
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 November 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02653
This article is part of the Research TopicFood Safety and Foodborne Pathogen – A Global Perspective on the Diversity, Combating Multidrug Resistance and ManagementView all 39 articles
Ground poultry is marketed as a healthier alternative to ground beef despite the fact that poultry is a major source of foodborne Salmonella. The objectives of this study were to determine the prevalence of Salmonella in Oklahoma retail ground poultry and to characterize representative isolates by serotyping, antimicrobial resistance, PFGE patterns, and large plasmid profiling. A total of 199 retail ground poultry samples (150 ground turkey and 49 ground chicken) were investigated. The overall prevalence of Salmonella in ground poultry was 41% (82/199), and the incidence in conventional samples (47%, 66/141) was higher than in organic samples (27%, 16/58). The prevalence of Salmonella in organic ground chicken and organic ground turkey was 33% (3/9) and 26% (13/49), respectively. Twenty six Salmonella isolates (19 conventional and 7 organic) were chosen for further characterization. The following six serotypes and number of isolates per serotype were identified as follows: Tennessee, 8; Saintpaul, 4; Senftenberg, 4; Anatum, 4 (one was Anatum_var._15+); Ouakam, 3; and Enteritidis, 3. Resistance to 16 tested antimicrobials was as follows: gentamycin, 100% (26/26); ceftiofur, 100% (26/26); amoxicillin/clavulanic acid, 96% (25/26); streptomycin, 92% (24/26); kanamycin, 88% (23/26); ampicillin, 85% (22/26); cephalothin, 81% (21/26); tetracycline, 35% (9/26); sulfisoxazole, 27% (7/26); nalidixic acid, 15% (4/26); and cefoxitin, 15% (4/26). All isolates were susceptible to amikacin, chloramphenicol, ceftriaxone, and trimethoprim/sulfamethoxazole. All screened isolates were multidrug resistant (MDR) and showed resistance to 4–10 antimicrobials; isolates from organic sources showed resistance to 5–7 antimicrobials. PFGE was successful in clustering the Salmonella isolates into distinct clusters that each represented one serotype. PFGE was also used to investigate the presence of large plasmids using S1 nuclease digestion. A total of 8/26 (31%) Salmonella isolates contained a ∼100 Kb plasmid that was present in all Anatum and Ouakam isolates. In conclusion, the presence of multidrug resistant Salmonella with various serotypes, PFGE profiles, and large plasmids in ground poultry stresses the importance of seeking novel interventions to reduce the risk of this foodborne pathogen. Multidrug resistance (MDR) is considered a high additional risk and continued surveillance at the retail level could minimize the risk for the consumer.
Nontyphoidal Salmonella spp. is the primary bacterial pathogen causing foodborne illness and the leading cause of hospitalization among the top five foodborne pathogens in the United States (Scallan et al., 2011). Contaminated meats are the major foodborne sources of Salmonella, which has been recovered and characterized from retail beef, pork, bison, chicken, and turkey meats in several countries worldwide (Li et al., 2006; Cetinkaya et al., 2008; Nde et al., 2008; Ammari et al., 2009; Yang et al., 2010; Tafida et al., 2013; Maka et al., 2014; Sallam et al., 2014; Aslam et al., 2012; Soufi et al., 2012; Thai et al., 2012). Consumption of ground poultry has increased in the last few years, partially because it is marketed as a healthier alternative to ground beef. However, ground poultry, particularly ground turkey and chicken, is often contaminated with Salmonella (White et al., 2001; Fakhr et al., 2006a; Erol et al., 2013; Cui et al., 2015). A large, multistate-outbreak caused by an antimicrobial-resistant Salmonella enterica subsp. enterica serovar Heidelberg occurred in 2011 from the consumption of contaminated ground turkey and resulted in one death (Folster et al., 2012). Three other multistate-outbreaks caused by Salmonella Heidelberg occurred between 2013 and 2014 that were linked to chicken consumption (CDC, 2013a, 2014a,b; Gieraltowski et al., 2016). An outbreak of Salmonella enterica serovar Stanley infections associated with turkey meat was reported in 10 European countries between 2011 and early 2013 (Kinross et al., 2014). Comparative genomic analysis using Whole Genome Sequencing revealed that the S. Heidelberg isolates in the 2011 ground turkey outbreak clustered together when compared to isolates from human, animal, and retail meat sources (Hoffmann et al., 2014). Using an experimental oral challenge experiment in turkey, a recent study showed that the Salmonella isolate causing the 2011 outbreak was high in cecal colonization, dissemination to internal organs, and tissue deposition (Nair et al., 2018). Recently, a food-grade essential oil from pimento leaves was shown to reduce attachment of the 2011 S. Heidelberg isolate to turkey skin (Nair and Johny, 2017). By testing the host transcriptional response, a recent study showed that young commercial turkeys are susceptible to colonization by S. Heidelberg isolated from the 2011 ground turkey outbreak (Bearson et al., 2017).
The presence of antimicrobial resistant Salmonella in retail meats, particularly in poultry, is a major risk to the treatment of foodborne illnesses caused by this bacterial pathogen (Antunes et al., 2016; Chai et al., 2017). The presence of multidrug resistant (MDR), nontyphoidal Salmonella in retail meats has been reported in several studies (Cetinkaya et al., 2008; Zhao et al., 2008; Yildirim et al., 2011; Thai et al., 2012; Van et al., 2012; Maka et al., 2014; Yang et al., 2014; Iwamoto et al., 2017; Clothier et al., 2018). Most of the antimicrobial resistance genes in Salmonella are carried on conjugative plasmids that facilitate transfer between different isolates (Jones and Stanley, 1992; Rotgers and Casadesüs, 1999; Carattoli, 2003; Rychlik et al., 2006). Conjugation experiments showed that 95% of the β–lactamase genes (blaCMY) in Salmonella are plasmid-encoded (Folster et al., 2011). Quinolone resistance genes were also plasmid-borne in Salmonella isolated from human cases in the United States (Sjölund-Karlsson et al., 2010). Three emerging European clones of Salmonella enterica subsp. enterica serovar Typhimurium circulating in Europe were found to harbor MDR plasmids that encode additional virulence functions (García et al., 2014). Plasmid profiling is often used in epidemiological studies related to surveillance of disease outbreaks and in tracing the dissemination of antibiotic resistance (Mayer, 1988).
Pulsed field gel electrophoresis (PFGE) is considered the gold standard in typing Salmonella and is known for its ability to discriminate isolates and for tracking the source of outbreaks (Tenover et al., 1995; Fakhr et al., 2005; Foley et al., 2006, 2009; Folster et al., 2012). PFGE profiling has been used with relative success as a method to identify Salmonella serotypes (Gaul et al., 2007; Zou et al., 2010). A meta-analysis of PFGE fingerprints based on a constructed Salmonella database of 45,923 PFGE patterns indicated the presence of serotype-specific patterns that may potentially reduce the need to perform the laborious, traditional serotyping (Zou et al., 2013).
Despite the risk associated with the consumption of ground poultry contaminated with Salmonella, studies investigating the prevalence and characterization of Salmonella in retail ground poultry are relatively scarce. The objectives of this study were to determine the prevalence of Salmonella in retail ground poultry sold in the Tulsa, Oklahoma area and to characterize a selected number of the recovered strains by serotyping, antimicrobial resistance screening, plasmid profiling, and PFGE.
Conventional methods were used to isolate Salmonella from ground turkey as described previously (Fakhr et al., 2006a; Nde et al., 2008). In the summer of 2009, 199 samples of ground poultry meat (150 and 49 from turkey and chicken, respectively) representing five brands were purchased at six retail stores representing six supermarket chains in Tulsa, Oklahoma. Ground poultry samples were stored in chilled containers, and transported to the laboratory within 4 h. Each sample (25 g) was subjected to a pre-enrichment step by combining it with 225 mL of Buffered Peptone Water (BPW) (EMD, Gibbstown, NJ, United States) in sterile plastic bags (VWR Scientific, Radnor, PA, United States); the samples was massaged briefly by hand for 5 min. The pre-enrichment rinsate was then incubated at 37°C for 24 h. To selectively enrich for Salmonella, 0.1 and 0.5 mL of each pre-enrichment broth sample was transferred to 10 mL of Rappaport-Vassiliadis broth (RVB; Difco, Becton Dickinson, Sparks, MD, United States) and tetrathionate broth (TTB; Difco, Becton Dickinson, Sparks, MD, United States), respectively, and incubated at 42°C for 24 h. The pre-enrichment broths of duplicate samples were then artificially spiked with 10 μL of an overnight broth of two Salmonella strains (one H2S-positive and one H2S-negative); these served as positive controls. After selective enrichment was completed, a loopful of broth contains each of the enriched samples, including the two artificially-spiked Salmonella positive controls, were inoculated by dilution-streaking onto two selective agar media, XLT4 (Difco, Becton Dickinson, Sparks, MD, United States) and Brilliant Green Sulfide (BGS) (Difco, Becton Dickinson, Sparks, MD, United States) and incubated at 37°C for 24 h. The identity of 4–6 suspected Salmonella colonies from each sample were confirmed biochemically by dilution streaking onto Triple Sugar Iron Agar (TSI) (Difco, Becton Dickinson, Sparks, MD, United States) and Lysine Iron Agar slants (Difco, Becton Dickinson, Sparks, MD, United States) and incubated at 37°C for 24 h. Suspected Salmonella isolates were subjected to molecular confirmation by PCR using invA as described below.
The invA gene was amplified using the following PCR primers: forward, 5′- GTGAAATTATCGCCACGTTCGGGCAA-3′; and reverse, 5′- TCATCGCACCGTCAAAGGAACC-3′ as described previously (Rahn et al., 1992). PCR was conducted in 25 μL reaction volumes containing the following: 12.5 μL GoTaq® Green Master Mix (Promega, Madison, WI, United States), 3.5 μL sterile water (Promega, Madison, WI, United States), 1 μL (25 pmol) of each primer (IDT, Coralville, IA, United States), and 3 μL of template DNA. The cycling conditions were as follows: (1) 95°C for 5 min; (2) 94°C for 1 min; (3) 55°C for 1 min; (4) 72°C for 1 min; and (5) 72°C for 10 min. Steps 2 through 4 were repeated for 35 cycles. PCR products were subjected to agarose gel electrophoresis, and a 1 kb plus DNA ladder (Bioneer, Alameda, CA, United States) was used as a molecular marker. Gel images were taken using a Bio-Rad Gel DocTM XR UV gel documentation system (Bio-Rad, Hercules, CA, United States). The presence of the 284 bp invA PCR product was considered to be positive for Salmonella molecular identification. Once confirmed as Salmonella, one isolate was kept as a representative for each ground poultry sample and further characterized by serotyping, antibiotic resistance profile, PFGE, and plasmid content.
Salmonella isolates selected for serotyping were given a serial designation from GP001 to GP023 and from GP025 to GP027. Isolates were sent to the National Veterinary Service Laboratory (NVSL) in Ames, Iowa, for serotyping.
Salmonella isolates were subjected to antimicrobial resistance profiling using the following 16 antimicrobials: cefoxitin (FOX), amikacin (AMI), chloramphenicol (CHL), tetracycline (TET), ceftriaxone (CTR), amoxicillin/clavulanic acid (AMC), ciprofloxacin (CIP), gentamycin (GEN), nalidixic acid (NAL), ceftiofur (TIO), sulfisoxazole (FIS), trimethoprim/sulfamethoxazole (SXT), cephalothin (CEP), kanamycin (KAN), ampicillin (AMP), and streptomycin (STR). Isolates were grown on Mueller-Hinton (MH) agar (Difco) and incubated for 24 h at 37°C. Cultures were then added to Mueller-Hinton broth (Difco), and the turbidity was adjusted to a 0.5 McFarland standard, and inoculated onto 6-inch MH agar plates supplemented with the appropriate antimicrobials. Multiple antibiotic concentrations were tested including the breakpoint established for each antimicrobial according to the Clinical and Laboratory Standards Institute (CLSI) (Cockerill, 2011). The ranges of the concentrations used and the breakpoint of each of the 16 antimicrobials tested in this study were detailed previously (Fakhr et al., 2006b). Plates were then incubated at 37°C for 48 h; results were read for growth or no growth and denoted as resistant or susceptible, respectively, according to the breakpoints for each antimicrobial.
Plug preparation for PFGE profiling was performed according to the PulseNet protocol and conditions established by the CDC (CDC, 2013b). Slices of the prepared PFGE plugs (2-mm wide) were incubated with XbaI (Promega, Madison, WI, United States) at a concentration of 50 U/plug for 3 h at 37°C. Plug slices were then inserted into the wells of 1% Seakem Gold Agarose gels. XbaI-digested Salmonella serovar Braenderup H9812 was used as a sizing marker. PFGE was conducted in a CHEF Mapper PFGE system (Bio-Rad, Hercules, CA, United States) for 18 h following the electrophoresis conditions established for Salmonella by the PulseNet protocol; these included an initial switch time of 2.16 s, and a final switch time of 63.8 s (CDC, 2013b). After electrophoresis, gel images were captured using a Bio-Rad Gel DocTM XR UV gel documentation system (Bio-Rad, Hercules, CA, United States). Images were then imported and analyzed using the BioNumerics software v. 6.6 (Applied Maths, Austin, TX, United States). Similarity analysis and the banding patterns were analyzed using the Dice coefficient and clustered using the unweighted pair group method with arithmetic mean (UPGMA) and a 1.5% band tolerance.
Screening of large plasmids was performed by PFGE as described previously (Barton et al., 1995; Marasini and Fakhr, 2014). The PFGE plugs were prepared as described above; thin slices were cut and digested with S1 nuclease (17 IU/plug) for 45 min at 37°C to linearize the plasmids. Plug slices were then inserted into the wells of 1% Seakem Gold Agarose gels, and XbaI-digested Salmonella serovar Braenderup H9812 was used as a sizing marker. PFGE was conducted using the CHEF Mapper PFGE system for 16 h using the conditions established for Salmonella by the PulseNet protocol (CDC, 2013b).
Large plasmids detected by PFGE were isolated by alkaline lysis using the Qiagen Miniprep kit and protocols established for Gram-negative bacteria (Qiagen Inc., Valencia, CA, United States). Isolated plasmids were analyzed by electrophoresis in 0.8% agarose gels at 120 V for 2 h. Gels were stained with ethidium bromide for 45 min, and images were captured using the Bio-Rad gel documentation system. DNA markers for sizing included plasmids preps of E. coli strains NCTC 50192 and NCTC 50193 and the 1 Kb plus DNA ladder (Bioneer). The isolated plasmids were also digested with EcoRI and HindIII (Promega, Madison, WI, United States) and subjected to agarose gel electrophoresis to determine variable restriction patterns.
A total of 199 retail ground poultry samples were investigated in this study (Table 1). Although only 6% (9/150) of the ground turkey samples were organic, all 49 ground chicken samples were organic. The overall prevalence of Salmonella in ground poultry was 41% (82/199), whereas the prevalence in conventional samples (47%; 66/141) was higher than organic samples (27%; 16/58) (Table 1). The prevalence of Salmonella in organic ground chicken was 26% (13/49), whereas the incidence in ground turkey was 47% (66/141) and 33% (3/9) for conventional and organic samples, respectively (Table 1).
To reduce the cost, twenty six isolates representing unique Salmonella-positive ground poultry samples (19 conventional, 7 organic) were selected for further characterization by serotyping, antibiotic resistance profiling, PFGE, and plasmid profiling. The twenty six isolates were carefully chosen to fairly represent the eighty two positive samples in this study in regards to variation in the collection and expiration date, brand, supermarket chain and location, and meat source (ground turkey or ground chicken). Six serotypes were identified: Tennessee (8 isolates), Saintpaul (4 isolates), Senftenberg (4 isolates), Anatum (4 isolates, including one Anatum_var._15+), Ouakam (3 isolates), and Enteritidis (3 isolates) (Table 2). Serotypes Saintpaul, Ouakam, and Anatum were detected in conventional ground turkey, but not in organic ground chicken; the latter contained serotypes Tennessee (n = 4), Enteritidis (n = 2), and Senftenberg (n = 1) (Table 2).
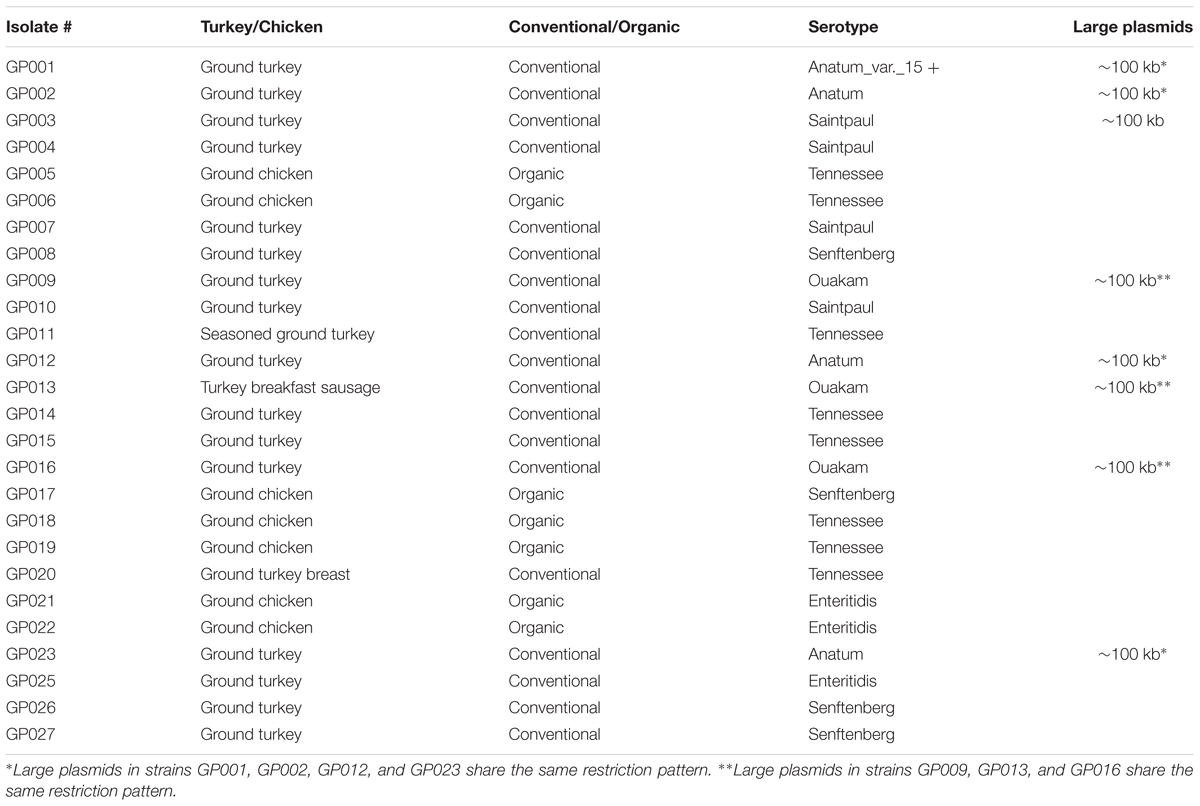
TABLE 2. Ground poultry sources, serotypes and large plasmid profiles of the 26 Salmonella isolates characterized in this study.
The 26 serotyped Salmonella isolates were subjected to antibiotic resistance profiling to 16 antimicrobials (Figure 1). All 26 isolates were resistant to both gentamycin and ceftiofur. Resistance to the remaining antimicrobials was as follows: amoxicillin/clavulanic acid, 96% (25/26); streptomycin, 92% (24/26); kanamycin, 88% (23/26); ampicillin, 85% (22/26); cephalothin, 81% (21/26); tetracycline, 35% (9/26 ), sulfisoxazole 27% (7/26); nalidixic acid 15% (4/26), and cefoxitin, 15% (4/26). All isolates were susceptible to amikacin, chloramphenicol, ceftriaxone, and trimethoprim/sulfamethoxazole. All 26 tested isolates were multidrug-resistant (MDR) and exhibited resistance to 4–10 antimicrobials (Figure 1). Isolates from organic sources also exhibited MDR to 5–7 antimicrobials. Sulfisoxazole resistance was observed only in the Anatum and Ouakam serotypes. Although there was some variability, some antibiotic profiles were common among a particular serotype (Figure 1).
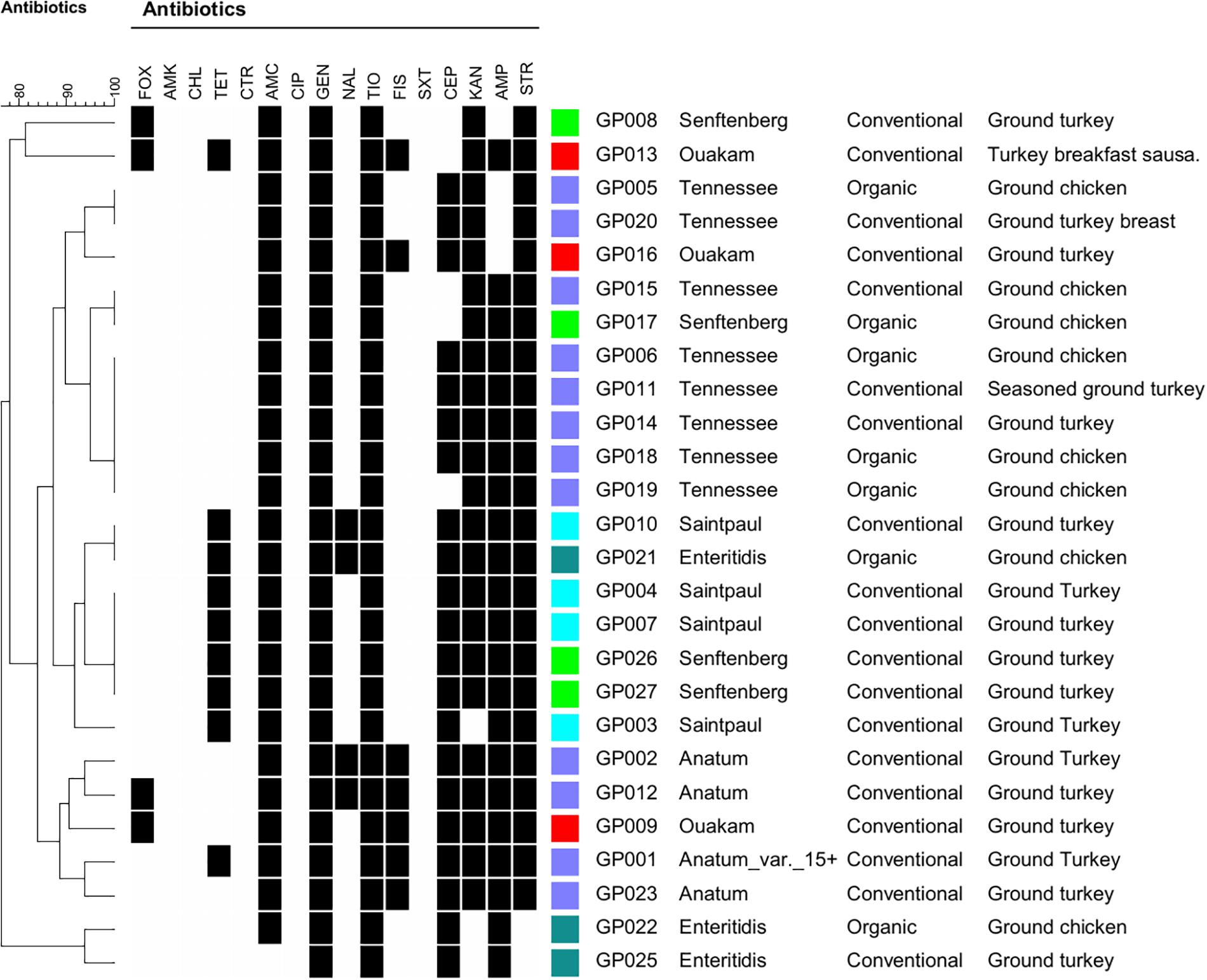
FIGURE 1. Dendrogram of 26 Salmonella strains showing antibiotic resistance profiles, serotypes, and sources of ground poultry. Clustering was based on antibiotic resistance profiling, and the dendogram was created using BioNumerics software. Black squares indicate resistance. Abbreviations: FOX, cefoxitin; AMK, amikacin; CHL, chloramphenicol: TET, tetracycline; CTR, ceftiaxone; AMC, amoxicillin-clavulanic acid; CIP, ciprofloxac; GEN, gentamicin; NAL, nalidixic acid; TIO, ceftiofur; FIS, sulfisoxazole; SXT, trimethoprim/sulfamethoxazole; CEP, cephalothin, KAN, kanamycin; AMP, ampicillin; STR, streptomycin.
All 26 serotyped Salmonella isolates were subjected to PFGE to determine XbaI restriction patterns. Although the four Saintpaul isolates were non-typable by XbaI-mediated PFGE, the remaining 22 isolates representing the other five serotypes were successfully analyzed (Figure 2). PFGE grouped the 22 Salmonella isolates into five distinct clusters each representing one of the following five serotypes: Enteritidis, Senftenberg, Ouakam, Anatum (including the Anatum_var._15+), and Tennessee.
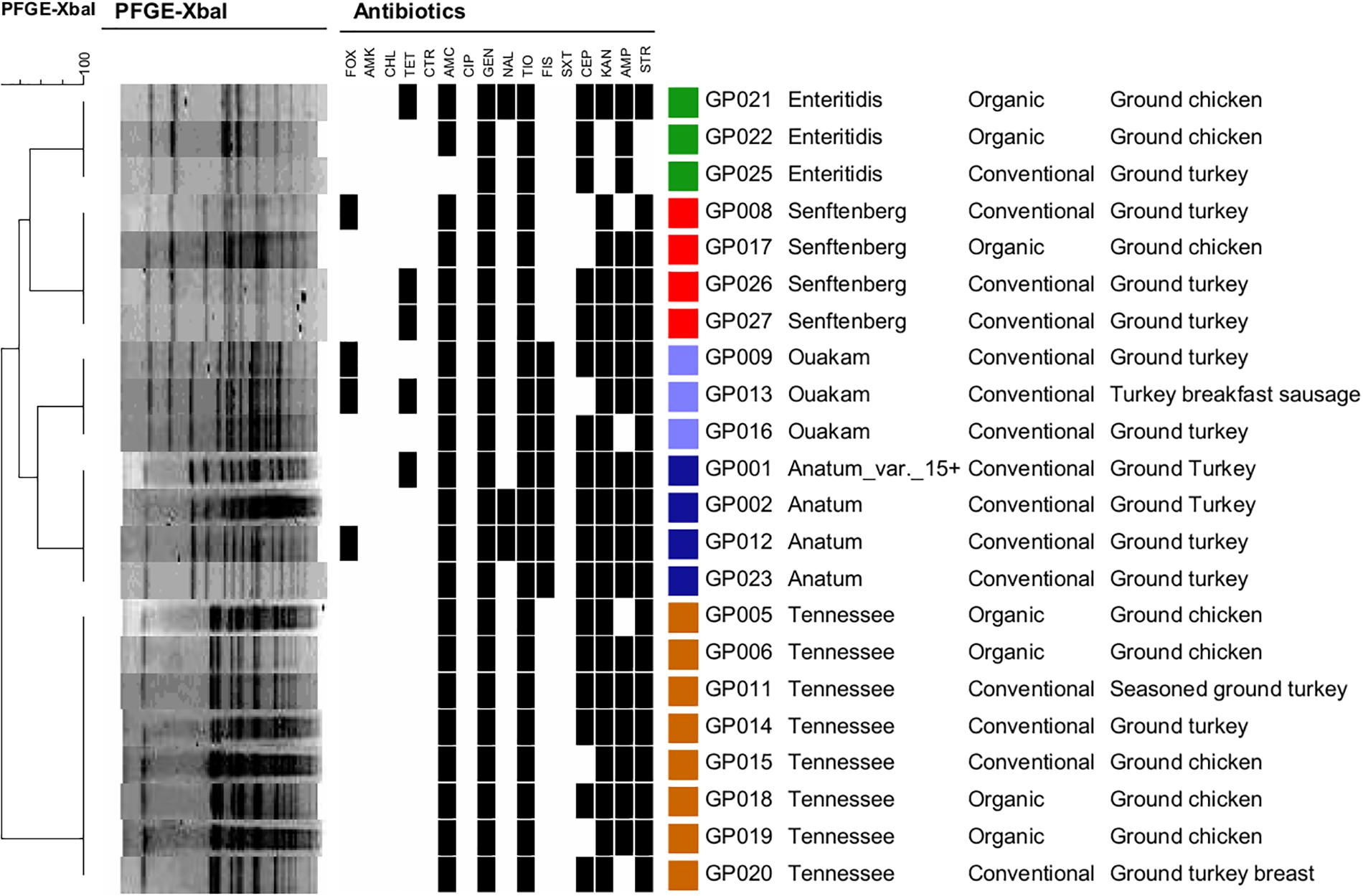
FIGURE 2. Dendogram and pulsed field gel electrophoresis (PFGE) profiling of 22 Salmonella isolates using XbaI. Antibiotic resistance, serotypes, and the ground poultry sources are shown for the 22 PFGE-typable Salmonella strains. Similarity analysis was performed using the Dice coefficient, and clustering was performed using UPGMA based on PFGE profiles. Black squares indicate resistance. FOX, cefoxitin; AMK, amikacin; CHL, chloramphenicol: TET, tetracycline; CTR, ceftiaxone; AMC, amoxicillin-clavulanic acid; CIP, ciprofloxac; GEN, gentamicin; NAL, nalidixic acid; TIO, ceftiofur; FIS, sulfisoxazole; SXT, trimethoprim/sulfamethoxazole; CEP, cephalothin, KAN, kanamycin; AMP, ampicillin; STR, streptomycin.
PFGE was used to investigate the presence of large plasmids in the 26 serotyped isolates using S1 nuclease digestion. Eight of the 26 Salmonella isolates contained a large ∼100 Kb plasmid (Table 2). The four serotype Anatum strains, including the Anatum_var._15 + isolate, contained a ∼100 Kb plasmid, as did the three Ouakam isolates and one of the four Saintpaul isolates (Table 2). A PFGE gel showing the 100 Kb plasmid in one of the Salmonella Ouakam isolates is presented in Figure 3 (lane 3). All eight isolates harboring large plasmids were isolated from conventional samples. Restriction digestion analysis using EcoRI and/or HindIII revealed that the large plasmids harbored by the four Anatum isolates had similar restriction patterns (Table 2). Likewise, similar restriction patterns were obtained for large plasmids of the three Ouakam isolates (Table 2).
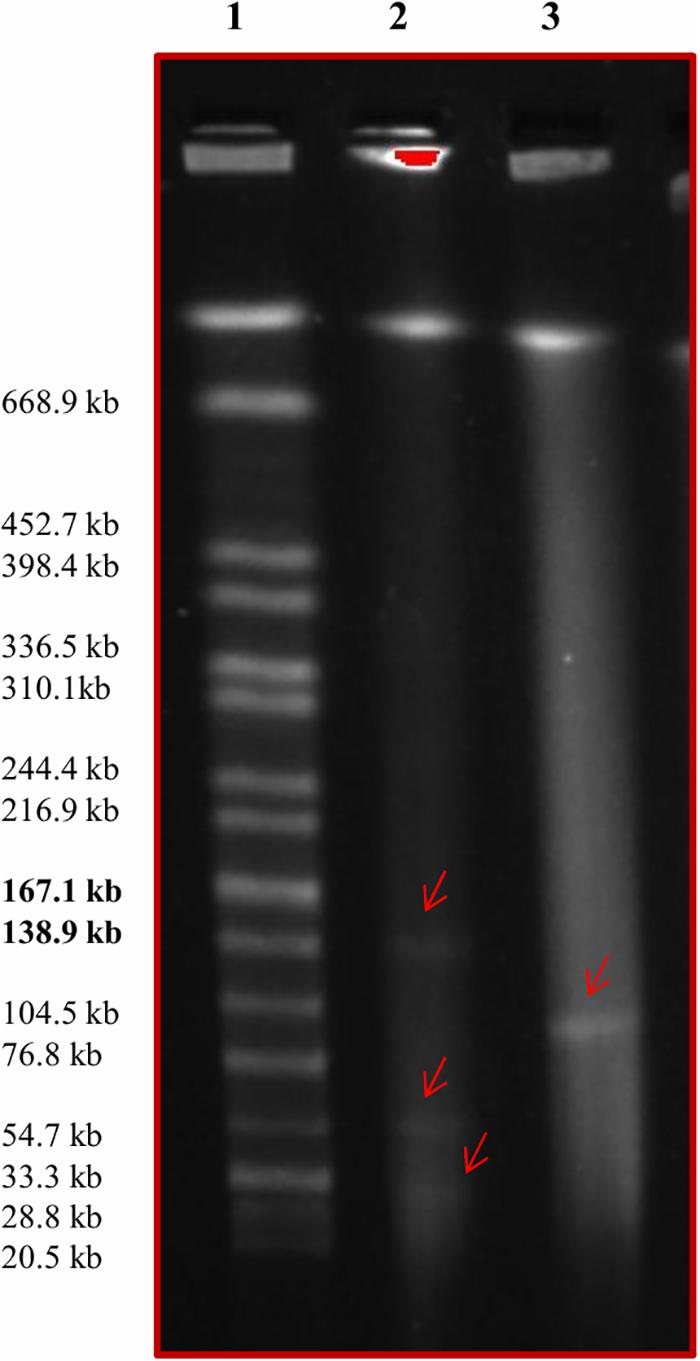
FIGURE 3. Pulsed field gel electrophoresis image showing the presence of large plasmids. Lane 1 contains the Salmonella serovar Braenderup H9812 molecular marker. Lane 2 contains E. coli NCTC 50192 treated with S1 nuclease; the three large plasmids (147, 63, and 43.5 kb) are indicated with red arrows. Lane 3 contains Salmonella Ouakam strain GP016 treated with S1 nuclease; the red arrow indicates the presence of a ∼100 kb plasmid.
Salmonella was recovered from 41% (82/199) of ground poultry samples collected in this study, which stresses the importance of monitoring this foodborne pathogen at the retail level. This result is similar to another study where Salmonella was recovered from 40% (30/74) of ground turkey samples in Fargo, North Dakota (Fakhr et al., 2006b). In a larger study conducted by researchers at the FDA, Salmonella prevalence was 52% in ground turkey after screening 1,499 ground turkey samples collected from grocery stores in several FoodNet sites across the United States (Zhao et al., 2006). A recent study indicated that the high incidence of Salmonella in turkey neck skin may predict a flock with greater potential for Salmonella contamination of ground turkey (Cui et al., 2015). Another study found that preharvest screening of Salmonella using a rapid protocol could potentially reduce Salmonella in ground turkey meat and possibly decrease future salmonellosis outbreaks (Evans et al., 2015). Recently, the Salmonella lytic bacteriophage preparation (SalmoFresh) was investigated for efficacy in reducing Salmonella populations on turkey breast cutlets and ground turkey (Sharma et al., 2015). While the bacteriophage preparation was effective in reducing Salmonella on turkey breast cutlets, it did not reduce the incidence of Salmonella Heidelberg in ground turkey (Sharma et al., 2015).
The detection of six serotypes in the 26 Salmonella-positive isolates indicates a high level of Salmonella diversity in ground turkey. This variability was also observed in other studies where different serotypes were detected depending on the geographic location and the date when studies were conducted (Fakhr et al., 2006b; Erol et al., 2013). All isolates were susceptible to amikacin, chloramphenicol, trimethoprim/sulfamethoxazole, and ceftriaxone; the latter is particularly important because ceftriaxone is the drug of choice for treating salmonellosis in children (White et al., 2001; Iwamoto et al., 2017). A recent study examining NARMS data between 1996 and 2013 showed that ceftriaxone resistance in Salmonella isolated from humans correlated with resistance in retail meats and food animals in the United States (Iwamoto et al., 2017). In the present study, the high percentages of resistance for gentamycin (100%), ceftiofur (100%), amoxicillin/clavulanic acid (96%), streptomycin (92%), kanamycin (88%), ampicillin (85%), and cephalothin (81%) is alarming. Similar high percentages of resistance ranging from 91.6 to 100% to several of these antimicrobials were reported in Salmonella isolated from chicken meat and giblets collected from Mansoura, Egypt (Abd-Elghany et al., 2015). The high incidence of resistance to aminoglycoside and β-lactam antibiotics is coincident with high prevalence of S. enterica resistance to these two classes of antibiotics in food animals (Foley and Lynne, 2008; Frye and Jackson, 2013). Ceftiofur has been used to prevent the death of 1-day old turkey poults, and its use in animal feed might select for the acquisition of plasmids with antibiotic resistance (Wittum, 2012). In our study, the moderate level of resistance in screened Salmonella isolates to tetracycline (35%), sulfisoxazole (27%), nalidixic acid (15%), and cefoxitin (15%) has been documented in other studies (Aslam et al., 2012; Thai et al., 2012; Nisar et al., 2017). A recent study analyzed the surveillance data of 18 years on antimicrobial resistance profiling showed higher level of resistance of chicken breast isolates toward third-generation cephalosporins and tetracyclines when compared to human isolates (Paudyal et al., 2018). It is noteworthy that all 26 isolates in this study, including those isolated from organic sources, exhibited multidrug resistance (MDR). The presence of MDR Salmonella in retail meats in the United States, Canada, and the European Union is well- established (White et al., 2001; Zhao et al., 2006; Aslam et al., 2012; Florez-Cuadrado et al., 2018). In a recent study, 36% of Salmonella isolates were multidrug resistant to two to five antimicrobials despite being isolated from a antimicrobials free turkey production facility (Sanad et al., 2016). In a large study conducted in Spain, 41% of Salmonella isolates from meat products was resistant to three or more antibiotics (Doménech et al., 2015). The high number of MDR Salmonella detected in retail meats sold in Oklahoma is not surprising since previous studies have shown the high incidence of MDR Campylobacter spp. and Staphylococcus aureus in Oklahoma retail meats (Noormohamed and Fakhr, 2012, 2013, 2014; Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b).
In this study, PFGE successfully grouped Salmonella isolates into distinct clusters that represented individual serotypes. PFGE previously showed discriminative ability for some Salmonella serotypes and antimicrobial resistance profiles (Fakhr et al., 2006b; Zhao et al., 2006). PFGE profiling was considered a possible alternative for identification of some Salmonella serotypes (Gaul et al., 2007; Zou et al., 2010, 2013). In a recent study, PFGE showed that Salmonella Heidelberg isolates from turkeys were more genetically diverse than those isolated from chickens (Nisar et al., 2017). In this study, PFGE in combination with S1 nuclease digestion enabled successful detection of large plasmids in 8/26 (31%) of the Salmonella isolates. A large plasmid of ∼100 Kb was detected in all Anatum and Ouakam isolates and one Saintpaul isolate; furthermore, the Anatum and Ouakam were the only isolates with sulfisoxazole resistance, which might indicate a role for these plasmids in mediating resistance to this antimicrobial. Recently, we released the whole genome sequences of three isolates described in this study including Salmonella Ouakam, Anatum, and Anatum var. 15+; these isolates harbored large plasmids of 109,715, 112,176, and 112,176 bp, respectively (Marasini et al., 2016a,b). The large plasmids in the Anatum and Anatum_var._15+ isolates were identical in size, which is consistent with the restriction patterns observed in this study. The large plasmids in Salmonella are known to encode genes for virulence and MDR, and their conjugative properties facilitates dissemination of virulence and antimicrobial resistance (Carattoli, 2003; Rychlik et al., 2006; Sajid and Schwarz, 2009; Folster et al., 2011; García et al., 2014).
In conclusion, the presence of MDR Salmonella with various serotypes, PFGE profiles, and large plasmids in ground poultry is alarming. Intervention strategies to reduce this important foodborne pathogen in retail meats are imperative, particularly in ground turkey. While ground poultry is being marketed as a healthier alternative to ground beef, consumers should apply strict food safety practices when handling ground turkey and consider cooking the meat thoroughly. The high prevalence of Salmonella strains recovered in this study with resistance to several antimicrobials can complicate the treatment of salmonellosis and increase the risk of this human illness. This is particularly critical for treating children with salmonellosis, since ceftriaxone is the drug of choice for pediatric salmonellosis and resistance to this compound would derail the efficacy of this antibiotic.
MF performed the research design and provided the laboratory supplies. AG, U-AS, KH, and MF carried out the bench work and data analysis. MF and AG prepared the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the University of Tulsa for granting KH a Tulsa Undergraduate Research Challenge (TURC) grant.
Abdalrahman, L., and Fakhr, M. (2015). Incidence, antimicrobial susceptibility, and toxin genes possession screening of Staphylococcus aureus in retail chicken livers and gizzards. Foods 4, 115–129. doi: 10.3390/foods4020115
Abdalrahman, L. S., Stanley, A., Wells, H., and Fakhr, M. K. (2015a). Isolation, virulence, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin sensitive Staphylococcus aureus (MSSA) strains from Oklahoma retail poultry meats. Int. J. Environ. Res. Public Health 12, 6148–6161. doi: 10.3390/ijerph120606148
Abdalrahman, L. S., Wells, H., and Fakhr, M. (2015b). Staphylococcus aureus is more prevalent in retail beef livers than in pork and other beef cuts. Pathogens 4, 182–198. doi: 10.3390/pathogens4020182
Abd-Elghany, S. M., Sallam, K. I., Abd-Elkhalek, A., and Tamura, T. (2015). Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 143, 997–1003. doi: 10.1017/S0950268814001708
Ammari, S., Laglaoui, A., En-Nanei, L., Bertrand, S., Wildemauwe, C., Barrijal, S., et al. (2009). Isolation, drug resistance and molecular characterization of Salmonella isolates in northern Morocco. J. Infect. Dev. Ctries. 3, 41–49. doi: 10.3855/jidc.104
Antunes, P., Mourão, J., Campos, J., and Peixe, L. (2016). Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 110–121. doi: 10.1016/j.cmi.2015.12.004
Aslam, M., Checkley, S., Avery, B., Chalmers, G., Bohaychuk, V., Gensler, G., et al. (2012). Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 32, 110–117. doi: 10.1016/j.fm.2012.04.017
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Bearson, B. L., Bearson, S. M. D., Looft, T., Cai, G., and Shippy, D. C. (2017). Characterization of a multidrug-resistant Salmonella enterica serovar Heidelberg outbreak strain in commercial turkeys: colonization, transmission, and host transcriptional response. Front. Vet. Sci. 4:156. doi: 10.3389/fvets.2017.00156
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5, 113–122.
CDC (2013a). CDC - Salmonella Heidelberg Infections Linked to Chicken - Salmonella - Feb, 2013. Available at: https://www.cdc.gov/salmonella/heidelberg-02-13/index.html [accessed September 19, 2018].
CDC (2013b). Standard operating procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non - O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. E. coli Salmonella Detect. Meat 157, 1–13. doi: 10.1089/fpd.2006.3.59
CDC (2014a). Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Foster Farms Brand Chicken (Final Update), 1–18. Available at: https://www.cdc.gov/salmonella/heidelberg-10-13/index.html [accessed September 19, 2018].
CDC (2014b). Salmonella Heidelberg Infections Linked to Tyson Brand Mechanically Separated Chicken at a Correctional Facility | February 2014 | Salmonella | CDC. Available at: https://www.cdc.gov/salmonella/heidelberg-01-14/index.html [accessed September 19, 2018].
Cetinkaya, F., Cibik, R., Ece Soyutemiz, G., Ozakin, C., Kayali, R., and Levent, B. (2008). Shigella and Salmonella contamination in various foodstuffs in Turkey. Food Control 19, 1059–1063. doi: 10.1016/j.foodcont.2007.11.004
Chai, S. J., Cole, D., Nisler, A., and Mahon, B. E. (2017). Poultry: the most common food in outbreaks with known pathogens, United States, 1998-2012. Epidemiol. Infect. 145, 316–325. doi: 10.1017/S0950268816002375
Clothier, K. A., Kim, P., Mete, A., and Hill, A. E. (2018). Frequency, serotype distribution, and antimicrobial susceptibility patterns of Salmonella in small poultry flocks in California. J. Vet. Diagn. Investig. 30, 471–475. doi: 10.1177/1040638718755418
Cockerill, F. (2011). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement, Vol. 31. Wayne, PA: CLSI, 1–172.
Cui, Y., Guran, H. S., Harrison, M. A., Hofacre, C. L., and Alali, W. Q. (2015). Salmonella levels in turkey neck skins, drumstick bones, and spleens in relation to ground turkey. J. Food Prot. 78, 1945–1953. doi: 10.4315/0362-028X.JFP-15-240
Doménech, E., Jiménez-Belenguer, A., Pérez, R., Ferrús, M. A., and Escriche, I. (2015). Risk characterization of antimicrobial resistance of Salmonella in meat products. Food Control 57, 18–23. doi: 10.1016/j.foodcont.2015.04.001
Erol, I., Goncuoglu, M., Ayaz, N. D., Ellerbroek, L., Bilir Ormanci, F. S., and Iseri Kangal, O. (2013). Serotype distribution of Salmonella isolates from turkey ground meat and meat parts. Biomed Res. Int. 2013:281591. doi: 10.1155/2013/281591
Evans, N. P., Evans, R. D., Regalado, J., Sullivan, J. F., Dutta, V., Elvinger, F., et al. (2015). Preharvest Salmonella detection for evaluation of fresh ground poultry product contamination. J. Food Prot. 78, 1266–1271. doi: 10.4315/0362-028X.JFP-14-509
Fakhr, M. K., McEvoy, J. M., Sherwood, J. S., and Logue, C. M. (2006a). Adding a selective enrichment step to the iQ-CheckTM real-time PCR improves the detection of Salmonella in naturally contaminated retail turkey meat products. Lett. Appl. Microbiol. 43, 78–83. doi: 10.1111/j.1472-765X.2006.01903.x
Fakhr, M. K., Sherwood, J. S., Thorsness, J., and Logue, C. M. (2006b). Molecular characterization and antibiotic resistance profiling of Salmonella isolated from retail Turkey meat products. Foodborne Pathog. Dis. 3, 366–374. doi: 10.1089/fpd.2006.3.366
Fakhr, M. K., Nolan, L. K., and Logue, C. M. (2005). Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar typhimurium. J. Clin. Microbiol. 43, 2215–2219. doi: 10.1128/JCM.43.5.2215-2219.2005
Florez-Cuadrado, D., Moreno, M. A., Ugarte-Ruíz, M., and Domínguez, L. (2018). Antimicrobial resistance in the food chain in the European union. Adv. Food Nutr. Res. 86, 115–136. doi: 10.1016/bs.afnr.2018.04.004
Foley, S. L., and Lynne, A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86(Suppl. 14), E173–E187. doi: 10.2527/jas.2007-0447
Foley, S. L., Lynne, A. M., and Nayak, R. (2009). Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect. Genet. Evol. 9, 430–440. doi: 10.1016/j.meegid.2009.03.004
Foley, S. L., White, D. G., McDermott, P. F., Walker, R. D., Rhodes, B., Fedorka-Cray, P. J., et al. (2006). Comparison of subtyping methods for differentiating Salmonella enterica serovar typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 44, 3569–3577. doi: 10.1128/JCM.00745-06
Folster, J. P., Pecic, G., McCullough, A., Rickert, R., and Whichard, J. M. (2011). Characterization of bla CMY -encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog. Dis. 8, 1289–1294. doi: 10.1089/fpd.2011.0944
Folster, J. P., Pecic, G., Rickert, R., Taylor, J., Zhao, S., Fedorka-Cray, P. J., et al. (2012). Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground Turkey-associated outbreak in the United States in 2011. Antimicrob. Agents Chemother. 56, 3465–3466. doi: 10.1128/AAC.00201-12
Frye, J. G., and Jackson, C. R. (2013). Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front. Microbiol. 4:135. doi: 10.3389/fmicb.2013.00135
García, P., Hopkins, K. L., García, V., Beutlich, J., Mendoza, M. C., Threlfall, J., et al. (2014). Diversity of plasmids encoding virulence and resistance functions in Salmonella enterica subsp. enterica serovar typhimurium monophasic variant 4,[5],12:i: strains circulating in Europe. PLoS One 9:e89635. doi: 10.1371/journal.pone.0089635
Gaul, S. B., Wedel, S., Erdman, M. M., Harris, D. L., Harris, I. T., Ferris, K. E., et al. (2007). Use of pulsed-field gel electrophoresis of conserved XbaI fragments for identification of swine Salmonella serotypes. J. Clin. Microbiol. 45, 472–476. doi: 10.1128/JCM.00962-06
Gieraltowski, L., Higa, J., Peralta, V., Green, A., Schwensohn, C., Rosen, H., et al. (2016). National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 11:e0162369. doi: 10.1371/journal.pone.0162369
Hoffmann, M., Zhao, S., Pettengill, J., Luo, Y., Monday, S. R., Abbott, J., et al. (2014). Comparative genomic analysis and virulence differences in closely related Salmonella enterica serotype Heidelberg isolates from humans, retail meats, and animals. Genome Biol. Evol. 6, 1046–1068. doi: 10.1093/gbe/evu079
Iwamoto, M., Reynolds, J., Karp, B. E., Tate, H., Fedorka-Cray, P. J., Plumblee, J. R., et al. (2017). Ceftriaxone-resistant nontyphoidal Salmonella from humans, retail meats, and food animals in the United States, 1996–2013. Foodborne Pathog. Dis. 14, 74–83. doi: 10.1089/fpd.2016.2180
Jones, C., and Stanley, J. (1992). Salmonella plasmids of the pre-antibiotic era. J. Gen. Microbiol. 138, 189–197. doi: 10.1099/00221287-138-1-189
Kinross, P., van Alphen, L., Martinez Urtaza, J., Struelens, M., Takkinen, J., Coulombier, D., et al. (2014). Multidisciplinary investigation of a multicountry outbreak of Salmonella stanley infections associated with Turkey meat in the European Union, August 2011 to January 2013. Euro Surveill. 19:20801. doi: 10.2807/1560-7917.ES2014.19.19.20801
Li, Q., Skyberg, J. A., Fakhr, M. K., Sherwood, J. S., Nolan, L. K., and Logue, C. M. (2006). Antimicrobial susceptibility and characterization of Salmonella isolates from processed bison carcasses. Appl. Environ. Microbiol. 72, 3046–3049. doi: 10.1128/AEM.72.4.3046-3049.2006
Maka,Ł., Maćkiw, E., Ściezyńska, H., Pawłowska, K., and Popowska, M. (2014). Antimicrobial susceptibility of Salmonella strains isolated from retail meat products in Poland between 2008 and 2012. Food Control 36, 199–204. doi: 10.1016/j.foodcont.2013.08.025
Marasini, D., Abo-Shama, U. H., and Fakhr, M. K. (2016a). Complete genome sequences of Salmonella enterica serovars anatum and anatum var. 15 +, isolated from retail ground turkey. Genome Announc. 4:e01619-15. doi: 10.1128/genomeA.01619-15
Marasini, D., Abo-Shama, U. H., and Fakhr, M. K. (2016b). Whole-genome sequencing of Salmonella enterica subsp. enterica serovar Ouakam isolated from ground turkey. Genome Announc. 4:e01618-15. doi: 10.1128/genomeA.01618-15
Marasini, D., and Fakhr, M. (2014). Exploring PFGE for detecting large plasmids in Campylobacter jejuni and Campylobacter coli isolated from various retail meats. Pathogens 3, 833–844. doi: 10.3390/pathogens3040833
Mayer, L. W. (1988). Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin. Microbiol. Rev. 1, 228–243. doi: 10.1128/CMR.1.2.228
Nair, D. V. T., and Johny, A. K. (2017). Food grade Pimenta leaf essential oil reduces the attachment of Salmonella enterica Heidelberg (2011 Ground Turkey Outbreak Isolate) on to Turkey Skin. Front. Microbiol. 8:2328. doi: 10.3389/fmicb.2017.02328
Nair, D. V. T., Thomas, J. V., Noll, S., Porter, R., and Johny, A. K. (2018). Effect of various inoculum levels of multidrug-resistant Salmonella enterica serovar Heidelberg (2011 Ground Turkey Outbreak Isolate) on cecal colonization, dissemination to internal organs, and deposition in skeletal muscles of commercial Turkeys after e. Front. Microbiol. 8:2680. doi: 10.3389/fmicb.2017.02680
Nde, C. W., Fakhr, M. K., Doetkott, C., and Logue, C. M. (2008). An evaluation of conventional culture, invA PCR, and the real-time PCR iQ-check kit as detection tools for Salmonella in naturally contaminated premarket and retail turkey. J. Food Prot. 71, 386–391. doi: 10.4315/0362-028X-71.2.386
Nisar, M., Kassem, I. I., Rajashekara, G., Goyal, S. M., Lauer, D., Voss, S., et al. (2017). Genotypic relatedness and antimicrobial resistance of Salmonella Heidelberg isolated from chickens and turkeys in the midwestern United States. J. Vet. Diagn. Investig. 29, 370–375. doi: 10.1177/1040638717690784
Noormohamed, A., and Fakhr, M. (2013). A higher prevalence rate of campylobacter in retail beef livers compared to other beef and pork meat cuts. Int. J. Environ. Res. Public Health 10, 2058–2068. doi: 10.3390/ijerph10052058
Noormohamed, A., and Fakhr, M. K. (2012). Incidence and antimicrobial resistance profiling of campylobacter in retail chicken livers and gizzards. Foodborne Pathog. Dis. 9, 617–624. doi: 10.1089/fpd.2011.1074
Noormohamed, A., and Fakhr, M. K. (2014). Prevalence and antimicrobial susceptibility of Campylobacter spp. in Oklahoma conventional and organic retail poultry. Open Microbiol. J. 8, 130–137. doi: 10.2174/1874285801408010130
Paudyal, N., Pan, H., Li, X., Fang, W., and Yue, M. (2018). Antibiotic resistance in Salmonella enteritidis isolates recovered from chicken, chicken breast, and humans through national antimicrobial resistance monitoring system between 1996 and 2014. Foodborne Pathog. Dis. doi: 10.1089/fpd.2017.2402 [Epub ahead of print].
Rahn, K., De Grandis, S. A., Clarke, R. C., McEwen, S. A., Galán, J. E., Ginocchio, C., et al. (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6, 271–279. doi: 10.1016/0890-8508(92)90002-F
Rotgers, R., and Casadesüs, J. (1999). The virulence plasmids of Salmonella. Int. Microbiol. 2, 177–184. doi: 10.2436/im.v2i3.9210
Rychlik, I., Gregorova, D., and Hradecka, H. (2006). Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112, 1–10. doi: 10.1016/j.vetmic.2005.10.030
Sajid, S. U., and Schwarz, S. (2009). Plasmid fingerprinting and virulence gene detection among indigenous strains of Salmonella enterica serovar Enteritidis. J. Ayub Med. Coll. 21, 83–86.
Sallam, K. I., Mohammed, M. A., Hassan, M. A., and Tamura, T. (2014). Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Control 38, 209–214. doi: 10.1016/j.foodcont.2013.10.027
Sanad, Y. M., Johnson, K., Park, S. H., Han, J., Deck, J., Foley, S. L., et al. (2016). Molecular characterization of Salmonella enterica serovars isolated from a turkey production facility in the absence of selective antimicrobial pressure. Foodborne Pathog. Dis. 13, 80–87. doi: 10.1089/fpd.2015.2002
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Sharma, C. S., Dhakal, J., and Nannapaneni, R. (2015). Efficacy of lytic bacteriophage preparation in reducing Salmonella in vitro, on turkey breast cutlets, and on ground turkey. J. Food Prot. 78, 1357–1362. doi: 10.4315/0362-028X.JFP-14-585
Sjölund-Karlsson, M., Howie, R., Rickert, R., Krueger, A., Tran, T. T., Zhao, S., et al. (2010). Plasmid-mediated quinolone resistance among non-typhi Salmonella enterica isolates, USA. Emerg. Infect. Dis. 16, 1789–1791. doi: 10.3201/eid1611.100464
Soufi, L., Sáenz, Y., de Toro, M., Salah Abbassi, M., Rojo-Bezares, B., Vinué, L., et al. (2012). Phenotypic and genotypic characterization of Salmonella enterica recovered from poultry meat in Tunisia and identification of new genetic traits. Vector Borne Zoonotic Dis. 12, 10–16. doi: 10.1089/vbz.2011.0667
Tafida, S. Y., Kabir, J., Kwaga, J. K. P., Bello, M., Umoh, V. J., Yakubu, S. E., et al. (2013). Occurrence of Salmonella in retail beef and related meat products in Zaria, Nigeria. Food Control 32, 119–124. doi: 10.1016/j.foodcont.2012.11.005
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Thai, T. H., Hirai, T., Lan, N. T., and Yamaguchi, R. (2012). Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. 156, 147–151. doi: 10.1016/j.ijfoodmicro.2012.03.016
Van, T. T. H., Nguyen, H. N. K., Smooker, P. M., and Coloe, P. J. (2012). The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia. Int. J. Food Microbiol. 154, 98–106. doi: 10.1016/j.ijfoodmicro.2011.12.032
White, D. G., Zhao, S., Sudler, R., Ayers, S., Friedman, S., Chen, S., et al. (2001). The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 345, 1147–1154. doi: 10.1056/NEJMoa010315
Wittum, T. E. (2012). The challenge of regulating agricultural ceftiofur use to slow the emergence of resistance to extended-spectrum cephalosporins. Appl. Environ. Microbiol. 78, 7819–7821. doi: 10.1128/AEM.01967-12
Yang, B., Qu, D., Zhang, X., Shen, J., Cui, S., Shi, Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 141, 63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015
Yang, B., Wang, Q., Cui, S., Wang, Y., Shi, C., Xia, X., et al. (2014). Characterization of extended-spectrum beta-lactamases-producing Salmonella strains isolated from retail foods in Shaanxi and Henan Province, China. Food Microbiol. 42, 14–18. doi: 10.1016/j.fm.2014.02.003
Yildirim, Y., Gonulalan, Z., Pamuk, S., and Ertas, N. (2011). Incidence and antibiotic resistance of Salmonella spp. on raw chicken carcasses. Food Res. Int. 44, 725–728. doi: 10.1016/j.foodres.2010.12.040
Zhao, S., Dermott, P. F. M. C., Friedman, S., Abbott, J., Ayers, S., Glenn, A., et al. (2006). Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog. Dis. 3, 106–117. doi: 10.1089/fpd.2006.3.106
Zhao, S., White, D. G., Friedman, S. L., Glenn, A., Blickenstaff, K., Ayers, S. L., et al. (2008). Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl. Environ. Microbiol. 74, 6656–6662. doi: 10.1128/AEM.01249-08
Zou, W., Chen, H. C., Hise, K. B., Tang, H., Foley, S. L., Meehan, J., et al. (2013). Meta-analysis of pulsed-field gel electrophoresis fingerprints based on a constructed Salmonella database. PLoS One 8:e59224. doi: 10.1371/journal.pone.0059224
Keywords: Salmonella, serotyping, antimicrobial resistance, PFGE, plasmids, ground poultry
Citation: Gad AH, Abo-Shama UH, Harclerode KK and Fakhr MK (2018) Prevalence, Serotyping, Molecular Typing, and Antimicrobial Resistance of Salmonella Isolated From Conventional and Organic Retail Ground Poultry. Front. Microbiol. 9:2653. doi: 10.3389/fmicb.2018.02653
Received: 15 June 2018; Accepted: 17 October 2018;
Published: 05 November 2018.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Jianmin Zhang, South China Agricultural University, ChinaCopyright © 2018 Gad, Abo-Shama, Harclerode and Fakhr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed K. Fakhr, bW9oYW1lZC1mYWtockB1dHVsc2EuZWR1
†Present address: Ahmed H. Gad, Northeastern State University, Broken Arrow, OK, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.