
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 14 August 2018
Sec. Virology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01866
This article is part of the Research Topic Innate Immunity Evasion by Picornaviruses View all 9 articles
As a major component of immunological defense against a great variety of pathogens, innate immunity is capable of activating the adaptive immune system. Viruses are a type of pathogen that proliferate parasitically in cells and have multiple strategies to escape from host immune pressure. Here, we review recent studies of the strategies and mechanisms by which enteroviruses evade innate immune monitoring.
Immunological defense mechanisms in vertebrates against various pathogens in the environment comprise innate immunity and adaptive immunity (Medzhitov and Janeway, 1997a,b; Hoebe et al., 2004). Innate immunity is capable of providing a persistent and non-specific physiological response to interrupt pathogenic invasion through a series of activated molecules and immune cells, including macrophages, neutrophils, natural killer (NK) cells, dendritic cells (DCs), and innate lymphoid cells (ILCs) (Medzhitov and Janeway, 1997a; Kleinnijenhuis et al., 2012). These cells are usually located in skin and mucosal tissues and are recruited to sites of pathogen infection by immunological signaling molecules secreted from infected or stimulated epithelia that recognize various pathogen-associated molecular patterns (PAMPs) through pattern-recognition receptors (PRRs) expressed on the cell membrane and cellular organelles (Modlin, 2012; Kawamura et al., 2014; Matejuk, 2018). This process initiates the activation of the immune system, which includes the expression of the first and second levels of immune-activating molecules, such as cytokines or chemokines, and the activation of T and B cells (Medzhitov and Janeway, 1997a; Kawamura et al., 2014). Innate immunity is the primary activator of the adaptive immune response.
Viruses, which are important vertebrate pathogens, show characteristic parasitic proliferation that depends on restrictive host cells and the development of systematic immune evasion mechanisms under host immune pressure over the long course of viral evolution (Finlay and McFadden, 2006; Ressing et al., 2015; Schulz and Mossman, 2016). These mechanisms include various virus-encoded molecules that are capable of targeting different immune receptors and signaling molecules and targeting functional innate and/or adaptive immune molecules to block the immune response; these interactions between viral and host components might lead to clinical and pathological outcomes in the body (Ploegh, 1998; Medzhitov, 2001). As a typical example of immune evasion, human immunodeficiency virus (HIV) uses five mechanisms to escape immune monitoring, including antigen evasion due to high variation in its genome (Borrow et al., 1997), evasion through integration of viral genes into the host genome (Lusic and Siliciano, 2017), inaccurate recognition of cytotoxic T lymphocytes (CTLs) on viral antigenic epitopes due to conformational changes in antigenic structure (Evans et al., 1999; Leviyang and Ganusov, 2015), structural mutability of major histocompatibility complex (MHC)-antigen epitope complexes by antigenic variation (Collins and Baltimore, 1999; Gorin et al., 2017), and lethal damage to CD4+ T cells caused by toxic viral proteins (Piguet and Trono, 2001; Sugden et al., 2017). These mechanisms are based on the tendency of RNA viruses to accumulate mutations in their genomes.
Enterovirus (EV) is a group of the Picornavirus family of non-enveloped, positive-stranded RNA viruses that contains more than 100 members, including poliovirus, coxsackievirus, echovirus, and many common enteric viral pathogens (Jubelt and Lipton, 2014). Humans are a natural host of enteroviruses (Solomon et al., 2010) and are usually infected through the fecal-oral and aerosol-respiratory routes, followed by viral shedding, as observed in throat and/or fecal swab assays several days after infection (Verboon-Maciolek et al., 2003; Han et al., 2010; Li et al., 2013). Clinical symptoms include fever, weakness, lethargy and diarrhea, and severe infections are associated with complications such as lesions and organ failure, including of the central nervous system, heart and liver, all of which may be observed in children (Lake et al., 1976; Wang et al., 1999; Verboon-Maciolek et al., 2008; Ooi et al., 2010). The high morbidity of enterovirus infections is notable, even though it is lower than that of fulminating infections. Severe infection with an enterovirus such as poliovirus was once a nightmare in the history of childhood infectious diseases due to irreversible paralysis, and the results of comprehensive studies of inactivated and attenuated poliovirus vaccines were a highly successful historic achievement (Chumakov et al., 2007; Reynolds, 2007). Although it is reasonable for humankind to be proud of this work, the occurrence of vaccine-associated paralysis poliomyelitis (VAPP) and vaccine-derived poliovirus (VDPV) suggests that immune evasion is an important biological characteristic of enteroviruses (Kew et al., 2004; Kew et al., 2005; Minor, 2009). In addition, epidemiological analyses have provided data showing constant pandemic infection with enteroviruses, such as human foot-hand-mouth disease and viral cardiomyopathy (Blomberg et al., 1974; Wenner, 1982; Ooi et al., 2010). In this case, prophylactic or curative treatments of diseases induced by enteroviruses should be developed based on an understanding of viral immune evasion strategies during infection.
To further understand the immunological escape characteristics of enteroviruses, especially their interactions with innate immunity, a brief review is presented here based on our knowledge and experience in enterovirus studies.
An immunological study indicated that host cells usually identify external pathogens through their PRRs, which recognize PAMPs (Medzhitov and Janeway, 1997a; Saito and Gale, 2007; Stuart et al., 2013). Five PRRs, include toll-like receptors (TLRs) (Athman and Philpott, 2004; Saito and Gale, 2007; Kawai and Akira, 2010), RIG-I-like receptors (RIG-I) (Thompson and Locarnini, 2007; Fredericksen et al., 2008; Wilkins and Gale, 2010), NOD-like receptors (NLRs) (Kanneganti, 2010; Wilkins and Gale, 2010; Kufer and Sansonetti, 2011), AIM2-like receptors (ALRs) (Yang et al., 2015), and the cGAS/STING system (Cai et al., 2014; Ma and Damania, 2016), have been shown to recognize viral PAMPs. TLRs, which include 10 members in humans and 13 in mice, were the first group of PRRs identified and are responsible for sensing invasive pathogenic stimuli signals (Meylan et al., 2006; Sansonetti, 2006). TLRs are evolutionally conserved proteins that are distributed extracellularly or intracellularly in various cells, especially immune cells and epithelial cells, highlighting their importance in the process of host antiviral immunity (Anders and Schlondorff, 2007). TLR-3, -4, -7, -8, -9 are involved in antiviral immunity, and after binding by viral fragments of DNA or RNA and proteins, these TLRs are capable of initiating conformational change in the structure of extracellular and cytoplasmic domains to recruit adaptor molecules in their signaling pathways, which sequentially activate the core component NF-κB and AP-1 motif in the innate immune response (O’Neill et al., 2013; Kawamura et al., 2014). As part of the same mechanism, the other PRRs, including RIG-I and NLRs, are also responsible for sensing intracellular viral pathogenic signals and transferring the stimulus to the NF-κB motif (Jeong and Lee, 2011; Oviedo-Boyso et al., 2014). This comprehensive process has been identified as a key step for innate immunity and cooperation (Meylan et al., 2006; Hansen et al., 2011). This cooperation is modulated systematically and closely with pathogenic stimulation and the release of signaling molecules for the formation of adaptive immunity.
After breaking through the physical barrier of the skin, the first priority of enteroviruses is to avoid recognition by the innate immune system. Evasion is very difficult for viruses because of the wide distribution of TLRs on the surfaces of more than 20 types of immune cells, especially innate immune cells, including monocytes, NK cells, macrophages, ILCs, DCs (Fearon and Locksley, 1996; Geissmann et al., 2010; Galli et al., 2011), and adaptive immune cells, including T and B lymphocytes and even epithelial cells (Medzhitov and Janeway, 1997a; Schleimer et al., 2007; Hammad and Lambrecht, 2008). Five members of the TLR family are involved in the recognition of viral PAMPs (Stuart et al., 2013; Yang et al., 2015; Ma and Damania, 2016). To escape recognition by TLRs and find shelter for replication, enteroviruses use various means, which are described below (Figure 1).
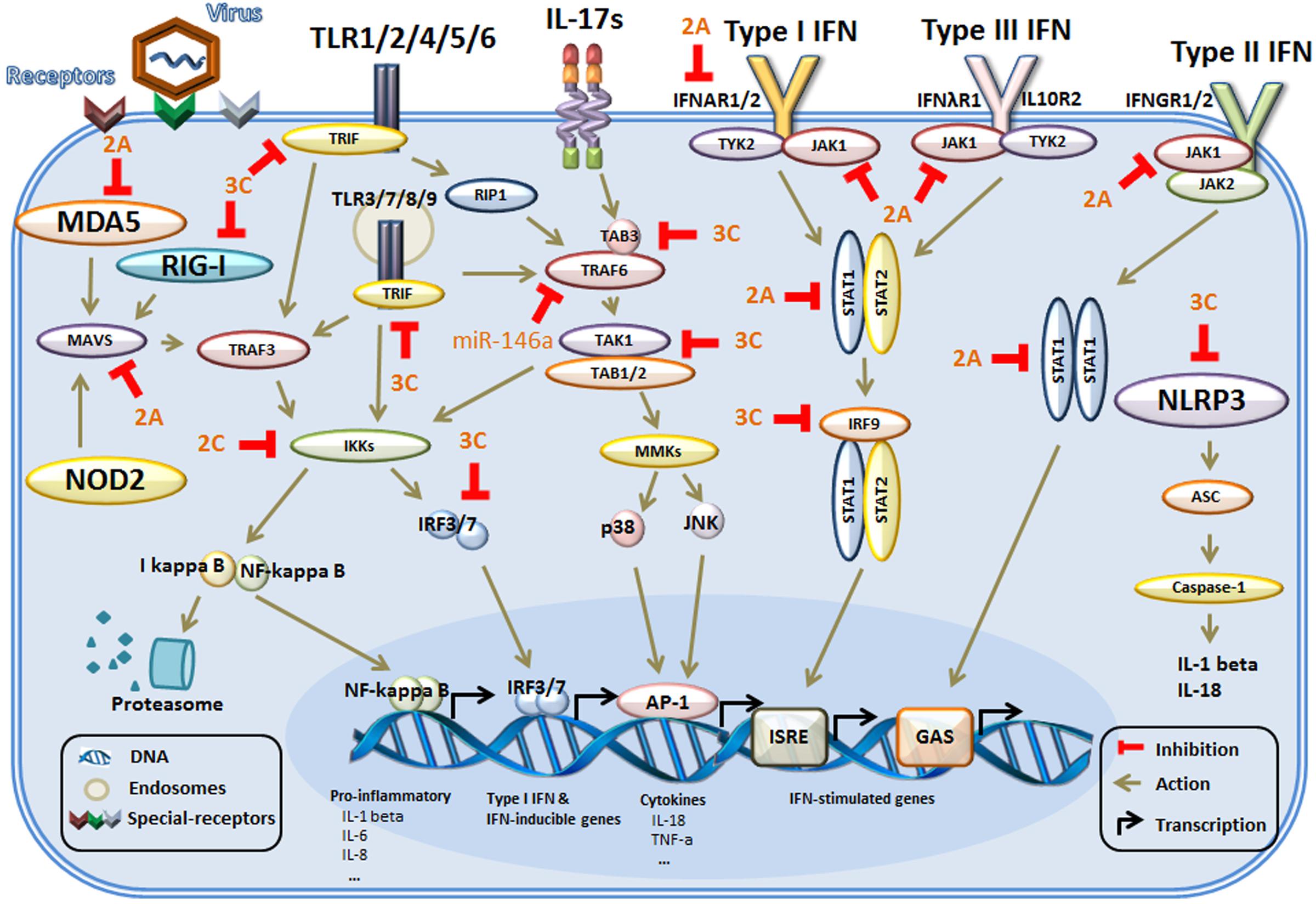
FIGURE 1. The main mechanism of innate immunity evasion by enteroviruses in host cells. Host cells identify external pathogens through their PRRs, including TLRs, RIG-I, and NLRs. The PRRs initiate conformational changes and recruit adaptor molecules in their signaling pathways. Then, the downstream signals are propagated through the activation of core components, such as TRAF6 and the IKK complex, culminating in the activation of transcription factors, which regulate the production of genes associated with the innate immune response. In addition, during binding of IFNs to the IFN receptors, multiple signaling pathways are activated. Then, activation of the kinases and downstream formation of various STAT complexes mediates IFN-dependent gene transcription of IFN-stimulated genes. The adapter proteins and key kinases that mediate these signaling pathways are targeted by viral proteins (such as 2A, 2C, and 3C) or microRNAs (miR-146a). TLRs, toll-like receptors; RIG-I, RIG-I-like receptors; NLRs, NOD-like receptors; NOD2, nucleotide binding oligomerization domain 2; MDA5, melanoma differentiation gene 5; MAVS, mitochondrial antiviral signaling protein; NLRP3, NOD-like receptor 3; TIRAP, toll-interleukin 1 receptor (TIR) domain-containing adaptor protein; TRIF, TIRAP inducing IFN-β; IB, inhibitor of NF-κB; IKKs, IB kinases; STAT, signal transducer and activator of transcription factor; TRAF6, tumor-necrosis-factor- receptor-associated factor 6; JNK, c-Jun N-terminal kinase; IRFs, interferon-regulatory factors; ASC, apoptosis-associated speck-like protein containing a CARD (caspase recruitment domain); ISRE, IFN-stimulated response element); GAS, IFN-γ activation sequence.
Usually, various physiological receptors are expressed in cells for transduction of specific signals and other functions (Aplin et al., 1998; Medzhitov, 2001; Lefkowitz and Shenoy, 2005). Viruses strategically use these receptor molecules that are expressed physiologically in the cell membrane to mediate their entry (Doranz et al., 1996; Agnello et al., 1999; Parker et al., 2001). Many cellular receptors, especially those expressed in immune cells to regulate immune function, are enterovirus-specific receptors (Canton et al., 2013). Previously, CD155 (also called nectin-like molecule, among other names) was found to interact specifically with poliovirus as its receptor (Belnap et al., 2000b; He et al., 2000) and function in the cell-mediated immune response via binding to CD226 (Lozano et al., 2012, 2013). Scavenger receptor class B member 2 (SCARB2) interacts with enterovirus type 71 (EV71), which mediates its entry into epithelial cells and some immune cells expressing this protein (Yamayoshi et al., 2009). The human T cell immunoglobulin and mucin domain 1 (Tim1) protein were identified as specific receptors for hepatitis A virus (HAV) (McIntire et al., 2003; Kim et al., 2011). Accumulative studies suggest that many proteins that are expressed on the surfaces of epithelial and immune cells that play different cellular biological roles, including adhesion, migration, and immune regulation, are also capable of binding to members of the enterovirus family (Rossmann et al., 2002). P-selectin glycoprotein ligand-1 (PSGL-1) is capable of enhancing EV71 and Coxsackievirus A type 16 (CA16) entry into cells after interaction with SCARB2 and PSGL-1 (Nishimura et al., 2010; Nishimura and Shimizu, 2012). Neuron-specific intercellular adhesion molecule 5 (ICAM-5/telencephalin) assists enterovirus 68 (EV68) binding and entry into cells (Wei et al., 2016). Additionally, SA-a-2,3-Gal and SA-linked O-glycan of sialylated glycans can act as receptors to co-mediate infection by Coxsackievirus A type 24 (CA24) and EV71 (Yang et al., 2009; Mistry et al., 2011).
Due to the distributions of these molecules on the surfaces of various cells, the binding of enteroviruses to these molecules not only mediates viral entry into cells but also accelerates viral evasion of recognition by the innate immune system.
As a basic immune defense function, immune cells are capable of initiating internalization processes to clear pathogens by recognizing pathogen PAMPs and damage-associated molecular patterns (DAMPs) to aggregate phagosomes (Mellman et al., 1986; Michallet et al., 2013). However, this immunological strategy is also utilized by viruses for entry into cells; for example, poliovirus and HAV change the conformation of the 160S capsid, leading to the formation of a cell-entry intermediate (Bergmann et al., 1997; Belnap et al., 2000a) known as the 135S or A-particle that subtly changes the pH environment in the cellular phagosome (Curry et al., 1996; Huang et al., 2000). This process helps the virus release its genome into the cytosol for replication. CA9 employs a similar strategy by using the cellular protein GRP78, a member of the heat shock protein 70 (HSP70) family, to assist MHC I in mediating virus internalization (Triantafilou et al., 2002). Echo 1 is capable of utilizing cellular multivesicular bodies (MVBs) as carriers for internalization into cells (Karjalainen et al., 2011). All of these data suggest that this is a strategy used by members of the enterovirus family to evade recognition by the innate immune system.
Although most members of the enterovirus family cause cytopathic infection in various cells, they still require new tactics to elude innate immune monitoring in vitro after avoiding extracellular monitoring (Figure 1).
Immunological studies have shown that when a TLR successfully recognizes a pathogenic molecule, it initiates a conformational change signal and gradually activates each molecule in the signaling pathway for signal transfer (West et al., 2006). Furthermore, the signals transferred from this pathway are able to activate the core component of innate immunity, namely, the NF-κB transcriptional complex, which enables the expression of various immune functional factors, including IFN-α, -β, -γ and other cytokines or chemokines (Muzio et al., 2000; West et al., 2006). These factors and the immune cells they activate have systematic antiviral effects in vitro and enable the restriction of viral proliferation (West et al., 2006). Enteroviruses encode specific proteins that interact with key molecules in this signaling pathway and interfere with signal transduction for the immune response. The non-structural EV71 component protein 3C is a typical example; it is capable of hydrolyzing specific peptide bonds in viral or cellular proteins, not only at the main site, Cys/Ser (or Gly), but also at other cleavage sites in a few cases (Laitinen et al., 2016), potentially leading to the inactivation of several proteins in the TLR pathway, including TRIF, TAK1, and IRF7 (Cui et al., 2011; Lei et al., 2011, 2013, 2014). Obviously, the result of this process is inhibition of NF-κB and AP-1 transcriptional systems modulated by MMKs or IRF3/7, leading to inhibited expression of sets of genes, such as those of inflammatory factors, cytokines, and the interferon (IFN) family, which is related to innate immunity (Baeuerle and Baichwal, 1997; Zeytun et al., 2007). Another EV71 protein, 2C, is also involved in viral interference with the TLR pathway by blocking IKKα/β, which impacts the NF-κB and IRF3/7 transcriptional systems (Zheng et al., 2011; Li et al., 2016). Additionally, EV71 enables RNA interference with the TLR pathway, in which an miRNA is capable of inhibiting TRAF6 and reducing the efficiency of the innate response in vitro (Ho et al., 2014; Hu et al., 2016). HAV, poliovirus, and CB3 are all capable of targeting similar proteins in the TLR pathway using similar mechanisms (Mukherjee et al., 2011; Qu et al., 2011).
As the major adaptor in the RIG-I pathway of mammalian cells, RIG-I is a key component of virus recognition in this pathway and interacts with viral RNA containing the 5′-ppp terminal structure (Takeuchi and Akira, 2008; Schmidt et al., 2009; Wang et al., 2010). This feature allows RIG-I to effectively recognize members of the Orthomyxoviridae, Paramyxoviridae, and Flaviviridae families (Yoneyama et al., 2004; Kato et al., 2006; Yoneyama and Fujita, 2009). Another key adaptor, MDA5, is thought to recognize long viral double-stranded RNA molecules (Takeuchi and Akira, 2008), although its mechanism is unclear. MDA5 sensitively detects members of the Picornaviridae and Coronaviridae families (Yoneyama and Fujita, 2009). To evade immune monitoring by the RIG-I pathway, enteroviruses encode a small protein, Vpg, that binds to the 5’-ppp end of its RNA genome and allows the RNA to escape molecular recognition by RIG-I (Yoneyama and Fujita, 2009). In addition, viruses induce the formation of stress granule (SG)-like aggregates, termed antiviral SG (avSG), in the cytoplasm to escape recognition by MDA5 (Yoneyama et al., 2015). Furthermore, enteroviruses might utilize various encoded proteins to block or interrupt the RIG-I pathway, such as the 3C protein, which is encoded by poliovirus and EV71 and is capable of cleaving RIG-I (Barral et al., 2009; Feng et al., 2014). The cleavage of MDA5 is enabled via EV71-encoded non-structural protein 2A or the proteasome and caspases induced by poliovirus (Barral et al., 2007; Feng et al., 2014). During EV71, coxsackieviruses and polio infection, host MAVS/IPS-1 is cleaved by viral proteins (Mukherjee et al., 2011; Feng et al., 2014; Kotla and Gustin, 2015; Lind et al., 2016).
The NLR family can be divided into the NOD, NLRP, and IPAF subfamilies based on the type of central NACHT domain, which is responsible for activation and oligomerization (Schroder and Tschopp, 2010).
NOD2 recognizes peptidoglycan motifs from bacterial cells and intracellular muramyl dipeptides (Girardin et al., 2003). NOD2 signaling is disrupted when its downstream signaling partially overlaps with that of the RIG-I pathway (Ramos and Gale, 2011). The immune gene transcription system mediated by NF-κb and IRF3/7 is perturbed, leading to changes in the innate immune response after the EV71-encoded 2A protein inhibits MAVS/IPS-1, the 2C protein inhibits IKKs, and the 3C protein inhibits IRF3/7 (Lei et al., 2013; Feng et al., 2014; Li et al., 2016).
The NLRP3 inflammasome, which is a critical component of innate immunity that senses various pathogens and DAMPs (Schroder and Tschopp, 2010), is capable of controlling the maturation and secretion of the proinflammatory cytokines IL-1β and IL-18 (Schmidt and Lenz, 2012). NLRP3 induces an effective immune response to defend the host from EV71 and polio infection, whereas the NLRP3 inflammasome is regulated via viral proteins produced by enteroviruses (Ito et al., 2012; Wang et al., 2015). An in vitro cellular study indicated that inflammasome activation is suppressed by the cleavage of NLRP3 by EV71-encoded 2A and 3C proteases (Wang et al., 2015; Yu et al., 2017). Additionally, this study demonstrated that the EV71 3C protease interacts with NLRP3 and suppresses the secretion of IL-1β in mammals, which affects the innate immune response (Wang et al., 2015). Non-structural protein 2B of poliovirus and EV71 can induce redistribution of NLRP3, thus affecting activation of innate immunity (Ito et al., 2012).
As a proinflammatory cytokine, IL-17 is produced by innate immune T helper cells (Th 17 cells) and is capable of inducing downstream transcription and expression of immune genes based on activation of the NF-κb and MAPK pathways (Hata et al., 2002; Korn et al., 2009). Thus, EV71 infection regulates core processing of the IL-17 pathway because it overlaps with TLRs and the RIG-I pathway, as described above.
Major histocompatibility complex, a cell surface protein, is essential for recognizing and triggering the T cell immune response by binding to peptides cleaved from pathogens (Hughes and Nei, 1988). The MHC family is divided into three subgroups (MHC class I, II, and III) based on structure, function and distribution. Class I MHC (MHC I) occurs on all nucleated cells, and class II occurs on only professional antigen-presenting cells (APCs) and is directly related to virus infection and antigen presentation (Hughes and Nei, 1988; Inaba et al., 1993). By presenting virus-specific peptide segments on the cell surface, MHC initiates the activation of CTLs to kill infected cells, prevent virus replication and release, and thus assist in the elimination of virus from the host (Schwartz, 1985).
Enteroviruses have different strategies for suppressing the presentation of peptides by MHC at different stages for evasion from CTL recognition as follows.
A typical example is Coxsackievirus B3, which contains at least three different viral proteins, including viral protein 3A, which interrupts the transportation of complexes to the Golgi apparatus (Wessels et al., 2005), as well as 2B and 2BC, which activate autophagy to rapidly remove proteins from the cell surface to inhibit the presentation of MHC I on the cell surface for host immune evasion (Cornell et al., 2007). This virus not only interferes with the presentation of MHC I on the cell surface but also induces the production of an efficient inhibitor of IFNγ-induced MHC II expression-IK, leading to the inhibition of MHC II presentation (Park et al., 2013). Similarly, poliovirus utilizes the 3A protein to suppress MHC I-dependent antigen presentation (Deitz et al., 2000). Additionally, MHC II deficiency is the most common diagnosis (54%) in enterovirus-positive patients (Driss et al., 2012). Although the reason and mechanism by which the EV71 virus renders MHC II ineffective during infection is not yet clear, all evidence suggests that enteroviruses are capable of evading the innate immune response in specific ways.
Interferons are a group of active proteins that function as broad-spectrum antivirals (Tomoda et al., 2001; Liu et al., 2012). Instead of directly killing or inhibiting viruses, IFNs activate receptors on the cell surface to produce antiviral proteins to eradicate viruses (Fensterl and Sen, 2009). In addition, IFNs are capable of activating immune cells, including NK cells, macrophages and T lymphocytes, and increasing host defenses by regulating the immune system (Djeu et al., 1979; Murray, 1988; Le Page et al., 2000). IFNs can be divided into three classes, type I IFN, type II IFN, and type III IFN, based on amino acid structure, antigenicity, and secretory cells (Chelbi-Alix and Wietzerbin, 2007).
In response to virus-specific antigen molecules (DNA or RNA), innate immune cell receptors are able to activate the transcription factor IRF3/7 and induce type I IFN through signaling cascades (Sato et al., 2000). Enteroviruses can impede core components of these pathways with non-structural proteins (e.g., 2A, 2C, 3C) or microRNAs (e.g., miR-146a) to prevent type I IFN production (Lei et al., 2010; Ho et al., 2014).
Type II (γ) IFN is released by APCs (Frucht et al., 2001). There is no obvious evidence that enterovirus directly inhibits IFN-γ production.
The antiviral function of the IFN pathway is realized by binding to specific cell surface receptors. Receptors that can recognize IFNs are divided into three types: type I IFN receptors consist of IFNAR1 and IFNAR2 (de Weerd et al., 2007), type II IFN receptors consist of IFNGR1 and IFNGR2 (Schroder et al., 2004), and type III IFN receptors consist of IFNλR1 and IL10R2 (Zhou et al., 2011). Signaling cascades are activated downstream to regulate the innate immune response after IFN receptors interact with specific IFN signaling molecules. Enteroviruses have evolved mechanisms to escape this complex but effective innate immune response at different stages by interacting with different core molecules. EV71, for example, is capable of regulating signaling pathways at the initial cell surface signaling stage by cleaving eIF4G to prevent IFNAR1 production of the type I IFN receptor via viral protein 2A to reduce the expression of the type I IFN receptor and disrupt the type I IFN pathway (Lu et al., 2012). Moreover, viral protein 2A blocks phosphorylation of JAK1, TYK2, STAT1, and STAT2, members of the JAK-STAT pathway, resulting in attenuation of immune signaling (Liu et al., 2014). Furthermore, the cleavage of the IRF9 protein by viral protein 3C further inhibits JAK-STAT signaling and eventually affects the transcription of IFN-induced genes to resist activation of the innate immune response (Hung et al., 2011).
Innate lymphoid cells are a recently discovered subset of lymphocytes that lack antigen-specific B or T cell receptors because of the absence of the recombination activating gene (Rag) (Moretta and Locatelli, 2016). ILCs are widely found in tissues and organs, particularly in mucosal tissues and lymphoid organs (Spits et al., 2013). ILCs are usually activated within hours after infection and play an important role in modulating innate and adaptive immune responses (Gasteiger and Rudensky, 2014). Based on their function in immunity against pathogens, ILCs respond to the initial invasion of pathogens and mediate innate immune signaling to activate adaptive immunity (Spits et al., 2013). Currently, much concern about ILCs focuses on understanding the role of ILCs in assisting the induction of specific immune responses.
Our research investigating the role of ILCs in enterovirus infection, including CA16, EV71, and CA10, has provided interesting data. These viruses show similar pathogenic characteristics of infection of the respiratory or gut epithelium followed by typical viremia and viral proliferation in various organs, leading to clinical pathogenic processes characterized by blisters on the hands, feet and mouth associated with other clinical manifestations, such as fever and flu-like symptoms (Liu et al., 2011; Wang et al., 2017). However, the immunity induced during infection with these viruses presents obvious differences in efficacy, especially their clinical protective efficacy against viral attack (Li et al., 2011; Wang et al., 2017). Our recent unpublished work has suggested that CA16-infected epithelial cells play a role in initiating the transduction of antigenic stimulus signals from local tissue to ILCs and activation of these ILCs, which were found to be co-localized with viral antigen in tissues. Similarly, EV71 infection is co-localized with ILC2 in respiratory mucosal tissue, in contrast to the co-localization of CA16 infection with ILC1 and 3. Although our data do not clearly explain the relationships between viral infection and these differences in localization and the induced immune response, the elucidation of the significance of the interactions between enteroviruses and ILCs might provide novel insights.
In summary, the data obtained to date suggest that interactions between enteroviruses and the immune system are complicated and that enteroviruses employ various ingenious tactics by targeting different molecules in different pathways. To understand the systematic process of virus infection, more work is still needed.
YZ and QL wrote this review. JL modified the manuscript. YZ wrote the first draft.
This work was supported by CAMS Initiative for Innovative Medicine (CAMS-I2M-1-019), the State Project for Essential Drug Research and Development (2017ZX09307013-2 and 2016ZX09101120-2), the National Natural Science Foundation of China (31700931), Medical reserve talents of Yunnan Province Health and Family Planning (H-201620), and Major science and technology special projects of Yunnan Province (2017ZF020, 2016ZF002, 2014HB066, and 2017FB018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Agnello, V., Abel, G., Elfahal, M., Knight, G. B., and Zhang, Q. X. (1999). Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 12766–12771. doi: 10.1073/pnas.96.22.12766
Anders, H. J., and Schlondorff, D. (2007). Toll-like receptors: emerging concepts in kidney disease. Curr. Opin. Nephrol. Hypertens. 16, 177–183. doi: 10.1097/MNH.0b013e32803fb767
Aplin, A. E., Howe, A., Alahari, S. K., and Juliano, R. L. (1998). Signal transduction and signal modulation by cell adhesion receptors: the role of integrins, cadherins, immunoglobulin-cell adhesion molecules, and selectins. Pharmacol. Rev. 50, 197–263.
Athman, R., and Philpott, D. (2004). Innate immunity via Toll-like receptors and Nod proteins. Curr. Opin. Microbiol. 7, 25–32. doi: 10.1016/j.mib.2003.12.013
Baeuerle, P. A., and Baichwal, V. R. (1997). NF-kappa B as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv. Immunol. 65, 111–137. doi: 10.1016/S0065-2776(08)60742-7
Barral, P. M., Morrison, J. M., Drahos, J., Gupta, P., Sarkar, D., Fisher, P. B., et al. (2007). MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81, 3677–3684. doi: 10.1128/JVI.01360-06
Barral, P. M., Sarkar, D., Fisher, P. B., and Racaniello, V. R. (2009). RIG-I is cleaved during picornavirus infection. Virology 391, 171–176. doi: 10.1016/j.virol.2009.06.045
Belnap, D. M., Filman, D. J., Trus, B. L., Cheng, N., Booy, F. P., Conway, J. F., et al. (2000a). Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74, 1342–1354.
Belnap, D. M., McDermott, B. M. Jr., Filman, D. J., Cheng, N., Trus, B. L., Zuccola, H. J., et al. (2000b). Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. U.S.A. 97, 73–78.
Bergmann, E. M., Mosimann, S. C., Chernaia, M. M., Malcolm, B. A., and James, M. N. (1997). The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71, 2436–2448.
Blomberg, J., Lycke, E., Ahlfors, K., Johnsson, T., Wolontis, S., and von Zeipel, G. (1974). Letter: new enterovirus type associated with epidemic of aseptic meningitis and-or hand, foot, and mouth disease. Lancet 2:112. doi: 10.1016/S0140-6736(74)91684-5
Borrow, P., Lewicki, H., Wei, X., Horwitz, M. S., Peffer, N., Meyers, H., et al. (1997). Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3, 205–211. doi: 10.1038/nm0297-205
Cai, X., Chiu, Y. H., and Chen, Z. J. (2014). The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 54, 289–296. doi: 10.1016/j.molcel.2014.03.040
Canton, J., Neculai, D., and Grinstein, S. (2013). Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634. doi: 10.1038/nri3515
Chelbi-Alix, M. K., and Wietzerbin, J. (2007). Interferon, a growing cytokine family: 50 years of interferon research. Biochimie 89, 713–718. doi: 10.1016/j.biochi.2007.05.001
Chumakov, K., Ehrenfeld, E., Wimmer, E., and Agol, V. I. (2007). Vaccination against polio should not be stopped. Nat. Rev. Microbiol. 5, 952–958. doi: 10.1038/nrmicro1769
Collins, K. L., and Baltimore, D. (1999). HIV’s evasion of the cellular immune response. Immunol. Rev. 168, 65–74. doi: 10.1111/j.1600-065X.1999.tb01283.x
Cornell, C. T., Kiosses, W. B., Harkins, S., and Whitton, J. L. (2007). Coxsackievirus B3 proteins directionally complement each other to downregulate surface major histocompatibility complex class I. J. Virol. 81, 6785–6797. doi: 10.1128/JVI.00198-07
Cui, S., Wang, J., Fan, T., Qin, B., Guo, L., Lei, X., et al. (2011). Crystal structure of human enterovirus 71 3C protease. J. Mol. Biol. 408, 449–461. doi: 10.1016/j.jmb.2011.03.007
Curry, S., Chow, M., and Hogle, J. M. (1996). The poliovirus 135S particle is infectious. J. Virol. 70, 7125–7131.
de Weerd, N. A., Samarajiwa, S. A., and Hertzog, P. J. (2007). Type I interferon receptors: biochemistry and biological functions. J. Biol. Chem. 282, 20053–20057. doi: 10.1074/jbc.R700006200
Deitz, S. B., Dodd, D. A., Cooper, S., Parham, P., and Kirkegaard, K. (2000). MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. U.S.A. 97, 13790–13795. doi: 10.1073/pnas.250483097
Djeu, J. Y., Heinbaugh, J. A., Holden, H. T., and Herberman, R. B. (1979). Augmentation of mouse natural killer cell activity by interferon and interferon inducers. J. Immunol. 122, 175–181.
Doranz, B. J., Rucker, J., Yi, Y., Smyth, R. J., Samson, M., Peiper, S. C., et al. (1996). A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85, 1149–1158. doi: 10.1016/S0092-8674(00)81314-8
Driss, N., Ben-Mustapha, I., Mellouli, F., Ben Yahia, A., Touzi, H., Bejaoui, M., et al. (2012). High susceptibility for enterovirus infection and virus excretion features in Tunisian patients with primary immunodeficiencies. Clin. Vaccine Immunol. 19, 1684–1689. doi: 10.1128/CVI.00293-12
Evans, D. T., O’Connor, D. H., Jing, P., Dzuris, J. L., Sidney, J., da Silva, J., et al. (1999). Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5, 1270–1276. doi: 10.1038/15224
Fearon, D. T., and Locksley, R. M. (1996). The instructive role of innate immunity in the acquired immune response. Science 272, 50–53. doi: 10.1126/science.272.5258.50
Feng, Q., Langereis, M. A., Lork, M., Nguyen, M., Hato, S. V., Lanke, K., et al. (2014). Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 88, 3369–3378. doi: 10.1128/JVI.02712-13
Fensterl, V., and Sen, G. C. (2009). Interferons and viral infections. Biofactors 35, 14–20. doi: 10.1002/biof.6
Finlay, B. B., and McFadden, G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. doi: 10.1016/j.cell.2006.01.034
Fredericksen, B. L., Keller, B. C., Fornek, J., Katze, M. G., and Gale, M. Jr. (2008). Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82, 609–616. doi: 10.1128/JVI.01305-07
Frucht, D. M., Fukao, T., Bogdan, C., Schindler, H., O’Shea, J. J., and Koyasu, S. (2001). IFN-gamma production by antigen-presenting cells: mechanisms emerge. Trends Immunol. 22, 556–560. doi: 10.1016/S1471-4906(01)02005-1
Galli, S. J., Borregaard, N., and Wynn, T. A. (2011). Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat. Immunol. 12, 1035–1044. doi: 10.1038/ni.2109
Gasteiger, G., and Rudensky, A. Y. (2014). Interactions between innate and adaptive lymphocytes. Nat. Rev. Immunol. 14, 631–639. doi: 10.1038/nri3726
Geissmann, F., Manz, M. G., Jung, S., Sieweke, M. H., Merad, M., and Ley, K. (2010). Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661. doi: 10.1126/science.1178331
Girardin, S. E., Boneca, I. G., Viala, J., Chamaillard, M., Labigne, A., Thomas, G., et al. (2003). Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872. doi: 10.1074/jbc.C200651200
Gorin, A. M., Du, Y., Liu, F. Y., Zhang, T. H., Ng, H. L., Hofmann, C., et al. (2017). HIV-1 epitopes presented by MHC class I types associated with superior immune containment of viremia have highly constrained fitness landscapes. PLoS Pathog. 13:e1006541. doi: 10.1371/journal.ppat.1006541
Hammad, H., and Lambrecht, B. N. (2008). Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat. Rev. Immunol. 8, 193–204. doi: 10.1038/nri2275
Han, J., Ma, X. J., Wan, J. F., Liu, Y. H., Han, Y. L., Chen, C., et al. (2010). Long persistence of EV71 specific nucleotides in respiratory and feces samples of the patients with Hand-Foot-Mouth Disease after recovery. BMC Infect. Dis. 10:178. doi: 10.1186/1471-2334-10-178
Hansen, J. D., Vojtech, L. N., and Laing, K. J. (2011). Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev. Comp. Immunol. 35, 886–897. doi: 10.1016/j.dci.2011.01.008
Hata, K., Andoh, A., Shimada, M., Fujino, S., Bamba, S., Araki, Y., et al. (2002). IL-17 stimulates inflammatory responses via NF-kappaB and MAP kinase pathways in human colonic myofibroblasts. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G1035–G1044. doi: 10.1152/ajpgi.00494.2001
He, Y., Bowman, V. D., Mueller, S., Bator, C. M., Bella, J., Peng, X., et al. (2000). Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. U.S.A. 97, 79–84. doi: 10.1073/pnas.97.1.79
Ho, B. C., Yu, I. S., Lu, L. F., Rudensky, A., Chen, H. Y., Tsai, C. W., et al. (2014). Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon. Nat. Commun. 5:3344. doi: 10.1038/ncomms4344
Hoebe, K., Janssen, E., and Beutler, B. (2004). The interface between innate and adaptive immunity. Nat. Immunol. 5, 971–974. doi: 10.1038/ni1004-971
Hu, Y., Song, J., Liu, L., Li, J., Tang, B., Wang, J., et al. (2016). Different microRNA alterations contribute to diverse outcomes following EV71 and CA16 infections: Insights from high-throughput sequencing in rhesus monkey peripheral blood mononuclear cells. Int. J. Biochem. Cell Biol 81(Pt A), 20–31. doi: 10.1016/j.biocel.2016.10.011
Huang, Y., Hogle, J. M., and Chow, M. (2000). Is the 135S poliovirus particle an intermediate during cell entry? J. Virol. 74, 8757–8761. doi: 10.1128/JVI.74.18.8757-8761.2000
Hughes, A. L., and Nei, M. (1988). Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170. doi: 10.1038/335167a0
Hung, H. C., Wang, H. C., Shih, S. R., Teng, I. F., Tseng, C. P., and Hsu, J. T. (2011). Synergistic inhibition of enterovirus 71 replication by interferon and rupintrivir. J. Infect. Dis. 203, 1784–1790. doi: 10.1093/infdis/jir174
Inaba, K., Inaba, M., Deguchi, M., Hagi, K., Yasumizu, R., Ikehara, S., et al. (1993). Granulocytes, macrophages, and dendritic cells arise from a common major histocompatibility complex class II-negative progenitor in mouse bone marrow. Proc. Natl. Acad. Sci. U.S.A. 90, 3038–3042. doi: 10.1073/pnas.90.7.3038
Ito, M., Yanagi, Y., and Ichinohe, T. (2012). Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 8:e1002857. doi: 10.1371/journal.ppat.1002857
Jeong, E., and Lee, J. Y. (2011). Intrinsic and extrinsic regulation of innate immune receptors. Yonsei Med. J. 52, 379–392. doi: 10.3349/ymj.2011.52.3.429
Jubelt, B., and Lipton, H. L. (2014). Enterovirus/picornavirus infections. Handb. Clin. Neurol. 123, 379–416. doi: 10.1016/B978-0-444-53488-0.00018-3
Kanneganti, T. D. (2010). Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10, 688–698. doi: 10.1038/nri2851
Karjalainen, M., Rintanen, N., Lehkonen, M., Kallio, K., Maki, A., Hellstrom, K., et al. (2011). Echovirus 1 infection depends on biogenesis of novel multivesicular bodies. Cell Microbiol. 13, 1975–1995. doi: 10.1111/j.1462-5822.2011.01685.x
Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. doi: 10.1038/nature04734
Kawai, T., and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863
Kawamura, T., Ogawa, Y., Aoki, R., and Shimada, S. (2014). Innate and intrinsic antiviral immunity in skin. J. Dermatol. Sci. 75, 159–166. doi: 10.1016/j.jdermsci.2014.05.004
Kew, O. M., Sutter, R. W., de Gourville, E. M., Dowdle, W. R., and Pallansch, M. A. (2005). Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59, 587–635. doi: 10.1146/annurev.micro.58.030603.123625
Kew, O. M., Wright, P. F., Agol, V. I., Delpeyroux, F., Shimizu, H., Nathanson, N., et al. (2004). Circulating vaccine-derived polioviruses: current state of knowledge. Bull. World Health Organ. 82, 16–23.
Kim, H. Y., Eyheramonho, M. B., Pichavant, M., Gonzalez Cambaceres, C., Matangkasombut, P., Cervio, G., et al. (2011). A polymorphism in TIM1 is associated with susceptibility to severe hepatitis A virus infection in humans. J. Clin. Invest. 121, 1111–1118. doi: 10.1172/JCI44182
Kleinnijenhuis, J., Quintin, J., Preijers, F., Joosten, L. A., Ifrim, D. C., Saeed, S., et al. (2012). Bacille calmette-guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 17537–17542. doi: 10.1073/pnas.1202870109
Korn, T., Bettelli, E., Oukka, M., and Kuchroo, V. K. (2009). IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517. doi: 10.1146/annurev.immunol.021908.132710
Kotla, S., and Gustin, K. E. (2015). Proteolysis of MDA5 and IPS-1 is not required for inhibition of the type I IFN response by poliovirus. Virol. J. 12:158. doi: 10.1186/s12985-015-0393-2
Kufer, T. A., and Sansonetti, P. J. (2011). NLR functions beyond pathogen recognition. Nat. Immunol. 12, 121–128. doi: 10.1038/ni.1985
Laitinen, O. H., Svedin, E., Kapell, S., Nurminen, A., Hytonen, V. P., and Flodstrom-Tullberg, M. (2016). Enteroviral proteases: structure, host interactions and pathogenicity. Rev. Med. Virol. 26, 251–267. doi: 10.1002/rmv.1883
Lake, A. M., Lauer, B. A., Clark, J. C., Wesenberg, R. L., and McIntosh, K. (1976). Enterovirus infections in neonates. J. Pediatr. 89, 787–791. doi: 10.1016/S0022-3476(76)80808-6
Le Page, C., Genin, P., Baines, M. G., and Hiscott, J. (2000). Interferon activation and innate immunity. Rev. Immunogenet. 2, 374–386.
Lefkowitz, R. J., and Shenoy, S. K. (2005). Transduction of receptor signals by beta-arrestins. Science 308, 512–517. doi: 10.1126/science.1109237
Lei, X., Han, N., Xiao, X., Jin, Q., He, B., and Wang, J. (2014). Enterovirus 71 3C inhibits cytokine expression through cleavage of the TAK1/TAB1/TAB2/TAB3 complex. J. Virol. 88, 9830–9841. doi: 10.1128/JVI.01425-14
Lei, X., Liu, X., Ma, Y., Sun, Z., Yang, Y., Jin, Q., et al. (2010). The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J. Virol. 84, 8051–8061. doi: 10.1128/JVI.02491-09
Lei, X., Sun, Z., Liu, X., Jin, Q., He, B., and Wang, J. (2011). Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J. Virol. 85, 8811–8818. doi: 10.1128/JVI.00447-11
Lei, X., Xiao, X., Xue, Q., Jin, Q., He, B., and Wang, J. (2013). Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 87, 1690–1698. doi: 10.1128/JVI.01855-12
Leviyang, S., and Ganusov, V. V. (2015). Broad CTL response in early HIV infection drives multiple concurrent CTL escapes. PLoS Comput. Biol. 11:e1004492. doi: 10.1371/journal.pcbi.1004492
Li, J., Lin, C., Qu, M., Li, X., Gao, Z., Zhang, X., et al. (2013). Excretion of enterovirus 71 in persons infected with hand, foot and mouth disease. Virol. J. 10:31. doi: 10.1186/1743-422X-10-31
Li, L., Liu, S., Dong, C., Zhao, H., Wang, J., Zhang, Y., et al. (2011). Immunoprotection of inactivated EV71 vaccine against enterovirus in neonatal rhesus monkey. Sci. Chin. Life Sci. 41, 439–448. doi: 10.1360/052011-114
Li, Q., Zheng, Z., Liu, Y., Zhang, Z., Liu, Q., Meng, J., et al. (2016). 2C proteins of enteroviruses suppress IKKbeta phosphorylation by recruiting protein phosphatase 1. J. Virol. 90, 5141–5151. doi: 10.1128/JVI.03021-15
Lind, K., Svedin, E., Domsgen, E., Kapell, S., Laitinen, O., Moll, M., et al. (2016). Coxsackievirus counters the host innate immune response by blocking type III interferon expression. J. Gen. Virol. 97, 1–12. doi: 10.1099/jgv.0.000443
Liu, L., Zhao, H., Zhang, Y., Wang, J., Che, Y., Dong, C., et al. (2011). Neonatal rhesus monkey is a potential animal model for studying pathogenesis of EV71 infection. Virology 412, 91–100. doi: 10.1016/j.virol.2010.12.058
Liu, M. Q., Zhou, D. J., Wang, X., Zhou, W., Ye, L., Li, J. L., et al. (2012). IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One 7:e35902. doi: 10.1371/journal.pone.0035902
Liu, Y., Zhang, Z., Zhao, X., Yu, R., Zhang, X., Wu, S., et al. (2014). Enterovirus 71 inhibits cellular type I interferon signaling by downregulating JAK1 protein expression. Viral Immunol. 27, 267–276. doi: 10.1089/vim.2013.0127
Lozano, E., Dominguez-Villar, M., Kuchroo, V., and Hafler, D. A. (2012). The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 188, 3869–3875. doi: 10.4049/jimmunol.1103627
Lozano, E., Joller, N., Cao, Y., Kuchroo, V. K., and Hafler, D. A. (2013). The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J. Immunol. 191, 3673–3680. doi: 10.4049/jimmunol.1300945
Lu, J., Yi, L., Zhao, J., Yu, J., Chen, Y., Lin, M. C., et al. (2012). Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J. Virol. 86, 3767–3776. doi: 10.1128/JVI.06687-11
Lusic, M., and Siliciano, R. F. (2017). Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 15, 69–82. doi: 10.1038/nrmicro.2016.162
Ma, Z., and Damania, B. (2016). The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19, 150–158. doi: 10.1016/j.chom.2016.01.010
Matejuk, A. (2018). Skin immunity. Arch. Immunol. Ther. Exp. 66, 45–54. doi: 10.1007/s00005-017-0477-3
McIntire, J. J., Umetsu, S. E., Macaubas, C., Hoyte, E. G., Cinnioglu, C., Cavalli-Sforza, L. L., et al. (2003). Immunology: hepatitis A virus link to atopic disease. Nature 425:576. doi: 10.1038/425576a
Medzhitov, R. (2001). Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145. doi: 10.1038/35100529
Medzhitov, R., and Janeway, C. A. Jr. (1997a). Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol. 9, 4–9. doi: 10.1016/S0952-7915(97)80152-5
Medzhitov, R., and Janeway, C. A. Jr. (1997b). Innate immunity: the virtues of a nonclonal system of recognition. Cell 91, 295–298. doi: 10.1016/S0092-8674(00)80412-2
Mellman, I., Fuchs, R., and Helenius, A. (1986). Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55, 663–700. doi: 10.1146/annurev.bi.55.070186.003311
Meylan, E., Tschopp, J., and Karin, M. (2006). Intracellular pattern recognition receptors in the host response. Nature 442, 39–44. doi: 10.1038/nature04946
Michallet, M. C., Rota, G., Maslowski, K., and Guarda, G. (2013). Innate receptors for adaptive immunity. Curr. Opin. Microbiol. 16, 296–302. doi: 10.1016/j.mib.2013.04.003
Minor, P. (2009). Vaccine-derived poliovirus (VDPV): impact on poliomyelitis eradication. Vaccine 27, 2649–2652. doi: 10.1016/j.vaccine.2009.02.071
Mistry, N., Inoue, H., Jamshidi, F., Storm, R. J., Oberste, M. S., and Arnberg, N. (2011). Coxsackievirus A24 variant uses sialic acid-containing O-linked glycoconjugates as cellular receptors on human ocular cells. J. Virol. 85, 11283–11290. doi: 10.1128/JVI.05597-11
Modlin, R. L. (2012). Innate immunity: ignored for decades, but not forgotten. J. Invest. Dermatol. 132(3 Pt 2), 882–886. doi: 10.1038/jid.2011.373
Moretta, L., and Locatelli, F. (2016). Innate lymphoid cells in normal and disease: an introductory overview. Immunol. Lett. 179:1. doi: 10.1016/j.imlet.2016.07.008
Mukherjee, A., Morosky, S. A., Delorme-Axford, E., Dybdahl-Sissoko, N., Oberste, M. S., Wang, T., et al. (2011). The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 7:e1001311. doi: 10.1371/journal.ppat.1001311
Murray, H. W. (1988). Interferon-gamma, the activated macrophage, and host defense against microbial challenge. Ann. Intern. Med. 108, 595–608. doi: 10.7326/0003-4819-108-4-595
Muzio, M., Polentarutti, N., Bosisio, D., Manoj Kumar, P. P., and Mantovani, A. (2000). Toll-like receptor family and signalling pathway. Biochem. Soc. Trans. 28, 563–566. doi: 10.1042/bst0280563
Nishimura, Y., and Shimizu, H. (2012). Cellular receptors for human enterovirus species a. Front. Microbiol. 3:105. doi: 10.3389/fmicb.2012.00105
Nishimura, Y., Wakita, T., and Shimizu, H. (2010). Tyrosine sulfation of the amino terminus of PSGL-1 is critical for enterovirus 71 infection. PLoS Pathog. 6:e1001174. doi: 10.1371/journal.ppat.1001174
O’Neill, L. A., Golenbock, D., and Bowie, A. G. (2013). The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 13, 453–460. doi: 10.1038/nri3446
Ooi, M. H., Wong, S. C., Lewthwaite, P., Cardosa, M. J., and Solomon, T. (2010). Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 9, 1097–1105. doi: 10.1016/S1474-4422(10)70209-X
Oviedo-Boyso, J., Bravo-Patino, A., and Baizabal-Aguirre, V. M. (2014). Collaborative action of Toll-like and NOD-like receptors as modulators of the inflammatory response to pathogenic bacteria. Mediators Inflamm. 2014:432785. doi: 10.1155/2014/432785
Park, H. L., Kim, Y. J., Na, H. N., Park, M. Y., Kim, J. Y., Yun, C. W., et al. (2013). IK induced by coxsackievirus B3 infection transiently downregulates expression of MHC class II through increasing cAMP. Viral Immunol. 26, 13–24. doi: 10.1089/vim.2012.0054
Parker, J. S., Murphy, W. J., Wang, D., O’Brien, S. J., and Parrish, C. R. (2001). Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75, 3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001
Piguet, V., and Trono, D. (2001). Living in oblivion: HIV immune evasion. Semin. Immunol. 13, 51–57. doi: 10.1006/smim.2000.0295
Ploegh, H. L. (1998). Viral strategies of immune evasion. Science 280, 248–253. doi: 10.1126/science.280.5361.248
Qu, L., Feng, Z., Yamane, D., Liang, Y., Lanford, R. E., Li, K., et al. (2011). Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 7:e1002169. doi: 10.1371/journal.ppat.1002169
Ramos, H. J., and Gale, M. Jr. (2011). RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 1, 167–176. doi: 10.1016/j.coviro.2011.04.004
Ressing, M. E., van Gent, M., Gram, A. M., Hooykaas, M. J., Piersma, S. J., and Wiertz, E. J. (2015). Immune evasion by Epstein-Barr virus. Curr. Top. Microbiol. Immunol 391, 355–381. doi: 10.1007/978-3-319-22834-1_12
Rossmann, M. G., He, Y., and Kuhn, R. J. (2002). Picornavirus-receptor interactions. Trends Microbiol. 10, 324–331. doi: 10.1016/S0966-842X(02)02383-1
Saito, T., and Gale, M. Jr. (2007). Principles of intracellular viral recognition. Curr. Opin. Immunol. 19, 17–23. doi: 10.1016/j.coi.2006.11.003
Sansonetti, P. J. (2006). The innate signaling of dangers and the dangers of innate signaling. Nat. Immunol. 7, 1237–1242. doi: 10.1038/ni1420
Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., et al. (2000). Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13, 539–548. doi: 10.1016/S1074-7613(00)00053-4
Schleimer, R. P., Kato, A., Kern, R., Kuperman, D., and Avila, P. C. (2007). Epithelium: at the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 120, 1279–1284. doi: 10.1016/j.jaci.2007.08.046
Schmidt, A., Schwerd, T., Hamm, W., Hellmuth, J. C., Cui, S., Wenzel, M., et al. (2009). 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072. doi: 10.1073/pnas.0900971106
Schmidt, R. L., and Lenz, L. L. (2012). Distinct licensing of IL-18 and IL-1beta secretion in response to NLRP3 inflammasome activation. PLoS One 7:e45186. doi: 10.1371/journal.pone.0045186
Schroder, K., Hertzog, P. J., Ravasi, T., and Hume, D. A. (2004). Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189. doi: 10.1189/jlb.0603252
Schroder, K., and Tschopp, J. (2010). The inflammasomes. Cell 140, 821–832. doi: 10.1016/j.cell.2010.01.040
Schulz, K. S., and Mossman, K. L. (2016). Viral Evasion Strategies in Type I IFN Signaling - A Summary of Recent Developments. Front. Immunol. 7:498. doi: 10.3389/fimmu.2016.00498
Schwartz, R. H. (1985). T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility complex. Annu. Rev. Immunol. 3, 237–261. doi: 10.1146/annurev.iy.03.040185.001321
Solomon, T., Lewthwaite, P., Perera, D., Cardosa, M. J., McMinn, P., and Ooi, M. H. (2010). Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10, 778–790. doi: 10.1016/S1473-3099(10)70194-8
Spits, H., Artis, D., Colonna, M., Diefenbach, A., Di Santo, J. P., Eberl, G., et al. (2013). Innate lymphoid cells–a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149. doi: 10.1038/nri3365
Stuart, L. M., Paquette, N., and Boyer, L. (2013). Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat. Rev. Immunol. 13, 199–206. doi: 10.1038/nri3398
Sugden, S. M., Pham, T. N., and Cohen, E. A. (2017). HIV-1 Vpu downmodulates ICAM-1 expression, resulting in decreased killing of infected CD4(+) T cells by NK Cells. J. Virol. 91, e2442–e2416. doi: 10.1128/JVI.02442-16
Takeuchi, O., and Akira, S. (2008). MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20, 17–22. doi: 10.1016/j.coi.2008.01.002
Thompson, A. J., and Locarnini, S. A. (2007). Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85, 435–445. doi: 10.1038/sj.icb.7100100
Tomoda, A., Shiraishi, S., Hosoya, M., Hamada, A., and Miike, T. (2001). Combined treatment with interferon-alpha and ribavirin for subacute sclerosing panencephalitis. Pediatr. Neurol. 24, 54–59. doi: 10.1016/S0887-8994(00)00233-2
Triantafilou, K., Fradelizi, D., Wilson, K., and Triantafilou, M. (2002). GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 76, 633–643. doi: 10.1128/JVI.76.2.633-643.2002
Verboon-Maciolek, M. A., Krediet, T. G., Gerards, L. J., de Vries, L. S., Groenendaal, F., and van Loon, A. M. (2008). Severe neonatal parechovirus infection and similarity with enterovirus infection. Pediatr. Infect. Dis. J. 27, 241–245. doi: 10.1097/INF.0b013e31815c1b07
Verboon-Maciolek, M. A., Nijhuis, M., van Loon, A. M., van Maarssenveen, N., van Wieringen, H., Pekelharing-Berghuis, M. A., et al. (2003). Diagnosis of enterovirus infection in the first 2 months of life by real-time polymerase chain reaction. Clin. Infect. Dis. 37, 1–6. doi: 10.1086/375222
Wang, H., Lei, X., Xiao, X., Yang, C., Lu, W., Huang, Z., et al. (2015). Reciprocal Regulation between Enterovirus 71 and the NLRP3 Inflammasome. Cell Rep. 12, 42–48. doi: 10.1016/j.celrep.2015.05.047
Wang, J., Zhang, Y., Zhang, X., Hu, Y., Dong, C., Liu, L., et al. (2017). Pathologic and immunologic characteristics of coxsackievirus A16 infection in rhesus macaques. Virology 500, 198–208. doi: 10.1016/j.virol.2016.10.031
Wang, S. M., Liu, C. C., Tseng, H. W., Wang, J. R., Huang, C. C., Chen, Y. J., et al. (1999). Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin. Infect. Dis. 29, 184–190. doi: 10.1086/520149
Wang, Y., Ludwig, J., Schuberth, C., Goldeck, M., Schlee, M., Li, H., et al. (2010). Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 17, 781–787. doi: 10.1038/nsmb.1863
Wei, W., Guo, H., Chang, J., Yu, Y., Liu, G., Zhang, N., et al. (2016). ICAM-5/Telencephalin Is a Functional Entry Receptor for Enterovirus D68. Cell Host Microbe 20, 631–641. doi: 10.1016/j.chom.2016.09.013
Wessels, E., Duijsings, D., Notebaart, R. A., Melchers, W. J., and van Kuppeveld, F. J. (2005). A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-golgi transport. J. Virol. 79, 5163–5173. doi: 10.1128/JVI.79.8.5163-5173.2005
West, A. P., Koblansky, A. A., and Ghosh, S. (2006). Recognition and signaling by toll-like receptors. Annu. Rev. Cell Dev. Biol. 22, 409–437. doi: 10.1146/annurev.cellbio.21.122303.115827
Wilkins, C., and Gale, M. Jr. (2010). Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22, 41–47. doi: 10.1016/j.coi.2009.12.003
Yamayoshi, S., Yamashita, Y., Li, J., Hanagata, N., Minowa, T., Takemura, T., et al. (2009). Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801. doi: 10.1038/nm.1992
Yang, B., Chuang, H., and Yang, K. D. (2009). Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol. J. 6, 141. doi: 10.1186/1743-422X-6-141
Yang, J., Zhao, Y., and Shao, F. (2015). Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 32, 78–83. doi: 10.1016/j.coi.2015.01.007
Yoneyama, M., and Fujita, T. (2009). RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227, 54–65. doi: 10.1111/j.1600-065X.2008.00727.x
Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., et al. (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737. doi: 10.1038/ni1087
Yoneyama, M., Onomoto, K., Jogi, M., Akaboshi, T., and Fujita, T. (2015). Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32, 48–53. doi: 10.1016/j.coi.2014.12.012
Yu, J., Wu, Y., and Wang, J. (2017). Activation and Role of NACHT, LRR, and PYD Domains-Containing Protein 3 Inflammasome in RNA Viral Infection. Front. Immunol. 8:1420. doi: 10.3389/fimmu.2017.01420
Zeytun, A., van Velkinburgh, J. C., Pardington, P. E., Cary, R. R., and Gupta, G. (2007). Pathogen-specific innate immune response. Adv. Exp. Med. Biol. 598, 342–357. doi: 10.1007/978-0-387-71767-8_24
Zheng, Z., Li, H., Zhang, Z., Meng, J., Mao, D., Bai, B., et al. (2011). Enterovirus 71 2C protein inhibits TNF-alpha-mediated activation of NF-kappaB by suppressing IkappaB kinase beta phosphorylation. J. Immunol. 187, 2202–2212. doi: 10.4049/jimmunol.1100285
Keywords: innate immunity, enterovirus, immune evasion, Picornaviridae, innate lymphoid cells (ILCs)
Citation: Zhang Y, Li J and Li Q (2018) Immune Evasion of Enteroviruses Under Innate Immune Monitoring. Front. Microbiol. 9:1866. doi: 10.3389/fmicb.2018.01866
Received: 29 April 2018; Accepted: 25 July 2018;
Published: 14 August 2018.
Edited by:
Zhiyong Li, Lanzhou Veterinary Research Institute (CAAS), ChinaReviewed by:
Jianwei Wang, Chinese Academy of Medical Sciences, ChinaCopyright © 2018 Zhang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qihan Li, bGlxaWhhbkBpbWJjYW1zLmNvbS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.