
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 21 September 2018
Sec. Evolutionary and Genomic Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02066
This article is part of the Research Topic Horizontal Gene Transfer and Genetic Diversity in Bacteria View all 20 articles
 Insha Sultan1†
Insha Sultan1† Safikur Rahman2†
Safikur Rahman2† Arif Tasleem Jan3*
Arif Tasleem Jan3* Mohammad Tahir Siddiqui1
Mohammad Tahir Siddiqui1 Aftab Hossain Mondal1
Aftab Hossain Mondal1 Qazi Mohd Rizwanul Haq1*
Qazi Mohd Rizwanul Haq1*History of mankind is regarded as struggle against infectious diseases. Rather than observing the withering away of bacterial diseases, antibiotic resistance has emerged as a serious global health concern. Medium of antibiotic resistance in bacteria varies greatly and comprises of target protection, target substitution, antibiotic detoxification and block of intracellular antibiotic accumulation. Further aggravation to prevailing situation arose on observing bacteria gradually becoming resistant to different classes of antibiotics through acquisition of resistance genes from same and different genera of bacteria. Attributing bacteria with feature of better adaptability, dispersal of antibiotic resistance genes to minimize effects of antibiotics by various means including horizontal gene transfer (conjugation, transformation, and transduction), Mobile genetic elements (plasmids, transposons, insertion sequences, integrons, and integrative-conjugative elements) and bacterial toxin-antitoxin system led to speedy bloom of antibiotic resistance amongst bacteria. Proficiency of bacteria to obtain resistance genes generated an unpleasant situation; a grave, but a lot unacknowledged, feature of resistance gene transfer.
Antibiotics, representing both naturally as well as chemically synthesized entities, emerged as a powerful tool in counteracting infectious diseases, following serendipitous discovery of penicillin from Penicillium notatum by Alexander Fleming in 1928. Widespread usage of antibiotics that imposes strong selection pressure for resistance development (ability to withstand effects of antibiotics) took a strong grip over the health care system globally as concerns regarding resistance to available drug regime restrict therapeutic options available to treat the disease. Emergence of resistance at rapid pace made the pathogens well-fit and well-adapted, resulting in causing serious life threatening complications as we lack robust drugs to curb the menace of multidrug resistance. Growing menace of antibiotic resistance is inevitable fallout of the introduction of new antibiotics aimed at long-term efficacy in the treatment of infectious diseases. Deteriorating public health ensuing emergence among pathogenic and commensal bacteria of resistance, illustrates a grave predicament globally (Bennett, 2008). Steadily increase in the development of resistance among bacteria thwarts current treatment regimes in hospitals and community settings. Through each passing day, treatments of infectious diseases require administration of high doses of antibiotics and longer stay in hospital. Widening gap between lean productions of drugs increases need of either to rejuvenate the drying antibacterial pipelines or design innovative strategies to combat bacterial antibiotic resistance. The present review analyses development of resistance and focusing on the factors that regulate acquisition of resistant determinants.
Antibiotics are the agents used commonly in the treatment and prevention of infections. Owing to their structure and degree of affinity to target sites, they are classified into Penicillin's, Cephalosporins, Tetracyclines, Aminoglycosides, Macrolides, Sulfonamides, Quinolones, Diaminopyrimidines, Polymyxin and Carbapenems (Sengupta et al., 2013; Bi et al., 2015; Liu et al., 2016). Being specific in their effect toward different bacterial species, antibiotics culminates them either by: (i) affecting cell wall synthesis (β-lactams), (ii) by targeting protein synthesis machinery via, interaction with ribosomal subunits (Tetracycline, Chloromphenicol, Aminoglycosides etc), (iii) disrupting with nucleic acid machinery (Rifampcin, Fluoroquinolones), (iv) interfering with metabolic pathways (Folic acid analogs, sulfonamides), and (v) by disrupting bacterial membrane structure (Polymyxins; Walsh, 2010; Table 1).
The main aminoglycoside resistance mechanism involves modification of the enzymes. Three major classes of proteins are classified in accordance with the kind of modification: AAC (acetyltransferases) which are AAC(1), AAC(2), AAC(3), and AAC(6); ANT (nucleotidyl transferases or adenyl transferases), which includes five nucleotidyl transferases: ANT(2), ANT(3), ANT(4), ANT(6), and ANT(9), and APH (phosphotransferases) which includes seven phosphotransferases: APH(2), APH(3), APH(3), APH(4), APH(6), APH(7), and APH(9) (Kotra et al., 2000; Ramirez and Tolmansky, 2010).
Resistance is acquired through production of β-lactamases, like extended-spectrum β-lactamases (ESBLs), ESBL genes (blaCTX-M, blaSHV, blaTEM) plasmid-mediated AmpC enzymes, and carbapenem-hydrolyzing β-lactamases (carbapenemases). Though, Stenotrophomonas maltophilia have endogenous metallo β-lactamases (MBL) L1 that makes it resistant to carbapenems (Sánchez, 2015). Carbapenem resistance among gram positive bacteria is acquired by mutations in the penicillin binding proteins (PBPs). However, in gram negative bacteria, lower penetration of the drug through decrease in the expression of outer membrane porin proteins such as OprD of Pseudomonas aeruginosa (Bonomo and Szabo, 2006). A tripartite efflux pump that causes exclusion of carbapenems from periplasmic space, adds to carbapenems resistance (Schweizer, 2003). Additionally, carbapenemases also contributes to carbapenem resistance (Poirel et al., 2007; Walsh, 2010). The main efficient carbapenemases responsible for carbapenem hydrolysis and its geographical dissemination are KPC, VIM, IMP, NDM, and OXA-48 types (Poirel et al., 2010; Nordmann et al., 2012). In a plasmid of K. pneumoniae HS11286 strain it was seen that deletion of bla KPC−2 abolished resistance toward carbapenem (cefoxitin, ceftazidime), and exhibited dose-dependent susceptibility toward cefepime supporting that bla KPC−2 is a key factor for the resistance toward cephalosporins and carbapenems in K. pneumoniae (Bi et al., 2015).
It acts as a broad spectrum antibiotic resistance mechanism for chloramphenicol involves enzymatic inactivation via acetylation mediated by chloramphenicol acetyltransferases (CATs) (Schwarz et al., 2004; Wright, 2005). Apart from enzyme inactivation chloramphenicol resistance mechanisms, also involves inactivation by phosphotransferases, target site mutation, permeability barriers and efflux pumps (Schwarz et al., 2004).
The vancomycin resistance originated from the production of modified peptidoglycan precursor, d-Ala–d-Lac (VanA, VanB, and VanD) or d-Ala–d-Ser (VanC, VanE, and VanG), to which glycopeptides display diminished binding affinities. The vanA and vanB operons are positioned on plasmids as well as on chromosome; whereas the vanC1, vanC2/3, vanD, vanE, and vanG solely show their presence on chromosomes (Klare et al., 2003; Depardieu et al., 2007).
Though resistance mechanism for quinolone was found restricted to chromosomes, three plasmid-mediated resistance mechanisms have also been reported (Courvalin, 2008; Martinez-Martinez et al., 2008). The chromosome-encoded resistance produce a declined outer-membrane permeability linked with porin loss, while over expression of the naturally existing efflux pumps create mutations in the molecular targets, DNA gyrase and topoisomerase IV (Hooper, 2000; Jacoby, 2005). Mutations were found occurring at quinolone resistance determining regions (QRDR) in the genes gyrA, gyrB, parC, and parE; which program the subunits of DNA gyrase and topoisomerase IV. Despite the fact that qnr determinant is the first recognized plasmid-mediated quinolone resistance gene, five new lineage of qnr genes have been accounted: qnrA, qnrB, qnrC, qnrD, and qnrS. Second kind of plasmid positioned quinolone resistant gene is a cr variant of aac(6)-Ib, that is aac(6)-Ib-cr, encoding aminoglycoside acetyl transferase (Park et al., 2006; Strahilevitz et al., 2009). The third means of resistance involves qepA, a plasmid-mediated efflux pump along with its E. coli derivative QepA2 (Cattoir et al., 2008), is able to expel hydrophilic fluoroquinolones, e.g., ciprofloxacin (Perichon et al., 2007).
Sulfonamide resistance in chromosome appears through mutations in the folP gene, encoding dihydropteroate synthase (DHPS; Grape, 2006). Acquired sulfonamide resistance was identified in the 1960s, and the plasmid-mediated genes sul1 and sul2 were described after 1980s (Swedberg and Sköld, 1983; Rådström and Swedberg, 1988). In addition a third plamid mediated gene sul3 has also been recognized (Perreten and Boerlin, 2003).
Mechanisms of resistance for tetracycline hold three key strategies: energy-dependent efflux pumps (ABC efflux pumps) ribosomal protection proteins {RPPs, Tet(O)} or enzymatic inactivation (TetX; Roberts, 2002).
Modifications in the lpxA, lpxC, and lpxD genes of A. baumannii result in neutralization of lipid A biosynthesis, causing total loss of LPS leading to a loss of the polymyxin target (Moffatt et al., 2010). Polymyxin resistance is controlled by two-component systems PhoP/PhoQ and PmrA/PmrB (Olaitan et al., 2014), which react to cation (calcium, iron, and magnesium) concentrations and pH variations. These systems are concerned in the alterations of LPS resulting in polymyxin resistance. Several molecular mechanisms have been associated with colistin resistance in Gram-negative bacteria, like modifications with PmrA/PmrB, PhoP/PhoQ, ParR/ParS, ColR/ColS, and CprR/CprS two-component systems and alterations in the mgrB gene, that codes for negative regulator of PhoPQ. Addition of cationic groups on lipid A due to mutations creat less anionic lipid A ultimately causing less fixation of polymyxins. The polymyxins remains one of the last classes of antibiotics in which resistance is not known to spread from cell to cell via plasmid mediated. There is a current report of plasmid mediated colistin resistance in china designated as mcr-1 gene (Liu et al., 2016) which is also reported closely in five continents viz, Asia, Europe, Africa, North America, and South America (Schwarz and Johnson, 2016).
Antibiotics do not, in themselves, cause resistance but frequent and high exposure of antibiotics to bacteria creates a selection pressure which triggers resistance strategies of bacteria. Acquirement of resistance genes has been viewed as main donor in favor of the extensive dispersal and increase in antimicrobial resistance through horizontal transfer involving MGEs (Xu et al., 2011). Their presence on mobile genetic elements facilitate transfer to un-related bacteria in a process referred to as horizontal gene transfer (HGT) via, conjugation, transduction, or transformation (Aminov and Mackie, 2007; Martinez, 2008). Transformation involves movement of cellular DNA among closely linked bacteria, imparted by chromosomal set of proteins that occur in naturally transformable bacteria. Conjugation needs autonomously replicating genetic elements known as conjugative plasmids that cause movement of plasmid from the donor cell to a recipient cell that is devoid of it. Transduction involves transfer of DNA facilitated by bacteriophages, constituted host DNA in their capsid and insert this DNA into a new host, where it combines with cellular chromosome and is inherited (Frost et al., 2005). Movement of genes confers new metabolic capabilities to the recipient, thereby helps them in their adaptation to new ecological niches. Resistance to antibiotics conferred by chromosomal or mobile genetic elements, is achieved by following strategies: (i) reduction of membrane permeability to antibiotics either by decreasing uptake or increasing efflux, (ii) drug inactivation either by hydrolysis or by modification, (iii) alteration in drug target and decreased binding permeability, and (iv) mutation (Walsh, 2000; Figure 1).
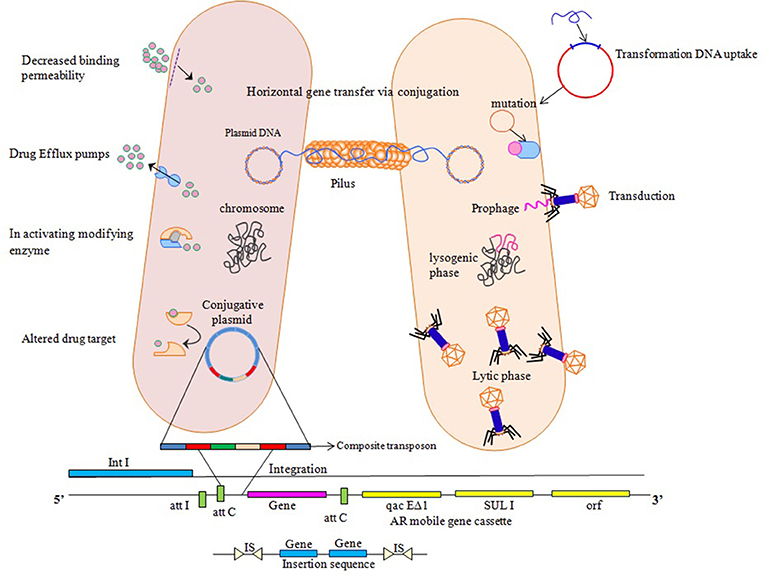
Figure 1. Various ways of resistance mechanisms to counteract effect of antibiotics. Horizontal Gene transfer facilitates transfer and exchange of genetic material among bacterial cells. Transformation involves direct uptake of genetic material from the surrounding by competent recipient having chromosomal set of proteins. Transduction involves DNA insertion into chromosome as prophage which then replicates, packages host DNA alone or in combination with the host cell chromosome. Conjugative plasmids utilize a protein structure pilus to make a link with the recipient cell so as to move them into the recipient cell that ultimately transfers the copy of entire bacterial chromosome, multicopy plasmid or a small portion to a recipient cell, where these genetic elements insert into the chromosome or replicate independently if compatible with the inhabitant plasmids. Integrons use site specific recombination mechanism where it provides a promoter for gene cassettes to exchange and disseminate. Transposons and insertion sequences insert into new sites on the chromosome or plasmids by non-homologous recombination and increase the copy number of transferred genes giving rise to chromosomal mutations, deletions and rearrangements.
Bacteria appeared on this planet billion of years ago, so have their skills sharpened due to genomic flexibility at shielding themselves from toxic chemicals. Bacteria are well-known potent originators for the dissemination of antimicrobial-resistant genetic apparatus (Woodford et al., 2011). They are competent to offer secured platform for the upholding and transmission of genes accountable for antimicrobial resistance as part of mobile genetic elements (MGE; plasmids, transposons and integrons). The transposons and integrons, owing to their genomic plasticity have contributed a great deal to the fitness quotient and robustness of bacteria to survive in varying environments. Integrons, typically transported by plasmids or enclosed in transposons, performing the task of resistance gene dissemination plays an important role in the revealing of Super Bugs (Xu et al., 2011). Since its earliest assessment in 1989 (Stokes and Hall, 1989) molecular mechanisms involved in the mobility of integrons, their excision and integration for gene cassettes, is currently being scrutinized (Hall et al., 1999; Mazel, 2006). Establishing role of MGEs in genomic evolution justifies the predictions of Barbara McClintok that transposons play a major role in the genomic diversity and evolution. Owing to their capacity to relocate between host genomes, MGEs play a vital function of acting as vehicles for resistance gene acquisition and their successive propagation.
Plasmids that mediate horizontal movement of plasmid-borne genes are accountable for global spread of resistance (Carattoli, 2013). Resistance plasmids (attributing resistance to commonly used antibiotics) are mostly conjugative; additional are mobilizable. Conjugative plasmids display both broad (no host restriction within the division) and narrow (shifting limited to small number of related bacterial groups) host range. Genes acquired through homologous recombination, integration and excision from the host chromosome relocate from donor to recipient cells by conjugation. These type of plasmid-encoded complexes help the contributor by attaching to promissing recipient that lead to the generation of secured association, required prior to the relocation of DNA. Plasmids that fails to get reloctaed by this approach are transferred by conjugative elements subsequent to the development of transitory or steady fusions called cointegrates. Plasmids also encourage cell contact development through production of pheromone influenced microfibrillar exterior covering substances. Mobilizable plasmids carry DNA transfer genes essential for structure of all or element of the relaxasome, but are deficient of genes essential for mating pore formation. They have a capacity to use conjugative plasmids for horizontal spread; these are immobile in cells which are short of mobile elements carrying compatible mating-pore genes. The majority of known mobilizable plasmids utilize conjugative element mating-pores by expressing their own relaxase (Mob) which works on the plasmid's cognate oriT (Joshua et al., 2016). Current studies have revealed that plasmid transfer can also take place even when the mobilizing plasmid and the plasmid being mobilized are in two diverse bacterial cells (Andersen and Sandaa, 1994; Sia et al., 1996). This type of recruitment, in which a contributor strain possessing a self-transmissible plasmid is getting a second plasmid from a receiver strain, is known as retrotransfer. Retrotransfer take place through two stages; (1) self-transmissible plasmid move from the contributor to the receiver, and (2) mobilization of plasmid from receiver back to the contributor (Ankenbauer, 1997). As the capacity of a self-transmissible plasmid to promote the acquirement of novel plasmids by its bacterial host possibly grant a benefit to the contributor bacterium, it can be said retrotransfer may play a significant job in the progression of plasmid transfer system.
Transphylum mobilization events that incorporate elements from entirely diverse phylogenetic group of bacteria, underscores broad range of interactive capacity originated in gene relocation elements. Besides being recognized through PCR amplification of identified incompatibility groups (Götz et al., 1996), mobilizing plasmids are identified by conducting a triparental mating among E. coli having a mobilizable IncQ plasmid, a recipient lacking plasmid and amalgam of local soil or marine bacteria (Top et al., 1994). Endurance of plasmids in natural isolates and their perceptible firmness in absence of antibiotics, opposes a common thought that in non-existence of selection pressure plasmids additional gene transport factors are easily lost. The multitude of antibiotic-resistant strains in environmental milieu where bacteria apparently do not appear to be in touch with antibiotics, suggest that resistance genes can also be firmly retained even in the paucity of antibiotic selection (Andersen and Sandaa, 1994; McKeon et al., 1995; Calva et al., 1996). As plasmids exhibit remarkable property of crossing species borders effortlessly (Hatch and Michael, 2011) co-mobilization of resistance genes aggravates furthermore clinical crisis.
Transposable elements (TE) are the DNA sequences that provide flexibility to the genome (Archana et al., 2013). Being proficient to alter their position, they are able to alter their genetic background along with that they change the genetic setting of the locus, where they get inserted (Wicker et al., 2007; Shapiro, 2010). Based on their role in identification and recombination of particular sequences, TEs are categorized into two classes; composite transposons (Class I; holding a range of resistance genes which possess identical structural and functional characteristics, but small DNA homology) and complex transposons (class II; constituting three dissimilar but interrelated families; Tn3, Tn21 and Tn2501; Schmitt, 1986; Wiedemann et al., 1986; Lafond et al., 1989). Some of the composite transposons in gram negative bacteria are Tn5, Tn9, Tn10, Tn903, Tn1525, and Tn2350 and among gram positive bacteria are Tn4001 and Tn4003. Compared to Tn1, Tn3, Tn21, Tn501, Tn1721, and Tn3926 found among gram negative bacteria, gram positive bacteria encompass Tn551, Tn917 and Tn4451 complex resistance transposons. These components are possessed with the capability to progress both intra and inter-molecularly which means they can jump within a DNA molecule or from one DNA molecule to another (Bennett, 2008). Tn21 being majorly studied, bear OXA (a carbapenems, possess oxacillinase activity) and PSE (β-lactam gene Pseudomonas specific enzyme) determinants that makes them resistant to aminoglycoside antibiotics. Tn21 also show resistance toward mercury compounds (Brown et al., 1986) and trimethoprim imparted by dhfr II and V (Sundström et al., 1988). Class I or retro-transposons work by copying RNA from DNA by transcription and RNA to DNA by reverse transcription; thereby get inserted into the genome at a diverse location (Kapitonov and Jurka, 2008). Acting in cut and paste manner, class II transposons does not involve RNA intermediate (Wicker et al., 2007). These, transposases create staggered cut at specific site, creating sticky ends; following its transposition to the aimed site, generally followed by target site duplication and construction of short direct repeat at insertion sites (Madigan et al., 2006). Though transposons provide antibiotic resistance due to the existence of an extra gene on a plasmid, there are chances that transposons can jump from chromosomal DNA to plasmid DNA and vice versa for development of resistance (Wagner, 2006).
Insertion Sequences (ISs; size < 2.5 kb) are basic form of mobile genetic elements disseminated in bacteria. ISs are contemplated as non-complex bacterial mobile DNA taking into account their structure (Allaaeddin El et al., 2013). They include more than 19 families, having dissimilar size (Wagner et al., 2007). ISs include an open reading frame that codes for a transposase enzyme, surrounded by inverted repeat sequences of 10–40 base pairs at both ends. The transposase enzyme cuts target DNA and inserts the IS due to possible association with the inverted repeat sequences. Exhibiting fondness toward AT-rich region of DNA, higher chances of undergoing homologous recombination, creates variety of possibilities such as deletions, inversions and duplications. There are evidences that when two identical IS elements surround a region of DNA, a composite transposon is produced, and the total interceded DNA flanked by the terminal inverted repeats get mobilized by one or both of the IS coded transposases (Ochi et al., 2009; Gyles and Boerlin, 2014).
Integrons attribute a great deal to the fitness quotient and robustness of bacteria to survive in varying environments. Harboring resistance determinants such as antibiotic resistance genes, their mobilization as part of chromosomes and plasmids and integration far off from their origin confer resistance to antimicrobials. Their categorization is based on amino acid sequences of integrase IntI; those carrying IntI1 are referred as class 1, IntI2 as class 2, IntI3 as class 3 and so on. Integrase IntI1, IntI2, and IntI3 were found associated with mobile genetic elements, while IntI4 was found linked with chromosomal integrons.
Class I integrons are found associated with the acquisition and mobilization of antibiotic resistance genes. Originated from Tn402, they are composed of two sequence; 5′ conserved sequence (5′CS) representing an integrase gene and a 3′ conserved sequence (3′CS) encoding quaternary ammonium compound resistance gene (qacΔE1) and sulfonamide resistance gene (sul1), respectively (Cambray et al., 2010). With three recombination sites (attI1, attC and secondary site), expression of captured gene cassettes acquired via site-specific recombination is driven by a promoter located in the 5′-conserved segment (5′-CS) region (Collis et al., 1993). Class 1 integrons are associated with a variety of resistance gene cassettes, but most integrons contain an aadA resistance determinant, encoding streptomycin-spectinomycin resistance. Trimethoprim resistance determinants are also detected frequently (Fluit and Schmitz, 2004; Mazel, 2006; Cambray et al., 2010). Showing prevalence of 22–59%, its localization is reported among diverse groups of Gram negative bacteria; Escherichia, Klebsiella, Aeromonas, Enterobacter, Providencia, Mycobacterium, Burkholderia, Alcaligenes, Campylobacter, Citrobacter, Stenotrophomonas, Acinetobacter, Pseudomonas, Salmonella, Serratia, Vibrio, and Shigella (Ramírez et al., 2005; Crowley et al., 2008; Partridge et al., 2009; Xu et al., 2009, 2011). Gram positive bacteria; Enterococcus, Corynebacterium, Streptococcus, Brevibacterium, Aerococcus, and Staphylococcus show high prevalence of aadA and dfrA gene cassettes (Nandi et al., 2004; Xu et al., 2010; Veise et al., 2013).
Class 2 integrons associated with the Tn7 transposon family (Tn1825, Tn1826, and Tn4132), carry a recombination site attI2 and promoter within these transposons (Xu et al., 2009). Its 3′ conserved segment (3′-CS) contains 5 tns genes (tnsA, tnsB, tnsC, tnsD and tnsE) associated with movement and preferential insertion at unique site within bacterial chromosomes (Hansson et al., 2002; Labbate et al., 2009). The amino-acid sequences coded by intI2 gene show <50% homology with intI1, and its non-functionality was found attributed by replacement of glutamic acid with a termination codon (amino acid 179) that leads to production of a shorter and inactive polypeptide (Barlow and Gobius, 2006). The classic structure of class 2 integrons contain a range of gene cassettes, including streptothricin acetyltransferase (sat1), adenyltransferase (aadA1), dihydrofolate reductase (dfrA1; Hansson et al., 2002; Xu et al., 2009). Class 2 integrons have been reported among Salmonella, Enterobacteriaceae, Acinetobacter, and Psuedomonas (Machado et al., 2008; Vinué et al., 2008; Macedo-Viñas et al., 2009; Ozgumus et al., 2009; Xu et al., 2009, 2010, 2011).
Class 3 integrons (IntI3) create excision of integrated cassettes and integration of circularized cassettes into the attI3 site with a considerably lower recombination than that observed with IntI1 (Arakawa et al., 1995). This class of integron was firstly identified from Serratia marcescens in 1993, and then found associated with blaGES-1 from Klebsiella pneumoniae strain FFUL 22K. Class 3 integron containing blaGES-1 within the IncQ plasmid was also found in E. coli (Collis et al., 2002). Occurence of class 3 integrons associated with IMP-1 metallo-beta-lactamase is limited to Acinetobacter, Alcaligenes, Citrobacter, Escherichia, Klebsiella, Pseudomonas, Salmonella, and Serratia (Arakawa et al., 1995; Rowe-Magnus et al., 1999, 2001; Ploy et al., 2003).
Class 4 integrons are distinguished from other Resistance Integrons (RIs) by two key features; incorporation of hundreds of cassettes (V. cholerae, 216 unidentified genes in an array of 179 cassettes, holding roughly 3% of the genome) and the high similarity between the attC sites of those assembled cassettes (Poirel et al., 2010). Class 4 integrons carry gene cassettes for antibiotics chloramphenicol and fosfomycin (Fluit and Schmitz, 2004). Inspite of huge array of cassettes, identification of class 4 integron has been limited within members of Pseudomonas, Xanthomonas, Shewanella, Vibrionaceae, and other proteobacteria (Clark et al., 2000; Rowe-Magnus et al., 2001).
Integrative and conjugative elements (ICEs) were first of all anticipated by Burrus et al. (2002) are different mobile elements found in both Gram-positive and Gram-negative bacteria. These are self transmissible integrative elements that code for complete match of conjugation apparatus. ICEs can integrate into and excise from a host chromosome. These versatile entities support their own mobilization facilitating horizontal transfer of antibiotic-resistant genes, virulence factors and various bacterial traits. ICEs possess three genetic modules: (i) integration and excision module; (ii) conjugation module; and (iii) regulation module. These modules contain different array of genes that code for proteins operating by distinct mechanisms. ICEs contain a gene encoding an integrase (Int) that promotes site-specific integration and excision of the element, frequently into a unique site on the chromosome of the host organism (Boyd et al., 2009). Some ICEs bear maintenance modules such as toxin–antitoxin systems (Wozniak and Waldor, 2009) and additional partition systems that guarantee thriving vertical inheritance of these elements. In contrast to plasmids, ICEs, are not found in extrachromosomal state, because they lack autonomous replication, the first known MGEs with ICE-like properties were Tn916 in Enterococcus faecalis and CTnDOT in Bacteriodes thetaiotaomicron; Franke and Clewell, 1981; Shoemaker et al., 1989). Bacteroides CTnDOT promote dissemination of antibiotic-resistant genes (Whittle et al., 2002). ICEs are distinguished by element-specific properties although they possess a general life cycle and modular structure. Apart from resistance to antibiotics (Böltner et al., 2002; Whittle et al., 2002) ICEs show a extensive collection of phenotypes on their hosts, including resistance for heavy metals (Böltner et al., 2002; Davies et al., 2009) and the power to degrade aromatic compounds (Ravatn et al., 1998). In addition, complex traits such as the capacity to inhabit a eukaryotic host (Sullivan and Ronson, 1998) fix nitrogen (Sullivan and Ronson, 1998) or encourage virulence and biofilm development have been recognized (Drenkard and Ausubel, 2002; He et al., 2004; Davies et al., 2009) the connection between ICEs and the propagation of antibiotic resistance genes in some pathogens show that these mobile elements have considerable clinical significance (Hochhut et al., 2001; Whittle et al., 2002; Mohd-Zain et al., 2004). ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/) is an integrated database that provides comprehensive information about integrative and conjugative elements (ICEs) found in bacteria (Bi et al., 2012).
Toxin-antitoxin (TA) systems, initially identified as plasmid addiction modules, are plentiful in the chromosomes of most free-living bacteria (Xie et al., 2018). TA systems provide endurance to bacterial populations in conditions of stress like nutrient deprivation or antibiotic pressure (Harms et al., 2016). Generally TA systems are made of a stable toxin and a labile antitoxin coded by a bicistronic locus (Lee and Lee, 2016). Toxin genes encode for proteins, while the antitoxin genes code for RNAs or antitoxin proteins, classifying them as type I–VI TA loci (Gerdes and Maisonneuve, 2012; Chan et al., 2016; Page and Peti, 2016) categorized due to mechanisms applied by the antitoxins to counteract the actions of the toxins. In TA systems I-VI product of the toxin gene is typically a protein, whereas the antitoxin gene is either a non-coding RNA among TA I and III or a low-molecular-weight protein in TA systems II, IV, V and VI. Toxins work on diverse targets to distress various cellular processes such as DNA replication, cell wall synthesis or protein synthesis. Amongst six types of TA system, type II is broadly studied, due to great quantity and high quality of freely accessible data. Currently two open-access bioinformatics resources in the field of type II TA loci, the online tool RASTA (Sevin and Barloy-Hubler, 2007) and the web-based database TADB (Shao et al., 2011) are available.
Presently, a new toxin is reported that contains a Gcn5-related N-acetyltransferase (GNAT) domain that transfers the acetyl group from acetyl coenzyme A (Acetyl Co~A) to the amine group of tRNAs (Jurenas et al., 2017b; Van Melderen and Wood, 2017) resulting in acetylation of tRNAs following inhibition of translation in bacterial cells. Similarly TacT of Salmonella enterica Typhimurium (Cheverton et al., 2016) and AtaT of Escherichia coli O157:H7 (Jurenas et al., 2017a), also transfer the acetyl group from acetyl coenzyme A to the amine group of the tRNAs, In stress environment, the intensity of the alarmone molecule, (p)ppGpp, is amplified, which activates a particular proteinase that degrades the antitoxin by proteolytic cleavage, thus permitting the toxin to stop cell expansion. The free toxins then cause the dormant state of bacterial cells (persister cells) which can encourage bacterial tolerance to antibiotics (Gerdes and Maisonneuve, 2012). Persisters are a fraction of bacterial cells in the culture that survive through a prolonged antibiotic treatment. Many studies have shown that toxins of diverse chromosomal TA systems encourage the development of persister cells. GNAT-RHH TA system is a newly exposed approach of bacterial cells to support persister cell formation by disturbing tRNA functions (Jurenas et al., 2017b), in S. enterica over expression of the TacT causes drug tolerance (Cheverton et al., 2016). Chromosomal type II TA loci have been reported to be activated by environmental stress (Li et al., 2016). To assess if antibiotic stress would stimulate the transcription of kacAT operon, exponential phase HS11286-RR2 cells were checked with different antibiotics at the minimum inhibitory concentration (MIC). Transcript levels of the toxin gene, kacT, were quantified by RT-qPCR at various time points after antibiotic challenge. RT-qPCR results showed that the exposure to meropenem or tigecycline antibiotics caused 10-fold or 40-fold increase in kacT transcription levels (Qian et al., 2018).
Klebsiella pneumoniae faces a wide diversity of environmental conditions, including antibiotic stress. Fifteen pairs of putative type II TA loci are detected on the K. pneumoniae HS11286 chromosome. Activation of the toxin plays an important role in bacterial multidrug tolerance (Harms et al., 2016). The chromosomally encoded kacAT bicistronic operon of K. pneumoniae HS11286 is a functional GNAT-RHH TA locus with kacA encoding the cognate antitoxin to the toxic product of kacT. (Hall et al., 2017). Over expression of KacT inhibited K. pneumoniae cell growth and resulted in dormant cell formation. Crystal structure analyses show that the KacT toxin adapts a typical GNAT-fold, which may confer the same catalytic mechanism as the one revealed for TacT of S. enterica (Cheverton et al., 2016). It may bind cellular tRNAs via its positive groove and transfer the acetyl group from AcCoA to tRNAs halting translation leading to cell arrest.
Emerging antibiotic resistance is a problem of global magnitude. Confronted by increasing amounts of antibiotics use, emergence and dissemination of antibiotic resistant strains has compromised therapeutic potential of antibiotics. Of the different strategies adopted, techniques that materialize ideally include; (1) Designing antimicrobial peptides (AMPs; Bacteriocins, Cathelicidins, Microcins, etc.) with broader spectrum of targets (Gordya et al., 2017), (2) Phage therapy (exploiting phages such as OMKO1, wksl3 and Φ1 to kill antibiotic-resistant bacteria; Chan et al., 2016), (3) combination therapy (using combination of antibiotics e.g., colistin in association with tigecycline, aminoglycoside, meropenem, etc or combination of inhibitor and antibiotic such as Augmention i.e., combination of clavulanate and amoxicillin; Soudeiha et al., 2017), (4) Delivery of drugs, AMPs and essential oils as nanoparticles (NPs) for sustained and controlled release (AgNPs of penicillin G, amoxicillin, erythromycin, and vancomycin; Wang et al., 2017), (5) Liposomes as drug targeting vehicles (Poerio et al., 2017), (6) Use of natural compounds such as Flavonoids, Alkaloids, Coumarins, etc. (Chandra et al., 2017), and (7) Modification of antimicrobials e.g., Plazomicin (ACHN-490); derivative of sisomicin produced by addition of a hydroxy-aminobutyric acid substituent at position 1 and a hydroxyethyl substituent at position 6 (Lopez-Diaz et al., 2017; Table 2). Other approaches include use of genomics to find out new bacterial targets and optimization of newer approaches that target bacterial pathogens while exerting selection for reduced pathogenesis, if bacteria evolve resistance to therapeutic intervention. Additionally, strategies such as designing molecules that can block bacterial attachment to surfaces and target bacterial virulence factors along with contribution to protect through production of inactivating antibodies, seems other suitable options to over the menace of drug resistance.
Bacterial infections continue to be one of the leading causes of morbidity and mortality worldwide. Fallout of excessive and imprudent antibiotic use, widespread dissemination of resistant determinants as part of MGEs has increased the rate of resistance development. Being capable to relocate between host genomes, they act as vehicles for resistance gene acquisition and their successive propagation. Thorough molecular studies have identified several mechanisms in microbes to attain the antimicrobial resistance. Among these mechanisms, plasmids, transposons, insertion sequences, integrons, ICEs and bacterial Toxin Anti-toxin systems have exposed how and why resistance has attained alarming stage. The possibility for recombination of genes from different bacterial populations is huge and it seems that it doesn't take bacteria much time to acquire the genetic resources to flourish in surroundings that would have otherwise hindered it's growth. Occurring with increasing frequency, resistance limits therapeutic option, resulting in the cases where certain human infections cannot be treated. Pertinently, where there is stiff resistance on the implementation of evidence-based clinical practice, scientists of the health care organizations are still searching as how to keep pace with the demand of actionable knowledge. This adverse condition of antimicrobial resistance demands the rejuvenation of dried pipeline for the development of new and efficient drugs to treat the deadly infection. With a goal to get hold of the menace of antibiotic resistance, it seems essential for everybody to have some basic knowledge about the systems in order to ensure optimal use of antibiotics from the surrounding milieu, to slow down the development of antibiotic-resistant superbugs.
QH and AJ conceived the idea. IS, SR and AJ contributed to writing of the manuscript. MS and AM contributed to reference and graphics section.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author extends thanks to colleagues from Department of Biosciences, Jamia Millia Islamia and School of Biosciences and Biotechnology, Baba Ghulam Shah Badshah University, for criticism that helped to improve the quality of contents in the perspective of broader audience. Authors also acknowledge help from Dr. Waseem Ahmad Wani, who helped with the designing of antibiotic structures. AJ would like to thank University Grants Commission (UGC), New Delhi for startup.
Aldred, K. J., Kerns, R. J., and Osheroff, N. (2014). Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574. doi: 10.1021/bi5000564
Allaaeddin El, S., Timothey, R. W., and Chouchani, C. (2013). Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in gram-negative bacteria. Crit. Rev. Microbiol. 39, 113–122. doi: 10.3109/1040841X.2012.691870
Aminov, R. I., and Mackie, R. I. (2007). Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161. doi: 10.1111/j.1574-6968.2007.00757.x
Andersen, S. R., and Sandaa, R. A. (1994). Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 60, 908–912.
Andriole, V. T. (2005). The quinolones: past, present, and future. Clin. Infect. Dis. 41, S113–S119. doi: 10.1086/428051
Ankenbauer, R. G. (1997). Reassessing forty years of genetic doctrine: retrotransfer and conjugation. Genetics 145, 543–549.
Arakawa, Y., Murakami, M., Suzuki, K., Ito, H., Wacharotayankun, R., Ohsuka, S., et al. (1995). A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39, 1612–1615. doi: 10.1128/AAC.39.7.1612
Archana, I., Elie, B., Esam, I. A., Alaaeddin, A. E. S., Hani Mutlak, A. H., Ishtiaq, Q., et al. (2013). Transposable elements in Escherichia coli antimicrobial resistance. Adv. Biosci. Biotechnol. 4, 415–423. doi: 10.4236/abb.2013.43A055
Bae, S., Kim, M. C., Park, S. J., Kim, H. S., Sung, H., Kim, M. N., et al. (2016). In vitro synergistic activity of antimicrobial agents in combination against clinical isolates of colistin-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 6774–6779. doi: 10.1128/AAC.00839-16
Barlow, R. S., and Gobius, K. S. (2006). Diverse class 2 integrons in bacteria from beef cattle sources. J. Antimicrob. Chemother. 58, 1133–1138. doi: 10.1093/jac/dkl423
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153, S347–S357. doi: 10.1038/sj.bjp.0707607
Beyth, N., Houri-Haddad, Y., Domb, A., Khan, W., and Hazan, R. (2015). Alternative antimicrobial approach: nano-antimicrobial materials. Evid. Based Complement Alternat. Med. 2015:246012. doi: 10.1155/2015/246012
Bi, D., Jiang, X., Sheng, Z. K., Ngmenterebo, D., Tai, C., Wang, M., et al. (2015). Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a resistance-disarmed' model organism. J. Antimicrob. Chemother. 70, 2770–2774. doi: 10.1093/jac/dkv204
Bi, D., Xu, Z., Harrison, E. M., Tai, C., Wei, Y., He, X., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40, D621–D626. doi: 10.1093/nar/gkr846
Biswas, S., Brunel, J. M., Dubus, J. C., Reynaud-Gaubert, M., and Rolain, J. M. (2012). Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78
Böltner, D., MacMahon, C., Pembroke, J. T., Strike, P., and Osborn, A. M. (2002). R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184, 5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002
Bonfiglio, G., Russo, G., and Nicoletti, G. (2002). Recent developments in carbapenems. Expert Opin. Investig Drugs 11, 529–544. doi: 10.1517/13543784.11.4.529
Bonomo, R. A., and Szabo, D. (2006). Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43, S49–56. doi: 10.1086/504477
Boyd, E. F., Almagro-Moreno, S., and Parent, M. A. (2009). Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17, 47–53. doi: 10.1016/j.tim.2008.11.003
Brown, N. L., Misra, T. K., Winnie, J. N., Schmidt, A., Seiff, M., and Silver, S. (1986). The nucleotide sequence of the mercuric resistance operons of plasmid RlOO and transposon TnSOZ; further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 202, 143–151. doi: 10.1007/BF00330531
Burrus, V., Pavlovic, G., Decaris, B., and Guedon, G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46, 601–610. doi: 10.1046/j.1365-2958.2002.03191.x
Calva, J. J., Niebla-Perez, A., Rodriguez-Lemoine, V., Santos, J. I., and Amabile-Cuevas, C. F. (1996). “Antibiotic usage and antibiotic resistance in Latin America,” in Antibiotic Resistance: From Molecular Basics to Therapeutic Options, ed C. F. Amabile-Cuevas (Austin, TX: R. G. Landes Co.), 78–97.
Cambray, G., Guerout, A. M., and Mazel, D. (2010). Integrons. Annu. Rev. Genet. 44, 141–166. doi: 10.1146/annurev-genet-102209-163504
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Cattoir, V., Poirel, L., and Nordmann, P. (2008). Plasmid-Mediated Quinolone Resistance Pump QepA2 in an Escherichia coli Isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804. doi: 10.1128/AAC.00638-08
Chan, B. K., Sistrom, M., Wertz, J. E., Kortright, K. E., Narayan, D., and Turner, P. E. (2016). Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6:26717. doi: 10.1038/srep26717
Chandra, H., Bishnoi, P., Yadav, A., Patni, B., Mishra, A. P., and Nautiyal, A. R. (2017). Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials-a review. Plants 6:16. doi: 10.3390/plants6020016
Cheverton, A. M., Gollan, B., Przydacz, M., Wong, C. T., Mylona, A., Hare, S. A., et al. (2016). A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 63, 86–96. doi: 10.1016/j.molcel.2016.05.002
Cho, J. H., Sung, B. H., and Kim, S. C. (2009). Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim. Biophys. Acta 1788, 1564–1569. doi: 10.1016/j.bbamem.2008.10.025
Cirioni, O., Silvestri, C., Ghiselli, R., Orlando, F., Riva, A., Mocchegiani, F., et al. (2008). Protective effects of the combination of alpha-helical antimicrobial peptides and rifampicin in three rat models of Pseudomonas aeruginosa infection. J. Antimicrob. Chemother. 62, 1332–1338. doi: 10.1093/jac/dkn393
Clark, C. A., Purins, L., Kaewrakon, P., Focareta, T., and Manning, P. A. (2000). The Vibrio cholerae O1 chromosomal integron. Microbiology 146, 2605–2612. doi: 10.1099/00221287-146-10-2605
Collis, C. M., Grammaticopoulos, G., Briton, J., Stokes, H. W., and Hall, R. M. (1993). Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9, 41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x
Collis, C. M., Kim, M. J., Partridge, S. R., Stokes, H. W., and Hall, R. M. (2002). Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 84, 3017–3126. doi: 10.1128/JB.184.11.3017-3026.2002
Courvalin, P. (2008). New plasmid mediated resistances to antimicrobial agents. Arch. Microbiol. 189, 289–291. doi: 10.1007/s00203-007-0331-9
Crowley, D., Cryan, B., and Lucey, B. (2008). First detection of a class 2 integron among clinical isolates of Serratia marcescens. Br. J. Biomed. Sci. 65, 86–99. doi: 10.1080/09674845.2008.11732803
Davies, M. R., Shera, J., Van Domselaar, G. H., Sriprakash, K. S., and McMillan, D. J. (2009). A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other β-hemolytic streptococci. J. Bacteriol. 191, 2257–2265. doi: 10.1128/JB.01624-08
de Leeuw, E., Li, C., Zeng, P., Li, C., Diepeveen-de Buin, M., Lu, W. Y., et al. (2010). Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 584, 1543–1548. doi: 10.1016/j.febslet.2010.03.004.
Depardieu, F., Podglajen, I., Leclercq, R., Collatz, E., and Courvalin, P. (2007). Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20, 79–114. doi: 10.1186/s12941-015-0100-6
Drenkard, E., and Ausubel, F. M. (2002). Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743. doi: 10.1038/416740a
Emmerson, A. M., and Jones, A. M. (2003). The quinolones: decades of development and use. J. Antimicrob. Chemother. 51, 13–20. doi: 10.1093/jac/dkg208
Fluit, A. C., and Schmitz, F. J. (2004). Resistance integrons and super-integrons. Clin. Microbiol. Infect. 10, 272–288. doi: 10.1111/j.1198-743X.2004.00858.x
Franci, G., Falanga, A., Galdiero, S., Palomba, L., Rai, M., Morelli, G., and Galdiero, M. (2015). Silver nanoparticles as potential antibacterial agents. Molecules 20, 8856–8874. doi: 10.3390/molecules20058856
Franke, A. E., and Clewell, D. B. (1981). Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145, 494–502.
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
García, A., Bocanegra-Garcia, V., Palma-Nicolas, J. P., and Rivera, G. (2012). Recent advances in antitubercular natural products. Eur. J. Med. Chem. 49, 1–23. doi: 10.1016/j.ejmech.2011.12.029
Gerdes, K., and Maisonneuve, E. (2012). Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66, 103–123. doi: 10.1146/annurev-micro-092611-150159
Goossens, H., Ferech, M., Vander, S. R., and Elseviers, M. (2005). Outpatient antibiotic use in europe and association with resistance: a cross-national database study. Lancet 365, 579–587. doi: 10.1016/S0140-6736(05)17907-0
Gordya, N., Yakovlev, A., Kruglikova, A., Tulin, D., Potolitsina, E., Suborova, T., et al. (2017). Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: calliphora vicina medicinal maggots. PLoS ONE 12:e0173559. doi: 10.1371/journal.pone.0173559
Götz, A., Pukall, R., Smit, E., Tietze, E., Prager, R., Tschape, H., et al. (1996). Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62, 2621–2628.
Grape, M. (2006). Molecular Basis for Trimethoprim and Sulphonamide Resistance in Gram Negative Pathogens. Ph.D. thesis, Karolinska Institutet, Stockholm.
Gyles, C., and Boerlin, P. (2014). Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 51, 328–340. doi: 10.1177/0300985813511131
Hall, A. M. J., Gollan, B., and Helaine, S. (2017). Toxin–antitoxin systems: reversible toxicity. Curr. Opin. Microbiol. 36, 102–110. doi: 10.1016/j.mib.2017.02.003
Hall, R. M., Collis, C. M., Kim, M. J., Partridge, S. R., Recchia, G. D., and Stokes, H. W. (1999). Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870, 68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x
Hansson, K., Sundstrom, L., Pelletier, A., and Roy, P. H. (2002). Intl2 integron integrase in Tn7. J. Bacteriol. 184, 1712–1721. doi: 10.1128/JB.184.6.1712-1721.2002
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268
Hassan, M., Kjos, M., Nes, I. F., Diep, D. B., and Lotfipour, F. (2012). Natural antimicrobial peptides from bacteria:characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 113, 723–736. doi: 10.1111/j.1365-2672.2012.05338.x
Hatch, W. S., and Michael, R. G. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. doi: 10.1111/j.1574-6976.2011.00273.x
He, J., Baldini, R. L., Déziel, E., Saucier, M., Zhang, Q., Liberati, N. T., et al. (2004). The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl Acad. Sci. U.S.A. 101, 2530–2535. doi: 10.1073/pnas.0304622101
Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R., and Waldor, M. K. (2001). Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45, 2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001
Hooper, D. C. (2000). Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31, S24–8. doi: 10.1086/314056
Jacob, J. T., and Gaynes, R. P. (2010). Emerging trends in antibiotic use in US Hospitals: quality, quantification and stewardship. Expert Rev. Anti Infect. Ther. 8, 893–902. doi: 10.1586/eri.10.73
Jacoby, G. A. (2005). Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41, S120–S126. doi: 10.1086/428052
Joshua, P. R., Stephen, M. K., Riley, J. T. M., Karina, Y. E., Karina, J. P., Quang, T. N., et al. (2016). An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 6:e1208317. doi: 10.1080/2159256X.2016.1208317
Jurenas, D., Chatterjee, S., Konijnenberg, A., Sobott, F., Droogmans, L., Garcia-Pino, A., et al. (2017a). AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet. Nat. Chem. Biol. 13, 640–646. doi: 10.1038/nchembio.2346
Jurenas, D., Garcia-Pino, A., and Van Melderen, L. (2017b). Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity. Plasmid 93, 30–35. doi: 10.1016/j.plasmid.2017.08.005
Kaneider, N. C., Djanani, A., and Wiedermann, C. J. (2007). Heparan sulfate proteoglycan-involving immunomodulation by cathelicidin antimicrobial peptides LL-37 and PR-39. Sci. World J. 7, 1832–1838. doi: 10.1100/tsw.2007.285
Kapitonov, V. V., and Jurka, J. (2008). A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 9, 411–2. doi: 10.1038/nrg2165-c1
Kaur, K., Tarassova, O., Dangeti, R. V., Azmi, S., Wishart, D., McMullen, L., et al. (2016). Characterization of a highly potent antimicrobial peptide microcin N from uropathogenic Escherichia coli. FEMS Microbiol. Lett. 363:11. doi: 10.1093/femsle/fnw095
Klare, I., Konstabel, C., Badstubner, D., Werner, G., and Witte, W. (2003). Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 88, 269–290. doi: 10.1016/S0168-1605(03)00190-9
Kluytmans, J., van Belkum, A., and Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520.
Kościuczuk, E. M., Lisowski, P., Jarczak, J., Strzałkowska, N., Jozwik, A., Horbanczuk, J., et al. (2012). Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39, 10957–10970. doi: 10.1007/s11033-012-1997-x
Kotra, L. P., Haddad, J., and Mobashery, S. (2000). Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44, 3249–3256. doi: 10.1128/AAC.44.12.3249-3256.2000
Labbate, M., Case, R. J., and Stokes, H. W. (2009). The integron/gene cassette system: an active player in bacterial adaptation. Methods Mol. Biol. 532, 103–125. doi: 10.1007/978-1-60327-853-9_6
Lafond, M., Couture, F., Vezina, G., and Levesque, R. C. (1989). Evolutionary perspectives on multiresistance β-lactamase transposons. J. Bacteriol. 171, 6423–6429. doi: 10.1128/jb.171.12.6423-6429.1989
Lee, K. Y., and Lee, B. J. (2016). Structure, biology, and therapeutic application of toxin–antitoxin systems in pathogenic bacteria. Toxin 8:305. doi: 10.3390/toxins8100305
Lehrer, R. I., Barton, A., Daher, K. A., Harwig, S. S., Ganz, T., and Selsted, M. E. (1989). Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Invest. 8, 553–561. doi: 10.1172/JCI114198
Li, P., Tai, C., Deng, Z., Gan, J., Oggioni, M. R., and Ou, H. Y. (2016). Identification and characterization of chromosomal relBE toxin-antitoxin locus in Streptomyces cattleya DSM46488. Sci. Rep. 6:32047. doi: 10.1038/srep32047
Lina, G., Piemont, Y., Godail-Gamot, F., Bes, M., Peter, M. O., Gauduchon, V., et al. (1999). Involvement of panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and Pneumonia. Clin. Infect. Dis. 29, 1128–1132. doi: 10.1086/313461
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lopez-Diaz, M. C., Culebras, E., Rodriguez-Avial, I., Rios, E., Vinuela-Prieto, J. M., Picazo, J. J., et al. (2017). Plazomicin activity against 346 extended-spectrum β-lactamase/Amp C producing Escherichia coli urinary isolates in relation to aminoglycoside modifying enzymes. Antimicrob. Agents Chemother. 61, e02454–e02516. doi: 10.1128/AAC.02454-16
Macedo-Viñas, M., Cordeiro, N. F., Bado, I., Herrera-Leon, S., Vola, M., Robino, L., et al. (2009). Surveillance of antibiotic resistance evolution and detection of class 1 and 2 integrons in human isolates of multi-resistant Salmonella Typhimurium obtained in Uruguay between 1976 and 2000. Int. J. Infect. Dis. 13, 342–348. doi: 10.1016/j.ijid.2008.07.012
Machado, E., Coque, T. M., Canton, R., Sousa, J. C., and Peixe, L. (2008). Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 62, 296–302. doi: 10.1093/jac/dkn179
Madigan, M. T., Martinko, J. M., and Brock, T. D. (2006). Brock Biology of Microorganisms. Upper Saddle River, NJ: Pearson Prentice Hall.
Maria-Neto, S., Caroline de Almeida, K., Macedo, M. L. R., and Franco, O. L. (2015). Understanding bacterial resistance to antimicrobial peptides: From the surface to deep inside. Biochim. Biophys. Acta 1848, 3078–3088. doi: 10.1016/j.bbamem.2015.02.017
Martinez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 18, 365–7. doi: 10.1126/science.1159483
Martinez-Martinez, L., Cano, M. E., Rodriguez-Martinez, R. M., Calvo, J., and Pascual, A. (2008). Plasmid mediated quinolone resistance. Expert Rev. Anti Infect. Ther. 6, 685–711. doi: 10.1586/14787210.6.5.685
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
McKeon, D. M., Calabrese, J. P., and Bissonette, G. K. (1995). Antibiotic resistant gram-negative bacteria in rural ground water supplies. Water Res. 29, 1902–1908. doi: 10.1016/0043-1354(95)00013-B
Meletis, G. (2016). Carbapenem resistance: overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 3, 15–21. doi: 10.1177/2049936115621709
Mitscher, L. A. (2005). Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem. Rev. 105, 559–592. doi: 10.1021/cr030101q
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D. F., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of Lipopolysaccharide production. Antimicrobial. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Mohd-Zain, Z., Turner, S. L., Cerdeño-Tárraga, A. M., Lilley, A. K., Inzana, T. J., Duncan, A. J., et al. (2004). Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186, 8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004
Nag, O. K., and Awasthi, V. (2013). Surface engineering of liposomes for stealth behavior. Pharmaceutics 5, 542–569. doi: 10.3390/pharmaceutics5040542
Nandi, S., Maurer, J. J., Hofacre, C., and Summers, A. O. (2004). Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci. U.S.A. 101, 7118–7122. doi: 10.1073/pnas.0306466101
Nocek, B., Tikhonov, A., Babnigg, G., Gu, M., Zhou, M., Makarova, K. S., et al. (2012). A Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis. J. Mol. Biol. 420, 366–383. doi: 10.1016/j.jmb.2102.04.011
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Ochi, S., Shimizu, T., and Ohtani, K. (2009). Nucleotide sequence analysis of the enterotoxigenic Escherichia coli Ent plasmid. DNA Res.16, 299–309. doi: 10.1093/dnares/dsp015
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Ozgumus, O. B., Sandalli, C., Sevim, A., Celik-Sevim, E., and Sivri, N. (2009). Class 1 and class 2 integrons and plasmid-mediated antibiotic resistance in coliforms isolated from ten rivers in northern Turkey. J. Microbiol. 47, 19–27. doi: 10.1007/s12275-008-0206-z
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., and Hooper, D. C. (2006). Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06 .
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Perichon, B., Courvalin, P., and Galimand, M. (2007). Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51, 2464–2469. doi: 10.1128/AAC.00143-07
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47, 1169–1172. doi: 10.1128/AAC.47.3.1169-1172.2003
Ploy, M. C., Chainier, D., Tran Thi, N. H., Poilane, I., Cruaud, P., Denis, F., et al. (2003). Integron-associated antibiotic resistance in Salmonella enterica serovar typhi from Asia. Antimicrob. Agents Chemother. 47, 1427–1429. doi: 10.1128/AAC.47.4.1427-1429.2003
Poerio, N., Bugli, F., Taus, F., Santucci, M. B., Rodolfo, C., Cecconi, F., et al. (2017). Liposomes loaded with bioactive lipids enhance antibacterial innate immunity irrespective of drug resistance. Sci. Rep. 7:45120. doi: 10.1038/srep45120
Poirel, L., Carattoli, A., Bernabeu, S., Bruderer, T., Frei, R., and Nordmann, P. (2010). A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J. Antimicrob. Chemother. 65, 1594–1598. doi: 10.1093/jac/dkq166
Poirel, L., Pitout, J. D., and Nordmann, P. (2007). Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2, 501–512. doi: 10.2217/17460913.2.5.501
Projan, S. J. (2003). Why is big pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6, 427–430. doi: 10.1016/j.mib.2003.08.003
Qian, H., Yao, Q., Tai, C., Deng, Z., Gan, J., and Ou, H. Y. (2018). Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae. Mol. Microbiol. 108, 336–349. doi: 10.1111/mmi.13934
Rådström, P., and Swedberg, G. (1988). RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32, 1684–1692. doi: 10.1128/AAC.32.11.1684
Ramirez, M. S., and Tolmansky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ramírez, M. S., Vargas, L. J., Cagnoni, V., and Tokumoto, M. (2005). Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob. Agents Chemother. 49, 4418–4420. doi: 10.1128/AAC.49.10.4418-4420.2005
Ravatn, R., Studer, S., Springael, D., Zehnder, A. J., and van der Meer, J. R. (1998). Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180, 4360–4369.
Rebuffat, S. (2012). Microcins in action: amazing defence strategies of Enterobacteria. Biochem. Soc. Trans. 40, 1456–1462. doi: 10.1042/BST20120183
Reynolds, P. E. (1989). Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8, 943–950.
Richard, J. F., and Yitzhak, Tor. (2014). Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 6, 25–64. doi: 10.4137/PMC.S14459
Roberts, M. C. (2002). Resistance to tetracycline, macrolidelincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechnol. 20, 261–284. doi: 10.1385/MB:20:3:261
Rowe-Magnus, D. A., Guerout, A. M., and Mazel, D. (1999). Super-integrons. Res. Microbiol. 150, 641–651. doi: 10.1016/S0923-2508(99)00127-8
Rowe-Magnus, D. A., Guerout, A. M., Ploncard, P., Dychinco, B., Davies, J., and Mazel, D. (2001). The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci, U.S.A. 98, 652–657. doi: 10.1073/pnas.98.2.652
Rukavina, Z., and Vanic, Z. (2016). Current trends in development of liposomes for targeting bacterial biofilms. Pharmaceutics 8:E18. doi: 10.3390/pharmaceutics8020018
Sánchez, M. (2015). Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 6:658. doi: 10.3389/fmicb.2015.00658
Savoia, D. (2012). Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol. 7, 979–90. doi: 10.2217/fmb.12.68
Schaack, S., Gilbert, C., and Feschotte, C. (2010). Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 25, 537–546. doi: 10.1016/j.tree.2010.06.001
Schmitt, R. (1986). Molecular biology of transposable elements. J. Antimicrob. Chemother. 18, 25–34. doi: 10.1093/jac/18.Supplement_C.25
Schwarz, S., and Johnson, A. P. (2016). Transferable resistance to colistin: a new but old threat. J. Antimicrob. Chemother. 71:206670. doi: 10.1093/jac/dkw274
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Schweizer, H. (2003). Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 2, 48–62.
Sengupta, S., Chattopadhyay, M. K., and Grossart, H.P. (2013). The multifaceted roles of antibiotics and antib iotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Sevin, E. W., and Barloy-Hubler, F. (2007). RASTA-bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 8:R155. doi: 10.1186/gb-2007-8-8-r155
Shao, Y., Harrison, E. M., Bi, D., Tai, C., He, X., Ou, H. Y., et al. (2011). TADB: a web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, 606–611. doi: 10.1093/nar/gkq908
Shapiro, J. A. (2010). Mobile DNA and evolution in the 21st century. Mob. DNA 1:4. doi: 10.1186/1759-8753-1-4
Shoemaker, N. B., Barber, R. D., and Salyers, A. A. (1989). Cloning and characterization of a Bacteroidesconjugal tetracycline erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 171, 1294–1302. doi: 10.1128/jb.171.3.1294-1302.1989
Sia, E. A., Kuehner, D. M., and Figurski, D. H. (1996). Mechanism of retrotransfer in conjugation: prior transfer of the conjugative plasmid is required. J. Bacteriol. 178, 1457–1464. doi: 10.1128/jb.178.5.1457-1464.1996
Soudeiha, M. A. H., Dahdouh, E. A., Azar, E., Sarkis, D. K., and Daoud, Z. (2017). In vitro evaluation of the colistin-carbapenem combination in clinical isolates of a baumannii using the checkerboard, etest, and time-kill curve techniques. Front. Cell. Infect. Microbiol. 7:209. doi: 10.3389/fcimb.2017.00209
Spellberg, B., Guidos, R., Gilbert, D., Bradley, J., Boucher, H. W., Scheld, W. M., et al. (2008). The epidemic of antibiotic-resistant infections: a call to action for the medical community from the infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164. doi: 10.1086/524891
Stokes, H. W., and Hall, R. M. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3, 1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Sullivan, J. T., and Ronson, C. W. (1998). Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl Acad.Sci. U.S.A. 95, 5145–5149. doi: 10.1073/pnas.95.9.5145
Sundström, L., Radstrom, P., Swedberg, G., and Skold, O. (1988). Site specific recombination promotes linkage between trimethoprim and sulfonamide-resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213, 191–201. doi: 10.1007/BF00339581
Swedberg, G., and Sköld, O. (1983). Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. J. Bacteriol. 153, 1228–1237.
Thangamani, S., Mohammad, H., Abushahba, M. F. N., Sobreira, T. J. P., Hedrick, V. E., Paul, L. N., et al. (2016). Antibacterial activity and mechanism of action of auranofin against multi-drug resistant bacterial pathogens. Sci. Rep. 6:22571. doi: 10.1038/srep22571
Tillotson, G. S., and Theriault, N. (2013). New and alternative approaches to tackling antibiotic resistance. F1000Prime Rep. 5:51. doi: 10.12703/P5-51
Top, E. I., De Smet, W., Verstraete, R., Dijkmans, R, and Mergeay, M. (1994). Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl. Environ. Microbiol. 60, 831–839.
Tsodikova, S. G., and Labby, K. J. (2016). Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Med. Chemcomm. 7, 11–27. doi: 10.1039/C5MD00344J
Van Melderen, L., and Wood, T. K. (2017). Commentary: what is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 8:191. doi: 10.3389/fmicb.2017.00191
Veise, P., Ramazanzadeh, R., Khiababi, Z., Derakhshi, B., and Amirmozafari, N. (2013). Identification of class I integrons gene in Staphylococcus strains isolated from clinical samples. Cell Biol. 1, 24–27. doi: 10.11648/j.cb.20130103.11
Velkov, T., Thompson, P. E., Nation, R. L., and Li, J. (2010). Structure activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916. doi: 10.1021/jm900999h
Vinué, L., Saenz, Y., Somalo, S., Escudero, E., Moreno, M. A., Ruiz-Larrea, F., et al. (2008). Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62, 934–937. doi: 10.1093/jac/dkn331
Wagner, A. (2006). Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol. Biol. Evol. 23, 723–733. doi: 10.1093/molbev/msj085
Wagner, A., Lewis, C., and Bichsel, M. (2007). A survey of bacterial insertion sequences using IScan. Nucleic Acids Res. 35, 5284–5293. doi: 10.1093/nar/gkm597
Walsh, C. (2000). Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781. doi: 10.1038/35021219
Walsh, T. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36, S8–S14. doi: 10.1016/S0924-8579(10)70004-2
Wang, L., Hu, C., and Shao, L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 12, 1227–1249. doi: 10.2147/IJN.S121956
Whittle, G., Shoemaker, N. B., and Salyers, A. A. (2002). The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell Mol. Life Sci. 59, 2044–2054. doi: 10.1007/s000180200004
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–82. doi: 10.1038/nrg2165
Wiedemann, B., Meyer, J. F., and Ziihlsdorf, M. T. (1986). Insertions of resistance genes into Tn21-like transposons. J. Antimicrob. Chemother. 18, 85–92. doi: 10.1093/jac/18.Supplement_C.85
Woodford, N., Turton, J. F., and Livermore, D. M. (2011). Multi resistant Gram negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Wozniak, R. A., and Waldor, M. K. (2009). A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. doi: 10.1371/journal.pgen.1000439
Wright, G. D. (2005). Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57, 1451–1470. doi: 10.1016/j.addr.2005.04.002
Xie, Y., Wei, Y., Shen, Y., Li, X., Zhou, H., Tai, C., et al. (2018). TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 46, D749–D753. doi: 10.1093/nar/gkx1033
Xiea, Y., Fleminga, E., Chena, J. L., and Elmorea, D. E. (2011). Effect of proline position on the antimicrobial mechanism of buforin II. Peptides 32, 677–682. doi: 10.1016/j.peptides.2011.01.010
Xu, H., Broersma, K., Miao, V., and Davies, J. (2011). Class 1 and class 2 integrons in multidrug-resistant gram-negative bacteria isolated from the Salmon River, British Columbia. Can. J. Microbiol. 57, 460–470. doi: 10.1139/w11-029
Xu, Z., Li, L., Shirtliff, M., Alam, M., Yamasaki, S., and Shi, L. (2009). Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J. Clin. Microbiol. 47, 230–234. doi: 10.1128/JCM.02027-08
Xu, Z., Li, L., Shirtliff, M., Peters, B., Peng, Y., Alam, M., et al. (2010). First report of class 2 integron in clinical Enterococcus faecalis and class 1 integron in Enterococcus faecium in South China. Diag. Microbiol. Infect. Dis. 68, 315–317. doi: 10.1016/j.diagmicrobio.2010.05.014
Yu, Z., Qin, W., Lin, J., Fang, S., and Qiu, J. (2015). Antibacterial mechanisms of polymyxin and bacterial resistance. BioMed Res. Int. 11:679109. doi: 10.1155/2015/679109
Zanetti, M. (2004). Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75, 39–48. doi: 10.1189/jlb.0403147
Keywords: antibiotics, bacteria, bacterial resistance, diseases, health care
Citation: Sultan I, Rahman S, Jan AT, Siddiqui MT, Mondal AH and Haq QMR (2018) Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 9:2066. doi: 10.3389/fmicb.2018.02066
Received: 23 March 2018; Accepted: 13 August 2018;
Published: 21 September 2018.
Edited by:
Zhenyu Xie, Hainan University, ChinaReviewed by:
Hong-Yu Ou, Shanghai Jiao Tong University, ChinaCopyright © 2018 Sultan, Rahman, Jan, Siddiqui, Mondal and Haq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arif Tasleem Jan, YXRhc2xlZW1AZ21haWwuY29t
Qazi Mohd Rizwanul Haq, aGFxcW1yQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.