
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 December 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03042
This article is part of the Research Topic Engineering the Microbial Platform for the Production of Biologics and Small-Molecule Medicines View all 17 articles
 Qinying Peng
Qinying Peng Guixi Gao
Guixi Gao Jin Lü
Jin Lü Qingshan Long
Qingshan Long Xuefei Chen
Xuefei Chen Fei Zhang
Fei Zhang Min Xu
Min Xu Kai Liu
Kai Liu Yemin Wang
Yemin Wang Zixin Deng
Zixin Deng Zhiyong Li*
Zhiyong Li* Meifeng Tao*
Meifeng Tao*Streptomyces lividans is a suitable host for the heterologous expression of biosynthetic gene clusters (BGCs) from actinomycetes to discover “cryptic” secondary metabolites. To improve the heterologous expression of BGCs, herein we optimized S. lividans strain SBT5 via the stepwise integration of three global regulatory genes and two codon-optimized multi-drug efflux pump genes and deletion of a negative regulatory gene, yielding four engineered strains. All optimization steps were observed to promote the heterologous production of polyketides, non-ribosomal peptides, and hybrid antibiotics. The production increments of these optimization steps were additional, so that the antibiotic yields were several times or even dozens of times higher than the parent strain SBT5 when the final optimized strain, S. lividans LJ1018, was used as the heterologous expression host. The heterologous production of these antibiotics in S. lividans LJ1018 and GX28 was also much higher than in the strains from which the BGCs were isolated. S. lividans LJ1018 and GX28 markedly promoted the heterologous production of secondary metabolites, without requiring manipulation of gene expression components such as promoters on individual gene clusters. Therefore, these strains are well-suited as heterologous expression hosts for secondary metabolic BGCs. In addition, we successfully conducted high-throughput library expression and functional screening (LEXAS) of one bacterial artificial chromosome library and two cosmid libraries of three Streptomyces genomes using S. lividans GX28 as the library-expression host. The LEXAS experiments identified clones carrying intact BGCs sufficient for the heterologous production of piericidin A1, murayaquinone, actinomycin D, and dehydrorabelomycin. Notably, due to lower antibiotic production, the piericidin A1 BGC had been overlooked in a previous LEXAS screening using S. lividans SBT5 as the expression host. These results demonstrate the feasibility and superiority of S. lividans GX28 as a host for high-throughput screening of genomic libraries to mine cryptic BGCs and bioactive compounds.
Microbial secondary metabolites display tremendous diversity in chemical structure and bioactivity and play an important role in drug discovery and development (Cragg and Newman, 2013). In recent years, the exploitation of the potential of “cryptic” biosynthesis, in the form of biosynthetic gene clusters (BGCs), in microbial genomes has become the focus of natural product research (Scherlach and Hertweck, 2009). Heterologous expression of BGCs is now an important technology for genome mining, biosynthetic study, and metabolic engineering. In addition, genome mining and biosynthetic studies on slow-growing or uncultured microorganisms can only be carried out through heterologous expression (Ongley et al., 2013). Many Streptomyces strains have the advantages of rapid growth and simple genetic manipulation. Moreover, as Streptomyces spp. and closely related actinomycetes are rich in secondary metabolite resources and therefore have the ability to provide precursors and cofactors required for efficient biosynthesis, engineered Streptomyces strains are highly suitable hosts for the heterologous expression of BGCs (Martinez et al., 2004; Wenzel and Müller, 2005). Excellent hosts must also be able to express all of the enzymes of the candidate biosynthetic pathway efficiently, including gene transcription, translation, and post-translational modifications, in order to successfully produce the corresponding compounds (Baltz, 2010). Ideally, endogenous secondary metabolic pathways should also be deleted to make a clean metabolic background and avoid substrate competition between endogenous and heterologous pathways (Komatsu et al., 2010). Such optimized hosts include Streptomyces coelicolor M1152, M1154, Streptomyces avermitilis, and Streptomyces albus, which has a naturally minimized genome (Gomez-Escribano and Bibb, 2011; Komatsu et al., 2013; Kallifidas et al., 2018).
Streptomyces lividans, a species closely related to S. coelicolor, has additional advantages as an expression host since it does not restrict (cleave) exogenous methylated DNA, whereas most actinomycetes such as S. coelicolor and S. avermitilis cleave methylated plasmid DNA from most Escherichia coli strains (MacNeil, 1988). Furthermore, when used as a recipient for E. coli–Streptomyces intergeneric conjugation, S. lividans exhibits a high efficiency of conjugative transfer, and this advantage is particularly important for experiments that require high-throughput transfer of arrayed library clones for screening genomic or metagenomic libraries by the function of unknown compounds (Wang et al., 2000). Indeed, an S. lividans TK24-derived strain was chosen as the expression host for the expression and screening of a metagenomic BAC library (Martinez et al., 2004). On the other hand, the wild-type strains of S. lividans, such as strain 1326, have disadvantages as they contain endogenous BGCs for secondary metabolites, such as act (for actinorhodin), red (for streptorubin or undecylprodigiosin) (Liu et al., 2017), and cda [for calcium-dependent antibiotic (CDA)]. Of more concern, such wild-type strains do not produce corresponding antibiotics under most culture conditions, implying that at least these BGCs are silent in the wild-type host (Hu et al., 2002).
Streptomyces lividans TK24 is a spontaneous rpsL[K88E] mutant that increases the production of actinorhodin; rpsL[K88E] has been shown to induce global upregulation of secondary metabolite biosynthesis (Shima et al., 1996; Okamoto-Hosoya, 2003). Additionally, the global regulatory genes afsRScla from S. clavuligerus significantly promote the synthesis of actinorhodin, streptorubin, and CDA in S. lividans TK24 (Chen et al., 2012). On the basis of the above findings, we knocked out the act, red, and cda BGCs from S. lividans TK24 and inserted 1–2 copies of afsRScla, obtaining S. lividans SBT5 and SBT18 (Bai et al., 2014; Xu et al., 2016). Using these strains as expression hosts, Xu et al. (2016) optimized the previous functional genomic screening protocol (Martinez et al., 2004) and developed a library heterologous expression and function-directed screening system (LEXAS) for the screening of BGCs. LEXAS has facilitated the activation of cryptic BGCs and helped in mining compounds and new BGCs using the genomic libraries of Streptomyces spp. (Gao et al., 2017; Zheng et al., 2017; Chen et al., 2018).
In this study, in order to further improve the expression efficiency of heterologous BGCs and improve the screening efficiency of biologically active natural products by LEXAS technology, we optimized S. lividans SBT5 using a number of global positive and negative regulatory genes and genes encoding drug efflux pumps. We also demonstrated the superiority of the new strains in expression of heterologous BGCs and for LEXAS screening of cosmid and BAC libraries.
Streptomyces lividans SBT5 [S. lividans TK24 ΔactΔredKL ΔcdaPS3-SLI3600::afsRScla] (Bai et al., 2014) was used as the parent strain to construct optimized hosts for the heterologous expression of BGCs. Streptomyces griseoruber Sgr29 and Streptomyces galtieri Sag48 were isolated from Shennongjia (Eastern Hubei, China) forest soil (China Center for Type CultureCollection, CCTCC). Streptomyces parvulus 10 was isolated from the marine sponge Phyllospongia foliascens collected from Yongxing Island (South China Sea). S. lividans TK24 was used to construct the wblAsl knockout plasmid, and S. coelicolor M1154 (Gomez-Escribano and Bibb, 2011) was used as the cloning template for nusGsc. Mannitol soy flour agar (MS) was used for sporulation of Streptomyces spp. Liquid culture was performed in TSBY medium containing 3% tryptone soy broth medium, 0.5% yeast extract, and 10.3% sucrose (Kieser et al., 2000). Agar media YBP (Ou et al., 2009), R3 (Shima et al., 1996), GYM (Ochi, 1987), No18 and No24 (Farnet et al., 2008) were used for the fermentation of Streptomyces. Streptomyces cultures were grown and fermented at 30°C. To determine growth curves for recombinant strains of S. lividans, strains were cultured in baffled flasks at 30°C with TSBY liquid medium (30 mL/flask).
Escherichia coli strains, Staphylococcus aureus CICC 10201, and Bacillus mycoides were cultured in Luria-Bertani (LB) medium at 37°C. E. coli XL1-Blue (Stratagene) was used as the host for cosmid library construction, and E. coli DH10B (Invitrogen) was used for general cloning, plasmid maintenance, and as host for a BAC library. E. coli ET12567/pUB307 was used as a helper strain mediating tri-parental E. coli–Streptomyces intergeneric conjugation (MacNeil et al., 1992). S. aureus CICC 10201, B. mycoides, and Saccharomyces sake were used as indicator strains in the bioassay experiments.
pJTU2554 (Li et al., 2008) is a pSET152-derived, triplet COS site-bearing vector used to construct genomic cosmid libraries of S. galtieri Sag48 and S. parvulus 10. SuperCos 1 (Stratagene) was used to construct the genomic cosmid library of S. lividans. pHL921 (Xu et al., 2016) was the vector for the genomic BAC library of S. griseoruber Sgr29. pJTU2554-, pHL931-, and pHL921-derived clones carry the attP and int loci of the Streptomyces temperate phage ΦC31, and therefore can integrate into Streptomyces genomes at the attB site (Combes et al., 2002). pMS82, which bears the integration site attP and int loci of the Streptomyces temperate phage ΦBT1, was used as an integrative vector to carry genes of interest into the chromosome of S. lividans (Gregory et al., 2003). pUB307 is an RK2-derived, self-mobilizable plasmid that facilitates the intergeneric conjugation of oriTRK2-plasmids from E. coli to Streptomyces (Bennett et al., 1977). pIJ773 and pIJ778 were used as templates for PCR amplification of aac(3)IV and aadA resistance markers, respectively (Gust et al., 2003). pHL851 (Chen et al., 2012) was the source of afsRScla.
The actinomycin D standard was purchased from Aladdin Bio-Chem Technology, Co. Standard compounds of murayaquinone, hybrubin A, dehydrorabelomycin, and piericidin A1 were prepared as described (Liu et al., 2012, 2018; Zhao et al., 2016; Gao et al., 2017).
A 1.4 kb fragment containing nusGsc was amplified by PCR using S. coelicolor M1154 genomic DNA as template and primers nusG-R (5′-CTAGTTCTTCTGGATCTGGTGCTTG-3′) and nusG-F (5′-GTGACGGACGCCGTGGGCTCCA-3′), and then cut with XbaI and ligated with pMS82 to yield pJTU6725. Two codon-optimized genes, mdfAco and lmrAco, were synthesized based on the protein sequences of MdfA of Enterobacteriaceae (AFH35853) and LmrA of Lactococcus lactis subsp. cremoris MG1363 (CAL98427.1), respectively, with the codon usage table of S. coelicolor, and following the Codon Adaptation Tool1. The mdfAco gene has an overall GC content of 67% and a GC content of 99.5% for the third codon position. The lmrAco gene has an overall GC content of 65% and a GC content of 99.5% for the third codon position. The optimized lmrAco-mdfAco DNA fragment was synthesized at HongXun Biotechnology, Co., Ltd., and ligated into pJTU6725 to construct pJTU6727. The paired genes afsRScla (SCLAV_3382, SCLAV_3383) and the upstream ermE∗ promoter in pHL851 were cloned into pJTU6727 to construct pJTU6728. The plasmids pJTU6725, pJTU6727, and pJTU6728 retain the hygromycin resistance gene (hyg), origin of transfer (oriTRK2), and the attP and int loci of Streptomyces phage ΦBT1 from the integrative vector pMS82.
The genomic cosmid library of S. lividans TK24 was constructed using XL1-Blue as a host and SuperCos 1 as a vector according to the standard protocol (Kieser et al., 2000). The S. lividans TK24 genomic DNA was extracted and partially digested with Sau3AI. The 40–60 kb fragments were isolated by pulsed-field gel electrophoresis (PFGE), dephosphorylated, and ligated into the SuperCos 1 vector. The ligation product was packaged with λ phage packaging protein, transferred into E. coli XL1-Blue, and transformants were selected by kanamycin. The cosmid clones were extracted and digested with EcoRI and BamHI to verify that the average inserted exogenous fragment size was 39 kb. The genomic cosmid libraries of S. galtieri Sag48 and S. parvulus 10 were constructed using E. coli XL1-blue MR/pUZ8002 as a host and pJTU2554 as a vector as described (Chen et al., 2012).
The genomic cosmid library of S. lividans TK24 was screened by PCR amplification using primers wblA-F (5′-CGTCCTCAACTGGCGGCGGTGAAT-3′) and wblA-R (5′-GGCCCCTGATCCGGCCTCGGGGCT-3′), and the cosmid clone 10H1 containing wblAsl was obtained. The wblAsl gene knockout plasmid was then constructed using 10H1 according to a PCR-targeting protocol (Gust et al., 2003). Firstly, the wblAsl in 10H1 was replaced by an aac(3)IV cassette amplified from pSET152 by λ-Red recombination. The resulting plasmid 10H1-ΔwblA::aac(3)IV was then transformed into E. coli DH5α containing the recombinant plasmid BT340, which expresses the FLP recombinase gene, to remove the aac(3)IV cassette by FLP recombination (Gust et al., 2003), yielding 10H1-ΔwblA. The bla (ampicillin resistance gene) on the backbone of 10H1-ΔwblA was then replaced by the aac(3)IV-oriT cassette amplified from pIJ773 by λ-Red recombination, resulting in the wblAsl knockout plasmid pHLJ42. The wblAsl gene in S. lividans SBT5 was deleted by homologous recombination between pHLJ42 and the chromosomal DNA. pHLJ42 contains 25.7 and 14.7 kb regions of S. lividans chromosomal DNA flanking either side of the mutated wblAsl locus. When pHLJ42, which does not contain an autonomous replication region or integration locus, was introduced into S. lividans SBT5 by conjugation, the apramycin-resistant (AprR) exconjugant should be a single-crossover mutant. To identify double-crossover mutants, the offspring colonies from the single-crossover mutant were screened for the loss of apramycin resistance, indicating the loss of aac(3)IV. A double-crossover mutant strain S. lividans SBT5ΔwblA, i.e., a wblAsl mutant, was confirmed by PCR (herein renamed S. lividans LJ101).
Conjugation using mycelia was conducted following literature (Du et al., 2012). S. lividans LJ1018 was grown in 30 mL TSB liquid medium in a baffled flask, shaking at 180 rpm, 28°C for 48 h. The mycelia was collected by centrifugation at 5,000 rpm and washed with equal volume of 10% of glycerol once and 2× YT twice. Then 0.6 mL of washed mycelia was resuspended in 0.3 mL of 2× YT in an Eppendorf tube, mixed with 0.3 mL exponential phase donor E. coli cells. The mixture was spun at 5,000 rpm for 10 s, and the precipitate was spread on MS agar plate.
The high-throughput, tri-parental E. coli–Streptomyces conjugation of an arrayed genomic library, high-throughput fermentation, and bioactivity assay were carried out according to the LEXAS procedure (Xu et al., 2016). Cosmid or BAC libraries in E. coli DH10B in the format of 96-well plates were used as the arrayed donors for conjugation. Spores of S. lividans GX28 were used as recipients, and E. coli ET12567/pUB307 was used as the helper strain. The E. coli strains containing cosmid or BAC clones were cultured in LB liquid medium (150 μL/well), supplemented with apramycin, at 37°C overnight, and then transferred to antibiotic-free LB, cultured for 4–6 h until the optical density at 600 nm (OD600) reached 0.4 to 0.6. E. coli ET12567/pUB307 (helper strain) was cultured in LB (120 mL/library) containing a final concentration of 50 μg/mL chloramphenicol at 37°C until the OD600 was between 0.4 and 0.6. The cells were then collected by centrifugation and resuspended in 20 mL LB medium. Next, 20 μL of ET12567/pUB307 was pipetted into each well of the 96-well plates in which the BAC/cosmid library was inoculated, and the plates were shaken on a rotary shaker at 200 rpm for 5 min to allow thorough mixing. S. lividans GX28 (recipient) was grown on MS sporulation medium for 5–6 days at 30°C. The fresh spores were collected and resuspended in 4 mL of 2× YT medium, heat-shocked at 50°C for 10 min, and then spread on MS plates supplemented with Mg2+ (20 mM). The donor-helper E. coli mixtures were replicated from 96-well plates onto spore-coated MS plates using a 48-pin replicator. After incubation at 30°C for 12 to 16 h, the MS plates were covered with apramycin and trimethoprim to final concentrations of 50 μg/mL to inhibit the E. coli strains. The exconjugants were cultured for another 4–6 days and then replicated to MS plates containing final concentrations of 50 μg/mL apramycin and 25 μg/mL nalidixic acid to remove the residual E. coli. The S. lividans GX28 exconjugants were fermented and subjected to high-throughput screening based on antibacterial activity. The S. lividans GX28 exconjugants of libraries were replicated to the agar fermentation media YBP, R3, GYM, No18, and No24 by replicator and cultured at 30°C for 7 days. The surface of the fermentation media were covered with soft agar premixed with indicator bacteria, and the inhibition zones produced by heterologous expression of the active compounds were observed after 1–2 days of incubation.
The sequences of both ends of the inserts in BAC clones were determined with primers pHL921F (5′-ATGTTTTTCGTCTCAGCC-3′) and pHL921R (5′-CCTTTAGTTGTTCCTTTC-3′). The end sequences of cosmid clones were determined with primers pJTU2554F (TGTAAAACGACGGCCAGT) and pJTU2554F (GGCACCTGTCCTACGAGTTG). The DNA end sequences were then mapped to the genomic sequences. The DNA sequences of BAC or cosmid inserts were submitted to antiSMASH2 for the analysis of secondary metabolic BGCs.
Actinorhodin was isolated and measured using a published method (Bystrykh et al., 1996). The S. lividans strains were cultured on solid YBP medium for 84 h, 500 mg agar culture was taken from each plates, 500 μL of 1 M NaOH was added, followed by crushing using a homogenizer (5,000 rpm, 15 s; twice). The samples were centrifuged at 12,000 × g, 5 min and the absorbance of the supernatants was measured at 633 nm. The isolation and analysis of piericidin A1, murayaquinone, dehydrorabelomycin, and actinomycin D, followed a similar approach as the following. Fermented culture (40 mL) was extracted three times with ethylacetate (150 mL). The combined extracts were concentrated on a rotary evaporator (Buchi R210) at 37°C and then dissolved in 1 mL methanol. The crude extract (20 μL) was filtered and injected onto a C18 reversed-phase column (Agilent Zorbax ODS C18, 5 μm, 4.6 by 250 mm) and analyzed by high performance liquid chromatography (HPLC) in the Agilent 1260 HPLC system using mobile phase A (H2O supplemented with 0.1% formic acid) and mobile phase B (acetonitrile) at a flow rate of 0.6 mL/min. The elution procedure was: 0–2 min, 5% B (and 95%A); 2–25 min, 5–40% B; 25–35 min, 40–100% B; 35–40 min, 100% B; 40–45 min, 100–5% B; 45–55 min, 5% B.
The isolation and identification of hybrubin A was carried out as described (Zhao et al., 2016). Hybrubin A was eluted using the following HPLC conditions: mobile phase A was H2O (supplemented with 0.1% formic acid), mobile phase B was methanol; flow rate of 0.6 mL/min; 0 min, 40% B; 5–15 min, 65–80% B; 15–20 min, 80–100% B; 20–25 min, 100% B; 25–26 min, 100–40% B; 26–35 min, 40% B.
Agilent G6530 HR ESI-QTOF mass spectrometry equipped with Agilent 1260 HPLC system was used to identify piericidin A1, dehydrorabelomycin, murayaquinone, actinomycin D, and hybrubins.
The nusGsc gene of S. coelicolor A3(2) encodes an anti-terminator that is functionally conserved in prokaryotes, eukaryotes, and archaea (Mason and Greenblatt, 1991; Burmann et al., 2010). For cloning this gene with its native promoter, we amplified a 1.4 kb fragment containing the nusGSC coding region and the 536 bp upstream region by PCR, and then the fragment was ligated into pMS82 to yield the integrative plasmid pJTU6725 (Figure 1A). pJTU6725 was conjugated to S. lividans SBT5 to generate S. lividans GX25, in which the plasmid is integrated into the genome at the attBΦBT1 site.
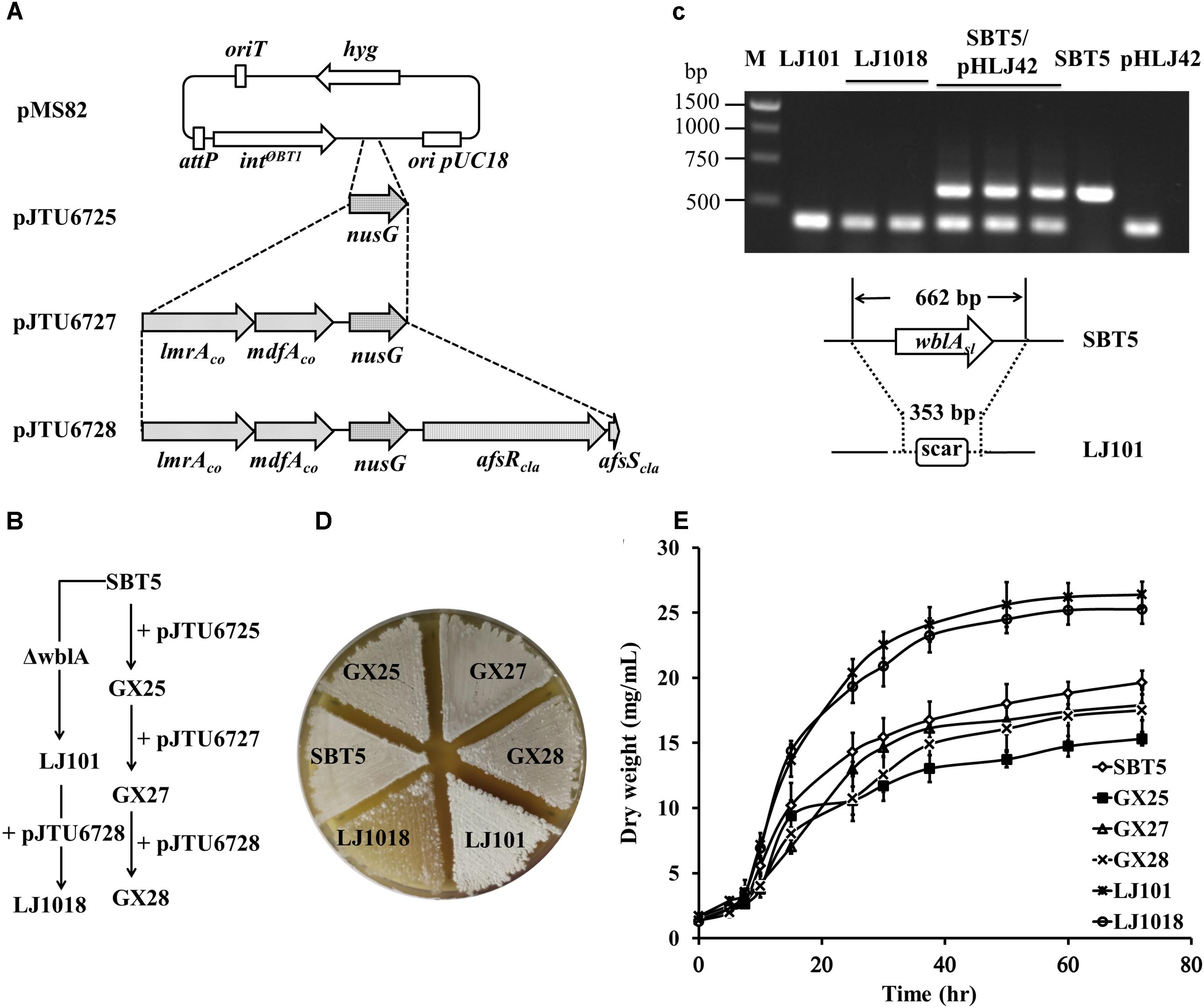
Figure 1. Engineering of Streptomyces lividans strains. (A) Integrative plasmids derived from pMS82 carrying regulatory genes and codon-optimized multidrug resistance transporter genes. (B) Construction of the engineered S. lividans strains from SBT5. (C) PCR confirmation of the wblAsl deletion mutants LJ101 and LJ1018. M, 1 kb ladder. pHLJ42, cosmid containing a deleted wblAsl locus; SBT5/pHLJ42, single-crossover mutant; SBT5, the parent strain. (D) Growth of the engineered strains and SBT5 on MS medium at 30°C for 72 h. LJ101 is white, and LJ1018 is bald. (E) Biomass accumulation of the engineered S. lividans strains and SBT5 over 72 h of cultivation in TSBY liquid medium. Spores (or mycelium of LJ101 and LJ1018) were pre-cultured on TSBY at 30°C for 48 h. An aliquot of the resultant vegetative culture was diluted to 100-fold by 30 mL TSBY and shaken at 180 rpm, 30°C. A 1 mL culture sample was taken and centrifuged for 10 min at 12,000 rpm. Supernatants were discarded, and the pellet was dried at 80°C for 48 h and weighed.
The lmrA gene in Lactococcus lactis subsp. cremoris MG1363 encodes a multidrug resistance ABC transporter ATP-binding and permease protein (van Veen et al., 1996), and the mdfA gene in Escherichia coli K-12 encodes a multidrug efflux transporter protein. Both lmrA and mdfA confer hosts with resistance to a variety of antibiotics by heterologous expression (Edgar and Bibi, 1997). The G+C contents of the original lmrA and mdfA genes were 39.0 and 52.4%, respectively. For the expression of these two multidrug resistance genes in the high G+C content genome of S. lividans, we synthesized the codon-optimized twin gene cassette lmrAco-mdfAco based on the protein sequences and the degenerate codon usage table of the S. coelicolor genome. The promoter of the non-ribosomal peptide synthase (NRPS) gene cdaPS1 from the CDA BGC was placed upstream of lmrAco to control the expression of lmrAco and mdfAco. The previously reported production of CDA in afsRScla-carrying S. lividans strains suggested that the PcdaPS1 promoter has been activated (Chen et al., 2012; Bai et al., 2014). The synthetic operon PcdaPS1-lmrAco-mdfAco was ligated to pJTU6725 to construct pJTU6727 (Figure 1A). The integrative plasmid pJTU6727 was conjugated to S. lividans SBT5 to yield S. lividans GX27.
The global transcriptional regulator AfsR/Scla from S. clavuligerus ATCC 27064 (NRRL3585) increased the production of actinorhodin and CDA in S. lividans TK24 (Chen et al., 2012). We cloned afsR/Scla and the ermE∗ promoter from pHL851 into pJTU6727 to construct pJTU6728 (Figure 1A), which was conjugated to S. lividans SBT5 to construct S. lividans GX28 (Figure 1B).
The gene wblAsl (SLIV_20395) of S. lividans TK24 encodes a global transcriptional regulator of the WhiB family (Yu et al., 2014) and has 99% similarity to S. coelicolor wblAsc (SCO3579). To knock out wblAsl in S. lividans SBT5, the cosmid clone 10H1 containing wblAsl was obtained from a genomic cosmid library of S. lividans TK24. An in-frame deletion was made in wblAsl on 10H1 to construct the gene knockout vector pHLJ42, which contains wblAsl flanking sequences of 25.7 and 14.7 kb for homologous recombination. The wblAsl-knockout strain LJ101 was constructed using pHL42 via homologous recombination and confirmed by PCR (Figure 1C). Compared with the parental strain SBT5, S. lividans LJ101 exhibited a “white” phenotype (Figure 1D): no spore pigment was produced and the white aerial hyphae did not develop into spores. This indicated that wblAsl plays an important role in aerial hyphae development similar to the wblA from S. coelicolor A3(2) (Fowler-Goldsworthy et al., 2011). pJTU6728 was conjugated to S. lividans LJ101 to yield S. lividans LJ1018, which displayed a “bald” phenotype with only sparse white mycelium (Figure 1D).
Growth curves indicated that the introduction of pJTU6725, pJTU6727, and pJTU6728 into S. lividans SBT5 did not significantly affect the growth and biomass accumulation of the host strain. However, the biomass of the wblAsl deletion strains S. lividans LJ101 and S. lividans LJ1018 was significantly improved. The dry weight of the two strains was 1.6 times higher than that of S. lividans GX28 after 72 h of culture (Figure 1E; p < 0.0001).
To test the ability of the engineered S. lividans strains to express polyketide BGCs, we expressed the actinorhodin BGC using S. lividans GX25, S. lividans GX27, S. lividans GX28, S. lividans LJ1018, and the parent strain S. lividans SBT5 as the expression hosts. The act BGC is a 22 kb type II polyketide synthase (PKS) BGC, and actinorhodin (Figure 2A) is a pH-sensitive, pigmented aromatic polyketide antibiotic that is red at acidic pH and blue at alkali pH. Plasmid pMM1 (45 kb) carrying the complete act BGC (Zhou et al., 2012) was introduced into the S. lividans series of hosts by conjugation. High conjugation frequencies, ca. 10-2/cfu, were observed when S. lividans GX25, S. lividans GX27, S. lividans GX28, and S. lividans SBT5 were used. Because S. lividans LJ1018 is deficient in sporulation, mycelium was used as the recipient for conjugation, and 100s of exconjugants were obtained on each conjugation plate, with a conjugation frequency of around 10-6/cfu. After fermentation in YBP medium for 72 h, the blue color of actinorhodin was observed due to the heterologous expression of the act BGC in the exconjugants. Observation of the color of the YBP fermentation medium revealed that the heterologous expression of actinorhodin in the optimized hosts S. lividans GX25, GX27, GX28, and LJ1018 progressively increased compared to levels in SBT5 (Figure 2B). The yield of actinorhodin of GX25/pMM1 was 1.3 times higher than that of SBT5/pMM1 (p < 0.001), and the yields of actinorhodin in S. lividans GX27/pMM1, GX28/pMM1, and LJ1018/pMM1 were 12.8, 21.6, and 23.3 times higher than that of SBT5/pMM1, respectively (p < 0.0001; Figure 2C), indicating that the addition of nusGsc, the drug efflux pump genes, and afsRScla and the knockout of wblAsl in SBT5 up-regulated the production of actinorhodin.
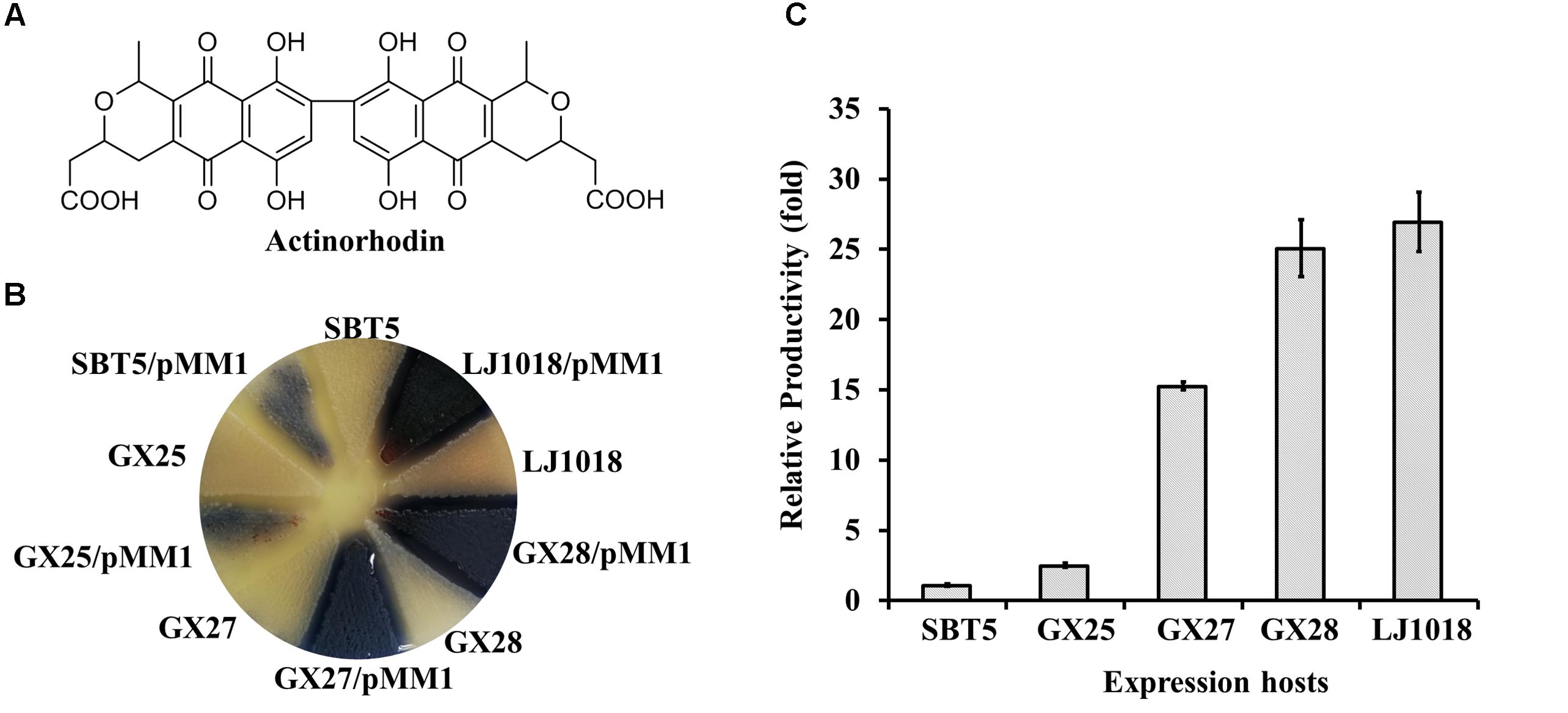
Figure 2. Heterologous expression of actinorhodin by the engineered S. lividans strains carrying the S. coelicolor actinorhodin BGC on pMM1. (A) Structure of actinorhodin. (B) Heterologous expression of the actinorhodin BGC in S. lividans strains on YBP agar medium. The top of the culture plate after 72 h fermentation is shown. pMM1, plasmid carrying the S. coelicolor actinorhodin BGC. (C) Quantification of actinorhodin production by various expression hosts carrying pMM1 on YBP medium. The productivity related to S. lividans SBT5/pMM1 was present. Data are from three biological replicates.
Murayaquinone is a tricyclic, angular aromatic polyketide 9,10-phenanthraquinone antibiotic produced by a type II PKS pathway (Figure 3A, Gao et al., 2017). The murayaquinone BGC is about 56 kb and was cloned into BAC clone 3B4. 3B4 was obtained by screening the genomic BAC library of S. griseoruber Sgr29 using S. lividans SBT5 as the high-throughput heterologous expression host, conferring the exconjugants with antibacterial activity against S. aureus (Gao et al., 2017). The exconjugants carrying 3B4 were fermented on solidified media No18 and No24, and the crude extracts were analyzed by HPLC. Three new peaks were observed on the HPLC trace of S. lividans GX28/3B4 fermented on No18 medium, with the same retention time as the standard samples of murayaquinone and murayalactone 1, 2 (Figure 3B). Murayalactone 1 and 2 were the main products on No18 medium, while murayaquinone was the main product on No24 medium. The identity of murayaquinone and murayalactones isolated from S. lividans GX28/3B4 was further confirmed by high-resolution mass spectrometry (HR-MS) (Figure 3C). However, murayaquinone and murayalactones were not detectable by HPLC from the S. griseoruber Sgr29 fermentation culture (Figure 3B), which is consistent with the literature. To compare the production of murayaquinone, all exconjugants were fermented on No24, and the areas of murayaquinone peaks in the HPLC traces were measured. The yield of murayaquinone was extremely low (about 0.11 mg/L) in the parent host SBT5/3B4 but was significantly increased in all engineered hosts (p < 0.05). The yields of murayaquinone in S. lividans GX28/3B4 and S. lividans LJ1018/3B4 were much higher than that of the original host SBT5/3B4 (74 and 96 times higher, respectively, p < 0.0001), and the yield of murayaquinone in S. lividans LJ1018/3B4 was 10.6 mg/L (Figure 3D).
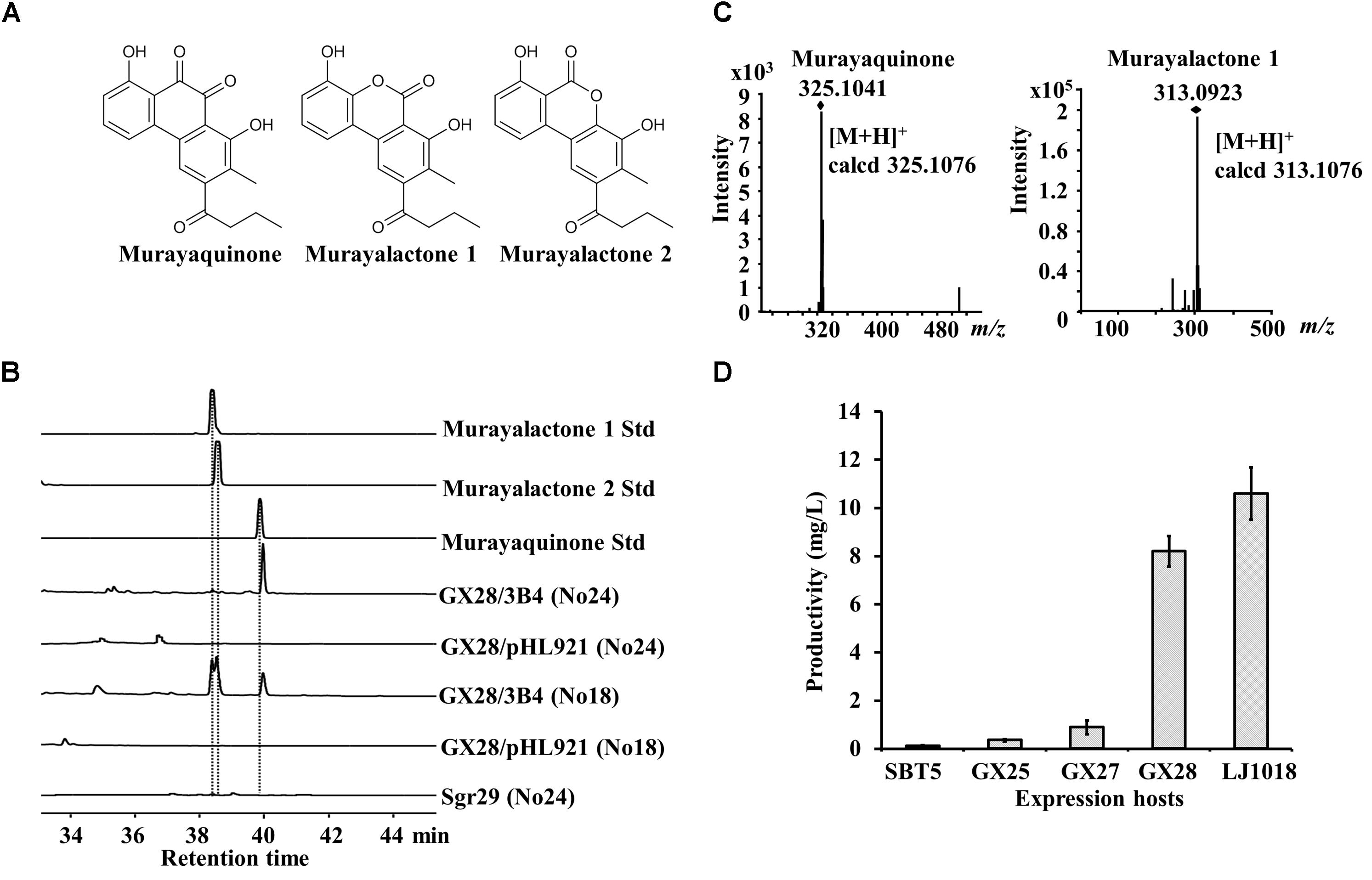
Figure 3. Heterologous expression of the murayaquinone BGC in engineered S. lividans strains. (A) Structure of murayaquinone and murayalactones. (B) HPLC analysis of the exconjugants carrying the murayaquinone BGC from BAC 3B4 and of the original strain Streptomyces griseoruber Sgr29. The absorbance was measured at 350 nm. No18 and No24 agar media were used for fermentation. 3B4, a BAC clone containing the murayaquinone BGC. No murayaquinone or murayalactones were detected from S. griseoruber Sgr29. (C) HR-MS spectrum of murayaquinone and murayalactone 1 isolated from S. lividans GX28/3B4. The spectra of murayalactone 1 and 2 are identical. (D) Quantification of murayaquinone production on No24 medium by engineered S. lividans strains carrying 3B4. Data are from three biological replicates.
The hbn BGC from Streptomyces variabilis Snt24 is a small PKS-NRPS hybrid BGC responsible for the biosynthesis of 5-ethylidenetetramic acid (ETA); the truncated red pathway in S. lividans SBT5 synthesizes 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC), and condensation of ETA with MBC produces the “non-natural” red compounds named hybrubins (Zhao et al., 2016). pZZL3 is an integrative plasmid containing a 13 kb hbn BGC cloned from the S. variabilis Snt24 genome. The heterologous expression of hbn BGC in S. lividans SBT5 led to the production of the red-pigmented secondary metabolites hybrubin A-C (Figure 4A, Zhao et al., 2016). When the pZZL3-carrying exconjugants of S. lividans SBT5, GX25, GX27, GX28, and LJ1018 were fermented with R3 medium, red pigment was observed in the crude extract whereas the vector control did not produce red pigment (Figure 4B). HPLC analysis indicated that hybrubins A-C were produced and that hybrubin A was the main component (Figure 4C). The identity of hybrubin A was confirmed by HR-ESI-MS (Figure 4D). The relative yield of hybrubin A was evaluated based on the HPLC peak area. The yield of hybrubin A in GX25/pZZL3 and GX27/pZZL3 was slightly higher than in SBT5/pZZL3 (2.2 and 2.5 times, respectively, p < 0.05), whereas the yield in GX28/pZZL3 and LJ1018/pZZL3 was greatly increased, reaching 13 times and 29 times the yield in SBT5/pZZL3, respectively (Figure 4E).
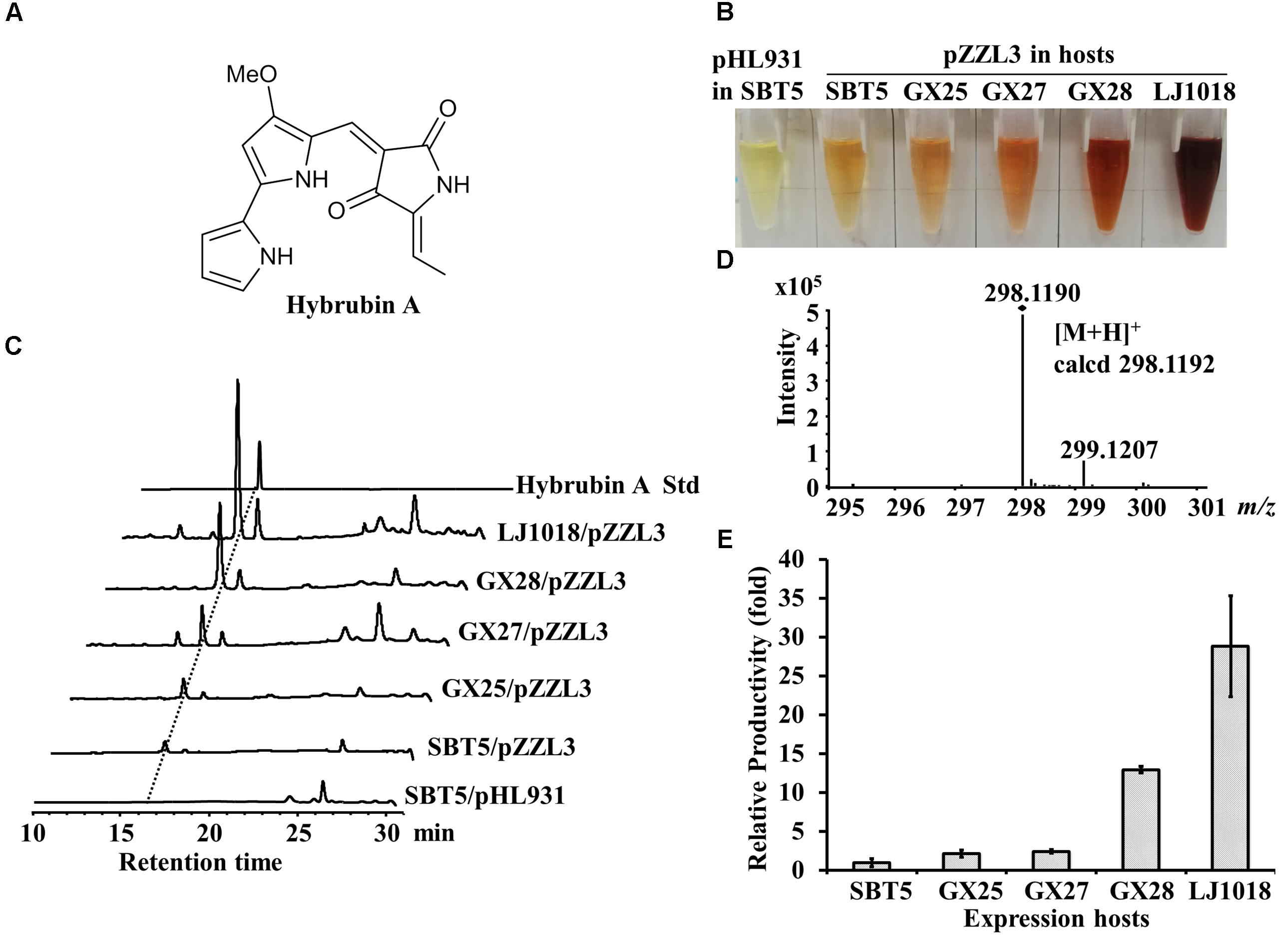
Figure 4. Heterologous expression of hybrubin A by engineered S. lividans strains carrying pZZL3. (A) Structure of hybrubin A. (B) Ethyl acetate crude extracts of the exconjugants carrying pZZL3 fermented on R3 medium. pZZL3, a plasmid containing the tetramic acid (ETA) BGC; pHL931, the empty vector control. Red pigmented hybrubin A was observed in extracts of the pZZL3-carrying exconjugants. The vector control did not produce red pigment. (C) HPLC analysis of hybrubin A production by S. lividans strains. (D) HR-MS spectrum of hybrubin A isolated from S. lividans GX28/pZZL3. (E) Quantification of the heterologous expression of hybrubin A in R3 liquid medium. Yields of hybrubin A from the optimized hosts S. lividans GX28 and LJ1018 were much higher than from the original host S. lividans SBT5. Data are from three biological replicates.
We tested the ability of the engineered strain S. lividans GX28 to serve as a host for LEXAS screening of antibiotics and their corresponding BGCs, using the S. griseoruber Sgr29 genomic BAC library, which contains 912 arrayed clones with an average insertion size of about 100 kb (Gao et al., 2017). S. lividans SBT5 had been used as a host for the high-throughput heterologous expression in a previous screening, and seven positive BAC clones with S. aureus resistance were obtained from this genomic BAC library, three of which contained the murayaquinone BGC (Gao et al., 2017). Using S. lividans GX28 as the expression host, nine new S. aureus-resistant positive BAC clones were obtained, five of which (4E6, 4F9, 1H5, 2D3, and 4G10) shared overlapping DNA regions (Figure 5A). The termini of these five BACs were sequenced with primers pHL921-F/R, and then the sequences were aligned with the S. griseoruber Sgr29 genomic sequence. The five BAC plasmids were found to have a 98 kb overlapping region, and analysis of this region by AntiSMASH revealed that it contains a 50 kb piericidin A1 BGC, which included six type I polyketide synthase (PKSI) genes and five post-modification genes highly homologous to piericidin A1 BGC genes in S. piomogeues. Piericidin A1 is an α-pyridone antibiotic (Figure 5B) that inhibits the mitochondrial respiratory chain and NADH-ubiquinone oxidase and exhibits weak antimicrobial and antitumor activities (Liu et al., 2012; Chen et al., 2014). To verify the function of the piericidin A1 BGC, one of the BAC clones, 1H5, was transferred to the expression host S. lividans GX28, and the exconjugants was fermented with R3 medium. The fermented culture of GX28/1H5 had inhibitory activity against B. mycoides, whereas the empty vector control (S. lividans GX28/pHL921) did not produce an inhibition zone (Figure 5C). HPLC and HR-MS analysis indicated that piericidin A1 was produced by GX28/1H5 and S. griseoruber Sgr29 (Figures 5D,E).
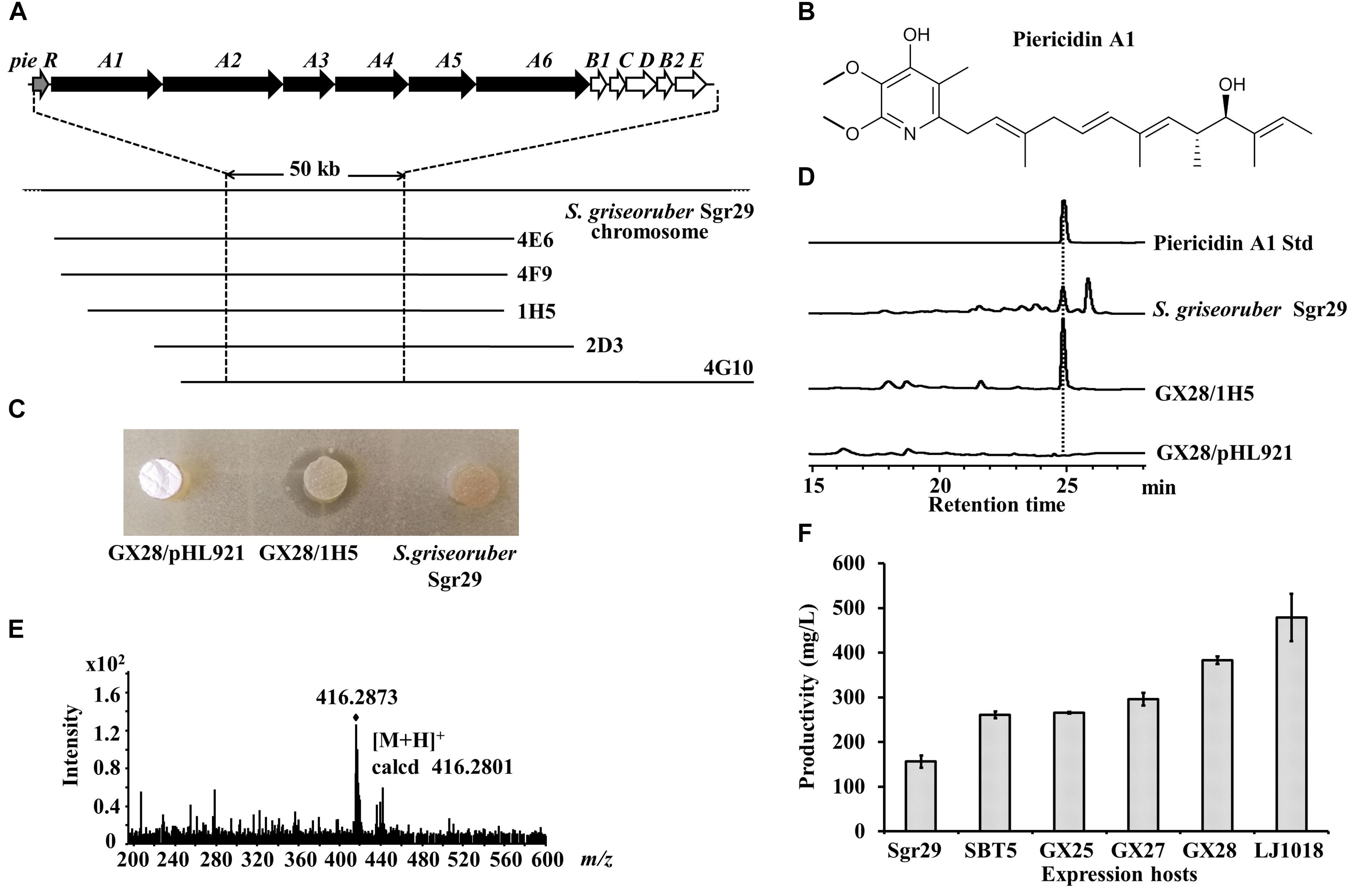
Figure 5. Identification of piericidin A1 and the pie BGC by LEXAS screening of the S. griseoruber Sgr29 BAC genomic library using S. lividans GX28 as host. (A) Overlapping map of the five BAC clones containing the 50 kb piericidin A1 BGC. Thick arrows on the top line denote genes of the piericidin biosynthetic pathway. (B) Structure of piericidin A1. (C) Bioassay of the exconjugant S. lividans GX28/1H5 against Bacillus mycoides. Plugs of fermented culture were placed on the surface of agar medium inoculated with B. mycoides. The bioassay plate was incubated for 24 h at 37°C. A zone of inhibition was observed around the plug of GX28/1H5. No antibacterial activity was observed from S. griseoruber Sgr29 or the vector control. (D) HPLC analysis of the ethyl acetate extracts of fermented cultures of S. lividans GX28/1H5, the vector control, and S. griseoruber Sgr29. The absorbance was measured at 254 nm. (E) HR-MS spectrum of piericidin A1 isolated from S. lividans GX28/1H5. (F) Quantification of the production of piericidin A1 by Sgr29 and the 1H5-carrying exconjugants in five expression hosts on R3 agar medium.
To detect the yield of piericidin A1 in different expression hosts, 1H5 was transferred into the five S. lividans hosts for heterologous expression, and the resulting exconjugants and the natural strain S. griseoruber Sgr29 were fermented. HPLC analysis showed that, although S. griseoruber Sgr29 produced high levels of piericidin A1 (156.6 mg/L), the yields resulting from heterologous expression in the S. lividans hosts were significantly higher (p < 0.001). The yield of piericidin A1 from S. lividans GX28/1H5 was 2.4 times that of S. griseoruber Sgr29, and the yield from S. lividans LJ1018/1H5 was even higher, at 3.1 times the yield from S. griseoruber Sgr29 and reaching 478 mg/L (Figure 5F).
We constructed a genomic cosmid library of S. galtieri Sag48, a species isolated from forest soil by CCTCC, and performed LEXAS screening using S. lividans GX28 as the high-throughput heterologous expression host. The LEXAS screening identified an exconjugant displaying weak inhibition activity against B. mycoides and which contained cosmid plasmid 8F5. Sequencing analysis revealed that 8F5 has a 32 kb insertion sequence containing 27 genes having high level of similarity (81–95%) to the alpA-alpW genes in the type II polyketide BGC of kinamycin from S. ambofaciens (Figure 6A). The complete kinamycin BGC is 63 kb and cannot be packaged into a single cosmid clone (Wang et al., 2015; Liu et al., 2018). Although cosmid 8F5 contains PKS genes (alpABC) and early modification genes for the synthesis of kinamycin intermediates, it does not contain other genes required for the synthesis of the final product (i.e., kinamycin). To analyze the metabolites produced via this cosmid, 8F5 and the vector pJTU2554 were introduced into the five S. lividans hosts by conjugation, and the resulting exconjugants and the natural strain S. galtieri Sag48 were fermented on No18 agar plates. Extracts of the fermented cultures were analyzed by HPLC. The crude extract of S. lividans LJ1018/8F5 produced an absorption peak at 39 min, which was not produced by S. galtieri Sag48 and the vector control strain S. lividans LJ1018/pJTU2554 (Figure 6B). The compound was detected by LC-MS, and its molecular weight, with an m/z value of 321.0710, was consistent with that of dehydrorabelomycin (m/z of [M+H]+ calcd. 321.0763) (Figures 6C,D), which is an intermediate of the kinamycin biosynthetic pathway.
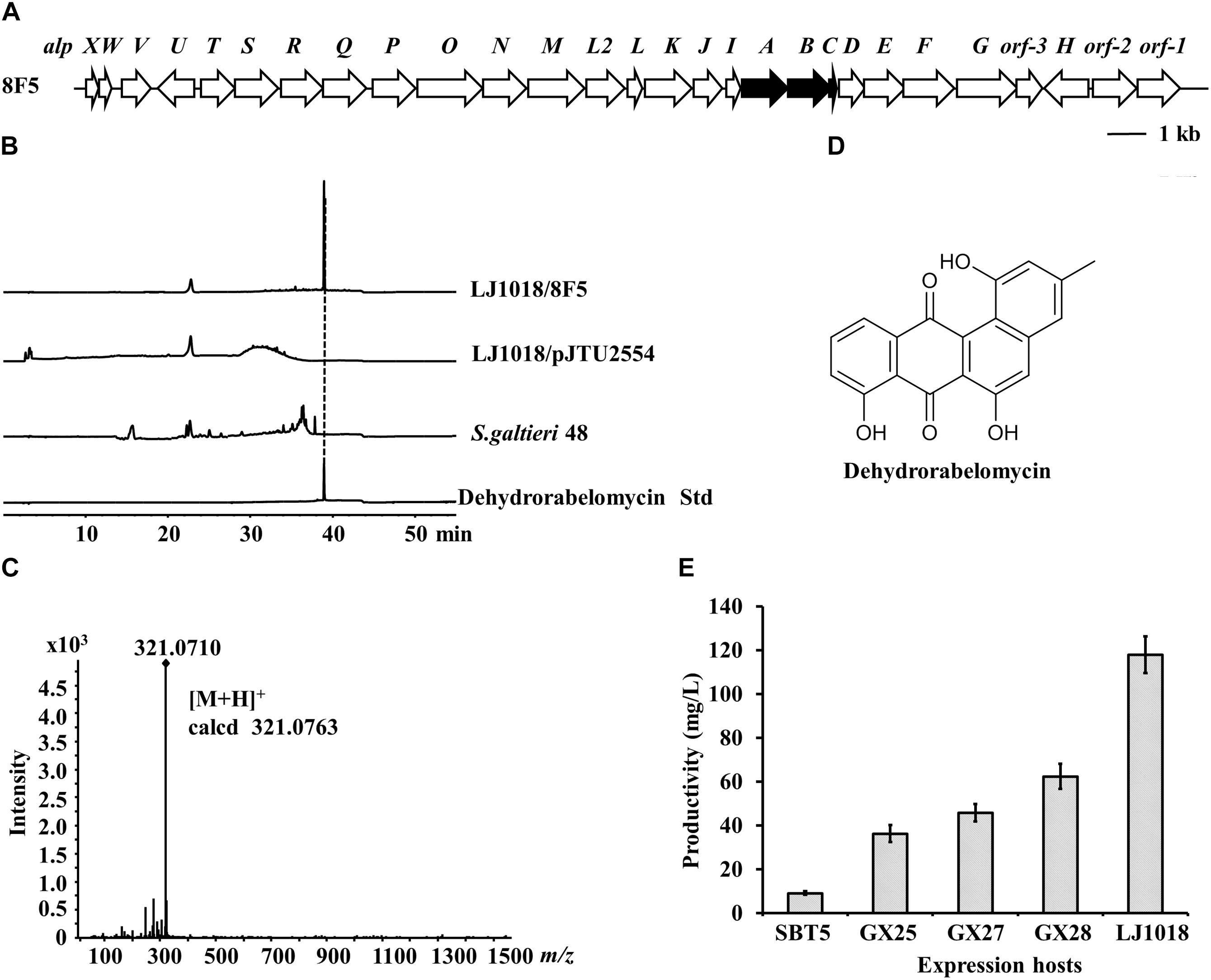
Figure 6. Identification of dehydrorabelomycin and its BGC by LEXAS screening of the Streptomyces galtieri Sag48 genomic cosmid library using S. lividans GX28 as host. (A) Gene organization of the dehydrorabelomycin BGC in cosmid 8F5. The minimal pks genes are black. (B) HPLC analysis of ethylacetate extracts of fermented cultures. The absorbance was measured at 350 nm. (C) HR-MS spectrum of dehydrorabelomycin isolated from S. lividans LJ1018/8F5. (D) Structure of dehydrorabelomycin. (E) Quantification of the production of dehydrorabelomycin by 8F5-carrying exconjugants in five expression hosts on R3 agar medium.
Quantitative comparison of dehydrorabelomycin production indicated that S. lividans GX28/8F5 and LJ1018/8F5 yielded levels 6.7 times and 12.7 times, respectively, the amount produced by the original host SBT5/8F5. The highest yield of dehydrorabelomycin was produced by S. lividans LJ1018/8F5, reaching a level of 118.0 mg/L (Figure 6E).
Streptomyces parvulus 10 was isolated from the marine sponge Carteriospongia foliascens collected from the South China Sea by the Zhiyong Li Group. We constructed a cosmid library of the S. parvulus 10 genome and used S. lividans GX28 as the heterologous expression host for high-throughput library screening. An exconjugant exhibiting S. aureus inhibitory activity was observed. Sequencing analysis of the corresponding cosmid, 5H11, revealed that it contains an NRPS BGC (Figure 7A) with 16 genes highly similar (78–95% similarity) to genes of the actinomycin C of Streptomyces anulatus (Keller et al., 2010). The compound produced by S. lividans GX28/5H11 was determined to be actinomycin D (Figure 7B) by HPLC and LC-MS (m/z of [M+H]+ obsd. 1255.6359, calcd. 1255.6363) (Figures 7C,D).
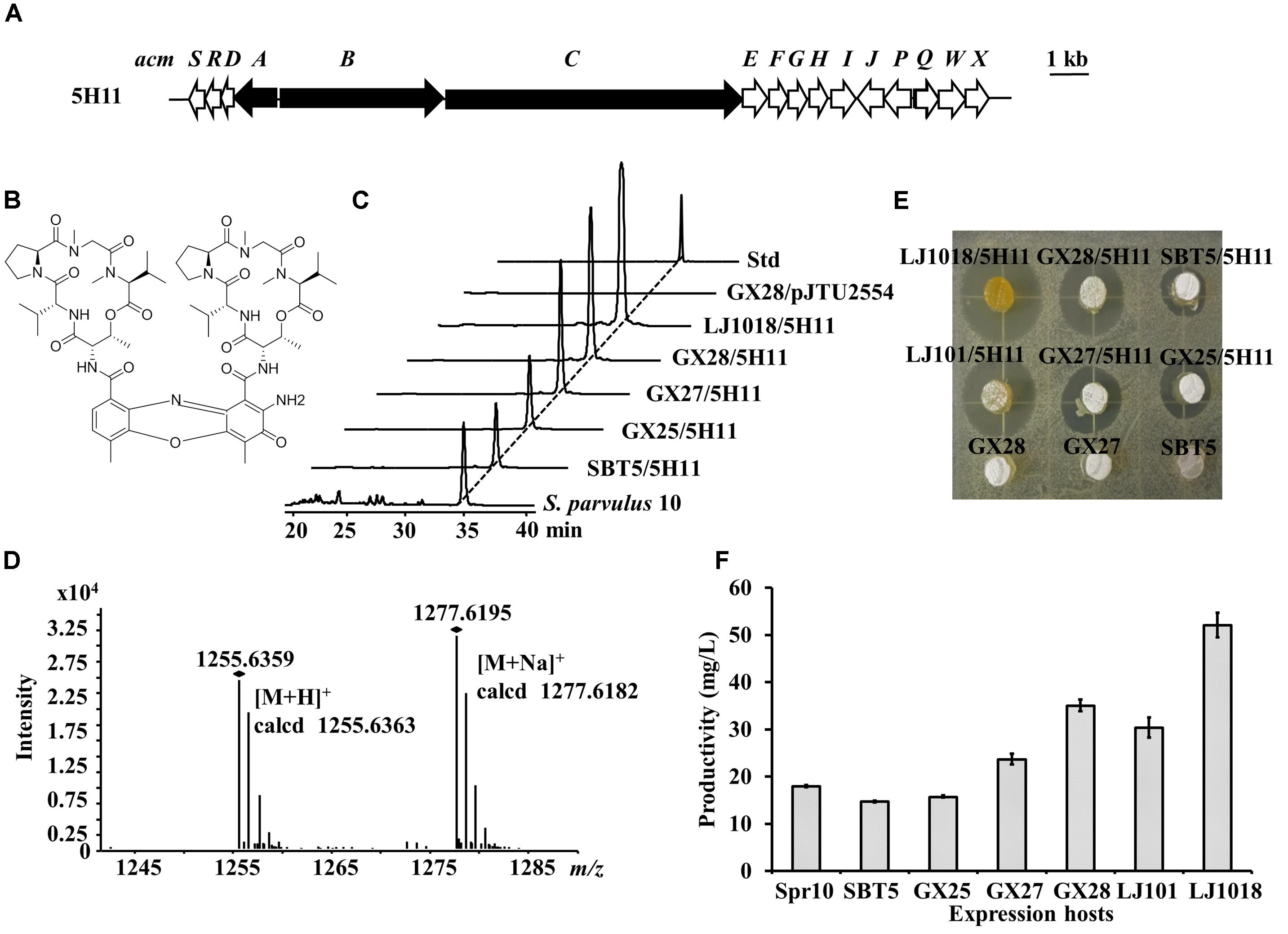
Figure 7. Identification of actinomycin D and its BGC by LEXAS screening of the Streptomyces parvulus 10 genomic cosmid library using S. lividans GX28 as host. (A) Gene organization of the actinomycin D BGC in cosmid 5H11. The PKS genes are black. (B) Structure of actinomycin D. (C) HPLC analysis of the extracts of fermented cultures of S. lividans exconjugants carrying 5H11. The absorbance was measured at 440 nm. Std, actinomycin D standard. (D) HR-MS spectrum of actinomycin D isolated from S. lividans GX28/5H11. (E) Bioassay against B. mycoides to detect actinomycin D production by 5H11 exconjugants. Agar plugs of fermented cultures of 5H11-containing S. lividans exconjugants were placed on LB agar pre-spread with B. mycoides. The plates were incubated for 12 h at 37°C for observing zones of inhibition. (F) Quantification of the production of actinomycin D by S. parvulus 10 and the 5H11-carrying exconjugants in six expression hosts on R3 agar medium. Spr10, S. parvulus 10.
To compare the heterologous expression of actinomycin BGC in different hosts, 5H11 was transferred to the five S. lividans hosts, and the exconjugants were fermented in YBP medium. After 4 days of fermentation, the bioactivity test indicated that the inhibition zones produced by the exconjugants of the newly engineered S. lividans hosts were larger than for the parental strain. The S. lividans LJ1018/5H11 fermented culture displayed the largest inhibition zone against B. mycoides (Figure 7E). HPLC quantitative determination confirmed that the production of actinomycin D in S. lividans strains increased in turn (p < 0.01 or p < 0.001), i.e., the production in LJ1018/5H11 > GX28/5H11 > GX27/5H11 > GX25/5H11 > SBT5/5H11 (Figure 7F). LJ1018/5H11 was capable of producing 52.1 mg/L actinomycin D, which was 3.5 times the level produced by SBT5/5H11. The native strain S. parvulus 10 also synthesized actinomycin D (18.0 mg/L). The actinomycin D yields of strains GX27/5H11, GX28/5H11, and LJ1018/5H11 were 32% (p < 0.0001), 95% (p < 0.0001), and 190% (p < 0.0001) higher than that of S. parvulus 10, respectively.
Whether a host can effectively express all the essential genes of a given heterologous synthetic pathway is key to the success of the heterologous production of secondary metabolites. When high-throughput heterologous expression methods are used to screen metagenomic or genomic libraries (Baltz, 2008), ideally overall gene expression should be improved by manipulating global regulatory genes in the expression host, rather than by attempting to modify all of the individual promoters within BGCs, a potentially complex and cumbersome task (Chen et al., 2010) and one not possible with previously unknown BGCs. Although previously engineered hosts, such as S. coelicolor, have altered global regulatory genes to promote the expression of BGCs, due to the restriction of methylated DNA and the slightly lower frequency of conjugative transfer (MacNeil, 1988), these strains are not well-suited to be LEXAS high-throughput screening hosts (Chen et al., 2012).
Streptomyces lividans has the advantage of high frequency of conjugative transfer and no restriction on exogenous methylated DNA (Martinez et al., 2004), and S. lividans TK24 strain itself contains an rpsL[K88E] mutation that promotes gene expression (Ochi, 2007). We previously added 1–2 copies of the global regulatory gene afsRScla to the TK24 genome, and the resultant strains indeed contributed significantly to the establishment of a high-throughput library expression and screening system (LEXAS) (Xu et al., 2016). To further optimize the S. lividans host and the LEXAS system, in this study we continued to optimize S. lividans with global regulatory genes, including nusGsc and wblAsl, as well as drug efflux pump genes, in addition to afsRScla.
Many antibiotic BGCs carry export genes, such as actII-ORF2 in the actinorhodin BGC (Fernándezmoreno et al., 1991) and rifP in the rifamycin BGC (August et al., 1998). These efflux pumps secrete the antibiotics out of the cell, thereby reducing the feedback inhibition of the end-products on the biosynthetic enzymes, while increasing the self-tolerance to the antibiotics. Therefore, overexpression of antibiotic efflux pumps is helpful when engineering strains to increase the production of antibiotics of interest (Qiu et al., 2011). MdfA of E. coli is a multi-drug transporter of the major facilitator superfamily (Sigal et al., 2006); it has a broad-spectrum recognition and efflux function for toxic compounds and enhances the tolerance of the host strains to natural or synthetic antibiotics such as daunomycin, rifampin, puromycin, aminoglycoside antibiotics, and quinolones (Edgar and Bibi, 1997). LmrA of Lactococcus lactis subsp. cremoris MG136362 belongs to a family of multidrug resistance ABC (ATP-binding cassette) transporters driven by ATP hydrolysis (van Veen et al., 1996; Wilkens, 2015), and its sequence is highly similar to that of the multi-drug resistance export pump P-glycoprotein (MDR) in mammals (Margolles et al., 1999). LmrA and MDR1 increased the tolerance of bacterial cells to compounds such as daunomycin, ethidium, rhodamine 6G, and tetraphenylphosphonium (van Veen et al., 1996). We added two codon-optimized efflux pump-encoding genes, mdfAco and lrmAco, into S. lividans GX25 to construct GX27. Our quantitative data on heterologous expression suggested that the introduction of these two efflux pump genes significantly increased the yield of four antibiotics, including actinorhodin, dehydrorabelomycin, piericidin A1, and actinomycin D, demonstrating that it is applicable to use multi-drug transporters for the general improvement of antibiotics production in heterologous hosts.
The second group of ideal engineering targets are global regulators. NusG, the regulator of the NusG-like family, functions as an RNAP processivity clamp and is the only anti-terminator factor conserved among the kingdoms of prokaryotes, eukaryotes, and archaea (Burmann et al., 2010). Behnken et al. (2012) used the constitutive strong promoter Pthl to increase the expression level of nusG, thereby successfully activating the originally silenced polythioamides BGC in the genome of the anaerobic bacterium Clostridium cellulolyticum and unveiling seven new compounds. In this study, we inserted S. coelicolor nusGsc into the S. lividans SBT5 and used the resultant GX25 as a heterologous host to express six different types of antibiotic BGCs, the production of five out of six antibiotics increased significantly (p < 0.05). Similarly, additional copy of the positive regulatory gene afsRcla and sigma factor-like gene afsScla increased the production of six antibiotics significantly. These results suggest that global positive regulatory genes like nusGsc and afsRScla are applicable for improving the heterologous expression of PKS, NRPS, and NRPS-PKS BGCs.
WblA is a global negative regulator unique to actinomycetes (Kang et al., 2007) and has obvious sequence similarity to the developmental differentiation factor WhiB (Chater et al., 2000). In many actinomycetes, knocking out wblA significantly improved antibiotic biosynthesis in the mutant strains (Kang et al., 2007; Noh et al., 2010; Rabyk et al., 2011; Nah et al., 2012; Yu et al., 2014). The molecular mechanism by which WblA negatively regulates antibiotic synthesis remains unclear. We knocked out wblAsl in S. lividans GX28 to obtain LJ1018, which led to significant increases in the production of hybrubins, dehydrorabelomycins, and actinomycin D (p < 0.05), and slight increases of the three antibiotics actinorhodin, murayaquinone, and piericidin A. However, the mutant strains S. lividans LJ1018 and LJ101 that we constructed do not produce spores. This is not surprised since WblA plays an important role in the formation of aerial hyphae in Streptomyces (Fowler-Goldsworthy et al., 2011). After knocking out wblA in S. coelicolor and S. chattanoogensis L10, no spores were formed on the aerial hyphae (Yu et al., 2014). As a consequence, we had to use mycelium instead of spores as the recipient during conjugation transfer, which reduced the frequency of conjugation sharply. Nevertheless, this characteristic did not affect the introduction of target BGCs into the host for heterologous expression, since dozens to 100s of exconjugants could be obtained for each mycelium conjugation in our laboratory. However, when we attempted to use S. lividans LJ1018 mycelium as LEXAS host for high-throughput expression of arrayed cosmid libraries and BAC libraries, only sporadic exconjugants emerged, so LJ1018 is not suitable as a host for high-throughput heterologous expression of arrayed libraries.
In contrast, strains GX25, GX27, and GX28 still produce abundant spores. Both high-throughput and conventional conjugation transfer worked as efficiently as with the parental strain SBT5. When screening the two cosmid libraries (from S. galtieri Sag48 and S. parvulus 10) using GX28 as the high-throughput expression host, 3948 out of 4032 cosmid clones (98%) yielded exconjugants, and when we screened a BAC library using GX28, 818 out of the 912 clones (93%) produced exconjugants. BGCs producing dehydrorabelomycin and actinomycin were identified from the cosmid libraries. Five clones containing the complete piericidin A1 BGC, in addition to clones carrying the murayaquinone BGC, were identified from the S. griseoruber Sgr29 genomic BAC library. Notably, these piericidin BGC clones had been overlooked during the previous screening using SBT5 as a host (Gao et al., 2017). Indeed, the corresponding SBT5 exconjugants did not show significant antibacterial activity, since no zone of inhibition was produced. Our genomic screening results demonstrate that the GX28 strain is an excellent expression host for the LEXAS procedure to screen for functional BGCs in arrayed cosmid libraries and BAC libraries, and also demonstrates the superiority of GX28 as a heterologous expression host.
In summary, S. lividans provides excellent host strains for high-throughput screening of genomes (such as LEXAS screening) due to its rapid growth, abundant sporulation, high frequency of conjugation transfer, no methylation restriction on methylated DNA, and efficient expression of heterologous BGCs after rational engineering. By sequentially engineering global regulatory genes and multi-drug transporters, we obtained four engineered strains of S. lividans, which in turn increased the yield of multiple synthesized antibiotics that involve PKS, NRPS, and PKS-NRPS hybrid pathways. Since cryptic BGCs in microorganisms usually encode new or unknown biosynthetic pathways, it is very difficult to specifically engineer pathway-specific regulatory factors or to appropriately modify promoters. Our optimized host GX28 produces high yields of antibiotics and does not require one-by-one modification of the promoters in a BGC of interest, so it is an excellent heterologous expression host for LEXAS high-throughput screening of cosmid or BAC libraries for the discovery of new, previously silenced BGCs and corresponding compounds. In addition, our study has revealed that the positive regulatory genes nusGsc and afsRScla, the negative regulatory gene wblAsl, and efflux pump genes, which are not regulatory genes by definition, have synergistic effects on the synthesis of antibiotics when they are combined in one host. The yields of the tested antibiotics were increased several times, even dozens of times in the case of hybrubins, over yields from the parent strain, SBT5. Furthermore, since the strain engineering conducted here mainly utilizes plasmid integration, these plasmids, especially pJTU6728, which integrates nusG, afsRScla, and efflux pump genes, can be used for the engineering of other strains for high-yield production of antibiotics in future. Therefore, the strains we have generated and the approaches we have used should aid in the identification of new BGCs and in optimizing the production of secondary metabolites of clinical and industrial value.
MT, ZD, and ZL were responsible for the original concept and designed the experiments. MT and YW analyzed the data. QP, GG, JL, QL, XC, FZ, MX, KL, and YW performed the experimental work. QP and MT wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the Chinese Ministry of Science and Technology through a China-Australia Joint Grant (Grant No. 2016YFE0101000), the National Natural Science Foundation of China (Grant No. 31770036), the Science and Technology Commission of Shanghai Municipality (Grant No. 15JC1400401), and the National Key Research and Development Program of China (Grant No. 2018YFC030900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Songwang Hou and Prof. Tianshen Tao for gifts of bacterial strains.
August, P. R., Tang, L., Yoon, Y. J., Ning, S., Müller, R., Yu, T. W., et al. (1998). Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei, s699. Chem. Biol. 5, 69–79. doi: 10.1016/S1074-5521(98)90141-7
Bai, T., Yu, Y., Xu, Z., and Tao, M. (2014). Construction of Streptomyces lividans SBT5 as an efficient heterologous expression host. J. Huazhong Agric. Univ. 33, 1–6.
Baltz, R. (2008). Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8, 557–563. doi: 10.1016/j.coph.2008.04.008
Baltz, R. H. (2010). Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 37, 759–772. doi: 10.1007/s10295-010-0730-9
Behnken, S., Lincke, T., Kloss, F., Ishida, K., and Hertweck, C. (2012). Antiterminator-mediated unveiling of cryptic polythioamides in an anaerobic bacterium. Angew. Chem. Int. Ed. 51, 2425–2428. doi: 10.1002/anie.201108214
Bennett, P. M., Grinsted, J., and Richmond, M. H. (1977). Transposition of TnA does not generate deletions. Mol. Gen. Genet. 154, 205–211. doi: 10.1007/BF00330839
Burmann, B. M., Schweimer, K., Luo, X., Wahl, M. C., Stitt, B. L., Gottesman, M. E., et al. (2010). A NusE: NusG complex links transcription and translation. Science 328, 501–504. doi: 10.1126/science.1184953
Bystrykh, L. V., Fernández-Moreno, M. A., Herrema, J. K., Malpartida, F., Hopwood, D. A., and Dijkhuizen, L. (1996). Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J. Bacteriol. 178, 2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996
Chater, K. F., Soliveri, J. A., Gomez, J., and Bishai, W. R. (2000). Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146, 333–343. doi: 10.1099/00221287-146-2-333
Chen, L., Wang, Y., Guo, H., Xu, M., Deng, Z., and Tao, M. (2012). High-throughput screening for Streptomyces antibiotic biosynthesis activators. Appl. Environ. Microbiol. 78, 4526–4528. doi: 10.1128/AEM.00348-12
Chen, X., Xu, M., Lü, J., Xu, J., Wang, Y., Lin, S., et al. (2018). Biosynthesis of tropolones in Streptomyces spp: interweaving biosynthesis and degradation of phenylacetic acid and hydroxylations on tropone ring. Appl. Environ. Microbiol. doi: 10.1128/AEM.00349-18 [Epub ahead of print].
Chen, Y., Smanski, M. J., and Shen, B. (2010). Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl. Microbiol. Biotechnol. 86, 19–25. doi: 10.1007/s00253-009-2428-3
Chen, Y., Zhang, W., Zhu, Y., Zhang, Q., Tian, X., Zhang, S., et al. (2014). Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis. Org. Lett. 16, 736–739. doi: 10.1021/ol4034176
Combes, P., Till, R., Bee, S., and Smith, M. C. M. (2002). The Streptomyces genome contains multiple pseudo-attB sites for the C31-encoded site-specific recombination system. J. Bacteriol. 184, 5746–5752. doi: 10.1128/JB.184.20.5746-5752.2002
Cragg, G. M., and Newman, D. J. (2013). Natural products: a continuing source of novel drug leads. Biochim. Biophys. Acta 1830, 3670–3695. doi: 10.1016/j.bbagen.2013.02.008
Du, L., Liu, R. H., Ying, L., and Zhao, G. R. (2012). An efficient intergeneric conjugation of DNA from Escherichia coli to mycelia of the lincomycin-producer Streptomyces lincolnensis. Int. J. Mol. Sci. 13, 4797–4806. doi: 10.3390/ijms13044797
Edgar, R., and Bibi, E. (1997). MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179, 2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997
Farnet, C. M., Mcalpine, J. B., Bachmann, B. O., Staffa, A., and Zazopoulos, E. (2008). System, knowledge repository and computer-readable medium for identifying a secondary metabolite from a microorganism. US Patent No 20,080,010,025. Toronto: Thallion Pharmaceuticals Inc.
Fernándezmoreno, M. A., Caballero, J., Hopwood, D. A., and Malpartida, F. (1991). The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66, 769–780. doi: 10.1016/0092-8674(91)90120-N
Fowler-Goldsworthy, K., Gust, B., Mouz, S., Chandra, G., Findlay, K. C., and Chater, K. F. (2011). The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157, 1312–1328. doi: 10.1099/mic.0.047555-0
Gao, G., Liu, X., Xu, M., Wang, Y., Zhang, F., Xu, L., et al. (2017). Formation of an angular aromatic polyketide from a linear anthrene precursor via oxidative rearrangement. Cell Chem. Biol. 24, 881–891.e4. doi: 10.1016/j.chembiol.2017.06.008
Gomez-Escribano, J. P., and Bibb, M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters: Streptomyces host for heterologous expression of gene clusters. Microb. Biotechnol. 4, 207–215. doi: 10.1111/j.1751-7915.2010.00219.x
Gregory, M. A., Till, R., and Smith, M. C. M. (2003). Integration site for Streptomyces phage BT1 and development of site-specific integrating vectors. J. Bacteriol. 185, 5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546. doi: 10.1073/pnas.0337542100
Hu, H., Zhang, Q., and Ochi, K. (2002). Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase subunit) of Streptomyces lividans. J. Bacteriol. 184, 3984–3991. doi: 10.1128/JB.184.14.3984-3991.2002
Kallifidas, D., Jiang, G., Ding, Y., and Luesch, H. (2018). Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters. Microb. Cell Factories 17:25. doi: 10.1186/s12934-018-0874-2
Kang, S.-H., Huang, J., Lee, H.-N., Hur, Y.-A., Cohen, S. N., and Kim, E.-S. (2007). Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J. Bacteriol. 189, 4315–4319. doi: 10.1128/JB.01789-06
Keller, U., Lang, M., Crnovcic, I., Pfennig, F., and Schauwecker, F. (2010). The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 192, 2583–2595. doi: 10.1128/JB.01526-09
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: The John Innes Foundation.
Komatsu, M., Komatsu, K., Koiwai, H., Yamada, Y., Kozone, I., Izumikawa, M., et al. (2013). Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 2, 384–396. doi: 10.1021/sb3001003
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. doi: 10.1073/pnas.0914833107
Li, L., Xu, Z., Xu, X., Wu, J., Zhang, Y., He, X., et al. (2008). The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5’-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. ChemBioChem 9, 1286–1294. doi: 10.1002/cbic.200800008
Liu, P., Zhu, H., Zheng, G., Jiang, W., and Lu, Y. (2017). Metabolic engineering of Streptomyces coelicolor for enhanced prodigiosins (RED) production. Sci. China Life Sci. 60, 948–957. doi: 10.1007/s11427-017-9117-x
Liu, Q., Yao, F., Chooi, Y. H., Kang, Q., Xu, W., Li, Y., et al. (2012). Elucidation of piericidin A1 biosynthetic locus revealed a thioesterase-dependent mechanism of α-pyridone ring formation. Chem. Biol. 19, 243–253. doi: 10.1016/j.chembiol.2011.12.018
Liu, X., Liu, D., Xu, M., Tao, M., Bai, L., Deng, Z., et al. (2018). Reconstitution of kinamycin biosynthesis within the heterologous host Streptomyces albus J1074. J. Nat. Prod. 81, 72–77. doi: 10.1021/acs.jnatprod.7b00652
MacNeil, D. J. (1988). Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J. Bacteriol. 170, 5607–5612. doi: 10.1128/jb.170.12.5607-5612.1988
MacNeil, D. J., Gewain, K. M., Ruby, C. L., Dezeny, G., Gibbons, P. H., and MacNeil, T. (1992). Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68. doi: 10.1016/0378-1119(92)90603-M
Margolles, A., Putman, M., van Veen, H. W., and Konings, W. N. (1999). The purified and functionally reconstituted multidrug transporter LmrA of Lactococcus lactis mediates the transbilayer movement of specific fluorescent phospholipids. Biochemistry 38, 16298–16306. doi: 10.1021/bi990855s
Martinez, A., Kolvek, S. J., Yip, C. L. T., Hopke, J., Brown, K. A., MacNeil, I. A., et al. (2004). Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl. Environ. Microbiol. 70, 2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004
Mason, S. W., and Greenblatt, J. (1991). Assembly of transcription elongation complexes containing the N protein of phage and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5, 1504–1512.
Nah, J.-H., Park, S.-H., Yoon, H.-M., Choi, S.-S., Lee, C.-H., and Kim, E.-S. (2012). Identification and characterization of wblA-dependent tmcT regulation during tautomycetin biosynthesis in Streptomyces sp. CK4412. Biotechnol. Adv. 30, 202–209. doi: 10.1016/j.biotechadv.2011.05.004
Noh, J.-H., Kim, S.-H., Lee, H.-N., Lee, S. Y., and Kim, E.-S. (2010). Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Appl. Microbiol. Biotechnol. 86, 1145–1153. doi: 10.1007/s00253-009-2391-z
Ochi, K. (1987). Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 169, 3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987
Ochi, K. (2007). From microbial differentiation to ribosome engineering. Biosci. Biotechnol. Biochem. 71, 1373–1386. doi: 10.1271/bbb.70007
Okamoto-Hosoya, Y. (2003). An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 149, 3299–3309. doi: 10.1099/mic.0.26490-0
Ongley, S. E., Bian, X., Neilan, B. A., and Müller, R. (2013). Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 30, 1121–1138. doi: 10.1039/c3np70034h
Ou, X., Zhang, B., Zhang, L., Zhao, G., and Ding, X. (2009). Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl. Environ. Microbiol. 75, 2158–2165. doi: 10.1128/AEM.02209-08
Qiu, J., Zhuo, Y., Zhu, D., Zhou, X., Zhang, L., Bai, L., et al. (2011). Overexpression of the ABC transporter AvtAB increases avermectin production in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 92, 337–345. doi: 10.1007/s00253-011-3439-4
Rabyk, M., Ostash, B., Rebets, Y., Walker, S., and Fedorenko, V. (2011). Streptomyces ghanaensis pleiotropic regulatory gene wblAgh influences morphogenesis and moenomycin production. Biotechnol. Lett. 33, 2481–2486. doi: 10.1007/s10529-011-0728-z
Scherlach, K., and Hertweck, C. (2009). Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760. doi: 10.1039/b821578b
Shima, J., Hesketh, A., Okamoto, S., Kawamoto, S., and Ochi, K. (1996). Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178, 7276–7284. doi: 10.1128/jb.178.24.7276-7284.1996
Sigal, N., Cohen-Karni, D., Siemion, S., and Bibi, E. (2006). MdfA from Escherichia coli, a model protein for studying secondary multidrug transport. J. Mol. Microbiol. Biotechnol. 11, 308–317. doi: 10.1159/000095633
van Veen, H. W., Venema, K., Bolhuis, H., Oussenko, I., Kok, J., Poolman, B., et al. (1996). Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. U.S.A. 93, 10668–10672. doi: 10.1073/pnas.93.20.10668
Wang, B., Ren, J., Li, L., Guo, F., Pan, G., Ai, G., et al. (2015). Kinamycin biosynthesis employs a conserved pair of oxidases for B-ring contraction. Chem. Commun. 51, 8845–8848. doi: 10.1039/C5CC01986A
Wang, G.-Y.-S., Graziani, E., Waters, B., Pan, W., Li, X., McDermott, J., et al. (2000). Novel natural products from soil DNA libraries in a Streptomycete host. Org. Lett. 2, 2401–2404. doi: 10.1021/ol005860z
Wenzel, S. C., and Müller, R. (2005). Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 16, 594–606. doi: 10.1016/j.copbio.2005.10.001
Wilkens, S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep. 7:14. doi: 10.12703/P7-14
Xu, M., Wang, Y., Zhao, Z., Gao, G., Huang, S.-X., Kang, Q., et al. (2016). Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl. Environ. Microbiol. 82, 5795–5805. doi: 10.1128/AEM.01383-16
Yu, P., Liu, S.-P., Bu, Q.-T., Zhou, Z.-X., Zhu, Z.-H., Huang, F.-L., et al. (2014). WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Appl. Environ. Microbiol. 80, 6879–6887. doi: 10.1128/AEM.01849-14
Zhao, Z., Shi, T., Xu, M., Brock, N. L., Zhao, Y.-L., Wang, Y., et al. (2016). Hybrubins: bipyrrole tetramic acids obtained by crosstalk between a truncated undecylprodigiosin pathway and heterologous tetramic acid biosynthetic genes. Org. Lett. 18, 572–575. doi: 10.1021/acs.orglett.5b03609
Zheng, X., Cheng, Q., Yao, F., Wang, X., Kong, L., Cao, B., et al. (2017). Biosynthesis of the pyrrolidine protein synthesis inhibitor anisomycin involves novel gene ensemble and cryptic biosynthetic steps. Proc. Natl. Acad. Sci. U.S.A. 114, 4135–4140. doi: 10.1073/pnas.1701361114
Keywords: optimal hosts, global regulatory genes, heterologous expression, biosynthetic gene clusters (BGCs), secondary metabolites, library expression and function-directed screening system (LEXAS)
Citation: Peng Q, Gao G, Lü J, Long Q, Chen X, Zhang F, Xu M, Liu K, Wang Y, Deng Z, Li Z and Tao M (2018) Engineered Streptomyces lividans Strains for Optimal Identification and Expression of Cryptic Biosynthetic Gene Clusters. Front. Microbiol. 9:3042. doi: 10.3389/fmicb.2018.03042
Received: 07 October 2018; Accepted: 26 November 2018;
Published: 10 December 2018.
Edited by:
Dipesh Dhakal, Sun Moon University, South KoreaReviewed by:
Hiroyuki Morita, University of Toyama, JapanCopyright © 2018 Peng, Gao, Lü, Long, Chen, Zhang, Xu, Liu, Wang, Deng, Li and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Li, enlsaUBzanR1LmVkdS5jbg== Meifeng Tao, dGFvX21laWZlbmdAc2p0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.