
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 September 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02197
This article is part of the Research Topic Advances in Understanding and Controlling Bacteria-Material Interactions, with a Focus on how Bacteria Sense a Surface View all 5 articles
Escherichia coli swarm on semi-solid surfaces with the aid of flagella. It has been hypothesized that swarmer cells overcome the increased viscous drag near surfaces by developing higher flagellar thrust and by promoting surface wetness with the aid of a flagellar switch. The switch enables reversals between clockwise (CW) and counterclockwise (CCW) directions of rotation of the flagellar motor. Here, we measured the behavior of flagellar motors in swarmer cells. Results indicated that although the torque was similar to that in planktonic cells, the tendency to rotate CCW was higher in swarmer cells. This suggested that swarmers likely have a smaller pool of phosphorylated CheY. Results further indicated that the upregulation of the flagellin gene was not critical for flagellar thrust or swarming. Consistent with earlier reports, moisture added to the swarm surface restored swarming in a CCW-only mutant, but not in a FliG mutant that rotated motors CW-only (FliGCW). Fluorescence assays revealed that FliGCW cells grown on agar surfaces carried fewer flagella than planktonic FliGCW cells. The surface-dependent reduction in flagella correlated with a reduction in the number of putative flagellar preassemblies. These results hint toward a possibility that the conformational dynamics of switch proteins play a role in the proper assembly of flagellar complexes and flagellar export, thereby aiding bacterial swarming.
Swarming in flagellated bacteria is a type of surface-dependent motility that is marked by rapid and coordinated movements of groups of cells in coherent structures and swirling patterns on top of semi-solid surfaces (Henrichsen, 1972). Swarming has been implicated in several types of infections (Mobley and Belas, 1995; Callegan et al., 2006; Yang et al., 2017), as well as elevated antibiotic resistance (Kim and Surette, 2003; Kim et al., 2003; Overhage et al., 2008; Butler et al., 2010). The initiation of swarming is preceded by the arrival of planktonic cells on a surface – in a laboratory setting, this involves the inoculation of planktonic cells on agar substrates (Hughes and Kearns, 2017). Subsequently, the planktonic cells transition into a swarmer state (Belas, 2014). The swarmer state is often accompanied by, depending on the bacterial species, elongation of the cell body, expression of more number of flagella, multi-nucleation, and secretion of polysaccharides that promote surface motility (Harshey, 2003; Darnton et al., 2010; Kearns, 2010). In Escherichia coli, swarmer cells reportedly double their lengths but maintain similar flagellar numbers per unit surface area as the planktonic cells. Due to the proximity of the swarmer cells to a solid surface, the viscous drag experienced is likely high (Lauga et al., 2006; Zhang et al., 2009). It is unclear how cells overcome surface drag but one possibility is that motors in swarming bacteria adapt to produce higher torque than motors in planktonic cells (Tuson et al., 2013), although this remains untested. Another way that cells might overcome the drag is with the aid of the flagellar switch – it is known that the ability of the motor to switch its direction of rotation between clockwise (CW) and counterclockwise (CCW) is vital for swarming. Mutants that rotate motors CCW-only or CW-only fail to swarm whereas those that are able to switch their motors are able to swarm (Harshey and Matsuyama, 1994; Burkart et al., 1997). It has been proposed that switching aids in the lubrication of the surface by extracting water from the underlying agar, and possibly helps liberate cells from the secreted LPS (lipopolysaccharides), thereby facilitating cell movement (Wang et al., 2005; Mariconda et al., 2006). Much of this remains unexplained and these are unlikely to be the only mechanisms of drag reduction employed by swarming bacteria (Chen et al., 2007).
The flagellar motor rotates with the aid of stator units. Experiments in planktonic cells have shown that motors recruit stator units in greater numbers when the viscous loads are higher (Lele et al., 2013; Tipping et al., 2013; Terahara et al., 2017). Mechano-sensitive stator recruitment is one way to adapt torque in response to increased viscous drag near surfaces (Chawla et al., 2017). However, motor torque in swarming bacteria has never been measured. Torque enables the rotation of extracellular filaments, thereby resulting in a thrust on the cell body. Proper motor and filament assembly is, therefore, the key in coupling torque and cell propulsion. The initial steps in motor assembly include the formation of the FliG, FliM, and FliN complexes that form the flagellar switch (Schuster and Khan, 1994; Zhao et al., 1996; Macnab, 2003; Li and Sourjik, 2011). Concurrently, parts of the flagellar export apparatus begin to form with the final components, FliI, FliH, and FliJ, assembling prior to the formation of the flagellar hook (Kojima and Blair, 2004; Fukumura et al., 2017). The export complexes associate with FliN through the export apparatus component, FliH (McMurry et al., 2006; Paul et al., 2006; Minamino, 2018). Following the assembly of the rod and the flagellar hook, with a remarkable precision (Homma et al., 1990; Cohen et al., 2017), the anti-sigma factor FlgM is exported. This activates FliA, resulting in the transcription of class 3 genes including the flagellin protein, FliC, which forms the extracellular filament (Gillen and Hughes, 1991; Calvo and Kearns, 2015).
Reversals in the direction of rotation of the extracellular filament are mediated by the binding of an intracellular response regulator, CheY-P, to FliM and FliN, which promotes CW rotation in an otherwise CCW-rotary motor (Sarkar et al., 2010; Pandini et al., 2016). Biasing of the flagellar rotation in either the CCW or CW direction is known to cause remodeling of the flagellar switch complex, independent of CheY-P binding (Lele et al., 2012). The number of FliM and FliN subunits is fewer in motors that are locked in the CW direction and higher in the motors that are locked in the CCW direction (Paul et al., 2011b; Lele et al., 2012, 2015b; Branch et al., 2014; Delalez et al., 2014). Estimates indicate that there are numerous putative flagellar preassemblies in a planktonic cell (Sourjik and Berg, 2000; Delalez et al., 2010), although only three to four exhibit complete flagellar assembly. It is unclear if there is a role for the remodeling of FliM and FliN in swarming, however, deficiencies in the flagellar switch assembly do hamper filament formation (Konishi et al., 2009).
Here, we characterized torque generation and flagellar switching in swarmer cells of E. coli. Our measurements with wild-type E. coli indicated that the magnitude of torque generated in swarmer cells was similar to that in planktonic cells. However, the CWbias (fraction of time that the motors rotate CW) was much lower in swarmer cells compared to that in planktonic cells. This reduction was dramatic, with ∼30% of the swarmer cells rotating CCW only. Despite the preference for CCW rotation in the swarmer state, the wild-type strain swarmed on agar substrates, whereas a CCW-only strain was unable. In agreement with earlier reports (Wang et al., 2005), the CCW-only strain could swarm in the presence of additional moisture on the agar surface. Experiments further indicated that transcriptional upregulation of the flagellin gene was neither critical for developing adequate flagellar thrust in the swarmer state nor for swarming. However, a CW-only mutant that rotates its motors exclusively CW due to a mutation in fliG (Togashi et al., 1997) could not swarm irrespective of the surface conditions. Using fluorescence visualization techniques, we found that the FliGCW planktonic cells expressed fewer filaments compared to their wild-type planktonic counterparts. The number of FliGCW filaments was even lower when the strain was grown on agar surfaces, which correlated with a reduction in the number of putative preassembled flagellar complexes. It is possible that flagellar assemblies in agar-grown cells are influenced, at least partially, by the conformations of the monomeric FliG subunits.
Bacterial strains were derived from either RP437 or AW405 parent E. coli strains (Table 1). All plasmids were prepared with pTrc99A vector backbone, unless otherwise noted. Chromosomal alterations were achieved with the λ-red mediated homologous recombination technique (Datsenko and Wanner, 2000).
Overnight cultures were grown from isolated colonies in 5 mL of Tryptone Broth (TB) at 30 °C. Day cultures were grown by diluting 100 μL of overnight culture in 10 mL of fresh TB at 33°C to OD600∼0.5. Swarm-agar plates (Peptone, 10 g/L; NaCl, 5 g/L; beef extract, 3 g/L; 0.45% Eiken Agar, 0.5% Glucose) were prepared fresh, poured, and allowed to dry for 20 min open-faced on the work bench prior to inoculation with the strain of interest (2 μL, overnight culture grown at 30°C). Motility buffer (0.01 M phosphate buffer, 0.067 M NaCl, 10-4 M EDTA, 0.01 M sodium lactate and 1 μM methionine, pH∼7.0) was employed in motility assays.
Cells were prepared for tethered cell-assays by washing several times in motility buffer followed by shearing of the flagella, as described previously (Ford et al., 2017). Flagellar tethering to beads or coverslips was achieved with a sticky fliC mutation (Scharf et al., 1998). Cell rotation was imaged and recorded on a Nikon Eclipse Ti-E with a 20× phase objective or a Nikon Optiphot with a 40× phase objective at ∼60 fps with a CCD camera (DCC1545M-GL, Thorlabs Inc). Bead rotation was imaged on a Nikon Optiphot with a 60× phase objective coupled to a photomultiplier setup (Yuan et al., 2010; Ford et al., 2017).
Swarm assays were carried out in an environmental chamber (ETS Model 5472, Electro-Tech Systems, Inc) that allowed for a precise control over humidity and temperature. Swarm plates were incubated for 8–10 h following inoculation at 75% relative humidity and 30°C, and then allowed to dry open faced for 2–4 h to increase the density of growth before imaging with a gel imager (ChemiDoc Touch Imaging System, BioRad). To add moisture to the agar surface, plates were sprayed with water from a spray bottle until the agar surface was visibly wet. The surface was allowed to dry with the lid open for 1 min before inoculation. The amount of water added in such an approach was 0.38 ± 0.03 mL. As an additional check, swarming was visually confirmed by observing colony movement within a region of the swarm plate with a 40× phase objective on a Nikon Optiphot.
Planktonic cells were diluted in either fresh TB (1:10 dilution) or suspended in motility buffer. The dilute suspension was observed in a standard flow cell and cell motion was recorded away from either surface (coverslip or the microscope slide) at ∼60 fps with a CCD camera (DCC1545M-GL, Thorlabs Inc.) In the case of swarmer cells, cells were recovered from swarm plates by gently pouring 10 mL of motility buffer on the agar surface and swirling to dislodge surface-associated cells. Experiments were also done where cells were recovered specifically from the leading edge of the swarm and from the center of the colony; however, the CWbias was similar in cells recovered from the two areas. The supernatant was collected in a Falcon tube and introduced in a flow cell for observation under a Nikon Optiphot microscope.
A Nikon Ti-E microscope with a 100 mW, 514-nm laser (Cobalt Fandango) focused in the back focal plane of a 60× TIRF objective was used to generate evanescent fields. An Andor iXon DU897 camera was used for capturing TIRF (total internal reflection fluorescence) images while a CCD camera (DCC1545M-GL, Thorlabs Inc.) was used for capturing phase contrast images. Strains carrying fliCcys were labeled with maleimide dye as described elsewhere (Blair et al., 2008). The fliM-eYFP-fliM internal fusion was gifted to us by the Berg lab. The allele carried a [Gly Gly][YFPSer…YFPLys][Ser Gly Gly] insertion between codons 15 and 16 of fliM. Tethered motor assays indicated that the fusion motors were fluorescent and functional.
Optical traps were generated with a 976-nm laser (Azurlight ALS-IR-976-10-I-SF) by overfilling the back-aperture of a 60× objective. Optically trapped beads or cell bodies were then used to physically interrupt the rotation of tethered cells by moving the trapped object in the path of a rotating cell. The trap strength was adequate to prevent rotation of all tethered cells. When the traps were turned off, the cell body was free to rotate again.
Prior to extraction, bacterial strains were grown in fresh liquid swarm medium at 30°C or grown on solid swarm plates as previously described. All swarm assays for directionally biased cells were carried out by adding moisture and cells were only collected from plates that indicated successful swarming. Cell suspensions were centrifuged at 1000 g for 3 min, re-suspended in RNAlater®, and stored at 4°C for 1 day, after which time cells were transferred to -80°C for storage. RNA was extracted using an illustra RNAspin Mini Kit manufactured by GE Life Sciences. RNA was quantified using a NanoDrop 2000c (ThermoFisher Scientific) and a sample was run on a gel to check for ribosomal bands. The RNA was then converted into cDNA using qScript cDNA SuperMix manufactured by Quantabio. Separate reactions were carried out for the gene of interest –fliC (5′GCACCACCAGCATCGTT TGTAGTT3′) and the housekeeping gene –gapA (5′ACCGGTAGAAGACGGGATGATGTT3′).
A LightCycler® 96 Real-Time PCR System (Roche) was employed for real-time PCR. Each reaction (20 μL) was carried out in a 96-well optical grade PCR plate, sealed with optical sealing tape. Amplifications were carried out using SYBR® Green JumpStartTM Taq ReadyMixTM manufactured by Sigma-Aldrich, 2 μL of cDNA, and 250 nM of each primer (see primer information in Supplementary Table S1). Three biological and two technical replicates were carried out for each strain in planktonic and swarming cells. Relative quantification was performed by the ΔΔCT method.
Videos of tethered cells were analyzed with custom-written codes in MATLAB to find the rotational speed as a function of time (Lele et al., 2015a). Time-averaged CWbias values were determined for each tethered motor over the duration of 1–2 min. Mean speeds for cells were determined from Gaussian fits to speed distributions.
Most cells swam in straight lines for limited time periods in the liquid medium. For each cell, the frames over which straight line motion was observed were averaged which resulted in a single image with bright streaks on a gray background. The corresponding length of the straight line intensity profile was determined and divided by the period of observation to obtain swimming speed.
All statistical analyses were performed with the Student’s T-test. Results with p < 0.05 were considered statistically significant.
Previous research indicated that the flagellar switch is able to adapt to mechanical stimuli, although the mechanisms are presently unknown (Fahrner et al., 2003; Lele et al., 2013). To determine how cells adapt flagellar motor functions in order to continue their surface existence, we characterized motor behavior in swarmer cells. We inoculated overnight cultures of a wild-type strain (RP437 background) that carried a sticky fliC allele on standard swarm agar plates. The colonies swarmed and were recovered from the plates several hours later, as discussed in Section “Materials and Methods.” The swarmer cells were then sheared and washed in motility buffer, before tethering to glass surfaces. The CWBias (fraction of time that motors rotate CW) was quantitatively determined from digital recordings of cell rotation with custom-written MATLAB codes (Lele and Berg, 2015). Reversal frequencies were also determined. As shown in Figure 1A, the average CWbias in swarmer cells (left panel) was lower than that in the corresponding planktonic cells (right panel). The distribution in the case of the former was skewed toward a CWbias ∼0, with 36% of cells rotating CCW only. The reversal rates further illustrated this disparity. Figure 1B indicates that the reversal rates were lower in the case of the swarmer cells (left panel) compared to those observed in the planktonic cells (right panel). These observations are consistent with a recent work that observed lower tumbling frequencies in swarmer cells in comparison to planktonic cells grown in a liquid medium (Turner et al., 2016). This result is unexpected considering that switching of the flagella has been reported to be crucial for swarming (Burkart et al., 1997). The lower CWbias likely indicates that the pool of phosphorylated CheY may be reduced in swarmer cells in comparison to planktonic cells, although differential acetylation levels could also play a role (Liarzi et al., 2010).
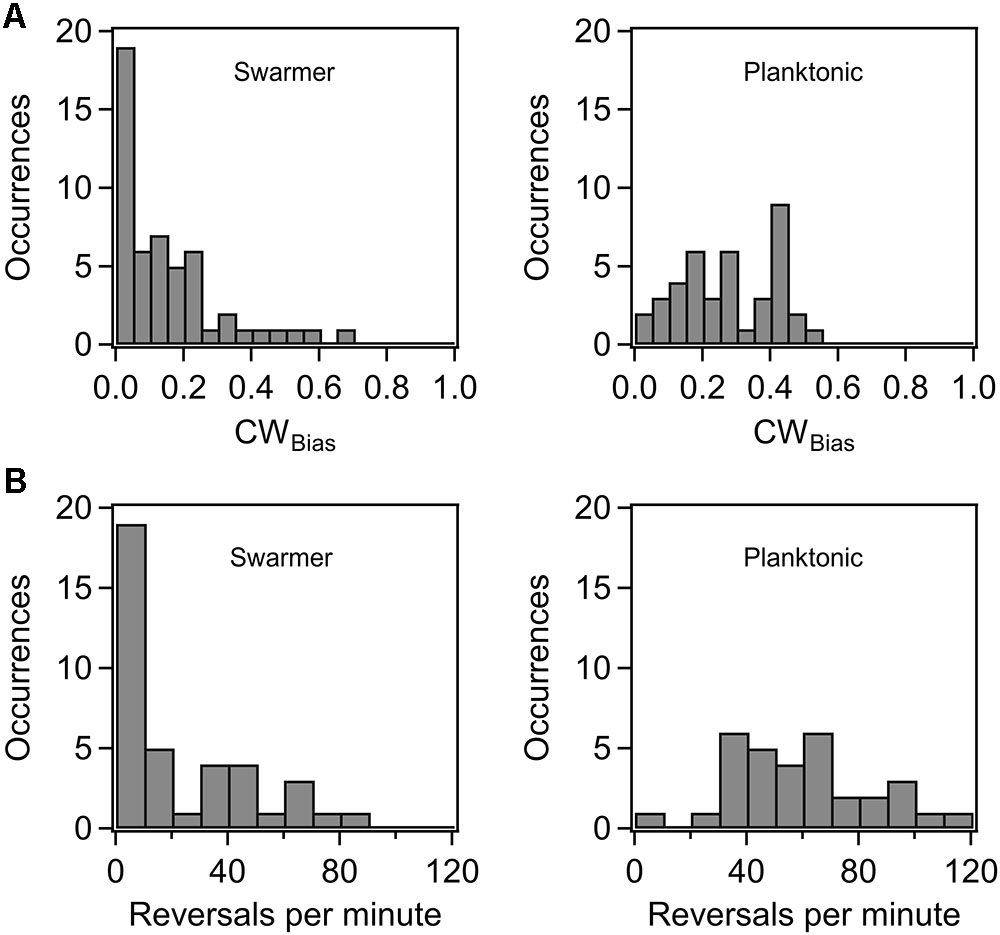
FIGURE 1. (A) Probability of CW rotation in wild-type swarmers and planktonic cells. The mean CWBias was 0.27 ± 0.02 (n = 40 motors) for the wild-type planktonic cells and 0.15 ± 0.02 (n = 52 motors) for the wild-type swarmer cells. The difference in the mean CWBias for planktonic and swarmer cells was significant (p < 0.05). (B) Distribution of motor reversals observed in wild-type swarmer and planktonic cells. The mean reversal rates were 22.4 and 59.9 reversals/min, respectively. The difference in the means was significant (p < 0.05).
To test the role of the flagellar switch in swarming, several directionally biased mutant strains were constructed from the parent AW405 and RP437 wild-type strains. In general, two types of strains were employed in swarm-experiments: a strain lacking cheY in which motors rotated exclusively CCW (CCW strain) and a fliG mutant strain in which motors rotated exclusively CW (FliGCW strain) because the FliG subunits were locked in the CW conformation (Togashi et al., 1997; Lele and Berg, 2015). In addition, a strain lacking cheR-cheB-cheZ was also constructed in which motors rotated predominantly CW (CW strain) due to an excess of [CheY-P] (Sourjik and Berg, 2002). As anticipated, all the directionally biased mutants failed to swarm in a standard swarm assay, unlike the wild-type strains. Swarming was restored in the CCW strain when CheY was expressed from an inducible plasmid. In agreement with earlier reports (Wang et al., 2005), it was possible to restore swarming, at least partially, in the CCW and CW strains by moistening the agar surface with water (see Section “Materials and Methods”; Supplementary Figure S1). Next, the CCW strain was transformed with an inducible vector from which a constitutively active form of CheY (CheYD13KY106W) was expressed (Scharf et al., 1998). Background expression was adequate to predispose tethered cells in this strain to rotate CW-only. It was possible to restore swarming partially in this strain by adding moisture. Conversely, swarming could not be restored in the FliGCW strain. These results suggested that the ability to swarm was not completely inhibited by switch inactivation in strains that carried the wild-type fliG allele. However, the CW conformation of the monomeric FliG subunits in the FliGCW strain probably precluded swarming.
In order to determine if the loss in swarming ability in the directionally biased mutants was, in part, due to an emergent deficiency in torque generation following surface inoculation, we measured the speed of rotation in tethered cells that had been recovered from agar substrates. Based on the size of the cells and the mean speeds, we calculated the average torque generated in the CCW and CW strains (see Section “Materials and Methods”). As indicated in Figure 2A, there were no significant differences in the mean torques generated in the planktonic and the agar-grown cells, irrespective of whether they belonged to the CCW or CW strain. The same was true in the case of the wild-type planktonic and swarmer cells (data not shown). In the FliGCW strain, we measured the mean torque over a range of viscous loads. The viscous load was varied by employing latex beads of different sizes (2, 1, and 0.75 μm) and rotation rates were measured via a photomultiplier-based high speed tracking technique (Yuan et al., 2010; Ford et al., 2017). Torque was calculated from rotation speeds and bead sizes (Ryu et al., 2000). As shown in Figure 2B, the differences between the mean torque generated by the FliGCW motors and that developed by the CW and CCW motors were statistically insignificant at high loads (load = 147.10 pN⋅nm⋅s). At lower loads (19.31 pN⋅nm⋅s), these differences were significant – FliGCW motors produced ∼15% less torque than the CW motors. At the lowest loads, we employed (8.55 pN⋅nm⋅s), the difference between the average torque in FliGCW motors and that in the CW strain was again insignificant, whereas that between the FliGCW motors and the CCW strain was significant, as expected from the anisotropy in torque generation in the two directions of motor rotation (Yuan et al., 2010; Lele et al., 2015b). To assess if the complete inhibition of swarming in the FliGCW strains arose due to such minor variations in torque, we transformed the FliGCW strain with an inducible plasmid carrying the motAmotB genes. Higher expression of MotA–MotB levels increased the average torque in the FliGCW strain to a level that was similar to that in the CCW strain (load = 19.31 pN⋅nm⋅s, Figure 2C). Yet, the same level of induction failed to restore swarming in the FliGCW strain. Together, the results indicated that swarmers are unlikely to develop higher flagellar power compared to planktonic cells at a given viscous load, in order to compensate for the increased surface drag.
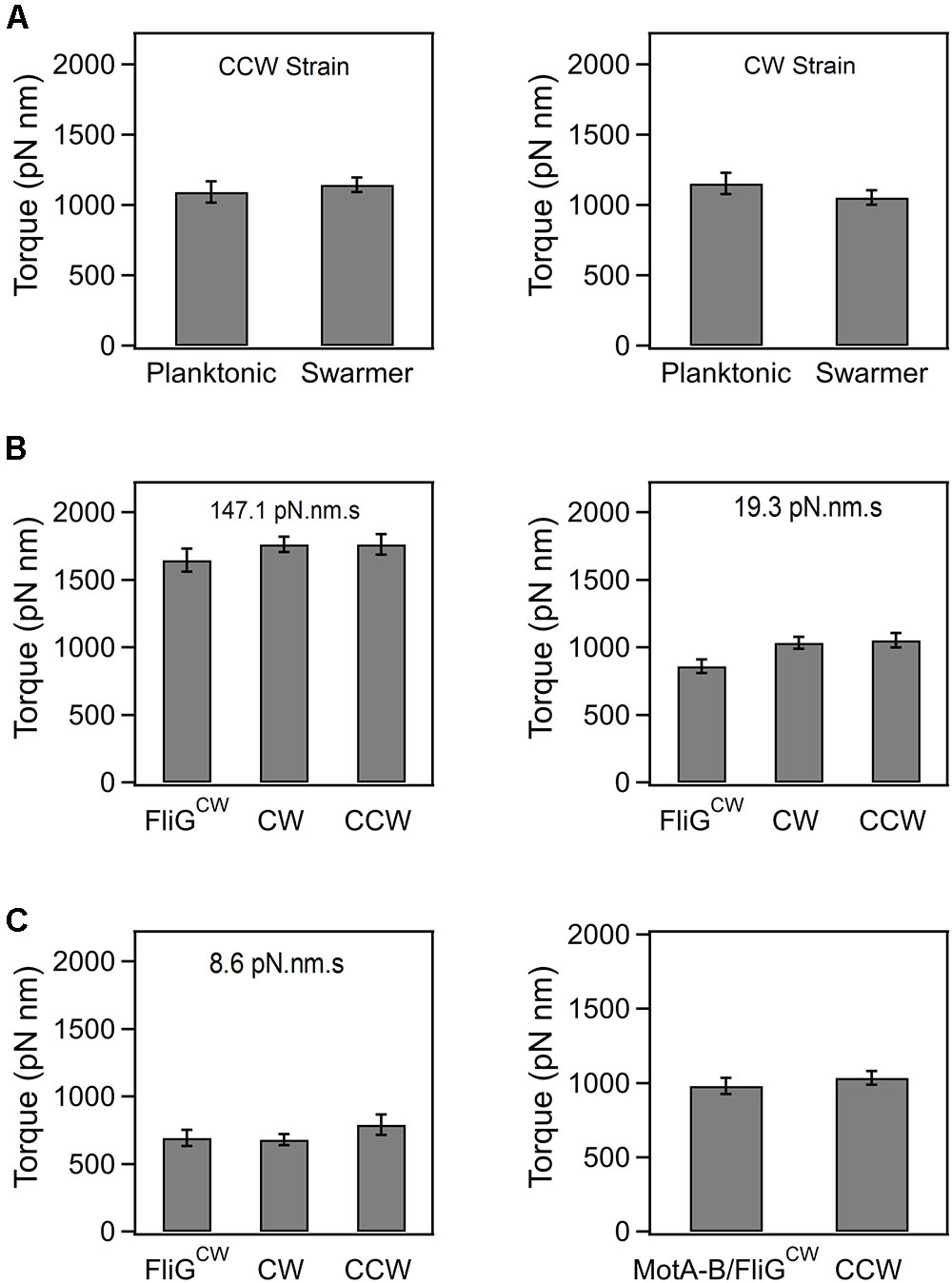
FIGURE 2. (A) Average torque generated in planktonic and swarming cells belonging to the CCW and CW strains. The average torque was 1154.9 ± 75.4 pN⋅nm (n = 35 motors, swarmer CCW strain) and 1121.7 ± 57.9 pN⋅nm (n = 32 motors, planktonic CCW strain). Differences in the mean torques were not significant (p > 0.05). The average torque was 1093 ± 81.5 pN⋅nm (n = 35 motors, swarmer CW strain) and 1144.5 ± 129.1 pN⋅nm (n = 16 motors, planktonic CW strain). Differences in the mean torques were not significant (p > 0.05). (B) Torque generated by the FliGCW strain in comparison to that in CCW and CW strains. At high loads (147.1 pN⋅nm⋅s), the differences in means were statistically insignificant. At medium loads (19.3 pN⋅nm⋅s), the differences in the mean torque in the FliGCW strain and the other strains were significant. At lower loads (8.6 pN⋅nm⋅s), the difference in the mean torque in the FliGCW strain and the CCW strain was significant. (C) The slight degradation in the torque in the FliGCW strain seen in (B) was remedied by expressing extra copies of MotA and MotB subunits in the strain from an inducible plasmid. The difference in mean torques in this strain and the CCW strain was insignificant at a load of 19.3 pN⋅nm⋅s. All plots indicate standard error.
Although our data suggests that differential torque generation is not the reason for the loss of swarming in directionally biased mutants, a reduction in flagellar thrust could play a role. Flagellar thrust depends on a rich interplay between the polymorphic form, arc lengths of the filament, filament numbers, and the ability to form tight flagellar bundles. Rather than making independent measurements of each of these factors, we opted to measure swimming speeds of the cells. The speeds encompass each of the key factors that influence motility, including cell lengths, enabling comparisons between different types of cells. To do this, swimming speeds were measured in the planktonic and agar-grown cultures in all the aforementioned strains. Agar-grown cells were recovered by adding and gently swirling motility buffer on the swarm substrates. Motility was subsequently recorded in standard flow cells and quantitatively analyzed; the data are shown in Table 2. Speeds measured in strains that were able to swarm on agar and were able to swim in motility buffer have been labeled as “swarmer speeds.” Speeds measured in strains that did not swarm but were able to swim in motility buffer have been labeled as “non-swarmer speeds.” Speeds measured in strains that were grown in liquid media have been labeled “planktonic speeds.” There was no significant difference in the mean speeds measured in planktonic cells belonging to the directionally biased strains carrying the native fliG allele and the wild-type cells. However, the difference in the mean speeds in planktonic cells belonging to the FliGCW and the wild-type strains was statistically significant; FliGCW swam at ∼33% lower speed. This indicated degradation in the flagellar thrust in FliGCW planktonic cells. In the case of cells recovered from the agar surfaces, the differences between the mean speeds in FliGCW and the wild type were further amplified. The wild-type swarmer cells experienced an increased flagellar thrust by ∼ 9% relative to the wild-type planktonic cells. By contrast, the agar-grown FliGCW cells were mostly non-motile. Among the ones that exhibited motility, the average swimming speed was ∼1/2 the speed of the wild-type swarmers. In comparison, CCW and wild-type swarmer speeds were similar but the CCW non-swarmer speed was ∼20% lower than the wild-type swarmer speed. This suggested that the inhibition of swarming in the CCW mutant (in the absence of added moisture) was partially attributable to reduced flagellar thrust. Considering that the flagellar motor torque is not deficient, this indicated a degradation in either the flagellar lengths or the number of flagella in the directionally biased mutant. In the case of FliGCW, this deficiency was extreme. In the wild type, there was no significant difference in the swimming speeds of swarmers in the presence or absence of added moisture on the agar surface.
To determine why the flagellar thrust in the FliGCW strain was dramatically lower in comparison to other strains, we employed fluorescence visualization techniques and measured the number of filaments in planktonic as well as agar-grown cells. Filaments carrying cysteine residues were labeled with a maleimide-based fluorescent dye and visualized as detailed in Section “Materials and Methods.” The filament numbers were manually counted from the fluorescence microscopy images. The average number of filaments per cell in the planktonic FliGCW strain was determined to be 3 ± 1 (Figure 3A). The average number in the planktonic wild-type cells was 4 ± 1 (not shown). Thus, there was ∼25% drop in the number of flagella per cell in the planktonic FliGCW cells. This was consistent with a 33% decrease in swimming speeds in the planktonic FliGCW cells. In comparison, the distribution of the number of filaments per cell in the agar-grown FliGCW strain was skewed toward zero (Figure 3A). Of the 200 cells analyzed, 115 cells (57.5%) appeared to carry no filaments. In the cells that had visible flagella, the average number of filaments per cell was 1.6 ± 0.1. In wild-type swarmer cells, only 12 cells out of 115 appeared to carry no filaments. In the cells that had visible flagella, the average number of filaments per cell was 3.0 ± 0.2 (see Supplementary Figure S2), in agreement with recent measurements of wild-type E. coli swarmers by Turner et al. (2016). The reduction in the number of visible filaments in the FliGCW cells was correlated with the complete loss of swarming ability. Additionally, upon visual inspection of these filaments, filaments appeared to be shorter than those found in wild-type cells.
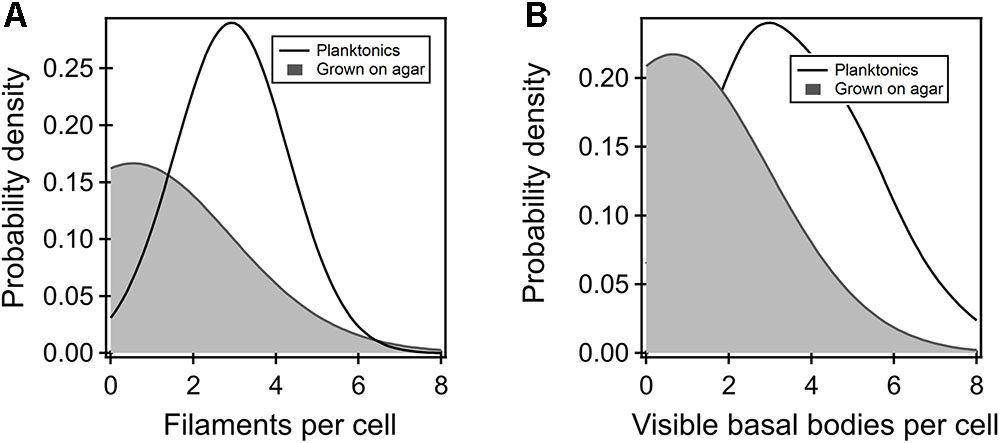
FIGURE 3. (A) Kernel density estimates of the flagellar filaments per cell in the planktonic (black curve) and agar-grown FliGCW strain (gray-shaded region). The difference in means was significant. Filaments were not observed in more than 57% of the agar-grown FliGCW cells. The average filament number observed in the planktonic cells was lower than that observed in wild-type planktonic cells (4 ± 1). (B) The number of putative flagellar preassemblies were determined from TIRF measurements. There were fewer such bodies in the agar-grown FliGCW cells (number of preassemblies = 0.64 ± 0.16, n = 28 cells) relative to the planktonic FliGCW cells (number of preassemblies = 3.72 ± 0.27, n = 29 cells), as seen from the kernel density estimates from the raw data. The difference in the means was significant (p < 0.05).
One explanation for the lower filament numbers in the FliGCW strain, planktonic or otherwise, could be that the flagellum is more susceptible to shear, similar to the reported propensity of the flagella to shear in fliL mutants of Salmonella enterica (Attmannspacher et al., 2008). To test this hypothesis, we employed optical traps to stall tethered motors of the FliGCW strain. Stalling ensures that the flagellar motor delivers the maximum possible force on the rotor (Ryu et al., 2000), thereby subjecting the flagellum to high physiologically relevant shearing forces. Several tethered motors were stalled for approximately 10 min (n = 10 cells), as described in Section “Materials and Methods.” Shearing of the flagellum was expected to be detectable through the detachment of the cell from the surface during stalling or through the loss of the ability to rotate following the removal of the optically trapped bodies. In all of the experiments, flagellar motors in the FliGCW strain remained tethered and functional even after trap removal. This indicated that the FliG mutation is unlikely to result in flagella that are easily sheared under high viscous loads.
We tested whether incomplete or inefficient assembly of the export apparatus could be compensated for by increasing the expression levels of the export ATPase, FliI, or the levels of FlhA. The latter is a part of the export gate of the type 3 secretion system and forms a dock for FliI (Abrusci et al., 2012). FliI overexpression especially has been shown to partially restore flagellation in strains carrying partial deletions in the switch proteins (Konishi et al., 2009; Erhardt and Hughes, 2010). However, overexpression of these two proteins from inducible plasmids failed to restore swarming in the FliGCW strain (Supplementary Figure S3). Similarly, overexpression of FliF and FliGCW also failed to elicit swarming in that strain.
The reduced flagellar thrust and filament numbers could arise due to a reduction in the flagellin levels in the directionally biased mutants. To test this, qPCR experiments were performed as detailed in Section “Materials and Methods.” Briefly, the flagellin gene was selected based on earlier reports that fliC was one of the only genes that is differentially regulated in wild-type swarmers in Salmonella typhimurium (Wang et al., 2004). Comparisons were made between the transcriptional levels of fliC in the planktonic and swarmer cell types for each of the three strains: the wild-type, the CCW, and the FliGCW strain. In the case of the CCW and the FliGCW strain, the cells were recovered from agar substrates that had been treated with water to increase moisture. Swarming was observed in the former but not in the latter strain. FliC mRNA levels were upregulated by twofold in wild-type swarmers, but there was no significant change in the two directionally biased strains (Supplementary Table S2). It was interesting to note that although the flagellin gene was not significantly upregulated in the agar-grown CCW cells, unlike the wild-type, swarming was not inhibited. Flagellar thrust was not diminished in CCW swarmers either. We did not observe a downregulation in flagellin transcription in the agar-grown FliGCW cells (relative to the planktonic cells) despite a clear reduction in the number of filaments. These observations were further supported by swarm experiments where flgM was deleted in the FliGCW strain. The anti-sigma factor, FlgM, binds to FliA and prevents the transcription of class 3 genes. Inefficient functioning of the export apparatus could result in decreased FlgM export, preventing the transcription of flagellar genes. The ΔflgM fliGCW strain, however, failed to swarm; a wild-type strain deleted for flgM retained its swarming ability. Prior observations also suggest that the deletion of flgM in a CCW strain did not restore swarming (Wang et al., 2005). Thus, a reduction in the expression of flagellar genes due to the inactivation of FliA by un-exported FlgM is unlikely to be the reason for the loss of swarming in the directionally biased mutants. Instead, the data pointed to inefficient flagellar assembly as the key problem.
Previous in vivo TIRF measurements with fluorescent fusions of FliM indicate that several putative flagellar preassemblies can be found throughout the cell membrane in planktonic cells of E. coli, although only three to four exist as complete flagella (Sourjik and Berg, 2000; Delalez et al., 2010). These assemblies are identifiable as fluorescent foci (Li and Sourjik, 2011). In each functional motor, there are 34–45 molecules of FliM subunits (Park et al., 2006; Delalez et al., 2010; Paul et al., 2011a; Lele et al., 2012). Remodeling of FliM is dependent on the direction of rotation of the motor and not on the interactions with CheY-P per se (Lele et al., 2012, 2015b). The assembly of FliN is proportionate to that of FliM (Branch et al., 2014; Delalez et al., 2014). The assembly of FliM is not disrupted in the FliGCW mutant; previous observations indicate that the number of FliM subunits per motor in the FliGCW strain is quantitatively similar to that observed in wild-type motors that rotate CW-only due to an excess pool of phosphorylated CheY (Lele et al., 2012; Supplementary Figure S4). Furthermore, incomplete or deficient FliM assembly is known to degrade motor torque (Tang and Blair, 1995), whereas our torque measurements (Figures 2B,C) indicate little or no degradation in torque in FliGCW motors, over the range of viscous loads studied here. It is likely, then, that the reduction in the number of filaments in planktonic cells of the FliGCW strain, relative to the wild-type, is not due to a defective C-ring assembly. Rather, the reduction in filaments could be due to subtle changes in the interactions of the export apparatus with FliG–FliM–FliN protomers that have adopted a locked CW conformation.
Next, we attempted to determine whether the dramatic reduction in the number of filaments in FliGCW cells grown on agar surfaces, relative to the planktonic FliGCW cells, was entirely due to deficient flagellar export or whether the locked conformations of the FliG monomers inhibited the assembly of putative flagellar complexes on agar substrates. For this purpose, a FliGCW strain was constructed that carried a genomic fliM-eYFP-fliM allele (Section “Materials and Methods”). Motors in this strain were functional and rotated CW-only when tethered. The strain was grown in liquid media and on agar surfaces. TIRF visualization enabled quantitative determination of the number of putative flagellar complexes with custom-written MATLAB codes for fluorescent foci detection (Lele et al., 2012). The distribution of the number of foci detected per cell is indicated in Figure 3B for the planktonic and agar-grown FliGCW cells. The average numbers of foci observed here are fewer than those reported earlier (Delalez et al., 2010). This is likely because the TIRF field employed only allows visualization of ∼1/6th of the total volume of the cell body. As can be seen, there were less than half the number of foci in the agar-grown cells (n = 28 cells) when compared with the planktonic cells (n = 29 cells). This is consistent with our observations of very few filaments in the agar-grown FliGCW cells, and the loss of swarming in this strain. To further test this notion, we attempted to construct the corresponding wild-type control for fluorescence assays. However, that strain was unable to swarm and as a result, was not employed in further experimentation. Nonetheless, to test whether the FliGCW mutation, rather than the CW-locked conformation of the FliG monomers, was responsible for the reduction in the foci on agar, we constructed and tested a strain that carried the native fliG and the fliM-eYFP-fliM alleles on its chromosome, and an excess of the constitutively active CheY variant. Motors in this strain rotated CW-only and the strain did not swarm. A similar reduction in the number of foci in agar-grown cells was observed in comparison to the corresponding planktonic cells (Supplementary Figure S5). This suggested that it is not the FliGCW mutation per se that interfered with the assembly of the putative flagellar assemblies in cells grown on agar surfaces, but possibly the lack of conformational transitions in the switch protomers. These experiments provide a measure of support to the notions that in cells that fail to swarm on agar, the number of putative flagellar preassemblies is decreased relative to that in the planktonic cells, and that the export of flagellar proteins is influenced by the conformations of the switch protomers.
Our results suggest that the flagellar motors in swarming cells of E. coli do not develop a higher power relative to the planktonic motors, for a given viscous load. This is in contrast to species such as Pseudomonas aeruginosa, where cells employ a specialized set of stator proteins that are capable of developing higher power when the cell finds itself near a surface (Kuchma et al., 2015). Unexpectedly, the CWbias was lower in swarmer cells compared to that in planktonic cells, in fact, one out of three swarmer cells showed no inclination to reverse the direction of flagellar rotation. This is likely due to a smaller pool of phosphorylated CheY in the swarmer cells. This may be a consequence of an overall reduction in the chemotaxis protein abundances, or a reduction in the sensitivity of the flagellar switch. In any scenario, the reduction in reversal rates and CWbias is at odds with the swarm assays that clearly emphasize the role of flagellar switching in swarming. We propose, then, that it is not switching per se, but rather some associated property of the wild type that enables swarming.
Our experiments also indicate that the transcriptional upregulation of the flagellin gene was neither critical for developing adequate flagellar thrust in the swarmer state nor for swarming. This conclusion was derived from the observation that the CCW strain did not upregulate the expression of the flagellin gene, but was still able to swarm, provided that moisture was added. Swarmer cells belonging to the CCW strain were also able to generate similar flagellar thrusts as the wild-type swarmer cells. This suggested that the upregulation of fliC expression might simply be a consequence of agar-based growth of the wild-type strain. Transcriptional activity was also unchanged between the planktonic and swarmer cells belonging to the FliGCW strain. The deletion of flgM did not restore swarming in this strain. Considering that this strain was severely deficient in producing flagella on the agar substrate, this further suggested that the extreme defects in flagellar production on agar were unlikely due to reduced fliC expression. Optical tweezer experiments ruled out the possibility that the reduction in flagellar numbers in the FliGCW strain was due to shearing near the agar surface. The fliGCW mutation in planktonic cells of E. coli did not interfere with the assembly and functioning of the flagellar C-rings, as evidenced by previous measurements of FliM assembly in a FliGCW strain (Lele et al., 2012), and the torque measurements in the present work (Figures 2B,C). This suggested that the CW-locked conformation of the FliG–FliM–FliN protomers, rather than assembly defects, affected flagellar export in this strain. Finally, we also found evidence that the assembly of putative FliM assemblies in the FliGCW cells was inhibited on agar surfaces, relative to the planktonic state. This decrease correlated with reduced flagellar numbers on agar surfaces, which was most likely responsible for the degradation in flagellar thrust and the loss of swarming. However, the corresponding wild-type fusion strain failed to swarm, which prevented us from determining if a high density of preassemblies was maintained in wild-type swarmers. Based on the available data, we speculate that flagellin export depends on the switch-activity, and not just assembly, and that the stochastic transitions in the conformations of switch protomers likely help anneal putative preassemblies. Under the conditions of increased shear near agar surfaces, locked protomeric conformations might result in inadequate assembly and inefficient export of flagellar substrates.
KF, JA, AN, and MJ performed the experiments and analyses. KF, AN, and PL developed the experimental setups and designed the research. KF, JA, and PL wrote the manuscript. All authors reviewed the manuscript.
This research was funded by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM123085. KF was supported by NSF-GRP, DGE 1252521. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation and the National Institute of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge helpful advice by Dr. Michael Manson. We are also grateful to Dr. Howard Berg for strains and plasmids.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02197/full#supplementary-material
Abrusci, P., Vergara-Irigaray, M., Johnson, S., Beeby, M. D., Hendrixson, D. R., Roversi, P., et al. (2012). Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104. doi: 10.1038/nsmb.2452
Attmannspacher U., Scharf, B. E., and Harshey, R. M. (2008). FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Molx. Microbiol. 68, 328–341. doi: 10.1111/j.1365-2958.2008.06170.284590
Belas, R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. doi: 10.1016/j.tim.2014.05.002
Blair, K. M., Turner, L., Winkelman, J. T., Berg, H. C., and Kearns, D. B. (2008). A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320, 1636–1638. doi: 10.1126/science.1157877
Branch, R. W., Sayegh, M. N., Shen, C., Nathan, V. S. J., and Berg, H. C. (2014). Adaptive remodelling by FliN in the bacterial rotary motor. J. Mol. Biol. 426, 3314–3324. doi: 10.1016/j.jmb.2014.07.009
Burkart, M., Toguchi, A., Harshey, R. M., (1997). The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 2568–2573. doi: 10.1073/pnas.95.5.2568
Butler, M. T., Wang, Q., and Harshey, R. M. (2010). Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U.S.A. 107, 3776–3781. doi: 10.1073/pnas.0910934107
Callegan, M. C., Novosad, B. D., Ramirez, R., Ghelardi, E., and Senesi, S. (2006). Role of swarming migration in the pathogenesis of Bacillus Endophthalmitis. Invest. Ophthalmol. Vis. Sci. 47, 4461–4467. doi: 10.1167/iovs.06-0301
Calvo, R. A., and Kearns, D. B. (2015). FlgM Is secreted by the flagellar export apparatus in Bacillus subtilis. J. Bacteriol. 197, 81–91. doi: 10.1128/JB.02324-14
Chawla, R., Ford, K. M., and Lele, P. P. (2017). Torque, but not FliL, regulates mechanosensitive flagellar motor-function. Sci. Rep. 7:5565. doi: 10.1038/s41598-017-05521-8
Chen, B. G., Turner, L., and Berg, H. C. (2007). The wetting agent required for swarming in Salmonella enterica serovar typhimurium is not a surfactant. J. Bacteriol. 189, 8750–8753. doi: 10.1128/JB.01109-07
Cohen, E. J., Ferreira, J. L., Ladinsky, M. S., Beeby, M., and Hughes, K. T. (2017). Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356, 197–200. doi: 10.1126/science.aam6512
Darnton, N. C., Turner, L., Rojevsky, S., and Berg, H. C. (2010). Dynamics of bacterial swarming. Biophys. J. 98, 2082–2090. doi: 10.1016/j.bpj.2010.01.053
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Delalez, N. J., Berry, R. M., and Armitage, J. P. (2014). Stoichiometry and turnover of the bacterial flagellar switch protein FliN. mBio 5:e01216-14. doi: 10.1128/mBio.01216-14
Delalez, N. J., Wadhams, G. H., Rosser, G., Xue, Q. A., Brown, M. T., Dobbie, I. M., et al. (2010). Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. U.S.A. 107, 11347–11351. doi: 10.1073/pnas.1000284107
Erhardt, M., and Hughes, K. T. (2010). C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol. Microbiol. 75, 376–393. doi: 10.1111/j.1365-2958.2009.06973.x
Fahrner, K. A., Ryu, W. S., and Berg, H. C. (2003). Biomechanics: bacterial flagellar switching under load. Nature 423:938. doi: 10.1038/423938a
Ford, K. M., Chawla, R., and Lele, P. P. (2017). Biophysical characterization of flagellar motor functions. J. Vis. Exp. 119:e55240. doi: 10.3791/55240
Fukumura, T., Makino, F., Dietsche, T., Kinoshita, M., Kato, T., Wagner, S., et al. (2017). Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 15:e2002281. doi: 10.1371/journal.pbio.2002281
Gillen, K. L., and Hughes, K. T. (1991). Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 173, 2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991
Harshey, R. M. (2003). Bacterial Motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Harshey, R. M., and Matsuyama, T. (1994). Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U.S.A. 91, 8631–8635. doi: 10.1073/pnas.91.18.8631
Henrichsen, J. (1972). Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36, 478–503.
Homma, M., Kutsukake, K., Hasebe, M., Iino, T., and Macnab, R. M. (1990). FlgB, FlgC, FlgF and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211, 465–477. doi: 10.1016/0022-2836(90)90365-S
Hughes, A. C., and Kearns, D. B. (2017). “Swimming, swarming and sliding motility in bacillus subtilis. Bacillus,” in Cellular and Molecular Biology, 3 Edn, ed. P. L. Graumann (Poole: Caister Academic Press), 415–438. doi: 10.21775/9781910190579-14
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kim, W., Killam, T., Sood, V., and Surette, M. G. (2003). Swarm-cell differentiation in Salmonellaenterica Serovar Typhimurium results in elevated resistance to multiple antibiotics. J. Bacteriol. 185, 3111–3117. doi: 10.1128/JB.185.10.3111-3117.2003
Kim, W., and Surette, M. G. (2003). Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol. Proc. Online 5, 189–196. doi: 10.1251/bpo61
Kojima, S., and Blair, D. F. (2004). The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233, 93–134. doi: 10.1016/S0074-7696(04)33003-2
Konishi, M., Kanbe, M., McMurry, J. L., and Aizawa, S. I. (2009). Flagellar formation in C-ring-defective mutants by overproduction of FliI, the atpase specific for flagellar type III secretion. J. Bacteriol. 191, 6186–6191. doi: 10.1128/JB.00601-09
Kuchma, S. L., Delalez, N. J., Filkins, L. M., Snavely, E. A., Armitage, J. P., and O’Toole, G. A. (2015). Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J. Bacteriol. 197, 420–430. doi: 10.1128/JB.02130-14
Lauga, E., DiLuzio, W. R., Whitesides, G. M., and Stone, H. A. (2006). Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 90, 400–412. doi: 10.1529/biophysj.105.069401
Lele, P. P., and Berg, H. C. (2015). Switching of bacterial flagellar motors triggered by mutant flig. Biophys. J. 108, 1275–1280. doi: 10.1016/j.bpj.2015.02.004
Lele, P. P., Branch, R. W., Nathan, V. S. J., and Berg, H. C. (2012). Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc. Natl. Acad. Sci. U.S.A. 109, 20018–20022. doi: 10.1073/pnas.1212327109
Lele, P. P., Hosu, B. G., and Berg, H. C. (2013). Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 110, 11839–11844. doi: 10.1073/pnas.1305885110
Lele, P. P., Roland, T., Shrivastava, A., Chen, Y., and Berg, H. C. (2015a). The flagellar motor of Caulobacter crescentus generates more torque when a cell swims backwards. Nat. Phys. 12, 175–178.
Lele, P. P., Shrivastava, A., Roland, T., and Berg, H. C. (2015b). Response thresholds in bacterial chemotaxis. Science Advances 1, e1500299. doi: 10.1126/sciadv.1500299
Li, H., and Sourjik, V. (2011). Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80, 886–899. doi: 10.1111/j.1365-2958.2011.07557.x
Liarzi, O., Barak, R., Bronner, V., Dines, M., Sagi, Y., Shainskaya, A., et al. (2010). Acetylation represses the binding of CheY to its target proteins. Mol. Microbiol. 76, 932–943. doi: 10.1111/j.1365-2958.2010.07148.x
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Mariconda, S., Wang, Q., and Harshey, R. M. (2006). A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60, 1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x
McMurry, J. L., Murphy, J. W., and González-Pedrajo, B. (2006). The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45, 11790–11798. doi: 10.1021/bi0605890
Minamino, T. (2018). Hierarchical protein export mechanism of the bacterial flagellar type III protein export apparatus. FEMS Microbiol. Lett. 365:fny117. doi: 10.1093/femsle/fny117
Mobley, H. L. T., and Belas, R. (1995). Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3, 280–284. doi: 10.1016/S0966-842X(00)88945-3
Overhage, J., Bains, M., Brazas, M. D., and Hancock, R. E. (2008). Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190, 2671–2679. doi: 10.1128/JB.01659-07
Pandini, A., Morcos, F., and Khan, S. (2016). The gearbox of the bacterial flagellar motor switch. Structure 24, 1209–1220. doi: 10.1016/j.str.2016.05.012
Park, S. Y., Lowder, B., Bilwes, A. M., Blair, D. F., and Crane, B. R. (2006). Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. U.S.A. 103, 11886–11891. doi: 10.1073/pnas.0602811103
Paul, K., Brunstetter, D., Titen, S., and Blair, D. F. (2011a). A molecular mechanism of direction switching in the flagellar motor of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 17171–17176. doi: 10.1073/pnas.1110111108
Paul, K., Gonzalez-Bonet, G., Bilwes, A. M., Crane, B. R., and Blair, D. (2011b). Architecture of the flagellar rotor. EMBO J. 30, 2962–2971. doi: 10.1038/emboj.2011.188
Paul, K., Harmon, J. G., and Blair, D. F. (2006). Mutational Analysis of the Flagellar Rotor Protein FliN: Identification of Surfaces Important for Flagellar Assembly and Switching. J. Bacteriol. 188, 5240–5248. doi: 10.1128/JB.00110-06
Ryu, W. S., Berry, R. M., and Berg, H. C. (2000). Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447. doi: 10.1038/35000233
Sarkar, M. K., Paul, K., and Blair, D. (2010). Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 9370–9375. doi: 10.1073/pnas.1000935107
Scharf, B. E., Fahrner, K. A., Turner, L., and Berg, H. C. (1998). Control of direction of flagellar rotation in bacterial?chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 95, 201–206. doi: 10.1073/pnas.95.1.201
Schuster, S. C., and Khan, S. (1994). The bacterial flagellar motor. Annu. Rev. Biophys. Biomol. Struct. 23, 509–539. doi: 10.1146/annurev.bb.23.060194.002453
Sourjik, V., and Berg, H. C. (2000). Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37, 740–751. doi: 10.1046/j.1365-2958.2000.02044.x
Sourjik, V., and Berg, H. C. (2002). Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 99, 12669–12674. doi: 10.1073/pnas.192463199
Tang, H., and Blair, D. F. (1995). Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J. Bacteriol. 177, 3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995
Terahara, N., Noguchi, Y., Nakamura, S., Kami-ike, N., Ito, M., Namba, K., et al. (2017). Load- and polysaccharide-dependent activation of the Na+-type MotPS stator in the Bacillus subtilis flagellar motor. Sci. Rep. 7:46081. doi: 10.1038/srep46081
Tipping, M. J., Delalez, N. J., Lim, R., Berry, R. M., and Armitage, J. P. (2013). Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13
Togashi, F., Yamaguchi, S., Kihara, M., Aizawa, S. I., and Macnab, R. M. (1997). An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179, 2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997
Turner, L., Ping, L., Neubauer, M., and Berg, H. C. (2016). Visualizing flagella while tracking bacteria. Biophys. J. 111, 630–639. doi: 10.1016/j.bpj.2016.05.053
Tuson, H. H., Copeland, M. F., Carey, S., Sacotte, R., and Weibel, D. B. (2013). Flagellum density regulates Proteus mirabilis swarmer cell motility in viscous environments. J. Bacteriol. 195, 368–377. doi: 10.1128/JB.01537-12
Wang, Q., Frye, J. G., McClelland, M., and Harshey, R. M. (2004). Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52, 169–187. doi: 10.1111/j.1365-2958.2003.03977.x
Wang, Q., Suzuki, A., Mariconda, S., Porwollik, S., and Harshey, R. M. (2005). Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24, 2034–2042.
Yang, A., Tang, W. S., Si, T., and Tang, J. X. (2017). Influence of physical effects on the swarming motility of Pseudomonas aeruginosa. Biophys. J. 112, 1462–1471. doi: 10.1016/j.bpj.2017.02.019
Yuan, J., Fahrner, K. A., Turner, L., and Berg, H. C. (2010). Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc Natl Acad Sci U.S.A. 107, 12846–12849. doi: 10.1073/pnas.1007333107
Zhang, H. P., Be’er, A., Smith, R. S., Florin, E.-L., and Swinney, H. L. (2009). Swarming dynamics in bacterial colonies. Europhys. Lett. 87:48011. doi: 10.1209/0295-5075/87/48011
Keywords: flagellar motor, motility, FliG, optical tweezers, chemotaxis
Citation: Ford KM, Antani JD, Nagarajan A, Johnson MM and Lele PP (2018) Switching and Torque Generation in Swarming E. coli. Front. Microbiol. 9:2197. doi: 10.3389/fmicb.2018.02197
Received: 15 June 2018; Accepted: 28 August 2018;
Published: 18 September 2018.
Edited by:
Dacheng Ren, Syracuse University, United StatesReviewed by:
Florian Altegoer, Philipps-Universität Marburg, GermanyCopyright © 2018 Ford, Antani, Nagarajan, Johnson and Lele. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pushkar P. Lele, cGxlbGVAdGFtdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.