
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 30 August 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02015
This article is part of the Research TopicExtracellular Vesicles Exported by MicroorganismsView all 14 articles
 Antoine Malabirade1†
Antoine Malabirade1† Janine Habier1†
Janine Habier1† Anna Heintz-Buschart1‡
Anna Heintz-Buschart1‡ Patrick May1
Patrick May1 Julien Godet2
Julien Godet2 Rashi Halder1
Rashi Halder1 Alton Etheridge3
Alton Etheridge3 David Galas3
David Galas3 Paul Wilmes1*
Paul Wilmes1* Joëlle V. Fritz1*‡
Joëlle V. Fritz1*‡Bacterial outer membrane vesicles (OMVs), as well as OMV-associated small RNAs, have been demonstrated to play a role in host–pathogen interactions. The presence of larger RNA transcripts in OMVs has been less studied and their potential role in host–pathogen interactions remains largely unknown. Here we analyze RNA from OMVs secreted by Salmonella enterica serovar Typhimurium (S. Typhimurium) cultured under different conditions, which mimic host–pathogen interactions. S. Typhimurium was grown to exponential and stationary growth phases in minimal growth control medium (phosphate-carbon-nitrogen, PCN), as well as in acidic and phosphate-depleted PCN, comparable to the macrophage environment and inducing therefore the expression of Salmonella pathogenicity island 2 (SPI-2) genes. Moreover, Salmonella pathogenicity island 1 (SPI-1), which is required for virulence during the intestinal phase of infection, was induced by culturing S. Typhimurium to the stationary phase in Lysogeny Broth (LB). For each condition, we identified OMV-associated RNAs that are enriched in the extracellular environment relative to the intracellular space. All RNA classes could be observed, but a vast majority of rRNA was exported in all conditions in variable proportions with a notable decrease in LB SPI-1 inducing media. Several mRNAs and ncRNAs were specifically enriched in/on OMVs dependent on the growth conditions. Important to note is that some RNAs showed identical read coverage profiles intracellularly and extracellularly, whereas distinct coverage patterns were observed for other transcripts, suggesting a specific processing or degradation. Moreover, PCR experiments confirmed that distinct RNAs were present in or on OMVs as full-length transcripts (IsrB-1/2; IsrA; ffs; SsrS; CsrC; pSLT035; 10Sa; rnpB; STM0277; sseB; STM0972; STM2606), whereas others seemed to be rather present in a processed or degraded form. Finally, we show by a digestion protection assay that OMVs are able to prevent enzymatic degradation of given full-length transcripts (SsrS, CsrC, 10Sa, and rnpB). In summary, we show that OMV-associated RNA is clearly different in distinct culture conditions and that at least a fraction of the extracellular RNA is associated as a full-length transcripts with OMVs, indicating that some RNAs are protected by OMVs and thereby leaving open the possibility that those might be functionally active.
The secretion of biomolecules into the environment is a common feature of living cells, as organisms can communicate, eliminate undesired products, protect themselves or mediate pathogenicity by the means of export. A wide variety of secretion pathways exist, such as direct export, encapsulation or vesicle formation. Gram-negative bacteria possess several of them, with exports of molecules through the passage across their membranes and cell walls or by the release of outer membrane vesicles (OMVs). OMVs are 50–250 nm spherical particles derived from the outer membrane of Gram-negative bacteria, containing a wide range of molecules. Their release is a phenomenon observed constitutively in many Gram-negative bacteria, including Salmonella (Schwechheimer and Kuehn, 2015). OMV formation is still poorly understood, even though recent advances have highlighted the importance of the LPS synthesis pathway (Schwechheimer et al., 2013; Bonnington and Kuehn, 2016; Elhenawy et al., 2016). Environmental conditions such as oxygen or iron availability, growth phase and media composition influence vesicle biogenesis (Orench-Rivera and Kuehn, 2016). Therefore, to shed light on the identification of OMV-associated molecules it becomes essential to study the content of OMVs originating from different culture conditions.
Outer membrane vesicles’ physiological roles are broad, ranging from functions such as nutrient acquisition, sharing of resistance, stress response or biofilm reinforcement, to more offensive roles like toxin concentration, delivery of virulence factors or modulation of host immune defenses (Manning and Kuehn, 2013; Jan, 2017). Their role and importance in intra- or interspecies communication and in pathogenicity is of growing interest (Schwechheimer and Kuehn, 2015; Jan, 2017; Lynch and Alegado, 2017). Whereas the type III, type IV, and type VI secretion systems imply direct contact between bacteria and their host to allow the release of molecules into the host cytosol, the release of OMVs provides a way to transfer bacterial cargo without being in close contact with the host and thus OMVs can act as long distance carriers (Jan, 2017; Tashiro et al., 2017). In this context, the classification of OMVs as a type 0 secretion system has been proposed recently (Guerrero-Mandujano et al., 2017). OMV delivery to their target occurs by different means, such as membrane fusion with the host or a variety of endocytosis mechanisms (O’Donoghue and Krachler, 2016). OMV-associated biomolecules mainly consist of the periplasmic content, but also contain outer membrane proteins such as OmpA (Bai et al., 2014) as well as a variety of lipids, nucleic acids, proteins and metabolites from the cytoplasm (Jan, 2017).
Over the past few years, several studies on mammalian cells suggest that the horizontal transfer of secreted RNAs occurs between cells and this points to a novel role for these molecules in intercellular and interspecies communication [summarized in the following review: (Fritz et al., 2016)]. So far, intercellular transfer of extracellular RNA molecules has not only been shown in mammals but also in fungi and in bacteria. Indeed, a plant-residing fungal pathogen has been shown to be able to transfer specific RNA effectors into its host cells to suppress host immunity (Weiberg et al., 2013). Moreover, extracellular RNA secreted by Listeria spp. are key components for developing immunity against the bacterial infection (Abdullah et al., 2012) and extracellular RNA fragments of Mycobacterium tuberculosis have been shown to be able to induce early apoptosis in human monocytes (Obregón-Henao et al., 2012). Finally, an OMV-associated small RNA has been demonstrated to participate in Pseudomonas pathogenicity by reducing the host immune response (Koeppen et al., 2016). Thus, for the moment only a few examples of interspecies communication via extracellular RNA are known, but the fact that such pathways have been identified indicates that communication via extracellular RNAs seem to be species-conserved and could be a widespread and important mechanism.
Generally, it is assumed that by using a microvesicle-dependent active trafficking system, secreted RNAs can be delivered into recipient cells where they function analogous to endogenous RNAs, and take part in regulating multiple target genes or signaling events in intra- and interspecies communication [reviewed in: (Chen et al., 2012)]. Interestingly, OMVs contain a wide range of RNAs including ribosomal RNA (rRNA), transfer RNA (tRNA), messenger RNA (mRNA), and small non-coding RNA (sRNA) (Ghosal et al., 2015; Ho et al., 2015; Sjöström et al., 2015; Blenkiron et al., 2016; Choi et al., 2017; Soares et al., 2017; Dauros-Singorenko et al., 2018), which indicates that OMVs are important to study in the context of extracellular RNA communication.
To study OMV-associated RNA in the context of host–pathogen interaction, S. Typhimurium seems to be a good model as Salmonella does secrete OMVs (Schwechheimer and Kuehn, 2015) and it has been shown that some in vitro culture conditions are able to mimic different steps of the Salmonella life cycle (Ibarra et al., 2010; Kröger et al., 2013). Salmonella encounters different environments during its life cycle and exhibits different behaviors in each infection step. Thereby, one distinguishes between the invasive state where bacteria are extracellular, in the gut, generally flagellated, and the intracellular state when Salmonella has entered the epithelium and then propagates to macrophages or dendritic cells to replicate (Fabrega and Vila, 2013; LaRock et al., 2015). Depending on the environment, different expression profiles are generated by a complex regulatory network of virulence factors, encoded in several of the so-called Salmonella Pathogenicity Islands (SPI), on the chromosome and on the pSLT virulence plasmid (Yoon et al., 2009; Hébrard et al., 2011; Fabrega and Vila, 2013). SPI-1 genes, which are triggered when Salmonella encounters epithelial cells, can be activated when Salmonella is grown to a high optical density (OD) within Lysogeny Broth (LB) (Ibarra et al., 2010; Kröger et al., 2013). Similarly, the infection of macrophages by Salmonella, where SPI-2 genes are required, can be partially reproduced by a specific minimal medium with low pH and low phosphate availability (Beuzón et al., 1999; Srikumar et al., 2015). Salmonella RNA expression during host–pathogen interactions is a dynamic process, with significant modulation as the infection progresses (Hautefort et al., 2008; Ortega et al., 2012; Kröger et al., 2013; Avital et al., 2017).
In this study, we aimed to characterize the RNA cargo of Salmonella Typhimurium OMVs through OMV isolation and RNA sequencing, in different culture conditions representative of some of the environments encountered by the bacteria during the infection process.
Salmonella enterica subsp. enterica serovar Typhimurium LT2 (henceforth referred to as S. Typhimurium) preculture triplicates were grown overnight in control (PCN), SPI-2 inducing (SPI-2ind) or SPI-1 inducing (SPI-1ind) conditions at 37°C and 160 Rotations Per Minute (RPM). Control conditions consisted of PCN buffer (pH 7.4, 80 mM MOPS, 50 mM NaCl, 25 mM K2HPO4, 22.2 mM glucose, 15 mM NH4Cl, 4 mM Tricine, 1 mM MgSO4, 376 μM K2SO4, 100 μM FeCl3, 10 μM CaCl2, 800 nM MnCl2, 300 nM CoCl2, 100 nM CuSo4, 10 nM Na2MoO4, 10 nM Na2SeO3, 4 nM boric acid, and 1 nm ZnSO4). SPI-2ind conditions (inducing Salmonella Pathogenicity Island 2) consisted of PCN buffer with lower pH (5.8, 80 mM MES replacing MOPS) and low inorganic phosphate content (0.4 mM K2HPO4) (Löber et al., 2006; Kröger et al., 2013). SPI-1ind conditions (inducing Salmonella Pathogenicity Island 1) consisted of LB supplemented to 10 g.l-1 NaCl [according to Miller, (Miller, 1972)] grown until early stationary phase (Kröger et al., 2013). 1.5 l cultures in the same respective conditions were inoculated the next day and grown until the desired growth phase (i.e., log phase with an OD600nm of 0.4 for Control Low OD and SPI-2ind Low OD; and stationary phase with an OD600nm 0.8 for SPI-2ind High OD or 1.6 for Control High OD and SPI-1ind). At this step, a viability check was included for all the different culture conditions to ensure that purified OMVs would not be contaminated with membrane debris originating from lysed cells (LIVE/DEAD BacLight Bacterial Viability Kit, Invitrogen). Supplementary Figure S1 shows that hardly any dead bacteria were apparent for any of the culture conditions tested within this study.
Outer membrane vesicles extraction was performed as described previously (Habier et al., 2018). Briefly, bacteria, grown as described above, were centrifuged (4700 g, 10 min, 4°C) and the pellet was stored at -80°C in order to be able to extract intracellular RNA at a later time point. The intracellular pellet was conserved without any RNA preservation agent. 109 cells were used for extraction of intracellular RNAs. The whole 1,5 l supernatant was filtered using a 0.22 μm filter before being concentrated by ultrafiltration using tangential flow (Quixstand Benchtop System with 100-kDa hollow-fiber membrane). The collected OMVs-enriched liquid was then ultracentrifuged using a precooled Beckman ultracentrifuge with a 90Ti rotor during 3 h at 150,000 g and 4°C to obtain crude OMVs (before density gradient separation). After resuspension of the crude OMVs in 500 μl of buffer (50 mM Hepes, 150 mM NaCl and pH 6.8), pure OMVs were isolated by iodixanol gradient (Optiprep, Sigma) using a precooled Beckman ultracentrifuge with a swinging bucket rotor (SW40 Ti) during 16 h at 100,000 g and 4°C. Continuous gradients were built with a bottom-up approach, loading vesicles in the 2 ml bottom fraction containing 45% w/v iodixanol, which was overlaid successively by 2 ml of 40, 35, 30, 25, and 20% w/v iodixanol fractions. After overnight (16 h) ultracentrifugation, 12 fractions of 1 ml were collected from the top low-density fractions to the bottom high-density fractions. The OMV-containing fractions were identified by subjecting 12.5 μl of each of the 12 isolated iodixanol fractions to SDS-PAGE followed by Western-Blot with an anti-OmpA IgG (Biorbyt) (for an example see Supplementary Figure S2A). Once identified, the remaining 987,5 μl of each OMV-containing fraction (positive for OmpA) was pooled and subjected to Trichloroacetic Acid precipitation (final concentration 10% w/v) followed by centrifugation during 30 min at 17136 g and 4°C. The pellet was resuspended in 450 μl ice-cold 80% acetone before a further centrifugation step for 30 min at 17136 g and 4°C. After acetone evaporation, the pellet was resuspended in 100 μl of buffer (50 mM Hepes, 150 mM NaCl, pH 6.8). Vesicles were treated with freshly prepared lysozyme (from chicken-egg white, Sigma-Aldrich, final concentration 1 μg.ml-1), even though this step might be superfluous as lysozyme digest sugars, but we aimed treating intracellular and extracellular samples as similarly as possible in order to be able to compare the different samples later on. RNA extraction was then performed with the All-in-One Purification Kit (Norgen), allowing the separation of the RNA within a small RNA (<200 nt) and a large RNA (>200 nt) fraction. The “large RNA fraction” (>200 nt) has been used for RNA-Seq, even though our data clearly indicates that all small RNAs (except tRNAs) can be resolved by RNA-Seq within the “large RNA fraction” (Figures 3, 11). OMV-associated RNA was further concentrated using a Zymo Research RNA clean up and concentrator kit (including On-column DNaseI treatment) up to a volume of 6 μl (corresponding to 1.5 l of original culture). The extraction of intracellular RNA was also performed with the All-in-One Purification Kit (Norgen), followed by a DNaseI treatment. 1 μl of RNA was quality-checked with an Agilent 2100 Bioanalyzer. Detected RNAs were longer than 25 nucleotides in length (see Supplementary Figure S3).
The presence of OMVs after density gradient separation was confirmed by Transmission Electron Microscopy (TEM). For this purpose, the SPI-1ind condition has been used. OMV purification was performed as stated before but each of the 12 obtained iodixanol fractions obtained after gradient ultracentrifugation was fixed with 4% w/v paraformaldehyde during 10 min. OMVs were then rinsed with 8 ml of buffer (50 mM Hepes, 150 mM NaCl and pH 6.8) and pelleted using a precooled Beckman ultracentrifuge with a 90Ti rotor during 3 h at 150,000 g at 4°C. The pellet was finally resuspended in 100 μl of the same buffer. A sample of the crude OMV preparation was also observed by TEM.
A 5 μl drop of each sample was deposited on a glow-discharged carbon-coated Electron Microscopy (EM) copper grid (400 mesh square grid, EMS). After 5 min of application, the excess of sample was blotted out using a Whatman filter paper and then the grid was washed with water three times to remove any buffer salts, which would react with uranyl acetate used for negative staining. To perform negative staining, 5 μl of uranyl acetate solution (2%) was applied on the grids. After 30 s of incubation, the excess of uranyl acetate was blotted out and then the grids were kept in a dry, dark, dust-free environment until observation with the electron microscope.
The EM grids were mounted on to a room temperature-equilibrated holder and subsequently introduced into a FEI Tecnai 20 transmission electron microscope (FEI Eindhoven Holland). Images (2048 pixels × 2048 pixels) were acquired using a US1000 camera (Gatan) at 50000× (nominal magnification, the corresponding pixel size was 0.21 nm). OMVs were found in gradient fractions where OmpA was detected by Western-blot, demonstrating that the OmpA screening is a suitable method for identifying OMV-enriched iodixanol gradient fractions (see Supplementary Figure S2).
In order to have an estimate of the amount of OMVs secreted by S. Typhimurium, OMVs were concentrated from a 0.75 l culture in SPI-1ind condition by tangential flow as explained previously. Vesicles were subsequently fixed with 4% w/v paraformaldehyde during 10 min, rinsed with 8 ml of buffer (50 mM Hepes, 150 mM NaCl and pH 6.8) and pelleted using a precooled Beckman ultracentrifuge with a 90Ti rotor during 3 h at 150 000 g at 4°C. The pellet was finally resuspended in 500 μl of the same buffer. A portion of the culture was plated on LB agar plates to determine CFU.ml-1 (Colony Forming Units). Quantification was not performed on samples subjected to density gradient separation, as it is not possible to obtain the dilution factor of this technique, which is needed to obtain OMVs/CFU in the original culture.
Quantification of OMVs was achieved by Fluorescence Correlation Spectroscopy (FCS). OMVs were stained with 100 nM of Nile Red lipophilic fluorescent dye (Sigma-Aldrich). Measurements were performed on a two-photon platform, as previously described (Didier et al., 2009). Briefly, two photon excitation is provided by an InSight DeepSee Laser (Spectra Physics). Pulses of about 100 fs were produced at a wavelength of 830 nm. FCS measurements were performed on an Olympus- I×70 inverted microscope. The laser beam was focused into the sample by a water-immersion objective (60× 1.2 NA Olympus) generating a diffraction-limited focal spot of about 0.6 μm in diameter (∼0.3 μm3). The measurements were carried out in a 8 Well Lab-Tek II cover glass system. The sample fluorescence was collected through the same objective and directed by a dichroic mirror (COWL 750 nm; Coherent) to the lateral output of the microscope. After rejection of residual IR light by a short pass filter (E700SP, Chroma), the emitted photons were focused by a 200 mm achromatic lens on the 64 μm core of a multimode optical fiber coupled to an avalanche photodiode (SPCM-AQR-14-FC, EG&G). The detected signal was correlated online by an ALV-5000E correlator (ALV, Germany) to generate autocorrelation curves G(τ). The analysis of G(τ) can provide information about the underlying mechanisms responsible for the intensity fluctuations such as number and diffusion of the particles. For an ideal case of freely diffusing monodisperse fluorescent particles undergoing a triplet-like blinking process in a Gaussian excitation volume, the correlation function, G(τ), calculated from the fluorescence fluctuations can be fitted according to:
where τd is the apparent diffusion time (a parameter that is inversely related to the diffusion constant of the molecule), N is the mean number of molecules within the excitation volume, S is the ratio between the axial and lateral radii of the sample volume (S ∼ 5), ft is the mean fraction of fluorophores in their dark state and τt is the triplet state lifetime. Using carboxytetramethylrhodamine (TMR) in water as a reference (DTMR = 421 μm2.s-1), the diffusion coefficient, Dexp, of the labeled OMVs was calculated by:
where τd (TMR) and τd (sample) are the measured correlation times for TMR and the sample, respectively. Typical data recording times were 10 min. Quantification results gave us an estimate of 8.51 OMVs/CFU in SPI-1ind condition (see Supplementary Figure S4).
RNA sequencing was performed for Low OD and High OD samples in two distinct sequencing facilities, with minor changes indicated below. 1 μg of intracellular RNA was subjected to rRNA depletion using the Ribo-Zero Magnetic Kit (Gram-negative Bacteria) (Illumina, MRZB 12424), to obtain a sufficient sequencing depth. For better insight in the composition of OMV-associated RNAs, the rRNA depletion step was omitted for OMV libraries. RNA-Sequencing libraries were prepared using the TruSeq Stranded mRNA LT kit (Illumina, used for High OD samples) or the KAPA stranded RNA kit (Kapa Biosystems, used for Low OD samples) without mRNA pull down steps according to the manufacturer’s protocol. The insert size and the quality of libraries were checked with an Agilent 2100 Bioanalyzer and quantified using the Qubit dsDNA HS assay kit. Libraries were diluted to 4 nM each, pooled, denatured and sequenced 2 × 80 (Low OD) or 2 × 75 (High OD) cycles on a NextSeq500 (Illumina) according to the manufacturer’s instructions. For each experimental condition, three biological replicates were sequenced. The raw sequence libraries are deposited in the BioProject database under the experiment accession number PRJNA438234, available at https://www.ncbi.nlm.nih.gov/bioproject/438234.
Raw RNA 75 nt or 80 nt paired-end sequencing reads from each condition were first trimmed and quality filtered using the trim-fastq.pl script from PoPoolation package with the following parameters – quality-threshold 20 – fastq-type sanger – min-length 35 (Kofler et al., 2011). The Salmonella genome and plasmid sequences and annotations (in GFF3 format) were downloaded from NCBI (genome accession number AE006468.2 and plasmid accession number AE006471.2). Samtools were used to merge the bam files (Li et al., 2009). Tophat2 was used to map the trimmed paired-end and single-end reads to the Salmonella genome (Kim et al., 2013). Moreover, small RNA sequences from (Srikumar et al., 2015) were mapped to the LT2 genome. Only sequences that had 100% sequence identity over the full length on LT2 strain were retained. Expression analysis by counting reads per genomic feature from the Salmonella annotation file was performed using the featureCounts tool from the Subread package (Liao et al., 2014). Read counts per genomic feature were subsequently subjected to statistical analysis with R. All libraries were depleted of rRNA in silico to allow intracellular/extracellular comparisons, except if stated otherwise.
Normalization among samples and conditions was performed using the DESeq2 package (Love et al., 2014), i.e., results were normalized for library sizes but not for gene length as the purpose of our analysis was to compare the expression of identical genes in different conditions. Moreover, rRNA was removed in silico from all datasets (otherwise stated) to avoid potential biases in the comparisons between intracellular and OMV-associated RNAs as they were experimentally removed from intracellular fractions only. Principal component analyses (PCA) were obtained with the plotPCA function from DESeq2. Analysis of variance (ANOVA) was performed with subsequent post hoc analysis to test if significant differences in relative amounts of exported RNA classes between the different culture conditions exist, using Tukey’s range test from the multcomp package (Hothorn et al., 2008) with FDR-adjusted p-values (Benjamini–Hochberg procedure). To produce the scatterplots in Figure 7, sum normalization was performed using the decostand function from the vegan package (Oksanen et al., 2018).
Exported RNAs associated to OMVs were filtered for a normalized count over 1, averaged from biological triplicates. Enriched RNAs associated to OMVs were determined using DESeq2 (Love et al., 2014), calculating the differential presence between intracellular and OMV-associated fractions for each condition. Transcripts with an average normalized number of extracellular reads over 5 for a given set of triplicates, a log2 Fold Change(OMV/intracellular) over 2, and a corresponding FDR-adjusted p-value under 0.05 were considered as enriched.
RNA coverage from sequencing data was visualized with the Integrative Genomics Viewer software (v2.4.8) using default parameters (Robinson et al., 2011).
Figure 1 was generated using the heatmap.2 function from the gplots package, scaled by row. Figure 8 was generated using the UpSet R package (Conway et al., 2017). Other graphs were obtained with ggplot2 (Wickham, 2009).
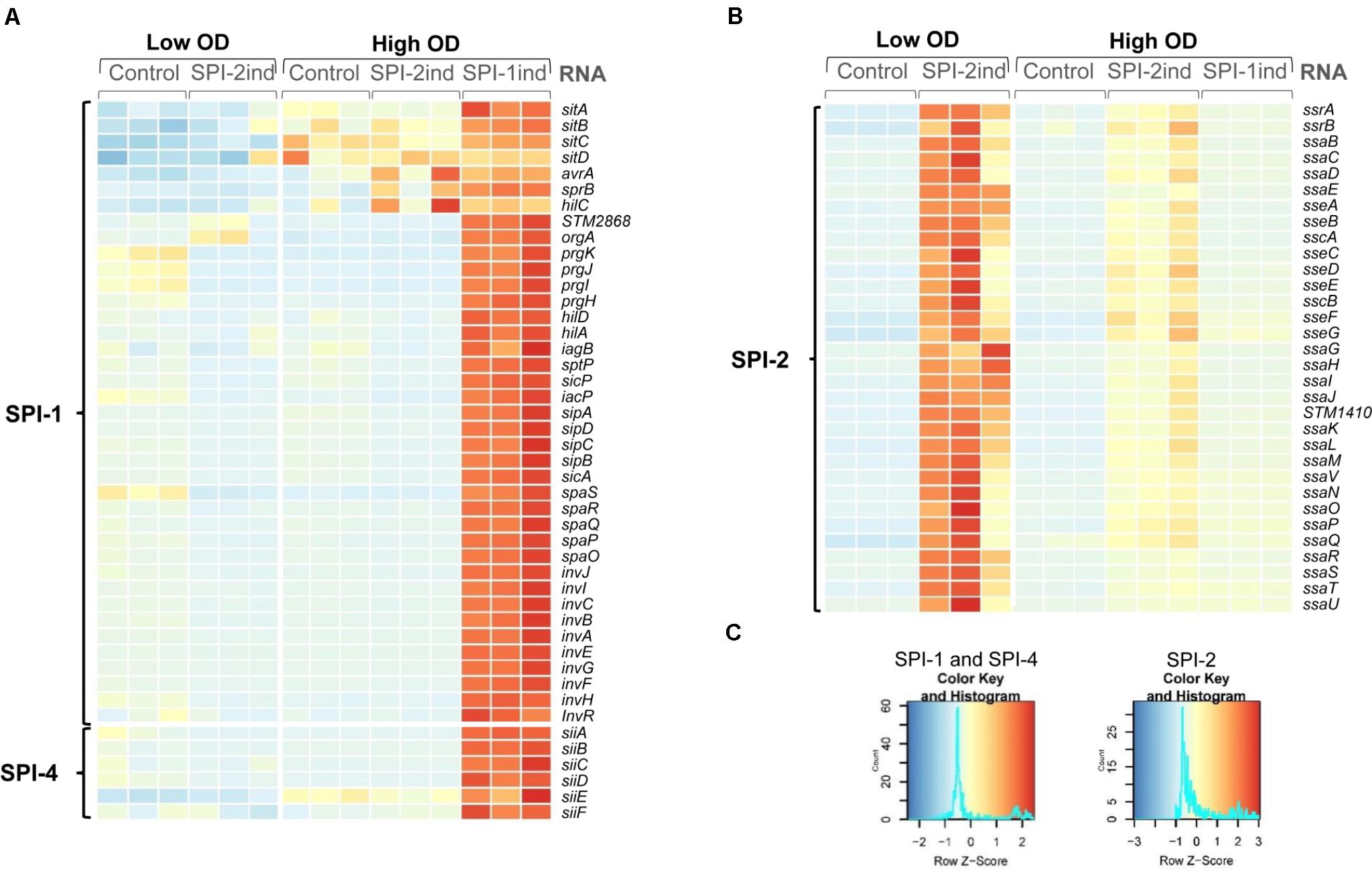
FIGURE 1. Activation of SPI-1 and SPI-2 by different in vitro culture conditions. Heatmaps showing relative expression of SPI-encoded genes in different conditions and biological replicates used in the study. RNA-Seq data of the intracellular fractions were normalized using DESeq2. Red or blue colors indicate a higher or lower expression, respectively. (A) Bacteria grown in SPI-1ind medium (LB) until stationary phase (OD600nm = 1.6) express SPI-1 and SPI-4 genes whereas growth in SPI-2ind medium or in control condition (PCN) represses these Pathogenicity Islands as expected. (B) For SPI-2 genes, the in vitro conditions with low pH and limited phosphate availability clearly trigger activation of the island in low OD, and to a lower extent in high OD. (C) Color keys and histograms for the heatmaps shown in (A,B).
To confirm results obtained by RNA sequencing and to show that RNAs of interest are present in or on OMVs in full-length, PCR validation was performed for distinct RNAs (Supplementary Dataset C) on independent replicates from high OD conditions.
New S. Typhimurium intracellular samples and purified OMV fractions were obtained from SPI-1ind, SPI-2ind, and Control conditions at high OD, with two biological replicates per condition. Intracellular and OMV-associated RNA was obtained as previously described with the following differences: the initial culture volume was 0.75 l per replicate instead of 1.5 l; RNA extractions were performed using TRIzol Reagent (Invitrogen); cDNA libraries were prepared from 1 μl of OMV-associated RNA or 100 ng of intracellular RNA with the SuperScript III First-Strand Synthesis System (Invitrogen) using random hexamer primers according to the manufacturer instructions and without further cDNA fragmentation step.
Polymerase Chain Reaction was performed using Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Scientific). A set of primers was designed to amplify the totality of the targeted Open Reading Frame (from start to stop codon), or the totality of the selected ncRNAs (list of targets and primers available in Supplementary Dataset C). 0.5 μl of cDNA library was used for each 20 μl PCR reaction as template, together with 1 μM of primers. Intracellular libraries were used as positive control, and cDNA synthesis reactions were performed on OMV-associated RNA without adding reverse transcriptase (RT) as a negative control to check for the presence of residual genomic DNA. Three different Touch-Down PCR programs were used, depending on the target length and differed only by their extension time (15 s per cycle for small targets under 500 bp, 45 s for medium targets between 500 and 1500 bp, and 70 s for large targets above 1500 bp). The 3 first cycles used an annealing temperature of 72°C during 7 s, which was lowered by 3°C steps every 3 cycles until reaching 57°C for the last 20 cycles. This approach was used to ensure a good selectivity without the need of further optimization. The size of PCR products was checked by running 10 μl of PCR reactions on an agarose gel prepared with 3% w/v TopVision Agarose (Thermo Scientific) in Tris-Borate-EDTA buffer (89 mM Tris, 89 mM Boric acid, 2 mM EDTA, pH = 8) for small size products or on a 1% w/v agarose gel in Tris-Acetate-EDTA buffer (40mM Tris, 20 mM acetic acid, 1 mM EDTA, pH = 8) for large size products, using the MassRuler DNA Ladder Mix (Thermo Scientific) as a size guide. In the case of 3% gels, the agarose solution was autoclaved to ensure an optimal dissolution before pouring the gel. Electrophoresis was run at 130 V during 200 min (3% gels) or 100 V during 100 min (1% gel) and DNA was stained with ethidium bromide. Pictures were acquired with a Canon EOS 1200D camera coupled to a BioDocAnalyzer (Biometra).
In addition to the PCR, qPCR was used to confirm for some RNAs their higher signal in RT-positives (+RT) libraries compared to RT-negative (-RT) controls, using absolute comparison between Ct values from both - and + RT conditions. As cDNA synthesis was performed with an identical protocol and the same quantity of template and as all qPCR runs had been performed on the same plate within the same instrument, we assumed that a greater Ct for a given gene in the +RT library compared to the -RT control is indicative for the presence of the targeted RNA. qPCR reactions were performed on technical duplicates for each biological replicate, using PerfeCTa SYBR® Green SuperMix (QuantaBio). The primers used are listed in Supplementary Dataset C. 0.75 μl of cDNA was used as a template for each 10 μl reaction, which was carried out with 100 nM of primers. Cycling conditions were as follow: initial denaturation 5 min at 95°C, 45 cycles of 15 s at 95°C, 30 s at the adapted annealing temperature (see Supplementary Dataset C), 30 s at 72°C, and a melting curve consisting of 5 s at 95°C and 1 min at 55°C. A first gradient run was performed on intracellular cDNA synthesis to select the optimal annealing temperature and to ensure that no coproducts were formed by analyzing the melting curves. The data was treated and analyzed with the LightCycler480 software to determine the Ct. Results with Ct equal or above 40 were considered as negative. Only technical duplicates with less than 5% variation were considered as reliable. For two given technical duplicates, if the difference between the average of +RT Ct and the average of -RT Ct is equal or superior to 3, the difference is considered to be significant (eight times more cDNA than residual gDNA in the sample).
To analyze if some nucleoprotein aggregates were co-purified with our OMV preparations, we performed a digestion protection assay (Shelke et al., 2014). OMV isolation was performed from the SPI-1ind condition as described in Section “OMV Purification and RNA Extraction,” except that a supplemental enzymatic step was added before density gradient ultracentrifugation. The crude OMV preparation was divided into four fractions. One was kept untouched (Untreated). The second was subjected to the same protocol as well as the third and fourth fraction without any enzyme added (Control). The third was digested by RNaseA (QIAGEN, 7000 U/ml) at 0.1 μg.μl-1 and 37°C during 20 min. The fourth was digested by ProteinaseK (Thermo Scientific, >600 U/ml) at 0.05 μg.μl-1 and 37°C during 30 min, followed by proteinase inactivation (adding 5 mM phenylmethylsulfonyl, Sigma-Aldrich) and RNAseA digestion. The protocol was then followed until the cDNA libraries from the OMV-associated RNAs were obtained. End-point PCR was then performed with the same primers and conditions used previously for SsrS, CsrC, 10Sa, and rnpB genes. Resulting samples were deposited on a 3% w/v agarose gel in TBE buffer, subjected to electrophoresis and stained with Ethidium Bromide. The efficiency of RNase digestion was confirmed using an intracellular RNA extract (10 ng) subjected to the same protocol (Supplementary Figures S5B,C). The efficiency of proteinase digestion was confirmed on an intracellular protein extract (5 μg), which had been digested under the same conditions (Supplementary Figure S5D).
Outer membrane vesicles originating from S. Typhimurium cultured in five distinct conditions, with three biological replicates per condition, were purified and the associated RNAs were sequenced, together with their respective intracellular counterparts.
RNA sequencing (RNA-Seq) was performed on the RNA isolated from OMVs secreted by S. Typhimurium in order to study their RNA composition. A significant number of transcripts was found in OMVs under all conditions (control Low and High OD, SPI-2ind Low and High OD, SPI-1ind High OD). Reads were mapped to the S. Typhimurium LT2 genome and pSLT plasmid and annotated using NCBI, and sRNA supplemental annotations from (Srikumar et al., 2015). Basic statistics of the RNA-Seq data are displayed in Supplementary Dataset A for low and high OD samples. Briefly, intracellular samples had roughly all around 10 million reads mapped to the target genome. For OMV samples, the number of reads and mapping percentages were lower, especially for SPI-1ind samples. The main contaminating RNAs mapped to Agrobacterium, Rhizobium, and Achromobacter taxa, which are common contaminants originating from column purification kits, when dealing with low abundance samples (Susilawati et al., 2016; Heintz-Buschart et al., 2018). These contaminating RNAs were thus added during the isolation protocol of RNAs and were originally not present in the S. Typhimurium cultures. As they did not originate from Salmonella, they were not taken into account for any further analysis. However, all OMV libraries were composed of reads mapping to most of the annotated genes in the Salmonella LT2 genome, leading to a mean of 3601 ± 545 expressed genes with an average count over 5 per condition (see Supplementary Figure S6A). As our annotated genome comprised 4964 genes, it means that on average, around 73% of the genes were expressed and identified in/on OMVs, even if it does not imply that these were native transcripts.
To confirm that the specific in vitro culture conditions chosen to mimic different life cycle steps of S. Typhimurium during host–pathogen interaction effectively induced the expression of the desired Salmonella pathogenicity island (SPI), we looked at the intracellular expression of the different SPI genes. As expected, SPI-1 expression was increased in SPI-1ind high OD conditions (Figures 1A,C), in agreement with previous findings (Kröger et al., 2013). The activation of SPI-4, which is controlled by SPI-1, was also observed. Concerning SPI-2, the expression of the pathogenicity island was favored in SPI-2ind conditions especially at low OD (Figures 1B,C). Residual expression was also present in stationary phase for the same medium (Figures 1B,C).
In order to acquire a broad view of the RNA expression profiles in the different samples, we performed Principal Component Analyses (PCA) on low and high OD samples (Figures 2A,B). The results confirm distinct clustering of the intracellular samples and, to a lesser extent, the OMVs triplicates. However, a general impression emerges from this data: most of the variance in these profiles is between the intracellular and OMV samples, which clearly points to a selective packaging of the exported RNAs. A difference is also observed in the high OD samples between SPI-1ind OMVs versus SPI-2ind OMVs triplicates. It is notable that these are conditions that mimic different infection steps (Figure 2B). Moreover, when only RNAs (rRNA included) associated with OMVs within SPI-1ind and SPI-2ind conditions were used for a PCA, we could observe that 79% of the variance was explained by the growth conditions (Supplementary Figure S7A). When only intracellular RNAs from these same conditions were compared, 81% of the variance was explained (Supplementary Figure S7B). Thus, the OMV-RNA complement reflects the adaptation of S. Typhimurium to its environment.
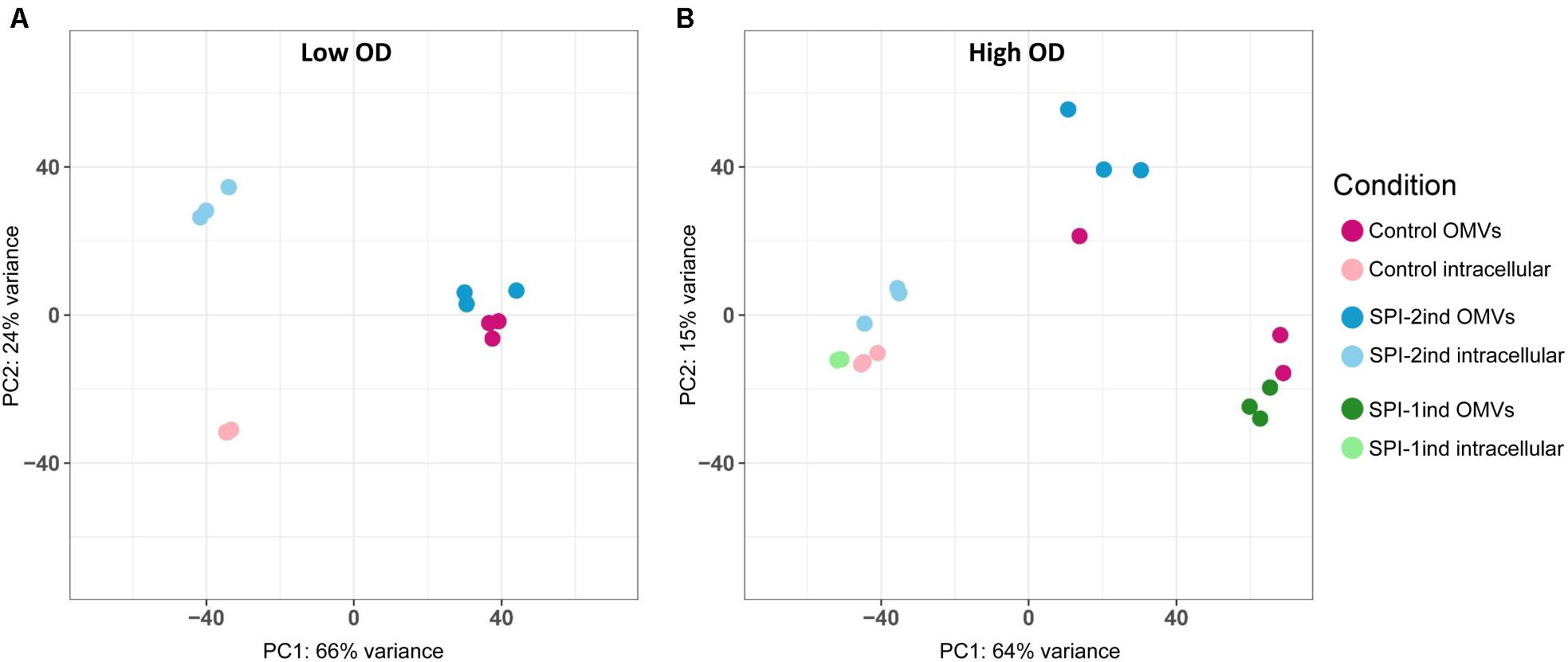
FIGURE 2. Principal component analysis. (A) Low OD samples. (B) High OD samples. Both intracellular and OMV-associated fractions are displayed. Normalized counts obtained with the DESeq2 package were used as input, omitting ribosomal RNAs, which were experimentally depleted for intracellular fractions, and removed in silico for OMV fractions. OMV and intracellular fractions are displayed in dark and light colors, respectively.
When analyzing the RNA classes present in OMV-associated fractions, it became clear that every class of transcript is exported, including rRNAs, mRNAs, tRNAs and others ncRNAs (Figure 3 and Supplementary Dataset B). One can observe that for all but the OMVs originating from SPI-1ind, the vast majority of exported RNAs were rRNAs, as it has already been observed in other species [e.g., in Escherichia coli, see (Blenkiron et al., 2016)]. However, in the case of SPI-1ind grown bacteria, the proportion of rRNA associated to OMVs was much lower, down to only 12.1 ± 15.2%. Concomitantly, we observed an increase in the mRNA relative amount up to 85.9 ± 15.1%. Despite the variance observed between triplicates, both of these values were significantly different from their respective counterpart in other conditions as determined by Tukey’s range test (Figure 3). Thus, it seems that the balance between rRNAs and mRNAs depends on the environmental conditions (pseudogenes followed the same trend than mRNAs). This result needs to be put in perspective, taking into account the lower sequencing depth obtained for the SPI-1ind OMV samples (Supplementary Dataset A). On the contrary, no significant differences were detected concerning tmRNA, tRNAs, and others ncRNAs, which appear to have been exported the same way in all conditions (see all ANOVA and Tukey’s test results in Supplementary Dataset B). Concerning tRNAs, we found a limited relative amount of tRNAs (<0.2% for all conditions). These were probably exported, but mainly found in the small RNA fraction (<50 bp) and therefore would have probably been more visible in a small RNA-Seq library, which was not included within the present study (Ghosal et al., 2015; Blenkiron et al., 2016).
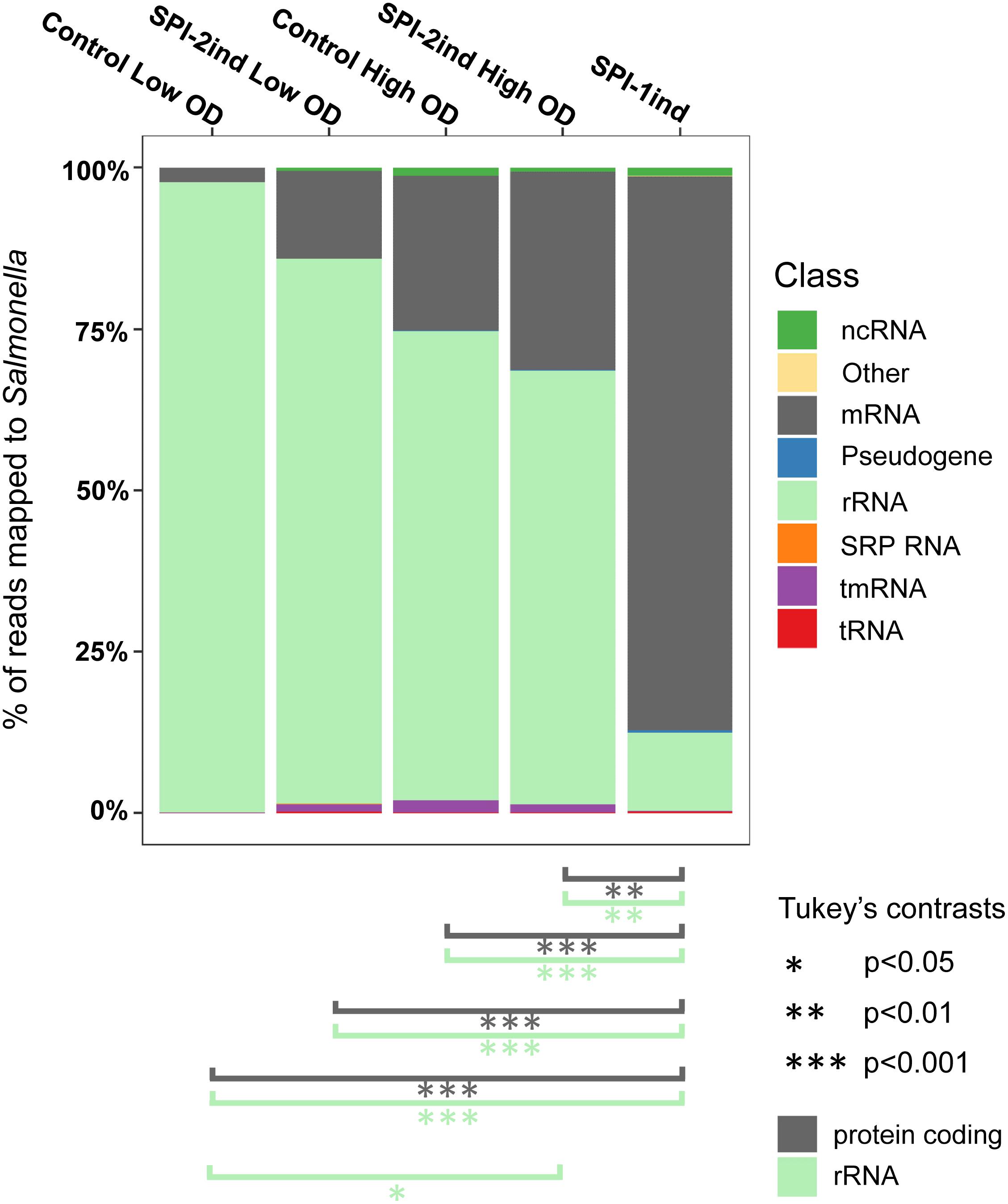
FIGURE 3. Relative composition of the OMVs RNA cargo for each culture condition. Individual colored bars represent the relative amount of each RNA class in the respective OMV RNA-Seq data (average of the biological triplicates). Number of reads have been normalized with DESeq2. RNA classes are defined from the Salmonella LT2 genome annotation (NCBI accession number AE006468.2) or pSLT plasmid annotation (NCBI accession number AE006471.2), with supplemental small RNA annotations from Srikumar et al. (2015). The existence of significant proportion differences among conditions for a specific RNA class was assessed by one-way ANOVA and F-test. Significant differences for rRNA and mRNA classes between conditions are indicated under the graph, determined by Tukey’s range test. Further details for other RNA classes can be found in Supplementary Dataset B.
Most of the existing studies on OMV-associated RNAs have emphasized non-coding small and micro RNAs in cell-to-cell information transfer (Gong et al., 2011; Dauros-Singorenko et al., 2018; González Plaza, 2018). Concerning longer transcripts, both their biological pertinence and integrity must be questioned. As we found a good proportion of reads mapping to larger RNAs in our samples, we investigated the apparent state of degradation of some OMV-associated RNAs.
To obtain a first insight into the apparent state of degradation of exported transcripts, we investigated the sequencing coverage of various genes in the intracellular and OMV-associated RNA-Seq data. Results show a variety of coverage profiles, falling into three broad categories. The first group consisted of transcripts showing identical patterns between intracellular and OMV-associated RNAs, and included 10Sa, CsrB, CsrC, rnpB, and SsrS (visible in Figure 4 and Supplementary Figure S8A). For these RNAs, the identity was almost constant through all media conditions tested. The 16S and 23S rRNA also felt into that group with few changes between intracellular and OMV-specific coverages (intracellular rRNA remaining after experimental rRNA depletion was sufficient to obtain a good coverage, see the example of rrlA in Supplementary Figure S8B). The second group of transcripts was fully covered by sequencing, but with sometimes very different read densities between the two fractions. For instance, it comprised ffs (except for SPI-2ind LowOD where patterns were identical), gogB in High OD, IsrA in SPI-1ind and Control High OD conditions, IsrB-1/2, nanH, pagC, pipA, sipADCB in SPI-1ind and Control HighOD, slrP, pSLT035, sseC, sseI, tae4 (STM0277), STM0289, STM0972 (sopD2). Some examples are displayed in Supplementary Figures S8B,C. These differences between intracellular and extracellular fractions might be explained by the low abundance of some RNA in the extracellular fraction, but drastic changes could also come from preferential export of processed RNAs or degradation products. The third category consisted of RNAs poorly or not detected in OMVs, meaning they were heavily degraded or their export was almost inexistent.
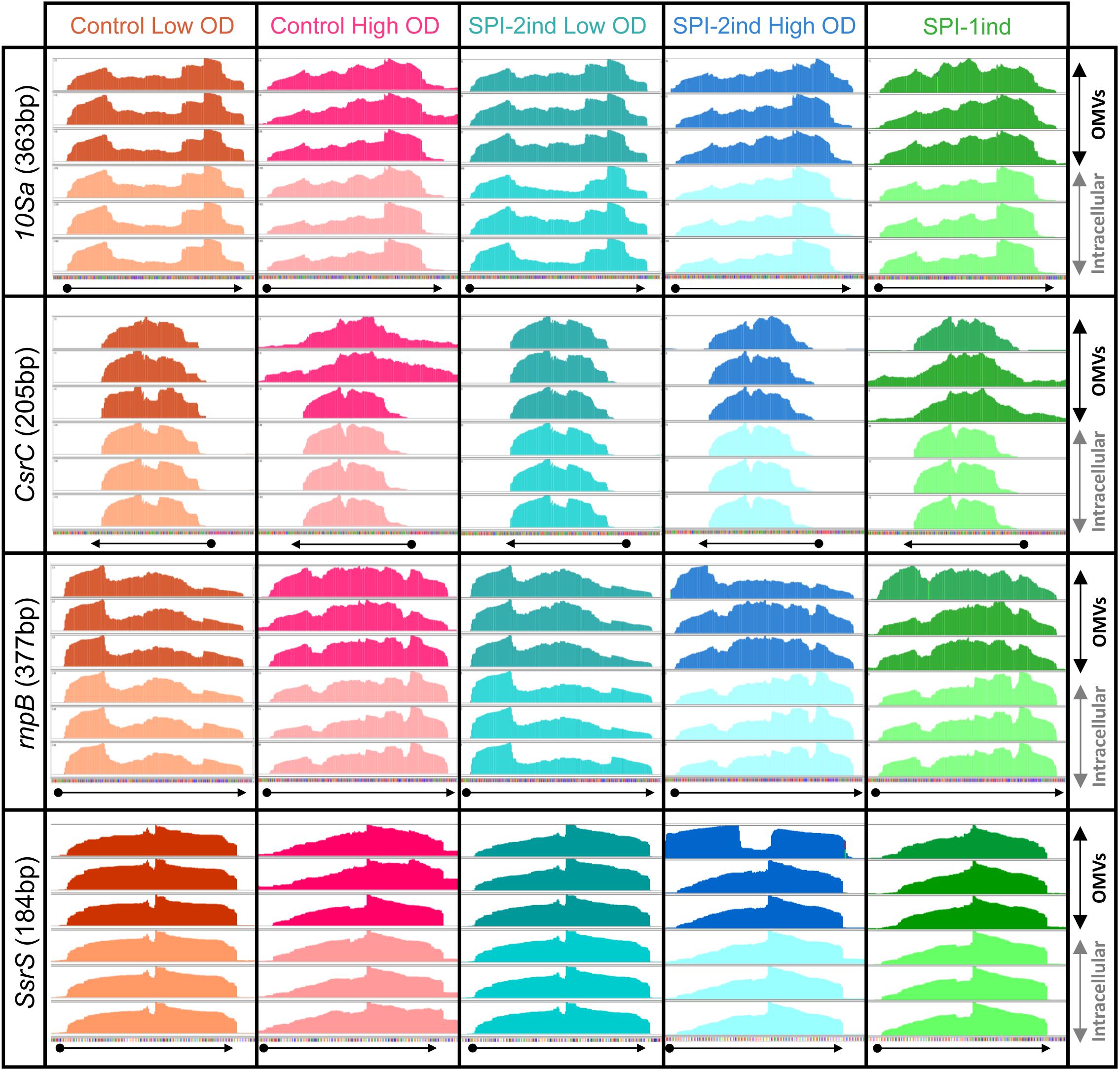
FIGURE 4. Visualization of intracellular and OMV-related read coverage plots of distinct sRNAs in distinct culture conditions. RNA coverage from sequencing data was visualized with the Integrative Genomics Viewer software (v2.4.8) using default parameters (Robinson et al., 2011). Each plot represents the raw number of reads mapped along the observed sequence, automatically scaled relatively to the highest number of read existing in this portion of the genome. The three plots in dark colors at the top of each window represents the sequencing coverage of OMV-associated fractions, in biological triplicate for each condition. On the contrary, the three plots in lighter colors at the bottom of each window show the coverage for the same RNA but from the corresponding intracellular fractions. The arrows under each window precise the genes positions and orientations, according to the data extracted from the Salmonella LT2 genome annotation (NCBI accession number AE006468.2) or pSLT plasmid annotation (NCBI accession number AE006471.2). The identical coverage patterns observed in both fractions for the displayed genes stand for a native export through OMVs without specific processing or degradation.
As some exported RNAs appeared to be complete transcripts according to their coverage profiles, we designed a set of primers to validate their integrity experimentally by PCR amplification. The full coding-sequence of 20 mRNAs and the totality of 10 ncRNAs sequences were targeted and amplifications were performed on high OD condition samples (Supplementary Dataset C). 12 RNAs were found to be amplified in their full-length from our cDNA libraries in at least one condition: 7 ncRNAs (IsrB-1/2, ffs, IsrA, SsrS, CsrC, 10Sa, rnpB) and 5 mRNAs [pSLT035, STM0277 also called tae4, sseB, STM0972 (sopD2), STM2606] (Figure 5).
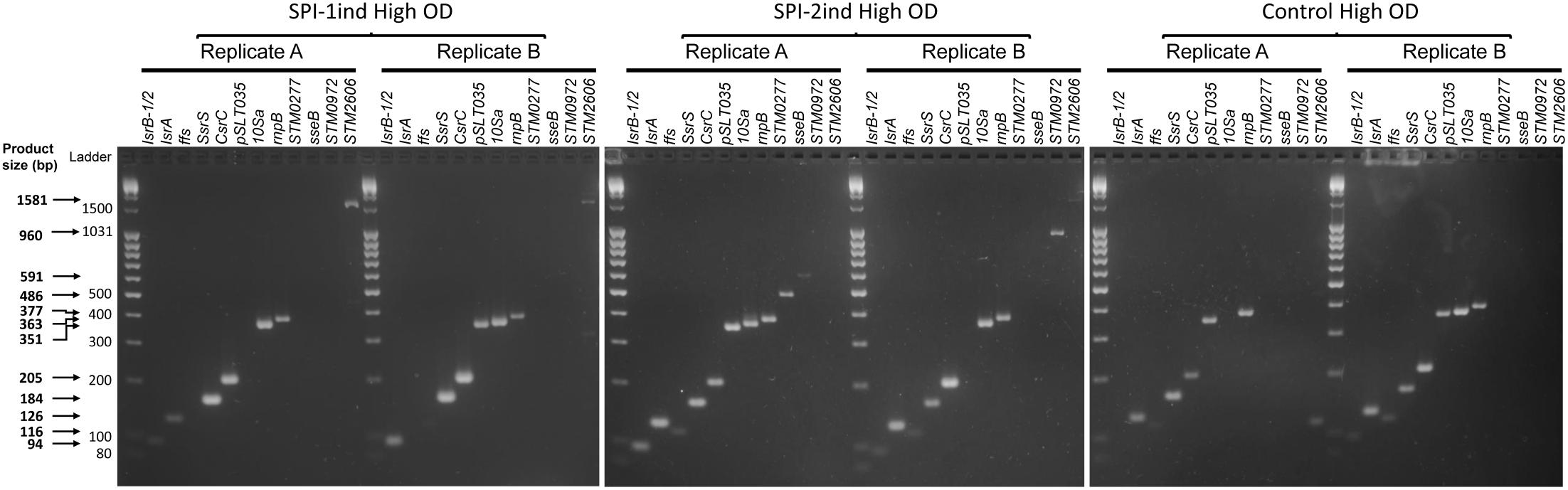
FIGURE 5. Integrity analysis of selected High OD exported RNAs by full-length RT-PCR. After OMVs isolation and RNA extraction as described in Section “Materials and Methods,” cDNA synthesis was performed. End-point PCR was then conducted with primers flanking the coding sequence (mRNA) or the full sequence (ncRNA) of 30 selected RNAs. Resulting samples were deposited on a 3% w/v agarose gel in TBE buffer, subjected to electrophoresis and stained with Ethidium Bromide. Only transcripts that were amplified as full-length amplicons are shown (12 out of 30 selected RNAs that were tested). Two biological replicates for SPI-1ind, SPI-2ind High OD and Control High OD conditions are visualized. Primers can be seen in Supplementary Dataset C, –RT controls are displayed in Supplementary Figure S9 and a positive control of full-length amplifications performed on intracellular RNAs is visible in Supplementary Figure S10.
Full-length mRNAs were amplified in only one out of the two replicates analyzed (for instance, sseB and STM0972 in the case of SPI-2ind High OD). STM0277 was also detected in SPI-2ind High OD but not in SPI-1ind, whereas a greater number of reads was detected in OMVs from the later by sequencing. Interestingly, we observed a tendency toward the amplification of the smaller sized RNAs compared to the larger ones (Figure 5 and Supplementary Dataset C). This trend might be due to a higher degradation predisposition of larger RNAs compared to small RNAs, which are usually more structured and therefore more stable. Especially, protein-associated ncRNAs such as ffs (SRP RNA binding to Ffh and routing nascent proteins to the membrane), rnpB (RNA catalytic component of the RNaseP ribozyme involved in pre-tRNAs maturation), SsrS (6S RNA that binds and regulates the σ70 transcription factor) or 10Sa (tmRNA associated to SmpB) were remarkably conserved in OMVs, in complete agreement with the RNA sequencing results (Figures 4, 5 and Supplementary Figure S8A). Fragments originating from tmRNA, SRP-RNA or 6S RNA were already reported to be present in the extracellular complement of E. coli, but completeness was not assessed (Ghosal et al., 2015).
Hence, one may consider that the presence of these RNAs in our samples might be due to a “contamination” with nucleoprotein complexes occurring during the purification process of the vesicles. This issue has already been identified in the Extracellular Vesicle field, resulting in artificial enrichment of the OMV-associated cargo with protein-associated RNAs (Mateescu et al., 2017). Therefore, we performed an RNase and proteinase protection assay on OMVs isolated from SPI-1ind condition (Figure 6; -RT controls are represented in Supplementary Figure S5A). As neither RNase nor proteinase followed by RNase treatment prevented RT-PCR on SsrS, CsrC, 10Sa, and rnpB, we assume that these ncRNAs were packaged inside the OMVs instead of being passively enriched during vesicles purification. However, this association might also occur when these are bound to their partner proteins explaining their stability. Note that in the same digestion conditions, intracellular RNA or protein extract was almost completely digested and only rnpB gave a low signal by RT-PCR after RNase treatment (Supplementary Figures S5B,C).
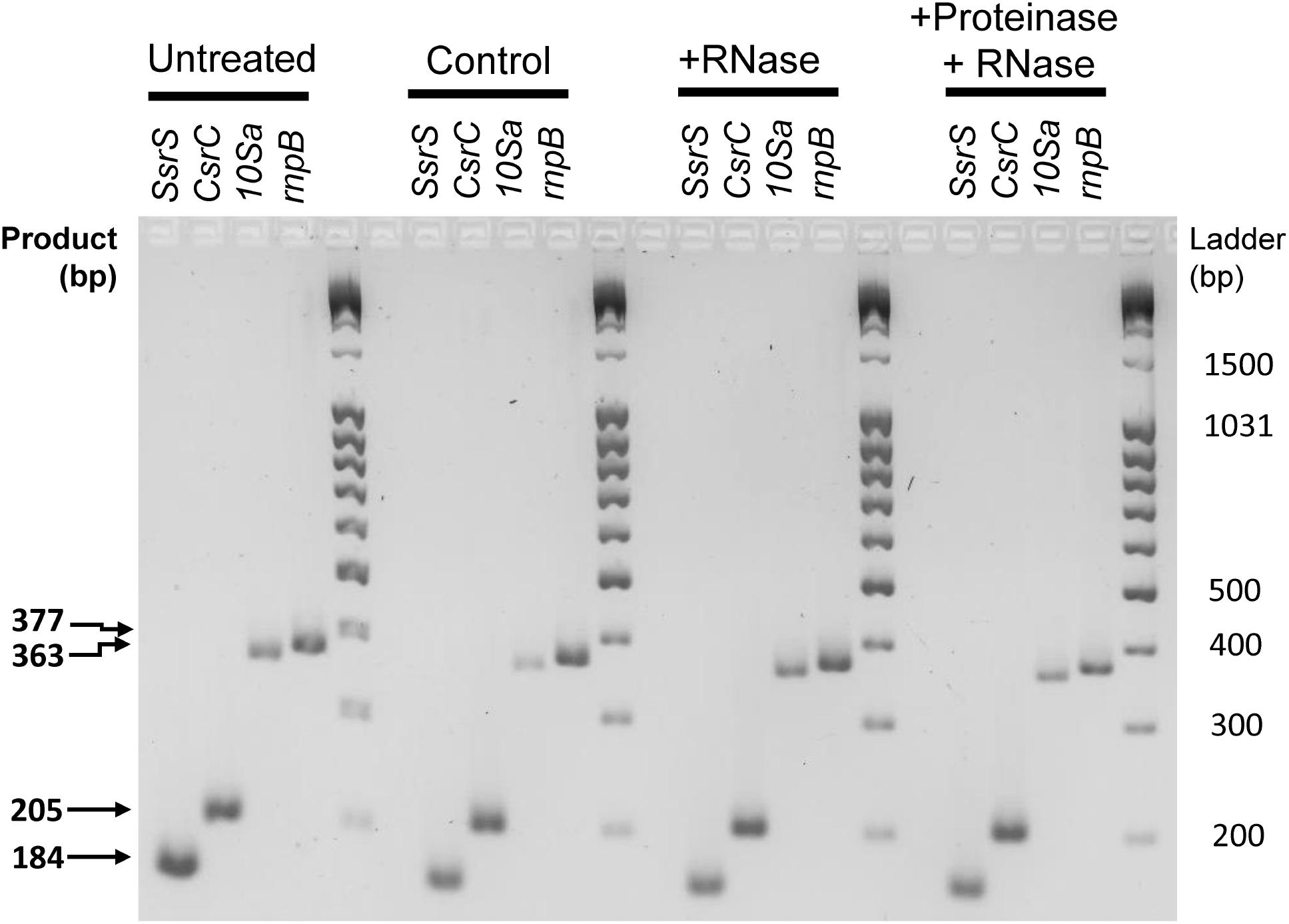
FIGURE 6. RNase protection assay of selected protein-associated sRNAs isolated from OMVs. OMV isolation and enzymatic digestions were performed from SPI-1ind condition as described in Section “Materials and Methods (see section “OMV Purification and RNA Extraction”).” Crude OMV preparation was divided in four fractions. One was kept untouched (Untreated). The second was subjected to the same protocol than the third and fourth fraction without any enzyme added (Control). The third was digested by RNaseA. The fourth was digested by ProteinaseK followed by proteinase inactivation and RNAseA digestion. The protocol was then followed as described in Section “OMV Purification and RNA Extraction” until cDNA libraries from OMV-associated RNAs were obtained End-point PCR was then conducted with the same primers and conditions used previously, for SsrS, CsrC, 10Sa, and rnpB genes. Resulting samples were deposited on a 3% w/v agarose gel in TBE buffer, subjected to electrophoresis and stained with Ethidium Bromide. Most of the signal is still present after enzymatic digestion, showing that the RNAs are protected by the vesicle membrane. –RT controls and positive controls for enzymatic digestions can be seen in Supplementary Figure S5.
Some -RT controls showed a remaining DNA contamination (Supplementary Figure S9) despite the DNAseI digestion step before cDNA synthesis. Therefore, we performed qPCR on +RT and -RT samples for several genes and confirmed that the amount of cDNA was greater in +RT than in -RT for ffs, CsrC, rnpB, sseB, and STM0972 (sopD2) (Table 1). For sseB and STM0972, both replicates were positive contrary to the full-length PCR, indicating that the transcripts were present but not in their native form or the end-point PCR was not sensitive enough to detect them on a gel.
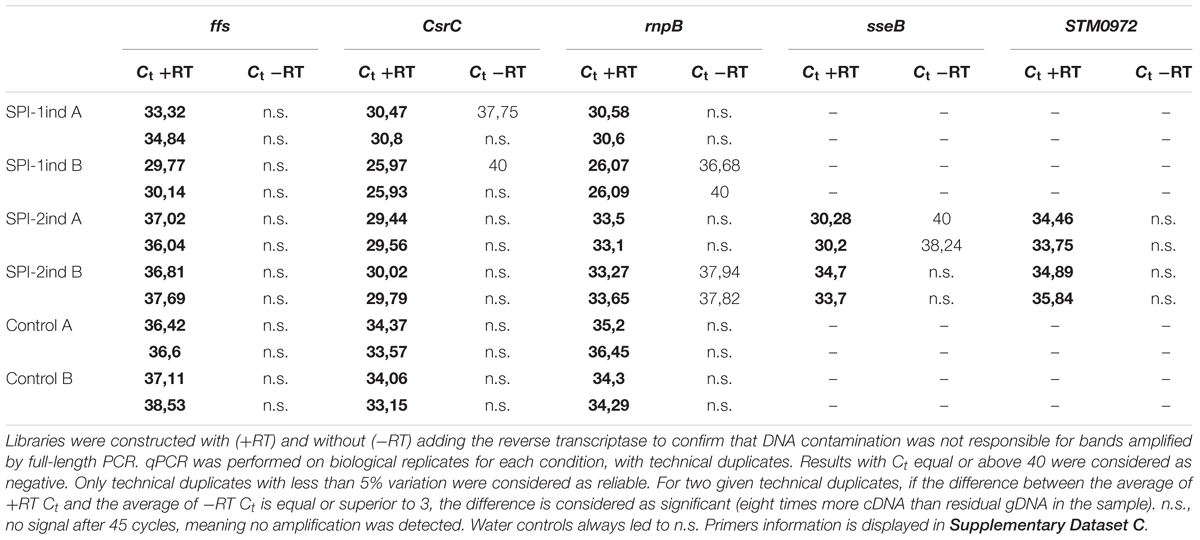
TABLE 1. q-RT-PCR Ct comparison for five selected genes between cDNA libraries generated from OMV-extracted RNA isolated from SPI-1ind, SPI-2ind High OD and Control High OD conditions.
In contrast to the genes highlighted above, no full-length transcripts associated to OMVs could be detected for the following genes: STnc440, STM0278, STM1026, pagC, spvD, STM0292, ydgH, sseI, AmgR, sipC, nanH, gogB, sseC, sipB, sipA, STM0289, slrP, and STM0291 (Supplementary Dataset C).
We could show that different culture conditions induce different OMV-associated RNA export. In order to study the RNA cargo present in the vesicles in detail, we performed pairwise comparisons between intracellular and OMV fractions among our five culture conditions. Simple scatterplots of normalized counts already provided a good view of the existence of transcripts that were enriched in/on OMVs compared to the intracellular fraction (Figure 7). In this context, it is important to note that we did not have the possibility to distinguish between full-length or processed RNAs which remain functional and degradation products for each individual transcript enriched in/on OMVs as described and explained in section “Native and Processed and/or Degraded RNA Co-exist Among OMV-Associated RNAs.” As, however, this remained true for all the different culture conditions, we performed pairwise comparisons between intracellular and OMV-associated RNAs among our five culture conditions, in order to obtain a global overview of RNA sequences which were differentially expressed among the different culture media. The RNAs that were enriched in/on OMVs compared to their intracellular counterpart were mainly coding sequences and to a lesser extent, ncRNAs (Figure 7, in black and green, respectively). On the contrary, tRNAs seemed more internalized for the studied transcript libraries (Figure 7, red). Note that rRNAs are not displayed here as the intracellular samples had been experimentally depleted of the latter and thus intracellular versus OMV RNA sample comparisons became impossible. The pattern was clearly dependent on the condition, with a wider spread in high OD conditions and especially in SPI-1ind (Figures 7C–F). Interestingly, the translation rescuing RNA tmRNA (10Sa), which was experimentally proven to be packaged as full-length transcript (Figures 5, 6) was always highly represented in both fractions. As 10Sa binds other proteins for its activity, and among them elongation factors or ribosomal proteins, its OMV association could be favored by the presence of the elongation factor EF-G in the vesicles (Bai et al., 2014; Himeno et al., 2014). However, no trace of its main protein partner SmpB has been reported to date.
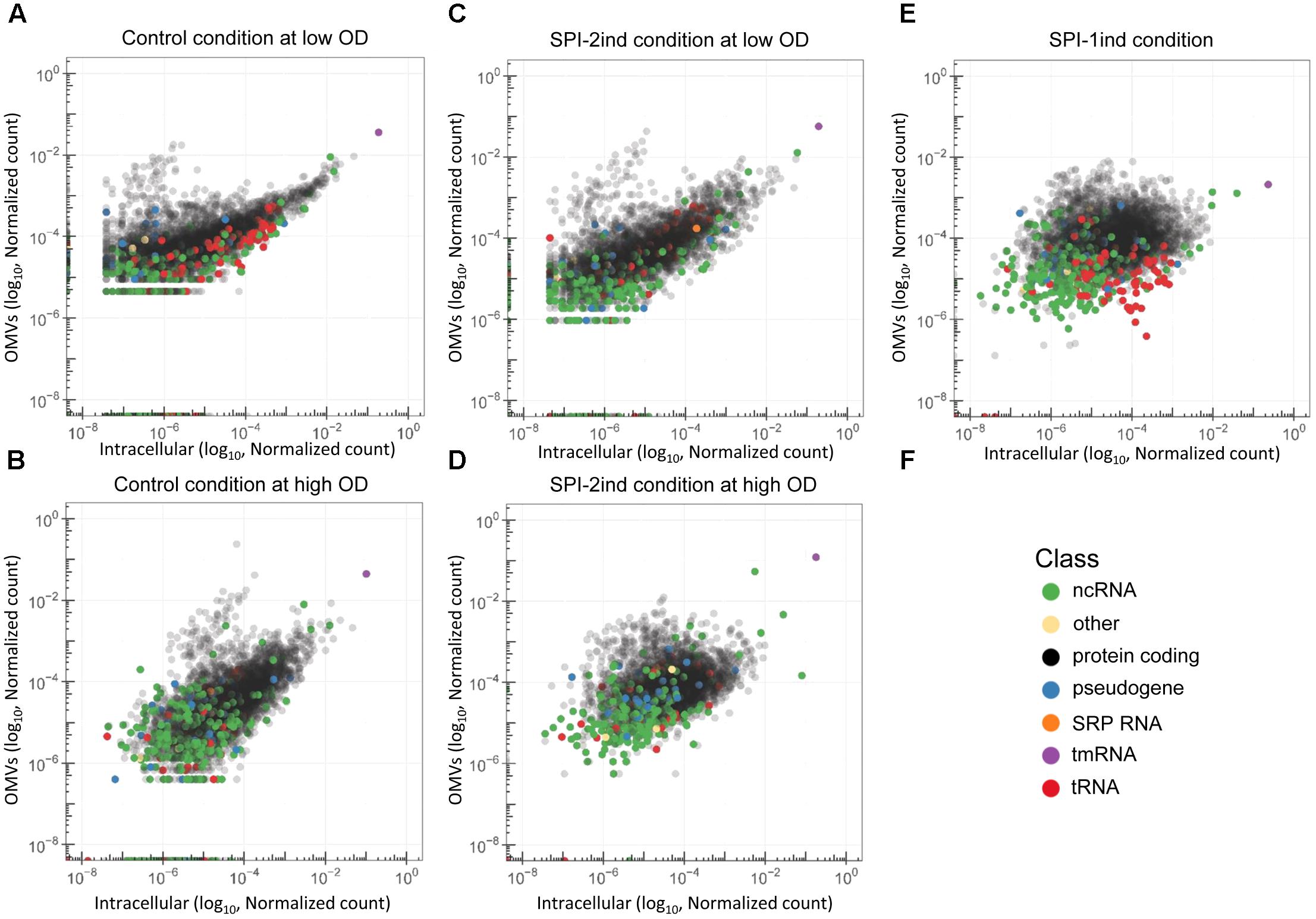
FIGURE 7. Differential abundance of RNAs in OMVs and their intracellular counterparts. (A–E) Scatterplots showing the reads repartition within a given growth condition. Each dot represents the relative abundance of an expressed gene in or attached to the OMVs (vertical axis) and in the intracellular (horizontal axis) fraction for each growth condition used in this study. RNA annotation from the Salmonella LT2 genome (NCBI accession number AE006468.2) or pSLT plasmid (NCBI accession number AE006471.2) is precized by a color code. Number of reads were normalized by sum normalization for each condition using the decostand function from the vegan R package. Ribosomal RNAs, which were experimentally depleted for intracellular fractions, and removed in silico for OMV fractions, are omitted. (F) Color legend used to underline the RNA class of each annotated transcript in the graphs.
In order to define more precisely the OMV-enriched transcripts in comparison to their intracellular counterpart, a differential expression analysis with the DESeq2 package (Love et al., 2014) was performed. We chose to select RNAs with a log2 Fold Change (logFC) (OMV/intracellular) ≥ 2, a p-value < 0.05 (Welch’s t-test), and an averaged number of reads per condition ≥ 5 in order to ensure a stringent selection of “enriched RNAs.” Henceforth, all RNAs that fulfill the aforementioned criteria are referred to as enriched RNAs, whereas we refer to exported RNAs when a given RNA is associated with OMVs, but not necessarily enriched compared to its intracellular counterpart. In each media condition, a significant number of transcripts was indeed selectively exported (Figure 8). In total, we found 1151 enriched transcripts in Control Low OD condition, 763 in SPI-2ind Low OD conditions, 881 in Control High OD condition, 643 in SPI-2ind High OD condition and 1196 in SPI-1ind condition (detailed lists are available in Supplementary Dataset D). A smaller core set of 193 RNAs was enriched in all conditions tested, with the notable absence of ncRNAs in this set, these being mainly found in condition-specific groups. However, these group sizes were very low compared to the totality of the RNA cargo, which originated from 4670 genes in total, from which 2217 were found in all the conditions tested (Supplementary Figure S6A). This difference may underline the dual nature of OMV-RNA association, reflecting the intracellular state of the transcriptome through global packaging, but also containing specific sets of read fragments mapping to distinct RNAs that depend or not on the current environmental state.
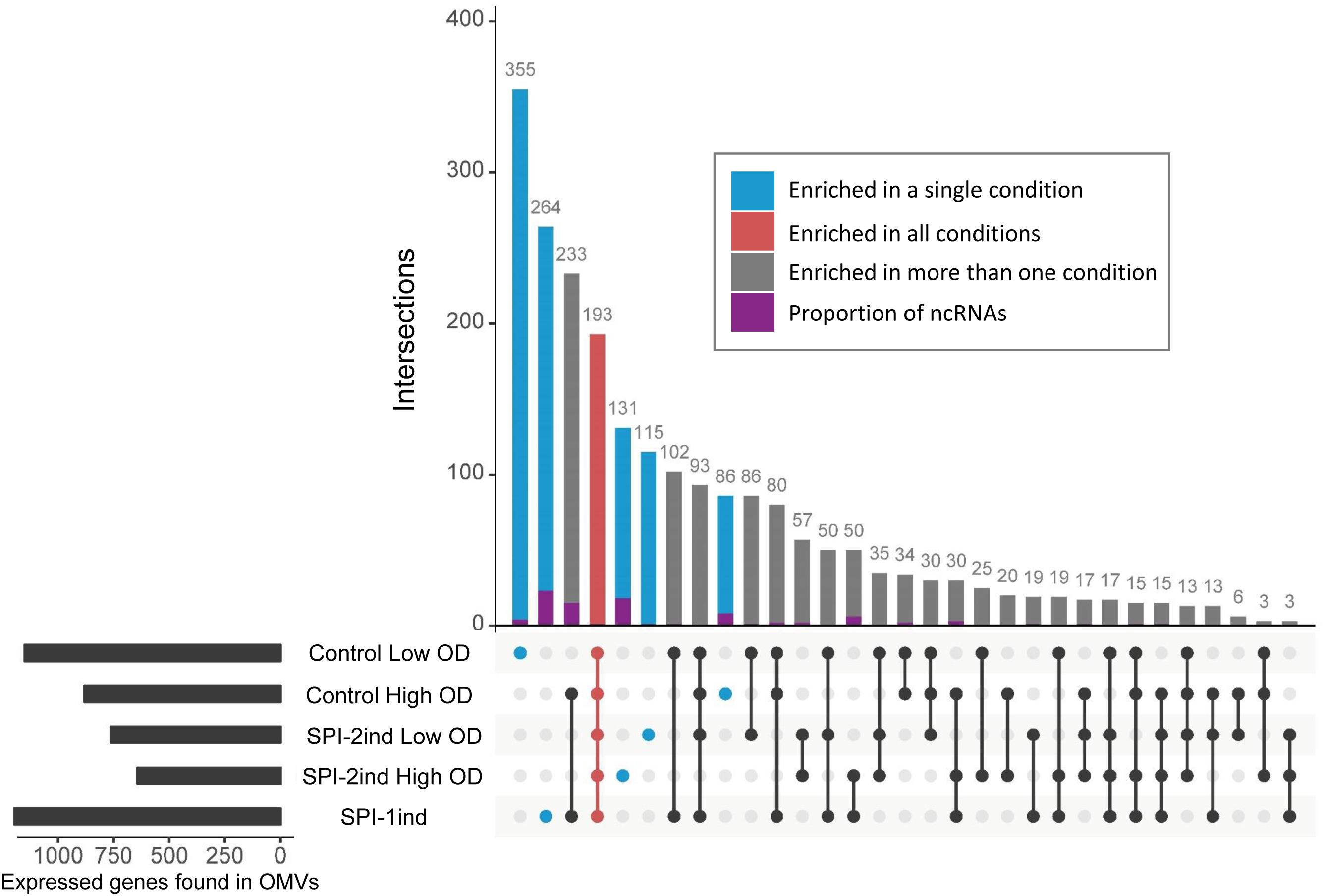
FIGURE 8. Repartition of enriched RNAs in/on OMVs isolated from the different analyzed in vitro culture conditions. UpSet plot representing the number of enriched RNAs shared between different conditions. One unit represents one expressed gene. The graph was generated using the UpSet R package. Enriched RNAs were determined using DESeq2, calculating differential presence between intracellular and OMV-associated fractions for each condition. Expressed genes with an average number of reads over 5 for a given set of triplicates, a log2 Fold Change over 2, and a corresponding adjusted p-value under 0.05 were selected. ncRNA class attribution is defined from the Salmonella LT2 genome annotation (NCBI accession number AE006468.2) or pSLT plasmid annotation (NCBI accession number AE006471.2). Moreover, small RNA annotations from Srikumar et al. (2015) were mapped to the LT2 genome. Only sequences that had 100% sequence identity over the full length on LT2 strain were kept. Identical datasets were used for the generation of Figure 8 and Supplementary Figure S6B.
As we validated the association of several full-length virulence-related mRNAs with OMVs (Figure 5, Table 1 and Supplementary Dataset C) and as most of the effectors were covered over the whole length of their gene (Supplementary Figures S11A–E), we looked at the more than 40 effectors known to be secreted by Salmonella during invasion and infection (Niemann et al., 2011; Yoon et al., 2011a; Kidwai et al., 2013; LaRock et al., 2015). Intracellular expression, OMV- exported and enriched effectors are displayed in Figure 9. As expected, the intracellular expression of secreted molecules was linked to their known export route: SPI-1 translocated effectors were more expressed in non-SPI-2ind conditions and vice-versa (Figure 9A). However, even if almost all effectors’ mRNAs were found associated with OMVs in every condition, the OMV export route appeared to be more important when the corresponding T3SS (Type 3 Secretion System) was absent (Figure 9B). This was confirmed by the analysis of enriched effectors RNAs, where a distinct pattern did exist (Figure 9C and Supplementary Dataset E): SPI-1 translocated effectors seemed enriched in/on vesicles produced in SPI-2ind conditions (i.e., sip genes), whereas SPI-2 translocated ones were enriched in/on OMVs originating from non-SPI-2ind conditions, i.e., Control and SPI-1ind (i.e., some sse transcripts). Interestingly, we also identified prophage-encoded effectors. For instance, GogB is a secreted effector originating from Gifsy-1 prophage that can be translocated through both the SPI-1 and SPI-2 T3SS, but which is mainly expressed under SPI-2 control by ssrB (Coombes et al., 2005). It has an anti-inflammatory effect by interacting with ubiquitin ligase in the host, limiting tissue damage during infection (Pilar et al., 2012). It was also associated with OMVs in all conditions and singularly enriched in the controls and mainly SPI-1ind (logFC = 6.39, p < 0.001), emphasizing its multiple roles in pathogenicity as this protein might be involved in regulating immune processes from an early stage of the host–pathogen interaction. One can also note the stable presence of transcripts of T3SS-independent effectors such as ydgH (or sssB) and STM0082 (or srfN) (Eletsky et al., 2014). No enrichment of T3SS-independent effectors was observed in OMVs isolated from SPI-2ind conditions even though their intracellular expression is increased in acidic medium and they are suspected to be exported through OMVs to the host cell (Figure 9B, Yoon et al., 2011b).
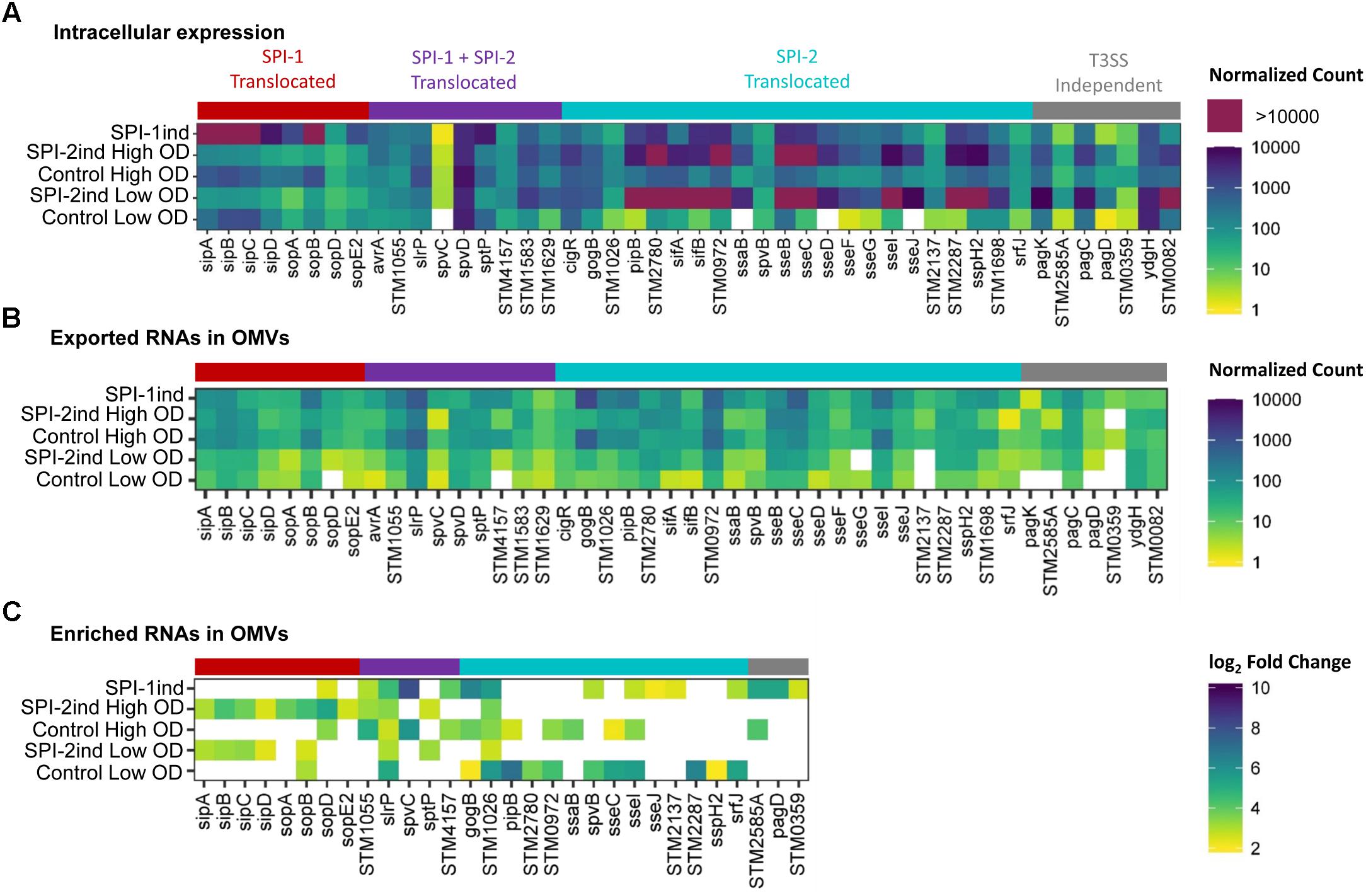
FIGURE 9. Intracellular expression, export and enrichment of OMV-associated effectors mRNAs. List of translocated effectors involved in virulence and infection of the host by Salmonella, regrouped by their known export route (Niemann et al., 2011; Kidwai et al., 2013; LaRock et al., 2015). T3SS, Type 3 Secretion System. (A,B) Mean count among triplicates in intracellular (A) or in OMV fractions (B) is displayed as a colored square, from low (yellow) to high count (dark blue). Counts above the upper limit of the scale are colored in purple. Counts were normalized using DESeq2 (Love et al., 2014), and filtered for a minimum averaged count of one read per biological triplicate. (C) Enriched RNAs associated to OMVs were determined using DESeq2, calculating differential presence between intracellular and OMV-associated fractions for each condition. Expressed genes with an average number of reads over 5 for a given set of triplicates, a log2 Fold Change over 2, and a corresponding adjusted p-value under 0.05 were selected. The log2 Fold Change is displayed as colored squares from 2 (yellow) to 10 (dark blue).
Interestingly, a number of SPI-6 mRNAs were found to be exported and enriched in/on OMVs (Figure 10). In this case, an increased intracellular expression caused a greater export by vesicles, as it was clearly visible in Control conditions at high OD and SPI-1ind. The in vitro expression level of SPI-6 is generally low, as reported earlier (Mulder et al., 2012). However, OMV-association apparently occurred in all conditions, and to a higher extent in non-SPI-2ind conditions (Figure 10B). Interestingly, the same was observed concerning the enrichment in OMVs, showing a correlated expression and enrichment in vesicles for most of the SPI-6 transcribed RNAs (Figure 10C and Supplementary Dataset F). The coverage data of the mapped reads was relatively constant between intracellular and OMV-associated fractions (Supplementary Figures S12A,B).
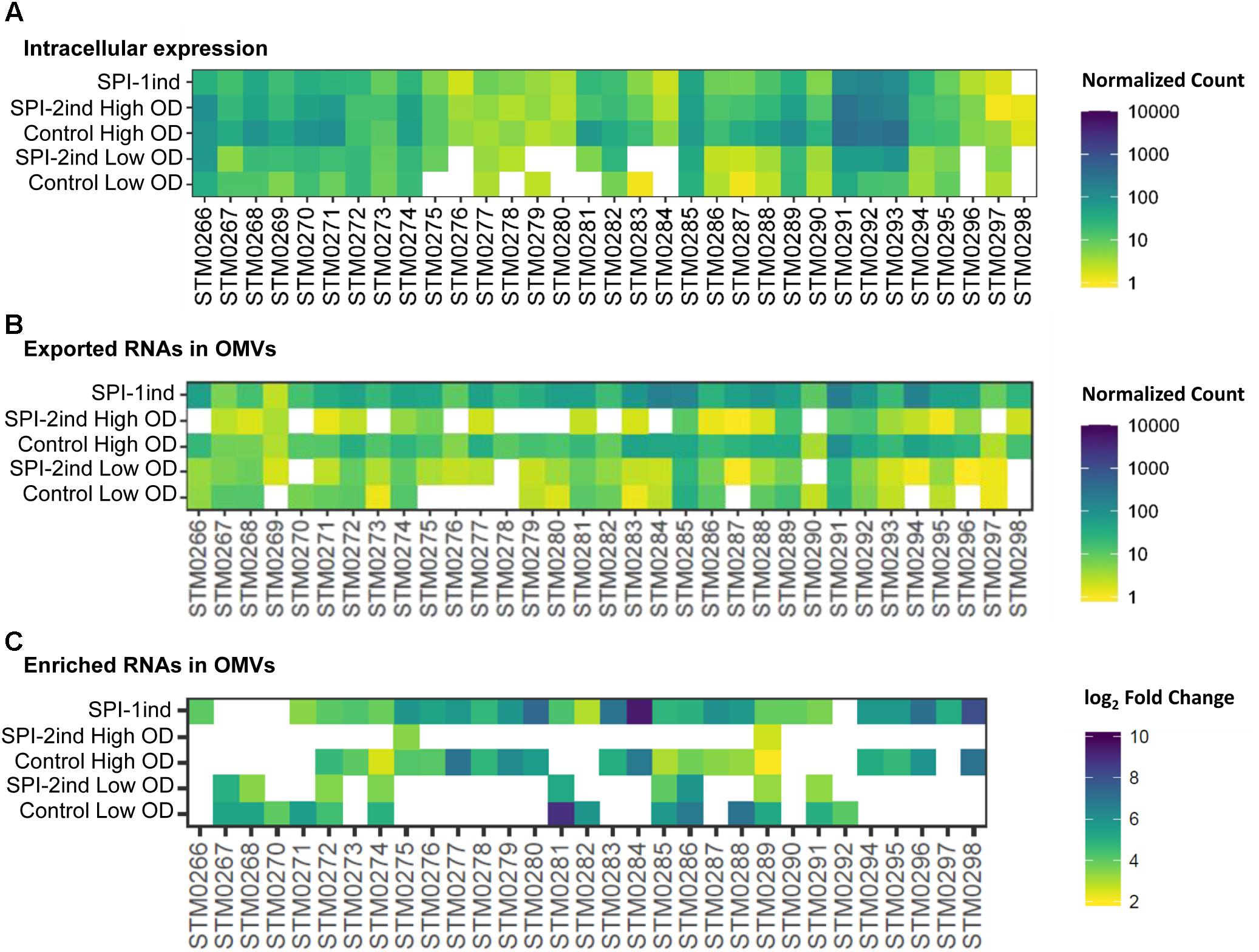
FIGURE 10. Intracellular expression, export and enrichment of SPI-6 encoded mRNAs. For each enriched SPI-6 mRNA, the intracellular expression, the presence and the enrichment in OMVs (if applicable) are displayed. (A,B) Mean count among triplicates in intracellular (A) or OMV fractions (B) is displayed as a colored square, from low (yellow) to high count (dark blue). The counts were normalized using DESeq2 and filtered for a minimum averaged count of one read per biological triplicate. (C) Enriched RNAs associated to OMVs were determined using DESeq2, calculating differential presence between intracellular and OMV-associated fractions for each condition. Expressed genes with an average number of reads over 5 for a given set of triplicates, a log2 Fold Change over 2, and a corresponding adjusted p-value under 0.05 were selected. The log2 Fold Change is displayed as colored squares from 2 (yellow) to 10 (dark blue).
SPI-6 in S. Typhimurium encodes for a Type 6 Secretion System (T6SS) that is involved in eukaryotic cell infection (Mulder et al., 2012). However, the role of the T6SS is not limited to virulence toward eukaryotes. Accumulating evidence demonstrates that the T6SS is mainly involved in interbacterial intoxication, targeting competitors by contact-dependent transport of bacteriocins from a donor cell to a recipient cell (Durand et al., 2014; Russell et al., 2014). Interestingly, one can note that the peptidoglycan hydrolase mRNA tae4 (STM0277: validated as being present as a full-length transcript within OMVs: Figure 5) and its cognate immunity-protein mRNA tai4 (STM0278) were only moderately present in the intracellular fraction (Figure 10A) under the conditions studied, but their mRNAs were enriched in/on OMVs purified in SPI-1ind and Control conditions at high OD (Figure 10C). Another pair of toxin/antitoxin encoded by the SPI-6 is rhs (STM0291 and STM0292)/rhsI (STM0293). Rhs has a putative nuclease activity and was expressed in the intracellular case together with RhsI (Figure 10A). Both of their mRNAs were found associated with OMVs but only rhs was slightly enriched in several conditions (Figures 10B,C).
As we could experimentally validate the presence of several full-length sRNAs (Figure 5 and Supplementary Dataset C) and as we have seen in Figure 8 that a smaller proportion of these latter was also enriched in/on OMVs in a condition-dependent manner, we analyzed in more detail these sRNAs (Figure 11 and Supplementary Dataset G). The majority of them was found in the SPI-2ind and SPI-1ind cases, thus linked to virulence-like conditions. An enrichment in/on OMVs was mainly observed in high OD conditions; with only a few transcripts enriched at low OD. If these were exported in their native form through OMVs, they might be involved in pathway regulation in surrounding cells up-taking OMVs.
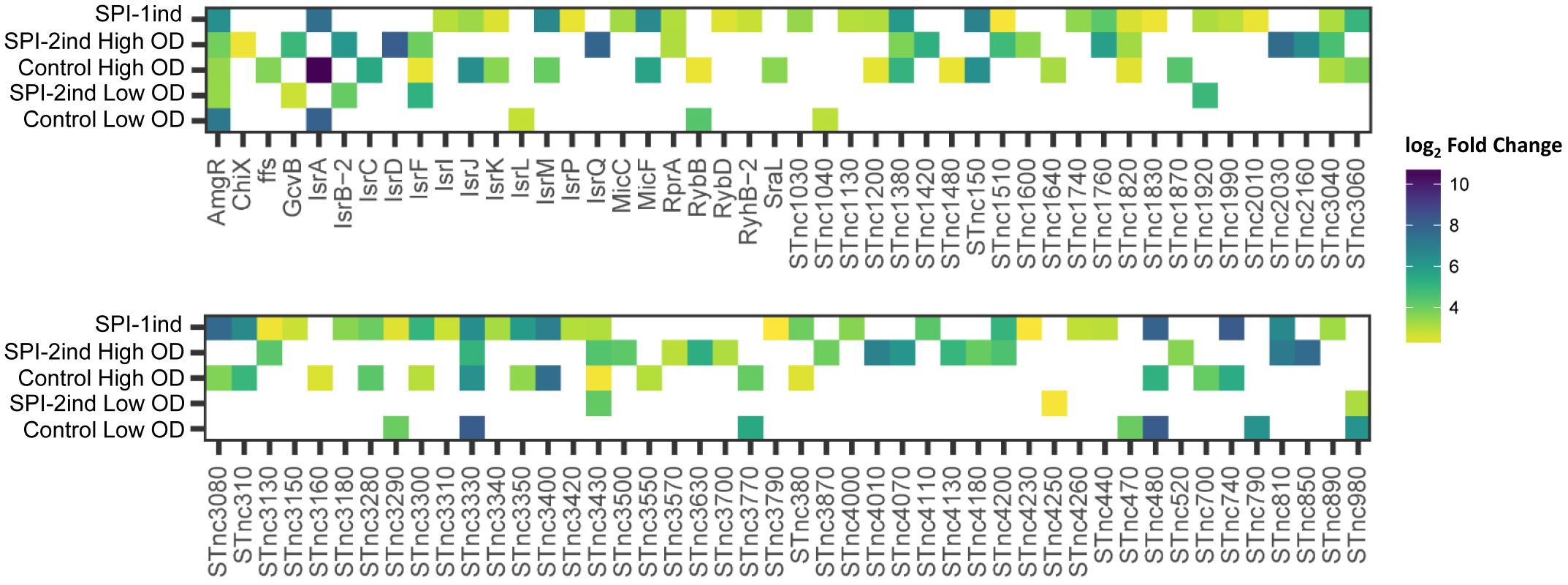
FIGURE 11. List of enriched sRNAs in/on OMVs isolated from the different culture conditions. For each enriched sRNA, the log2 Fold Enrichment in the OMV fraction is displayed as a colored square for the corresponding culture condition, from 2 (yellow) to 10 (dark blue). Enriched sRNAs associated to OMVs for each condition were determined using DESeq2 (Love et al., 2014), calculating differential presence between intracellular and OMV-associated fractions for each condition. Transcripts with an average number of reads over 5 for a given set of triplicates, a log2 Fold Change over 2, and a corresponding adjusted p-value under 0.05 were selected.
Traces of almost all pathogenicity-island encoded sRNA were found in OMVs (Supplementary Figure S13). Among them, one can note the presence of IsrC, triggering the degradation of the mgrA mRNA, involved in Salmonella survival in macrophages (Padalon-Brauch et al., 2008). It was expressed in our control (PCN) as reported earlier (Hébrard et al., 2012). We found that it was also enriched in/on OMVs generated in Control High OD condition, with a logFC = 5.45 (p < 0.001). No trace of it was found in/on OMVs from SPI-2ind conditions (Supplementary Figure S13). IsrM is known to be expressed in gut tract conditions in vitro (Hébrard et al., 2012). We found the transcript enriched in the control and SPI-1ind at high OD, but not in SPI-2ind. This sRNA is targeting hilE mRNA, modulating SPI-1 expression and repressing sopA. ΔisrM strains are less invasive and have a reduced growth rate in host cells or the gut tract (Hautefort et al., 2008; Gong et al., 2011). IsrJ is a trans-acting sRNA that was enriched in the control at high OD and SPI-1ind condition. It is upregulated in conditions which promote invasion of epithelial cells and affects the translocation efficiency of virulence-associated effector proteins into non-phagocytic cells (Padalon-Brauch et al., 2008).
Some core-genome sRNA were also found to be enriched. For example, MicF which is a Hfq-dependent trans-acting sRNA that regulates the synthesis of a lipid A modifying enzyme (Papenfort and Vogel, 2010) was enriched in SPI-1ind OMVs. Finally, GcvB was enriched in SPI-2ind conditions only, even if it was found associated to OMVs in all conditions. This sRNA is controlling amino-acids uptake and has also a demonstrated positive effect on bacteria resistance to low pH in E. coli, which is a physiological condition encountered by Salmonella in macrophages (Jin et al., 2009; Sharma et al., 2011).
Others sRNAs involved in virulence were also found in/on OMVs, but without specific enrichment regarding their intracellular presence. For example, CrsB and CsrC (the later validated experimentally to be internalized as a full-length transcript in OMVs by PCR: Figures 5, 6, Table 1, and Supplementary Dataset C) were among the most exported sRNAs through OMVs, and the most abundant intracellular sRNAs (Supplementary Figure S13). These sRNAs control the translational regulator CsrA that regulates SPI-1 genes and the expression of flagella components, playing a central role in virulence (Fortune et al., 2006; Timmermans and Van Melderen, 2010). The 6S RNA, SsrS (validated experimentally to be internalized as a full-length transcript to OMVs by PCR: Figures 5, 6, Table 1, and Supplementary Dataset C), was also highly expressed inside the cells and associated with OMVs. It is necessary for acidic-environment adaptation and epithelial cell invasion (Ren et al., 2017). STnc1480 is a PhoP/Q and SlyA regulated sRNA whose role during the intracellular life of Salmonella has been suggested, and was found in OMVs in a high amount (Colgan et al., 2016). SgrS, a repressor of sugar uptake that also represses the SopD secreted virulence factor (Papenfort et al., 2012; Balasubramanian and Vanderpool, 2013), was also exported in all conditions.
This study provides a first global picture of the RNA cargo (>25 nt) associated with S. Typhimurium OMVs released under different in vitro culture conditions. These conditions mimic various environments encountered by Salmonella in its life cycle through the invasion and infection steps from the gut to macrophages. Even if this in vitro model does not fully reproduce the in vivo environment, a significant number of common pathways are induced, and among them a number of pathogenicity islands and virulence factors (Kröger et al., 2013; Srikumar et al., 2015). RNA was isolated from density gradient purified vesicles, with samples positive for a known outer membrane protein marker (OmpA), and lacking any obvious non-OMV structures like pili and flagellae, as judged by examination of TEM images (Supplementary Figure S2). These contaminating structures are present in crude OMV preparations (Supplementary Figure S2B). We cannot rule out the co-purification of nucleoprotein aggregates, however, these should be resistant to RNase A and the combination of RNase A and proteinase K. As the understanding of microbial membrane vesicles advances, we expect the validation of purity markers will provide the appropriate tools for even more stringent quality control.
The global quantity of OMVs and associated RNAs was not studied in this work. From western-blots, we could see differences in OmpA quantities between conditions. These do not automatically reflect differences in OMV secretion but can also originate from a change in the composition of the outer membrane. However, several studies already demonstrated the variation of membrane vesicles secretion induced by different cultivation conditions (Tashiro et al., 2010; McCaig et al., 2013). The global quantity of associated RNA depending on conditions did not clearly vary on our Bioanalyzer experiments, but quantification of such small amounts is challenging and should be addressed for further investigations (Aranda et al., 2009; Mateescu et al., 2017).
In accordance with several other studies, the majority of OMV-exported large RNAs were ribosomal RNAs (Dauros-Singorenko et al., 2018). It is interesting that these have been identified as a major immunostimulatory component for bacterial RNA recognition by dendritic cells and macrophages (Eberle et al., 2009; Li and Chen, 2012). They have also been reported to trigger a response from the human NLRP3 inflammasome in macrophages, as other types of RNAs do, thereby being part of the host–pathogen interactions during infection (Sha et al., 2014).
Other typically abundant ncRNAs, i.e., tRNAs were found associated with OMVs only at low levels in our data. This differs from previous observations on E. coli where they were among the most exported OMV-RNAs (Ghosal et al., 2015; Blenkiron et al., 2016). These differences might be due to the fact that we did not produce a small RNA sequencing library in the present study, which would probably better resolve the tRNA complement. As it has been demonstrated for E. coli, OMV-small-RNA libraries are highly enriched with fragmented tRNAs, largely at 15–50 nucleotides in length (Ghosal et al., 2015; Blenkiron et al., 2016).
In our data, the second most commonly found RNA type in OMVs were messenger RNAs, after rRNAs. The mRNA population included the majority of Salmonella’s expressed genes, however, there was a core population of transcripts that seemed always to be selected for export and enriched in the OMV RNA cargo. Some of them may originate from the OMV formation mechanism: one could expect some messengers coding for OMV-formation related proteins to be packaged at the same time as their product, as translation can occur in a spatial-dependent manner in bacteria (Nevo-Dinur et al., 2012; Buskila et al., 2014). However, RNA localization was also regulated independently from translation for some transcripts, suggesting a possible OMV-targeting process for specific mRNAs (Nevo-Dinur et al., 2011). Our data suggest that this differential packaging is adapted to environmental conditions and may be relevant for Salmonella’s virulence, as some populations of enriched mRNAs which were present in the RNA cargo were specific to each culture condition and contained pathogenicity-related mRNAs. This differs from previous findings in UPEC (Blenkiron et al., 2016). A common finding was the presence of prophage-originating RNAs, which are also known to represent pathogenicity-related regions. Type III secretion systems in E. coli are also connected to a vast network of phage-encoded genes which in turn are crucial for the evolution of pathogenesis (Tobe et al., 2006). This makes sense since phages are reported to be paramount in the evolution of bacterial pathogens by participating in horizontal gene transfer (Brussow et al., 2004). It is also important to note that the phage transcripts that we identified in our OMV RNA-Seq were encoded by temperate phages (phages that replicate using both lytic and lysogenic cycles) (Frye et al., 2005). We did not observe many bacterial cell lysis during our different culture conditions (Supplementary Figure S1), indicating that most of the phage-encoded RNA identified in our RNA-Seq data is originating from prophages. Nevertheless, we cannot exclude that the purification method used to purify OMVs did result in a co-purification of some active Salmonella’s phages, as gradient purification is sometimes also used for phage isolation (Effantin et al., 2010). As the phage capsid usually protects phage DNA from DNases and releases its content upon heating, “phage contamination” could also occur during cDNA library preparation (Owen et al., 2017). This should be verified in depth in upcoming studies as phage-related transcripts are enriched in/on OMVs purified in all tested conditions.
It is interesting to note that some OMV-associated mRNAs were enriched in/on vesicles compared to their intracellular fractions, suggesting that this export is not a passive but rather an active, regulated process. The purpose of such an operation and the underlying mechanism remain obscure. Read coverage of transcripts varied consequently depending on the exported RNA, sometimes showing an almost identical pattern to the coverage observed for the intracellular transcript, or on the contrary adopting distinct coverage profiles suggesting a specific processing or degradation. This dual nature was confirmed by PCR experiments in which we identified a number of native full-length RNAs associated with OMVs but failed to validate other predicted full-length transcripts. Whether this degradation or processing happens inside the cell or is ongoing during the long vesicle isolation procedure is an open question and further work should address this topic. However, we confirmed the presence of several full-length RNAs in independent replicates and by different techniques, showing that at least a fraction of RNAs are constantly associated and protected by OMVs.
Hence some OMV-associated RNAs might have a functional potential. This should be verified in future studies. Indeed, full-length mRNAs could potentially be translated by surrounding bacteria to which OMVs may be delivered. It could be a way to help members of the population by providing mRNAs even before a specific transcription activation signal has been received by the receptor cell, offering a quick response to a changing environment sensed by the exporting cell. RNA transfer between cells has been observed for exosome shuttles (Valadi et al., 2007). However, no comparable findings exist for OMV RNAs so far, even if evidence exists that OMV cargo in general is exchanged between bacteria [see for instance transfer of Pseudomonas Quinone Signal in Pseudomonas aeruginosa, (Mashburn and Whiteley, 2005)]. Bacterial exchange of OMVs is a preferential process, with different affinity and delivery efficiency for each receiving species (Tashiro et al., 2017). On the contrary, in the case of competing strains or host cells, these delivered mRNAs may “hack” the target’s translation machinery by producing virulence factors directly inside them, being part of the invasion toolkit of Salmonella enterica. In that sense, bacterial mRNA translation by mammalian cells has already been observed (Schoen et al., 2005). However, in this case prokaryotic RNAs were optimized for this purpose. For native mRNAs lacking eukaryotic nuclear processing like 5′ capping or the lack of internal ribosome entry sites (IRES), this process is likely to be impossible (Pelletier and Sonenberg, 1988; Pisarev et al., 2005).
sRNAs also populated the enriched transcripts which were associated with OMVs and their over-representation compared to the intracellular environment was mainly condition-dependent. However, it is difficult to see a relationship between the known implication of some sRNAs in virulence and the corresponding enrichment in vesicles. For instance, STnc440, STnc470, STnc3750, and IsrH are highly expressed in the Salmonella Containing Vacuole (Srikumar et al., 2015) but not specifically enriched in/on OMVs from SPI-2ind. A proportion of small RNAs originating from E. coli OMVs has been successfully aligned with the human genome in a way resembling eukaryotes’ long non-coding RNAs, which could also be the case in Salmonella with potential implications in host–pathogen communication (Celluzzi and Masotti, 2016). The interplay between the levels of regulatory sRNAs and their mRNA targets, together with their concomitant association to OMVs, would be an interesting point to look at in future work in order to be able to understand why given sRNAs are released via OMVs.
The confirmed presence of many full-length protein-associated RNAs in OMVs remains to be clarified. Whereas the export of SRP RNA might be explained by a side-effect of its location (proximity to the cytoplasmic membrane), the strong presence of others is mysterious. Interestingly, most of them are linked to virulence and resistance of Salmonella to external stresses to some extent. For instance, 10Sa is involved in the regulation of specific genes linked to virulence and may take part in conferring resistance to the oxidative stress encountered throughout infection by preventing the buildup of abnormal proteins which may be toxic to the cell (Julio et al., 2000). Another interesting moiety is the 6S RNA (SsrS) which has been shown to be crucial for resistance and adaptation to acidic environment encountered during invasion (Ren et al., 2017). As we demonstrated that these molecules were inside OMVs rather than bound on the surface for SsrS, CsrC, 10Sa, and rnpB, questions arise on the possible targeted mechanisms leading to packaging.
Finally, in OMVs isolated from Borrelia burgdorferi cultures, an enrichment of plasmid-encoded RNAs over chromosome-encoded transcripts has been recently shown (Malge et al., 2018). We validated the presence of a full-length plasmid-encoded transcript (pSLT035) in Salmonella-derived OMVs but in general we could not observe an enrichment of plasmid-encoded RNAs over chromosome-encoded transcripts associated with Salmonella OMVs (data not shown).
In summary, this work provides a first insight into the differential association of RNAs (>25 nt) with Salmonella-derived OMVs and emphasizes the impact of the bacteria-encountered environmental conditions on the RNA content of the vesicles. Much more work is needed to fully understand the underlying mechanisms and physiological processes that lead to this export, which results in the release of many virulence-associated RNAs and could have important consequences in host–pathogen interplay during infection. Whether this adaptable RNA export is a mechanism for targeted transfer or a way for cells to eliminate undesirable RNAs remains to be clarified. However, the confirmation of the export of full-length transcripts opens the door to numerous possible functional implications in cell-to-cell communication.
JF and PW conceived the original idea. JH, JF, and AM designed the experiments. JH, JF, AM, AH-B, JG, RH, AE, and DG performed the experiments. AH-B, PM, AM, and JF analyzed the data. AM and JF wrote the manuscript. All authors analyzed results, commented on and edited the manuscript.
This present work was financed by the Luxembourg National Research Fund (FNR) (CORE Junior/14/BM/8066232) to JF and (CORE/15/BM/10404093) and (CORE/16/BM/11276306) to PW and by a grant from the NIH Common Fund Extracellular RNA Communication Consortium (1U01HL126496), as well as a Baylor subaward (5U54DA036134) to DG.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the scientists and technical staff of the Luxembourg Centre for Systems Biomedicine and the HPC facilities of the University of Luxembourg where in silico analyses presented in this paper were carried (Varrette et al., 2014). We are grateful to the whole Eco-Systems Biology group, as well as to Esther Nolte-’t Hoen for fruitful discussions. We also acknowledge the support and the use of resources of the French Infrastructure for Integrated Structural Biology FRISBI ANR-10-INBS-05 and of Instruct-ERIC for TEM imaging.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02015/full#supplementary-material
Abdullah, Z., Schlee, M., Roth, S., Mraheil, M. A., Barchet, W., Böttcher, J., et al. (2012). RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31, 4153–4164. doi: 10.1038/emboj.2012.274
Aranda, R., Dineen, S. M., Craig, R. L., Guerrieri, R. A., and Robertson, J. M. (2009). Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 10(4) concentration range. Anal. Biochem. 387, 122–127. doi: 10.1016/j.ab.2009.01.003
Avital, G., Avraham, R., Fan, A., Hashimshony, T., Hung, D. T., and Yanai, I. (2017). scDual-Seq: mapping the gene regulatory program of Salmonella infection by host and pathogen single-cell RNA-sequencing. Genome Biol. 18:200. doi: 10.1186/s13059-017-1340-x
Bai, J., Kim, S. I., Ryu, S., and Yoon, H. (2014). identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infect. Immun. 82, 4001–4010. doi: 10.1128/IAI.01416-13
Balasubramanian, D., and Vanderpool, C. K. (2013). Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J. Bacteriol. 195, 4620–4630. doi: 10.1128/JB.00586-13
Beuzón, C. R., Banks, G., Deiwick, J., Hensel, M., and Holden, D. W. (1999). pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33, 806–816. doi: 10.1046/j.1365-2958.1999.01527.x
Blenkiron, C., Simonov, D., Muthukaruppan, A., Tsai, P., Dauros, P., Green, S., et al. (2016). Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS One 11:e0160440. doi: 10.1371/journal.pone.0160440
Bonnington, K. E., and Kuehn, M. J. (2016). Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in salmonella during environmental transitions. mBio 7:e01532-16. doi: 10.1128/mBio.01532-16
Brussow, H., Canchaya, C., and Hardt, W.-D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Buskila, A. A., Kannaiah, S., and Amster-Choder, O. (2014). RNA localization in bacteria. RNA Biol. 11, 1051–1060. doi: 10.4161/rna.36135
Celluzzi, A., and Masotti, A. (2016). How our other genome controls our epi-genome. Trends Microbiol. 24, 777–787. doi: 10.1016/j.tim.2016.05.005
Chen, X., Liang, H., Zhang, J., Zen, K., and Zhang, C.-Y. (2012). Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 22, 125–132. doi: 10.1016/j.tcb.2011.12.001
Choi, J.-W., Kim, S.-C., Hong, S.-H., and Lee, H.-J. (2017). Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 96, 458–466. doi: 10.1177/0022034516685071
Colgan, A. M., Kröger, C., Diard, M., Hardt, W.-D., Puente, J. L., Sivasankaran, S. K., et al. (2016). The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12:e1006258. doi: 10.1371/journal.pgen.1006258
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364
Coombes, B. K., Wickham, M. E., Brown, N. F., Lemire, S., Bossi, L., Hsiao, W. W. L., et al. (2005). Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar Typhimurium with autonomous expression from its associated phage. J. Mol. Biol. 348, 817–830. doi: 10.1016/j.jmb.2005.03.024
Dauros-Singorenko, P., Blenkiron, C., Phillips, A., and Swift, S. (2018). The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 365:fny023. doi: 10.1093/femsle/fny023
Didier, P., Godet, J., and Mély, Y. (2009). Two-photon two-focus fluorescence correlation spectroscopy with a tunable distance between the excitation volumes. J. Fluoresc. 19, 561–565. doi: 10.1007/s10895-008-0424-0
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Eberle, F., Sirin, M., Binder, M., and Dalpke, A. H. (2009). Bacterial RNA is recognized by different sets of immunoreceptors. Eur. J. Immunol. 39, 2537–2547. doi: 10.1002/eji.200838978
Effantin, G., Figueroa-Bossi, N., Schoehn, G., Bossi, L., and Conway, J. F. (2010). The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology 402, 355–365. doi: 10.1016/j.virol.2010.03.041
Eletsky, A., Michalska, K., Houliston, S., Zhang, Q., Daily, M. D., Xu, X., et al. (2014). Structural and functional characterization of DUF1471 domains of Salmonella proteins SrfN, YdgH/SssB, and YahO. PLoS One 9:e101787. doi: 10.1371/journal.pone.0101787
Elhenawy, W., Bording-Jorgensen, M., Valguarnera, E., Haurat, M. F., Wine, E., and Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940-16. doi: 10.1128/mBio.00940-16
Fabrega, A., and Vila, J. (2013). Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/CMR.00066-12
Fortune, D. R., Suyemoto, M., and Altier, C. (2006). Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar typhimurium. Infect. Immun. 74, 331–339. doi: 10.1128/IAI.74.1.331-339.2006
Fritz, J. V., Heintz-Buschart, A., Ghosal, A., Wampach, L., Etheridge, A., Galas, D., et al. (2016). Sources and functions of extracellular small RNAs in human circulation. Annu. Rev. Nutr. 36, 301–336. doi: 10.1146/annurev-nutr-071715-050711
Frye, J. G., Porwollik, S., Blackmer, F., Cheng, P., and McClelland, M. (2005). Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 187, 1485–1492. doi: 10.1128/JB.187.4.1485-1492.2005
Ghosal, A., Upadhyaya, B. B., Fritz, J. V., Heintz-Buschart, A., Desai, M. S., Yusuf, D., et al. (2015). The extracellular RNA complement of Escherichia coli. Microbiologyopen 4, 252–266. doi: 10.1002/mbo3.235
Gong, H., Vu, G.-P., Bai, Y., Chan, E., Wu, R., Yang, E., et al. (2011). A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 7:e1002120. doi: 10.1371/journal.ppat.1002120
González Plaza, J. J. (2018). Small RNAs in cell-to-cell communications during bacterial infection. FEMS Microbiol. Lett. 365:fny024. doi: 10.1093/femsle/fny024
Guerrero-Mandujano, A., Hernández-Cortez, C., Ibarra, J. A., and Castro-Escarpulli, G. (2017). The outer membrane vesicles: secretion system type zero. Traffic 18, 425–432. doi: 10.1111/tra.12488
Habier, J., May, P., Heintz-Buschart, A., Ghosal, A., Wienecke-Baldacchino, A. K., Nolte-’t Hoen, E. N. M., et al. (2018). Extraction and analysis of RNA isolated from pure bacteria-derived outer membrane vesicles. Methods Mol. Biol. 1737, 213–230. doi: 10.1007/978-1-4939-7634-8_13
Hautefort, I., Thompson, A., Eriksson-Ygberg, S., Parker, M. L., Lucchini, S., Danino, V., et al. (2008). During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10, 958–984. doi: 10.1111/j.1462-5822.2007.01099.x
Hébrard, M., Kröger, C., Sivasankaran, S. K., Händler, K., and Hinton, J. C. (2011). The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium. Curr. Opin. Biotechnol. 22, 200–210. doi: 10.1016/j.copbio.2011.02.007
Hébrard, M., Kröger, C., Srikumar, S., Colgan, A., Händler, K., and Hinton, J. C. D. (2012). sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol. 9, 437–445. doi: 10.4161/rna.20480
Heintz-Buschart, A., Yusuf, D., Kaysen, A., Etheridge, A., Fritz, J. V., May, P., et al. (2018). Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 16:52. doi: 10.1186/s12915-018-0522-7
Himeno, H., Kurita, D., and Muto, A. (2014). tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front. Genet. 5:66. doi: 10.3389/fgene.2014.00066
Ho, M.-H., Chen, C.-H., Goodwin, J. S., Wang, B.-Y., and Xie, H. (2015). Functional advantages of Porphyromonas gingivalis vesicles. PLoS One 10:e0123448. doi: 10.1371/journal.pone.0123448
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biom. J. 50, 346–363. doi: 10.1002/bimj.200810425
Ibarra, J. A., Knodler, L. A., Sturdevant, D. E., Virtaneva, K., Carmody, A. B., Fischer, E. R., et al. (2010). Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella–host cell interactions in vitro. Microbiology 156, 1120–1133. doi: 10.1099/mic.0.032896-0
Jan, A. T. (2017). Outer Membrane Vesicles (OMVs) of gram-negative bacteria: a perspective update. Front. Microbiol. 8:1053. doi: 10.3389/fmicb.2017.01053
Jin, Y., Watt, R. M., Danchin, A., and Huang, J. (2009). Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10:165. doi: 10.1186/1471-2164-10-165
Julio, S. M., Heithoff, D. M., and Mahan, M. J. (2000). ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol. 182, 1558–1563. doi: 10.1128/JB.182.6.1558-1563.2000
Kidwai, A. S., Mushamiri, I., Niemann, G. S., Brown, R. N., Adkins, J. N., and Heffron, F. (2013). Diverse secreted effectors are required for Salmonella persistence in a mouse infection model. PLoS One 8:e70753. doi: 10.1371/journal.pone.0070753
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 12:e1005672. doi: 10.1371/journal.ppat.1005672
Kofler, R., Orozco-terWengel, P., De Maio, N., Pandey, R. V., Nolte, V., Futschik, A., et al. (2011). PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6:e15925. doi: 10.1371/journal.pone.0015925
Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlöf, D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14, 683–695. doi: 10.1016/j.chom.2013.11.010
LaRock, D. L., Chaudhary, A., and Miller, S. I. (2015). Salmonella interactions with host processes. Nat. Rev. Microbiol. 13, 191–205. doi: 10.1038/nrmicro3420
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, X.-D., and Chen, Z. J. (2012). Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife 1:e00102. doi: 10.7554/eLife.00102
Liao, Y., Smyth, G. K., and Shi, W. (2014). Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Löber, S., Jäckel, D., Kaiser, N., and Hensel, M. (2006). Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296, 435–447. doi: 10.1016/j.ijmm.2006.05.001
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lynch, J. B., and Alegado, R. A. (2017). Spheres of hope, packets of doom: the good and bad of outer membrane vesicles in interspecies and ecological dynamics. J. Bacteriol. 199:e00012-17. doi: 10.1128/JB.00012-17
Malge, A., Ghai, V., Reddy, P. J., Baxter, D., Kim, T.-K., Moritz, R. L., et al. (2018). mRNA transcript distribution bias between Borrelia burgdorferi bacteria and their outer membrane vesicles. FEMS Microbiol. Lett. 365:fny135. doi: 10.1093/femsle/fny135
Manning, A. J., and Kuehn, M. J. (2013). Functional advantages conferred by extracellular prokaryotic membrane vesicles. J. Mol. Microbiol. Biotechnol. 23, 131–141. doi: 10.1159/000346548
Mashburn, L. M., and Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. doi: 10.1038/nature03925
Mateescu, B., Kowal, E. J. K., van Balkom, B. W. M., Bartel, S., Bhattacharyya, S. N., Buzás, E. I., et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J. Extracell. Vesicles 6:1286095. doi: 10.1080/20013078.2017.1286095
McCaig, W. D., Koller, A., and Thanassi, D. G. (2013). Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 195, 1120–1132. doi: 10.1128/JB.02007-12
Miller, J. (ed.) (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Mulder, D. T., Cooper, C. A., and Coombes, B. K. (2012). Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect. Immun. 80, 1996–2007. doi: 10.1128/IAI.06205-11
Nevo-Dinur, K., Govindarajan, S., and Amster-Choder, O. (2012). Subcellular localization of RNA and proteins in prokaryotes. Trends Genet. 28, 314–322. doi: 10.1016/j.tig.2012.03.008
Nevo-Dinur, K., Nussbaum-Shochat, A., Ben-Yehuda, S., and Amster-Choder, O. (2011). Translation-independent localization of mRNA in E. coli. Science 331, 1081–1084. doi: 10.1126/science.1195691
Niemann, G. S., Brown, R. N., Gustin, J. K., Stufkens, A., Shaikh-Kidwai, A. S., Li, J., et al. (2011). Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43. doi: 10.1128/IAI.00771-10
Obregón-Henao, A., Duque-Correa, M. A., Rojas, M., García, L. F., Brennan, P. J., Ortiz, B. L., et al. (2012). Stable extracellular RNA fragments of Mycobacterium tuberculosis induce early apoptosis in human monocytes via a caspase-8 dependent mechanism. PLoS One 7:e29970. doi: 10.1371/journal.pone.0029970
O’Donoghue, E. J., and Krachler, A. M. (2016). Mechanisms of outer membrane vesicle entry into host cells: microreview - OMV entry into host cells. Cell. Microbiol. 18, 1508–1517. doi: 10.1111/cmi.12655
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). Vegan: Community Ecology Package R package version 2.5-2. Available at: https://CRAN.R-project.org/package=vegan
Orench-Rivera, N., and Kuehn, M. J. (2016). Environmentally controlled bacterial vesicle-mediated export: environmentally controlled bacterial vesicle-mediated export. Cell. Microbiol. 18, 1525–1536. doi: 10.1111/cmi.12676
Ortega,Á.D., Gonzalo-Asensio, J., and García-del Portillo, F. (2012). Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol. 9, 469–488. doi: 10.4161/rna.19317
Owen, S. V., Wenner, N., Canals, R., Makumi, A., Hammarlöf, D. L., Gordon, M. A., et al. (2017). Characterization of the prophage repertoire of African Salmonella Typhimurium ST313 reveals high levels of spontaneous induction of novel phage BTP1. Front. Microbiol. 8:235. doi: 10.3389/fmicb.2017.00235
Padalon-Brauch, G., Hershberg, R., Elgrably-Weiss, M., Baruch, K., Rosenshine, I., Margalit, H., et al. (2008). Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 36, 1913–1927. doi: 10.1093/nar/gkn050
Papenfort, K., Podkaminski, D., Hinton, J. C. D., and Vogel, J. (2012). The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl. Acad. Sci. U.S.A. 109, E757–E764. doi: 10.1073/pnas.1119414109
Papenfort, K., and Vogel, J. (2010). Regulatory RNA in bacterial pathogens. Cell Host Microbe 8, 116–127. doi: 10.1016/j.chom.2010.06.008
Pelletier, J., and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. doi: 10.1038/334320a0
Pilar, A. V. C., Reid-Yu, S. A., Cooper, C. A., Mulder, D. T., and Coombes, B. K. (2012). GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathog. 8:e1002773. doi: 10.1371/journal.ppat.1002773
Pisarev, A. V., Shirokikh, N. E., and Hellen, C. U. T. (2005). Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 328, 589–605. doi: 10.1016/j.crvi.2005.02.004
Ren, J., Sang, Y., Qin, R., Cui, Z., and Yao, Y.-F. (2017). 6S RNA is involved in acid resistance and invasion of epithelial cells in Salmonella enterica serovar Typhimurium. Future Microbiol. 12, 1045–1057. doi: 10.2217/fmb-2017-0055
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Schoen, C., Kolb-Mäurer, A., Geginat, G., Löffler, D., Bergmann, B., Stritzker, J., et al. (2005). Bacterial delivery of functional messenger RNA to mammalian cells: bacterial delivery of mRNA to mammalian cells. Cell. Microbiol. 7, 709–724. doi: 10.1111/j.1462-5822.2005.00507.x
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Schwechheimer, C., Sullivan, C. J., and Kuehn, M. J. (2013). Envelope control of outer membrane vesicle production in gram-negative bacteria. Biochemistry 52, 3031–3040. doi: 10.1021/bi400164t
Sha, W., Mitoma, H., Hanabuchi, S., Bao, M., Weng, L., Sugimoto, N., et al. (2014). Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc. Natl. Acad. Sci. U.S.A. 111, 16059–16064. doi: 10.1073/pnas.1412487111
Sharma, C. M., Papenfort, K., Pernitzsch, S. R., Mollenkopf, H.-J., Hinton, J. C. D., and Vogel, J. (2011). Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 81, 1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x
Shelke, G. V., Lässer, C., Gho, Y. S., and Lötvall, J. (2014). Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 3:24783. doi: 10.3402/jev.v3.24783
Sjöström, A. E., Sandblad, L., Uhlin, B. E., and Wai, S. N. (2015). Membrane vesicle-mediated release of bacterial RNA. Sci. Rep. 5:15329. doi: 10.1038/srep15329
Soares, R. P., Xander, P., Costa, A. O., Marcilla, A., Menezes-Neto, A., Del Portillo, H., et al. (2017). Highlights of the São Paulo ISEV workshop on extracellular vesicles in cross-kingdom communication. J. Extracell. Vesicles 6, 1407213. doi: 10.1080/20013078.2017.1407213
Srikumar, S., Kröger, C., Hébrard, M., Colgan, A., Owen, S. V., Sivasankaran, S. K., et al. (2015). RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11:e1005262. doi: 10.1371/journal.ppat.1005262
Susilawati, T. N., Jex, A. R., Cantacessi, C., Pearson, M., Navarro, S., Susianto, A., et al. (2016). Deep sequencing approach for investigating infectious agents causing fever. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1137–1149. doi: 10.1007/s10096-016-2644-6
Tashiro, Y., Hasegawa, Y., Shintani, M., Takaki, K., Ohkuma, M., Kimbara, K., et al. (2017). Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front. Microbiol. 8:571. doi: 10.3389/fmicb.2017.00571
Tashiro, Y., Ichikawa, S., Shimizu, M., Toyofuku, M., Takaya, N., Nakajima-Kambe, T., et al. (2010). Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76, 3732–3739. doi: 10.1128/AEM.02794-09
Timmermans, J., and Van Melderen, L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67, 2897–2908. doi: 10.1007/s00018-010-0381-z
Tobe, T., Beatson, S. A., Taniguchi, H., Abe, H., Bailey, C. M., Fivian, A., et al. (2006). An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U.S.A. 103, 14941–14946. doi: 10.1073/pnas.0604891103
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. doi: 10.1038/ncb1596
Varrette, S., Bouvry, P., Cartiaux, H., and Georgatos, F. (2014). “Management of an academic hpc cluster: The ul experience,” in Proceedings of the 2014 International Conference On. IEEE High Performance Computing & Simulation (HPCS), (Luxembourg: University of Luxembourg), 959–967. doi: 10.1109/HPCSim.2014.6903792
Weiberg, A., Wang, M., Lin, F.-M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis, Use R!. New York, NY: Springer-Verlag. doi: 10.1007/978-0-387-98141-3
Yoon, H., Ansong, C., Adkins, J. N., and Heffron, F. (2011a). Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79, 2182–2192. doi: 10.1128/IAI.01277-10
Yoon, H., Ansong, C., McDermott, J. E., Gritsenko, M., Smith, R. D., Heffron, F., et al. (2011b). Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella. BMC Syst. Biol. 5:100. doi: 10.1186/1752-0509-5-100
Keywords: outer membrane vesicle (OMV), RNA export, Salmonella enterica pathogenicity, host–pathogen interaction, SPI-1, SPI-2, virulence, sRNA
Citation: Malabirade A, Habier J, Heintz-Buschart A, May P, Godet J, Halder R, Etheridge A, Galas D, Wilmes P and Fritz JV (2018) The RNA Complement of Outer Membrane Vesicles From Salmonella enterica Serovar Typhimurium Under Distinct Culture Conditions. Front. Microbiol. 9:2015. doi: 10.3389/fmicb.2018.02015
Received: 15 March 2018; Accepted: 09 August 2018;
Published: 30 August 2018.
Edited by:
Rosana Puccia, Federal University of São Paulo, BrazilReviewed by:
Kai Papenfort, Ludwig-Maximilians-Universität München, GermanyCopyright © 2018 Malabirade, Habier, Heintz-Buschart, May, Godet, Halder, Etheridge, Galas, Wilmes and Fritz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul Wilmes, cGF1bC53aWxtZXNAdW5pLmx1 Joëlle V. Fritz, am9lbGxlLnBlbm55LWZyaXR6QGV4dC51bmkubHU=
†These authors have contributed equally to this work
‡Present address: Anna Heintz-Buschart, Department of Soil Ecology, Helmholtz-Centre for Environmental Research - UFZ, Halle, Germany; German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany Joëlle V. Fritz, Centre Hospitalier de Luxembourg, Luxembourg, Luxembourg
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.