- Department for Sustainable Food Process–DiSTAS, Università Cattolica del Sacro Cuore, Piacenza, Italy
Lactobacillus casei/Lactobacillus paracasei group of species contains strains adapted to a wide range of environments, from dairy products to intestinal tract of animals and fermented vegetables. Understanding the gene acquisitions and losses that induced such different adaptations, implies a comparison between complete genomes, since evolutionary differences spread on the whole sequence. This study compared 12 complete genomes of L. casei/paracasei dairy-niche isolates and 7 genomes of L. casei/paracasei isolated from other habitats (i.e., corn silage, human intestine, sauerkraut, beef, congee). Phylogenetic tree construction and average nucleotide identity (ANI) metric showed a clustering of the two dairy L. casei strains ATCC393 and LC5, indicating a lower genetic relatedness in comparison to the other strains. Genomic analysis revealed a core of 313 genes shared by dairy and non-dairy Lactic Acid bacteria (LAB), within a pan-genome of 9,462 genes. Functional category analyses highlighted the evolutionary genes decay of dairy isolates, particularly considering carbohydrates and amino acids metabolisms. Specifically, dairy L. casei/paracasei strains lost the ability to metabolize myo-inositol and taurine (i.e., iol and tau gene clusters). However, gene acquisitions by dairy strains were also highlighted, mostly related to defense mechanisms and host-pathogen interactions (i.e., yueB, esaA, and sle1).
This study aimed to be a preliminary investigation on dairy and non-dairy marker genes that could be further characterized for probiotics or food applications.
Introduction
Strains allotted to Lactobacillus casei/Lactobacillus paracasei group of species have been isolated from a range of ecological niches such as the different sites of the human body, breeding animals' intestinal tract and dairy environment, sourdough bread starter, and fermented vegetables, as well as from plants (Smokvina et al., 2013). A wide number of strain belonging to these two species are also commercially exploited as dairy starters, probiotics for humans and animals.
The wealth of data created by comparative genomics is allowing the identification of the core and of pan-genomes in a significant number of Lactic Acid bacteria (LAB) (Wassenaar and Lukjancenko, 2014; Sun et al., 2015; Zheng et al., 2015), allowing scientists to describe an updated evolutionary history of these bacteria and to identify genes responsible of their adaptation to an impressive number of ecological niches.
Dairy lactobacilli have been described by Stefanovic et al. (2017) as “niche specialist” (bacteria able to live in a limited number of environments) in opposition to “niche generalist” (bacteria able to live in a large number of environments) and their niche adaption have been related to their genomes enriched with sugar amino acid transportation systems as well as an enhanced proteolytic ability. In addition, dairy-adapted lactobacilli have been shown to harbor substantial gene decay, limiting their ability to survive in niche different from milk and dairy.
A relevant number of 100 Lb. rhamnosus genomes have been compared by Douillard et al. (2013) for their niche adaptability. They were able to cluster together most of the dairy isolates, while intestinal and probiotic strains grouped together with other human isolates. Combining phenotypic and genomic data authors were able to identify two clearly distinguishable geno-phenotypes. The “dairy group” was characterized by the absence of SpaCBA pili, lactose, maltose, and rhamnose metabolism, while the “intestinal group” harbored genes encoding for bile resistance, pili, and L-fucose utilization, traits believed to be relevant for intestinal tract survival.
On the contrary, a massive comparative genomic analysis performed by Smokvina et al. (2013) on 34 L. paracasei strains allowed authors to identify over 4,200 ortholog groups that comprise the pan-genome of this species, while they discover that factors generally associated with animal host-microbe interactions such as pili, cell-envelope proteinase, hydrolases p40 and p75 are part of the L. paracasei core genome present in all analyzed strains. What they called the variome consists mainly of hypothetical proteins, phages, plasmids, transposon/conjugative elements, and known functions such as sugar metabolism, cell-surface proteins, transporters. They concluded that “no specific genes or gene clusters were found to correlate with strain origin of isolation.”
A Japanese group (Toh et al., 2013) published a paper focused on the genomic adaptation of the Lactobacillus casei, in which genomes of L. casei and L. paracasei were compared, including in the survey also the well-known probiotic strain ATCC 53103 (LGG) belonging to the L. rhamnosus species. Their comparative genomic analysis identified 1,682 core genes and a wide synteny in the considered strains. However, beside this synteny, authors identified a large presence of Genomic Islands (GI) in these strains, many of which located at the genetic same loci, suggesting the presence, in the chromosomes of the L. casei group, of a number of hypervariable regions at the same loci. These GI could explain the adaptation to each ecological niche.
More recently, Stefanovic and McAuliffe (2018) performed a comparative genomic and metabolic analysis of three Lactobacillus paracasei strains, all of them isolated from the dairy environment. These authors were unable to identify a specific set of genes related to this niche, while their findings suggested a high degree of heterogeneity among strains of the species of Lactobacillus paracasei even if isolated from the same ecological niche.
The potential of comparative genomics as applied to the elucidation of the niche adaptation mechanisms of the L. casei/paracasei groups of species seems, as suggested by the above cited papers, still to be fully exploited.
The current study aimed to evaluate further genomic features of species belonging to L. casei/paracasei groups, to identify discriminant traits between dairy and non-dairy niche isolates. This goal was reached by focusing on complete genomes only, since evolutionary differences rely on the whole sequence and not only on gene clusters (Inglin et al., 2018). Moreover, it was shown that incomplete genomes hugely affect the core-genome identification within a species, thus high-quality input sequences are fundamental to define a reliable genomic core (Inglin et al., 2018).
Furthermore, the secretion of the lactococcal bacteriocin Lactococcin G (Moll et al., 1996) was phenotypically tested, since a dedicated ABC transporter gene (lagD) was identified in some of the strains investigated.
Materials and Methods
Downloading of Publicly Available Complete Genomes and Quality Control
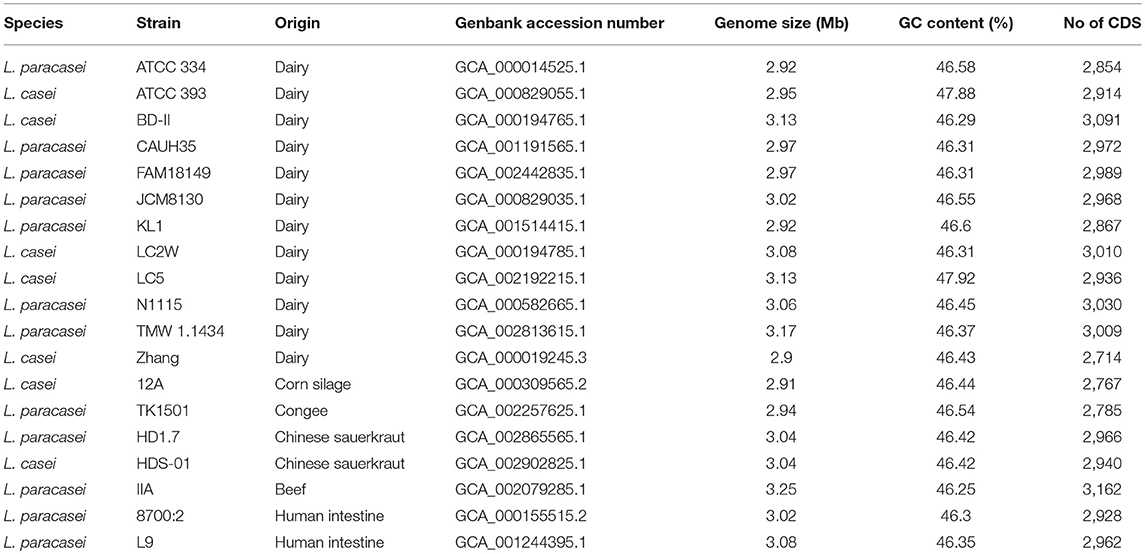
All the complete genomes of L. casei/paracasei available on the NCBI database were downloaded and the last update was done on 16 April 2018. The quality of the 19 genomes and their GC content were evaluated using Quast 4.6 (Gurevich et al., 2013), retaining high quality genomes, with N75 values >10 kbp and undetermined bases (N) per 100 kbp < 500, as suggested by Wuyts et al. (2017).
Comparative Genomic Analyses
Genes were reannotated using Prokka (v1.12; Seemann, 2014) with the genus option. ANIb calculation (Richter and Rossello-Mora, 2009) was performed using pyani (v0.2.7) to evaluate distances between all L. casei/paracasei genomes.
Pan-genome analyses were carried out with Roary (v3.11.2; Page et al., 2015) by setting strict parameters: blastp identity ≥95% and gene isolates threshold ≥99% to consider a gene as conserved. The output core gene alignment obtained with Roary was used in RAxML (v8.2.4) to build a maximum-likelihood phylogenetic tree. Genomes were clustered in R (R Development Core Team, 2008) based on the gene presence–absence distance matrix calculated with the Manhattan method. The output was then visualized by using heatmap.2 from the gplots package (Warnes et al., 2016). Orthogroups (OGs) were inferred by means of OrthoFinder (v2.2.3; Emms and Kelly, 2015) and the gene content analysis was performed by mapping the orthogroups to the optimized eggNOG bacterial database with eggNOG-mapper (v1.0.3; Huerta-Cepas et al., 2016). Statistical analysis for each functional category was carried out by performing non-metric multidimensional scaling, (NMDS) on the orthogroups count profiles for dairy and non-dairy genomes, by using the metaMDS function from the vegan package in R (Oksanen et al., 2016).
Phenotypic Evaluation of Lactococcin G Expression in Selected Lactobacilli
Gene expression of the bacteriocin Lactococcin G was evaluated by testing the growth of Lactococcus lactis subsp. lactis MG1614 in presence of a Lactobacillus paracasei strain containing a complete lagD gene (Lactobacillus paracasei 8700:2) and a strain with a fragmented lagD gene (Lactobacillus paracasei ATCC 334). All the strains were grown at 37°C in MRS agar plates and anaerobic conditions.
Results and Discussion
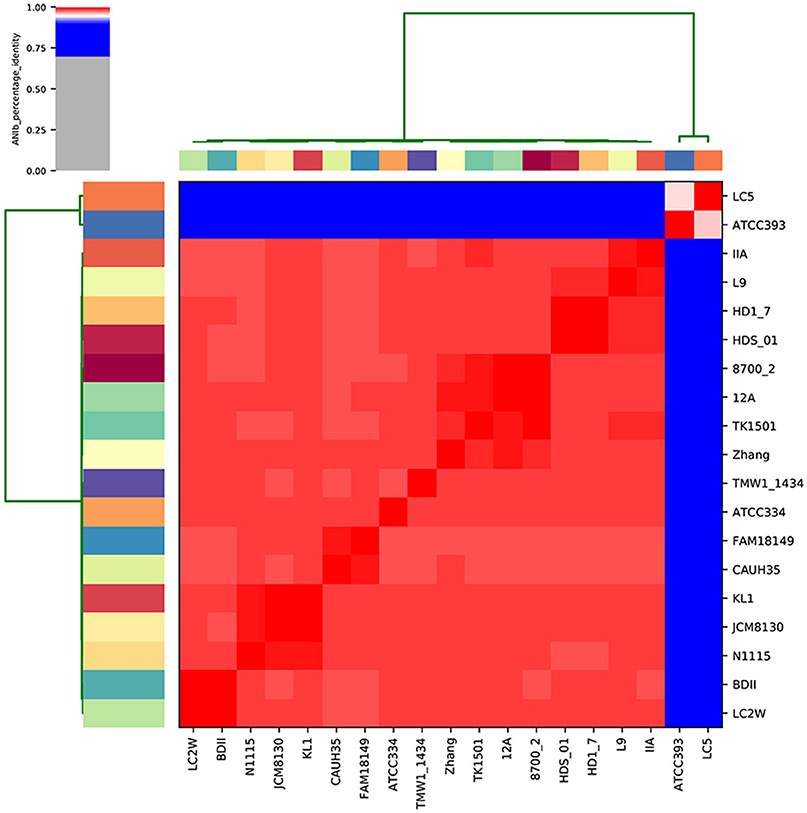
To evaluate what made L. casei/paracasei strains adapted to dairy niches (D) rather than non-dairy environments (ND), a genomic comparison was carried out on 19 publicly available complete genomes. After genomes quality checking and re-annotation, it was possible to notice that there were no appreciable differences in genomes size, GC content, and number of coding sequences (CDS) between D and ND strains (Table 1), indicating low intraspecies variation. To detect strains boundaries, pairwise genome comparisons were performed using average nucleotide identity (ANI) metrics. Particularly, ANIb distance between L. casei/paracasei strains was calculated (Figure 1) and it was evidenced the presence of two distinct clades: one represented by the dairy L. casei strains ATCC 393 and LC5, and the other including the remaining 17 genomes of both L. casei/paracasei strains. However, a deeper investigation of the pan-genome of such isolates is needed to better reveal the genic losses and acquisitions which led to different niches colonization.
Core- and Pan-Genome Analyses on Complete L. Casei/Paracasei Strains
The presence/absence of core and accessory genes identified by Roary (Supplementary Material, Data set A) additionally revealed the lower genetic relatedness of the dairy L. casei strains, ATCC393 and LC5, in relation to the others. The peculiarity of these two strains, which share a high genome similarity, has been previously highlighted (Kang et al., 2017). Therefore, ATCC393 and LC5 were included in the maximum-likelihood phylogenetic tree to serve as an outgroup (Figure 2). From the inferred tree, it was possible to notice the grouping of the strains in four clades (excluding the outgroup). Specifically, ND strains were separated from D strains, except for the dairy isolate L. casei Zhang. Moreover, the D strain L. paracasei ATCC334 is represented by a single subtree separated from the other dairy niche adapted genomes. Interestingly, based on the tree inference, it is also highlighted that dairy strains originated from two distinct evolutionary events (Figure 2). The core gene alignment, on which relied the phylogenetic tree, included 313 marker genes present in 99% of the genomes investigated and with a blastp identity over 95%. Besides core genes, shell and cloud genes were also identified (3,573 and 5,576, respectively), completing a pan-genome of 9,462 genes. The 98% of the genes identified considering all genomes were clustered into 3,727 orthogroups by Orthofinder (Emms and Kelly, 2015), from which 1,791 were defined as core orthogroups (containing all species; Table 2). These orthogroups were mapped to the bacterial eggNOG database (Huerta-Cepas et al., 2016) to evaluate the main functional COG categories related to the pan-genome (Supplementary Material, Data set B). The major fraction of orthogroups (28%) were grouped under the category of “unknown function” (S). This assignment is in line with previous findings (Smokvina et al., 2013; Wuyts et al., 2017), indicating the need for further improvements in functional gene prediction. However, 13% of the OGs were grouped into the “Carbohydrate transport and metabolism” category (G), followed by “transcription” activities (K), “amino acid transport and metabolism” (E) and “inorganic ion transport and metabolism” (P; 8, 7, and 5% OGs, respectively). Interestingly, 4% of the OGs were included into the V category of “Defense mechanisms.” However, it is worth to highlight that considering the dairy and non-dairy isolates separately, the percentage of OGs grouped in the COG categories related to metabolisms was identical (Figure 3). This outcome suggests that the adaptation to the dairy environment of D strains was not reached toward metabolic simplification, but it mostly relied on a process of gene losses and acquisitions. The NMDS analysis on the orthogroups gene counts for each functional category additionally highlighted the orthogroup composition overlapping for almost all categories of dairy and non-dairy niche isolates (Figure 4). This finding is in line with a previous study (Wuyts et al., 2017), except for the U category representing the intracellular trafficking and secretion systems, which exhibited a higher separation between D and ND strains.

Figure 2. Maximum-likelihood phylogenetic tree of L. casei/paracasei strains, dairy and non-dairy niche isolates (indicated with colored dots). *Outgroup represented by L. casei ATCC393 and L. casei LC5. Four clades of strains (1–4) were highlighted.
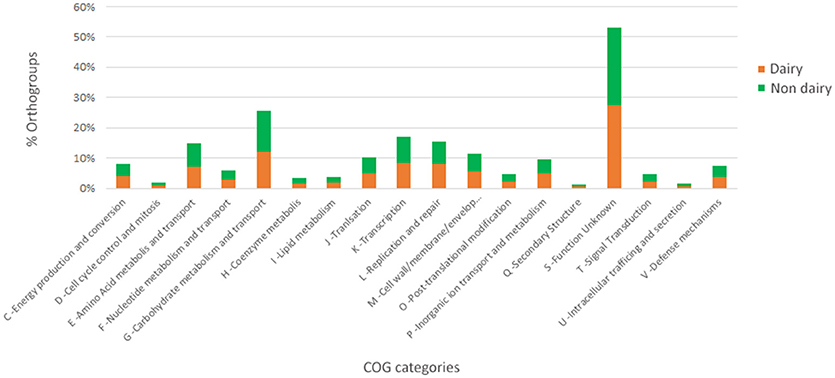
Figure 3. Functional COG categories related to the pan-genome of dairy and non-dairy L. casei/paracasei strains.
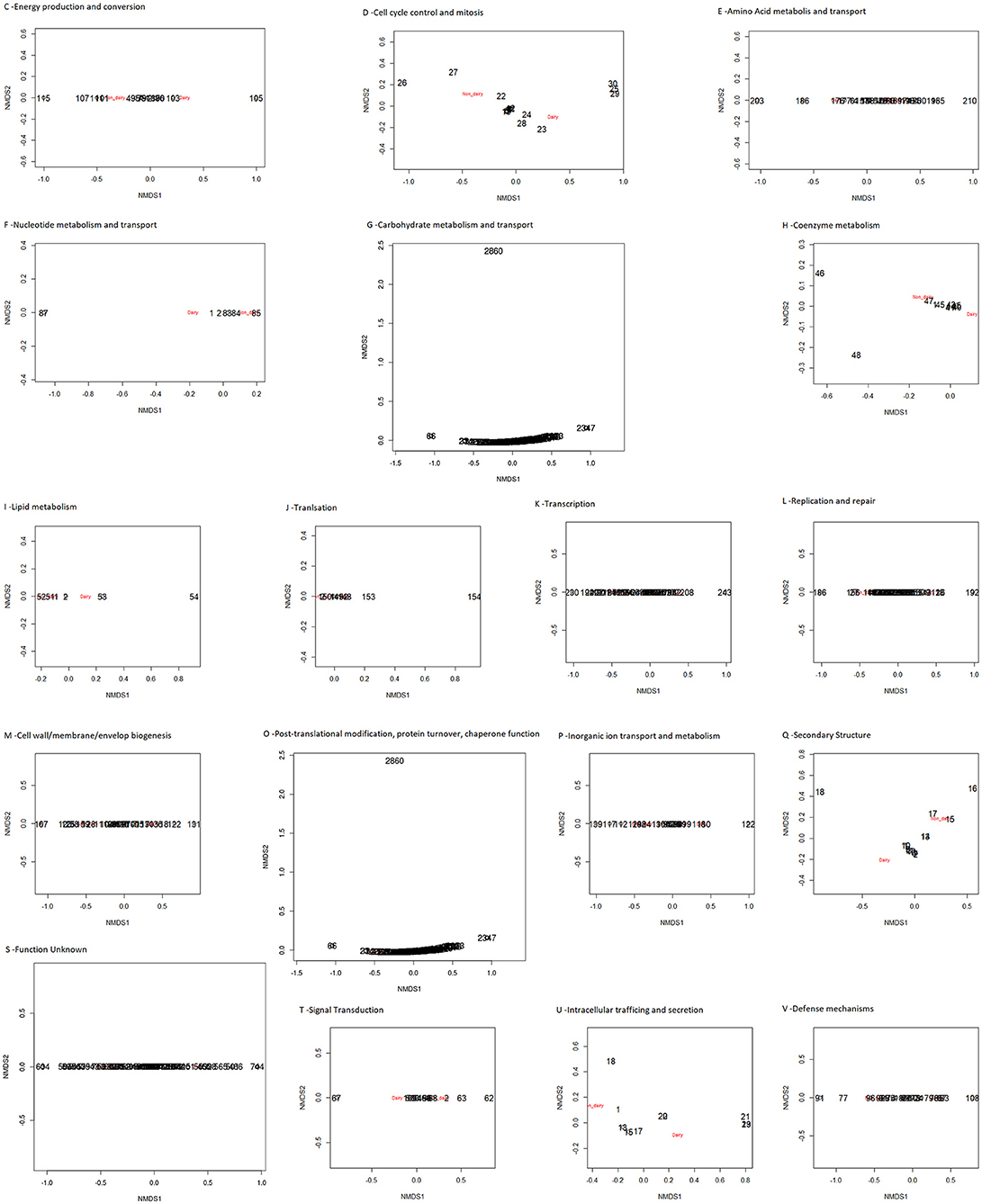
Figure 4. NMDS of the predicted functional capacity per niche (dairy and non-dairy), considering each COG category separately.
The variable genome content (i.e., variome) of the investigated strains was additionally analyzed by using the gene presence/absence matrix to build a “pan-genome” tree (Figure 5). From the hierarchical clustering it was possible to classify the OGs of genes in three clusters, whereas, at strain level, four clades were identified (excluding the outgroup). Thus, the variome-based tree recognized the same numbers of clades as the core-genome-based tree (Figure 2), but it presented a slightly different strain distribution. This outcome indicates that the discrimination between dairy and non-dairy niche adapted genomes mostly rely on the core-genome content.
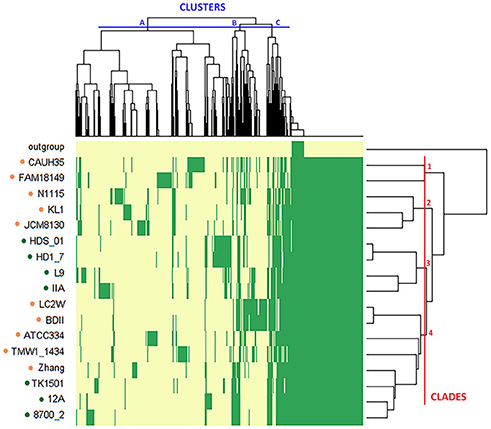
Figure 5. Hierarchical map and clustering of the 19 L. casei/paracasei genomes based on presence–absence of genes (yellow: absence, green: presence). The niche from which the strains were isolated is indicated by the colored dots (orange: dairy, green: non-dairy). The outgroup includes the two dairy isolated L. casei strains ATCC393 and LC5. OGs of genes are grouped in three clusters (A–C), while strains are grouped in four clades (1–4).
Considering the KEGG categories of the pan-genome (Supplementary Material, Data set B), many genes encoded for ABC transporters and, particularly, for amino acids translocation (i.e., MetQIN for methionine, LivFGHKL for branched-chain amino acids, TcyKLMN for cystein, GlnHPQ for glutamine, ProVWX for glycin/proline, LysXY for lysine), phosphate/phosphonate (PstABCS/PhnCDE), oligopeptides (OppABCDF), cations and organic ions (i.e., manganese, zinc, alkanesulfonates), oligo and monosaccharides (galactose oligomers/maltooligosaccharides, melibiose, ribose/xylose), and for glycerol-3-phosphate (UgpABCE). Moreover, there were found 65 genes related to phosphotransferase systems (PTS), namely N-acetylgalactosamine, alpha/beta-glucoside, cellobiose, fructose, galactitol, glucitol, lactose, mannose, mannitol, sucrose, sorbose, trehalose, ascorbate, maltose, galactose, 2-O-A-mannosyl-D-glycerate-specific components. Interestingly, there were also found 12 genes encoding for penicillin-binding protein (Pbp) under the category of peptidoglycan synthesis. Among these genes there was pbp3, which has been suggested to be one of the targets of intestinal bile acids, by functioning as cholate resistance factor (Hattori et al., 2017). Concerning the two-component systems (TCS) category, 68 genes have been identified, among which bacitracin-related components proteins (braERS, bceB). TCS enable LAB responding to many change/stress conditions, as it was elucidated by Alcántara et al. for Lactobacillus casei BL23 (Alcántara et al., 2011).
COG and KEGG functional categories were further investigated by exploring the presence/absence of specific genes of interest in each strain (Supplementary Material, Data set B) within the two clusters of L. casei/paracasei (dairy niche isolates and other niches adapted LAB) to find putative marker genes for such clades.
Carbohydrate Transport and Metabolism
All ND strains (including both L. casei/paracasei strains) exhibit a cluster of iol genes (iolABCDEGJTX) involved in the myo-Inositol (MI) transport and catabolic pathway (Yebra et al., 2007). As it was found in B. subtilis, the iol cluster is regulated by the catabolite control protein A (CcpA) and the corepressor phosphocarrier protein HPr (PtsH; Miwa and Fujita, 2001), which were both found in addition to the iol genes. These findings suggest an adaptation of certain LAB to non-dairy niches by acquiring new genes to metabolize other compounds than lactose, as it was found in Lactobacillus casei BL23 by Yebra et al. (2007).
Moreover, all ND strains contain the gene coding for 2-keto-3-deoxy-6-phosphogluconate aldolase (kdgA), which catalyze a step of the galacturonate metabolism from pectin degradation (Slováková et al., 2002). In addition, three of these strains (L. casei 12A, L. paracasei 8700:2, and TK1501) contain the whole gene cluster kdgKAT, indicating an adaptation to plant carbohydrates. A similar set of genes has been identified in two L. lactis strains isolated from beans and cress (Siezen et al., 2008), indicating an evolutionary adaptation to plant ecological niches.
The absence of two genes in four out of seven NDs were instead highlighted in comparison to D strains, which preferentially exhibited ugl and hepC genes responsible of GAGs (glycosaminoglycans) hydrolysis (i.e., chondroitin and heparin-sulfate). GAGs can interfere with cell-cell adhesion, as it was pointed out by Martín et al. working on HeLa cells and a Lactobacillus salivarius strain (Martín et al., 2013).
Amino Acid Transport and Metabolism
ND strains showed the absence of some gene clusters related to amino acid metabolism. For instance, the yke genes cluster (ykeMNOP) was mainly identified in D strains. The high fraction of proteins in dairy products (mostly caseins) could be related to the maintenance of these genes, which are putatively responsible of amino acids import and hydrolyzation. In addition, the presence of a spermidine/putrescine-binding periplasmic protein (PotD) was identified among the dairy-niche isolates (i.e., CAUH35, FAM18149, JCM8130, KL1, N1115, TMW1_1434). Putrescine is one of the most abundant biogenic amine found in dairy products, and its production was found to be strain-specific, relying on horizontal gene transfer acquisition (Linares et al., 2011).
Interestingly, ND strains contain the genes tauA and tauB encoding for two proteins (periplasmatic and ATP-binding, respectively) involved in taurine uptake under sulfate or cysteine starvation conditions (Javaux et al., 2007). The previously tested high affinity for the substrate of TauA protein (Javaux et al., 2007) can be applied in biological dosage assays of this compound.
Defense Mechanisms
ESX secretion system protein (YueB) and ESAT-6 secretion accessory factor (EsaA) were only present in D strains. These proteins belong to the so called “type VII secretion systems” or T7S system, in Gram-positive bacteria (Abdallah et al., 2007). In B. subtilis, the yueB gene has been identified as phage receptor and homologs of this protein have been also found in other species, like S. aureus (São-José et al., 2004, 2006). Most of the proteins belonging to the ESX/T7S system are involved in host–pathogen interactions but a further characterization of these secretion systems is still needed (Simeone et al., 2009).
Interestingly, a N-acetylmuramyl-L-alanine amidase gene (sle1) coding for a peptidoglycan hydrolase (PGH) was only present in D strains. This protein was found to be involved in cell separation of Staphylococcus aureus (Kajimura et al., 2005). However, it is known that the anti-inflammatory properties of probiotic lactobacilli often rely on their PGHs activities (Frirdich and Gaynor, 2013), resulting in enzymes of high interest.
The presence of the pgcA gene (phosphoglucomutase) in all ND strains but only in 4 out of 12 D strains was also highlighted. This gene is required for the synthesis of the membrane glycolipid diglucosyl-diacylglycerol (Glc2-DAG), whose linkage to lipoteichoic acid (LTA) seems to be a key point during S. aureus infection (Gründling and Schneewind, 2007). Furthermore, it is noteworthy the presence of the lactococcin A secretion protein (LcnD) in all strains but ATCC393 and LC5 (both dairy isolates). LcnD is an accessory membrane protein which enables the secretion of Lactococcin A (LcnA), a bacteriocin produced by some strains of Lactococcus lactis (Stoddard et al., 1992). On the contrary, the lactococcin G ABC transporter (LagD) is specifically included in all NDs strains. The presence of the putative lactococcin G transporter gene can suggest also the inclusion of the bacteriocin gene itself. This hypothesis was verified by carrying out phenotypic trials on selected Lactobacillus strains to evaluate its expression.
Lactococcin G Expression Trial
The secretion of Lactococcin G was tested by co-plating L. lactis MG1614 by inclusion and by spotting on the surface the putative bacteriocin producer L. paracasei 8700:2 and the non-producer (L. casei ATCC 334) for comparison. It was possible to appreciate an inhibition ring in proximity of the non-dairy L. paracasei 8700:2 (Figure 6). This data aims to be a preliminary finding for further investigations to support the expression of Lactococcin G gene among the proteins secreted by some strains of L. casei/paracasei.
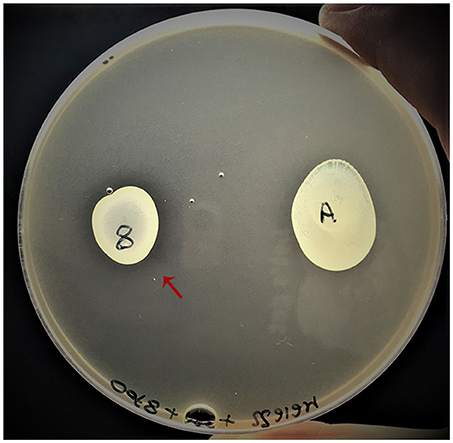
Figure 6. Phenotypic trial of Lactococcin G expression. The putative producer L. paracasei 8700:2 (indicated with the number “8”) is highlighted by a red arrow and the negative control is represented by L. casei ATCC334 (indicated with the letter “A”).
Conclusion
This study highlighted the origin of L. casei/paracasei dairy-isolated strains from two distinct evolutionary events. The variome-based analyses indicated that the discrimination between dairy and non-dairy niche adapted genomes mostly rely on the core-genome content, suggesting the presence of marker genes in dairy-niche isolated strains of L. casei/paracasei. These genetic signatures relied on the presence/absence of specific genes in relation to non-dairy L. casei/paracasei strains, supporting the occurrence of both gene gains and losses during the evolution of these LAB. Specifically, dairy-niche isolates lost many genes included in carbohydrates and amino acids metabolisms, such as the ability to utilize myo-inositol and taurine as carbon and sulfate sources, respectively. However, gene acquisitions have been also highlighted, especially regarding defense mechanisms. Dairy isolates indeed presented two genes (yueB and esaA) belonging to the type VII secretion systems and a peptidoglycan hydrolase (Sle1). These proteins are involved with host–pathogen interactions and further characterization should be done for probiotics development and food applications.
Author Contributions
AF performed bioinformatic analyses and wrote the manuscript. CZ supervised analyses and revised the manuscript. LM designed, supervised analyses, and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Dr. Irene Falasconi, Paolo Bellassi, and Ernesto Brambilla for the technical guidance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02611/full#supplementary-material
References
Abdallah, A. M., Gey van Pittius, N. C., Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M., et al. (2007). Type VII secretion–mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891. doi: 10.1038/nrmicro1773
Alcántara, C., Revilla-Guarinos, A., and Zúiga, M. (2011). Influence of two-component signal transduction systems of Lactobacillus casei BL23 on tolerance to stress conditions. Appl. Environ. Microbiol. 77, 1516–1519. doi: 10.1128/AEM.02176-10
Douillard, F. P., Ribbera, A., Kant, R., Pietil,ä, T. E., Järvinen, H. M., Messing, M., et al. (2013). Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 9:e1003683. doi: 10.1371/journal.pgen.1003683
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Frirdich, E., and Gaynor, E. C. (2013). Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr. Opin. Microbiol. 16, 767–778. doi: 10.1016/j.mib.2013.09.005
Gründling, A., and Schneewind, O. (2007). Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J. Bacteriol. 189, 2521–2530. doi: 10.1128/JB.01683-06
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hattori, M., Torres, G. A., Tanaka, N., Okada, S., and Endo, A. (2017). Detection and analysis of Lactobacillus paracasei penicillin-binding proteins revealed the presence of cholate-sensitive penicillin-binding protein 3 and an elongated cell shape in a cholate-sensitive strain. Biosci. Microbiota Food Health 36, 65–72. doi: 10.12938/bmfh.16-019
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). EGGNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Inglin, R. C., Meile, L., and Stevens, M. J. A. (2018). Clustering of pan- and core-genome of Lactobacillus provides novel evolutionary insights for differentiation. BMC Genomics 19:284. doi: 10.1186/s12864-018-4601-5
Javaux, C., Joris, B., and De Witte, P. (2007). Functional characteristics of TauA binding protein from TauABC Escherichia coli system. Protein J. 26, 231–238. doi: 10.1007/s10930-006-9064-x
Kajimura, J., Fujiwara, T., Yamada, S., Suzawa, Y., Nishida, T., Oyamada, Y., et al. (2005). Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol.Microbiol. 58, 1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x
Kang, J., Chung, W. H., Lim, T. J., Whon, T. W., Lim, S., Nam, Y., and Do (2017). Complete genome sequence of Lactobacillus casei LC5, a potential probiotics for atopic dermatitis. Front. Immunol. 8:413. doi: 10.3389/fimmu.2017.00413
Linares, D. M., Martln, M. C., Ladero, V., Alvarez, M. A., and Fernández, M. (2011). Biogenic amines in dairy products. Crit. Rev. Food Sci. Nutr. 51, 691–703. doi: 10.1080/10408398.2011.582813
Martín, R., Martín, C., Escobedo, S., Suárez, J. E., and Quirós, L. M. (2013). Surface glycosaminoglycans mediate adherence between HeLa cells and Lactobacillus salivarius Lv72. BMC Microbiol. 13:210. doi: 10.1186/1471-2180-13-210
Miwa, Y., and Fujita, Y. (2001). Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 183, 5877–5884. doi: 10.1128/JB.183.20.5877-5884.2001
Moll, G., Ubbink-Kok, T., Hildeng-Hauge, H., Nissen-Meyer, J., Nes, I. F., Konings, W. N., et al. (1996). Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J. Bacteriol. 178, 600–605. doi: 10.1128/jb.178.3.600-605.1996
Oksanen, J., Blanchet, F., Kindt, R., Legendre, P., and O'Hara, R. (2016). Vegan: community ecology package. R Packag. 2.3–3. doi: 10.4135/9781412971874.n145
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
São-José, C., Baptista, C., and Santos, M. A. (2004). Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186, 8337–8346. doi: 10.1128/JB.186.24.8337-8346.2004
São-José, C., Lhuillier, S., Lurz, R., Melki, R., Lepault, J., Santos, M. A., et al. (2006). The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281, 11464–11470. doi: 10.1074/jbc.M513625200
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siezen, R. J., Starrenburg, M. J. C., Boekhorst, J., Renckens, B., Molenaar, D., and Van Hylckama Vlieg, J. E. T. (2008). Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74, 424–436. doi: 10.1128/AEM.01850-07
Simeone, R., Bottai, D., and Brosch, R. (2009). ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12, 4–10. doi: 10.1016/j.mib.2008.11.003
Slováková, L., Dusková, D., and Marounek, M. (2002). Fermentation of pectin and glucose, and activityof pectin-degrading enzymes in the rabbit caecalbacterium Bifidobacterium pseudolongum. 35, 126–160. doi: 10.1046/j.1472-765X.2002.01159.x
Smokvina, T., Wels, M., Polka, J., Chervaux, C., Brisse, S., Boekhorst, J., et al. (2013). Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS ONE 8:e68731. doi: 10.1371/journal.pone.0068731
Stefanovic, E., Fitzgerald, G., and McAuliffe, O. (2017). Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol. 61, 33–49. doi: 10.1016/j.fm.2016.08.009
Stefanovic, E., and McAuliffe, O. (2018). Comparative genomic and metabolic analysis of three Lactobacillus paracasei cheese isolates reveals considerable genomic differences in strains from the same niche. BMC Genomics. 19:205. doi: 10.1186/s12864-018-4586-0
Stoddard, G. W., Petzel, J. P., Van Belkum, M. J., Kok, J., and McKay, L. L. (1992). Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl. Environ. Microbiol. 58, 1952–1961.
Sun, Z., Harris, H. M. B., McCann, A., Guo, C., Argimón, S., Zhang, W., et al. (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6:8322. doi: 10.1038/ncomms9322
Toh, H., Oshima, K., Nakano, A., Takahata, M., Murakami, M., Takaki, T., et al. (2013). Genomic adaptation of the Lactobacillus casei group. PLoS ONE 8:e75073. doi: 10.1371/journal.pone.0075073
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W. H. A., Lumley, T., et al. (2016). Package “gplots”: various R programming tools for plotting data. R Packag. version 2.17.10. doi: 10.1111/j.0022-3646.1997.00569.x
Wassenaar, T. M., and Lukjancenko, O. (2014). “Comparative genomics of Lactobacillus and other LAB,” in Lactic Acid Bacteria: Biodiversity and Taxonomy, eds W. H. Holzapfel and B. J. B. Wood (Chichester: Wiley-Blackwell), 55–69.
Wuyts, S., Wittouck, S., De Boeck, I., Allonsius, C. N., Pasolli, E., Segata, N., et al. (2017). Large-scale phylogenomics of the Lactobacillus casei group highlights taxonomic inconsistencies and reveals novel clade-associated features. mSystems 2:e00061–17. doi: 10.1128/mSystems.00061-17
Yebra, M. J., Zúñiga, M., Beaufils, S., Pérez-Martínez, G., Deutscher, J., and Monedero, V. (2007). Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl. Environ. Microbiol. 73, 3850–3858. doi: 10.1128/AEM.00243-07
Keywords: lactic acid bacteria, Lactobacillus casei group, comparative genomics, pan genome, niche adaptation
Citation: Fontana A, Zacconi C and Morelli L (2018) Genetic Signatures of Dairy Lactobacillus casei Group. Front. Microbiol. 9:2611. doi: 10.3389/fmicb.2018.02611
Received: 29 May 2018; Accepted: 12 October 2018;
Published: 30 October 2018.
Edited by:
Sarah Lebeer, University of Antwerp, BelgiumReviewed by:
Giuseppe Spano, University of Foggia, ItalyMichiel Wels, NIZO Food Research BV, Netherlands
Copyright © 2018 Fontana, Zacconi and Morelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Morelli, bG9yZW56by5tb3JlbGxpQHVuaWNhdHQuaXQ=
 Alessandra Fontana
Alessandra Fontana Carla Zacconi
Carla Zacconi Lorenzo Morelli
Lorenzo Morelli