- 1Laboratory of Veterinary Pharmacology, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2LABioMed at Harbor-UCLA Medical Center, Torrance, CA, United States
- 3Geffen School of Medicine at UCLA, Los Angeles, CA, United States
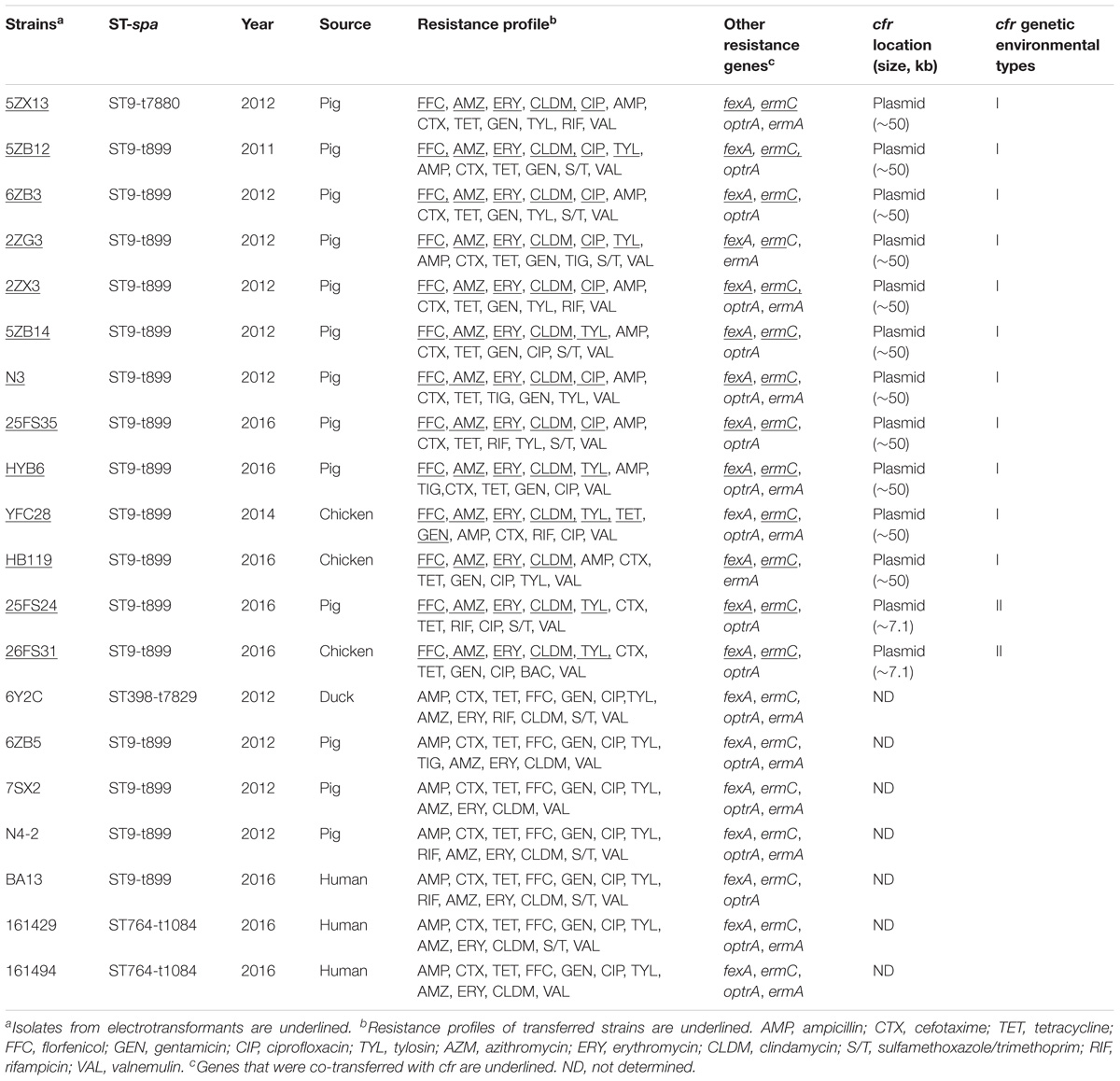
We investigated cfr-positive and -negative MRSA strains isolated from animals and humans in different geographical areas of China, from 2011 to 2016. Twenty cfr-positive strains (15.6%) were identified from 128 MRSA strains including 17 from food animals and three from humans. The resistance rates and prevalence of the tested antibiotic resistance genes (ARGs) in the cfr-positive MRSA isolates were higher than that in the cfr-negative MRSA isolates. All cfr-positive MRSA isolates were co-carrying fexA and ermC, and had significantly higher optrA incidence rate vs. the cfr-negative isolates (P < 0.05). In addition, multilocus sequence typing (MLST) assays showed that ST9 and spa-type t899 were the most prevalent ST and spa types in the study strains. However, all of the 20 cfr-positive and 10 randomly selected cfr-negative MRSA isolates were clonally unrelated as determined by pulsed-field gel electrophoresis (PFGE) analyses. Importantly, the cfr gene was successfully transferred to a recipient Staphylococcus aureus strain RN4220 from 13 of the 20 cfr-positive MRSA isolates by electroporation. Among these 13 cfr-positive MRSA isolates, two different genetic contexts surrounding cfr were determined and each was associated with one type of cfr-carrying plasmids. Of note, the predominant genetic context of cfr was found to be a Tn558 variant and locate on large plasmids (∼50 kb) co-harboring fexA in 11 of the 13 MRSA isolates. Furthermore, the cfr gene was also identified on small plasmids (∼ 7.1 kb) that co-carried ermC in two of the 13 MRSA isolates. Our results demonstrated a high occurrence of multi-drug resistance in cfr-positive MRSA isolates, and the spread of cfr might be attributed to horizontal dissemination of similar cfr-carrying transposons and plasmids.
Introduction
The chloramphenicol–florfenicol resistance (cfr) gene encodes a methyltransferase that modifies position A-2503 in bacterial 23S rRNA and confers resistance to five classes of antibiotics (phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A) (Long et al., 2006; Morales et al., 2010). These antibiotics have been widely used for the treatment of infections in human and animal (Inkster et al., 2017; Li J. et al., 2017). Since the first identification of the cfr gene in Staphylococcus sciuri isolates in 2000, it has been subsequently found in Enterococcus spp., Bacillus spp., Streptococcus suis, Proteus vulgaris, and Escherichia coli (Long et al., 2006; Wang et al., 2011, 2012a,b). In China, most cfr-positive isolates were derived from domestic animals (mainly pigs). In addition, plasmids and insertion sequences were implicated in cfr gene dissemination between species and genera (Shen et al., 2013).
Methicillin-resistant Staphylococcus aureus (MRSA) can cause a wide range of infections, including skin and soft-tissue infections as well as endocarditis and respiratory tract infections (Marshall and McBryde, 2014; Rodvold and McConeghy, 2014). Hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA) are the primary origins for infections in humans (Woodford and Livermore, 2009). However, livestock-acquired MRSA (LA-MRSA) have been identified in pigs, ducks, poultry, and rats (Voss et al., 2005; Wulf et al., 2006; de Neeling et al., 2007; van de Giessen et al., 2009). Importantly, LA-MRSA containing the plasmid-borne cfr gene has been identified in infections of farmers suggesting zoonotic transmission (Wulf et al., 2006; Cui et al., 2009).
In this study, we investigated the epidemiological characteristics and dissemination of the cfr gene in clinical MRSA isolates from animal and human sources. We compared the phenotypic and genotypic profiles of cfr-positive MRSA strains with cfr-negative MRSA strains.
Materials and Methods
In total, 128 MRSA strains were isolated from pigs, chickens, and ducks in 10 different regions of China and from clinical patients in two different hospitals in Guangzhou, China during 2011–2016. All MRSA isolates were confirmed by MALDI-TOF/MS system (Shimadzu-Biotech, Japan), multiplex PCR amplification, and DNA sequencing of the mecA gene.
Minimum inhibitory concentrations (MIC) were determined using a standard agar dilution method -CLSI M100-S28 and VET01-A4/VET01-S2. The tested antibiotics were phenicols (florfenicol), lincosamides (clindamycin), oxazolidinones (linezolid), pleuromutilins (valnemulin), β-lactams (ampicillin and cefotaxime), macrolides (tylosin, azithromycin, and erythromycin) and ciprofloxacin, gentamycin, tetracycline, rifampicin, trimethoprim-sulfamethoxazole, vancomycin, and daptomycin. The MIC breakpoints of each antibiotic against MRSA were used as recommended by the current CLSI guidance (Clinical and Laboratory Standards Institute [CLSI], 2013, 2018). S. aureus ATCC 29213 was used as a quality control strain.
Detection of Resistance Genes
The presence of the cfr gene in the MRSA strains was determined with PCR as described previously (Kehrenberg and Schwarz, 2006). Other genes that encoded resistance to phenicols (fexA), lincomycin [lnu(A), (F)], oxazolidinones (optrA), pleuromutilins (vgaAV), macrolide–lincosamide–streptogramin B (ermA-C), macrolides (ereA-B), tetracycline [tet(A), (C), (L), (M), and (K)]. and aminoglycosides [aac(3′)-Ia, aac(3′)-IIc, aadA1, aadB, aph(3′)-II, aph(3′)-IV, aph(4′)-Ia, and aac(6′)-Ib] were identified by PCR using gene-specific primers (Supplementary Table S1).
Molecular Typing
Genetic diversity of cfr-positive and -negative MRSA isolates was determined by SmaI pulsed-field gel electrophoresis (PFGE) (Tenover et al., 1995). Comparison of PFGE patterns was performed with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms were generated using Dice similarity coefficient and analogical values to categorize identical PFGE types cut-offs were fixed at 100%. Further determinations of clonality were performed by multilocus sequence typing (MLST) and spa typing as described previously.1,2 Salmonella enterica serotype Braenderup H9812 DNA was used as a molecular size marker (Tenover et al., 1995).
Transformation of cfr Gene and Determination of cfr Location
Plasmid DNA from cfr-positive MRSA strains was extracted using a Qiagen Prep Plasmid Midi Kit (Qiagen, Hilden, Germany) and transferred into a recipient S. aureus strain RN4220 by electroporation using Gene Pulser apparatus (Bio-Rad, Hercules, CA, United States). Electrotransformants were selected on brain heart infusion (BHI) agar containing 8 μg/mL of florfenicol. The presence of cfr was further confirmed by PCR (Kehrenberg et al., 2009). To determine the location of cfr gene, DNA was separated by PFGE after treatment with S1 nuclease (Takara, Dalian, China) and plasmids carrying cfr were identified by Southern blot hybridization using a digoxigenin-labeled cfr probe (Roche, Mannheim, Germany) according to the manufacturer’s instruction.
Genetic Environment of cfr Gene
The genetic environment surrounding cfr was determined by PCR mapping, inverse PCR, and sequencing (Wang et al., 2015). The primers used to determine the regions upstream and downstream of cfr gene and reference sequences containing the cfr gene used for PCR mapping are listed in Supplementary Table S2. The obtained DNA sequences were analyzed using BLAST,3 and then compared to those deposited in GenBank.
Statistical Analyses
Statistical significance for the comparison of prevalence data and proportions was determined using a χ2 test. P < 0.05 was consideredto be statistical significant.
Results
Antimicrobial Susceptibility and Presence of Resistance Genes
We demonstrated that >80% of the 128 MRSA isolates were resistant to all tested antimicrobial agents with the exception of sulfamethozaxole/trimethoprim (43.8%), rifampicin (21.1%), linezolid (1.56%), vancomycin (0%), and daptomycin (0%) (Figure 1). Importantly, resistance rates in cfr-positive strains were higher than in cfr-negative strains for sulfamethozaxole/trimethoprim (60 vs. 40.7%) and rifampicin (30 vs. 19.4%) (Figure 1). In addition, the proportion of isolates with increased linezolid MIC (≥2 μg/mL) was significantly higher in the cfr-positive MRSA vs. the cfr-negative MRSA (40 vs. 6.5%, P < 0.001) (Supplementary Table S3).
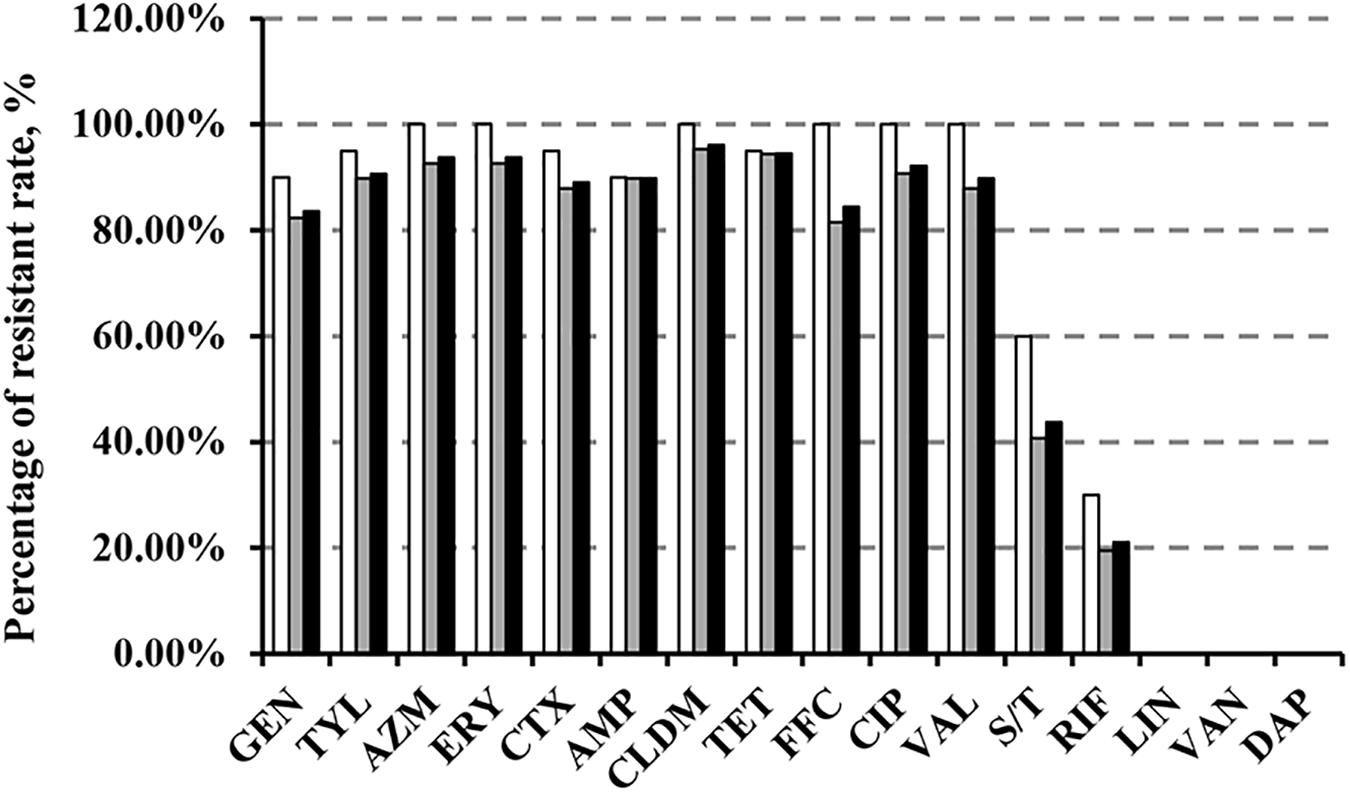
FIGURE 1. Antibiotic resistance in MRSA strains from animals and humans. GEN, gentamicin; TYL, tylosin; AZM, azithromycin; ERY, erythromycin; CTX, cefotaxime; AMP, ampicillin; CLDM, clindamycin; TET, tetracycline; FFC, florfenicol; CIP, ciprofloxacin; VAL, valnemulin; S/T, sulfamethoxazole/trimethoprim; RIF, rifampicin; LIN, linezolid; VAN, vancomycin; DAP, daptomycin.
In addition, 20 of the 128 MRSA strains (15.6%) harbored the cfr gene, and included 13 isolates from pigs (10.2%), three from chickens (2.3%), one from duck (0.8%), and three from humans (2.3%). Interestingly, all of the cfr-positive MRSA strains also carried the fexA, ermC, ereA, and aadA1 genes (Table 1). In addition, the prevalence of all the other tested antibiotic resistance genes (ARGs) was higher in the cfr-positive MRSA isolates than in cfr-negative MRSA isolates, especially for the optrA, ereB, aac (3′)-IIc, and aph (3′)-IV genes (P < 0.05; Figure 2).
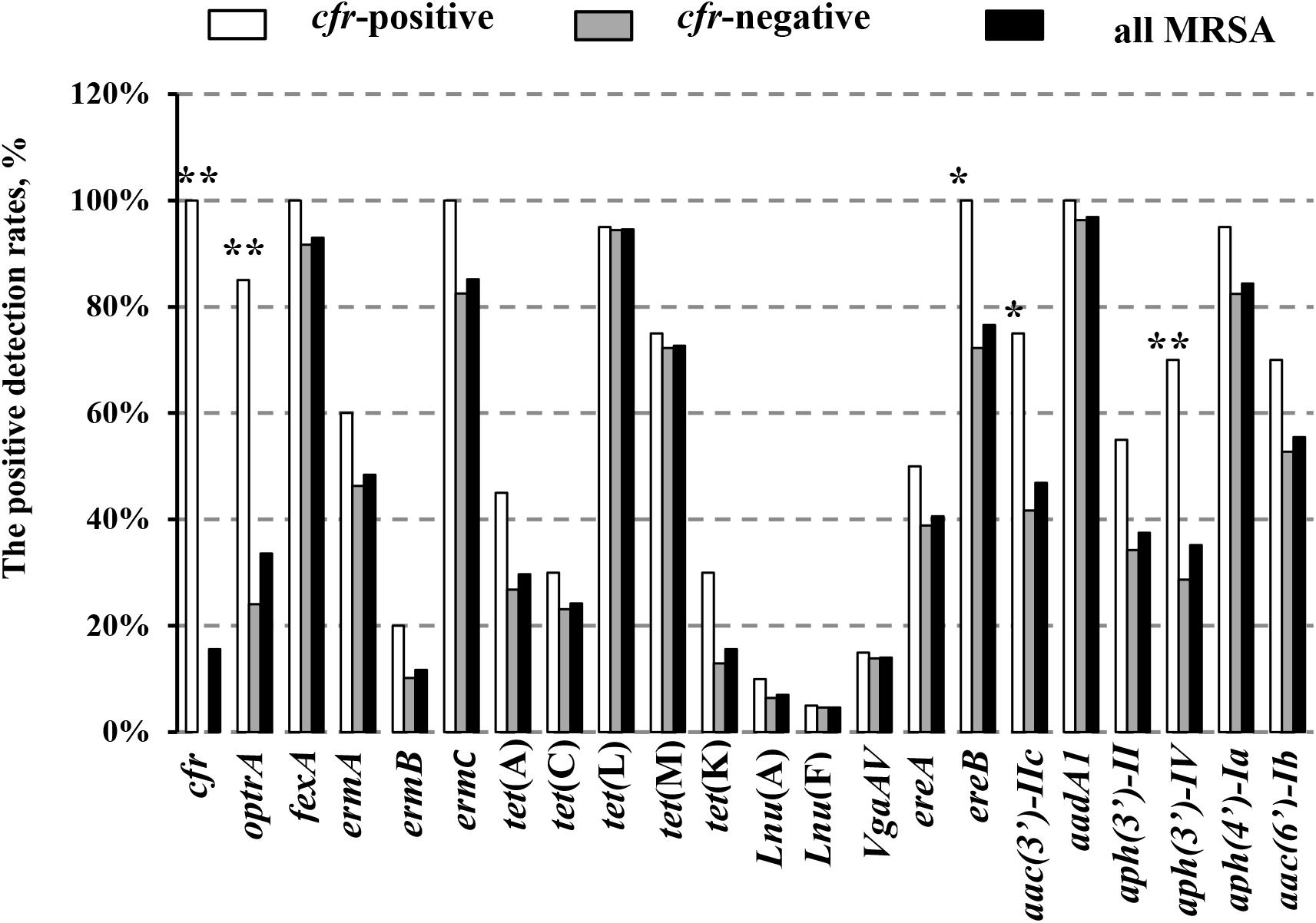
FIGURE 2. Positive detection rates of genes related to antibiotic resistance in the study MRSA strains. Detection rates between the cfr-positive and cfr-negative MRSA strains were determined using the χ2 test. ∗P < 0.05; ∗∗P < 0.01 cfr-positive MRSA strains vs. cfr-negative MRSA strains.
Molecular Typing
The 128 MRSA strains contained eight ST types and seven spa types, and ST9 (82.0%, 105/128) and spa type t899 (80.5%, 103/128) were predominated. In the cfr-positive MRSA isolates, three ST types and four spa types were observed, and ST9 (85%, 17/20) and spa type t899 (75%, 15/20) were also the most prevalent of these types. We observed 12 different profiles using a combination of MLST and spa typing in the 128 MRSA isolates. ST9-t899 (78.9%, 101/128) and ST764-t1084 (6.3%, 8/128) were the most and second most ST-spa types, respectively (Supplementary Table S4).
We also found 20 different PFGE profiles in the cfr-positive and 10 cfr-negative MRSA isolates (Figure 3). PFGE analysis suggested that MRSA isolates in the current study were epidemiologically unrelated clones.
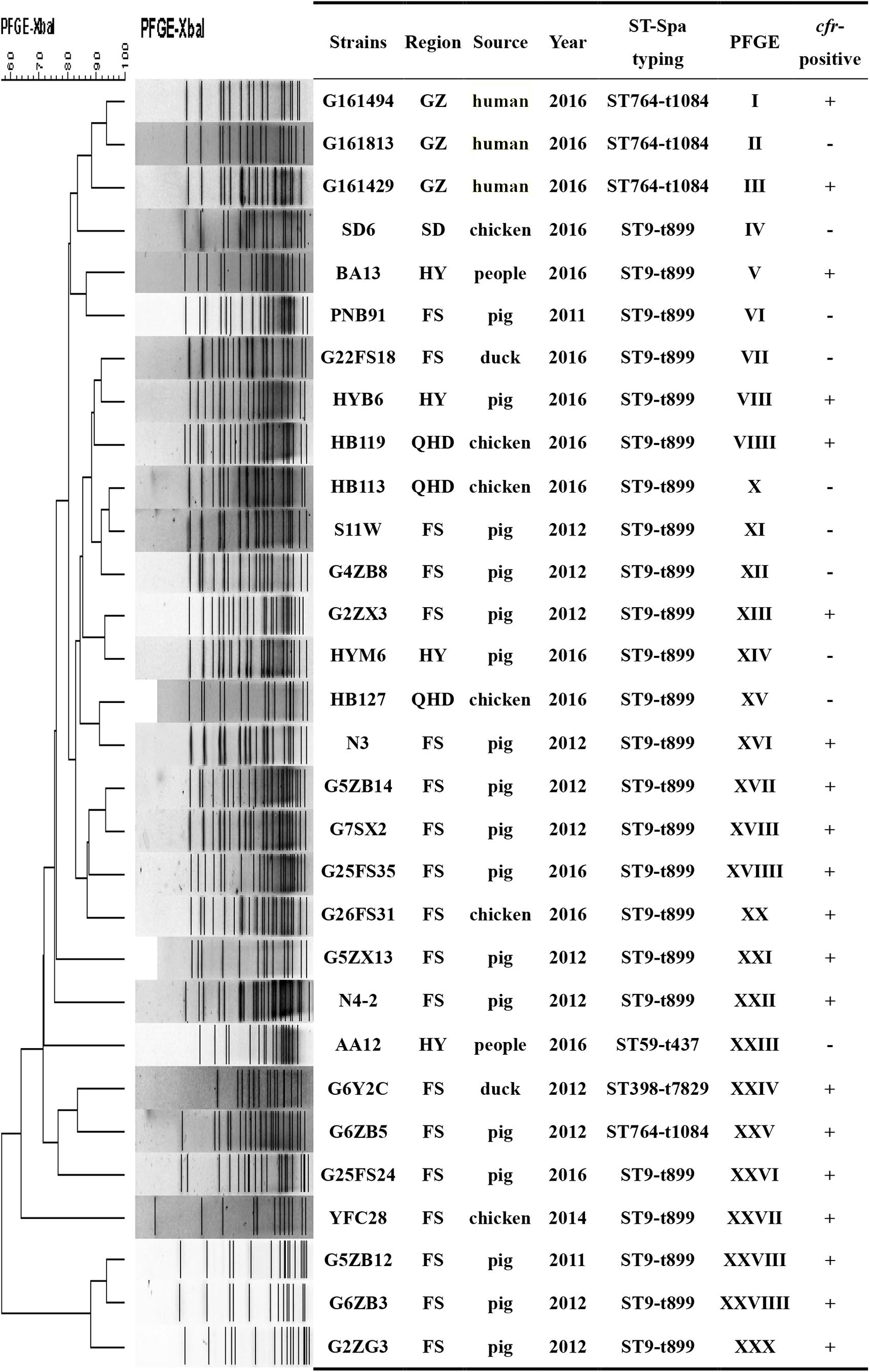
FIGURE 3. PFGE fingerprint patterns of SmaI-digested total DNA preparations from 20 MRSA strains harboring cfr and 10 cfr-negatives MRSA strains. A similarity cutoff of 100% was used to identify a PFGE cluster. Guangzhou (GZ), Qinhuangdao (QHD), Foshan (FS), Shandong (SD), Huadu (HD), and Heyuan (HY). “+”, cfr-positive; “–” <LIST>cfr-negative.
Transfer of cfr and Plasmids Analyses
The cfr gene from 13 of the 20 cfr-positive MRSA isolates were successfully transferred to a recipient strain (S. aureus RN4220) and showed 4- to 64-fold increases in the MICs of florfenicol as compared with the recipient strain lacking the cfr gene. In addition, cfr gene transfenerated strains were resistant to erythromycin, azithromycin, and clindamycin. Co-transfer of cfr with fexA and ermC genes were found in eight of 13 electrotransformants. S1-PFGE and Southern blot hybridizations revealed that the cfr genes were located on plasmids with sizes of 50 kb (n = 11) or 7.1 kb (n = 2) (Table 1).
Genetic Environment of cfr Gene
The genomic structure surrounding cfr in the 13 cfr-carrying electrotransformants showed two different genetic contexts. Type I was the most common structure observed in 11 of 13 among which the cfr gene was located on ∼ 50 kb plasmids. The 9,880 bp cfr-containing regions comprised a truncated tnpA (DeltatnpA), istA, istB, cfr, tnpB, tnpC, orf138, and fexA. This was a Tn558 variant with a 5′ deletion of tnpB by insertion of the IS21-558 element (istA-B) and cfr in the same orientation. Type II was similar to that in plasmid pHNCR35 (KF861983), pSS-02 (JX827253), pHK01 (KC820816), and pSA737 (KC206006). Type II was found in two electrotransformants among which the complete nucleotide sequences of 7,057-bp circular plasmids harboring cfr (p26FS31 and p25FS24) were obtained. A plasmid comparison based on a BLAST query revealed that p26FS31 and p25FS24 were identical to plasmid pSS-03 (JQ219851) and pHNLKJC2 (KF751701). These plasmids consisted of five open reading frames (ORF) (rep-Deltapre/mob-cfr-pre/mob-ermC) (Figure 4).
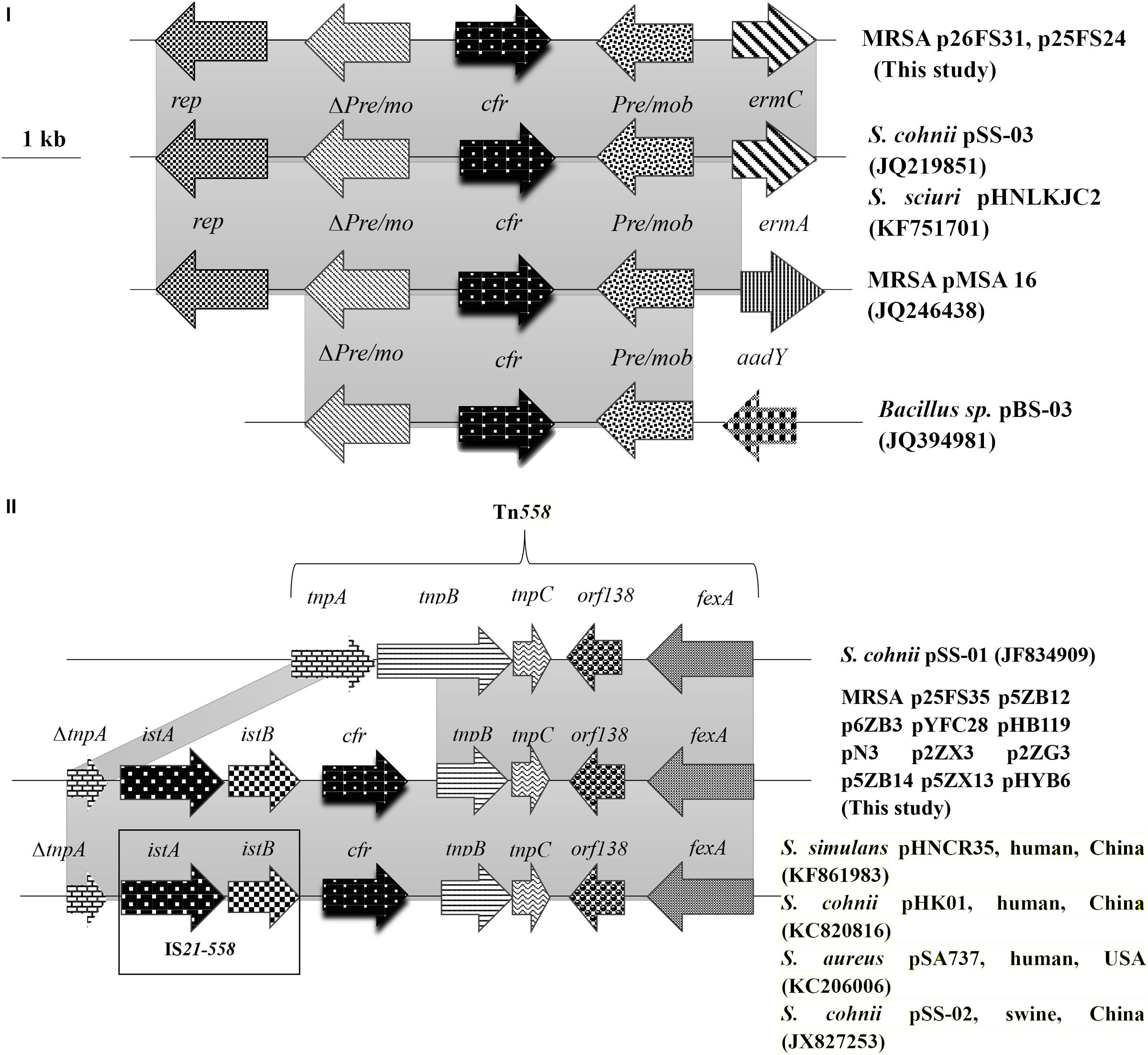
FIGURE 4. The genetic context surrounding the cfr gene in plasmids and their structural comparison with plasmids possessing have >98% similarity. The arrows indicate the positions and directions of the transcription of each gene. Gray shaded regions indicate homology >98%. “Delta” represents a truncated gene.
Discussion
In this study, we investigated the prevalence of cfr in 128 MRSA strains isolated from animals and humans in China. Our study showed a significantly higher positive rate of cfr in LA-MRSA strains from animals (15.17%) than that recently reported in domestic studies (1.11–3.46%) (Li et al., 2015; Li J. et al., 2017). In addition, cfr was also present in one MRSA isolate from domestic duck. To the best of our knowledge, this is the first report on the cfr gene in MRSA strains from waterfowl. This finding may implicate a recent and rapid dissemination process of cfr in MRSA strains from different food animals in China. Moreover, the prevalence of cfr in MRSA strains from humans (2.34%) was higher in the current study than that previously reported for clinical patients (0.30%) (Cai et al., 2015), but lower in isolates from a teaching hospital in a different region of China (9.38%) (Tian et al., 2014).
Most of the cfr-positive MRSA strains in the current study presented a multidrug-resistant phenotype and harbored diverse ARGs. These observations were similar to the high occurrence of multidrug resistance previously reported in cfr-positive MRSA isolates from swine farms and retail meat in China (Zeng et al., 2014; Li J. et al., 2017). In addition, the cfr gene has been reported to be associated with oxazolidinone resistance in several studies (Schwarz et al., 2000; Shen et al., 2013), but it only mediated low levels of resistance to this antibiotic class. In the current study, we found that the proportion of MRSA isolates with increased linezolid MICs in the cfr-positive MRSA strains was higher than in the cfr-negative MRSA strains. Interestingly, we also determined that the majority of cfr-positive MRSA isolates harbored optrA, which is in agreement with previous reports suggesting that optrA and cfr coexist (Li et al., 2016; Fan et al., 2017). In addition to optrA, our study cfr-positive MRSA isolates also co-carried fexA and ermC, which is also consistent with previous studies (Liu .X et al., 2017). This linked the cotransmission of fexA and ermA-C with cfr gene in diverse plasmids from coagulase-negative Staphylococci as well as Enterobacteriaceae of different origins (Wang et al., 2012a, 2013; Ye et al., 2015). Moreover, we observed different ratios of ermA, ermB, and ermC in our study strains that may be related to the location of the genes. For instance, ermA and ermB are primary chromosomal genes, while ermC gene is often plasmid-borne (Schwarz et al., 2011; Kadlec et al., 2012). The ermB was present in a minority of our strains, while ermA and ermC were frequent in MRSA strains (Lina et al., 1999; Liu H. et al., 2017). Furthermore, we found that the majority of the cfr genes were located on plasmids. Therefore, these factors may have influenced on the high ratio of ermC as we observed in the current studies.
Among all the study MRSA isolates, ST9 and t899 were the most prevalent ST and spa types, respectively. ST9 was reported as the predominant ST type in S. aureus isolates from animals in China (Cui et al., 2009), and sporadically occurred in Canada, England, Germany, and the United States (Mulders et al., 2010; Fessler et al., 2011; Dhup et al., 2015). In other and our current study, ST9 in S. aureus isolates were also found in farmers (Fessler et al., 2011; Dhup et al., 2015; Sun et al., 2015). Emergence of the cfr gene in the prevalent ST9 MRSA isolates from pigs and pig-handlers would probably extend the potential reservoirs and expand the risk to human health (Ye et al., 2015; Yan et al., 2016). Since the ST398 was first identified in pigs and pig farmers in 2005 (Armand-Lefevre et al., 2005), it has become the most prevalent MLST-type in LA-MRSA in the United States and Europe (Armand-Lefevre et al., 2005; Cuny et al., 2010; Antoci et al., 2013). More importantly, the ST398 LA-MRSA carrying the cfr gene has been detected in Korea and other countries (Kadlec et al., 2012; Moon et al., 2015). Despite the wide and rapid dissemination of cfr gene in S. aureus isolates in China, to date, cfr was only identified in ST398 MRSA isolates from pigs (Li W. et al., 2017).
In the current study, we also found a cfr-positive ST398 MRSA strain isolated from a duck indicating a possibility of widespread dissemination of the cfr-harboring ST398 LA-MRSA clone in China. In addition, all of the three cfr-positive MRSA isolates from patients were identified as ST764, the increased prevalent hybrid variant of the ST5 HA-MRSA lineage with the arginine catabolic mobile element (ACME) in China, Japan, and other Asian areas (Otsuka et al., 2012; Nakaminami et al., 2014; Wang et al., 2016). These results indicated that cfr-positive MRSA isolates from animals and humans belonged to different ST types and were probably from epidemiologically unrelated MRSA clones.
In the MRSA isolates from food animals, the cfr genes were primarily located on two types of transferable plasmids with sizes of ∼ 50 and ∼ 7.1 kb. Two different genetic contexts surrounding cfr were found, and each was associated with one type of cfr-carrying plasmid. The predominant genetic context of cfr was found to be a Tn558 variant in the large plasmids that co-carried fexA. This suggested that the acquisition of cfr could be involved in IS21-558 mediated recombination. Importantly, the Tn558 variant also occurred in Bacillus, S. sciuri, Staphylococcus simulans, and MRSA isolates from humans and swine (Wang et al., 2015; Li J. et al., 2017).
In addition, we also found the cfr gene on small plasmids that co-carried ermC in MRSA isolates from food animals.These small plasmids were also identified in Staphylococcus and Bacillus species isolates from pigs (Wang et al., 2012c). The high similarity of the genetic environment of cfr among diverse MRSA strains and sources indicated that horizontal transmission mediated by plasmids and transposons played a significant role in dissemination of cfr.
Conclusion
Our studies demonstrated higher antibiotic resistance rates in the cfr-positive vs. -negative MRSA isolates. Horizontal transmission mediated by plasmids and transposons likely played an important role in co-dissemination of cfr with fexA and ermC. The transmission of similar cfr-carrying transposons and plasmids from diverse bacteria species and origins requires continued investigation.
Ethics Statement
All procedure of strain isolation from animals was approved by the South China Agriculture University (SCAU) Animal Ethics Committee and conducted in strict accordance with technical guidelines for isolation and identification of animal-origin Staphylococcus aureus (DB51/T 2363-2017), as issued by the Quality and Technical Supervision Bureau of China, and in accordance with the SCAU Institutional Animal Care and Use Committee guidelines. The owner of farms from which animal-related samples were taken gave permission for their animals to be used in this study. All strains with human-origin were kindly provided by the Third Affiliated Hospital of Sun Yat-sen University and Guangdong Second Traditional Chinese Medicine Hospital, and the isolation procedure was in accordance with their Institutional Strain Isolation guidelines.
Author Contributions
Y-HL and Y-QX designed and organized the study. S-ML did the research. J-HD, F-RL, H-QL, Y-TW, and W-QG did the assisted help. LL, L-XF, X-PL, and JS analyzed the data. S-ML and Y-FZ wrote the paper.
Funding
This work was supported by the International Cooperation of Science and Technology Planning Project of Guangdong Province, China (2016A050502046) and the National Key Research and Development Program of China (2016YFD0501300).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to acknowledge the Third Affiliated Hospital of Sun Yat-sen University and Guangdong Second Traditional Chinese Medicine Hospital for providing clinical S. aureus isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02925/full#supplementary-material
Footnotes
- ^ http://saureus.mlst.net/
- ^ http://www.spaserver.ridom.de/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Antoci, E., Pinzone, M. R., Nunnari, G., Stefani, S., and Cacopardo, B. (2013). Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among subjects working on bovine dairy farms. Infez. Med. 21, 125–129.
Armand-Lefevre, L., Ruimy, R., and Andremont, A. (2005). Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11, 711–714. doi: 10.3201/eid1105.040866
Cai, J. C., Hu, Y. Y., Zhou, H. W., Chen, G. X., and Zhang, R. (2015). Dissemination of the same cfr-carrying plasmid among methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates in China. Antimicrob. Agents Chemother. 59, 3669–3671. doi: 10.1128/AAC.04580-14
Clinical and Laboratory Standards Institute [CLSI] (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests Forbacteria Isolated from Animals; Approved Standard. VET01-A4/VET01-S2. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Document M100-S28. Wayne, PA: CLSI.
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, Y., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Cuny, C., Friedrich, A., Kozytska, S., Layer, F., Nubel, U., Ohlsen, K., et al. (2010). Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300, 109–117. doi: 10.1016/j.ijmm.2009.11.002
de Neeling, A. J., Van Den Broek, M. J., Spalburg, E. C., Van Santen-Verheuvel, M. G., Dam-Deisz, W. D., Boshuizen, H. C., et al. (2007). High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122, 366–372. doi: 10.1016/j.vetmic.2007.01.027
Dhup, V., Kearns, A. M., Pichon, B., and Foster, H. A. (2015). First report of identification of livestock-associated MRSA ST9 in retail meat in England. Epidemiol. Infect. 143, 2989–2992. doi: 10.1017/S0950268815000126
Fan, R., Li, D., Fessler, A. T., Wu, C., Schwarz, S., and Wang, Y. (2017). Distribution of optrA and cfr in florfenicol-resistant Staphylococcus sciuri of pig origin. Vet. Microbiol. 210, 43–48. doi: 10.1016/j.vetmic.2017.07.030
Fessler, A. T., Kadlec, K., Hassel, M., Hauschild, T., Eidam, C., Ehricht, R., et al. (2011). Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 77, 7151–7157. doi: 10.1128/AEM.00561-11
Inkster, T., Coia, J., Meunier, D., Doumith, M., Martin, K., Pike, R., et al. (2017). First outbreak of colonization by linezolid- and glycopeptide-resistant Enterococcus faecium harbouring the cfr gene in a UK nephrology unit. J. Hosp. Infect. 97, 397–402. doi: 10.1016/j.jhin.2017.07.003
Kadlec, K., Fessler, A. T., Hauschild, T., and Schwarz, S. (2012). Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 745–755. doi: 10.1111/j.1469-0691.2012.03842.x
Kehrenberg, C., Cuny, C., Strommenger, B., Schwarz, S., and Witte, W. (2009). Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53, 779–781. doi: 10.1128/AAC.01376-08
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Li, D., Wang, Y., Schwarz, S., Cai, J., Fan, R., Li, J., et al. (2016). Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 71, 1474–1478. doi: 10.1093/jac/dkw040
Li, D., Wu, C., Wang, Y., Fan, R., Schwarz, S., and Zhang, S. (2015). Identification of multiresistance gene cfr in methicillin-resistant Staphylococcus aureus from pigs: plasmid location and integration into a Staphylococcal cassette chromosome mec complex. Antimicrob. Agents Chemother. 59, 3641–3644. doi: 10.1128/AAC.00500-15
Li, J., Jiang, N., Ke, Y., Fessler, A. T., Wang, Y., Schwarz, S., et al. (2017). Characterization of pig-associated methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 201, 183–187. doi: 10.1016/j.vetmic.2017.01.017
Li, W., Liu, J. H., Zhang, X. F., Wang, J., Ma, Z. B., Chen, L., et al. (2017). Emergence of methicillin-resistant Staphylococcus aureus ST398 in pigs in China. Int. J. Antimicrob. Agents 51, 275–276. doi: 10.1016/j.ijantimicag.2017.10.013
Lina, G., Quaglia, A., Reverdy, M. E., Leclercq, R., Vandenesch, F., and Etienne, J. (1999). Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among Staphylococci. Antimicrob. Agents Chemother. 43, 1062–1066. doi: 10.1128/AAC.43.5.1062
Liu, H., Li, S., Meng, L., Dong, L., Zhao, S., Lan, X., et al. (2017). Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J. Dairy Sci. 100, 8796–8803. doi: 10.3168/jds.2017-13370
Liu, X., Deng, S., Huang, J., Huang, Y., Zhang, Y., Yan, Q., et al. (2017). Dissemination of macrolides, fusidic acid and mupirocin resistance among Staphylococcus aureus clinical isolates. Oncotarget 8, 58086–58097. doi: 10.18632/oncotarget.19491
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Marshall, C., and McBryde, E. (2014). The role of Staphylococcus aureus carriage in the pathogenesis of bloodstream infection. BMC Res. Notes 7:428. doi: 10.1186/1756-0500-7-428
Moon, D. C., Tamang, M. D., Nam, H. M., Jeong, J. H., Jang, G. C., Jung, S. C., et al. (2015). Identification of livestock-associated methicillin-resistant Staphylococcus aureus isolates in Korea and molecular comparison between isolates from animal carcasses and slaughterhouse workers. Foodborne Pathog. Dis. 12, 327–334. doi: 10.1089/fpd.2014.1868
Morales, G., Picazo, J. J., Baos, E., Candel, F. J., Arribi, A., Pelaez, B., et al. (2010). Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50, 821–825. doi: 10.1086/650574
Mulders, M. N., Haenen, A. P., Geenen, P. L., Vesseur, P. C., Poldervaart, E. S., Bosch, T., et al. (2010). Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in the Netherlands. Epidemiol. Infect. 138, 743–755. doi: 10.1017/S0950268810000075
Nakaminami, H., Noguchi, N., Ito, A., Ikeda, M., Utsumi, K., Maruyama, H., et al. (2014). Characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals in Tokyo, Japan. J. Infect. Chemother. 20, 512–515. doi: 10.1016/j.jiac.2014.03.006
Otsuka, T., Zaraket, H., Fujii, K., Masuda, Y., Komiyama, K., Ishikawa, Y., et al. (2012). Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from children in a community with low antimicrobial pressure in Japan. Jpn. J. Infect. Dis. 65, 483–488. doi: 10.7883/yoken.65.483
Rodvold, K. A., and McConeghy, K. W. (2014). Methicillin-resistant Staphylococcus aureus therapy: past, present, and future. Clin. Infect. Dis. 58(Suppl. 1), S20–S27. doi: 10.1093/cid/cit614
Schwarz, S., Fessler, A. T., Hauschild, T., Kehrenberg, C., and Kadlec, K. (2011). Plasmid-mediated resistance to protein biosynthesis inhibitors in Staphylococci. Ann. N. Y. Acad. Sci. 1241, 82–103. doi: 10.1111/j.1749-6632.2011.06275.x
Schwarz, S., Werckenthin, C., and Kehrenberg, C. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000
Shen, J., Wang, Y., and Schwarz, S. (2013). Presence and dissemination of the multiresistance gene cfr in gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092
Sun, J., Yang, M., Sreevatsan, S., and Davies, P. R. (2015). Prevalence and characterization of Staphylococcus aureus in growing pigs in the USA. PLoS One 10:e0143670. doi: 10.1371/journal.pone.0143670
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tian, Y., Li, T., Zhu, Y., Wang, B., Zou, X., and Li, M. (2014). Mechanisms of linezolid resistance in staphylococci and enterococci isolated from two teaching hospitals in Shanghai, China. BMC Microbiol. 14:292. doi: 10.1186/s12866-014-0292-5
van de Giessen, A. W., Van Santen-Verheuvel, M. G., Hengeveld, P. D., Bosch, T., Broens, E. M., and Reusken, C. B. (2009). Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev. Vet. Med. 91, 270–273. doi: 10.1016/j.prevetmed.2009.05.016
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966. doi: 10.3201/eid1112.050428
Wang, J., Lin, D. C., Guo, X. M., Wei, H. K., Liu, X. Q., Chen, X. J., et al. (2015). Distribution of the multidrug resistance gene cfr in staphylococcus isolates from pigs, workers, and the environment of a hog market and a slaughterhouse in Guangzhou, China. Foodborne Pathog. Dis. 12, 598–605. doi: 10.1089/fpd.2014.1891
Wang, R., Wan, H. Y., Shi, G. C., Li, M., Han, L. Z., Jin, X. Y., et al. (2016). Gene typing and antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from lower respiratory tract at two hospitals in Shanghai. Zhonghua Jie He He Hu Xi Za Zhi 39, 286–290. doi: 10.3760/cma.j.issn.1001-0939.2016.04.007
Wang, Y., He, T., Schwarz, S., Zhao, Q., Shen, Z., Wu, C., et al. (2013). Multidrug resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int. J. Med. Microbiol. 303, 84–87. doi: 10.1016/j.ijmm.2012.12.004
Wang, Y., He, T., Schwarz, S., Zhou, D., Shen, Z., Wu, C., et al. (2012a). Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 67, 1094–1098. doi: 10.1093/jac/dks020
Wang, Y., Wang, Y., Schwarz, S., Shen, Z., Zhou, N., Lin, J., et al. (2012b). Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J. Antimicrob. Chemother. 67, 1824–1827. doi: 10.1093/jac/dks163
Wang, Y., Zhang, W., Wang, J., Wu, C., Shen, Z., Fu, X., et al. (2012c). Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56, 1485–1490. doi: 10.1128/AAC.05827-11
Wang, Y., Wang, Y., Wu, C. M., Schwarz, S., Shen, Z., Zhang, W., et al. (2011). Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66, 2521–2526. doi: 10.1093/jac/dkr322
Woodford, N., and Livermore, D. M. (2009). Infections caused by gram-positive bacteria: a review of the global challenge. J. Infect. 59(Suppl. 1), S4–S16. doi: 10.1016/S0163-4453(09)60003-7
Wulf, M., Van Nes, A., Eikelenboom-Boskamp, A., De Vries, J., Melchers, W., Klaassen, C., et al. (2006). Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, the Netherlands. Emerg. Infect. Dis. 12, 1939–1941. doi: 10.3201/eid1212.060355
Yan, X., Li, Z., Chlebowicz, M. A., Tao, X., Ni, M., Hu, Y., et al. (2016). Genetic features of livestock-associated Staphylococcus aureus ST9 isolates from Chinese pigs that carry the lsa(E) gene for quinupristin/dalfopristin resistance. Int. J. Med. Microbiol. 306, 722–729. doi: 10.1016/j.ijmm.2016.08.001
Ye, X., Liu, W., Fan, Y., Wang, X., Zhou, J., Yao, Z., et al. (2015). Frequency-risk and duration-risk relations between occupational livestock contact and methicillin-resistant Staphylococcus aureus carriage among workers in Guangdong, China. Am. J. Infect. Control 43, 676–681. doi: 10.1016/j.ajic.2015.03.026
Keywords: cfr, MRSA, multi-drug resistance, plasmid, food animals
Citation: Li S-M, Zhou Y-F, Li L, Fang L-X, Duan J-H, Liu F-R, Liang H-Q, Wu Y-T, Gu W-Q, Liao X-P, Sun J, Xiong Y-Q and Liu Y-H (2018) Characterization of the Multi-Drug Resistance Gene cfr in Methicillin-Resistant Staphylococcus aureus (MRSA) Strains Isolated From Animals and Humans in China. Front. Microbiol. 9:2925. doi: 10.3389/fmicb.2018.02925
Received: 26 February 2018; Accepted: 14 November 2018;
Published: 27 November 2018.
Edited by:
Patrick Rik Butaye, Ross University School of Veterinary Medicine, Saint Kitts and NevisReviewed by:
Xu Jia, Chengde Medical College, ChinaKristina Kadlec, Friedrich Loeffler Institut, Germany
Copyright © 2018 Li, Zhou, Li, Fang, Duan, Liu, Liang, Wu, Gu, Liao, Sun, Xiong and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Qiong Xiong, eXhpb25nQHVjbGEuZWR1 Ya-Hong Liu, bHloQHNjYXUuZWR1LmNu
 Shu-Min Li1
Shu-Min Li1 Yu-Feng Zhou
Yu-Feng Zhou Liang Li
Liang Li Xiao-Ping Liao
Xiao-Ping Liao Jian Sun
Jian Sun Yan-Qiong Xiong
Yan-Qiong Xiong Ya-Hong Liu
Ya-Hong Liu