
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Microbiol. , 09 October 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02341
This article is part of the Research Topic Bacterial Cell Wall Structure and Dynamics View all 15 articles
 Antony T. Vincent1,2
Antony T. Vincent1,2 Sammy Nyongesa1
Sammy Nyongesa1 Isabelle Morneau3
Isabelle Morneau3 Michael B. Reed2,4,5
Michael B. Reed2,4,5 Elitza I. Tocheva3
Elitza I. Tocheva3 Frederic J. Veyrier1,2*
Frederic J. Veyrier1,2*Mycobacteria are well known for their taxonomic diversity, their impact on global health, and for their atypical cell wall and envelope. In addition to a cytoplasmic membrane and a peptidoglycan layer, the cell envelope of members of the order Corynebacteriales, which include Mycobacterium tuberculosis, also have an arabinogalactan layer connecting the peptidoglycan to an outer membrane, the so-called “mycomembrane.” This unusual cell envelope composition of mycobacteria is of prime importance for several physiological processes such as protection from external stresses and for virulence. Although there have been recent breakthroughs in the elucidation of the composition and organization of this cell envelope, its evolutionary origin remains a mystery. In this perspectives article, the characteristics of the cell envelope of mycobacteria with respect to other actinobacteria will be dissected through a molecular evolution framework in order to provide a panoramic view of the evolutionary pathways that appear to be at the origin of this unique cell envelope. In combination with a robust molecular phylogeny, we have assembled a gene matrix based on the presence or absence of key determinants of cell envelope biogenesis in the Actinobacteria phylum. We present several evolutionary scenarios regarding the origin of the mycomembrane. In light of the data presented here, we also propose a novel alternative hypothesis whereby the stepwise acquisition of core enzymatic functions may have allowed the sequential remodeling of the external cell membrane during the evolution of Actinobacteria and has led to the unique mycomembrane of slow-growing mycobacteria as we know it today.
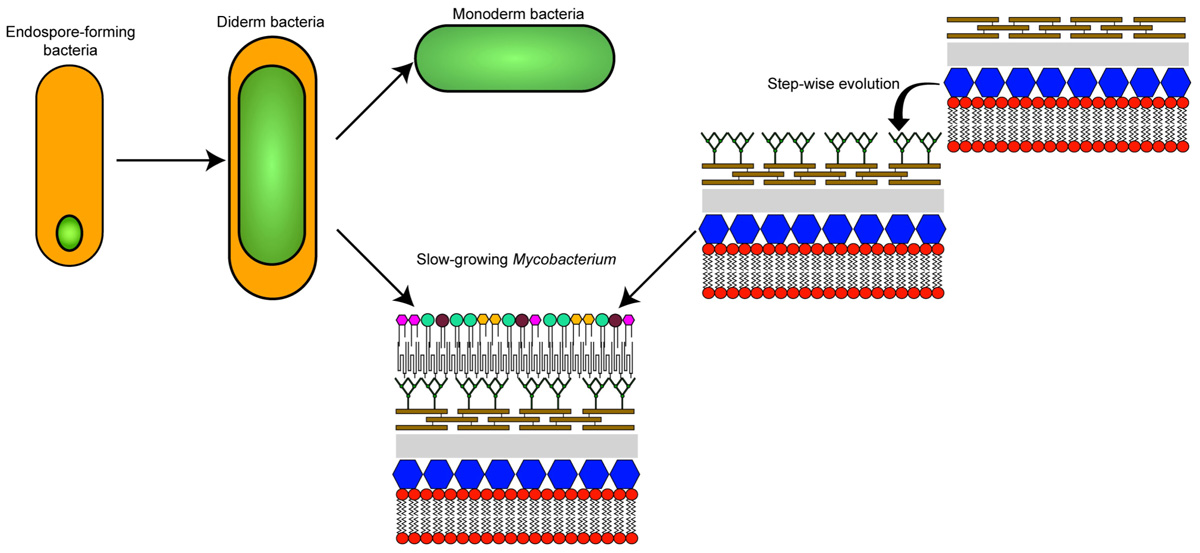
GRAPHICAL ABSTRACT The different theories on the evolutionary origin of the mycobacterial cell envelope.
The Actinobacteria phylum of Gram-positive bacteria forms an extremely diverse group that includes several species that have evolved specific symbioses (commensal or parasitic) with a wide range of hosts including numerous mammals. For example, certain species from the genera Mycobacterium and Nocardia are pathogenic while others, belonging to the genus Bifidobacterium, are part of the normal gut microbial flora and are known to have a beneficial and important effect on human health (Barka et al., 2016; O’Callaghan and van Sinderen, 2016). Several Actinobacteria are also involved in the production of antibiotic compounds (e.g., Streptomyces sp.), amino acids (e.g., Corynebacterium sp.), biofuels, and other bioproducts (Becker and Wittmann, 2016; Lewin et al., 2016).
One of the most studied bacteria from the Actinobacteria phylum is Mycobacterium tuberculosis, the etiological agent of tuberculosis, a disease that causes significant morbidity and mortality. It is a leading cause of death worldwide making its control a top priority for the World Health Organization (Pai et al., 2016; Friedrich, 2017). Several bacteria from the Corynebacteriales order, that includes M. tuberculosis, are studied for having an atypical structural characteristic: the presence of a so-called “mycomembrane” that, in an organizational sense, is believed to resemble the outer membrane of typical Gram-negative bacteria (Zuber et al., 2008; Touchette and Seeliger, 2017; Figure 1). This mycomembrane is limited to members of the Corynebacteriales with some species-specific variation (Goodfellow and Jones, 2015; Mohammadipanah and Dehhaghi, 2017). Besides the components common to other Gram-positive bacteria, Corynebacteriales also have a layer of arabinogalactan attached to the peptidoglycan layer and to the inner leaflet of the mycolic-acid containing mycomembrane. This unusual structure, along with some phylogenetic ambiguity, have led certain authors to suggest that M. tuberculosis has more in common with Gram-negative bacteria than with their Gram-positive relatives (Fu and Fu-Liu, 2002). In the 1970s, two distinct cell envelope cleavage planes were recorded for freeze-etched mycobacteria (Barksdale and Kim, 1977). These findings contributed to the original proposal for a mycobacterial outer membrane. Further unequivocal evidence was provided by labeling with selective fluorescent probes (Christensen et al., 1999). In fact, the debate about the existence and composition of the mycomembrane was partially resolved only 10 years ago when it was visualized by cryo-electron microscopy of vitreous sections (CEMOVIS) (Hoffmann et al., 2008; Zuber et al., 2008). The reasons for the evolution of this membrane are still not totally clear, although we know that it is important for several aspects of the virulence and intrinsic antibiotic resistance of pathogenic species such as M. tuberculosis (Forrellad et al., 2013; Becker and Sander, 2016). In addition, as this membrane (along with the rest of the cell envelope) is at the frontline of environmental interactions, it is expected that differences in the constitution of the mycomembrane are associated with adaptation to specific environments or ecological niches (Veyrier et al., 2009).
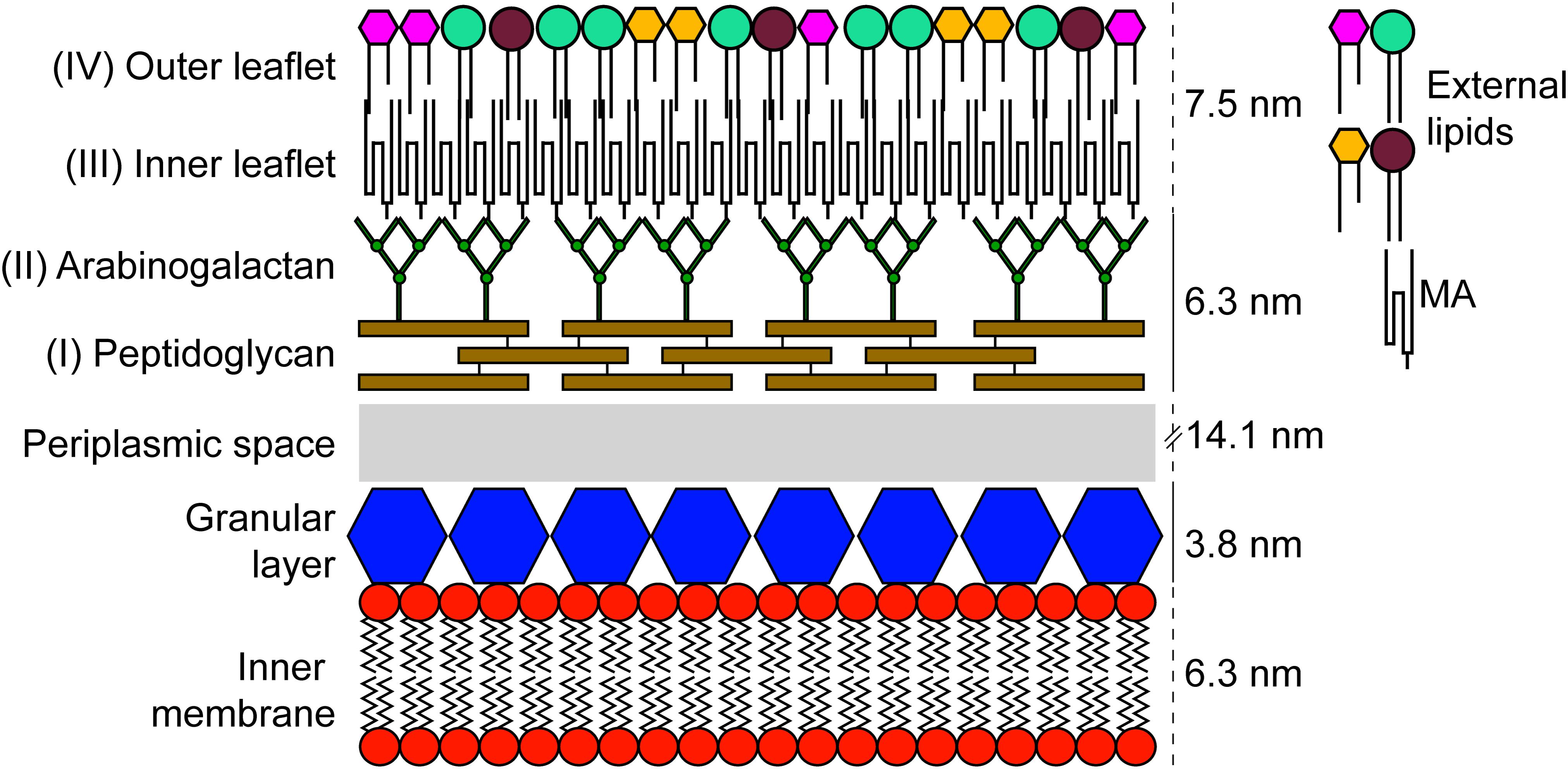
FIGURE 1. Schematic representation of the cell envelope found in members of the order Corynebacteriales. The components were scaled up using results obtained by CEMOVIS for M. bovis BCG (Zuber et al., 2008).
In this era of large-scale DNA sequencing (Vincent et al., 2017), we can now investigate and compare the genomic sequences of bacteria at an unprecedented rate (Land et al., 2015) and infer the key steps that have led to the development and modification of the various subcomponents of bacterial cells. In recent years, numerous actinobacterial genomes have been sequenced including a broad representation of each genus. As expected, analysis of these data revealed a great heterogeneity in terms of the many biological functions associated with these bacteria (Ventura et al., 2007; Gomez-Escribano et al., 2016). Based on this large amount of sequence information, it is now possible to carry out an evolutionary analysis of the cell envelope in order to dissect the genetic events that have led to its development. Here, we describe the different layers of the cell envelope for the Actinobacteria phylum with a specific emphasis on species that harbor the mycomembrane. Although a number of mysteries still remain, our goal for this article is to provide new perspectives on the evolutionary path and benefits surrounding the unique membrane features of this important group of bacteria.
By using CEMOVIS, it is possible to visualize a “granular layer” in between the plasma membrane and the peptidoglycan layer of Mycobacterium bovis BCG, Mycobacterium smegmatis, and Corynebacterium glutamicum (Zuber et al., 2008). A previous study made on other Gram-positive bacteria has shown that the granular layer is possibly linked to the plasma membrane and composed of penicillin-binding proteins, lipoproteins, and lipoteichoic acids (or teichuronic acid in some species; for review see Tul’skaya et al., 2011) (Zuber et al., 2006). Although, it seems this structure is common to several Gram-positive bacteria, the function and its precise composition are poorly characterized.
The cell envelope of Actinobacteria is composed of a layer of peptidoglycan that provides essential functions such as rigidity and helps to maintain an optimal osmotic stability (Vollmer et al., 2008; Jankute et al., 2015). Although peptidoglycan is common amongst bacteria, there are many subtle differences in its composition that have been used in the past as a way to identify distinct species in the context of chemotaxonomy (Lechevalier and Lechevalier, 1970). The standard peptidoglycan consists of short peptides and glycan strands that are composed of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues linked byβ-1→4 bonds. The third amino acid in the peptide stem is usually meso-diaminopimelic acid (meso-DAP) for Gram-negative and L-lysine for Gram-positive bacteria (Egan et al., 2017). In Actinobacteria, this amino acid is variable with numerous species (including the Corynebacteriales order) that harbor meso-DAP. This property is potentially due to the species-specific affinity of the ligase MurE (UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelate ligase) for meso-DAP (Hammes et al., 1977; Basavannacharya et al., 2010). Furthermore, differences in these third amino-acids were used as a marker for Actinobacteria cell wall types II, III, and IV (Lechevalier and Lechevalier, 1970).
In addition to having meso-DAP, the peptidoglycan of members of the Corynebacteriales order has another major distinction: a portion of MurNAc molecules is oxidized to become N-glycolylmuramic acid (MurNGlyc). This characteristic, in addition to playing an important role in the structure of peptidoglycan, has been suggested to increase resistance to lysozyme and β-lactam antibiotics (Raymond et al., 2005). The hydroxylase responsible for this modification is encoded by the gene namH (Rv3818; as per the reference M. tuberculosis H37Rv genome) (Raymond et al., 2005; Coulombe et al., 2009; Hansen et al., 2014). As expected, this gene is found in several members of the Corynebacteriales order (Figure 2). However, it appears to be absent from the genomes of genera such as Hoyosella and in those that are the most “basal” of the order: Corynebacterium, Turicella, Lawsonella, Dietzia, and Segniliparus. Interestingly, a few other genera in the Actinobacteria phylum, but not in the Corynebacteriales, also have a putative namH homolog (Figure 2). This is the case for Stackebrandtia (Glycomycetales), Micromonospora (Micromonosporales), and Salinispora (Micromonosporales) that form a discrete sub-lineage and have been described to harbor a cell wall with MurNGlyc (Parte et al., 2012). As an exception, a few scattered species (such as Catenulispora acidiphila and Rubrobacter radiotolerans) possess a homologous copy of NamH, although to the best of our knowledge there is no evidence for the presence of MurNGlyc in their peptidoglycan (Parte et al., 2012).
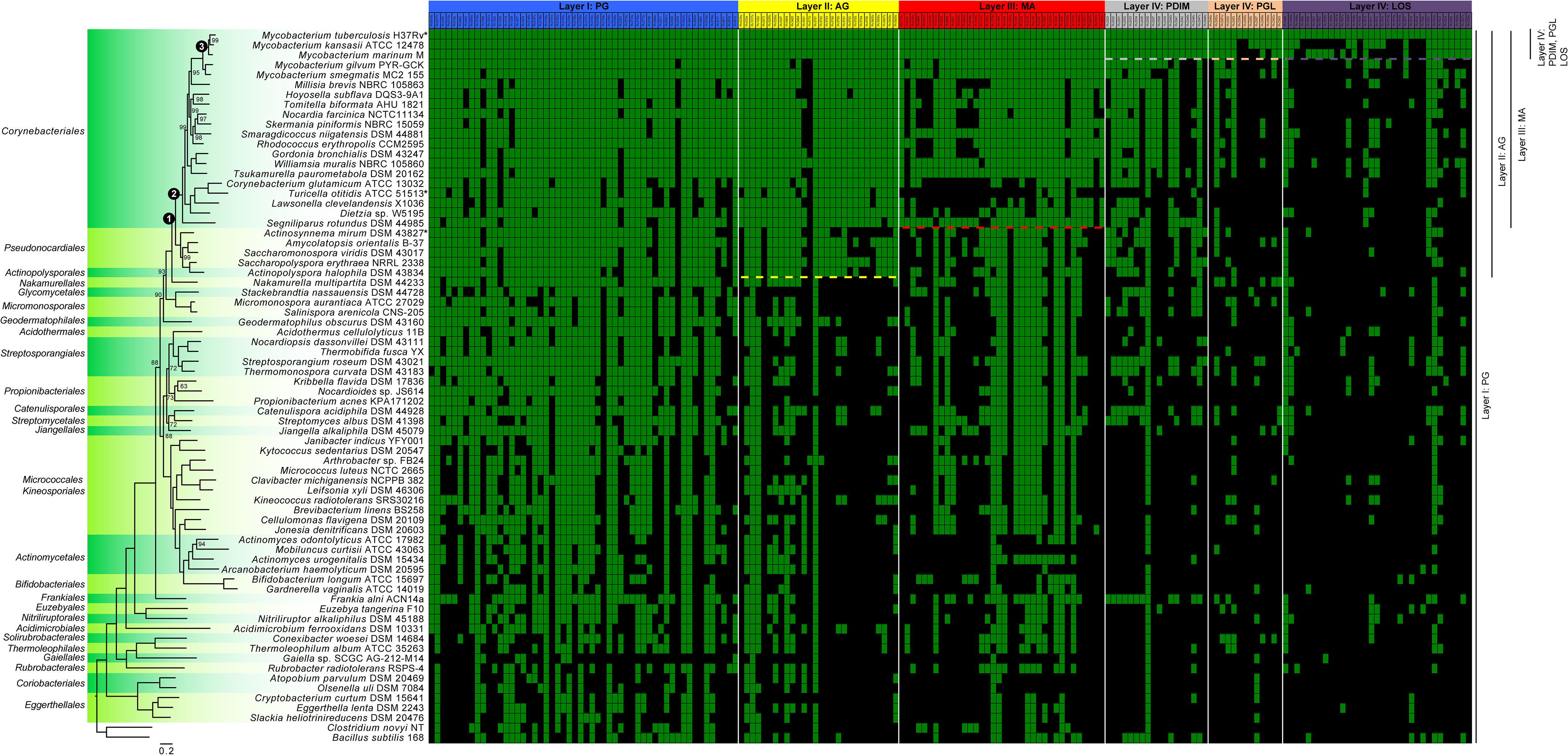
FIGURE 2. Distribution within the Actinobacteria phylum of genes involved in the synthesis of components of the cell envelope of M. tuberculosis. The 72 genome sequences (Supplementary Table S1), comprising at least one species per order, were annotated using PROKKA version 1.12 (Seemann, 2014). The phylogenetic tree was generated from 86 sequences inferred as orthologous by GET_HOMOLOGUES version 20180313 (Contreras-Moreira and Vinuesa, 2013). The sequences were aligned using MAFFT version 7.397 (Katoh and Standley, 2013) and concatenated and partitioned using AMAS (Borowiec, 2016). Finally, the best evolutionary model for each partition was found by IQ-TREE version 1.6.3 (Kalyaanamoorthy et al., 2017) and maximum-likelihood phylogenetic analysis was also performed using IQ-TREE (Nguyen et al., 2015) using 10,000 ultrafast bootstrap replicates (Hoang et al., 2018). Only bootstrap values under 100 are shown on the tree. Nodes that are discussed in the article are highlighted with a numbered black circle. The green and black squares of the matrix highlight genes that are present or absent, respectively. Individual genes (Supplementary Table S2) were considered to be present when they had a sequence similarity ≥50% relative to M. tuberculosis [an e-value cut-off of 1e-10 has also been applied in TBLASTN version 2.7.1 (Altschul et al., 1997)]. The raw data, containing the similarity values, is presented in the Supplementary Table S2 for clarity. The “∗” represent micro-evolution of specific strains (described in the corresponding sections of the article). Dashed lines represent delineation, described in the literature, of the presence/absence of the specific layers also depicted at the right side of the figure. Of note, although the phylogenetic position of Gaiella sp. SCGC AG-212-M14 is expected for a member of the order Gaiellales, the size of the assembly (<1 Mb) coming from a single cell amplified by WGA-X, suggests that the sequence of this strain is likely incomplete.
In addition to its distinct composition, the peptidoglycan of Corynebacteriales is also the point of attachment of arabinogalactan, a highly branched heteropolysaccharide composed of galactose and arabinose in a furanoid form (Figure 1; McNeil et al., 1987). It is estimated that approximately 10% of the MurNAc residues of the peptidoglycan are covalently linked to arabinogalactan [α-L-rhamnopyranose-(1→3-α-D-GlycNAc-(1→P))] (McNeil et al., 1990). Although this linkage is unique, it resembles the one that connects cell wall teichoic acids to peptidoglycan in other Gram-positive species. This similarity led to the discovery of a phosphotransferase of the LytR-CpsA-Psr family, named Lcp1, which is encoded by the Rv3267 gene and plays a leading role in linking arabinogalactan to peptidoglycan in M. tuberculosis (Baumgart et al., 2016; Grzegorzewicz et al., 2016; Harrison et al., 2016).
In terms of its distribution within the Actinobacteria phylum, arabinogalactan is produced exclusively by the members of the Corynebacteriales order and in some species from related orders such as Pseudonocardiales and Actinopolysporales (Goodfellow et al., 1984) [with the exception of Actinosynnema mirum (Hasegawa et al., 1978)]. These latter two orders possess a typical type IV cell wall (Lechevalier and Lechevalier, 1970), although they lack mycolic acids (Ludwig et al., 2015). The global distribution of genes implicated in the synthesis and transport of arabinogalactan is well correlated with the fact that these species produce arabinogalactan (see Figure 2). Nevertheless, a deeper investigation of these latter two orders will be required to exclude possible exceptions or misinterpretation.
The inner leaflet of the external mycomembrane is homogeneous and is mainly composed of mycolic acids (MAs) – long-chain fatty acids that are exclusive to the order Corynebacteriales (Bansal-Mutalik and Nikaido, 2014; Marrakchi et al., 2014). On one hand, MAs form a barrier to hydrophilic molecules including several antibiotics (Liu et al., 1995), while on the other hand, they are also the target of some first- and second-line anti-TB drugs including isoniazid (Jackson et al., 2013; Marrakchi et al., 2014). In an evolutionary context, MAs play a prominent role in taxonomic differentiation within the Corynebacteriales order. In this sense, there is a great diversity in the number of carbons and functional groups comprising the MAs, thus making it possible to use them as a chemotaxonomic marker to differentiate between genera and also at the species level (Minnikin et al., 1984; Barry et al., 1998; Song et al., 2009; Marrakchi et al., 2014).
As reviewed elsewhere (Marrakchi et al., 2014), the raw materials of MAs, fatty acids, are produced by the combination of two fatty acid synthases (FAS-I and FAS-II). The first, FAS-I, is a eukaryotic-like protein narrowly distributed in bacteria (Cabruja et al., 2017) and encoded by the fas gene (Rv2524c). FAS-I produces acyl-CoAs with a bimodal distribution of C16-C18 and C24-C26. FAS-II, on the other hand, is far more common across bacterial species (Cabruja et al., 2017) and begins with the formation of β-ketoacyl-ACP by Claisen condensation of malonyl-ACP [produced from malonyl-CoA by MtFabD (Rv2243)], with acyl-CoA (the product of FAS-I), through the action of MtFabH (Rv0533). Other enzymes such as HadA (Rv0635), HadB (Rv0636), HadC (Rv0637), InhA (Rv1484), KasA (Rv2245), and KasB (Rv2246) are involved in the subsequent steps of elongation and maturation of fatty acids by FAS-II. The final MA structure is produced though several crucial steps that include, among others, activation (FadD32 and Rv3801c), condensation (Pks13 and Rv3800c) and reduction (CmrA and Rv2509).
Although the presence of MAs is well-conserved throughout the Corynebacteriales, there are some exceptions. For example, basal species of Corynebacteriales (such as Corynebacterium, Dietzia, Lawsonella, and Turicella) that form a monophyletic group, lack multiple genes confirming the well described diversity in length and composition of MA. It has already been reported that some species of Corynebacterium possess two FAS-I genes (such as C. glutamicum), while some do not have the genes encoding for the typical FAS-II machinery (Radmacher et al., 2005). Interestingly, Corynebacterium cannot elongate MAs (Radmacher et al., 2005; Burkovski, 2013), which is consistent with the lack of more than 50% of genes implicated in this pathway.
Turicella otitidis possesses almost none of the genes involved in MA biosynthesis (see “∗” in Figure 2). This species is clearly in the order Corynebacteriales based on its genome sequence (Baek et al., 2018). However, a recent study suggests that the genome of T. otitidis has lost the key genes involved in MA synthesis (Baek et al., 2018). This result corroborates the fact that this bacterium does not produce MAs which is exceptional for a bacterium of the order Corynebacteriales and this microevolution has made its positioning within this order rather difficult (Funke et al., 1994). Interestingly, this species still produces arabinogalactan (Renaud et al., 1996).
While the composition of the inner leaflet of the mycomembrane is homogenous, the outer leaflet is highly heterogeneous and consists of lipids, lipoglycans, and proteins (Chiaradia et al., 2017). Due to its complexity and species-specific constitution, it remains poorly characterized with the exception of the pathogen M. tuberculosis and closely related slow-growing mycobacteria. In these bacteria, several lipids are associated with the external leaflet including – but not limited to – phthiocerol dimycocerosate (PDIM), phenolic glycolipid (PGL, phenolphthiocerol-based glycolipids that share a similar long-chain fatty acid backbone with PDIM), and lipooligosaccharides (LOS). These components seem to be important virulence factors (Reed et al., 2004; Forrellad et al., 2013) and have been shown to be involved in the infection of macrophages as well as in escape from the immune system (Cambier et al., 2014). The PDIM and PGL-associated genes (Rv2928 to Rv2963), are all co-located within the same cluster of the chromosome (Goude and Parish, 2008; Supplementary Table S2). LOS are common to several slow growing mycobacteria (including “M. canettii” from the M. tuberculosis complex) although they are not produced by M. tuberculosis due to the loss of Pks5.1 and truncation of PapA4. The remaining genes implicated in this pathway are present (Figure 2) which suggests micro-evolution in this species has led to the loss of LOS production (Boritsch et al., 2016; Brennan, 2016). This apparent evolutionary change correlates with a marked increase in whole cell hydrophobicity and enhanced aerosol transmission of current TB strains (Jankute et al., 2017).
The external layer of the mycomembrane also contains trehalose monomycolate – TMM, and trehalose dimycolate – TDM (also called “cord factor”) that also have crucial functions in the regulation of the host-symbiont relationship (Hunter et al., 2006). These molecules consist of glucose disaccharides (α-D-glucopyranosyl-α-D-glucopyranoside) esterified with MAs. TMM is generated in the cytoplasm, whereupon MmpL3 (Rv0206c) subsequently transports this molecule to the periplasm (Grzegorzewicz et al., 2016). Finally, the accepted model (for a review see Marrakchi et al., 2014) is that TMM in the periplasm serves as the MA donor for the mycolylation of arabinogalactan, and is also processed to give TDM. Both reactions involve the Ag85 enzyme complex (Belisle et al., 1997). Three genes (Rv3803c, Rv1886c, and Rv0129c) encode for the Ag85 complex and they are only present in the MA-positive species as seen in Figure 2. As TMM and TDM rely on the presence of MAs and Ag85, their distribution is therefore restricted to MA positive species. Indeed, they have been isolated from Mycobacterium, Corynebacterium, Nocardia, and Rhodococcus (Noll et al., 1956; Ioneda et al., 1970; Jean-Claude et al., 1976; Mompon et al., 1978; Lopes Silva et al., 1979; Pommier and Michel, 1979; Rapp et al., 1979; Batrakov et al., 1981). However, further characterization is necessary for other MA-positive species (such as Segniliparus).
The biological functions and molecular mechanisms surrounding the biosynthesis of the mycomembrane present in the Corynebacteriales are being uncovered slowly, but surely. However, the precise origin of this feature that is unique amongst members of the Actinobacteria (otherwise composed essentially of monoderm bacteria) continues to be a mystery. Two main theories can potentially explain the biogenesis of the mycomembrane: (1) that it arose via the remodeling of an already existing outer membrane, or (2) that it has a de novo origin (Graphical Abstract).
The first theory was proposed based on the observation that a double membrane is generated as a byproduct of endospore formation in bacteria (although endospores have not been found in Actinobacteria) (Tocheva et al., 2011; Tocheva et al., 2013). Assuming that the primordial cell was monoderm, a double membraned cell could have appeared via the retention of the second spore membrane, thus giving rise to a diderm common ancestor of all bacteria. This theory implies a bottleneck that selected for this ancestor but also that the current monoderm species appeared by independent loss of the outer membrane. In order to generate a mycomembrane, a significant re-engineering of the gene content (difficult to assess due to gene erosion over time) would have occurred in Corynebacteriales and could account for the atypical envelope characteristics when compared to other diderm species (Tocheva et al., 2016). This theory remains an open question in the field (Sutcliffe and Dover, 2016).
The second theory proposes that the double bacterial membrane is a homoplastic character (i.e., a similar trait from different evolutionary origins) that has evolved several times independently in a functionally convergent manner. With regard to the Actinobacteria phylum, the double membrane could have appeared “recently” by successive horizontal acquisition of genes allowing a step-by-step construction of the cell envelope that is present in Mycobacterium.
The distribution of the major genes implicated in the synthesis of the different layers of the mycomembrane is presented for the Actinobacteria in Figure 2. This figure needs to be interpreted carefully as the function of the different orthologous proteins has not been proven to be conserved across all the listed species. Nevertheless, one can observe a “phylogenetic gradient” in terms of the distribution of genes involved in the construction of the cell-envelope from inside to outside (Figure 2). In addition, one can observe a step-wise distribution in the evolution of the layers comprising the cell envelope. Both the gene content and the cell wall characteristics are well correlated. When investigating the genes involved in peptidoglycan synthesis throughout the genomes of the Actinobacteria, it can be observed that they are broadly distributed. On the other hand, arabinogalactan (and the genes implicated in this pathway) are restricted to bacteria that have diverged after the evolutionary node that we called “Node 1” (Figure 2). The MAs are even more narrowly distributed within bacteria that diverged after Node 2 (if we exclude the polyvalent FAS-II machinery). Finally, the genes associated with synthesis of the external leaflet are present in a sublineage-specific manner within the slow-growing mycobacteria that diverged after Node 3. This step-wise gradient of gene acquisition would support the theory in which the sequential gain-of-function has led to the successive evolution of the outer-membrane components in the ancestors of modern M. tuberculosis. The first step in this pathway appears to be the ability to attach and produce arabinogalactan that, in turn, supports the addition of the mycomembrane. Within this scenario, the final layer that corresponds to the external leaflet would have evolved most recently in a species-specific manner.
In this article, M. tuberculosis (and other slow growing mycobacteria) are used as our end point as they are the best characterized. However, we expect that with further characterization of the cell envelope of other Corynebacteriales and identification of the genes involved in their synthesis, we will obtain a similar general pattern. This will be particularly interesting for the highly heterogeneous outer leaflet. In fact, Corynebacteriales colonize multiple ecological niches from extreme and diverse habitats including soil and the human microbiome (Ramakrishnan et al., 2013; Goodfellow and Jones, 2015). This diversity implies a specific adaptation for individual species. This phenomenon would involve a distinctive selection of cell envelope constituents and, more specifically, for the external leaflet of the outer-membrane that is in immediate contact with the environment. For example, Figure 2 shows that several genes involved in PDIM formation are present in the genomes of members of the Corynebacteriales order that are assumed to be PDIM-negative. This is especially true for M. gilvum, which is a rapid-growing mycobacterium (Wee et al., 2017). It is important to note that a number of genes involved in PDIM and PGL synthesis are polyketide synthases, which are versatile enzymes involved in the production of various natural products (Quadri, 2014). The question remains whether these genes serve in the production of molecules localized in the outer leaflet of “DIM-negative” bacteria that may have a common origin with PDIM as we know it in the pathogenic slow-growing mycobacteria.
Regardless of whether the double membrane of Corynebacteriales represents a relict sharing a common origin with the double membrane present in typical Gram-negative species, or is the result of a more recent adaptation, it will undoubtedly be interesting and informative to attempt to experimentally reconstruct the double membrane of mycobacteria starting from an Actinobacteria species possessing a single membrane (such as Turicella sp.). This daring experiment will teach us more about the mechanism of biogenesis of this double membrane and, perhaps, even inform on its molecular evolution and the natural selection of such an adaptation. In doing so, we could mimic the putative evolutionary process that has led to the Mycobacterium membrane as well as identify the functional differences linked to possible species-specific adaptation of the mycomembrane. This work may also highlight important enzyme functions that can be specifically targeted in vaccination or chemotherapeutic approaches aimed at killing pathogenic species such as M. tuberculosis.
ATV and FJV conceived and designed the experiments. ATV, FJV, MR, ET, SN, and IM contributed to the writing and editing of the manuscript. All authors read and approved the final manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) under Grant RGPIN-2016-04940. ATV received a Postdoctoral Fellowship from the NSERC. FJV is a research scholar of the Fonds de Recherche du Québec – Santé.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02341/full#supplementary-material
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Baek, I., Kim, M., Lee, I., Na, S. I., Goodfellow, M., and Chun, J. (2018). Phylogeny trumps chemotaxonomy: a case study involving Turicella otitidis. Front. Microbiol. 9:834. doi: 10.3389/fmicb.2018.00834
Bansal-Mutalik, R., and Nikaido, H. (2014). Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963. doi: 10.1073/pnas.1403078111
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Barry, C. E., Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A., et al. (1998). Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37, 143–179.
Basavannacharya, C., Robertson, G., Munshi, T., Keep, N. H., and Bhakta, S. (2010). ATP-dependent MurE ligase in Mycobacterium tuberculosis: biochemical and structural characterisation. Tuberculosis 90, 16–24. doi: 10.1016/j.tube.2009.10.007
Batrakov, S. G., Rozynov, B. V., Koronelli, T. V., and Bergelson, L. D. (1981). Two novel types of trehalose lipids. Chem. Phys. Lipids 29, 241–266. doi: 10.1016/0009-3084(81)90055-4
Baumgart, M., Schubert, K., Bramkamp, M., and Frunzke, J. (2016). Impact of LytR-CpsA-Psr proteins on cell wall biosynthesis in Corynebacterium glutamicum. J. Bacteriol. 198, 3045–3059. doi: 10.1128/JB.00406-16
Becker, J., and Wittmann, C. (2016). “Industrial microorganisms: Corynebacterium glutamicum,” in Industrial Biotechnology: Microorganisms, Vol. 1, eds C. Wittmann and J. C. Liao (Hoboken, NJ: John Wiley & Sons), 183–220. doi: 10.1002/9783527807796.ch6
Becker, K., and Sander, P. (2016). Mycobacterium tuberculosis lipoproteins in virulence and immunity - fighting with a double-edged sword. FEBS Lett. 590, 3800–3819. doi: 10.1002/1873-3468.12273
Belisle, J. T., Vissa, V. D., Sievert, T., Takayama, K., Brennan, P. J., and Besra, G. S. (1997). Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422.
Boritsch, E. C., Frigui, W., Cascioferro, A., Malaga, W., Etienne, G., Laval, F., et al. (2016). pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat. Microbiol. 1:15019. doi: 10.1038/nmicrobiol.2015.19
Borowiec, M. L. (2016). AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4:e1660. doi: 10.7717/peerj.1660
Brennan, P. J. (2016). Bacterial evolution: emergence of virulence in TB. Nat. Microbiol. 1:15031. doi: 10.1038/nmicrobiol.2015.31
Burkovski, A. (2013). Cell envelope of corynebacteria: structure and influence on pathogenicity. ISRN Microbiol. 2013:935736. doi: 10.1155/2013/935736
Cabruja, M., Mondino, S., Tsai, Y. T., Lara, J., Gramajo, H., and Gago, G. (2017). A conditional mutant of the fatty acid synthase unveils unexpected cross talks in mycobacterial lipid metabolism. Open Biol. 7:160277. doi: 10.1098/rsob.160277
Cambier, C. J., Takaki, K. K., Larson, R. P., Hernandez, R. E., Tobin, D. M., Urdahl, K. B., et al. (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222. doi: 10.1038/nature12799
Chiaradia, L., Lefebvre, C., Parra, J., Marcoux, J., Burlet-Schiltz, O., Etienne, G., et al. (2017). Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci. Rep. 7:12807. doi: 10.1038/s41598-017-12718-4
Christensen, H., Garton, N. J., Horobin, R. W., Minnikin, D. E., and Barer, M. R. (1999). Lipid domains of mycobacteria studied with fluorescent molecular probes. Mol. Microbiol. 31, 1561–1572.
Contreras-Moreira, B., and Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
Coulombe, F., Divangahi, M., Veyrier, F., De Leseleuc, L., Gleason, J. L., Yang, Y., et al. (2009). Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 206, 1709–1716. doi: 10.1084/jem.20081779
Egan, A. J., Cleverley, R. M., Peters, K., Lewis, R. J., and Vollmer, W. (2017). Regulation of bacterial cell wall growth. FEBS J. 284, 851–867. doi: 10.1111/febs.13959
Forrellad, M. A., Klepp, L. I., Gioffre, A., Sabio, Y., Garcia, J., Morbidoni, H. R., et al. (2013). Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4, 3–66. doi: 10.4161/viru.22329
Fu, L. M., and Fu-Liu, C. S. (2002). Is Mycobacterium tuberculosis a closer relative to Gram-positive or Gram-negative bacterial pathogens? Tuberculosis 82, 85–90.
Funke, G., Stubbs, S., Altwegg, M., Carlotti, A., and Collins, M. D. (1994). Turicella otitidis gen. nov., sp. nov., a coryneform bacterium isolated from patients with otitis media. Int. J. Syst. Bacteriol. 44, 270–273. doi: 10.1099/00207713-44-2-270
Gomez-Escribano, J. P., Alt, S., and Bibb, M. J. (2016). Next generation sequencing of Actinobacteria for the discovery of novel natural products. Mar. Drugs 14:E78. doi: 10.3390/md14040078
Goodfellow, M., and Jones, A. L. (2015). “Corynebacteriales ord. nov,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds W. B. Whitman, F. Rainey, P. Kämpfer, M. Trujillo, J. Chun, P. DeVos, et al. (Hoboken, NJ: Wiley), doi: 10.1002/9781118960608.obm00009
Goodfellow, M., Mordarski, M., and Williams, S. T. (1984). The Biology of the Actinomycetes. Cambridge, MA: Academic Press.
Goude, R., and Parish, T. (2008). The genetics of cell wall biosynthesis in Mycobacterium tuberculosis. Future Microbiol. 3, 299–313. doi: 10.2217/17460913.3.3.299
Grzegorzewicz, A. E., De Sousa-D’auria, C., Mcneil, M. R., Huc-Claustre, E., Jones, V., Petit, C., et al. (2016). Assembling of the Mycobacterium tuberculosis cell wall core. J. Biol. Chem. 291, 18867–18879. doi: 10.1074/jbc.M116.739227
Hammes, W. P., Neukam, R., and Kandler, O. (1977). On the specificity of the uridine diphospho-N-acetylmuramyl-alanyl-D-glutamic acid: diamino acid ligase of Bifidobacterium globosum. Arch. Microbiol. 115, 95–102.
Hansen, J. M., Golchin, S. A., Veyrier, F. J., Domenech, P., Boneca, I. G., Azad, A. K., et al. (2014). N-glycolylated peptidoglycan contributes to the immunogenicity but not pathogenicity of Mycobacterium tuberculosis. J. Infect. Dis. 209, 1045–1054. doi: 10.1093/infdis/jit622
Harrison, J., Lloyd, G., Joe, M., Lowary, T. L., Reynolds, E., Walters-Morgan, H., et al. (2016). Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. MBio 7:e00972-16. doi: 10.1128/mBio.00972-16
Hasegawa, T., Lechevalier, M. P., and Lechevalier, H. A. (1978). New genus of the Actinomycetales: Actinosynnema gen. nov. Int. J. Syst. Evol. Microbiol. 28, 304–310. doi: 10.1099/00207713-28-2-304
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M., and Engelhardt, H. (2008). Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967. doi: 10.1073/pnas.0709530105
Hunter, R. L., Olsen, M. R., Jagannath, C., and Actor, J. K. (2006). Multiple roles of cord factor in the pathogenesis of primary, secondary, and cavitary tuberculosis, including a revised description of the pathology of secondary disease. Ann. Clin. Lab. Sci. 36, 371–386.
Ioneda, T., Lederer, E., and Rozanis, J. (1970). Sur la structure des diesters de tréhalose (“cord factors”) produits par Nocardia asteroides et Nocardia rhodochrous. Chem. Phys. Lipids 4, 375–392. doi: 10.1016/0009-3084(70)90037-X
Jackson, M., Mcneil, M. R., and Brennan, P. J. (2013). Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol. 8, 855–875. doi: 10.2217/fmb.13.52
Jankute, M., Cox, J. A., Harrison, J., and Besra, G. S. (2015). Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423. doi: 10.1146/annurev-micro-091014-104121
Jankute, M., Nataraj, V., Lee, O. Y., Wu, H. H. T., Ridell, M., Garton, N. J., et al. (2017). The role of hydrophobicity in tuberculosis evolution and pathogenicity. Sci. Rep. 7:1315. doi: 10.1038/s41598-017-01501-0
Jean-Claude, P., Lacave, C., Ahibo-Coffy, A., and Savagnac, A. (1976). Séparation et étude structurale des espéces moléculaires de monomycolates et de dimycolates de α-d-tréhalose présents chez Mycobacterium phlei. Eur. J. Biochem. 63, 543–552. doi: 10.1111/j.1432-1033.1976.tb10258.x
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Land, M., Hauser, L., Jun, S. R., Nookaew, I., Leuze, M. R., Ahn, T. H., et al. (2015). Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. doi: 10.1007/s10142-015-0433-4
Lechevalier, M. P., and Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443. doi: 10.1099/00207713-20-4-435
Lewin, G. R., Carlos, C., Chevrette, M. G., Horn, H. A., Mcdonald, B. R., Stankey, R. J., et al. (2016). Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 70, 235–254. doi: 10.1146/annurev-micro-102215-095748
Liu, J., Rosenberg, E. Y., and Nikaido, H. (1995). Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. U.S.A. 92, 11254–11258.
Lopes Silva, C., Gesztesi, J. L., and Ioneda, T. (1979). Thehalose mycolates from Nocardia asteroides, Nocardia farcinica, Gordona lentifragmenta and Gordona bronchialis. Chem. Phys. Lipids 24, 17–25. doi: 10.1016/0009-3084(79)90092-6
Ludwig, W., Euzéby, J., Schumann, P., Busse, H., Trujillo, M. E., Kämpfer, P., et al. (2015). “Road map of the phylum Actinobacteria,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds W. B. Whitman, F. Rainey, P. Kämpfer, M. Trujillo, J. Chun, P. DeVos, et al. (Hoboken, NJ: Wiley), doi: 10.1002/9781118960608.bm00029
Marrakchi, H., Laneelle, M. A., and Daffe, M. (2014). Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85. doi: 10.1016/j.chembiol.2013.11.011
McNeil, M., Daffe, M., and Brennan, P. J. (1990). Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 265, 18200–18206.
McNeil, M., Wallner, S. J., Hunter, S. W., and Brennan, P. J. (1987). Demonstration that the galactosyl and arabinosyl residues in the cell-wall arabinogalactan of Mycobacterium leprae and Mycobacterium tuberculosis are furanoid. Carbohydr. Res. 166, 299–308.
Minnikin, D. E., Minnikin, S. M., Parlett, J. H., Goodfellow, M., and Magnusson, M. (1984). Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 139, 225–231.
Mohammadipanah, F., and Dehhaghi, M. (2017). “Classification and taxonomy of Actinobacteria,” in Biology and Biotechnology of Actinobacteria, eds J. Wink, F. Mohammadipanah, and J. Hamedi (Cham: Springer International Publishing), 51–77.
Mompon, B., Federici, C., Toubiana, R., and Lederer, E. (1978). Isolation and structural determination of a “cord-factor” (trehalose 6,6’ dimycolate) from Mycobacterium smegmatis. Chem. Phys. Lipids 21, 97–101. doi: 10.1016/0009-3084(78)90057-9
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Noll, H., Bloch, H., Asselineau, J., and Lederer, E. (1956). The chemical structure of the cord factor of Mycobacterium tuberculosis. Biochim. Biophys. Acta 20, 299–309. doi: 10.1016/0006-3002(56)90289-X
O’Callaghan, A., and van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7:925. doi: 10.3389/fmicb.2016.00925
Pai, M., Behr, M. A., Dowdy, D., Dheda, K., Divangahi, M., Boehme, C. C., et al. (2016). Tuberculosis. Nat. Rev. Dis. Primers 2:16076. doi: 10.1038/nrdp.2016.76
Parte, A., Whitman, W. B., Goodfellow, M., Kämpfer, P., Busse, H. J., Trujillo, M. E., et al. (2012). Bergey’s Manual of Systematic Bacteriology: The Actinobacteria, Vol. 5. New York, NY: Springer.
Pommier, M. T., and Michel, G. (1979). Glycolipides des nocardiae. Isolement et caractérisation de mononocardomycolates et de dinocardomycolates de tréhalose dans Nocardia caviae. Chem. Phys. Lipids 24, 149–155. doi: 10.1016/0009-3084(79)90084-7
Quadri, L. E. (2014). Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit. Rev. Biochem. Mol. Biol. 49, 179–211. doi: 10.3109/10409238.2014.896859
Radmacher, E., Alderwick, L. J., Besra, G. S., Brown, A. K., Gibson, K. J., Sahm, H., et al. (2005). Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum. Microbiology 151, 2421–2427. doi: 10.1099/mic.0.28012-0
Ramakrishnan, V. R., Feazel, L. M., Gitomer, S. A., Ir, D., Robertson, C. E., and Frank, D. N. (2013). The microbiome of the middle meatus in healthy adults. PLoS One 8:e85507. doi: 10.1371/journal.pone.0085507
Rapp, P., Bock, H., Wray, V., and Wagner, F. (1979). Formation, isolation and characterization of trehalose dimycolates from Rhodococcus erythropolis grown on N-Alkanes. Microbiology 115, 491–503. doi: 10.1099/00221287-115-2-491
Raymond, J. B., Mahapatra, S., Crick, D. C., and Pavelka, M. S. Jr. (2005). Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280, 326–333. doi: 10.1074/jbc.M411006200
Reed, M. B., Domenech, P., Manca, C., Su, H., Barczak, A. K., Kreiswirth, B. N., et al. (2004). A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84–87. doi: 10.1038/nature02837
Renaud, F. N., Gregory, A., Barreau, C., Aubel, D., and Freney, J. (1996). Identification of Turicella otitidis isolated from a patient with otorrhea associated with surgery: differentiation from Corynebacterium afermentans and Corynebacterium auris. J. Clin. Microbiol. 34, 2625–2627.
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Song, S. H., Park, K. U., Lee, J. H., Kim, E. C., Kim, J. Q., and Song, J. (2009). Electrospray ionization-tandem mass spectrometry analysis of the mycolic acid profiles for the identification of common clinical isolates of mycobacterial species. J. Microbiol. Methods 77, 165–177. doi: 10.1016/j.mimet.2009.01.023
Sutcliffe, I. C., and Dover, L. G. (2016). Comment on Tocheva et al. ”Sporulation, bacterial cell envelopes and the origin of life”. Nat. Rev. Microbiol. 14:600. doi: 10.1038/nrmicro.2016.113
Tocheva, E. I., Lopez-Garrido, J., Hughes, H. V., Fredlund, J., Kuru, E., Vannieuwenhze, M. S., et al. (2013). Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 88, 673–686. doi: 10.1111/mmi.12201
Tocheva, E. I., Matson, E. G., Morris, D. M., Moussavi, F., Leadbetter, J. R., and Jensen, G. J. (2011). Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146, 799–812. doi: 10.1016/j.cell.2011.07.029
Tocheva, E. I., Ortega, D. R., and Jensen, G. J. (2016). Sporulation, bacterial cell envelopes and the origin of life. Nat. Rev. Microbiol. 14, 535–542. doi: 10.1038/nrmicro.2016.85
Touchette, M. H., and Seeliger, J. C. (2017). Transport of outer membrane lipids in mycobacteria. Biochim. Biophys. Acta 1862, 1340–1354. doi: 10.1016/j.bbalip.2017.01.005
Tul’skaya, E. M., Shashkov, A. S., Streshinskaya, G. M., Senchenkova, S. N., Potekhina, N. V., Kozlova, Y. I., et al. (2011). Teichuronic and teichulosonic acids of actinomycetes. Biochemistry 76, 736–744. doi: 10.1134/S0006297911070030
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Veyrier, F., Pletzer, D., Turenne, C., and Behr, M. A. (2009). Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol. Biol. 9:196. doi: 10.1186/1471-2148-9-196
Vincent, A. T., Derome, N., Boyle, B., Culley, A. I., and Charette, S. J. (2017). Next-generation sequencing (NGS) in the microbiological world: how to make the most of your money. J. Microbiol. Methods 138, 60–71. doi: 10.1016/j.mimet.2016.02.016
Vollmer, W., Blanot, D., and De Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Wee, W. Y., Dutta, A., and Choo, S. W. (2017). Comparative genome analyses of mycobacteria give better insights into their evolution. PLoS One 12:e0172831. doi: 10.1371/journal.pone.0172831
Zuber, B., Chami, M., Houssin, C., Dubochet, J., Griffiths, G., and Daffe, M. (2008). Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680. doi: 10.1128/JB.01919-07
Zuber, B., Haenni, M., Ribeiro, T., Minnig, K., Lopes, F., Moreillon, P., et al. (2006). Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 188, 6652–6660. doi: 10.1128/JB.00391-06
Keywords: cell envelope, Actinobacteria, Mycobacterium, evolution, genomics
Citation: Vincent AT, Nyongesa S, Morneau I, Reed MB, Tocheva EI and Veyrier FJ (2018) The Mycobacterial Cell Envelope: A Relict From the Past or the Result of Recent Evolution?. Front. Microbiol. 9:2341. doi: 10.3389/fmicb.2018.02341
Received: 27 June 2018; Accepted: 12 September 2018;
Published: 09 October 2018.
Edited by:
Christoph Mayer, Eberhard Karls Universität Tübingen, GermanyReviewed by:
Andreas Burkovski, Friedrich-Alexander-Universität Erlangen-Nürnberg, GermanyCopyright © 2018 Vincent, Nyongesa, Morneau, Reed, Tocheva and Veyrier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frederic J. Veyrier, ZnJlZGVyaWMudmV5cmllckBpYWYuaW5ycy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.