
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02754
 Ting Huang1,2,3,4
Ting Huang1,2,3,4 Biao Gao1,2,3,4
Biao Gao1,2,3,4 Wen-Lu Chen1,2,3,4
Wen-Lu Chen1,2,3,4 Rong Xiang1,2,3,4
Rong Xiang1,2,3,4 Ming-Gui Yuan1,2,3,4
Ming-Gui Yuan1,2,3,4 Zhi-Hong Xu1,2,3,4
Zhi-Hong Xu1,2,3,4 Xin-Yu Peng1,2,3,4*
Xin-Yu Peng1,2,3,4*Necrotic enteritis (NE) caused by Clostridium perfringens is responsible for huge financial losses in the poultry industry annually. A diet highly supplemented with fishmeal is one factor predisposing chickens to the development of clinical NE. However, the effects of fishmeal-rich diets on the gut microbiota and immune response in chickens with C. perfringens challenge over the long-term are not well-understood. Here, a chicken NE model was established in which chickens were fed high fishmeal diet and subsequently infected with C. perfringens (FM/CP). Two control groups of chickens, one that was not infected and had a high fishmeal feeding (FM) and another group only infected with C. perfringens with basic diets (CP), were used as comparators. We analyzed the gut microbiota and immune response of the three groups at the age of 20, 24 [1 day post-infection (dpi)] and 30 days (7 dpi) using 16S rDNA sequencing and real-time PCR, respectively. We found that the composition of the gut microbiota had significant shifted in both the CP and FM/CP groups, although the CP group did not have intestinal lesions. The structure of the gut microbiota in C. perfringens-challenged chickens, independent of a high fishmeal diet, had the tendency to return to their non-infection state if the chickens no longer received C. perfringens challenge. Gut microbiota variation with time in challenged chickens with high fishmeal diet feeding was superimposed upon that of non-infected chickens with high fishmeal feeding. For the immune response, the relative expression of IL-8 in the ileum was significantly higher in infected chickens independent of high fishmeal feeding than in non-infected chickens. However, the expression of alpha 1-acid glycoprotein (AGP) and serum amyloid A (SAA) genes in chicken liver were significantly increased in FM/CP compared to the other groups. In conclusion, high fishmeal feeding induced significant changes to the structure of chicken gut microbiota over time and such changes provided an opening for C. perfringens infection to progress to NE. The relative expression of AGP and SAA in liver tissue may be used as diagnostic biomarkers for poultry NE but such an indication requires further investigation.
Necrotic enteritis (NE) is intestinal disease of poultry caused by Clostridium perfringens (C. perfringens) common in domestic poultry-producing countries and poses a risk to animal health. This disease has been documented to cause a loss of more than US $2 billion per year in the international poultry industry, with an average loss of $0.05 per chicken (Kaldhusdal et al., 1999; Van der Sluis, 2000). Recently, the true cost of NE to the producer has been elevated to US $6 billion in 2015, with a cost of more than $0.062 per chicken (Wade Ben, 2015).
Clostridium perfringens infection alone is not enough to induce NE. Predisposing factors are necessary to produce special conditions to induce disease. Usually, the third to fourth week of a chicken’s life provide the most susceptible conditions for the development of NE. Using high-protein diets containing fishmeal are one predisposing factor that have been extensively studied in experimental NE models (Wu et al., 2010; Fernandes da Costa et al., 2013). Now we know that high fishmeal diets have an abundancy of available nutrients which can promote C. perfringens proliferation (Drew et al., 2004). It has been also reported that fishmeal feeding alone as well as the combination of fishmeal feeding and C. perfringens infection could induce a significant shift in chicken gut microbiota (Stanley et al., 2014; Wu et al., 2014). However, it is not clear how such a high-protein diet promotes NE.
The expression of several different genes has been found to be associated with C. perfringens infection. Two receptors of intestinal epithelial cells, nucleotide-binding oligomerization domain (NOD) and Toll-like receptor (TLR) 2, were shown to be activated by peptidoglycan (PGN) (Wu et al., 2007; Lin et al., 2011). Moreover, PGN is the main cell wall component of C. perfringens which may activate these two receptors. However, the recognition of C. perfringens and the following immune responses are not well-understood. In addition, acute phase protein (APP) expression could be affected by microbial infection. Five APP have been found to have different expression in diseased chickens, alpha 1-acid glycoprotein (AGP), serum amyloid A (SAA), PIT54, ovotrasferrin (OVT), and C-reactive protein (CRP) (O’Reilly and Eckersall, 2014). It has been reported that different bacterial or viral infections could elevate corresponding APPs levels. For example, OVT was overexpressed in chickens infected with Escherichia coli (Rath et al., 2009). Therefore, the APPs may provide a target for poultry infection disease diagnostics.
The aim of the present study was to compare changes to the microbiota in chickens exposed to high fishmeal diets as well as C. perfringens challenge over time. Combination of these two factors were used to gain a better understanding of how high fishmeal affects the microbiome and the immune response in chickens with C. perfringens infection.
One-day old Ross 308 commercial chickens (negative for C. perfringens) were purchased from a hatchery in Guangzhou City, Guangdong Province, China. They were kept in pens for 14 days and then were randomly assigned on the basis of weight to one of three experimental groups. Food and water were available ad libitum and the chickens were maintained with 12 h light/dark cycles. Animal trials were conducted at the poultry house located at the Institute of Animal Health, Guangdong Academy of Agricultural Sciences. Three groups (60 chickens per group) were designed: (1) a chicken NE model in which chickens were fed high fishmeal diet and subsequently infected with C. perfringens (FM/CP), (2) chickens without additional infection subject to a high fishmeal feeding (FM), and (3) chickens infected with C. perfringens under basic diet (CP). From day of age 1 to 14, all groups were fed an antibiotic-free chicken basic diet (Supplementary Table S1). From day 15 to 30, group FM and FM/CP were switched to a high-protein diet (Supplementary Table S1) and group CP was kept on the same diet as before. Group FM/CP and CP were orally gavaged in the corresponding diets twice per day with actively growing concentration of 7–9 × 108 cfu/ml C. perfringens cpnetBF (Stanley et al., 2012) on days 21–23. The strain C. perfringens cpnetBF carries the netb gene isolated from intestines of broiler chickens with necrotic enteritis in Guangdong Province, China. The strain was stored at -80°C in fluid thioglycollate broth (FT, Beckson, Dickinson and Company) supplemented with 30% glycerol. None of the chickens died during the experiment. This animal experimental design is shown in Figure 1.
Seven chickens from each group were euthanized by cervical dislocation for sampling at day of age 20, 24, and 30. Jejunum–ileum content samples were collected for subsequent DNA extraction. Ileal and liver tissues were collected for subsequent RNA isolation. Ileal tissues were collected from approximately 3 cm distal to Meckel’s diverticulum. The collected tissue were immediately frozen in liquid nitrogen.
All the experimental procedures involving chickens were approved by the guidelines and regulations of Guangdong Academy of Agricultural Sciences [No. SYXK (yue)-20171211].
The lesion scores of the small intestine (duodenum to ileum) were observed from 10 replications for three different groups as previous study (Keyburn et al., 2006): (0) no gross lesions; (1) thin or friable walls; (2) focal necrosis or ulceration (one to five foci); (3) focal necrosis or ulceration (6 to 15 foci); (4) focal necrosis or ulceration (16 or more foci); (5) patches of necrosis 2–3 cm long; (6) diffused necrosis typical of field cases.
DNA from chickens’ jejunum–ileum content contents was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. Total DNA was quantified using a NanoDrop® ND-2000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). The instrument measures absorbance at 260 nm (A260) to quantify DNA in samples, at 280 nm (A280) to verify protein contamination and at 230 nm (A230) for determining contamination by phenol. Only DNA samples with A260/A280 ratio as 1.7 and A260/A230 > 1.8 were used for further analysis (Li et al., 2014). The extracts were stored at -20°C until use (Singh et al., 2015).
Quantitative PCR reaction was performed with intestinal content DNA temple in triplicate using SYBR Premix Ex Taq (TAKARA Bio, Otsu, Japan). The C. perfringens genes values were normalize to the 16S rRNA gene. The relative abundance of C. perfringens in the intestine were calculated based on the value of the 16S rRNA gene using 2-ΔCt method. The primers used for qPCR of C. perfringens and 16S rRNA are showed in Supplementary Table S2.
The V3+V4 hypervariable regions of 16S rDNA were PCR amplified from microbial DNA harvested from seven replications for three different groups at days 20, 24, 30 (Abrahamsson et al., 2014; Jakobsson et al., 2014). The gene-specific sequences for the 16S V3 and V4 region were performed by using primers 341F 5′-CCTACGGGNGGCWGCAG-3′ and 805R 5′-GACTACHVGGGTATCTAATCC-3′ (Abrahamsson et al., 2014; Jakobsson et al., 2014). The PCR conditions were as follows: one pre-denaturation cycle at 94°C for 4 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 45 s, and elongation at 72°C for 30 s, and one post-elongation cycle at 72°C for 5 min. The PCR amplicons were separated on 0.8% agarose gels and then extracted using QIAEX II gel extraction kit (Qiagen) according to the handbook. Only PCR products without primer dimers and contaminant bands were used for sequencing. Amplicons were purified using AMPure X using the manufacturer’s instructions (Beckman Coulter, Mississauga, ON, Canada). Bar-coded V3 and V4 amplicons were sequenced using the 2 × 300 paired-end method by Illumina MiSeq with a seven-cycle index read. Sequences processing was performed using QIIME (version 1.6.0) to get clean data. Sequences with an average Phred (Q) score lower than 30, with ambiguous bases or homopolymer runs exceeding 6 bp, primer mismatches or sequence lengths shorter than 100 bp were removed. The consensus sequence was generated by FLASH (Fast length Adjustment of Short reads, v1.2.11) as following: only sequences with an overlap longer than 10 bp and without any mismatches were assembled according to their overlap sequences. Reads that could not be assembled were discarded. Barcode and sequencing primers were trimmed from the assembled sequence. The high quality paired-end reads were combined to tags based on overlap. The tags were clustered to operational taxonomic unit (OTU) by software USEARCH (v7.0.1090). OTU representative sequences were taxonomically classified using Ribosomal Database Project (RDP) Classifier v2.2 trained on the Greengenes database.
Using the EZNA® Total RNA Isolation Kit (Omega Bio-Tek), RNA was isolate from ileal and liver tissue according to the manufacturer’s protocol. RNA was eluted in DEPC-treated water and stored at -80°C. Total RNA was quantified using a NanoDrop® ND-2000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States).
Reverse transcription was performed with M-MLV Frist Strand cDNA Synthesis Kit (Omega Bio-Tek), following the manufacturer’s protocol. Quantitative PCR reaction was performed with cDNA temple in triplicate using SYBR Premix Ex Taq (TAKARA Bio, Otsu, Japan). The target genes values were normalize to the housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The relative mRNA level of each target gene were calculated based on the expression of the GAPDH using 2-ΔCt method. The primers used for qPCR of interleukin-8 (IL-8), tumor necrosis factor α (TNF-α), Toll-like receptor 2 (TLR2), nucleotide-binding oligomerization domain 1 (NOD1), alpha1-acid glycoprotein (AGP), serum amyloid A (SAA), C- reactive protein (CRP), PIT54, ovotransferrin (OVT) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are showed in Supplementary Table S2.
The relative expression of mRNA, the relative abundance of C. perfringens and the alpha diversity indices of Shannon and abundance-based coverage (ACE) among the three groups were tested using single-factor analysis of molecular variance ANOVA with a p-value of < 0.05 considered to represent significance. Genus and species abundance were compared using Kruskal–Wallis test with Benjamini–Hochberg p-value correction.
No intestinal lesions were observed in unchallenged chickens (Figure 2). In the challenged groups, only the group FM/CP had obvious lesions at 1 day post-infection (dpi). At 7 dpi, all the challenged chickens showed no intestinal lesions.
All chickens were negative for C. perfringens until experimental challenging and group FM remained detection negative for C. perfringens throughout the study (Figure 3). The relative abundance of C. perfringens in chickens with FM/CP was significantly higher than group CP at 1 dpi (p < 0.05). However, at 7 dpi, there was no significant difference between group FM/CP and CP.
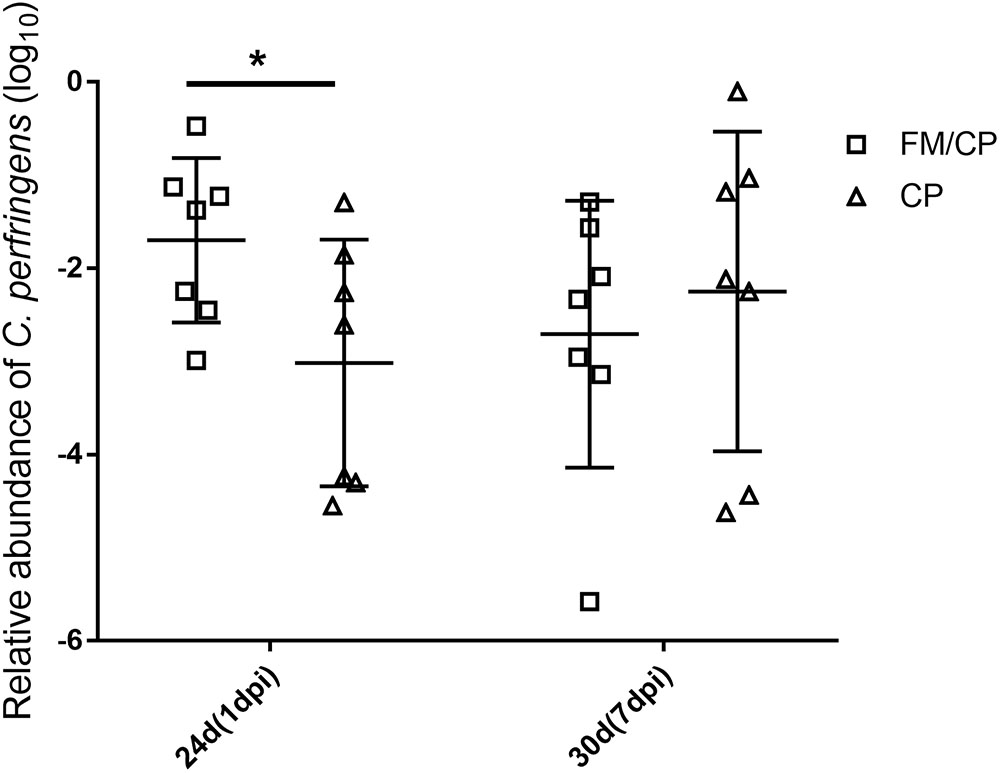
FIGURE 3. Viable relative abundance of Clostridium perfringens in the jejunum–ileum lumen. FM/CP and CP groups at day 20 and group FM remained negative for C. perfringens throughout the study. No significant differences between group FM/CP and CP at day 30 were observed (∗p > 0.05; ANOVA).
In order to investigate the effect of high fishmeal diet on the intestine and liver immune function in C. perfringens challenged chickens, the relative expression genes of cytokines, chemokine, Toll-like and NOD receptor and APPs was evaluated using the qPCR method (Figures 4, 5). At -1 dpi, the relative expression of all the detected immune genes had no significant difference among the three groups in ileum and liver tissue except the relative expression of OVT in liver and IL-8 in ileum. Moreover, the relative expression of TNF-α, AGP, CRP, and OVT in ileum tissue as well as PIT54 and CRP in liver tissue were not significantly different among the three groups at each time point. At 1 dpi, the relative expression of IL-8 in the ileum was significantly higher in infected chickens independent of high fishmeal diet as compared to non-infected chickens (p < 0.05). In addition, the relative expression of TNF-α, NOD1, AGP, SAA, CRP, and OVT had similar results as IL-8 expression among the three groups in ileum at same time points, without significant difference (p > 0.05). For liver tissue, the relative expression of AGP and SAA genes were significantly higher in group FM/CP compared to those in group FM and CP at 1 dpi. At 7 dpi, no significant changes were observed among three groups in liver and ileum tissue except IL-8, NOD1, and SAA in the ileum.
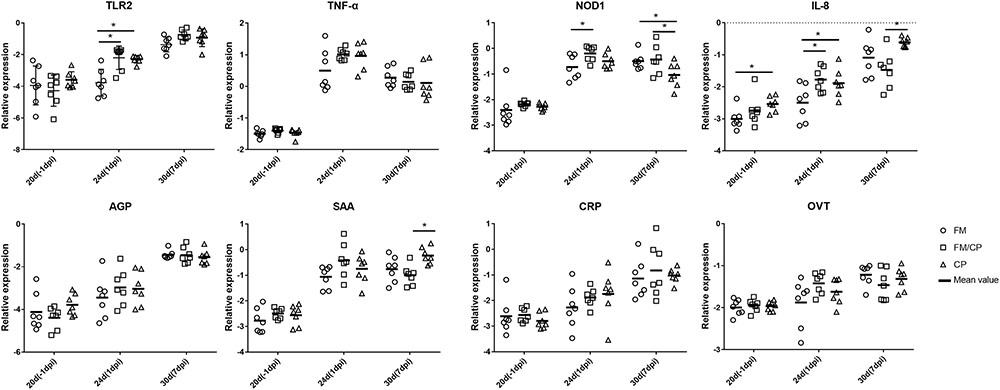
FIGURE 4. Relative change in expression of immune genes in ileum tissue. Relative gene expression represented as log10. Significant difference among three groups are indicated by an asterisk (ANOVA, ∗p < 0.05) for the expression of each genes at the corresponding time point. The Ct values of PIT54 in three groups was below the detection level. IL, interleukin; TNF-α, tumor necrosis factor α; TLR, toll-like receptor; NOD, nucleotide-binding oligomerization domain; AGP, alpha 1- acid glycoprotein; SAA, serum amyloid A; CRP, C- reactive protein; OVT, ovotransferrin; dpi, day post-infection.
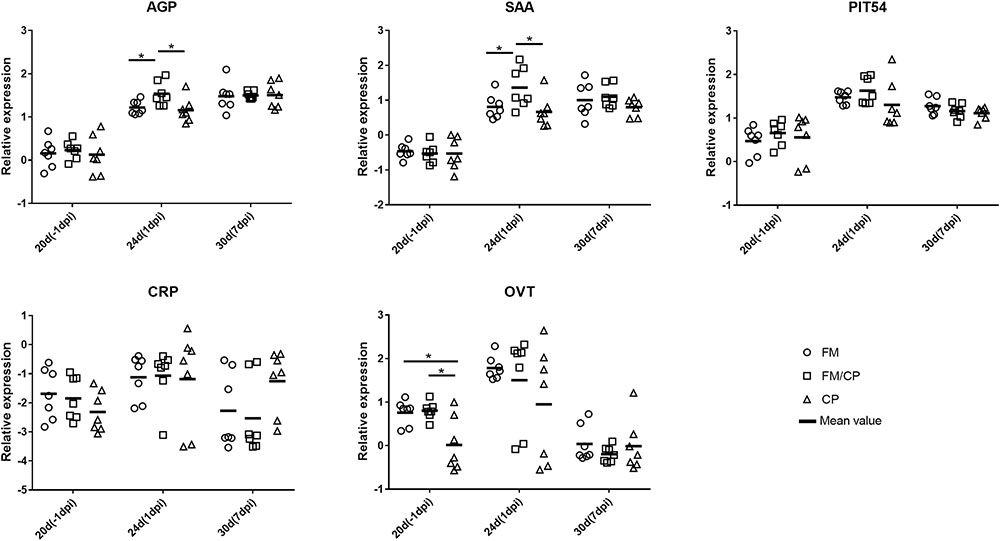
FIGURE 5. Relative change in expression of immune genes in liver tissue. Relative gene expression represented as log10. Significant differences among the three groups are indicated by an asterisk (ANOVA, ∗p < 0.05) for the expression of each gene at the corresponding time point. AGP, alpha 1- acid glycoprotein; SAA, serum amyloid A; CRP, C- reactive protein; OVT, ovotransferrin; dpi, day post-infection.
Shannon and ACE indices were used to measure the alpha diversity (evenness and richness) of the gut microbiota. The FM group did not have significant changes in alpha diversity of microbiota in chickens from day of age 20 to 30 (p > 0.05, Figure 6). After challenging, the ACE indices in the group FM/CP were significantly lower at 1 dpi compared to that at -1 and 7 dpi. The FM/CP group had significantly lower Shannon indices at the challenge stage than at the stage baseline while the group CP showed the opposite.
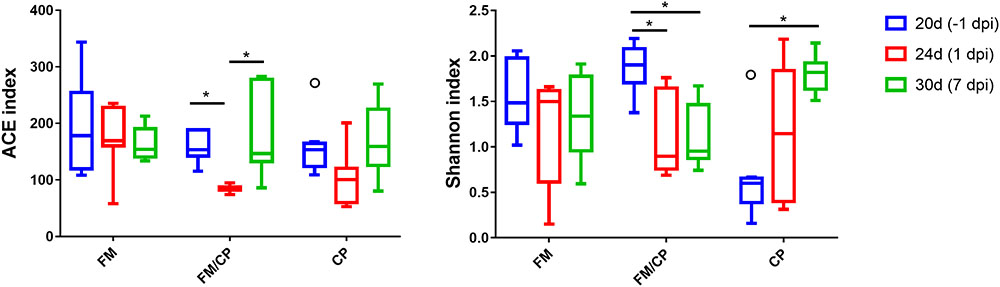
FIGURE 6. Estimates of alpha diversity for the gut microbiota from intestine content. ∗p < 0.05, ANOVA test.
Principal coordinate analysis (PCoA) of Bray–Curtis indices demonstrated the difference in bacteria composition among the three experimental groups at age days 20, 24, and 30 (-1, 1, and 7 dpi, Figure 7). Before challenging, the structure of gut microbiota in group FM and FM/CP at -1 dpi was significantly different from stage-matched chickens in group CP. After challenging, the gut bacteria composition of group FM/CP and CP at 1 and 7 dpi were significantly different from corresponding group at -1 dpi. At 1 dpi, the PCoA plot showed clustering the gut bacteria with respect to three different groups. At 7 dpi, the gut bacterial composition of chickens in the group FM/CP and CP both had the tendency to return to an unchallenged stage.
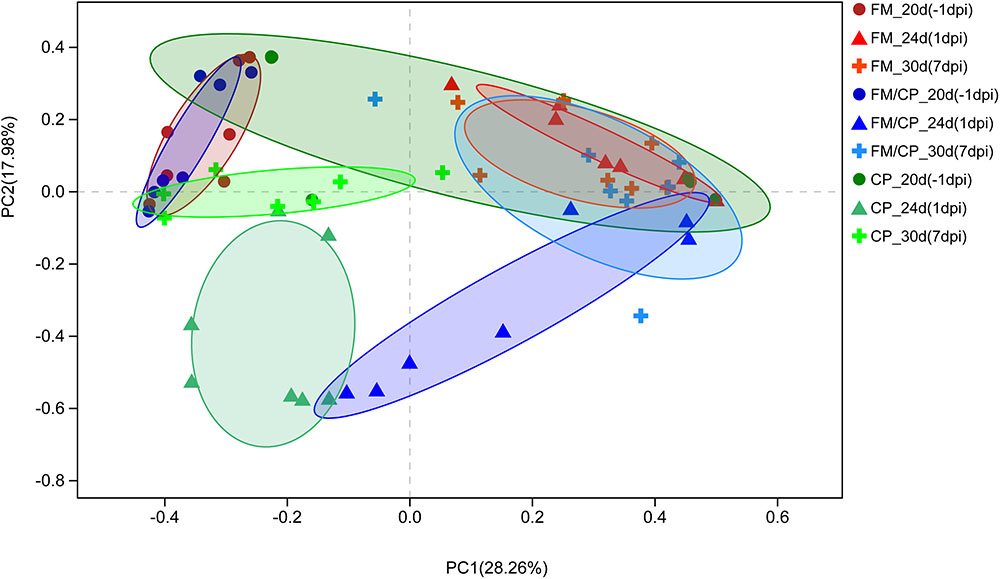
FIGURE 7. Principal coordinate analysis (PCoA) plot of Bray–Curtis indices for the gut microbiota of three different groups. The different degree red, blue, and green indicated group FM, FM/CP, and CP respectively. Circle: day 20 (–1 dpi); Triangle: day 24 (1 dpi); Plus sign: day 30 (7 dpi).
The results of comparison of microbiota in each group showed that the relative abundance of genus Candidatus Arthromitus in the group FM and FM/CP was significantly lower at day 30 than that at day 20 (p < 0.05, Figure 8A), but group CP had no significant change. The unclassified_p_Firmicutes bacterial strain found in group FM/CP and FM was significantly higher at day 24 than that at day 20, moreover, this genus in the group FM/CP was significantly higher compared to that in the group FM at day 24. The relative abundance of Enterococcus in the group FM/CP and CP at 1 dpi was significantly lower than that at -1 dpi whereas the genus Clostridium sensu stricto 1 showed the opposite result (p < 0.05).
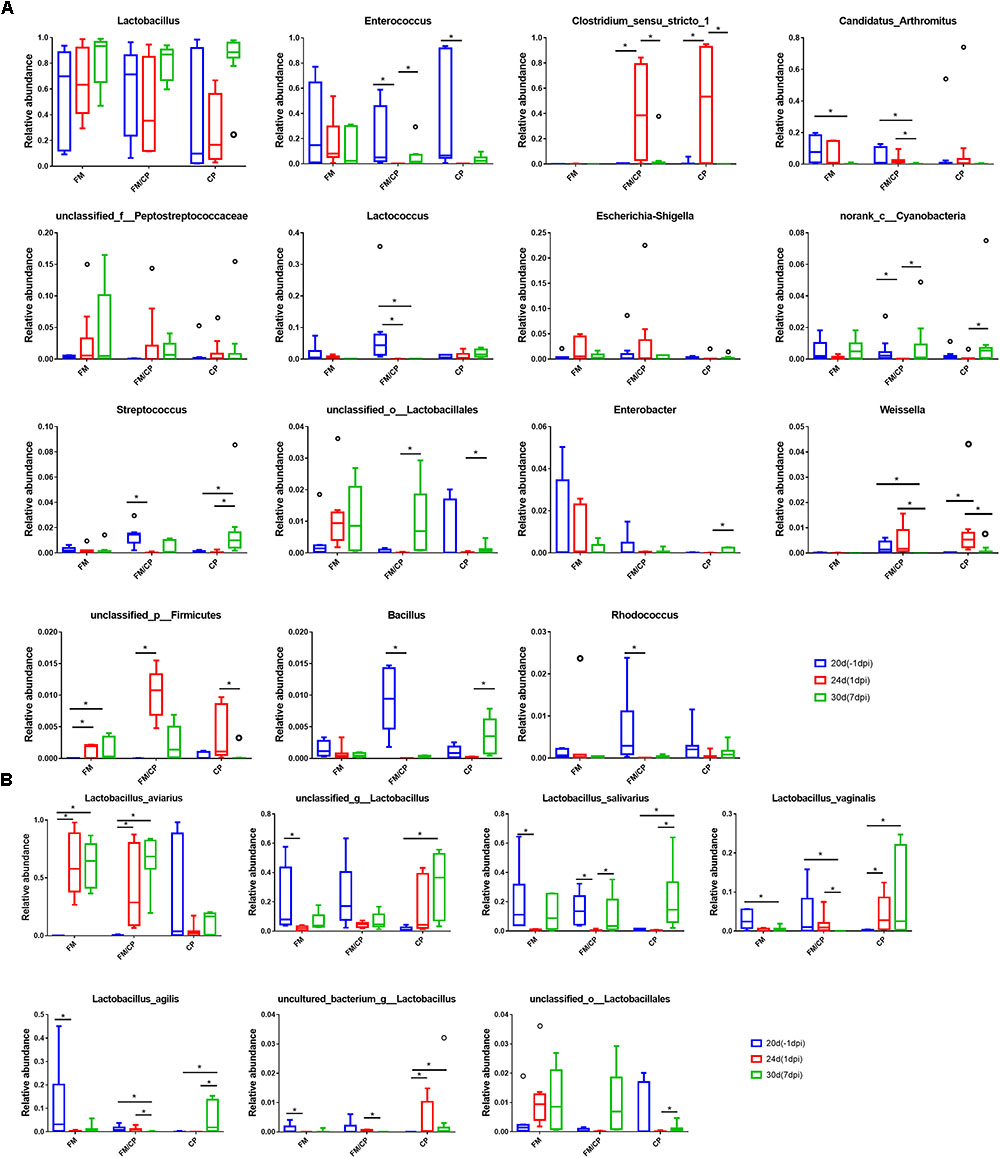
FIGURE 8. Comparison of bacterial composition using relative abundance. At the genus (A) and species (B) level. ∗p < 0.05, Kruskal–Wallis test.
Although the total of genus Lactobacillus was predominant in the chickens gut bacterial composition were not significantly shifted from day 20 to day 30 in group FM, FM/CP and CP respectively (p > 0.05, Figure 8A), some species of Lactobacillus had significant changes (Figure 8B). The relative abundance of Lactobacillus aviarius (L. aviarius) in group FM and FM/CP at days 24 and 30 were significantly higher compared to day 20 (p < 0.05). Additionally, L. aviarius become the dominant species of Lactobacillus in group FM and FM/CP at day 24. All raw sequence were submitted to the Sequence Read Archive database at NCBI (Accession No. SRP159590).
It has been previously shown that C. perfringens challenge can shift the structure of gut microbiota in chickens (Stanley et al., 2012). However, the effect of fishmeal-rich diets on gut bacterial composition and the immune response during the development of NE have not been thoroughly investigated. To determine these effects, we studied the community structures and the relative expression of immune related genes in NE models with high fishmeal diet.
It was previously reported that long-term feeding of a high-protein diet could decrease the expression of TNF-α (Kitabchi et al., 2013) and feeding with a high fishmeal could increase the AGP concentration in the plasma of chickens (Takahashi et al., 2007). However, in this study, at day of age 20, we observed that the high fishmeal did not affect the expression of TNF-α in ileum or AGP in liver of non-infected chickens, but a high fishmeal diet did reduce the expression of IL-8 in ileum and increase the expression of OVT in the liver.
Remarkably, the recognition of C. perfringens by NOD and TLR2 receptor has been shown to lead to activation of downstream signaling pathways to induce chemokine (e.g., IL-8) and cytokine (e.g., TNF-α) expression (Girardin et al., 2003; Lu et al., 2009). Eimeria maxima/C. perfringens co-infection and C. perfringens infection alone with high fishmeal diet could up-regulated the IL-8 and TNF-α transcript level in chickens respectively (Lee et al., 2013; Li et al., 2018). Moreover, E. maxima/C. perfringens challenge significantly increased the expression of NOD1 gene in the intestine (Zhang et al., 2017). However, the TNF-α expression in chicken intestinal epithelial cells was not significantly affected by CP in vitro (Guo et al., 2015). In this study, we observed that C. perfringens infection independent of high fishmeal diet significantly increased the expression of TLR2, NOD1 and IL-8 in intestine tissue at 1 dpi, suggesting that C. perfringens infection independent of high fishmeal diet induced the expression of the above genes to increase.
Relative APP expression was higher in the liver compared with ileum, consistent with the previous report that the expression of APP genes was higher in liver than other tissue (Marques et al., 2017). AGP is a highly glycosylated protein secreted by hepatocytes (Murata et al., 2004). It plays important role in the early stage of inflammation and infection in chickens. For example, Escherichia coli as well as Salmonella typhimurium could significantly increase AGP expression (Takahashi et al., 1998; Adler et al., 2001). Moreover, Chamanza et al. (1999) demonstrated that perhaps SAA is a reliable APP for diagnosing inflammatory lesions in chickens. The expression of SAA also could be increased by bacterial and viral infection such as with Pasteurella multocida, Staphylococcus aureus and infectious bronchitis virus (Chamanza et al., 1999; Kovacs et al., 2007; Asasi et al., 2013). In our study, C. perfringens challenge with a high fishmeal diet, rather than infection alone, resulted in a higher intestinal lesion score and increased the relative abundance of C. perfringens at 1 dpi that was accompanied by an increase in AGP and SAA increase in liver. This suggested that the effect on the level of the AGP and SAA expression in response to the C. perfringens infection was dependent on a high fishmeal diet.
With regard to gut bacterial structure, the PCoA plot of Bray–Curtis indices demonstrated that chickens with high fishmeal diets were clustered and separated from the basic diet feeding chickens at day 20. This was consistent with other studies that showed high fishmeal diets had significant effects on the structure of the chickens gut microbiota (Stanley et al., 2014; Wu et al., 2014). Notably, with prolonged high fishmeal diet, PCoA showed a partition of chickens independent of C. perfringens challenge between early (20 days) and later (24 and 30 days) phases of the experiment. We hypothesized that the structure of the chicken gut microbiota undergoes transition with exposure to the high fishmeal over time and less shifting occurs after reaching a certain composition. Studies about the relationship between exposure time and fishmeal diet and the gut microbiota in chickens are rare and thus require further investigation.
In addition, C. perfringens challenge destabilized chickens gut microbiota independent of high fishmeal diet, and the structure of chicken gut bacteria had the tendency to return to the non-infection state when the chickens did not keep C. perfringens challenge. Our results were different from a study showed the stability of chickens intestinal microbiota is not influenced by the C. perfringens challenge alone (Stanley et al., 2014). Furthermore, in this study, the gut bacterial structure of group FM/CP were different from that of group CP at 1 dpi, which is corresponding to our results showed that C. perfringens-challenged chickens with high fishmeal feeding demonstrated significantly higher C. perfringens colonization level and higher lesion score compared to challenged alone chickens.
For gut bacterial composition, Candidatus Arthromitus (C. Arthromitus) are commensal bacteria in the family Lachnospiraceae which belong to the segmented filamentous bacteria (SFB) group (Thompson et al., 2012b). These bacteria can attach to the intestinal epithelium and modulated the host immune system (Thompson et al., 2012a). We observed that the high fishmeal could decrease abundance of C. Arthromitus however C. perfringens challenge alone did not affect the abundance of these organisms. A decreased fraction of C. Arthromitus was detected in the gut of broiler chickens with the combination E. maxima and C. perfringens or E. maxima alone infection (Stanley et al., 2014; Kim et al., 2015). Previous studies also reported that the genus Lactobacillus was predominant in chicken gut microbiota and the total genus Lactobacillus was not perturbed in chickens following high fishmeal diet feeding or C. perfringens challenge as also shown in the present study (Stanley et al., 2014; Yan et al., 2017). L. aviarius was one of the predominant Lactobacillus species in chickens found by Wang et al. (2014). Similarly, we found that the increase of L. aviarius in high fishmeal feeding chickens independent of C. perfringens challenge in this study was the result of high fishmeal diet and not C. perfringens challenge.
Although C. perfringens challenge alone did not cause intestine lesions, it shifted the structure of the chicken gut microbiota. After infection, both C. perfringens challenge with a high fishmeal diet and C. perfringens challenge alone had the tendency to return to the non-infection stage when the chickens did not have persistent C. perfringens challenge. The temporal shift of gut microbiota structure in C. perfringens-challenged chickens with high fishmeal diet were superimposed upon the chickens with high fishmeal diet alone. Finally, we found that the expression of AGP and SAA in chicken liver tissue may be able to provide an assessment of poultry NE but such an application requires further investigation.
X-YP and Z-HX conceived of this study and participated in its design and coordination. TH designed the experiment, drafted the manuscript, and participated in the statistical analyses. TH, BG, W-LC, M-GY, and RX carried out animal experiments. TH and BG carried out the amplicon sequencing and qPCR. All authors read and approved the final manuscript.
This study was supported by the Special Fund for Agro-Scientific Research in the Public Interest Ministry of Agriculture of the People’s Republic of China (Grant No. 201303040-16), Science and Technology Planning Project of Guangdong Province, China (Grant No. 2014A020208133), and Agricultural Science and Technology Innovation and Major Projects of Guangdong Province (2018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02754/full#supplementary-material
Abrahamsson, T. R., Jakobsson, H. E., Andersson, A. F., Bjorksten, B., Engstrand, L., and Jenmalm, M. C. (2014). Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 44, 842–850. doi: 10.1111/cea.12253
Adler, K. L., Peng, P. H., Peng, R. K., and Klasing, K. C. (2001). The kinetics of hemopexin and alpha1-acid glycoprotein levels induced by injection of inflammatory agents in chickens. Avian Dis. 45, 289–296. doi: 10.2307/1592967
Asasi, K., Mohammadi, A., Boroomand, Z., Hosseinian, S. A., and Nazifi, S. (2013). Changes of several acute phase factors in broiler chickens in response to infectious bronchitis virus infection. Poult. Sci. 92, 1989–1996. doi: 10.3382/ps.2012-02902
Chamanza, R., Toussaint, M. J., van Ederen, A. M., van Veen, L., Hulskamp-Koch, C., and Fabri, T. H. (1999). Serum amyloid A and transferrin in chicken. A preliminary investigation of using acute-phase variables to assess diseases in chickens. Vet. Q. 21, 158–162. doi: 10.1080/01652176.1999.9695012
Drew, M. D., Syed, N. A., Goldade, B. G., Laarveld, B., and Van Kessel, A. G. (2004). Effects of dietary protein source and level on intestinal populations of clostridium perfringens in broiler chickens. Poult. Sci. 83, 414–420. doi: 10.1093/ps/83.3.414
Fernandes da Costa, S. P., Mot, D., Bokori-Brown, M., Savva, C. G., Basak, A. K., Van Immerseel, F., et al. (2013). Protection against avian necrotic enteritis after immunisation with NetB genetic or formaldehyde toxoids. Vaccine 31, 4003–4008. doi: 10.1016/j.vaccine.2013.05.063
Girardin, S. E., Travassos, L. H., Herve, M., Blanot, D., Boneca, I. G., Philpott, D. J., et al. (2003). Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708. doi: 10.1074/jbc.M307198200
Guo, S., Li, C., Liu, D., and Guo, Y. (2015). Inflammatory responses to a Clostridium perfringens type A strain and alpha-toxin in primary intestinal epithelial cells of chicken embryos. Avian. Pathol. 44, 81–91. doi: 10.1080/03079457.2015.1005573
Jakobsson, H. E., Abrahamsson, T. R., Jenmalm, M. C., Harris, K., Quince, C., Jernberg, C., et al. (2014). Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63, 559–566. doi: 10.1136/gutjnl-2012-303249
Kaldhusdal, M., Hofshagen, M., Lovland, A., Langstrand, H., and Redhead, K. (1999). Necrotic enteritis challenge models with broiler chickens raised on litter: evaluation of preconditions, Clostridium perfringens strains and outcome variables. FEMS Immunol. Med. Microbiol. 24, 337–343. doi: 10.1111/j.1574-695X.1999.tb01303.x
Keyburn, A. L., Sheedy, S. A., Ford, M. E., Williamson, M. M., Awad, M. M., Rood, J. I., et al. (2006). Alpha-toxin of clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74, 6496–6500. doi: 10.1128/IAI.00806-06
Kim, J. E., Lillehoj, H. S., Hong, Y. H., Kim, G. B., Lee, S. H., Lillehoj, E. P., et al. (2015). Dietary Capsicum and Curcuma longa oleoresins increase intestinal microbiome and necrotic enteritis in three commercial broiler breeds. Res. Vet. Sci. 102, 150–158. doi: 10.1016/j.rvsc.2015.07.022
Kitabchi, A. E., McDaniel, K. A., Wan, J. Y., Tylavsky, F. A., Jacovino, C. A., Sands, C. W., et al. (2013). Effects of high-protein versus high-carbohydrate diets on markers of beta-cell function, oxidative stress, lipid peroxidation, proinflammatory cytokines, and adipokines in obese, premenopausal women without diabetes: a randomized controlled trial. Diabetes Care 36, 1919–1925. doi: 10.2337/dc12-1912
Kovacs, B. M., Toussaint, M. J., Gruys, E., Fabian, I. B., Szilagyi, L., Janan, J., et al. (2007). Evaluation of goose serum amyloid a acute phase response by enzyme-linked immunosorbent assay. Acta Vet. Hung. 55, 349–357. doi: 10.1556/AVet.55.2007.3.9
Lee, S. H., Lillehoj, H. S., Jang, S. I., Lillehoj, E. P., Min, W., and Bravo, D. M. (2013). Dietary supplementation of young broiler chickens with Capsicum and turmeric oleoresins increases resistance to necrotic enteritis. Br. J. Nutr. 110, 840–847. doi: 10.1017/S0007114512006083
Li, Y., He, J., He, Z., Zhou, Y., Yuan, M., Xu, X., et al. (2014). Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 8, 1879–1891. doi: 10.1038/ismej.2014.28
Li, Z., Wang, W., Liu, D., and Guo, Y. (2018). Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 9, 25. doi: 10.1186/s40104-018-0243-3
Lin, H. Y., Tang, C. H., Chen, J. H., Chuang, J. Y., Huang, S. M., Tan, T. W., et al. (2011). Peptidoglycan induces interleukin-6 expression through the TLR2 receptor, JNK, c-Jun, and AP-1 pathways in microglia. J. Cell. Physiol. 226, 1573–1582. doi: 10.1002/jcp.22489
Lu, Y., Sarson, A. J., Gong, J., Zhou, H., Zhu, W., Kang, Z., et al. (2009). Expression profiles of genes in Toll-like receptor-mediated signaling of broilers infected with clostridium perfringens. Clin. Vaccine Immunol. 16, 1639–1647. doi: 10.1128/CVI.00254-09
Marques, A. T., Nordio, L., Lecchi, C., Grilli, G., Giudice, C., and Ceciliani, F. (2017). Widespread extrahepatic expression of acute-phase proteins in healthy chicken (Gallus gallus) tissues. Vet. Immunol. Immunopathol. 190, 10–17. doi: 10.1016/j.vetimm.2017.06.006
Murata, H., Shimada, N., and Yoshioka, M. (2004). Current research on acute phase proteins in veterinary diagnosis: an overview. Vet. J. 168, 28–40. doi: 10.1016/S1090-0233(03)00119-9
O’Reilly, E. L., and Eckersall, P. D. (2014). Acute phase proteins: a review of their function, behaviour and measurement in chickens. Worlds Poul. Sci. J. 70, 27–44. doi: 10.1017/s0043933914000038
Rath, N. C., Anthony, N. B., Kannan, L., Huff, W. E., Huff, G. R., Chapman, H. D., et al. (2009). Serum ovotransferrin as a biomarker of inflammatory diseases in chickens. Poult. Sci. 88, 2069–2074. doi: 10.3382/ps.2009-00076
Singh, P., Teal, T. K., Marsh, T. L., Tiedje, J. M., Mosci, R., Jernigan, K., et al. (2015). Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3, 45. doi: 10.1186/s40168-015-0109-2
Stanley, D., Keyburn, A. L., Denman, S. E., and Moore, R. J. (2012). Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet. Microbiol. 159, 155–162. doi: 10.1016/j.vetmic.2012.03.032
Stanley, D., Wu, S. B., Rodgers, N., Swick, R. A., and Moore, R. J. (2014). Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PLoS One 9:e104739. doi: 10.1371/journal.pone.0104739
Takahashi, K., Miyake, N., Ohta, T., Akiba, Y., and Tamura, K. (1998). Changes in plasma alpha 1-acid glycoprotein concentration and selected immune response in broiler chickens injected with Escherichia coli lipopolysaccharide. Br. Poult. Sci. 39, 152–155. doi: 10.1080/00071669889547
Takahashi, K., Yodogawa, S., Akiba, Y., and Tamura, K. (2007). Effect of dietary protein concentration on responses to Escherichia coli endotoxin in broiler chickens. Br. J. Nutr. 74, 173. doi: 10.1079/bjn19950121
Thompson, C. L., Mikaelyan, A., and Brune, A. (2012a). Immune-modulating gut symbionts are not “Candidatus Arthromitus”. Mucosal Immunol. 6, 200. doi: 10.1038/mi.2012.91
Thompson, C. L., Vier, R., Mikaelyan, A., Wienemann, T., and Brune, A. (2012b). ‘Candidatus Arthromitus’ revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ. Microbiol. 14, 1454–1465. doi: 10.1111/j.1462-2920.2012.02731.x
Van der Sluis, W. (2000). Clostridial enteritis is an often underestimated problem. World Poultry 16, 42–43.
Wang, L., Fang, M., Hu, Y., Yang, Y., Yang, M., and Chen, Y. (2014). Characterization of the most abundant Lactobacillus species in chicken gastrointestinal tract and potential use as probiotics for genetic engineering. Acta Biochim. Biophys. Sin. 46, 612–619. doi: 10.1093/abbs/gmu037
Wu, L., Feng, B. S., He, S. H., Zheng, P. Y., Croitoru, K., and Yang, P. C. (2007). Bacterial peptidoglycan breaks down intestinal tolerance via mast cell activation: the role of TLR2 and NOD2. Immunol. Cell Biol. 85, 538–545. doi: 10.1038/sj.icb.7100079
Wu, S. B., Rodgers, N., and Choct, M. (2010). Optimized necrotic enteritis model producing clinical and subclinical infection of Clostridium perfringens in broiler chickens. Avian Dis. 54, 1058–1065. doi: 10.1637/9338-032910-Reg.1
Wu, S. B., Stanley, D., Rodgers, N., Swick, R. A., and Moore, R. J. (2014). Two necrotic enteritis predisposing factors, dietary fishmeal and Eimeria infection, induce large changes in the caecal microbiota of broiler chickens. Vet. Microbiol. 169, 188–197. doi: 10.1016/j.vetmic.2014.01.007
Yan, W., Sun, C., Yuan, J., and Yang, N. (2017). Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci. Rep. 7:45308. doi: 10.1038/srep45308
Keywords: Clostridium perfringens, high fishmeal diet, gut microbiota, immune response, chickens
Citation: Huang T, Gao B, Chen W-L, Xiang R, Yuan M-G, Xu Z-H and Peng X-Y (2018) Temporal Effects of High Fishmeal Diet on Gut Microbiota and Immune Response in Clostridium perfringens-Challenged Chickens. Front. Microbiol. 9:2754. doi: 10.3389/fmicb.2018.02754
Received: 13 September 2018; Accepted: 29 October 2018;
Published: 13 November 2018.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Yewande Fasina, North Carolina Agricultural and Technical State University, United StatesCopyright © 2018 Huang, Gao, Chen, Xiang, Yuan, Xu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Yu Peng, cC14eUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.