- 1Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 2Department of Molecular Science and Technology, Department of Applied Chemistry and Biological Engineering, Ajou University, Suwon, South Korea
- 3Center for Food and Bioconvergence, Seoul National University, Seoul, South Korea
Enterohemorrhagic Escherichia coli (EHEC) is a foodborne pathogen that poses a serious threat to humans. Although EHEC is problematic mainly in food products containing meat, recent studies have revealed that many EHEC-associated foodborne outbreaks were attributable to spoiled produce such as sprouts and green leafy vegetables. To understand how EHEC adapts to the environment in fresh produce, we exposed the EHEC isolate FORC_035 to canola spouts (Brassica napus) and water dropwort (Oenanthe javanica) and profiled the transcriptome of this pathogen at 1 and 3 h after incubation with the plant materials. Transcriptome analysis revealed that the expression of genes associated with iron uptake were down-regulated during adaptation to plant tissues. A mutant strain lacking entB, presumably defective in enterobactin biosynthesis, had growth defects in co-culture with water dropwort, and the defective phenotype was complemented by the addition of ferric ion. Furthermore, gallium treatment to block iron uptake inhibited bacterial growth on water dropwort and also hampered biofilm formation. Taken together, these results indicate that iron uptake is essential for the fitness of EHEC in plants and that gallium can be used to prevent the growth of this pathogen in fresh produce.
Introduction
Enterohemorrhagic Escherichia coli (EHEC) is a virulent foodborne pathogen that causes outbreaks associated with serious health problems worldwide (Sperandio and Nguyen, 2012). It causes bloody diarrhea, abdominal cramps, and hemolytic uremic syndrome in human, which may be life-threatening, especially in young children and the elderly (Karmali et al., 1983; Karmali, 1989; Griffin and Tauxe, 1991). EHEC is commonly found in animal-based foods made with beef, pork, or poultry (Meng and Doyle, 1997). However, fresh produce (e.g., lettuce, spinach, alfalfa sprouts) has been identified as another notable source of EHEC outbreaks (Taormina et al., 1999; Sivapalasingam et al., 2004). The increase in number of outbreaks associated with fresh produce has been attributed in part to increased consumption of fruits and vegetables in recent decades (Lynch et al., 2009). Furthermore, fresh produce is generally consumed without thermal treatment, which is likely to aggravate the occurrence of produce-associated foodborne diseases (Herman et al., 2015).
A number of studies have shown that many foodborne pathogens can survive and replicate in plants, taking advantage of plant-derived amino acids and carbon compounds as nutrients (Cooley et al., 2003; Jablasone et al., 2005; Schikora et al., 2008; Deering et al., 2012). The interactions between bacterial pathogens and their host plants in the context of unfavorable plant conditions including desiccation, UV radiation, and plant immune responses have also been investigated extensively (Shi et al., 2007; Erickson, 2012; Fink et al., 2012). Interestingly, bacterial determinants required for persistence in plants are variable depending on bacterial genomic features and reservoir host plants (O’brien and Lindow, 1989; Heaton and Jones, 2008; Junker et al., 2011). Therefore, to decipher the mechanisms of persistence and replication of foodborne pathogens in plants, the behavior of diverse bacterial pathogens in contact with a variety of plants should be documented. From a food safety and hygiene perspective, although any produce has the potential to carry foodborne pathogens in the field or during post-harvest management, it is critical to understand bacterial survival strategies in fresh produce, which is consumed without thermal treatments.
Recent studies of plant–microbe interactions have been biased toward lettuce, spinach, and tomato, which are associated with foodborne disease outbreaks (Carey et al., 2009; Kyle et al., 2010). To extend our understanding of the interactions between foodborne bacteria and plants, we investigated canola sprouts (Brassica napus) and water dropwort (Oenanthe javanica) in this study. Sprouted seeds of plants like canola are cultivated using warm water and therefore provide favorable niches for growth of pathogens such as Salmonella enterica and E. coli O157:H7 (Andrews et al., 1982; Taormina et al., 1999; Erickson, 2012). A multistate outbreak of EHEC occurred in the United States in 1997 because of alfalfa sprout contamination (Breuer et al., 2001). Water dropworts are members of the genus Oenanthe in the plant family Apiaceae and the species O. javanica is mostly consumed raw in eastern Asia. Water dropwort is usually cultivated in flooded soil and is prone to contamination with sewage containing bacterial pathogens.
Transcriptome analysis can lead to a systematic understanding of bacterial responses to environmental changes such as contact with plant tissues (Schenk et al., 2012). In this study, to gain insight into the interaction between fresh produce and EHEC, we analyzed the transcriptome of EHEC FORC_035 when exposed to canola sprouts and water dropwort using an RNA-Sequencing (RNA-Seq) approach, as this provides higher resolution data than microarrays or related techniques (Matkovich et al., 2010). FORC_035, a strain isolated from kimchi in South Korea, possesses multiple genes encoding virulence factors including Stx2 and hemolysin. It causes attaching and effacing (A/E) lesions, and has the potential to cause severe diseases in humans when consumed in the form of contaminated fresh produce. Comprehensive transcriptome analysis of FORC_035 in contact with plants provided informative data for unraveling the interaction between pathogens and plants and revealed that iron acquisition was critical for EHEC survival in plants. Iron is an essential element for all living organisms, including bacteria. Therefore, bacteria possess a variety of complex systems for iron acquisition and utilization (Doherty, 2007). Among multiple iron uptake systems, siderophore-mediated iron acquisition is the most common form in bacteria (Visca et al., 2002). E. coli has been reported to utilize diverse forms of siderophores such as ferrichrome, enterobactin, yersiniabactin, and catecholate that are transported through highly specific proteins encoded by fhu operon, fep operon, ybt operon, and fiu/cir/iroN, respectively (Braun, 2003). The role of iron in bacterial survival and growth has been extensively investigated in bacterial pathogens that encounter iron starvation stress upon entering their hosts (Skaar, 2010). As a critical growth determinant in plants, we further investigated the role of iron in E. coli adaptation to plant environments.
Materials and Methods
Bacterial Strains and Culture Conditions
The EHEC isolate strain FORC_035 and isogenic mutant derivatives used in this study are described in Table 1. FORC_035 is EHEC (O8:H30) producing Stx2 and hemolysin E. FORC_035 strain was isolated from kimchi by the Incheon Institute of Health and Environment (Incheon, South Korea). The genome sequence of FORC_035 was deposited in the database of the Foodborne pathogen Omics Research Center1. Unless stated, all bacterial strains were grown in Luria-Bertani (LB) medium at 37°C, and the following antibiotics were added if needed: kanamycin (25 μg/mL) and ampicillin (100 μg/mL). The selective medium of tryptone bile X-glucuronide (TBX, Oxoid, Cambridge, United Kingdom) was used to enumerate FORC_035 populations. As a nutrient-limitation condition, M9 minimal medium supplemented with 0.4% glucose was used. Fe2(SO4)3 and Ga(NO3)3 (Sigma-Aldrich, St. Louis, MO, United States) at pH 7.0 in sterile water were added to bacterial cultures to examine the effect of iron on bacterial growth.
To study the effect of two siderophores, enterobactin and yersiniabactin, ΔentB (deletion mutant of entB encoding enterobactin synthase component B) and ΔybtS (deletion mutant of ybtS encoding yersiniabactin biosynthesis salicylate synthase) were constructed. An isogenic mutant of FORC_035, ΔentB, was constructed using the λ Red recombination system. PCR products containing a kanamycin resistance gene were generated using pKD13 as a template (Datsenko and Wanner, 2000; Murphy and Campellone, 2003). For homologous recombination, PCR primers were designed to introduce nucleotides identical to the flanking regions of entB at each end of the PCR products (Supplementary Table S1). FORC_035 strain harboring the Red helper plasmid pKD46 was grown in the presence of L-arabinose (50 mM) at 30°C with shaking (220 rpm) until the OD600 reached 0.6. Bacterial cells were then centrifuged (10,000 × g, 5 min, 4°C), resuspended in ice-cold water, and transformed by electroporation (Bio-Rad Laboratories, Hercules, CA, United States). Recombinant cells were selected by kanamycin resistance (Jandu et al., 2009), and subjected to diagnostic PCR using the primers listed in Supplementary Table S1. The kanamycin resistance cassette was subsequently removed using the FLP recombinase of pCP20 (Cherepanov and Wackernagel, 1995). A ΔybtS mutant strain was also constructed in the same way using ybtS-Lambda-F/ybtS-Lambda-R primer set (Supplementary Table S1).
Preparation of Canola Sprouts and Water Dropwort
Canola sprouts (Brassica napus) and water dropwort (Oenanthe javanica) were purchased from commercial markets in Seoul, South Korea. To prepare water dropwort, only stems around 15 cm in length were cut into pieces, because these are more likely to be contaminated by bacteria than other parts of the plant. Five grams of each plant were washed using 50 mL of sterile water three times with agitation (220 rmp) for 10 min each time prior to contact with bacteria.
In planta Assay
To prepare bacterial total RNA after contact with plants, the FORC_035 strain was cultured in LB until OD600 reached 1.0, then bacterial cells were washed twice using M9 minimal medium and resuspended in fresh M9 minimal medium (15 mL) at 109 CFU/mL. Five-gram of canola sprouts or water dropwort was added to the bacterial culture and cells were incubated at 37°C with shaking at 220 rpm. The bacterial culture grown in the same condition without adding plants in 20 mL M9 minimal medium was used as a control. At 1- and 3-h post-inoculation, total RNAs were prepared in biological duplicate.
Total RNA Extraction, Sequencing, and Analysis
One hundred microliter of bacterial cultures was treated with RNAprotect bacterial reagent (Qiagen, Hilden, Germany) to quench RNA degradation, and bacterial total RNAs were isolated using an RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. RNA samples were treated with Ambion Turbo DNA-freeTM (Ambion, Austin, TX, United States) to remove all genomic DNA. The quantity and quality of total RNA samples were examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States), and RNA samples with RNA integrity numbers (RIN) larger than 9 were used for further analysis (Schroeder et al., 2006). Total RNA samples were stored at −81°C until use.
Five micrograms of total RNA from each sample was used as a starting material and subjected to an rRNA-removal process based on the subtractive hybridization/bead capture system of the Ribo-Zero kit (Epicentre Biotechnologies, Madison, WI, United States). Purified RNA samples were used for mRNA-Seq library construction using the Illumina TruSeq RNA Sample Preparation kit v.2 (Illumina, San Diego, CA, United States). RNA-Seq was performed by two runs of Illumina Hiseq to generate single-end-reads around 100 bp in length. All RNA-Seq data analyzed in this study, including whole transcriptome profiles, are provided as supplementary information files (Supplementary Tables S2, S3) and deposited in the Foodborne pathogen Omics Research Center2. Using CLRNAseq program (Chunlab, Seoul, South Korea), sequencing reads were mapped to the FORC_035 genome sequence and normalized. Normalization methods employed in RNA-Seq analysis included Reads Per Kilobase of transcript per Million mapped reads (RPKM), Relative Log Expression (RLE), and Trimmed Mean of M-value (TMM) (Robinson and Oshlack, 2010; Dillies et al., 2013; Risso et al., 2014) (Supplementary Table S4). Because the coefficient of variation (CV) value for the TMM method was lower than those of RPKM and RLE, TMM was used for the normalization of expression level of genes. The p-value was calculated using edgeR and the [fold change] value was calculated as [TMMFreshproduce/TMMControl]. For further experiments, differentially expressed genes (DEGs) with an absolute log2 [fold change] larger than 2 were filtered and visualized using the CLRNAseq program (Chunlab, Seoul, South Korea). Clusters of orthologous groups (COG) analysis (Tatusov et al., 1997) was used for functional grouping of all genes of EHEC FORC_035. The proportion of DEGs in each functional group was calculated.
Heat Map Generation
Heat maps were drawn to graphically visualize global response patterns. Log2 [fold change] values were used to determine relative gene expression between FORC_035 only and FORC_035 co-cultured with canola sprouts and water dropwort. Heat maps and hierarchical clusters were then generated using Gitools v2.2.2 (Perez-Llamas and Lopez-Bigas, 2011).
Quantitative Real-Time RT-PCR (qRT-PCR)
The expression level of specific genes was validated using qRT-PCR as described in Yoo et al. (2017). For all results of qRT-PCR, GAPDH was used as normalization control. The primers used for the detection of genes are listed in Supplementary Table S5.
Construction of pEntB Plasmid
To construct pEntB plasmid expressing entB gene, pUHE21-2lacIq plasmid was used (Soncini et al., 1995). The entB gene was amplified by PCR using the primer set of entB-F-pUHE and entB-R-pUHE (Supplementary Table S1). The purified PCR product was inserted between the BamHI and HindIII sites of the vector.
Bacterial Cell Count Assay for Growth Comparison
Bacterial cells cultured overnight were washed with M9 minimal media and added in 15 mL of fresh M9 minimal media at a 1:100 ratio with or without 5 g of fresh produce. Bacterial viability was examined by plating on TBX selective agar and represented as log(N/N0) (CFU/mL) values, where N0 is the initial number of cells at 0 h and N is the number of cells after 5 h incubation. Ferric sulfate or gallium nitrate at indicated concentrations was added to M9 minimal media when required. For strains harboring empty plasmid or pEntB plasmid, 50 μM of IPTG was used for induction. All experiments were performed in biological triplicate. P-value was calculated by Student t-test.
Biofilm Reduction Assay
Bacterial cells cultivated in LB medium overnight were diluted into fresh LB medium (200 μL) at a 1:100 ratio in a polystyrene 96-well plate (SPL, Seoul, South Korea), and grown at 37°C for 48 h without shaking. Ga(NO3)3 dissolved in sterile water was added at different concentrations when needed. Considering no-observable-adverse-effect level (NOAEL) and acceptable-daily-intake (ADI) values of gallium, Ga(NO3)3 was used up to 1 mM (Gómez et al., 1992). Biofilm formation was quantified by staining with 0.1% crystal violet for 5 min followed by washing with phosphate buffer (0.1 M, pH 7.4). All dye attached to the biofilm was dissolved with 200 μL of 33% glacial acetic acid, and OD570 was measured to quantify the total biofilm mass (Vikram et al., 2010). The assay was performed in biological triplicate, and the p-value was calculated by Student t-test.
Results and Discussion
Comprehensive Transcriptome Analysis of FORC_035 Cultivated With Canola Sprouts or Water Dropwort
Bacterial transcriptome analysis is an effective approach to understand the interactions between foodborne pathogens and plants (Kyle et al., 2010; Fink et al., 2012; Landstorfer et al., 2014). In this study, the transcriptome of FORC_035 grown with canola sprouts or water dropwort was analyzed using RNA-Seq technology. Bacterial growth was increased in the presence of plant tissues (Figure 1), which was consistent with a previous study showing that bacteria could take advantage of plants-derived nutrients for their growth (Cooley et al., 2003; Jablasone et al., 2005; Schikora et al., 2008; Deering et al., 2012). To understand the bacterial response and adaptations in contact with plants, FORC_035 cells were harvested at 1 and 3 h post-inoculation and their RNAs were isolated for RNA-Seq analysis. RNA-Seq data were acquired, mapped, and normalized as described in the methods (Supplementary Tables S2–S4). After mRNA expression levels were compared between plant-exposed samples and not-exposed samples, genes with a log2 (fold change) greater than 2 or less than −2 were selected and considered to be DEGs (Supplementary Figure S1).
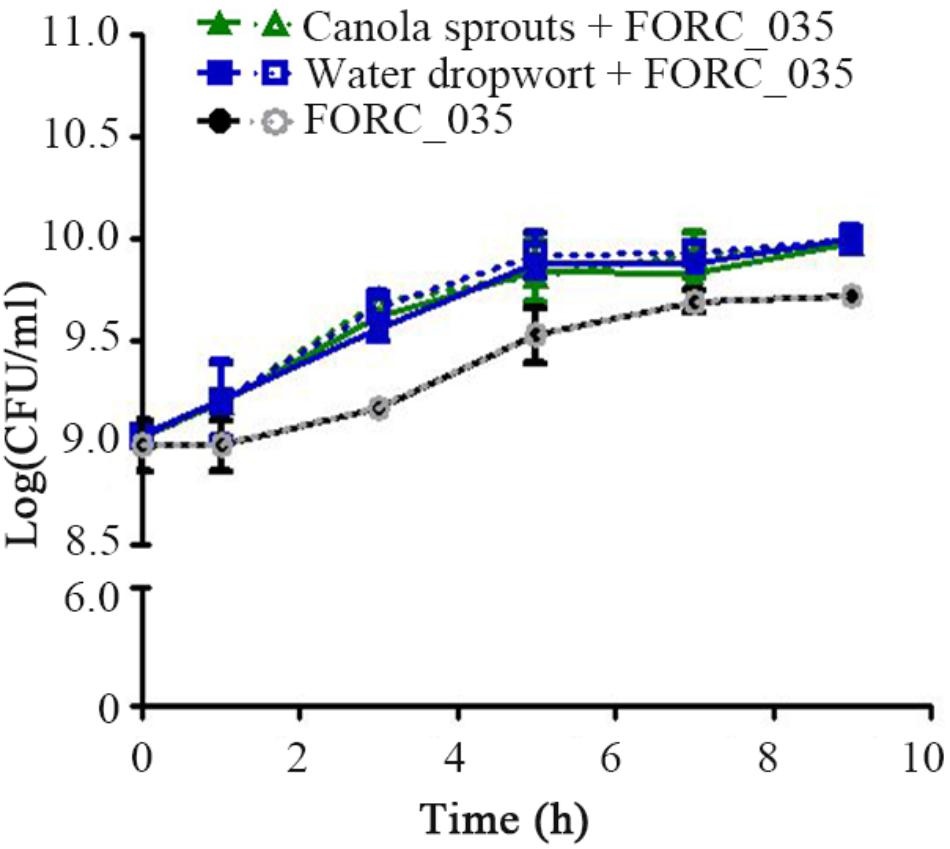
FIGURE 1. Growth of FORC_035 in M9 minimal media with or without fresh produce. Bacterial cells were inoculated at 109 CFU/mL to fresh M9 minimal media containing canola sprouts or water dropwort, respectively. As a control, FORC_035 was cultivated in M9 minimal medium without plant material. In order to discriminate E. coli among indigenous bacteria of plant tissues, culture suspension was diluted and spread onto E. coli selective TBX agar (dotted line) and LB agar (solid line), simultaneously. When only plants were incubated, growth of E. coli was not detected (Supplementary Figure S2). Each symbol indicates the mean value from triplicate measurements.
To delineate the physiological changes that occurred in the bacterial cells during contact with plants, 4,977 genes of FORC_035 were categorized according to clusters of orthologous groups (COG) (Tatusov et al., 1997) and the proportion of DEGs in each functional group was calculated (Figure 2). In FORC_035 co-cultured with canola sprouts, the expression of 8.14% (405/4,977) and 12.54% (624/4,977) genes changed significantly at 1 and 3 h, respectively. In cells exposed to water dropwort, 5.65% (281/4,977) and 5.67% (282/4,977) of genes exhibited significant differences in their expression at 1 and 3 h, respectively. In both of canola sprouts and water dropwort, co-cultivation for 1 h induced significant expression changes in genes associated with translation, ribosomal structure, and biogenesis (30.56% for canola sprouts and 19.44% for water dropwort), cell motility (20.79% for canola sprouts and 10.89% for water dropwort), energy production and conversion (14.98% for canola sprouts and 10.10% for water dropwort), and carbohydrate transport and metabolism (12.70% for canola sprouts and 9.26% for water dropwort). Meanwhile at 3 h post-inoculation, groups of genes related to secondary metabolites biosynthesis, transport and catabolism (42.55% for canola sprouts and 10.64% for water dropwort), nucleotide transport and metabolism (31.00% for canola sprouts and 11.00% for water dropwort), and energy production and conversion (27.36% for canola sprouts and 19.54% for water dropwort) showed significant differences in their expression in common. Functional groups of genes that showed significant expression changes upon contact with both plants were graphically compared at 1 and 3 h based on COG (Figures 2C,D).
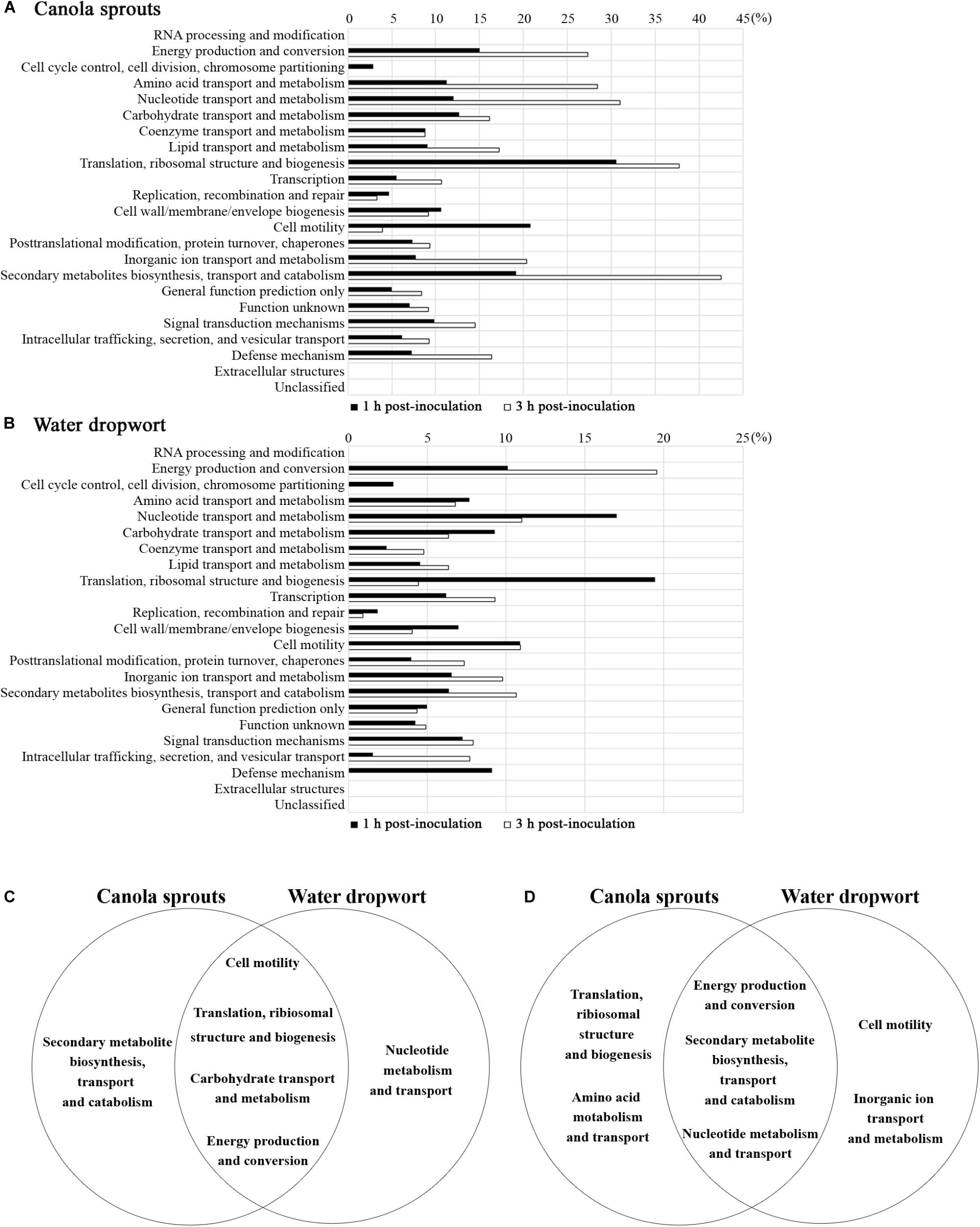
FIGURE 2. Functional categorization of genes that were differentially regulated upon contact with plants. (A,B) Genes with log2 [fold change] ≥ 2 or log2 [fold change] ≤ –2 in response to canola sprouts (A) or water dropwort (B) were grouped based on COG. Bars represent the percentages of increased or decreased genes in a given category after 1 h of exposure ( ) or 3 h of exposure (
) or 3 h of exposure ( ). (C,D) Venn diagram illustrating functional groups with significant expression changes in contact with plants. Five top functional groups showing differential expressions in response to each plant were sorted at 1 h (C) and 3 h (D) post-inoculation. Functional groups identified in both of plants in common were placed in the intersections of the sets.
). (C,D) Venn diagram illustrating functional groups with significant expression changes in contact with plants. Five top functional groups showing differential expressions in response to each plant were sorted at 1 h (C) and 3 h (D) post-inoculation. Functional groups identified in both of plants in common were placed in the intersections of the sets.
RNA-Seq Analysis Identifies Genes Differentially Expressed in Response to Canola Sprouts or Water Dropwort
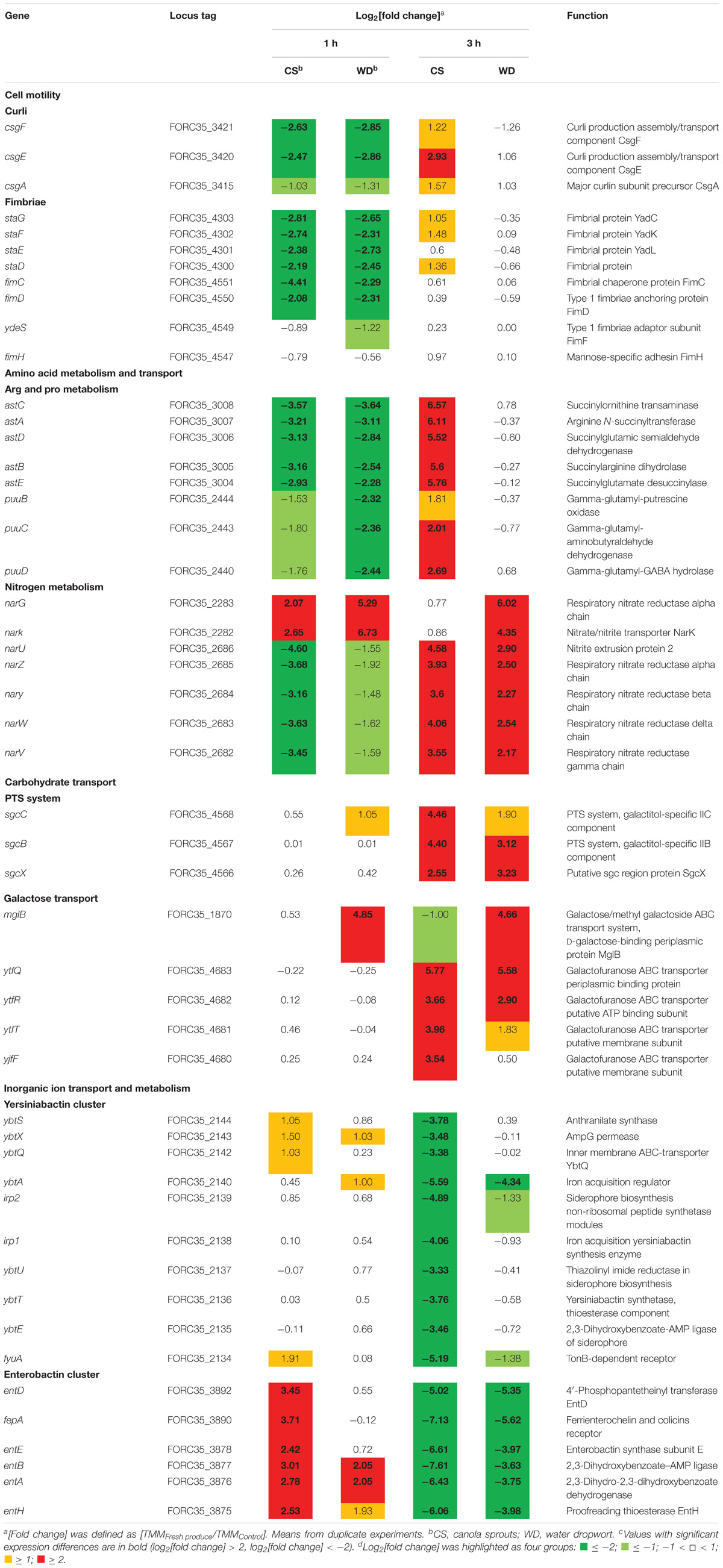
Based on their predicted functions, genes up- or down-regulated as a cluster or an operon in response to plant tissues were further defined, because these genes might be involved in bacterial adaptation to plant environments. Genes specifically regulated by exposure to fresh produce and their functions are presented in Table 2. Representative genes from each group were validated by qRT-PCR (Supplementary Figure S3).
(i) Adhesion
In this study, genes encoding various types of fimbriae, including curli and type 1 fimbriae, were down-regulated upon contact with canola sprouts and water dropwort (Table 2). Bacteria switch between planktonic and sessile states depending on environmental conditions. When they encounter stresses such as nutrient limitation, they adhere to surfaces and develop extracellular polymeric substances (EPSs) to form a bacterial aggregation called a biofilm (Oh et al., 2007), which enables them to resist various stresses and acquire scarce nutrients (Enos and Taylor, 1996). Bacteria utilize diverse cell surface appendages, including fimbriae, lipopolysaccharides, O-antigen capsule, flagellae, and non-fimbrial adhesions for initial attachment to surfaces (Kierek-Pearson and Karatan, 2005). For example, enteric pathogens such as E. coli and Salmonella enterica possess several different kinds of fimbrial structures and exploit them to colonize salad and alfalfa sprouts (Barak et al., 2005; Fink et al., 2012). In contrast, bacteria favor a planktonic lifestyle under nutrient-rich conditions (Marshall et al., 1971; Brown et al., 1977). Therefore, consistent with the increased growth of FORC_035 in the presence of plants (Figure 1), the down-regulation of fimbriae genes in response to plants implies that free-living cells are preferred due to the abundance of nutrients exuded from plant tissues.
(ii) Amino Acid and Nitrogen Metabolism
In accordance with the profound expression changes of genes associated with energy production and conversion (Figure 2), numerous genes important for amino acid and nitrogen metabolism showed significantly altered expression upon contact with plants, intimating the efflux of nutrients from co-inoculated plant tissues.
For example, genes in the ast and puu operons, which are required for arginine and proline metabolism, respectively, tended to be induced during adaptation in plant-supplemented environments (Table 2): the log2 [fold change] was higher at 3 h than at 1 h in both plants. Arginine succinyltransferase (AST) pathway comprises an ast operon that allows utilization of arginine as a nitrogen source in E. coli (Schneider et al., 1998). Genes in the puu operon encode enzymes required for degradation of putrescine, a major polyamine molecule involved in a variety of biological processes, including growth rate control (Tweeddale et al., 1998). The induction of genes involved in arginine and putrescine degradation during bacterial adaptation to plant tissues might be indicative of abundant nitrogen during early exposure, but the depletion of preferable nitrogen sources due to bacterial overgrowth in the medium at later time points. In a similar context, considering that canola sprouts are enriched in arginine and proline (Chung et al., 1989), the overproduction of AST enzymes in canola sprout-supplemented medium at 3 h might be a bacterial strategy to exploit plant arginine as an alternative nitrogen source. Bacteria utilize putrescine not only as a sole nitrogen source, but also as a signaling molecule in response to unfavorable conditions such as oxidative stress, which is a plausible stress in the context of contact with plant tissues (Schneider and Reitzer, 2012). Interestingly, arginine can be converted to putrescine, suggesting that the two pathways are closely related (Schneider et al., 2013).
Bacteria colonizing plants are able to scavenge nitrate, a source of nitrogen in plants (Mantelin and Touraine, 2004), and utilize it as an alternative electron acceptor for energy generation (Bonnefoy and Demoss, 1994). E. coli produces two different membrane-bound respiratory nitrate reductases, nitrate reductase A (NRA) and nitrate reductase Z (NRZ), for dissimilative nitrate reduction (Bonnefoy and Demoss, 1994). Transcriptome analysis revealed that the expression of genes encoding NRA (narG and narK) and NRZ (narUZYWV) were both significantly altered during adaptation to a plant environment (Table 2); there was an increase in narGK expression at both time points, while narUZYWV expression decreased at 1 h but increased at 3 h. These results suggest that FORC_035 might operate an aerobic respiration system for shorter exposure times to plant material, but convert to anaerobic respiration for longer exposure times, using nitrate leaked from plant tissues as an electron acceptor.
(iii) Carbohydrate Transport
Given the role of sugars and carbohydrates as carbon sources, E. coli employs a range of different transporter systems for efficient sugar acquisition, including the phosphotransferase system (PTS) and ATP-binding cassette (ABC) transporter system (Braun, 2003). The genes sgcCBX, mglB, ytfQRT, and ytfF were up-regulated during FORC_035 adaptation to both plant types (Table 2). Genes of the sgc operon encode a PTS and are involved in uptake and breakdown of pentose and pentitol (Reizer et al., 1996; Tchieu et al., 2001). Products of mglB, ytfQRT, and ytfF comprise ABC transporters for galactopyranose (mglB) or galactofuranose (ytfQRT and ytfF) (Horler et al., 2009). When preferred carbon sources such as glucose are in limited supply, bacteria undergo catabolite repression to consume secondary carbon sources and improve their fitness in response to changes in nutrients (Stülke and Hillen, 1999). E. coli deprived of glucose is known to up-regulate the expression of genes required for uptake of alternative sugars such as galactose, ribose, and mannose (Busby and Kolb, 1996; Raman et al., 2005). Up-regulation of genes involved in diverse carbohydrate transport at 3 h post-inoculation in this study suggests that FORC_035 might experience nutrient starvation at 3 h after contact with plants and switch metabolic pathways to utilize alternative carbon sources abundant in plant tissues.
(iv) Iron Acquisition
Iron is a critical nutrient for bacterial growth and also important for virulence in pathogenic bacteria (Miethke and Marahiel, 2007). Therefore, bacteria possess many different iron transport systems (Porcheron et al., 2013). E. coli also exploits a variety of chelating compounds for iron uptake and siderophores are the primary chelators responsible for ferric iron acquisition (Miethke and Marahiel, 2007). E. coli produces redundant siderophores including enterobactin, salmochelin, and aerobactin. In addition, yersiniabactin, a bacterial siderophore found in various pathogenic Yersinia species, is also present in several pathogenic E. coli strains (Garcia et al., 2011; Johnstone and Nolan, 2015). The complex of ferric ion and siderophore is actively transported across the outer membrane using ATP hydrolysis mediated by the TonB-ExbB-ExbD system (Braun, 2003). Once in the periplasm, periplasmic binding proteins (PBPs) capture ferric-siderophores and deliver them to cytoplasmic membrane ABC transporters to release ferric iron into the cytosol for subsequent utilization as a nutrient (Braun, 2003). Transcriptome analysis revealed a significant change in the expression of genes associated with iron uptake. In particular, the expression of genes required for the production of yersiniabactin and enterobactin siderophores declined during bacterial adaptation to plant tissues (Table 2). Taking into account the high binding affinities of siderophores for ferric iron under iron-deficient conditions, decreased expression of siderophore genes at 3 h post-inoculation might be indicative of an abundant supply of iron from plant exudates.
Enterobactin Is Required for Survival of FORC_035 in Water Dropwort
Iron is an essential nutrient involved in a variety of biological processes in bacteria, including DNA synthesis, electron transport system, ATP synthesis, and oxygen transport (Andrews et al., 2003; Krewulak and Vogel, 2008). Multiple genes relevant to iron uptake showed an opposing expression tendency between 1 and 3 h post-inoculation; up-regulation at 1 h versus down-regulation at 3 h (Table 2 and Supplementary Figure S4). The expression levels were quantified by qRT-PCR, supporting consistency with the RNA-Seq results (Supplementary Figure S5). A similar gene expression pattern was observed when bacterial cells were cultured at room temperature (25°C) in the presence of fresh produce (Supplementary Figures S6, S7). These results suggest that FORC_035 might encounter a dynamic change in iron abundance during its adaptation to the plant environment and control its iron uptake ability to improve its fitness. Especially, genes associated with two siderophores, enterobactin and yersiniabactin, were down-regulated by 3 log-fold or more when bacterial cells were exposed to fresh produce for 3 h (Table 2). Considering the indispensable roles of iron in numerous biological processes, we evaluated the importance of iron uptake via enterobactin and yersiniabactin during bacterial adaptation to the plants by comparing the growth of ΔentB and ΔybtS strains in the presence and absence of plant tissues. Deletion of entB did not compromise bacterial growth significantly in M9 minimal medium supplemented with canola sprouts. Taking into account the observation that co-culturing with plant tissues stimulated FORC_035 to alter transcription of diverse genes required for iron transport, including those involved in ferrous ion and ferric citrate transport (Supplementary Figure S4), we reasoned that the ΔentB strain might be able to circumvent the lack of enterobactin by activating iron transport systems other than enterobactin to acquire iron released from canola sprouts. However, lack of entB dampened bacterial growth significantly in the presence of water dropwort, which has a lower iron content than sprouts (Park and Kim, 1996) (Figure 3A and Supplementary Figure S8). This result suggests that the ΔentB strain suffered from a shortage of iron when interacting with water dropwort due to its lack of enterobactin. To test this possibility, ferric sulfate was added to the culture medium at 10 μM as an iron supplement and bacterial growth was compared. As expected, addition of ferric ion complemented the growth defect of the ΔentB strain in water dropwort-containing medium (Figure 3A). Moreover, the growth defect was complemented with trans-encoded entB by the pEntB plasmid (Figure 3B), indicating that iron uptake via enterobactin is required for the growth of bacterial cells exposed to water dropwort. On the other hand, the mutation on yersiniabactin showed no defects in bacterial growth. These results suggested that enterobactin plays a greater role than yersiniabactin does for iron acquisition. Differential contribution to bacterial growth by enterobactin and yersiniabactin has been reported previously in Klebsiella pneumonia (Lawlor et al., 2007).
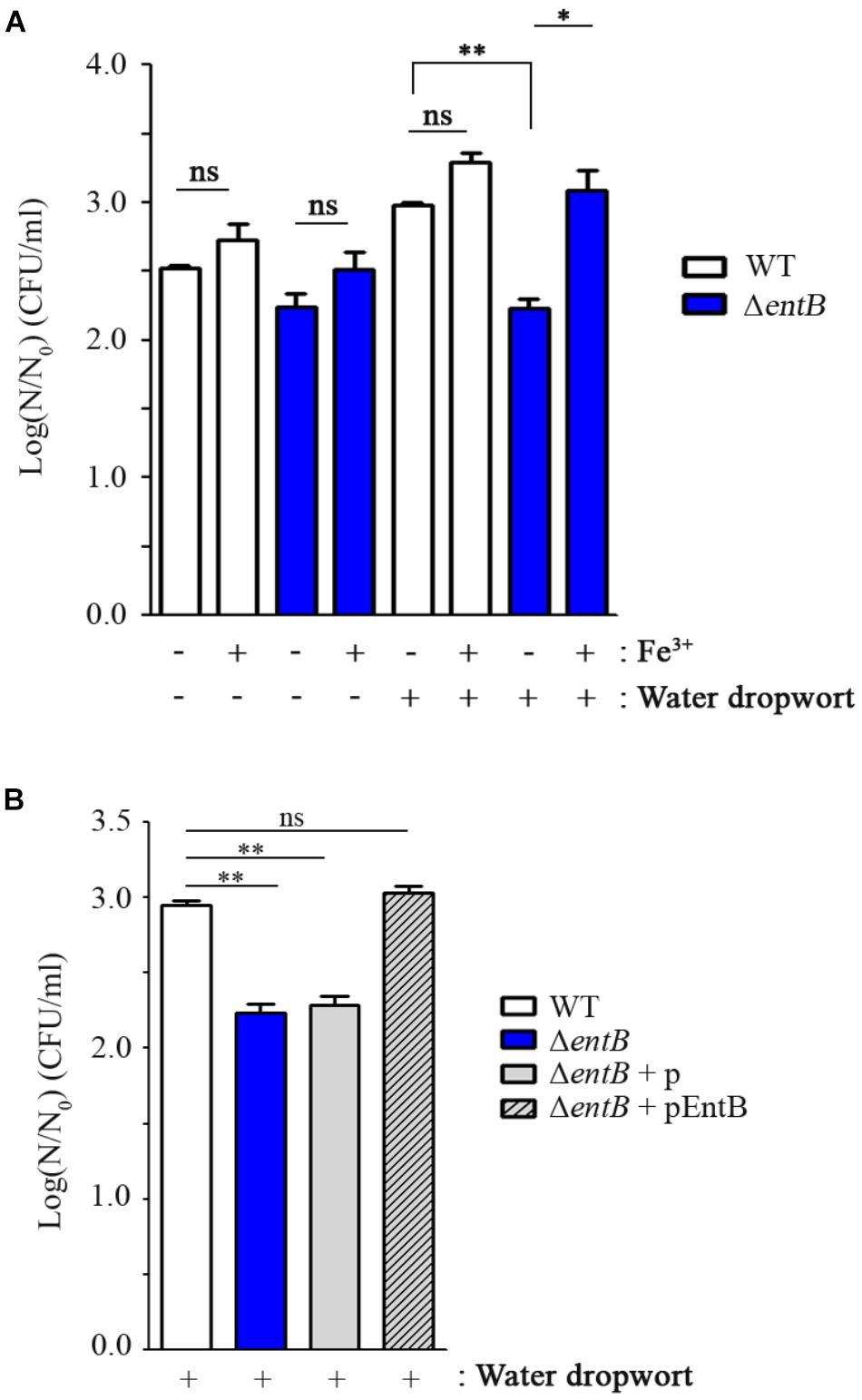
FIGURE 3. Comparison of bacterial growth between FORC_035 wild-type (WT) and ΔentB mutant strains. (A) Ferric sulfate (10 μM) was added to M9 minimal media with or without water dropwort. (B) Four strains of WT, ΔentB, and ΔentB with either empty plasmid or pEntB were cultivated in the presence of water dropwort. pUHE21-2lacIq plasmid was used and 50 μM of IPTG was used for induction. In both graphs, live bacterial numbers were measured in M9 minimal medium broth supplemented with water dropwort after a 5-h incubation. Viability of bacterial cells was measured by comparing log(N/N0) values, where N0 is the initial number of cells at 0 h and N is the number of cells after a 5-h incubation. The t-test was used to evaluate the significance of differences in viability, and significance is indicated as follows: ∗P < 0.05; ∗∗P < 0.01; ns, not significant.
Gallium as an Iron Mimetic Inhibits FORC_035 Growth and Biofilm Formation
The transition metal gallium (Ga3+), which has an ionic radius nearly identical to iron (Fe3+), can act as a “Trojan horse” to disturb bacterial iron uptake, as many biologic systems, including bacteria, cannot distinguish between Ga3+ and Fe3+ (Chitambar and Narasimhan, 1991; Kaneko et al., 2007). Taking into account that the ΔentB strain suffering from iron shortage due to its lack of enterobactin slowed down its growth when in contact with water dropwort, disturbance of cellular iron uptake by the addition of gallium could be a promising strategy to control the growth and survival of EHEC FORC_035 on water dropwort. To examine this possibility, FORC_035 in M9 minimal medium containing water dropwort was supplemented with Ga(NO3)3 at 1 mM and its growth was evaluated (Figure 4). FORC_035 co-cultured with water dropwort multiplied faster than FORC_035 grown in the absence of plant tissues, but the addition of gallium led to a 1.7 log reduction in bacterial numbers, indicating that gallium had antimicrobial activity against EHEC FORC_035. Gallium had a growth inhibitory effect regardless of the presence of water dropwort, but gallium exerted greater growth inhibition when bacteria were in contact with water dropwort tissues than when they were not (Figure 4). The greater growth inhibition of gallium in the presence of water dropwort was probably because the growth-promoting effect of the plant (Figure 1) was abolished by gallium, which functioned as a competitive antagonist of iron in the plant-supplemented environment. Antimicrobial effect of gallium has been observed in other bacteria as well, including Pseudomonas aeruginosa and Mycobacterium tuberculosis wherein gallium inhibits cellular Fe-dependent metabolic pathways by substituting Fe3+ (Olakanmi et al., 2000; Kaneko et al., 2007). In mammalian cells, Ga3+ makes up Ga-transferrin complex instead of Fe3+, suggesting the possibility that gallium exerts its role by combining with iron-chelating proteins.
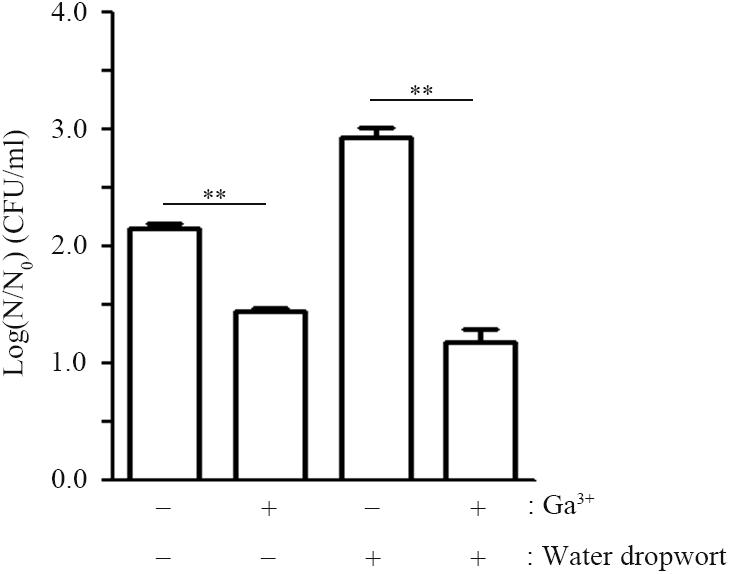
FIGURE 4. Effect of gallium nitrate on the growth of FORC_035. Wild-type FORC_035 was cultivated in M9 minimal medium broth with or without water dropwort and Ga(NO3)3 was added to the medium at 1 mM. Bacterial numbers were estimated at 9 h post-inoculation. Note that log(N/N0) is plotted on y-axis, where N0 is the initial number of cells at 0 h and N is the number of cells after a 9-h incubation. Asterisks indicate significant differences (∗∗P < 0.01).
Biofilm formation is achieved through a complex process influenced by multiple environmental signals. Among diverse signals, iron can also regulate biofilm formation and this regulation depends on bacterial species. The availability of iron influences the ability of bacteria to form biofilms effectively; high concentrations of iron promote bacterial biofilm development in E. coli and P. aeruginosa (Banin et al., 2005; Wu and Outten, 2009). Therefore, the influence of gallium on bacterial biofilm formation was investigated. The presence of Ga(NO3)3 at 10 μM inhibited biofilm formation by threefold without changing the growth of FORC_035 (Figure 5). The inhibitory effect of gallium on biofilm formation was not influenced by temperature changes (Supplementary Figure S9). Biofilms enable bacteria to resist stressful conditions such as disinfectants and the defense responses of plants and animals, which makes biofilms an important issue to deal with in food hygiene (Srey et al., 2013). Gallium was approved for hypercalcemia treatment by the FDA, and has low toxicity at appropriate doses (Chitambar and Narasimhan, 1991; Gómez et al., 1992). The inhibitory effects of gallium on bacterial growth and biofilm formation suggest that gallium can potentially be used as an antimicrobial agent.
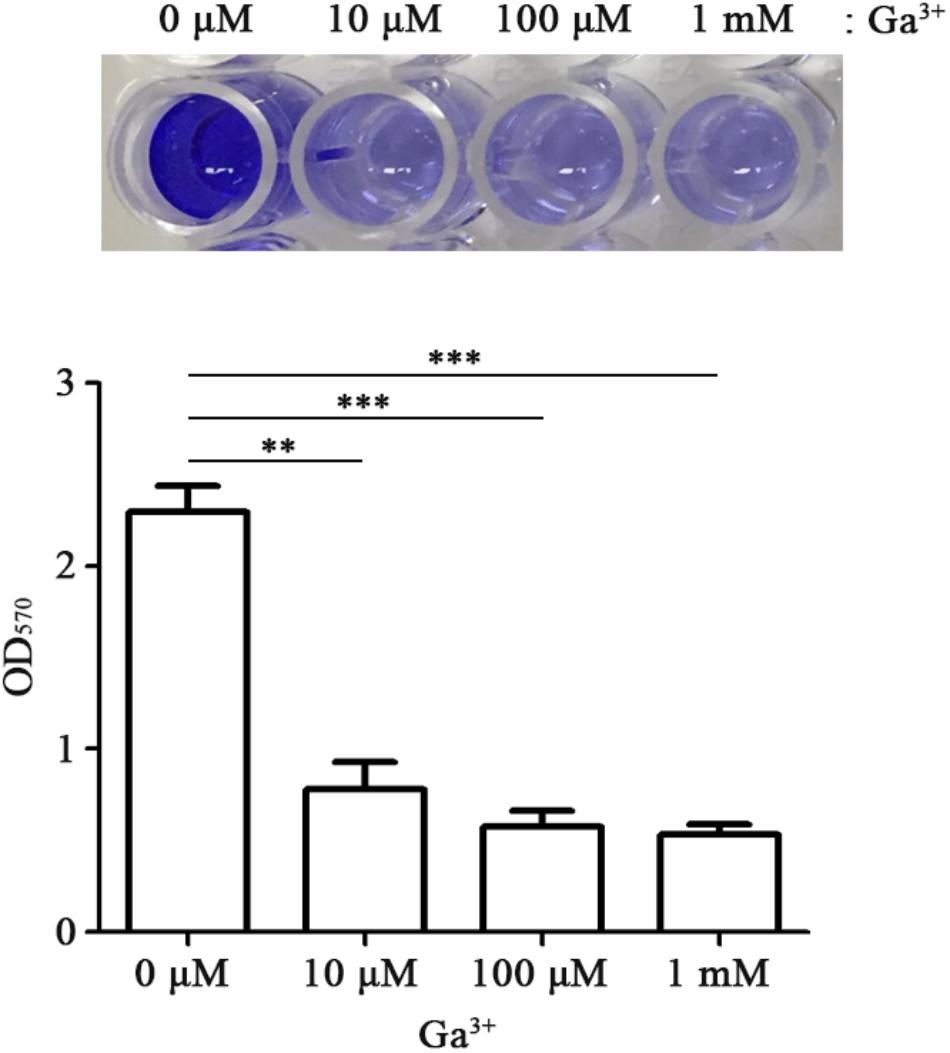
FIGURE 5. Gallium inhibits biofilm formation of FORC_035. FORC_035 was statically cultivated in LB medium in polystyrene 96-well plates for 48 h. Gallium was added at the indicated concentrations. Biofilm was stained using 0.1% crystal violet (Top) and OD570 was measured (Bottom). Asterisks indicate significant differences (∗∗P < 0.01; ∗∗∗P < 0.001).
Conclusion
In conclusion, the transcriptomes of FORC_035 in contact with canola sprouts and water dropwort provided insights into the overall bacterial transcriptional response to plants. There were significant changes in the expression of genes associated with fitness and survival upon co-culture with plant materials. In particular, genes required for iron uptake were found to be critical determinants of the adaptation of EHEC FORC_035 to plants.
Data Availability
The database of the Foodborne pathogen Omics Research Center (http://forcdb.snu.ac.kr) in which the datasets for this manuscript were deposited are not publicly available due to policy of the institution. Requests to access the datasets should be directed to SR.
Author Contributions
HN, HY, and SR conceived and designed the experiments. HN, YK, and DK performed the experiments. HN, YK, DK, and HY analyzed the data and wrote the paper. HY and SR revised the manuscript.
Funding
This study was supported by a grant (14162MFDS972) from the Ministry of Food and Drug Safety, South Korea, in 2018. This work was also supported by the BK21 Plus Program of the Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02397/full#supplementary-material
Footnotes
References
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Andrews, W., Mislivec, P., Wilson, C., Bruce, V., Poelma, P., Gibson, R., et al. (1982). Microbial hazards associated with bean sprouting. J. Assoc. Off. Anal. Chem. 65, 241–248.
Banin, E., Vasil, M. L., and Greenberg, E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081. doi: 10.1073/pnas.0504266102
Barak, J. D., Gorski, L., Naraghi-Arani, P., and Charkowski, A. O. (2005). Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71, 5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005
Bonnefoy, V., and Demoss, J. A. (1994). Nitrate reductases in Escherichia coli. Antonie Van Leeuwenhoek 66, 47–56. doi: 10.1007/BF00871632
Breuer, T., Benkel, D. H., Shapiro, R. L., Hall, W. N., Winnett, M. M., Linn, M. J., et al. (2001). A multistate outbreak of Escherichia coli O157: H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerg. Infect. Dis. 7, 977–982. doi: 10.3201/eid0706.010609
Brown, C., Ellwood, D., and Hunter, J. (1977). Growth of bacteria at surfaces: influence of nutrient limitation. FEMS Microbiol. Lett. 1, 163–166. doi: 10.1111/j.1574-6968.1977.tb00605.x
Busby, S., and Kolb, A. (1996). “The CAP modulon,” in Regulation of Gene Expression in Escherichia coli, eds E. C. C. Lin and A. S. Lynch (Berlin: Springer), 255–279. doi: 10.1007/978-1-4684-8601-8_12
Carey, C. M., Kostrzynska, M., and Thompson, S. (2009). Escherichia coli O157: H7 stress and virulence gene expression on Romaine lettuce using comparative real-time PCR. J. Microbiol. Methods 77, 235–242. doi: 10.1016/j.mimet.2009.02.010
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: Tc R and Km R cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-A
Chitambar, C. R., and Narasimhan, J. (1991). Targeting iron-dependent DNA synthesis with gallium and transferrin-gallium. Pathobiology 59, 3–10. doi: 10.1159/000163609
Chung, T., Nwokolo, E., and Sim, J. (1989). Compositional and digestibility changes in sprouted barley and canola seeds. Plant Foods Hum. Nutr. 39, 267–278. doi: 10.1007/BF01091937
Cooley, M. B., Miller, W. G., and Mandrell, R. E. (2003). Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157: H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69, 4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Deering, A. J., Mauer, L. J., and Pruitt, R. E. (2012). Internalization of E. coli O157: H7 and Salmonella spp. in plants: a review. Food Res. Int. 45, 567–575. doi: 10.1016/j.foodres.2011.06.058
Dillies, M.-A., Rau, A., Aubert, J., Hennequet-Antier, C., Jeanmougin, M., Servant, N., et al. (2013). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 14, 671–683. doi: 10.1093/bib/bbs046
Doherty, C. P. (2007). Host-pathogen interactions: the role of iron. J. Nutr. 137, 1341–1344. doi: 10.1093/jn/137.5.1341
Enos, D., and Taylor, S. (1996). Influence of sulfate-reducing bacteria on alloy 625 and austenitic stainless steel weldments. Corrosion 52, 831–842. doi: 10.5006/1.3292075
Erickson, M. C. (2012). Internalization of fresh produce by foodborne pathogens. Annu. Rev. Food Sci. Technol. 3, 283–310. doi: 10.1146/annurev-food-022811-101211
Fink, R. C., Black, E. P., Hou, Z., Sugawara, M., Sadowsky, M. J., and Diez-Gonzalez, F. (2012). Transcriptional responses of Escherichia coli K-12 and O157: H7 associated with lettuce leaves. Appl. Environ. Microbiol. 78, 1752–1764. doi: 10.1128/AEM.07454-11
Garcia, E. C., Brumbaugh, A. R., and Mobley, H. L. (2011). Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79, 1225–1235. doi: 10.1128/IAI.01222-10
Gómez, M., Sánchez, D. J., Domingo, J. L., and Corbella, J. (1992). Developmental toxicity evaluation of gallium nitrate in mice. Arch. Toxicol. 66, 188–192. doi: 10.1007/BF01974013
Griffin, P. M., and Tauxe, R. V. (1991). The epidemiology of infections caused by Escherichia coli O157: H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13, 60–98. doi: 10.1093/oxfordjournals.epirev.a036079
Heaton, J. C., and Jones, K. (2008). Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J. Appl. Microbiol. 104, 613–626. doi: 10.1111/j.1365-2672.2007.03587.x
Herman, K., Hall, A., and Gould, L. (2015). Outbreaks attributed to fresh leafy vegetables, United States, 1973–2012. Epidemiol. Infect. 143, 3011–3021. doi: 10.1017/S0950268815000047
Horler, R. S., Müller, A., Williamson, D. C., Potts, J. R., Wilson, K. S., and Thomas, G. H. (2009). Furanose-specific sugar transport characterization of a bacterial galactofuranose-binding protein. J. Biol. Chem. 284, 31156–31163. doi: 10.1074/jbc.M109.054296
Jablasone, J., Warriner, K., and Griffiths, M. (2005). Interactions of Escherichia coli O157: H7, Salmonella typhimurium and Listeria monocytogenes plants cultivated in a gnotobiotic system. Int. J. Food Microbiol. 99, 7–18. doi: 10.1016/j.ijfoodmicro.2004.06.011
Jandu, N., Ho, N. K., Donato, K. A., Karmali, M. A., Mascarenhas, M., Duffy, S. P., et al. (2009). Enterohemorrhagic Escherichia coli O157: H7 gene expression profiling in response to growth in the presence of host epithelia. PLoS One 4:e4889. doi: 10.1371/journal.pone.0004889
Johnstone, T. C., and Nolan, E. M. (2015). Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 44, 6320–6339. doi: 10.1039/c4dt03559c
Junker, R. R., Loewel, C., Gross, R., Dötterl, S., Keller, A., and Blüthgen, N. (2011). Composition of epiphytic bacterial communities differs on petals and leaves. Plant Biol. 13, 918–924. doi: 10.1111/j.1438-8677.2011.00454.x
Kaneko, Y., Thoendel, M., Olakanmi, O., Britigan, B. E., and Singh, P. K. (2007). The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117, 877–888. doi: 10.1172/JCI30783
Karmali, M., Petric, M., Steele, B., and Lim, C. (1983). Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 321, 619–620. doi: 10.1016/S0140-6736(83)91795-6
Karmali, M. A. (1989). Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2, 15–38. doi: 10.1128/CMR.2.1.15
Kierek-Pearson, K., and Karatan, E. (2005). Biofilm development in bacteria. Adv. Appl. Microbiol. 57, 79–111. doi: 10.1016/S0065-2164(05)57003-5
Krewulak, K. D., and Vogel, H. J. (2008). Structural biology of bacterial iron uptake. Biochimica Biophysica Acta 1778, 1781–1804. doi: 10.1016/j.bbamem.2007.07.026
Kyle, J. L., Parker, C. T., Goudeau, D., and Brandl, M. T. (2010). Transcriptome analysis of Escherichia coli O157: H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 76, 1375–1387. doi: 10.1128/AEM.02461-09
Landstorfer, R., Simon, S., Schober, S., Keim, D., Scherer, S., and Neuhaus, K. (2014). Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157: H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15:353. doi: 10.1186/1471-2164-15-353
Lawlor, M. S., O’connor, C., and Miller, V. L. (2007). Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75, 1463–1472. doi: 10.1128/IAI.00372-06
Lynch, M. F., Tauxe, R. V., and Hedberg, C. W. (2009). The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137, 307–315. doi: 10.1017/S0950268808001969
Mantelin, S., and Touraine, B. (2004). Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J. Exp. Bot. 55, 27–34. doi: 10.1093/jxb/erh010
Marshall, K., Stout, R., and Mitchell, R. (1971). Mechanism of the initial events in the sorption of marine bacteria to surfaces. Microbiology 68,337–348.
Matkovich, S. J., Zhang, Y., Van Booven, D. J., and Dorn, G. W. (2010). Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators. Circ. Res. 106, 1459–1467. doi: 10.1161/CIRCRESAHA.110.217513
Meng, J., and Doyle, M. (1997). Emerging issues in microbiological food safety. Annu. Rev. Nutr. 17, 255–275. doi: 10.1146/annurev.nutr.17.1.255
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/MMBR.00012-07
Murphy, K. C., and Campellone, K. G. (2003). Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11.
O’brien, R., and Lindow, S. (1989). Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology 79, 619–627. doi: 10.1094/Phyto-79-619
Oh, Y., Jo, W., Yang, Y., and Park, S. (2007). Influence of culture conditions on Escherichia coli O157: H7 biofilm formation by atomic force microscopy. Ultramicroscopy 107, 869–874. doi: 10.1016/j.ultramic.2007.01.021
Olakanmi, O., Britigan, B. E., and Schlesinger, L. S. (2000). Gallium disrupts iron metabolism of mycobacteria residing within human macrophages. Infect. Immun. 68, 5619–5627. doi: 10.1128/IAI.68.10.5619-5627.2000
Park, Y., and Kim, Y. (1996). Study on the nutrient composition of hydroponic water dropwort (Oenanthe stolonifera DC). Food Chem. 57, 79–79. doi: 10.1016/0308-8146(96)89021-2
Perez-Llamas, C., and Lopez-Bigas, N. (2011). Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One 6:e19541. doi: 10.1371/journal.pone.0019541
Porcheron, G., Garénaux, A., Proulx, J., Sabri, M., and Dozois, C. M. (2013). Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 3:90. doi: 10.3389/fcimb.2013.00090
Raman, B., Nandakumar, M., Muthuvijayan, V., and Marten, M. R. (2005). Proteome analysis to assess physiological changes in Escherichia coli grown under glucose-limited fed-batch conditions. Biotechnol. Bioeng. 92, 384–392. doi: 10.1002/bit.20570
Reizer, J., Ramseier, T. M., Reizer, A., Charbit, A., and Saier, M. H. Jr. (1996). Novel phosphotransferase genes revealed by bacterial genome sequencing: a gene cluster encoding a putative N-acetylgalactosamine metabolic pathway in Escherichia coli. Microbiology 142, 231–250. doi: 10.1099/13500872-142-2-231
Risso, D., Ngai, J., Speed, T. P., and Dudoit, S. (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. doi: 10.1038/nbt.2931
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Schenk, P. M., Carvalhais, L. C., and Kazan, K. (2012). Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trends Biotechnol. 30, 177–184. doi: 10.1016/j.tibtech.2011.11.002
Schikora, A., Carreri, A., Charpentier, E., and Hirt, H. (2008). The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One 3:e2279. doi: 10.1371/journal.pone.0002279
Schneider, B. L., Hernandez, V. J., and Reitzer, L. (2013). Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol. Microbiol. 88, 537–550. doi: 10.1111/mmi.12207
Schneider, B. L., Kiupakis, A. K., and Reitzer, L. J. (1998). Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180, 4278–4286.
Schneider, B. L., and Reitzer, L. (2012). Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J. Bacteriol. 194, 4080–4088. doi: 10.1128/JB.05063-11
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-3
Shi, X., Namvar, A., Kostrzynska, M., Hora, R., and Warriner, K. (2007). Persistence and growth of different Salmonella serovars on pre-and postharvest tomatoes. J. Food Prot. 70, 2725–2731. doi: 10.4315/0362-028X-70.12.2725
Sivapalasingam, S., Friedman, C. R., Cohen, L., and Tauxe, R. V. (2004). Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67, 2342–2353. doi: 10.4315/0362-028X-67.10.2342
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Soncini, F. C., Véscovi, E. G., and Groisman, E. A. (1995). Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177, 4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995
Sperandio, V., and Nguyen, Y. (2012). Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2:90. doi: 10.3389/fcimb.2012.00090
Srey, S., Jahid, I. K., and Ha, S.-D. (2013). Biofilm formation in food industries: a food safety concern. Food Control 31, 572–585. doi: 10.1016/j.foodcont.2012.12.001
Stülke, J., and Hillen, W. (1999). Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2, 195–201. doi: 10.1016/S1369-5274(99)80034-4
Taormina, P. J., Beuchat, L. R., and Slutsker, L. (1999). Infections associated with eating seed sprouts: an international concern. Emerg. Infect. Dis. 5, 626–634. doi: 10.3201/eid0505.990503
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Tchieu, J. H., Norris, V., Edwards, J. S., and Saier, M. H. Jr. (2001). The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3, 329–346.
Tweeddale, H., Notley-Mcrobb, L., and Ferenci, T. (1998). Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180,5109–5116.
Vikram, A., Jesudhasan, P. R., Jayaprakasha, G., Pillai, B., and Patil, B. S. (2010). Grapefruit bioactive limonoids modulate E. coli O157: H7 TTSS and biofilm. Int. J. Food Microbiol. 140, 109–116. doi: 10.1016/j.ijfoodmicro.2010.04.012
Visca, P., Leoni, L., Wilson, M. J., and Lamont, I. L. (2002). Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45, 1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x
Wu, Y., and Outten, F. W. (2009). IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J. Bacteriol. 191, 1248–1257. doi: 10.1128/JB.01086-08
Keywords: enterohemorrhagic Escherichia coli FORC_035, canola sprouts, water dropwort, iron acquisition, gallium
Citation: Na H, Kim Y, Kim D, Yoon H and Ryu S (2018) Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli FORC_035 Reveals the Essential Role of Iron Acquisition for Survival in Canola Sprouts and Water Dropwort. Front. Microbiol. 9:2397. doi: 10.3389/fmicb.2018.02397
Received: 17 May 2018; Accepted: 19 September 2018;
Published: 08 October 2018.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jincai Ma, Jilin University, ChinaYanhong Liu, United States Department of Agriculture, United States
Nora Lía Padola, National University of Central Buenos Aires, Argentina
Copyright © 2018 Na, Kim, Kim, Yoon and Ryu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangryeol Ryu, c2FuZ3J5dUBzbnUuYWMua3I=
†These authors have contributed equally to this work
 Hongjun Na
Hongjun Na Yeonkyung Kim
Yeonkyung Kim Dajeong Kim
Dajeong Kim Hyunjin Yoon
Hyunjin Yoon Sangryeol Ryu
Sangryeol Ryu