- Department of Genetics and Applied Microbiology, University of Debrecen, Debrecen, Hungary
Over the last one and a half decade, interspecies hybridisation has gained continuously increasing attention as a breeding technique suitable for transferring of genetic information between Saccharomyces species and mixing of their gene pools without genetic engineering. The hybrids frequently show positive transgressive phenotypes. Segregation of the hybrid genome results in mosaic (chimeric) strains that can outperform both the parents and the hybrids or exhibit novel positive phenotypic properties. Mitotic segregation can take place during the vegetative propagation of the sterile allodiploid hybrid cells. Meiotic segregation becomes possible after genome duplication (tetraploidisation) if it is followed by break-down of sterility. The allotetraploid cells are seemingly fertile because they form viable spores. But because of the autodiploidisation of the meiosis, sterile allodiploid spores are produced and thus the hybrid genome does not segregate (the second sterility barrier). However, malsegregation of MAT-carrying chromosomes in one of the subgenomes during allotetraploid meiosis (loss of MAT heterozygosity) results in fertile alloaneuploid spores. The breakdown of (the second) sterility barrier is followed by the loss of additional chromosomes in rapid succession and recombination between the subgenomes. The process (genome autoreduction in meiosis or GARMe) chimerises the genome and generates strains with chimeric (mosaic) genomes composed of various combinations of the genes of the parental strains. Since one of the subgenomes is preferentially reduced, the outcome is usually a strain having an (almost) complete genome from one parent and only a few genes or mosaics from the genome of the other parent. The fertility of the spores produced during GARMe provides possibilities also for introgressive backcrossing with one or the other parental strain, but genome chimerisation and gene transfer through series of backcrosses always with the same parent is likely to be less efficient than through meiotic or mitotic genome autoreduction. Hybridisation and the evolution of the hybrid genome (resizing and chimerisation) have been exploited in the improvement of industrial strains and applied to the breeding of new strains for specific purposes. Lists of successful projects are shown and certain major trends are discussed.
Introduction
Strains of Saccharomyces cerevisiae, the major yeast used in fermentation technologies (Tuite and Oliver, 1991) show high genetic and phenotypic diversity. The favorable properties of the different strains can be brought together by hybridisation. The hybrids not only show transgressive phenotypes but also produce mitotic and meiotic segregants of diverse combinations of the traits of the hybridizing partners. Strains of the other species of the genus can have additional favorable properties but their combination with those of the S. cerevisiae strains is hampered by the postzygotic reproductive isolation of the species manifested as hybrid sterility. The interspecies hybrids of the Saccharomyces species are viable but either do not produce gametes (ascospores) or if they do so, the viability of the gametes is extremely low. This sterility barrier keeps the species biologically isolated but the isolation is not absolute. The hybrid genomes can change and certain types of changes make the barrier permeable. In a previous review a model was proposed to integrate these postzygotic events into a coherent system based on what was then known (Sipiczki, 2008). According to the model, the hybrid genome undergoes a gradual size reduction by losing chromosomes, either in the course of vegetative propagation of the allodiploid hybrid cells or during allotetraploid meiosis which takes place upon spontaneous genome duplication. Concomitantly with size reduction the subgenomes can interact and recombine. The “stabilized” outcomes of these processes are recombinant haploids and aneuploids, in fact strains with chimeric (mosaic) genomes. Hybridisation and postzygotic genome chimerisation can be observed in the laboratory but can take place also in natural habitats as demonstrated by the occurrence of chimerised genomes in strains isolated from yeast communities fermenting beverages. Over the past 10 years, considerable progress has been made in the investigation of hybrid sterility, the breakdown of the sterility barrier, and the mechanisms underlying the postzygotic reduction and chimerisation of the hybrid genome. These processes and their exploitation in the improvement of industrial Saccharomyces strains, as a non-GMO alternative of targeted genetic manipulation, are the subjects of this review. A review of this length cannot be comprehensive and thus it will not cover the hybrid species, the natural “hybrid strains” and the evolutionary aspects of hybridisation. The reader interested in the developments in these fields can consult review papers (e.g., Sipiczki, 2008; Louis, 2011; Albertin and Marullo, 2012; Morales and Dujon, 2012; Dujon and Louis, 2017; Krogerus et al., 2017a; Bisson, 2017; Gibson et al., 2017; Guillamón and Barrio, 2017; Lopandic, 2018) published elsewhere. Given that certain genetic terms are often inconsistently used in the literature, a section will address terminological issues.
Taxonomy of Saccharomyces (Saccharomyces sensu stricto)
The taxonomy of Saccharomyces changed many times in the history of the genus. van der Walt (1970) separated the highly fermenting species from the rest of the genus and proposed the name Saccharomyces sensu stricto complex for them. Since then the species not included in this group (sensu lato) were transferred to other genera, so the name “sensu stricto complex” has become obsolete. Yeast taxonomy currently accepts 7 “natural,” “clean” or “single-genome-based” [S. arboricolus (S. arboricola), S. cariocanus, S. cerevisiae, S. kudriavzevii, S. mikatae, S. paradoxus, and S. uvarum (S. bayanus var. uvarum)] and 2 “hybrid” [S. pastorianus/carlsbergensis and S. bayanus (S. bayanus var. bayanus)] Saccharomyces species (Vaughan-Martini and Martini, 2011). Since 2011 two new Saccharomyces species S. eubayanus and S. jurei were described. However, this classification is in contradiction with the results of whole-genome sequencing. Whole-genome analysis has shown that Saccharomyces cariocanus should be considered to be more a S. paradoxus variant as this species is reproductively isolated from Saccharomyces paradoxus by four translocations but not by sequence (reviewed in Borneman and Pretorius, 2015; Dujon and Louis, 2017; Nguyen and Boekhout, 2017). But it does not fit with mtDNA gene order, which is considered as a species-specific feature. The S. cariocanus mtDNA is not syntenic to that of S. paradoxus and therefore S. cariocanus should be designated as a separate species (Sulo et al., 2017).
The hybrid species (S. bayanus, and S. pastorianus/ carlsbergensis) occurring almost exclusively in beer fermentation are highly heterogeneous in genome structure and the proportion of the genomic mosaics originating from the assumed parental species (e.g., Nguyen and Gaillardin, 2005; Rainieri et al., 2006; Dunn and Sherlock, 2008; Nakao et al., 2009; Libkind et al., 2011; Nguyen et al., 2011; Walther et al., 2014; Pérez-Través et al., 2014a; Van den Broek et al., 2015). In addition to these hybrid species, many other Saccharomyces strains have been identified mainly among wine yeasts that contained mosaics of foreign origin in their genomes. These strains usually referred to as “hybrids” are not considered distinct taxonomic entities (for reviews, see Sipiczki, 2008; Albertin and Marullo, 2012; Morales and Dujon, 2012; Dujon and Louis, 2017; Lopandic, 2018). There is no consensus about where to draw the line between the hybrid species and the interspecies hybrids. Both groups have diverse chimeric genomes.
There is considerable confusion also around the taxonomic identity of S. bayanus and S. uvarum. Both species have been described in the 19th century but because of their proposed merger under the name S. bayanus some three decades ago (Vaughan-Martini and Kurtzman, 1985), the name S. uvarum has become sparsely used in the literature until recently. However, the molecular genetic and genomic analyses clearly separated two groups, a clean and a hybrid group, among the strains that had been assigned to S. bayanus over the last 30 years and led to the reinstatement of the species S. uvarum for the clean group (Nguyen and Gaillardin, 2005; Nguyen et al., 2011). As most S. bayanus strains isolated from substrates not related to beer production have turned out in molecular tests to be conspecific with the type strain of S. uvarum, the name S. bayanus will be restricted only to brewery yeasts in this review.
All discrepancies in fungal taxonomy come from the three species concepts that have been applied to species delimitation: the classical morphology/physiology-based, the biological and the evolutionary/phylogenetic species concepts. The classical concept is difficult to apply generally in Saccharomyces taxonomy because of the strain variation in the utilization of different substrates and the absence of distinctive morphological properties. The biological species concept, which defines a species as a group of actually or potentially inbreeding natural population reproductively isolated from other such groups (Mayr, 1940), was introduced to Saccharomyces taxonomy by Naumov (1987) and successfully applied to the description of novel species and to the determination of the taxonomic affiliation of novel isolates. This approach is based on the observation that the hybrids of the Saccharomyces strains are viable, but only hybrids of conspecific strains can produce viable spores at frequencies higher than 1%. However, this method is impractical in large-scale screening and routine testing of taxonomic affiliation of larger numbers of isolates. The phylogenetic concept defines a species as a monophyletic group of organisms sharing molecular characters that derive from a common ancestor (Moore et al., 2011). The phylogenetic relationships of yeast strains can be examined by comparing the sequences of their evolutionary conserved genes or non-coding chromosomal segments such as the D1/D2 domains of the genes encoding the large-subunit (LSU) rRNA molecules and the internal transcribed segments (ITS), two components of the repeats of the rDNA arrays. A recent study of 9,000 yeast isolates of the CBS collection (Vu et al., 2016) found that strains differing by more than 0.49% in their D1/D2 domains and by more than 1.59% in their ITS segments are usually not conspecific. The “natural” Saccharomyces species can be separated with these criteria. The “hybrid species” S. pastorianus/carlsbergensis and S. bayanus are difficult to distinguish from the “natural species” because certain their strains show no sexual activity (untestable for biological isolation) and/or have rDNA sequences of different “natural” species in heterozygous constitution (e.g., Nguyen and Gaillardin, 2005; Rainieri et al., 2006; Nguyen and Boekhout, 2017). Nevertheless their type strains sporulate and it is possible to mate them e.g., to S. cerevisiae (Špírek et al., 2014). The pan-genome analysis (e.g., Tettelin et al., 2005; Dunn et al., 2012) of large numbers of strains of all species of the genus may shed new light on the problem of species boundaries.
Terminological Difficulties
Authenticity of Species
The widespread view is that the Saccharomyces species have the same number (16) of mostly syntenic nuclear chromosomes (e.g., Fischer et al., 2000; Kellis et al., 2003; Scannell et al., 2011; Dunn et al., 2013; Liti et al., 2013; Baker et al., 2015). This view based on the comparison of a limited number of structurally assembled genome sequences is at odds with the high intraspecies diversity of karyotypes and genome structures observed in S. cerevisiae and S. uvarum isolates (e.g., Johnston and Mortimer, 1986; Bakalinsky and Snow, 1990; Yamamoto et al., 1991; Adams et al., 1992; Bidenne et al., 1992; Kishimoto and Goto, 1995; Miklos et al., 1997; Sipiczki, 2002; Csoma et al., 2010; Leducq et al., 2016; Nguyen and Boekhout, 2017; Albertin et al., 2018). If this diversity is not taken into account, the interpretation of the results of the hybridisation experiments can easily lead to disputable conclusions. Perhaps it is more accurate to conceive each species as a population of diverse strains that share certain species-specific attributes but may show diversity in other properties and also in genome structure. For example, it does not seem substantiated to declare a sequenced genome “the S. cerevisiae genome” and another sequenced genome “the S. uvarum genome” and claim that these two species differ in specific translocations or inversions that occurred a certain time ago. This claim is correct if only two strains are compared. However, other strains of these species show more or different structural genome differences including mosaics of foreign origin attributable to more recent events.
Hybrid – Mosaic – Genetically Admixed – Chimera – Evolved Hybrid
Further confusion can be caused by the presence of genes of different species in the genome. Genomes containing foreign genes were found in many strains isolated from yeast populations of fermenting beverages or constructed in laboratories. Many isolates identified as S. cerevisiae (for reviews, see e.g., Sipiczki, 2008; Morales and Dujon, 2012; Lopandic, 2018) or S. uvarum (e.g., Almeida et al., 2014; Albertin et al., 2018) turned out to have genes of foreign origin (alien genes) in their genomes. It can be assumed that, when subjected to molecular testing, many more wine and ale strains of these species will turn out to have genomic mosaics acquired from other species. The question arises as to whether the presence of such alien genes is characteristic of all yeasts fermenting alcoholic beverages as a consequence of “domestication.” These strains pose a terminological problem. They can be considered conspecific with one or the other species or can be regarded as interspecies hybrids. The latter term has become widely used even for strains in which only one or two foreign genes were detected. The application of terms like “natural S. cerevisiae × S. kudriavzevii hybrid” or “natural S. cerevisiae × S. kudriavzevii × S. uvarum hybrid” to such strains can be confusing because it conceals the exact genetic identity of the strains and may lead to the erroneous assumption that these strains have no taxonomic affinities. Certain authors (e.g., Groth et al., 1999; Belloch et al., 2009; Sipiczki, 2011) tried to resolve the confusion by applying the term “chimera” or “genomic chimera” (used in other groups of organisms; e.g., Jansen et al., 2011; Watanabe et al., 2012; Pryszcz et al., 2014) to Saccharomyces strains having mosaics of different origin in their genomes. Unfortunately, this has not become a widely used practice. Nevertheless, for the sake of ambiguity, the term chimera (chimeric) will be used in this review to distinguish strains having mosaic genome structures from “true” hybrids. Natural true hybrids having complete genomes of two species were rarely found (e.g., Masneuf et al., 1998; Naumov et al., 2000; Borneman et al., 2012). A good alternative for “chimeric” and “mosaic” can be “admixed” (e.g., Tilakaratna and Bensasson, 2017).
In the case of the hybrids constructed in laboratories, the terminological diversity is not smaller. The most frequently used names (synonyms) are synthetic, artificial, constructed, laboratory-constructed, newly formed, de novo and experimental hybrids. Derivatives of the hybrids which usually have reduced and chimeric genomes are often called “evolved hybrids” instead of the more accurate “segregants.”
HTG – Introgression – Genetic Admixture – Genome Autoreduction
Formally, the foreign genes/mosaics are horizontally transferred segments of the genome(s) of the donor species to the chimeric (“hybrid”) strains of the acceptor species. Their origin can be traced back by searching databases for similar sequences, but the mechanism of the transfer cannot be reconstructed by sequence analysis. In spite of having no real clues to the mechanism, most authors regard them introgressed sequences without considering other possible transfer mechanisms (e.g., Liti et al., 2006; Muller and McCusker, 2009; Louis, 2011; Dunn et al., 2013; Boynton and Greig, 2014; Marsit and Dequin, 2015; Hou et al., 2016; Dujon and Louis, 2017; Guillamón and Barrio, 2017; Peris et al., 2017; Albertin et al., 2018). Hittinger et al. (2015) assumed that S. cerevisiae gains genes from other Saccharomyces species by introgression whereas from non-Saccharomyces species by horizontal gene transfer (HTG). This distinction was used in other works as well (e.g., Legras et al., 2018). Other authors use the latter term also for gene transfer between Saccharomyces species (Marinoni et al., 1999). Automatic attribution of all gene flow events within the genus to introgression appears to be a disputable oversimplification of a complex issue. By definition, introgression (or “introgressive hybridisation”) incorporates genes from one entity (species) into the gene pool of a second, divergent entity (species) via hybridisation and backcrossing of the hybrid with the latter entity (Anderson and Hubricht, 1938). But the hybrids of the Saccharomyces species are sterile, either produce no spores (no gametes suitable for backcrossing with the parent) at all, or if they sporulate, the frequency of the viable, usually highly aneuploid spores is very low and these spores are frequently unable to mate (e.g., Gjermansen and Sigsgaard, 1981; Zambonelli et al., 1993; Hawthorne and Philippsen, 1994; Kishimoto, 1994; Hunter et al., 1996; Marinoni et al., 1999; Liti et al., 2006; Kunicka-Styczyńska and Rajkowska, 2011; Pfliegler et al., 2012). Several recent works pointed out the conflict between the hypothesized (exclusive) role of introgression in mixing of genetic information of Saccharomyces species and its low chance of occurrance due to the sterility of the hybrids (e.g., Morales and Dujon, 2012; Marsit and Dequin, 2015; Dujon and Louis, 2017). Perhaps the more general term “genetic admixture” would be more accurate than “introgression” when the mechanism of the gene transfer is not known.
The alternative of introgressive backcrosses is the gradual genome reduction and chimerisation in meiotic and/or mitotic divisions, which will be referred to as GARMe and GARMi in the forthcoming sections. GARMe (Genome Autoreduction in Meiosis) generates chimeric genomes in series of successive meiotic divisions after the breakdown of the sterility barrier upon tetraploidisation (Karanyicz et al., 2017), whereas GARMi (Genome Autoreduction in Mitosis) is its counterpart operating during vegetative (mitotic) propagation of the hybrid cells. Both mechanisms are described in the forthcoming sections. Neither involves introgressive backcrosses and thus it is misleading to call their outcomes introgressions. Occasionally, the latter process is also called introgression (e.g., Dunn et al., 2013) despite the fact that it does not involve sexual interactions.
Fertility – Sterility
A strain is fertile if it produces viable meiospores (ascospores) that can act as gametes capable of fusion (conjugation) with other meiospores. As the Saccharomyces meiospores can propagate vegetatively and produce clones of cells (spore clones), the ability to mate as a criterion of fertility applies also to these cells (“propagating gametes”). As discussed in the previous paragraph, the allodiploid interspecies hybrid is sterile because its cells neither produce viable spores (spore viability is usually much lower than 1%; e.g., Naumov et al., 2000; Delneri et al., 2003) nor mate with other cells. The allotetraploid hybrid is different because its cells are defective only in mating. The allotetraploid cells can sporulate and produce viable spores (e.g., Banno and Kaneko, 1989). However, the spores (spore clones) are usually sterile because they are usually allodiploid like the allodiploid hybrids (e.g., Pfliegler et al., 2012; Karanyicz et al., 2017) (for a detailed description, see the section on the second sterility barrier). Allotetraploid meiosis produces also alloaneuploid spores which can be sterile or fertile, depending on which chromosomes are missing (Antunovics et al., 2005; Pfliegler et al., 2012). If nullisomic for one of the MAT-carrying parental chromosomes, the alloaneuploids can mate and form alloaneuploid zygotes capable of sporulation (see section on GARMe). Several studies reported on interspecies Saccharomyces hybrids producing viable spores but often without providing information on the frequency of such spores, and/or on the ploidy of the hybrids and their spores (e.g., Greig et al., 2002b; Lee et al., 2008; Chou et al., 2010; Verspohl et al., 2018).
An important condition for viable spore production is homothallism, allowing the mating type switch in haploid/alloaneuploid cells after almost every division. Therefore, progeny of interspecies hybrids between different Saccharomyces species is able to establish fertile lines after sporulation and self-fertilization, provided they lose MAT heterozygosity (see section on the break-down of sterility barrier). In case of different biological species spores of allodiploid hybrids are rarely viable and only about 0.1–1% germinate and proliferate into visible colonies, from which many produce immortal lines (e.g., Naumov et al., 2000; Delneri et al., 2003 and many others).
Generation Terminology
In the Mendelian terminology the pairing (mating) individuals constitute the P (parental) generation, their descendants (hybrids) are the first filial generation (F1), and the descendants of the pairing (or self-pollinating) F1 individuals are the F2 generation. For pairing, the sexually propagating animals and plants produce gametes by meiosis which then either fuse with other gametes to form zygotes (fertilization) or die. In contrast to animals and plants, the products of the yeast meiosis (meiospores, ascospores, basidiospores) do not die if they cannot mate. Instead, they germinate and form clones of vegetatively propagating cells. For these clones of “propagating gametes” there is no Mendelian term. In this review, they will be referred to as F1 spore clones. If the F1 spore is fertile and homothallic, the cells of its vegetative clone can switch mating types and then can mate with other cells of the same clone (selfing, autofertilisation) (Pfliegler et al., 2012). The resulting zygotes will propagate vegetatively, and can be considered F2 generation (equivalents of the F2 generation of self-pollinating plants). Their spores (gametes) are the F2 spores and the cells produced by the F2 spores are the F2 spore clones. If the F2 spore clone is homothallic, its cells can conjugate with each other and produce the F3 generation. This nomenclature will be used in this review. It may differ from those used in certain other works.
The Complexity of (Allodiploid) Hybrid Sterility
Strains of the natural (“clean” or “single-genome-based”) Saccharomyces species are usually homothallic (for a review, see Rainieri et al., 2003). Their genomes are diploid or aneuploid and heterozygous for the mating-type alleles. MATa/MATalpha heterozygosity makes the cells unable to conjugate by repressing the mating program. However, it allows meiosis and sporulation in response to proper external signals (e.g., starvation). Meiosis produces haploid meiospores that have only one MAT locus, either MATa or MATalpha. The loss of MAT heterozygosity blocks the meiotic program and concomitantly abolishes the block of the mating program. So the spores can mate with other mating-competent conspecific spores and thus can act as gametes to form autodiploid zygotes. The zygotes then produce mitotically propagating autodiploid cells, equivalents of the somatic cells of higher eukaryotes. Having a MATa/MATalpha genotype, these cells are also sporogenic and can produce viable gametes (ascospores). For a review of the genetic determination of sexual processes of Saccharomyces strains, see Herskowitz (1988).
Mating can take place also between non-conspecific Saccharomyces spores. The resulting allodiploid zygotes are viable and produce vegetative cells much like the autodiploid zygotes. But in contrast to those, the allodiploid cells cannot sporulate or if they do so, their spores are very rarely viable (e.g., Gjermansen and Sigsgaard, 1981; Zambonelli et al., 1993; Hawthorne and Philippsen, 1994; Kishimoto, 1994; Hunter et al., 1996; Marinoni et al., 1999; Liti et al., 2006; Kunicka-Styczyńska and Rajkowska, 2011; Pfliegler et al., 2012). This phenomenon called hybrid sterility or hybrid incompatibility plays an essential role in the biological isolation of the Saccharomyces species because it impairs the competitiveness of the hybrids under unfavorable natural conditions (for explanation, see section the bad and good sides of sterility) and prevents the recombination of their gene pools.
What is the genetic basis of hybrid sterility in Saccharomyces?
Four major hybrid sterility mechanisms have been proposed:
Interactions of Incompatibility Genes
Although nucleo-mitochondrial incompatibility is apparently the main reason for divergence of species, it is not involved substantially in the sterility of interspecific hybrids, because they contain both copies of incompatibility genes. Debilitated nucleo-mitochondrial communication has been reported only in haploid cybrids with reduced respiration or in non-respiring chimeras containing mitochondria from one partner as well as a set of original chromosomes, where one or two were replaced with their counterpart from second partner. However, these cells can mate and the respiration is rescued (Lee et al., 2008; Chou et al., 2010; Špírek et al., 2014). In principle, other forms of incompatibility with adverse effect on spore formation are also conceivable. For example, the spores could be killed by the interactions of recessive or dominant chromosomal incompatibility (speciation) genes. The aforementioned sporulation proficiency of the allotetraploid hybrids rules out the possibility that dominant incompatibility genes cause the sterility of allodiploids (Greig et al., 2002a). Efforts to find recessive incompatibility genes were unsuccessful (Greig, 2007; Kao et al., 2010). However, a recent computer simulation raised the possibility that two- and multilocus incompatibilities with incomplete penetrance could operate in S. cerevisiae × S. paradoxus hybrids (Li et al., 2013). Such interactions might also affect sporulation but incomplete penetrance is likely to cause only slight reduction of spore viability.
Chromosome Rearrangement
Many of the spores from yeast hybrids are unviable, because they do not contain a complete genome’s worth of genes. In the case of single chromosomal translocation, 50% of the gametes will lack part of one of the translocated chromosome and die (Delneri et al., 2003).
Misexpression of Meiotic Genes
Swain Lenz et al. (2014) found that the meiotic gene expression program proceeds more rapidly in S. cerevisiae × S. paradoxus hybrids than in the parents, and the change in the timing results in a heterochronic pattern of misexpression during midmeiosis. The authors hypothesized that the temporal changes might compromise the efficiency of meiosis. In a different study the fast evolution of the meiosis-related genes was hypothesized to account for hybrid sterility but the hypothesis was not tested experimentally (Xu and He, 2011). These models are also difficult to reconcile with the high spore viability of allotetraploids.
Aberrant Chromosomal Behavior in Meiosis and Antirecombination
Numerous early studies have shown that the sterility of the hybrids of different plant and animal species is due to inadequate or deficient chromosome pairing during meiosis (e.g., Walters, 1958; John and Weissman, 1977; Gangadevi et al., 1985). Similar pairing aberrations were later observed also in hybrids of Saccharomyces species (Lorenz et al., 2002), shown to hamper chromosome segregation (Hawthorne and Philippsen, 1994; Sebastiani et al., 2002; Boynton and Greig, 2014) and implicated in hybrid sterility (e.g., Ryu et al., 1998; Delneri et al., 2003).
Chromosome alignment requires DNA strand exchange between similar segments of the pairing chromosomes (as part of recombination) (Sansam and Pezza, 2015). If the sequence similarity is low or restricted to shorter segments (e.g., between homeologous chromosomes of the subgenomes in the alloploid hybrid), there are fewer opportunities for strand exchange and thus the chromosomes cannot be properly aligned. Because of the sequence differences, strand exchange creates mainly heteroduplexes with mismatched nucleotides. Mismatches are targets for the mismatch repair machinery. This repair process can eliminate heteroduplexes and thus impair the pairing of the homeologous chromosomes (antirecombinational effect). The improvement in spore viability observed by Hunter et al. (1996) after the attenuation of the mismatch repair in S. cerevisiae and S. paradoxus hybrids might have been due to the elimination of fewer heteroduplexes (reduction of the antirecombinational effect). Until recently, mismatch repair was regarded a major player in the sterility barrier. However, recent results revealed that structural differences between the chromosomes alone are sufficient for reproductive isolation (Leducq et al., 2016). Besides, a recent work found that karyotype engineering can lead to reproductive isolation and eight chromosome–chromosome fusion events suffice to isolate a S. cerevisiae strain reproductively from other conspecific strains (Luo et al., 2018).
If the homeologous chromosomes differ in synteny, aberrant bivalents (syntenic segments having different positions in the homeologous chromosomes pair) (e.g., Karanyicz et al., 2017) and even partial multivalents (syntenic segments located on non-homeologous chromosome pairs) (Lorenz et al., 2002) can be formed. The meiotic spindle apparatus then cannot handle correctly the poorly and chaotically synapsed chromosomes and most probably either collapses (no sporulation) or forms nullisomic spores lacking essential genes and aneuploid spores with non-functional combinations of parental chromosomes (dead spores) (Hawthorne and Philippsen, 1994; Boynton et al., 2018). Thus, the probability of the formation of viable spores is very small and the proportion of haploids among them can be extremely low. Interestingly, in certain works, all viable spores were considered haploid without being subjected to any ploidy tests (Greig et al., 2002b; Lee et al., 2008; Chou et al., 2010). This, however, seems to be a disputable oversimplification in view of a recent estimation of alloaneuploid formation during allodiploid meiosis performed by the same authors (Boynton et al., 2018). Their model is difficult to reconcile with the results of Xu and He (2011), who found that the vast majority of the viable spores of their synthetic S. cerevisiae × S. paradoxus allodiploid had either S. cerevisiae or S. paradoxus haploid genomes. The reason for the discrepancy is not clear. Nevertheless, the production of spores with parental genomes would be a dead end on the road toward introgression and other modes of genome chimerisation.
The Second (Yeast-Specific) Sterility Barrier Operating in Allotetraploids
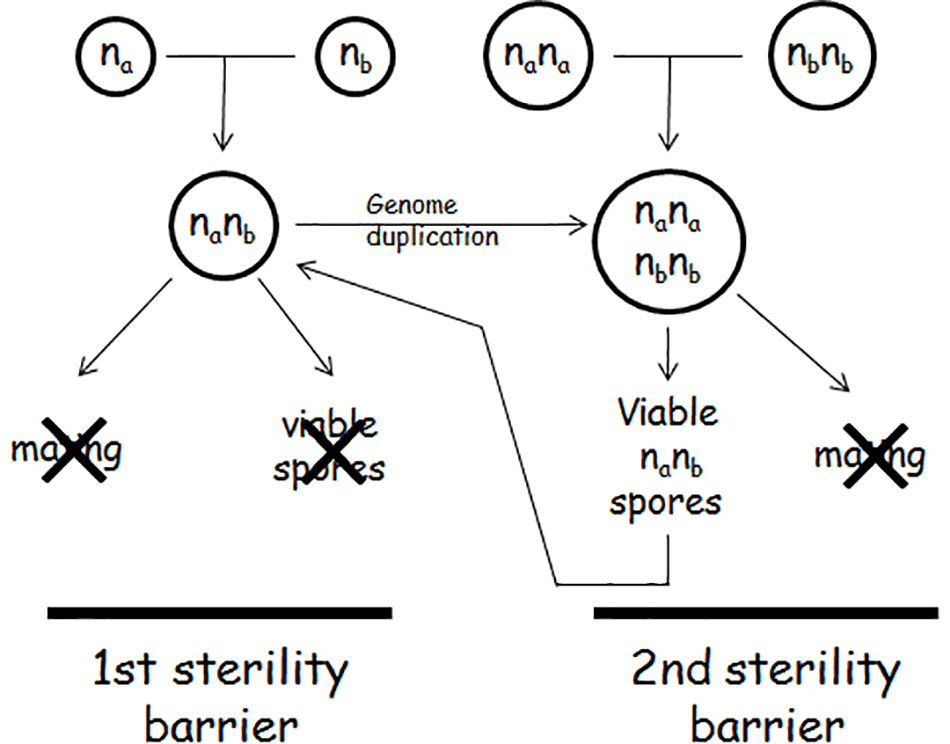
In plants, the sterile allodiploid hybrids become fertile when they duplicate their genomes because in the allotetraploid genomes each chromosome has a homologous partner to pair with, and thus a normal meiosis can take place. Upon genome duplication the hybrids produce viable and functional allodiploid gametes (for a review, see Ramsey and Schemske, 2002). A seemingly similar process takes place also in Saccharomyces interspecies hybrids but with a different outcome. If the hybridisation of two Saccharomyces species is followed by genome doubling or the mating cells were diploid (rare event), the hybrid becomes able to form viable spores (Cummings and Fogel, 1978; Banno and Kaneko, 1989; Marinoni et al., 1999; Johnston et al., 2000; Naumov et al., 2000; Greig et al., 2002a; Sebastiani et al., 2002; Antunovics et al., 2005; Pfliegler et al., 2012; Karanyicz et al., 2017). This observation led to the widespread view that genome duplication restores fertility in Saccharomyces interspecies hybrids as well. However, where investigated, these spores turned out to possess allodiploid chromosomal sets and thus represented a return to the ancestral sterile allodiploidy (Sebastiani et al., 2002; Pfliegler et al., 2012; Karanyicz et al., 2017) (Figure 1). Their sterility makes them different from the gametes of the allotetraploid plants which are fertile. Another difference from the plant allodiploid gametes is that the yeast allodiploid gametes (F1 spores) can propagate vegetatively and produce allodiploid F1 spore clones. The third difference is that the cells of these clones can sporulate. But like the spores of the allodiploid hybrids, the spores of the allodiploid F1 spore clones are dead. The inability of the F1 spore clones of the allotetraploid cells to mate and produce viable spores is the second sterility barrier in the biological isolation of the Saccharomyces species (Pfliegler et al., 2012). Thus, in contrast to the plant interspecies hybrids, genome duplication does not restore fertility in the Saccharomyces hybrids, it only produces non-functional gametes that can propagate vegetatively but cannot fertilize (conjugate).
The Second Sterility Barrier Is Maintained by Autodiploidisation During the Allotetraploid Meiosis
The pairing of a chromosome with its homologous partner (autosyndesis) in the allotetraploid meiosis excludes its pairing with a chromosome from the other subgenome (allosyndesis) (Ramsey and Schemske, 2002; Moore, 2002). Thus, each diploid subgenome performs meiosis without interaction with the companion subgenome and then transmits its halved (haploid) sets of chromosomes into each of the four gametes. This mode of meiotic division is referred to as autodiploidisation of the allotetraploid genome in plant genetics (Hutchinson et al., 1983). The investigation of the viable spores produced by allotetraploid Saccharomyces hybrids revealed that the meiosis of the allotetraploid yeasts is also autodiploidised (Pfliegler et al., 2012; Karanyicz et al., 2017). Like the gametes of the allotetraploid plants, the spores of the allotetraploid yeasts are allodiploid with a single set of chromosomes in both subgenomes. Each subgenome contains a MAT-carrying chromosome but their MAT alleles are different because the diploid subgenomes of the allotetraploid were homozygous for different alleles: one was MATa/MATa the other was MATalpha/MATalpha (because hybridisation can take place between cells of opposite mating types and mating-type switching is then repressed by the MAT heterozygosity; Herskowitz, 1988). The simultaneous presence of both MAT alleles makes the spores (and their vegetative progeny) of the allotetraploid hybrid unable to mate but able to launch the meiotic program, although the spores produced are not viable (Pfliegler et al., 2012). This is exactly what characterizes the allodiploid hybrids. Thus, after genome duplication and a round of successful meiosis the hybrid returns to the sterile allodiploid state (Figure 1). This mode of meiosis and chromosomes segregation is very different from those characteristic of autotetraploids. In an autotetraploid S. cerevisiae cell any chromosome can freely pair with any of the other three homologous copies regardless of their origin. As a result, many allodiploid spores will be homozygous for the mating type (MATa/MATa or MATalpha/MATalpha) and capable of mating. Neither the heterozygous spores are sterile because their vegetative descendants produce viable haploid spores (e.g., Hilger, 1973; Al Safadi et al., 2010).
Escape From the Biological Isolation: Breakdown of the (Second) Sterility Barrier by Loss of Mat Heterozygosity
The allotetraploid meiosis turned out to be prone to errors at the distribution of the chromosomes to the spores. It was noticed that certain asci of certain allotetraploids contained two types of spores, usually in 2:2 proportion. Two spores were sterile, and two spores formed clones of cells producing viable spores (Antunovics et al., 2005; Pfliegler et al., 2012; Karanyicz et al., 2017). The latter type lacked the MAT-carrying chromosome in one of the subgenomes. This chromosome (designated III in S. cerevisiae and III, 3 or 2 in other species, depending on the numbering system used) is known to be the least stable chromosome even in autoploid S. cerevisiae strains (Kumaran et al., 2013). Having only one MAT allele (being hemizygous at the MAT locus), the cells of these clones can (in contrast to the clones of the MATa/MATalpha allodiploid spores) switch their mating types and then conjugate with each other to form uniparentally disomic allotetraploid zygotes. This can take place because the loss of mating-type heterozygosity reactivates both the mating-type switching machinery and the mating program (Herskowitz, 1988). The intraclonal conjugation (selfing, autofertilisation) converts the F1 spore clone to an F2 population of cells having allotetraploid genomes nullisomic for the lost MAT-carrying chromosome of one subgenome and disomic for the MAT-carrying chromosome of the other subgenome. In these cells autosyndetic (homologous) chromosome pairing can take place, and thus the meiosis produces fertile alloaneuploid F2 spores. The cells of the F2 spore clones then form a fertile F3 generation and so on. These observations demonstrate that the loss of MAT heterozygosity by the loss of the MAT-carrying chromosomes in one of the subgenomes abolishes the sterility of the alloploid hybrid (Figure 2). The conjugation-proficient spore clones of the tetraploid S. cerevisiae × S eubayanus hybrids used by Krogerus et al. (2017b) for hybridisation with a S. cerevisiae strain might have been this sort of fertile alloaneuploids. The cost of the breakdown of the sterility barrier is the destabilization of the hybrid genome. The loss of the MAT-carrying chromosome converts the euploid genome into aneuploid after which additional chromosomes can be lost in rapid succession (see next section).
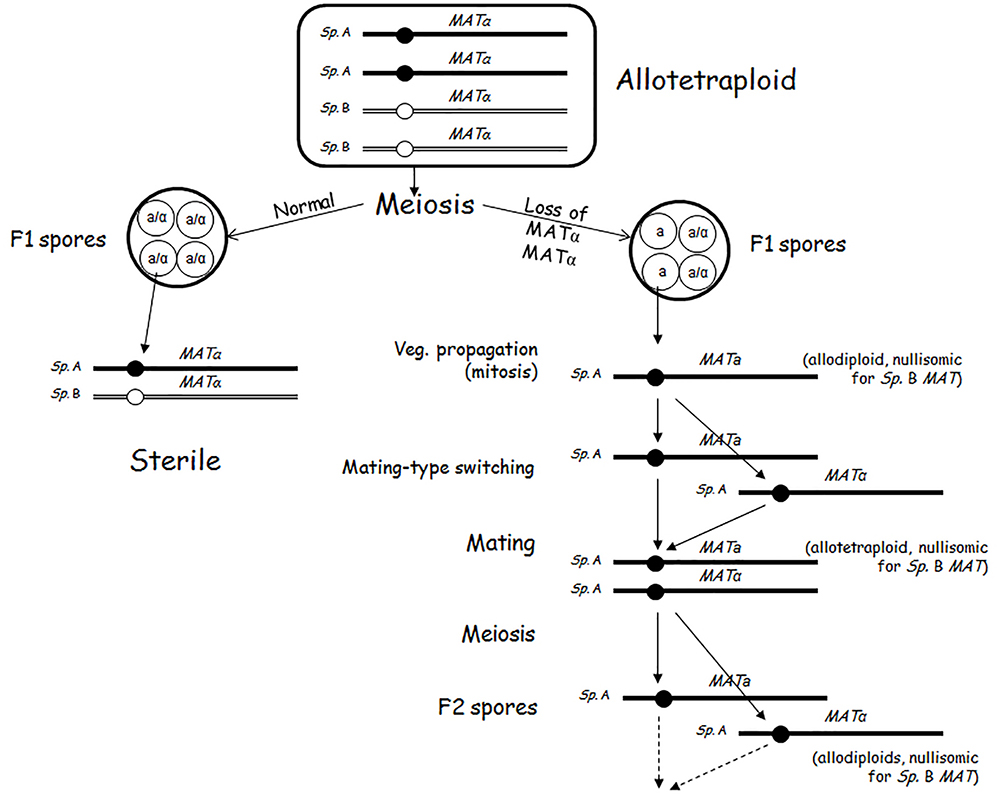
Figure 2. Breakdown of the sterility barrier by malsegregation of the MAT-carrying chromosomes of one subgenome in allotetraploid meiosis.
In principle, conjugation-proficient spores can occur also among the few viable aneuploid spores produced by allodiploid hybrids provided they have only one MAT-carrying chromosome. The fertile “haploid” spores described in Greig et al. (2002b) and in Lee et al. (2008) could have been such segregants but because their ploidy was not examined, the possibility cannot be excluded that they were allodiploid spores nullisomic for one parental MAT-carrying chromosome produced by cells of duplicated (allotetraploid) genomes.
Chimerisation of the Hybrid Genome in Meiotic Divisions: Introgression Versus Garme
The fertile alloaneuploid spores produced after the loss of MAT heterozygosity can conjugate with any fertile gamete of any other strain, also with the spores of one or the other parental species which opens the way for introgression. The caveat is that the products of these backcrosses will have low fertility due to their unbalanced sets of chromosomes and MAT heterozygosity. In an early work Sebastiani et al. (2002) observed conjugation between a few F1 spore clones of allotetraploids and the parental strains, but the hybrids obtained from the backcross produced mainly dead spores. de Barros Lopes et al. (2002) raised the possibility that introgression might take place by serial matings of the vegetative cells of the sterile hybrid with cells of one of the parental species [by the process called rare mating (Gunge and Nakatomi, 1972)]. Lee et al. (2008) managed to backcross the vegetative and autodiploidised (via self-fertilization) descendants of the spores of a S. cerevisiae × S bayanus (uvarum?) hybrid to the S. cerevisiae parent several times successively. Because of the very concise description of certain parts of the experimental procedure, it is impossible to resolve the inconsistency between the assumable MATa/MATalpha genotype (mating program repressed) of the cells and their ability to mate. It is conceivable that the spore clones contained conjugation-proficient segregants that had lost one of the parental MAT-carrying chromosomes. Nevertheless, the transfer of genetic information between Saccharomyces species via introgressive backcrosses has not been verified yet experimentally.
A recently described alternative process leading to chimerised genomes is GARMe (Genome Autoreduction in Meiosis) (Karanyicz et al., 2017) (Figure 3). It is a progressive process starting with the breakdown of the sterility barrier by the malsegregation of the MAT-carrying chromosomes during the allotetraploid meiosis. The loss of a pair of MAT-carrying chromosomes destabilizes the genome and additional pairs of autosyndetically paired chromosomes can malsegregate in the meiotic divisions of the consecutive filial generations of autofertilised spore clones. The genome gradually becomes smaller at each meiotic division, with one partner’s chromosomes being lost preferentially. Occasionally, recombination between chromosomes of the subgenomes can also take place (Antunovics et al., 2005; Karanyicz et al., 2017; Krogerus et al., 2017b), most probably due to allosyndetic interactions between similar segments of homeologous chromosomes (Lorenz et al., 2002). During GARMe, the hybrid genome is gradually transformed into various genomic chimeras comprising predominantly chromosomes of one species but containing also mosaics from the genome of the other species (Antunovics et al., 2005; Pfliegler et al., 2012, 2014; Lopandic et al., 2016; Karanyicz et al., 2017). The process gradually leads to chimeric genomes similar to those found in “natural hybrids” (in fact chimeras) that have only a few genes from the donor genome (see above). The differences between the hybrids created by hybridizing conjugation-proficient spore clones of an allotetraploid S. cerevisiae × S. eubayanus hybrid and a S. cerevisiae strain (Krogerus et al., 2017b) might be attributed to different chromosome losses in the spores.
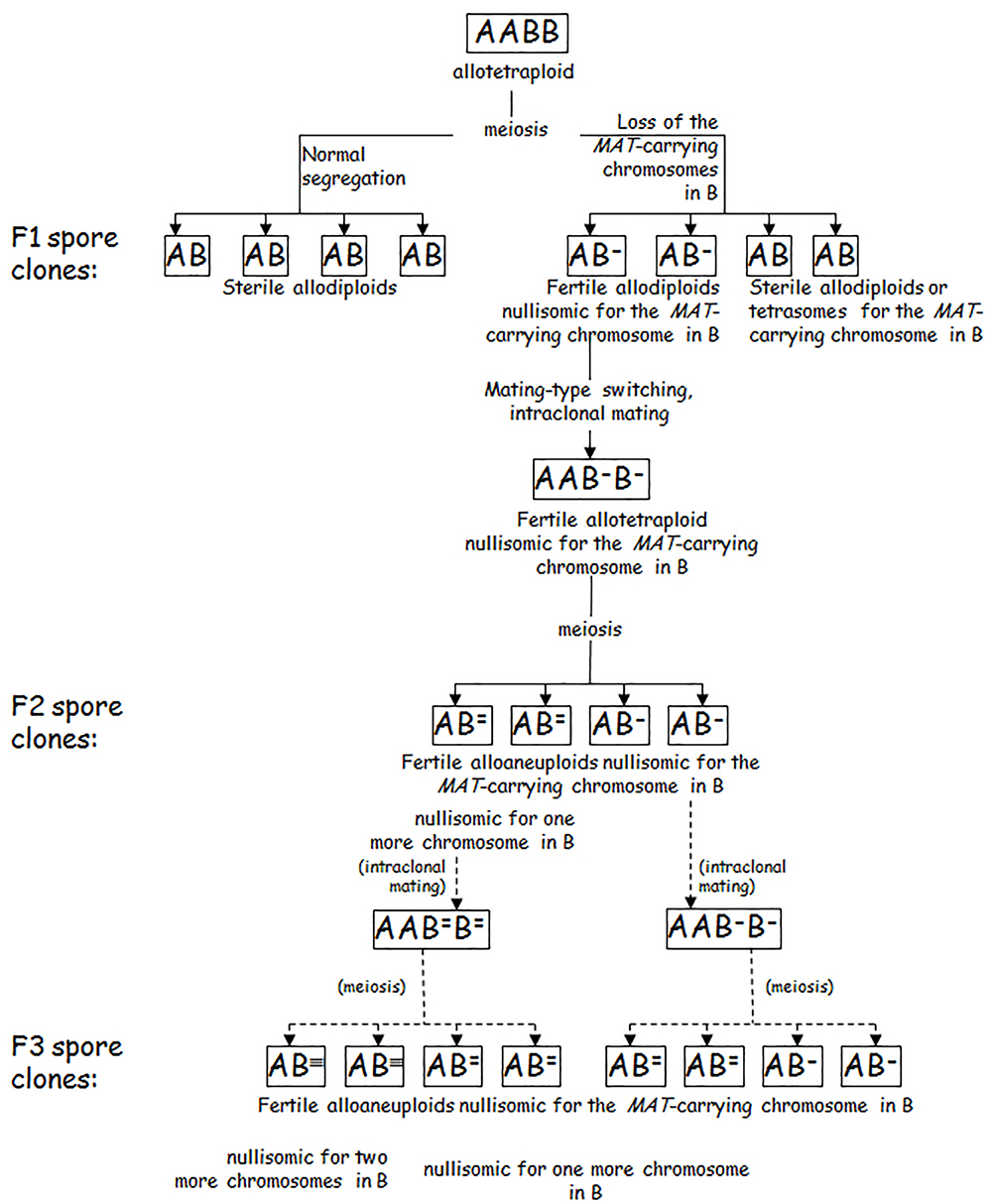
Figure 3. Genome autoreduction in meiosis (GARMe). A and B: parental genomes. The superscript hyphens represent lost chromosomes. Although all hyphens are shown over B, chromosomes can be lost from either subgenome. Chimeric genomes are produced when chromosomes are lost from both subgenomes, and allosyndetic interactions (recombination) take place between the subgenomes during meiosis.
Alteration of the Hybrid Genome During Vegetative (Mitotic) Propagation (Hybrid Evolution, Garmi)
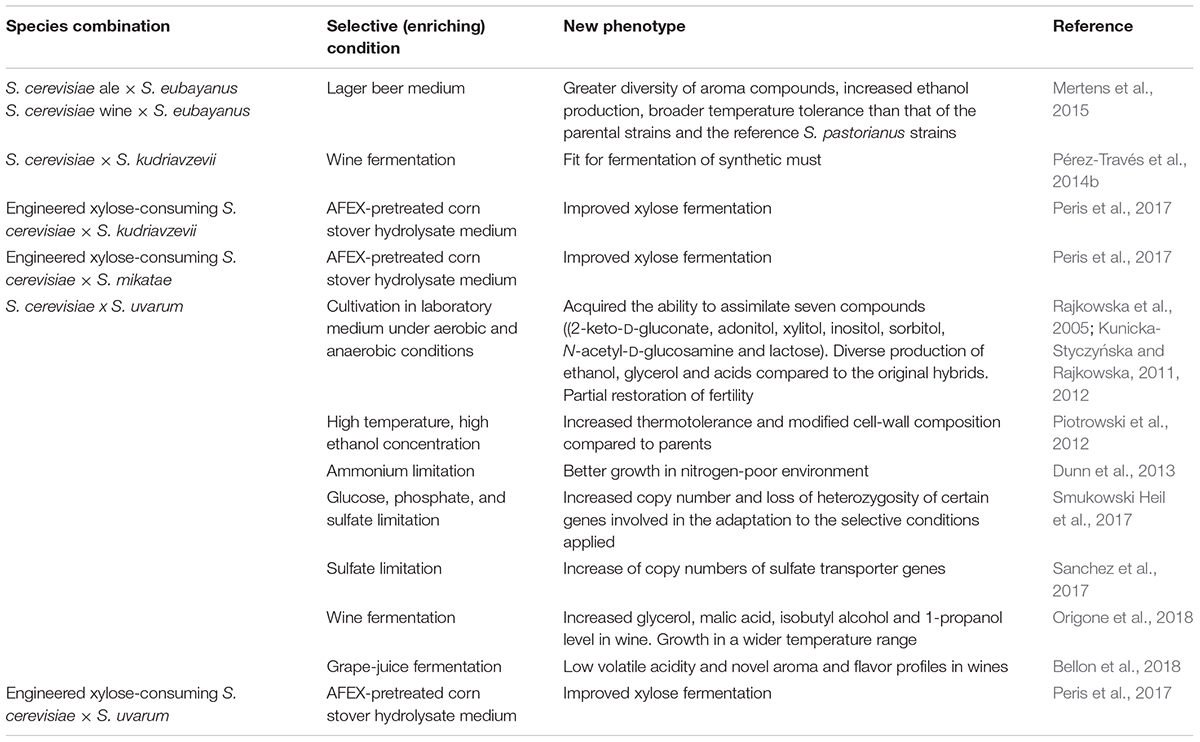
Vegetatively propagating diploid S. cerevisiae cells can lose chromosomes by rare spontaneous chromosome non-disjunction during mitosis. The event generates two aneuploid daughter cells one of which is monosomic, the other being trisomic (Parry and Zimmerman, 1976). Aneuploidy destabilizes the genome and thus initiates further karyotype changes (e.g., Parry and Cox, 1970; Zhu et al., 2012). As predicted in the model proposed a decade ago (Sipiczki, 2008), malsegregation leading to aneuploidy can occur also in vegetatively propagating populations of allodiploid hybrid cells. Recent studies confirmed the prediction. It was found that alloploid Saccharomyces hybrids can lose chromosomes during vegetative propagation, which can also be accompanied by other forms of structural rearrangement in the genome (e.g., Kunicka-Styczyńska and Rajkowska, 2011; Piotrowski et al., 2012; Dunn et al., 2013; Bellon et al., 2013, 2015, 2018; Pérez-Través et al., 2014b; Mertens et al., 2015; Lopandic et al., 2016; Krogerus et al., 2017b; Peris et al., 2017). Such derivatives (frequently called “evolved hybrids”) constitute minor components of the population of the hybrid cells but can become relatively more abundant if the hybrid culture is cultivated under enriching conditions favoring their propagation for longer periods of time (for a recent review, see Lopandic, 2018). Various “enriching” or selective conditions have been applied to obtain strains with specific properties such as anaerobiosis (Kunicka-Styczyńska and Rajkowska, 2011), high temperature and alcohol content (Piotrowski et al., 2012), ammonium limitation (Dunn et al., 2013), glucose limitation, phosphate limitation and sulfate limitation (Smukowski Heil et al., 2017) sulfate limitation (Sanchez et al., 2017), grape must fermentation (Pérez-Través et al., 2012, 2014b; Bellon et al., 2013, 2018; Origone et al., 2018), fermentation at high sugar concentrations (Lopandic et al., 2016), cultivation in lager beer medium (Mertens et al., 2015), in xylose fermentation medium (Peris et al., 2017), etc.
The most frequently detected genome modifications were losses of chromosomes and changes of the genome size. An extreme example is the loss of the almost entire S. uvarum subgenome during culturing of the S. cerevisiae x S. uvarum hybrid at increased temperature (31–46.5°C) (Piotrowski et al., 2012). Preferential alterations and reduction of one subgenome was reported several times. For example Bellon et al. (2018) also detected chromosomal loss and much more rearrangements in the S. uvarum subgenome than in the S. cerevisiae subgenome during must fermentation. Lopandic et al. (2016) observed chromosome loss in the S. uvarum subgenome of two F1 spore clones of a S. cerevisiae x S. uvarum allotetraploid and in the S. kudriavzevii subgenome of F1 spore clones of a S. cerevisiae x S. kudriavzevii allotetraploid hybrid during experimental wine fermentation.
In addition to aneuploidisation of the genome, also size changes of certain chromosomes were observed (e.g., Kunicka-Styczyńska and Rajkowska, 2011; Lopandic et al., 2016; Bellon et al., 2018) which indicates that various forms of recombination (e.g., exchange of chromosomal arms, translocation) can occur too. Other experiments demonstrated that recombination can take place also at the level of individual genes. Dunn et al. (2013) detected frequent recombination events between the MEP2 genes of the partner genomes in vegetatively propagating S. cerevisiae × S. uvarum hybrids. The observed genomic changes and interactions between the subgenomes in vegetatively propagating hybrids strongly suggest that the hybrid genome can be reduced and chimerised not only by GARMe but also during vegetative propagation without introgressive backcrosses. To the analogy of GARMe, this process can be called GARMi (Genome Autoreduction in Mitosis).
Different selective conditions can enrich derivatives of different phenotypes from the same hybrid (e.g., Piotrowski et al., 2012). When there is no selective pressure, the culture can segregate into subpopulations differing in genome size and chromosomal constitution even in the same culturing conditions (e.g., Mertens et al., 2015). The phenotypic diversity found in a recent study among hybrids obtained by hybridisation of the same pair of isogenic strains (Verspohl et al., 2018) might have been the consequence of different spontaneous genome rearrangements in the individual hybrid lines during their maintenance in non-selective culturing media.
The overgrowth of the unchanged hybrid cells by a segregant is a sort of adaptive evolution at population level because the cells of the segregant (evolved hybrid) are more fit than the original hybrid under the applied culturing conditions. The winner then remains stable, thus the process can also be regarded genome stabilization (e.g., Piotrowski et al., 2012; Pérez-Través et al., 2014b; Krogerus et al., 2017b; Bellon et al., 2018) but cultivation of the “stabilized” strain under different conditions would show whether its genome is stable indeed.
The aneuploid spore clones produced after the breakdown of the sterility barrier also change during vegetative propagation. Lopandic et al. (2016) found extensive segregation in F1 spore clones of S. cerevisiae × S. uvarum and S. cerevisiae × S. kudriavzevii hybrids, whereas Krogerus et al. (2017b) observed less drastic changes in S. cerevisiae × S. eubayanus hybrids. In both cases the non-cerevisiae subgenome was less stable.
When interpreting the changes of the hybrid genome during vegetative propagation, it has to be borne in mind that occasional meiotic divisions may also contribute to the process despite the adverse cultivation conditions for sporulation. Mortimer et al. (1994) observed sporulation in certain wine yeast strains even in sugar-rich media and Sebastiani et al. (2002) found that S. cerevisiae × S. uvarum hybrids could form asci on YPD (yeast-peptone-glucose medium).
In principle, GARMi can also break down the sterility barrier, if the hybrid cell loses the MAT-carrying chromosome in one of the subgenomes (loss of MAT heterozygosity). Kunicka-Styczyńska and Rajkowska (2011) found fertile segregants in cultures of sterile S. cerevisiae × S. uvarum hybrids after 50–80 generations of mitotically dividing cells.
Why Is the Postzygotic Genome Reduction Asymmetric?
As shown above, one of the subgenomes usually undergoes a faster and more extensive reduction both in the meiotic (GARMe) and in the mitotic (GARMi) divisions. The less stable subgenome in meiosis was that of S. uvarum in the S. cerevisiae × S. uvarum hybrids (Antunovics et al., 2005; Pfliegler et al., 2012; Piotrowski et al., 2012), S. eubayanus in the S. cerevisiae × S. eubayanus hybrids (Krogerus et al., 2017b), S. kudriavzevii in the S. cerevisiae × S. kudriavzevii (Pfliegler et al., 2014; Lopandic et al., 2016) and S. kudriavzevii × S. uvarum hybrids (Karanyicz et al., 2017). The mechanisms underlying the unequal reduction of the subgenomes (concerted loss of chromosomes in one of the subgenomes) are unknown but a hypothetical model was recently proposed to explain the asymmetry (Karanyicz et al., 2017). The model is based on the assumption that the loss of a chromosome is unlikely to affect physically the segregation of other chromosomes. More plausibly, it creates a situation in which the subsequent loss of certain chromosomes of the other subgenome would have deleterious effects. For example, when the fertility is restored by the loss of the MAT-carrying chromosome of one of the subgenomes, all genes located on that chromosome are lost. The absence of these genes has no discernible effect on viability because their orthologs in the companion subgenome can substitute them. Most of the orthologs are in the MAT-carrying chromosome of the partner subgenome but some of them are on different, non-homeologous chromosomes [e.g., as shown in S. kudriavzevii × S. uvarum hybrids (Karanyicz et al., 2017)]. If one of these non-homeologous chromosomes is then lost during the next meiosis, the orthologues located on it will be eliminated from the genome. If any of them performs an essential function in the life of the cell, the spore lacking this chromosome will die. The dead spores in the F1 and F2 tetrads (Pfliegler et al., 2012; Karanyicz et al., 2017) can be the products of lethal combinations of chromosome losses in the subgenomes. The loss of another chromosome from the subgenome from which the MAT-carrying chromosome was lost is not entailed with similar risk but can render additional chromosomes of the partner subgenome essential for viability. Thus, the spores that lose chromosomes from the same subgenome have better chances to remain viable. Because of the differences in gene contents even between chromosomes considered homeologous, random losses of chromosomes are likely to eliminate essential genes and thus lead to non-functional genomes. Retaining mainly the chromosomes of one subgenome and reducing preferentially the other subgenome also prevents eventual clashes of the different regulatory networks of the hybridized species (e.g., Tirosh et al., 2010; Schraiber et al., 2013; Metzger et al., 2017). It is pertinent to mention here that structural and regulatory incompatibilities have been assumed to shape the highly diverse alloaneuploid genomes in the S. pastorianus lager strains (for a review, see Krogerus et al., 2017a). The question remains as to why the same subgenome loses its MAT chromosomes in different hybrids of the same species pair. A similar model can be proposed for the postzygotic genome evolution during the vegetative propagation of the hybrid cells.
Biotechnological Aspects
The combination of the gene pools of two or even more Saccharomyces species by natural mating of cells and the postzygotic chimerisation of the hybrid genomes by natural meiotic and mitotic segregation have great potentials in breeding of novel production strains for food industry. These processes generate a great diversity of new phenotypes without producing GMO strains with targeted genetic modifications. Both hybridisation and segregation are natural processes that take place in the nature without human intervention. The role of the human activity is to guide these processes in experimental conditions so that new phenotypes useful in fermentation and food industry can emerge.
The hybrids differ in phenotype from the parental species, but the differences can be both favorable and unfavorable. During the chimerisation process the nascent hybrid can get rid of “bad” genes or alleles, retain and combine their “good” counterparts and create new regulatory networks allowing better adaptation to technological environments. From the technological point of view, the allodiploid hybrids and the chimerised end products of the postzygotic genome evolution are better suited to the technological demands because their genomes are more stable than those of the intermediate forms (Figure 4). Examples of successful applications of these processes are shown in Tables 1–3, and certain major conclusions are discussed in the following sections.
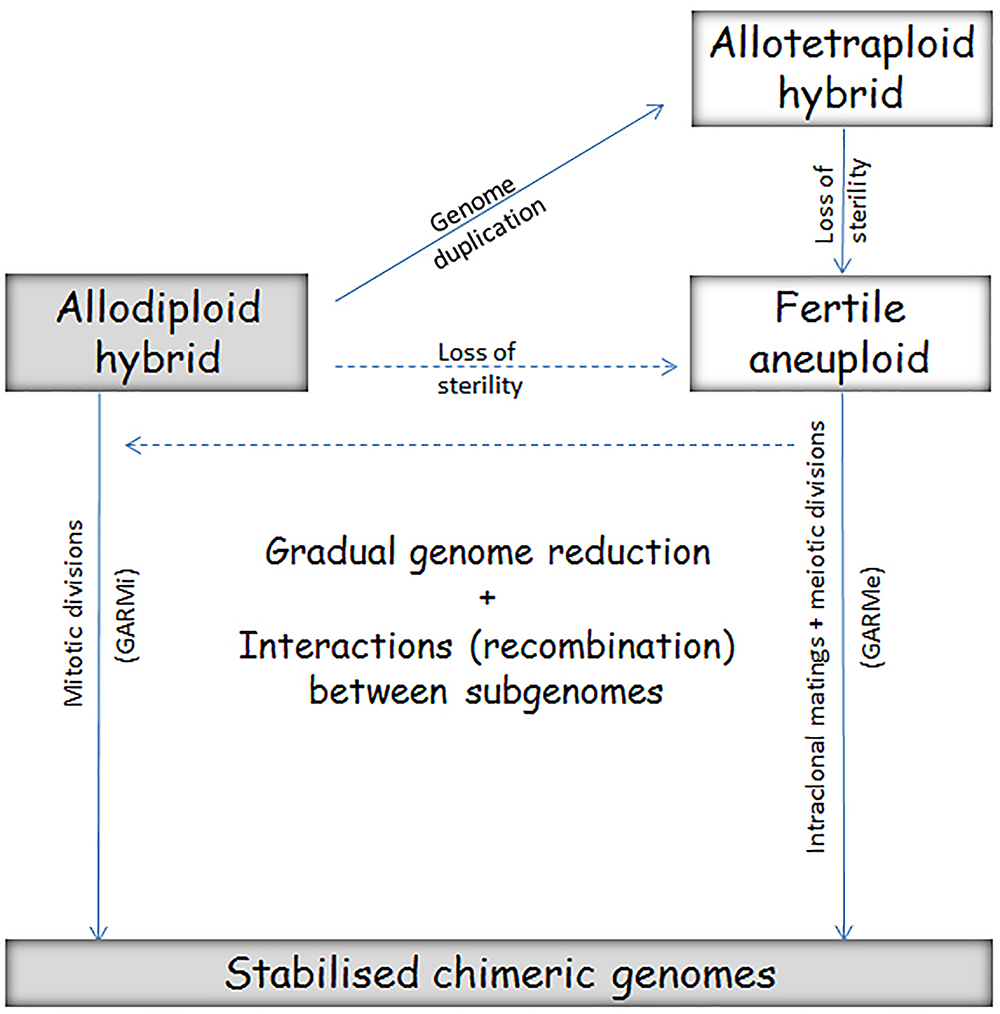
Figure 4. Two pathways of conversion of the interspecies hybrid into chimeric strains through genetically unstable intermediates. The conversion by serial biased mitotic segregation and recombination of chromosomes during vegetative propagation (GARMi) is exploited in the breeding strategy called adaptive evolution. As the outcomes are chimeric strains (“evolved hybrids”) that are genetically stable under the selective conditions that preferentially promoted their growth, the process is also called genetic stabilization of the hybrid. The alternative pathway leads to similar chimeric strains through biased segregation and recombination in a series of successive meiotic divisions (GARMe). The key events in this process are genome duplication, breakdown of sterility and autofertilisation (self-conjugation/mating) in the spore clones. Switching from one pathway to the other is possible (broken arrows). The fertile spores produced during GARMe can mate also with other fertile spores and cells of parental strains (a possibility for introgression). The allodiploid hybrids and the final chimeric strains are more suitable for biotechnological applications than the forms being in intermediary stages because the latter are less stable and may change unpredictably during propagation in the technology.
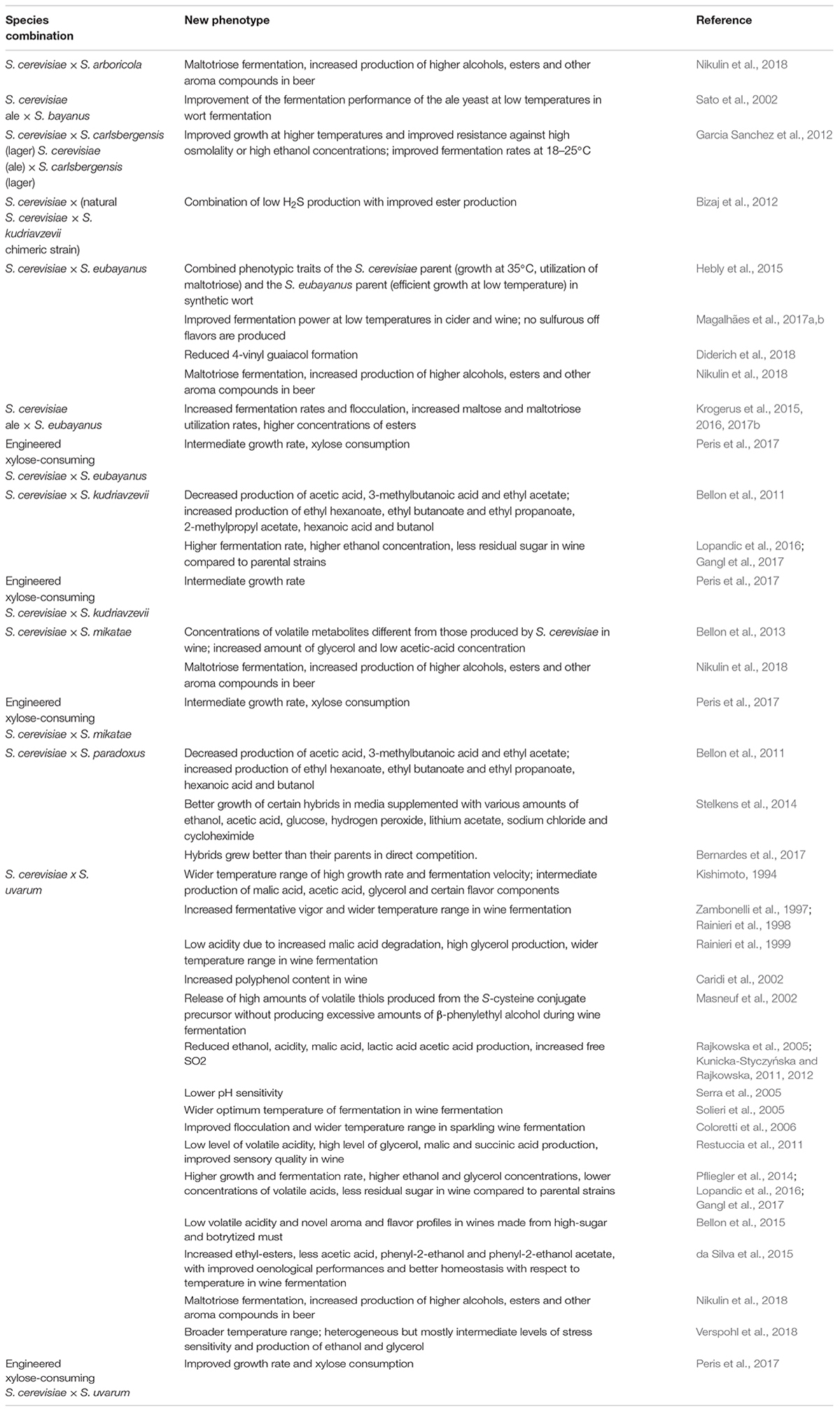
Hybridisation Generates Both Favorable and Unfavorable Phenotypes
Hybrids of different species usually show phenotypes, referred to as transgressive phenotypes that differ from those of their parents. Transgressive phenotypes can be products of additive effects of the orthologous parental genes, epistatic interactions and/or novel regulatory networks for biochemical processes not easily predicted from the properties of the parental strains. The transgressive phenotypes can be either positive or negative in terms of fitness or technological usefulness. While many of these phenotypes are of no obvious value for food industry, there are a number of documented cases where novel beneficial traits have appeared in the hybrids. These hybrids can outperform their parental strains in one or more properties of technological relevance. Such positive examples are listed in Table 1. The most frequently observed positive traits are faster fermentation rates in wider temperature ranges, more efficient sugar utilization, better stress tolerance, broader or better aroma profiles (increased complexity of the sensory properties), reduced production of acetic acid, SH2 and other compounds with adverse effects on the quality of the fermented products. Some of these traits confer a competitive fitness advantage to the hybrid cells in the fermentation environment. The positive transgression, the superior performance of the hybrid compared to either of its parent is frequently referred to as heterosis or hybrid vigor (e.g., Krogerus et al., 2017b). Negative transgressive phenotypes are rarely published but one must be aware of the risk of fitness reduction and the production of undesirable off-flavors (Steensels et al., 2014; Krogerus et al., 2016).
Hybridisation of the Same Parents Can Result in Diverse Hybrids
Hybrids of a pair of strains frequently show phenotypic diversity (e.g., Kunicka-Styczyńska and Rajkowska, 2011; Piotrowski et al., 2012; Lopandic et al., 2016). This phenomenon can be attributed to intragenomic heterogeneity of the parental strains. Wine and beer strains are usually highly heterozygous, frequently also aneuploid, and thus produce spores with diverse combinations of alleles and copy numbers of chromosomes (e.g., Johnston et al., 2000; Sipiczki et al., 2001, 2004; Steensels et al., 2014). Spores (spore clones) of different genomes form genetically and phenotypically different hybrids. Another reason for the heterogeneity can be the inherent instability of the hybrid genomes. If the hybrids of the same pair of parents are cultured for longer periods of time in non-selective laboratory media, they can become different due to random mitotic or meiotic segregation events as discussed above. Hybrid diversity reduces the predictability of the outcome of the breeding program but extends the range of phenotypes from which the breeder can choose.
The Good and Bad Sides of Sterility
The inability of the allodiploid hybrids to produce viable spores has two important consequences of practical relevance. On the one hand, it keeps the genome stabile by preventing meiotic segregation; on the other hand it reduces the survival chances of the hybrid under stress conditions. Since allodiploid meiosis is abortive, the hybrid genome can change only by mitotic segregation, which is a much slower process than meiotic segregation (see above). Genetic stability is particularly important in the brewing industry where a yeast culture is reused multiple times (for a review, see Gibson et al., 2017). In the brewing technology the negative side of hybrid sterility, the high mortality of hybrids under stress conditions does not cause problems because the environment does not change much. But in natural wine-making, the fermenting yeast populations are suddenly exposed to adverse conditions after the completion of fermentation and have to withstand multiple stresses until the next vintage season. Wine yeasts can survive these periods on the winery equipment (Rosini, 1984; Martini, 2003), in vineyard soil (Cordero-Bueso et al., 2011) or in mummified grape berries (Sipiczki, 2016), where the ability to produce ascospores greatly increases their chances of survival. Spores are not only gametes but also dormant resting cells resistant to many stress conditions deleterious to vegetative cells (Honigberg, 2016). The hybrids are easily selected out from the population because they have only vegetative cells that die under conditions which the spores of the natural strains withstand. The lack of viable spores is a severe disadvantage that may account (synergistically with other factors such as genetic instability) for the rare occurrence of true hybrids in the nature. In inoculated wine fermentation the sterility of the starter yeast strain is irrelevant.
Postzygotic Genomic Changes Broaden the Phenotypic Diversity
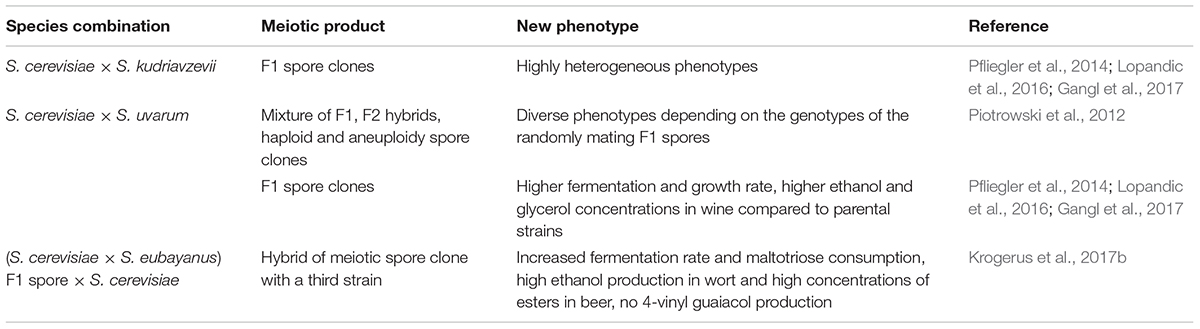
The inherent instability of interspecific yeast hybrids can be exploited for obtaining segregants (evolved hybrids) of chimeric genomes that display phenotypes outside the range of variation observed in the parents and the hybrids. The changes of the hybrid genome during the vegetative propagation of the cells (GARMi) are most probably spontaneous random events that take place independently of the culturing conditions. However, by culturing the population in a specific medium (e.g., high sugar content, low concentration of nitrogen sources) or under specific conditions (e.g., low or high temperature), the segregant having the best suited phenotype gradually overgrows the unchanged hybrid cells and the other segregants in the population (adaptive evolution). Examples of successful application of this experimental approach are listed in Table 2. Meiotic division also generates segregants with valuable novel phenotypes, provided that the hybrid can produce viable spores. Allotetraploids form viable spores but because of the autodiploidisation of the meiosis (see above) most spores are allodiploid having complete parental chromosomal sets. These are unlikely to show much difference in phenotype from the “ancestral” hybrid. But the alloaneuploid spores produced during GARMe can have chimerised genomes that express favorable novel phenotypes. Table 3 shows examples of strain improvement by generating spore clones from alloploid hybrids. The diversity can be further broadened by crossing the fertile spore clones with each other or with cells of different strains (e.g., Piotrowski et al., 2012). The drawback of strain improvement by meiotic segregation of the hybrids is the high instability of the fertile spore clones. Attempts have been made to obtain stabilized derivatives of spore clones by propagating them under technological conditions (e.g., Lopandic et al., 2016).
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This study was supported by the grant K-124417 provided by the National Research, Development and Innovation Office of Hungary.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, J., Puskas-Rozsa, S., Simlar, J., and Wilke, C. M. (1992). Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr. Genet. 22, 13–19. doi: 10.1007/BF00351736
Al Safadi, R., Weiss-Gayet, M., Briolay, J., and Aigle, M. (2010). A polyploid population of Saccharomyces cerevisiae with separate sexes (dioecy). FEMS Yeast Res. 10, 757–768. doi: 10.1111/j.1567-1364.2010.00660.x
Albertin, W., Chernova, M., Durrens, P., Guichoux, E., Sherman, D. J., Masneuf-Pomarede, I., et al. (2018). Many interspecific chromosomal introgressions are highly prevalent in holarctic Saccharomyces uvarum strains found in human-related fermentations. Yeast 35, 141–156. doi: 10.1002/yea.3248
Albertin, W., and Marullo, P. (2012). Polyploidy in fungi: evolution after whole-genome duplication. Proc. Biol. Sci. 279, 2497–2509. doi: 10.1098/rspb.2012.0434
Almeida, P., Gonçalves, C., Teixeira, S., Libkind, D., Bontrager, M., Masneuf-Pomarède, I., et al. (2014). A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5:5044. doi: 10.1038/ncomms5044
Anderson, E., and Hubricht, L. (1938). Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 25, 396–402. doi: 10.1002/j.1537-2197.1938.tb09237.x
Antunovics, Z., Nguyen, H. V., Gaillardin, C., and Sipiczki, M. (2005). Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1141–1150. doi: 10.1016/j.femsyr.2005.04.008
Bakalinsky, A. T., and Snow, R. (1990). The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6, 367–382. doi: 10.1002/yea.320060503
Baker, E., Wang, B., Bellora, N., Peris, D., Hulfachor, A. B., Koshalek, J. A., et al. (2015). The genome sequence of Saccharomyces eubayanus and the domestication of later brewing yeasts. Mol. Biol. Evol. 32, 2818–2831. doi: 10.1093/molbev/msv168
Banno, I., and Kaneko, Y. (1989). A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast 5,S373–S377.
Belloch, C., Pérez-Torrado, R., González, S. S., Pérez-Ortín, J. E., García-Martínez, J., Querol, A., et al. (2009). Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl. Environ. Microbiol. 75, 2534–2544. doi: 10.1128/AEM.02282-08
Bellon, J. R., Eglinton, J. M., Siebert, T. E., Pollnitz, A. P., Rose, L., de Barros Lopes, M., et al. (2011). Newly generated interspecific wine yeast hybrids introduce flavor and aroma diversity to wines. Appl. Microbiol. Biotechnol. 91, 603–612. doi: 10.1007/s00253-011-3294-3
Bellon, J. R., Ford, C. M., Borneman, A. R., and Chambers, P. J. (2018). A novel approach to isolating improved industrial interspecific wine yeasts using chromosomal mutations as potential markers for increased fitness. Front. Microbiol. 9:1442. doi: 10.3389/fmicb.2018.01442
Bellon, J. R., Schmid, F., Capone, D. L., Dunn, B. L., and Chambers, P. J. (2013). Introducing a new breed of wine yeast: interspecific hybridization between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae. PLoS One 8:e62053. doi: 10.1371/journal.pone.0062053
Bellon, J. R., Yang, F., Day, M. P., Inglis, D. L., and Chambers, P. J. (2015). Designing and creating Saccharomyces interspecific hybrids for improved, industry relevant, phenotypes. Appl. Microbiol. Biotechnol. 99, 8597–8609. doi: 10.1007/s00253-015-6737-4
Bernardes, J. P., Stelkens, R. B., and Greig, D. (2017). Heterosis in hybrids within and between yeast species. J. Evol. Biol. 30, 538–548. doi: 10.1111/jeb.13023
Bidenne, C., Blondin, B., Dequin, S., and Vezinhet, F. (1992). Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr. Genet. 22, 1–7. doi: 10.1007/BF00351734
Bisson, L. F. (2017). Yeast hybrids in winemaking. Catalyst 1, 27–34. doi: 10.5344/catalyst.2016.16001
Bizaj, E., Cordente, A. G., Bellon, J. R., Raspor, P., Curtin, C. D., and Pretorius, I. S. (2012). A breeding strategy to harness flavor diversity of Saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res. 12, 456–465. doi: 10.1111/j.1567-1364.2012.00797.x
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2012). The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 12, 88–96. doi: 10.1111/j.1567-1364.2011.00773.x
Borneman, A. R., and Pretorius, I. S. (2015). Genomic insights into the Saccharomyces sensu stricto complex. Genetics 199, 281–291. doi: 10.1534/genetics.114.173633
Boynton, P. J., and Greig, D. (2014). The ecology and evolution of non-domesticated Saccharomyces species. Yeast 31, 449–462.
Boynton, P. J., Janzen, T., and Greig, D. (2018). Modeling the contributions of chromosome segregation errors and aneuploidy to Saccharomyces hybrid sterility. Yeast 35, 85–98. doi: 10.1002/yea.3282
Caridi, A., Cufari, A., and Ramondino, D. (2002). Winemaking from Gaglioppo grapes with hybrid strains of Saccharomyces. Folia Microbiol. 47, 407–408. doi: 10.1007/BF02818698
Chou, J. Y., Hung, Y. S., Lin, K. H., Lee, H. Y., and Leu, J. Y. (2010). Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 8:e1000432. doi: 10.1371/journal.pbio.1000432
Coloretti, F., Zambonelli, C., and Tini, V. (2006). Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol. 23, 672–676. doi: 10.1016/j.fm.2005.11.002
Cordero-Bueso, G., Arroyo, T., Serrano, A., Tello, J., Aporta, I., Vélez, M. D., et al. (2011). Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 145, 132–139. doi: 10.1016/j.ijfoodmicro.2010.11.040
Csoma, H., Zakany, N., Capece, A., Romano, P., and Sipiczki, M. (2010). Biological diversity of Saccharomyces yeasts of spontaneously fermenting wines in four wine regions: comparative genotypic and phenotypic analysis. Int. J. Food Microbiol. 140, 239–248. doi: 10.1016/j.ijfoodmicro.2010.03.024
Cummings, J., and Fogel, S. (1978). Genetic homology of wine yeasts with Saccharomyces cerevisiae. J. Inst. Brew. 84, 267–270. doi: 10.1002/j.2050-0416.1978.tb03885.x
da Silva, T., Albertin, W., Dillmann, C., Bely, M., la Guerche, S., Giraud, C., et al. (2015). Hybridization within Saccharomyces genus results in homoeostasis and phenotypic novelty in winemaking conditions. PLoS One 10:e0123834. doi: 10.1371/journal.pone.0123834
de Barros Lopes, M., Bellon, J. R., Shirly, N. J., and Ganter, P. F. (2002). Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1, 323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x
Delneri, D., Colson, I., Grammenoudi, S., Roberts, I. N., Louis, E. J., and Oliver, S. G. (2003). Engineering evolution to study speciation in yeasts. Nature 422, 68–72. doi: 10.1038/nature01418
Diderich, J. A., Weening, S. M., van den Broek, M., Pronk, J. T., and Daran, J. G. (2018). Selection of Pof- Saccharomyces eubayanus variants for the construction of S. cerevisiae × S. eubayanus hybrids with reduced 4-vinyl guaiacol formation. Front. Microbiol. 9:1640. doi: 10.3389/fmicb.2018.01640
Dujon, B. A., and Louis, E. J. (2017). Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 206, 717–750. doi: 10.1534/genetics.116.199216
Dunn, B., Paulish, T., Stanbery, A., Piotrowski, J., Koniges, G., Kroll, E., et al. (2013). Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgressions. PLoS Genet. 9:e1003366. doi: 10.1371/journal.pgen.1003366
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908–924. doi: 10.1101/gr.130310.111
Dunn, B., and Sherlock, G. (2008). Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 18, 1610–1623. doi: 10.1101/gr.076075.108
Fischer, G., James, S. A., Roberts, I. N., Oliver, S. G., and Louis, E. J. (2000). Chromosomal evolution in Saccharomyces. Nature 405, 451–454. doi: 10.1038/35013058
Gangadevi, T., Rao, P. N., Rao, B. H., and Satyanarayana, K. V. (1985). A study of morphology, cytology and sterility in interspecific hybrids and amphidiploids of Nicotiana knightiana X N. umbratica. Theor. Appl. Genet. 70, 330–332. doi: 10.1007/BF00304921
Gangl, H., Tiefenbrunner, W., Pfliegler, W. P., Sipiczki, M., Leitner, G., Tscheik, G., et al. (2017). Influence of artificial interspecies yeast hybrids and their F1 offspring on the aroma profile of wine. Mitt. Klosterneuburg 67, 68–83.
Garcia Sanchez, R., Solodovnikova, N., and Wendland, J. (2012). Breeding of lager yeast with Saccharomyces cerevisiae improves stress resistance and fermentation performance. Yeast 29, 343–355. doi: 10.1002/yea.2914
Gibson, B., Geertman, J. A., Hittinger, C. T., Krogerus, K., Libkind, D., Louis, E. J., et al. (2017). New yeasts-new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 17:fox038. doi: 10.1093/femsyr/fox038
Gjermansen, C., and Sigsgaard, P. (1981). Construction of a hybrid brewing strain of Saccharomyces carlsbergensis by mating of meiotic segregants. Carlsberg Res. Commun. 46, 1–11. doi: 10.1007/BF02906193
Greig, D. (2007). A screen for recessive speciation genes expressed in the gametes of F1 hybrid yeast. PLoS Genet. 3:e21. doi: 10.1371/journal.pgen.0030021
Greig, D., Borts, R. H., Louis, E. J., and Travisano, M. (2002a). Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. B 269, 1167–1171. doi: 10.1098/rspb.2002.1989
Greig, D., Louis, E. J., Borts, R. H., and Travisano, M. (2002b). Hybrid speciation in experimental populations of yeast. Science 298, 1773–1775. doi: 10.1126/science.1076374
Groth, C., Hansen, J., and Piskur, J. (1999). A natural chimeric yeast containing genetic material from three species. Int. J. Syst. Bacteriol. 4, 1933–1938. doi: 10.1099/00207713-49-4-1933
Guillamón, J. M., and Barrio, E. (2017). Genetic polymorphism in wine yeasts: mechanisms and methods for its detection. Front. Microbiol. 8:806. doi: 10.3389/fmicb.2017.00806
Gunge, N., and Nakatomi, Y. (1972). Genetic mechanisms of rare matings of the yeast Saccharomyces cerevisiae heterozygous for mating type. Genetics 70, 41–58.
Hawthorne, D., and Philippsen, P. (1994). Genetic and molecular analysis of hybrids in the genus Saccharomyces involving S. cerevisiae, S. uvarum and a new species, S. douglasii. Yeast 10, 1285–1296. doi: 10.1002/yea.320101005
Hebly, M., Brickwedde, A., Bolat, I., Driessen, M. R., de Hulster, E. A., van den Broek, M., et al. (2015). S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15:fov005. doi: 10.1093/femsyr/fov005
Herskowitz, I. (1988). Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52, 536–553.
Hilger, F. (1973). Construction and analysis of tetraploid yeast sets for gene dosage studies. J. Microbiol. 75, 23–31. doi: 10.1099/00221287-75-1-23
Hittinger, C. T., Rokas, A., Bai, F. Y., Boekhout, T., Gonçalves, P., Jeffries, T. W., et al. (2015). Genomics and the making of yeast biodiversity. Curr. Opin. Genet. Dev. 35, 100–109. doi: 10.1016/j.gde.2015.10.008
Honigberg, S. M. (2016). Similar environments but diverse fates: responses of budding yeast to nutrient deprivation. Microb. Cell 3, 302–328. doi: 10.15698/mic2016.08.516
Hou, J., Fournier, T., and Schacherer, J. (2016). Species-wide survey reveals the various flavors of intraspecific reproductive isolation in yeast. FEMS Yeast Res. 16:fow048. doi: 10.1093/femsyr/fow048
Hunter, N., Chambers, S. R., Louis, E. J., and Borts, R. H. (1996). The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15, 1726–1733. doi: 10.1002/j.1460-2075.1996.tb00518.x
Hutchinson, E. S., Price, S. C., Kahier, A. L., Morris, M. I., and Allard, R. W. (1983). An experimental verification of segregation theory in a diploidized tetraploild: esterase loci in Avena barbata. J. Hered. 74, 381–383. doi: 10.1093/oxfordjournals.jhered.a109816
Jansen, A. M., Hall, L. J., Clare, S., Goulding, D., Holt, K. E., Grant, A. J., et al. (2011). A Salmonella typhimurium-typhi genomic chimera: a model to study Vi polysaccharide capsule function in vivo. PLoS Pathog. 7:e1002131. doi: 10.1371/journal.ppat.1002131
John, B., and Weissman, D. B. (1977). Cytogenetic components of reproductive isolation in Trimerotropis thalassica and T. occidentalis. Chromosoma 60, 187–203. doi: 10.1007/BF00288465
Johnston, J. R., Baccari, C., and Mortimer, R. K. (2000). Genotypic characterization of strains of commercial wine yeasts by tetrad analysis. Res. Microbiol. 151, 583–590. doi: 10.1016/S0923-2508(00)00228-X
Johnston, J. R., and Mortimer, R. K. (1986). Electrophoretic karyotyping of laboratory and commercial strains of Saccharomyces and other yeasts. Int. J. Syst. Bacteriol. 36, 569–572. doi: 10.1099/00207713-36-4-569
Kao, K. C., Schwartz, K., and Sherlock, G. (2010). A genome-wide analysis reveals no nuclear Dobzhansky-Muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS Genet. 6:e1001038. doi: 10.1371/journal.pgen.1001038
Karanyicz, E., Antunovics, Z., Kallai, Z., and Sipiczki, M. (2017). Non-introgressive genome chimerisation by malsegregation in autodiploidised allotetraploids during meiosis of Saccharomyces kudriavzevii x Saccharomyces uvarum hybrids. Appl. Microbiol. Biotechnol. 101, 4617–4633. doi: 10.1007/s00253-017-8274-9
Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E. S. (2003). Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254. doi: 10.1038/nature01644
Kishimoto, M. (1994). Fermentation Characteristics of hybrids between the cryophilic wine yeast Saccharomyces bayanus and the mesophilic wine yeast Saccharomyces cerevisiae. J. Ferment. Bioeng. 77, 432–435. doi: 10.1016/0922-338X(94)90019-1
Kishimoto, M., and Goto, S. (1995). Growth temperatures and electrophoretic karyotyping as tools for practical discrimination of Saccharomyces bayanus and Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 41, 239–247. doi: 10.2323/jgam.41.239
Krogerus, K., Arvas, M., De Chiara, M., Magalhães, F., Mattinen, L., Oja, M., et al. (2016). Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl. Microbiol. Biotechnol. 100, 7203–7222. doi: 10.1007/s00253-016-7588-3
Krogerus, K., Magalhães, F., Vidgren, V., and Gibson, B. (2015). New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 42, 769–778. doi: 10.1007/s10295-015-1597-6
Krogerus, K., Magalhães, F., Vidgren, V., and Gibson, B. (2017a). Novel brewing yeast hybrids: creation and application. Appl. Microbiol. Biotechnol. 101, 65–78. doi: 10.1007/s00253-016-8007-5
Krogerus, K., Seppänen-Laakso, T., Castillo, S., and Gibson, B. (2017b). Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb. Cell Fact. 16:66. doi: 10.1186/s12934-017-0679-8
Kumaran, R., Yang, S. Y., and Leu, J. Y. (2013). Characterization of chromosome stability in diploid, polyploid and hybrid yeast cells. PLoS One 8:e68094. doi: 10.1371/journal.pone.0068094
Kunicka-Styczyńska, A., and Rajkowska, K. (2011). Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J. Appl. Microbiol. 110, 1538–1549. doi: 10.1111/j.1365-2672.2011.05009.x
Kunicka-Styczyńska, A., and Rajkowska, K. (2012). Fermentative stability of wine yeast Saccharomyces sensu stricto complex and their hybrids. Food Technol. Biotechnol. 50, 222–229. doi: 10.1111/j.1365-2672.2011.05009.x
Leducq, J. B., Nielly-Thibault, L., Charron, G., Eberlein, C., Verta, J. P., Samani, P., et al. (2016). Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat. Microbiol. 1:15003. doi: 10.1038/nmicrobiol.2015.3
Lee, H., Chou, J., Cheong, L., Chang, N. H., Yang, S. Y., and Leu, J. Y. (2008). Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073. doi: 10.1016/j.cell.2008.10.047
Legras, J. L., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Li, C., Wang, Z., and Zhang, J. (2013). Toward genome-wide identification of Bateson-Dobzhansky-Muller incompatibilities in yeast: a simulation study. Genome Biol. Evol. 5, 1261–1272. doi: 10.1093/gbe/evt091
Libkind, D., Hittinger, C. T., Valério, E., Gonçalves, C., Dover, J., Johnston, M., et al. (2011). Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 14539–14544. doi: 10.1073/pnas.1105430108
Liti, G., Ba, A. N. N., Blythe, M., Müller, C. A., Bergström, A., Cubillos, F. A., et al. (2013). High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome. BMC Genomics 14:69. doi: 10.1186/1471-2164-14-69
Liti, G., Barton, D. B., and Louis, E. J. (2006). Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850. doi: 10.1534/genetics.106.062166
Lopandic, K. (2018). Saccharomyces interspecies hybrids as model organisms for studying yeast adaptation to stressful environments. Yeast 35, 21–38. doi: 10.1002/yea.3294
Lopandic, K., Pfliegler, W., Tiefenbrunner, W., Gangl, H., Sipiczki, M., and Sterflinger, K. (2016). Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation. Appl. Microbiol. Biotechnol. 100,6331–6343. doi: 10.1007/s00253-016-7481-0
Lorenz, A., Fuchs, J., Trelles-Sticken, E., Scherthan, H., and Loidl, J. (2002). Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae x S. paradoxus hybrids. J. Cell Sci. 115, 3829–3835. doi: 10.1242/jcs.00066
Louis, E. J. (2011). Population genomics and speciation in yeasts. Fungal Biol. Rev. 25, 136–142. doi: 10.1016/j.fbr.2011.06.001
Luo, J., Sun, X., Cormack, B. P., and Boeke, J. D. (2018). Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 560, 392–396. doi: 10.1038/s41586-018-0374-x
Magalhães, F., Krogerus, K., Castillo, S., Ortiz-Julien, A., Dequin, S., and Gibson, B. (2017a). Exploring the potential of Saccharomyces eubayanus as a parent for new interspecies hybrid strains in winemaking. FEMS Yeast Res. 17:fox049. doi: 10.1093/femsyr/fox049
Magalhães, F., Krogerus, K., Vidgren, V., Sandell, M., and Gibson, B. (2017b). Improved cider fermentation performance and quality with newly generated Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. J. Ind. Microbiol. Biotechnol. 44, 1203–1213. doi: 10.1007/s10295-017-1947-7
Marinoni, G., Manuel, M., Petersen, R. F., Hvidtfeldt, J., Sulo, P., and Piskur, J. (1999). Horizontal transfer of genetic material among Saccharomyces yeasts. J. Bacteriol. 181, 6488–6496.
Marsit, S., and Dequin, S. (2015). Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res. 15:fov067. doi: 10.1093/femsyr/fov067
Martini, A. (2003). Biotechnology of natural and winery-associated strains of Saccharomyces cerevisiae. Int. Microbiol. 6, 207–209. doi: 10.1007/s10123-003-0135-y
Masneuf, I., Hansen, J., Groth, C., Piškur, J., and Dubourdieu, D. (1998). New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64, 3887–3892.
Masneuf, I., Murat, M. L., Naumov, G. I., Tominaga, T., and Dubourdieu, D. (2002). Hybrids Saccharomyces cerevisiae x Saccharomyces bayanus var. uvarum having a high liberating ability of some sulfur varietal aromas of Vitis vinifera Sauvignon blanc wines. J. Int. Sci. Vigne Vin 36, 205–212.
Mertens, S., Steensels, J., Saels, V., De Rouck, G., Aerts, G., and Verstrepen, K. J. (2015). A large set of new created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 81, 8202–8214. doi: 10.1128/AEM.02464-15
Metzger, B. P. H., Wittkopp, P. J., and Coolon, J. D. (2017). Evolutionary dynamics of regulatory changes underlying gene expression divergence among Saccharomyces species. Genome Biol. Evol. 9, 843–854. doi: 10.1093/gbe/evx035
Miklos, I., Varga, T., Nagy, A., and Sipiczki, M. (1997). Genome instability and chromosomal rearrangements in a heterothallic wine yeast. J. Basic Microbiol. 37, 345–354. doi: 10.1002/jobm.3620370507
Moore, D., Robson, G. D., and Trinci, A. P. J. (2011). 21st Century Guidebook to Fungi. Cambridge: Cambridge University Press. doi: 10.1017/CBO9780511977022
Moore, G. (2002). Meiosis in allopolyploids – the importance of ‘Teflon’ chromosomes. Trends Genet. 18, 456–463. doi: 10.1016/S0168-9525(02)02730-0
Morales, L., and Dujon, B. (2012). Evolutionary role of interspecies hybridization and genetic exchanges in yeast. Microbiol. Mol. Biol. Rev. 76, 721–739. doi: 10.1128/MMBR.00022-12
Mortimer, R. K., Romano, P., Suzzi, G., and Polsinelli, M. (1994). Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10, 1543–1552. doi: 10.1002/yea.320101203
Muller, L. A., and McCusker, J. H. (2009). A multispecies-based taxonomic microarray reveals interspecies hybridization and introgression in Saccharomyces cerevisiae. FEMS Yeast Res. 9, 143–152. doi: 10.1111/j.1567-1364.2008.00464.x
Nakao, Y., Kanamori, T., Itoh, T., Kodama, Y., Rainieri, S., Nakamura, N., et al. (2009). Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 16, 115–129. doi: 10.1093/dnares/dsp003
Naumov, G. I. (1987). Genetic basis for classification and identification of the ascomycetous yeasts. Stud. Mycol. 30, 469–475.
Naumov, G. I., Naumova, E. S., Masneuf, I., Aigle, M., Kondratieva, V. I., and Dubourdieu, D. (2000). Natural polyploidisation of some cultured yeast Saccharomyces sensu stricto: auto- and allotetraploidy. Syst. Appl. Microbiol. 23, 442–449. doi: 10.1016/S0723-2020(00)80076-4
Nguyen, H. V., and Boekhout, T. (2017). Characterization of Saccharomyces uvarum (Beijerinck, 1898) and related hybrids: assessment of molecular markers that predict the parent and hybrid genomes and a proposal to name yeast hybrids. FEMS Yeast Res. 17:fox014. doi: 10.1093/femsyr/fox014
Nguyen, H. V., and Gaillardin, C. (2005). Evolutionary relationships between the former species Saccharomyces uvarum and the hybrids Saccharomyces bayanus and Saccharomyces pastorianus; reinstatement of Saccharomyces uvarum (Beijerinck) as a distinct species. FEMS Yeast Res. 5, 471–483. doi: 10.1016/j.femsyr.2004.12.004
Nguyen, H. V., Legras, J., Neuvéglise, C., and Gaillardin, C. (2011). Deciphering the hybridisation history leading to the lager lineage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380. PLoS One 6:e25821. doi: 10.1371/journal.pone.0025821
Nikulin, J., Krogerus, K., and Gibson, B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. doi: 10.1002/yea.3246
Origone, A. C., Rodríguez, M. E., Oteiza, J. M., Querol, A., and Lopes, C. A. (2018). Saccharomyces cerevisiae × Saccharomyces uvarum hybrids generated under different conditions share similar winemaking features. Yeast 35, 157–171. doi: 10.1002/yea.3295
Parry, E. M., and Cox, B. S. (1970). The tolerance of aneuploidy in yeast. Genet. Res. 16, 333–340. doi: 10.1017/S0016672300002597
Parry, J. M., and Zimmerman, F. K. (1976). The detection of monosomic colonies produced by mitotic chromosome non-disjunction in the yeast Saccharomyces cerevisiae. Mutat. Res. 36, 49–66. doi: 10.1016/0027-5107(76)90020-8
Pérez-Través, L., Lopes, C. A., Barrio, E., and Querol, A. (2014a). Stabilization process in Saccharomyces intra and interspecific hybrids in fermentative conditions. Int. Microbiol. 17, 213–224. doi: 10.2436/20.1501.01.224
Pérez-Través, L., Lopes, C. A., Querol, A., and Barrio, E. (2014b). On the complexity of the Saccharomyces bayanus taxon: hybridization and potential hybrid speciation. PLoS One 9:e93729. doi: 10.1371/journal.pone.0093729
Pérez-Través, L., Lopes, C. A., Barrio, E., and Querol, A. (2012). Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int. J. Food Microbiol. 156, 102–111. doi: 10.1016/j.ijfoodmicro.2012.03.008
Peris, D., Arias, A., Orlić, S., Belloch, C., Pérez-Través, L., Querol, A., et al. (2017). Mitochondrial introgression suggests extensive ancestral hybridization events among Saccharomyces species. Mol. Phylogenet. Evol. 108, 49–60. doi: 10.1016/j.ympev.2017.02.008
Pfliegler, W. P., Antunovics, Z., and Sipiczki, M. (2012). Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res. 12, 703–718. doi: 10.1111/j.1567-1364.2012.00820.x
Pfliegler, W. P., Atanasova, L., Karanyicz, E., Sipiczki, M., Bond, U., Druzhinina, I. S., et al. (2014). Generation of new genotypic and phenotypic features in artificial and natural hybrids. Food Technol. Biotechnol. 52, 46–57.
Piotrowski, J. S., Nagarajan, S., Kroll, E., Stanbery, A., Chiotti, K. E., Kruckeberg, A. L., et al. (2012). Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol. Biol. 12:46. doi: 10.1186/1471-2148-12-46
Pryszcz, L. P., Nemeth, T., Gacser, A., and Gabaldon, T. (2014). Genome comparison of Candida orthopsilosis clinical strains reveals the existence of hybrids between two distinct subspecies. Genome Biol. Evol. 6, 1069–1078. doi: 10.1093/gbe/evu082
Rainieri, S., Giudici, P., and Zambonelli, C. (1998). Oenological properties of Saccharomyces bayanus and Saccharomyces cerevisiae interspecific hybrids. Food Technol. Biotechnol. 36, 51–53.
Rainieri, S., Kodama, Y., Kaneko, Y., Mikata, K., Nakao, Y., and Ashikari, T. (2006). Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 72, 3968–3974. doi: 10.1128/AEM.02769-05
Rainieri, S., Zambonelli, C., and Kaneko, Y. (2003). Saccharomyces sensu stricto: systematics, genetic diversity and evolution. J. Biosci. Bioeng. 96, 1–9. doi: 10.1016/S1389-1723(03)90089-2
Rainieri, S., Zambonelli, C., Tini, V., Castellari, L., and Giudici, P. (1999). Oenological properties of an interspecific Saccharomyces hybrid. S. Afr. J. Enol. Vitie. 20, 47–52.
Rajkowska, K., Kunicka, A., Cebula, B., Robak, T., and Smolewski, P. (2005). Characterization of wine yeast hybrids. Folia Univ. Agric. Stetin Sci. Aliment. 246, 241–254.
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639. doi: 10.1146/annurev.ecolsys.33.010802.150437
Restuccia, C., Muratore, G., Muccilli, S., Randazzo, C. L., Caggia, C., Mazzaglia, A., et al. (2011). Saccharomyces hybrids as a tool for improving the quality of moscato Di siracusa DOC wine. Ital. J. Food Sci. 23, 28–35.
Rosini, G. (1984). Assessment of dominance of added yeast in wine fermentation and origin of Saccharomyces cerevisiae in wine-making. J. Gen. Appl. Microbiol. 30, 249–256. doi: 10.2323/jgam.30.249
Ryu, S. L., Murooka, Y., and Kaneko, Y. (1998). Reciprocal translocation at duplicated RPL2 loci might cause speciation of Saccharomyces bayanus and Saccharomyces cerevisiae. Curr. Genet. 33, 345–351. doi: 10.1007/s002940050346
Sanchez, M. R., Miller, A. W., Liachko, I., Sunshine, A. B., Lynch, B., Huang, M., et al. (2017). Differential paralog divergence modulates genome evolution across yeast species. PLoS Genet. 13:e1006585. doi: 10.1371/journal.pgen.1006585
Sansam, C. L., and Pezza, R. J. (2015). Connecting by breaking and repairing: mechanisms of DNA strand exchange in meiotic recombination. FEBS J. 282, 2444–2457. doi: 10.1111/febs.13317
Sato, M., Kishimoto, M., Watari, J., and Takashio, M. (2002). Breeding of brewer’s yeast by hybridisation between a top-fermenting yeast Saccharomyces cerevisiae and a cryophilic yeast Saccharomyces bayanus. J. Biosci. Bioeng. 93, 509–511. doi: 10.1016/S1389-1723(02)80101-3
Scannell, D. R., Zill, O. A., Rokas, A., Payen, C., Dunham, M. J., Eisen, M. B., et al. (2011). The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1, 11–25. doi: 10.1534/g3.111.000273
Schraiber, J. G., Mostovoy, Y., Hsu, T. Y., and Brem, R. B. (2013). Inferring evolutionary histories of pathway regulation from transcriptional profiling data. PLoS Comput. Biol. 9:e1003255. doi: 10.1371/journal.pcbi.1003255
Sebastiani, F., Barberio, C., Casalone, E., Cavalieri, D., and Polsinelli, M. (2002). Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res. Microbiol. 153, 53–58. doi: 10.1016/S0923-2508(01)01286-4
Serra, A., Strehaiano, P., and Taillandier, P. (2005). Influence of temperature and pH on Saccharomyces bayanus var. uvarum growth: impact of a wine yeast interspecific hybridization on these parameters. Int. J. Food Microbiol. 104, 257–265. doi: 10.1016/j.ijfoodmicro.2005.03.006
Sipiczki, M. (2002). “Taxonomic and physiological diversity of Saccharomyces bayanus,” in Biodiversity and Biotechnology of Wine Yeasts, ed. M. Ciani (Kerala: Research Signpost), 53–69.
Sipiczki, M. (2008). Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 8, 996–1007. doi: 10.1111/j.1567-1364.2008.00369.x
Sipiczki, M. (2011). Diversity, variability and fast adaptive evolution of the wine yeast (Saccharomyces cerevisiae) genome - a review. Ann. Microbiol. 61, 85–93. doi: 10.1007/s13213-010-0086-4
Sipiczki, M. (2016). Overwintering of vineyard yeasts: survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 7:212. doi: 10.3389/fmicb.2016.00212
Sipiczki, M., Romano, P., Capece, A., and Paraggio, M. (2004). Genetic segregation of natural Saccharomyces cerevisiae strains derived from spontaneous fermentation of Aglianico wine. J. Appl. Microbiol. 96, 1169–1175. doi: 10.1111/j.1365-2672.2004.02254.x
Sipiczki, M., Romano, P., Lipani, G., Miklos, I., and Antunovics, Z. (2001). Analysis of yeasts derived from natural fermentation in a Tokaj winery. Antonie Van Leeuwenhoek 79, 97–105. doi: 10.1023/A:1010249408975
Smukowski Heil, C. S., DeSevo, C. G., Pai, D. A., Tucker, C. M., Hoang, M. L., and Dunham, M. J. (2017). Loss of heterozygosity drives adaptation in hybrid yeast. Mol. Biol. Evol. 34, 1596–1612. doi: 10.1093/molbev/msx098
Solieri, L., Gullo, M., De Vero, L., Antúnez, O., Pérez-Ortín, J. E., and Giudici, P. (2005). Homogeneity of interspecific hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum by phenotypic and transcriptional analysis. Int. J. Biotechnol. Biochem. 1, 11–21.
Špírek, M., Poláková, S., Jatzová, K., and Sulo, P. (2014). Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5:454. doi: 10.3389/fgene.2014.00454
Steensels, J., Meersman, E., Snock, T., Saels, V., and Verstrepen, K. (2014). Large scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975. doi: 10.1128/AEM.02235-14
Stelkens, R. B., Brockhurst, M. A., Hurst, G. D., Miller, E. L., and Greig, D. (2014). The effect of hybrid transgression on environmental tolerance in experimental yeast crosses. J. Evol. Biol. 27, 2507–2519. doi: 10.1111/jeb.12494
Sulo, P., Szaboova, D., Bielik, P., Polakova, S., Soltys, K., Jatzova, K., et al. (2017). The evolutionary history of Saccharomyces species inferred from completed mitochondrial genomes and revision in the ’yeast mitochondrial genetic code’. DNA Res. 24, 571–583. doi: 10.1093/dnares/dsx026
Swain Lenz, D., Riles, L., and Fay, J. C. (2014). Heterochronic meiotic misexpression in an interspecific yeast hybrid. Mol. Biol. Evol. 31, 1333–1342. doi: 10.1093/molbev/msu098
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Tilakaratna, V., and Bensasson, D. (2017). Habitat predicts levels of genetic admixture in Saccharomyces cerevisiae. G3 7, 2919–2929. doi: 10.1534/g3.117.041806
Tirosh, I., Sigal, N., and Barkai, N. (2010). Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol. Syst. Biol. 6:365. doi: 10.1038/msb.2010.20
Tuite, M. F., and Oliver, G. O. (1991). Saccharomyces. Biotechnology Handbooks, Vol. 4. New York, NY: Springer. doi: 10.1007/978-1-4899-2641-8
Van den Broek, M., Bolat, I., Nijkamp, J., Ramos, E., Luttik, M., Koopman, F., et al. (2015). Chromosomal copy number variation in Saccharomyces pastorianus evidence for extensive genome dynamics in industrial lager brewing strains. Appl. Environ. Microbiol. 81, 6253–6267. doi: 10.1128/AEM.01263-15
van der Walt, J. P. (1970). “Saccharomyces (Meyen) emend. Reess,” in The Yeasts, A Taxonomic Study, ed. J. Lodder (Amsterdam: North-Holland), 555–718.
Vaughan-Martini, A., and Kurtzman, C. P. (1985). Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int. J. Syst. Bacteriol. 35, 508–511. doi: 10.1099/00207713-35-4-508
Vaughan-Martini, A., and Martini, A. (2011). “Saccharomyces meyen ex Reess (1870),” in The Yeasts, A Taxonomic Study, ed. J. Lodder (Amsterdam: North-Holland), 733–746.
Verspohl, A., Pignedoli, S., and Giudici, P. (2018). The inheritance of mitochondrial DNA in interspecific Saccharomyces hybrids and their properties in winemaking. Yeast 35, 173–187. doi: 10.1002/yea.3288
Vu, D., Groenewald, M., Szoke, S., Cardinali, G., Eberhardt, U., Stielow, B., et al. (2016). DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Walters, M. S. (1958). Aberrant chromosome movement and spindle formation in meiosis of Bromus hybrids: an interpretation of spindle organization. Am. J. Bot. 45, 271–289. doi: 10.1002/j.1537-2197.1958.tb13127.x
Walther, A., Hesselbart, A., and Wendland, J. (2014). Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 4, 783–793. doi: 10.1534/g3.113.010090
Watanabe, S., Shiwa, Y., Itaya, M., and Yoshikawa, H. (2012). Complete sequence of the first chimera genome constructed by cloning the whole genome of Synechocystis strain PCC6803 into the Bacillus subtilis 168 genome. J. Bacteriol. 194:7007. doi: 10.1128/JB.01798-12
Xu, M., and He, X. (2011). Genetic incompatibility dampens hybrid fertility more than hybrid viability: yeast as a case study. PLoS One 6:e18341. doi: 10.1371/journal.pone.0018341
Yamamoto, N., Amemiya, H., Yokomori, Y., Shimizu, K., and Totosuka, A. (1991). Electrophoretic karyotypes of wine yeasts. Am. J. Enol. Vitic. 42, 358–363.
Zambonelli, C., Passarelli, P., Rainieri, S., Bertolini, L., Giudici, P., and Castellari, L. (1997). Technological properties and temperature response of interspecific Saccharomyces hybrids. J. Sci. Food Agric. 74, 7–12. doi: 10.1002/(SICI)1097-0010(199705)74:1<7::AID-JSFA753>3.0.CO;2-X
Zambonelli, C., Passarelli, P., Rainieri, S., and Giudici, P. (1993). Taxonomic and technological implications of sterility in hybrids from cryotolerant and non cryotolerant Saccharomyces strains. Ann. Microbiol. 43, 217–223.
Keywords: interspecies hybridisation, sterility, alloploid, meiosis, genome chimerisation, strain improvement, fermentation, yeast
Citation: Sipiczki M (2018) Interspecies Hybridisation and Genome Chimerisation in Saccharomyces: Combining of Gene Pools of Species and Its Biotechnological Perspectives. Front. Microbiol. 9:3071. doi: 10.3389/fmicb.2018.03071
Received: 12 September 2018; Accepted: 28 November 2018;
Published: 11 December 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Pavol Sulo, Comenius University, SlovakiaFederico Sebastiani, Istituto per la Protezione Sostenibile delle Piante (IPSP), Italy
Copyright © 2018 Sipiczki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias Sipiczki, Z2VjZWxhQHBvc3Quc2s=; bGlwb3Z5QGdteC5jb20=; bGlwb3Z5QHRpZ3Jpcy51bmlkZWIuaHU=
 Matthias Sipiczki
Matthias Sipiczki