- 1Graduate Interdisciplinary Program in Entomology and Insect Science, Tucson, AZ, United States
- 2Department of Entomology, University of Arizona, Tucson, AZ, United States
- 3Center for Insect Science, Department of Entomology, University of Arizona, Tucson, AZ, United States
Bombardier beetles (Carabidae: Brachininae) are well known for their unique explosive defensive chemistry. These beetles are found in riparian corridors throughout the American Southwest, where they commonly form large diurnal multispecies aggregations in moist areas under rocks, in crevices, and in leaf litter. Using high throughput 16S amplicon sequencing, we provide the first microbiome survey of a bombardier beetle, Brachinus elongatulus, collected from two sites in Arizona. Two bacterial taxa were present in all individuals sampled: Enterococcus and Dysgonomonas. Enterococcus has been implicated in the production of fecal aggregation pheromone components, which have been shown to regulate aggregation in the German cockroach; it is possible that Enterococcus plays a similar role in Brachinus. Dysgonomonas was found in all the secretory cells of the defensive system and gut samples. Additional studies are needed to determine if these microbes play a role in these beetles’ unique chemical defense. Results also show that the majority of B. elongatulus individuals collected from both sites were infected with Spiroplasma. Many Spiroplasma are intracellular, vertically transmitted insect symbionts that may manipulate host reproduction (e.g., cause male-killing) or provide resistance to nematodes and/or parasitoid wasps. Defensive protection could be especially beneficial to B. elongatulus, which are frequently parasitized by horsehair worms (Nematomorpha). In sum, findings suggest several testable hypotheses on the effects bacteria may have on bombardier beetle behavior and physiology.
Introduction
Members of the genus Brachinus (Coleoptera: Brachininae) belong to a group of carabids known as bombardier beetles, which produce noxious exothermic defensive sprays to deter their predators (Arndt et al., 2015) (Figure 1A). They produce defensive chemicals with a pair of pygidial glands (Figure 2). Each gland system consists of secretory cells, a long collecting duct, a reservoir chamber, and a heavily sclerotized reaction chamber. Hydroquinones and hydrogen peroxide are transferred through the collecting ducts and stored in the reservoir chamber. When the beetles are threatened, muscles surrounding the reservoir chamber contract sending these chemical precursors through a one-way valve to mix with catalases and peroxidases in the reaction chamber creating defensive benzoquinones in an exothermic reaction just before they are expelled from the body in an audible explosion (Schildknecht and Holoubek, 1961; Aneshansley et al., 1983). This intricate mechanism is broadly acknowledged as one of the most complex and sophisticated defensive systems known in the animal kingdom (Eisner, 2003).

FIGURE 1. (A) Habitus of Brachinus elongatulus (UAIC1004027), United States: Arizona, Madera Canyon. (B) Multispecies aggregation of carabid beetles photographed in the laboratory after lifting a piece of wood under which the beetles had settled. Individuals were collected along the San Pedro River and are members of the following species: Brachinus favicollis, B. elongatulus, B. phaeocerus, B. hirsutus, B. lateralis, B. mexicanus, Chlaenius ruficauda, C. cumatilis, and Agonum sp.
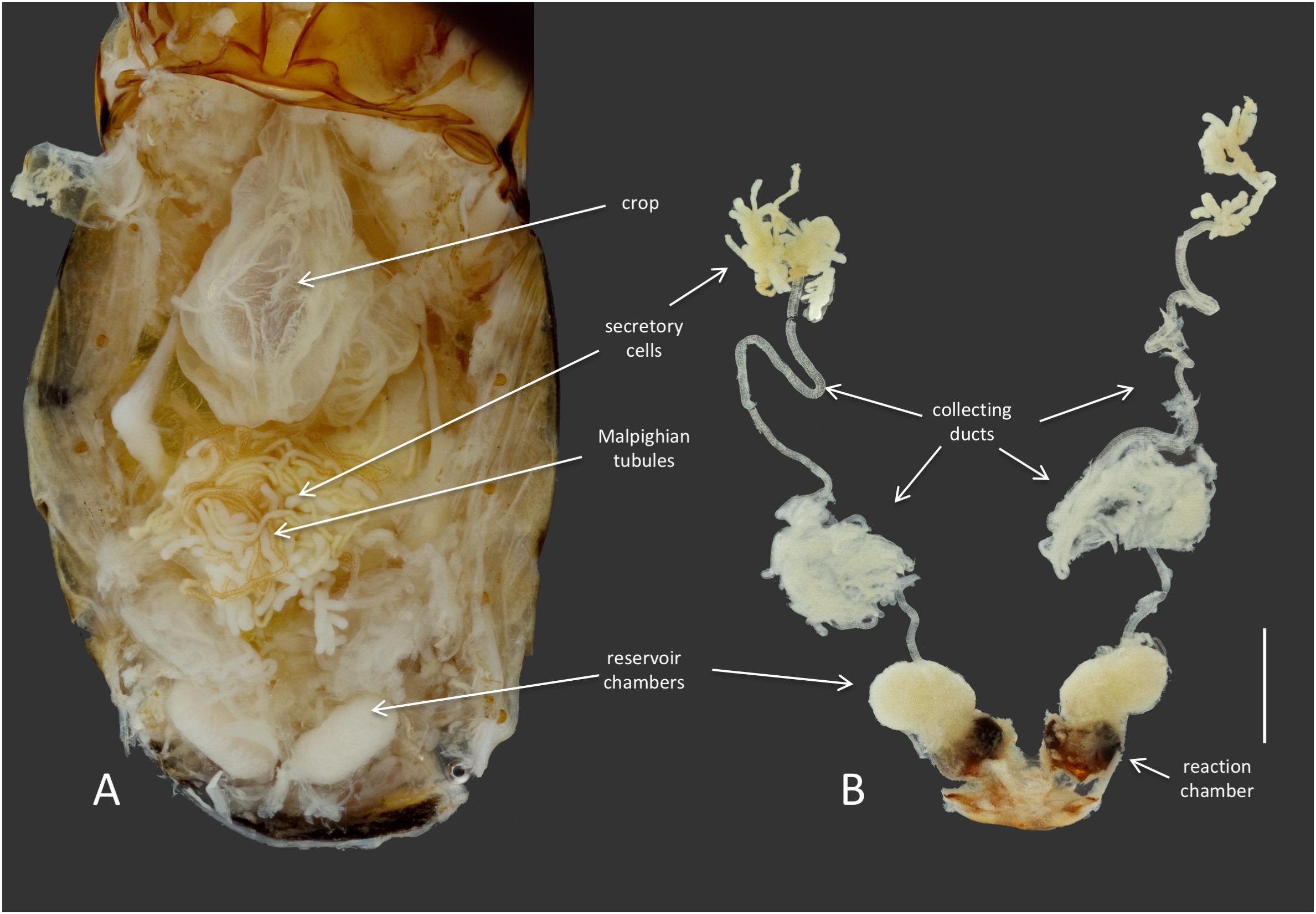
FIGURE 2. Anatomy of the defensive system of B. elongatulus and its orientation inside of the abdomen. (A) Dorsal view of the abdomen of B. elongatulus, tergites removed. (B) Paired defensive gland system dissected from the abdomen. Note that the secretory cells of the defensive system are intertwined with the Malpighian tubules of the digestive system. This bundle lays dorsal to the midgut. Scale bar = 1 mm.
Adult Brachinus elongatulus are found in moist areas under rocks, in crevices and in leaf litter along riparian corridors in Southeastern Arizona. Females oviposit in the moist mud or gravel along the water’s edge. After hatching, the triungulan larva seeks out a pupa of water beetle (Hydrophilidae, Dysticidae, Gyrinidae) which it consumes as it undergoes full larval development (Erwin, 1967; Saska and Honek, 2004). Due to living in close proximity to streams, adult Brachinus have often been found to be infected with parasitic horsehair worms (Nematomorpha) (McManus and Moore, personal observation) (Figure 3).

FIGURE 3. Nematomorphoran infection of Brachinus elongatulus. The wings have been removed from the beetle and a horsehair worm can be seen coiled inside the abdomen beneath the abdominal tergites. Another single horsehair worm, removed from another beetle’s abdomen, surrounds the beetle.
During the day, adult Brachinus are gregarious and commonly form diurnal multispecies aggregations that may contain many species of Brachinus as well as members of other carabid genera, such as Agonum, Chlaenius, Platynus, and Galerita (Schaller et al., in press) (Figure 1B). Adults forage at night scavenging on decaying organic matter and acting as opportunistic predators before forming a new aggregation by dawn. Similar to the behavior of some of their gyrinid hosts, Brachinus do not return to their aggregation of origin after dispersing at night (Heinrich and Vogt, 1980; Moore, personal observation). Aggregations provide a variety of benefits to the participating individuals, such as providing protection by deterring predators, facilitating mating, and allowing chemically defended organisms to conserve energy (Machado and Vasconcelos, 1998).
Cues involved in the formation of these aggregations are unknown but are thought to be chemical and possibly tactile in nature (Wautier, 1971; Schaller et al., in press). Multispecies aggregations are extremely rare among arthropods with only a few cases being reported in harvestmen (Archnida: Opiliones), net-winged beetles (Coleoptera: Lycidae) and whirligig beetles (Coleoptera: Gyrinidae) (Eisner et al., 1962; Heinrich and Vogt, 1980; Machado and Vasconcelos, 1998).
Other gregarious insects, such as German cockroaches (Ectobiidae), firebrats (Lepismatidae) and desert locusts (Acrididae), have been shown to aggregate in response to volatile fecal pheromones produced by their gut microbiota (Dillion et al., 2000; Woodbury and Gries, 2013; Wada-Katsumata et al., 2015). Digestive microbes in termites have also been implicated in providing cues for nest mate recognition (Minkley et al., 2006). To date the role of microbiota in influencing Brachinus aggregation behavior is unknown.
The present study is the first exploration into the bacterial associates of a bombardier beetle and the first survey of a carabid beetle microbiome using high throughput sequencing methods. Previous studies have examined the bacterial communities present in the digestive tracts of three species of carabid beetles—Harpalus pensylvanicus, Anisodactylus sanctaecrucis (both of which specialize on eating seeds) and Poecilus chalcites, a predator of living arthropods. Both of these studies used terminal restriction fragment length polymorphism (tRFLP) in tandem with 16S rRNA gene cloning (Lundgren et al., 2007; Lehman et al., 2009). The objectives of our study were to (1) characterize the microbial communities of B. elongatulus using high-throughput 16S amplicon sequencing, (2) determine if there is a core set of sequence variants common across individuals from two localities, (3) determine if B. elongatulus hosts any potential obligate or facultative endosymbionts and (4) lay the groundwork for future investigations into how the microbiome of B. elongatulus might shape their behavior and/or physiology.
Materials and Methods
Insect Collection and Dissections
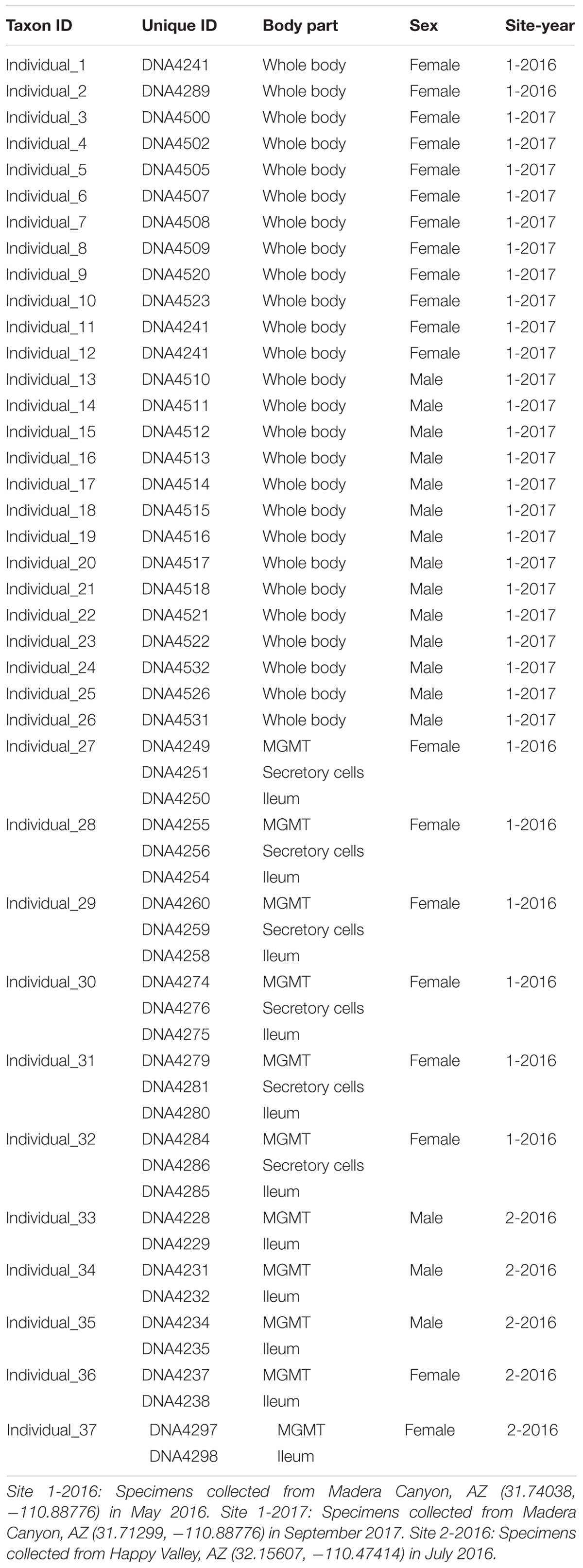
Brachinus elongatulus were collected from two sites in Southeastern Arizona. Thirty-two specimens were sampled from Madera Canyon (Santa Rita Mountains, Santa Cruz Co.) (Site 1) and 5 specimens were sampled from Happy Valley (Rincon Mountains, Pima Co.) (Site 2) (Table 1). All specimens were kept alive and transferred back to the lab for dissections. All dissections were performed within 24 h of collection. Specimens were briefly preserved in 100% ethanol and surfaced sterilized using an 8% bleach solution followed by a 1-min wash in phosphate-buffered saline (PBS) and a 1-min wash in 100% ethanol just prior to dissection. Each dissection was made using flame-sterilized forceps and flame-sterilized glass petri dishes. Details on the individuals and tissues sampled are presented in Table 1. For 26 specimens from Site 1, the whole bodies, minus the hindwings and elytra, were processed. For an additional six specimens from Site 1, the mid-gut and Malpighian tubules (MGMT), ileum, and the secretory cells of the defensive system were dissected out of the abdomen and processed separately. For the five specimens from Site 2, the MGMT and ileum were dissected out of the abdomen and processed separately.
DNA Extraction, PCR Amplification, and High-Throughput Sequencing
Dissected tissues and whole bodies were placed in individual 1.5 mL Eppendorf tubes containing lysozyme buffer [180 μl for tissues and 360 μl for whole bodies: 20 mM Tris-HCl (pH 8.0), 2mM EDTA, 1.2% Triton X, 10 mg ml−1 lysozyme]. Tissues were ground using sterile DNase/RNase free pestles. Samples were then incubated at 37°C for 90 min. DNA was precipitated and purified following the Qiagen Blood and Tissue Kit protocol for gram-positive bacteria. Using a two-step protocol, the bacterial hypervariable V3-V4 domain of 16S rDNA was PCR amplified using the primers 341F and 785R (Klindworth et al., 2013). The thermocycler protocol was denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 60 s, and extension at 72°C for 90 s, with a final extension of 72°C for 10 min (Gilbert et al., 2014). PCR products were then purified using QIAquick PCR Purification Kit (Qiagen). In a second PCR unique barcodes were ligated to the amplified region of interest (Hamady et al., 2008). The thermocycler protocol was denaturation at 95°C for 3 min followed by 8 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 68°C for 50 s, with a final extension of 68°C for 10 min. After the second PCR, products were cleaned with magnetic beads (Sera-Mag SpeedBeads, GE Healthcare 65152105050250) and the final concentration of each sample was quantified fluorometrically with a 96-well plate reader (Qubit dsDNA HS kit, Thermo Fisher Q32854) (Rohland and Reich, 2012). The bacterial library was sequenced on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) at The University of Arizona Genomics Core (Tucson, AZ, United States) with 300 × 2 chemistry.
Illumina Sequence Data Processing
After trimming primer sequences, DADA2 was used to identify sequence variants from raw Illumina paired-end reads (Callahan et al., 2016). DADA2 uses an error model that incorporates quantitative abundances to infer the true abundances of every unique bacterial sequence; it is effectively the same as 100% OTUs but with error removed. Forward and reverse reads were quality filtered, de-replicated, merged, and bimeras were removed. The resulting de novo amplicon sequence variants (ASVs) were assigned taxonomic classifications using a native implementation of the RDP classifier (Wang et al., 2007) against version 128 of Silva taxonomic training data formatted for DADA2 (Quast et al., 2013). The R package “phyloseq” (McMurdie and Holmes, 2013) was used to organize, visualize and analyze data. Analyses of whole body individuals from Site 1 samples were rarified to 1669 reads per tissue type. For the analysis of differences in microbial community in organs systems within Site 1 reads were rarified to 1772. For analysis of microbial communities in MGMT and ileum samples from Site 2 reads were rarified to 4468. For comparison of MGMT samples between Site 1 and Site 2 reads were rarified to 3714. For comparison of ileum samples between Site 1 and Site 2 reads were rarified to 1796. All rarefaction depths employed in our analyses adequately sampled these microbial communities (Supplementary Figure S1).
Statistical Analysis
Using the R package “vegan” (Oksanen et al., 2018), non-metric multi-dimensional scaling (NMDS) scaling plot analyses of microbial communities was performed using Bray-Curtis dissimilarity. Permutational analysis of variance (PERMANOVA) was run to test whether within-group distances were significantly different from the between-group distances. PERMANOVA testing was run using the Adonis command. FDR-corrected multiple pairwise t-tests were ran in the “phyloseq” (McMurdie and Holmes, 2013) R package using the mt() command.
Spiroplasma Survey Using PCR
DNA extractions from various tissue types of thirty-one previously collected B. elongatulus specimens (see Supplementary Table S1) were screened for Spiroplasma using Spiroplasma-diagnostic primers 23f (5′-CTCAGGATGAACGCTGGCGGCAT-3′) and TKSSsp (5′-TAGCCGTGGCTTTCTGGTAA-3′) producing a 410 base pair fragment of the V1-V2 region of 16S rDNA. This primer pair was selected for its ability to amplify almost all Spiroplasma, including male-killing and non-male killing strains (Watts et al., 2009). However, these primers can amplify other types of bacteria as well (Watts et al., 2009). The thermocycler protocol was denaturation at 94°C for 2 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 90 s, with a final extension of 72°C for 5 min. PCR products were sequenced with an ABI 3730 at The University of Arizona Genomics Core (Tucson, AZ, United States).
Molecular Phylogenetic Inference of Spiroplasma
We used 16S rDNA to reconstruct the phylogeny of Spiroplasma. Five sequences from the Spiroplasma PCR survey (410 bp fragment of the V1–V2 region), four ASVs identified as Spiroplasma from the Illumina survey (427 bp fragment of the V3–V4 region), two sequences from transcriptomes of Brachinus elongatulus from Madera Canyon (available in the Moore lab) (692 bp fragment of the V1–V4 region), and 113 full length 16S Tenericutes sequences from NCBI (DiBlasi et al., 2011; Benson et al., 2012) were used to infer the evolutionary relationships of Spiroplasma species.
See Supplementary Table S2 for accession numbers. The sequences were aligned using MAFFT v 7.310 (Katoh and Standley, 2013) with default settings in Mesquite (Maddison and Maddison, 2017). The aligned matrix was trimmed to the V1–V4 region, resulting in an alignment of 719 bp. The best fitting model of nucleotide evolution was selected with PartitionFinder (Lanfear et al., 2012). The maximum likelihood phylogeny was inferred with the GTR + Gamma model of nucleotide evolution using RAxML v8.2.8 (Stamatakis, 2014) on CIPRES Science Gateway portal (Miller et al., 2010). 500 search replicates were conducted to find the maximum likelihood tree. Clade support was conducted using rapid bootstrapping with a subsequent ML search letting RAxML halt bootstrapping automatically (using MRE-based bootstopping criterion).
Results
Data Summary of the MiSeq Analysis
A total of 864,299 rDNA V3–V4 reads were obtained from 54 unique samples comprised of either whole-body individuals or a specific tissue dissected from an individual specimen (Table 1). After quality control filtering and merging of paired-end reads 424,425 sequences remained with an average read length of 422 bases. The DADA2 algorithm identified 248 unique ASVs with an average of 26 ASVs per sample. The 54 libraries ranged in size from 1,669 to 54,077 sequences.
Bacterial Taxa Associated With Brachinus elongatulus
Site 1: Whole-Body Samples
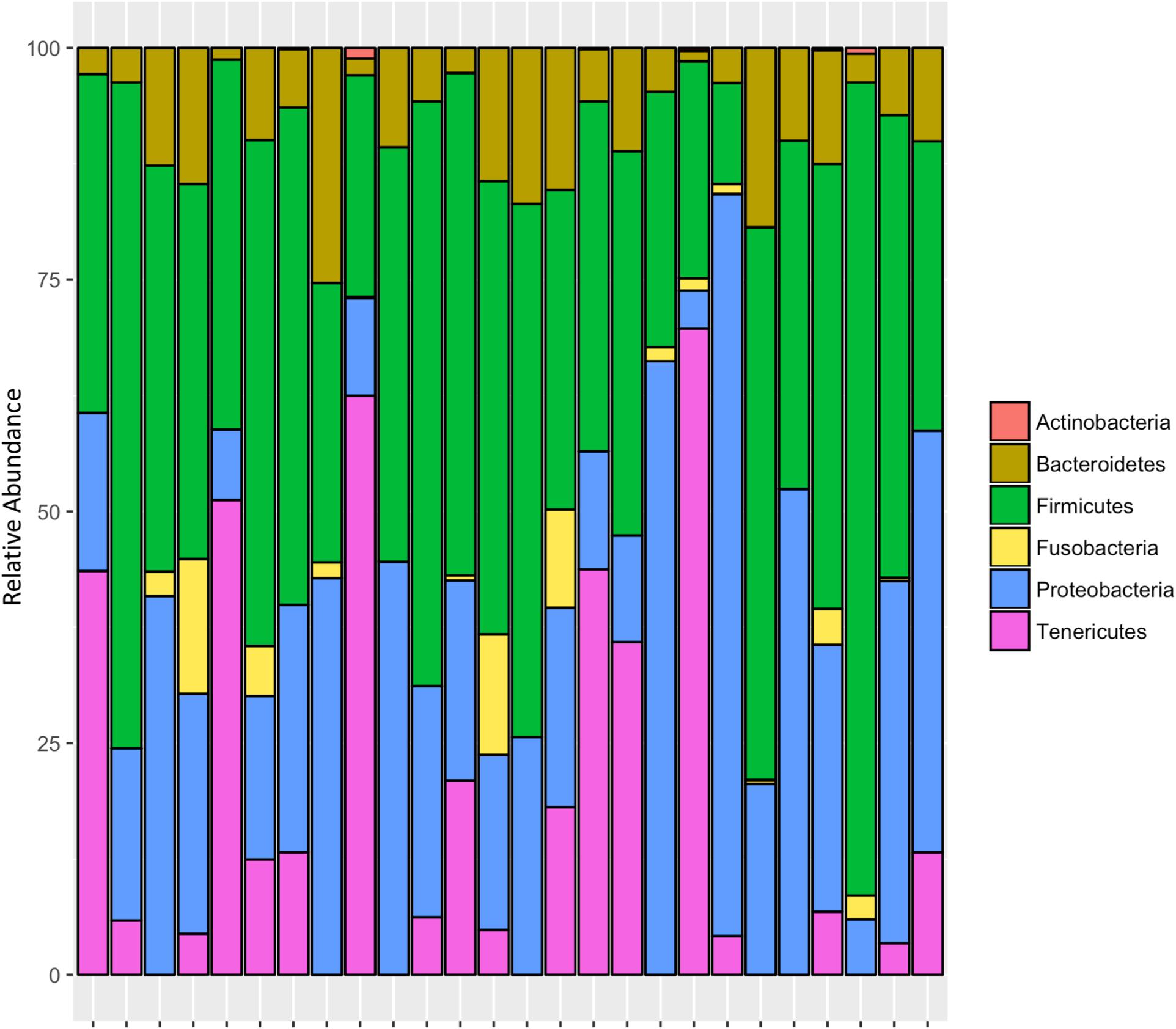
After rarifying to 1669 reads per sample, a total of 165 bacterial ASVs across six bacterial phyla were detected in the whole-body samples from Site 1 (Madera Canyon). The bacterial phyla with greater than one percent of total reads across all B. elongatulus whole-body individuals were Firmicutes (44%), Proteobacteria (28%), Tenericutes (16%), Bacteroidetes (9%) and Fusobacteria (2%) (Figure 4). The top 20 most abundant ASVs are shown in Table 2. See Supplementary Table S3 for a full list of ASVs. There were no significant differences in bacterial communities based on sex (PERMANOVA, R2 = 0.034, p = 0.58) (Supplementary Figure S2).
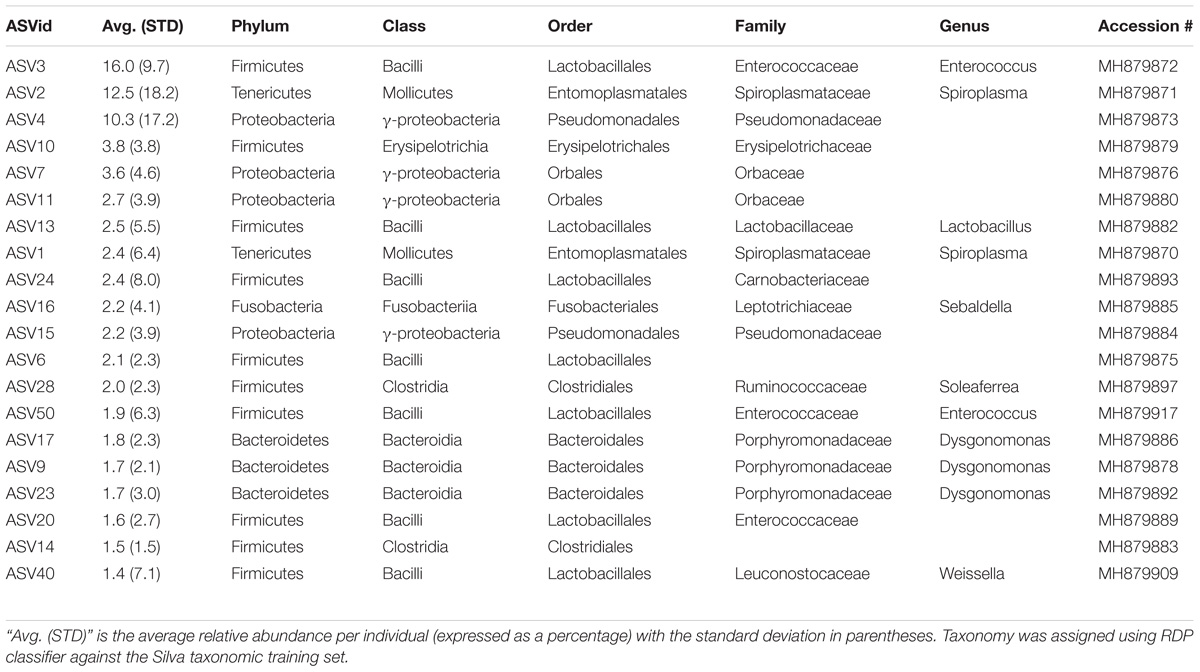
Six genera were present in at least 50% of the samples from Madera Canyon (Table 3). Enterococcus and Dysgonomonas were present in all 26 whole-body samples. There were no significant differences in Dysgonomonas variation between sexes (t-test, FDR-corrected p = 1.0, Supplementary Figure S3). There were also no significant differences in Enterococcus variation between sexes (t-test, FDR-corrected p = 1.0, Supplementary Figure S3). Spiroplasma was present in 69% of the individuals, Candidatus Soleaferrea and Sulfurospirillum were present in 65% of the samples, and Sebaldella was present in 58% of samples.
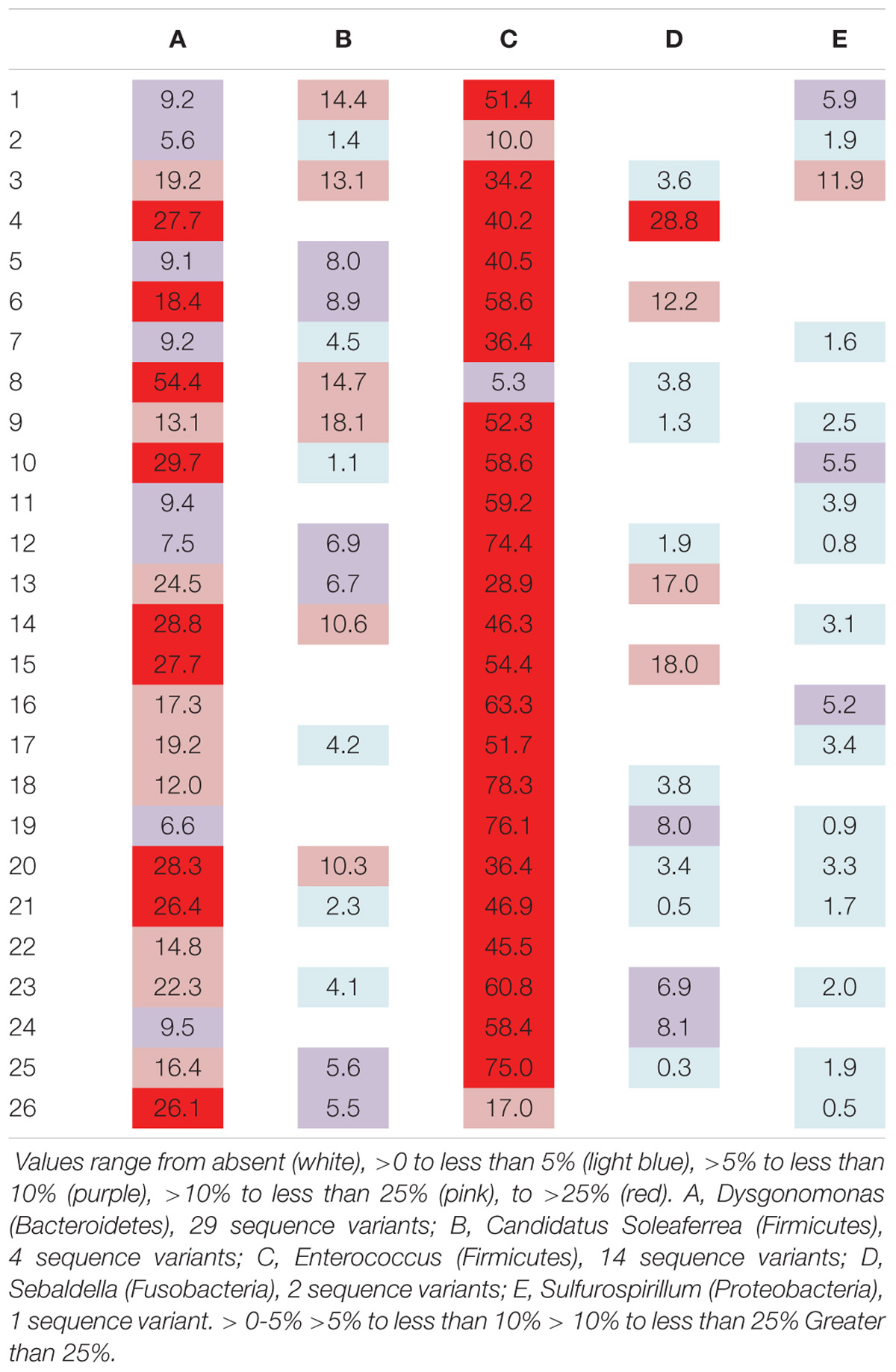
TABLE 3. Relative abundance of reads per individual for genera found in at least 50% of the individuals from Site 1 (Madera Canyon, AZ, United States).
Site 1: Gut Tissues and Secretory Cells of the Defensive System
We asked whether the bacterial community differed among organ systems sampled from Site 1. Samples were rarified to 1772 reads per tissue type. PERMANOVA testing resulted in no significant difference in overall bacterial communities between the Malpighian tubules and midgut (MGMT), ileum, and secretory cells of the defensive system (PERMANOVA, R2 = 0.13, P = 0.29; Supplementary Figure S4). FDR-corrected pairwise t-tests also revealed no significant differences in ASV abundance between tissue types. While it is possible that bacterial communities do not vary between these organ systems, it is also possible that the small sample size did not allow us to detect differences in community composition.
There was a significant difference in overall bacterial communities between MGMT and ileum (PERMANOVA, R2 = 0.15, P = 0.02, Supplementary Figure S5A). FDR-corrected multiple pairwise t-tests revealed no genera that significantly varied between the tissues. One MGMT sample (DNA4284) was comprised of 100% Spiroplasma reads; after removing this sample, there was no longer a significant difference between the two tissue types (PERMANOVA, R2 = 0.14, P = 0.06, Supplementary Figure S5B).
MGMT samples had an average of 20 ASVs per tissue type. The bacterial phyla with greater than one percent of total reads across MGMT samples at Site 1 were Firmicutes (44%), Tenericutes (26%), Proteobacteria (22%), Bacteroidetes (4%), and Fusobacteria (4%) (Figure 5A). See Supplementary Table S4 for the top 10 ASVs. The genera Enterococcus, Dysgonomonas, and Weisella were present in four of the five MGMT samples (the fifth sample was composed of 100% Spiroplasma reads).
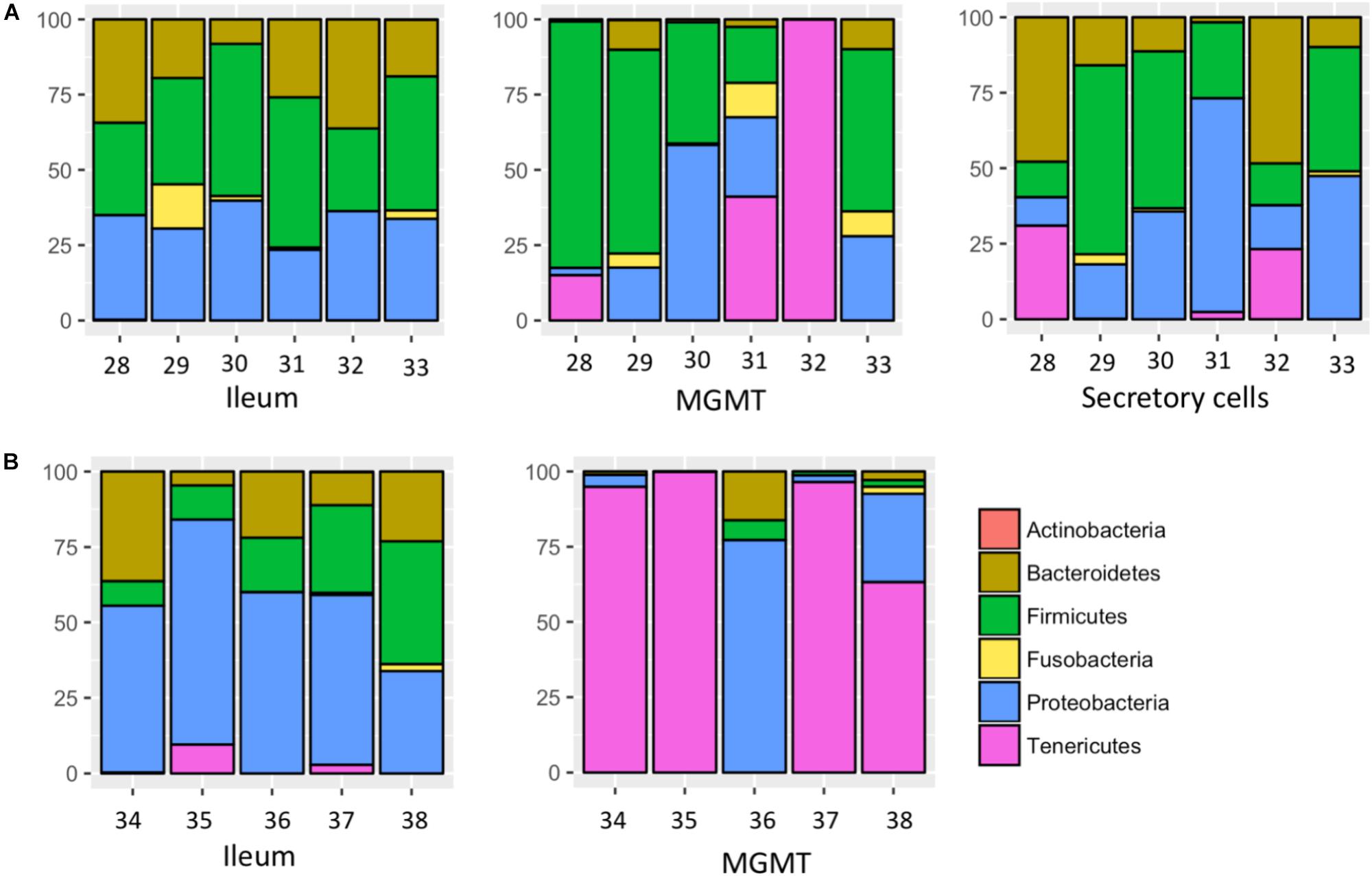
FIGURE 5. Relative abundance of bacterial phyla in individual tissues from Site 1 (A) Madera Canyon, AZ and Site 2 (B) Happy Valley, AZ. The Taxon ID number is on the x-axis.
The ileum samples had an average of 37 ASVs per tissue type. The bacterial phyla with greater than one percent of total reads across ileum samples Site 1 were Firmicutes (40%), Proteobacteria (33%), Bacteroidetes (24%) and Fusobacteria (3%) (Figure 5A). See Supplementary Table S5 for the top 10 ASVs. For the ileum samples, the genera Candidatus_Soleaferrea, Dysgonomonas, Enterococcus, including ASV3, and Qingshengfania were present in all six samples. For the secretory cell samples, the genera Dysgonomonas, including ASV9, and Enterococcus were present in all samples.
The secretory cell samples had an average of 32 ASVs per tissue type. The bacterial phyla with greater than one percent of total reads across secretory cell samples Site 1 were Firmicutes (34%), Proteobacteria (33%), Bacteroidetes (22%), Tenericutes (9%) and Fusobacteria (1%) (Figure 5A). See Supplementary Table S6 for the top 10 ASVs.
Of note, the genus Dysgonomonas was the only genus present in all 6 samples of the secretory cells of the defensive system and in all gut samples.
Site 2: Gut Tissues
We asked whether the bacterial community differed among the tissue types sampled from Site 2 (Happy Valley). Samples were rarified to 4468 reads per tissue type. PERMANOVA testing resulted in no significant difference in overall bacterial communities between the MGMT and ileum (PERMANOVA, R2 = 0.19, P = 1.09).
The five MGMT samples had an average of 10 ASVs per sample. The bacterial phyla with greater than one percent of total reads across MGMT samples Site 2 were Tenericutes (71%), Proteobacteria (23%), Bacteroidetes (4%), and Firmicutes (2%) (Figure 5B). See Supplementary Table S7 for the top 10 ASVs for each tissue type No genera were present in every sample.
The five ileum samples had an average of 25 ASVs per sample. The bacterial phyla with greater than one percent of total reads across ileum samples Site 2 were Proteobacteria (56%), Firmicutes (21%), Bacteroidetes (19%), and Tenericutes (3%), (Figure 5B). See Supplementary Table S8 for the top 10 sequence variants for each tissue type. Dysgonomonas, Enterococcus, including ASV3, and Pseudomonadaceae (ASV4) were present in all ileum samples.
Comparison of Gut Tissues Between Site 1 and Site 2
There were no significant differences in overall bacterial communities in MGMT samples between Site 1 and Site 2 (PERMANOVA, R2 = 0.13, P = 0.13; Supplementary Figure S6). There were also no significant differences in overall bacterial communities ileum samples between Site 1 and Site 2 (PERMANOVA, R2 = 0.14, P = 0.06; Supplementary Figure S6).
The genera Dysgonomonas and Enterococcus were present in all ileum samples from both locations. Spiroplasma infections were present in individuals from both sites.
Differences Between Tissue Types Are Consistent With or Without Spiroplasma
Since our data suggest that Spiroplasma may be symbionts or pathogens of Brachinus (see Discussion), we ran all analyses comparing bacterial communities between tissue types twice: once including all Spiroplasma reads (the tests reported above), and again omitting all Spiroplasma reads to see if the infection was obscuring any signal from the rest of the microbial community. All test results were qualitatively identical, with one small exception: at Site 1, MGMT and ileum differed in community composition when Spiroplasma was removed and all samples were included in the test. This difference was driven by the same outlier sample that previously harbored 100% Spiroplasma reads.
Prevalence of Spiroplasma in Brachinus elongatulus
Spiroplasma (phylum Tenericutes) made up 16.2% of the total reads across 26 whole-body individuals. Four ASVs were identified as Spiroplasma. All four ASVs were found in individuals from Madera Canyon (ASV1, ASV2, ASV5, ASV36) and three ASVs were found in gut tissues from Happy Valley (ASV1, ASV2, ASV5). In both locations Spiroplasma was found in both males and females (Figure 6B). There were no significant differences in Spiroplasma between sexes (Supplementary Figure S7). Some individuals were infected with one sequence variant, while other individuals were infected with two or three ASVs. None of the individuals were infected with all four.
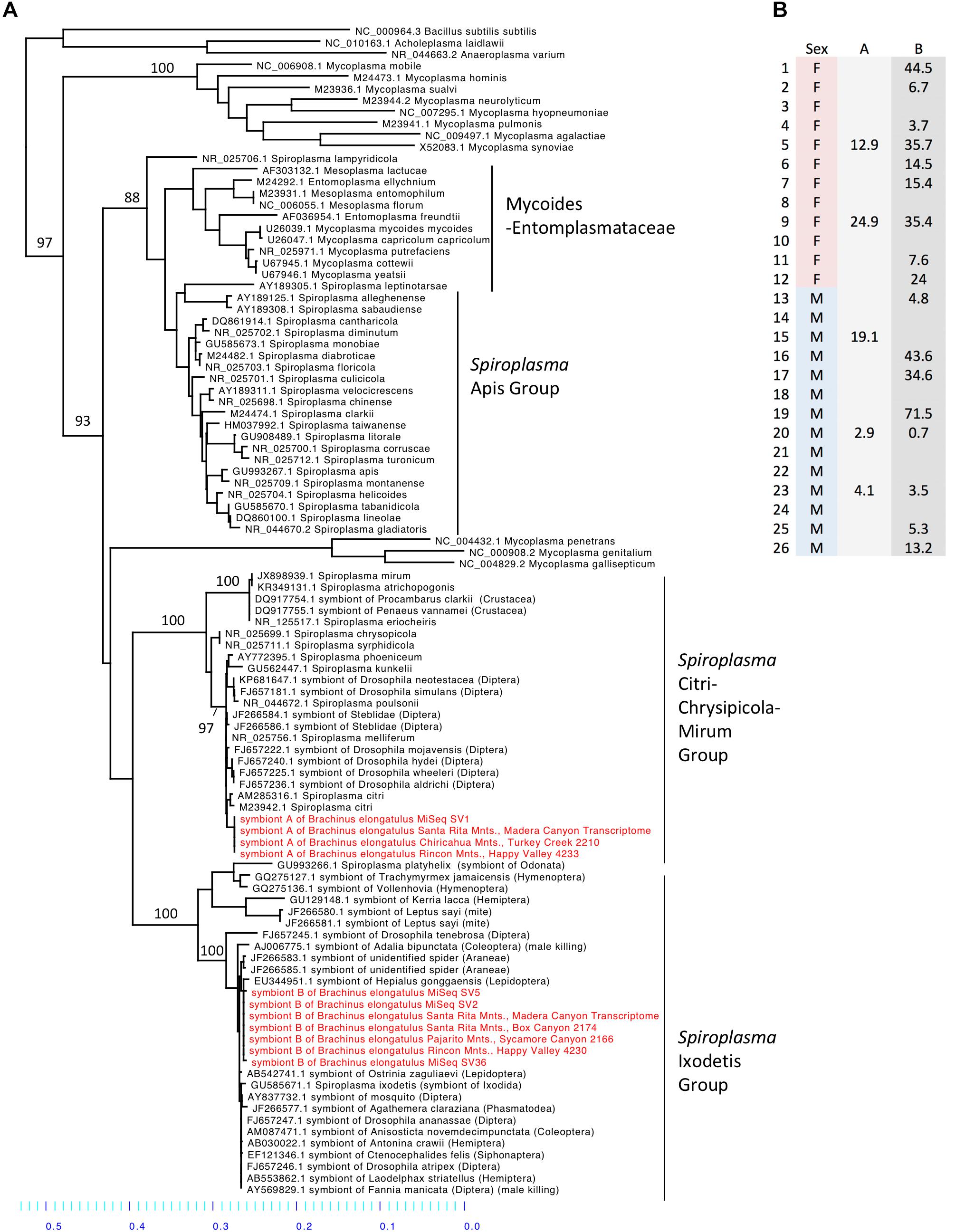
FIGURE 6. (A) Molecular phylogeny of Spiroplasma. Sequences from Brachinus (MiSeq, PCR survey and transcriptome survey) fall out in two clades (Ixodetis and CCM). (B) Relative abundance of Spiroplasma in individuals from Site 1. Column A is the relative abundance of Spiroplasma symbiont A (svl) belonging to the CCM. Column B is the relative abundance of Spiroplasma symbiont B (sv2, sv5, sv36) belonging to the Ixodetis clade.
Of the 31 B. elongatulus surveyed with Spiroplasma specific primers, 29 samples yielded bands and were sent for Sanger sequencing. Twelve samples produced clean sequences. Of those, nine matched Spiroplasma and three matched Streptococcus or Pseudomonas in BLAST searches. Sequences from 17 samples were too noisy to identify, which could suggest that more than one type of bacteria was present (Spiroplasma or otherwise). The PCR survey detected the presence of Spiroplasma in B. elongatulus at an additional three localities (Chiricahua Mountains, Turkey Creek, AZ, Santa Rita Mountains, Box Canyon, AZ, and Parajito Mountains, Sycamore Canyon, AZ) (PCR Table)
Molecular Phylogenetic Inference of Spiroplasma
Sequences from all of our surveys of Brachinus fell out into two Spiroplasma clades: the Ixodetis clade, and the Citri-Chrysipicola-Mirum (CCM) clade (Figure 6A and Supplementary Figure S8). The phylogeny indicates there are at least two species of Spiroplasma in the Brachinus elongatulus populations in Southern Arizona.
Discussion
Our research on the microbiome of B. elongatulus reveals a bacterial community similar to other insects’ microbiota in phylum composition and diversity. In particular, the microbiota of B. elongatulus is broadly similar to the microbiota of previously studied carabid beetles. However, our data also show that (1) Enterococcus is present in all individuals from two locations, (2) the acid producing Dysgonomonas genus is present in all individuals from both locations, and (3) Spiroplasma, a common facultative endosymbiont, is present in the majority of individuals from both locations.
Comparison to Other Insects’ Microbiomes
Among the most abundant phyla present in B. elongatulus the Proteobacteria, Firmicutes and Bacteroidetes are also the predominant bacterial phyla detected in gut samples and whole-body samples across several families of Coleoptera (Jones et al., 2013; Yun et al., 2014). In addition, Fusobacteria have been reported in the digestive tract of the predaceous carabid, P. chalcites (Lehman et al., 2009). This phylum of bacteria has also been found to be a common gut symbiont of carnivorous birds (Roggenbuck et al., 2014) and is a common hindgut symbiont of cockroaches (Schauer et al., 2012). Fusobacteria have also been found in low abundance in the midgut of several species of mosquitos (Muturi et al., 2017) and in the guts of the dipteran Bactrocera minax (Wang et al., 2014), although the functions of these gut bacteria are not known.
Of the six genera present in 50% or more of B. elongatulus surveyed, five genera have known associations with insects and, in particular, four of the five genera have associations with fungus-growing termites. A species of Sebaldella, S. termitidis, has been isolated and characterized from the gut of a termite (Harmon-Smith et al., 2010). The closest BLAST hit for the Sulfurospirillum sequence variant is to an uncultured epsilon-proteobacterium isolated from a gut homogenate of fungus-growing termite, Macrotermes gilvus. The closest BLAST hit for all four Candidatus Soleaferrea sequence variants is to an uncultured bacterium isolated from the hindgut of cranefly larvae (Tipula abdominalis). Only a few species of insects, none of which are carabids, have well characterized microbiomes, therefore, these genera may be more common than we know and/or be more closely related to symbionts of beetles or other non-termite insects that have yet to be surveyed.
Some insects host only a few bacterial species in their guts; for example, fewer than 10 species comprise the core microbiomes of honeybees, reed beetles and fruit flies (Engel and Moran, 2013). Other insects, such as the cockroach and termite, regularly host over 1000 OTUs (operational taxonomic units; roughly equivalent to species) in their gut (Tinker and Ottesen, 2016). B. elongatulus had an overall average of 15 ASVs in MGMT samples and an overall average of 31 ASVs in ileum samples. B. elongatulus hosts a low bacterial species diversity roughly comparable to that of honeybees and fruit flies. The microbial gut associates of Brachinus follow the trend of other insects in that the hindgut hosts more diversity than the midgut. The midgut is often a more hostile environment for microbes to reside due to the suite of digestive and immunological enzymes produced by the midgut epithelium (Douglas, 2015). The ileum environment is often more suitable for microbes, which may benefit from the ion and metabolite rich filtrate delivered into the hindgut by the Malpighian tubules.
Previous studies (Lundgren et al., 2007; Lehman et al., 2009) have examined the microbial communities present in digestive tracts of three species of carabid beetles with different diets: two granivores- Harpalus pensylvanucus and Ansiodactylus sanctaecrucis and one predator, Poecilus chalcites. All three species, like B. elongatulus, had an abundance of Proteobacteria. The community of B. elongatulus described here and the other predator, P. chalcites had the most similar bacterial community compositions with the most abundant OTUs being represented by Firmicutes, Fusobacteria, Proteobacteria and Bacteroidetes. The genera Enterococcus and Weissella were also identified in P. chalcites. It is impossible to more comprehensively compare the microbiomes of these other carabids to that of B. elongatulus due to the differences in experimental approach.
The Genus Enterococcus Was Present in All Individuals
Enterococcus and six other genera of the lactic-acid bacterial order Lactobacillales were present in B. elongatulus. Interestingly, a single ASV of Enterococcus (ASV3) was present in every ileum sample from both sites and in 88% of whole body samples from Site 1. Enterococcus is a common inhabitant of the digestive tract of mammals, birds, reptiles, and insects. It has been found across a broad range of insect orders with the most prevalent species studied being E. faecalis, E. faecium, E. casseliflavus (Martin and Mundt, 1972) and most recently E. termitis, which was isolated from the gut of a termite (Svec et al., 2006). E. faecalis has been shown to support digestion in the carabid, Harpalus pensylvanicus (Schmid et al., 2014). E. casseliflavus has been implicated in assisting Lepidopteran larvae metabolize toxic compounds ingested from plants, for example by forming a monospecific biofilm in the foregut of Hyles euphorbiae (Vilanova et al., 2016).
Presence of a single ASV in most individuals from two localities suggests the potential for a specific symbiotic association between Brachinus and this Enterococcus species. Our data cannot confirm this, nor can they indicate the potential function of such a relationship. However, some species of these bacteria have been implicated in the production of volatile carboxylic acids (VCAs) that act as fecal aggregation pheromone components, which regulate aggregation in the German cockroach (Wada-Katsumata et al., 2015). Cockroach feces containing isolates of Enterococcus avium were found to have a significant effect on German cockroach aggregation compared to axenic cockroach feces. Isolates of Enterobacter cloacae were found to cause an aggregation response in the firebrat, Thermobia domestica (Woodbury and Gries, 2013). Brachinus are notable for their multispecies aggregations and it remains unknown what triggers them. Future research should investigate whether volatiles produced by Enterococcus induce aggregation, not only in B. elongatulus, but also in other Brachinus species.
The Genus Dysgonomonas Was Present in All Individuals
The genus Dysgonomonas was present in all individuals surveyed although, unlike Enterococcus, there was not a single common ASV. Indeed, Dysgonomonas accounted for the most sequence variants assigned to a single genus (28 ASVs). Dysgonomonas has been found in the digestive tracts of several insects with varying diets, such as fungus-growing termites (Macrotermes annadalei), subterranean termites Reticulitermes speratus, and several Drosophila populations (Chandler et al., 2011; Yang et al., 2014; Pramono et al., 2015). Within Coleoptera, Dysgonomonas has been found to be highly abundant in two species of dung beetle (Scarabaeidae) with different dietary lifestyles- detritivore, Pachysoma endroedyi, and dry-dung-feeding, P. striatum (Franzini et al., 2016). Dysgonomonas has also been found in the digestive tracts of red palm weevil larvae, Rhynchophorus ferrugineus (Dryophthoridae) (Muhammad et al., 2017). Two species of Dysgonomonas from the guts of the termites have been characterized and both species ferment glucose and xylan as a sole carbon source to produce acetic acid as the major end product (Yang et al., 2014; Pramono et al., 2015). Other species of Dysgonomonas have been found to produce lactic acid, propionic acid, succinic acid, and/or acetic acid as the major end product of glucose fermentation (Sakamoto, 2014).
Dysgonomonas was found in all of the Brachinus digestive tract samples, but more interestingly it was the only genus present in all six defensive secretory cell samples. Additional studies are needed to determine if these microbes play a role in these beetles’ unique chemical defense. However, we noted that Brachinus has a unique anatomical association between the defensive and digestive systems, that has apparently not been reported from other carabid beetles. The secretory cells of the defensive system are always intertwined with the Malpighian tubules of the digestive system (Figure 2).
It seems that proximity to the digestive and defensive systems is important. In fact, during dissections of individuals with late-stage nematomorphan infections, we have observed the secretory cells adhered to the ileum. In these cases, the Malpighian tubules were no longer present, likely having been consumed by the parasite. The secretory cells of the defensive system, which are thought to produce precursors of quinones used in bombardier beetle defense, have a high density of mitochondria and are very metabolically active (Di Giulio et al., 2015). It could be that the association with the digestive system is important solely for detoxification. It is possible that the association is also important for secretory cells to obtain nutrients and/or building blocks for quinone synthesis. The close association between the defensive system and the Malpighian tubules is most likely due to the need for detoxification. Our data cannot confirm the involvement of microbes in these processes, more studies are needed to determine if microbes play a role.
Spiroplasma Was Present in the Majority of Individuals
Spiroplasma accounted for 16.2% of the microbiota of B. elongatulus, and the genus was present in 70% of individuals surveyed. In B. elongatulus, we hypothesize that at least one species of Spiroplasma is an intracellular symbiont and/or present in the hemolymph because the bacterium was successfully amplified from the muscle tissue in an individual leg. Spiroplasma are estimated to infect at least 7% of all insects (Duron et al., 2008). Spiroplasma have been found to have commensal, mutualistic and/or pathogenic relationships with their plant and arthropod hosts (e.g., crustaceans, arachnids, insects). Insects most commonly infected with Spiroplasma include species of Odonata, Hemiptera, Hymenoptera, Diptera, Lepidoptera and Coleoptera (Cisak et al., 2015). In Coleoptera, three species of Spiroplasma have been characterized from members of Scarabaeidae, Lampyridae, and Cantharidae (Whitcomb et al., 1993; Hackett et al., 1996). Spiroplasma has also been found in Tenebrionidae and Coccinellidae (Tinsley and Majerus, 2006; Wang and Zhang, 2015). Different species of Spiroplasma have been shown (1) to be pathogens of honeybees (Schwarz et al., 2014), (2) to cause reproductive manipulation in planthoppers and Drosophila (Haselkorn, 2010; Sanada-Morimura et al., 2013), (3) to provide host defense against nematode-induced sterilization in Drosophila neotestacea (Jaenike et al., 2010) and parasitoid wasps (Xie et al., 2014), and (4) there are many Spiroplasma-host interactions in a wide variety of insects, crustaceans, and arachnids that are not well characterized (Cisak et al., 2015). Future research is needed to determine whether Spiroplasma are mutualists, pathogens, or commensals in B. elongatulus.
Spiroplasma sequences from the MiSeq, PCR and transcriptome surveys fell out in two separate clades (Ixodetis and CCM), therefore it appears at least two species are present. It was not possible to derive the function of these Spiroplasma species based on their positions in the phylogeny. The CCM clade contains plant and crustacean pathogens, male-killing species of dipteran hosts, as well as a recently identified species that encodes a ribosome-inactivating protein (RIP) that has defensive effects against nematode infection in Drosophila neotestacea (Hamilton et al., 2016). The Ixodetis clade houses male-killing species of coleopteran, dipteran and lepidopteran hosts, defensive species that protect Drosophila against parasitoid wasps (Xie et al., 2014) and aphids against pathogenic fungi (Lukasik et al., 2013), and this clade also house a suite of commensal species and species with unknown function (DiBlasi et al., 2011).
We found Brachinus that were doubly (N = 7) infected with Spiroplasma ASVs. Co-infection by two or more species of Spiroplasma in arthropods hosts has also been reported in Hymenoptera. Two pathogenic species of Spiroplasma belonging to the Apis clade, S. apis and S. melliferum, have been found to co-infect honeybee colonies in North and South America (Schwarz et al., 2014). Due to the presence of Spiroplasma in male and female B. elongatulus, we hypothesize our Spiroplasma species do not cause male killing (Figure 6B), parthenogenesis or feminization. Because of the difficulty in rearing these beetles in captivity and performing controlled matings with newly enclosed adults (the larvae are parasitoids of diving beetles), we are unlikely to be able to rule out cytoplasmic incompatibility (CI), although CI has not yet been associated with Spiroplasma. A more likely possibility is a defensive role. Brachinus elongatulus live in riparian areas and are commonly found infected with nematomorpha (McManus and Moore, personal observation). Further investigation is needed to determine if one or both of the species of Spiroplasma confer defense against helminth parasite infection.
Conclusion
This study paves the way for further investigations into the roles of microorganisms in the behavior, immune function, and physiology of B. elongatulus. In particular, our results suggest that a single sequence variant of Enterococcus, the genus Dysgonomonas, and the genus Spiroplasma may have specialized relationships with Brachinus that merit further investigation. To resolve the number of Spiroplasma infecting B. elongatulus and to derive their function, genome sequencing from both cultivable and uncultivable associates would provide important insight. Fluorescence in situ hybridization (FISH) could provide insight into localizing and confirming the presence of Dysgonomonas. Microbial cultures of Brachinus feces, treatment of beetles with antibiotics and follow-up behavioral studies could also provide insight into the potential roles bacteria may be playing in Brachinus aggregation behavior.
Data Accessibility
Sequences and associated metadata are archived under NCBI BioProject PRJNA489455.
Author Contributions
RM designed the experiments, performed the fieldwork, processed the samples, analyzed the data, and prepared the manuscript. AR processed the samples, analyzed the data, and prepared the manuscript. WM designed the experiments, processed the samples, analyzed the data, and prepared the manuscript.
Funding
This work was supported by University of the Arizona, Office for Research and Discovery, Faculty Seed Grants Program, Microbiome Discovery and its Influence on Beetle-Ant Symbiosis (to WM and RM). Supported by National Science Foundation Award 1556813 to WM; and U.S. National Institutes of Health PERT Training Grant to the Center for Insect Science at the University of Arizona (K12GM000708) to AR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank J. Schaller and A. Yanahan for assistance in collecting beetles, A. Gill for providing primers and guidance for a preliminary study and M. Hunter and B. Hurwitz for discussion in developing the study and insightful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02361/full#supplementary-material
References
Aneshansley, D. J., Jones, T. H., Alsop, D., and Meinwald, J., and Eisner, T. (1983). Thermal concomitants and biochemistry of the explosive discharge mechanism of some little known bombardier beetles. Experientia 39, 366–368. doi: 10.1007/BF01963125
Arndt, E. M., Moore, W., Wah-Keat, L., and Ortiz, C. (2015). Mechanistic origins of bombardier beetle (Brachinini) explosion-induced defensive spray. Science 348, 563–567. doi: 10.1126/science.1261166
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A., and Wheeler, D. L. (2012). GenBank. Nucleic Acids Res. 40, D48–D53. doi: 10.1093/nar/gkr1202
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/NMETH.3869
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7:e1002272. doi: 10.1371/journal.pgen.1002272
Cisak, E., Wójcik-Fatla, A., Zając, V., Sawczyn, A., Sroka, J., and Dutkiewicz, J. (2015). Spiroplasma – an emerging arthropod-borne pathogen? Ann. Agric. Environ. Med. 22, 589–593. doi: 10.5604/12321966.1185758
Di Giulio, A., Muzzi, M., and Romani, R. (2015). Functional anatomy of the explosive defensive system of bombardier beetles (Coleoptera, Carabidae, Brachininae). Arthropod. Struct. Dev. 44, 468–490. doi: 10.1016/j.asd.2015.08.013
DiBlasi, E., Morse, S., Mayberry, J. R., Avila, L. J., Morando, M., and Dittmar, K. (2011). New Spiroplasma in parasitic Leptus mites and their Agathemera walking stick hosts from Argentina. J. Invertebr. Pathol. 107, 225–228. doi: 10.1016/j.jip.2011.05.013
Dillion, R. J., Vennard, C. T., and Charnley, A. K. (2000). Exploitation of gut bacteria in the locust. Nature 403:851. doi: 10.1038/35002669
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Duron, O., Didier, B., Sébastien, B., Bellamy, L., Zhou, L., and Engelstädter, J., (2008). The diversity of reproductive parasites among arthropods: wolbachia do not walk alone. BMC Biol. 6:27. doi: 10.1186/1741-7007-6-27
Eisner, T., Kafatos, F. C., and Linsley, E. G. (1962). Lycid predation by mimetic adult Cerambycidae (Coleoptera). Evolution 16, 316–324. doi: 10.1111/j.1558-5646.1962.tb03223.x
Engel, P., and Moran, N. (2013). The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Erwin, T. L. (1967). Bombardier beetles (Coleoptera, Carabidae) of North America: part II. biology and behavior of Brachinus pallidus in California. Coleopt. Bull. 21, 41–55.
Franzini, P. Z. N., Ramond, J. B., Scholtz, C. H., Sole, C. L., Ronca, S., and Cowan, D. A. (2016). The gut microbiomes of two Pachysoma MacLeay desert dung beetle species (Coleoptera: Scarabaeidae: Scarabaeinae) feeding on different diets. PLoS One 11:8. doi: 10.1371/journal.pone.0161118
Gilbert, J. A., Jansson, J. K., and Knight, R. (2014). The earth microbiome project: successes and aspirations. BMC Biol. 12:69. doi: 10.1186/s12915-014-0069-1
Hackett, K. J., Whitcomb, R. F., French, F. E., Tully, J. G., Gasparich, G. E., Rose, D. L., et al. (1996). Spiroplasma corruscae sp. nov., from a firefly beetle (Coleoptera: Lampyridae) and tabanid flies (Diptera: Tabanidae). Int. J. Syst. Bacteriol. 46, 947–950. doi: 10.1099/00207713-46-4-947
Hamady, M., Walker, J. J., Harris, J. K., Gold, N. J., and Knight, R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:3. doi: 10.1038/nmeth.1184
Hamilton, P. T., Peng, F., Boulanger, M. J., and Perlman, S. J. (2016). A ribosome-inactivating protein in a Drosophilia defensive symbiont. Proc. Natl. Acad. Sci. U.S.A. 113, 350–355. doi: 10.1073/pnas.1518648113
Harmon-Smith, M., Celia, L., Chertkov, O., Lapidus, A., Copeland, A., Glavina Del Rio, G., et al. (2010). Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300T). Stand. Genomic Sci. 2, 220–227. doi: 10.4056/sigs.811799
Haselkorn, T. (2010). The Spiroplasma heritable bacterial endosymbiont of Drosophila. Fly 4, 80–87. doi: 10.4161/fly.4.1.10883
Heinrich, B., and Vogt, F. D. (1980). Aggregation and foraging behavior of whirligig beetles (Gyrinidae). Behav. Ecol. Sociobiol. 7, 179–186. doi: 10.1007/BF00299362
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of Drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Jones, R. T., Sanchez, L. G., and Fierer, N. (2013). A cross-taxon analysis of insect-associated bacterial diversity. PLoS One 8:4. doi: 10.1371/journal.pone.0061218
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvement in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lehman, R. M., Lundgren, J. G., and Petzke, L. M. (2009). Bacterial communities associated with the digestive tract of the predatory ground beetle, Poecilus chalcites, and their modification by laboratory rearing and antibiotic treatment. Microb. Ecol. 57, 349–358. doi: 10.1007/s00248-008-9415-6
Lukasik, P., Guo, H., van Asch, M., Ferrari, J., and Godfray, H. C. (2013). Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J. Evol. Biol. 26, 2654–2661. doi: 10.1111/jeb.12260
Lundgren, J. G., Lehman, R. M., and Chee-Sanford, J. (2007). Bacterial communities within digestive tracts of ground beetles (Coleoptera: Carabidae). Ann. Entomol. Soc. Am. 100, 275–282. doi: 10.1007/s00248-008-9415-6
Machado, G., and Vasconcelos, C. H. F. (1998). Multi-species aggregations in neotropical harvestmen (Opiliones, Gonyleptidae). J. Arachn. 26, 389–391.
Maddison, W. P., and Maddison, D. R. (2017). Mesquite: A Modular System for Evolutionary Analysis, Version 3.2. Available at: http://mesquiteproject.org
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:4. doi: 10.1371/journal.pone.0061217
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8. doi: 10.1109/GCE.2010.5676129
Minkley, N., Fujita, A., Brune, A., and Kirchner, W. H. (2006). Nest specificity of bacterial community in termite guts (Hodotermes mossambicus). Insect Soc. 53, 339–344. doi: 10.1007/s00040-006-0878-5
Muhammad, A., Fang, Y., Hou, Y., and Shi, Z. (2017). The gut entomotype of red palm weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae) and their effect on host nutrition metabolism. Front. Microbiol. 8:2291. doi: 10.3389/fmicb.2017.02291
Muturi, E. J., Ramirez, J. L., Rooney, A. P., and Kim, C. H. (2017). Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 11:2. doi: 10.1371/journal.pntd.0005377
Oksanen, J., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). vegan: Community Ecology Package. R package version 2.4-6. Available at: https://CRAN.R-project.org/package=vegan
Pramono, A. K., Sakamoto, M., Iino, T., Hongoh, Y., and Ohkuma, M. (2015). Dysgonomonas termitidis sp. nov., isolated from the gut of the subterranean termite Reticulitermes speratus. Int. J. Syst. Evol. Microbiol. 65, 681–685. doi: 10.1099/ijs.0.070391-0
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Roggenbuck, M., Bœrholm Schnell, I., Blom, N., Bœlum, J., Frost Bertelsen, M., Sicheritz-Pontén, T., et al. (2014). The microbiome of New World vultures. Nat. Commun. 5:5498. doi: 10.1038/ncomms6498
Rohland, N., and Reich, D. (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946. doi: 10.1101/gr.128124.111
Sakamoto, M. (2014). “The family porphyromonadaceae,” in The Prokaryotes- Other Major Linesges of Bacteria and Archaea, eds E. Rosenberg et al. (Berlin: Springer), 811–822. doi: 10.1007/978-3-642-38954-2_132
Sanada-Morimura, S., Matsumura, M., and Noda, H. (2013). Male-killing caused by a Spiroplasma symbiont in the small brown Planthopper, Laodelphax striarellus. J. Hered. 104, 821–829. doi: 10.1093/jhered/est052
Saska, P., and Honek, A. (2004). Development of the beetle parasitoids, Brachinus explodens and B. crepitans (Coleoptera:Carabidae). J. Zool. Lond. 262, 29–36. doi: 10.1017/S0952836903004412
Schaller, J. C., Davidowitz, G., Papaj, D., Smith, R. L., and Moore, W. (in press). Molecular phylogeny, ecology and multispecies aggregation behaviour of bombardier beetles in Arizona. PLoS One
Schauer, C., Thompson, C. L., and Brune, A. (2012). The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl. Environ. Microbiol. 78, 2758–2767. doi: 10.1128/AEM.07788-11
Schildknecht, H., and Holoubek, K. (1961). Die Bombardierkäfers. Agnew. Chem. 69:62. doi: 10.1002/ange.19570690115
Schmid, R. B., Lehman, R. M., Brözel, V. S., and Lundgren, J. G. (2014). An indigenous gut bacterium, Enterococcus faecalis (Lactobacillales, Enterococcaceae), increases seed consumption by Harpalus pensylvanicus (Coleoptera: Carabidae). FLA Entomol. 2, 575–584. doi: 10.1653/024.097.0232
Schwarz, R. S., Teixeira, E. W., Tauber, J. P., Birke, J. M., Martins, M. F., Fonseca, I., et al. (2014). Honey bee colonies act as reservoirs for two Spiroplasma facultative symbionts and incur complex, multiyear infection dynamics. MicrobiologyOpen 3, 341–355. doi: 10.1002/mbo3.172
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Svec, P., Vancanneyt, M., Sedlácek, I., Naser, S. M., Snauwaert, C., Lefebvre, K., et al. (2006). Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. Int. J. Syst. Evol. Microbiol. 56, 577–581. doi: 10.1099/ijs.0.63937-0
Tinker, K. A., and Ottesen, E. A. (2016). The core gut microbiome of the American cockraoch, Periplaneta americana, is stable and resilent to dietary shifts. Appl. Environ. Microbiol. 82, 6603–6610. doi: 10.1128/AEM.01837-16
Tinsley, M. C., and Majerus, M. E. (2006). A new male-killing parasitism: Spiroplasma bacteria infect ladybird beetle Anisosticta novemdecimpunctata (Coleoptera: Coccinellidae). Parasitology 132, 757–765. doi: 10.1017/S0031182005009789
Vilanova, C., Baixeras, J., Latorre, A., and Porcar, M. (2016). The generalist inside the specialist: gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. Microbiol. 7:1005. doi: 10.3389/ficb.2016.01005
Wada-Katsumata, A., Zurek, L., Nalyanya, G., Roelofs, W. L., Zhang, A., and Schal, C. (2015). Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. U.S.A. 112, 15678–15683. doi: 10.1073/pnas.1504031112
Wang, A., Yao, Z., Zheng, W., and Zhang, H. (2014). Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing. PLoS One 9:e106988. doi: 10.1371/journal.pone.0106988
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Y., and Zhang, Y. (2015). Investigation of gut-associated bacteria in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae using culture-dependent and DGGE methods. Ann. Entomol. Soc. Am. 108, 941–949. doi: 10.1093/aesa/sav079
Watts, T., Haselkorn, T. S., Moran, N. A., and Markow, T. A. (2009). Variable incidence of Spiroplasma infections in natural populations of Drosophila species. PLoS One 4:e5703. doi: 10.1371/journal.pone.0005703
Wautier, V. (1971). Un phénomène social chez les Coléoptères : le grégarisme des Brachinus (Caraboïdea Brachinidae). Insectes Soc. 18, 1–84.
Whitcomb, R. F., Chastel, C., Abalain-Colloc, M., Stevens, C., Tully, J. G., Rose, D. L., et al. (1993). Spiroplasma cantharicola sp. nov., from cantharid beetles (Coleoptera: Cantharidae). Int. J. Syst. Bacteriol. 43, 421–424. doi: 10.1099/00207713-43-3-421
Woodbury, N., and Gries, G. (2013). Firebrats, Thermobia domestica, aggregate in response to the microbes Enterobacter cloacae and Mycotypha microspora. Entomol. Exp. Appl. 147, 154–159. doi: 10.1111/eea.12054
Xie, J., Butler, S., Sanchez, G., and Mateos, M. (2014). Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity 112, 399–408. doi: 10.1038/hdy.2013.118
Yang, Y. J., Zhang, N., Ji, S. Q., Lan, X., Shen, Y. L., Li, F. L., et al. (2014). Dysgonomonas macrotermitis sp. nov., isolated from the hindgut of a fungus-growing termite. Int. J. Syst. Evol. Microbiol. 64, 2956–2961. doi: 10.1099/ijs.0.061739-0
Keywords: 16s rDNA, Spiroplasma, bombardier beetle, Brachinus elongatulus, microbiome, Nematomorpha
Citation: McManus R, Ravenscraft A and Moore W (2018) Bacterial Associates of a Gregarious Riparian Beetle With Explosive Defensive Chemistry. Front. Microbiol. 9:2361. doi: 10.3389/fmicb.2018.02361
Received: 28 June 2018; Accepted: 14 September 2018;
Published: 05 October 2018.
Edited by:
Joerg Graf, University of Connecticut, United StatesReviewed by:
Panagiotis Sapountzis, University of Copenhagen, DenmarkSandra Breum Andersen, University of Oxford, United Kingdom
Copyright © 2018 McManus, Ravenscraft and Moore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reilly McManus, cm1jbWFudXMxQGVtYWlsLmFyaXpvbmEuZWR1
 Reilly McManus
Reilly McManus Alison Ravenscraft
Alison Ravenscraft Wendy Moore
Wendy Moore