- Institut de Recherche pour le Développement, Assistance Publique – Hôpitaux de Marseille, Microbes, Evolution, Phylogeny and Infection, and Institut Hospitalo-Universitaire – Méditerranée Infection, Aix-Marseille Université, Marseille, France
Acanthamoeba are ubiquitous phagocytes predominant in soil and water which can ingest many microbes. Giant viruses of amoebae are listed among the Acanthamoeba-resisting microorganisms. Their sympatric lifestyle within amoebae is suspected to promote lateral nucleotide sequence transfers. Some Acanthamoeba species have shown differences in their susceptibility to giant viruses. Until recently, only the genome of a single Acanthamoeba castellanii Neff was available. We analyzed the draft genome sequences of Acanthamoeba polyphaga through several approaches, including comparative genomics, phylogeny, and sequence networks, with the aim of detecting putative nucleotide sequence exchanges with giant viruses. We identified a putative sequence trafficking between this Acanthamoeba species and giant viruses, with 366 genes best matching with viral genes. Among viruses, Pandoraviruses provided the greatest number of best hits with 117 (32%) for A. polyphaga. Then, genes from mimiviruses, Mollivirus sibericum, marseilleviruses, and Pithovirus sibericum were best hits in 67 (18%), 35 (9%), 24 (7%), and 2 (0.5%) cases, respectively. Phylogenetic reconstructions showed in a few cases that the most parsimonious evolutionary scenarios were a transfer of gene sequences from giant viruses to A. polyphaga. Nevertheless, in most cases, phylogenies were inconclusive regarding the sense of the sequence flow. The number and nature of putative nucleotide sequence transfers between A. polyphaga, and A. castellanii ATCC 50370 on the one hand, and pandoraviruses, mimiviruses and marseilleviruses on the other hand were analyzed. The results showed a lower number of differences within the same giant viral family compared to between different giant virus families. The evolution of 10 scaffolds that were identified among the 14 Acanthamoeba sp. draft genome sequences and that harbored ≥ 3 genes best matching with viruses showed a conservation of these scaffolds and their 46 viral genes in A. polyphaga, A. castellanii ATCC 50370 and A. pearcei. In contrast, the number of conserved genes decreased for other Acanthamoeba species, and none of these 46 genes were present in three of them. Overall, this work opens up several potential avenues for future studies on the interactions between Acanthamoeba species and giant viruses.
Introduction
Acanthamoeba spp. (Eukaryota, Amoebozoa, Acanthamoebidae) are among the most predominant protozoa in soil and water (Rodríguez-Zaragoza, 1994). These amoebae are found in natural or artificial habitats, mostly humid ones, such as the marine environment, sediments, salt lakes, cooling towers, stagnant water, treatment plant sewage, drinking water, or soil (Rodríguez-Zaragoza, 1994). Their ubiquity in water and soil promotes contacts with animals including humans (Sherr and Sherr, 2002; Marciano-cabral and Cabral, 2003). Moreover, Acanthamoeba spp. are phagocytic protists that can ingest all particles with a size > 0.5 μm, which includes notably bacteria and fungi (Raoult and Boyer, 2010). Whereas most of these microorganisms are degraded post-internalization, some are able to survive and multiply (Raoult and Boyer, 2010). They are known as amoeba-resisting microorganisms (ARMs) (Greub and Raoult, 2004). Examples of ARMs include human pathogens such as Legionella pneumophila or Mycobacterium sp., which also resist degradation by macrophages (Greub and Raoult, 2004; Salah et al., 2009).
Due to their giant size, giant viruses of amoebae are ARMs that multiply inside Acanthamoeba polyphaga and A. castellanii (Colson et al., 2017b). These amoebae have been used in laboratory settings for giant virus isolation during the last 14 years since the discovery of their first representative, Mimivirus (La Scola et al., 2003; Raoult et al., 2004; Colson et al., 2017a). Mimivirus led to the creation of the family Mimiviridae and to the discovery of many other giant viruses of amoebae, as well as virophages that replicate in mimivirus factories, and transpovirons integrated in mimivirus genomes (Colson et al., 2017a). Until now, three new families of amoeba-infecting viruses have been described, which include Mimiviridae, Marseilleviridae, and Lavidaviridae, as well as seven new putative lineages consisting in pandoraviruses, pithoviruses, faustoviruses, Mollivirus sibericum, Kaumoebavirus, cedratviruses and Pacmanvirus (Colson et al., 2017b). These giant viruses are commonly found in environmental water and soil and have a broad geographical distribution. They differ from classical viruses and have a complexity similar to that of other microbes. Emblematically, the virions are visible with an optical microscope and can reach 1.5 μm in size, and their genomes harbor between 444 (for a marseillevirus) and 2,544 (for a pandoravirus) genes (Raoult et al., 2004; Colson et al., 2017a).
The hosting by Acanthamoeba spp. of several ARMs living sympatrically confers to these microorganisms increased opportunities to exchange sequences between each other and with the amoebal host (Greub and Raoult, 2004; Raoult and Boyer, 2010; Bertelli and Greub, 2012). This is suspected to promote the broad mosaicism and large size of the genomes of giant viruses of amoebae that are in close vicinity with other viruses and microorganisms inside Acanthamoeba. It has also been observed that bacteria and viruses living in sympatry in Acanthamoeba harbored larger genomes than their closest relatives with an allopatric lifestyle (Raoult and Boyer, 2010). Such sequence exchanges may allow accumulating a substantial gene armory to multiply and compete with other amoeba-resisting microorganisms (Boyer et al., 2011). Consistently, culturing Mimivirus in allopatric conditions on ARM-free Acanthamoeba led to a 16% reduction of the viral genome after 100 passages (Boyer et al., 2011).
According to recent findings, the species A. castellanii and A. polyphaga, which were those used to isolate giant viruses of amoebae, have different levels of tolerance to viruses from different or even similar lineages (Dornas et al., 2015). We observed that some virus lineages relied on specific Acanthamoeba species for their replication. For example, pandoraviruses and pithoviruses were only isolated on A. castellanii and faustoviruses were only isolated on Vermamoeba vermiformis (Khalil et al., 2016; Reteno et al., 2015). Moreover, different mimivirus isolates were obtained from the same sample with different Acanthamoeba species (Dornas et al., 2015). These data raise some questions about the relationship between Acanthamoeba species and giant viruses of amoebae.
Giant viruses of amoebae have raised a radically new issue regarding their genomic content. Since the description of the Mimivirus, the question of the origin of their genes has arisen. From the onset, genes corresponding to nucleotide sequences putatively transferred from amoebae to viruses were identified, and it was proposed that giant viruses were essentially bags of exogenous genes (Filée et al., 2007; Moreira and Brochier-Armanet, 2008). This assertion is only partially true, since several genes were suspected to be shared by several giant viruses, and the proportion of ORFan genes, for which no source is identified, remains very high in giant viruses (Colson et al., 2017b). Nucleotide sequences from bacterial viruses (bacteriophages), viruses of viruses (virophages), archaeal viruses, or eukaryotic viruses can possibly be transferred inside their host genomes. Regarding amoebae, at present, nucleotide sequence transfers are mostly studied by comparing their genomes to those of giant viruses. A. castellanii was the only Acanthamoeba species for which we had draft genome sequences (from 2010 to 2015), which was completed using transcriptomic data (Clarke et al., 2013). The study of this genome led to infer sequence exchanges with other eukaryotes and with archaea, bacteria, and viruses of the proposed order Megavirales, which comprises the formerly described Nucleocytoplasmic large DNA viruses (NCLDVs) and giant viruses of amoebae (Yutin et al., 2009; Colson et al., 2013). In this work, we sought to detect and characterize putative nucleotide sequence transfers involving giant virus genomes and the draft genome sequences of a second Acanthamoeba species, A. polyphaga, which has been one of the most used to isolate giant viruses of amoebae.
Materials and Methods
Gene Content of Giant Viruses
The study of the gene trafficking between A. polyphaga and giant viruses was carried out by using genes of all annotated genomes described for these viruses at the time of our analysis (including mimiviriruses, marseilleviruses, pandoraviruses, Pithovirus sibericum, Mollivirus sibericum, faustoviruses, phycodnaviruses, ascoviruses, iridoviruses, asfarviruses and poxviruses) (Supplementary Table S1).
Draft Genome Sequences of Acanthamoeba polyphaga and Other Acanthamoeba Species
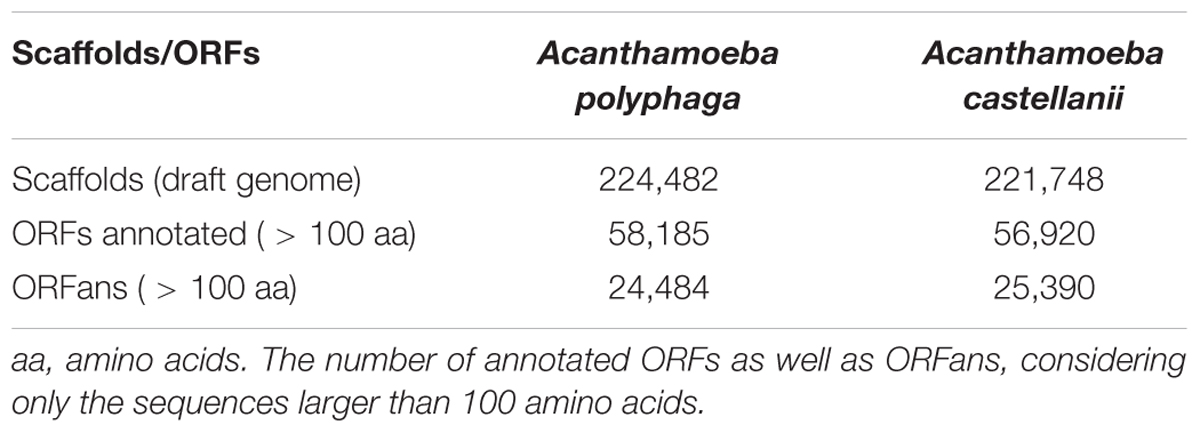
The draft genome of A. polyphaga ATCC 30872 is publicly available on the NCBI website1 (accession: PRJEB7687). It is part of the project «Phylogenomics of Acanthamoeba species» (Institute of Integrative Biology, University of Liverpool), along with the draft genomes of 13 other Acanthamoeba species. The A. polyphaga ATCC 30872 draft genome contains 224,482 scaffolds with a total length of 120.6 megabases (Mb). The 14,974 genes identified in the genome of A. castellanii Neff with the support of transcriptomic data (Clarke et al., 2013) were used for comparative genomic analyses.
Optimization of Assembly of the Acanthamoeba Draft Genome Sequences
We used the CLC Genomics Workbench software (version 7.5)2 to reassemble the draft genome sequences of A. polyphaga and some other Acanthamoeba species, including another A. castellanii strain (ATCC 50370). The default kmer size of 50 was used.
Gene Prediction, Functional Annotation, and Analysis of Taxonomical Distribution
Gene prediction for the draft genome sequences of A. polyphaga and other Acanthamoeba species was performed using the Prodigal program, as we searched for sequences best matching those of giant viruses. This tool identifies ribosome binding sites to localize translation initiation positions and localizes precisely the 3′ end of each gene (Hyatt et al., 2010). The hits identified as those best matching with giant virus genes were then checked and compared with the 47,246 genes predicted for A. polyphaga using GeneMarkES, a program developed specifically for eukaryotes (Lomsadze et al., 2005). For the functional annotation, sequences homologous with ORFs predicted from non-redundant scaffolds were searched for in the NCBI GenBank protein sequence database (nr) using the BLASTp program (Altschul et al., 1990). In addition, ORFs were identified through BLASTp searches (with an e-value threshold of 0.1) in the database of Clusters of Orthologous Groups of proteins (COGs) of the NCBI (Tatusov, 2000). Taxonomical origins were determined using MEGAN6 (Huson et al., 2016).
Comparative Genomic Analyses
Predicted protein sequences of A. polyphaga were compared with those from giant viruses and those predicted from the draft genome sequences of the 13 other species of Acanthamoeba (Supplementary Table S2). The orthologous genes (≥100 amino acids) of A. polyphaga and the other Acanthamoeba were identified using the Proteinortho program (Lechner et al., 2011). Phylogenetic reconstructions were then performed using two sets of aligned sequences. First, 16 draft genome sequences of Acanthamoeba strains classified in 14 species including two A. polyphaga strains and two A. castellanii strains were aligned by using the progressive Mauve program (Darling et al., 2010). Second, the 18S ribosomal DNA sequences from 33 different Acanthamoeba strains were retrieved, including those of the 16 Acanthamoeba strains classified in 14 Acanthamoeba species. The 18S ribosomal DNA sequences were obtained by sequencing the complete 18S ribosomal DNA from strains available in our laboratory, and for non-available strains, the sequences were retrieved from the NCBI GenBank database or directly from the Acanthamoeba draft genome sequences. Nucleotide sequences alignments were performed with the MUSCLE program (Edgar, 2004). Phylogenetic trees were constructed using FastTree (Price et al., 2010).
We investigated more specifically any possible occurrences of horizontal gene transfers (HGT), i.e., the gene trafficking between this amoeba and giant viruses. A. polyphaga proteins which had as significant hit a giant virus sequence were used as queries to search into the NCBI GenBank non-redundant protein sequence database (nr). Phylogenetic analyses were performed to confirm suspicion of HGT for the genes showing the highest level of sequence similarity with a viral homolog. Amino acid sequences alignments were performed with the MUSCLE program. Phylogenetic trees were constructed using FastTree. Ancestral major capsid protein (MCP) sequences were predicted using the MEGA6 program3. Additionally, similarity searches were performed by using the tBLASTn program for all ORFs of giant viruses, virophages and transpovirons against the draft genome sequences of A. polyphaga. The following criteria were used: a percentage of amino acid identity ≥ 30%; an e-value ≤ 1e-2; and a percentage of coverage of aligned sequences ≥ 30%. Finally, results from similarity searches were formatted to create networks of gene trafficking using the Cytoscape tool (Smoot et al., 2011). This software was also used to generate a network between protein sequences from two giant viruses of amoebae, Pandoravirus dulcis and Pandoravirus salinus, and from the draft genome sequences of A. polyphaga and A. castellanii ATCC 50370. Finally, a ‘rhizome’ of genes was determined for a few A. polyphaga genes whose best hit was a giant virus. This information was obtained by performing BLASTp searches with fragments of this gene obtained by fenestrating its amino acid sequence with a window of 40 amino acids and a sliding step of 20 amino acids. The representation of the mosaicism of these genes was built using the Circos tool4.
We identified 115 ORFs (i) best matching with a giant virus gene, (ii) larger than 100 amino acids, and (iii) present in scaffolds that harbor a majority of ‘non-viral’ ORFs. For these 115 ORFs, we performed BLASTp searches against the GenBank protein sequence database nr and a tBLASTn search against all 14 Acanthamoeba draft genome sequences. Then, we merged the results of these two BLAST searches by creating a database with significant sequence hits (e-value < 1e-4, length of query and subject sequence alignments > 100 amino acids), and performed another BLASTp search for the 115 ORFs against this database. We thereafter examined if best hits were exclusively or in majority sequences from giant viruses or Acanthamoeba spp. or both and performed a phylogenetic reconstruction to determine the sense of the nucleotide sequence transfers between these organisms.
Synteny Analysis in the Draft Genome Sequences of the 14 Acanthamoeba Species of A. polyphaga Genes for Which the Best Match Is a Giant Virus Gene
Synteny preservation of genes from A. polyphaga for which the best match is a giant virus gene was evaluated in the draft genome sequences available for the 13 other Acanthamoeba species. For this purpose, we selected 10 A. polyphaga scaffolds that carried at least 3 such genes, then searched for scaffolds harboring similar genomic sequences in other Acanthamoeba species and strains. Finally, we determined whether A. polyphaga genes for which the best match is a giant virus gene were present in the genome scaffolds from other species and strains, and whether these genes were in synteny in the different Acanthamoeba draft genome sequences.
Results
Improvement of the Assembly of the Acanthamoeba Draft Genome Sequences
The estimated size of the A. polyphaga draft genome was 120.6 Mb, compared to 115.3 Mb for the draft genome sequences of A. castellanii ATCC 50370, and 41 Mb for the A. castellanii Neff genome (Clarke et al., 2013). Using CLC Genomics Workbench, the number of contigs for the A. polyphaga draft genome sequences was reduced from 224,482 to 56,709. Contig number reduction was in the same order of magnitude for A. castellanii ATCC 50370 (from 221,748 to 56,469) (Table 1). We obtained a statistically significant reduction in the average number (±standard deviation) of contigs for the 14 draft genome sequences of Acanthamoeba (p < 1e-3, ANOVA test) (Supplementary Figure S1).
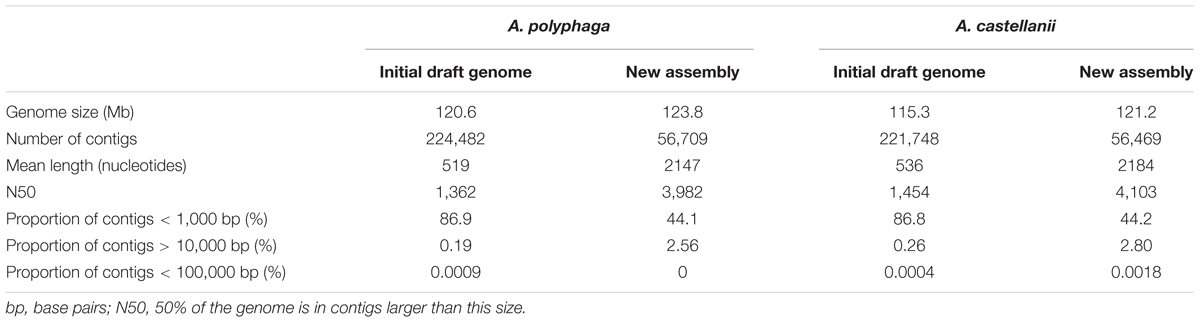
TABLE 1. Assembly statistics for the draft genome sequences of the two species A. polyphaga and A. castellanii using the CLC software.
Acanthamoeba polyphaga Gene Content and Comparison With the Gene Content of Other Acanthamoeba Species
Gene prediction performed for the A. polyphaga draft genome sequences detected a substantial number of ORFs (equal to 310,496) shorter than 50 amino acids (but greater than 35 amino acids). The number of predicted ORFs with a size comprised between 50 and 100 amino acids or larger than 100 amino acids was also considerable, being of 223,728 and 97,092, respectively (Supplementary Table S2). Comparison with the A. castellanii Neff gene set showed that 97.2% of its genes were detected in the A. polyphaga draft genome sequences. The same proportion (97.2%) of A. castellanii Neff genes was detected in the draft genome sequences of A. castellanii ATCC 50370. These results indicate that a large majority of A. castellanii Neff transcribed genes are present in these Acanthamoeba sp. strains (Supplementary Table S3). The 421 A. castellanii Neff genes that were not detected in the A. polyphaga draft genome sequences included mostly genes encoding hypothetical proteins [383 genes (91%)]. Other genes included proteins encoding the six MCP described in the A. castellanii Neff genome (Maumus and Blanc, 2016), three NAD-dependent epimerase/dehydratase family proteins, two HEAT repeat domains containing proteins, two Rho termination factor domain containing proteins, a chitin synthase, and a putative autophagy protein (Supplementary Table S4). In addition, 48,583 (78.6%) of the 61,786 orthologous groups of genes found in the A. polyphaga draft genome sequences were also detected in the A. castellanii ATCC 50370 draft genome sequences (Supplementary Figure S2). The number of non-ORFan genes and ORFan genes larger than 100 amino acids in the A. polyphaga draft genome sequences was equal to 58,185 (70.4%) and 24,484 (29.6%), respectively (Table 2). Similar proportions were found for the draft genome sequences of A. castellanii ATCC 50370, with 56,920 ORFan genes (69.2%) and 25,390 non-ORFan genes (30.8%).
Phylogenetic reconstruction based on 18S ribosomal genes (Supplementary Figure S3) and the tree based on similarities between the 14 draft genome sequences and the genome of A. castellanii Neff (Supplementary Figure S4) showed that the A. castellanii ATCC 50370 and Neff are not clustered together. In contrast, the A. castellanii ATCC 50370 is clustered with A. polyphaga and A. pearci, A. pearci being the isolate closest to A. polyphaga. These findings suggest that the genomes of A. castellanii Neff and those of A. castellanii ATCC 50370 belong to different species. We further checked for similarities between 18S ribosomal DNA sequences from the draft genome sequences of A. polyphaga analyzed here and the sequence AY026244 from A. polyphaga ATCC 30872. We observed that 18S ribosomal DNA sequences from both strains were not clustered together, which questions the accuracy of the identification of one or both genomes.
Taxonomical Distribution of Acanthamoeba polyphaga Genes and Possible Gene Trafficking Between Acanthamoeba spp. and Giant Viruses
The taxonomical distribution of the best BLAST hits obtained for the A. polyphaga proteins indicated that 43% belong to Amoebozoa (98% of them belonging to A. castellanii Neff), 3% belong to eukaryotes other than Amoebozoa, 3% belong to bacteria, 3% to archaea, and 51% were identified to be ORFans. A total of 366 genes (0.07%) had viral genes as best match, which suggests an important gene trafficking between amoebae and their infecting viruses (Figures 1, 2A). A total of 41 (11%) of the 366 viral genes in A. polyphaga were found to match with genes transcribed in A. castellanii Neff. The functions of these 366 A. polyphaga genes are mostly related to replication, recombination and repair [COG category L (18%)]; then signal transduction [T (16%)]; general function [R (15%)]; post-translational modification [O (12%)]; transcription [K (9%)]; and molecule transport and metabolism [E, F, G, H and P (17%)] (Table 3). Most of these 366 genes belong to giant viruses of the proposed order Megavirales. Pandoraviruses were the giant viruses that provided the greatest number of best hits with 117 (32%) for A. polyphaga. Then, genes from mimiviruses, Mollivirus sibericum, marseilleviruses, and Pithovirus sibericum, were best hits in 67 (18%), 37 (10%), 24 (7%), and 2 (0.5%) cases, respectively. Among other viral genes, there were 34 genes from phycodnaviruses. Among other viral genes, there were 29 genes from phycodnaviruses. A similar number of viral genes (356) was identified in the draft genome sequences of A. polyphaga than in those of A. castellanii ATCC 50370 (Supplementary Table S5). The 47,246 genes predicted for A. polyphaga by using the GeneMarkES program were compared to the 366 genes best matching with virus genes, and this comparison showed that coverage inferior to 50% was only observed for 25 genes (6.8%), while 67 genes did not match with any of the available 47,246 genes. A same analysis was specifically carried out using only available ORFomes from all giant viruses isolated on Acanthamoeba spp., allowing the identification of 1,797 genes in the A. polyphaga draft genome sequences (1.9% of the 97,092 ORFs larger than 100 amino acids) with a giant viral homolog. In accordance with our previous findings, pandoravirus genes were the most abundant of these giant viral homologous genes, before mimiviruses, marseilleviruses, other giant viruses of amoebae, and phycodnaviruses. A substantial level of gene trafficking between A. polyphaga and giant viruses was illustrated by a sequence network that made it possible to observe the number and the nature of genes involved in putative nucleotide sequence transfers (Figure 2B).
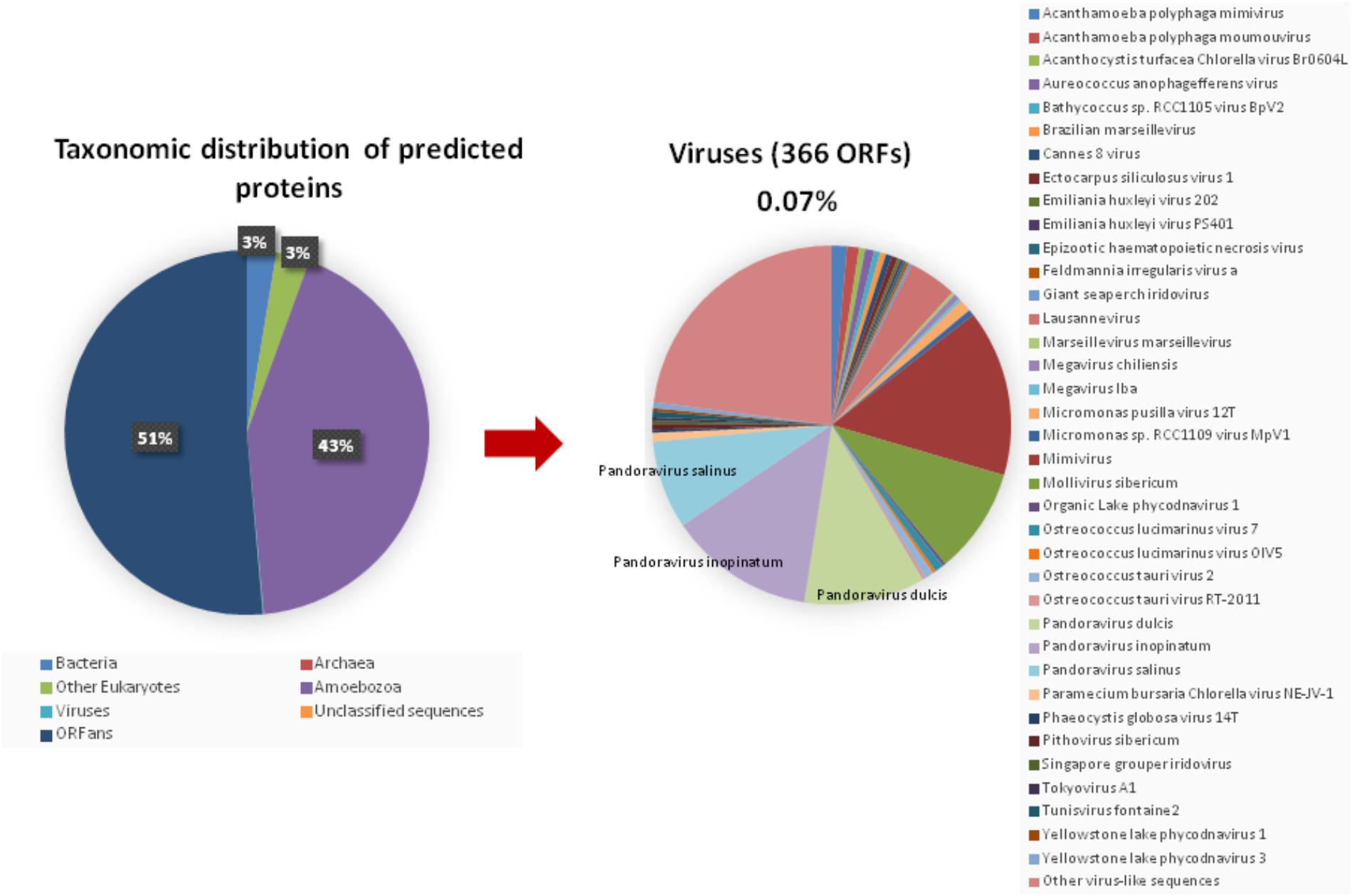
FIGURE 1. Taxonomic distribution of the predicted proteins in A. polyphaga. (Left) All predicted proedicted proteins; (right) viral proteins. Other virus-like sequences than giant viruses are represented, encompassing the viral families or groups: Herpesviridae, Baculoviridae, Podoviridae, Nimaviridae, haloviruses, Dicistroviridae, Paramyxoviridae, Totiviridae, and Retroviridae, and Haloviruses.
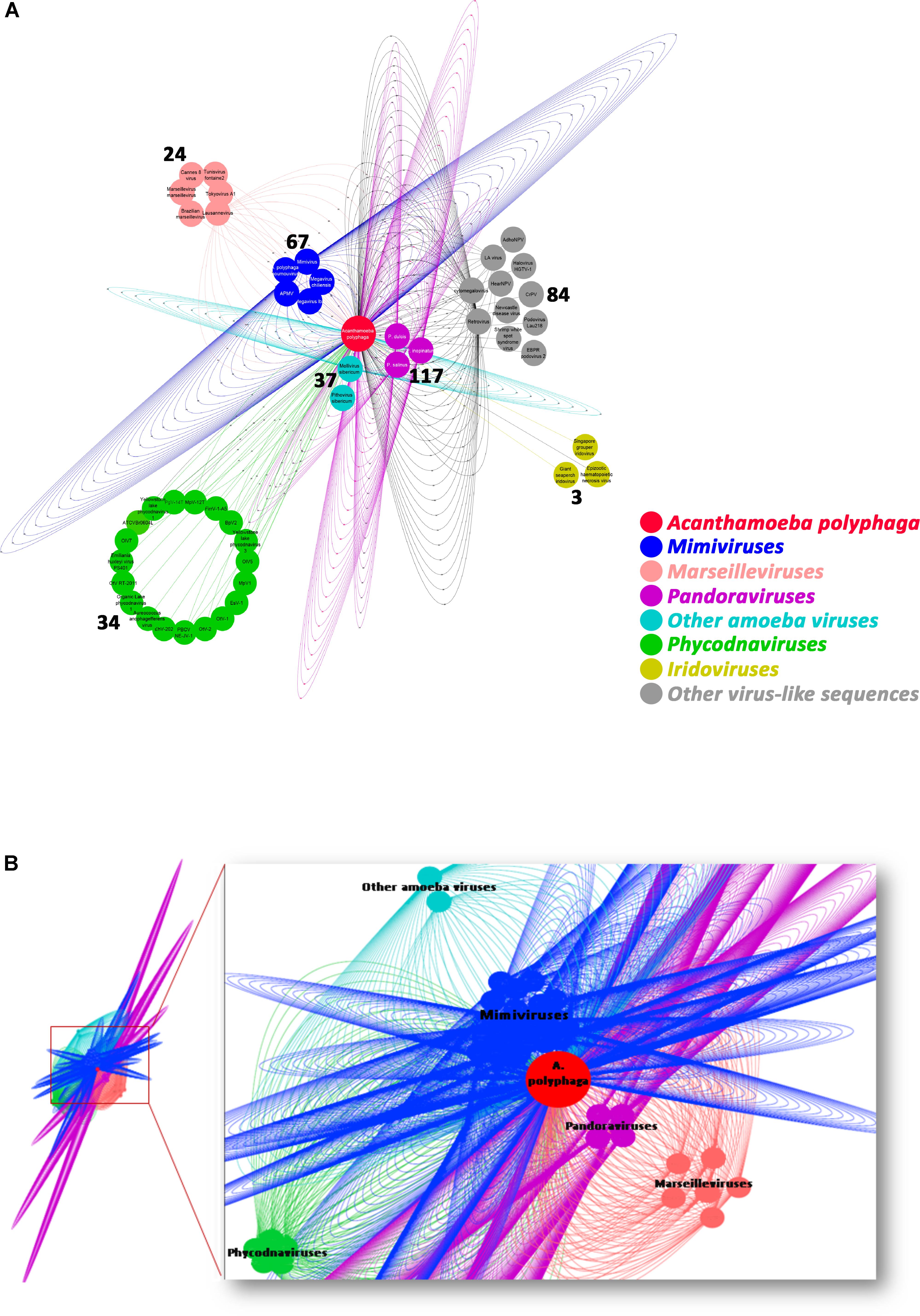
FIGURE 2. Network of viral genes in A. polyphaga (A) and gene trafficking between A. polyphaga and giant viruses (B). (A) genes were classified regarding their viral families, and the number of exchanged genes was indicated for each group of viruses; (B) the 1,797 genes were classified with respect to their viral families, and represented by a different color: pandoraviruses in pink; mimiviruses in dark blue; marseilleviruses in light orange; light blue includes other amoeba viruses (Mollivirus sibericum and Pithovirus sibericum); and phycodnaviruses in green.
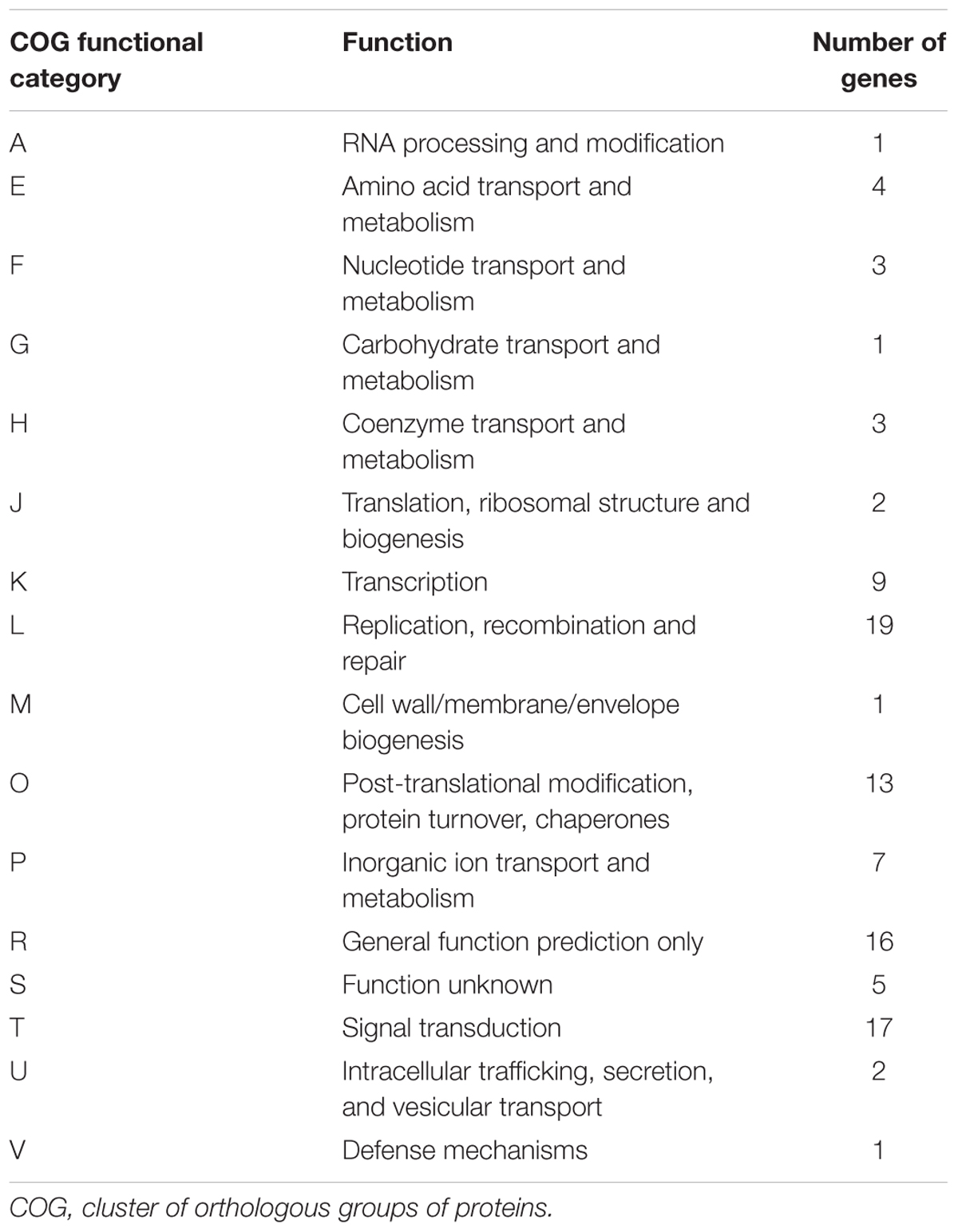
TABLE 3. Distribution of the genes predicted from the draft genome sequences of A. polyphaga with a viral gene as best hit in the functional “COG” categories.
A total of 262 A. polyphaga ORFs had as best hit a coding sequence of a giant virus of amoeba, among which 134 are larger than 100 amino acids and 115 are present in scaffolds harboring a majority of ‘non-viral’ ORFs. For these 115 ORFs, results from BLAST searches against the non-redundant GenBank protein sequence database and all 14 Acanthamoeba draft genome sequences showed different patterns of best hits, including sequence sets comprising a majority of sequences from giant viruses or from Acanthamoeba spp. Subsequent phylogenetic reconstructions enabled us to infer that the most parsimonious evolutionary scenario was, in at least three cases, a gene sequence transfer from giant viruses of amoebae to Acanthamoeba spp., although alternative scenarios could not be ruled out (Figures 3A–C). Also, in at least three cases, gene sequence transfer was supposed to have occurred in the opposite way, from Acanthamoeba spp. to giant viruses of amoebae (Figures 3D–F). Nevertheless, phylogenies were most often inconclusive regarding the putative sense of the gene flow (Supplementary Figure S5). We analyzed further two cases for which the putative sense of the gene sequence transfer was from giant viruses to amoebae and two cases for which the putative sense of the gene sequence transfer was from amoebae to giant viruses. We searched for the most similar sequences for short fragments of these genes. We found that the best hits for these fragments were organisms that belonged to different cellular domains (Eukarya, Bacteria, or Archaea), or were of putative viral origin (Figure 4). This gene sequence mosaicism was observed in all four cases, although with level differences. This indicates that sequence mosaicism may also occur within genes, and may challenge the interpretation of gene-based phylogeny.

FIGURE 3. Phylogenetic trees for six examples of putative viral proteins in A. polyphaga. Gene sequence transfer was inferred from the comparison between annotated sequences with a putative viral origin and their best hits as well as their homologs in the other Acanthamoeba genomes. The way of the transfer was supposed to be from giant viruses to amoebae (a–c) or from amoebae to giant viruses (d–f). Trees are unrooted. In red: A. polyphaga gene; in blue: the viral homolog; in orange homologs from other Acanthamoeba species; in black: homologs from other organisms.
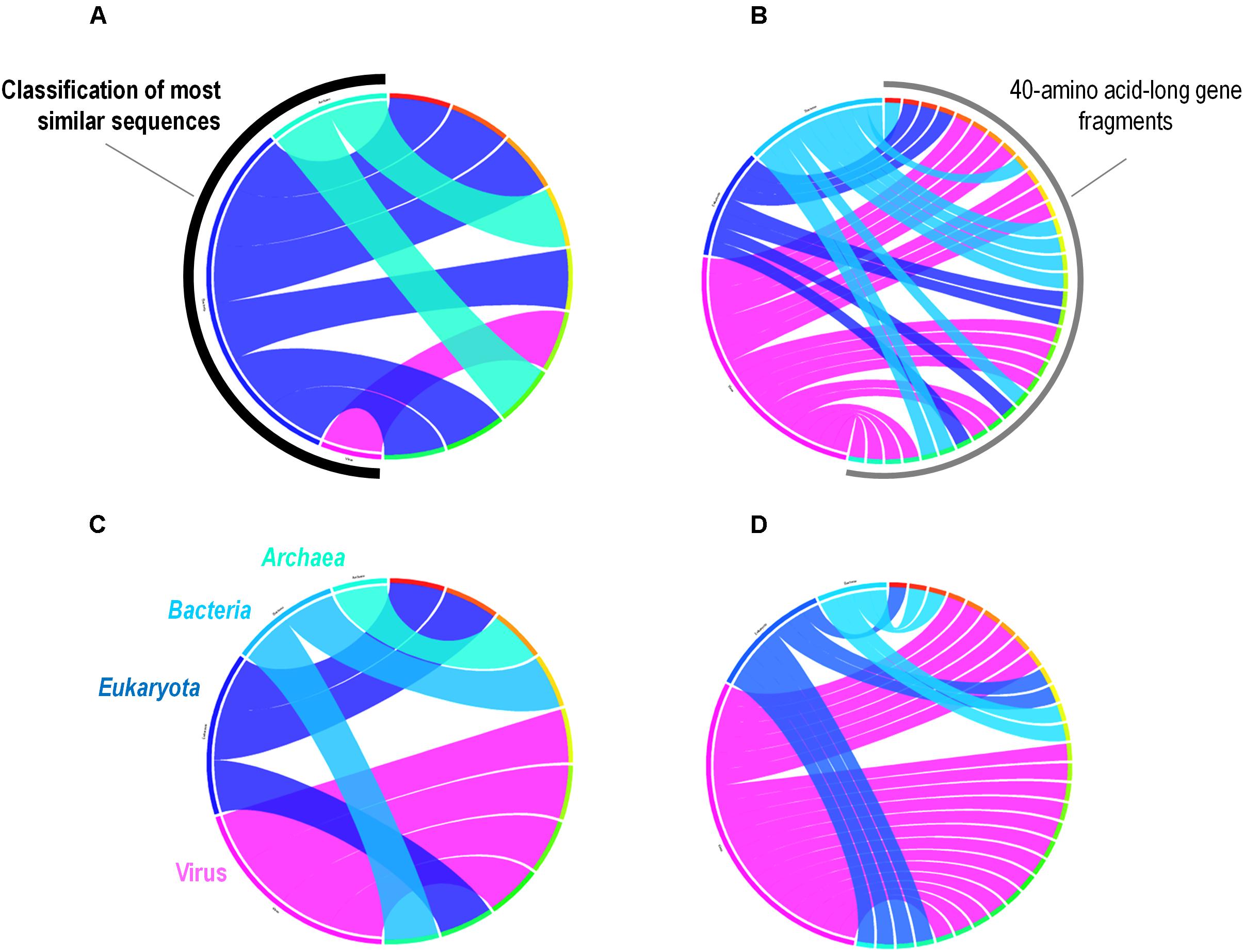
FIGURE 4. Rhizomes of A. polyphaga gene fragments illustrative of the mosaicism of the genes. Forty amino acid-long fragments of four A. polyphaga genes either supposed to have been transferred from viruses to amoebae (A,B) or in the opposite way (C,D) were linked to their most similar sequences in the NCBI GenBank protein sequence database according to the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi), classified according to their belonging to viruses, eukaryotes, bacteria or archaea, and integrated in a circular gene data visualization. The figures were performed using the CIRCOS online tool (http://mkweb.bcgsc.ca/tableviewer/visualize/).
As previous culture isolation experiments suggested different levels of permissivity to giant viruses according to the Acanthamoeba species (Dornas et al., 2015), sequences detected in A. polyphaga and A. castellanii ATCC 50370 that were homologous with giant viral genes were compared (Supplementary Table S6 and Supplementary Figure S6). This showed that the numbers and sets of viral genes that are homologs of genes present in these two Acanthamoeba species differ between giant viral families. In contrast, for a given viral family, a majority of genes found as best matches of genes from these two Acanthamoeba species were conserved in different viruses. Nonetheless, in some viruses, we identified genes that are homologous with only one of the two Acanthamoeba species. For example, homologs of some mimivirus genes were specifically found in A. polyphaga (Supplementary Table S6). Furthermore, analysis of the presence and conservation of these genes in the draft genome sequences of the other Acanthamoeba species showed that some were present in a majority of these genomes, being only absent in those of two or three other Acanthamoeba species. For instance, Pandoravirus salinus gene Ps_2278 was only absent in A. castellanii ATCC 50370 and A. healyi, whereas Pandoravirus dulcis gene Pd_13-16 was absent in A. polyphaga, A. mauritaniensis, and A. pearcei. In contrast, no giant virus homologous genes were found in the draft genome sequences of a substantial number of Acanthamoeba species, including pandoravirus genes Ps_1170 and Pd_589. In addition, the same gene in the draft genome sequences of A. castellanii ATCC 50370 was homologous with both Pandoravirus salinus gene Ps_2319 and Pandoravirus dulcis gene Pd_1426 (Supplementary Table S7).
Acanthamoeba polyphaga is one of the Acanthamoeba species for which no MCP homologs were detected. In contrast, eight MCP homologs were found in the draft genome sequences of other Acanthamoeba species including A. lenticulata, A. lugdunensis, A. quina, A. healyi, and A. mauritaniensis (Supplementary Figure S7 and Supplementary Table S8), as previously described (Maumus and Blanc, 2016). In addition, A. castellanii Neff was previously found to harbor six genes encoding MCPs of giant viruses (Maumus and Blanc, 2016). A phylogenetic analysis showed that three of these sequences, including MCP homolog 1, MCP homolog 2, and iridovirus MCP homolog 2, were clustered. Moreover, the iridovirus homolog 1 sequence is clustered with a sequence from Mollivirus sibericum, the nearest neighbor of the pandoraviruses for which no gene encoding a capsid protein has been identified (Legendre et al., 2015) (Supplementary Figure S8a). A BLAST search was performed against the NCBI non-redundant protein sequence database. The query used was the ancestral sequence inferred for the MCP protein detected in the draft genome sequences of the different Acanthamoeba species and the MCP of Mollivirus sibericum. This search retrieved MCP-encoding sequences from phycodnaviruses, which are DNA viruses that infect algae and are classified with giant viruses of Acanthamoeba in the proposed order Megavirales (Supplementary Figure 8b). The G + C content was homogenous along the Acanthamoeba scaffolds carrying these virus-related genes.
Synteny of Acanthamoeba polyphaga Genes With Viral Genes as Best Hits in the Draft Genome Sequences of the 16 Acanthamoeba spp.
The evolution of the 10 genomic regions identified among the draft genome sequences of Acanthamoeba spp. and carrying the highest number of genes best matching with viruses (at least 3 genes) was analyzed. This showed that the three amoebal species A. polyphaga ATCC30872, A. castellanii ATCC 50370 and A. pearcei all conserved the 10 scaffolds that harbored in totality 46 viral genes. In addition, this analysis revealed a shared synteny with similar contents and co-localization of the viral-related genes in these three Acanthamoeba species. In contrast, for A. quina, A. lugdunensis, A. mauritaniensis, A. rhysodes, A. palestinensis, A. healyi, A. lenticulata, and A. royreba, the number of conserved genes was 24, 11, 10, 10, 8, 4, 4, and 3, respectively. In addition, none of these 46 genes was found in A. culbertsoni, A. astronyxis and A. divionenesis (Figure 5). The tree based on similarities between the 14 draft genome sequences and the genomes of A. castellanii Neff and A. polyphaga Linc-AP1 showed that the pattern of conservation of these genes homologous with viral genes in the draft genome sequences of the different Acanthamoeba species was congruent with the phylogenomic analyses since they displayed the same distribution into the groups of species.
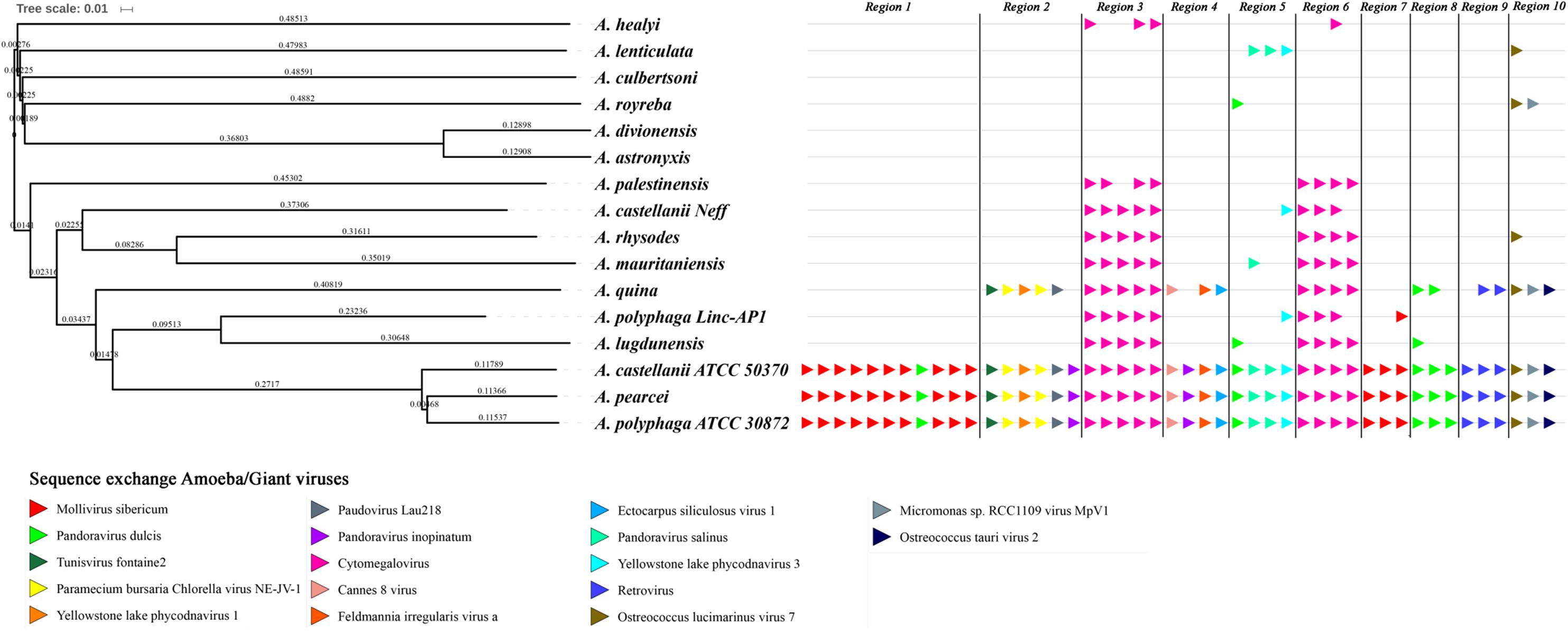
FIGURE 5. Comparison between gene synteny for genes with giant virus genes as best match in the genome sequence of 16 Acanthamoeba isolates classified in 14 species, and genome tree built for these Acanthamoeba species. Phylogenetic tree using the complete draft genome sequences of the 16 Acanthamoeba strains was represented aside the synteny distribution of genes best matching with giant virus genes in 10 selected genomic regions from Acanthamoeba spp. Phylogenetic reconstructions were performed using the alignment of 16 draft genome sequences of Acanthamoeba strains classified in 14 species including two A. polyphaga strains and two A. castellanii strains, by using the progressive Mauve program (Darling et al., 2010). Pointing triangles on the right part of the Figure correspond to viral genes; viruses are represented by different colors.
Discussion
We give here new insights into the possible interactions between Acanthamoeba species and giant viruses. Only one A. castellanii genome has been analyzed so far (Clarke et al., 2013). Our analyses primarily focused on A. polyphaga, for which the body of data regarding the isolation and propagation by culture of giant viruses is the greatest among amoebal species (along with that of A. castellanii). Great similarities were suggested between A. polyphaga and A. castellanii Neff, as A. polyphaga was found to harbor the majority of the 14,974 genes previously predicted through genome and transcriptome sequencing in A. castellanii Neff (Clarke et al., 2013). In contrast, it is worthy to note that A. castellanii Neff and A. castellanii ATCC 50370, whose draft genome sequences were analyzed here, did not cluster together based on our phylogenetic analyses. Furthermore, there is possibly an incorrect identification of sequences presented as originating from ATCC 30872 but belonging to A. polyphaga species. Indeed, the 18S ribosomal DNA previously described (AY026244) and part of the draft genome sequences analyzed here were not the closest among sequences from different Acanthamoeba species, and were not clustered in the phylogenetic analysis. These issues deserve clarification in future studies.
More than half of the genes predicted from the A. polyphaga draft genome sequences analyzed here had no homologs in the NCBI non-redundant sequence database. This suggests that a large part of the genome sequence of this amoeba remains unknown. Yet, a significant proportion of the predicted ORFs contained less than 100 codons. They should be considered carefully, especially when identified as ORFans or annotated as hypothetical proteins. Among annotated genes, the vast majority had homologs in amoebae, and 3% had homologs in other eukaryotes or in bacteria. Finally, a small proportion of annotated genes was mostly related to viral sequences, among which a majority belonged to the three pandoraviruses described to date and to Mollivirus sibericum.
The presence of sequences homologous with coding sequences of giant viruses of amoebae in eukaryotic genomes has been described by several teams (Filée, 2014; Maumus et al., 2014; Blanc et al., 2015; Sharma et al., 2016; Maumus and Blanc, 2016; Gallot-Lavallée and Blanc, 2017). Notably, MCPs were recently unexpectedly described in some Acanthamoeba species (Clarke et al., 2013; Maumus and Blanc, 2016; Gallot-Lavallée and Blanc, 2017). Congruently, our analysis showed that these sequences comprised two groups: firstly, the sequence of the “iridovirus-like” MCP of A. castellanii Neff and a sequence of Mollivirus sibericum, and, secondly, the other sequences which are related to those of a phycodnavirus (genus Raphidovirus), Heterosigma akashiwo virus 01. The G+C content was found to be homogeneous along the Acanthamoeba scaffolds carrying these genes best matching MCPs. However, this does not completely rule out the possibility that these sequences are the result of an exchange of sequences between these organisms because the GC% in some giant viruses is relatively close to that of their host, for example for pandoraviruses and A. castellanii (60.6% vs. 58.3%) (Antwerpen et al., 2015). These results also confirm previous observations concerning the presence of homologous sequences of MCPs in the genome of some eukaryotes (Maumus et al., 2014). Thus, these data suggest nucleotide sequence transfers between giant viruses and Acanthamoeba, and raise the question of the significance of homologies between genes present in giant virus and Acanthamoeba genomes. One hypothesis is that capsid proteins may be involved in defense mechanisms. Interestingly, it has been described that, in addition to their function as capsid proteins, the MCPs of totivirus, a RNA virus, was also able to inactivate the host mRNAs by eliminating their 5’-cap (Koonin, 2010).
Preliminary analysis of possible nucleotide sequence transfers within the A. polyphaga genome showed that there are 366 genes that could have been exchanged between this amoeba and viruses. We compared this gene set with the set of 267 genes recently described in A. castellanii Neff as putatively exchanged with viruses (Maumus and Blanc, 2016). We found that among these 366 A. polyphaga genes, only 30 (8.2%) were shared with the 267 genes identified in A. castellanii Neff as putatively exchanged with viruses. In addition, only 39 (11.0%) among the 356 genes which had as best hit a coding sequence of viruses in the draft genome sequences of A. castellanii ATCC 50370 were shared with the 267 genes described by Maumus and Blanc (Maumus and Blanc, 2016). A total of 11% of the 366 genes were matching with genes transcribed in A. castellanii Neff (Clarke et al., 2013), suggesting that they might be transcribed and play yet unknown roles. Some of these differences might be explained by differences between the sets of giant virus genomes available at the time when the different analyses were performed, as giant virus diversity expands considerably (Maumus and Blanc, 2016; Colson et al., 2017b). In addition, differences in parameters used for gene prediction and annotation and BLAST searches are likely to generate discrepant results. It should be also taken into account that the amoebal genomes analyzed here were non-assembled draft genome sequences. Among giant viruses which infect Acanthamoeba spp., pandoraviruses were found to share the highest number of genes with A. polyphaga, before mimiviruses and marseilleviruses, with 117, 65 and 24 putatively exchanged genes, respectively. This might suggests a co-evolution of A. polyphaga with pandoraviruses, but it is worth considering that this Acanthamoeba species was far less permissive to pandoraviruses than A. castellanii (Dornas et al., 2015). In addition, there is little evidence of gene exchanges with other viruses, including phycodnaviruses. The sense of the gene nucleotide sequence transfers remained undetermined in a large majority of cases. This is due to an insufficient number of matches, or an absence of significantly delineated cluster in the phylogenetic trees. Nevertheless, for some of the phylogenies, a transfer from Acanthamoeba to giant viruses was the most parsimonious evolutionary hypothesis, indicating that giant viruses are not only bags of genes but contribute to the gene sequence flow. Estimating times of divergence might be helpful in more extensive analyses that would be conducted on giant viruses of amoebae and several Acanthamoeba species in order to try inferring the sense of gene sequence transfers between giant viruses and their amoebal hosts. Besides, the analyses of fragments of A. polyphaga genes that had a giant virus sequence as best hit showed a different, more complex pattern of best hits compared to the analysis of the whole genes, with mosaics of sequences from eukaryotes, bacteria, archaea and giant viruses as best matches. A similar pattern has also been illustrated by fragmenting a gene encoding an aminoacyl-tRNA synthetase in Klosneuvirus, a mimivirus relative (Abrahao et al., 2018). These findings may extend to genes the paradigm that no single tree can define the mosaic origin of genomes, which may result from nucleotide sequence transfers rather than from gene transfers (Dagan and Martin, 2006; Merhej et al., 2011). These mosaic patterns that affect the sequences of genes may represent a pitfall for the robustness of phylogenetic analyses and inference of the putative way of nucleotide sequence transfer. This further suggests that the vision of species in Darwin’s tree of life is rather outdated, and that each organism has a complex family tree that is a testimony of its chimerical origin and is better represented in the form of a rhizome than of a tree (Raoult, 2010; Merhej et al., 2011). Overall, the global analysis of all giant virus homologs to A. polyphaga predicted genes potentially demonstrates the complexity of the putative gene trafficking between this amoeba and giant viruses, with 1,797 genes involved, although only 366 A. polyphaga genes have viral genes as best matches. This might further suggest intermediate interactions with organisms other than viruses. Moreover, these results highlight the fact that the gene flow was not a one way mechanism and likely resulted from the sympatric lifestyle of giant viruses in amoebae (Moliner et al., 2010).
The comparisons of possible gene sequence transfers between the two species A. polyphaga and A. castellanii ATCC 50370 and some representatives of giant viruses (pandoraviruses, Acanthamoeba polyphaga mimivirus and Marseillevirus) shows that the differences regarding the number and the nature of potentially transferred genes remain limited within the same viral family. However, the number and the nature of potentially transferred genes were found to be more variable when considering different families of giant viruses. These observations might be explained by differences in the frequency of interactions between some Acanthamoeba species and some giant viruses, and between genes used by both the amoebae and the giant viruses to deal with these interactions. The recent study by Clarke et al. (2013) showed that the plasticity of protists living in community with microbes is as important as that of bacteria with the same lifestyle. This supports the hypothesis that an essential explanation of the chimerism of the genomes of organisms and micro-organisms that live sympatrically relies on this lifestyle, and possibly on their genomic plasticity according to their phylogenetic origin (eukaryotic, bacterial, archaeal or viral). These observations may also reflect the time of sequence exchange through the evolutionary course of both giant viruses and amoeba species. Phylogenetic and synteny analyses of viral genes performed in the present study suggest that the sequence exchanges between Acanthamoeba species and giant viruses occurred along the Acanthamoeba speciation events and evolved through species-specific events. Among the Acanthamoeba species, A. castellanii ATCC 50370 and A. pearcei were found to be the most structurally conserved with A. polyphaga regarding their content of genes homologous with viral genes. These results did follow the expectation that decreased phylogenetic distance would correspond to increased levels of genome preservation.
In summary, this work opens up several potential avenues for future works on the interactions between Acanthamoeba species and giant viruses. The annotation of all Acanthamoeba species and their accurate identification are important tasks for a greater understanding of why some amoebae are more susceptible than others to giant viruses, and possibly to other microorganisms. Thus, the considerable diversity of gene repertoires among Acanthamoeba species might lead to differences regarding potential interactions with giant viruses. The characterization of the genes present and absent in the different species of Acanthamoeba could be performed and correlated with the observed phenotypic differences that need to be studied more extensively.
Author Contributions
PC and BLS designed the study. NC, PC, and AL performed the experiments. NC, PC, AL, PP, DR, and BLS analyzed the data. All the authors contributed to manuscript redaction and review.
Funding
This work was supported by a grant from the French State managed by the National Research Agency under the “Investissements d’avenir (Investments for the Future)” program with the reference ANR-10-IAHU-03 (Méditerranée Infection). NC was financially supported through a grant from the Infectiopole Sud foundation. This work was supported by Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02098/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/bioproject/
- ^ https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/
- ^ http://www.megasoftware.net/
- ^ http://circos.ca/
References
Abrahao, J., Silva, L., Santos Silva, L., Bou Khalil, J. Y., Rodrigues, R., Arantes, T., et al. (2018). Tailed giant tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9:749. doi: 10.1038/s41467-018-03168-3161
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-80362
Antwerpen, M. H., Georgi, E., Zoeller, L., Woelfel, R., Stoecker, K., and Scheid, P. (2015). Whole-genome sequencing of a Pandoravirus isolated from keratitis-inducing Acanthamoeba. Genome Announc. 3:e00136-15. doi: 10.1128/genomeA.00136-15
Bertelli, C., and Greub, G. (2012). Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front. Cell. Infect. Microbiol. 2:110. doi: 10.3389/fcimb.2012.00110
Blanc, G., Gallot-Lavallée, L., and Maumus, F. (2015). Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc. Natl. Acad. Sci. U.S.A. 112, E5318–E5326. doi: 10.1073/pnas.1506469112
Boyer, M., Azza, S., Barrassi, L., Klose, T., Campocasso, A., Pagnier, I., et al. (2011). Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. 108, 10296–10301. doi: 10.1073/pnas.1101118108
Clarke, M., Lohan, A. J., Liu, B., Lagkouvardos, I., Roy, S., Zafar, N., et al. (2013). Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14:R11. doi: 10.1186/gb-2013-14-2-r11
Colson, P., De Lamballerie, X., Yutin, N., Asgari, S., Bigot, Y., Bideshi, D. K., et al. (2013). “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 158, 2517–2521. doi: 10.1007/s00705-013-1768-1766
Colson, P., La Scola, B., Levasseur, A., Caetano-Anolles, G., and Raoult, D. (2017a). Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 15, 243–254. doi: 10.1038/nrmicro.2016.197
Colson, P., La Scola, B., and Raoult, D. (2017b). Giant viruses of amoebae: a journey through innovative research and paradigm changes. Annu. Rev. Virol. 4, 61–85. doi: 10.1146/annurev-virology-101416-141816
Dagan, T., and Martin, W. (2006). The tree of one percent. Genome Biol. 7:118. doi: 10.1186/gb-2006-7-10-118
Darling, A. E., Mau, B., and Perna, N. T. (2010). Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
Dornas, F. P., Khalil, J. Y. B., Pagnier, I., and Raoult, D. (2015). Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 6:1086. doi: 10.3389/fmicb.2015.01086
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Filée, J. (2014). Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: the visible part of the iceberg? Virology 466–467, 53–59. doi: 10.1016/j.virol.2014.06.004
Filée, J., Siguier, P., and Chandler, M. (2007). I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 1, 10–15. doi: 10.1016/j.tig.2006.11.002
Gallot-Lavallée, L., and Blanc, G. (2017). A glimpse of nucleo-cytoplasmic large DNA virus biodiversity through the eukaryotic genomics window. Viruses 9:17. doi: 10.3390/v9010017
Greub, G., and Raoult, D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433. doi: 10.1128/CMR.17.2.413
Huson, D. H., Beier, S., Flade, I., Górska, A., and El-hadidi, M. (2016). MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12:e1004957. doi: 10.1371/journal.pcbi.1004957
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Khalil, J. Y. B., Andreani, J., and La Scola, B. (2016). Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 31, 80–87. doi: 10.1016/j.mib.2016.03.004
Koonin, E. V. (2010). Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 8:2. doi: 10.1186/1741-7007-8-2
La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., et al. (2003). A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867
Lechner, M., Findeiß, S., Steiner, L., Marz, M., Stadler, P. F., and Prohaska, S. J. (2011). Proteinortho:detection of (co-) orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124
Legendre, M., Lartigue, A., Bertaux, L., Jeudy, S., Bartoli, J., Lescot, M., et al. (2015). In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U.S.A. 112, E5327–E5335. doi: 10.1073/pnas.1510795112
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, Y. O., and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506. doi: 10.1093/nar/gki937
Marciano-cabral, F., and Cabral, G. (2003). Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16, 273–307. doi: 10.1128/CMR.16.2.273
Maumus, F., and Blanc, G. (2016). Study of gene trafficking between Acanthamoeba and giant viruses suggests an undiscovered family of amoeba-infecting viruses. Genome Biol. Evol. 8, 3351–3363. doi: 10.1093/gbe/evw260
Maumus, F., Epert, A., Nogué, F., and Blanc, G. (2014). Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 5:4268. doi: 10.1038/ncomms5268
Merhej, V., Notredame, C., Royer-Carenzi, M., Pontarotti, P., and Raoult, D. (2011). The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Mol. Biol. Evol. 28, 3213–3223. doi: 10.1093/molbev/msr239
Moliner, C., Fournier, P. E., and Raoult, D. (2010). Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34, 281–294. doi: 10.1111/j.1574-6976.2009.00209.x
Moreira, D., and Brochier-Armanet, C. (2008). Giant viruses, giant chimeras: the multiple evolutionary histories of mimivirus genes. BMC Evol. Biol. 8:12. doi: 10.1186/1471-2148-8-12
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Raoult, D. (2010). The post-Darwinist rhizome of life. Lancet 375, 104–105. doi: 10.1016/S0140-6736(09)61958-61959
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., et al. (2004). The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350. doi: 10.1126/science.1101485
Raoult, D., and Boyer, M. (2010). Amoebae as genitors and reservoirs of giant viruses. Intervirology 53, 321–329. doi: 10.1159/000312917
Reteno, D. G., Benamar, S., Khalil, J. B., Andreani, J., Armstrong, N., Klose, T., et al. (2015). Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 89, 6585–6594. doi: 10.1128/JVI.00115-115
Rodríguez-Zaragoza, S. (1994). Ecology of free-living amoebae. Crit. Rev. Microbiol. 20, 225–241. doi: 10.3109/10408419409114556
Salah, I. B., Ghigo, E., and Drancourt, M. (2009). Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin. Microbiol. Infect. 15, 894–905. doi: 10.1111/j.1469-0691.2009.03011.x
Sharma, V., Colson, P., Pontarotti, P., and Raoult, D. (2016). Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr. Opin. Microbiol. 31, 16–24. doi: 10.1016/j.mib.2015.12.010
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308. doi: 10.1023/A:1020591307260
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P., and Ideker, T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. doi: 10.1093/bioinformatics/btq675
Tatusov, R. L. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Keywords: Acanthamoeba polyphaga, Acanthamoeba, giant viruses, mimivirus, draft genome sequences, horizontal gene transfer, nucleotide sequence transfer
Citation: Chelkha N, Levasseur A, Pontarotti P, Raoult D, La Scola B and Colson P (2018) A Phylogenomic Study of Acanthamoeba polyphaga Draft Genome Sequences Suggests Genetic Exchanges With Giant Viruses. Front. Microbiol. 9:2098. doi: 10.3389/fmicb.2018.02098
Received: 14 June 2018; Accepted: 16 August 2018;
Published: 06 September 2018.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Kiran Kondabagil, Indian Institute of Technology Bombay, IndiaCarsten Balczun, Bundeswehrkrankenhaus Koblenz, Germany
Erna Geessien Kroon, Universidade Federal de Minas Gerais (UFMG), Brazil
Copyright © 2018 Chelkha, Levasseur, Pontarotti, Raoult, La Scola and Colson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Colson, cGhpbGlwcGUuY29sc29uQHVuaXYtYW11LmZy
 Nisrine Chelkha
Nisrine Chelkha Anthony Levasseur
Anthony Levasseur Pierre Pontarotti
Pierre Pontarotti Didier Raoult
Didier Raoult Bernard La Scola
Bernard La Scola Philippe Colson
Philippe Colson