
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 17 August 2018
Sec. Virology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01865
This article is part of the Research TopicInnate Immunity Evasion by Picornaviruses View all 9 articles
 Xin Cao1,2,3†
Xin Cao1,2,3† Yu-jia Xue1,2†
Yu-jia Xue1,2† Jiang-long Du1,2
Jiang-long Du1,2 Qiang Xu1,2
Qiang Xu1,2 Xue-cai Yang1,2
Xue-cai Yang1,2 Yan Zeng3
Yan Zeng3 Bo-bo Wang1,2
Bo-bo Wang1,2 Hai-zhen Wang4
Hai-zhen Wang4 Jing Liu5
Jing Liu5 Kui-zheng Cai1,2*
Kui-zheng Cai1,2* Zhong-ren Ma1,2*
Zhong-ren Ma1,2*Hepatitis A virus (HAV) belongs to the family Picornaviridae. It is the pathogen of acute viral hepatitis caused by fecal-oral transmission. RNA viruses are sensed by pathogen-associated pattern recognition receptors (PRRs) such as Toll-like receptor 3 (TLR3), retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated gene 5 (MDA5). PRR activation leads to production of type 1 interferon (IFN-α/β), serving as the first line of defense against viruses. However, HAV has developed various strategies to compromise the innate immune system and promote viral propagation within the host cells. The long coevolution of HAV in hosts has prompted the development of effective immune antagonism strategies that actively fight against host antiviral responses. Proteases encoded by HAV can cleave the mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, VISA, or Cardif), TIR domain- containing adaptor inducing IFN-β (TRIF, also known as TICAM-1) and nuclear factor-κB (NF-κB) essential modulator (NEMO), which are key adaptor proteins in RIG-I-like receptor (RLR), TLR3 and NF-κB signaling, respectively. In this mini-review, we summarize all the recent progress on the interaction between HAV and the host, especially focusing on how HAV abrogates the antiviral effects of the innate immune system.
HAV is a positive-strand RNA virus lacking a lipid envelope. As a member of the Picornaviridae family, it has strong tropism for the human hepatocyte like hepatitis C virus (HCV). It is the sole member of the genus Hepatovirus. Enveloped HAV (eHAV) were formed by hijacking cellular membranes, thereby virion were protected from antibody-mediated neutralization (Feng et al., 2013). The HAV genome is ~7,500 nucleotides in length and contains a 5′-untranslated region (UTR), a single open reading frame (ORF) and a 3′-UTR with a polyadenosine tail. The large single open-reading frame encodes a polyprotein that consists of the structural genes (VP1 to VP4, i.e., P1 segment) and the non-structural genes (2A−2C, i.e., P2 segment and 3A−3D, i.e., P3 segment) (Martin and Lemon, 2006; Debing et al., 2014). The investigators use high-resolution X-ray structures to observe HAV mature virus and empty particle. The structures of the two particles are very similar, both forming an icosahedral protein capsid. However, the HAV mature virus has the small viral protein VP4, whereas the empty particle contains only the uncleaved precursor, VP0 (Wang et al., 2015b). A putative receptor for HAV, the HAV cellular receptor (HAVcr-1), is an integral membrane mucin-like glycoprotein with unknown natural function and was identified in African green monkey kidney cells (AGMK) (Kaplan et al., 1996). The human homolog (HuHAVcr-1), otherwise known as T-cell immunoglobulin and mucin-containing domain protein 1 (Tim-1), was also identified and characterized as a human HAV receptor (Feigelstock et al., 1998). In addition, Tim-3 and HAV-specific immunoglobulin A (IgA)-HAVcr-1 association facilitated virus entry into target cells (Dotzauer et al., 2000; Sui et al., 2006; Tami et al., 2007).
In this review, we summarize recent advances in the understanding of induction and suppression of innate antiviral responses by the hepatitis A virus.
The large ORF encodes a single ~2230 amino acid polyprotein that is cleaved into 10 mature proteins primarily by a single virally encoded proteinase, 3Cpro and unknown cellular protease (Schultheiss et al., 1994; Gosert et al., 1996; Debing et al., 2014). Six non-structural proteins, each involved in replication of the HAV RNA, are derived from the P2 and P3 segments of the polyprotein: 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol. Interestingly, 3A is targeted to mitochondrial membranes (Yang et al., 2007). 3Cpro is a cysteine protease, while 3Dpol is the viral RNA-dependent RNA polymerase and the catalytic core of the replicase complex (McKnight and Lemon, 2018). The 3ABC processing intermediate is unique among picornaviruses in that it is relatively stable and has distinct activities in particle assembly (Probst et al., 1998). Processing at the 3CD site is more efficient than at 3AB and 3BC, and 3CD is proteolytically active like 3ABC (Probst et al., 1998).
Hepatocytes, the primary cell type targeted by HAV for infection, express both retinoic acid activated gene I (RIG-I)-like RNA helicases (RLRs) and Toll-like receptors (TLRs) (Li et al., 2005). However, the nucleotide-binding domain and leucine-rich repeat (NLR) proteins play an extensive role in the innate immune and inflammatory reactions, and have not been well-studied in the liver and hepatitis virus infection (Qu and Lemon, 2010). RIG-I, melanoma differentiation- associated gene 5 (MDA5) and Toll-like receptor 3 (TLR3) are pathogen-associated pattern recognition receptors, which identify the presence of RNA viruses and stimulate signaling pathways that lead to induction of an antiviral state (Meylan and Tschopp, 2006). RIG-I seems to be the major recognition receptor that recognizes the 5′-triphosphate group (5′-ppp) and blunt end of short (low molecular weight) RNAs with high affinity (Hornung et al., 2006; Pichlmair et al., 2006; Jiang et al., 2011; Luo et al., 2011). In contrast, MDA5 appears to be the major recognition receptor that senses the internal duplex structure of long (high-molecular-weight) double-stranded (ds)RNAs with a weaker affinity (Takeuchi and Akira, 2010; Kato et al., 2011). LGP2 (Laboratory of Genetics and Physiology 2), a cytoplasmic DExH helicase that shares the domain structure of RIG-I and MDA-5 with the exception of the caspase-recruitment domain (CARD), has been reported to exert both positive and negative effects on RIG-I and MDA5 regulation in different cell types in response to different viruses (Venkataraman et al., 2007; Satoh et al., 2010; Kato et al., 2011; Childs et al., 2012; Malur et al., 2012). TLR3 contains an extracellular leucine-rich repeat (LRR) motif, a transmembrane (TM) domain and an intracellular Toll and IL-1R (TIR) domain (Bell et al., 2003; Leulier and Lemaitre, 2008). Compared with short dsRNAs, long dsRNAs are more potent inducers of TLR3 signaling (Bouteiller et al., 2005; Okahira et al., 2005; Leonard et al., 2008; Liu et al., 2008; Pirher et al., 2008; Botos et al., 2009). Currently known RNA-sensing pathways are summarized in Figure 1. MDA5 and TLR3 is likely to sense the genomic RNA of HAV. As the HAV genome possesses the covalently linked 5′ VPg, it is not likely to be sensed by RIG-I, which generally recognizes RNAs with free 5′ triphosphate (Qu and Lemon, 2010). The function of LGP2 during HAV infection was not investigated in details. Changes in the intrahepatic transcriptome during acute HAV infection in experimentally infected chimpanzees were observed by microarray assay. With the exception of CXCL10 (IP10) and interferon-stimulated gene 20 kDa protein (ISG20), both of which are dually regulated by IFN-α/β and IFN-γ, low-level ISG induction is restricted to the first weeks of infection and subsides before peak HAV RNA abundance in the liver (Lanford et al., 2011). The early type I IFN response diminishes before peak replication of HAV within the liver and the onset of liver injury, when a type II IFN (IFN-γ) response becomes evident (Lanford et al., 2011; Zhou et al., 2012; Feng et al., 2015).
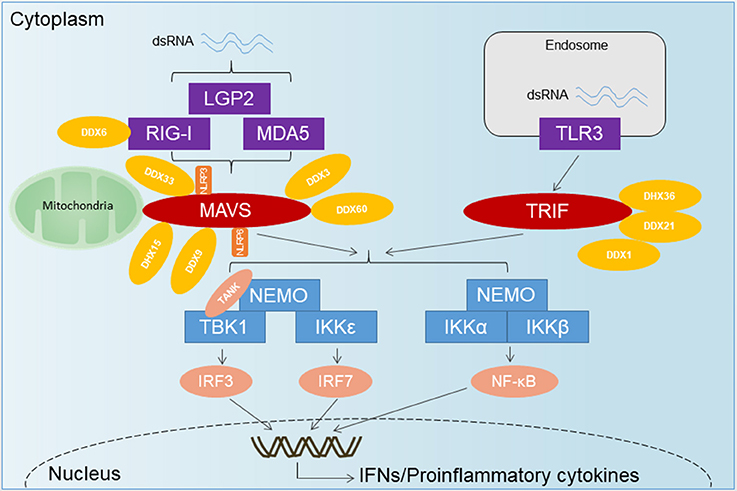
Figure 1. The overview of RNA-sensing pathway. TLR3 senses dsRNA and utilizes the adaptor TRIF to activate IRF3 and NF-κB. Upon recognition of dsRNA, RLRs are recruited by the adaptor MAVS located on the outer membrane of the mitochondria, leading to the activation of several transcription factors including IRF3, IRF7, and NF-κB. DExD/H-Box helicases other than the RLRs and some RNA-binding proteins have emerged as important for innate immune signaling and control of virus infection. DDX1-DDX21-DHX36 forms a complex with TRIF. DDX3 has been shown to associate with RIG-I, MDA5, and MAVS. DDX60 has been shown to bind RIG-I, whereas DHX9 has been shown to interact with MAVS. DHX33 has also been shown to bind MAVS, as well as the NLR NLRP3, to induce inflammasome assembly. DHX15 may serve as a sensor for viral RNA in the cytosol to signal NLRP6-mediated interferon responses in a MAVS-dependent manner, independent of inflammasome formation.
Plasmacytoid dendritic cells (pDCs) are “professional” type I IFN-producer cells that play a central role in host antiviral immunity (Gilliet et al., 2008; Reizis et al., 2011). pDCs mainly sense viruses via endosomal TLR7 and TLR9 (Lund et al., 2003), but they can also sense viral nucleic acids in the cytosol (Feng et al., 2015). pDCs require either close contact with cells infected with HAV or exposure to concentrated culture supernatants for IFN-α production. Enveloped virions (eHAV), and not viral RNA exosomes, are responsible for IFN-α induction. Although membrane envelopment protects HAV against neutralizing antibodies, it also facilitates an early but limited detection of HAV infection by pDCs (Feng et al., 2015). During acute hepatitis A (AHA), non-HAV-specific memory CD8+ T cells are activated by the IL-15 produced by HAV-infected cells. These CD8+ T cells exert innate-like cytotoxicity by activating receptors NKG2D and NKp30 without TCR engagement. Innate-like cytotoxicity of CD8+ T cells is associated with liver injury in AHA (Kim et al., 2018).
HAV was shown to restrain double-stranded RNA (dsRNA)-induced IFNβ gene expression by intervening in RIG-I-mediated IRF3 activation (Brack et al., 2002; Fensterl et al., 2005). Both RIG-I and MDA-5 employ an adaptor protein called MAVS that is localized to the outer mitochondrial membrane via a C-terminal transmembrane domain (Seth et al., 2005). Once activation by RIG-I or MDA-5, MAVS recruits and activates TANK-binding kinase 1 (TBK1) and NF-κB kinase ε (IKKε). TBK1 and IKKε are both accountable for the phosphorylation of IFN regulatory factor 3 (IRF-3), eventually causing IRF-3 dimerization, nuclear translocation, and induction of IFNβ transcription.
HAV proteins 3ABCpro and 2B have been described to interfere with MAVS, thereby disturbing the innate cellular antiviral defense mechanism (Yang et al., 2007; Paulmann et al., 2008). The 3ABC cleavage of MAVS requires both the protease activity of 3Cpro and a transmembrane domain in 3A that targets 3ABC to the mitochondria (Yang et al., 2007). The non-structural HAV 2B protein partially colocalizes with MAVS and interferes with the activities of MAVS and the TBK1/IKKε kinases (Paulmann et al., 2008). However, the exact mechanism of this protein still needs to be investigated in detail.
The TLR3 signaling pathway is mediated exclusively by the TRIF adapter, which is recruited to TLR3 by the interaction between the TIR domains of the two molecules (Oshiumi et al., 2003; Yamamoto et al., 2003). TRIF is proteolytically cleaved by 3CD but not by the mature 3Cpro protease or the 3ABC precursor that degrades MAVS. 3CD-mediated degradation of TRIF depends on both the cysteine protease activity of 3Cpro and the downstream 3Dpol sequence but not 3Dpol polymerase activity (Qu et al., 2011).
NEMO has been shown to link the TLR3-IRF3 pathway (Zhao et al., 2007). 3Cpro-mediated proteolytic cleavage of NEMO is directly involved in inhibition of IFN-β transcription (Wang et al., 2014).
The key innate immune signaling proteins degraded by HAV proteases are illustrated in Figure 2.
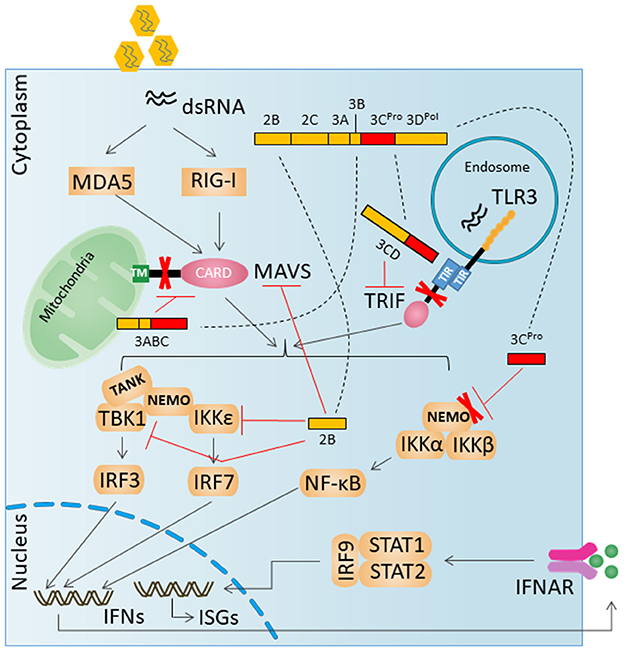
Figure 2. HAV evades innate antiviral responses by degradation key innate immune signaling proteins. A virally encoded, catalytically active polyprotein processing intermediate, 3ABC, degrades MAVS. A second catalytically active 3Cpro processing intermediate, 3CD, cleaves the TLR3 adaptor protein TRIF. 3Cpro can mediate the degradation of NEMO.
Nod-like receptors (NLRs) can play an important role in the host response to infections by RNA viruses by both promoting and suppressing innate immunity and inflammation (Wen et al., 2013; Jha and Pan-Yun Ting, 2015). Of note, activated RIG-I has been shown to associate with apoptosis-associated speck-like protein containing CARD (ASC) to form an NLR-independent RIG-I inflammasome complex that induces caspase-1 activation, leading to IL-1β and IL-18 release (Poeck et al., 2010; Pothlichet et al., 2013). Kupffer cells and monocytes express high levels of NLRs, which play important roles in mediating inflammatory responses and modulating liver injury (Dixon et al., 2013). Exposure to either eHAV or HAV neither initiates nor blocks NLRP3 inflammasome assembly or IL-1β secretion by THP-1 cells, a human monocyte cell line derived from an acute monocytic leukemia (Feng and Lemon, 2018). Nonetheless, NLRX1 positively regulates very early (3 h) RLR-induced cytokine responses to HAV in PH5CH8 cells, which are T antigen-transformed adult human hepatocytes. NLRX1 promotes IL-6 and other early cytokine responses by inhibiting activation of the dsRNA-induced PKR. Suppression of PKR activation allows for early, virus-induced increases in synthesis of the IRF1 protein, which plays a key role in regulating these cytokine responses in hepatocytes (Feng et al., 2017).
The host range of HAV is believed to be restricted to humans and non-human primates. The range of the HAV host species mainly depends on its capacity to evade MAVS- mediated type I IFN responses, which have revealed an unexpected role for MAVS signaling in virus-mediated liver injury (Hirai-Yuki et al., 2016). Type I IFNs, but not type II IFNs, are a major barrier for cross-species infection by HAV. MAVS-dependent, RLR-induced IFN responses play a much more important role in restricting HAV replication than TLR3 in vivo, at least in mice, despite the fact that HAV targets adaptors in both signaling pathways for degradation (Feng and Lemon, 2018). Since the sequences targeted in human MAVS and TRIF are not conserved in small mammals, the failure of HAV to infect these species could derive from inability to disrupt IFN responses (Hirai-Yuki et al., 2016). RLR signaling through MAVS in the mitochondria and mitochondria-associated membranes (MAM) results in the expression of both type I and type III interferons, whereas RLR signaling through MAVS in the peroxisome induces the expression of type III interferon alone (Odendall et al., 2014; Bender et al., 2015; Chow et al., 2018). Although type I IFNs clearly play a pivotal role in restricting HAV infection in mice (Hirai-Yuki et al., 2016), type III IFNs are predominantly detected in cultured human hepatocytes infected with HAV (Feng and Lemon, 2018). Human hepatocytes produce and respond robustly to type III interferon as noted above, but mouse hepatocytes express negligible amounts of the receptor for type III IFN and respond poorly to type III interferon (Hermant et al., 2014; Chow et al., 2018). Studies are needed in Ifnlr1−/− mice lacking functional expression of the type III IFN receptor.
Some DExD/H-Box Helicases may act as bona fide RNA sensors, while others may instead act as accessory factors required to promote innate immune signaling through one of the aforementioned RNA-sensing pathways (Figure 2). DDX3 has been shown to bind poly(I:C), VSV RNA, and abortive RNA products from HIV replication (Gringhuis et al., 2017). DHX9 was proposed as an RNA sensor during viral infection in myeloid dendritic cells (Zhang et al., 2011b). A cytosolic complex formed by DDX1, DDX21, and DHX36 has been reported to sense dsRNA in cDCs in order to induce interferons in response to poly(I:C), IAV, and reovirus (Zhang et al., 2011a). DHX33 may act as a sensor for cytosolic PAMP RNA and activate the NLRP3 inflammasome (Mitoma et al., 2013). DHX15 may diverge in its signaling of innate immune responses in different cell types (Lu et al., 2014; Mosallanejad et al., 2014; Wang et al., 2015a). DDX60 has been shown to be required for RIG-I- mediated signaling in response to dsRNA or virus infection (Miyashita et al., 2011; Oshiumi et al., 2015). Small nuclear ribonucleoprotein U5 subunit 200 protein (SNRNP200), typically involved in spliceosome processes, associates with TBK1 to activate IRF3 during SeV infection (Tremblay et al., 2017). The interaction between HAV and these DExD/H-Box Helicases needs to be investigated in detail in future research.
When activated by binding to dsRNA, oligoadenylate synthetase (OAS) catalyzes the conversion of ATP into 2′-5′-linked oligoadenylates (2–5A), which in turn become second messengers that bind to and activate RNase L. RNase L functions to cleave ssRNA, and the products of this cleavage can cooperate to trigger RIG-I- and MDA5-dependent interferon induction (Malathi et al., 2007, 2010). Protein kinase R (PKR) functions to suppress translation in virus-infected cells by inhibiting eukaryotic translation initiation factor 2A (eIF2A) (Hull and Bevilacqua, 2016). The interferon-induced protein with tetratricopeptide repeats (IFIT) family of proteins can inhibit viral translation and sequester viral RNA (Fensterl and Sen, 2015). LRRFIP1 activates β-catenin to enhance transcriptional activation of Ifnb1 (Yang et al., 2010). A DNA-dependent activator of IRFs (DAI), otherwise known as ZBP1/DLM-1, can activate RIPK1/3/ MLKL (Thapa et al., 2016) and the NLRP3 inflammasome (Kuriakose et al., 2016). High-mobility-group box (HMGB) proteins have been implicated in the signaling of interferon and proinflammatory responses to control virus infection (Ugrinova and Pasheva, 2017). The interplay between some RNA-binding proteins and RLRs pathways is shown in Figure 2. The effect of the aforementioned RNA-binding proteins on HAV hasn't been studied systematically. It would be intriguing to further investigate the function of these RNA-binding proteins during HAV infection.
In HAV-infected cells, viral dsRNA replication intermediates are sensed by cytosolic RLRs (RIG-I and MDA5) as well as endosomal TLR3. However, adaptor proteins MAVS and TRIF and the IκB kinase complex's regulatory subunit, NEMO, are degraded by viral proteinases. This disrupts signals extending from RLRs and TLR3 such that little or no activated IRF3 and NF-κB reach responsive promoters in the nucleus, and therefore, little or no IFNs are produced. Despite many interesting features, HAV remains a largely understudied virus on a fundamental biological level. Future studies on the interactions between HAV and the host immunological system will shed light on the pathogenesis and therapeutic approaches of this virus.
XC, YX, and JD wrote the review. QX, XY, YZ, BW, HW, JL, KC, and ZM participated in the conception and design of the review. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31700763, 81760287), the Science and Technology Fund Program of Gansu Province for Young Investigators (17JR5RA277), Gansu Provincial Science and Technology Grant (1504WKCA094), Innovative Research Team in University (IRT_17R88), Ministry of Science and Technology Assistance Project Grant (KY201501005), the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SKLVEB2016KFKT013), the Open Fund of the Ministry of Education Key Laboratory of Molecular Microbiology and Technology, Nankai University, the Central Universities deriving from the Northwest Minzu University (31920170165), and the Introduction of Talent Research Projects from the Northwest Minzu University (xbmuyjrcs201611).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with several of the authors XC and YZ.
Bell, J. K., Mullen, G. E., Leifer, C. A., Mazzoni, A., Davies, D. R., and Segal, D. M. (2003). Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24, 528–533. doi: 10.1016/S1471-4906(03)00242-4
Bender, S., Reuter, A., Eberle, F., Einhorn, E., Binder, M., and Bartenschlager, R. (2015). Activation of type i and iii interferon response by mitochondrial and peroxisomal mavs and inhibition by hepatitis c virus. Cytokine 76, 85–85. doi: 10.1371/journal.ppat.1005264
Botos, I., Liu, L., Wang, Y., Segal, D. M., and Davies, D. R. (2009). The toll-like receptor 3:dsRNA signaling complex. Biochim. Biophys. Acta 1789, 667–674. doi: 10.1016/j.bbagrm.2009.06.005
Bouteiller, O., Merck, E., Hasan, U. A., Hubac, S., Benguigui, B., Trinchieri, G., et al. (2005). Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J. Biol. Chem. 280, 38133–38145. doi: 10.1074/jbc.M507163200
Brack, K., Berk, I., Magulski, T., Lederer, J., Dotzauer, A., and Vallbracht, A. (2002). HepatitisAvirus inhibits cellular antiviral defense mechanisms induced by double-stranded RNA. J. Virol. 76, 11920–11930. doi: 10.1128/JVI.76.23.11920-11930.2002
Childs, K., Randall, R., and Goodbourn, S. (2012). Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 86, 3411–3421. doi: 10.1128/JVI.06405-11
Chow, K. T., Gale, M. Jr., and Loo, Y. M. (2018). RIG-I and Other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 36:667–694. doi: 10.1146/annurev-immunol-042617-053309
Debing, Y., Neyts, J., and Thibaut, H. J. (2014). Molecular biology and inhibitors of hepatitis a virus. Med. Res. Rev. 34, 895–917. doi: 10.1002/med.21292
Dixon, L. J., Barnes, M., Tang, H., Pritchard, M. T., and Nagy, L. E. (2013). Kupffer cells in the liver. Compr. Physiol. 3, 785–797. doi: 10.1002/cphy.c120026
Dotzauer, A., Gebhardt, U., Bieback, K., Göttke, U., Kracke, A., Mages, J., et al. (2000). Hepatitis A virus-specific immunoglobulin A mediates infection of hepatocytes with hepatitis A virus via the asialoglycoprotein receptor. J. Virol. 74, 10950–10957. doi: 10.1128/JVI.74.23.10950-10957.2000
Feigelstock, D., Thompson, P., Mattoo, P., Zhang, Y., and Kaplan, G. G. (1998).The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 72, 6621–6628.
Feng, H., Lenarcic, E. M., Yamane, D., Wauthier, E., Mo, J., Guo, H., et al. (2017). NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat. Immunol. 18, 1299–1309. doi: 10.1038/ni.3853
Feng, Z., Hensley, L., McKnight, K. L., Hu, F., Madden, V., Ping, L., et al. (2013). A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371. doi: 10.1038/nature12029
Feng, Z., and Lemon, S. M. (2018). Innate immunity to enteric hepatitis viruses. Cold Spring Harb. Perspect. Med. 23:a033464. doi: 10.1101/cshperspect.a033464
Feng, Z., Li, Y., McKnight, K. L., Hensley, L., Lanford, R. E., Walker, C. M., et al. (2015). Human pDCs preferentially sense enveloped hepatitis A virions. J. Clin. Invest. 125, 169–176. doi: 10.1172/JCI77527
Fensterl, V., Grotheer, D., Berk, I., Schlemminger, S., Vallbracht, A., and Dotzauer, A. (2005). Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J. Virol. 79, 10968–10977. doi: 10.1128/JVI.79.17.10968-10977.2005
Fensterl, V., and Sen, G. C. (2015). Interferon-induced ifit proteins: their role in viral pathogenesis. J. Virol. 89, 2462–2468. doi: 10.1128/JVI.02744-14
Gilliet, M., Cao, W., and Liu, Y. J. (2008). Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8, 594–606. doi: 10.1038/nri2358
Gosert, R., Cassinotti, P., Siegl, G., and Weitz, M. (1996). Identification of hepatitis A virus non-structural protein 2B and its release by the major virus protease 3C. J. Gen. Virol. 77, 247–255. doi: 10.1099/0022-1317-77-2-247
Gringhuis, S. I., Hertoghs, N., Kaptein, T. M., Zijlstrawillems, E. M., Sarramiforooshani, R., Sprokholt, J. K., et al. (2017). Hiv-1 blocks the signaling adaptor mavs to evade antiviral host defense after sensing of abortive hiv-1 rna by the host helicase ddx3. Nat. Immunol. 18, 225–235. doi: 10.1038/ni.3647
Hermant, P., Demarez, C., Mahlakõiv, T., Staeheli, P., Meuleman, P., and Michiels, T. (2014). Human but not mouse hepatocytes respond to interferon-lambda in vivo. PLoS ONE 9:e87906. doi: 10.1371/journal.pone.0087906
Hirai-Yuki, A., Hensley, L., McGivern, D. R., González-López, O., Das, A., Feng, H., et al. (2016). MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353, 1541–1545. doi: 10.1126/science.aaf8325
Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., et al. (2006). 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. doi: 10.1126/science.1132505
Hull, C. M., and Bevilacqua, P. C. (2016). Discriminating self and non-self by rna: roles for rna structure, misfolding, and modification in regulating the innate immune sensor pkr. Acc. Chem. Res. 49:1242. doi: 10.1021/acs.accounts.6b00151
Jha, S., and Pan-Yun Ting, J (2015). Holding the inflammatory system in check: NLRs keep it cool. F1000prime Rep. 7:15. doi: 10.12703/P7-15
Jiang, F., Ramanathan, A., Miller, M. T., Tang, G. Q., Gale, M. Jr, Patel, S. S., et al. (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427. doi: 10.1038/nature10537
Kaplan, G., Totsuka, A., Thompson, P., Akatsuka, T., Moritsugu, Y., and Feinstone, S. M. (1996). Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 15, 4282–4296. doi: 10.1002/j.1460-2075.1996.tb00803.x
Kato, H., Takahasi, K., and Fujita, T. (2011). RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243, 91–98. doi: 10.1111/j.1600-065X.2011.01052.x
Kim, J., Chang, D. Y., Lee, H. W., Lee, H., Kim, J. H., Sung, P. S., et al. (2018). Innate-like Cytotoxic Function of Bystander-Activated CD8+ T cells is associated with liver injury in acute hepatitis A. Immunity 48, 161–173.e5. doi: 10.1016/j.immuni.2017.11.025
Kuriakose, T., Man, S. M., Malireddi, R. K., Karki, R., Kesavardhana, S., Place, D. E., et al. (2016). Zbp1/dai is an innate sensor of influenza virus triggering the nlrp3 inflammasome and programmed cell death pathways. Sci. Immunol. 1:aag2045. doi: 10.1126/sciimmunol.aag2045
Lanford, R. E., Feng, Z., Chavez, D., Guerra, B., Brasky, K. M., Zhou, Y., et al. (2011). Acute hepatitis Avirus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 11223–11228. doi: 10.1073/pnas.1101939108
Leonard, J. N., Ghirlando, R., Askins, J., Bell, J. K., Margulies, D. H., Davies, D. R., et al. (2008). The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 258–263. doi: 10.1073/pnas.0710779105
Leulier, F., and Lemaitre, B. (2008). Toll-like receptors — taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178. doi: 10.1038/nrg2303
Li, K., Chen, Z., Kato, N., Gale, M. Jr, and Lemon, S. M. (2005). Distinct poly-I:C and virus-activated interferon signaling pathways in hepatocytes. J. Biol. Chem. 280, 16739–16747. doi: 10.1074/jbc.M414139200
Liu, L., Botos, I., Wang, Y., Leonard, J. N., Shiloach, J., Segal, D. M., et al. (2008). Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320, 379–381. doi: 10.1126/science.1155406
Lu, H., Lu, N., Weng, L., Yuan, B., Liu, Y. J., and Zhang, Z. (2014). Dhx15 senses double-stranded rna in myeloid dendritic cells. J. Immunol. 193, 1364–1372. doi: 10.4049/jimmunol.1303322
Lund, J., Sato, A., Akira, S., Medzhitov, R., and Iwasaki, A. (2003). Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198, 513–520. doi: 10.1084/jem.20030162
Luo, D., Ding, S. C., Vela, A., Kohlway, A., Lindenbach, B. D., and Pyle, A. M. (2011). Structural insights into RNA recognition by RIG-I. Cell 147, 409–422. doi: 10.1016/j.cell.2011.09.023
Malathi, K., Dong, B., Michael Gale, J., and Silverman, R. H. (2007). Small self-rna generated by rnase l amplifies antiviral innate immunity. Nature 448:816. doi: 10.1038/nature06042
Malathi, K., Saito, T., Crochet, N., Barton, D. J., Gale, M.Jr, and Silverman, R. H. (2010). Rnase l releases a small rna from hcv rna that refolds into a potent pamp. RNA Public. RNA Soc. 16, 2108–2119. doi: 10.1261/rna.2244210
Malur, M., Gale, M. Jr, and Krug, R. M. (2012). LGP2 downregulates interferon production during infection with seasonal human influenza A viruses that activate interferon regulatory factor 3. J. Virol. 86, 10733–10738. doi: 10.1128/JVI.00510-12
Martin, A., and Lemon, S. M. (2006). Hepatitis a virus: from discovery to vaccines. Hepatology 43, S164–S172. doi: 10.1002/hep.21052
McKnight, K. L., and Lemon, S. M. (2018). Hepatitis A virus genome organization and replication strategy. Cold Spring Harb. Perspect. Med. 2:a033480. doi: 10.1101/cshperspect.a033480
Meylan, E., and Tschopp, J. (2006). Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell. 22, 561–569. doi: 10.1016/j.molcel.2006.05.012
Mitoma, H., Hanabuchi, S., Kim, T., Bao, M., Zhang, Z., Sugimoto, N., et al. (2013). The dhx33 rna helicase senses cytosolic rna and activates the nlrp3 inflammasome. Immunity 39:123. doi: 10.1016/j.immuni.2013.07.001
Miyashita, M., Oshiumi, H., Matsumoto, M., and Seya, T. (2011). Ddx60, a dexd/h box helicase, is a novel antiviral factor promoting rig-i-like receptor-mediated signaling. Mol. Cell. Biol. 31, 3802–3819. doi: 10.1128/MCB.01368-10
Mosallanejad, K., Sekine, Y., Ishikura-Kinoshita, S., Kumagai, K., Nagano, T., Matsuzawa, A., et al. (2014). The deah-box rna helicase dhx15 activates nf-κb and mapk signaling downstream of mavs during antiviral responses. Sci. Signal. 7:ra40. doi: 10.1126/scisignal.2004841
Odendall, C., Dixit, E., Stavru, F., Bierne, H., Franz, K. M., Durbin, A. F., et al. (2014). Diverse intracellular pathogens activate type iii interferon expression from peroxisomes. Nat. Immunol. 15, 717–726. doi: 10.1038/ni.2915
Okahira, S., Nishikawa, F., Nishikawa, S., Akazawa, T., Seya, T., and Matsumoto, M. (2005). Interferon-beta induction through toll-like receptor 3 depends on double-stranded RNA structure. DNA Cell Biol. 24, 614–623. doi: 10.1089/dna.2005.24.614
Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T., and Seya, T. (2003). TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4, 161–167. doi: 10.1038/ni886
Oshiumi, H., Miyashita, M., Okamoto, M., Okabe, M., Matsumoto, M., et al. (2015). Ddx60 is involved in rig-i-dependent and independent antiviral responses, and its function is attenuated by virus-induced egfr activation. Cell Rep. 11, 1193–1207. doi: 10.1016/j.celrep.2015.04.047
Paulmann, D., Magulski, T., Schwarz, R., Heitmann, L., Flehmig, B., Vallbracht, A., et al. (2008). Hepatitis A virus protein 2B suppresses beta interferon (IFN) gene transcription by interfering with IFN regulatory factor 3 activation. J. Gen. Virol. 89:1593–1604. doi: 10.1099/vir.0.83521-0
Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., et al. (2006). RIG-I- mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314, 997–1001. doi: 10.1126/science.1132998
Pirher, N., Ivicak, K., Pohar, J., Bencina, M., and Jerala, R. (2008). A second binding site for double- stranded RNA in TLR3 and consequences for interferon activation. Nat. Struct. Mol. Biol. 15, 761–763. doi: 10.1038/nsmb.1453
Poeck, H., Bscheider, M., Gross, O., Finger, K., Roth, S., Rebsamen, M., et al. (2010). Recognition of rna virus by rig-i results in activation of card9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11, 63–69. doi: 10.1038/ni.1824
Pothlichet, J., Meunier, I., Davis, B. K., Ting, J. P., Skamene, E., Von, M. V., et al. (2013). Type i ifn triggers rig-i/tlr3/nlrp3-dependent inflammasome activation in influenza a virus infected cells. PLoS Pathog. 9:e1003256. doi: 10.1371/journal.ppat.1003256
Probst, C., Jecht, M., and Gauss-Muller, V. (1998). Processing of proteinase precursors and their effect on hepatitis A virus particle formation. J. Virol. 72, 8013–8020.
Qu, L., Feng, Z., Yamane, D., Liang, Y., Lanford, R. E., Li, K., et al. (2011). Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A virus protease- polymerase processing intermediate, 3CD. PLoS Pathog. 7:e1002169. doi: 10.1371/journal.ppat.1002169
Qu, L., and Lemon, S. M. (2010). Hepatitis A and Hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses. Semin. Liver Dis. 30, 319–332. doi: 10.1055/s-0030-1267534.
Reizis, B., Bunin, A., Ghosh, H. S., Lewis, K. L., and Sisirak, V. (2011). Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 29, 163–183. doi: 10.1146/annurev-immunol-031210-101345
Satoh, T., Kato, H., Kumagai, Y., Yoneyama, M., Sato, S., Matsushita, K., et al. (2010). LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U.S.A. 107, 1512–1517. doi: 10.1073/pnas.0912986107
Schultheiss, T., Kusov, Y. Y., and Gauss-Müller, V. (1994). Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology 198, 275–281. doi: 10.1006/viro.1994.1030
Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682. doi: 10.1016/j.cell.2005.08.012
Sui, L., Zhang, W., Chen, Y., Zheng, Y., Wan, T., Zhang, W., et al. (2006). Human membrane protein Tim-3 facilitates hepatitis A virus entry into target cells. Int. J. Mol. Med. 17, 1093–1099. doi: 10.3892/ijmm.17.6.1093
Takeuchi, O., and Akira, S. (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Tami, C., Silberstein, E., Manangeeswaran, M., Freeman, G. J., Umetsu, S. E., DeKruyff, R. H., et al. (2007). Immunoglobulin A (IgA) is a natural ligand of hepatitis A virus cellular receptor 1 (HAVCR1), and the association of IgA with HAVCR1 enhances virus-receptor interactions. J. Virol. 81, 3437–3446. doi: 10.1128/JVI.01585-06
Thapa, R. J., Ingram, J. P., Ragan, K. B., Nogusa, S., Boyd, D. F., Benitez, A. A., et al. (2016). Dai senses influenza a virus genomic rna and activates ripk3-dependent cell death. Cell Host Microbe. 20, 674–681. doi: 10.1016/j.chom.2016.09.014
Tremblay, N., Baril, M., Chatelchaix, L., Essaad, S., Park, A. Y., Koenekoop, R. K., et al. (2017). Spliceosome snrnp200 promotes viral rna sensing and irf3 activation of antiviral response. PLoS Pathog. 13:e1005772. doi: 10.1371/journal.ppat.1005772
Ugrinova, I., and Pasheva, E. (2017). Hmgb1 protein: a therapeutic target inside and outside the cell. Adv. Protein Chem. Struct. Biol. 107:37. doi: 10.1016/bs.apcsb.2016.10.001
Venkataraman, T., Valdes, M., Elsby, R., Kakuta, S., Caceres, G., Saijo, S., et al. (2007). Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178, 6444–6455. doi: 10.4049/jimmunol.178.10.6444
Wang, D., Fang, L., Wei, D., Zhang, H., Luo, R., Chen, H., et al. (2014). Hepatitis A Virus 3C Protease Cleaves NEMO To Impair Induction of Beta Interferon. J. Virol. 88, 10252–10258. doi: 10.1128/JVI.00869-14
Wang, P., Zhu, S., Yang, L., Cui, S., Pan, W., Jackson, R., et al. (2015a). Nlrp6 regulates intestinal antiviral innate immunity. Science 350:826. doi: 10.1126/science.aab3145
Wang, X., Ren, J., Gao, Q., Hu, Z., Sun, Y., Li, X., et al. (2015b). Hepatitis A virus and the origins of picornaviruses. Nature 517, 85–88. doi: 10.1038/nature13806
Wen, H., Miao, E. A., and Ting, J. P. (2013). New mechanisms of nod-like receptor-associated inflammasome activation. Immunity 39, 432–441. doi: 10.1016/j.immuni
Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., et al. (2003). Role of adaptor TRIF in the MyD88- independent toll-like receptor signaling pathway. Science 301, 640–643. doi: 10.1126/science.1087262
Yang, P., An, H., Liu, X., Wen, M., Zheng, Y., Rui, Y., et al. (2010). The cytosolic nucleic acid sensor lrrfip1 mediates the production of type i interferon via a β-catenin-dependent pathway. Nat. Immunol. 11, 487–494. doi: 10.1038/ni.1876
Yang, Y., Liang, Y., Qu, L., Chen, Z., Yi, M., Li, K., et al. (2007). Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U.S.A. 104, 7253–7258. doi: 10.1073/pnas.0611506104
Zhang, Z., Kim, T., Bao, M., Facchinetti, V., Jung, S. Y., Ghaffari, A. A., et al. (2011a). Ddx1, ddx21, and dhx36 helicases form a complex with the adaptor molecule trif to sense dsrna in dendritic cells. Immunity 34, 866–878. doi: 10.1016/j.immuni.2011.03.027
Zhang, Z., Yuan, B., Lu, N., Facchinetti, V., and Liu, Y. J. (2011b). Dhx9 pairs with ips-1 to sense double-stranded rna in myeloid dendritic cells. J. Immunol. 187:4501. doi: 10.4049/jimmunol.1101307
Zhao, T., Yang, L., Sun, Q., Arguello, M., Ballard, D. W., Hiscott, J., et al. (2007). The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat. Immunol. 8, 592–600. doi: 10.1038/ni1465
Keywords: HAV, MAVS, TRIF, NEMO, type 1 interferon
Citation: Cao X, Xue Y, Du J, Xu Q, Yang X, Zeng Y, Wang B, Wang H, Liu J, Cai K and Ma Z (2018) Induction and Suppression of Innate Antiviral Responses by Hepatitis A Virus. Front. Microbiol. 9:1865. doi: 10.3389/fmicb.2018.01865
Received: 29 April 2018; Accepted: 25 July 2018;
Published: 17 August 2018.
Edited by:
Zhiyong Li, Lanzhou Veterinary Research Institute (CAAS), ChinaReviewed by:
Koji Ishii, National Institute of Infectious Diseases (NIID), JapanCopyright © 2018 Cao, Xue, Du, Xu, Yang, Zeng, Wang, Wang, Liu, Cai and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kui-zheng Cai, Y2t6MDAwQDEyNi5jb20=
Zhong-ren Ma, bXpyQHhibXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.