- 1Divison of Crop Protection and Plant Health, Crop Research Institute, Prague, Czechia
- 2Department of Medical Chemistry and Clinical Biochemistry, Second Faculty of Medicine, Motol University Hospital, Charles University, Prague, Czechia
- 3Department of Zoology, Faculty of Science, Charles University, Prague, Czechia
- 4Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI, United States
- 5Institute of Biology, University of Tyumen, Tyumen, Russia
Background: Tyrophagus putrescentiae is a ubiquitous mite species in soil, stored products and house dust and infests food and causes allergies in people. T. putrescentiae populations harbor different bacterial communities, including intracellular symbionts and gut bacteria. The spread of microorganisms via the fecal pellets of T. putrescentiae is a possibility that has not been studied in detail but may be an important means by which gut bacteria colonize subsequent generations of mites. Feces in soil may be a vector for the spread of microorganisms.
Methods: Extracts from used mite culture medium (i.e., residual food, mite feces, and dead mite bodies) were used as a source of feces-inhabiting microorganisms as food for the mites. Two T. putrescentiae populations (L and P) were used for experiments, and they hosted the intracellular bacteria Cardinium and Wolbachia, respectively. The effects of the fecal fraction on respiration in a mite microcosm, mite nutrient contents, population growth and microbiome composition were evaluated.
Results: Feces from the P population comprised more than 90% Bartonella-like sequences. Feces from the L population feces hosted Staphylococcus, Virgibacillus, Brevibacterium, Enterobacteriaceae, and Bacillus. The mites from the P population, but not the L population, exhibited increased bacterial respiration in the microcosms in comparison to no-mite controls. Both L- and P-feces extracts had an inhibitory effect on the respiration of the microcosms, indicating antagonistic interactions within feces-associated bacteria. The mite microbiomes were resistant to the acquisition of new bacterial species from the feces, but their bacterial profiles were affected. Feeding of P mites on P-feces-enriched diets resulted in an increase in Bartonella abundance from 6 to 20% of the total bacterial sequences and a decrease in Bacillus abundance. The population growth was fivefold accelerated on P-feces extracts in comparison to the control.
Conclusion: The mite microbiome, to a certain extent, resists the acquisition of new bacteria when mites are fed on feces of the same species. However, a Bartonella-like bacteria-feces-enriched diet seems to be beneficial for mite populations with symbiotic Bartonella-like bacteria. Coprophagy on the feces of its own population may be a mechanism of bacterial acquisition in T. putrescentiae.
Introduction
Bacterial symbionts can directly or indirectly affect the interaction of the host populations with other species within a community (Ferrari and Vavre, 2011). Many arthropod species are infected by more than one symbiont (Ferrari and Vavre, 2011). For example, spider mite (Tetranychus phaselus) populations can host multiple simultaneous infections by Cardinium and two distinct lineages of Wolbachia (Zhao et al., 2013). This complexity may generate a meta-community structure of host-associated microbiota. Members of stored food astigmatid mites (Acari: Astigmata) have variable microbiomes, not only among different species (Hubert et al., 2016b) but also among different populations of a single species (Zindel et al., 2013; Erban et al., 2017). For example, among five observed populations of mold mite [Tyrophagus putrescentiae (Schrank, 1781)], two populations hosted the intracellular bacterial symbiont Cardinium in combination with Bacillus cereus, Staphylococcus, and Enterobacteriaceae; another two populations hosted Wolbachia in combination with Solitalea-like, Blattabacterium-like and Bartonella-like bacteria; and the remaining population lacked the intracellular symbionts but was inhabited by B. cereus, Staphylococcus, and Bartonella-like bacteria (Erban et al., 2016a; Hubert et al., 2016a). These populations also differed in fitness, measured as the nutrient composition of their bodies and population growth (Erban et al., 2017).
The mite T. putrescentiae is a ubiquitous species living in various natural habitats, such as soil, that decomposes plant materials and vertebrate nests; T. putrescentiae is also very common in human-created habitats, infesting various commodities, such as wheat, oil seeds, cheese, dried ham, dried fruits, mushrooms and grain debris (Hughes, 1976), as well as dog food (Brazis et al., 2008) and fungal, plant and insect cultures in laboratories (Walter et al., 1986; Duek et al., 2001). Moreover, Tyrophagus mites have been reported to feed on nematodes (Walter et al., 1986; Abou El-Atta and Osman, 2016), and related species, namely, T. similis and T. curvipenis, can feed on plant leaves in greenhouses (Fain and Fauvel, 1993; Jung et al., 2010). These mites can spread dangerous fungi (e.g., those developing on grain) by carrying fungal spores on their bodies, in the digestive system, or in feces (Griffiths et al., 1959). T. putrescentiae is common and the second most important species (after pyroglyphid house dust mites) among house dust mites and causes allergies in people and domestic animals (Arlian et al., 1984).
The hypothesis that bacterial metacommunities influence the host food specialization in mites is linked to fungal chitin digestion and utilization by symbiotic bacteria (Smrz and Catska, 2010). Chitin-digesting enzymes produced by symbiotic bacteria have been suggested to break up structural chitin from the cell walls of filamentous fungi, which can make up a substantial part of the mites’ diet (Smrz and Catska, 1989; Erban et al., 2016b). Depending on the fungal diet, T. putrescentiae can have different communities of cultivable chitinolytic bacteria, Pseudomonas, Brevundimonas, and Stenotrophomonas (Smrz et al., 1991; Smrz and Trelova, 1995; Smrz and Catska, 2010). Culturing different bacterial taxa from T. putrescentiae fed with various diet regimes provides the first proof of bacterially assisted feeding of mites and the first evidence that bacterial communities differ among different mite diets/habitats (Smrz, 2003; Smrz and Catska, 2010). Based on these findings, one can expect different bacterial communities within a single host mite species with different diets. However, this hypothesis is not supported by manipulative experiments using diet switching (Erban et al., 2017).
Transmission of intracellular symbiotic bacteria was suggested to be vertical from mother to offspring since known (Cardinium and Wolbachia) and suspected (Solitalea-like, Blattabacterium-like) intracellular symbionts were detected inside the surface-sterilized eggs of mites (Erban et al., 2016a; Hubert et al., 2016a). Coprophagy, namely, feeding on feces, is a strategy for the acquisition of bacterial communities from feces employed by various arthropods (Nalepa et al., 2001; Engel and Moran, 2013). Although astigmatid mites do not exhibit social behavior, they naturally form aggregations on food, resulting in colonies with high densities of mite individuals (Kuwahara, 2004). In cockroaches, bacteria in feces are responsible for their host aggregation, and cockroaches are thought to acquire their gut microbiota via coprophagy (Wada-Katsumata et al., 2015). Mites’ feces contain a nitrogen waste product, guanine, which is a mite attractant (kairomone) (Levinson et al., 1991b). Mites migrate into feces-rich habitats and aggregate at these sites (Levinson et al., 1991a). In addition to guanine, astigmatid mites exhibit advanced chemical communication via various volatile secretions of the opisthosomal glands, including aggregation pheromones, which signal food availability and the presence of the same-species individuals. Massive aggregations of mites on, for example, cheese (Robertson, 1946) or dog food, are well known (Baker and Swan, 2013). Under such conditions, mite-infested habitats become covered with their feces, and feeding on the feces or feces-contaminated food may enable the acquisition of gut microbiota by mites. Mite feces are small and lightweight and may be carried by air currents along with their contents. The realization that mites’ feces serve as a vector for microorganisms is important because this strategy could be exploited for the dissemination of bacteria and fungi in soil (Griffiths et al., 1959; Brasier, 1978).
In this study, we selected two populations of T. putrescentiae (L and P) with different microbiomes and population growth dynamics (Erban et al., 2016a, 2017). The microbiome of L population was dominated by Cardinium (71% of total sequences), Staphylococcus (10%), B. cereus (10%) and Enterobacteriaceae with low similarity to Xenorhabdus (7%). In contrast, in the microbiome of P population, Wolbachia was the most prevalent (61%), followed by Bartonella-like (17%) and Solitalea-like species (21%) (Erban et al., 2016a, 2017). Growth medium after mite cultivation, namely, the leftover diet plus dead mite bodies and feces, was used as a source of microorganisms. Using both populations of T. putrescentiae, we conducted the experiments to assess the following factors: (i) microcosm experiments, in which respiration was measured in the microcosms with and without mites on mite feces-treated and control diets; (ii) the influence of feeding on a feces-enriched diet on mite fitness, measured as the amount of nutrients in the mite bodies and mite population growth; and (iii) the influence of feeding on the feces-enriched diet on the microbiome composition.
Materials and Methods
Experimental Mites and Spent Growth Medium for Experiments
Two populations of T. putrescentiae (Schrank, 1781) were used for the experiments: the L population (strain) originated from mites sampled in a grain store in Bustehrad, Bohemia, Czechia, by E. Zdarkova in 1996; the P population was obtained from specimens collected on dog food by T. Phillips in Manhattan, KS, United States, in 2014. The mites were mass-reared as previously described (Erban et al., 2016b), with the L population reared on a stored product mite wheat-derived diet (SPMd) (Erban and Hubert, 2008) and the P population on reared on ground dog kernels (DDFd) (Rybanska et al., 2016). The diet was heated at 70°C for 30 min and rehydrated (Hubert et al., 2012). Mites were removed from the cup and culture chamber surfaces and used for experiments. The spent growth medium in the chambers was used for the initial feces extract after 60 days of mite cultivation.
Experimental Design and Samples
Feces extracts were obtained from the mite-rearing chambers for L and P populations, i.e., LE and PE extracts, with six independent replicates per population (Figure 1). The chambers containing the mites and SPGM were washed with 300 mL of sterile phosphate-buffered saline (PBST – 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl) with 0.05% (v/v) Tween® 20 detergent (Sigma-Aldrich, Saint Louis, MO, United States). The extract was transferred from the six chambers to a beaker and passed through a polyamide fiber mesh with a mesh size of 41 to 420 μm (Silk & Progress, Brnenec, Czechia) using a vacuum pump. The filtered supernatant was centrifuged (CL31R, Thermo Fisher Scientific, Waltham, MA, United States) at 845 × g for 5 min, and the supernatant was removed. The pellet was rinsed by resuspending in PBST and then centrifuged twice. The final pellet was resuspended in 10 mL of PBST. In the case of initial extract, the extracts were pooled per population for DNA extraction and diet inoculation. The extracts were applied to a powdered dog kernel; a diet lacking any extract was used in pure form as the control. Ground dog kernels (10 g) (Friskies Junior Life Plus Nutrition, Nestle Purina, Buk, Hungary) (Rybanska et al., 2016) were mixed with 4 mL of the appropriate extract; the mixtures were then vacuum dried and remoistened as previously described (Hubert et al., 2005).
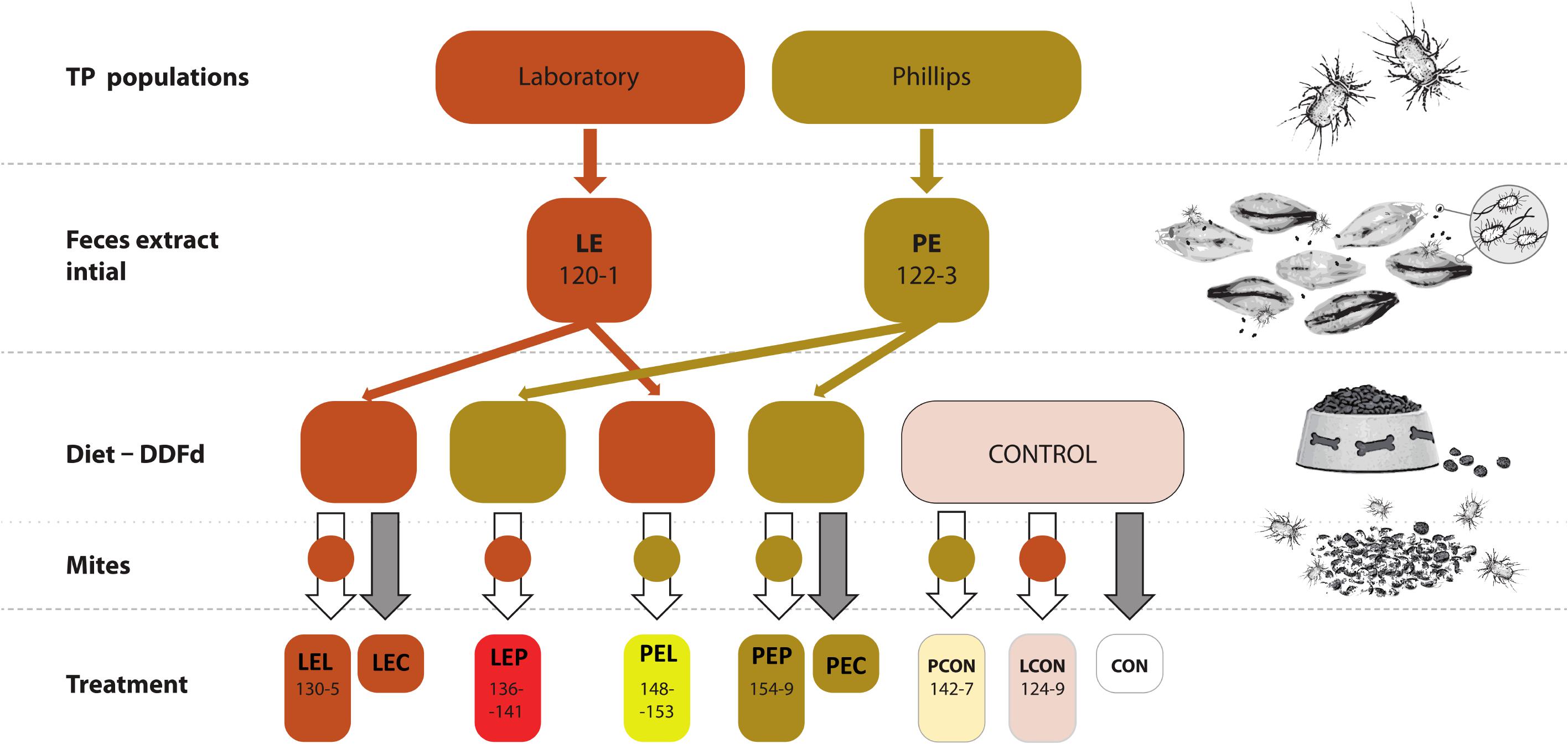
FIGURE 1. Experimental design for manipulative crosswise experiments with Tyrophagus putrescentiae and feces fraction. Two populations of mites (and P) were used to obtain initial extracts, which were added to the mite diets. The numbers indicate the numbers of biosamples in GenBank (Supplementary Table S1). CON, control diet without mites; LCON, control diet with mites from Laboratory population; LE, feces extract from the Laboratory population; LEC, diet treated with feces extract of mites from the Laboratory population, no mites were added; LEL, diet treated with feces extract from Laboratory population of mites with Laboratory population mites added; LEP, diet treated with feces extract from the Phillips population of mites with Laboratory population mites added; PCON, control diet with mites from the Phillips population mites; PE, feces extract from Phillips population mites; PEC, diet treated with feces extract of mites from the Phillips population mites, no mites were added; PEL, diet treated with feces extract from the laboratory population of mites with Phillips population mites added; PEP, diet treated with feces extract from the Phillips population of mites with Phillips mites added; DDFd, diet of mites from ground dog kernels.
Mites were introduced to the diets as follows: LCON or PCON – L or P mites fed on the control diet without any feces extracts; LEL or PEL – L or P mites fed on L mite feces extracts; LEP or PEP – L or P mites fed on P mite feces extracts. After the incubation period, the mites were removed from the rearing chambers and used for experiments. For the respiration assay, additional control treatments were prepared to include L and P-feces extract-treated diet without any mites for LEC and PEC, respectively, and control diet without mites for CON.
The Effect of Feces Addition on Microcosm Respiration
Carbon dioxide (CO2) production in a microcosm consisting of mites and their diet can provide an indirect estimate of the mite feeding effect on the microbial metabolism (Hanlon and Anderson, 1979; Bengtsson and Rundgren, 1983; Siepel and Maaskamp, 1994). As shown in a preliminary experiment, mite respiration has a negligible influence on the experimental results. We conducted a respiration assay to compare CO2 production by diets with and without the addition of feces and mites. The following experiments were performed: CON – microcosms with diet only, no mites or feces extracts were added; microcosms with diet treated by feces extract (LE or PE) and no mites added; microcosms with mites and no feces extract treatment (LCON and PCON); and mites with feces extract-treated diet (LEL, LEP, PEL, PEP).
An infrared gas analyzer (IRGA) based on the Gascard II infrared card (Edinburgh Sensors, Livingston, SCT, United Kingdom) and a respiration apparatus (Cat No: RP1LP; Qubit systems, Kingston, ON, Canada) were used as described previously (Hubert et al., 2010). The apparatus measured CO2 injected from the syringe manifold. Data were collected using LabPro Interface (Cat No: C410; Qubit) and processed with LabPro 3 software (Vernier Software & Technology, Beaverton, OR, United States). The experimental design is described in Figure 1. Feces-treated and control diets were weighed to 0.01 ± 0.001 g on a Mettler AE 240 microbalance (Mettler-Toledo, Columbus, OH, United States), transferred to an Iwaki chamber to which 50 mites were added, and incubated for 7 days at 85% humidity in darkness. After incubation, the diet and mites were weighed and transferred to the syringe in the respiration manifold. The aperture of the syringe was filled with a piece of filter paper to prevent movement of the mites into the apparatus. The manifold was connected to the apparatus, and the air in the syringe was replaced with moistened synthetic gas (80% N2 and 20% O2, Cat No: GA231; Linde Gas, Prague, Czechia). The volume of the gas in every syringe was 0.6 mL. The control syringes did not contain any diets or mites. The manifolds were incubated for 2 h in an ES-500 thermostat (Trigon Plus, Cestlice, Czechia) at 25 ± 0.1°C. After the incubation, the respiration manifold was connected to the respiration apparatus, and the CO2 content was measured immediately via the application of 0.5 mL of gas from the syringe at 120-s intervals. Logger Pro software (Vernier, Beaverton, OR, United States) recorded the concentration of CO2 for 120 s. The concentration was checked 180 times per second, and 21,600 observations were collected. The total volume of CO2 in 0.5 mL of injected air was calculated by comparing the total sums of the IRGA–CO2 signals (integrals of the concentration × time curve) minus the average of the sums of the signals from the control syringes. The CO2 produced by the chamber over 2 h of incubation was recalculated to μL of CO2 per g of fresh weight for 1 h. Respiration data did not meet normality and were therefore analyzed using the Kruskal–Wallis test with a Dunn post hoc comparison and Bonferroni corrections.
The Effect of Feces Addition on Mite Nutrient Contents and Population Growth
The estimation included analyses of nutrient contents in the bodies of mites, including protein, saccharide, lipid and glycogen, and a growth test. The samples included the following treatments: LEL, LEP, PEL, PEP, PCON, and LCON.
Two sets of mite culture samples were prepared. For the population growth test, 10 individuals per chamber were used, with six replicates per treatment; the experiment followed a previously described protocol (Erban et al., 2017). Although the mites underwent sexual reproduction, they were not sexed in the experiment. Separately, 100 unsexed mites were added to chambers in six replicates per treatment and cultivated for 21 days. For nutrient content analysis and DNA extraction, mites were collected from the chamber surface and plug as described above, transferred to 1.5-mL Eppendorf tubes, and weighed using a microbalance (MS Mettler-Toledo, Greifensee, Switzerland) to obtain 0.05 ± 0.01-g wet-weight samples (Erban et al., 2016a).
Nutrient content analyses were based on the Kaufmann method (Kaufmann, 2014) with modifications (Erban et al., 2017). The design included six biological and two technical replicates per treatment (2 samples per chamber). The data were processed using a previously described protocol (Erban et al., 2017). Data were analyzed with the Kruskal–Wallis test using a Dunn post hoc comparison and Bonferroni correction.
The growth test began by introducing 10 adult unsexed mites to treated diets; 10 replicates were performed. The mites were transferred to Iwaki flasks with 0.01 ± 0.001 g of diet and maintained under conditions described for mite cultivation (Erban et al., 2017). The experiment was terminated after 21 days, as described above, and the mites were counted under a dissecting microscope (Erban et al., 2016b). Data were analyzed with a Kruskal–Wallis test.
The Effect of Feces on the Mite Gut Contents
Approximately 100 individuals per treatment described above were removed, added to modified Bouin–Dubosque fluid (Smrz, 1989) and used for histological observations. The mites were transferred to paraffin and sectioned using a Microm HM 200 ErgoStar Microtome (Carl Zeiss, Jena, Germany) according to a previously described protocol (Erban et al., 2016a). Slide sections were stained with Mann–Domenici stains (Exbrayat, 2013) for bacterial visualization (Supplementary Figure S1). At least 15 specimens per treatment were observed using an Axioskop compound microscope equipped with a digital camera and AxioVision software (Carl Zeiss). Gut terminology followed that used for Acarus siro (Sobotnik et al., 2008).
The Effect of Feces on the Mite Microbiome
The microbiome was characterized for feces extract LE and PE, mites originated from the control cultures (LCON, PCON), and feces extract-treated diets (LEL, LEP, PEL, and PEP) (Figure 1). Samples used for mite DNA extraction originated from the same cultures as the samples used for nutrient contents experiments (see above) but were processed independently. Mites were surface cleaned, and the DNA was extracted according to a previously described protocol (Erban et al., 2016a). DNA was also extracted from 3 mL of feces extracts. DNA was isolated using a Wizard Genomic DNA Purification Kit (Cat No. A1125, Promega, Madison, WI, United States) and stored in a freezer at -28°C. We extracted DNA from every mite sample for a total of six replicates (rearing chambers) per population and two technical replicates from pooled extracts from the six rearing chambers.
DNA samples were sent to MR DNA1 (Shallowater, TX, United States) for sequencing of the V1–V3 portion of the 16S rRNA gene with the universal primers 27Fmod and 519Rmod (Supplementary Table S1); the Illumina MiSeq platform was used with the bTEFAP® process (Chiodini et al., 2015). The forward and reverse read lengths were 300 bp. The sequences were deposited in GenBank as SUB2874221 and bioproject PRJNA394876. Lists of samples and barcodes are provided in Supplementary Table S1.
Sequencing reads were processed using MOTHUR v.1.37.1 software according to the MiSeq standard operating procedure (Schloss et al., 2009; Kozich et al., 2013) and UPARSE (Edgar, 2013). Sequences were processed as previously described (Erban et al., 2017). OTUs were identified using Ribosomal Database Project (Cole et al., 2014) training set no. 15 available from UPARSE (Edgar, 2013). Chimeras were removed via alignment to the SILVA reference database (Quast et al., 2013) and UCHIME (Edgar et al., 2011), and representative sequences for OTUs were processed using the BLASTn program on the NCBI platform (Altschul et al., 1997). The resulting identifications were based on both RDP and GenBank (Supplementary Table S3). We identified Cardinium, Solitalea-like and Bartonella-like OTUs by comparing representative sequences to previously obtained near-full-length 16S rRNA cloned sequences from TP (Erban et al., 2016a), as previously described (Erban et al., 2017). Taxonomical membership and abundance of bacterial OTUs in the samples were visually summarized in Krona (Ondov et al., 2011). Data standardization was based on the subsample created in MOTHUR using 23,081 sequences. The subsampled dataset (Supplementary Table S4) was applied for all further statistical analyses using MOTHUR and later using PAST 3.06 software (Hammer et al., 2001) and R software using the “vegan” R package (Oksanen et al., 2016).
Illumina amplicon analyses: The effects of population and treatment on OTU distribution were tested by performing two-way and one-way permutational multivariate analyses of variance (PERMANOVAs) using Jaccard and Bray–Curtis dissimilarity. The Jaccard index is based on the presence and absence of OTUs, while Bray–Curtis dissimilarity uses relative OTU abundance data in non-Euclidean space (Oksanen et al., 2016). In the one-way analysis, we compared the differences between treatments using Bonferroni-corrected P-values. Visualization was carried out for the dbRDA (redundancy analysis) models based on the Bray–Curtis matrix of the “vegan” package; OTU abundance data were log-transformed (log2) (Anderson et al., 2006). Environmental variables were selected based on the forward.sel function of the package “adespatial” (Dray et al., 2017). METASTATS (White et al., 2009) was applied to describe the effects of treatment on OTUs for each population separately in MOTHUR.
Quantitative polymerase chain reaction (qPCR) analyses were carried out using a StepOnePlusTM Real-Time PCR System (Life Technologies, Grand Island, NY, United States) in 96-well plates with GoTaq qPCR Master Mix (Promega). SYBR Green (Bio-Rad Laboratories, Veenendaal, Netherlands) was used as a double-stranded DNA (dsDNA) binding dye. The protocol and primers were described previously (Erban et al., 2017); see Supplementary Table S2. We employed a design consisting of six biological replicates, each of which was analyzed with two technical replicates. The data describing the numbers of gene copies amplified by universal and specific primers were log-transformed (log10) and analyzed by PERMANOVA as described above using the Gower distance (Hammer et al., 2001). Finally, the data were separated for populations and taxa and subjected to the Kruskal–Wallis test using treatment as the comparison of treatments.
Results
The Effect of Feces Extracts on Microcosm Respiration
Microcosm respiration was measured on diets without feces extracts with and without mites (CON, LCON, and PCON). CO2 production differed significantly between the microcosms based on the control diet without mites and control diet with L mites (CON and LCON) vs. control diet with P mites (PCON) (Figure 1). Almost 60-fold higher CO2 production was observed in the control microcosms with P mites (PCON) than in the control diet without any mites (CON). No difference in the respiration of microcosms of L mites (LCON) and the control without mites was observed (CON). These results indicate that the activity of P mites accelerated microbial metabolism. When a feces extract was added into a microcosm, the CO2 production differed among the populations of mites in the microcosm. The respiration of microcosms containing L population did not differ from those without mites (i.e., LEC, LEL, and LEP). The respiration of microcosm with P mites (PEP and PEL) was three times higher than that without mites (PEC); no differences in respiration between the microcosms consisting of feces extract-treated diets without mites was found (PEC and LEC). The respiration of microcosms based on P-feces extract-treated diet and P mites (PEP) was lower than in the control experiments when P mites were fed a diet without the extract (PCON). Thus, both the feces extracts inhibited the interaction between mites and bacteria, resulting in a lower microbial respiration.
The Effect of Feces Extracts on the Mite Nutrient Contents and Population Growth
The addition of feces extract to the diet did not influence the nutrient contents in the bodies of L and P mites (Supplementary Table S5). Although there were differences among treatments in lipid, saccharide and glycogen contents, post hoc comparison revealed that none of the experiments involving extract treatment (LEL, LEP, PEL, PEP) differed from the controls (LCON or PCON). There was a single exception, in which the protein content in PEP was threefold lower than that in PCON.
Population growth experiments revealed that in the control diets without any feces extracts (LCON and PCON), the growth of P mites was 1.5 higher than that in L mites. The feces extract from the L population stimulated the growth of P mites but not L mites, while P-feces extract stimulated both L and P mites. The growth of L mites was threefold higher on P extract-treated diet than on the control (LEP and LCON). The growth of P mites was fourfold greater than in the control (PEP and PCON) (Figure 1).
The Effect of Feces Extract on the Gut Contents of Mites
Histological analyses did not detect fragments of fungal mycelium in the mite gut. There was a highly intense mesodermal cell secretion in the anterior gut, i.e., the ventriculus and the caecum (Supplementary Figure S1A). A typical food bolus (Supplementary Figure S1B) contained fragmented pieces of plant material and debris from proliferating mesodeal cells (Supplementary Figures S1B,E). Bacteria were observed attached to the surface of the food bolus (Supplementary Figure S1C) or were present in the gut lumen (Supplementary Figure S1F). There were no differences in the contents of food boli among the control mites and mites originated from feces extract-treated diets. A single exception was the food boli contained bacteria in specimens from the L population treated with P-feces extract (Figure 2). Unlike previous observations (Erban et al., 2016a), none of the specimens observed contained bacteriocytes in the fat tissues; guanine deposits in the fat tissues were observed only in specimens from the L population reared on P-feces extract-treated diet.
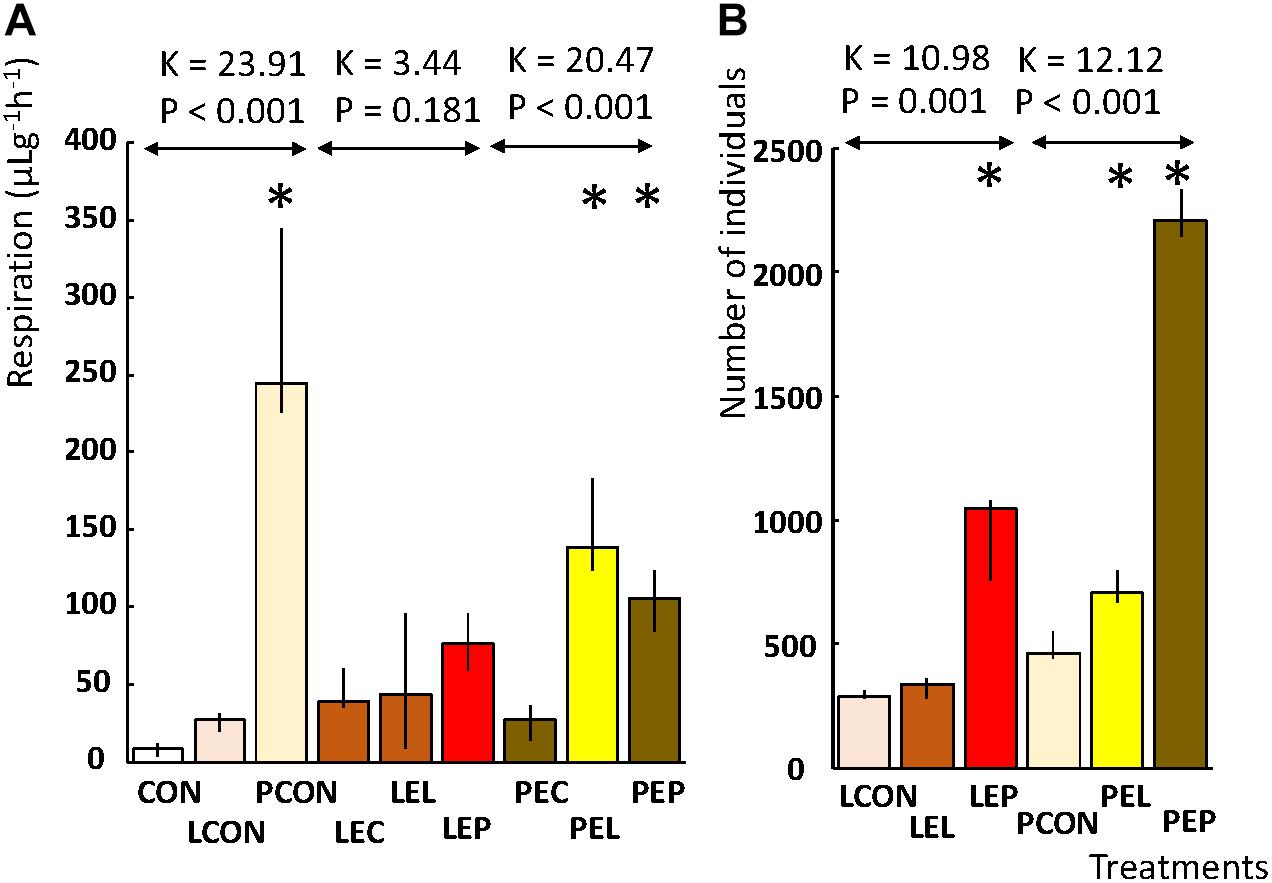
FIGURE 2. Effects of feces extract addition on the diet on the respiration and population growth of T. putrescentiae in a manipulative crosswise experiment. The columns are medians, and bars represent interquartile range. (A) Respiration is expressed as μg of CO2 per g of weight per hour. (B) Population growth was observed as the final number of individuals after 21 days from a starting population of 10 individuals; an asterisk indicates a significant difference compared to the control treatment. CON, control diet without mites; LCON, control diet with mites from the laboratory population; PCON, control diet with mites from the Phillips population; LEC, diet treated with feces extract of mites from the Laboratory population, no mites were added; PEC, diet treated with feces extract of mites from the Phillips population, no mites were added; LEL, diet treated with feces extract from the Laboratory population of mites, with laboratory mites added; LEP, diet treated with feces extract from the Phillips population of mites, with Laboratory population of mites added; PEL, diet treated with feces extract from the Laboratory population of mites, with Phillips mites added; LEP, diet treated with feces extract from the Phillips population of mites, with Phillips mites added.
Does Mite Feeding on Feces Affect Their Microbiomes?
The feces extract-treated diet had no effect on the presence/absence of OTUs in the microbiome but did influence the OTU profiles in the microbiome in both the standardized Illumina dataset and the qPCR dataset (Table 1). In the L population, feeding of mites on the diet treated by PE feces extract significantly (P < 0.05) altered the microbiome profile, while mite feeding on the diet treated by LE extract did not result in significant changes in the microbiome compared to that from the control (Table 1). For the P population, feeding of mites on the diets treated with both extracts (LE and PE) resulted in a significant difference in the microbial profiles from the control. In the qPCR dataset, feeding of the two populations on both feces extract-treated diets resulted in a difference in the numbers of copies of observed bacterial taxa compared to the control.
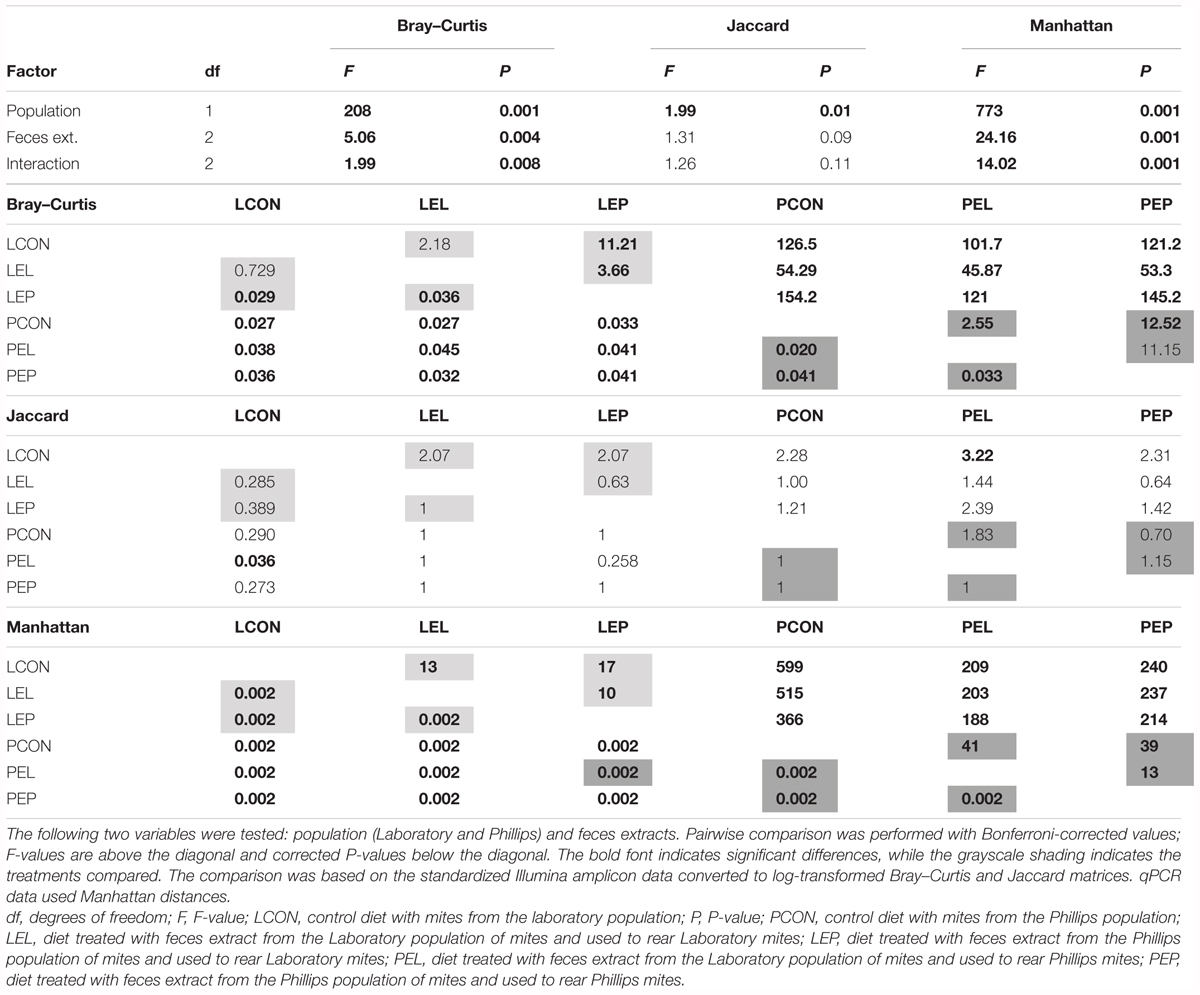
TABLE 1. Comparison (PERMANOVA) of the microbiomes of the two populations of Tyrophagus putrescentiae in the manipulative experiment.
The log2-transformed OTU profiles based on Bray–Curtis distance were used to construct a dbRDA model (F = 31.92; P < 0.001; R2 = 0.87), which explained 85% of the total variance in the dataset. After forward selection of environmental variables, P population, diet treatments by both feces extracts, and population growth were significantly correlated with the model, whereas nutrient content in the bodies of mites was not. The populations were separated by the first axis and the treatments by the second axis (Figure 3A). Another dbRDA model was constructed based on qPCR data using Manhattan distances (F = 132.92; P < 0.001; R2 = 0.90), which explained 80% of the total variance. After forward variable selection, the following variables had significant effects on the model: saccharides, P population, diet treatments with both feces extracts (PE and LE) and population growth. Mite populations and treatments were separated by the first and second axes, respectively (Figure 3B).
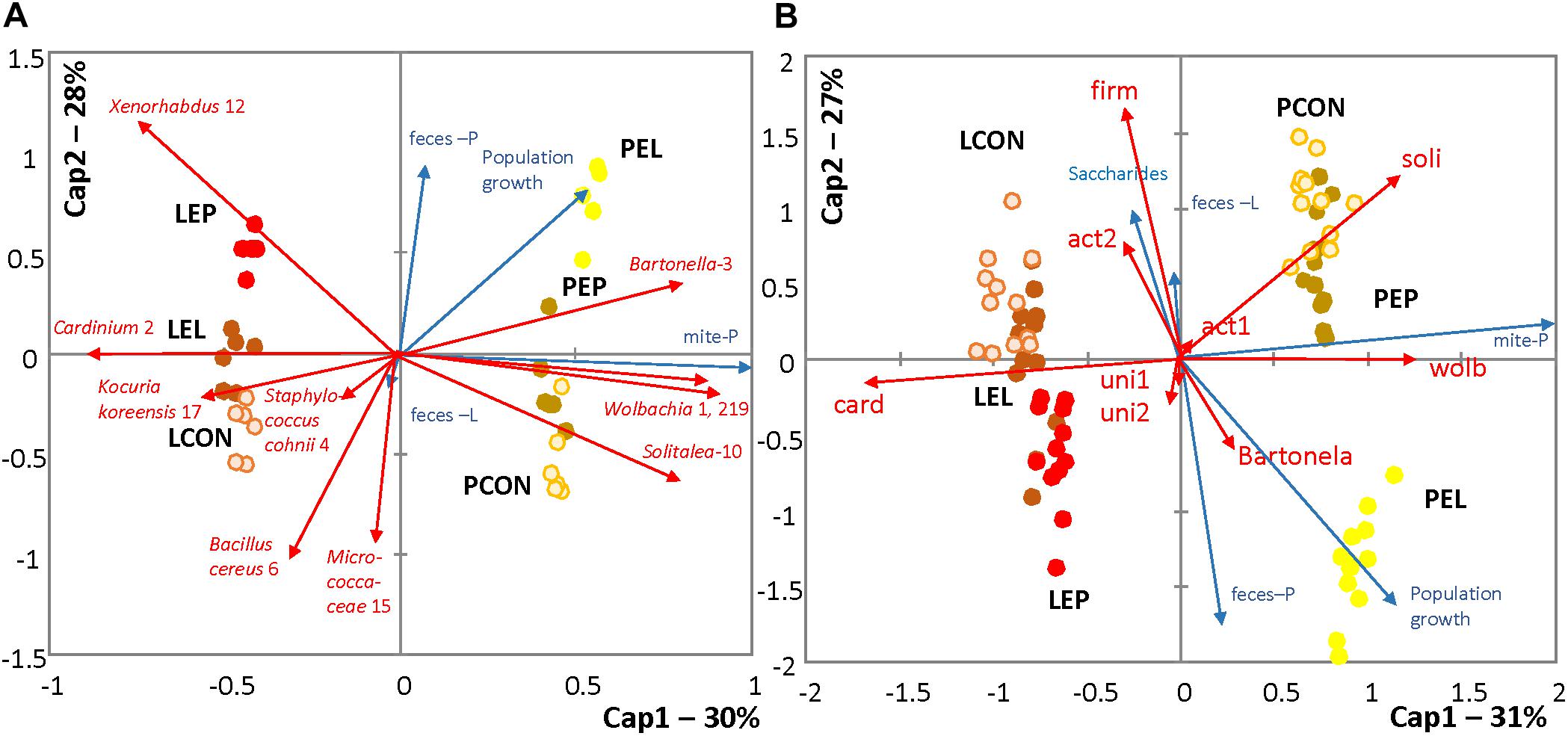
FIGURE 3. RDA triplots showing distances among profiles of the T. putrescentiae microbiomes under different treatments; environmental variables with a significant effect on the models are shown. (A) Comparison of the effects of different treatments for standardizing the Illumina dataset; the relative abundance of sequences was log-transformed (log2) and converted to a Bray–Curtis distance matrix. (B) Comparison of the absolute numbers of sequences obtained using species-specific and universal primers (Supplementary Table S1); the data were log-transformed (log10) and converted to a Manhattan distance matrix. LCON, control diet with mites from the laboratory population; PCON, control diet with mites from the Phillips population; LEC, diet treated with feces extract of mites from the Laboratory population, no mites were added; PEC, diet treated with feces extract of mites from the Phillips population, no mites were added; LEL, diet treated with feces extract from the Laboratory population of mites, with laboratory mites added; LEP, diet treated with feces extract from the Phillips population of mites, with Laboratory population of mites added; PEL, diet treated with feces extract from the Laboratory population of mites, with Phillips mites added; LEP, diet treated with feces extract from the Phillips population of mites, with Phillips mites added.
METASTATS analyses were used to identify which OTUs were responsible for the differences in the microbiomes (Tables 2, 3). The feces extract from the L population was composed mainly of Staphylococcus cohnii (OTU4), Brevibacterium avium (OTU20) and Virgibacillus halotolerans (OTU22) (Figure 4), but the relative numbers of these OTUs in both microbiomes of mites fed on extract-treated diet were not significantly different from those of the control treatments. The differences in L mites fed on L and P extract-treated diet compared to the control were as follows: an increase in Enterobacteriaceae with low similarity to Xenorhabdus (OTU12) and a decrease in Kocuria koreensis (OTU17) (Supplementary Figure S2). Micrococcaceae (OTU15) decreased in P mite feces-enriched diet (Figure 3A). The observed decrease in Actinomycetes of the Illumina profile was in agreement with the qPCR data (Figure 3B). The abundance of Cardinium, a dominant taxon in the Illumina profile (OTU2) (Figure 4), was not influenced by mite feeding on feces-treated diets, as indicated both by qPCR and Illumina amplicon datasets (Supplementary Table S6).
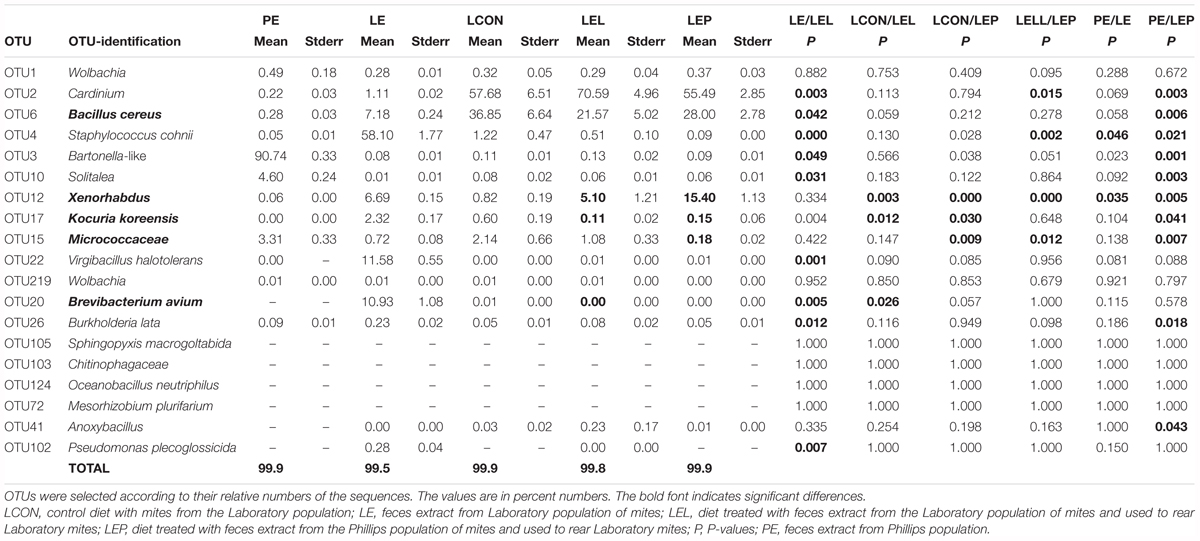
TABLE 2. Comparison of the effect of mite feces extract on the microbiome profile of T. putrescentiae Laboratory population based on METASTATS analyses of the standardized Illumina amplicon data set.
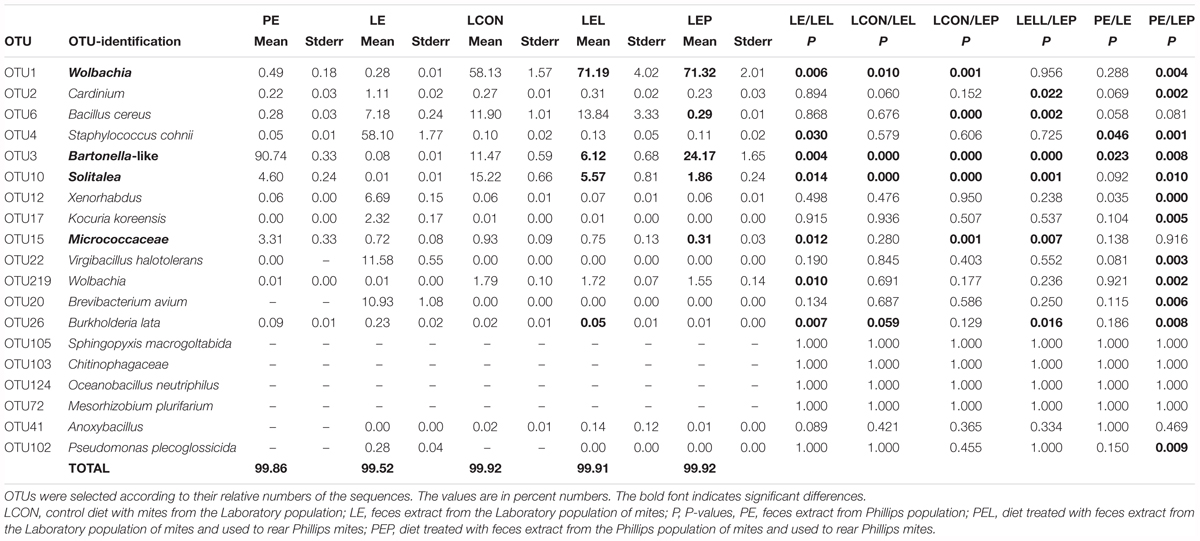
TABLE 3. Comparison of the effect of feces extract on the microbiome profile of T. putrescentiae Phillips population based on METASTATS analyses of the standardized Illumina amplicon data set.
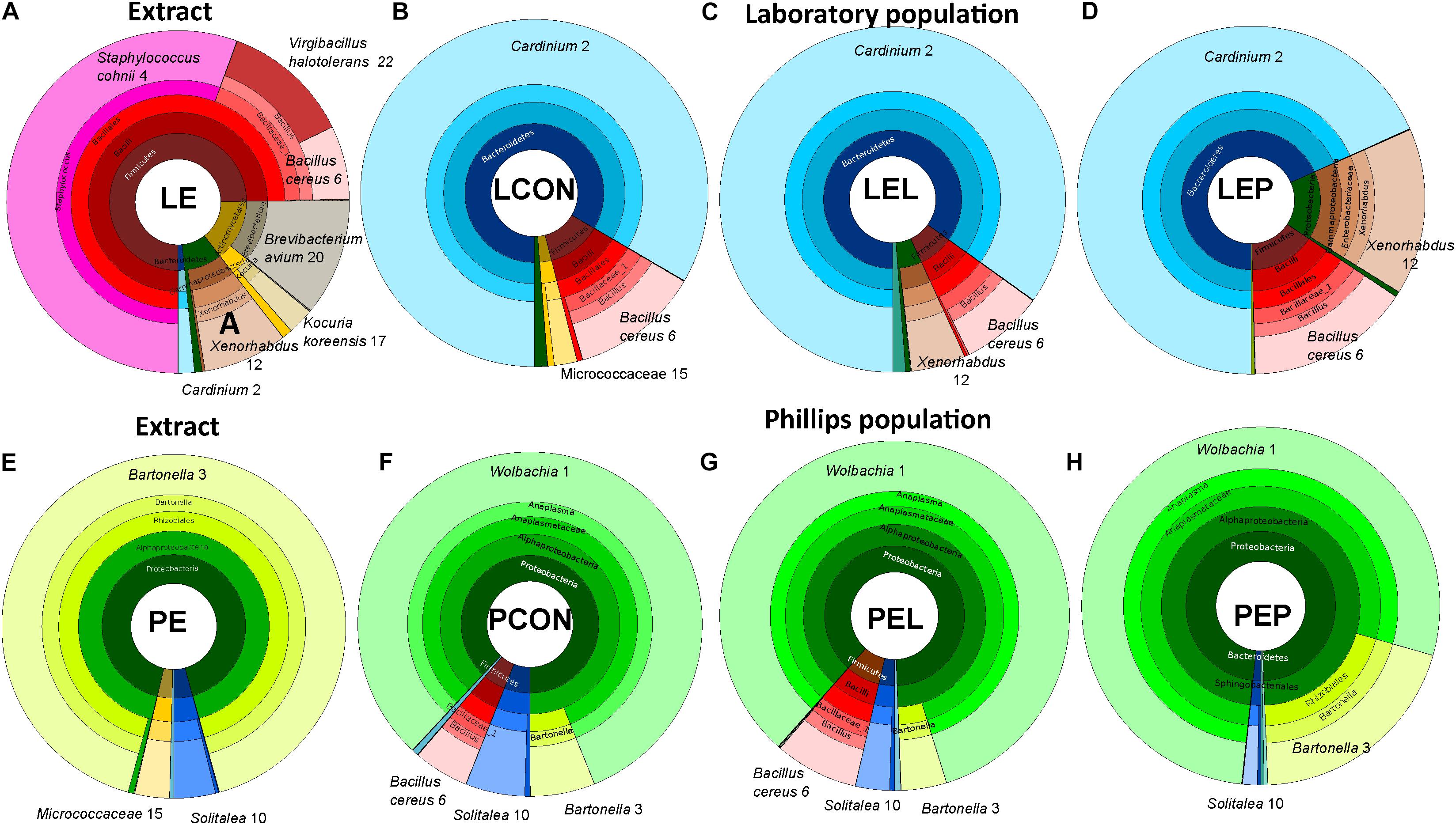
FIGURE 4. Krona visualization of microbiomes of the two populations of T. putrescentiae in the manipulative experiment based on feeding of mites on a diet enriched with mite feces; (A) bacterial composition of the feces extract form the Laboratory population (LE); (B) microbiome of the Laboratory population of mites fed on the control diet without any extract (LCON); (C) microbiome of the Laboratory population of mites fed on the Laboratory mites feces extract added to the diet (LEL); (D) microbiome of the Laboratory population fed on Phillips mites feces extract added to the diet (LEP); (E) bacterial composition of the feces extract from the Phillips population (PE); (F) microbiome of the Phillips population fed on the control diet without any extract added (PCON); (G) microbiome of the Phillips population fed on Laboratory mites feces extract added to the diet (PEL); (H) microbiome of the Phillips population feeding on Phillips mites feces extract added to the diet (PEP).
The feces extract from P mites consisted of 91% Bartonella (OTU3), and the diet containing PE resulted in an increase in the relative and absolute abundance of Bartonella in the microbiome of the mites fed on P mites feces extract-treated diet (Figure 3A). In comparison to the microbiome of mites fed on control diet, the increase in Bartonella in mites reared on P-feces extract-treated diet was from 6 to 20% of the total number of sequences (Figure 4). qPCR data (Figure 4) also supported the increase in the abundance of Bartonella in the mites fed on P extract-treated diet. The Wolbachia (OTU1) profile increased in the microbiomes of mites fed on feces-enriched diets (Figures 3A,B). In contrast, the relative and absolute abundance of Solitalea-like bacteria decreased (Figure 4) in the microbiome of mites fed on P extract-treated diet in comparison to mites fed on the control diet.
Discussion
The Effect of Feces on Respiration of Microcosms With and Without Mites
Mites and their symbiotic microorganisms depend on each other, as evidenced by the increases in microbial metabolism in microcosm experiments (Hanlon and Anderson, 1979; Bengtsson and Rundgren, 1983; Siepel and Maaskamp, 1994). It is believed that the grazing of microarthropods at optimal densities accelerates microbial metabolism (Hanlon and Anderson, 1979; Bengtsson and Rundgren, 1983; Siepel and Maaskamp, 1994). Our model organism, T. putrescentiae, feeds on various plant materials, fungi, fungal mycelium and spores and, sometimes, on nematodes, all of which are clearly recognizable in the gut (Smrz et al., 2016). However, the respiration observed in our study was linked to bacterial metabolism because fungi were not present in the gut of mites, and fungal mycelium was not observed in the treated diet.
A new aspect of our study is the finding that different populations of T. putrescentiae can have different effects on microbial respiration, namely, the feeding of P mites accelerated CO2 production more than that of L mites in microcosm experiments. Surprisingly, the highest respiration rate was observed with the control diet (not treated with feces) after 7 days with 50 individuals of TP. The explanation is that the control diet lacked microorganisms due to pasteurization and that the mites added to this diet also inoculated a new microbial community via transfer on their bodies and feces. This microbial community has no competition with the residential bacteria. The high level of CO2 production in the microcosms was caused by rapid microbial growth and the rapid metabolism of available nutrients. Because the presence of mites appears to be more important than the presence of feces extracts, it is possible that the main interaction depends more on direct feeding by the mite than on the vectoring of bacteria. Although the “feces extracts” used here may not have been representative of the mite gut microbiome, as the old culture medium was washed and filtered, they still may represent a good source of feces microbiota. Nonetheless, we suggest that the bacterial taxa with the ability to colonize plant material in the diets remained, as indicated by the increase in respiration in the microcosms with feces treatment in comparison to the control without mites.
The Effect of Feces Addition to the Diet on the Nutrient Contents in Mites and Population Growth
Mite population growth rate was suggested to be a suitable fitness indicator (Matsumoto, 1965). There were significant changes in the population growth among treatments and populations in this study. The changes in nutrients contents (lipid, glycogen, saccharide, and protein) in the mite bodies were low among the treatments. However, saccharide content was a significant factor affecting OTU distribution as evidenced by the RDA model with forward variable selection (Figure 3B).
The response of the populations of T. putrescentiae to feces extract addition to the diet was different. Additionally, the effect of feces extract on mite population growth differed by the origin of feces (L and P populations). The highest increase in the population growth was in P mites reared on P mite feces extract-treated diets (PEP). This increase was correlated with a positive increase in Bartonella-like bacteria and a decrease in B. cereus in the microbiome profiles. The addition of B. cereus to the diet decreased the population growth of T. putrescentiae, indicating an antagonistic effect of this bacterium on mites (Erban et al., 2016b). However, feces extracts from L mites slowly increased the growth of P mites (PEL); P mite feces extract slowly increased the growth of L mites (LEP) without any correlation with the abundance of Bartonella. Our explanation of these observations is that the strong bacterial growth in these experiments provided nutrients to the mites. Long-term laboratory cultivation of mites can lead to inbreeding (Wolska, 1980), which can influence mite physiology. This finding may explain the lower population growth of the L mites than of the P mites because L mites have been reared in laboratory culture for almost 30 years. Low genetic variation is indicative of inbreeding. The finding of similar ITS and CO1 haplotypes (Erban et al., 2016a) is indicative of the genetic equivalency of the cultures; thus, potential differences in response to environmental factors due to different genetic makeups can probably be ruled out.
The observed stimulatory effect of feces on the population growth may offer an explanation for the density-dependent population growth observed previously in A. siro (Pekar and Hubert, 2008). The rate of the increase in the A. siro populations was higher when starting with 100 than when starting with 50 individuals (Pekar and Hubert, 2008). Thus, the higher number of feces produced by a denser mite population enhances population growth.
The Effect of Feces Extract Addition on the Intracellular Bacterial Symbionts of Mites
We observed changes in the profile of certain bacterial taxa among the treatments in the Illumina dataset (barcoded tag sequencing) and the qPCR dataset, which gives the absolute numbers of bacterial sequences. It is likely that certain differences in the microbiome profiles among treatments resulted from neutral processes (Burns et al., 2016) and may be interpreted as random changes in intracellular symbiont abundance (Blattabacterium-like, Solitalea-like, Wolbachia, and Cardinium).
The two populations of TP differ in the profiles of intracellular symbionts, i.e., the microbiome of the L population contains 65–85% Cardinium sequences, while the P population contains 78–84% of Wolbachia sequences. Both of these bacteria are known to affect the sexual reproduction of hosts (Werren, 1997; Zchori-Fein and Perlman, 2004). As reproductive parasites, these bacteria can occupy the same niche in different mite populations, i.e., in the reproductive organs, as in spider mites (Zhao et al., 2013), and may indicate competitive exclusion of Wolbachia/Cardinium. Wolbachia is able to affect the microbial species present in the fruit fly gut (Drosophila melanogaster) (Simhadri et al., 2017). Both intracellular symbiotic bacteria can prevent new invaders from persisting in the mite host. However, in a previous experiment, a TP population lacking intracellular symbionts did not differ from mite populations infested with intracellular symbionts (Wolbachia and Cardinium) in response to the diet perturbation (Erban et al., 2017). The present study was based on mite population-level-associated bacteria screening; the situation at the level of individuals may be different and should be studied in the future.
The Effect of Feces Extract Addition on the Bartonella-Like Bacteria
Systematic (non-random) effects in the microbiome profiles were observed for Bartonella-like bacteria, which represented 90% of the sequences in P mite feces extracts. However, the relative and absolute numbers of the Bartonella-like bacteria increased in L mites after feeding on P mite feces extract-treated diet. This bacterium was present at almost 10-fold lower levels in the L microbiome than in the P microbiome. For other core taxa of the microbiome, we did not observe increases correlated with feeding on the diet treated by P mite feces extract. Thus, the change in the diet did not modulate the microbiome, as observed previously (Erban et al., 2017), even though the bacteria in feces can modulate the environment of mites.
Although we did not observe bacterial cross-contamination between the mite populations, there was a substantial horizontal transfer of Bartonella-like bacteria by P mites. Treatment of the diet with P mite feces extract increased the P mite fitness, and P mites increased microbial respiration. Feeding of P mites on the diet treated by P mite feces resulted in an increase in the proportion Bartonella-like sequences from 6 to 20% in the mite microbiome profile and accelerated mite population growth by fivefold compared to that on the untreated control.
Recent phylogenetic analyses of the 16S rRNA sequences of Bartonella-like bacteria (Hubert et al., 2017) have revealed that these microbes cluster outside of pathogenic Bartonella but together with tick-associated Bartonella tamiae (Billeter et al., 2008; Anderson et al., 2012) and form a sister group with B. apis (Kesnerova et al., 2016) together with unidentified sequences from human skin (Kong et al., 2012), ants (Bonasio et al., 2010), and stinkbugs (Matsuura et al., 2012). In ants, the bacteria have been suggested to fix atmospheric nitrogen and recycle insect waste products (Stoll et al., 2007; Anderson et al., 2012). A phylogenetic analysis of the genome of the non-pathogenic B. apis suggested its sister group relationship with B. tamiae (Segers et al., 2017). B. apis engages in an extracellular lifestyle and competes with other gut bacteria in nitrogen-limited plant diets (Segers et al., 2017). Our results provide evidence of an extracellular lifestyle of the Bartonella-like mite symbionts because these bacteria are present in relatively high numbers in the mite feces. B. apis possesses a conserved vitamin B12 biosynthesis pathway (Segers et al., 2017), and house dust mites require B vitamins for accelerated reproduction and growth (de Saint Georges-Gridelet, 1987). T. putrescentiae was observed feeding on crystalline B12 (Pankiewicz-Nowicka et al., 1986), indicating possible benefits to mite growth from the presence of B12 in Bartonella.
The related ant symbiotic bacterium Ca. Tokpelaia hoelldoblerii has the ability to recycle urea from uric acid (Neuvonen et al., 2016); however, some genes in this pathway are missing in B. apis, which vertically inherited the urease gene cluster to degrade urea into ammonia, which in turn is converted to glutamine and glutamate (Segers et al., 2017). Mites produce guanine as a nitrogen waste product, and guanine accumulation in fat tissues results in a pathological condition called white body symptoms. In this scenario, consumption of a nitrogen-rich diet results in accumulation of guanine crystals (Levinson et al., 1991a,b) in the fat tissue and damage to internal organs, such as the reproductive tract (Smrz and Catska, 1989; Smrz, 2003). Although we have no direct proof of the participation of Bartonella-like bacteria in the guanine recycling, six TP populations were compared, among which P mites showed low numbers of bodily guanine deposits (Erban et al., 2016a), corresponding to a high proportion of Bartonella-like bacteria in the microbiome profile. Although a comparison to the closely related symbiotic Bartonella indicated beneficial effects during the experiment, Bartonella did not exhibit proportional increases in the microbiome of L mites after treatment with PE.
Our experiments indicate that of the two predictors, the population origin and the diet, the former is the most important variable affecting the microbiome profiles. Our crosswise manipulation experiments using feces-enriched diet failed to significantly alter the microbiome composition and mite fitness. These results also highlight significant differences in the fitness of a single mite population in relation to the microbiome. Bartonella-like bacteria were not transferred between populations but were transferred within populations from adults to juveniles via feces. The presence of bacterial symbionts for recycling or synthesis of nitrogen compounds has not yet been proven. However, the Bartonella-like symbiont is a candidate for such an interaction, and further characterization of the genome of this bacterium is needed.
The Microbiome of T. putrescentiae Is Resistant to the Acquisition of New Bacteria From the Feces of Mites
The recent concepts of symbiotic association between host and microbes predict that the host microbiota strongly influences the physiology, anatomy, behavior, reproduction and fitness of the host (Bordenstein and Theis, 2015; Douglas and Werren, 2016). In insects, such differences have been linked to the diet (Chandler et al., 2011; Montagna et al., 2016; Chaturvedi et al., 2017) or population origin, i.e., field or laboratory (Bansal et al., 2014). This concept also includes differences in microbiomes within a species, which can be associated with the great plasticity in the ability to consume a variety of food types in T. putrescentiae. In this study, however, we found that feeding on feces did not change the presence/absence of bacteria in the microbiome of T. putrescentiae. It is not surprising that T. putrescentiae populations differ in their microbiome composition and fitness among populations (Erban et al., 2017), as was also confirmed in this study. Our experiments indicate the resistance of the microbiome to change in response to feeding on a diet containing feces extracts from different populations with different microbiomes. However, the feeding on feces resulted in changes of mite fitness, namely, accelerated population growth and changes in the abundance of certain bacterial species in the mite microbiomes.
Author’s Note
Prof. RNDr. Jaroslav Smrž, CSc., passed away suddenly on 11 August 2018 at the age of 67.
Author Contributions
JH, TE, JS, PK, and BS carried out the scientific writing. BS and JH performed the statistical analyses and bioinformatics. MN performed the experiments and the molecular biology. JH, PK, JS, and TE executed the experimental design. JH and JS executed the microanatomy.
Funding
This study was supported by project number GA15-09038S of the Czech Science Foundation (GA CR) and RO0418 by the Ministry of Agriculture of the Czech Republic institutional support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Martin Markovic for his technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02590/full#supplementary-material
Abbreviations
CON, control diet without mites; DDFd, diet of mites from ground dog kernels; L, Laboratory population of Tyrophagus putrescentiae; LCON, control diet with mites from the Laboratory population; LE, feces extract from the Laboratory population; LEC, diet treated with feces extract of mites from the Laboratory population, no mites were added; LEL, diet treated with feces extract from the Laboratory population of mites with Laboratory population mites added; LEP, diet treated with feces extract from the Phillips population of mites with Laboratory population mites added; P, Phillips population of T. putrescentiae; PCON, control diet with mites from the Phillips population; PE, feces extract from the Phillips population; PEC, diet treated with feces extract of mites from the Phillips population, no mites were added; PEL, diet treated with feces extract from the laboratory population of mites with Phillips mites added; PEP, diet treated with feces extract from the Phillips population of mites with Phillips mites added; SPGM, spent growth medium, including the remaining contents of the culture chamber after mite cultivation, containing the diets, debris of mite bodies and mite feces; SPMd, stored product mite diet; TP, T. putrescentiae.
Footnotes
References
Abou El-Atta, D. A., and Osman, M. A. (2016). Development and reproductive potential of Tyrophagus putrescentiae (Acari: Acaridae) on plant-parasitic nematodes and artificial diets. Exp. Appl. Acarol. 68, 477–483. doi: 10.1007/s10493-015-0002-5
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anderson, K. E., Russell, J. A., Moreau, C. S., Kautz, S., Sullam, K. E., Hu, Y., et al. (2012). Highly similar microbial communities are shared among related and trophically similar ant species. Mol. Ecol. 21, 2282–2296. doi: 10.1111/j.1365-294x.2011.05464.x
Anderson, M. J., Ellingsen, K. E., and McArdle, B. H. (2006). Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693. doi: 10.1111/j.1461-0248.2006.00926.x
Arlian, L. G., Geis, D. P., Vyszenski-Moher, D. L., Bernstein, I. L., and Gallagher, J. S. (1984). Antigenic and allergenic properties of the storage mite Tyrophagus putrescentiae. J. Allergy Clin. Immunol. 74, 166–171. doi: 10.1016/0091-6749(84)90281-1
Baker, A. S., and Swan, M. C. (2013). A puzzling domestic infestation of the storage mite Tyrophagus longior. J. Stored Prod. Res. 54, 64–66. doi: 10.1016/j.jspr.2013.05.004
Bansal, R., Mian, M. A. R., and Michel, A. P. (2014). Microbiome diversity of Aphis glycines with extensive superinfection in native and invasive populations. Environ. Microbiol. Rep. 6, 57–69. doi: 10.1111/1758-2229.12108
Bengtsson, G., and Rundgren, S. (1983). Respiration and growth of a fungus, Mortierella isabellina, in response to grazing by Onychiurus armatus (Collembola). Soil Biol. Biochem. 15, 469–473. doi: 10.1016/0038-0717(83)90013-5
Billeter, S. A., Miller, M. K., Breitschwerdt, E. B., and Levy, M. G. (2008). Detection of two Bartonella tamiae-like sequences in Amblyomma americanum (Acari: Ixodidae) using 16S-23S intergenic spacer region-specific primers. J. Med. Entomol. 45, 176–179. doi: 10.1093/jmedent/45.1.176
Bonasio, R., Zhang, G., Ye, C., Mutti, N. S., Fang, X., Qin, N., et al. (2010). Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071. doi: 10.1126/science.1192428
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Brasier, C. M. (1978). Mites and reproduction in Ceratocystis ulmi and other fungi. Trans. Br. Mycol. Soc. 70, 81–89. doi: 10.1016/S0007-1536(78)80175-2
Brazis, P., Serra, M., Selles, A., Dethioux, F., Biourge, V., and Puigdemont, A. (2008). Evaluation of storage mite contamination of commercial dry dog food. Vet. Dermatol. 19, 209–214. doi: 10.1111/j.1365-3164.2008.00676.x
Burns, A. R., Stephens, W. Z., Stagaman, K., Wong, S., Rawls, J. F., Guillemin, K., et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664. doi: 10.1038/ismej.2015.142
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7:e1002272. doi: 10.1371/journal.pgen.1002272
Chaturvedi, S., Rego, A., Lucas, L. K., and Gompert, Z. (2017). Sources of variation in the gut microbial community of Lycaeides melissa caterpillars. Sci. Rep. 7:11335. doi: 10.1038/s41598-017-11781-1
Chiodini, R. J., Dowd, S. E., Chamberlin, W. M., Galandiuk, S., Davis, B., and Glassing, A. (2015). Microbial population differentials between mucosal and submucosal intestinal tissues in advanced Crohn’s disease of the ileum. PLoS One 10:e0134382. doi: 10.1371/journal.pone.0134382
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
de Saint Georges-Gridelet, D. (1987). Vitamin requirements of the European house dust mite, Dermatophagoides pteronyssinus (Acari: Pyroglyphidae), in relation to its fungal association. J. Med. Entomol. 24, 408–411. doi: 10.1093/jmedent/24.4.408
Douglas, A. E., and Werren, J. H. (2016). Holes in the hologenome: why host–microbe symbioses are not holobionts. mBio 7:e02099. doi: 10.1128/mBio.02099-15
Dray, S., Bauman, D., Blanchet, G., Borcard, D., Clappe, S., Guenard, G., et al. (2017). Adespatial: Multivariate Multiscale Spatial Analysis. Available at: https://cran.r-project.org/web/packages/adespatial/ [accessed March 15, 2018].
Duek, L., Kaufman, G., Palevsky, E., and Berdicevsky, I. (2001). Mites in fungal cultures. Mycoses 44, 390–394. doi: 10.1046/j.1439-0507.2001.00684.x
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Erban, T., and Hubert, J. (2008). Digestive function of lysozyme in synanthropic acaridid mites enables utilization of bacteria as a food source. Exp. Appl. Acarol. 44, 199–212. doi: 10.1007/s10493-008-9138-x
Erban, T., Klimov, P. B., Smrz, J., Phillips, T. W., Nesvorna, M., Kopecky, J., et al. (2016a). Populations of stored product mite Tyrophagus putrescentiae differ in their bacterial communities. Front. Microbiol. 7:1046. doi: 10.3389/fmicb.2016.01046
Erban, T., Rybanska, D., Harant, K., Hortova, B., and Hubert, J. (2016b). Feces derived allergens of Tyrophagus putrescentiae reared on dried dog food and evidence of the strong nutritional interaction between the mite and Bacillus cereus producing protease bacillolysins and exo-chitinases. Front. Physiol. 7:53. doi: 10.3389/fphys.2016.00053
Erban, T., Ledvinka, O., Nesvorna, M., and Hubert, J. (2017). Experimental manipulation shows a greater influence of population than dietary perturbation on the microbiome of Tyrophagus putrescentiae. Appl. Environ. Microbiol. 83:e00128-17. doi: 10.1128/AEM.00128-17
Exbrayat, J.-M. (2013). Histochemical and Cytochemical Methods of Visualization, 1st Edn. Boca Raton, FL: CRC Press. doi: 10.1201/b14967
Fain, A., and Fauvel, G. (1993). Tyrophagus curvipenis n. sp. from an orchid cultivation in a greenhouse in Portugal (Acari: Acaridae). Int. J. Acarol. 19, 95–100. doi: 10.1080/01647959308683544
Ferrari, J., and Vavre, F. (2011). Bacterial symbionts in insects or the story of communities affecting communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1389–1400. doi: 10.1098/rstb.2010.0226
Griffiths, D. A., Hodson, A. C., and Christensen, C. M. (1959). Grain storage fungi associated with mites. J. Econ. Entomol. 52, 514–518. doi: 10.1093/jee/52.3.514
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9. Available at: http://palaeo-electronica.org/2001_1/past/issue1_01.htm [accessed March 15, 2018].
Hanlon, R. D. G., and Anderson, J. M. (1979). The effects of collembola grazing on microbial activity in decomposing leaf litter. Oecologia 38, 93–99. doi: 10.1007/BF00347827
Hubert, J., Doleckova-Maresova, L., Hyblova, J., Kudlikova, I., Stejskal, V., and Mares, M. (2005). In vitro and in vivo inhibition of alpha-amylases of stored-product mite Acarus siro. Exp. Appl. Acarol. 35, 281–291. doi: 10.1007/s10493-004-7834-8
Hubert, J., Erban, T., Kopecky, J., Sopko, B., Nesvorna, M., Lichovnikova, M., et al. (2017). Comparison of microbiomes between red poultry mite populations (Dermanyssus gallinae): predominance of Bartonella-like bacteria. Microb. Ecol. 74, 947–960. doi: 10.1007/s00248-017-0993-z
Hubert, J., Kopecky, J., Nesvorna, M., Perotti, M. A., and Erban, T. (2016a). Detection and localization of Solitalea-like and Cardinium bacteria in three Acarus siro populations (Astigmata: Acaridae). Exp. Appl. Acarol. 70, 309–327. doi: 10.1007/s10493-016-0080-z
Hubert, J., Kopecky, J., Sagova-Mareckova, M., Nesvorna, M., Zurek, L., and Erban, T. (2016b). Assessment of bacterial communities in thirteen species of laboratory-cultured domestic mites (Acari: Acaridida). J. Econ. Entomol. 109, 1887–1896. doi: 10.1093/jee/tow089
Hubert, J., Kopecky, J., Perotti, M. A., Nesvorna, M., Braig, H. R., Sagova-Mareckova, M., et al. (2012). Detection and identification of species-specific bacteria associated with synanthropic mites. Microb. Ecol. 63, 919–928. doi: 10.1007/s00248-011-9969-6
Hubert, J., Pekar, S., Nesvorna, M., and Sustr, V. (2010). Temperature preference and respiration of acaridid mites. J. Econ. Entomol. 103, 2249–2257. doi: 10.1603/ec10237
Hughes, A. M. (1976). The Mites of Stored Food and Houses: Technical Bulletin 9 of the Ministry of Agriculture, Fisheries and Food, 2nd Edn. London: Her Majesty’s Stationery Office.
Jung, J.-A., Cho, M.-R., Kim, H.-H., Kang, T.-J., Lee, J.-H., and Do, K.-R. (2010). Damages by Tyrophagus similis (Acari: Acaridae) in greenhouse spinach in Korea. Korea J. Appl. Entomol. 49, 429–432. doi: 10.5656/ksae.2010.49.4.429
Kaufmann, C. (2014). “Determination of lipid, glycogen and sugars in mosquitoes,” in MR4 Methods in Anopheles Research, 4th Edn, ed. M. Benedict (Manassas, VA: BEI Resources). Available at: https://www.beiresources.org/Publications/MethodsinAnophelesResearch.aspx [accessed March 15, 2018].
Kesnerova, L., Moritz, R., and Engel, P. (2016). Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 66, 414–421. doi: 10.1099/ijsem.0.000736
Kong, H. H., Oh, J., Deming, C., Conlan, S., Grice, E. A., Beatson, M. A., et al. (2012). Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859. doi: 10.1101/gr.131029.111
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kuwahara, Y. (2004). “Chemical ecology of astigmatid mites,” in Advances in Insect Chemical Ecology, eds R. T. Carde and J. G. Millar (Cambridge: Cambridge University Press), 76–109. doi: 10.1017/CBO9780511542664.004
Levinson, H. Z., Levinson, A. R., and Muller, K. (1991a). The adaptive function of ammonia and guanine in the biocoenotic association between ascomycetes and flour mites (Acarus siro L.). Naturwissenschaften 78, 174–176. doi: 10.1007/bf01136207
Levinson, H. Z., Levinson, A. R., and Muller, K. (1991b). Functional adaptation of two nitrogenous waste products in evoking attraction and aggregation of flour mites (Acarus siro L.). Anz. Schadlingskde. Pflanzenschutz Umweltschutz 64, 55–60. doi: 10.1007/bf01909743
Matsumoto, K. (1965). Studies on environmental factors for breeding of grain mites VII. Relationship between reproduction of mites and kind of carbohydrates in the diet. Med. Entomol. Zool. 16, 118–122. doi: 10.7601/mez.16.118 (in Japanese with English Summary).
Matsuura, Y., Kikuchi, Y., Meng, X. Y., Koga, R., and Fukatsu, T. (2012). Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl. Environ. Microbiol. 78, 4149–4156. doi: 10.1128/AEM.00673-12
Montagna, M., Mereghetti, V., Gargari, G., Guglielmetti, S., Faoro, F., Lozzia, G., et al. (2016). Evidence of a bacterial core in the stored products pest Plodia interpunctella: the influence of different diets. Environ. Microbiol. 18, 4961–4973. doi: 10.1111/1462-2920.13450
Nalepa, C. A., Bignell, D. E., and Bandi, C. (2001). Detritivory, coprophagy, and the evolution of digestive mutualisms in Dictyoptera. Insect. Soc. 48, 194–201. doi: 10.1007/pl00001767
Neuvonen, M.-M., Tamarit, D., Naslund, K., Liebig, J., Feldhaar, H., Moran, N. A., et al. (2016). The genome of Rhizobiales bacteria in predatory ants reveals urease gene functions but no genes for nitrogen fixation. Sci. Rep. 6:39197. doi: 10.1038/srep39197
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2016). Vegan: Community Ecology Package. R package Version 2.3–5. Available at: http://CRAN.R-project.org/package=vegan [accessed March 15, 2018].
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385
Pankiewicz-Nowicka, D., Boczek, J., and Davis, R. (1986). Attraction by selected organic compounds to Tyrophagus putrescentiae (Acari: Acaridae). Ann. Entomol. Soc. Am. 79, 293–299. doi: 10.1093/aesa/79.2.293
Pekar, S., and Hubert, J. (2008). Assessing biological control of Acarus siro by Cheyletus malaccensis under laboratory conditions: effect of temperatures and prey density. J. Stored Prod. Res. 44, 335–340. doi: 10.1016/j.jspr.2008.02.011
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Robertson, P. L. (1946). Tyroglyphid mites in stored products in New Zealand. Trans. R. Soc. N. Z. 76, 185–207.
Rybanska, D., Hubert, J., Markovic, M., and Erban, T. (2016). Dry dog food integrity and mite strain influence the density-dependent growth of the stored-product mite Tyrophagus putrescentiae (Acari: Acaridida). J. Econ. Entomol. 109, 454–460. doi: 10.1093/jee/tov298
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Segers, F. H. I. D., Kesnerova, L., Kosoy, M., and Engel, P. (2017). Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J. 11, 1232–1244. doi: 10.1038/ismej.2016.201
Siepel, H., and Maaskamp, F. (1994). Mites of different feeding guilds affect decomposition of organic matter. Soil Biol. Biochem. 26, 1389–1394. doi: 10.1016/0038-0717(94)90222-4
Simhadri, R. K., Fast, E. M., Guo, R., Schultz, M. J., Vaisman, N., Ortiz, L., et al. (2017). The gut commensal microbiome of Drosophila melanogaster is modified by the endosymbiont Wolbachia. mSphere 2:00287-17. doi: 10.1128/mSphere.00287-17
Smrz, J. (1989). Internal anatomy of Hypochthonius rufulus (Acari: Oribatida). J. Morphol. 200, 215–230. doi: 10.1002/jmor.1052000210
Smrz, J. (2003). Microanatomical and biological aspects of bacterial associations in Tyrophagus putrescentiae (Acari: Acaridida). Exp. Appl. Acarol. 31, 105–113. doi: 10.1023/B:APPA.0000005111.05959.d6
Smrz, J., and Catska, V. (1989). The effect of the consumption of some soil fungi on the internal microanatomy of the mite Tyrophagus putrescentiae (Schrank) (Acari. Acaridida). Acta Univ. Carol. Biol. 33, 81–93.
Smrz, J., and Catska, V. (2010). Mycophagous mites and their internal associated bacteria cooperate to digest chitin in soil. Symbiosis 52, 33–40. doi: 10.1007/s13199-010-0099-6
Smrz, J., Soukalova, H., Catska, V., and Hubert, J. (2016). Feeding patterns of Tyrophagus putrescentiae (Sarcoptiformes: Acaridae) indicate that mycophagy is not a single and homogeneous category of nutritional biology. J. Insect Sci. 16:94. doi: 10.1093/jisesa/iew070
Smrz, J., Svobodova, J., and Catska, V. (1991). Synergetic participation of Tyrophagus putrescentiae (Schrank) (Acari; Acaridida) and its associated bacteria on the destruction of some soil micromycetes. J. Appl. Entomol. 111, 206–210. doi: 10.1111/j.1439-0418.1991.tb00312.x
Smrz, J., and Trelova, M. (1995). The association of bacteria and some soil mites (Acari: Oribatida and Acaridida). Acta Zool. Fenn. 196, 120–123.
Sobotnik, J., Alberti, G., Weyda, F., and Hubert, J. (2008). Ultrastructure of the digestive tract in Acarus siro (Acari: Acaridida). J. Morphol. 269, 54–71. doi: 10.1002/jmor.10573
Stoll, S., Gadau, J., Gross, R., and Feldhaar, H. (2007). Bacterial microbiota associated with ants of the genus Tetraponera. Biol. J. Linn. Soc. 90, 399–412. doi: 10.1111/j.1095-8312.2006.00730.x
Wada-Katsumata, A., Zurek, L., Nalyanya, G., Roelofs, W. L., Zhang, A., and Schal, C. (2015). Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. U.S.A. 112, 15678–15683. doi: 10.1073/pnas.1504031112
Walter, D. E., Hudgens, R. A., and Freckman, D. W. (1986). Consumption of nematodes by fungivorous mites, Tyrophagus spp. (Acarina: Astigmata: Acaridae). Oecologia 70, 357–361. doi: 10.1007/bf00379497
Werren, J. H. (1997). Biology of Wolbachia. Annu. Rev. Entomol. 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
White, J. R., Nagarajan, N., and Pop, M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. doi: 10.1371/journal.pcbi.1000352
Wolska, K. J. (1980). Effect of inbreeding on quantitative features of copra mite Tyrophagus putrescentiae Schr., Acarina: Acaridae. Genet. Pol. 21, 291–307.
Zchori-Fein, E., and Perlman, S. J. (2004). Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 13, 2009–2016. doi: 10.1111/j.1365-294X.2004.02203.x
Zhao, D.-X., Chen, D.-S., Ge, C., Gotoh, T., and Hong, X.-Y. (2013). Multiple infections with Cardinium and two strains of Wolbachia in the spider mite Tetranychus phaselus Ehara: revealing new forces driving the spread of Wolbachia. PLoS One 8:e54964. doi: 10.1371/journal.pone.0054964
Keywords: Bartonella, feeding, feces, transmission, soil, fungi, bacteria, diets
Citation: Hubert J, Nesvorna M, Sopko B, Smrz J, Klimov P and Erban T (2018) Two Populations of Mites (Tyrophagus putrescentiae) Differ in Response to Feeding on Feces-Containing Diets. Front. Microbiol. 9:2590. doi: 10.3389/fmicb.2018.02590
Received: 11 June 2018; Accepted: 10 October 2018;
Published: 30 October 2018.
Edited by:
Suhelen Egan, University of New South Wales, AustraliaReviewed by:
Felicity Victoria Crotty, Royal Agricultural University, United KingdomThomas L. Kieft, New Mexico Institute of Mining and Technology, United States
Copyright © 2018 Hubert, Nesvorna, Sopko, Smrz, Klimov and Erban. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Hubert, aHViZXJ0QHZ1cnYuY3o=
 Jan Hubert
Jan Hubert Marta Nesvorna
Marta Nesvorna Bruno Sopko1,2
Bruno Sopko1,2 Pavel Klimov
Pavel Klimov Tomas Erban
Tomas Erban