
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 30 October 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02577
This article is part of the Research TopicMALDI-TOF MS Application for Susceptibility Testing of Microorganisms View all 12 articles
MALDI-TOF MS technology has made possible revolutionary advances in the diagnosis of infectious diseases. Besides allowing rapid and reliable identification of bacteria and fungi, this technology has been recently applied to the detection of antimicrobial resistance. Several approaches have been proposed and evaluated for application of MALDI-TOF MS to antimicrobial susceptibility testing of bacteria, and some of these have been or might be applied also to yeasts. In this context, the comparison of proteomic profiles of bacteria/yeasts incubated with or without antimicrobial drugs is a very promising method. Another recently proposed MALDI-TOF MS-based approach for antifungal susceptibility testing is the application of the semi-quantitative MALDI Biotyper antibiotic susceptibility test rapid assay, which was originally designed for antimicrobial susceptibility testing of bacteria, to yeast isolates. Increasingly effective and accurate MS tools and instruments as well as the possibility to optimize analytical parameter settings for targeted applications have generated an expanding area in the field of clinical microbiology diagnostics, paving the way for the development and/or optimization of rapid methods for antifungal susceptibility testing in the near future. In the present study, the state of the art of MALDI-TOF MS applications to antifungal susceptibility testing is reviewed, and cutting-edge developments are discussed, with a particular focus on methods allowing rapid detection of drug resistance in pathogenic fungi causing systemic mycoses.
During the last two decades, MALDI-TOF MS technology has rapidly evolved toward new applications in the field of clinical diagnostic microbiology, generating a novel, robust and accurate tool for rapid identification of bacteria and yeasts (Kliem and Sauer, 2012; Richter et al., 2012; Spanu et al., 2012; Szabados et al., 2012; Idelevich et al., 2014; Barnini et al., 2015; Florio et al., 2018b). In parallel, major efforts have been made to expand and apply the enormous potentialities of this technology to another important issue: testing resistance and, in some cases, susceptibility of microorganisms to antimicrobial drugs. The emergence and diffusion of bacterial and fungal pathogens resistant to multiple drugs and the high incidence of hospital-acquired bloodstream infections (BSIs) have made the purpose of developing new methods for rapid and reliable antimicrobial susceptibility testing an urgent issue. In this context, considerable efforts have been made in order to apply MALDI-TOF MS technology to the detection of antimicrobial resistance, especially for multi-drug resistant (MDR) microorganisms causing systemic infections (Nordmann et al., 2011; Cantòn et al., 2012; Patel and Bonomo, 2013; Lamoth et al., 2018), which represent a major concern for public health (Walsh, 2010; Walsh and Gamaletsou, 2013; Lee et al., 2016; Barchiesi et al., 2017). Several MALDI-TOF MS-based approaches have been attempted for the detection of antibiotic resistance in bacteria. These include: (i) the assessment of β-lactamase activity by MALDI-TOF MS detection of hydrolysis products of β-lactam/carbapenem antibiotics (Hooff et al., 2012; Hrabák et al., 2012; Jung et al., 2014; Oviano et al., 2016); (ii) identification in mass spectra of genes (Lau et al., 2014; Rodríguez-Baño et al., 2018) or peptides specifically involved in antibiotic resistance mechanisms (Gaibani et al., 2016; Chang et al., 2018); (iii) identification of biomarkers somehow correlated with, though not responsible for drug resistance, such as the phenol-soluble protein toxin (PSM-mec), produced by a subset of methicillin-resistant Staphylococcus aureus strains (Chatterjee et al., 2011), and detectable by MALDI-TOF as a 2415 ± 2.00 m/z peak (Rhoads et al., 2016); and (iv) assays based on discrimination of mass spectra of resistant from susceptible microbial isolates after exposure to breakpoint concentrations of antimicrobial agents directly on the target plate for MALDI-TOF MS analysis (Idelevich et al., 2017). This latter approach seems very promising since, theoretically, it might be applied to any microbial species and antimicrobial agent independently from the underlying resistance mechanisms, and it may be suitable for laboratory automation and simultaneous testing of a panel of different antimicrobial drugs.
As multi-drug resistance is an increasing widespread problem also in fungal infections, there is a pressing need for rapid methods allowing to obtain timely and reliable information on antifungal susceptibility/resistance of fungal infectious agents, especially those causing systemic mycoses. In spite of advances in diagnosis and treatment, the incidence of invasive fungal infections has dramatically increased during the last two decades (Barchiesi et al., 2016; Benedict et al., 2017; Cornely et al., 2017; Guo et al., 2017; Pana et al., 2017), with only a few classes of antifungal drugs being available (Pfaller, 2012; Pierce and Lopez-Ribot, 2013; Patil and Majumdar, 2017). The growing elderly population, frequently suffering from co-morbidities, high colonization rate by Candida albicans, immunosuppressive treatment after organ transplantation, and prolonged antibiotic therapies have substantially contributed to such an increase in fungal infections (Guimarães et al., 2012; Wang et al., 2014; Barchiesi et al., 2017). Although C. albicans remains the most frequently isolated species, other fungal pathogens have been isolated with remarkably increasing frequency; these include Candida glabrata (Cleveland et al., 2012; Pfaller et al., 2014), the emerging pathogen Candida auris (Bao et al., 2018; Kohlenberg et al., 2018; Kordalewska et al., 2018), Trichosporon spp. (Davies and Thornton, 2014), and filamentous fungi, such as Scedosporium spp., mucoralean fungi, and Fusarium spp. (Walsh and Gamaletsou, 2013; Davuodi et al., 2015).
Systemic infections sustained by Candida spp. as well as by other fungal pathogens are associated with mortality rates that can be higher than 60%, depending on the patient category (Kett et al., 2011; Kollef et al., 2012; Barchiesi et al., 2016, 2017; Guo et al., 2017). Since timely administration of effective antifungal therapy is of vital importance for the outcome of patients affected by these infections (Garey et al., 2006) and susceptibility profiles to antifungal agents vary greatly among fungi, rapid species identification and antifungal susceptibility testing (AFST) is fundamental to reduce mortality and improve patients’ outcome. MALDI-TOF MS, extensively used for identification of bacteria, has been increasingly proposed also for rapid identification of fungal pathogens directly in positive BCs (Ferreira et al., 2011; Yan et al., 2011; Spanu et al., 2012; Idelevich et al., 2014; Vecchione et al., 2018). Yeast identification by itself provides relevant clinical information since different fungal species may differ in virulence and drug resistance. For example, the antimicrobial susceptibility profile of Candida parapsilosis and C. glabrata can be quite different, with C. parapsilosis more frequently resistant to echinocandins and C. glabrata to azoles (Silva et al., 2012). Furthermore, invasive trichosporonosis is characterized by resistance to amphotericin and echinocandins, and poor prognosis (Miceli et al., 2011). Therefore, the ability to rapidly identify these yeasts may be useful to promptly streamline empirical antimicrobial therapy. However, the emergence and spread of MDR fungal pathogens (Lamoth et al., 2018) have posed a pressing need for rapid AFST. In general, echinocandins are widely used as empirical antifungal therapy for patients with candidemia, at least until AFST results become available; when resistance is detected, treatment with echinocandins needs to be immediately streamlined. Therefore, a rapid method for AFST may be of vital importance to achieve a favorable outcome, especially in critically ill patients. To this purpose, the application of MALDI-TOF MS to AFST has been attempted in the last few years (Vella et al., 2013; Saracli et al., 2015; Vatanshenassan et al., 2018). In this context, two methods have been proposed, which revealed potential for the development of rapid assays based on MALDI-TOF MS. The first is based on the comparative analysis of mass spectra of fungal isolates exposed to different concentrations of antifungal drugs (Vella et al., 2013, 2017; Saracli et al., 2015). The second relies on the application and optimization of the MALDI Biotyper antibiotic susceptibility test rapid assay (MBT ASTRA), originally developed for rapid antibiotic susceptibility testing in bacteria (Sparbier et al., 2016), to AFST (Vatanshenassan et al., 2018).
The present review provides a synthetic, updated overview of the proposed methods based on MALDI-TOF MS and aimed at yielding rapid and accurate information regarding antimicrobial resistance of clinically relevant fungi.
A method has been proposed for the detection of resistance to specific antifungal agents that relies on the comparison of MALDI-TOF MS spectra of yeast strains after incubation with high, intermediate or null antifungal concentrations (De Carolis et al., 2012; Vella et al., 2013, 2017; Saracli et al., 2015; Sanguinetti and Posteraro, 2016). In particular, MALDI-TOF MS spectra of yeast isolates after exposure to high, intermediate or null drug concentrations are used to generate a composite correlation index (CCI). The intermediate drug concentration is the minimal concentration of the compound able to induce a detectable proteome change in the mass spectrum of a susceptible strain. When a susceptible strain is tested, the mass spectrum at the intermediate drug concentration is more similar to that at the high concentration; therefore, the CCI of the intermediate/null concentration is lower than that of the intermediate/high concentration. This method has been used to test the resistance of C. albicans, C. tropicalis, and C. glabrata to triazole drugs. The results showed essential agreement ranging between 54 and 97% for the MALDI-TOF method in comparison to conventional AFST. The wide variability in essential agreement was dependent on both the triazole drug and Candida species tested (Saracli et al., 2015). A limit of this method was that the time saving over the conventional AFST method was of modest entity (overnight versus 24 h).
In another study based on the same approach (Vella et al., 2013), resistance to caspofungin could be detected in 10/11 (90.6%) C. albicans clinical isolates after an incubation of only 3 h, and 51/51 (100%) isolates were correctly classified as susceptible. The single very major error (false susceptibility) related to an isolate with a low level of caspofungin resistance. Caspofungin selectively targets the fungal cell wall by inhibiting 1,3-β-glucan synthase. Due to its high efficacy and reduced cases of side effects and adverse events in comparison with other antifungal agents, caspofungin has become a front-line agent in the treatment of candidemia and other invasive fungal infections (Patil and Majumdar, 2017). However, resistance to caspofungin can arise because of mutations in the FSK1 gene in C. albicans (Balashov et al., 2006), and FSK1 and FSK2 genes in C. glabrata (Pham et al., 2014). Due to the emergence of drug resistant yeast strains (Chen et al., 2011), rapid detection of caspofungin resistance in Candida spp. clinical isolates is essential for timely and appropriate treatment of systemic fungal infections.
Recently, the same method based on detection of changes in the protein spectrum after a 3-h incubation with antifungal drugs was used to test a panel of 80 C. glabrata clinical isolates against anidulafungin or fluconazole (Vella et al., 2017). In comparison to the reference method recommended by the Clinical and Laboratory Standards Institute [CLSI], 2012), 58/58 (100%) susceptible isolates were classified as susceptible, and 11/22 (50.0%) isolates in the resistant category were classified as resistant to anidulafungin. For fluconazole, 40/41 (97.6%) susceptible isolates were classified as susceptible, and 37/39 (94.9%) isolates in the resistant category were classified as resistant. Of interest, when the assay was repeated for the 11 resistant isolates giving very major error with longer incubation times (6, 9, and 12 h), only two errors for anidulafungin remained unresolved; with a 15-h incubation, 100% essential agreement was reached with both the antifungal drugs. Therefore, incubation time was a critical factor to achieve maximum accuracy and reliability of this assay, especially for anidulafungin.
In a recent study (Paul et al., 2018), an assay based on CCI analysis was used to evaluate fluconazole resistance in 15 fluconazole resistant (MICs ranging from 16 to 128 μg/mL) and 19 fluconazole susceptible C. tropicalis isolates (MIC ≤ 1 μg/mL) in comparison with the reference CLSI microdilution method (Clinical and Laboratory Standards Institute [CLSI], 2012). In India, C. tropicalis is the commonest agent causing candidemia, and fluconazole resistant strains have been increasingly isolated (Chakrabarti et al., 2015). Therefore, a rapid method allowing the detection of fluconazole resistance in C. tropicalis strains grown in BCs may be very helpful to promptly streamline antifungal therapy. The authors reported that spectral changes were detectable by visual inspection soon after 4-h exposure to high (128 μg/mL), intermediate (1 μg/mL) or null concentrations of fluconazole for all C. tropicalis isolates. For software-based analysis, the incubation time was optimized at 5 h with 4 μg/mL as intermediate drug concentration. Among the 34 isolates tested, the minimal profile change concentrations (MPCCs), i.e., the minimum drug concentration at which an alteration of mass spectra can be detected, coincided with the MICs for 16 isolates (4 resistant, 12 susceptible), whereas the MPCC was one twofold dilution lower than the corresponding MIC in the remaining 18 isolates (11 resistant, 7 susceptible). Categorical and essential agreement were observed for all 34 isolates, indicating that CCI analysis might be used as a rapid screening method for fluconazole resistance in C. tropicalis clinical isolates.
In a recent proof of concept study (Vatanshenassan et al., 2018) the detection of C. albicans and C. glabrata strains resistant to caspofungin was evaluated by MBT ASTRA. This is a MALDI-TOF-based semi-quantitative technique, which was originally designed for rapid antibiotic susceptibility testing in bacteria. MBT ASTRA is a phenotypic assay comparing growth of microorganisms after incubation in the absence or in the presence of different concentrations of antimicrobial drugs. Cell growth is inferred from the comparison of the area under the curve (AUC) of the MALDI-TOF MS spectra for the different incubation setup (with or without antimicrobial drug). The relative growth (RG) is calculated for each concentration of antimicrobial agent as the ratio of the AUC observed with or without drug exposure, and a RG cutoff discriminating resistance from susceptibility is determined experimentally: strains showing RG above this cutoff are considered resistant, whereas those below the cutoff are considered susceptible. The MBT ASTRA results were compared with those obtained by the CLSI reference microdilution method (Clinical and Laboratory Standards Institute [CLSI], 2017) on 58 C. albicans and 57 C. glabrata clinical isolates. A categorical agreement of 100% was observed for 29 susceptible and 22 resistant strains of C. albicans. For seven C. albicans strains, insufficient growth was observed in the control setup (no caspofungin) and, therefore, these strains were excluded from further evaluation, resulting in a validity of the MBT ASTRA assay of 88%. For C. glabrata, 31 out of 33 strains categorized as resistant by the microdilution method resulted resistant also by MBT ASTRA, and four out of five susceptible strains were correctly detected. Eighteen out of 19 strains categorized as intermediate by the microdilution method resulted resistant by MBT ASTRA, while only one intermediate strain resulted susceptible. Therefore, a sensitivity of 94% and a specificity of 80% were observed using MBT ASTRA on C. glabrata strains. These results were obtained after a 6-h incubation of yeast cells with or without caspofungin. Time saving compared to the reference microdilution method was approximately 18 and 42 h for C. albicans and C. glabrata, respectively. The RG cutoff was set at 0.6 for both C. albicans and C. glabrata; breakpoint concentrations of 1 and 0.5 μg/mL caspofungin, discriminating susceptible from resistant strains, were determined for C. albicans and C. glabrata, respectively.
Overall, this approach seems promising, and it would be interesting to evaluate its performance in clinical settings, and the possibility to extend its application to the detection of resistance to other antifungal agents. In addition, it would be of great usefulness if it could be efficiently applied to yeasts directly recovered from positive BCs by lysis/centrifugation in Serum Separator Tubes (BD Vacutainer system) or using other suitable methods (Florio et al., 2018a). To this purpose, further studies might be necessary to optimize the analysis software and experimental conditions.
In this review, we summarized recently proposed methods and significant advances for rapid detection of antifungal resistance by MALDI-TOF MS with a particular focus on methods applicable to positive BCs, as schematically represented in Figure 1. The diagnosis of fungal BSIs by conventional methods is particularly time consuming, while in case of severe, life-threatening infections, such as systemic mycoses, timeliness and accuracy of test results are crucial factors for clinicians to decide and promptly administer an effective and targeted antifungal therapy. Conventional microdilution methods for AFST are generally very accurate but require long incubation times. Since mortality rates in systemic infections are strictly correlated with time between onset of symptoms and administration of effective therapy, rapid methods for AFST are strongly needed. Therefore, major research efforts have to be made to refine and optimize MALDI-TOF MS-based assays in order to obtain timely, accurate and reliable results. Because of the increasing diffusion of MDR fungal pathogens, methods that allow simultaneous susceptibility testing to different classes of antifungals are expected to represent a major focus of research in this field. Differently from bacteria, the existence of specific peaks associated with antimicrobial resistance in mass spectra of fungal pathogens has not been reported so far, leaving this possibility an open issue. In addition, the development of new analytical algorithms, automation of procedures, and optimization of assays are expected to expand and refine the clinical applications of MALDI-TOF MS technology to AFST in the near future.
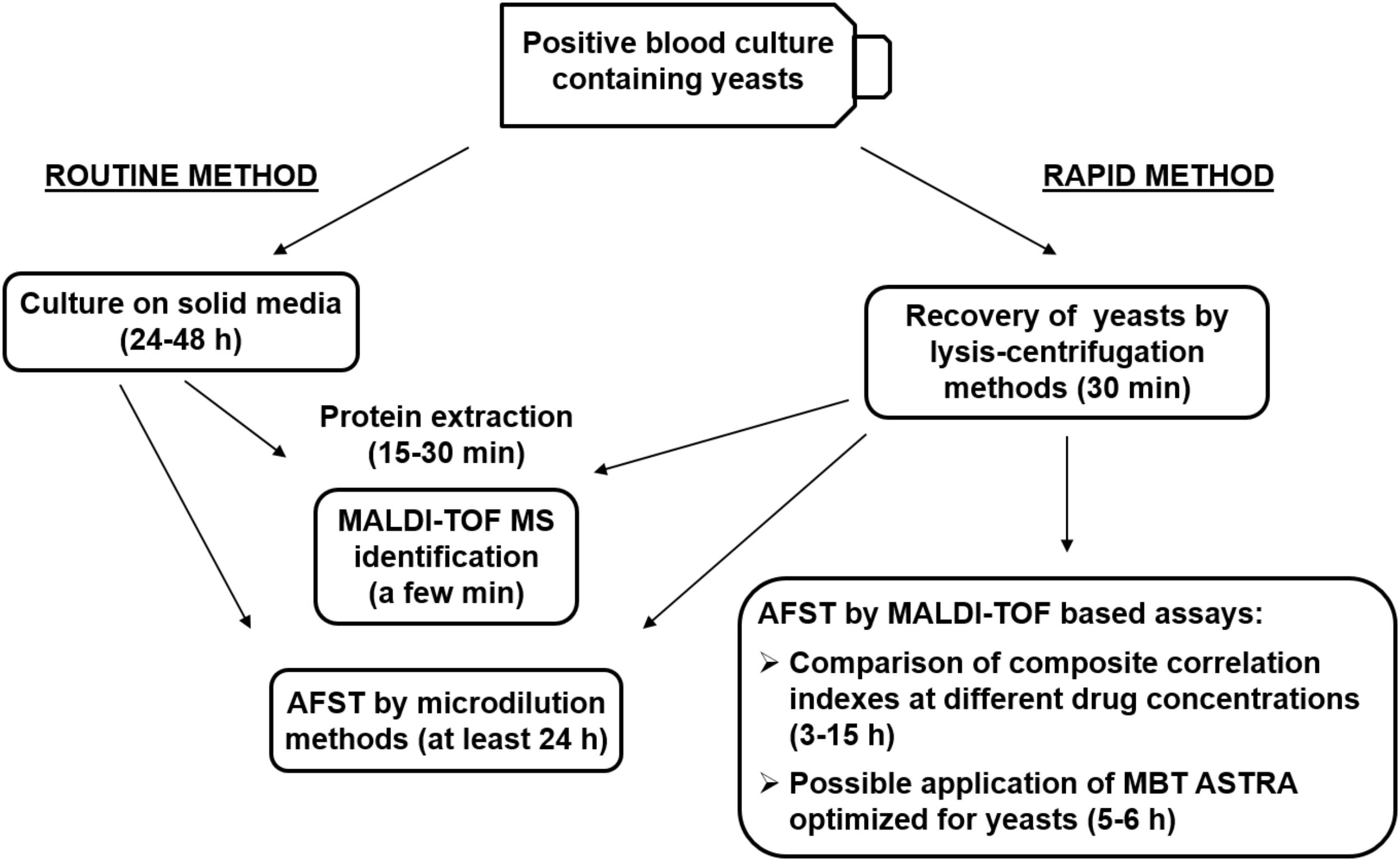
FIGURE 1. Schematic representation of the routine and rapid methods for identification and antifungal susceptibility testing (AFST) of yeasts grown in monomicrobial blood cultures, and possible applications of newly described methods. MBT ASTRA, MALDI Biotyper antibiotic susceptibility test rapid assay.
WF and AL contributed to the conception and design of the study. WF wrote the first draft of the manuscript. WF, AT, EG, and AL contributed to manuscript revision and read and approved the submitted version.
Financial support was obtained from the University of Pisa (Grant No. PRA_2017_18 n°18).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Balashov, S., Park, S., and Perlin, D. S. (2006). Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50, 2058–2063.
Bao, J. R., Master, R. N., Azad, K. N., Schwab, D. A., Clark, R. B., Jones, R. S., et al. (2018). Rapid, accurate identification of Candida auris by using a novel matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) database (library). J. Clin. Microbiol. 56:e01700-17. doi: 10.1128/JCM.01700-17
Barchiesi, F., Orsetti, E., Gesuita, R., Skrami, E., Manso, E., and The Candidemia Study Group (2016). Epidemiology, clinical characteristics, and outcome of candidemia in a tertiary referral center in Italy from 2010 to 2014. Infection 44, 205–213. doi: 10.1007/s15010-015-0845-z
Barchiesi, F., Orsetti, E., Mazzanti, S., Trave, F., Salvi, A., Nitti, C., et al. (2017). Candidemia in the elderly: what does it change? PLoS One 12:e0176576. doi: 10.1371/journal.pone.0176576
Barnini, S., Ghelardi, E., Brucculeri, V., Morici, P., and Lupetti, A. (2015). Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol. 15:124. doi: 10.1186/s12866-015-0459-8
Benedict, K., Richardson, M., Vallabhaneni, S., Jackson, B. R., and Chiller, T. (2017). Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect. Dis. 17, e403–e411. doi: 10.1016/S1473-3099(17)30443-7
Cantòn, R., Akòva, M., Carmeli, Y., Giske, C. G., Glupczynski, Y., Gniadkowski, M., et al. (2012). The European network on Carbapenemases. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18, 413–431. doi: 10.1111/j.1469-0691.2012.03821.x
Chakrabarti, A., Sood, P., Rudramurthy, S. M., Chen, S., Kaur, H., Capoor, M., et al. (2015). Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 41, 285–295. doi: 10.1007/s00134-014-3603-2
Chang, K.-C., Chung, C.-Y., Yeh, C.-H., Hsu, K.-H., Chin, Y.-C., Huang, S.-S., et al. (2018). Direct detection of carbapenemase-associated proteins of Acinetobacter baumannii using nanodiamonds coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Meth. 147, 36–42. doi: 10.1016/j.mimet.2018.02.014
Chatterjee, S. S., Chen, L., Joo, H. S., Cheung, G. Y., Kreiswirth, B. N., and Otto, M. (2011). Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS One 6:e28781. doi: 10.1371/journal.pone.0028781
Chen, S., Slavin, M. A., and Sorrell, T. C. (2011). Echinocandin antifungal drugs in fungal infections: a comparison. Drugs 71, 11–41. doi: 10.2165/11585270-000000000-00000
Cleveland, A. A., Farley, M. M., Harrison, L. H., Stein, B., Hollick, R., Lockhart, S. R., et al. (2012). Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin. Infect. Dis. 55, 1352–1361. doi: 10.1093/cid/cis697
Clinical and Laboratory Standards Institute [CLSI] (2012). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts Informational Supplement, 4th Edn. Wayne, PA: Clinical and Laboratory Standards Institue.
Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antifungal Susceptibility Testing of Yeasts; Approved Standard, CLSI Document M60, 1st Edn. Wayne, PA: Clinical and Laboratory Standards Institue.
Cornely, O. A., Lass-Flörl, C., Lagrou, K., Arsic-Arsenijevic, V., and Hoenigl, M. (2017). Improving outcome of fungal diseases - Guiding experts and patients towards excellence. Mycoses 60, 420–425. doi: 10.1111/myc.12628
Davies, G. E., and Thornton, C. R. (2014). Differentiation of the emerging human pathogens Trichosporon asahii and Trichosporon asteroides from other pathogenic yeasts and moulds by using species-specific monoclonal antibodies. PLoS One 9:e84789. doi: 10.1371/journal.pone.0084789
Davuodi, S., Manshadi, S. A., Salehi, M. R., Yazdi, F., Khazravi, M., and Fazli, J. T. (2015). Fatal cutaneous mucormycosis after kidney transplant. Exp. Clin. Transplant. 13, 82–85. doi: 10.6002/ect.2013.0216
De Carolis, E., Vella, A., Florio, A. R., Posteraro, P., Perlin, D. S., Sanguinetti, M., et al. (2012). Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol. 50, 2479–2483. doi: 10.1128/JCM.00224-12
Ferreira, L., Sanchez-Juanes, F., Porras-Guerra, I., Garcia-Garcia, M. I., Garcia-Sanchez, J. E., González-Buitrago, J. M., et al. (2011). Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17, 546–551. doi: 10.1111/j.1469-0691.2010.03257.x
Florio, W., Morici, P., Ghelardi, E., Barnini, S., and Lupetti, A. (2018a). Recent advances in the microbiological diagnosis of bloodstream infections. Crit. Rev. Microbiol. 44, 351–370. doi: 10.1080/1040841X.2017.1407745
Florio, W., Tavanti, A., Barnini, S., Ghelardi, E., and Lupetti, A. (2018b). Recent advances and ongoing challenges in the diagnosis of microbial infections by MALDI-TOF mass spectrometry. Front. Microbiol. 9:1097. doi: 10.3389/fmicb.2018.01097
Gaibani, P., Galea, A., Fagioni, M., Ambretti, S., Sambri, V., and Landini, M. P. (2016). Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of KPC-producing Klebsiella pneumoniae. J. Clin. Microbiol. 54, 2609–2613. doi: 10.1128/JCM.01242-16
Garey, K. W., Rege, M., Pai, M. P., Mingo, D. E., Suda, K. J., Turpin, R. S., et al. (2006). Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clint. Infect. Dis. 43, 25–31.
Guimarães, T., Nucci, M., Mendonça, J. S., Martinez, R., Brito, L. R., Silva, N., et al. (2012). Epidemiology and predictors of a poor outcome in elderly patients with candidemia. Int. J. Infect. Dis. 16, e442–e447. doi: 10.1016/j.ijid.2012.02.005
Guo, F., Yang, Y., Kang, Y., Zang, B., Cui, W., Qin, B., et al. (2017). China-SCAN Team. Invasive candidiasis in intensive care units in China: a multicentre prospective observational study. J. Antimicrob. Chemother. 68, 1660–1668. doi: 10.1093/jac/dkt083
Hooff, G. P., van Kampen, J. J., Meesters, R. J., van Belkum, A., Goessens, W. H., and Luider, T. M. (2012). Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J. Proteome Res. 11, 79–84. doi: 10.1021/pr200858r
Hrabák, J., Studentová, V., Walková, R., Zemlicková, H., Jakubu, V., Chudácková, E., et al. (2012). Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50, 2441–2443. doi: 10.1128/JCM.01002-12
Idelevich, E. A., Grunewald, C. M., Wüllenweber, J., and Becker, K. (2014). Rapid identification and susceptibility testing of Candida spp. from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS One 9:e114834. doi: 10.1371/journal.pone.0114834
Idelevich, E. A., Sparbier, K., Kostrzewa, M., and Becker, K. (2017). Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 24, 738–743. doi: 10.1016/j.cmi.2017.10.016
Jung, J. S., Popp, C., Sparbier, K., Lange, C., Kostrzewa, M., and Schubert, S. (2014). Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J. Clin. Microbiol. 52, 924–930. doi: 10.1128/JCM.02691-13
Kett, D. H., Azoulay, E., Echeverria, P. M., and Vincent, J. L. (2011). Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 39, 665–670. doi: 10.1097/CCM.0b013e318206c1ca
Kliem, M., and Sauer, S. (2012). The essence on mass spectrometry based microbial diagnostics. Curr. Opin. Microbiol. 15, 397–402. doi: 10.1016/j.mib.2012.02.006
Kohlenberg, A., Struelens, M. J., Monnet, D. L., Plachouras, D., and The Candida Auris Survey Collaborative Group (2018). Candida auris: epidemiological situation, laboratory capacity and preparedness in European Union and European Economic Area countries, 2013 to 2017. Euro Surveill. 23:18-00136. doi: 10.2807/1560-7917.ES.2018.23.13.18-00136
Kollef, M., Micek, S., Hampton, N., Doherty, J. A., and Kumar, A. (2012). Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin. Infect. Dis. 54, 1739–1746. doi: 10.1093/cid/cis305
Kordalewska, M., Lee, A., Park, S., Berrio, I., Chowdhary, A., Zhao, Y., et al. (2018). Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 62:e00238-18. doi: 10.1128/AAC.00238-18
Lamoth, F., Lockhart, S. R., Berkow, E. L., and Calandra, T. (2018). Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 73, i4–i13. doi: 10.1093/jac/dkx444
Lau, A. F., Wang, H., Weingarten, R. A., Drake, S. K., Suffredini, A. F., Garfield, M. K., et al. (2014). A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 52, 2804–2812. doi: 10.1128/JCM.00694-14
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Miceli, M. H., Diaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Oviano, M., Sparbier, K., Barba, M. J., Kostrzewa, M., and Bou, G. (2016). Universal protocol for the rapid automated detection of carbapenem-resistant Gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS). Int. J. Antimicrob. Agents 48, 655–660. doi: 10.1016/j.ijantimicag.2016.08.024
Pana, Z. D., Roilides, E., Warris, A., Groll, A. H., and Zaoutis, T. (2017). Epidemiology of invasive fungal disease in children. J. Pediatric Infect. Dis. Soc. 6(Suppl._1), S3–S11. doi: 10.1093/jpids/pix046
Patel, G., and Bonomo, R. A. (2013). “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. doi: 10.3389/fmicb.2013.00048
Patil, A., and Majumdar, S. (2017). Echinocandins in antifungal pharmacotherapy. J. Pharm. Pharmacol. 69, 1635–1660. doi: 10.1111/jphp.12780
Paul, S., Singh, P., Shamanth, A. S., Rudramurthy, S. M., Chakrabarti, A., and Ghosh, A. K. (2018). Rapid detection of fluconazole resistance in Candida tropicalis by MALDI-TOF MS. Med. Mycol. 56, 234–241. doi: 10.1093/mmy/myx042
Pfaller, M. A. (2012). Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125, S3–S13. doi: 10.1016/j.amjmed.2011.11.001
Pfaller, M. A., Andes, D. R., Diekema, D. J., Horn, D. L., Reboli, A. C., Rotstein, C., et al. (2014). Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS One 9:e101510. doi: 10.1371/journal.pone.0101510
Pham, C. D., Iqbal, N., Bolden, C. B., Kuykendall, R. J., Harrison, L. H., Farley, M. M., et al. (2014). Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 58, 4690–4696. doi: 10.1128/AAC.03255-14
Pierce, C. G., and Lopez-Ribot, J. L. (2013). Candidiasis drug discovery and development: new approaches targeting virulence for discovering and identifying new drugs. Expert Opin. Drug Discov. 8, 1117–1126. doi: 10.1517/17460441.2013.807245
Rhoads, D. D., Wang, H., Karichu, J., and Richter, S. S. (2016). The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 86, 257–261. doi: 10.1016/j.diagmicrobio.2016.08.001
Richter, C., Hollstein, S., Woloszyn, J., Kaase, M., Gatermann, S. G., and Szabados, F. (2012). Evaluation of species-specific score cut-off values for various Staphylococcus species using a MALDI Biotyper-based identification. J. Med. Microbiol. 61, 1409–1416. doi: 10.1099/jmm.0.042606-0
Rodríguez-Baño, J., Gutiérrez-Gutiérrez, B., Machuca, I., and Pascual, A. (2018). Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 31:e00079-17. doi: 10.1128/CMR.00079-17
Sanguinetti, M., and Posteraro, B. (2016). Mass spectrometry applications in microbiology beyond microbe identification: progress and potential. Expert Rev. Proteomics 13, 965–977. doi: 10.1080/14789450.2016.1231578
Saracli, M. A., Fothergill, A. W., Sutton, D. A., and Wiederhold, N. P. (2015). Detection of triazole resistance among Candida species by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). Med. Mycol. 53, 736–742. doi: 10.1093/mmy/myv046
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., and Azeredo, J. (2012). Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 36, 288–305. doi: 10.1111/j.1574-6976.2011.00278.x
Spanu, T., Posteraro, B., Fiori, B., D’Inzeo, T., Campoli, S., Ruggeri, A., et al. (2012). Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 50, 176–179. doi: 10.1128/JCM.05742-11
Sparbier, K., Schubert, S., and Kostrzewa, M. (2016). MBT-ASTRA: a suitable tool for fast antibiotic susceptibility testing? Methods 104, 48–54. doi: 10.1016/j.ymeth.2016.01.008
Szabados, F., Tix, H., Anders, A., Kaase, M., Gatermann, S. G., and Geis, G. (2012). Evaluation of species-specific score cutoff values of routinely isolated clinically relevant bacteria using a direct smear preparation for matrix-assisted laser desorption/ionization time-of flight mass spectrometry-based bacterial identification. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1109–1119. doi: 10.1007/s10096-011-1415-7
Vatanshenassan, M., Boekhout, T., Lass-Flörl, C., Lackner, M., Schubert, S., Kostrzewa, M., et al. (2018). Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)-based method to detect caspofungin resistance in Candida albicans and Candida glabrata. J. Clin. Microbiol. 56:e00420-18. doi: 10.1128/JCM.00420-18
Vecchione, A., Florio, W., Celandroni, F., Barnini, S., Lupetti, A., and Ghelardi, E. (2018). A rapid procedure for identification and antifungal susceptibility testing of yeasts from positive blood cultures. Front. Microbiol. 9:2400. doi: 10.3389/fmicb.2018.02400
Vella, A., De Carolis, E., Mello, E., Perlin, D. S., Sanglard, D., Sanguinetti, M., et al. (2017). Potential use of MALDI-TOF mass spectrometry for rapid detection of antifungal resistance in the human pathogen Candida glabrata. Sci. Rep. 7:9099. doi: 10.1038/s41598-017-09329-4
Vella, A., De Carolis, E., Vaccaro, L., Posteraro, P., Perlin, D. S., Kostrzewa, M., et al. (2013). Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization time-of- flight mass spectrometry analysis. J. Clin. Microbiol. 51, 2964–2969. doi: 10.1128/JCM.00903-13
Walsh, T. J., and Gamaletsou, M. N. (2013). Treatment of fungal disease in the setting of neutropenia. Hematology Am. Soc. Hematol. Educ. Program 2013, 423–427. doi: 10.1182/asheducation-2013.1.423
Walsh, T. R. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36, S8–S14. doi: 10.1016/S0924-8579(10)70004-2
Wang, H., Liu, N., Yin, M., Han, H., Yue, J., Zhang, F., et al. (2014). The epidemiology, antifungal use and risk factors of death in elderly patients with candidemia: a multicentre retrospective study. BMC Infect. Dis. 14:609. doi: 10.1186/s12879-014-0609-x
Yan, Y., He, Y., Maier, T., Quinn, C., Shi, G., Li, H., et al. (2011). Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization biotyper system. J. Clin. Microbiol. 49, 2528–2532. doi: 10.1128/JCM.00339-11
Keywords: MALDI-TOF mass spectrometry, antimicrobial resistance, antifungal susceptibility testing, blood culture, rapid AFST
Citation: Florio W, Tavanti A, Ghelardi E and Lupetti A (2018) MALDI-TOF MS Applications to the Detection of Antifungal Resistance: State of the Art and Future Perspectives. Front. Microbiol. 9:2577. doi: 10.3389/fmicb.2018.02577
Received: 21 June 2018; Accepted: 09 October 2018;
Published: 30 October 2018.
Edited by:
Karsten Becker, Universität Münster, GermanyReviewed by:
Jozsef Soki, University of Szeged, HungaryCopyright © 2018 Florio, Tavanti, Ghelardi and Lupetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Lupetti, YW50b25lbGxhLmx1cGV0dGlAbWVkLnVuaXBpLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.