- 1LOEWE Centre for Synthetic Microbiology-SYNMIKRO, Philipps-Universität Marburg, Marburg, Germany
- 2Manchester Institute of Biotechnology, The University of Manchester, Manchester, United Kingdom
- 3United States Army Medical Research Institute of Infectious Diseases, Frederick, MD, United States
- 4Defense Biological Product Assurance Office, Frederick, MD, United States
- 5The Tauri Group, LLC, Alexandria, VA, United States
Chromosomal inheritance in bacteria usually entails bidirectional replication of a single chromosome from a single origin into two copies and subsequent partitioning of one copy each into daughter cells upon cell division. However, the human pathogen Vibrio cholerae and other Vibrionaceae harbor two chromosomes, a large Chr1 and a small Chr2. Chr1 and Chr2 have different origins, an oriC-type origin and a P1 plasmid-type origin, respectively, driving the replication of respective chromosomes. Recently, we described naturally occurring exceptions to the two-chromosome rule of Vibrionaceae: i.e., Chr1 and Chr2 fused single chromosome V. cholerae strains, NSCV1 and NSCV2, in which both origins of replication are present. Using NSCV1 and NSCV2, here we tested whether two types of origins of replication can function simultaneously on the same chromosome or one or the other origin is silenced. We found that in NSCV1, both origins are active whereas in NSCV2 ori2 is silenced despite the fact that it is functional in an isolated context. The ori2 activity appears to be primarily determined by the copy number of the triggering site, crtS, which in turn is determined by its location with respect to ori1 and ori2 on the fused chromosome.
Introduction
The generally accepted paradigm of chromosome replication in bacteria is elucidated in Escherichia coli. Replication is initiated at a unique singular sequence, the origin of replication (oriC) by DnaA, proceeds bidirectionally along the chromosome and ends at the terminus diametrically opposite to oriC on the circular chromosome. In E. coli and related bacteria, immediate re-initiation of chromosome replication is hindered due to the hemi-methylated status of the sister chromosomes and sequestration of oriC by SeqA which has a high binding affinity to hemimethylated ori sequences (Lu et al., 1994; Slater et al., 1995; Waldminghaus and Skarstad, 2009). Most bacteria have single chromosomes and follow this general replication paradigm. However, about 10% of bacterial species have more than one chromosome and exhibit some deviation from this norm (Fournes et al., 2018). Among these, Vibrio cholerae with chromosome 1 (Chr1, ∼3 Mbps) and chromosome 2(Chr2, ∼1 Mbps) has served as a model system for studies pertaining to multi-chromosome replication mechanisms, and in recent years, an extensive body of information has been accumulated on various aspects of Chr1 and Chr2 replication (Egan et al., 2005; Jha et al., 2012; Val et al., 2014b; Espinosa et al., 2017; Ramachandran et al., 2017).
Chr1 in V. cholerae is similar to the E. coli chromosome in that the replication follows the same pattern: replication origin, ori1, contains multiple DnaA boxes, which are bound by DnaA that unwinds the DNA and initiate replication (Duigou et al., 2006). The similarity is so striking that V. cholerae ori1 can functionally substitute the E. coli replication origin oriC (Egan and Waldor, 2003; Demarre and Chattoraj, 2010; Koch et al., 2010; Kamp et al., 2013).
In contrast, the V. cholerae Chr2 appears to have an origin that resembles those of low copy number plasmids such as P1 and F (Fournes et al., 2018). The ori2 contains an array of repeats (iterons) where the Chr2 specific initiator protein, RctB, binds and unwinds the DNA for ori2 firing (Egan and Waldor, 2003; Duigou et al., 2008) but also exerts a form of negative regulation, termed ‘handcuffing,’ originally discovered in plasmids (Venkova-Canova and Chattoraj, 2011). Although ori2 has plasmid-like features, Chr2 resembles typical chromosomes in some respects: (1) Participation of SeqA and Dam in regulation of ori2 (Saint-Dic et al., 2008; Demarre and Chattoraj, 2010; Koch et al., 2010; Stokke et al., 2011). (2) Indispensability of Chr2, unlike plasmids, for cell survival because it harbors essential genes (Heidelberg et al., 2000; Kamp et al., 2013). (3) High level of coordination of replication between Chr1 and Chr2 in order to prevent over replication of Chr2 and ensure a guaranteed inheritance of a single copy of both chromosomes (Baek and Chattoraj, 2014; Val et al., 2016; Ramachandran et al., 2018). This raises the question on how coordination between Chr1 and Chr2 with respect to their timing of replication initiation is achieved given the disparity in their sizes and mechanisms of replication.
Chr1 replication is initiated at the onset of the replication period while initiation of Chr2 is delayed and occurs only when 2/3rd of Chr1 replication has been completed. Since Chr2 is 1/3rd the size of Chr1, both chromosomes consequently terminate their replication roughly at the same time (Rasmussen et al., 2007; Stokke et al., 2011). This termination synchrony appears not to be accidental but is selected for during evolution and is conserved within Vibrionaceae despite differing ratios of chromosome sizes (Kemter et al., 2018). This synchrony occurs through the crtS (Chr2 replication triggering Site) present on Chr1 that positively regulates ori2 initiation (Val et al., 2016). Translocation of the crtS locus on Chr1, either closer to ori1 or farther away, resulted in a corresponding shift in Chr2 initiation time as revealed by marker frequency analysis (Val et al., 2016), indicating that the native position of crtS sets the timing of Chr2 replication initiation such that its replication terminates synchronously with Chr1 (Kemter et al., 2018). The exact mechanism of crtS action remains to be elucidated but may include physical contacts between crtS and ori2 as well as sequestration of Chr2 replication initiator protein, RctB (Baek and Chattoraj, 2014; Val et al., 2016). Recently, the global transcription factor Lrp was shown to bind to the crtS site and to facilitate RctB binding (Ciaccia et al., 2018). Heterologous E. coli systems have been established based on ori2 mini-chromosomes demonstrating that crtS provided in trans increases mini chromosome copy number indicating a positive role played by crtS in ori2 firing (Baek and Chattoraj, 2014; Schallopp et al., 2017; de Lemos Martins et al., 2018). Recently, it was demonstrated that the copy number of crtS rather than the act of replicating the crtS is critical in the triggering of ori2 firing (de Lemos Martins et al., 2018; Ramachandran et al., 2018).
In order to assess the differential genetic requirements of Chr1 and Chr2 replication, an artificial single chromosome V. cholerae strain has been created by genetic engineering in which ori1 drives the replication of the fused chromosome (Val et al., 2012). In this strain, designated MCH1, the sequences to the left and right of ori2 were fused to the terminus of Chr1. In this arrangement the direction in which chromosome arms are replicated is conserved to minimize conflicts between DNA replication and transcription. The MCH1 strain was instrumental in establishing the essentiality of Dam methyltransferase in V. cholerae because of its role in ori2 function which was first shown by Demarre and Chattoraj (2010). This conclusion was further supported by the finding that depletion of Dam leads to spontaneous chromosomal fusion (Val et al., 2014a). In this case, the entire genome is replicated from ori1 which can tolerate the absence of Dam (Val et al., 2014a).
Recently, we described two naturally occurring V. cholerae strains (NSCV1 and NSCV2) containing both ori1 and ori2 on the same chromosome (Chapman et al., 2015; Xie et al., 2017). In these strains, Chr1 and Chr2 are fused at two different locations (Figure 1). The locations of relevant features such as ori1, ori2, and crtS sites are indicated in Figure 1A. In NSCV1, crtS is located about 670 kbs away from ori1 (Figure 1). It is similar to the standard two-chromosome reference strain N16961 with respect to distance, where crtS is located 695 kbs away from ori1. In NSCV2, the distance between ori1 and crtS is 1,566 kbs due to a large inversion that has occurred around the terminus region of the chromosome (Figure 1C).
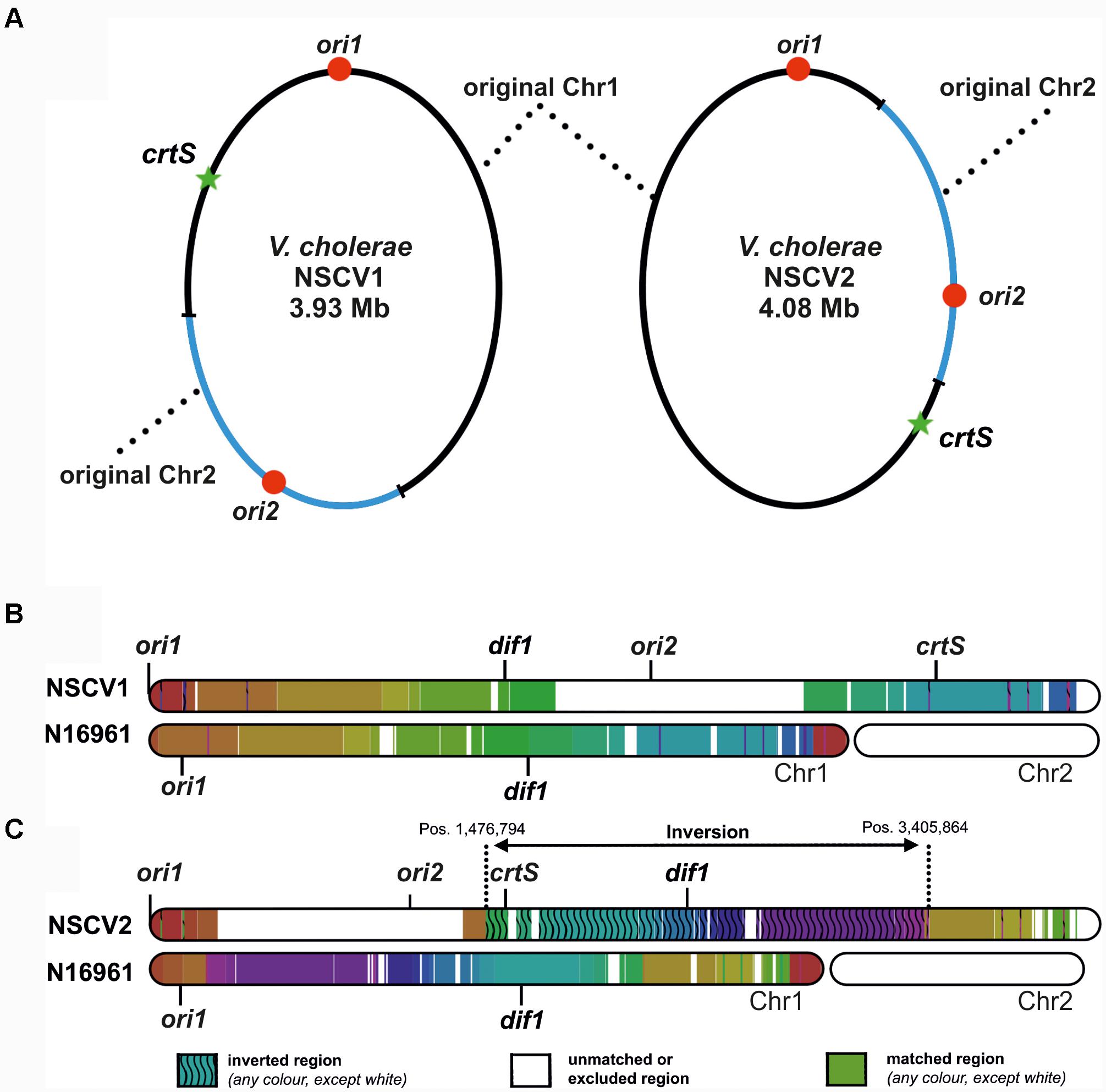
FIGURE 1. Genetic maps of V. cholerae NSCV1 and NSCV2 chromosomes depicting fusion junctions and other genomic features. (A) The fused chromosomes of strain NSCV1 (left) and NSCV2 (right) are shown with the original Chr1 and Chr2 in black and blue parts of the ring, respectively. The origins are indicated by red dots and the crtS sites by green stars. (B,C) Inversion analysis of V. cholerae NSCV1 and NSCV2 chromosomes using Smash tool (Pratas et al., 2015). The genome sequences were analyzed for sequence homology with the two-chromosome genome of strain N16961 as reference (accession numbers NC002505 and NC002506). Regions with identical information content are marked by the same color. Inverted regions are indicated by waved lines. White areas represent unmatched regions or Chr2.
This genomic architecture raised interesting questions about whether both origins are functional in the same cell or one or the other ori is silenced since in principle a single origin should suffice to replicate the fused chromosome. The strains also allowed us to ask if the chromosomal fusions are maintained without resorting to genome splitting which is the predominant genome configuration in Vibrionaceae. We found that in NSCV1 both origins are active in the same cell whereas in NSCV2 ori2 appears to be silent. Further, these chromosomes appear to be in a locked configuration since even after prolonged continuous growth they remain fused without splitting into two.
Results
Activity of ori1 and ori2 in V. cholerae NSCV Strains
In general, the replication in bacteria relies on one origin of replication for one replicon. Fusion of two replicons, such as seen in NSCV1 and NSCV2, would initially give rise to a chromosome with two functional replication origins where one could be superfluous. This raises the question of whether both, ori1 and ori2 are active in the fused chromosomes of NSCV1 and NSCV2, or only one of the origins is active and the other is silent. Inspection of the sequence of the origins and the replication initiator genes revealed no obvious mutational changes that could indicate non-functionality of one or the other of the origins in the two NSCV strains (Figure 2) (Xie et al., 2017). Compared to strain N16961, both strains possess complete ori1 sequences, with seven SNPs (NSCV1) and four SNPs (NSCV2) spanning the 474 bps long gidA-mioC intergenic region and none of the DnaA boxes were affected by mutations. The 5,656 bps long ori2 regions (including genes parB, parA, and rctB) are also intact, with 54 SNPs in NSCV1 and 57 SNPs in NSCV2. Notably, the RctB-binding iteron sequences are not affected by mutations.
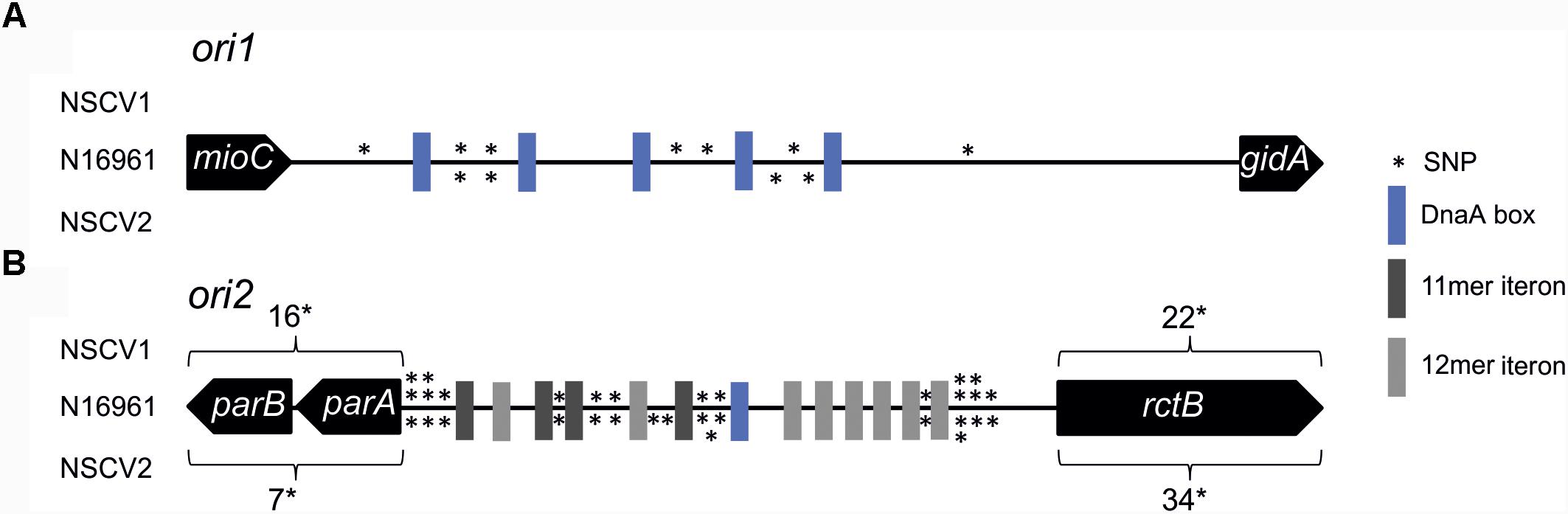
FIGURE 2. Single nucleotide polymorphism (SNP) analysis of replication origins of V. cholerae strains NSCV1 and NSCV2 in comparison to N16961. Genes are indicated as black arrows, RctB binding iterons in dark gray (11 mer iteron) and light gray (12 mer iteron), DnaA boxes in blue and SNPs as stars. The figure is not drawn to scale. Reference sequence: N16961 (Accession # NC002505 and NC002506). Apart from the highlighted SNPs, the sequences are identical. (A) Alignment of ori1. (B) Alignment of ori2.
To test the activity of replication origins experimentally, we carried out marker frequency analyses (MFA). It is known that actively growing cells have a higher copy number of ori proximal sequences compared to ori distal/ter proximal sequences. We employed next generation sequencing technology to obtain whole genome sequences of DNAs isolated from logarithmic and stationary phase cultures of NSCV1 and NSCV2 and analyzed the sequence data for marker frequency. Read data from stationary phase DNAs were used for normalization and read mapping plots were created with ori1 repositioned at the center of the plot to represent bidirectional replication as well as for easy visualization of ori activity (Figure 3). Both NSCV1 and NSCV2 exhibited a maximum copy number of reads close to ori1 and a decreasing gradient on either side of ori1 moving toward the terminus creating a tent-shape (Skovgaard et al., 2011), indicating an active ori1 and bidirectional replication in both strains. In strain NSCV1, a local higher marker frequency was observed at the ori2 position, indicating that ori2 also is active in this strain. In contrast, no such local peak in marker frequency was found at the ori2 position in NSCV2 consistent with a silent ori2. In addition, an almost 3X lower ori1/ter ratio was observed in NSCV2 compared to NSCV1 indicating less or no overlap of replication cycles. The rationale for this interpretation is as follows: On a replicon with overlapping replication, the ori/ter ratio would be four since replication is initiated twice before termination can occur. A replicon that initiates only once would consequently have an ori/ter ratio of two. Considering that the culture represents a mixed population of cells before and after termination an ori/ter ratio of higher than two, as in the case of NSCV1, indicates overlapping replication cycles while values less than two indicate no overlap of replication cycles.
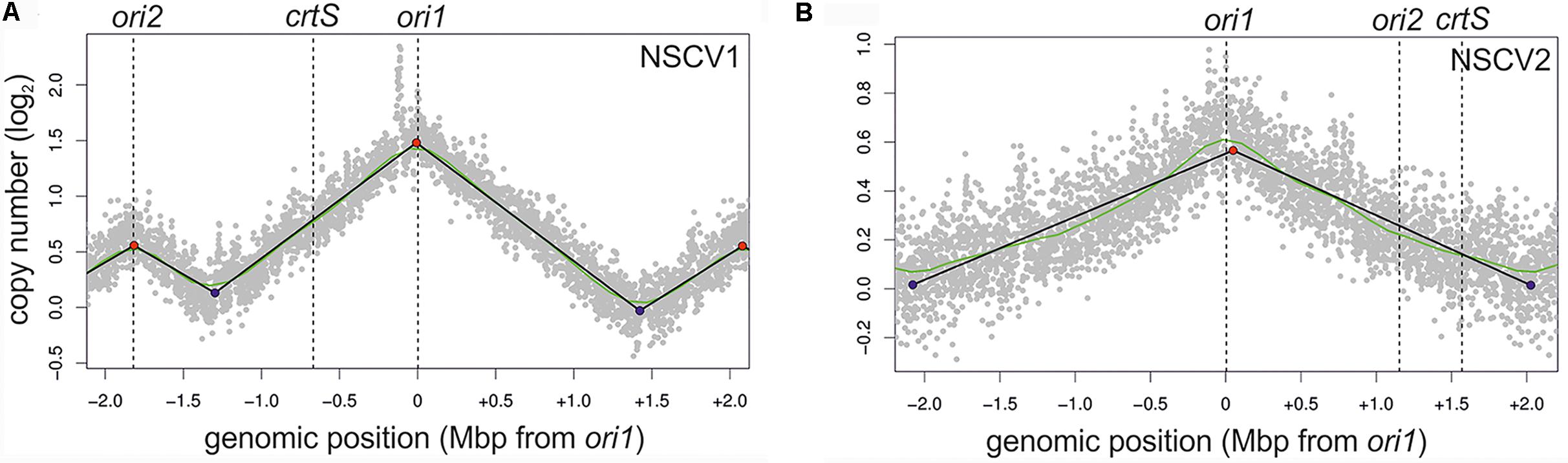
FIGURE 3. Marker frequency analysis (MFA) of V. cholerae NSCV1 (A) and NSCV2 (B) to assess origin activities. Profiles of genome-wide copy numbers based on Illumina sequencing and read mapping. Gray dots represent log numbers of normalized reads as mean values for 1 kbp windows relative to the stationary phase sample. The genome position is shown as the distance from ori1. Vertical dotted black lines mark the locations of replication origins and the crtS site. The solid black lines represent the fitting of regression lines and the green line corresponds to the Loess regression (F = 0.05). Maxima are highlighted by red and minima as blue dots. Plots of biological replicates are shown in Supplementary Figure S1.
NSCV1 and NSCV2 Carry a Functional ori2
The apparent inactivity of ori2 in strain NSCV2 raises the question of whether this origin of replication is functional but silenced or non-functional. To answer this question, we cloned the origins into a mini replicon and assessed independent replication in a plasmid backbone. Plasmid pMA135 carries oriR6K and can replicate conditionally in E. coli strains that provide in trans, the replication initiator protein, Pir, from a lambda prophage. In addition pMA135 can be transferred by conjugation from a donor to a recipient (Messerschmidt et al., 2015). In the absence of λpir, no exconjugants are obtained unless replication is driven by another fully functional origin of replication. The ori2 fragments of NSCV1 and NSCV2, including the core ori2 plus the genes rctB and parAB2, were cloned into pMA135 independently and the number of exconjugants was enumerated in an E. coli strain that does not contain λpir. The ori2 minichromosome constructs from all three strains (N16961, NSCV1, and NSCV2) yielded exconjugants at a frequency of about 1% (10-2) of the recipients, as did the positive control F plasmid ori (Table 1). Minichromosomes based on oriII of strain N16961 have been shown not to integrate into the E. coli chromosome (Messerschmidt et al., 2016). To test if the replicons based on NSCV oriII are also replicating autonomously, we performed a plasmid isolation procedure for five individual clones for each of the two tested NSCV replicons. In all cases, we were able to isolate the corresponding minichromosomes as evidenced by agarose gel electrophoresis (data not shown), verifying autonomous replication without integration into the primary chromosome. The oriR6K replicon by itself did not yield any exconjugant. We conclude that both NSCV1 and NSCV2 carry a functional ori2.
Genetic Stability of Chromosome Fusions in NSCV1 and NSCV2
An active ori2 in strain NSCV1 could potentially allow the chromosome fusion to be reversed. Similarly, the silent ori2 in NSCV2 might be inactive only in the context of a fused chromosome; in either case, splitting of Chr1 and Chr2 is conceivable. It was observed that V. cholerae with an artificially fused chromosome grew slower compared to the two-chromosome parental strain indicating a negative fitness burden on the bacterium (Val et al., 2012). Similarly, we observed an increased doubling time of strains NSCV1 (20 ± 0.5 min) and NSCV2 (29 ± 1.3 min) compared to the two-chromosome strain N16961 (16 ± 0.2 min) (Supplementary Table S4). In addition, microscopic examination showed that strain NSCV2 cells exhibited a distinct phenotype. While the two-chromosome reference strain N16961 and NSCV1 cells are comma-shaped as typical for V. cholerae, NSCV2 cells are much more curled and occasionally S-shaped (Figure 4). It is unknown whether this phenotype is related to the chromosome fusion.
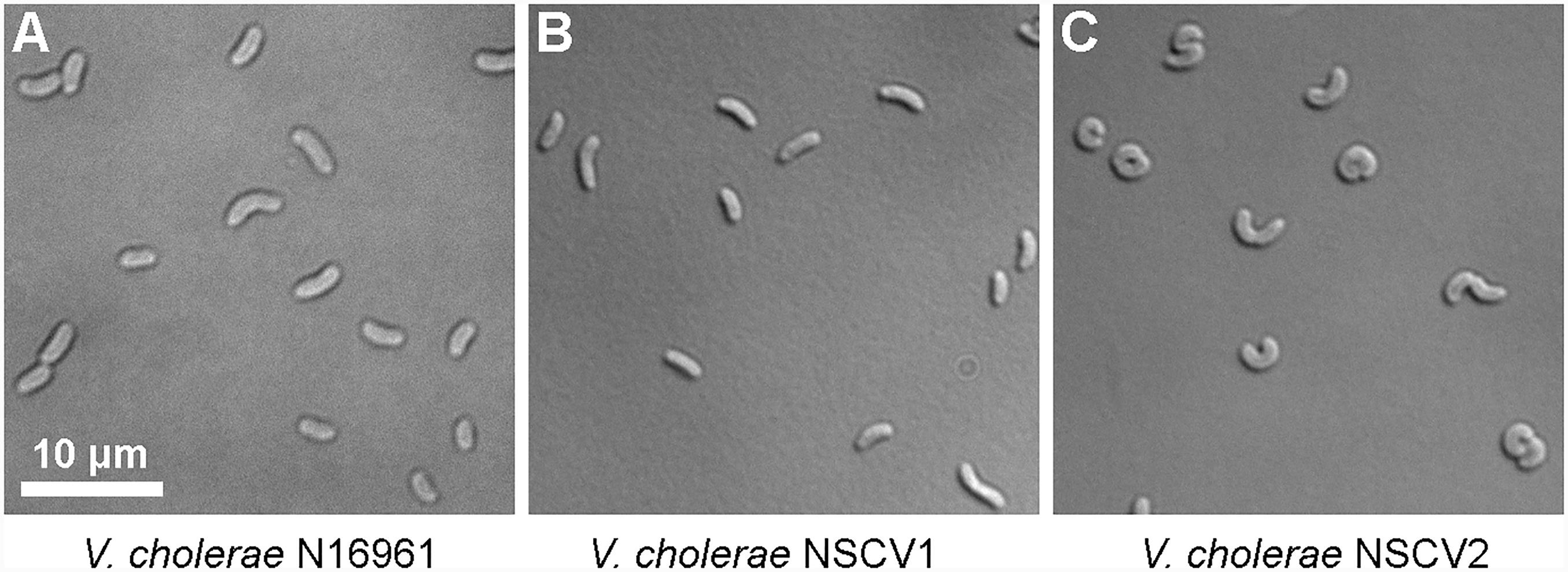
FIGURE 4. Differential interference contrast (DIC) microscopic images of cells of various V. cholerae strains growing in exponential phase. (A–C) N16961 represents the typical 2 chromosome V. cholerae and NSCV1 and NSCV2 are Chr1 and Chr2 fusion strains.
It is conceivable that the slower growth of the fused chromosome strains may have negative fitness value leading to instability, thus promoting genome splitting. On the other hand, if the fused chromosome were under a positive selection pressure to remain fused even at a greater cost in terms of slower growth rate or if they are locked in the fusion configuration due to genetic defects that prevent splitting, then the fusion would be stable even after long term continuous culturing. This led us to test whether the two NSCV strains potentially could revert back to a two-chromosome arrangement upon prolonged continuous culturing. If splitting of the fused chromosomes occurs and if this splitting leads to a fitness advantage due to increased growth rates one would expect two-chromosome clones to appear in a population of NSCV1 and NSCV2 and these clones could potentially replace the fused chromosome cells after prolonged growth by clonal expansion. To test this hypothesis, we cultured the two NSCV strains for 16 days in 100 ml of liquid medium with replenishment of fresh medium every 24 h resulting in approximately 160 generations of growth in total. To examine if the NSCV strains reverted back to a two-chromosome arrangement we isolated DNA from long-term grown cells and performed long-read DNA sequencing using the PacBio technology to be able to detect long range chromosomal rearrangements such as the chromosomal fusion junctions/or junctions (∼24–51 kbs) of the split chromosomes. A de novo genome assembly led to one single contig for both NSCV1 and NSCV2 reflecting the original one-chromosome configuration. In conclusion, the chromosome fusions in NSCV1 and NSCV2 appear to be stable and chromosome splitting is not a frequent event or the fused state is probably under positive selection pressure.
Replication Origins Are Active in an Engineered System Resembling the NSCV Arrangement
Why is ori2 of strain NSCV2 silenced while it is active in NSCV1? One obvious difference between the two strains is the differential positioning of the two replication origins ori1 and ori2 to one another. While the distance between ori1 and ori2 is 1.828 Mbps in NSCV1 it is only 1.118 Mbps in the genome of NSCV2.
To test experimentally, if the ori positioning of NSCV1 and NSCV2 influences the initiation outcome, we re-constructed the ori1 to ori2 arrangement in a genetically accessible system since NSCV1 and NSCV2 are recalcitrant for genetic manipulations. To accomplish this, we transferred a functional hapR to V. cholerae strain MCH1 to render it naturally competent (Lo Scrudato and Blokesch, 2012, 2013). Strain MCH1 was derived from the prototype V. cholerae strain N16961 by fusion of the two chromosomes with a deletion of ori2 (Val et al., 2012). We inserted a copy of ori2 including the flanking genes parAB and rctB into MCH1 at positions analogous to NSCV1 and NSCV2 with respect to the distance from ori1 giving rise to strains VC61 and VC62, respectively, and performed marker frequency analysis of exponentially grown cultures (Figure 5).
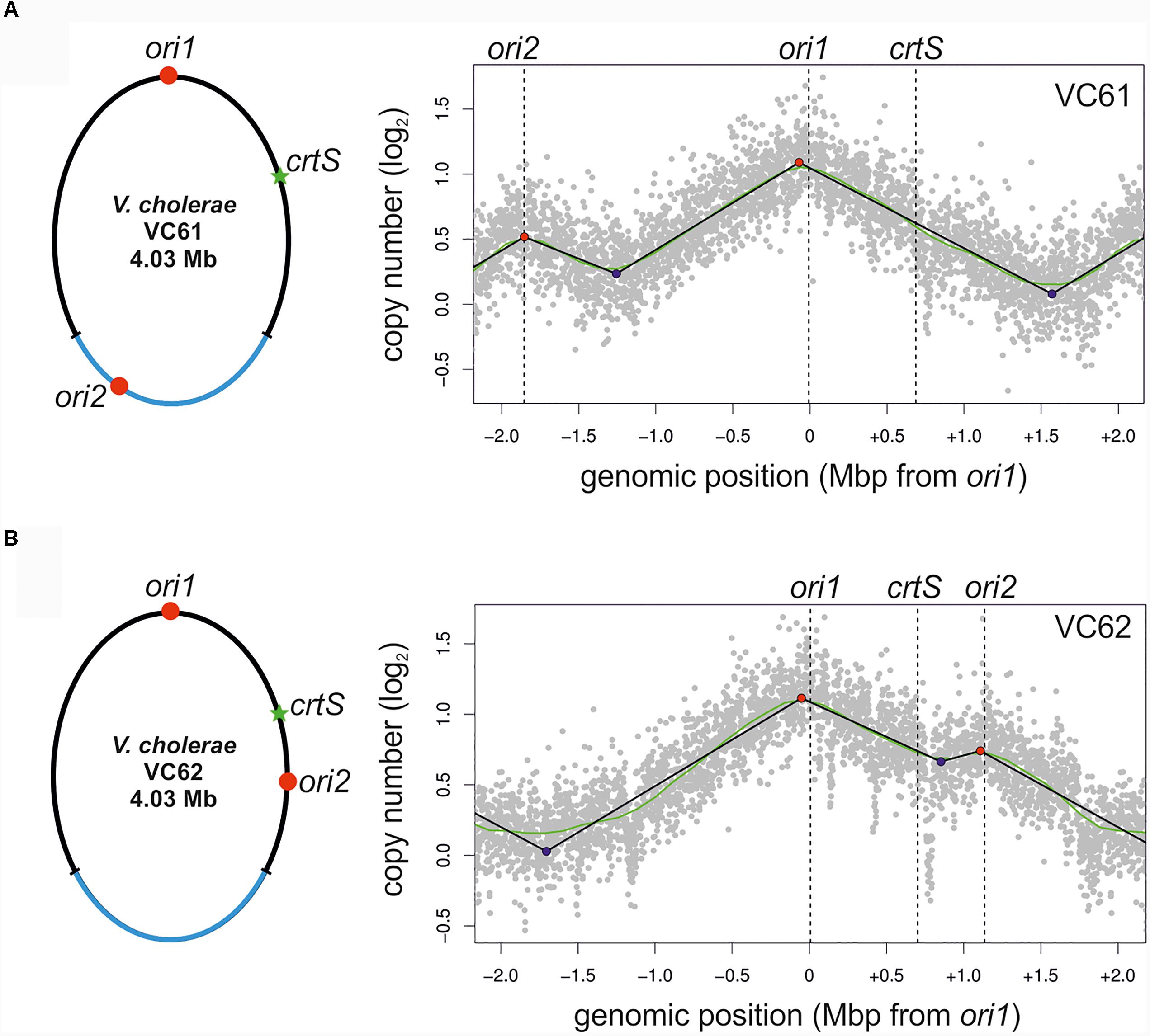
FIGURE 5. Marker frequency analysis of engineered single chromosome V. cholerae strains VC61 (A) and VC62 (B). Left panel: Genomic maps of VC61 (analogous to NSCV1) and VC62 (analogous to NSCV2) showing the respective locations of ori1, ori2, and crtS. Right panel: Profiles of genome-wide copy numbers based on Illumina sequencing. Gray dots represent log numbers of normalized reads as mean values for 1 kbp windows relative to the stationary phase sample. Vertical dotted black lines mark the locations of replication origins of replication and the crtS sites. The solid black lines represent the fitting of regression lines and the green line corresponds to the Loess regression (F = 0.05). Maxima are highlighted by red and minima as blue dots. Plots of biological replicates are shown in Supplementary Figure S1.
The MFA plot of strain VC61 resembled the pattern seen in NSCV1 (Figure 5A, compared to Figure 3A). In contrast, a clear local peak of marker frequency was seen at the ori2 position of strain VC62 (Figure 5B). Thus, even though the origin arrangement is similar in NSCV2 and VC62, the ori2 copy appears to be active only in the engineered strain VC62 and not in NSCV2. To test if the ori2 positioning at the NSCV2 position has more negative effect on growth compared to the ori2 insertion at the NSCV1 position we measured the doubling times of strains VC61 (26 ± 0.1 min) and VC62 (27 ± 0.5 min) (Supplementary Table S4). The difference in doubling time was only marginally affected suggesting that the differences in DNA replication between the two strains do not result in severe impairment in growth. We conclude that an ori2 insertion at positions analogous to those in the NSCV strains relative to ori1 can be active as determined by MFA.
Dam Methyltransferase Is Functional in NSCV Strains
We have shown above that ori2 in strain NSCV2 is functional but appears to be not active. One potential cause of ori2 silencing could be an inactive Dam methylation system. Methylation of the adenine within the Dam recognition site ‘GATC’ present at ori2 locus is a prerequisite for ori2 activity (Demarre and Chattoraj, 2010). Sequence analyses showed intact dam genes in both strains (Xie et al., 2017). The methylation status of GATC sites can be analyzed by the differential sensitivities of the genomic DNA to DpnI (cleaves only methylated/hemimethylated GATC sites), DpnII (cleaves only unmethylated GATC sites) and Sau3A1 (cleaves both unmethylated and methylated GATC sites) restriction enzymes.
Genomic DNAs of NSCV1 and NSCV2 were sensitive to DpnI and Sau3A1 and resistant to DpnII, indicative of full methylation at GATC sites (Figure 6). This result was further confirmed by the PacBio sequence data. In PacBio SMRT (Single Molecule Real Time) sequencing, presence of modified base (A in GATC) in the DNA template results in a delayed incorporation of the corresponding T nucleotide, i.e., longer inter-pulse duration (IPD) compared to template lacking the modification (Flusberg et al., 2010). These kinetic measurements create specific signatures for different types of base modifications. Analyses of the PacBio sequence data for modified bases indicated that both NSCV1 (38571/38572 sites) and NSCV2 (37573/37590) have fully methylated (>99.99%) GATC sites. We conclude that Dam is functional in both NSCV strains and a lack of methylation cannot explain the inactivity of ori2 in NSCV2.
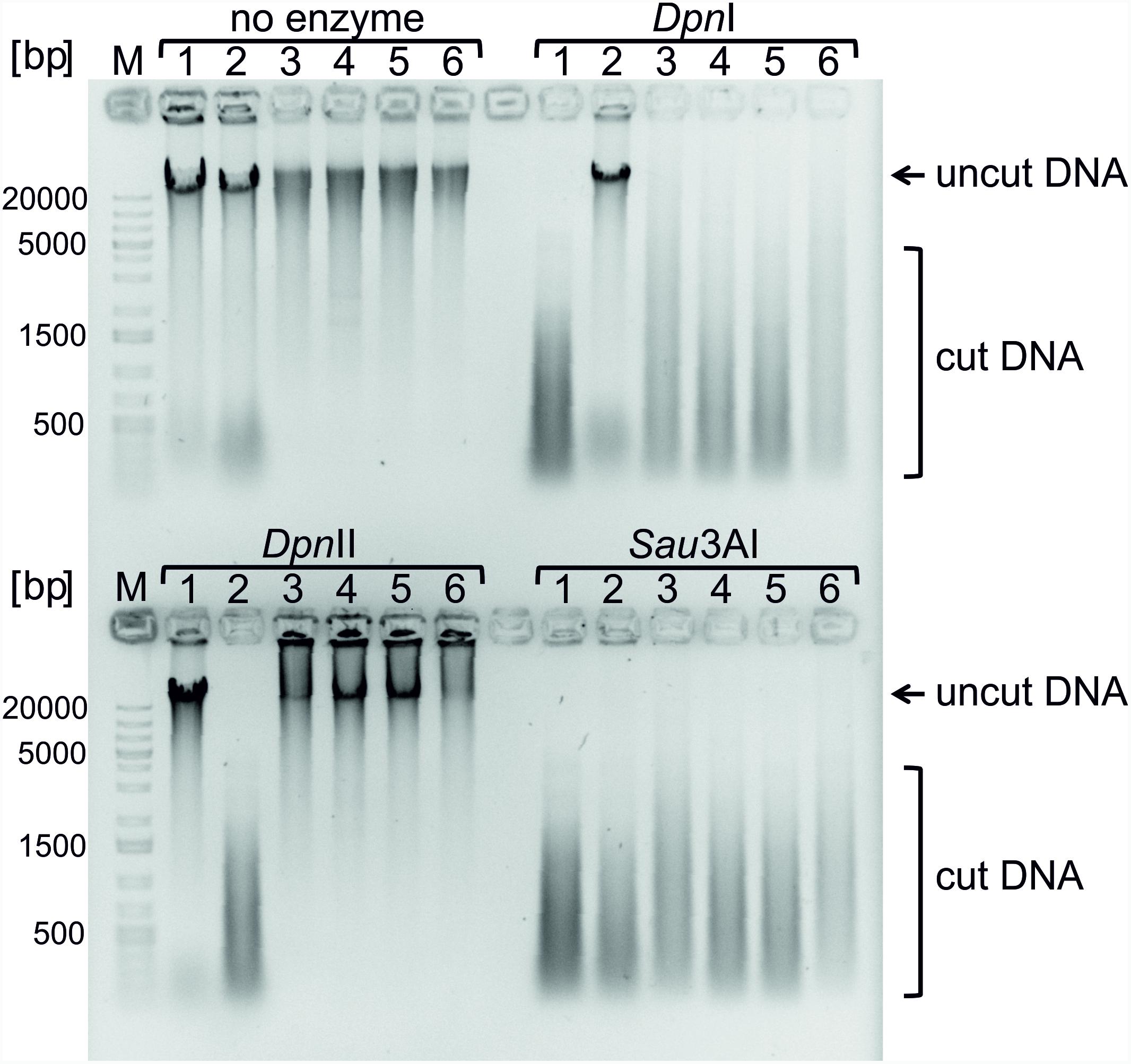
FIGURE 6. Testing Dam methylation sensitivities of genomic DNAs of strains V. cholerae NSCV1 and NSCV2. Restriction digestion of 1 μg of genomic DNA using the indicated enzymes was carried out and the various strains are as follows: 1. E. coli MG1655; 2. E. coli MG1655Δdam; 3 and 4. V. cholerae NSCV1; 5 and 6. V. cholerae NSCV2. DpnI cleaves methylated GATC sequences. DpnII cleaves only unmethylated GATC sequences. Sau3AI cleaves GATCs independent of the methylation state. DNA cleavage is evident from the disappearance of high molecular weight band and appearance of low molecular weight streak of DNA.
crtS Sites of NSCV Strains Are Functional
Another possibility for ori2 silencing in NSCV2 is through alterations of crtS activity. This short DNA sequence is found on Chr1 in all available whole genome sequences of two-chromosome strains of Vibrionaceae and its replication triggers ori2 firing on Chr2 (Baek and Chattoraj, 2014; Val et al., 2016; Kemter et al., 2018). Consequently, a non-functional crtS could lead to an inactive ori2. To decipher the functionality of crtS sites in NSCV1 and NSCV2 in silico, we extracted the respective sequences and aligned them to the consensus sequence we established recently (Figure 7A) (Kemter et al., 2018). Notably, all highly conserved parts of the crtS sequence are also conserved in the crtS sequences of NSCV1 and NSCV2 (Figure 7A). To test the functionality of crtS in the context of a fused chromosome experimentally, we deleted the crtS in strain VC62 (resulting in strain VC71) and performed marker frequency analysis (Figure 7B).
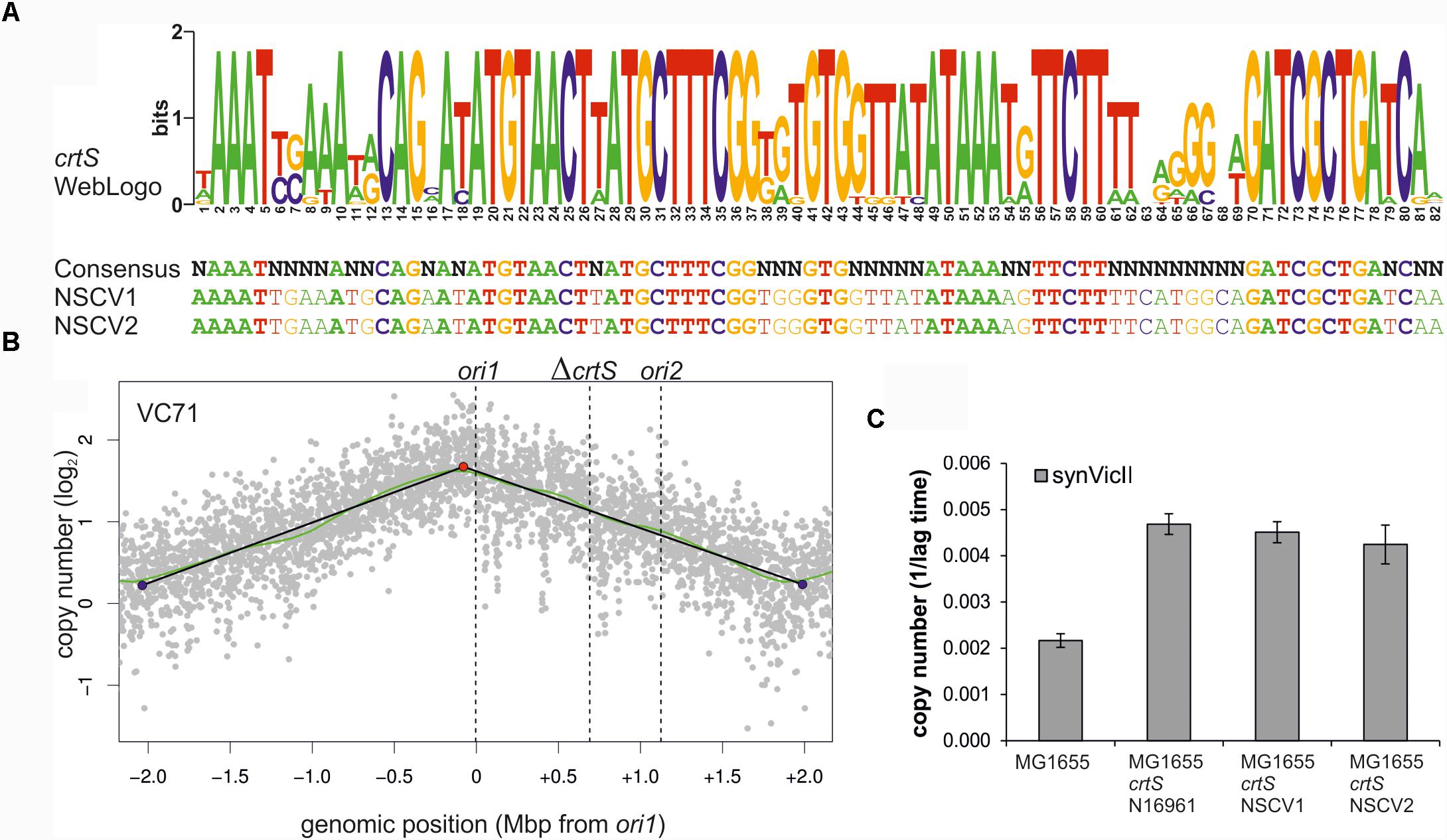
FIGURE 7. crtS sites of NSCV strains are functional. (A) An alignment of crtS sites from 13 sequenced Vibrio genomes was used to calculate a WebLogo (top panel). The height of the nucleotides in each position represents the measure of conservation. All positions with 100% sequence conservation are conserved in the crtS sequences of NSCV1 and NSCV2 as well (indicated in bold, lower panel) (Kemter et al., 2018). (B) Marker frequency analysis of V. cholerae strain VC71 lacking a functional crtS site. Gray dots represent log numbers of normalized reads as mean values for 1 kbp windows relative to the stationary phase sample. The genome position is shown as the distance from ori1. Vertical dotted black lines mark the locations of replication origins of replication and the crtS sites. The solid black lines represent the fitting of regression lines and the green line corresponds to the Loess regression (F = 0.05). Maxima are highlighted by red and minima as blue dots. Plots of biological replicates are shown in Supplementary Figure S1. (C) crtS sites of NSCV1 and NSCV2 increase the copy number of ori2-based mini-chromosomes in E. coli. E. coli strains harboring the ori2-based mini-chromosome synVicII and chromosomal insertions of crtS from different V. cholerae strains as indicated were grown in LB medium with 500 μg/ml ampicillin in a 96-well plate at 37°C. As strains with a lower replicon copy number have a longer lag period to initiate growth, the value 1 divided by the time to reach an OD600 ≥ 0.1 was used as measure for the replicon copy number. Values are the mean of three biological replicates with indicated standard deviation. Growing the strains in standard concentrations of ampicillin did not show any difference between wild type and crtS carrying strains as expected (data not shown).
As expected, no local peak of copy number increase was seen at the ori2 position of the ΔcrtS strain VC71 in contrast to the MFA of the parental strain VC62 (compare Figures 5B,7B) confirming the necessity of a functional crtS site for ori2 firing. Incidentally, the doubling time of this strain is not much different (25 ± 0.6 min) from that of VC61 and VC62 (Supplementary Table S4). To analyze the functionality of the crtS sites from NSCV1 and NSCV2, we inserted these sequences into the genome of E. coli and transformed the corresponding strains with the ori2-based minichromosome synVicII. The rationale behind this approach is the observation of a copy number increase of an ori2-based minichromosome in E. coli strains carrying a functional crtS site (Baek and Chattoraj, 2014; Kemter et al., 2018). The copy number of ori2 minichromosomes was measured in E. coli strains either carrying the crtS sites of strain N16961, NSCV1, NSCV2 or no crtS (Figure 7C). In this case, we tested the resistance level of the respective strains to ampicillin as an indirect measure of the minichromosome copy number as described previously (Schallopp et al., 2017). The crtS sites of both NSCV strains increased the copy number of the ori2 minichromosome compared to the strain lacking any crtS site and to a similar extent as the crtS site from the two-chromosome V. cholerae strain N16961 used as positive control (Figure 7C). We conclude that crtS sites in NSCV1 and NSCV2 are functional in a heterologous system and a defective crtS might not explain the silent ori2 in strain NSCV2.
Relative ori1, ori2, and crtS Locations Determine ori2 Activity in NSCV Strains
In a canonical two-chromosome V. cholerae, replication of the crtS on Chr1 triggers the initiation at ori2 on Chr2. This scenario can in principle be analogous in a fused chromosome such as NSCV1 in which ori1, ori2 and crtS are present on the same molecule. Here the crtS is located about 677 kbps away from ori1 and is replicated first with replication forks originating at ori1 which subsequently triggers ori2 firing, which is further downstream of crtS. The organization is different in strain NSCV2 where the crtS lies about 416 kbps downstream of ori2. Thus, in NSCV2, ori2 precedes crtS. This arrangement is the consequence of a large inversion of the Chr1 part of the genome (Figure 1). The replicative outcome of this arrangement is difficult to predict considering the known triggering role of crtS; i.e., ori2 being replicated first by replication forks originating at ori1, subsequently upon duplication of the crtS site. In other words, crtS can also retrospectively trigger replication of already replicated ori2 that crtS replication is supposed to trigger in the first place. To test this scenario, we moved the crtS site in strain VC62 to the position analogous to strain NSCV2 (resulting in strain VC73). VC73 did not exhibit any significant difference in doubling time compared to other strains (26 ± 0.2 min) (Supplementary Table S4). Respective MFA analysis revealed an active ori2, suggesting that an ori2 copy on a fused chromosome can be triggered by a crtS site located downstream relative to ori2, contrary to what was observed in NSCV2 (compare Figures 3B– 8). The peak at the ori2 position is not very strong but it is important to note that the fitting of regression lines is automated based on the maxima found within the mapped reads (Kemter et al., 2018). There is no pre-selection of ori positions and the fact that the peak is detected at the ori2 position by the computational fitting model implies that it is an active origin as also seen in the biological replicate (Supplementary Figure S1F).
Discussion
Vibrio cholerae has partitioned its genome between a true bacterial chromosome and a “domesticated” plasmid replicon (Heidelberg et al., 2000; Venkova-Canova and Chattoraj, 2011; Val et al., 2014b). Unlike most bacteria, this two-replicon arrangement is conserved within the family of Vibrionaceae (Okada et al., 2005; Val et al., 2014b; Ramachandran et al., 2017). It was postulated that the bipartite genome in Vibrio species enables varying the copy numbers of both chromosomes in a niche-specific manner under certain environmental conditions as an adaptation strategy (Heidelberg et al., 2000; Schoolnik and Yildiz, 2000; Srivastava and Chattoraj, 2007). Alternatively, the two-chromosome setup in Vibrio species can be considered as an adaptive feature that enables rapid genome duplication if multiple chromosomes are replicated simultaneously. The latter assumption fits with the observation that Vibrio species is one of the fastest growing bacteria known, a feature that has led to a recent proposal for using the non-pathogenic V. natriegens as the new workhorse for bioengineering (Weinstock et al., 2016; Dalia et al., 2017; Hoffart et al., 2017). However, it brings up the question of how the two replicons are coordinating their replication and regulate over or under replication in order to ensure inheritance of the just one copy of the full complement of the genome into daughter cells upon cell division. Furthermore, the evolutionary driving force (or forces) that led to the two-chromosome setup in Vibrionaceae remain speculative. One way to better understand the evolutionary significance of this unique feature of Vibrio species is to study naturally occurring exceptions to the two chromosome rule: strains which have evolved into a single chromosome by chromosomal fusion. We have previously identified two Natural Single Chromosome Vibrio (NSCV) cholerae strains (Chapman et al., 2015; Johnson et al., 2015; Xie et al., 2017) and in this study we addressed the functionality and activity of the two origins of replication on the same chromosome.
Fortuitously, the two NSCV strains have different fusion junctions and other genetic rearrangements that provided an opportunity to compare and contrast their ori functionality. We used marker frequency analysis as an indirect measure of ori activity and found ori1 to be active in both strains. In strain NSCV1, we detected an additional, replication activity peak at the ori2 locus indicating that this second origin is active as well. Since the MFA experiments are population based, we cannot exclude that some cells use ori1 only and other cells ori2 only to replicate the fused chromosome. However, we consider it more likely that both origins are firing within the same cell. Interestingly, the copy number ratio of ori2/ter was lower than for ori1/ter consistent with a time delayed initiation of ori2 on the fused chromosome. Such an origin initiation differential has also been observed in conventional two-chromosome Vibrio strains resulting in synchronous termination of replication of the two chromosomes despite their different sizes (Rasmussen et al., 2007; Stokke et al., 2011; Kemter et al., 2018). Conservation of the orchestrated replication timing of the two origins in NSCV1 indicates that the single-chromosome setup does not lead to any origin interference. In two-chromosome Vibrio species, replication of the crtS site located on Chr1 has been shown to trigger ori2 firing (Baek and Chattoraj, 2014; Val et al., 2016; Ramachandran et al., 2018). In V. cholerae N16961, the time between crtS replication and ori2 initiation corresponds to the time the replication fork needs to replicate about 200 kbps of DNA (Val et al., 2016). Considering a similar delay in strain NSCV1, ori2 would have fired long before replication fork originating at ori1 reaches it since the distance between crtS to ori2 is more than 1 Mbps. As a consequence, replication forks originating at ori1 and ori2 will meet at some position of the fused chromosome, the timing of which will dependent on ori1 location and its regulation just as it occurs in two-chromosome V. cholerae.
The scenario is much different in NSCV2. Here, ori2 is replicated before crtS by replication forks originating at ori1 (Figure 1A). Intuitively, this would lead to a chaotic replication perturbance because ori2 firing precedes crtS duplication. In the two-chromosome context, crtS duplication occurs first which then triggers ori2 firing. Hence, if replication of the crtS triggers ori2 initiation also in this genomic arrangement it would happen on two copies of the already replicated ori2. In addition, the replication forks coming from this newly initiated ori2 copies have the potential to replicate the crtS in just a few minutes because of their close proximity and could potentially lead to additional rounds of ori2 initiation and thus an uncontrolled ori2 firing. If this were to happen, not only replication control at ori2 would be severely perturbed but also ori1 functioning can be interfered because ori1 might be replicated passively from replication forks, coming from ori2. Interestingly, what we observed in strain NSCV2 is not a replication out of control but instead a simple silencing of ori2 activity. Our data clearly demonstrate that ori2 of NSCV2 is functional in an isolated context as shown by its ability to drive replication of a mini-chromosome in E. coli. In addition, critical factors involved in regulation of ori2 appear to be fully functional in NSCV2, namely the Dam methylation system and the crtS site. Paradoxically, ori2 is not used to initiate DNA replication in the fused chromosome strain, NSCV2.
A recent study offers a simple explanation for our observation on ori2 silencing in NSCV2 (Ramachandran et al., 2018). These authors showed that it is the doubling of the crtS dosage rather than the process of replicating the crtS that triggers ori2 initiation which results in an even number of crtS and ori2 copies. This tendency of the regulatory system to produce similar copy numbers of crtS and ori2 have also been found in engineered systems with multiple crtS sites (de Lemos Martins et al., 2018). If the crtS site is duplicated before ori2 as it is the case in two-chromosome V. cholerae strains, the ori2 copy number will be lower compared to crtS and consequently the initiation at ori2 will be triggered to restore crtS/ori2 copy number balance. If the crtS site is replicated after ori2 has been copied by replication forks coming from ori1 as in the case of NSCV2, there is no need to initiate at ori2 because the copy number of crtS and ori2 are in balance already. The lack of initiation at ori2 in NSCV2 is therefore fully consistant with and confirmatory to the findings of the aforementioned studies (de Lemos Martins et al., 2018; Ramachandran et al., 2018). However, in contrast to the expectation of the crtS-to-ori2 copy control model, we observed that ori2 is active in the engineered strain VC73 in which the crtS lies downstream of ori2 analogous to the arrangement in NSCV2 (Figure 8). However, it is important to note that although the positioning of ori1, ori2 and crtS are similar in strains NSCV2 and VC73, the genomic contexts of ori2 and crtS are entirely artificial compared to their native positions and furthermore, VC73 does not share the large chromosomal inversion found in NSCV2. More precisely, in VC73, ori2 lies in a region of the original Chr1 and crtS within a region of the original Chr2 (Figure 8, left panel). The discrepancy in ori2 firing between the naturally occurring NSCV2 and the engineered strain VC73 may therefore be explained by their different genomic context.
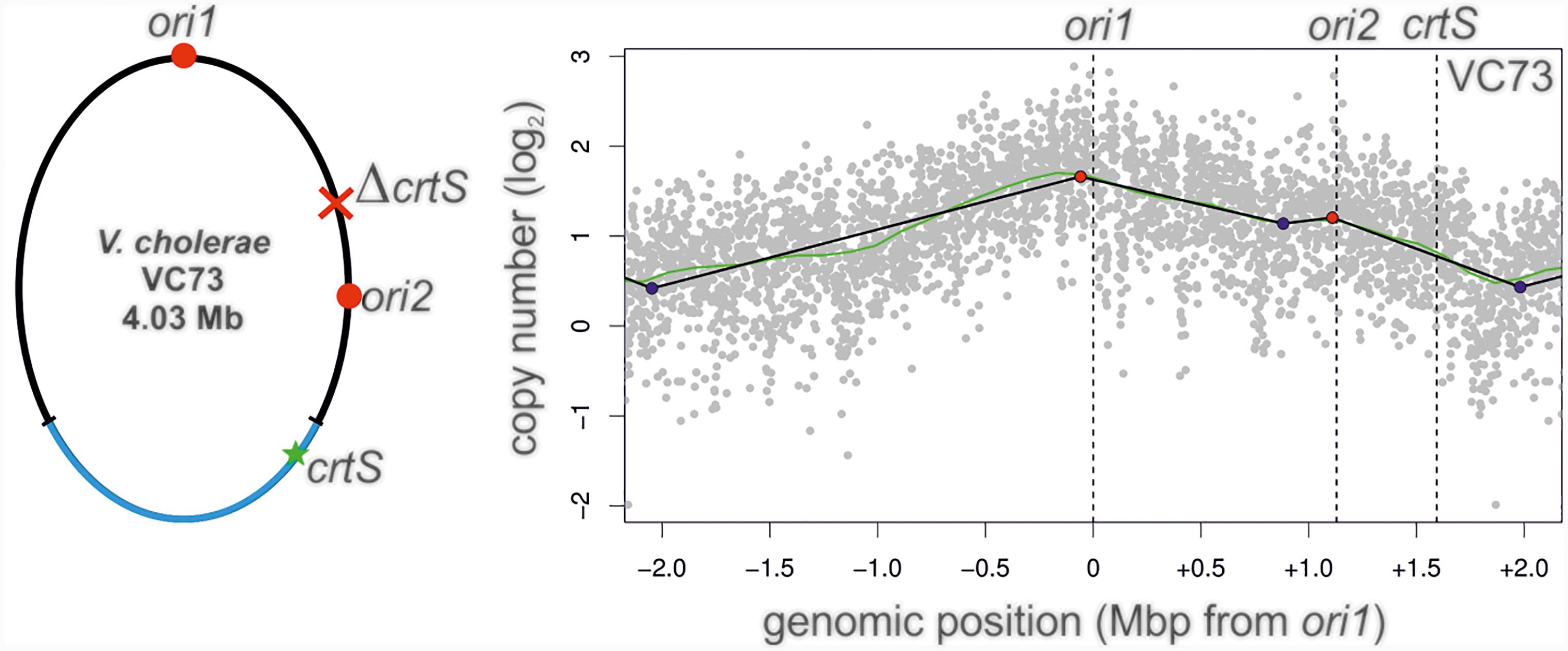
FIGURE 8. Marker frequency analysis of V. cholerae strain VC73. Left panel: Genomic map of VC73 showing the locations of ori1, ori2, and crtS. Right panel: Profile of genome-wide copy numbers. Gray dots represent log numbers of normalized reads as mean values for 1 kbp windows relative to the stationary phase sample. The genome position is shown as the distance from ori1. Vertical dotted black lines mark the locations of replication origins of replication and the crtS sites. The solid black lines represent the fitting of regression lines and the green line corresponds to the Loess regression (F = 0.05). Maxima are highlighted by red and minima as blue dots. Plots of biological replicates are shown in Supplementary Figure S1.
One interesting question that arises is if the two V. cholerae strains with fused chromosomes are evolutionarily stable. It could also be that the fusion occurred only transiently as an artifact of lab cultivation or during the original strain isolation from a patient sample. In fact, chromosome fusions in V. cholerae have been observed previously to occur frequently within a population but the two chromosome configuration seems to provide a selective advantage leading to rapid elimination of the fused chromosomes cells from the population (Val et al., 2014a). Based on this observation, we grew the NSCV strains for about 160 generations and expected a splitting of the fused chromosome that may provide a selective advantage thereby eliminating the cells with single chromosomes from the population. Contrary to the expectation, we found that cells retained the fused chromosomes indicating that they are in fact, locked in this configuration and not easily revertible. An alternative explanation would be that the fused state is under positive selection pressure. It remains to be seen what the significance of the fused chromosome status in NSCV1 and NSCV2 is, in contrast to vast majority of other V. cholerae strains where two chromosome status is the norm. In any case, the single-chromosome V. cholerae appears to be more frequent than expected as yet another NSCV strain was discovered recently (Yamamoto et al., 2018). We expect future studies of NSCV strains which are a deviation from the norm, would to lead to a better understanding of why most V. cholerae carry their genome split into two as the norm.
Materials and Methods
Strains, Plasmids, Oligonucleotides, and Growth Conditions
All strains, plasmids and oligonucleotides used in this study are listed in the Supplementary Tables S1–S3. Unless indicated otherwise, the bacterial cells were grown in LB medium at a temperature of 37°C. Antibiotic selection was performed at the following concentrations, if not indicated differently: Ampicillin 100 μg/ml, Kanamycin 35 μg/ml for E. coli and 70 μg/ml for V. cholerae, Spectinomycin 100 μg/ml, Gentamicin 20 μg/ml, Chloramphenicol 35 μg/ml. Where needed, diaminopimelic acid (DAP) was added to the medium at a concentration of 300 μM. To determine doubling times, cells were grown in LB medium in a 96-well plate at 37°C in a microplate reader (Infinite M200 pro multimode microplate reader, Tecan). OD600 was measured every 5 min for 18 h. Doubling times were calculated in exponential phase for OD600 values between 0.01 and 0.1. For continues cultivation of NSCV strains to investigate potential chromosome splitting strains were inoculated in the morning and grown in 100 ml liquid medium for 24 h. The next morning, new flasks were inoculated 1:1,000 from the previous cultures. After 4 working days (on day 5), 1 ml samples of the cultures were frozen at -80°C as glycerin stocks. Two days later, new flasks were inoculated from these glycerin stocks and the procedure was continued for a total of 16 days. Before sequencing, these were re-streaked on TCBS medium to verify that it is V. cholerae and to obtain single colonies. For each strain one single colony was used to inoculate a culture for DNA isolation and sequencing.
Construction of Replicons and Strains
NSCV minichromosomes were constructed by PCR amplifying the respective origins of replication using the primers 1227/1228 on V. cholerae NSCV1 genomic DNA for pMA739 or V. cholerae NSCV2 genomic DNA for pMA755. The vector pMA135 was digested with AscI and the PCR products were integrated in the linearized vector using Gibson Assembly (Gibson et al., 2009) used to transform E. coli WM3064. For construction of chromosomal integrations and deletions, integration cassettes were assembled by using the MoClo system as described previously (Weber et al., 2011; Milbredt et al., 2016; Schindler et al., 2016). The ori2 insertion cassettes on pMA735 and pMA736, as well as the crtS deletion and insertion cassettes on pMA748 and pMA749 were assembled by MoClo reactions using the plasmids indicated in Supplementary Table S2, which themselves were assembled by MoClo reactions of the respective PCR products into the respective backbones (primers, templates, and backbones indicated in Supplementary Table S2). The linear cassettes were released by restriction enzyme digestions with BsaI. Triparental mating was performed to deliver the plasmids pUXBF13 and pGP704-mTn7-hapR_ATN from E. coli S17-1 λpir to V. cholerae MCH1 (Bao et al., 1991; Meibom et al., 2005; Val et al., 2012). The created strain V. cholerae VC49 was naturally competent and by transforming it with the ori2 insertion cassettes released from pMA735 or pMA736, respectively, followed by transformation with pBR-flp and flippase reaction strains VC61 and VC62 were created (De Souza Silva and Blokesch, 2010). V. cholerae strain VC71 was constructed by deletion of the crtS sequence by transforming V. cholerae VC49 with the crtS deletion cassette from pMA748 and a flippase reaction to excise the resistance marker. Subsequently, strain VC71 was transformed with the crtS insertion cassette from pMA749 and the flippase reaction was performed to create strain VC73. To construct plasmid pMA449, crtS was amplified with primers 1439/1440 from gDNA of V. cholerae NSCV1 and for pMA450 with primers 1439/1440 from gDNA of V. cholerae NSCV2. PCR products were assembled in pMA349 by MoClo assembly as described (Schindler et al., 2016) and used to transform E. coli TOP10 cells. Details of further MoClo assemblies are provided in Supplementary Table S2. Assemblies were used to transform E. coli DH5α λpir. The derived integration cassettes were cut out with BsaI, integrated into the chromosome of E. coli AB330 and transferred to E. coli MG1655 per P1-transduction. FRT recombination was used to remove the resistance marker.
Sequence Comparison
Sequences from V. cholerae N16961, NSCV1 and NSCV2 were compared by multiple sequence alignment using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011) to find single nucleotide polymorphisms. To analyze the whole genomes of the V. cholerae strains NSCV1 and NSCV2 for sequence homology, the alignment free chromosome comparison tool SMASH (Pratas et al., 2015) was used with a minimum block size setting of 5,000 bp.
Preparation of Genomic DNA From Bacteria
The desired amount of culture (between 0.1 ml and 5 ml) was mixed 1:1 with ice-cold killing buffer and centrifuged at maximum speed and 4°C for 3 min. The pellet was resuspended in 300 μl TE, 40 μl SDS and 3 μl 0.5 M EDTA. After 5 min incubation at 65°C, 750 μl Isopropanol were added and the sample was centrifuged 5 min at maximum speed. The pellet was resuspended in 500 μl TE and 2 μl RNAse were added. After 30 min at 37°C, 2 μl of Proteinase K were added and incubated for another 15 min. The sample was purified via twofold Phenol-Chloroform extraction and precipitated over night at -20°C by mixing it with 1 ml pure ethanol and 40 μl 3 M sodium acetate.
The next day, DNA was spinned down at maximum speed for 10 min, the pellet was washed with 70% ethanol and centrifuged another 10 min. The pellet was dried and resuspended in 50 μl pure water.
Short Read Sequencing (Illumina MiSeq)
V. cholerae genomic DNA was isolated as described (Kemter et al., 2018) from various strains grown under different conditions (log phase vs. stationary) were quantitated using the Qubit fluorimeter (Thermo Fisher) and adjusted to 0.2 ng/μL with nuclease-free water. Sequencing libraries were prepared using the Illumina Nextera XT kit, processed and pooled according to manufacturer’s instructions (Illumina). The final, pooled sample was paired-end sequenced (2×300 bp) using the Illumina MiSeq with a v3 chemistry, 600 cycle kit. Post sequencing processing was performed the systems software packages and the final demultiplexed fastq reads produced by the instrument were used for MFA against the reference genome. Raw sequencing data are available on request.
Long Read Sequencing (PacBio)
Whole genome sequencing was performed on a Pacific Biosciences RSII platform. The sequencing library was prepared using the SMRTbellTM Template Prep Kit (Pacific Biosciences, Menlo Park, CA, United States) following manufacturer’s protocol. 5 μg of DNA was fragmented using gTUBE (Covaris Inc., Woburn, MA, United States) to ∼20 kb. After DNA damage repair and ends repair, blunt hairpin adapters were ligated to the template. Non-ligated products were digested by ExoIII and ExoVII exonucleases. Resulting SMRTbell template were purified with AMPure PB beads and size selected on BluePippin system (Sage Science, Beverly, MA, United States), using 0.75% dye-free agarose cassette, with 4–10 kb Hi-Pass protocol and lower cut set on 4 kb. Size selected purified libraries were quantified by Qubit dsDNA High Sensitivity assay. After primer annealing, and P6 polymerase binding, templates were bound to MagBeads for loading. Each sample was sequenced on two SMRT cells, using C4 sequencing kit and 360-min movies per SMRT cell. Presence of unidentified contaminant in two libraries (NSV2 16 days) inhibited sequencing reactions, which manifested in extremely low P1 (2%) and super short reads. Both libraries were subjected to the cleanup procedure that involves binding annealed SMRTbell libraries to magnetic beads, washing the bound annealed DNA SMRTbell templates to remove potential contaminants, and eluting the purified, annealed DNA SMRTbell templates from the magnetic beads. The purified SMRTbell templates were then re-quantified by Qubit and prepared for sequencing on the PacBio RSII according to the Binding Calculator. After cleanup procedure, PacBio RS II instrument sequencing yields were comparable to the other samples. Raw sequencing data are available on request.
Marker Frequency Analysis
Sequencing reads from a NGS were mapped to the respective genome using the program Geneious (Biomatters Ltd.; Kearse et al., 2012). Read densities were extracted and plotted using custom R scripts as described previously (Kemter et al., 2018).
Semiquantitative Conjugation
All used replicons possess an oriR6K and were conjugated from E. coli strain WM3064 to MG1655. For overnight cultures of donor and recipient strains the OD600 was determined and the amount of cells corresponding to 1 ml of OD600 = 1 was centrifuged 1 min at 13,000 ×g. The cells were washed twice in TBS and resuspended in 100 μl TBS. From each donor strain, 50 μl were mixed with 50 μl of the recipient strain and dropped on LB agar including DAP. After 6 h cells were scraped off the plate, washed twice in TBS. The total CFU of recipient cells was determined by plating dilutions on LB, while the CFU of plasmid bearing recipients was determined by plating the same dilutions on selective media. The selective CFU was then normalized to total CFU.
Microscopy
Differential interference contrast (DIC) microscopy was performed on 1% (w/v) agarose pads in PBS buffer using the Nikon Ti fluorescence microscope (100× objective, NA 1.45).
Quantification of Replicon Copy Number via Antibiotic Sensitivity
The copy-up effect of crtS was measured as described (Messerschmidt et al., 2016). Cells were grown in LB medium with either 100 or 500 μg/ml ampicillin at 37°C in 96-well plates in a microplate reader (Infinite M200 pro multimode microplate reader, Tecan). The main culture (150 μL) was inoculated 1:1,000 and growth curves recorded for 15 h. For better visualization, 1 divided by the time needed to reach an OD600 of 0.1 was defined as measure of the copy number.
Author Contributions
MB, DS, SS, and TW contributed to the conceptualization of the study. MB, DS, FK, MW, KC, GK, and GP contributed to the methodology. MB, DS, KC, and GK contributed to the software. MB, DS, FK, MW, KC, GK, GP, SS, and TW contributed to the formal analysis of the study. MB, DS, FK, MW, KC, GK, and GP executed the investigation for the study. MB, SS, and TW contributed to the writing of the original draft. SS and TW reviewed and edited the manuscript. DS, SS, and TW provided the supervision. SS, GP, and TW contributed to the funding acquisition.
Funding
This work was supported within the LOEWE program of the State of Hesse and a grant of the Deutsche Forschungsgemeinschaft (Grant No. WA2713/4-1). Funding for the sequencing work was provided by National Strategic Research Institute (NSRI, University of Nebraska) under the contract number FA4600-12-D-9000 – TOPR 0042 – TO0059).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all current and former members of the Waldminghaus lab for help and fruitful discussions. Nadine Schallopp is acknowledged for excellent technical assistance and Mehryad Mataei for experimental support. We are grateful to Melanie Blokesch for advice in V. cholerae genetics and for providing strains and plasmids and to Didier Mazel for providing V. cholerae strain MCH1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02932/full#supplementary-material
References
Baek, J. H., and Chattoraj, D. K. (2014). Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 10:e1004184. doi: 10.1371/journal.pgen.1004184
Bao, Y., Lies, D. P., Fu, H., and Roberts, G. P. (1991). An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109, 167–168. doi: 10.1016/0378-1119(91)90604-A
Chapman, C., Henry, M., Bishop-Lilly, K. A., Awosika, J., Briska, A., Ptashkin, R. N., et al. (2015). Scanning the landscape of genome architecture of non-O1 and non-O139 Vibrio cholerae by whole genome mapping reveals extensive population genetic diversity. PLoS One 10:e0120311. doi: 10.1371/journal.pone.0120311
Ciaccia, P. N., Ramachandran, R., and Chattoraj, D. K. (2018). A requirement for global transcription factor Lrp in licensing replication of Vibrio cholerae chromosome 2. Front. Microbiol. 9:2103. doi: 10.3389/fmicb.2018.02103
Dalia, T. N., Hayes, C. A., Stolyar, S., Marx, C. J., McKinlay, J. B., and Dalia, A. B. (2017). Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth. Biol. 6, 1650–1655. doi: 10.1021/acssynbio.7b00116
de Lemos Martins, F., Fournes, F., Mazzuoli, M. V., Mazel, D., and Val, M. E. (2018). Vibrio cholerae chromosome 2 copy number is controlled by the methylation-independent binding of its monomeric initiator to the chromosome 1 crtS site. Nucleic Acids Res. 46, 10145–10156. doi: 10.1093/nar/gky790
De Souza Silva, O., and Blokesch, M. (2010). Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64, 186–195. doi: 10.1016/j.plasmid.2010.08.001
Demarre, G., and Chattoraj, D. K. (2010). DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 6:e1000939. doi: 10.1371/journal.pgen.1000939
Duigou, S., Knudsen, K. G., Skovgaard, O., Egan, E. S., Lobner-Olesen, A., and Waldor, M. K. (2006). Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J. Bacteriol. 188, 6419–6424. doi: 10.1128/JB.00565-06
Duigou, S., Yamaichi, Y., and Waldor, M. K. (2008). ATP negatively regulates the initiator protein of Vibrio cholerae chromosome II replication. Proc. Natl. Acad. Sci. U.S.A. 105, 10577–10582. doi: 10.1073/pnas.0803904105
Egan, E. S., Fogel, M. A., and Waldor, M. K. (2005). Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol. Microbiol. 56, 1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x
Egan, E. S., and Waldor, M. K. (2003). Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114, 521–530. doi: 10.1016/S0092-8674(03)00611-1
Espinosa, E., Barre, F. X., and Galli, E. (2017). Coordination between replication, segregation and cell division in multi-chromosomal bacteria: lessons from Vibrio cholerae. Int. Microbiol. 20, 121–129. doi: 10.2436/20.1501.01.293
Flusberg, B. A., Webster, D. R., Lee, J. H., Travers, K. J., Olivares, E. C., Clark, T. A., et al. (2010). Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465. doi: 10.1038/nmeth.1459
Fournes, F., Val, M. E., Skovgaard, O., and Mazel, D. (2018). Replicate once per cell cycle: replication control of secondary chromosomes. Front. Microbiol. 9:1833. doi: 10.3389/fmicb.2018.01833
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Hoffart, E., Grenz, S., Lange, J., Nitschel, R., Muller, F., Schwentner, A., et al. (2017). High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microbiol. 83:e01614-17. doi: 10.1128/AEM.01614-17
Jha, J. K., Baek, J. H., Venkova-Canova, T., and Chattoraj, D. K. (2012). Chromosome dynamics in multichromosome bacteria. Biochim. Biophys. Acta 1819, 826–829. doi: 10.1016/j.bbagrm.2012.01.012
Johnson, S. L., Khiani, A., Bishop-Lilly, K. A., Chapman, C., Patel, M., Verratti, K., et al. (2015). Complete genome assemblies for two single-chromosome Vibrio cholerae Isolates, strains 1154-74 (Serogroup O49) and 10432-62 (Serogroup O27). Genome Announc. 3:e00462-15. doi: 10.1128/genomeA.00462-15
Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F., and Camilli, A. (2013). Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9:e1003800. doi: 10.1371/journal.ppat.1003800
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kemter, F. S., Messerschmidt, S. J., Schallopp, N., Sobetzko, P., Lang, E., Bunk, B., et al. (2018). Synchronous termination of replication of the two chromosomes is an evolutionary selected feature in Vibrionaceae. PLoS Genet. 14:e1007251. doi: 10.1371/journal.pgen.1007251
Koch, B., Ma, X., and Lobner-Olesen, A. (2010). Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation. J. Bacteriol. 192, 3903–3914. doi: 10.1128/JB.00311-10
Lo Scrudato, M., and Blokesch, M. (2012). The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 8:e1002778. doi: 10.1371/journal.pgen.1002778
Lo Scrudato, M., and Blokesch, M. (2013). A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 41, 3644–3658. doi: 10.1093/nar/gkt041
Lu, M., Campbell, J. L., Boye, E., and Kleckner, N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. doi: 10.1016/0092-8674(94)90156-2
Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C. Y., and Schoolnik, G. K. (2005). Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827. doi: 10.1126/science.1120096
Messerschmidt, S. J., Kemter, F. S., Schindler, D., and Waldminghaus, T. (2015). Synthetic secondary chromosomes in Escherichia coli based on the replication origin of chromosome II in Vibrio cholerae. Biotechnol. J. 10, 302–314. doi: 10.1002/biot.201400031
Messerschmidt, S. J., Schindler, D., Zumkeller, C. M., Kemter, F. S., Schallopp, N., and Waldminghaus, T. (2016). Optimization and characterization of the synthetic secondary chromosome synVicII in Escherichia coli. Front. Bioeng. Biotechnol. 4:96. doi: 10.3389/fbioe.2016.00096
Milbredt, S., Farmani, N., Sobetzko, P., and Waldminghaus, T. (2016). DNA replication in engineered Escherichia coli genomes with extra replication origins. ACS Synth. Biol. 5, 1167–1176. doi: 10.1021/acssynbio.6b00064
Okada, K., Iida, T., Kita-Tsukamoto, K., and Honda, T. (2005). Vibrios commonly possess two chromosomes. J. Bacteriol. 187, 752–757. doi: 10.1128/JB.187.2.752-757.2005
Pratas, D., Silva, R. M., Pinho, A. J., and Ferreira, P. J. (2015). An alignment-free method to find and visualise rearrangements between pairs of DNA sequences. Sci. Rep. 5:10203. doi: 10.1038/srep10203
Ramachandran, R., Ciaccia, P. N., Filsuf, T. A., Jha, J. K., and Chattoraj, D. K. (2018). Chromosome 1 licenses chromosome 2 replication in Vibrio cholerae by doubling the crtS gene dosage. PLoS Genet. 14:e1007426. doi: 10.1371/journal.pgen.1007426
Ramachandran, R., Jha, J., Paulsson, J., and Chattoraj, D. (2017). Random versus cell cycle-regulated replication initiation in bacteria: insights from studying Vibrio cholerae chromosome 2. Microbiol. Mol. Biol. Rev. 81, e00033-16. doi: 10.1128/MMBR.00033-16
Rasmussen, T., Jensen, R. B., and Skovgaard, O. (2007). The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 26, 3124–3131. doi: 10.1038/sj.emboj.7601747
Saint-Dic, D., Kehrl, J., Frushour, B., and Kahng, L. S. (2008). Excess SeqA leads to replication arrest and a cell division defect in Vibrio cholerae. J. Bacteriol. 190, 5870–5878. doi: 10.1128/JB.00479-08
Schallopp, N., Milbredt, S., Sperlea, T., Kemter, F. S., Bruhn, M., Schindler, D., et al. (2017). Establishing a system for testing replication inhibition of the Vibrio cholerae secondary chromosome in Escherichia coli. Antibiotics 7:E3. doi: 10.3390/antibiotics7010003
Schindler, D., Milbredt, S., Sperlea, T., and Waldminghaus, T. (2016). Design and assembly of DNA sequence libraries for chromosomal insertion in bacteria based on a set of modified MoClo vectors. ACS Synth. Biol. 5, 1362–1368. doi: 10.1021/acssynbio.6b00089
Schoolnik, G. K., and Yildiz, F. H. (2000). The complete genome sequence of Vibrio cholerae: a tale of two chromosomes and of two lifestyles. Genome Biol. 1, 1016.1–1016.3. doi: 10.1186/gb-2000-1-3-reviews1016
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Skovgaard, O., Bak, M., Lobner-Olesen, A., and Tommerup, N. (2011). Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 21, 1388–1393. doi: 10.1101/gr.117416.110
Slater, S., Wold, S., Lu, M., Boye, E., Skarstad, K., and Kleckner, N. (1995). E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82, 927–936. doi: 10.1016/0092-8674(95)90272-4
Srivastava, P., and Chattoraj, D. K. (2007). Selective chromosome amplification in Vibrio cholerae. Mol. Microbiol. 66, 1016–1028. doi: 10.1111/j.1365-2958.2007.05973.x
Stokke, C., Waldminghaus, T., and Skarstad, K. (2011). Replication patterns and organization of replication forks in Vibrio cholerae. Microbiology 157, 695–708. doi: 10.1099/mic.0.045112-0
Val, M. E., Kennedy, S. P., Soler-Bistue, A. J., Barbe, V., Bouchier, C., Ducos-Galand, M., et al. (2014a). Fuse or die: how to survive the loss of Dam in Vibrio cholerae. Mol. Microbiol. 91, 665–678. doi: 10.1111/mmi.12483
Val, M. E., Soler-Bistue, A., Bland, M. J., and Mazel, D. (2014b). Management of multipartite genomes: the Vibrio cholerae model. Curr. Opin. Microbiol. 22, 120–126. doi: 10.1016/j.mib.2014.10.003
Val, M. E., Marbouty, M., de Lemos Martins, F., Kennedy, S. P., Kemble, H., Bland, M. J., et al. (2016). A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci. Adv. 2:e1501914. doi: 10.1126/sciadv.1501914
Val, M. E., Skovgaard, O., Ducos-Galand, M., Bland, M. J., and Mazel, D. (2012). Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 8:e1002472. doi: 10.1371/journal.pgen.1002472
Venkova-Canova, T., and Chattoraj, D. K. (2011). Transition from a plasmid to a chromosomal mode of replication entails additional regulators. Proc. Natl. Acad. Sci. U.S.A. 108, 6199–6204. doi: 10.1073/pnas.1013244108
Waldminghaus, T., and Skarstad, K. (2009). The Escherichia coli SeqA protein. Plasmid 61, 141–150. doi: 10.1016/j.plasmid.2009.02.004
Weber, E., Engler, C., Gruetzner, R., Werner, S., and Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. doi: 10.1371/journal.pone.0016765
Weinstock, M. T., Hesek, E. D., Wilson, C. M., and Gibson, D. G. (2016). Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 13, 849–851. doi: 10.1038/nmeth.3970
Xie, G., Johnson, S. L., Davenport, K. W., Rajavel, M., Waldminghaus, T., Detter, J. C., et al. (2017). Exception to the rule: genomic characterization of naturally occurring unusual Vibrio cholerae strains with a single chromosome. Int. J. Genomics 2017:8724304. doi: 10.1155/2017/8724304
Keywords: DNA replication, secondary chromosome, plasmid, multipartite genome, replication initiation, pathogens, cholera
Citation: Bruhn M, Schindler D, Kemter FS, Wiley MR, Chase K, Koroleva GI, Palacios G, Sozhamannan S and Waldminghaus T (2018) Functionality of Two Origins of Replication in Vibrio cholerae Strains With a Single Chromosome. Front. Microbiol. 9:2932. doi: 10.3389/fmicb.2018.02932
Received: 28 September 2018; Accepted: 14 November 2018;
Published: 30 November 2018.
Edited by:
Alan Leonard, Florida Institute of Technology, United StatesReviewed by:
Dhruba Chattoraj, National Institutes of Health (NIH), United StatesOle Skovgaard, Roskilde University, Denmark
Gregory Marczynski, McGill University, Canada
Copyright © 2018 Bruhn, Schindler, Kemter, Wiley, Chase, Koroleva, Palacios, Sozhamannan and Waldminghaus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanmuga Sozhamannan, U2hhbm11Z2EuU296aGFtYW5uYW4uY3RyQG1haWwubWls Torsten Waldminghaus, VG9yc3Rlbi5XYWxkbWluZ2hhdXNAU1lOTUlLUk8uVW5pLU1hcmJ1cmcuZGU=
 Matthias Bruhn1
Matthias Bruhn1 Daniel Schindler
Daniel Schindler Gustavo Palacios
Gustavo Palacios Shanmuga Sozhamannan
Shanmuga Sozhamannan Torsten Waldminghaus
Torsten Waldminghaus