
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 December 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03032
This article is part of the Research Topic Proteomics for Studying Foodborne Microorganisms and their Impact on Food Quality and Human Health View all 10 articles
Alicyclobacillus acidoterrestris is a major putrefying bacterium that can cause pecuniary losses in the global juice industry. Current detection approaches are time-consuming and exhibit reduced specificity and sensitivity. In this study, an immunoproteomic approach was utilized to identify specific biomarkers from A. acidoterrestris for the development of new detection methods. Cell surface-associated proteins were extracted and separated by 2-D (two-dimensional) gel electrophoresis. Immunogenic proteins were detected by Western blot analysis using antisera against A. acidoterrestris. Twenty-two protein spots exhibiting immunogenicity were excised and eighteen of the associated spots were successfully identified by matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS). These proteins were observed to be involved in energy and carbohydrate metabolism, transmembrane transport, response to oxidative stress, polypeptide biosynthesis, and molecule binding activity. This is the first report detailing the identification of cell surface-associated antigens of A. acidoterrestris. The identified immunogenic proteins could serve as potential targets for the development of novel detection methods.
Alicyclobacillus acidoterrestris is a thermo-acidophilic, aerobic, spore-forming, Gram-positive bacterium which is capable of growing at a temperature range of 25–60°C, and a pH range of 2.5–6.0 (Chang et al., 2013). The spores that are produced by A. acidoterrestris can germinate and grow at pH < 4 and are capable of resisting heat treatment at 90°C for 16 to 23 min, a more stringent heat treatment regimen than the conventional pasteurization treatments used in juice processing (Steyn et al., 2011; Celenk and Ayse Handan, 2015). A. acidoterrestris can use vanillin and tyrosine as precursors in the synthesis of guaiacol, an organic compound that emits a “phenolic” odor (Uchida and Silva, 2017). The latter compound triggers spoilage of fruit juices and acidic beverages, resulting in significant economic losses to the juice industry (Oteiza et al., 2015; Fernandez et al., 2017). According to a survey conducted by the European Fruit Juice Association (AIJN) in 2005, about 45% of the 68 fruit processing industries experienced Alicyclobacillus related problems, including 33% undergoing problems more than once (Steyn et al., 2011). Current detection methods are either labor-intensive and time-consuming or highly technique-requiring (Chang and Kang, 2004; Concina et al., 2010; Pérez-Cacho et al., 2011; Wang et al., 2012). Although immunoassays have been developed for many years, the effectiveness of immunoassays largely depends on the quality of antibody. Bacteria with high homology could express similar antigens especially in gram-positive bacteria that teichoic-acid could serve as an important surface antigen in all species (Pasquina et al., 2013). So, finding species-specific biomarkers and preparing their corresponding antibodies could lead to the development of more accurate methods for the detection of A. acidoterrestris.
As a combination of immunology and proteomics, immunoproteomics has been successfully utilized in pathogenic microorganism for revealing the pathogenesis and elucidating novel diagnostic biomarkers (Hernandez-Haro et al., 2015; Ray et al., 2016; Chen et al., 2018). Thus, immunoproteomics can be used as a preliminary step for specific antibody and immunoassay development to facilitate the detection of foodborne microbiology accurately and rapidly possible (Xiong et al., 2012; Rasmussen et al., 2016). However, up until now limited research has been performed to investigate the use of this technique in detecting non-pathogenic bacteria capable of contaminating food matrices and products.
For most bacteria, the cell wall proteins are often exposed at the surface and exhibit strain specificity (Yu et al., 2012). Thus, based on these properties, cell surface exposed proteins have already served as drug targets and for vaccine and pathogen-specific immunoassay techniques development (Rodriguez-Ortega et al., 2006). The aim of this study was to resolve the cell wall proteins of A. acidoterrestris using an immunoproteomics approach to find species-specific biomarkers for immunodetection of this bacterium. Type strain A. acidoterrestris DSM3923 was used in this research. Cell wall proteins were extracted and separated by 2-D gel electrophoresis. Protein spots on gels exhibiting immunogenicity were identified and these proteins were chosen as biomarkers for future immunoassay development. We expect that we can monitor the A. acidoterrestris in real time from orchard to table in the future based on our findings. Therefore, it could reduce or prevent economic losses to the juice industry resulting from spoilage caused by metabolic products of A. acidoterrestris.
This study was carried out in accordance with the recommendations of the Animal Ethics Procedures and Guidelines of the People’s Republic of China. All efforts were exerted to minimize the suffering of animals. The animal experiments were approved by the Ethics Committee of Northwest A&F University.
Alicyclobacillus acidoterrestris DSM 3923 used in this study was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and stored at -80°C. The culture was grown in AAM broth (Yamazaki et al., 1996) with some modifications (yeast extract 2.0 g, glucose 2.0 g, (NH4)2SO4 0.4 g, MgSO47H2O 1.0 g, KH2PO4 1.2 g, CaCl2 0.38 g, distilled water 1000 mL, pH 4.0) on a shaker at 45°C.
Antisera against whole A. acidoterrestris DSM 3923 cells were obtained by immunizing rabbits as previously described (Wang et al., 2012). Briefly, two New Zealand white male rabbits (Xi’an Jiaotong University Health Science Center, China) were subcutaneously immunized with formaldehyde-inactivated A. acidoterrestris DSM 3923 at a dose of 1 × 108 CFU/rabbit mixed with an equal volume of Freund’s complete adjuvant (Sigma, United States). Four booster immunizations were administered every 2 weeks using the same concentration of bacterial cells mixed with Freund’s incomplete adjuvant (Sigma, United States). One week after the final booster immunization, the rabbits were anesthetized with absolute ether and sacrificed to collect blood samples. The blood samples were incubated at 37°C for 1–2 h followed by overnight incubation at 4°C. The samples were subsequently centrifuged at 4500 g for 15–20 min at 4°C and the sera were collected and divided into 1 mL-aliquots and stored at -20°C until further required. Next, the isolated antisera were purified by MabSelect SuRe (GE Healthcare, United States) and stored at -20°C until further required.
Cell wall proteins from A. acidoterrestris DSM 3923 were isolated according to a method (with some modifications) published by Siegel et al. (1981). Briefly, A. acidoterrestris cells grown to the log phase (OD600 = 0.5) were harvested by centrifugation at 10,000 g for 10 min at 4°C. The cells were subsequently washed three times with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) to remove the excess culture medium. The resultant pellets were suspended in extraction buffer (30 mM Tris–HCl (pH 7.5), 3 mM MgCl2, 25% sucrose, 125 U/mL mutanolysin, 400 U/mL lysozyme) containing a protease inhibitor cocktail (Roche, Switzerland) and incubated at 37°C for 90 min. The protoplast fraction was removed by centrifugation at 10,000 g for 10 min at 4°C and the supernatant containing the cell wall proteins was collected and filtered through a 0.22 μm R.C membrane. The cell wall proteins were precipitated using trichloroacetic acid (TCA)/acetone (Wu et al., 2014). Prechilled (-20°C) 10% TCA/acetone containing 0.1% Dithiothreitol (DTT) was added to the resultant supernatant. Proteins were allowed to precipitate for 1 h at -20°C. The precipitate was washed three times with cold (-20°C) acetone containing 0.1% DTT to remove TCA. The final pellet was air-dried and solubilized in rehydration buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 65 mM DTT and 0.2% (v/v) Bio-Lyte pH range 3–10 (Bio-Rad, United States)]. Protein concentration was determined by the Bradford assay.
For the first dimension, isoelectric focusing (IEF) was performed in a PROTEANTM IEF cell (Bio-Rad, United States). Cell wall protein samples (approximately 400 μg each) were suspended in rehydration buffer containing 0.001% (w/v) bromophenol blue and loaded onto the IEF focusing tray with ReadyStripTM IPG Strips (17 cm, pH 4–7). The samples were subsequently rehydrated overnight (12 h) at 20°C, and 50 V. IEF was carried out using the following conditions: 250 V for 30 min, 1000 V for 1 h, 8500 V for 5 h and final focusing at 8500 V for a total of 60,000 Vh. The current was limited to 50 μA per IPG strip, and the temperature was kept at 20°C for all of the focusing steps. After the IEF, the strip was equilibrated for 15 min with 5 mL of equilibration buffer (6 M urea, 2% (w/v) SDS, 0.375 M Tris-HCl (pH 8.8), 20% (v/v) glycerol) containing 2% (w/v) DTT and another 15 min in the same buffer containing 2.5% iodoacetamide. Finally, the IPG strip was dipped into running buffer (25 mM Tris base, 192 mM glycine, 0.1% (w/v) SDS, pH 8.3), positioned on top of a 12% polyacrylamide gel and sealed in place with 0.5% agarose. The second-dimension electrophoresis was carried out in PROTEAN II XL Cell (Bio-Rad, United States). The resultant gel was run at 10 mA for 30 min and 25 mA until the bromophenol dye reached the bottom of the gel. Following electrophoresis, the gel was stained using Coomassie Brilliant Blue G-250 and scanned using a GS-800TM Calibrated Densitometer (Bio-Rad, United States). Three independent replicates were performed for each experiment.
Proteins in the gel were transferred onto a 0.45 μm Immobilon-P polyvinylidene difluoride (PVDF) membrane (Merck Millipore, United States) using a Trans-Blot®SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad, United States) at 25 V for 30 min with transfer buffer (25 mM Tris base, 192 mM glycine, 0.02% (w/v) SDS, 20% (v/v) methanol). Next, the membrane was rinsed three times with TBST (20 mM Tris base, 137 mM NaCl, 0.1% Tween-20, pH 7.5) for 5 min and blocked with 5% skimmed milk in TBST for 2 h at room temperature. The membrane was subsequently incubated with 1:2000 diluted antisera at 4°C overnight. Next, the membrane was rinsed five times with TBST for 8 min and incubated with a 1:10000 dilution of goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson, United States) for 1 h at room temperature. Finally, the membrane was washed five times with TBST for 8 min and the excess buffer was dried with filter paper. Immunoreactive spots on the membrane were detected by chemiluminescence (PierceTM ECL Western Blotting Substrate, Thermo Scientific, United States) according to the manufacturer’s instructions. The blot was visualized using a ChemiDoTM XRS+ system (Bio-Rad, United States).
In-gel digestion was performed as previously described (Shevchenko et al., 1996) with some modifications. Briefly, immunoreactive protein spots were selected and manually excised from stained gels. The gel spots were destained with 30% acetonitrile (ACN) containing 100 mM NH4HCO3 and dehydrated in 100% ACN. The liquid phase was removed. The gel pieces were completely dried in a vacuum centrifuge. Next, the gel pieces were reswollen with 10 ng/μL sequencing-grade trypsin at 4°C for 30 min; the pieces were subsequently incubated at 37°C overnight (approximately 20 h). Excess trypsin solution was removed and peptides were extracted three times with 60% ACN containing 0.1% trifluoroacetic acid (TFA). Then the extracts were pooled and lyophilized.
The lyophilized peptides were dissolved in 20% acetonitrile. The samples (1 μL each) were spotted on a MALDI target plate and allowed to air-dry. Next, 0.5 μL of matrix (5 mg/mL α-cyano-4-hydroxycinnamic acid diluted in 0.1% TFA and 50% ACN) were mixed with dried peptides and allowed to air-dry. Sample analysis was carried out using a 4800 Plus MALDI-TOF/TOF MS Analyzer (Applied Biosystems, United States) in positive ion reflector mode with 2 kV accelerating voltage and 355 nm Nd:YAG laser source. Positive ion mode and automatic acquisition mode were used for data collection. A peptide mass fingerprint (PMF) in the 800–4000 Da range was generated. Peaks with a signal-to-noise ratio greater than 50 were selected for MS/MS analysis using a collision energy of 2 kV.
Data searches were performed using MASCOT 2.2 (Matrix Science, United Kingdom) and Global Proteome Server Explorer (Applied Biosystems) protein identification. The parameters were set as follows: UniProt database, and species restriction to A. acidoterrestris; Enzyme, Trypsin; Fixed modifications, Carbamidomethyl (C)s; Dynamical modifications, Oxidation (M); Peptide Mass Tolerance, ± 100 ppm; Fragment Mass Tolerance, ± 0.4 Da; Peptide Charge State, 1+; Max Missed Cleavages, 1. GPS software-reported protein scores with confidence intervals (C.I.%) greater than 95% were considered as successful identifications for proteins. Protein–protein interaction was performed using STRING [version 10.51 (Szklarczyk et al., 2017)] and the protein network was constructed.
Eight identified antigens, namely, GADPH, Polyamine aminopropyl transferase, Succinate-CoA ligase [ADP-forming] subunit alpha, EF-Tu, 6PGD, Hypothetical protein N007_04435, SOD, AhpC were chosen for antigenicity veridiction. Genomic DNA of DSM 3923 were extracted by QIAamp DNA Mini Kit (Qiagen, Germany) following manufacturer’s instruction. PCR reactions were performed using T100TM Thermal Cycler (Bio-Rad, United States). The PCR products were purified by Gel Extraction Kit (Omega BIO-TEK, United States) before cloned into pET-28a (+) and pET-32a (+) vectors (Novagen, Germany). The constructs were sequenced by ABI 3730XL. Recombinant plasmids were transformed into E. coli BL21 (DE3) (InvitrogenTM, United States) competent cells for expression. The recombinant proteins were purified by Ni-NTA His Bind Resin (Merck Millipore, United States) and examined by 12% SDS–PAGE. Western blot was performed as described above, apart from the transfer parameters which were changed from 25 V for 30 min into 15 V for 15 min.
Cell wall proteins of A. acidoterrestris were extracted using enzymolysis and separated by 2-D gel electrophoresis. A total of more than 200 spots in the gel were clearly observed with a molecular mass range from 10 to 75 kDa and pH range from 4 to 7 as shown in Figure 1A. Protein spots were evenly scattered and most of them showed in small light dots. High abundance proteins (big spots) were predominantly distributed in the 10–35 kDa range and low abundance proteins (small spots) in the 35–75 kDa range.
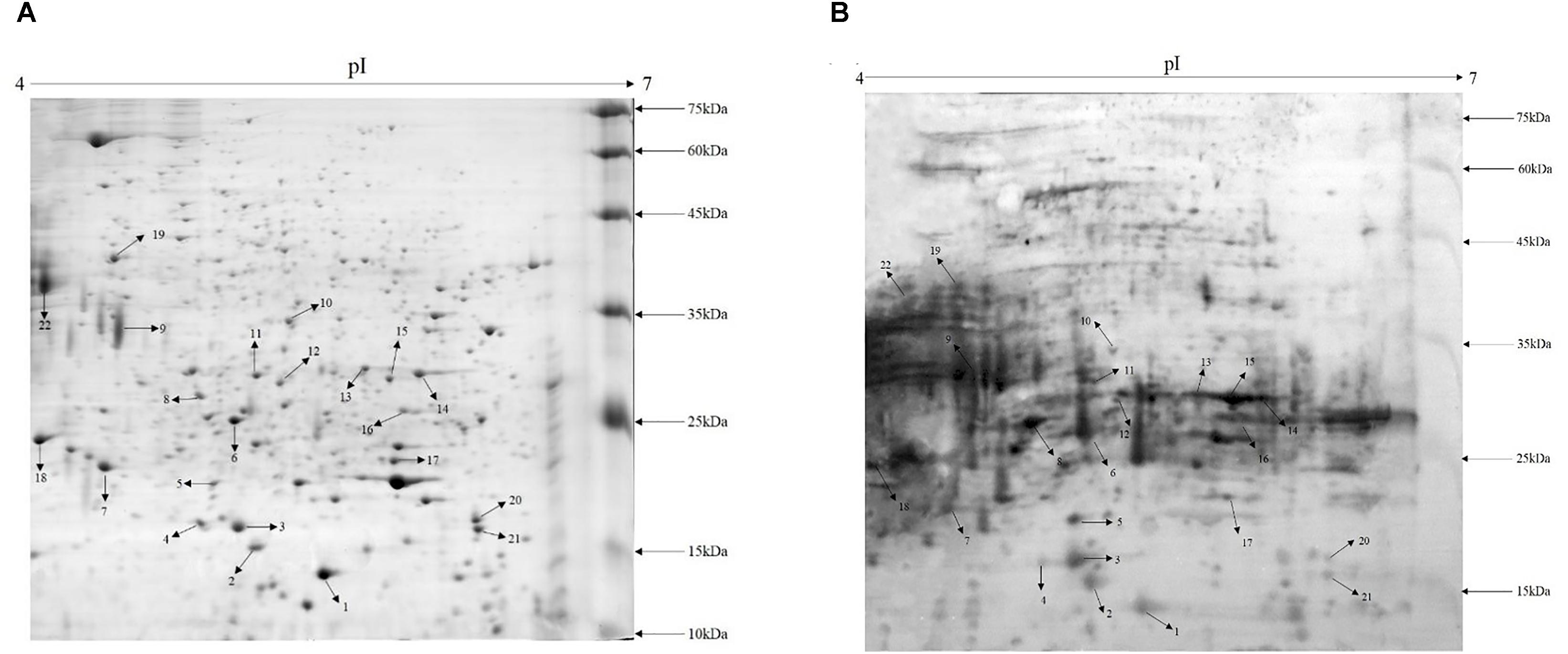
FIGURE 1. Proteome map and immunoblot profile of cell wall proteins of A. acidoterrestris. (A) 2-D map of cell wall proteins; (B) immunoblot analysis of cell wall proteins.
Cell wall extracts of A. acidoterrestris DSM 3923 separated by 2-D gel electrophoresis were blotted onto a PVDF membrane and visualized using chemiluminescence. Figure 1B shows that a large number of protein spots reacted with the antisera and the strongest reactors were mainly in the 15–35 kDa range. This analysis also indicated that, compared with the 2-D map, highly abundant proteins reacted strongly with antisera while those proteins that were less prevalent reacted with the antisera in a less intense fashion. Furthermore, several intense reaction spots that were not observed clearly on the gel were detected by Western blot, indicating that some proteins that were expressed at reduced levels but were exposed on the cell surface could potently stimulate host immune response (Wang et al., 2013).
Twenty-two immunoreactive spots recognized by antisera that correlated with relatively high abundance protein spots in the Coomassie-stained gel were picked for MALDI-TOF/TOF MS identification. All twenty-two immunoreactive spots were excised from the gel and eighteen spots were successfully identified (Table 1). Of the eighteen spots, eight were characterized as three kinds of hypothetical protein (N007_00440, N007_08970, N007_04435). Two spots were identified as the elongation factor Tu (EF-Tu), and the rest were classified as metabolic enzymes (Table 2). These proteins were mainly involved in transmembrane transport of matter, carbohydrate and energy metabolic process, chain elongation in polypeptide synthesis, maintaining redox homeostasis in respond to oxidative stress, and cell growth (Figure 2A and Table 2). Among the identified antigens, two were newly found in our study that never been reported in the previous researches. Five were identified as “common antigen” and the rest five were classified into “antigen been reported” (Figure 2B and Table 2). To further understand the property and correlation of immunogenic proteins, Protein–protein interaction prediction was performed using online database STRING. Figure 3 shows that seven proteins which involved in glycometabolism and oxidative stress could interact with each other. The three hypothetical proteins include two newly identified antigens showed no relationship with others.
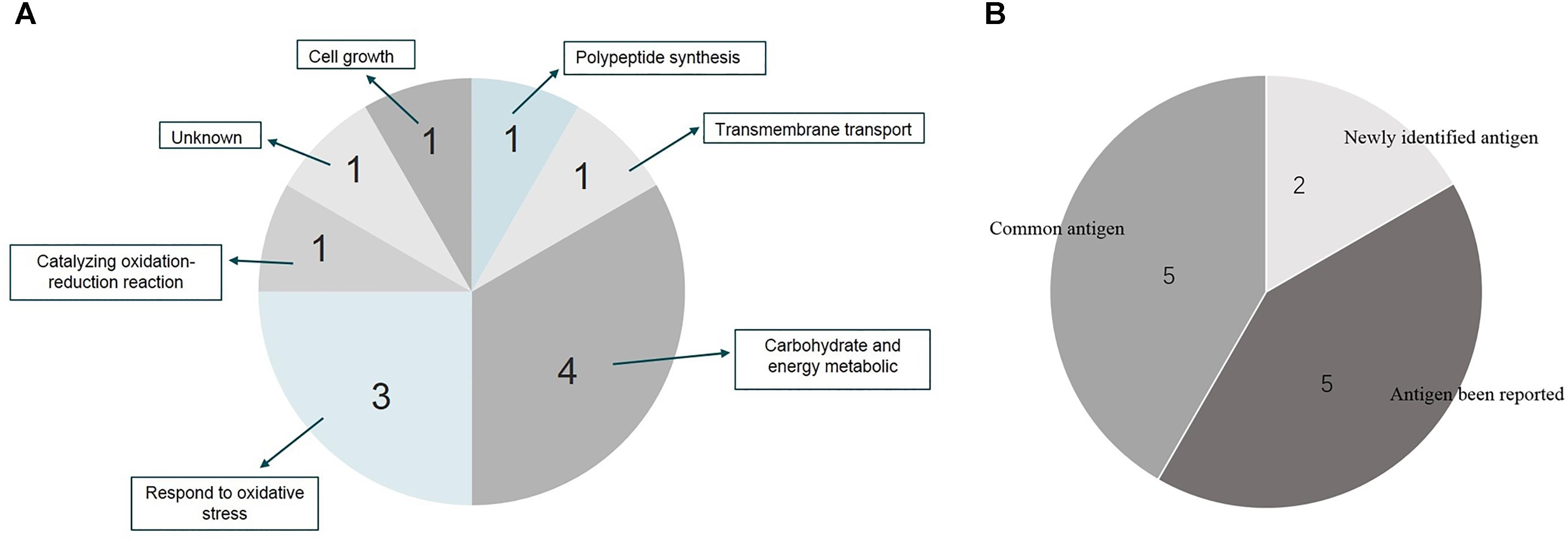
FIGURE 2. Distribution and function summary of identified antigens from A. acidoterrestris cell wall. (A) Function characterization of identified antigens; (B) antigen description of identified proteins.
To further evaluate the potential as biomarkers of the identified proteins, eight proteins were cloned and expressed in E. coli BL21 (DE3) cell. Figure 4A confirms that the recombinant proteins were successfully expressed. The molecule mass of recombinant proteins matched the data of MS analysis. Western blot demonstrates that the eight proteins could be recognized by anti-A. acidoterrestris antisera (Figure 4B).
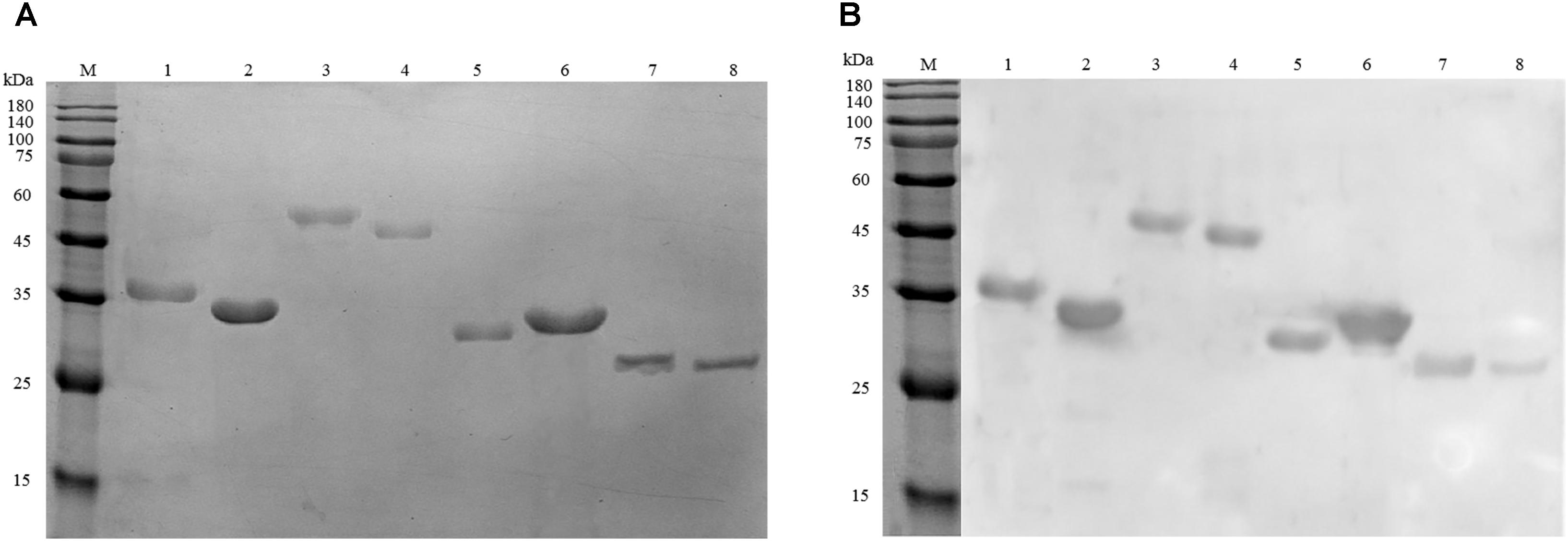
FIGURE 4. Analysis of recombinant antigen proteins. (A) SDS–PAGE of eight purified recombinant proteins. (B) Immunoreactivity analysis of recombinant proteins using anti-A. acidoterrestris polyclonal antibody. Lanes: M, pre-stain protein molecular weight marker (kDa). 1–8: GADPH, polyamine aminopropyl transferase, succinate-CoA ligase [ADP-forming] subunit alpha, EF-Tu, 6PGD, hypothetical protein N007_04435, SOD, AhpC.
Alicyclobacillus acidoterrestris is a thermo-acidophilic bacterial species that is capable of surviving conventional pasteurization procedures for fruit juices and acidic products. The presence of this bacterium results in product spoilage and economic losses (dos Anjos et al., 2016). However, existing detection methods used to identify this microorganism are not without problems (Chang et al., 2013). It has previously been suggested that bacterial cell wall-associated proteins could serve as potential diagnostic candidates for detection. However, the surface proteome of A. acidoterrestris cells has yet to be elucidated. Hence, in the present study, we utilized 2-D gel electrophoresis followed by Western blotting to elucidate new biomarkers for A. acidoterrestris.
The use of two kinds of cell wall hydrolases, lysozyme and mutanolysin, in the extraction of cell wall proteins, has facilitated the release proteins attached to the cell wall, thereby giving a more complete picture of the proteome. Figure 1A shows most of protein spots are small and faint, while only a few are large and of high intensity. In gram-positive bacteria, cell walls mainly contain peptidoglycan and teichoic acid,with less than 10% of the exported proteins covalently attached to the bacterial cell wall (Siegel et al., 2016). Thus, the cell wall proteome indicates that most proteins are expressed at low levels. However, a large number of spots could elicit strong reactions against the antisera including many spots which were not clearly observed on the gel. Overall, the data show that the cell wall proteins of A. acidoterrestris were successfully extracted and had a high affinity to the antisera. While these proteins were present at low levels, they were sufficiently exposed on the surface to elicit a strong antibody response (from the rabbits) (Wang et al., 2013).
Among the eighteen spots that were successfully identified, three proteins (Hypothetical protein N007_00440, Hypothetical protein N007_08970, Elongation factor Tu) were identified from two or more spots. The presence of the same protein in multiple spots was also observed in the proteomic study of Sporothrix schenckii (Messias et al., 2015). The existence of natural isoforms, post-translational modifications, or differential methods of sample preparation might explain these divergences (Wang et al., 2013).
Five of the identified proteins were recognized as “common antigen,” as these proteins have been widely reported in pathogenic bacteria. Typically, 6-phosphogluconate dehydrogenase (6PGD) (spot 21), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (spot 17) and malate dehydrogenase (MDH) (spot 10) which play a key role in the carbohydrate and energy metabolic pathway so that they can catalyze different substrates to maintain normal function of bacteria (Rendina et al., 1984; Sirover, 1997; Musrati et al., 1998). Consequently, these proteins are the most active in the cellular metabolism and have been identified as immunogenic proteins in many pathogenic bacteria (Severin et al., 2007; Wang et al., 2013; Elfaham et al., 2017). Superoxide dismutase (SOD) (spot 20) were also recognized by the antisera. It can convert superoxide (O2-) radicals into either ordinary molecular oxygen (O2) or hydrogen peroxide (H2O2) to protect cells from oxidative stress and contributes to the survival of some pathogenic bacteria in vivo (Zhang et al., 1991). Thus, it has served as important biomarker in Cronobacter sakazakii (Wang et al., 2013). Notably, Elongation factor Tu (EF-Tu) (spot 4, 6) were also identified in our study. This protein is mainly involved in chain elongation during polypeptide synthesis which could bind and transport of aminoacyl-tRNA to the ribosome and interact with GTP in the catalysis of GTP into GDP (Caldas et al., 1998). It is widely distributed in bacteria and has been identified as immunogen in many microorganisms (Fowsantear et al., 2014; Casas et al., 2017).
Five proteins were classified as “antigen been reported,” which means that these proteins have been identified as antigen candidates and served as biomarkers in diagnosis of pathogens in other studies (Stent et al., 2012; Kim et al., 2014; Gomes et al., 2016). These proteins also participate in the metabolic pathway to regulate the function of bacteria. Thiol peroxidase and alkyl hydroperoxide reductase subunit C (AhpC) (spot 5) reduce hydrogen peroxide and peroxynitrite to maintain homeostasis of bacteria and protect cell from oxidative damage (Delaunay et al., 2002; Zuo et al., 2014). They have been identified as important immunogens and vaccine candidates of Helicobacter pylori (Stent et al., 2012) and anthrax (Kim et al., 2014), respectively. Succinate-CoA ligase [ADP-forming] (spot 12), also named Succinyl-CoA synthetase, functions in substrate-level phosphorylation in the citric acid cycle (TCA), catalyzing succinate, CoA and ATP or GTP into Succinyl-CoA, phosphate and ADP or GDP, respectively. It has been suggested that this enzyme is a potential antigenic target that could be utilized for the development of a novel diagnostic approach for pathogenic bacterium detection Bartonella bacilliformis (Gomes et al., 2016). Moreover, an ATP-binding cassette (ABC) transporter substrate-binding protein analog, hypothetical protein N007_00440 (spot 1, 18, and 19) was recognized as an immunoreactive protein. This protein is mainly involved in transmembrane transport process and is capable of binding substrates outside of the cell and delivering them into cell (Schneider and Hunke, 1998). Members of this family are also involved in the antigen presentation and bacterial pathogenesis and were able to be used as targets to facilitate the detection of pathogenic bacteria (Rodriguez-Ortega et al., 2006; Chen et al., 2018). Polyamine aminopropyl transferase (spot 13) is involved in the first step of the sub-pathway of polyamine biosynthesis which is mainly function in the growth and function of a normal cell (Wallace et al., 2003; Belda-Palazon et al., 2012).
Two proteins were newly identified as immunogens. Protein spots 9, 14, 16, and 22 were identified as the same protein, hypothetical protein N007_08970. This protein was first identified and reported in our research. The structural property and functional information have not been clearly elucidated in any research up to now. Furthermore, homologs from other organisms eliciting immunoreactions have yet to be reported. Hypothetical protein N007_04435 (spot 11) was also first identified as a novel antigen in our study and predicted to be similar to aldo–keto reductases which are involved in the catalysis of a diverse range of substrates, including aliphatic and aromatic aldehydes, monosaccharides, steroids, prostaglandins, polycyclic aromatic hydrocarbons, and isoflavonoids (Hyndman et al., 2003). As for hypothetical protein N007_04435 (spot 11), homologs from other organisms eliciting similar immunoreactions have yet to be elucidated. However, Aldo–keto reductases have been described as therapeutic drug targets (Jez et al., 1997). Our present work demonstrates that this protein is easily recognizable by antibodies. The results support that these proteins are excellent candidates as biomarkers for the detection of A. acidoterrestris. The fact that no interaction partners were identified for either N007_08970 or N007_04435 (Figure 3), illustrates their novelty as antigens from a different perspective.
This is the first study to report the occurrence of specific antigens in the cell surface of A. acidoterrestris, a species that causes fruit juice contamination leading to significant economic losses. Hypothetical protein N007_08970 and Hypothetical protein N007_04435 are two antigens that were first identified as part of this study. The other antigens that were identified as part of this study are homologs of antigens reported in pathogenic species and could also act as biomarkers. We believe that the species-specific antigens identified in this study may be important in the development of methods for the accurate detection of A. acidoterrestris in fruit juices.
GADPH, polyamine aminopropyl transferase, succinate-CoA ligase [ADP-forming] subunit alpha, EF-Tu, 6PGD, hypothetical protein N007_04435, SOD and AhpC were selected to express as recombinant in E. coli to further evaluate the potential as molecular biomarkers for detection or antibody development. All selected proteins could react with antisera against A. acidoterrestris. This demonstrates that these proteins are suitable targets for future development of an immunological detection method for A. acidoterrestris.
YS conceived and designed the experiments, analyzed the data, and wrote the manuscript. YS and YZ conducted the experiments. YY and TY supervised all work. YS, JW, YY, and TY revised the manuscript.
This work was supported by the National Natural Science Foundation of China (31671866), the Scientific and Technology Cooperation Project in Hong Kong, Macau, and Taiwan of China (2015DFT30130), the Shaanxi Special Project of China (2016KTCQ03-12), and the National Basic Research Program of China (2013FY113400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Shanghai Hoogen Biotechnology Co., Ltd., for their technical help.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03032/full#supplementary-material
Belda-Palazon, B., Ruiz, L., Marti, E., Tarraga, S., Tiburcio, A. F., Culianez, F., et al. (2012). Aminopropyl transferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7:e46907. doi: 10.1371/journal.pone.0046907
Caldas, T. D., El Yaagoubi, A., and Richarme, G. (1998). Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 273, 11478–11482. doi: 10.1074/jbc.273.19.11478
Casas, V., Rodriguez-Asiain, A., Pinto-Llorente, R., Vadillo, S., Carrascal, M., and Abian, J. (2017). Brachyspira hyodysenteriae and B. pilosicoli proteins recognized by sera of challenged pigs. Front. Microbiol. 8:723. doi: 10.3389/fmicb.2017.00723
Celenk, M., and Ayse Handan, B. (2015). Effects of pomegranate and pomegranate-apple blend juices on the growth characteristics of Alicyclobacillus acidoterrestris DSM 3922 type strain vegetative cells and spores. Int. J. Food Microbiol. 200, 52–56. doi: 10.1016/j.ijfoodmicro.2015.01.019
Chang, S. S., and Kang, D. H. (2004). Alicyclobacillus spp. in the fruit juice industry: history, characteristics, and current isolation/detection procedures. Crit. Rev. Microbiol. 30, 55–74. doi: 10.1080/10408410490435089
Chang, S. S., Park, S. H., and Kang, D. H. (2013). Development of novel agar media for isolating guaiacol producing Alicyclobacillus spp. Int. J. Food Microbiol. 164, 1–6. doi: 10.1016/j.ijfoodmicro.2013.03.015
Chen, S., Hao, H., Zhao, P., Ji, W., Li, M., Liu, Y., et al. (2018). Differential immunoreactivity to bovine convalescent serum between mycoplasma bovis biofilms and planktonic cells revealed by comparative immunoproteomic analysis. Front. Microbiol. 9:379. doi: 10.3389/fmicb.2018.00379
Concina, I., Bornsek, M., Baccelliere, S., Falasconi, M., Gobbi, E., and Sberveglieri, G. (2010). Alicyclobacillus spp detection in soft drinks by electronic nose. Food Res. Int. 43, 2108–2114. doi: 10.1016/j.foodres.2010.07.012
Delaunay, A., Pflieger, D., Barrault, M. B., Vinh, J., and Toledano, M. B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481. doi: 10.1016/S0092-8674(02)01048-6
dos Anjos, M. M., da Silva, A. A., de Pascoli, I. C., Mikcha, J. M., Machinski, M. Jr., Peralta, R. M., et al. (2016). Antibacterial activity of papain and bromelain on Alicyclobacillus spp. Int. J. Food Microbiol. 216, 121–126. doi: 10.1016/j.ijfoodmicro.2015.10.007
Elfaham, M. H., Eissa, M. M., Igetei, J. E., Amer, E. I., Liddell, S., Elazzouni, M. Z., et al. (2017). Treatment of Schistosoma mansoni with miltefosine in vitro enhances serological recognition of defined worm surface antigens. PLoS Negl. Trop. Dis. 11:e0005853. doi: 10.1371/journal.pntd.0005853
Fernandez, P., Antonio Gabaldon, J., and Jesus Periago, M. (2017). Detection and quantification of Alicyclobacillus acidoterrestris by electrical impedance in apple juice. Food Microbiol. 68, 34–40. doi: 10.1016/j.fm.2017.06.016
Fowsantear, W., Argo, E., Pattinson, C., and Cash, P. (2014). Comparative proteomics of Helicobacter species: the discrimination of gastric and enterohepatic Helicobacter species. J. Proteomics 97, 245–255. doi: 10.1016/j.jprot.2013.07.016
Gomes, C., Palma, N., Pons, M. J., Magallon-Tejada, A., Sandoval, I., Tinco-Valdez, C., et al. (2016). Succinyl-CoA synthetase: new antigen candidate of Bartonella bacilliformis. PLoS Negl. Trop. Dis. 10:e0004989. doi: 10.1371/journal.pntd.0004989
Hernandez-Haro, C., Llopis, S., Molina, M., Monteoliva, L., and Gil, C. (2015). Immunoproteomic profiling of Saccharomyces cerevisiae systemic infection in a murine model. J. Proteomics 112, 14–26. doi: 10.1016/j.jprot.2014.08.008
Hyndman, D., Bauman, D. R., Heredia, V. V., and Penning, T. M. (2003). The aldo-keto reductase superfamily homepage. Chem. Biol. Interact 143-144, 621–631. doi: 10.1016/S0009-2797(02)00193-X
Jez, J. M., Bennett, M. J., Schlegel, B. P., Lewis, M., and Penning, T. M. (1997). Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 326(Pt 3), 625–636. doi: 10.1042/bj3260625
Kim, Y. H., Kim, K. A., Kim, Y. R., Choi, M. K., Kim, H. K., Choi, K. J., et al. (2014). Immunoproteomically identified GBAA_0345, alkyl hydroperoxide reductase subunit C is a potential target for multivalent anthrax vaccine. Proteomics 14, 93–104. doi: 10.1002/pmic.201200495
Messias, R. A., Kubitschek-Barreira, P. H., Ferreira, F. G., de Almeida, S. R., Lopes-Bezerra, L. M., and de Camargo, Z. P. (2015). Immunoproteomic analysis reveals a convergent humoral response signature in the Sporothrix schenckii complex. J. Proteomics 115, 8–22. doi: 10.1016/j.jprot.2014.11.013
Musrati, R. A., Kollarova, M., Mernik, N., and Mikulasova, D. (1998). Malate dehydrogenase: distribution, function and properties. Gen. Physiol. Biophys. 17, 193–210.
Oteiza, J. M., Soto, S., Alvarenga, V. O., Sant’Ana, A. S., and Gianuzzi, L. (2015). Fate of Alicyclobacillus spp. in enrichment broth and in juice concentrates. Int. J. Food Microbiol. 210, 73–78. doi: 10.1016/j.ijfoodmicro.2015.05.021
Pasquina, L. W., Maria, J. P. S., and Walker, S. (2013). Teichoic acid biosynthesis as an antibiotic target. Curr. Opin. Microbiol. 16, 531–537. doi: 10.1016/j.mib.2013.06.014
Pérez-Cacho, P. R., Danyluk, M. D., and Rouseff, R. (2011). GC–MS quantification and sensory thresholds of guaiacol in orange juice and its correlation with Alicyclobacillus spp. Food Chem. 129, 45–50. doi: 10.1016/j.foodchem.2011.04.014
Rasmussen, K. J., Mattsson, A. H., Pilely, K., Asferg, C. A., Ciofu, O., Vitved, L., et al. (2016). Proteome-wide antigen discovery of novel protective vaccine candidates against Staphylococcus aureus infection. Vaccine 34, 4602–4609. doi: 10.1016/j.vaccine.2016.07.016
Ray, A., Kinch, L. N., Santos, M. D. S., Grishin, N. V., Orth, K., and Salomon, D. (2016). Proteomics analysis reveals previously uncharacterized virulence factors in Vibrio proteolyticus. mBio 7:e01077-16. doi: 10.1128/mBio.01077-16
Rendina, A. R., Hermes, J. D., and Cleland, W. W. (1984). Use of multiple isotope effects to study the mechanism of 6-phosphogluconate dehydrogenase. Biochemistry 23, 6257–6262. doi: 10.1021/bi00320a056
Rodriguez-Ortega, M. J., Norais, N., Bensi, G., Liberatori, S., Capo, S., Mora, M., et al. (2006). Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191–197. doi: 10.1038/nbt1179
Schneider, E., and Hunke, S. (1998). ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22, 1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x
Severin, A., Nickbarg, E., Wooters, J., Quazi, S. A., Matsuka, Y. V., Murphy, E., et al. (2007). Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J. Bacteriol. 189, 1514–1522. doi: 10.1128/JB.01132-06
Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi: 10.1021/ac950914h
Siegel, J. L., Hurst, S. F., Liberman, E. S., Coleman, S. E., and Bleiweis, A. S. (1981). Mutanolysin-induced spheroplasts of Streptococcus mutants are true protoplasts. Infect. Immun. 31, 808–815.
Siegel, S. D., Liu, J., and Ton-That, H. (2016). Biogenesis of the Gram-positive bacterial cell envelope. Curr. Opin. Microbiol. 34, 31–37. doi: 10.1016/j.mib.2016.07.015
Sirover, M. A. (1997). Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell. Biochem. 66, 133–140. doi: 10.1002/(SICI)1097-4644(19970801)66:2<133::AID-JCB1>3.0.CO;2-R
Stent, A., Every, A. L., Ng, G. Z., Chionh, Y. T., Ong, L. S., Edwards, S. J., et al. (2012). Helicobacter pylori thiolperoxidase as a protective antigen in single- and multi-component vaccines. Vaccine 30, 7214–7220. doi: 10.1016/j.vaccine.2012.10.022
Steyn, C. E., Cameron, M., and Witthuhn, R. C. (2011). Occurrence of Alicyclobacillus in the fruit processing environment–a review. Int. J. Food Microbiol. 147, 1–11. doi: 10.1016/j.ijfoodmicro.2011.03.004
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Uchida, R., and Silva, F. V. M. (2017). Alicyclobacillus acidoterrestris spore inactivation by high pressure combined with mild heat: modeling the effects of temperature and soluble solids. Food Control 73, 426–432. doi: 10.1016/j.foodcont.2016.08.034
Wallace, H. M., Fraser, A. V., and Hughes, A. (2003). A perspective of polyamine metabolism. Biochem. J. 376(Pt 1), 1–14. doi: 10.1042/BJ20031327
Wang, J., Du, X. J., Lu, X. N., and Wang, S. (2013). Immunoproteomic identification of immunogenic proteins in Cronobacter sakazakii strain BAA-894. Appl. Microbiol. Biotechnol. 97, 2077–2091. doi: 10.1007/s00253-013-4720-5
Wang, Z., Yue, T., Yuan, Y., Cai, R., Guo, C., Wang, X., et al. (2012). Development of polyclonal antibody-based indirect enzyme-linked immunosorbent assay for the detection of Alicyclobacillus strains in apple juice. J. Food Sci. 77, M643–M649. doi: 10.1111/j.1750-3841.2012.02961.x
Wu, X., Xiong, E., Wang, W., Scali, M., and Cresti, M. (2014). Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 9, 362–374. doi: 10.1038/nprot.2014.022
Xiong, X., Wang, X., Wen, B., Graves, S., and Stenos, J. (2012). Potential serodiagnostic markers for Q fever identified in Coxiella burnetii by immunoproteomic and protein microarray approaches. BMC Microbiol. 12:35. doi: 10.1186/1471-2180-12-35
Yamazaki, K., Teduka, H., and Shinano, H. (1996). Isolation and identification of Alicyclobacillus acidoterrestris from acidic beverages. Biosci. Biotechnol. Biochem. 60, 543–545. doi: 10.1271/bbb.60.543
Yu, T., Xu, X., Peng, Y., Luo, Y., and Yang, K. (2012). Cell wall proteome of Clostridium thermocellum and detection of glycoproteins. Microbiol. Res. 167, 364–371. doi: 10.1016/j.micres.2012.02.006
Zhang, Y., Lathigra, R., Garbe, T., Catty, D., and Young, D. (1991). Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol. Microbiol. 5, 381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x
Keywords: immunoproteomics, immunogenic proteins, Alicyclobacillus acidoterrestris, cell surface proteins, biomarkers
Citation: Shi Y, Yue T, Zhang Y, Wei J and Yuan Y (2018) Surface Immunoproteomics Reveals Potential Biomarkers in Alicyclobacillus acidoterrestris. Front. Microbiol. 9:3032. doi: 10.3389/fmicb.2018.03032
Received: 12 July 2018; Accepted: 23 November 2018;
Published: 04 December 2018.
Edited by:
Sergio Uzzau, University of Sassari, ItalyReviewed by:
Nurul-Syakima Ab Mutalib, UKM Medical Molecular Biology Institute (UMBI), MalaysiaCopyright © 2018 Shi, Yue, Zhang, Wei and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yahong Yuan, eXloMzI0QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.