- 1Institute for Infectious Diseases, Faculty of Medicine, University of Bern, Bern, Switzerland
- 2Graduate School for Cellular and Biomedical Sciences, University of Bern, Bern, Switzerland
- 3Janssen Vaccines and Prevention, Leiden, Netherlands
- 4Julius Center, UMC Utrecht, Utrecht, Netherlands
- 5Proteomics and Mass Spectrometry Core Facility, Department for BioMedical Research (DBMR), University of Bern, Bern, Switzerland
The Ami-AliA/AliB oligopeptide permease of Streptococcus pneumoniae has been suggested to play a role in environmental sensing and colonisation of the nasopharynx by this human bacterial pathogen by binding peptides derived from bacterial neighbours of other species in the microbiota. Here, we investigated the effects of the peptide ligands of the permease’s substrate binding proteins AmiA, AliA, and AliB on pneumococcal phenotype. AmiA and AliA ligands reduced pneumococcal growth, increased biofilm production and reduced capsule size. In contrast, AliB ligand increased growth and greatly increased bacterial chain length. A decrease in transformation rate was observed in response to all three peptides. Changes in protein expression were also observed, particularly those associated with metabolism and cell wall synthesis. Understanding interspecies bacterial communication and its effect on development of colonising versus invasive phenotypes has the potential to reveal new targets to tackle and prevent pneumococcal infections.
Introduction
Streptococcus pneumoniae (pneumococcus) is one of the most extensively studied microorganisms, but this opportunistic pathogen remains a major causative agent of several human diseases including otitis media, pneumonia, sepsis and bacterial meningitis. However, the pneumococcus is more often a harmless inhabitant of the human nasopharynx, a niche it shares with many other microorganisms. Sensing of these microbial neighbours has been proposed to occur by binding of short peptides of these other bacterial species by S. pneumoniae via oligopeptide binding proteins of ATP-binding cassette (ABC) transporters (Hathaway et al., 2014). ABC transporters are also of interest as virulence factors since mutations in some have resulted in a reduction in pneumonia and bacteraemia in mouse models (Polissi et al., 1998; Lau et al., 2001).
One ABC transporter, the Ami-AliA/AliB permease, has been suggested to play a role in uptake of environmental signals and in competence for genetic transformation (Claverys et al., 2000). One of these permease’s oligopeptide binding proteins, AliB, has also been shown to aid in colonisation of the nasopharynx in a mouse model (Kerr et al., 2004). Previously, it has been shown that peptides found in other bacterial species are recognised by homologs of AliB: AliB-like ORF1 and ORF2 in non-encapsulated S. pneumoniae and that binding of the proteins to their peptide ligands affected pneumococcal phenotype and gene expression (Hathaway et al., 2014; Nasher et al., 2018a).
For the three substrate binding proteins of the Ami-AliA/AliB permease, we have shown recently that AmiA binds peptide AKTIKITQTR, matching an amino acid sequence found in 50S ribosomal subunit protein L30; AliA binds peptide FNEMQPIVDRQ, matching an amino acid sequence found in 30S ribosomal protein S20 and AliB binds peptide AIQSEKARKHN, matching an amino acid sequence found in 30S ribosomal protein S20 (Nasher et al., 2018b). These Ami-AliA/AliB peptide ligands are found in multiple bacterial species in the class of Gammaproteobacteria which includes common colonisers of the nostrils and nasopharynx (Pettigrew et al., 2012; Biesbroek et al., 2014; Mika et al., 2015; Teo et al., 2015). We therefore speculate that the pneumococcus binds these peptides to sense the presence and composition of its bacterial neighbours and adapt by altering its phenotype.
We determined here the effects of binding of AmiA, AliA, and AliB to their ligands on phenotypes associated with colonisation or virulence in wild type strains and mutants lacking the substrate binding proteins. Effects were found on growth, capsule size, chaining, transformation rate, and biofilm as well as on the pneumococcal proteome.
Materials and Methods
Ethics Statement
The human nasopharyngeal epithelial cell line Detroit 562 (CCL-138 was obtained from the American Type Culture Collection (ATCC)).
Clinical isolates of S. pneumoniae were selected from two nationwide surveillance programmes (Mühlemann et al., 2003; Kronenberg et al., 2006), from anonymous donors and have been used in previous studies (Hathaway et al., 2004, 2012).
Bacterial Culture and Strains
Strain D39 (serotype 2) pneumococci were used as the parental strain for the production of mutants. Construction of the individual, ΔAmiA, ΔAliA, ΔAliB, and triple mutant ΔAmiA/ΔAliA/ΔAliB was described previously (Alloing et al., 1994; Kerr et al., 2004). Clinical isolates 106.66 (serotype 6B) and 110.58 (non-encapsulated, classic lineage) (Hilty et al., 2014) were also used. Bacteria, stored at -80°C using Protect bacterial preservers (Technical Service Consultants, Heywood, United Kingdom), were grown on Columbia sheep blood agar (CSBA) plates at 37°C, 5% CO2. Overnight cultures were prepared with 3–10 colonies in 5 mL brain heart infusion broth (Becton Dickinson and Company, le Pont de Claix, France) containing 5% foetal calf serum (FCS; Biochrom KG, Berlin, Germany; BHI+FCS medium).
Growth Assay
Strains were streaked onto CSBA plates and incubated at 37°C in a 5% CO2-enriched atmosphere overnight then sub-cultured in BHI+FCS medium to OD600nm 0.5, centrifuged at 3000 ×g for 5 min and resuspended in 5 mL chemically defined medium (CDM) which contains 5.5 mM glucose (Schaffner et al., 2014). Growth was monitored in sterile flat-bottomed 96-well microtiter plates (Nunclon Surface, Nunc, Denmark) based on the method of Brewster (2003). In brief, 200 μL bacteria culture was grown per well at 37°C and OD450nm measured every 30 min using a VersaMax microplate reader (Molecular Devices) over 22 h with or without peptide ligands (synthesised by PolyPeptide Group, Strasbourg, France). The plate was shaken automatically for 5 s before each reading. The problem of condensation affecting the readings was avoided by pre-treating the lids of the 96-well plates with 3 mL 0.05% Triton X-100 in 20% ethanol and allowing them to air-dry before use (Hathaway et al., 2012).
Growth Competition Assay
D39 and its mutants ΔAmiA, ΔAliA, ΔAliB, and ΔAmiA/ΔAliA/ΔAliB were streaked onto CSBA plates and incubated at 37°C in a 5% CO2-enriched atmosphere overnight then subcultured in BHI+FCS medium to OD600nm 0.5, centrifuged at 3000 ×g for 5 min and resuspended in 5 mL CDM. 250 μL bacterial culture (125 μL of each of the two strains being compared) was transferred to 4.75 mL CDM pre-warmed to 37°C. The initial inoculum was plated out to check that there was a 1:1 mixture of the two bacterial strains. Peptide ligands were added to a final concentration of 0.5 mg/mL, and the culture incubated to an OD600nm 0.3. Serial dilutions in PBS were plated onto CSBA plates with and without antibiotics to differentiate between wild type (susceptible) and mutants (resistant) strains. ΔAmiA mutant was selected on 10 μg/mL of tetracycline, ΔAliB on 2 μg/mL of chloramphenicol, and both ΔAliA and triple mutant ΔAmiA/ΔAliA/ΔAliB on 1 μg/mL of erythromycin/mL. After overnight incubation, the numbers of colonies were counted and the colony forming units (CFU) for each strain calculated.
FITC-Dextran Exclusion Assay
Capsule thickness was determined by measuring the zone of exclusion of FITC-dextran as described previously (Gates et al., 2004; Hathaway et al., 2012), using FITC-dextran of 2000 kDa (Sigma-Aldrich) with the following difference, bacteria were cultured overnight in BHI+FCS medium until OD600nm = 0.5 then 100 μL subcultured into 2 mL of CDM with and without peptide ligands and grown to an OD600nm = 0.3. After centrifuging at 3000 ×g for 5 min the pellet was resuspended in 500 μL of PBS. 10 μL bacterial suspension was mixed with 2 μL FITC-dextran (10 mg/mL in water), pipetted onto a microscope slide and a coverslip applied firmly. The slides were viewed using a Zeiss Axio Imager M1 fluorescence microscope with a 100× objective and photographed by a Zeiss AxioCam HRc camera. The images were converted to grayscale and analysed with UTHSCSA ImageTool for Windows v3.0 (University of Texas Health Science Center, San Antonio, TX, United States) software. A bright field image was also photographed for each fluorescent image recorded in order to count the number of bacteria. Each image contained between 8 and 150 bacteria, five images were counted per group in each of three independent experiments making a total of 15 images per group. Each data point in Figure 3B represents one image.
Chain Formation
Bacteria were cultured overnight in BHI+FCS medium until OD600nm = 0.5 then 100 μL sub-cultured into 2 mL of CDM with and without peptide ligands and grown to an OD600nm = 0.3. After centrifuging at 3000 ×g for 5 min the pellet was resuspended in 500 μL of PBS, 25 μL of the suspension was put on microscope slides and Gram stained. The slides were viewed using a Zeiss Axio microscope with a 100× objective and photographed by a Zeiss AxioCam HRc camera. A bright field image was acquired to count the number of bacteria per chain. 5 to 78 bacterial chains were counted per image, five images were counted per group in each of three independent experiments making a total of 15 images per group. Each data point in Figure 4B represents one image.
Adherence to Human Epithelial Cell Line
Adherence to the nasopharyngeal epithelial cell line Detroit 562 (CCL-138, from the ATCC) was performed as described previously (Hathaway et al., 2004) with the following differences: A total of 4 × 105 cells/well were cultured in a 24-well tissue culture plate in Minimum Essential Medium+Earle’s salts (MEM) containing 10% FCS, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids and 1 mM sodium pyruvate (Gibco, United Kingdom) until reaching complete confluence at 37°C in 5% CO2. S. pneumoniae strains D39 and its mutants as well as strains 106.66 and 110.58 were grown in BHI+FCS to the logarithmic phase OD600nm = 0.4 and washed in MEM then resuspended to give 107 bacteria in 0.5 mL MEM, which was added per well of washed Detroit cells. The bacteria were centrifuged onto the cells at 390 ×g for 5 min at room temperature. After 1 h incubation of the cells with the bacteria at 37°C and five washes with PBS, 200 μL of trypsin was then added to each well and incubated for 5 min in 37°C to detach the cells. Adhered bacteria were recovered, serially diluted in 0.85% NaCl and plated onto CSBA. CFU/mL was then calculated after an overnight incubation at 37°C in 5% CO2.
Biofilm Assay
Biofilm assays were performed as previously described (Marks et al., 2012b) with the following modifications. A total of 4 × 105 Detroit cells/well in MEM salts containing 10% FCS, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids and 1 mM sodium pyruvate (Gibco, United Kingdom) were cultured in 24-well tissue culture plate until reaching complete confluence at 37°C in 5% CO2. Strains D39 and its mutants and strains 106.66 and 110.58 were grown in CDM to OD600nm 0.4 to give 105 bacteria in 0.5 mL per well. 5 μL salmon sperm DNA was added into each well and, where indicated, corresponding peptide ligands, followed by 16 h of static incubation at 37°C, 5% CO2. The medium containing the planktonic bacteria was removed and each well was washed twice with PBS taking care not to disturb the biofilm. 200 μL PBS was then added per well and the plate was sonicated for 6 s in a water bath and then the biofilm scraped thoroughly from the wells. The contents of the wells were recovered and the CFU determined by plating out serial dilutions CSBA and incubating overnight at 37°C.
Transformation Assay
To determine transformation rates of strain D39 and its mutants, strains 106.66 and 110.58, bacteria were grown to an OD600nm = 0.15 in BHI+FCS. A total of 0.5 mL of the culture was transferred to 9.5 mL CDM prewarmed to 30°C and incubated for 15 min at 30°C in a waterbath. Competence-stimulating peptide 1 (CSP 1; ERLSKFFRFILQRLL) for D39 and its mutants and CSP 2 (EMRISRIILDFLFLRKK) for 106.66 and 110.58 were added to final concentration 100 ng/mL with and without AmiA, AliA, and AliB peptide ligands to a final concentration of 0.5 mg/mL, the culture incubated for 15 min at 30°C. Chromosomal DNA, 1 μg, from streptomycin resistant strain 104.37 was added and the culture incubated for 60 min at 30°C, then 90 min at 37°C. Serial dilutions in PBS were plated onto CSBA plates with and without 200 μg/mL streptomycin. After overnight incubation, the numbers of colonies were counted and the transformation rate calculated.
Proteomic Analysis
Strain D39 and its mutants were cultured overnight in BHI+FCS medium until OD600nm = 0.5 then 100 μL sub-cultured into 10 mL of CDM and grown to an OD600nm = 0.2, the culture was split into two 5 mL aliquots and 0.5 mg/mL of peptides were added to one sample and incubated for 15 min at 37°C. Both cultures were centrifuged for 5 min at 4000 ×g and the pellets were resuspended in 50 mM Tris–HCl and 8 M urea and sonicated. Sample processing, Liquid Chromatography Mass Spectrometry (LC-MS/MS) and data interpretation was essentially done as described previously (Engel et al., 2014) with the following changes. LC-MS/MS analysis was carried out on an UltiMate 3000 Nano LC coupled to an Orbitrap Fusion Lumos instrument (Thermo Fisher Scientific) acquiring full MS scans in the m/z range 400–1400 in the orbitrap at resolution 120,000 with AGC set to 4e5 and maximal ion injection time of 50 ms. Peptide precursors with charge 2–8 were fragmented once in the ion trap then excluded for 30 s. The ion trap settings were data-dependent MS2 cycle time of 3 s, isolation width of 1.6 m/z, fragmentation HCD mode with 30% normalised collision energy, AGC of 1e4 with maximal ion injection time of 35 ms. The LC-MS/MS data was processed with MaxQuant (version 1.5.4.1) using default settings for peak detection, strict trypsin cleavage rule allowing for up to three missed cleavages, variable oxidation on methionine and acetylation of protein N-termini with strict carbamidomethylation of cysteines. Match between runs was activated with a retention time window of 0.7 min. The fragment spectra were interpreted with the S. pneumoniae strain D39 (serotype 2) ensemble database (version gca_d39.ASM1436v1). LFQ intensities were used for data analysis with imputation of missing values from the low end of the LOG2 transformed intensity distribution of each LC-MS/MS run using Perseus (version 1.5.5.3) as suggested by Lazar et al. (2016).
Statistics
For proteomics, differential protein expression analysis was performed with t-tests within Perseus, including a permutation-based false discovery rate estimation (cutoff at 5% and using S0 function set at 0.5) to correct for multiple testing. For other experiments two-way ANOVA or student t-tests were performed to obtain p-values using the software GraphPad Prism (Version 7, GraphPad Software, Inc.).
Results
Peptide Ligands Affect Pneumococcal Growth
Streptococcus pneumoniae strain D39 (serotype 2) and its mutants in which one or all three of the substrate-binding proteins have been deleted (ΔAmiA, ΔAliA, ΔAliB, and ΔAmiA/ΔAliA/ΔAliB) were grown in CDM in 96 well plates in the absence and presence (at concentrations of between 0.5 and 0.062 mg/mL) of AmiA, AliA, and AliB peptide ligands for 22 h.
For strain D39, AmiA, and AliA peptide ligands reduced growth significantly in a dose dependent way (Figures 1A,B) whilst AliB peptide ligand slightly reduced the lag phase and increased the maximum optical density (OD) (Figure 1C). To check that these effects were peptide specific, D39 was grown in the presence of peptides with single amino acid differences from peptides AliA and AliB (Figure 1D). (FSEMQPIVDRQ differs from AliA ligand at the second amino acid and AIQSEKRRKHN differs from AliB ligand at the seventh amino acid, underlined; a single amino acid version of AmiA peptide was insoluble and so could not be used in the growth assay). A single amino acid substitution caused AliA peptide to lose its ability to suppress growth and the substitution in the AliB peptide reduced its ability to boost growth response to the peptide.
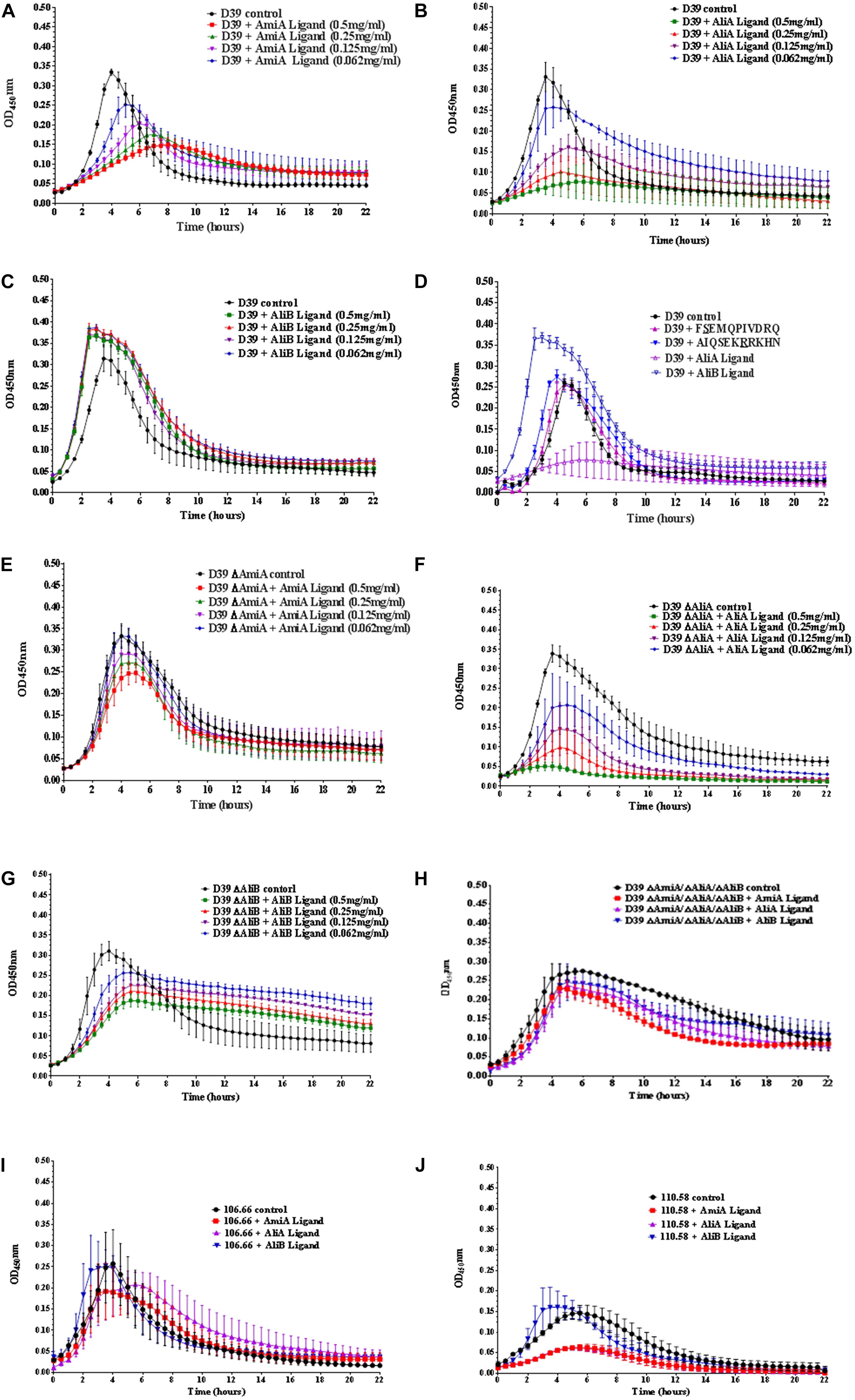
FIGURE 1. Growth curves of pneumococcal strain D39, its mutants and strains 106.66 and 110.58 in the presence and absence of the peptide ligands of AmiA, AliA, and AliB. Growth in chemically defined medium (CDM) was determined by measuring optical density over time for (A) wild type strain D39 in the absence (control) and presence of AmiA peptide ligand (at the different concentrations indicated), (B) D39 in the absence and presence of AliA ligand, (C) D39 in the absence and presence of AliB ligand, showing that AmiA and AliA ligands cause significant and dose dependent suppression of growth but AliB ligand does not. The effects of peptides on growth were specific as shown in panel (D) where D39 was grown in the absence and presence (0.5 mg/mL) of peptides with a single amino acid substitution: FSEMQPIVDRQ differs from AliA ligand at the second amino acid and AIQSEKRRKHN differs from AliB ligand at the seventh amino acid (underlined). Growth curves in the presence of the unmodified ligands have been included to allow direct comparison. AmiA peptide had much less suppressive effect on growth of the mutant lacking the AmiA receptor, D39 ΔAmiA (E). Whereas growth of the D39 ΔAliA mutant was suppressed by AliA peptide (F). Unlike the wild type D39 strain, growth of the D39 ΔAliB mutant was reduced by the AliB peptide (G). None of the three peptide ligands had a great effect on the triple mutant ΔAmiA/ΔAliA/ΔAliB (H). Suppression of growth by AmiA and AliA peptides but not AliB peptide was also seen in two clinical pneumococcal isolates: strain 106.66 (serotype 6B) (I) and non-encapsulated strain 110.58 (J) (Peptide concentrations of 0.5 mg/mL). Curves show the mean values of at least three independent experiments performed on different days, error bars equal SEM.
Less suppression of growth was seen by AmiA peptide in the ΔAmiA mutant supporting the idea that this is the specific receptor for the peptide (Figure 1E). In contrast, AliA peptide affects the ΔAliA mutant in the same way as it affects the D39 wild type as expected since AliA peptide is known to bind to AmiA receptor (Figure 1F; Nasher et al., 2018b). Unlike the wild type D39 strain, growth of the D39 ΔAliB mutant was reduced by the AliB peptide (Figure 1G). However, none of the three peptide ligands had a great effect on the triple mutant ΔAmiA/ΔAliA/ΔAliB indicating a redundancy of function between the three substrate binding proteins (Figure 1H).
To test whether the peptides had the same effect on growth for clinical isolates, we tested the effects on strain 106.66 (serotype 6B) and non-encapsulated strain 110.58 (Figures 1I,J). As for D39, AmiA and AliA peptide ligands reduced growth whilst AliB peptide ligand slightly shortened the lag phase for these clinical isolates.
AmiA and AliA peptides have a bacteriostatic, rather than bacteriolytic, effect as shown by plating out and counting CFU at different timepoints during exponential growth (Supplementary Figure S1).
Given the effects on growth by the peptides in Figure 1, next we determined whether possession of the oligopeptide binding proteins affected the outcome of a competition assay when wild type strain D39 and each of its mutants were cultured together in the presence of the peptides. Figure 2A shows that D39 is outcompeted by both the mutant lacking AmiA (ΔAmiA) and the triple mutant ΔAmiA/ΔAliA/ΔAliB when the AmiA peptide is present but not when the AmiA peptide is absent (Figure 2B). The same pattern is seen for AliA (Figure 2C) with mutants lacking the receptor for the peptide having a growth advantage in the competition assay when the peptide was present (Figure 2C) but not when the peptide was absent (Figure 2D). In contrast, the wild type D39 strain had a competitive advantage over the mutant lacking AliB (ΔAliB) and over the triple mutant (Figure 2E). D39 also had a growth advantage over the ΔAliB mutant in the absence of peptide, although to a lesser extent, and no advantage over the triple mutant (Figure 2F).
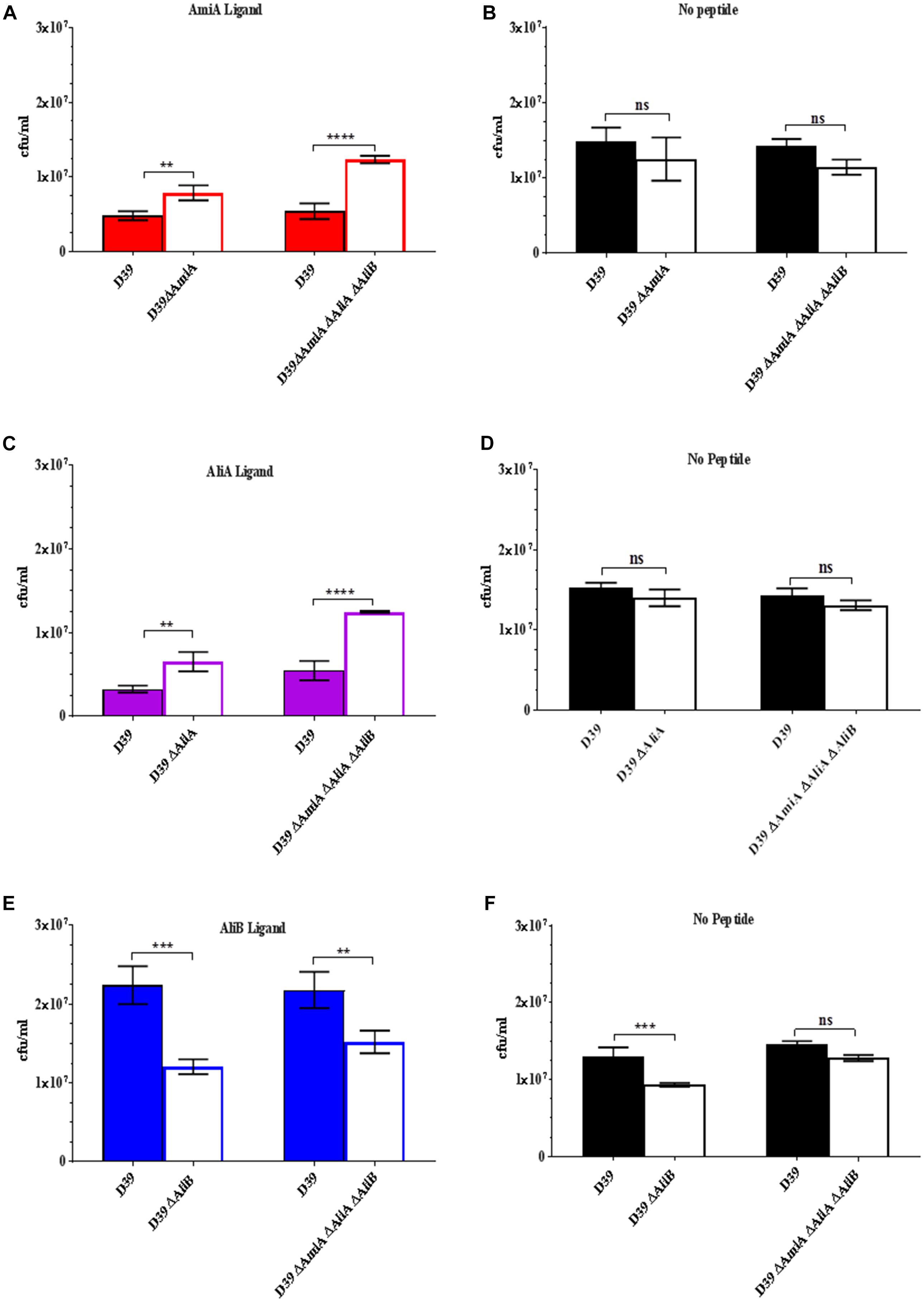
FIGURE 2. Competition assay between D39 and each of its mutants when cultured together in the absence and presence (0.5 mg/mL) of each of the peptides. Strain D39 was cultured together with mutant ΔAmiA or the triple mutant ΔAmiA/ΔAliA/ΔAliB in panel (A) the presence or (B) absence of AmiA ligand, with mutant ΔAliA or ΔAmiA/ΔAliA/ΔAliB in panel (C) the presence or (D) absence of AliA ligand and with mutant ΔAliB or the triple mutant ΔAmiA/ΔAliA/ΔAliB in panel (E) the presence or (F) absence of AliB ligand in CDM. When OD600nm reached 0.3, the bacteria were plated onto CSBA with and without antibiotics to differentiate between the wild type and mutants and CFU counted. Results are as the mean ± SD of three independent experiments, ∗∗p = 0.0021; ∗∗∗p = 0.00021; ∗∗∗∗p < 0.0001; ns, not significant.
(See Supplementary Figure S2 for all data points at OD600nm = 0.1, 0.2, and 0.3).
The results of the competition assays support those of the growth curves: peptides AmiA and AliA inhibit growth but AliB enhances growth.
Peptides Affect Capsule Thickness
Since the polysaccharide capsule is a major virulence factor of S. pneumoniae, we investigated whether any of the three peptides affected capsule thickness. Figure 3A shows an example of a FITC-dextran exclusion assay using strains D39 (serotype 2) and 106.66 (serotype 6B). For both strains there was a clear diminution of capsule size following incubation with AmiA peptide. The results for all three peptides for both strains are summarised in Figure 3B. The reduction in capsule thickness in response to AmiA peptide was significant for both strains. AliA peptide also reduced capsule thickness in both strains, although the difference was only significant for strain D39. In contrast, AliB peptide ligand caused a slight, non-significant, increase in capsule thickness in strain D39 only (Figure 3B). None of the three peptide ligands affected capsule thickness significantly in the triple mutant ΔAmiA/ΔAliA/ΔAliB (Supplementary Figure S3). In the absence of peptide ligands, mutants had thinner capsule than the parent strain D39. AliA peptide ligand did not affect capsule thickness in the ΔAliA mutant, nor did AliB peptide ligand affect capsule thickness in the ΔAliB mutant. AmiA peptide significantly increased capsule thickness in the ΔAmiA mutant but only to a value equivalent to the capsule thickness of D39 in the absence of peptide (Supplementary Figure S3).
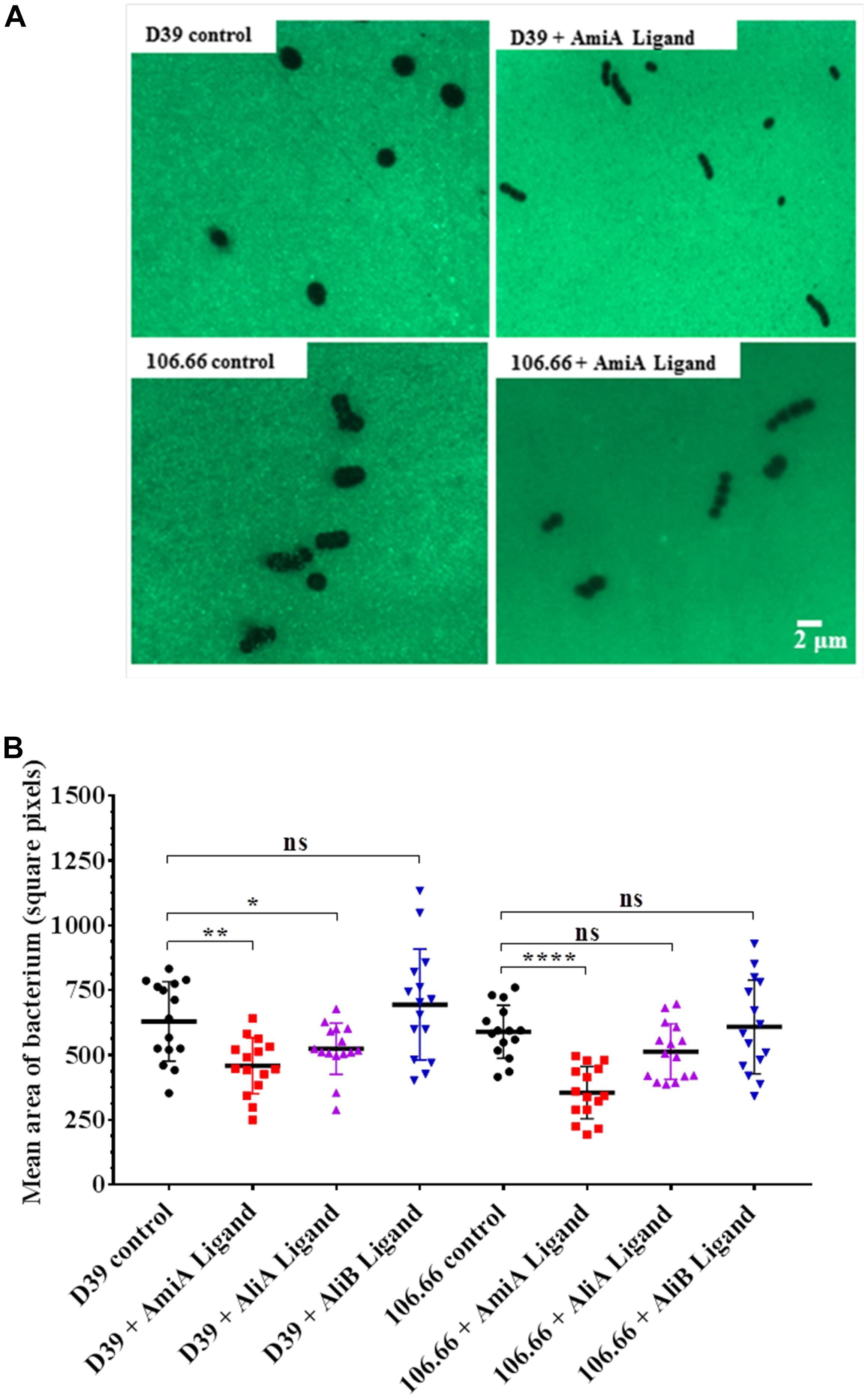
FIGURE 3. FITC-dextran exclusion assay to determine the effect of the peptides on capsule thickness. In CDM, bacteria were grown in the presence or absence of AmiA, AliA or AliB peptide ligands to OD600nm = 0.2. (A) A representative fluorescent image shows a visible reduction in capsule size following incubation with AmiA peptide ligand for both strains D39 and 106.66. (B) Quantification of results with all peptides presented as mean ± SD of mean area per bacterium in square pixels. Each symbol represents one image containing between 8 and 150 bacteria. ∗p = 0.0332; ∗∗p = 0.002; ∗∗∗∗p = 0.0001; ns, not significant.
Peptide Ligands Affect Chain Length of Encapsulated but Not Non-encapsulated Pneumococci
When performing the capsule thickness experiments, we noticed that the peptide ligands appeared to affect chain length of the encapsulated pneumococci. We therefore quantified the effects of the peptide ligands on chain length (Figure 4). All three peptide ligands increased the average number of bacteria per chain, but by far the greatest effect was seen in presence of AliB peptide ligand on strains D39 and 106.66 (Figure 4A). The peptides increased chain length in the encapsulated strains D39 (serotype 2) and 106.66 (serotype 6B) but not the non-encapsulated strain 110.58 (Figure 4B). None of the three peptides had any significant effect on chain length in any of the four mutant strains (Supplementary Figure S4). By replacing the AliB peptide by the one with the single amino acid substitution (AIQSEKRRKHN) we show that the effect on chain length is specific to the AliB peptide (Figure 4C).
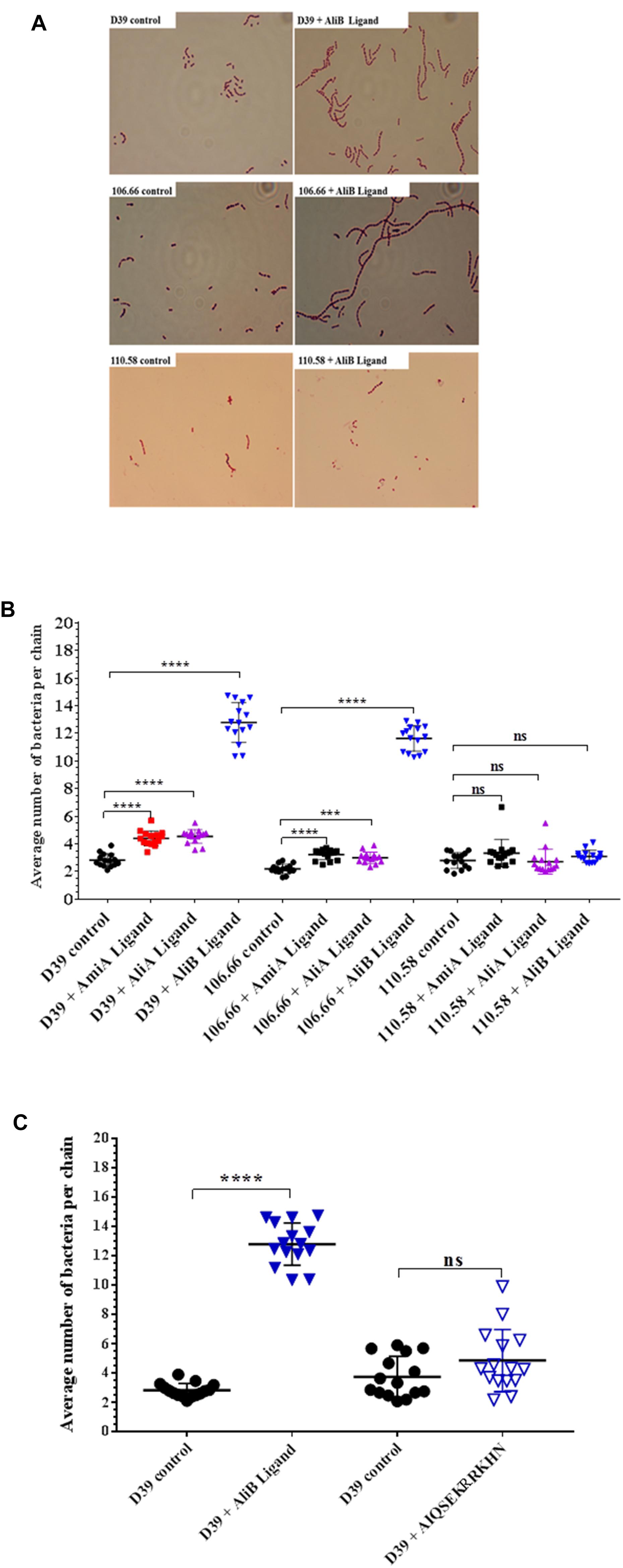
FIGURE 4. Effect of peptides on pneumococcal chain length. In CDM, strain D39 and its mutants, 106.66 and 110.58 were grown to OD600nm = 0.2 in the absence and presence of peptide ligands (0.5 mg/mL). (A) Representative brightfield images showing differences in chain length between controls and S. pneumoniae treated with AliB peptide ligand. (B) Quantification of results with all peptides and (C) comparison of effect of AliB ligand with that of a peptide with a single amino acid substitution (AIQSEKRRKHN) on chain length of strain D39. Results are presented as mean ± SD of average number of bacteria per chain. Each symbol represents an image containing between 5 and 78 chains. ∗∗∗p = 0.00021; ∗∗∗∗p = 0.0001; ns, not significant.
Peptides Affect Biofilm Formation
We found no effect of the peptide ligands on adherence (see Supplementary Figure S5) but nevertheless we looked for an effect on biofilm formation. Peptide ligands for AmiA and AliA significantly increased biofilm formation for all three strains; D39, 106.66, and 110.58 (Figure 5). AliB peptide ligand caused a significant increase in biofilm in strains D39 and 110.58, although to a lesser extent than the other two peptides, but not in strain 106.66 (Figure 5). AmiA peptide increased biofilm production in mutant ΔAmiA/ΔAliA/ΔAliB but not in mutant ΔAmiA. AliA and AliB peptide ligands caused no changes in biofilm formation in any of the mutants tested (Supplementary Figure S6).
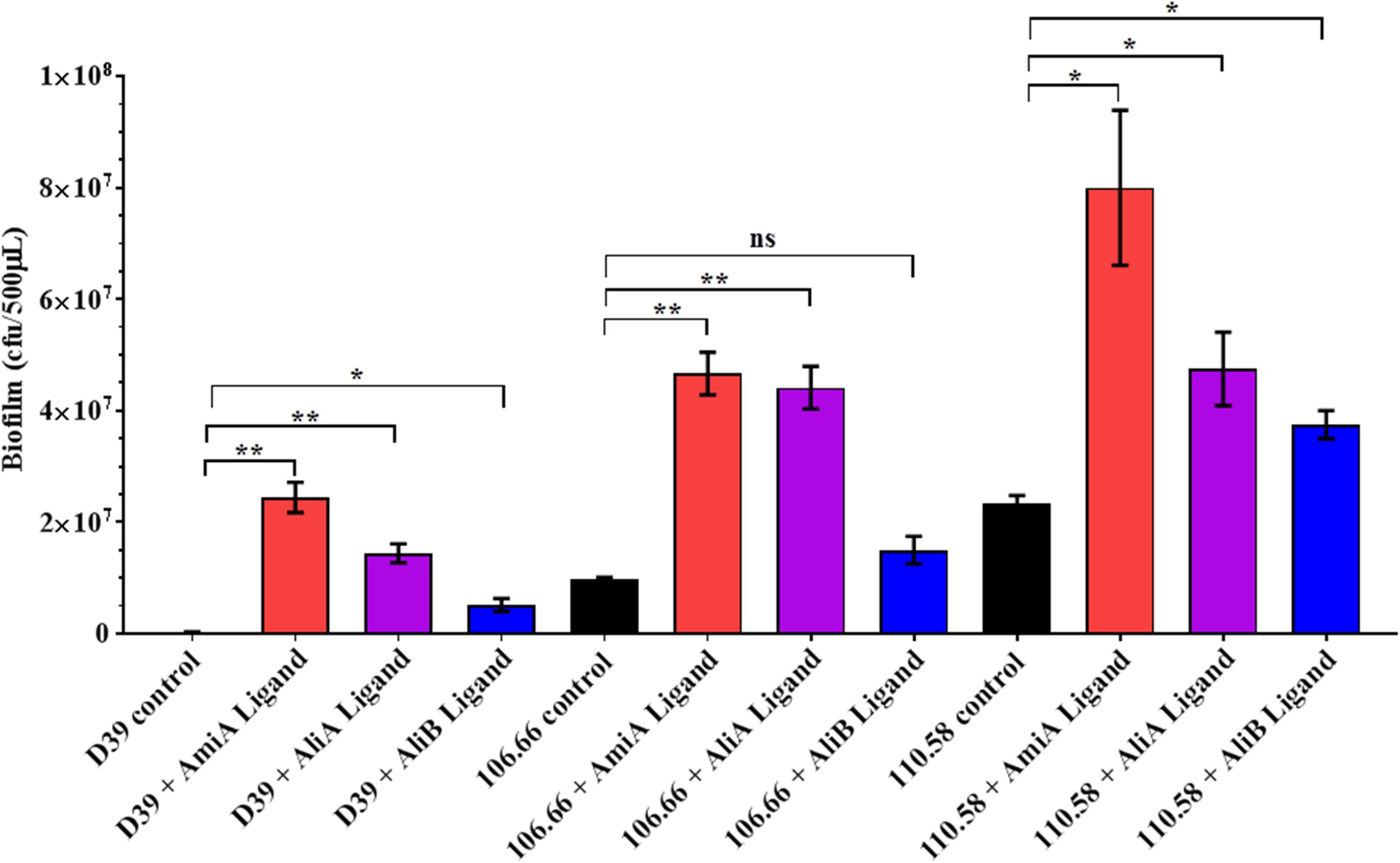
FIGURE 5. Effect of peptides on biofilm formation. Quantitative analysis of in vitro biofilm formation of strains D39, 106.66, and 110.58 on Detroit 562 nasopharyngeal epithelial cells in the absence and presence of peptide ligands (0.5 mg/mL) in CDM after 16 h of static incubation. Results are expressed as mean CFU/500 μL medium ± SD, ∗p = 0.0332; ∗∗p = 0.0021; ns, not significant.
Peptide Ligands Reduced Transformation Rate
Streptococcus pneumoniae is naturally competent for genetic transformation and being able to take up DNA in a hostile environment may confer a survival advantage. The effect of the peptides on CSP-mediated transformation rate was therefore tested in the strain D39, its mutants and strains 106.66 and 110.58. All three peptide ligands significantly reduced the transformation rate in strains D39, 106.66, and 110.58 (Figure 6). No significant effect was observed in the mutants (Supplementary Figure S7A). It must be noted that the peptide concentration is high (0.5 mg/mL) relative to the concentration of CSP (100 ng/mL) and that at such concentrations the AliA and AliB peptides with single amino acid substitutions and an irrelevant peptide reduced transformation rate (Supplementary Figure S7B), although in contrast to AmiA, AliA, and AliB, the results were not statistically significant.
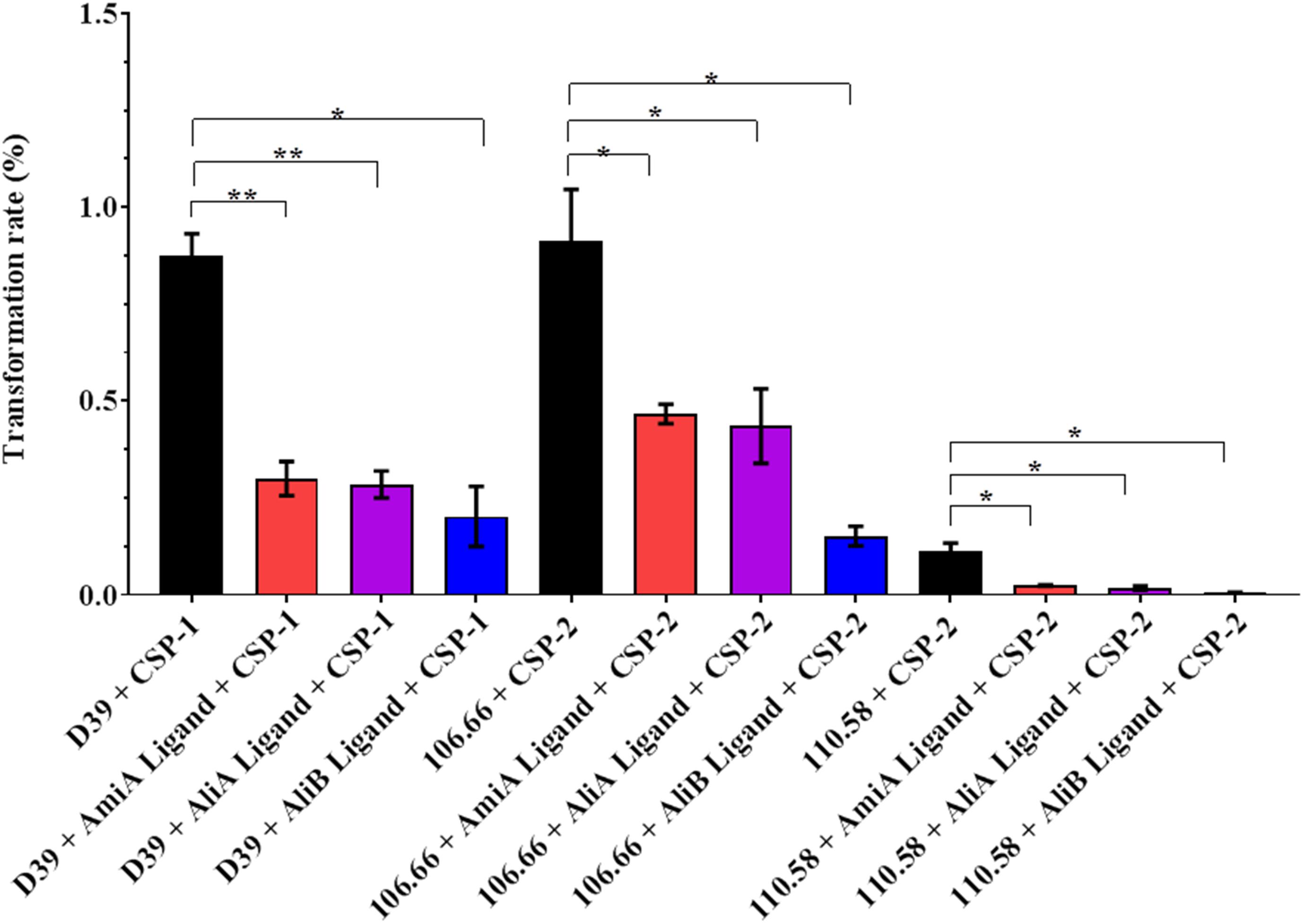
FIGURE 6. Effect of peptides on transformation rate. The rate of transformation to antibiotic resistance by the uptake of DNA from a resistant strain was determined for strains D39, 106.66, and 110.58 in the presence of the appropriate CSP (as indicated on the x-axis) and the absence and presence of the peptide ligands (0.5 mg/mL). Percentage transformation rates are the means ± SD of three independent experiments, ∗p = 0.0332; ∗∗p = 0.0021; ns, not significant.
Peptide Affects Protein Expression
To gain insight into how the peptides caused these phenotype changes, we also looked at the effect of AmiA, AliA, and AliB peptide ligands on the pneumococcal proteome. Proteomic analysis was performed by LC-MS/MS on strain D39 and its mutants ΔAmiA, ΔAliA, ΔAliB, and triple mutant ΔAmiAΔAliAΔAliB in the presence and absence of peptide ligands. Table 1 shows all proteins significantly upregulated and downregulated in the wild type following a 15 min exposure to each of the peptide ligands. Although there was some overlap of the effect on protein expression between the three peptides, we noted that only AliB peptide increased expression of alanine racemase, a protein that catalyses the racemisation of L-alanine and D-alanine. D-alanine is needed for biosynthesis of peptidoglycan in the cell wall, essential for AliB peptide-induced growth. AmiA peptide appeared to switch off expression of several proteins associated with metabolism such as Vex2, the ATP-binding protein of an ABC transporter for export of lipoproteins during cell division, Cof family protein involved in metabolism and RecN, a protein that repairs DNA during replication. Two proteins uniquely switched off by AliA were a replicative DNA helicase and penicillin-binding protein 2X which is required for building cell wall. These results seem to support the role of AmiA and AliA proteins in reducing growth. Of interest was also the CinA protein, which is associated with competence and was switched on by AmiA and AliA but not AliB.
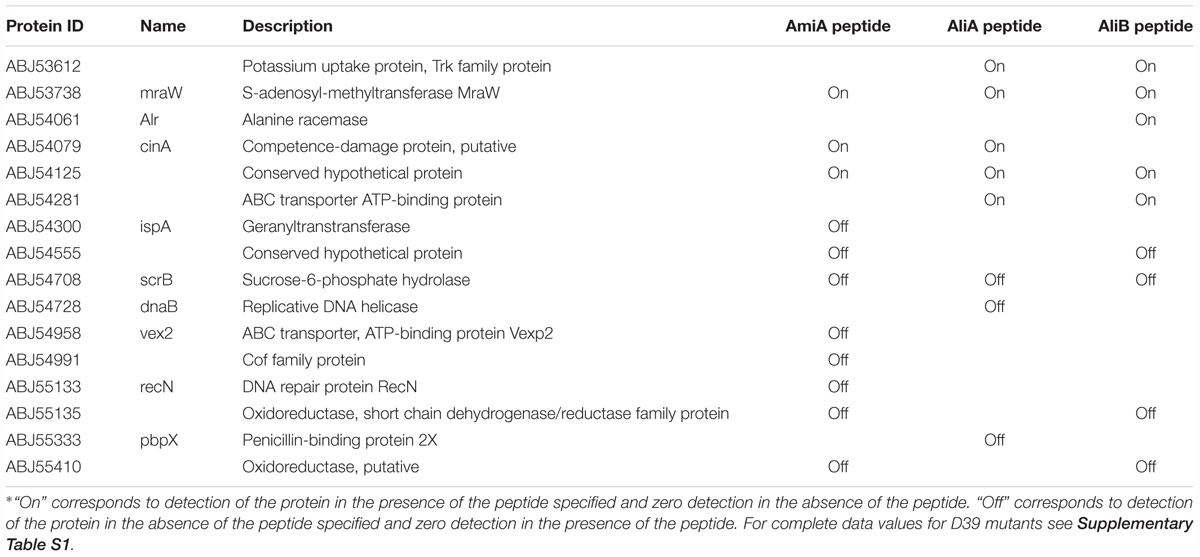
TABLE 1. All proteins significantly upregulated and downregulated in the wild type strain D39 following a 15 min exposure to each of the peptide ligands∗.
See Supplementary Table S1 for upregulated and downregulated proteins in the single and triple mutants in the presence of the peptides and Supplementary Table S2 for the complete set of proteins.
Discussion
We propose that there is interspecies communication between bacteria that live in the human nasopharynx and that detecting other bacterial species allows S. pneumoniae to adapt to its environment. Previously, we reported that S. pneumoniae AmiA, AliA, and AliB lipoproteins of an ABC transporter are able to bind peptides matching ribosomal proteins of other bacteria species including common nasopharynx colonisers (Nasher et al., 2018b). These proteins are encoded by genes of the core pneumococcal genome (Obert et al., 2006) and so are of relevance across the pneumococcal population. Here, we looked at what effects these novel peptide ligands have on the phenotype of the pneumococcus.
In the presence of AmiA and AliA peptide ligands, growth of all strains was greatly reduced; by contrast AliB peptide ligand slightly boosted growth. Steady but persistent growth may be an ideal strategy for a coloniser in an environment such as the nasopharynx where nutrient availability is limited. In contrast proliferation of the pneumococcus has been suggested to increase the risk of invasive disease (Albrich et al., 2012).
AmiA and AliA peptides, but not AliB peptide, decreased capsule thickness. The polysaccharide capsule is the most important virulence factor of pneumococci, playing a role in invasive disease (Kelly et al., 1994; Beech et al., 2018). The capsule prevents phagocytosis mediated by Fcγ receptors, complement receptors and non-opsonic receptor thus protecting the pneumococci from the host immune response (Kelly et al., 1994; Hardy et al., 2001; Hammerschmidt et al., 2005; Hyams et al., 2010, 2013; Geno et al., 2015). Additionally, and an increase in capsule size has been observed in systemic infections (Kim and Weiser, 1998; Kim et al., 1999). However, capsule is downregulated when pneumococci are in intimate contact with epithelial cells (Hammerschmidt et al., 2005), and a transparent phenotype that is associated with improved colonisation of the nasopharynx has a reduced expression of capsular polysaccharide (Weiser, 2010). We therefore propose that when S. pneumoniae comes into contact with AmiA ligand and AliA ligand, these peptides signal the pneumococci to downregulate capsule expression which may promote attachment to epithelial cells, the first step in colonisation. Colonisation involves biofilm formation and, whilst all three peptides increased the amount of biofilm, the effect was greatest for AmiA and AliA peptides. Biofilm formation may provide several advantages for the pneumococci including a protective environment in which the bacteria can upregulate factors that are involved in nutrient uptake from their environment while downregulating virulence factors involved in inflammation to evade host defence system (Blanchette-Cain et al., 2013; Qin et al., 2013). Several previous studies have found that capsule size is inversely associated with biofilm formation (Hall-Stoodley et al., 2008; Marks et al., 2012b; Junges et al., 2017). In contrast, proliferating bacteria are able to transition from colonising to invasive disease at a higher degree than biofilm forming bacteria (Trappetti et al., 2011; Marks et al., 2013) suggesting that AliB peptide might have a higher likelihood than the other two peptides of being associated with invasion. We found that peptide ligands for AmiA and AliA particularly increased biofilm formation, despite not increasing adherence to epithelial cells in our in vitro model, although others have found a correlation between adherence and biofilm formation (Konkel et al., 1997; Marks et al., 2012a; Sulaeman et al., 2012). It is possible that the peptide ligands enhance biofilm formation by another mechanism such as enhancing extracellular matrix formation.
AliB peptide ligand dramatically increased chain formation and length in the encapsulated strains but not in the non-encapsulated 110.58. We propose that increasing chain length may compensate for a thick capsule during colonisation as having a thicker capsule has also been reported to restrict adherence to human epithelial cells (Talbot et al., 1996; Adamou et al., 1998; Ring et al., 1998; Novick et al., 2017). Non-encapsulated pneumococci have an advantage in adherence and colonisation (Keller et al., 2016), and may therefore not require increase in chaining for effective colonisation. If a longer chain allows more secure attachment as suggested by a recent report (Rodriguez et al., 2012), the chances of progression to invasive disease may also be greater.
Our findings indicate that S. pneumoniae transformation rate can be influenced by other bacterial species. To our knowledge, there have not been any reports on the effect of other bacterial species that are common colonisers of the nasopharynx on the transformation rate of pneumococcus, although there are several studies that highlight that competence and transformation rate of the pneumococcus is affected by co-colonisation of multiple serotypes (Donati et al., 2010; Hiller et al., 2010; Croucher et al., 2011; Muzzi and Donati, 2011; Marks et al., 2012b). Transformation rate was decreased by all three peptides, despite the finding that only AmiA and AliA reduced capsule thickness, but the only competence-associated protein found in the proteomic studies was CinA which was switched on by AmiA and AliA but not AliB. Expression of this protein has been previously shown to be turned on during competence (Martin et al., 1995) but its role in transformation has not been confirmed (Mortier-Barrière et al., 1998). Since biofilm formation is generally associated with increased competence, further studies over a time course might be warranted to decipher the precise role of the peptides in competence.
Proteomic data indicated downregulation of some proteins involved in metabolism or cell division by AmiA peptide (Vex2, Cof family protein, RecN) and AliA (DnaB, PbpX). These results are compatible with the phenotypic data, where AmiA and AliA reduced growth and AliB increased both peptidoglycan production (Alr) and increased growth. Alr is an alanine racemase and uses a covalently bound pyridoxal 5′-phosphate cofactor to catalyse the racemisation of L-alanine and D-alanine. D-alanine is a necessary component of the peptidoglycan layer in bacterial cell walls (Lambert and Neuhaus, 1972; Milligan et al., 2007). In S. mutans, Alr has also been shown to be necessary for growth and competitiveness (Wei et al., 2016). However, proteomic data was not clear cut, for example MraW, involved in cell wall biogenesis was increased by all three peptides,
A limitation in this study was the incubation time of the pneumococcal strains and the peptide ligands which only provided a snapshot of the proteome profile. We cannot overlook the fact that we could have also missed regulation of many other essential proteins thought to be involved in pneumococcal pathogenesis. In addition, all our studies investigated the effects of one peptide ligand for each lipoprotein receptor but we do not exclude the possibility that other peptides may bind and that their effects on the pneumococcal phenotypes may be different from those found in the present study.
We have shown that AmiA, AliA, and AliB peptides, found in ribosomal proteins of other bacterial species, alter the phenotypes of S. pneumoniae. We propose that exploiting this knowledge could offer new targets in the prevention and treatment of pneumococcal diseases.
Author Contributions
LH conceived the study. FN wrote the first draft of the manuscript. FN, FA, and LH participated in design of the study. FN, SA, and FA performed the research. MH analyzed proteomic data. PH provided the strain D39 and mutants. All authors gave final approval for publication.
Funding
This work was supported by grant number 162808 from the Swiss National Science Foundation to LH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
PH is employed by the company Janssen Vaccines and Prevention.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Annelies Müller for her support, Sophie Braga, and Natasha Buchs for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03013/full#supplementary-material
References
Adamou, J. E., Wizemann, T. M., Barren, P., and Langermann, S. (1998). Adherence of Streptococcus pneumoniae to human bronchial epithelial cells (BEAS-2B). Infect. Immun. 66, 820–822.
Albrich, W. C., Madhi, S. A., Adrian, P. V., Van Niekerk, N., Mareletsi, T., Cutland, C., et al. (2012). Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin. Infect. Dis. 54, 601–609. doi: 10.1093/cid/cir859
Alloing, G., De Philip, P., and Claverys, J. P. (1994). Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the Gram-positive Streptococccus pneumoniae. J. Mol. Biol. 241, 44–58. doi: 10.1006/jmbi.1994.1472
Beech, J. P., Ho, B. D., Garriss, G., Oliveira, V., Henriques-Normark, B., and Tegenfeldt, J. O. (2018). Separation of pathogenic bacteria by chain length. Anal. Chim. Acta 1000, 223–231. doi: 10.1016/j.aca.2017.11.050
Biesbroek, G., Tsivtsivadze, E., Sanders, E. A., Montijn, R., Veenhoven, R. H., Keijser, B. J., et al. (2014). Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190, 1283–1292. doi: 10.1164/rccm.201407-1240OC
Blanchette-Cain, K., Hinojosa, C. A., Akula Suresh Babu, R., Lizcano, A., Gonzalez-Juarbe, N., Munoz-Almagro, C., et al. (2013). Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 4:e00745-13. doi: 10.1128/mBio.00745-13
Brewster, J. D. (2003). A simple micro-growth assay for enumerating bacteria. J. Microbiol. Methods 53, 77–86. doi: 10.1016/S0167-7012(02)00226-9
Claverys, J. P., Grossiord, B., and Alloing, G. (2000). Is the Ami-AliA/B oligopeptide permease of Streptococcus pneumoniae involved in sensing environmental conditions? Res. Microbiol. 151, 457–463.
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., Burton, J., Van Der Linden, M., et al. (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434. doi: 10.1126/science.1198545
Donati, C., Hiller, N. L., Tettelin, H., Muzzi, A., Croucher, N. J., Angiuoli, S. V., et al. (2010). Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11:R107. doi: 10.1186/gb-2010-11-10-r107
Engel, H., Mika, M., Denapaite, D., Hakenbeck, R., Mühlemann, K., Heller, M., et al. (2014). A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 3934–3941. doi: 10.1128/AAC.02547-14
Gates, M. A., Thorkildson, P., and Kozel, T. R. (2004). Molecular architecture of the Cryptococcus neoformans capsule. Mol. Microbiol. 52, 13–24. doi: 10.1111/j.1365-2958.2003.03957.x
Geno, K. A., Gilbert, G. L., Song, J. Y., Skovsted, I. C., Klugman, K. P., Jones, C., et al. (2015). Pneumococcal capsules and their types: past, present, and future. Clin. Microbiol. Rev. 28, 871–899. doi: 10.1128/CMR.00024-15
Hall-Stoodley, L., Nistico, L., Sambanthamoorthy, K., Dice, B., Nguyen, D., Mershon, W. J., et al. (2008). Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 8:173. doi: 10.1186/1471-2180-8-173
Hammerschmidt, S., Wolff, S., Hocke, A., Rosseau, S., Müller, E., and Rohde, M. (2005). Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73, 4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005
Hardy, G. G., Magee, A. D., Ventura, C. L., Caimano, M. J., and Yother, J. (2001). Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69, 2309–2317. doi: 10.1128/IAI.69.4.2309-2317.2001
Hathaway, L. J., Battig, P., Reber, S., Rotzetter, J. U., Aebi, S., Hauser, C., et al. (2014). Streptococcus pneumoniae detects and responds to foreign bacterial peptide fragments in its environment. Open Biol. 4:130224. doi: 10.1098/rsob.130224
Hathaway, L. J., Brugger, S. D., Morand, B., Bangert, M., Rotzetter, J. U., Hauser, C., et al. (2012). Capsule type of Streptococcus pneumoniae determines growth phenotype. PLoS Pathog. 8:e1002574. doi: 10.1371/journal.ppat.1002574
Hathaway, L. J., Stutzmann Meier, P., Battig, P., Aebi, S., and Muhlemann, K. (2004). A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J. Bacteriol. 186, 3721–3729. doi: 10.1128/JB.186.12.3721-3729.2004
Hiller, N. L., Ahmed, A., Powell, E., Martin, D. P., Eutsey, R., Earl, J., et al. (2010). Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6:e1001108. doi: 10.1371/journal.ppat.1001108
Hilty, M., Wuthrich, D., Salter, S. J., Engel, H., Campbell, S., Sa-Leao, R., et al. (2014). Global phylogenomic analysis of nonencapsulated Streptococcus pneumoniae reveals a deep-branching classic lineage that is distinct from multiple sporadic lineages. Genome Biol. Evol. 6, 3281–3294. doi: 10.1093/gbe/evu263
Hyams, C., Camberlein, E., Cohen, J. M., Bax, K., and Brown, J. S. (2010). The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78, 704–715. doi: 10.1128/IAI.00881-09
Hyams, C., Trzcinski, K., Camberlein, E., Weinberger, D. M., Chimalapati, S., Noursadeghi, M., et al. (2013). Streptococcus pneumoniae capsular serotype invasiveness correlates with the degree of factor H binding and opsonization with C3b/iC3b. Infect. Immun. 81, 354–363. doi: 10.1128/IAI.00862-12
Junges, R., Salvadori, G., Shekhar, S., Åmdal, H. A., Periselneris, J. N., Chen, T., et al. (2017). A quorum-sensing system that regulates Streptococcus pneumoniae biofilm formation and surface polysaccharide production. mSphere 2:e00324-17. doi: 10.1128/mSphere.00324-17
Keller, L. E., Robinson, D. A., and Mcdaniel, L. S. (2016). Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. mBio 7:e01792. doi: 10.1128/mBio.01792-15
Kelly, T., Dillard, J. P., and Yother, J. (1994). Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect. Immun. 62, 1813–1819.
Kerr, A. R., Adrian, P. V., Estevao, S., De Groot, R., Alloing, G., Claverys, J. P., et al. (2004). The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 72, 3902–3906. doi: 10.1128/IAI.72.7.3902-3906.2004
Kim, J. O., Romero-Steiner, S., Sørensen, U. B. S., Blom, J., Carvalho, M., Barnard, S., et al. (1999). Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 67, 2327–2333.
Kim, J. O., and Weiser, J. N. (1998). Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177, 368–377. doi: 10.1086/514205
Konkel, M. E., Garvis, S. G., Tipton, S. L., Anderson, D. E. Jr., and Cieplak, W. Jr., (1997). Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24, 953–963. doi: 10.1046/j.1365-2958.1997.4031771.x
Kronenberg, A., Zucs, P., Droz, S., and Muhlemann, K. (2006). Distribution and invasiveness of Streptococcus pneumoniae serotypes in Switzerland, a country with low antibiotic selection pressure, from 2001 to 2004. J. Clin. Microbiol. 44, 2032–2038. doi: 10.1128/JCM.00275-06
Lambert, M. P., and Neuhaus, F. C. (1972). Factors affecting the level of alanine racemase in Escherichia coli. J. Bacteriol. 109, 1156–1161.
Lau, G. W., Haataja, S., Lonetto, M., Kensit, S. E., Marra, A., Bryant, A. P., et al. (2001). A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40, 555–571. doi: 10.1046/j.1365-2958.2001.02335.x
Lazar, C., Gatto, L., Ferro, M., Bruley, C., and Burger, T. (2016). Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 15, 1116–1125. doi: 10.1021/acs.jproteome.5b00981
Marks, L. R., Davidson, B. A., Knight, P. R., and Hakansson, A. P. (2013). Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:e00438-13. doi: 10.1128/mBio.00438-13
Marks, L. R., Parameswaran, G. I., and Hakansson, A. P. (2012a). Pneumococcal Interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect. Immun. 80, 2744–2760. doi: 10.1128/IAI.00488-12
Marks, L. R., Reddinger, R. M., and Hakansson, A. P. (2012b). High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. mBio 3:e00200-12. doi: 10.1128/mBio.00200-12
Martin, B., Garcia, P., Castanié, M.-P., and Claverys, J.-P. (1995). The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15, 367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x
Mika, M., Mack, I., Korten, I., Qi, W., Aebi, S., Frey, U., et al. (2015). Dynamics of the nasal microbiota in infancy: a prospective cohort study. J. Allergy Clin. Immunol. 135, 905–912.e11. doi: 10.1016/j.jaci.2014.12.1909
Milligan, D. L., Tran, S. L., Strych, U., Cook, G. M., and Krause, K. L. (2007). The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of d-Alanine. J. Bacteriol. 189, 8381–8386. doi: 10.1128/JB.01201-07
Mortier-Barrière, I., De Saizieu, A., Claverys, J. P., and Martin, B. (1998). Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27, 159–170. doi: 10.1046/j.1365-2958.1998.00668.x
Mühlemann, K., Matter, H. C., Täuber, M. G., and Sentinel Working, G. (2003). Nationwide surveillance of nasopharyngeal Streptococcus pneumoniae isolates from children with respiratory infection, Switzerland, 1998–1999. J. Infect. Dis. 187, 589–596. doi: 10.1086/367994
Muzzi, A., and Donati, C. (2011). Population genetics and evolution of the pan-genome of Streptococcus pneumoniae. Int. J. Med. Microbiol. 301, 619–622. doi: 10.1016/j.ijmm.2011.09.008
Nasher, F., Förster, S., Yildirim, E. C., Grandgirard, D., Leib, S. L., Heller, M., et al. (2018a). Foreign peptide triggers boost in pneumococcal metabolism and growth. BMC Microbiol. 18:23. doi: 10.1186/s12866-018-1167-y
Nasher, F., Heller, M., and Hathaway, L. J. (2018b). Streptococcus pneumoniae proteins AmiA, AliA, and AliB Bind Peptides found in ribosomal proteins of other bacterial species. Front. Microbiol. 8:2688. doi: 10.3389/fmicb.2017.02688
Novick, S., Shagan, M., Blau, K., Lifshitz, S., Givon-Lavi, N., Grossman, N., et al. (2017). Adhesion and invasion of Streptococcus pneumoniae to primary and secondary respiratory epithelial cells. Mol. Med. Rep. 15, 65–74. doi: 10.3892/mmr.2016.5996
Obert, C., Sublett, J., Kaushal, D., Hinojosa, E., Barton, T., Tuomanen, E. I., et al. (2006). Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74, 4766–4777. doi: 10.1128/IAI.00316-06
Pettigrew, M. M., Laufer, A. S., Gent, J. F., Kong, Y., Fennie, K. P., and Metlay, J. P. (2012). Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78, 6262–6270. doi: 10.1128/AEM.01051-12
Polissi, A., Pontiggia, A., Feger, G., Altieri, M., Mottl, H., Ferrari, L., et al. (1998). Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629.
Qin, L., Kida, Y., Imamura, Y., Kuwano, K., and Watanabe, H. (2013). Impaired capsular polysaccharide is relevant to enhanced biofilm formation and lower virulence in Streptococcus pneumoniae. J. Infect. Chemother. 19, 261–271. doi: 10.1007/s10156-012-0495-3
Ring, A., Weiser, J. N., and Tuomanen, E. I. (1998). Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Invest. 102, 347–360. doi: 10.1172/JCI2406
Rodriguez, J. L., Dalia, A. B., and Weiser, J. N. (2012). Increased chain length promotes pneumococcal adherence and colonization. Infect. Immun. 80, 3454–3459. doi: 10.1128/IAI.00587-12
Schaffner, T. O., Hinds, J., Gould, K. A., Wüthrich, D., Bruggmann, R., Küffer, M., et al. (2014). A point mutation in cpsE renders Streptococcus pneumoniae nonencapsulated and enhances its growth, adherence and competence. BMC Microbiol. 14:210. doi: 10.1186/s12866-014-0210-x
Sulaeman, S., Hernould, M., Schaumann, A., Coquet, L., Bolla, J. M., De, E., et al. (2012). Enhanced adhesion of Campylobacter jejuni to abiotic surfaces is mediated by membrane proteins in oxygen-enriched conditions. PLoS One 7:e46402. doi: 10.1371/journal.pone.0046402
Talbot, U. M., Paton, A. W., and Paton, J. C. (1996). Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 64, 3772–3777.
Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715. doi: 10.1016/j.chom.2015.03.008
Trappetti, C., Ogunniyi, A. D., Oggioni, M. R., and Paton, J. C. (2011). Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One 6:e19844. doi: 10.1371/journal.pone.0019844
Wei, Y., Qiu, W., Zhou, X.-D., Zheng, X., Zhang, K.-K., Wang, S.-D., et al. (2016). Alanine racemase is essential for the growth and interspecies competitiveness of Streptococcus mutans. Int. J. Oral Sci. 8, 231–238. doi: 10.1038/ijos.2016.34
Keywords: Streptococcus pneumoniae, interspecies communications, growth, biofilm, capsule, chaining, ABC transporters
Citation: Nasher F, Aguilar F, Aebi S, Hermans PWM, Heller M and Hathaway LJ (2018) Peptide Ligands of AmiA, AliA, and AliB Proteins Determine Pneumococcal Phenotype. Front. Microbiol. 9:3013. doi: 10.3389/fmicb.2018.03013
Received: 24 September 2018; Accepted: 21 November 2018;
Published: 05 December 2018.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Chad W. Euler, Hunter College (CUNY), United StatesClaire Durmort, UMR5075 Institut de Biologie Structurale (IBS), France
Copyright © 2018 Nasher, Aguilar, Aebi, Hermans, Heller and Hathaway. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucy J. Hathaway, bHVjeS5oYXRoYXdheUBpZmlrLnVuaWJlLmNo
 Fauzy Nasher
Fauzy Nasher Fernando Aguilar
Fernando Aguilar Suzanne Aebi1
Suzanne Aebi1 Manfred Heller
Manfred Heller Lucy J. Hathaway
Lucy J. Hathaway