
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 October 2018
Sec. Extreme Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02458
This article is part of the Research TopicFor The Future of Alkaliphiles: 50th Anniversary Year Since the Rediscovery of Alkaliphiles by Dr. Koki Horikoshi View all 9 articles
 Luciana Terra1
Luciana Terra1 Paul J. Dyson1
Paul J. Dyson1 Matthew D. Hitchings1
Matthew D. Hitchings1 Liam Thomas1
Liam Thomas1 Alyaa Abdelhameed1
Alyaa Abdelhameed1 Ibrahim M. Banat2
Ibrahim M. Banat2 Salvatore A. Gazze1
Salvatore A. Gazze1 Dušica Vujaklija3
Dušica Vujaklija3 Paul D. Facey1
Paul D. Facey1 Lewis W. Francis1
Lewis W. Francis1 Gerry A. Quinn3*
Gerry A. Quinn3*In an effort to stem the rising tide of multi-resistant bacteria, researchers have turned to niche environments in the hope of discovering new varieties of antibiotics. We investigated an ethnopharmacological (cure) from an alkaline/radon soil in the area of Boho, in the Fermanagh Scarplands (N. Ireland) for the presence of Streptomyces, a well-known producer of antibiotics. From this soil we isolated a novel (closest relative 57% of genome relatedness) Streptomyces sp. capable of growth at high alkaline pH (10.5) and tolerant of gamma radiation to 4 kGy. Genomic sequencing identified many alkaline tolerance (antiporter/multi-resistance) genes compared to S. coelicolor M145 (at 3:1), hence we designated the strain Streptomyces sp. myrophorea, isolate McG1, from the Greek, myro (fragrance) and phorea (porter/carrier). In vitro tests demonstrated the ability of the Streptomyces sp. myrophorea, isolate McG1 to inhibit the growth of many strains of ESKAPE pathogens; most notably carbapenem-resistant Acinetobacter baumannii (a critical pathogen on the WHO priority list of antibiotic-resistant bacteria), vancomycin-resistant Enterococcus faecium, and methicillin-resistant Staphylococcus aureus (both listed as high priority pathogens). Further in silico prediction of antimicrobial potential of Streptomyces sp. myrophorea, isolate McG1 by anti-SMASH and RAST software identified many secondary metabolite and toxicity resistance gene clusters (45 and 27, respectively) as well as many antibiotic resistance genes potentially related to antibiotic production. Follow-up in vitro tests show that the Streptomyces sp. myrophorea, isolate McG1 was resistant to 28 out of 36 clinical antibiotics. Although not a comprehensive analysis, we think that some of the Boho soils’ reputed curative properties may be linked to the ability of Streptomyces sp. myrophorea, isolate McG1 to inhibit ESKAPE pathogens. More importantly, further analysis may elucidate other key components that could alleviate the tide of multi-resistant nosocomial infections.
The global increase in multi-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) has created an urgent need to develop replacement therapies. ESKAPE pathogens are responsible for the top 6 health care-associated infections (HAIs) and many have been identified as priority antibiotic-resistant bacteria (Jelic et al., 2016; Santajit and Indrawattana, 2016; Founou et al., 2017; Tacconelli et al., 2018). Infections with multi-resistant pathogens are extremely hard to treat and may spread throughout a hospital or community environment (Jelic et al., 2016; Santajit and Indrawattana, 2016). It was thought that developing new antibiotics from combinatorial chemistry would be able to eliminate these resistant bacteria. However, years of trials have not produced anything like the number of new drugs necessary to stem the tide of multi-resistant bacteria (Lewis, 2013). In addition, the production of new antibiotics tapered off in the early 1980s due to unfavorable market conditions which has led to a crisis in the supply line of new antibiotics (Santajit and Indrawattana, 2016; Founou et al., 2017). Normally, bacterial infections are treated with the simplest, most effective antibiotics, however, multi-resistant pathogens usually require treatment with higher tier antibiotics or antibiotics of last resort (Santajit and Indrawattana, 2016). There are no guarantees of success with these treatments and they can involve expensive and sometimes toxic chemotherapy. If all solutions fail, infections by ESKAPE pathogens can lead to death of the patient and a spread of multi-resistant strains. Antibacterial resistance has now been detected for nearly all new antibiotics, even those of last resort (Jelic et al., 2017; Li and Webster, 2018). As a consequence, the WHO have created an urgent priority list for discovery of new antibiotics (Tacconelli et al., 2018).
Current strategies to alleviate the shortage of new antibiotics have turned to niche environments such as deserts, thermal vents, and alkaline environments in the hope that they might produce exotic varieties of current antibiotics (Sato et al., 1983; Mao et al., 2007; Yucel and Yamac, 2010; Mohammadipanah and Wink, 2015). Alkaline environments in particular have proven to be a rich source of antibiotics, many derived from Streptomyces bacteria (Sato et al., 1983; Yucel and Yamac, 2010; Behroozian et al., 2016; Maciejewska et al., 2016). Together with other members of the phylum Actinobacteria, Actinomycetes, are responsible for the synthesis of more than half of modern medicines including antimicrobial, anti-cancerous (Noomnual et al., 2016), antiviral (Yokomizo et al., 1998), antifungal (Nguyen and Kim, 2015), and antiparasitic compounds (Procopio et al., 2012). In 1943, it was streptomycin (from Streptomyces) that was the great savior against the formally incurable scourge of tuberculosis (Schatz et al., 1944).
Another promising avenue of drug (re)discovery lies in the investigation of traditional medicines or ethnopharmacology. Although ancient medical traditions are well known in Chinese and Native American cultures, less is known about European folk medicines (Foley, 2015; Behroozian et al., 2016; Kung et al., 2018). One of the last vestiges of continuous ethnopharmacological culture can be found on the most westerly fringes of Europe, in rural locations on the island of Ireland (Foley, 2015). One such cure originates from a region of the West Fermanagh Scarplands known locally as Boho (pronounced Bo) in Northern Ireland. This cure is derived from an alkaline soil deposited in the late Pleistocene period (circa 9,126,000–11,700 years ago) on a bedrock of Carboniferous Dartry limestone (circa 335 million years ago) imparting an alkaline/high radon character to the soil (Brunton and Mason, 1979). Traditionally, this cure had been used to treat a variety of conditions from toothache to infections by placing a small portion of the soil wrapped in cloth next to the infection or underneath the users’ pillow for 9 days. The soil was then returned to the area of sampling. The exact specificity and origins of the cure are obscured by lack of documentation, however, some relatively recent written records remain, associating it with the grave of James McGirr, a cleric and healer who died in 1815 (Gallachair, 1975). Previous to this time the area had significance as an amphitheater for the Druids and a symbolic place for Neolithic peoples as evidenced by the nearby Reyfad stones (Halpin and Newman, 2009).
The purpose of this paper is to report the isolation of a novel alkaliphilic strain of Streptomyces from soil with antimicrobial activity against multiresistant ESKAPE pathogens which may have potential clinical applications.
The Boho soil sample was collected from an alkaline escarpment region (Latitude-54.364637°N Longitude-7.820939°W) at the Sacred Heart Church, in the townland of Toneel North, Boho, Fermanagh, United Kingdom (Donnelly et al., 2003) on the 28 July 2015. The test soil, which was pre-aliquoted into small cloth bags on site, was sampled with a sterile spatula. Instructions as to the traditional uses and practices with the soil are displayed on the door of an adjacent building. Approximately 25 g of this soil sample was collected in a sterile conical sample tube (50 ml) and dispatched to the laboratory for analysis. For laboratory analysis, 1 g of the Boho soil sample was diluted in 1 ml sterile water, vortexed, and cultured on International Streptomyces Project (ISP) 2 agar (1/5th strength), and Starch agar (1/5th strength) for the initial Streptomyces isolation.
Escherichia coli (ATCC, K12-MG1655) was provided by Dr. D. Zahradka, Ruđer Bošković Institute (RBI), Zagreb, Croatia.
Bacillus subtilis (strain 168) was provided by Dr. D. Vujaklija (RBI, Croatia).
ESKAPE pathogens listed below and other ATCC strains listed were provided by Dr. M. Jelić (Jelic et al., 2016, 2017).
Enterococcus faecium – strains: a, b, c (VRE), d, e, and f.
Staphylococcus aureus – strains: a, b (MRSA), c, d, and e (MRSA).
Klebsiella pneumoniae – strains: a, b, c, and d.
Acinetobacter baumannii – strains: a, b, c, d, and e.
Pseudomonas aeruginosa – strains: a, b, c, and ATCC 27853.
Enterobacter cloacae – strain: a.
The ESKAPE pathogens were clinical isolates collected through regular hospital activities. Species identification was performed using standard biochemical methods (tests) and the VITEK 2 system (bioMérieux, France) (Jelic et al., 2016, 2017). Isolates were assigned unique isolate IDs and subsequently anonymized with designations a, b, c, etc. (meaning the strains cannot be linked to patients in any identifiable manner) in accordance with European regulations. Stocks of original Streptomyces were frozen at -80°C in 18% glycerol after their initial isolation.
Microbiological media used in these experiments: ISP 2 media (1/5th strength: meaning 20% of standard ingredients except agar which was 2%) and Starch media (1/5th strength) for initial Streptomyces isolation. Soy Flour Mannitol (SFM) for Streptomyces sub culture and growth. Alkaline SFM for selection of alkaline tolerant Streptomyces (soy flour 10 g, mannitol 10 g, agar 20 g, CaCO3 1 g, humic acid 0.002 g, pH adjusted to 8.3 before sterilization). Blood agar was used for the isolation and cultivation of clinical isolates and Mueller-Hinton agar used for antimicrobial tests unless organisms specifically required enriched blood media.
For the determination of Streptomyces alkaline tolerance, we used ISP-2 media supplemented with Streptomyces minor elements solution [consisting of 0.1% (wt/vol) (each) of ZnSO4⋅7H2O, FeSO4⋅7H2O, MnCl2⋅4H2O, and CaCl2 anhydrous], 1% (wt/vol) glucose, and 0.02% (vol/vol) NaH2PO4–K2HPO4 buffer (0.1 M, pH 6.8). It was necessary to increase the concentration of agar to 4% after pH 13.2, and 6% after 13.4, due to the inability of lower agar concentrations to solidify. The pH of the agar was adjusted after sterilization (when the agar had cooled but was still liquid, i.e., 42–45°C) by the addition of appropriate volumes of filter sterilized sodium carbonate buffer (0.2 M Na2CO3 + 0.2 M NaH2CO3; pH 9.2 → 10.7) and potassium chloride/sodium hydroxide buffer (0.2 M KCl + 0.2 M NaOH; pH 11.6 → 13.0). The pH of the agar was checked immediately after the agar had set.
On the occasions when the robust growth of frozen stocks of Streptomyces isolates seemed to decline (as was observed from 2-year-old stocks, even when stored in glycerol at -80°C), stocks were revived by cultivation of the Streptomyces sp. myrophorea, isolate McG1 on ISP 2 agar (1/5th) supplemented with Streptomyces minor elements solution. In some cases, a mineral solution was made from alkaline soil by dissolving 1 g soil in 1 g water, vortexed (1-min), centrifuged (15,616 ×g, 10 min) to clear the supernatant and then added at a concentration of 0.5% (vol/vol) to pre-solidified agar. In both cases supplements were filter sterilized (syringe filter, 0.2 μm) prior to their addition to the agar to prevent precipitation of ferric compounds. Cores of this agar were used as negative controls in antimicrobial inhibition assays involving Mueller-Hinton agar. All reagents were supplied by Oxoid (Basingstoke, Hampshire, United Kingdom) except agar which was supplied by Melford (Melford, Suffolk, United Kingdom).
Atomic Force Microscopy (Bruker BioScope Catalyst; Bruker Instruments, Santa Barbara, CA, United States) was used to visualize Streptomyces hyphae growing on glass cover slips. The measurements were conducted in air, using TESPA cantilevers (Bruker Instruments, Santa Barbara, CA, United States) in Tapping Mode, with a nominal spring constant of 40 N/m and a nominal resonant frequency of 300 kHz. Off-line processing involved first-order plane fitting and flattening using the software Nan scope Analysis 1.50 (Bruker Instruments, Santa Barbara, CA, United States).
Soil pH was measured by dissolving 1 g of soil in 5 ml distilled water, shaking for 5 min and then waiting 1 h for the soil to settle (measuring in triplicate using a pH-meter; Mettler-Toledo Seven Compact, Leicester, United Kingdom). A similar procedure was used with pH paper, where 1 g of soil was dissolved in 2 ml distilled water, vortexed for 15 min, and applied in small aliquots to pH indicator strips (EMD Millipore ColorpHast, Burlington, MA, United States). The pH strips were accurate to 0.5 pH units.
To test the alkaline tolerance of bacteria, a pH gradient was established from pH 9.0 to 13.0 in ISP-2 by adding sterilized buffer as described above. The pH of agar surface was continuously monitored using flat pH sticks (Macherey-Nagel GmbH & Co. KG, Düren, Germany) as described (Jones et al., 2017). The pH strip accuracy was routinely tested against standard pH calibration solutions.
Streptomyces sp. myrophorea, isolate McG1 spore concentrations were estimated by cultivation of dilutions of the spore stock. A 1 ml spore suspension (estimated to be 1 × 105 spores/ml) received gamma radiation doses of 0.25, 0.5, 1.0, 2.0, 4.0, 10.0, 15.0, and 20.0 kiloGray (kGy) from a cobalt source (60Co, 8.2645 Gy/s). Spore suspensions were contained in 2 ml plastic microfuge tubes surrounded by a layer of ice inside a polystyrene ice container. After irradiation, aliquots of the spore suspensions were spread (100 μl) on SFM agar and cultivated for 2–3 weeks. The growth of one colony (colony-forming units = CFU) was interpreted to be the survival of one spore.
A standard agar overlay combined with an antibiotic assay was used to test the inhibitory potential of Streptomyces sp. myrophorea, isolate McG1 against ESKAPE pathogens (Nkanga and Hagedorn, 1978; Lehrer et al., 1991). Briefly, wells were made in a base layer (15 ml) of Mueller-Hinton agar. Control wells contained standard amounts of dissolved antibiotics were allowed to absorb into the agar over a period of 2 h. Once the wells were dry, an agar core of Streptomyces sp. myrophorea, isolate McG1 (cultivated for 9 days 20°C) was placed in an empty well in addition to a negative control of the original media without Streptomyces sp. myrophorea, isolate McG1. This base layer was then overlaid with 15 ml Mueller-Hinton agar (cooled to 43°C) that incorporating the test organisms (ESKAPE pathogens with a minimum 5 × 105 CFU/ml). Bacterial inhibition was indicated by a clear zone in the confluent growth of the test organism after overnight incubation.
Antibiotic sensitivity tests (antibiograms) for the Streptomyces sp. myrophorea, isolate McG1 and for ESKAPE pathogens followed the Kirby–Bauer protocol (Bauer et al., 1966). The resistance profiles (antibiograms) of clinical isolates (ESKAPE pathogens) were determined using standard concentrations of antibiotic impregnated discs (Oxoid) as proposed by the European Committee on Antimicrobial Susceptibility Tests (EUCAST) (Supplementary Table S1). Breakpoint tables for interpretation of MICs and zone diameters can be found in Version 8.0, 20181.
Cultures of Streptomyces sp. myrophorea, isolate McG1 were grown on 1/10th strength LB agar for DNA extraction using a Qiagen DNA mini kit (Qiagen, MD, United States) with the inclusion of a lysis step using lysozyme. Genomic DNA was prepared for sequencing using Qiagen FX and sequenced on an Illumina MiSeq platform using a 600 cycle V3 reagent kit.
Paired-end reads were subjected to quality filtering using the Trimmomatic tool (4 bp sliding window of Q20) (Bolger et al., 2014) prior to de novo genome assembly using SPAdes under default parameters (Bankevich et al., 2012). The genome assembly was assessed using QUAST (Gurevich et al., 2013) and annotated using Prokka (Seemann, 2014) and Rapid Annotation using Subsystem Technology (RAST)2 (Aziz et al., 2008).
In silico DNA–DNA hybridization was performed using the Genome–Genome Distance Calculator (GGDC) v 2.1 (Auch et al., 2010) using all Streptomyces genomes (868 sequences) as references. All genomes were downloaded from GenBank3.
Phylogenetic placement of Streptomyces sp. myrophorea, isolate McG1 was performed using PhyloPhlAn (Segata et al., 2013). Protein sequences from annotated Streptomyces genomes were retrieved autonomously from the GenBank FTP site using the term “Streptomyces” as a query. Ortholog identification and alignment was performed in Phylophlan. A maximum-likelihood phylogeny was reconstructed from the concatenated alignments in FastTree MP (JTT + CAT) implemented in the Cipres Science Gateway Server (Miller et al., 2010). The robustness of the phylogeny was assessed using 1000 bootstrap pseudoreplicates.
Gene clusters known to be involved in secondary metabolite biosynthesis, self-immunity, or resistance were identified using Antibiotics and Secondary Metabolite Analysis Shell (anti-SMASH) version 4.0.0 (Medema et al., 2011). The GenBank sequence file (from Prokka annotation) was submitted to the web interface selecting all extra features of annotation.
Aliquots (20 μl) of diluted soil samples were cultivated on several agars to select for Streptomyces including ISP2 (1/5th) and alkaline SFM (Figure 1). The original soil was returned to the sampling site as per local tradition.
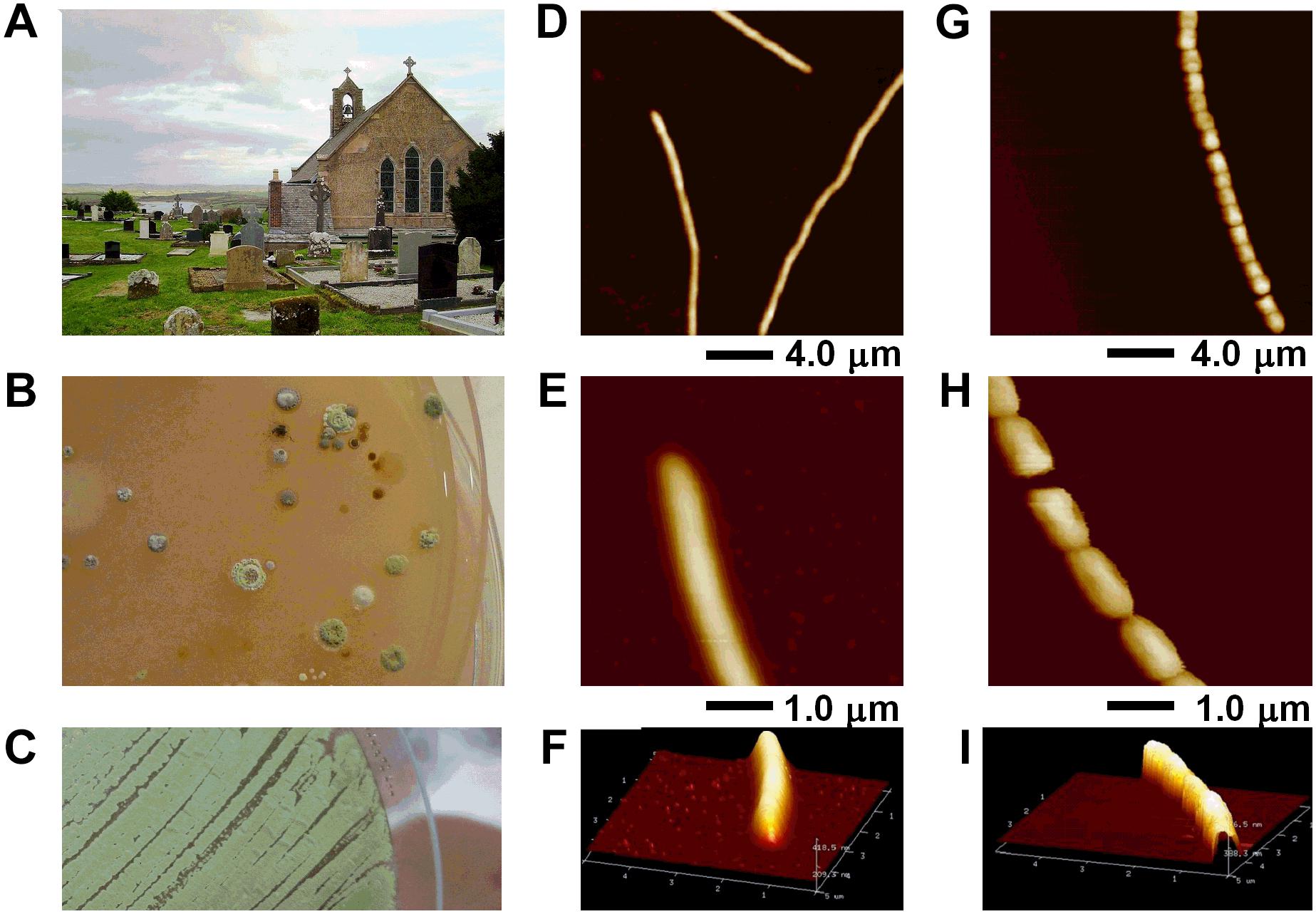
FIGURE 1. Streptomyces sp. myrophorea isolation from (A) Boho soil sample site, (B) selective enrichment agar, and (C) pure culture. (D–I) AFM data of Streptomyces sp. myrophorea, isolate McG1 grown on glass coverslips and imaged in air by Tapping Mode. Both vegetative (D–F) and sporulating (G–I) stages are shown. Hyphae overviews are shown in D and G, while higher resolution scans in E and F for vegetative hyphae, and in H and I for sporulating hyphae.
Preliminary screening of the Boho soil sample resulted in the isolation of eight (visually different) Streptomyces-like colony types as determined by colony morphology and growth characteristics. The Streptomyces isolate which had the most consistent inhibitory activity toward Gram-positive and Gram-negative bacteria (initially labeled as Streptomyces sp. myrophorea, isolate McG1) was selected for further characterization and testing (Figure 1).
Visually Streptomyces sp. myrophorea, isolate McG1 had (powdery) light to dark green colonies on SFM agar with green to very light green/white spores (Figures 1B,C). After a period of approximately 3–5 days, colonies emitted a distinctly “germaline” odor. Streptomyces sp. myrophorea, isolate McG1 appeared to be a non-motile, spore forming bacteria with very slender vegetative and aerial hyphae. Atomic force microscopy revealed that the bacterial hyphae were approximately 0.5–1.0 μm width, with spores in a linear conformation (17–20 spores) having a width of approximately 0.5–1.0 μm (Figures 1D–I).
The Boho soil sample had an average pH of 7.8 (mean ± standard deviation 0.35, by pH meter). Parallel measurements using pH strips indicated a wider range of pH from pH 7.8 to pH 8.5 (six readings at two different time points).
To measure the pH tolerance of Streptomyces sp. myrophorea, isolate McG1, a pH gradient was prepared as described. The pH levels of the agar surface were measured after it had set and every subsequent day after this using flat pH sticks (see the section “Materials and Methods”). The results revealed pronounced Streptomyces sp. myrophorea, isolate McG1 growth around pH 9.0 to pH 10 after 3–4 days. Growth was slower when bacteria were inoculated on agar that was prepared at pH 11.5 and took 5 days to be visible, however, during this time the pH of the agar also dropped to pH 10.5 (Figure 2A). In addition, we observed that Streptomyces sp. myrophorea, isolate McG1 inoculated at pH 12.2 or 13.0 could survive several days without prominent growth. As above, we noted that bacterial biomass became visible once the pH of the agar decreased to pH 10.5. In contrast S. coelicolor M145 was unable to grow in high alkaline conditions (pH 9 and above) implying that our new strain of Streptomyces was more alkaliphilic.
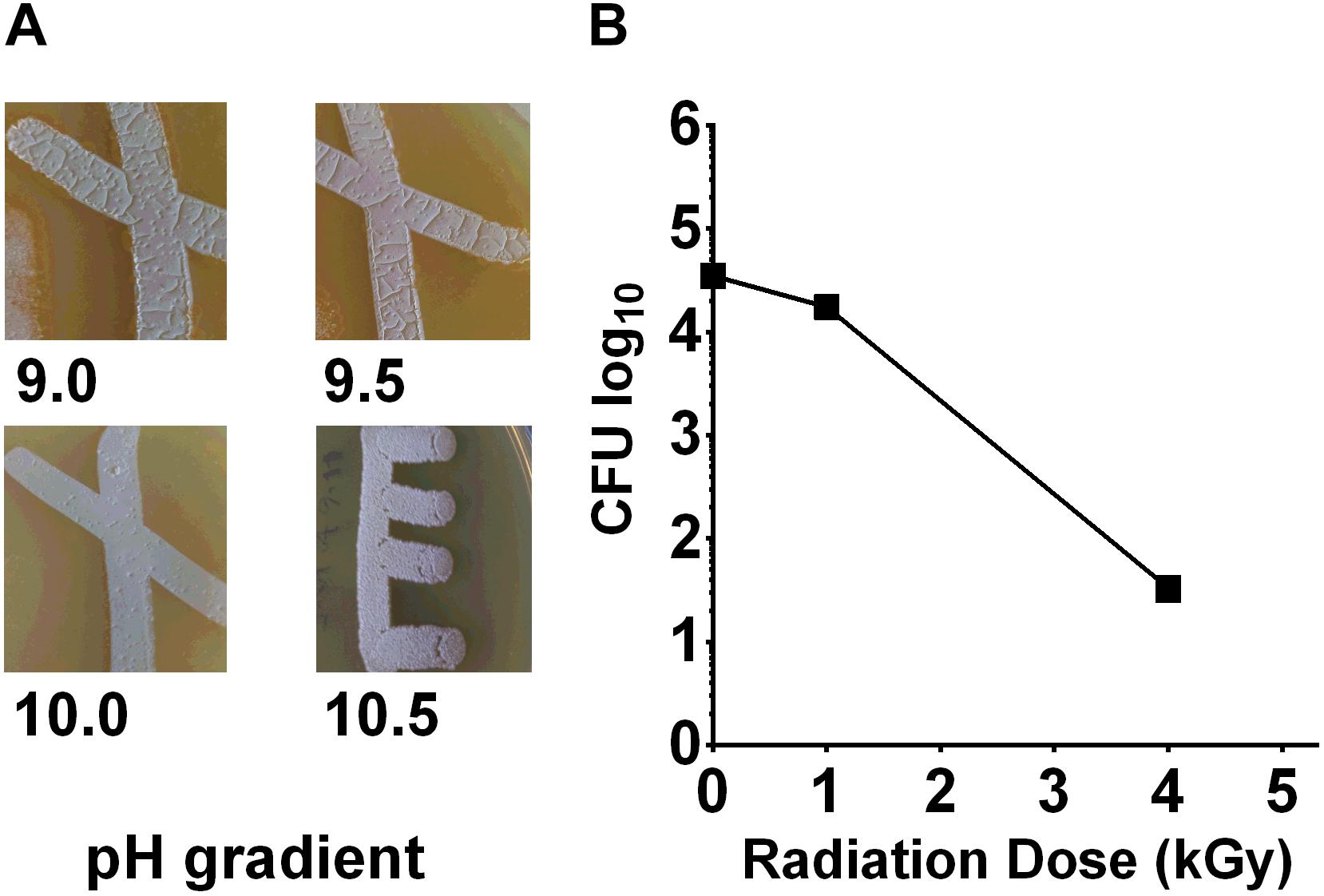
FIGURE 2. Streptomyces sp. myrophorea, isolate McG1 development on pH and irradiation gradients. (A) Streptomyces sp. myrophorea, isolate McG1 cultivated on ISP-2 agar under increasing pH. Images indicate final measurements, pH 9.0 (originally pH 9.5), pH 9.5 (originally pH 10.0), pH 10.0 (originally pH 10.7), and pH 10.5 (originally pH 11.6). Images are representative of three independent experiments. (B) Effect of γ-irradiation (60Co) on spore survival of Streptomyces sp. myrophorea, isolate McG1. All tests performed in triplicate. The variation in spore survival is indicated using standard error of the mean.
In contrast to the alkaline conditions, Streptomyces sp. myrophorea, isolate McG1 did not grow well under acidic conditions, the lowest pH for growth being pH 6.5.
Based on changes in pH during sterilization of media and between buffered and unbuffered media, we realize that it is very important to measure the pH of the agar surface throughout the bacterial growth cycle.
The alkaline environment of the Boho area is not the only characteristic which makes this region a niche habitat. The (limestone/shale) bedrock also releases radon gas which can be found at levels as high as 710 bq/m3 (domestic dwelling annual totals) in Boho and adjacent areas (Daraktchieva et al., 2015).
To test the effects of radiation on the survival of Streptomyces sp. myrophorea, isolate McG1, we subjected spore solutions to increasing levels of gamma radiation, i.e., spores were removed from a chamber after an exposure to absorbed doses of 0.25 → 20 kGy. After irradiation, aliquots (100 μl) of spores were spread evenly on diluted ISP2 agar in triplicate. The agar plates were then incubated at 4–10°C for 3 weeks before enumeration. Our measurements indicated that spores of Streptomyces sp. myrophorea, isolate McG1 were able to tolerate doses of 4 kGy of gamma irradiation and still remain viable (Figure 2B). In comparison, another Streptomyces, Streptomyces radiopugnans resists radiation expose of up to 15 kGy, whereas vegetative cells such as Deinococcus radiodurans tolerates an exposure of 12 kGy, E. coli 600 Gy, and human cells 4 Gy (Mao et al., 2007; Daly, 2012).
To test antimicrobial potential of Streptomyces sp. myrophorea, isolate McG1, agar cores from the Streptomyces were embedded in Mueller-Hinton test agar and overlaid with a suspension of ESKAPE pathogens. The pathogens were isolated from different hospitals in Croatia based on clinical antimicrobial susceptibility data. Controls consisted of ampicillin (Amp-20 μg), chloramphenicol (Cam-20 μg), ciprofloxacin (Cip-5 μg), gentamicin (Gen-30 μg), kanamycin (Kan-10 μg), streptogramin (10 μg), and ampicillin + sulbactam (Amp + Sulf-10 μg + 10 μg). An agar core from the original ISP2 supplemented agar was used as a negative control.
ESKAPE pathogens were considered susceptible (S) to antibiotics if the zone of inhibition was greater than 12 mm diameter; resistant (R), if zone was less than 8 mm radius and of intermediate status (I) if the zone was between 8 and 12 mm or there were a few colonies appearing in between the beginning of the zone and the edge of the antibiotic disc. A single mark indicates uniformity of result in a triplicate. Tests with variable results are indicated by the result of each replicate, i.e., S/R/S indicates the result of 1st/2nd/3rd test (Table 1).
ESKAPE pathogens are known to resist many clinical antibiotics including aminoglycosides, beta-lactams, carbapenems and glycopeptides (Founou et al., 2017). Our data on the incubation of ESKAPE pathogens reveal that Streptomyces sp. myrophorea, isolate McG1 was broadly inhibitory to both Gram-positive and Gram-negative bacteria. Specifically Streptomyces sp. myrophorea, isolate McG1 inhibited carbapenem-resistant A. baumannii [listed as a critical pathogen in the WHO priority pathogens list), vancomycin-resistant E. faecium, and methicillin-resistant S. aureus (which are both listed as high priority on the WHO pathogen list)] and K. pneumoniae (Figure 3 and Table 1). Some strains of E. faecium that could not be cultivated on Mueller-Hinton agar (they only grew on blood agar) as well as some strains of multi-resistant P. aeruginosa were resistant to Streptomyces sp. myrophorea, isolate McG1 (Table 1).
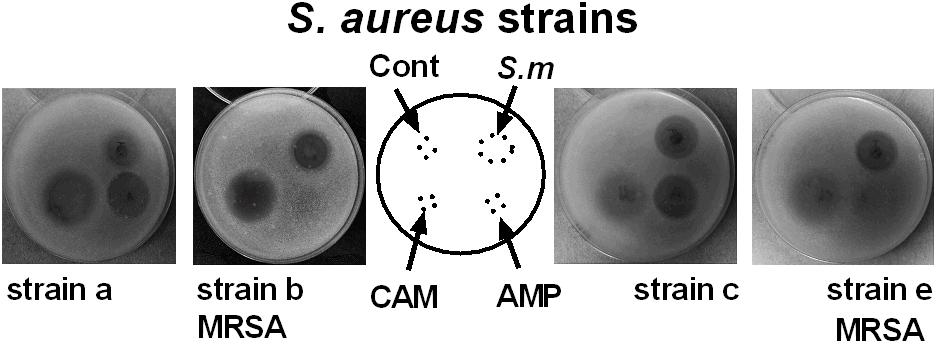
FIGURE 3. Inhibition of clinical strains of S. aureus by agar cores of Streptomyces sp. myrophorea, isolate McG1. S. aureus (SA) strains: SA strain a, SA strain b (MRSA, ciprofloxacin resistant), SA strain c and SA strain e (MRSA, ciprofloxacin sensitive) were cultivated on Mueller-Hinton agar together with agar cores of Streptomyces sp. myrophorea, isolate McG1 (S.m). Other test wells contained chloramphenicol (CAM-20 μg), ampicillin (AMP-20 μg), and a negative control of the original ISP-2 agar with mineral supplement (cont). Images are representative of three independent experiments.
Whole-genome sequencing of Streptomyces sp. myrophorea, isolate McG1 was performed using the Illumina MiSeq system. The total assembled size of the Streptomyces sp. myrophorea, isolate McG1 genome was almost 9 MB pairs with a GC content of 71.6%. Parameters predicted by Prokka and QUAST (Gurevich et al., 2013; Table 2).
The genome sequence was deposited in NCBI under the name of “Streptomyces sp. myrophorea, isolate McG1” (TaxID 2099643) or “Streptomyces sp. McG1,” Biosample accession number SAMN08518548, BioProject accession number PRJNA433829, Submission ID: SUB3653175, and Locus tag prefix: C4625. The project information is accessible on publication from http://www.ncbi.nlm.nih.gov/bioproject/433829.
Streptomyces sp. myrophorea, isolate McG1 was deposited with the National Collection of Type Cultures (NCTC 14177), United Kingdom and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) GmbH, Germany.
The top 12 matches against Streptomyces genomes and the intergenomic distances for in silico DNA–DNA hybridization (DDH) were calculated using GGDC (Table 3). The calculation of GGDC Formula 2 is independent of genome length (calculated by dividing found identities by high-scoring pairs and not by whole-sequence length). An absence of any DDH values over the recognized threshold of 70% revealed that our isolate was an uncharacterized species of Streptomyces.
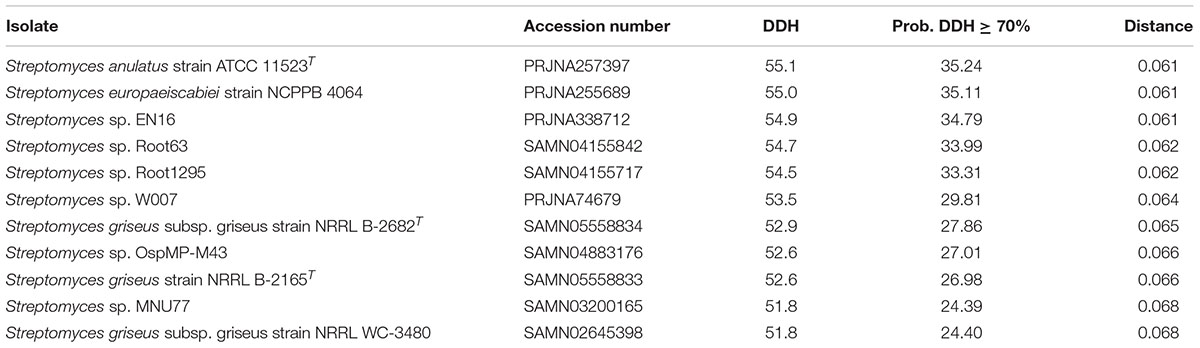
TABLE 3. The closest 12 matches to Streptomyces sp. myrophorea, isolate McG1 in NCBI (by GGDC) (Formula 2 statistics).
The ability of bacteria such as Streptomyces sp. myrophorea, isolate McG1 to tolerate high levels of alkalinity is often attributed to alkaline shock genes, proton antiporters (like nhaA), and multidrug resistance factors (like mdt/mdfA) (Krulwich et al., 2011; Holdsworth and Law, 2013). We identified the presence of these genes in the annotated genomes of Streptomyces sp. myrophorea, isolate McG1 and S. coelicolor M145 listing 25 of these genes in Streptomyces sp. myrophorea, isolate McG1 and 9 genes in the genome of S. coelicolor M145 (Table 4). Only Streptomyces sp. myrophorea, isolate McG1 possessed alkaline shock genes, aspartate/ammonium antiporters, and multidrug transporters of the type mdtH (Table 4). Hence, the Streptomyces sp. was referred to as strain myrophorea, isolate McG1, from the Greek myro (fragrance; the isolate emits a strong fragrant odor) and phorea (porter/carrier; in recognition of the number of predicted antiporter genes).
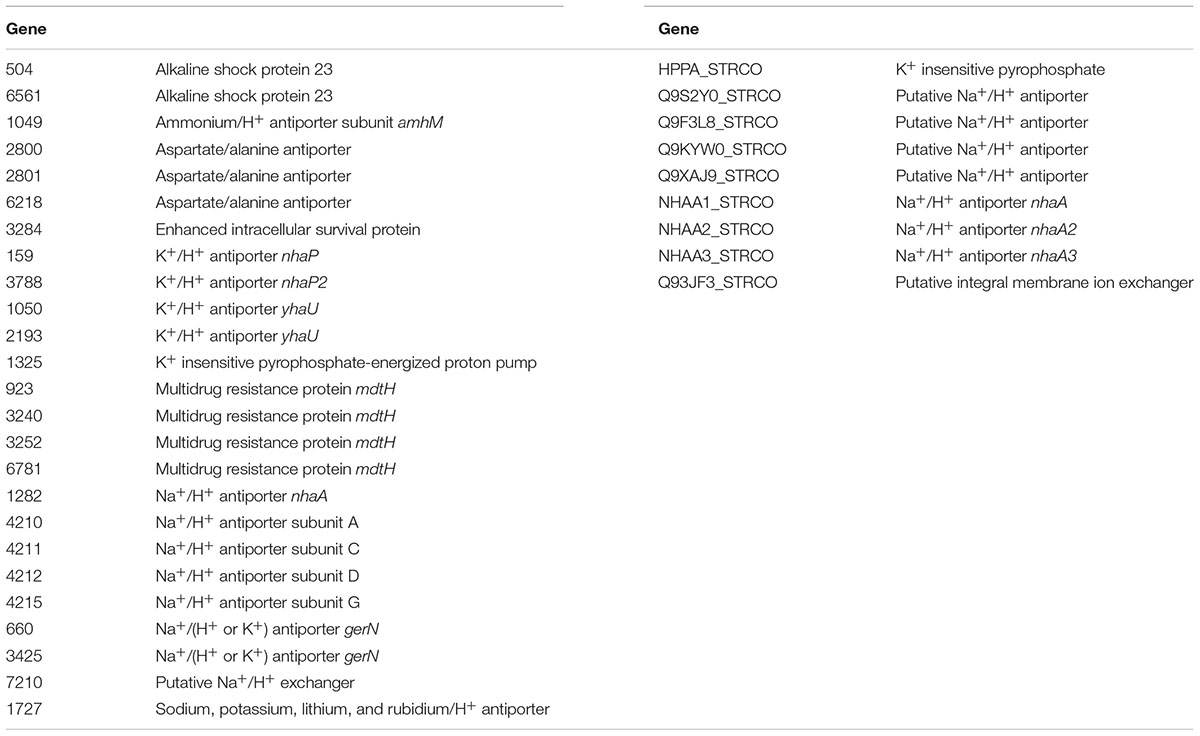
TABLE 4. List of alkaline tolerance genes identified in Streptomyces sp. myrophorea, isolate McG1 and S. coelicolor M145.
A similar identification of the number of DNA repair genes for Streptomyces sp. myrophorea, isolate McG1, and S. coelicolor M145 revealed little difference (Supplementary Table S2).
To infer the evolutionary history of Streptomyces sp. myrophorea, isolate McG1 a maximum-likelihood phylogeny was performed using a concatenation of 400 protein sequences (Figure 4).
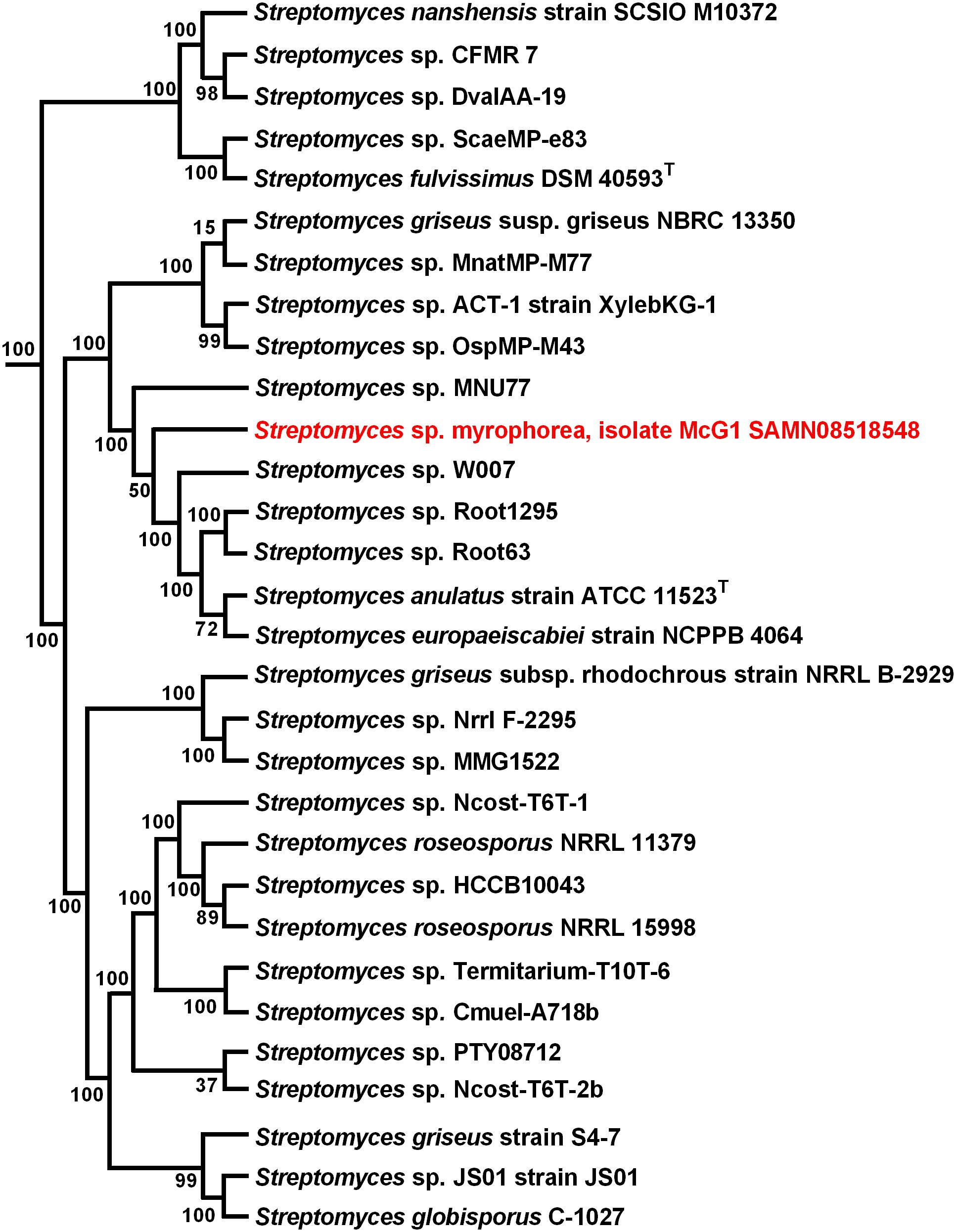
FIGURE 4. Maximum-likelihood phylogenetic tree of Streptomyces sp. myrophorea, isolate McG1. Tree shows the phylogenetic relationships between the closest GGDC matches; constructed with PhyloPhlAn. The phylogenetic tree was drawn in Fasttree from a concatenated alignment of 400 protein sequences in PhyloPhlAn. Numbers at nodes represent bootstrap values.
Potential antibiotic synthesis clusters from Streptomyces sp. myrophorea, isolate McG1 were predicted using anti-SMASH (Adamek et al., 2017). This revealed that Streptomyces sp. myrophorea, isolate McG1 possessed a total of 45 secondary metabolite biosynthesis gene clusters including multiple clusters with genes encoding the following secondary metabolite families: non-ribosomal peptide synthetase (NRPS) – 5 clusters; type I polyketide synthase (TI PKS) – 10 clusters; type III polyketide synthase (TIII PKS) – 2 clusters; terpenes – 5 clusters, lantipeptides – 3 clusters, and other biosynthesis genes clusters (BGCs) (Table 5).
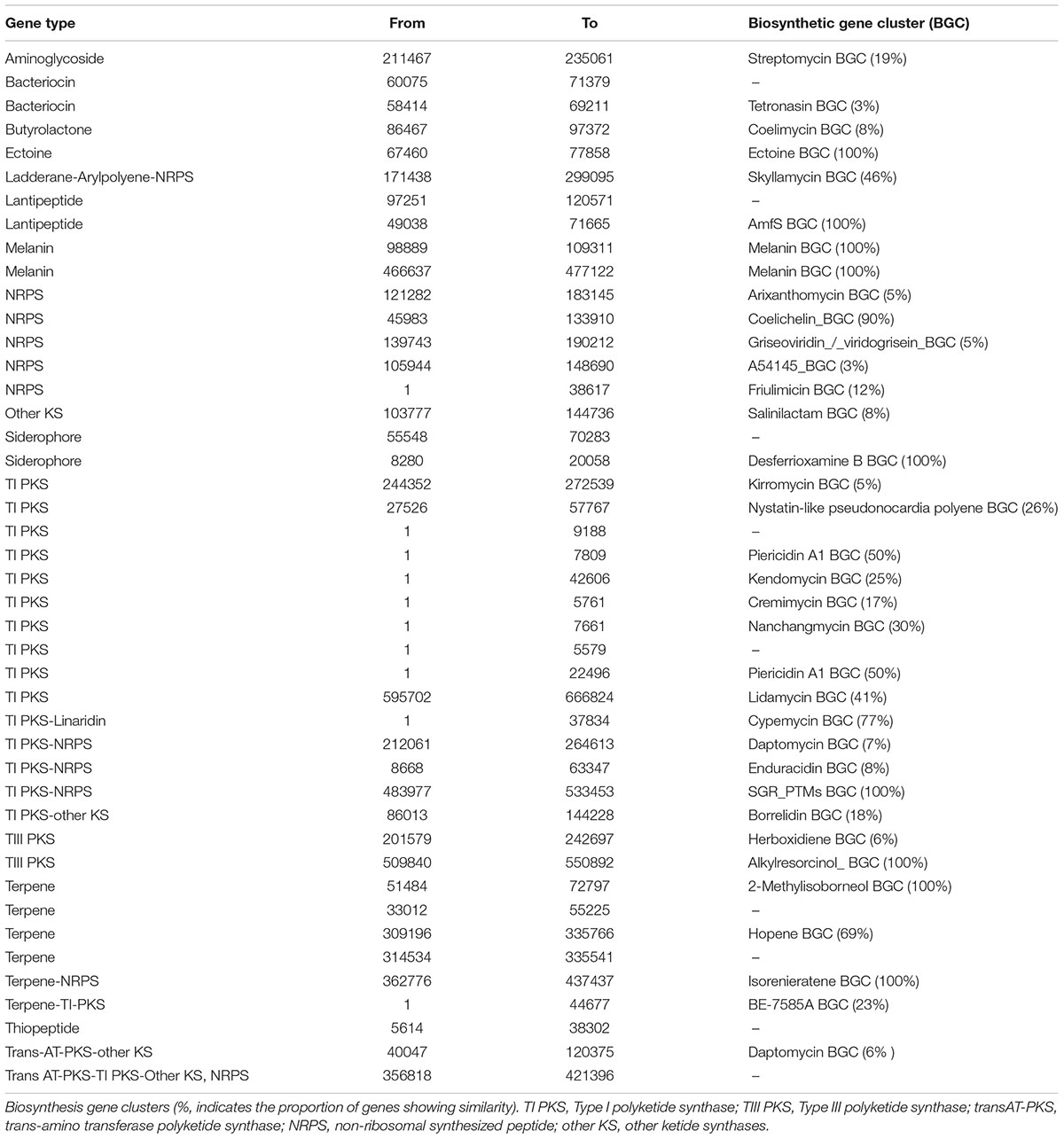
TABLE 5. The distribution of biosynthesis gene clusters in the Streptomyces sp. myrophorea, isolate McG1.
Antibiotic producing bacteria such as Streptomyces often require resistance genes to ameliorate the potentially toxic nature of their secondary metabolites. Such resistance elements can also be associated with antimicrobial biosynthesis gene clusters and can be used to predict potential antimicrobial synthesis (Nodwell, 2007). To better characterize these resistance elements, we cultivated Streptomyces sp. myrophorea, isolate McG1 in the presence of 36 different antibiotics (in triplicate). Streptomyces sp. myrophorea, isolate McG1 was resistant to 20 out of the 36 antibiotics tested after 2 days growth (Table 6 and Figure 5A). Resistance to a further eight antibiotics was visually apparent after a further 4 days, most notably in vancomycin, produced by Amycolatopsis orientalis; imipenim, a β-lactam stablized version of thienamycin, produced by Streptomyces cattleya and erythromycin, a macrolide antibiotic produced by Saccharopolyspora erythraea (Figure 5B).
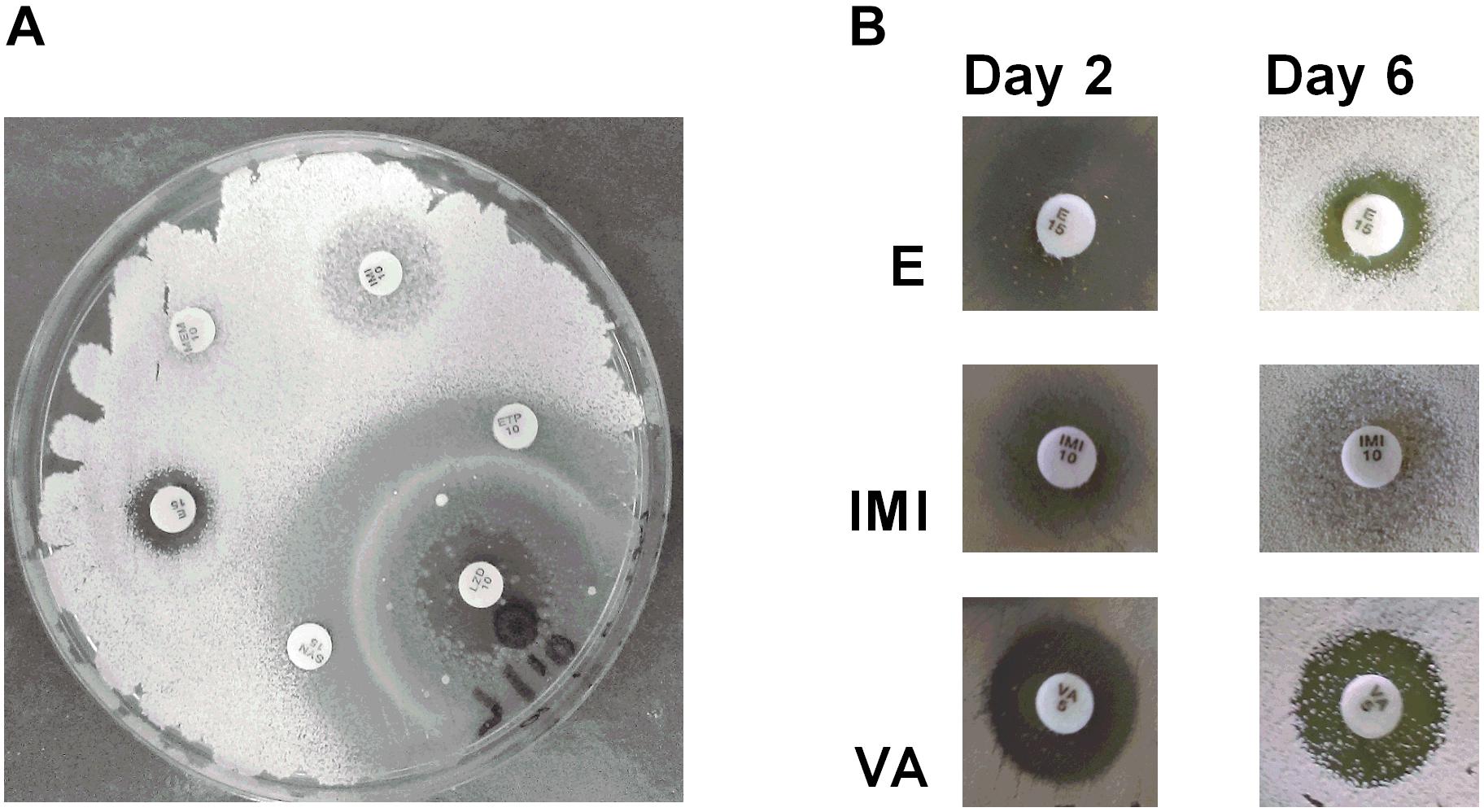
FIGURE 5. Antibiogram of Streptomyces sp. myrophorea, isolate McG1. (A) Streptomyces sp. myrophorea, isolate McG1 cultivated on ISP-2 agar (6 days) in the presence of an array of clinical antibiotics starting at the top the image and moving clockwise: imipenem (IMI 10), ertapenem (ETP 10), linezolid (LZD 10), quinpristin + dalropristin (SYN 15), erythromycin (E 15) and meropenem (MEM 10). (B) Delayed development of antibiotic resistance in Streptomyces sp. myrophorea, isolate McG1 only became apparent for some antibiotics after 6 days incubation in the case of erythromycin (E-15 μg), imipenem (IMI-10 μg), and vancomycin (VA-5 μg). The full antibiogram list is available in Table 6.
Antibiotic resistance gene clusters for Streptomyces sp. myrophorea, isolate McG1 were predicted in silico using anti-SMASH. Many copies of multiple antibiotic resistance elements were also predicted through RAST including β-lactamases (classes A and C), metal-dependant hydrolase of β-lactams (metallo β-lactamase L1), β-lactamase (cephalosporinase), and other penicillin binding proteins (Supplementary Table S3). In addition many metal resistance elements were identified (through RAST) to mercury, copper, cobalt, zinc/cadmium, and arsenic which have been linked to increased antibacterial resistance (Chenia and Jacobs, 2017) (Supplementary Table S3). It is also possible that resistance may also be mediated through other mechanisms such as general multi-resistance clusters.
We have isolated a novel species of Streptomyces from an alkaline/radon environment that inhibits the growth of many multiresistant ESKAPE pathogens. There have been several reports in recent years of the presence of Streptomyces and other organisms in alkaline environments that can tolerate high pH levels (Tiago et al., 2004; Yucel and Yamac, 2010; Janto et al., 2011; Maciejewska et al., 2016). Our original hypothesis presumed that the Boho soil sample most likely contained Streptomyces which may produce antibiotics given the alkaline nature of the environment (Kontro et al., 2005). However, we were surprised to find that our isolate, Streptomyces sp. myrophorea, isolate McG1 inhibited the growth of many of the multiresistant ESKAPE pathogens. Some of these bacteria have been listed in a recent WHO document on priority pathogens urgently requiring the development of new antibiotics, such as carbapenem-resistant A. baumannii (at the top of this list) classified as a critical priority pathogen and vancomycin-resistant Enterobacter faecium and methicillin-resistant S. aureus classified as high priority pathogens (Tacconelli et al., 2018).
We have not ascertained the active component(s) responsible for inhibition of ESKAPE pathogens by Streptomyces sp. myrophorea, isolate McG1 as yet but this forms part of our ongoing research. Many Streptomyces species have the capacity to produce multiple antibiotics whose composition and identity can vary from species to species (Watve et al., 2001; Procopio et al., 2012). Given that our isolate encodes many antimicrobial gene clusters, it is entirely possible that some of these are responsible for the inhibition of ESKAPE pathogens. Given that our species is also novel and has the capacity to inhibit many multi-resistant pathogens also raises the possibility that some of its inhibitory components could be novel.
Another unexpected finding of the research was that although the pH of the Boho soil sample was around pH 8; our species of Streptomyces was able to grow at pH 10.5. This suggested not only tolerance of high alkaline conditions but the capacity to grow and divide in an extreme environments. More detailed tests will have to be made to ascertain the specifics of these optimal growth conditions.
The mechanism by which organisms such as Streptomyces can ameliorate high alkaline conditions is thought to be a product of a group of pH homeostasis genes, some of which have been identified in other species as alkaline shock genes, multidrug resistance factors (mdt/mdfA) and proton antiporters (nhaA) (Krulwich et al., 2011; Holdsworth and Law, 2013). Alkaline tolerance is also linked to the production of antibiotics through the presence of multidrug resistance factors which are often associated with antimicrobial biosynthesis gene clusters (Nodwell, 2007).
In support of our results, genome sequencing of Streptomyces sp. myrophorea, isolate McG1 revealed that it contained more annotated alkaline tolerance genes than S. coelicolor M145 (at 25/9). Furthermore alkaline shock genes, ammonium and aspartate antiporters and multidrug resistance antiporters of the type mdtH were only identified in the Streptomyces sp. myrophorea, isolate McG1 genome and not in S. coelicolor M145 genome. Indeed, it was this abundance of porters and the fragrant smell of the Streptomyces that prompted us to refer to the strain as myrophorea, isolate McG1, myro (Greek for fragrance) and phorea (Greek for porter).
The sequencing of the Streptomyces sp. myrophorea, isolate McG1 genome also enabled the prediction of potential antibiotics through anti-Smash and RAST (Aziz et al., 2008; Medema et al., 2011). However, given the cryptic nature of many Streptomyces spp. metabolites, it cannot be assumed these antibiotics are produced until their products are identified. We were also able to predict many antibiotic resistance clusters through RAST which are often associated with antimicrobial biosynthesis gene clusters (Nodwell, 2007). Antimicrobial resistance predictions were followed by in vitro experiments on 36 clinical antibiotics. This data showed that Streptomyces sp. myrophorea, isolate McG1 was resistant to 28 out of 36 antibiotics. Specifically it was resistant to nearly all the β-lactams, with the exception of augmentin to which resistance developed after 6 days. The same pattern of delayed resistance was seen for glycopeptides, fluoroquinones (such as ciprofloxacin), and tetracyclines (such as tigecycline).
It is interesting that the ancient healers of Boho made a connection between alkaline soils (containing Streptomyces sp.) and (skin) infections. It has only recently been discovered that the pH of the specific infection sites can rise from normal skin pH about pH 5.5 to 8.5 and that bacterial biofilms, which can colonize wounds, can also reach similar pH levels (Schneider et al., 2007; Hostacka et al., 2010; Percival et al., 2014). Perhaps the indigenous people, who were undoubtedly in close proximity to the soil, noted (after many years) its curative properties under specific conditions. However, it is difficult to know the exact genesis of this cure because the previous occupants of the Boho site, the Druids, left no surviving records of healing and their Neolithic counterparts left only undeciphered carvings on some nearby stones. Anthropologically, the Boho folk tradition is similar to that of Kisameet Bay clay in Canada, another indigenous soil cure which was found to have inhibitory activity against a range of ESKAPE pathogens (Behroozian et al., 2016).
We have isolated and genome sequenced a novel alkaline and radio-tolerant species of Streptomyces from an ethnopharmacological soil cure; Streptomyces sp. myrophorea, isolate McG1. This Streptomyces sp. inhibits many multi-resistant ESKAPE pathogens including carbapenem-resistant A. baumannii (a critical priority species from the WHO priority list of antibiotic-resistant bacteria), vancomycin-resistant E. faecium, and methicillin-resistant S. aureus (listed as high priority by the WHO). Although not a complete elucidation of the antibacterial components of the Boho soil; we think that inhibition of such pathogens by Streptomyces sp. myrophorea, isolate McG1 may explain some of its reputed curative properties. It is hoped to further characterize some of these inhibitory components from Streptomyces sp. myrophorea, isolate McG1 and investigate the properties of other species contained in alkaline soil. We hope this will advance progress in stemming the tide of multi-resistant bacteria.
All datasets (GENERATED/ANALYZED) for this study are included in the manuscript and the Supplementary Files.
The idea for the isolation of extreme Streptomyces was an offshoot from the work of PD and LuT. Microbiology, strain growth, and testing were by GQ, LuT, DV, and IB. Genomic isolation and sequencing by AA and MH. Bioinformatics by PF, MH, PD, LiT, and AA. Imaging by SG and LF. Manuscript ideas and editing of relevant sections by all authors.
This study was supported in part by the Centre of Excellence for Bioprospecting, Ruđer Bošković Institute, Croatia, and from the kind donations of our collaborators at Swansea University and the University of Ulster.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge the help of reviewers of this manuscript. We would also like to thank Dr. Marko Jelić, Zagreb University Hospital for Infectious Diseases for the provision of the ESKAPE pathogens; Igor Sajko of the Radiation Chemistry and Dosimetry Lab at the RBI for his help in γ-irradiation; Dr. Petar Mitrikeski, Zelimira Filić, and Ela Šarić for help with pH measurements and imaging; Fermanagh local historian, F. McHugh, Parish of Botha, church curator Rev. J. McPhillips for background information and permissions; Mary McGurn, Pat McGurn, Mary Egan, and Bridget Quinn for knowledge of locations and customs; Vale Romani for illustrations and Vasilios Theocharidis for consultations on Greek etymology.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02458/full#supplementary-material.
Adamek, M., Spohn, M., Stegmann, E., and Ziemert, N. (2017). Mining bacterial genomes for secondary metabolite gene Clusters. Methods Mol. Biol. 1520, 23–47.
Auch, A. F., von Jan, M., Klenk, H.-P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Behroozian, S., Svensson, S. L., and Davies, J. (2016). Kisameet clay exhibits potent antibacterial activity against the eskape pathogens. mBio 7, e1842–e1815. doi: 10.1128/mBio.01842-15
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brunton, C. H. C., and Mason, T. R. (1979). Palaeoenvironments and correlation of the carboniferous rocks in West Fermanagh. Northern Ireland. Bull. Br. Mus. Nat. Hist. Geol. 32, 91–108.
Chenia, H. Y., and Jacobs, A. (2017). Antimicrobial resistance, heavy metal resistance and integron content in bacteria isolated from a South African tilapia aquaculture system. Dis. Aquat. Organ. 126, 199–209. doi: 10.3354/dao03173
Daly, M. J. (2012). Death by protein damage in irradiated cells. DNA Repair. 11, 12–21. doi: 10.1016/j.dnarep.2011.10.024
Daraktchieva, Z., Appleton, J. D., Rees, D. M., Adlam, K. A. M., Myers, A. H., Hodgson, S. A., et al. (2015). Radon in Northern Ireland: Indicative Atlas. Available at: https://www.gov.uk/government/ publications/radon-indicative-atlas-for-northern-ireland
Donnelly, C., MacDonald, P., Murphy, E., and Beer, N. (2003). Excavations at boho high cross, toneel north, county fermanagh. Ulst. J. Archaeol. 62, 121–142.
Foley, R. (2015). Indigenous narratives of health: (Re)placing folk-medicine within irish health histories. J. Med. Humanit. 36, 5–18. doi: 10.1007/s10912-014-9322-4
Founou, R. C., Founou, L. L., and Essack, S. Y. (2017). Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS One 12:e0189621. doi: 10.1371/journal.pone.0189621
Gallachair, P. Ó. (1975). Clogherici: a dictionary of the catholic clergy of the diocese of clogher (1535-1835) (continued). Clogher Rec. 8, 271–280. doi: 10.2307/27695713
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Halpin, A., and Newman, C. (2009). Ireland: An Oxford Archaeological Guide to Sites from Earliest Times to AD 1600. Oxford: Oxford Univ. Press.
Holdsworth, S. R., and Law, C. J. (2013). Multidrug resistance protein MdtM adds to the repertoire of antiporters involved in alkaline pH homeostasis in Escherichia coli. BMC Microbiol. 13:113. doi: 10.1186/1471-2180-13-113
Hostacka, A., Ciznar, I., and Stefkovicova, M. (2010). Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. 55, 75–78. doi: 10.1007/s12223-010-0012-y
Janto, B., Ahmed, A., Ito, M., Liu, J., Hicks, D. B., Pagni, S., et al. (2011). The genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external ph range from 7.5 to 11.4. Environ. Microbiol. 13, 3289–3309. doi: 10.1111/j.1462-2920.2011.02591.x
Jelic, M., Butic, I., Plecko, V., Cipris, I., Jajic, I., Bejuk, D., et al. (2016). KPC-Producing Klebsiella pneumoniae isolates in croatia: a nationwide survey. Microb. Drug Resist. 22, 662–667. doi: 10.1089/mdr.2015.0150
Jelic, M., Skrlin, J., Bejuk, D., Koscak, I., Butic, I., Guzvinec, M., et al. (2017). Characterization of isolates associated with emergence of OXA-48-producing Klebsiella pneumoniae in croatia. Microb. Drug Resist. 24, 973–979. doi: 10.1089/mdr.2017.0168
Jones, S. E., Ho, L., Rees, C. A., Hill, J. E., Nodwell, J. R., and Elliot, M. A. (2017). Streptomyces exploration is triggered by fungal interactions and volatile signals. eLife 6:e21738. doi: 10.7554/eLife.21738
Kontro, M., Lignell, U., Hirvonen, M.-R., and Nevalainen, A. (2005). pH effects on 10 Streptomyces spp. growth and sporulation depend on nutrients. Lett. Appl. Microbiol. 41, 32–38. doi: 10.1111/j.1472-765X.2005.01727.x
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kung, S. H., Lund, S., Murarka, A., McPhee, D., and Paddon, C. J. (2018). Approaches and recent developments for the commercial production of semi-synthetic artemisinin. Front. Plant Sci. 9:87. doi: 10.3389/fpls.2018.00087
Lehrer, R. I., Rosenman, M., Harwig, S. S., Jackson, R., and Eisenhauer, P. (1991). Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137, 167–173. doi: 10.1016/0022-1759(91)90021-7
Lewis, K. (2013). Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387. doi: 10.1038/nrd3975
Li, B., and Webster, T. J. (2018). Bacteria antibiotic resistance: new challenges and opportunities for implant-associated orthopaedic infections. J. Orthop. Res. 36, 22–32. doi: 10.1002/jor.23656
Maciejewska, M., Adam, D., Martinet, L., Naome, A., Calusinska, M., Delfosse, P., et al. (2016). A phenotypic and genotypic analysis of the antimicrobial potential of cultivable Streptomyces isolated from cave moonmilk deposits. Front. Microbiol. 7:1455. doi: 10.3389/fmicb.2016.01455
Mao, J., Tang, Q., Zhang, Z., Wang, W., Wei, D., Huang, Y., et al. (2007). Streptomyces radiopugnans sp. nov., a radiation-resistant actinomycete isolated from radiation-polluted soil in China. Int. J. Syst. Evol. Microbiol. 57,2578–2582. doi: 10.1099/ijs.0.65027-0
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. doi: 10.1093/nar/gkr466
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8. doi: 10.1109/GCE.2010.5676129
Mohammadipanah, F., and Wink, J. (2015). Actinobacteria from arid and desert habitats: diversity and biological activity. Front. Microbiol. 6:1541. doi: 10.3389/fmicb.2015.01541
Nguyen, T. M., and Kim, J. (2015). Antifungal and antibacterial activities of Streptomyces polymachus sp. nov. Isolated from soil. Int. J. Syst. Evol. Microbiol. 65, 2385–2390. doi: 10.1099/ijs.0.000268
Nkanga, E. J., and Hagedorn, C. (1978). Detection of antibiotic-producing streptomyces inhabiting forest soils. Antimicrob. Agents Chemother. 14, 51–59. doi: 10.1128/AAC.14.1.51
Nodwell, J. R. (2007). Novel links between antibiotic resistance and antibiotic production. J. Bacteriol. 189, 3683–3685. doi: 10.1128/JB.00356-07
Noomnual, S., Thasana, N., Sungkeeree, P., Mongkolsuk, S., and Loprasert, S. (2016). Streptanoate, a new anticancer butanoate from Streptomyces sp. DC3. J. Antibiot. 69, 124–127. doi: 10.1038/ja.2015.95
Percival, S. L., McCarty, S., Hunt, J. A., and Woods, E. J. (2014). The effects of pH on wound healing, biofilms, and antimicrobial efficacy. Wound Repair Regen. 22, 174–186. doi: 10.1111/wrr.12125
Procopio, R. E., Silva, I. R., Martins, M. K., Azevedo, J. L., and Araújo, J. M. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471. doi: 10.1016/j.bjid.2012.08.014
Santajit, S., and Indrawattana, N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res. Int. 2016:2475067. doi: 10.1155/2016/2475067
Sato, M., Beppu, T., and Arima, K. (1983). Studies on antibiotics produced at high alkaline pH. Agric. Biol. Chem. 47, 2019–2027. doi: 10.1080/00021369.1983.10865910
Schatz, A., Bugie, E., and Waksman, S. (1944). Streptomycin: a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Proc. Soc. Exp. Biol. Med. 55, 66–69. doi: 10.3181/00379727-55-14461
Schneider, L. A., Korber, A., Grabbe, S., and Dissemond, J. (2007). Influence of pH on wound-healing: a new perspective for wound-therapy? Arch. Dermatol. Res. 298, 413–420. doi: 10.1007/s00403-006-0713-x
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Segata, N., Bornigen, D., Morgan, X. C., and Huttenhower, C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/ncomms3304
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tiago, I., Chung, A. P., and Veríssimo, A. (2004). Bacterial diversity in a nonsaline alkaline environment: heterotrophic aerobic populations. Appl. Environ. Microbiol. 70, 7378–7387. doi: 10.1128/AEM.70.12.7378-7387.2004
Watve, M. G., Tickoo, R., Jog, M. M., and Bhole, B. D. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Yokomizo, K., Miyamoto, Y., Nagao, K., Kumagae, E., Habib, E. S., Suzuki, K., et al. (1998). Fattiviracin A1, a novel antiviral agent produced by Streptomyces microflavus strain No. 2445. II. Biological properties. J. Antibiot. 51, 1035–1039. doi: 10.7164/antibiotics.51.1035
Keywords: alkaliphile, antimicrobial, Streptomyces, ESKAPE pathogens, multi-resistant, ethnopharmacology
Citation: Terra L, Dyson PJ, Hitchings MD, Thomas L, Abdelhameed A, Banat IM, Gazze SA, Vujaklija D, Facey PD, Francis LW and Quinn GA (2018) A Novel Alkaliphilic Streptomyces Inhibits ESKAPE Pathogens. Front. Microbiol. 9:2458. doi: 10.3389/fmicb.2018.02458
Received: 17 May 2018; Accepted: 25 September 2018;
Published: 16 October 2018.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Saori Kosono, The University of Tokyo, JapanCopyright © 2018 Terra, Dyson, Hitchings, Thomas, Abdelhameed, Banat, Gazze, Vujaklija, Facey, Francis and Quinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerry A. Quinn, Z3F1aW5uQGlyYi5ocg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.