- 1College of Science and Engineering, James Cook University, Townsville, QLD, Australia
- 2Australian Institute of Marine Science, Townsville, QLD, Australia
- 3AIMS@JCU, Townsville, QLD, Australia
- 4ARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD, Australia
- 5Research School of Earth Sciences, The Australian National University, Canberra, ACT, Australia
- 6Department of Environmental Science and Policy, George Mason University, Fairfax, VA, United States
Ocean acidification (OA) as a result of increased anthropogenic CO2 input into the atmosphere carries consequences for all ocean life. Low pH can cause a shift in coral-associated microbial communities of pCO2-sensitive corals, however, it remains unknown whether the microbial community is also influenced in corals known to be more tolerant to high pCO2/low pH. This study profiles the bacterial communities associated with the tissues of the pCO2-tolerant coral, massive Porites spp., from two natural CO2 seep sites in Papua New Guinea. Amplicon sequencing of the hypervariable V3-V4 regions of the 16S rRNA gene revealed that microbial communities remained stable across CO2 seep sites (pH = 7.44–7.85) and adjacent control sites (ambient pH = 8.0–8.1). Microbial communities were more significantly influenced by reef location than pH, with the relative abundance of dominant microbial taxa differing between reefs. These results directly contrast with previous findings that increased CO2 has a strong effect on structuring microbial communities. The stable structure of microbial communities associated with the tissues of massive Porites spp. under high pCO2/low pH conditions confirms a high degree of tolerance by the whole Porites holobiont to OA, and suggest that pH tolerant corals such as Porites may dominate reef assemblages in an increasingly acidic ocean.
Introduction
Increasing levels of atmospheric carbon dioxide from anthropogenic emissions have led to higher CO2 concentrations in the surface seawaters of the oceans, changing seawater chemistry (Caldeira and Wickett, 2003; Doney et al., 2009). The chemical reaction between CO2 and water releases hydrogen ions (H+), thereby reducing pH (Doney et al., 2009). Seawater pH is defined by the negative base-10 logarithm of H+, thus a near 30% increase in H+ since the industrial revolution equates to a decrease of 0.1 pH units in global surface seawater (Rhein et al., 2013). Consequently, over the last 200 years, the average pH of global surface seawater has been reduced by ∼0.1 units to ∼8.1, and based on current CO2 emissions modeling, pH is predicted to decrease by a further 0.2–0.4 units by 2100. This decrease will lead to changes in ocean chemistry not seen for the past 23 million years (Rhein et al., 2013). Although seawater pH fluctuates (especially on coral reefs) with season, depth and productivity (Joint et al., 2011), the reduction in baseline pH levels, termed ocean acidification (OA), represents a growing concern for global ocean ecosystems (O’Brien et al., 2016).
Coral reefs have been a focal point for understanding the effects of OA, as skeletal growth (i.e., calcification) of scleractinian (reef-building) corals is progressively impeded at low pH (Gattuso et al., 1998). Reduced calcification in reef-building species poses a serious long-term threat to the future of coral reefs globally. A coral colony represents a meta-organism, a complex and interdependent association between a coral animal, endosymbiotic dinoflagellates of the family Symbiodiniaceae (LaJeunesse et al., 2018), and diverse members of the Bacteria, Archaea, Fungi, Protista, Apicomplexa and viruses (Rohwer et al., 2002; Bourne et al., 2016). Members of this inter-kingdom partnership are collectively referred to as the coral holobiont. The roles of bacteria within complex partnerships are becoming apparent, and it is now widely accepted that microbes in general are critical to the health and survival of multicellular organisms such as the coral holobiont (Rosenberg et al., 2007; McFall-Ngai et al., 2013).
Bacteria perform essential functions for the health of the coral host. For example, nitrogen-fixing bacteria in coral tissues are believed to help sustain the coral-Symbiodiniaceae relationship (Lesser et al., 2004; Lema et al., 2012), while carbon-fixing bacteria may provide an additional energy source when Symbiodiniaceae densities are low (Fine and Loya, 2002; Rosenberg et al., 2007). Antibiotic properties of bacteria within coral mucus are important for structuring coral-associated microbial communities, ensuring habitat niches for beneficial bacteria and preventing invasion by potentially opportunistic or pathogenic microorganisms (Ritchie, 2006). Metabolism of dimethylsulfoniopropionate (DMSP), which is produced by both the coral host and Symbiodiniaceae, provides antioxidants that protect coral tissues against harmful reactive oxygen species (Sunda et al., 2002; Raina et al., 2013). Under ideal conditions, microorganisms form communities that benefit the host animal through nutrient cycling and disease prevention. However, stressful environmental conditions can alter microbial community composition, which can negatively affect the fitness and survival of the holobiont.
Disruptions to microbial community composition and function are thought to adversely affect coral host fitness, and have been documented in response to a variety of environmental stressors, including temperature, eutrophication, pollution, and pH (Kline et al., 2006; Bourne et al., 2008; Webster et al., 2013, 2016; Ziegler et al., 2016). In general, perturbations resulting in dysbiotic community states are stochastic rather than deterministic, resulting in a variable and unstable microbiome (the aptly named Anna Karenina principle) (Zaneveld et al., 2017). In response to OA, aquaria-based studies have demonstrated that coral microbial communities generally shift toward an unhealthy state in response to low pH (Thurber et al., 2009; Meron et al., 2011). However, research has primarily focused on coral species that are sensitive to changes in pH (Fabricius et al., 2011; Strahl et al., 2015), and/or involve pH levels that are beyond what is predicted for the coming century (Thurber et al., 2009). Studies conducted in situ, where pH levels are more representative of future conditions, have produced variable results. For example, no change in coral bacterial communities was detected following translocation of two Mediterranean coral species along a natural pH gradient in the Gulf of Naples (Meron et al., 2012). Likewise, the endolithic community associated with massive Porites spp. did not change significantly between ambient and low pH sites found at naturally occurring CO2 seeps in Papua New Guinea (PNG) (Marcelino et al., 2017). Conversely, large shifts in the structure of tissue-associated bacterial communities were found in the corals Acropora millepora and Porites cylindrica across the same CO2 seep in PNG (Morrow et al., 2014).
Natural volcanic CO2 seeps have been used as experimental settings to assess the long-term effects of OA on marine organisms in situ due to their naturally high pCO2 levels (Hall-Spencer et al., 2008; Fabricius et al., 2011, 2014; Morrow et al., 2014). Within the D’Entrecasteaux Islands of the Milne Bay province of PNG, coral reefs are found within volcanic seeps that release 99% pure CO2 into the surrounding seawater. At these seeps, median pH levels range from 7.28–8.01 (Fabricius et al., 2014) and are within the range of “business-as-usual” predictions for the coming century (Rhein et al., 2013). Benthic invertebrate community structure, abundance and diversity are dramatically different at these seeps compared with nearby control sites exposed to the same temperatures, currents, geomorphology and light levels (Fabricius et al., 2011, 2014; Morrow et al., 2014). While the mechanism driving survival of communities at high pCO2 sites is unclear, evidence suggests that host-associated microbial communities resilient to low pH enable host fitness and survival (Morrow et al., 2014).
Some coral species, such as species of the genus Porites with massive morphology, appear tolerant to high pCO2 and are present in high abundance within the seep environments (Fabricius et al., 2011). While this coral maintains its physiological performance under low pH conditions (Edmunds et al., 2012; Strahl et al., 2015), and contains a stable endolithic community (Marcelino et al., 2017), it remains unknown whether microbial communities associated with the tissues of this pCO2-tolerant coral are impacted by low pH conditions. This study profiled the bacterial communities of massive Porites spp. at two CO2 seeps in PNG and compared microbial community profiles to corals from adjacent control sites. Through this comparison, we determined that these pCO2-tolerant corals can maintain more stable microbial communities than their pCO2-sensitive counterparts.
Materials and Methods
Study Site and Sample Collection
A total of 15 samples (n = 3 per site) were collected at two natural volcanic CO2 seep sites; Illi Illi near Upa-Upasina (herewith Illi), Normanby Island, and Dobu, Dobu Island, in the D’Entrecasteaux Islands, Milne Bay Province, Papua New Guinea (Figure 1). Seeps at both Illi (9.7625S, 150.8186E) and Dobu (9.7366S, 150.8691E) expel 99% pure CO2 into the water column, resulting in median ambient pH levels of 7.77 and 7.73, respectively. These pH levels are consistent with predictions for ocean chemistry over the next century. Control sites were less than 500 m away and have similar seawater temperature and salinity (Fabricius et al., 2011, 2014), with median ambient pH levels of 7.97 and 8.02 at Illi and Dobu, respectively (Fabricius et al., 2011).
FIGURE 1. Study site locations in Papua New Guinea (adapted from Fabricius et al., 2011).
Coral samples were collected on SCUBA at a depth of 3–5 m using a hammer and chisel in November 2014. Samples were collected from 3 colonies of massive Porites spp. at each of the two seeps and corresponding control sites, and at a second control site at Dobu. Samples were brought to the surface and briefly kept in running seawater (1.0 μm filtered) until being processed (<2 h). Coral tissues were rinsed with 0.22 μm filtered seawater and then removed from the skeleton using pressurized air (∼130–150 PSI) from a blow gun with sterile tips into a sterile bag (Whirlpak®, Nasco). Tissue blastate (2 mL) was pelleted in a fixed-angle centrifuge (20,000 × g for 20 min at 4°C; Beckman-Coulter, Brea, CA, United States), seawater supernatant decanted, tissue pellets snap frozen in liquid nitrogen, and stored at −80°C.
Coral Cover and Diversity
Coral cover and diversity were estimated at each site by placing five replicate 50 × 2 m belt transects parallel to shore at a depth of 3–7 m. A total of 25 photographs of 1 × 1 m quadrats were taken on alternating sides of the transect every 2 m along each transect. Photographs were analyzed using photoQuad (Trygonis and Sini, 2012) by overlaying 50 random points over the image and identifying the underlying coral to genus. Shannon’s diversity index (H) was calculated based on the number of coral genera for each transect. To determine if coral cover, the cover of massive Porites, and diversity varied significantly between seep and control sites, a generalized linear mixed effect model was performed using the package MASS (Venables and Ripley, 2002) in R Studio (R Core Team, 2016). The model nested seeps within reefs, and had transects as random factors, and a Poisson distribution was used for the percentage cover data.
Sample Processing and Sequencing of 16S rRNA Gene
Samples were thawed at 4°C and DNA was extracted using the PowerSoil DNA Isolation Kit, following the manufacturer’s instructions (MoBio Laboratories, Inc; Carlsbad, CA, United States), except a 10 min 65°C heating step was added prior to bead beating. Sequencing was completed at the Molecular Research DNA laboratory (MR DNA; Shallowater, TX, United States). The hypervariable V3-V4 regions were targeted using the 16S rRNA gene primer set, 341F (CCTACGGGNGGCWGCAG) and 785R (GACTACHVGGGTATCTAATCC) (Klindworth et al., 2013), in a 30 cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, United States) under the following conditions: 94°C for 3 min, 28 cycles of 94°C for 30 s, 53°C for 40 s and 72°C for 1 min, followed by a final elongation at 72°C for 5 min. After amplification, samples were pooled in equal proportions based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads and then used to prepare a DNA library by following the Illumina TruSeq DNA library preparation protocol. Sequencing was performed on the Illumina MiSeq platform (2 × 300 bp) following the manufacturer’s guidelines.
16S rRNA Gene Amplicon Analysis
Sequence reads of all samples were processed using QIIME2 (Caporaso et al., 2010). In summary, reads were demultiplexed and barcodes and primer regions were removed. Reads were filtered for quality and chimeric sequences using dada2 (Callahan et al., 2016). Taxonomic classification was assigned using a naïve Bayes classifier trained on the extracted region of interest from the SILVA 16S rRNA reference alignment [Release 132 (Quast et al., 2013)]. All sequences classified as chloroplast, mitochondria, or eukaryota were removed. The resulting amplicon sequence variant (ASV) table was used for statistical analysis in R Studio (R Core Team, 2016) using the packages vegan, car, VennDiagram, pheatmap, and IndicSpecies (De Caceres and Legendre, 2009; Chen and Boutros, 2011; Fox and Weisberg, 2011; Kolde, 2015; Oksanen et al., 2016).
The percent relative abundance of each microbial phylum present in each sample was calculated, and the top ten most abundant taxa were plotted. All additional analyses were conducted at the genus level. Alpha diversity was calculated using Shannon’s diversity index. Sequence count data were standardized with a Wisconsin double-standardization [i.e., each value is standardized by the column maximum followed by the row total (Bray and Curtis, 1957)]. Patterns in microbial community composition were visualized using non-parametric multidimensional scaling (NMDS) based on Bray–Curtis dissimilarity. Permutational multivariate analysis of variance (PERMANOVA) was used to identify differences in microbial community diversity as a function of location and CO2 treatment.
Core microbial taxa were identified as the genera occurring in 100% of samples across all sites. Indicator taxa were identified for each treatment (i.e., seep and control) and each location (i.e., Dobu and Illi) using the R package IndicSpecies (De Caceres and Legendre, 2009). Each microbial taxon was tested for significant correlation using 999 permutations. Sequence counts for the resulting indicator species were standardized to the number of total sequences per sample, and heatmaps were created to visualize relative abundance.
Modeling pH Using 14C
We utilized the 14C content in both the control and seep corals to estimate the seawater pH. Approximately 7 mg of coral powder was weighed into VacutainerTM blood vials. The samples were acidified under vacuum with 0.3 ml of 85% orthophosphoric acid at 70°C to evolve CO2 gas. The CO2 gas was cryogenically purified and converted to graphite using H2/Fe (Vogel et al., 1984). The 14C of the resulting graphite was measured on a Single Stage Accelerator Mass Spectrometer at the Australian National University (Fallon et al., 2010). Analytical results are reported as F14C using the Oxalic Acid-I, corrected for d13C and background subtracted using Carrera Marble (Supplementary Table S1 and Supplementary Equations). The pH values for seawater were modeled using CO2SYS (Pierrot et al., 2006) from in situ alkalinity and salinity measurements, high resolution IGOSS satellite temperature records and the calculated impacted dissolved inorganic carbon (DIC).
Results
Coral Cover and Diversity
There was no significant difference in total coral cover between seep and control sites (GLM; x2 = 2.6(1), p = 0.11; Figure 2A and Supplementary Figure S3). However, there was an increase in coral cover at the Dobu seep (37.93%) compared to the Dobu control sites (26.06%), largely due to a fivefold increase in cover of massive Porites spp., from 4.13% at the Dobu control site to 21.41% at Dobu seep. At Illi, percent cover of massive Porites spp. also increased from 7.89% cover at the control site to 19.28% at the seep. Across both reefs, percent cover of massive Porites spp. was significantly higher at the seep compared to the control sites (GLMM; x2 = 49.0(1), p < 0.001; Figure 2A). As a result, coral community diversity was significantly lower at the seep compared to the control sites at both reefs (ANOVA, F(2,17) = 7.356, p = 0.005; Figure 2B).
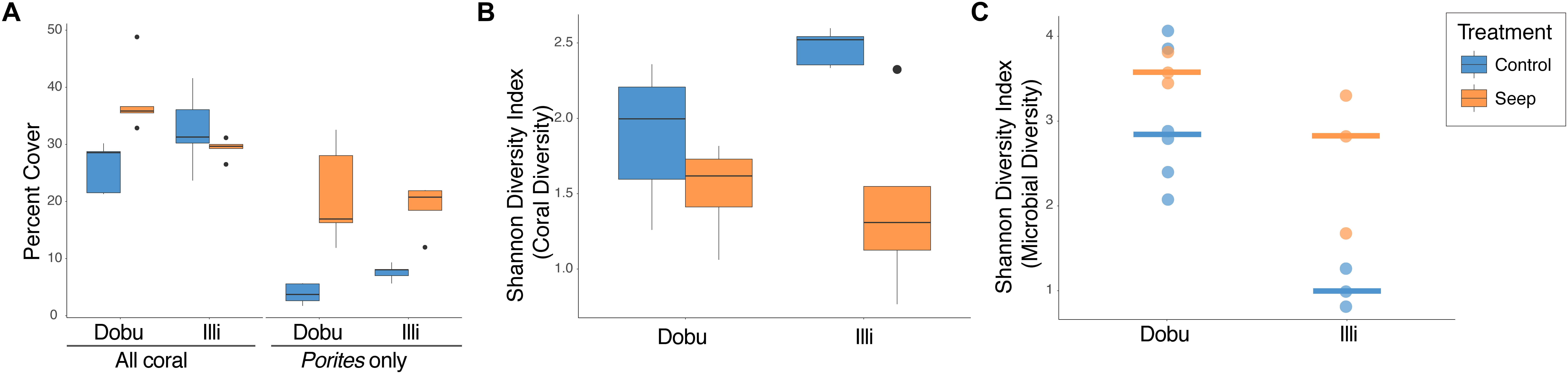
FIGURE 2. Percent total coral cover and massive Porites spp. cover (A); Shannon’s diversity index for scleractinian corals (B); and Shannon’s diversity index for microbial communities associated with massive Porites spp. (C) at control and seep sites at Dobu and Illi reefs.
Bacteria Community Diversity Comparisons
A total of 875,140 sequence reads were recovered from all 15 samples, though following filtering, only 119,332 high quality reads were subsequently used for taxonomic classification. A total of 537 bacterial genera (comprised of a total 683 sequence variants) were detected across all samples. Rarefaction analysis confirmed sampling depth was sufficient to estimate the total diversity of each sample, with species detection curves reaching an asymptote after a sequencing depth of approximately 450 reads (Supplementary Figure S1). Shannon’s diversity index of microbial communities associated with massive Porites spp. showed a significant interaction between locations and treatments (ANOVA; F(3,11) = 8.991, p = 0.003). Interestingly, mean alpha diversity was significantly lower at the Illi control reef (1.03 ± 0.13 SE) than the Illi seep (2.61 ± 0.48 SE; Figure 2C). At Dobu, microbial diversity was similar at control and seep sites (3.02 ± 0.32 and 3.62 ± 0.11, respectively; Figure 2C). Across all samples, community composition was significantly different between the two reefs (PERMANOVA; R2 = 0.19, p = 0.001; Figure 3A), but there was no significant difference between seep and control sites (PERMANOVA; R2 = 0.08, p = 0.23; Figure 3B). These results suggest that the greatest influence on massive Porites spp. microbial communities was the location of the reef, while the effect of higher pCO2 at the seeps had little influence at either reef. The samples from Dobu were not tightly clustered, suggesting a high degree of variability in microbial community composition (Figure 3). Conversely, the Illi samples were more tightly clustered, suggesting greater similarity in community composition.
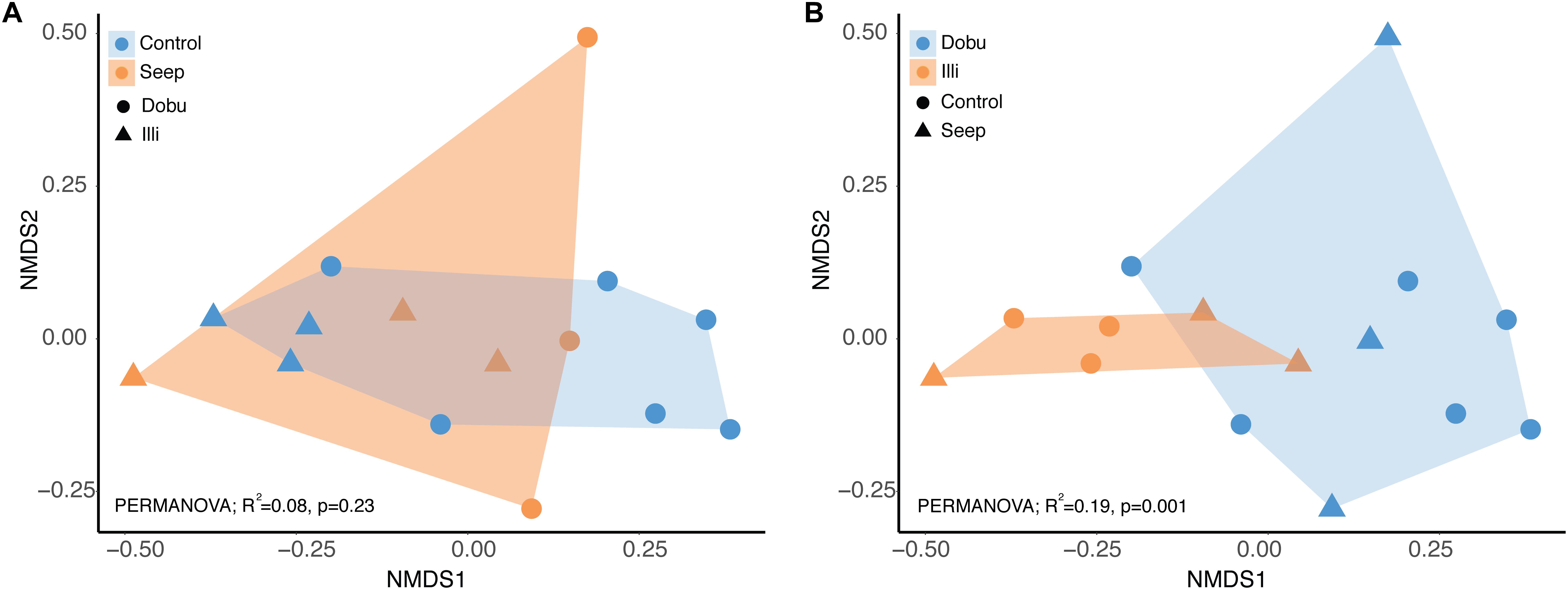
FIGURE 3. Microbial community composition visualized using NMDS. Two colorations of the same plot are shown to visualize the effect of treatment (A) or reef (B).
A total of 37 microbial phyla were identified within the 16S rRNA dataset. Sequences affiliated with the phylum Proteobacteria were the most abundant across all samples at both the seep and control sites, representing >93% of sequences from the Illi control site and between 29 and 82% of sequences from the Dobu control site (mean 94.7 ± 0.6 and 56.9 ± 9.6%, respectively; Figures 4, 5 and Supplementary Table S2). Two replicate samples from the Dobu control site with lower relative abundance of Proteobacteria displayed higher abundance (up to 45%) of sequences from the phylum Chlorobi (i.e., green sulfur bacteria; Figure 5). In general, both Illi and Dobu seep sites showed a lower abundance of Proteobacteria compared to the Illi control reef (Figures 4, 5). No trend was observed in the relative abundance of Proteobacteria between the Dobu control and seep sites (Figures 4, 5).
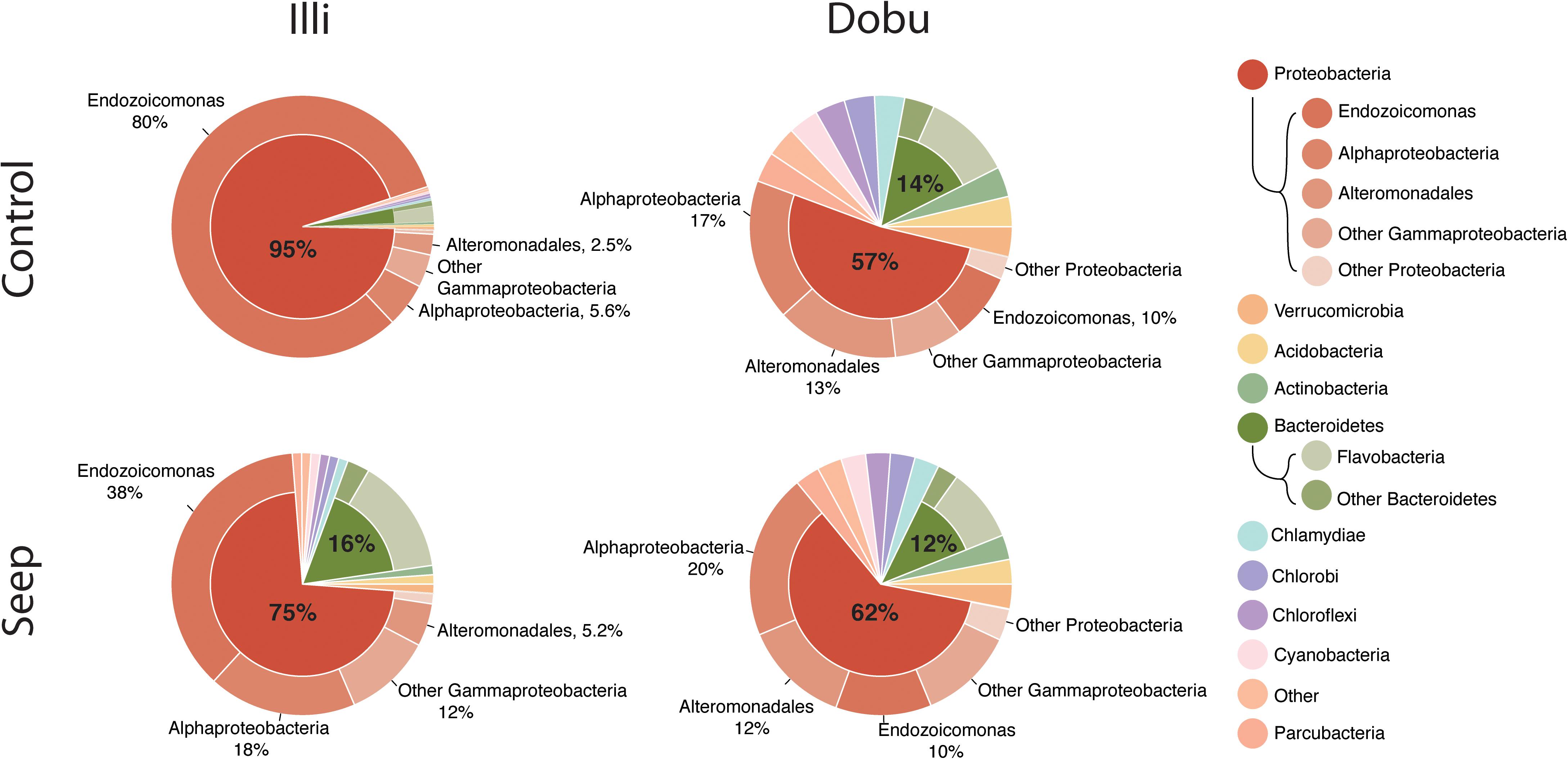
FIGURE 4. Ten most abundant microbial phyla associated with massive Porites. “Other” represents all microbes not in the top 10. The two most abundant phyla, Proteobacteria and Bacteroidetes, are split further into classes. Gammaproteobacteria are split further to illustrate the high relative abundance of the genus Endozoicomonas (outside ring). Percentages are mean proportion of total sequences.
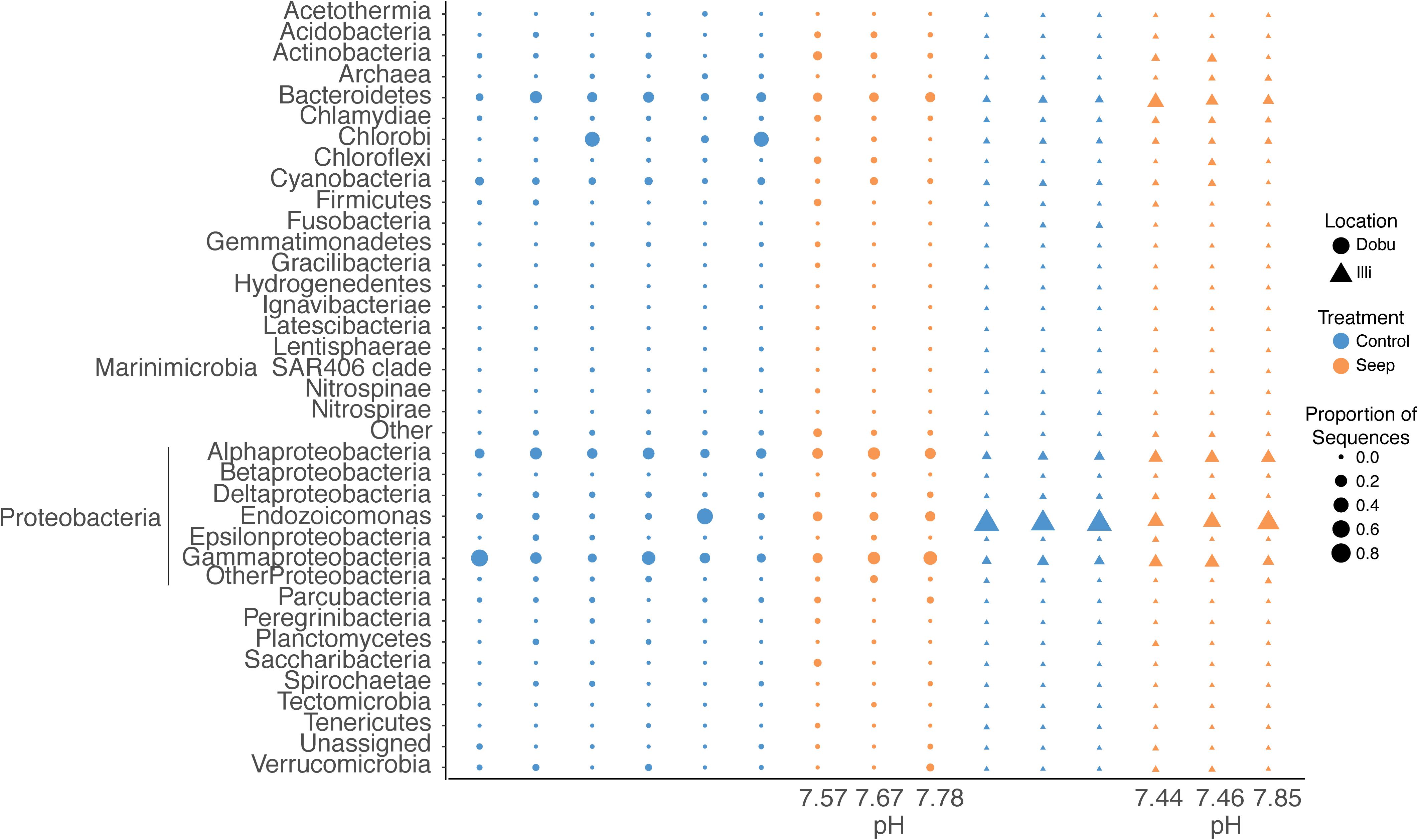
FIGURE 5. Relative abundance of the 37 microbial phyla associated with massive Porites spp. at seep and control sites at Illi and Dobu reefs. “Other” represents all that could not be classified below Kingdom level. “Archaea” represents all sequences affiliated to this domain. “Unassigned” represents all sequences that could not be assigned taxonomy. Sequences affiliated with Proteobacteria are split into classes (Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria, and Gammaproteobacteria) and genus (Endozoicomonas) to display higher resolution of dominant taxa associated with this phylum. Values below each column represent the pH value of each coral skeleton collected from the seep sites as analyzed by 14C chemistry.
Within the Proteobacteria, sequences associated with the genus Endozoicomonas and family Alteromonadaceae (class Gammaproteobacteria) and class Alphaproteobacteria made up the vast majority in all samples. At the Illi control site, all 3 samples were dominated by members affiliated with Endozoicomonas, comprising an average of 79.6 ± 2.9% of all taxa present (Figures 4, 5 and Supplementary Table S2). Members of the Endozoicomonas were also the dominant taxa at the Illi seep, however, their abundance was lower at 38.3 ± 10.2% of all sequences. At Dobu, the relative abundance of Endozoicomonas sequences was reduced compared to Illi (Figures 4, 5), but was similar across seep and control sites (10.4 ± 1.4 and 10.1 ± 7.7% of sequences, respectively).
Where relative abundance of Endozoicomonas was low, the relative abundance of sequences affiliated with the order Alteromonadales was generally higher. The Dobu control sites contained the lowest relative abundance of Endozoicomonas, and subsequently the greatest relative abundance of sequences related to Alteromonadales, with a mean of 13.4 ± 4.8% (ranging from 3–35.6%), compared to 11.6 ± 4.2% at the seep. Conversely, the seep site comprised slightly more sequences related to Alphaproteobacteria, with a mean of 20.4 ± 3%, compared to 17 ± 2.3% at the control. The Illi seep site contained a higher relative abundance of Alphaproteobacteria than the control, with a mean of 18.3 ± 0.4% compared to 5.6 ± 0.5%, respectively. Alteromonadales were present at low abundances in both Illi control and seep samples, with a mean of 2.5 ± 0.2 and 5.2 ± 1.2%, respectively. Overall, Alteromonadales and Alphaproteobacteria were present in marginally higher relative abundance at Dobu than Illi, regardless of treatment (Figure 4), suggesting that patterns in relative abundance are more strongly influenced by reef than by pH.
Patterns in the Core Microbiome and Indicator Species
The core microbiome and indicator taxa were investigated to determine if the high pCO2 environment at natural seeps may influence the presence of potentially beneficial bacterial taxa. Across all sites, 22 taxa (66,139 sequences) were present in every sample (Figure 6 and Supplementary Figure S2). The majority of core taxa were present in low abundances across all samples, with the genus Endozoicomonas being the most abundant, accounting for 33,974 sequences (51.4%), followed by Prosthecochloris spp. (class Chlorobia; 13.5%) and Marinobacteria spp. (order Alteromonadales; 9.6%). No trend in relative abundance was observed between seep and control sites across both reefs.
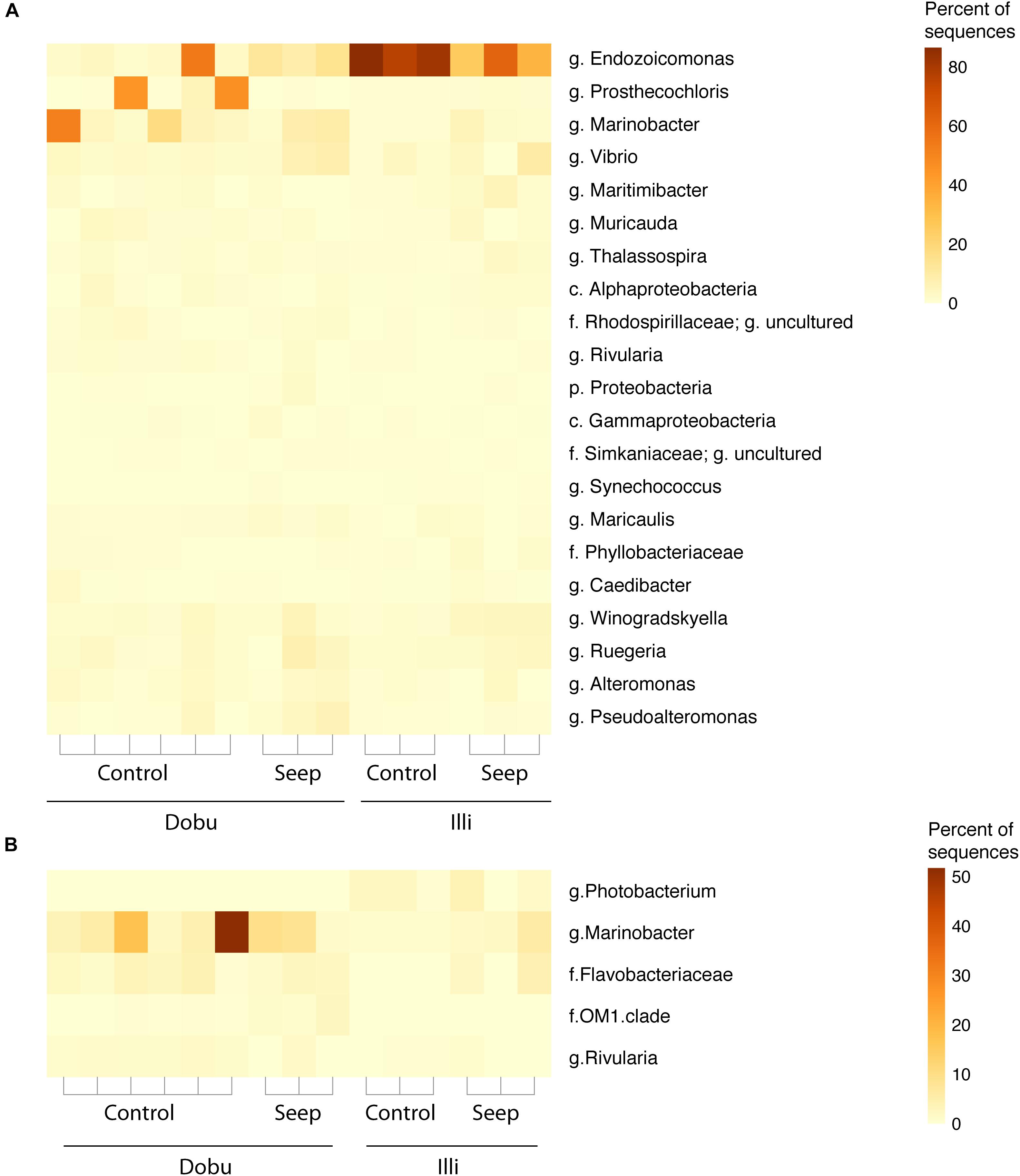
FIGURE 6. (A) Relative proportion of core microbes (the 22 taxa found in 100% of samples). (B) Relative proportion of indicator taxa (taxa found to be significantly correlated (p < 0.05) to each site). Taxa are identified to the lowest possible classification, where p. is phylum, c. is class, f. is family, and g. is genus.
Dobu Porites spp. supported three indicator taxa: the genus Marinobacter (p = 0.009), the family Flavobacteriaceae (p = 0.008), and the family OM1 clade of Acidimicrobiales (p = 0.016). The genus Marinobacter was the most abundant indicator species, representing a mean of 11.6% of sequences in Dobu samples. Only one taxon was found to be significantly indicative of Illi Porites spp.: the genus Photobacterium (c. Gammaproteobacteria, f. Vibrionaceae; p = 0.005) was present at low but consistent abundances, representing a mean of 1.8% of sequences from all Illi samples (Figure 6). The genus Rivularia was indicative of control sites (p = 0.013), but was present in very low abundances (mean of 0.9% of all control sample sequences). Notably, no indicator taxa were detected for seep sites.
pH Values Modeled Using 14C
A precise measurement of seawater pH was given for each coral living in the seep environment (Supplementary Table S1). Seawater pH was variable among colonies and dropped below what is predicted for the coming century. At the Dobu seep, pH varied from 7.67 to 7.78, while at the Illi seep pH varied from 7.44 to 7.85.
Discussion
The rising partial pressure of carbon dioxide (pCO2) in the oceans and subsequent OA poses a serious threat to marine life, particularly to calcifying organisms such as reef-building corals. However, corals show varying degrees of sensitivity to high pCO2 and some may even benefit from the change in ocean chemistry (Morrow et al., 2014; Strahl et al., 2015; Strahl et al., 2016). In the current study, we show that massive Porites corals that occur in high abundance at low pH/high pCO2 seep sites in PNG host robust bacterial communities that are stable across a broad range of environmental pH levels.
Massive Porites spp. Dominate CO2 Seeps in Papua New Guinea
The high abundance of massive Porites spp. at the PNG seeps (pH 7.44–7.85) in comparison to adjacent control sites demonstrates that Porites spp. may be able to withstand lower pH conditions than those predicted for the coming century (i.e., ∼7.8) (Rhein et al., 2013). The dominance of massive Porites spp. at the seeps led to a significant decrease in coral diversity in comparison to control sites. These findings are consistent with previous reports of coral community assemblages at the CO2 seeps of the D’Entrecasteaux Islands, in which structurally complex branching and tabular corals were found to dominate control sites, while massive, boulder formations dominated at the seeps (Fabricius et al., 2011; Morrow et al., 2014). Community changes resulting in a loss of topographical complexity have important impacts on the reef ecosystem as many coral reef organisms seek refuge in the complex habitat provided by branching and tabular coral species (Graham and Nash, 2013; Fabricius et al., 2014). Therefore, it is critical to understand what drives the success of massive Porites in low pH/high pCO2 environments. Evidence suggests that ecological success is linked to both physiological (Wall et al., 2016) and microbial mechanisms (Marcelino et al., 2017), with our data supporting the concept that Porites maintains a stable microbial community, even under extreme pCO2 conditions.
Response of Massive Porites spp. Microbial Communities to Low pH/High pCO2
The microbial communities of massive Porites were highly similar between colonies in volcanic seep habitats (pH 7.44–7.85) and nearby colonies living under ambient pH conditions (pH ∼8.1). With the exception of the high abundance of Endozoicomonas at the Illi control site, there were negligible differences in the relative abundance of dominant taxa between control and seep colonies, suggesting that the microbiome of Porites spp. is stable across pCO2 conditions. The major difference in microbial community was between the two reefs (i.e., Illi versus Dobu), and was largely characterized by increases in the abundance of Endozoicomonas and decreases in Chlorobi at Illi reef. Previous work investigating the endolithic community of Porites spp. at the PNG seeps found no significant difference in community composition between seep and control sites (Marcelino et al., 2017), suggesting the skeletal microbial community is robust to low pH conditions. Our results align with this previous work, demonstrating that the tissue-associated microbial community is also robust to high pCO2 conditions. Our findings that massive Porites show little change in microbial community structure across pH levels strongly contrasts with previous findings focused on pCO2-sensitive corals (Morrow et al., 2014). For instance, it was previously demonstrated that a significant decline in the abundance of the coral species A. millepora and P. cylindrica at CO2 seeps co-occurred with a substantial shift in associated microbial communities (Morrow et al., 2014). It may be hypothesized that one factor underpinning the tolerance of corals to a low pH environment could be the association with a microbial community that is robust toward changing pH levels.
An alternative hypothesis to explain the resilience of corals at seep sites is that the physiological performance of the coral host influences the diversity and stability of the associated microbial community. Recent work on massive Porites has shown that this coral is able to regulate and maintain its internal pH when the surrounding seawater pH is reduced to ∼7.9 (Wall et al., 2016). This method of pH regulation involves the active removal of protons (H+) from the calicoblastic space (Venn et al., 2013), however, it is unknown what energetic trade-offs may accompany this in massive Porites at the seeps. Massive Porites benefit from the high pCO2 environment at the PNG seeps through an increase in net photosynthesis (however, unaltered net calcification rates) in comparison to control sites (Strahl et al., 2015). In addition to providing added nutritional support, an increase in photosynthesis raises the internal pH of the coral tissues through the removal of CO2 (Kühl et al., 1995). Furthermore, tissue biomass, lipid, protein and tissue energy content, fatty acid content, pigment content, and oxidative stress parameters all remained unaffected by pCO2 up to 800 μatm (Strahl et al., 2015). Similarly, aquaria-based experiments have shown that calcification in massive Porites appears largely unaffected by increases in pCO2 (Anthony et al., 2008; Edmunds, 2011; Edmunds et al., 2012). Massive Porites have repeatedly been shown to be one of the most pCO2-tolerant coral taxa due to their ability to maintain higher internal pH values at the site of calcification than many other scleractinians (McCulloch et al., 2012). Therefore, through the combination of internal pH regulation and increased photosynthesis, massive Porites corals likely maintain a more stable pH, which in turn benefits the stability of their microbial assemblages.
This interpretation is consistent with previous investigations into the microbiology and physiology of the coral species A. millepora at the PNG CO2 seeps. The pCO2-sensitive species not only displayed a shift in microbial community structure at the seeps (Morrow et al., 2014), but also have significantly lower tissue biomass and lipid content in comparison to Porites (Strahl et al., 2015). Additionally, although daylight calcification rates and photosynthesis in A. millepora were unaffected, both dark and net calcification rates were significantly reduced (Strahl et al., 2015), and likely represent the coral’s inability to regulate internal pH, or a trade-off of resources away from processes such as calcification into pH regulation. It is possible that corals sensitive to changes in surrounding pH will divert resources away from key cellular metabolic processes, thereby changing the amount and composition of exudates and subsequently influencing the associated microbial consortia. Thus, the strong physiological performance of massive Porites at the seep potentially supports a stable microbial community, further increasing the resilience of massive Porites to low pH.
Response of Microbial Communities Associated With Massive Porites spp. to Reef Location
Reef location had the strongest influence on microbial communities associated with massive Porites. While the same dominant microbial taxa were present at both sites, changes in the relative abundance led to significant changes in overall community structure. Specifically, lower relative abundances of Endozoicomonas at Dobu reef were replaced with increased relative abundances of Alteromonadales and Chlorobi, and were largely responsible for the differences in microbiome diversity between reefs. In particular, the Illi control samples had the lowest microbial diversity due to the high abundance of Endozoicomonas at this site. Further site differences were highlighted by an indicator taxa analysis that identified the less abundant Marinobacter, Flavobacteriaceae and Acidimicrobiales as characteristic of the Dobu site. These changes were clearly reflected in the NMDS plot with multivariate data being clustered by reef location rather than CO2 treatment. Marinobacter are known to degrade some organic pollutants (Huu et al., 1999) (see below), and may indicate a difference in water quality between the two sites. However, previous measurements of seawater chemistry (excluding pH differences at the seeps) were consistent between the two reefs (Fabricius et al., 2014).
Illi and Dobu reefs are known to harbor different communities of zooplankton, where species abundances and biochemical composition vary significantly between the two reefs (Smith et al., 2016). This represents a potential difference in food quality for the coral populations, which can induce changes to the physiology of both the host and its algal symbionts (Houlbrèque and Ferrier-Pagès, 2009), and potentially its prokaryotic community as well. Differences in zooplankton communities between reefs highlight the possibility of a currently unknown factor affecting microbial communities between reefs at Illi and Dobu. However, it is not uncommon for microbial communities associated with coral to show spatial variation. Patterns in reef-specific coral-associated bacterial communities have previously been observed, with the same species of coral harboring different microbial communities in different locations (Littman et al., 2009; Morrow et al., 2012). It has also been speculated that variation in microbial community structure reflects differences in Symbiodiniaceae populations (Littman et al., 2010; Sogin et al., 2017), which may produce variable nutrient sources for associated microbes (Olson et al., 2009). However, it has been shown that Symbiodiniaceae community composition of massive Porites (and other corals) does not change between the locations in this study, nor do they change after life-long exposure to elevated CO2, (Noonan et al., 2013), and thus it is unlikely that microbial communities in the current study were affected by Symbiodiniaceae identity. Lastly, massive Porites includes multiple species that cannot be identified in the field. It is feasible that site differences in the microbial community are due to different species inhabiting different locations and future studies on this group of corals should incorporate molecular testing to resolve the taxonomy to species level. In the current study, it remains unclear what drives the differences between the two reefs microbial communities.
Core Microbes Associated With Massive Porites spp.
Endozoicomonas was the most common core taxa, and is considered a common marine symbiont, found in high abundance across many coral genera (Bayer et al., 2013b; Neave et al., 2016; Pogoreutz et al., 2018) and other marine invertebrates (Martínez-García et al., 2007; Jensen et al., 2010; Bayer et al., 2013a). The widespread association of Endozoicomonas with multiple taxa suggests that the genus has coevolved with marine invertebrates through a developed symbiotic relationship, similar to the relationship between corals and Symbiodiniaceae (Bayer et al., 2013a,b). Recent genomic work on Endozoicomonas highlighted the potential for important functional roles, including the transport of organic molecules between symbiont and host and the synthesis of amino acids (Neave et al., 2017). Due to these potentially fundamental roles of Endozoicomonas, this genus is emerging as a core symbiont in corals and other invertebrate taxa.
Sequences affiliated with both Marinobacter spp. (order Alteromonadales) and Prosthecochloris spp. (class Chlorobia) were also conserved across all samples but were far less abundant than Endozoicomonas. Interestingly, bacteria from both genera are able to grow optimally under low pH conditions (relative to ambient seawater pH), however, whether or not this confers any degree of tolerance to the coral holobiont under low pH is unknown (Gorlenko, 1970; Pringault et al., 1998; Duran, 2010). Bacterial strains from the genus Marinobacter are known for their ability to degrade hydrophobic organic compounds including some organic pollutants (Gauthier et al., 1992; Huu et al., 1999), and has recently been isolated from a Bryozoan sample, indicating its potential association with marine invertebrates (Romanenko et al., 2005). Direct contact with marine algae is known to transfer hydrophobic organic compounds to nearby corals, thereby stimulating microbial growth and creating a pathway for opportunistic pathogens (Barott and Rohwer, 2012). However, the role of Marinobacter within the coral holobiont is currently unknown.
Bacteria affiliated to Chlorobi have previously been found within the endolithic community of coral skeletons (Marcelino and Verbruggen, 2016; Yang et al., 2016; Marcelino et al., 2017), and have been previously discovered in the skeleton of massive Porites collected from the same seeps as the current study (Marcelino et al., 2017). Specifically, the green sulfur bacterium Prosthecochloris is likely an important member of the coral holobiont due to its role in nitrogen fixation, and has been found in the tissues of Porites lutea (Li et al., 2014) and dominates the green endolithic layer in the coral Isopora palifera (Yang et al., 2016). As Chlorobi appear to characterize the endolithic community, it is possible that sequences found within the tissues represent endolithic contamination when sampling. Nevertheless, it appears to be a recurrent member of the massive Porites holobiont community.
Conclusion
The conditions offered by natural CO2 seeps provide one of the best opportunities for assessing the long-term impact of OA on marine holobionts in situ. The pH levels experienced by the sampled corals at the volcanic seep in the D’Entrecasteaux Islands varied from 7.44 to 7.85, indicating that some of these corals experienced harsher OA conditions than those predicted for the next 100 years. This strengthens our findings that pCO2-tolerant corals are accompanied by a robust bacterial community, similar to what was found in their endolithic community (Marcelino et al., 2017). Coral-associated bacterial communities were stable across CO2 seep and control sites, contrasting with the shift previously observed in corals that were more pCO2-sensitive (Morrow et al., 2014). Instead, bacterial communities were mainly influenced by geographic site differences. These data, combined with results from previous studies of massive Porites at the PNG seeps, suggest that massive Porites are able to maintain and regulate internal pH in low pH/high pCO2 environments and subsequently provide a stable habitat for microbial partners. This potentially leads to a positive feedback for the resilience of this species at low pH, thereby contributing to its potential dominance and success in future OA scenarios.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive via accession number SRP148584.
Ethics Statement
This study was exempt from these requirements as research involving corals is not required to obtain ethics approval.
Author Contributions
DB, KM, PO, BW, and KF conceived, designed, and coordinated the study. KM collected specimens and field data. PO and KM carried out molecular lab work and photo analysis, SF carried out 14C lab work and analysis. HS completed molecular and statistical analysis. PO drafted the manuscript with input from all authors. All authors gave final approval for publication.
Funding
This research was funded by the Australian Institute of Marine Science (AIMS). Publication funding support provided by the College of Science and Engineering, James Cook University. PO was supported by AIMS@JCU.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Traditional Owners of Illi Illi Bwa Bwa and Dobu for allowing us to survey their reefs, and to the Councilors of the Dobu RLLG, the Milne Bay Province Research Committee, the National Research Institute and the Department of Environment and Conservation of Papua New Guinea for permits and logistic support. We would also like to thank Sara Bell (AIMS) for her assistance with molecular lab work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02621/full#supplementary-material
References
Anthony, K. R. N., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Barott, K. L., and Rohwer, F. (2012). Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. doi: 10.1016/j.tim.2012.08.004
Bayer, T., Arif, C., Ferrier-Pagès, C., Zoccola, D., Aranda, M., and Voolstra, C. R. (2013a). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 479, 75–84. doi: 10.3354/meps10197
Bayer, T., Neave, M. J., Alsheikh-Hussain, A., Aranda, M., Yum, L. K., Mincer, T., et al. (2013b). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Caldeira, K., and Wickett, M. E. (2003). Oceanography: anthropogenic carbon and ocean pH. Nature 425:365. doi: 10.1038/425365a
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35
De Caceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Doney, S. C., Fabry, V. J., Feely, R. A., and Kleypas, J. A. (2009). Ocean acidification: the other CO2 problem. Annu. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
Duran, R. (2010). “Marinobacter,” in Handbook of Hydrocarbon and Lipid Microbiology, eds K. N. Timmis, T. McGenity, J. R. Van Der Meer, and V. de Lorenzo (Berlin: Springer).
Edmunds, P. J. (2011). Zooplanktivory ameliorates the effects of ocean acidification on the reef coral Porites spp. Limnol. Oceanogr. 56, 2402–2410. doi: 10.4319/lo.2011.56.6.2402
Edmunds, P. J., Brown, D., and Moriarty, V. (2012). Interactive effects of ocean acidification and temperature on two scleractinian corals from Moorea, French Polynesia. Glob. Change Biol. 18, 2173–2183. doi: 10.1111/j.1365-2486.2012.02695.x
Fabricius, K. E., De’ath, G., Noonan, S., and Uthicke, S. (2014). Ecological effects of ocean acidification and habitat complexity on reef-associated macroinvertebrate communities. Proc. R. Soc. B 281:20132479. doi: 10.1098/rspb.2013.2479
Fabricius, K. E., Langdon, C., Uthicke, S., Humphrey, C., Noonan, S., De’ath, G., et al. (2011). Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat. Clim. Change 1, 165–169. doi: 10.1038/nclimate1122
Fallon, S. J., Fifield, L. K., and Chappell, J. M. (2010). The next chapter in radiocarbon dating at the Australian National University: status report on the single stage AMS. Nucl. Instrum. Methods Phys. Res. B 268, 898–901. doi: 10.1016/j.nimb.2009.10.059
Fine, M., and Loya, Y. (2002). Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc. R. Soc. Lond. B Biol. Sci. 269, 1205–1210. doi: 10.1098/rspb.2002.1983
Fox, J., and Weisberg, S. (2011). An R Companion to Applied Regression, 2nd Edn. Thousand Oaks CA: Sage.
Gattuso, J.-P., Frankignoulle, M., Bourge, I., Romaine, S., and Buddemeier, R. (1998). Effect of calcium carbonate saturation of seawater on coral calcification. Glob. Planet. Change 18, 37–46. doi: 10.1016/S0921-8181(98)00035-6
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Evol. Microbiol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Gorlenko, V. M. (1970). A new phototrophic green sulphur bacterium–Prosthecochloris aestuarii nov. gen. nov. spec. Z. Allg. Mikrobiol. 10, 147–149. doi: 10.1002/jobm.19700100207
Graham, N. A. J., and Nash, K. L. (2013). The importance of structural complexity in coral reef ecosystems. Coral Reefs 32, 315–326. doi: 10.1007/s00338-012-0984-y
Hall-Spencer, J. M., Rodolfo-Metalpa, R., Martin, S., Ransome, E., Fine, M., Turner, S. M., et al. (2008). Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 454, 96–99. doi: 10.1038/nature07051
Houlbrèque, F., and Ferrier-Pagès, C. (2009). Heterotrophy in tropical scleractinian corals. Biol. Rev. Camb. Philos. Soc. 84, 1–17. doi: 10.1111/j.1469-185X.2008.00058.x
Huu, N. B., Denner, E. B. M., Ha, D. T. C., Wanner, G., and Stan-Lotter, H. (1999). Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int. J. Syst. Evol. Microbiol. 49, 367–375. doi: 10.1099/00207713-49-2-367
Jensen, S., Duperron, S., Birkeland, N.-K., and Hovland, M. (2010). Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol. Ecol. 74, 523–533. doi: 10.1111/j.1574-6941.2010.00981.x
Joint, I., Doney, S. C., and Karl, D. M. (2011). Will ocean acidification affect marine microbes? ISME J. 5, 1–7. doi: 10.1038/ismej.2010.79
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kline, D. I., Kuntz, N. M., Breitbart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125. doi: 10.3354/meps314119
Kolde, R. (2015). Package “pheatmap.”. Available at: http://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf
Kühl, M., Cohen, Y., Dalsgaard, T., Jørgensen, B. B., and Revsbech, N. P. (1995). Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Ecol. Prog. Ser. 117, 159–172. doi: 10.3354/meps117159
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Li, J., Chen, Q., Long, L.-J., Dong, J.-D., Yang, J., and Zhang, S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4:7320. doi: 10.1038/srep07320
Littman, R. A., Bourne, D. G., and Willis, B. L. (2010). Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 19, 1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666.x
Marcelino, V. R., Morrow, K. M., van Oppen, M. J. H., Bourne, D. G., and Verbruggen, H. (2017). Diversity and stability of coral endolithic microbial communities at a naturally high pCO2 reef. Mol. Ecol. 26, 5344–5357. doi: 10.1111/mec.14268
Marcelino, V. R., and Verbruggen, H. (2016). Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci. Rep. 6:31508. doi: 10.1038/srep31508
Martínez-García, M., Díaz-Valdés, M., Wanner, G., Ramos-Esplá, A., and Antón, J. (2007). Microbial community associated with the colonial ascidian Cystodytes dellechiajei. Environ. Microbiol. 9, 521–534. doi: 10.1111/j.1462-2920.2006.01170.x
McCulloch, M., Falter, J., Trotter, J., and Montagna, P. (2012). Coral resilience to ocean acidification and global warming through pH up-regulation. Nat. Clim. Change 2, 623–627. doi: 10.1038/nclimate1473
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Meron, D., Atias, E., Kruh, L. I., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. doi: 10.1038/ismej.2010.102
Meron, D., Rodolfo-Metalpa, R., Cunning, R., Baker, A. C., Fine, M., and Banin, E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. doi: 10.1038/ismej.2012.19
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2014). Natural volcanic CO2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-12
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Noonan, S. H. C., Fabricius, K. E., and Humphrey, C. (2013). Symbiodinium community composition in scleractinian corals is not affected by life-long exposure to elevated carbon dioxide. PLoS One 8:e63985. doi: 10.1371/journal.pone.0063985
O’Brien, P. A., Morrow, K. M., Willis, B. L., and Bourne, D. G. (2016). Implications of ocean acidification for marine microorganisms from the free-living to the host-associated. Front. Mar. Sci. 3:47. doi: 10.3389/fmars.2016.00047
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Simpson, G. L., Solymos, P., et al. (2016). vegan: Community Ecology Package. Available at: https://CRAN.R-project.org/package=vegan
Olson, N. D., Ainsworth, T. D., Gates, R. D., and Takabayashi, M. (2009). Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371, 140–146. doi: 10.1016/j.jembe.2009.01.012
Pierrot, D., Lewis, D. E., and Wallace, D. W. R. (2006). MS Excel Program Developed for CO2 System Calculations. Oak Ridge, TN: Carbon Dioxide Information Analysis Center.
Pogoreutz, C., Rädecker, N., Cárdenas, A., Gärdes, A., Wild, C., and Voolstra, C. R. (2018). Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol. Evol. 8, 2240–2252. doi: 10.1002/ece3.3830
Pringault, O., Kühl, M., de Wit, R., and Caumette, P. (1998). Growth of green sulphur bacteria in experimental benthic oxygen, sulphide, pH and light gradients. Microbiology 144, 1051–1061. doi: 10.1099/00221287-144-4-1051
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raina, J.-B., Tapiolas, D. M., Forêt, S., Lutz, A., Abrego, D., Ceh, J., et al. (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502, 677–680. doi: 10.1038/nature12677
Rhein, M., Rintoul, S. R., Aoki, S., Campos, E., Chambers, D., Feely, R., et al. (2013). “Observations: ocean,” in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds T. F. Stocker, D. Qin, G.-K. Plattner, M. Tignor, S. K. Allen, J. Boschung, et al. (Cambridge: Cambridge University Press).
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Romanenko, L. A., Schumann, P., Rohde, M., Zhukova, N. V., Mikhailov, V. V., and Stackebrandt, E. (2005). Marinobacter bryozoorum sp. nov. and Marinobacter sediminum sp. nov., novel bacteria from the marine environment. Int. J. Syst. Evol. Microbiol. 55, 143–148. doi: 10.1099/ijs.0.63258-0
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Smith, J. N., De’ath, G., Richter, C., Cornils, A., Hall-Spencer, J. M., and Fabricius, K. E. (2016). Ocean acidification reduces demersal zooplankton that reside in tropical coral reefs. Nat. Clim. Change 6, 1124–1129. doi: 10.1038/nclimate3122
Sogin, E. M., Putnam, H. M., Nelson, C. E., Anderson, P., and Gates, R. D. (2017). Correspondence of coral holobiont metabolome with symbiotic bacteria, archaea and Symbiodinium communities. Environ. Microbiol. Rep. 9, 310–315. doi: 10.1111/1758-2229.12541
Strahl, J., Francis, D. S., Doyle, J., Humphrey, C., and Fabricius, K. E. (2016). Biochemical responses to ocean acidification contrast between tropical corals with high and low abundances at volcanic carbon dioxide seeps. ICES J. Mar. Sci. 73, 897–909. doi: 10.1093/icesjms/fsv194
Strahl, J., Stolz, I., Uthicke, S., Vogel, N., Noonan, S. H. C., and Fabricius, K. E. (2015). Physiological and ecological performance differs in four coral taxa at a volcanic carbon dioxide seep. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 184, 179–186. doi: 10.1016/j.cbpa.2015.02.018
Sunda, W., Kieber, D. J., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Trygonis, V., and Sini, M. (2012). photoQuad: a dedicated seabed image processing software, and a comparative error analysis of four photoquadrat methods. J. Exp. Mar. Biol. Ecol. 424, 99–108. doi: 10.1016/j.jembe.2012.04.018
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S, 4th Edn. New York, NY: Springer. doi: 10.1007/978-0-387-21706-2
Venn, A. A., Tambutté, E., Holcomb, M., Laurent, J., Allemand, D., and Tambutté, S. (2013). Impact of seawater acidification on pH at the tissue–skeleton interface and calcification in reef corals. Proc. Natl. Acad. Sci. U.S.A. 110, 1634–1639. doi: 10.1073/pnas.1216153110
Vogel, J. S., Southon, J. R., Nelson, D. E., and Brown, T. A. (1984). Performance of catalytically condensed carbon for use in accelerator mass spectrometry. Nucl. Instrum. Methods Phys. Res. B 5, 289–293. doi: 10.1016/0168-583X(84)90529-9
Wall, M., Fietzke, J., Schmidt, G. M., Fink, A., Hofmann, L. C., de Beer, D., et al. (2016). Internal pH regulation facilitates in situ long-term acclimation of massive corals to end-of-century carbon dioxide conditions. Sci. Rep. 6:30688. doi: 10.1038/srep30688
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Webster, N. S., Negri, A. P., Flores, F., Humphrey, C., Soo, R., Botté, E. S., et al. (2013). Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ. Microbiol. Rep. 5, 243–251. doi: 10.1111/1758-2229.12006
Yang, S.-H., Lee, S. T. M., Huang, C., Ching-Hung, T., Chiang, P., Chen, C., et al. (2016). Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol. Oceanogr. 61, 1078–1086. doi: 10.1002/lno.10277
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Keywords: ocean acidification, microbiome, coral, volcanic seep, Porites
Citation: O’Brien PA, Smith HA, Fallon S, Fabricius K, Willis BL, Morrow KM and Bourne DG (2018) Elevated CO2 Has Little Influence on the Bacterial Communities Associated With the pH-Tolerant Coral, Massive Porites spp. Front. Microbiol. 9:2621. doi: 10.3389/fmicb.2018.02621
Received: 20 July 2018; Accepted: 12 October 2018;
Published: 01 November 2018.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
John Everett Parkinson, Oregon State University, United StatesSen-Lin Tang, Biodiversity Research Center, Academia Sinica, Taiwan
Copyright © 2018 O’Brien, Smith, Fallon, Fabricius, Willis, Morrow and Bourne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David G. Bourne, ZGF2aWQuYm91cm5lQGpjdS5lZHUuYXU=
†These authors have contributed equally to this work
 Paul A. O’Brien
Paul A. O’Brien Hillary A. Smith
Hillary A. Smith Stewart Fallon5
Stewart Fallon5 Katharina Fabricius
Katharina Fabricius Bette L. Willis
Bette L. Willis Kathleen M. Morrow
Kathleen M. Morrow David G. Bourne
David G. Bourne