- 1Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 2Department of Microbiology, University of Georgia, Athens, GA, United States
- 3Odum School of Ecology, University of Georgia, Athens, GA, United States
Reforestation with different tree species could alter soil properties and in turn affect the bacterial community. However, the effects of long-term reforestation on bacterial community structure and diversity of subtropical forest soils are poorly understood. In the current study, we applied error-corrected barcoded pyrosequencing to characterize the differences in the soil bacterial community in a low mountain, subtropical forest subjected to reforestation. The communities were sampled in the summer and winter from a native broadleaved forest (BROAD-Nat) and two adjacent coniferous plantations, a Calocedrus formosana forest of 80 years (CONIF-80) and a Cunninghamia konishii forest of 40 years (CONIF-40). The soil bacterial communities among three forest types were dominated by Acidobacteria and Alphaproteobacteria. The distribution of abundant genera among communities was different. Based on the Shannon diversity index, the bacterial alpha diversity of CONIF-40 community was significantly higher than that in the CONIF-80 and BROAD-Nat soils. In both of the coniferous plantations, the soil bacterial diversity in summer was also higher than that in winter. Distribution of some abundant phylogenetic groups, K-shuff and redundancy analysis of beta diversity among communities showed that the bacterial structure of three soil communities differed between two seasons. These results suggest that seasonal differences influence the diversity and structure of bacterial soil communities and that the communities remain different even after a long period of reforestation.
Introduction
Soil bacteria play an important role in soil processes, including mineralization of organic matter and biogeochemical cycling of carbon and nitrogen (Bardgett et al., 2008; Chatterjee et al., 2008; Burton et al., 2010). The soil bacterial community can be influenced by soil properties (Stres et al., 2008; Ushio et al., 2008; Lauber et al., 2009). Different plant species can also result in distinct soil bacterial communities (Oh et al., 2012). Similarly, other environmental factors can play important roles, such as litter quality and root exudates (Quideau et al., 2001; Grayston and Prescott, 2005; Xu et al., 2008). Seasonal differences in temperature and precipitation also alter soil bacterial communities. For example, soil microbial phospholipid fatty acid (PLFA) composition associated with organic matter decomposition was significantly affected by temperature differences (Zhou et al., 2016). Multiple factors, including temperature, resulted in the complex responses of bamboo soil bacterial composition and diversity across altitudinal gradients (Lin et al., 2015). Previous studies also indicated that precipitation could affect bacterial communities indirectly by changing the soil moisture, which may have resulted in composition differences between perhumid forest sites with similar elevations but different precipitation (Lin et al., 2014).
The disturbances caused by forest management and other land uses are another important factor affecting soil microbes. They can alter soil characteristics and then influence the diversity and structure of the microbial community (Hartmann et al., 2014). For instance, the abundances of Proteobacteria and Firmicutes were significantly correlated with the disturbances of soil compaction associated with logging (Hartmann et al., 2014). Rainforest conversion to rubber plantation significantly increased the abundance of Chloroflexi in the bacterial community, while the most abundant phylum, Acidobacteria, was largely unchanged (Kerfahi et al., 2016). The proportion of some abundant bacterial groups differed between communities of native and regenerated perhumid montane forest soils (Lin et al., 2011b).
In the current study, we analyzed the differences of soil bacterial communities at a subtropical low mountain forest subjected to reforestation. This forest ecosystem is mainly covered by natural broadleaved forest. In the same region, the natural forest was replaced by a Calocedrus plantation 80 years ago and a Cunninghamia plantation 40 years ago. In a previous study using 16S ribosomal ribonucleic acid (rRNA) gene clone libraries and Sanger sequencing, we found that the reforestation with conifers altered the structure of the bacterial community and increased bacterial diversity (Lin et al., 2011a). In addition, another study indicated that the microbial biomass, PLFA content and soil enzymatic activities were higher in the summer than in the winter (Chang et al., 2016). However, the PLFA method cannot provide information about microbial populations at the genus or family level (Papadopoulou et al., 2011), and the low number of sequences from clone library only provided data on the abundant members of the community (Caporaso et al., 2011), leaving a gap in our understanding of the effects of season and disturbance on the soil bacterial community at this ecosystem. Hence, we applied the barcoded pyrosequencing method to obtain a more comprehensive survey of the soil bacterial communities. Compared to the clone library, the pyrosequencing method is less labor intensive and can provide between 4,500 and 52,000 unique sequences or operational taxonomic units (OTUs) in soils (Acosta-Martínez et al., 2010). Thus, it quickly provides much more comprehensive information about the microbial community structure. We hypothesized that both the change in management and seasonal differences would increase the soil bacterial diversity and alter the abundance of some bacterial taxa. The first objective of this study was to determine the soil bacterial diversity and structure in natural broadleaved forest compared to the coniferous plantation soils. Our second objective was to compare the effects of season on these soil bacterial communities.
Materials and Methods
Site Description and Soil Sampling
This study was conducted at Lienhuachi Experimental Forest, a subtropical low mountain area (about 700 m.a.s.l.) in central Taiwan (23°54′N, 120°54′E). It is a long-term ecological study site of low altitude forest ecosystem and encompasses 461 ha, with 261 ha covered by evergreen natural hardwood forest. Approximately 50% of the region has been replanted by coniferous plantations, including Calocedrus formosana and Cunninghamia konishii. The mean annual precipitation is about 2,200 mm. Nearly 90% of which occurs between March and September. The mean annual temperature is about 21°C (30°C in July and 10°C in January) (Lee et al., 2008). Soil samples were collected in July, 2010 (summer) and February, 2012 (winter). A native broadleaf forest (BROAD-Nat), and secondary coniferous Calocedrus plantation of 80 years (CONIF-80) and Cunninghamia plantation of 40 years (CONIF-40) were included for sampling. Each vegetation type was sampled at four (winter) or five (summer) 50 m × 50 m plots established 50 m apart along transect lines. The plots were marked to facilitate subsequent sample collections at the same location. After removing the surface litter, three subsamples at each plot were collected with a soil auger, 8 cm in diameter and 10 cm deep and pooled. The soil samples were passed through a 2 mm sieve, and a portion was stored at 4°C for soil biochemical assays. The remainder of the samples was freeze-dried and stored in -20°C for DNA extraction. The soils in sampling sites were moderately well-drained and classified as Typic Dystrudept (U.S. Soil Taxonomy) (Jien et al., 2009). The soils were strongly acidic, and the pH values of the surface soils ranged from 3.6 to 3.8. Other characteristics of each soil site are described in Chang et al. (2016).
DNA Extraction and Amplification of 16S rRNA Genes and Pyrosequencing
The soil communities DNA from 0.25 g of soil from each plot were extracted using the PowerSoilTM Soil DNA Isolation kit (MoBio Industries, Carlsbad, CA, United States) in accordance with manufacturer’s instructions. The V1 to V2 regions of the bacterial 16S rRNA gene were amplified using 27F and 338R primers (Lane, 1991). Polymerase chain reactions (PCRs) were performed as described previously (Lin et al., 2015). Secondary PCR (3 cycles rather than 20) was carried out to add the different barcodes for each sample (Lin et al., 2015). The unique and error-correcting barcodes facilitated sorting of sequences from a single pyrosequencing run (Hamady et al., 2008). Products were then purified using the PCR clean up system (Viogene BioTek Corp., New Taipei City, Taiwan). The qualities and concentrations of the purified barcoded PCR products were determined using a NanoDrop Spectrophotometer (Thermo Fisher Scientific). Amplicon pyrosequencing was performed by Mission Biotech (Taipei, Taiwan) using a 454/Roche GS-FLX Titanium Instrument (Roche, Branchburg, NJ, United States).
Pyrosequence Analyses
All the sequences were processed through the Ribosomal Data Project (RDP) pyrosequencing pipeline1 (RDP release 11.5; release date: 2016.09.30). Sequences were assigned to each sample by recognition of the barcode and trimmed to remove the barcodes, primers, and linker. The pyrosequences were then filtered, and those that did not have Ns, were more than 200 bp in length, were free of chimeras using Uchime (Edgar et al., 2011), and had quality scores >25 were used for further analyzes. The trimmed and filtered sequences used for the subsequent analyses were submitted to the Sequence Read Archives with the accession number of SRP136858. Taxonomic information was analyzed using the Naïve Bayesian rRNA classifier of the RDP with a confidence cutoff of 80% (Wang et al., 2007). OTUs were formed with an evolutionary distance of 0.03. Singletons that appeared only once in the data set were removed. To avoid measurement differences due to differences in sample size for estimation of the alpha and beta diversity (Roesch et al., 2007; Will et al., 2010), the sample size of each replicate was normalized to 1800 sequences by random selection from larger samples. Although this procedure reduced the number of rare OTUs analyzed, it was justified because the goals of these studies were to compare the effects of season and vegetation on the bacterial communities and not to capture all OTUs. To estimate bacterial alpha diversity, Shannon diversity indexes (Shannon and Weaver, 1963) were calculated based on the Complete Linkage Clustering data for OTUs with an evolutionary distance of 0.03. The “Venn” command of Mothur program (Schloss et al., 2009) was applied to visualize the shared OTUs among bacterial communities. The abundant genera and genus-level groups of three forest soil bacterial communities were identified with the Naïve Bayesian rRNA classifier of the RDP. The differences in distribution between the seasons were analyzed with one way ANOVA. The heat map was plotted with Heatmapper (Babicki et al., 2016). Community comparisons were further performed using K-shuff analysis (Jangid et al., 2016). Correlations (Spearman rank correlation ρ) between resemblance matrices of bacterial composition based on the distribution of OTUs, diversity, and soil properties were assessed using RELATE as implemented in the PRIMER V6 software (Clarke and Gorley, 2006). A permutation test (999 permutations) was applied to evaluate significance. The resemblance matrix was derived from normalized data. Under the null hypothesis that there is no relation between the two similarity matrices, ρ will be approximately zero. Redundancy analysis was carried out by using the vegan package in R v.3.2.1 to determine the relationship between the distribution of OTUs of bacterial community and soil properties.
Results
Bacterial Community Composition
Using barcoded pyrosequencing, the 16S rRNA gene sequences were retrieved from three forest soils in summer and winter to investigate the bacterial composition. About 12,300 to 24,000 pyrosequence reads were obtained from each soil communities (Supplementary Table S1). These sequences were dominated with Acidobacteria (42–48%) and Proteobacteria (30–33%) (Supplementary Figure S1). Actinobacteria was the third most abundant phyla, but accounted for less than 6% of the sequences. The proportion of other phyla, such as Bacteroidetes, Chloroflexi, and Firmicutes, were all less than 4% (Supplementary Figure S1). The Gp1, 2, and 3 were the most abundant groups of Acidobacteria, and the relative abundance of Gp2 was higher in winter (Table 1 and Figure 1). In contrast, Acidobacteria Gp3 was more abundant in summer. Within the Proteobacteria, Alphaproteobacteria was the most abundant group (17–22%), and Bradyrhizobium of the Rhizobiales was the most abundant genus, especially in the summer. The Beta-, Gamma- and Delta-proteobacteria were all only about 2–5% of sequences among the three communities in both seasons. In summary, although the same bacterial genera and genus-level groups were abundant under all the conditions examined, the relative abundances of some phylogenetic groups varied significantly with season.
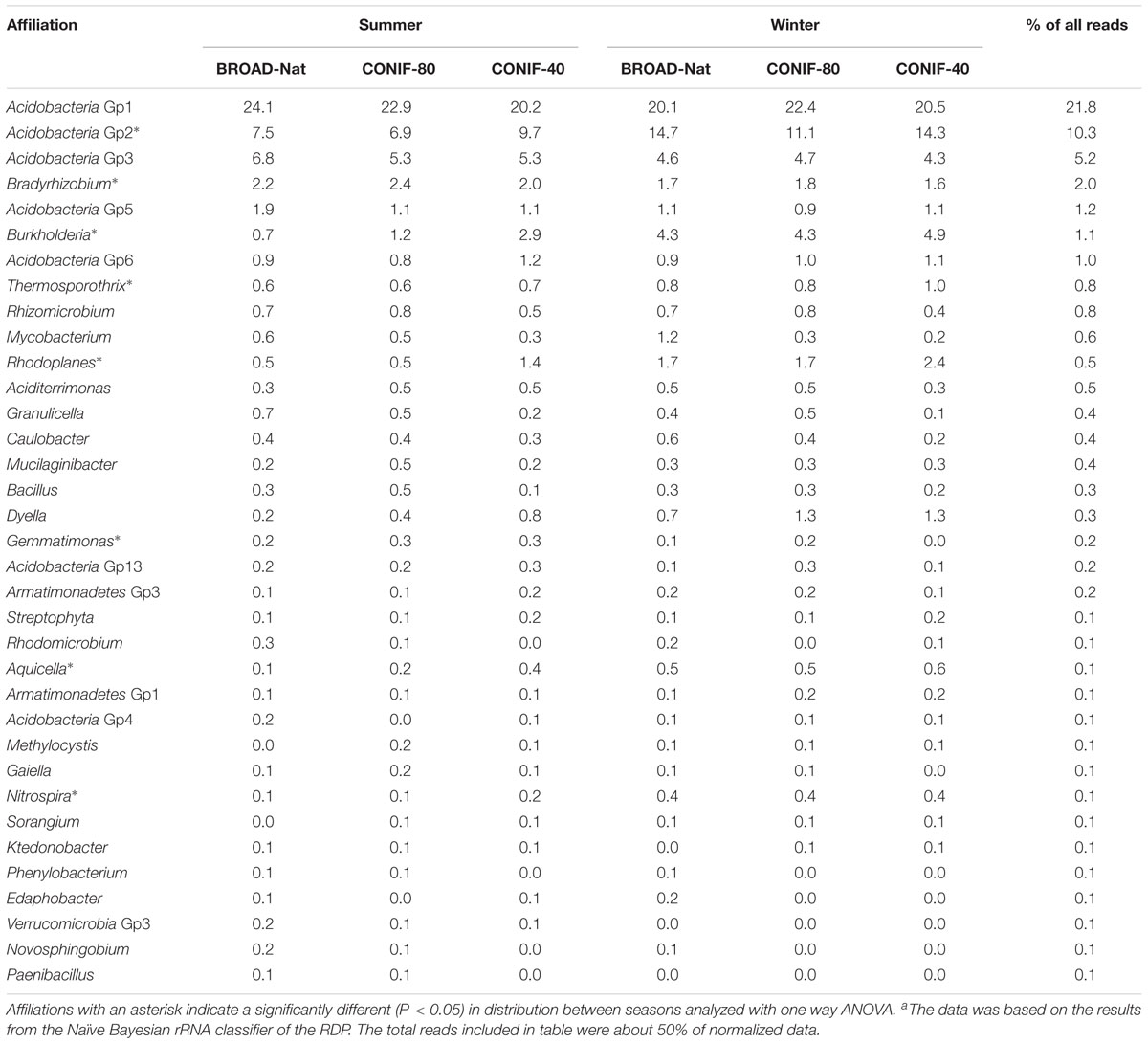
TABLE 1. The relative abundance (%) of some abundant genera and genus-level groups of three forest soil bacterial communitiesa.
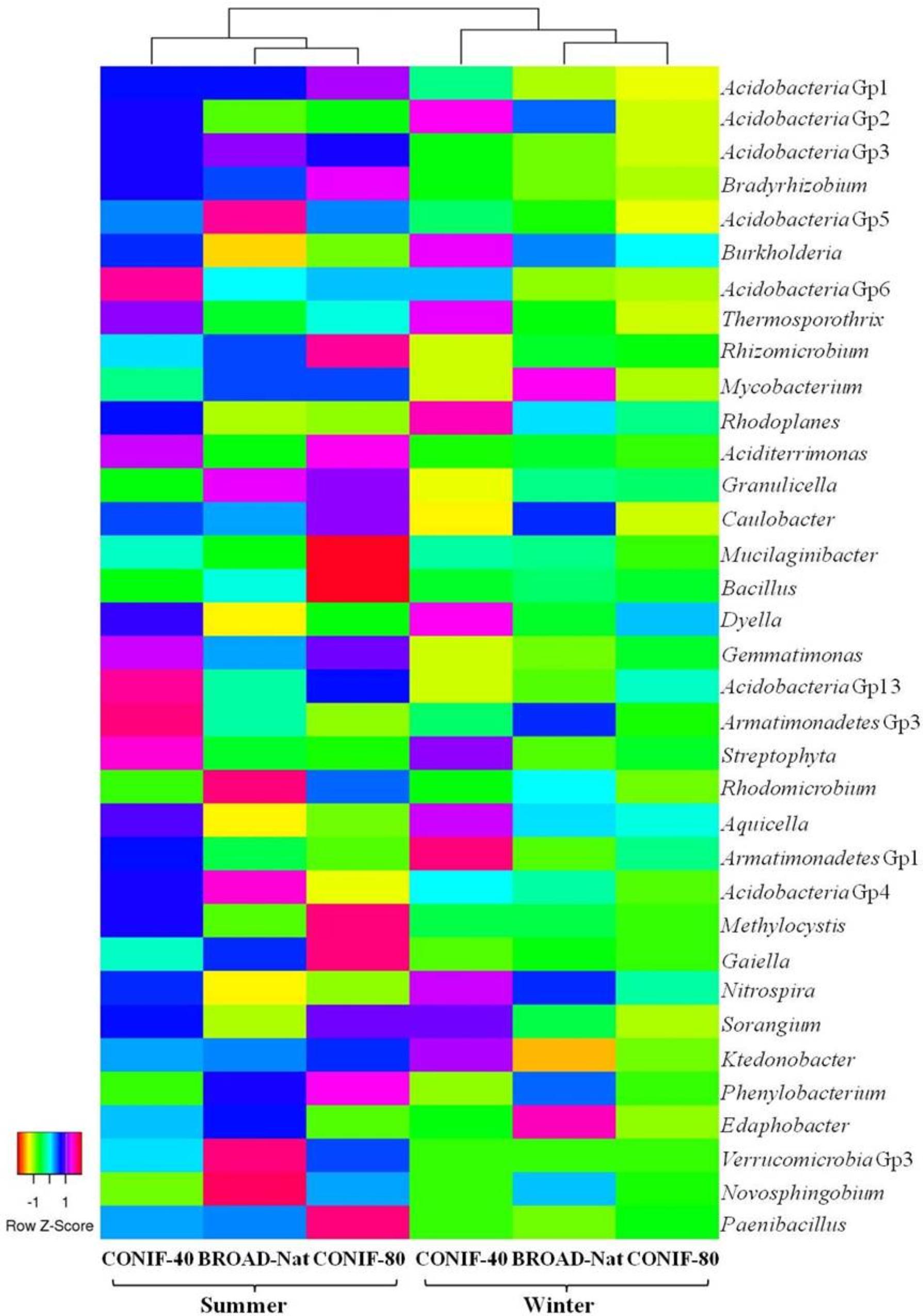
FIGURE 1. Heat map of the 35 most abundant genera or genus-level groups of three forest soil communities. Dendrogram of the Euclidean distances between the bacterial communities is shown at the top.
Similar conclusions were obtained by K-shuff analysis of the soil bacterial communities (Table 2). Within summer or winter, the community structures were significantly different between the three forest soils except for the comparison of CONIF-80 and CONIF-40 in summer. Moreover, the soil bacterial community of each forest type also differed significantly between two seasons. However, in all cases, the differences were relatively small, with Kcf values of less than 0.005.
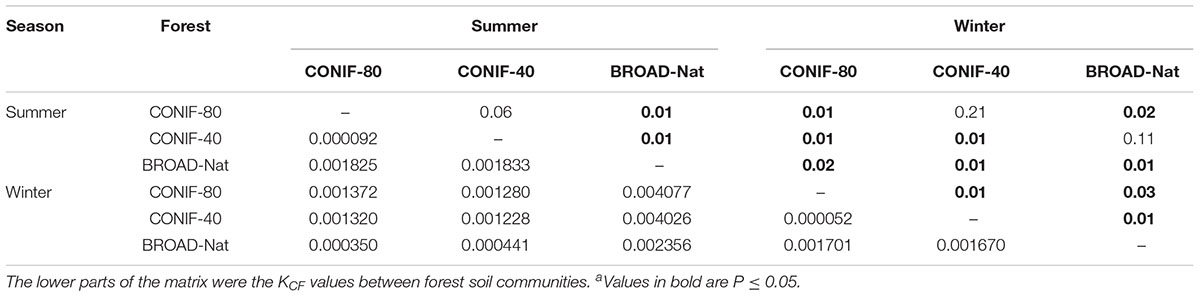
TABLE 2. Statistical significance (P-value) of differences among forest soil communities using K-shuff analysisa.
Bacterial Community Diversity
Although the major taxa present in all three forest soils were similar, the bacterial alpha diversity among three communities was different. Based on the Shannon diversity indexes calculated from the OTUs formed at an evolutionary distance <0.03 (about 97% sequence similarity), the CONIF-40 forest soil community was more diverse than both the CONIF-80 and BROAD-Nat communities in both the summer and winter (Figure 2). While the diversity of both the CONIF-40 and CONIF-80 forest communities appeared to be higher in the summer than the winter, this difference was not significant (Figure 2). Rarefaction curves supported these conclusions, and the slopes of the CONIF-40 curves were higher than the other communities in each season (Supplementary Figure S3). In addition, the rarefaction curves for the BROAD-Nat communities were nearly identical in both seasons.
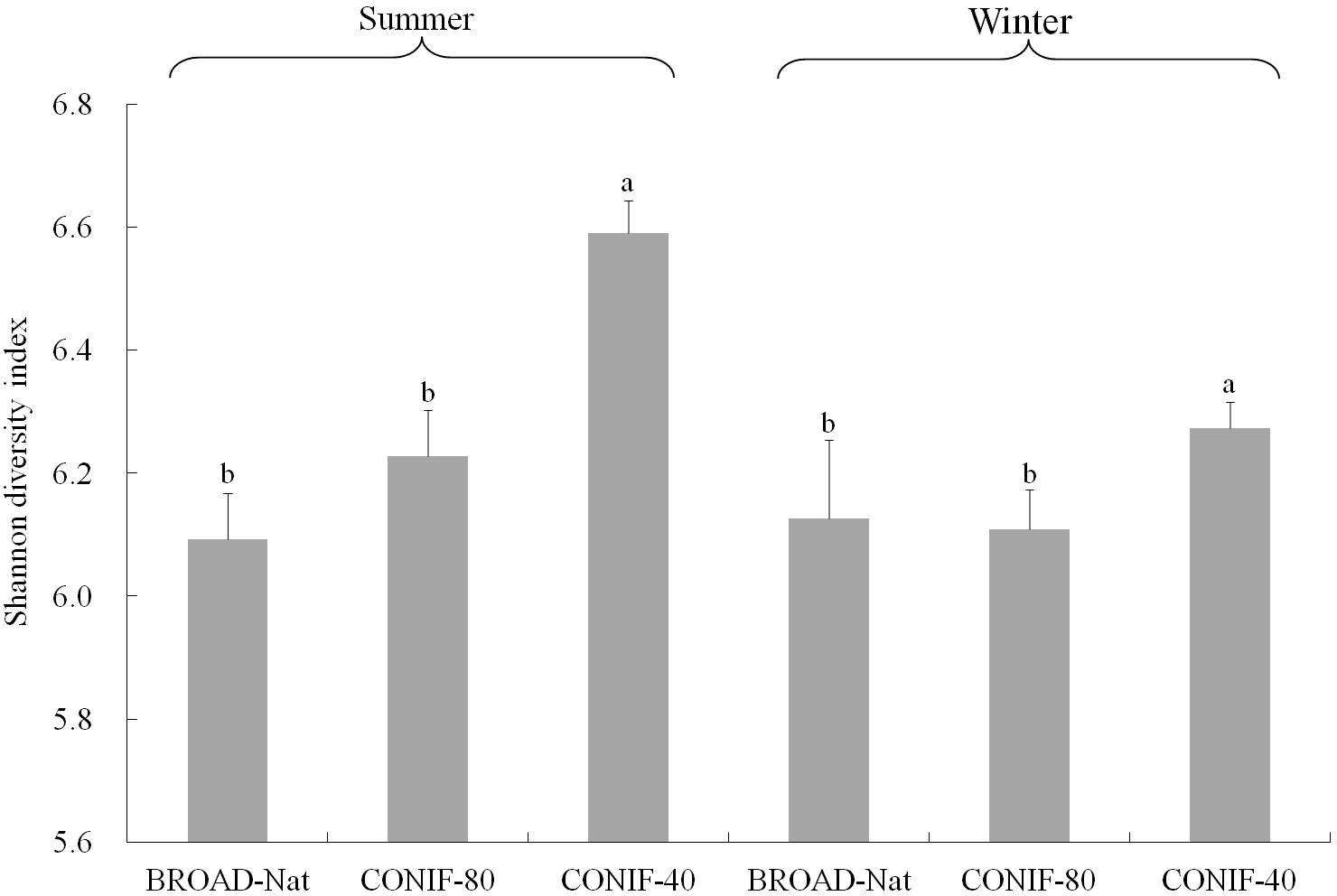
FIGURE 2. Shannon diversity index of three forest soil bacterial communities. OTUs were calculated at the 3% evolutionary distance. Bars of the same season with different letters indicate significant differences at P ≤ 0.05 analyzed with one way ANOVA. The bacterial diversity of the same forest types between two seasons was not significantly different.
Bacterial Community Comparison and Relationship With Soil Properties
While the same genera and genus-level groups were abundant in all the bacterial communities, there were significant variations in the relative abundance of some specific groups (Table 1). To examine the differences in more detail, the distributions of the OTUs with more than 10 reads in the normalized data set were visualized in Venn diagrams (Supplementary Figure S2). These OTUs represented 53–65% of the total number of reads. The minimum number of reads of 10 was chosen because the rarefaction curves indicated that the communities had not been completely sampled (Supplementary Figure S3), and OTUs with fewer reads might be found in only one community by chance even if it was evenly distributed throughout the samples. In all comparisons, either by management or season, nearly 70% of the OTUs were only found in one condition. Moreover, statistical analyses of the K-shuff results indicated that the communities were significantly different in both seasons (Supplementary Table S2). These results strongly supported the conclusion that the communities varied greatly with both season and management.
Based on the RELATE analysis, soil properties, including soil pH and organic carbon, were significantly correlated with the alpha diversity of bacterial communities (Table 3). Redundancy analysis was further performed based on the distribution of shared OTUs and the soil properties. The soil communities between two seasons, especially those in BROAD-Nat forests, formed separate clusters (Figure 3). This result supported the analyses of abundant genus-level groups and OTUs (Table 1), which reached similar conclusions. In addition, the results of Supplementary Table S2 also supported the conclusion that replicate samples mostly clustered together, as in Figure 3. The microbial biomass carbon was also positively associated with the BROAD-Nat and CONIF-80 forest communities in summer. The Org. C and TN showed positive associations with the summer CONIF-40 community, and the pH was positively associated with BROAD-Nat and CONIF-80 forest communities in winter (Figure 3).
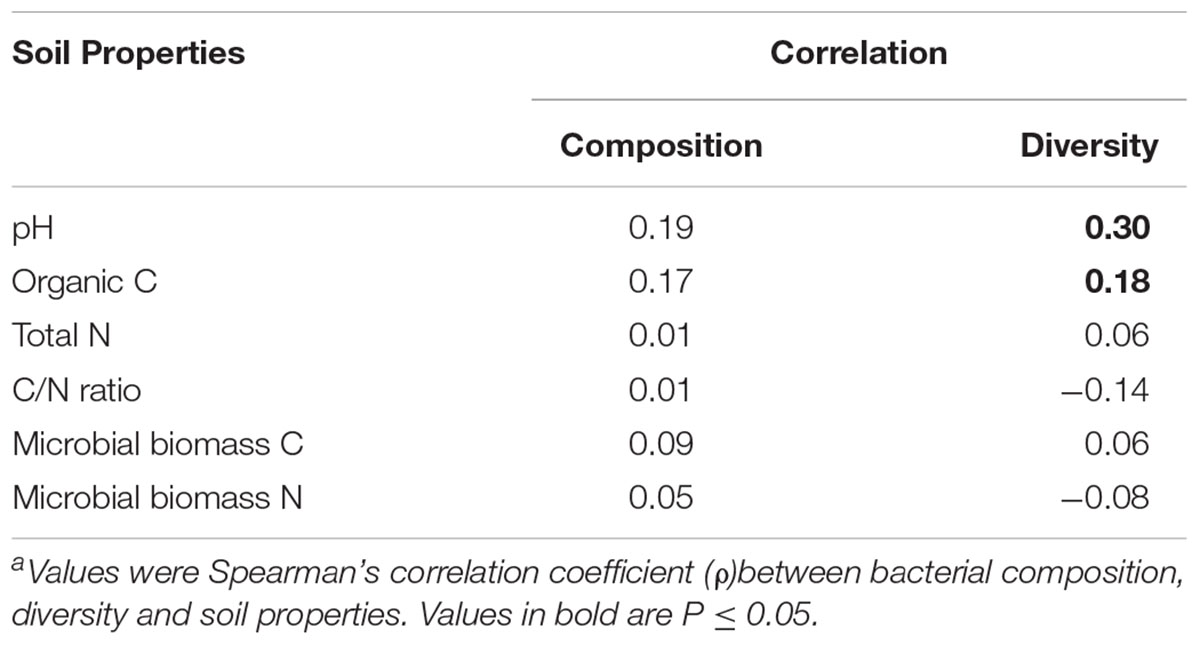
TABLE 3. Correlations of bacterial community composition based on the distribution of OTUs and Shannon diversity index with soil properties using the RELATE in the PRIMER V6 softwarea.
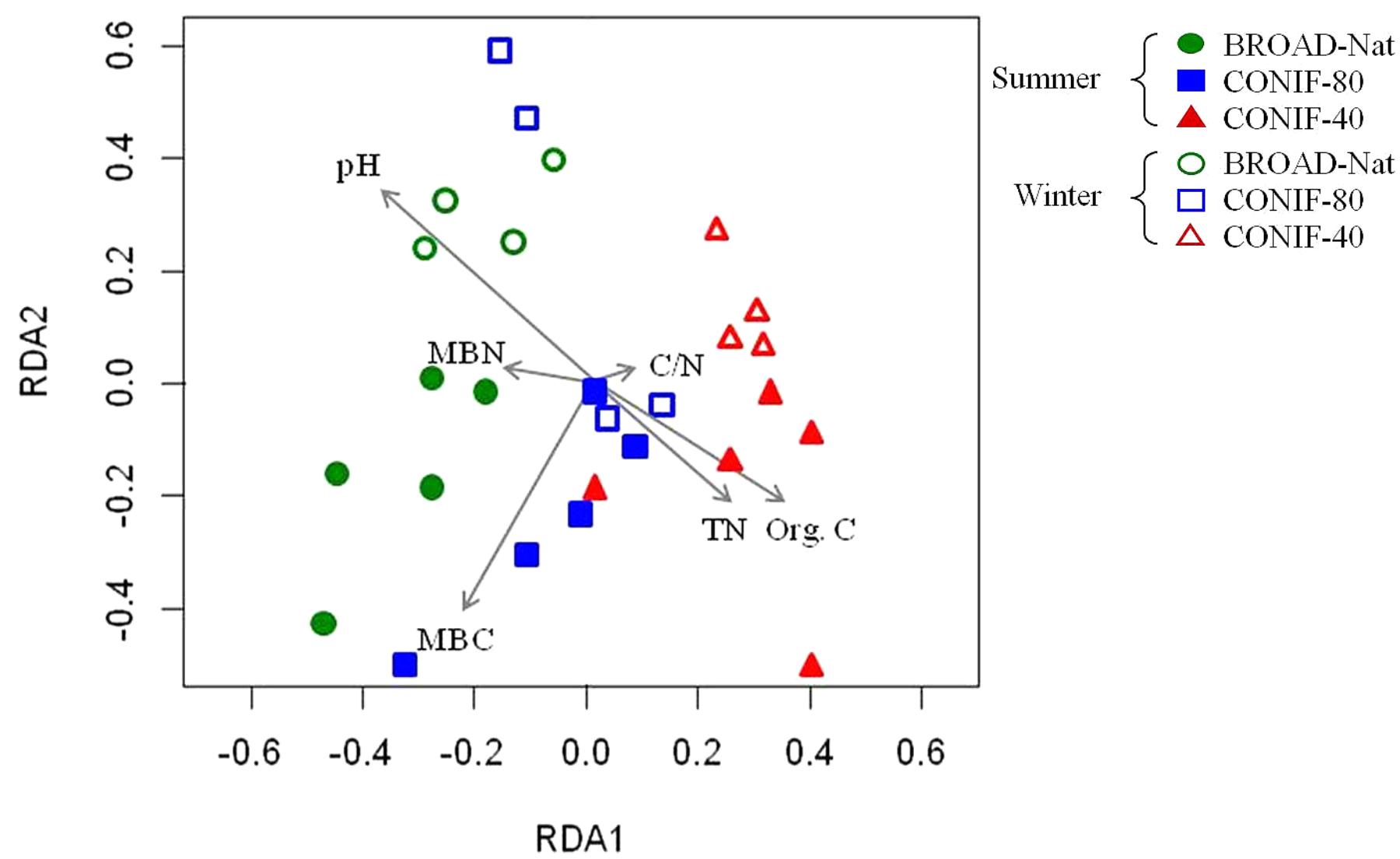
FIGURE 3. Redundancy analysis of bacterial community based on the distribution of OTUs among three forest soils in different seasons. Org. C, organic carbon; TN, total nitrogen; C/N, organic C/TN ratio; MBC, microbial biomass carbon; MBN, microbial biomass nitrogen.
Discussion
In the present study, the three bacterial communities were dominated by two phyla, Acidobacteria and Proteobacteria. With the other three less abundant groups, Actinobacteria, Bacteroidetes, and Firmicutes, they comprised more than 90% of the sequences in each of the communities. Using pyrosequencing, similar results have also been observed in other bacterial communities of forests (Lauber et al., 2009; Nemergut et al., 2010), grasslands (Will et al., 2010), and agricultural systems (Acosta-Martínez et al., 2008). These results indicated that a variety of soils were mainly composed of similar dominant bacterial groups, although the relative abundances of the taxa varied between the study sites.
The bacterial structure in natural broadleaved forest soils differed from those in secondary coniferous plantations. Several studies have shown that land use is one of the important factors to alter the soil bacterial structure (Deng et al., 2016; Wang et al., 2017). Our previous study at higher elevations also showed the bacterial structure changes between a natural broadleaved forest and reforested cedar plantation (Lin et al., 2017). Forest management in tree species changes could result in differences in litter chemistry and influence soil properties and microbial communities (Ushio et al., 2010). Soil nutrient availability is correlated with the bacterial community (Hofmann et al., 2016).
Our results revealed that the community diversity of coniferous CONIF-40 and perhaps the CONIF-80 plantation soils was higher than that in the original BROAD-Nat forest soils. Previous studies in different forest ecosystem also revealed that the disturbance of forest soils increased the diversity of microbial communities (Lin et al., 2011a, 2017). The disturbance of reforestation could disturb soil water content and nutrient conditions, which in turn could affect bacterial diversity (Jangid et al., 2011). Although the apparent diversity in the coniferous plantation soils was higher in the summer than the winter, this increase was not significant. Temperature and soil moisture are important factors that affect the soil bacterial community. A landscape-scale study in Scotland revealed that the soil bacterial composition was related to variation in precipitation (Nielsen et al., 2010). Differences in soil water content due to the temporal variability affected the soil bacterial alpha diversity (Rasche et al., 2011; Lauber et al., 2013). In this study site, precipitation is mainly between March and September. The temperature in summer could be up to 30°C and only 10°C in winter. Lower temperature and precipitation in winter could alter microbial activity (Sowerby et al., 2005) and shape the communities (Romanowicz et al., 2016), resulting in lower bacterial diversity in winter.
Using clone library method, a previous study at the same site also revealed the community differences between hardwood and coniferous plantations. However, the Proteobacteria comprised more than half of sequences, and the Gammaproteobacteria dominated within the Proteobacteria (Lin et al., 2011a). The results were quite different from the present study. In the present study, the soil samples were recollected and were different from those in the previous one. Besides sampling time, sequencing method could result in this difference. About 150 to 170 sequences per sample were used in the previous study or far less than the present one. The low number of sequences may have missed some parts of community, and 2,000 reads have been suggested as necessary to characterize soil microbial communities (Caporaso et al., 2011). In addition, an abundant Stenotrophomonas-related OTU was present in the previous study (Lin et al., 2011a). Less than five pyrosequences of this genus was retrieved in the present study. Thus, the specific cloning method may also have contributed to the differences.
In this study, Acidobacteria predominated in the communities of the three forests. The abundance is in accordance with findings of other studies of the composition of soil bacterial communities conducted in various environments, such as forest (Lauber et al., 2009), grassland (Will et al., 2010), and agricultural soils (Janssen, 2006). The Acidobacteria might also be metabolically active as well as numerically dominant in soils (Lee et al., 2008). They exhibit slow metabolic rates under low nutrient conditions and are versatile heterotrophs (Ward et al., 2009) and oligotrophs (Nemergut et al., 2010). The low soil pH value of this study site also facilitates the presence of Acidobacteria, and their distribution is negatively correlated with soil pH (Jones et al., 2009). According to the genomic and physiological data, Acidobacteria also well tolerate fluctuations in soil hydration (Ward et al., 2009). Moreover, some studies showed that Acidobacteria involve degradation of plant- and microorganism-based polysaccharides and are linked to the soil N availability (Fierer et al., 2007; Jones et al., 2009). In the present study, the abundant acidobacterial Gp1 and Gp2 were also reported as the two most abundant subdivisions of Acidobacteria in the tundra soil bacterial communities. They responded to environmental conditions differently, suggesting the wide functional diversity of these organisms (Männistö et al., 2013). Hence, based on their abundance, metabolic activity, and the presence in various soils, the Acidobacteria appear to play important roles in this forest ecosystem as well as in other environments.
Proteobacteria were abundant in the three soil communities. Several studies in other soils showed similar trends (Mendes et al., 2015; Montecchia et al., 2015). Members of this phylum possess an enormous range of morphological, physiological, and metabolic diversity and play essential roles in global carbon, nitrogen, and sulfur cycling (Spain et al., 2009). Among Proteobacteria, many alphaproteobacterial pyrosequences were related to the genus Bradyrhizobium of the Rhizobiales. Some species related to this genus are able to fix nitrogen and could contribute to the biogeochemical cycling in this ecosystem (Zhang and Xu, 2008; Yarwood et al., 2009). In addition, the members of Rhizobiales are considered a rhizospheric-plant promoting group (Fierer et al., 2007). Their abundance could indicate the essential roles in plant–microbe interactions and soil function, as well as in nitrogen fixation. Within Betaproteobacteria, the genus Burkholderia was also abundant and relatively higher in the winter communities. This group is a commonly part of rhizosphere soil communities. With functional diversity in nitrogen fixation and plant growth promotion (Coenye and Vandamme, 2003), it could play important role in maintaining stability in the soil community.
In the three communities, Actinobacteria accounted for less than 6% of the sequences. The abundance of Actinobacteria increases in soils with high organic matter and alkaline pH (Zhang et al., 2013; Li et al., 2016). The acidic pH value (pH < 4) of the study soils could result in lower abundance in Actinobacteria, since their relative abundance is positively correlate with soil pH (Lauber et al., 2009).
The phylum Chloroflexi also comprised about 3–4% of the three communities, with a relatively higher abundance in CONIF-40 soils. The species of this phylum are able to hydrolyze polysaccharides such as cellulose, xylan, and chitin (Yabe et al., 2010, 2016), suggesting their roles in degradation of plant residue.
Conclusion
Though similar in composition, the bacterial structure among forest types and seasons were significantly different. The composition of the communities differed, especially between seasons. The bacterial diversity was also higher in the community with a shorter reforestation time and in summer, except that in BROAD-Nat. The differences between two seasons could have resulted from differences in soil moisture and temperature. More environmental factors would need to be examined to completely elucidate the relationship between soil properties and soil bacterial communities and the effects of season on the soil bacterial communities at this site.
Author Contributions
Y-TL performed statistical analyses and wrote the manuscript. WW helped in analyzing and interpreting the data. WW, DC, and C-YC edited the manuscript. All authors read and approved the final manuscript.
Funding
The authors thank the grants awarded by Academia Sinica and the Ministry of Science and Technology of Taiwan (MOST 106-2313-B-001-002) for financially supporting this research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01968/full#supplementary-material
Footnotes
References
Acosta-Martínez, V., Dowd, S., Sun, Y., and Allen, V. (2008). Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40, 2762–2770. doi: 10.1016/j.soilbio.2008.07.022
Acosta-Martínez, V., Dowd, S., Sun, Y., Wester, D., and Allen, V. (2010). Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl. Soil Ecol. 45, 13–25. doi: 10.1016/j.apsoil.2010.01.005
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. doi: 10.1093/nar/gkw419
Bardgett, R. D., Freeman, C., and Ostle, N. J. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814. doi: 10.1038/ismej.2008.58
Burton, J., Chen, C., Xu, Z., and Ghadiri, H. (2010). Soil microbial biomass, activity and community composition in adjacent native and plantation forests of subtropical Australia. J. Soils Sediments 10, 1267–1277. doi: 10.1007/s11368-010-0238-y
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chang, E. H., Chen, T. H., Tian, G. L., Hsu, C. K., and Chiu, C. Y. (2016). Effect of 40 and 80 years of conifer regrowth on soil microbial activities and community structure in subtropical low mountain forests. Forests 7:244. doi: 10.1016/j.apsoil.2015.10.018
Chatterjee, A., Vance, G. F., Pendall, E., and Stahl, P. D. (2008). Timber harvesting alters soil carbon mineralization and microbial community structure in coniferous forests. Soil Biol. Biochem. 40, 1901–1907. doi: 10.1016/j.soilbio.2008.03.018
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Deng, Q., Cheng, X., Hui, D., Zhang, Q., Li, M., and Zhang, Q. (2016). Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in central China. Sci. Total Environ. 541, 230–237. doi: 10.1016/j.scitotenv.2015.09.080
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Grayston, S. J., and Prescott, C. E. (2005). Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 37, 1157–1167. doi: 10.1016/j.soilbio.2004.11.014
Hamady, M., Walker, J. J., Harris, J. K., Gold, N. J., and Knight, R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5, 235–237. doi: 10.1038/nmeth.1184
Hartmann, M., Niklaus, P. A., Zimmermann, S., Schmutz, S., Kremer, J., Abarenkov, K., et al. (2014). Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J. 8, 226–244. doi: 10.1038/ismej.2013.141
Hofmann, K., Lamprecht, A., Pauli, H., and Illmer, P. (2016). Distribution of prokaryotic abundance and microbial nutrient cycling across a high-alpine altitudinal gradient in the Austrian Central Alps is affected by vegetation, temperature, and soil nutrients. Microb. Ecol. 72, 704–716. doi: 10.1007/s00248-016-0803-z
Jangid, K., Kao, M. H., Lahamge, A., Williams, M. A., Rathbun, S. L., and Whitman, W. B. (2016). K-shuff: a novel algorithm for characterizing structural and compositional diversity in gene libraries. PLoS One 11:e0167634. doi: 10.1371/journal.pone.0167634
Jangid, K., Williams, M. A., Franzluebbers, A. J., Schmidt, T. M., Coleman, D. C., and Whitman, W. B. (2011). Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol. Biochem. 43, 2184–2193. doi: 10.1016/j.soilbio.2011.06.022
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006
Jien, S. H., Chen, T. H., Chiu, C. Y., and Nagatsuka, S. (2009). Relationships between soil mass movement and relief in humid subtropical low-elevation mountains. Soil Sci. 174, 563–573. doi: 10.1097/SS.0b013e3181bccf35
Jones, R. T., Robeson, M. S., Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. doi: 10.1038/ismej.2008.127
Kerfahi, D., Tripathi, B. M., Dong, K., Go, R., and Adams, J. M. (2016). Rainforest conversion to rubber plantation may not result in lower soil diversity of bacteria, fungi, and nematodes. Microb. Ecol. 72, 359–371. doi: 10.1007/s00248-016-0790-0
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackbrandt and M. Goodfellow (New York, NY: Wiley), 115–175.
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/aem.00335-09
Lauber, C. L., Ramirez, K. S., Aanderud, Z., Lennon, J., and Fierer, N. (2013). Temporal variability in soil microbial communities across land-use types. ISME J. 7, 1641–1650. doi: 10.1038/ismej.2013.50
Lee, M. F., Lin, T. C., Vadeboncoeur, M. A., and Hwong, J. L. (2008). Remote sensing assessment of forest damage in relation to the 1996 strong typhoon Herb at Lienhuachi Experimental Forest, Taiwan. For. Ecol. Manage. 255, 3297–3306. doi: 10.1016/j.foreco.2008.02.010
Li, X., Sun, M., Zhang, H., Xu, N., and Sun, G. (2016). Use of mulberry–soybean intercropping in salt–alkali soil impacts the diversity of the soil bacterial community. Microb. Biotechnol. 9, 293–304. doi: 10.1111/1751-7915.12342
Lin, Y. T., Hu, H. W., Whitman, W. B., Coleman, D. C., and Chiu, C. Y. (2014). Comparison of soil bacterial communities in a natural hardwood forest and coniferous plantations in perhumid subtropical low mountains. Bot. Stud. 55:50. doi: 10.1186/s40529-014-0050-x
Lin, Y. T., Jangid, K., Whitman, W. B., Coleman, D. C., and Chiu, C. Y. (2011a). Change in bacterial community structure in response to disturbance of natural hardwood and secondary coniferous forest soils in central Taiwan. Microb. Ecol. 61, 429–437. doi: 10.1007/s00248-010-9748-9
Lin, Y. T., Jangid, K., Whitman, W. B., Coleman, D. C., and Chiu, C. Y. (2011b). Soil bacterial communities in native and regenerated perhumid montane forests. Appl. Soil Ecol. 47, 111–118. doi: 10.1016/j.apsoil.2010.11.008
Lin, Y. T., Whitman, W. B., Coleman, D. C., Jien, S. H., and Chiu, C. Y. (2017). Cedar and bamboo plantations alter structure and diversity of the soil bacterial community from a hardwood forest in subtropical mountain. Appl. Soil Ecol. 112, 28–33. doi: 10.1016/j.apsoil.2017.01.001
Lin, Y. T., Whitman, W. B., Coleman, D. C., Shi, S. Y., Tang, S. L., and Chiu, C. Y. (2015). Changes of soil bacterial communities in bamboo plantations at different elevations. FEMS Microbiol. Ecol. 91:fiv033. doi: 10.1093/femsec/fiv033
Männistö, M. K., Kurhela, E., Tiirola, M., and Häggblom, M. M. (2013). Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol. Ecol. 84, 47–59. doi: 10.1111/1574-6941.12035
Mendes, L. W., de Lima Brossi, M. J., Kuramae, E. E., and Tsai, S. M. (2015). Land-use system shapes soil bacterial communities in Southeastern Amazon region. Appl. Soil Ecol. 95, 151–160. doi: 10.1016/j.apsoil.2015.06.005
Montecchia, M. S., Tosi, M., Soria, M. A., Vogrig, J. A., Sydorenko, O., and Correa, O. S. (2015). Pyrosequencing reveals changes in soil bacterial communities after conversion of Yungas forests to agriculture. PLoS One 10:e0119426. doi: 10.1371/journal.pone.0119426
Nemergut, D. R., Cleveland, C. C., Wieder, W. R., Washenberger, C. L., and Townsend, A. R. (2010). Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 42, 2153–2160. doi: 10.1016/j.soilbio.2010.08.011
Nielsen, U. N., Osler, G. H. R., Campbell, C. D., Burslem, D. F. R. P., and van der Wal, R. (2010). The influence of vegetation type, soil properties and precipitation on the composition of soil mite and microbial communities at the landscape scale. J. Biogeogr. 37, 1317–1328. doi: 10.1111/j.1365-2699.2010.02281.x
Oh, Y. M., Kim, M., Lee-Cruz, L., Lai-Hoe, A., Go, R., Ainuddin, N., et al. (2012). Distinctive bacterial communities in the rhizoplane of four tropical tree species. Microb. Ecol. 64, 1018–1027. doi: 10.1007/s00248-012-0082-2
Papadopoulou, E. S., Karpouzas, D. G., and Menkissoglu-Spiroudi, U. (2011). Extraction parameters significantly influence the quantity and the profile of PLFAs extracted from soils. Microb. Ecol. 62, 704–714. doi: 10.1007/s00248-011-9863-2
Quideau, S. A., Chadwick, O. A., Benesi, A., Graham, R. C., and Anderson, M. A. (2001). A direct link between forest vegetation type and soil organic matter composition. Geoderma 104, 41–60. doi: 10.1016/S0016-7061(01)00055-6
Rasche, F., Knapp, D., Kaiser, C., Koranda, M., Kitzler, B., Zechmeister-Boltenstern, S., et al. (2011). Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 5, 389–402. doi: 10.1038/ismej.2010.138
Roesch, L. R., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K. M., Kent, A. D., et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290. doi: 10.1038/ismej.2007.53
Romanowicz, K. J., Freedman, Z. B., Upchurch, R. A., Argiroff, W. A., and Zak, D. R. (2016). Active microorganisms in forest soils differ from the total community yet are shaped by the same environmental factors: the influence of pH and soil moisture. FEMS Microbiol. Ecol. 92:fiw149. doi: 10.1093/femsec/fiw149
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shannon, C. E., and Weaver, W. (1963). The Mathematical Theory of Communication. Urbana: University of Illinois Press.
Sowerby, A., Emmett, B., Beier, C., Tietema, A., Peñuelas, J., Estiarte, M., et al. (2005). Microbial community changes in healthland soil communities along a geographical gradient: interaction with climate change manipulations. Soil Biol. Biochem. 37, 1805–1813. doi: 10.1016/j.soilbio.2005.02.023
Spain, A. M., Krumholz, L. R., and Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi: 10.1038/ismej.2009.43
Stres, B., Danevčič, T., Pal, L., Fuka, M. M., Resman, L., Leskovec, S., et al. (2008). Influence of temperature and soil water content on bacterial, archaeal and denitrifying microbial communities in drained fen grassland soil microcosms. FEMS Microbiol. Ecol. 66, 110–122. doi: 10.1111/j.1574-6941.2008.00555.x
Ushio, M., Kitayama, K., and Balser, T. C. (2010). Tree species-mediated spatial patchiness of the composition of microbial community and physicochemical properties in the topsoils of a tropical montane forest. Soil Biol. Biochem. 42, 1588–1595. doi: 10.1016/j.soilbio.2010.05.035
Ushio, M., Wagai, R., Balser, T. C., and Kitayama, K. (2008). Variations in the soil microbial community composition of a tropical montane forest ecosystem: does tree species matter? Soil Biol. Biochem. 40, 2699–2702. doi: 10.1016/j.soilbio.2008.06.023
Wang, J., Ren, C., Cheng, H., Zou, Y., Bughio, M. A., and Li, Q. (2017). Conversion of rainforest into agroforestry and monoculture plantation in China: consequences for soil phosphorus forms and microbial community. Sci. Total. Environ. 595, 769–778. doi: 10.1016/j.scitotenv.2017.04.012
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Will, C., Thurmer, A., Wollherr, A., Nacke, H., Herold, N., Schrumpf, M., et al. (2010). Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 76, 6751–6759. doi: 10.1128/aem.01063-10
Xu, Z., Ward, S., Chen, C., Blumfield, T., Prasolova, N., and Liu, J. (2008). Soil carbon and nutrient pools, microbial properties and gross nitrogen transformations in adjacent natural forest and hoop pine plantations of subtropical Australia. J. Soils Sediments 8, 99–105. doi: 10.1065/jss2008.02.276
Yabe, S., Aiba, Y., Sakai, Y., Hazaka, M., and Yokota, A. (2010). Thermosporothrix hazakensis gen. nov., sp. nov., isolated from compost, description of Thermosporotrichaceae fam. nov. within the class Ktedonobacteria Cavaletti et al. 2007 and emended description of the class Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 60, 1794–1801. doi: 10.1099/ijs.0.018069-0
Yabe, S., Sakai, Y., and Yokota, A. (2016). Thermosporothrix narukonensis sp. nov., belonging to the class Ktedonobacteria, isolated from fallen leaves on geothermal soil, and emended description of the genus Thermosporothrix. Int. J. Syst. Evol. Microbiol. 66, 2152–2157. doi: 10.1099/ijsem.0.001004
Yarwood, S. A., Myrold, D. D., and Högberg, M. N. (2009). Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol. Ecol. 70, 151–162. doi: 10.1111/j.1574-6941.2009.00733.x
Zhang, L., and Xu, Z. (2008). Assessing bacterial diversity in soil. J. Soils Sediments 8, 379–388. doi: 10.1007/s11368-008-0043-z
Zhang, X., Chen, Q., and Han, X. (2013). Soil bacterial communities respond to mowing and nutrient addition in a steppe ecosystem. PLoS One 8:e84210. doi: 10.1371/journal.pone.0084210
Keywords: 16S rRNA genes, soil bacterial community, long-term reforestation, coniferous, broadleaved
Citation: Lin Y-T, Whitman WB, Coleman DC and Chiu C-Y (2018) Effects of Reforestation on the Structure and Diversity of Bacterial Communities in Subtropical Low Mountain Forest Soils. Front. Microbiol. 9:1968. doi: 10.3389/fmicb.2018.01968
Received: 23 January 2018; Accepted: 02 August 2018;
Published: 21 August 2018.
Edited by:
Gary M. King, Louisiana State University, United StatesReviewed by:
Hinsby Cadillo-Quiroz, Arizona State University, United StatesWei Shi, North Carolina State University, United States
Copyright © 2018 Lin, Whitman, Coleman and Chiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chih-Yu Chiu, Ym9jaGl1QHNpbmljYS5lZHUudHc=
 Yu-Te Lin
Yu-Te Lin William B. Whitman
William B. Whitman David C. Coleman
David C. Coleman Chih-Yu Chiu
Chih-Yu Chiu