- Molecular Plant Pathology and Fungal Biology Group, School of Biological Sciences, University of Bristol, Bristol, United Kingdom
Zymoseptoria tritici is the causal agent of septoria tritici blotch, a devastating fungal disease of wheat which can cause up to 40% yield loss. One of the ways in which Z. tritici spreads in the field is via rain splash-dispersed asexual pycnidiospores, however there is currently limited understanding of the genetic mechanisms governing the development of these propagules. In order to explore whether the existing models for conidiation in ascomycete fungi apply to Z. tritici, homologs to the well-characterized Aspergillus nidulans genes abacus (abaA), bristle (brlA), fluffy B (flbB), fluffy C (flbC), and stunted (stuA) were identified and knocked-out by Agrobacterium-mediated transformation. Although deletion of the ZtAbaA, ZtBrlA1, and ZtFlbB genes had no apparent effect on Z. tritici asexual sporulation or on pathogenicity, deletion of ZtFlbC or ZtBrlA2 resulted in mutants with reduced pycnidiospore production compared to the parental IPO323 strain. Deletion of ZtStuA gave non-pigmented mutants with altered vegetative growth and eliminated asexual sporulation and pathogenicity. These findings suggest that the well-established A. nidulans model of asexual sporulation is only partially applicable to Z. tritici, and that this pathogen likely uses additional, as yet uncharacterized genes to control asexual sporulation.
Introduction
The ascomycete fungus Zymoseptoria tritici (synonym Mycosphaerella graminicola) causes septoria tritici blotch, a major disease of wheat worldwide (Ponomarenko et al., 2011; Dean et al., 2012). There are currently no wheat varieties which are fully resistant to the fungus, so disease is often managed by fungicide application. It has been estimated that 70% of total annual fungicide usage in the European Union is used primarily against Z. tritici (Ponomarenko et al., 2011; Torriani et al., 2015), which highlights the importance of this pathogen.
In the field, Z. tritici is able to reproduce both sexually and asexually. Asexual reproduction occurs via the formation of asexual fruiting bodies (pycnidia) which produce the asexual spores (pycnidiospores).The pycnidia develop within the substomatal cavity of the infected wheat leaf and mature approximately 21–28 days after initial colonization by the fungus. These fruiting bodies appear macroscopically as dark brown dots against the pale necrotic lesions formed by the fungus (Kema et al., 1996; Dancer et al., 1999; Duncan and Howard, 2000).
Microscopically, pycnidia are typically 60–200 μm in diameter and subglobose in shape, with an ostiole below or protruding through the stomatal pore (Eyal et al., 1987; Kema et al., 1996). Each pycnidium produces an estimated 10,000 pycnidiospores, which are exuded through the ostiole during conditions of high humidity in a gelatinous matrix termed cirrhus (Eyal et al., 1987; Kema et al., 1996; Dancer et al., 1999; Duncan and Howard, 2000; Palmer and Skinner, 2002). The pycnidiospores are then transmitted to the leaves and stems of neighboring host plants by rain splash or direct mechanical contact (Lovell et al., 2004). It is not unusual for almost all substomatal cavities of a lesion to be colonized and contain mature pycnidia. As a result, massive asexual sporulation can occur from a single successful infection, allowing propagation and spread to adjacent plants.
Despite the global importance of this pathogen, to date there has been no detailed investigation into the key genes controlling asexual sporulation in Z. tritici. Previous studies focusing on other aspects of the pathogen's biology have occasionally reported defects in sporulation among mutant phenotypes following gene disruption. Examples include the Z. tritici Gα subunits MgGpa1, MgGpa3, and MgGpb1 mutants or the ZtWor1 deletion, all of which have reduced pathogenicity and are therefore unable to form mature pycnidia in planta (Mehrabi et al., 2009; Mirzadi Gohari et al., 2014). These results are typically obtained from in planta experiments where asexual sporulation has not been separated from virulence, so the impact on sporulation may be a by-product of a reduction in pathogenicity.
The most detailed and in-depth characterization of the genetic basis of asexual sporulation in an ascomycete species has been in the model fungus Aspergillus nidulans (reviewed in Adams et al., 1998; Etxebeste et al., 2010; Park and Yu, 2012). The key genes regulating asexual sporulation have been determined through mutant analysis, with clear phenotypes for each step in the process. In A. nidulans, asexual spores (termed conidia) are borne on stalk-like multicellular structures called conidiophores. Three central regulatory genes have been identified which together promote successful conidiophore development; bristle (which produces two overlapping transcripts, brlAα and brlAβ) (Adams et al., 1988; Prade and Timberlake, 1993), abacus (abaA) (Sewall et al., 1990; Andrianopoulos and Timberlake, 1994) and wet white (wetA) (Mirabito et al., 1989; Marshall and Timberlake, 1991).
The brlA gene is important for the transition from apical growth of the conidiophore stalk to vesicle formation. Null brlA mutants form bristle-like structures with elongated conidiophore stalks, about 20–30 times the normal length, that are unable to form the vesicle or the remaining structures (Clutterbuck, 1969; Adams et al., 1988). The abaA gene is activated by brlA and is required for phialide differentiation, the phialides being the progenitor cells from which conidia are produced. Mutants in this gene have normal conidiophore stalks but produce abacus-like structures on the vesicle (Clutterbuck, 1969; Sewall et al., 1990). Finally, the wetA gene is activated by abaA and has been shown to be involved in the proper synthesis of the conidial cell walls. Mutants in this gene produce colorless conidia which autolyse after a few days (Clutterbuck, 1969; Marshall and Timberlake, 1991). Upstream of the central regulatory genes are fluG, flbA, flbB, flbC, flbD, and flbE. These are all required for proper expression of brlA and for the initiation of conidiation. Deletion of these genes results in “fluffy” colonies of profuse aerial hyphae (Wieser et al., 1994). Other genes known to be involved in asexual sporulation in A. nidulans are the developmental modifiers medusa (medA) and stunted (stuA), which are involved in cell differentiation and patterning of the conidiophore (Clutterbuck, 1969; Martinelli, 1979; Miller et al., 1991, 1992; Adams et al., 1998).
Homologs to the genes regulating asexual sporulation in A. nidulans have been identified in the wider Dikarya, suggesting that a conserved set of genes may regulate the patterning of divergent fruiting body structures. For example, stuA is evolutionarily widely conserved and homologs with roles in sporulation have been identified in Ustilago maydis, Parastagonospora nodorum, Fusarium oxysporum, Aspergillus fumigatus, Magnaporthe oryzae, Acremonium chrysogenum, and Glomerella cingulata (Ohara and Tsuge, 2004; Sheppard et al., 2005; Tong et al., 2007; García-pedrajas et al., 2010; IpCho et al., 2010; Nishimura et al., 2014; Hu et al., 2015).
The objective of this study was to assess to what extent the A. nidulans model for regulation of conidiation holds true for Z. tritici. Potential homologs to the A. nidulans genes abaA, brlA, flbB, flbC, and stuA were selected for gene deletion studies in Z. tritici. Gene deletion plasmids were transformed into Z. tritici via Agrobacterium-mediated transformation, and the resulting knock-out mutants were characterized to assess differences in vegetative growth, pathogenicity, and the ability to sporulate asexually in vitro and in planta. The findings from this study shed light on some of the key genes required for asexual sporulation in Z. tritici.
Materials and Methods
Comparative Genomic Analyses in Z. tritici
Genes involved in asexual sporulation in the model fungus A. nidulans were identified through a literature search using the Web of SciencesTM (http://www.webofknowledge.com) and the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). The protein FASTA sequences were BLAST searched against the Z. tritici genome database (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html) using the tblastn and Filtered Models (transcripts) algorithms.
When more than one BLAST match occurred in Z. tritici, the sequence of the original query gene and the Z. tritici genes were aligned using Clustal X version 2.0 (Larkin et al., 2007). Molecular Evolutionary Genetics (MEGA) 6 software was then used to create neighbor-joining phylogenetic trees between the original sequence and the observed matches in Z. tritici.
Plasmid Construction
Knock-out plasmids were constructed using yeast-based homologous recombination. Primers for plasmid construction are listed in Supplementary Table 1. The plasmids consisted of a pCAMBIA0380YA (yeast-adapted) backbone, two 1.5 kb flanking regions and Hygromycin-trpC resistance cassette from pCB1003 (Carroll et al., 1994). The flanking regions and Hygromycin-trpC resistance cassette were amplified using Phusion® High-Fidelity DNA Polymerase (Thermo Scientific).
Plasmid DNA was recovered from Saccharomyces cerevisiae using the Zymoprep™ Yeast Plasmid Miniprep II kit (Zymo Research) following the manufacturer's instructions. The plasmids were then propagated in E. coli ccdB or DH5α cells, and isolated using the Gene JET Plasmid Miniprep Kit (Thermo Scientific) or Gene JET Plasmid Midiprep Kit (Thermo Scientific) following the manufacturer's instructions. Sequencing of plasmids was carried out by GATC Biotech using the LIGHTRUNTM sequencing service with primers listed in Supplementary Table 2.
Agrobacterium-Mediated Transformation
Knock-out plasmids were transformed into A. tumefaciens LBA1126 and AGL1 cells. Z. tritici IPO323 and Δku70 strains were transformed by Agrobacterium–mediated transformation after Derbyshire et al. (2015).
Confirmation of Knock-Out Mutants
Initial screening of knock-out mutants were confirmed by growth on YPDA supplemented with Hygromycin B and Timentin. Mutants were sub-cultured at least three times to single colonies, and successful knock-out of the candidate gene confirmed by double PCR. Fungal DNA was extracted using the protocol outlined in (Liu et al., 2000). Double PCR was carried out using two primer pairs; the first primer pair was designed to amplify the wild-type gene, and the second pair to amplify the Hygromycin-trpC resistance cassette. Primers used for knock-out confirmation are listed in Supplementary Table 3.
In vitro Experiments
Fungal isolates were cultured onto either Czapek Dox-V8 juice (CDV8) agar (46 g/L Czapek Dox agar, 200 ml/L V8® Original vegetable juice (Campbell's), 3 g/L calcium carbonate and 10 g/L technical agar), PDA (24 g/L potato dextrose broth and 20 g/L technical agar), YPDA agar (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose and 20 g/L technical agar) or wheat extract agar (37.5 g/L homogenized 21 day-old wheat leaves cv. Riband, 20 g/L technical agar). Cultures were incubated under white light or UV-A light (16:8 light:dark cycle) at 20°C for up to 28 days.
Z. tritici liquid cultures were grown by inoculating a 250 ml conical flask containing 50 ml PDB with a 10 μl loop of the fungus. The culture was incubated in a shaker at 200 rpm in the dark at 20°C for up to 10 days.
In planta Experiments
The attached leaf wheat inoculation procedure used was similar to that described previously by Keon et al. (2007). The susceptible wheat cultivar Riband was used for all experiments. Wheat seedlings were grown in a single line along the edge of a 9” plastic seed tray in Levington® “F2” peat-based compost. The seedlings were grown for about 21 days at an average of 20°C under a 16 h day length.
The first true leaf of wheat plants at growth stages 12–13 (Tottman, 1987) were then held adaxial side up on polystyrene blocks. Wheat leaves were inoculated with spores from seven day-old CDV8 plates suspended in filter-sterilized 0.1% Tween20 at a concentration of 4 × 106 spores/ml, using a cotton bud dipped in the suspension (Motteram et al., 2009) The spore suspension was swabbed over the leaf surface ten times. Infected plants were sealed inside a clear 40 μm thick autoclave bag for the initial 72 h of infection at 20°C under a 16 h day length in order to maintain high relative humidity. Following this period of time, the bags were removed and the infected plants remained at conditions of 20°C and a 16 h day length.
Virulence of Z. tritici knock-out mutants was assessed by monitoring and recording disease progression every 2–3 days. Disease symptoms on the infected leaves were scored from 1 to 5 using a modified version of the scale outlined in Skinner (2001).
Infected and control leaves were collected at 28 days post infection to compare pycnidia and pycnidiospore production. Leaf sections 4 cm in length were taken starting at the base of the lesion, and subjected to 100% humidity for 48 h to induce release of pycnidiospores. The leaf sections were then immersed in 2 ml SDW and vortexed for 15 s. Ten μl of the suspension was immediately pipetted onto a haemocytometer to count the number of spores released from the pycnidia. This procedure was used for 10 leaf sections per strain of Z. tritici.
In order to assess pycnidia number, a 1 cm section was also taken from each leaf, and the area of the leaf was measured using ImageJ software. The number of pycnidia were counted using a dissection microscope, and used to calculate the number of pycnidia per square millimeter (mm2).
Microscopy Stains
Light microscopy was used to assess differences in Z. tritici vegetative hyphal morphology, pycnidiosospore number and pycnidiospore morphology between the knock-out mutants and parental IPO323 or Δku70 strain. Z. tritici vegetative hyphae were obtained from liquid PDB cultures. Pycnidiospores were harvested from the infected leaves collected at 28 dpi.
Specimens were prepared by putting 5 μl of lactophenol cotton blue stain on a glass microscope slide, followed by 5 μl of the fungal cell suspension and a cover slip. The specimen was left to stand for 1 min before microscope analysis.
Results
Identification of Zymoseptoria tritici Sporulation Genes Using Aspergillus nidulans Homologs
Homologs to the A. nidulans genes abaA, brlA, flbB, and stuA were identified in Z. tritici by BLAST search using the A. nidulans protein sequences and tblastn searched against the Filtered Models (transcripts) of the Z. tritici genome database (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html). A single homolog was obtained in Z. tritici for abaA, however the brlA, flbB, and stuA genes all gave multiple significant matches (Table 1).
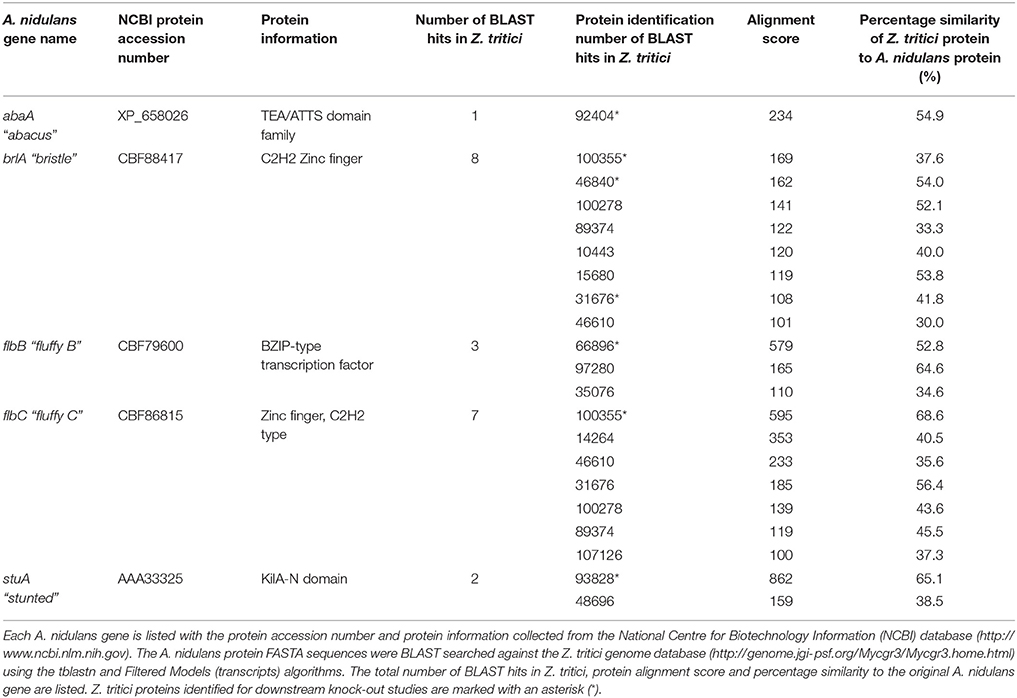
Table 1. Summary of the Aspergillus nidulans sporulation genes abaA, brlA, flbB, flbC, and stuA, and all potential homologs identified in Zymoseptoria tritici.
In order to allow confident identification of the most likely homolog in Z. tritici, phylogenetic trees using known sequences for each protein family from A. nidulans and the wider Dikarya were used to infer relationships. Where possible, RNAseq data available in the literature were used to validate the Z. tritici homologs selected (Yang et al., 2013; Rudd et al., 2015). During the search for brlA, the Z. tritici protein number 100355 was identified as a match to both brlA and the flbC proteins (Figure 1). In addition, one of the matches identified, ZtBrlA2, was identified as synonymous with the previously identified MGSTE12 (Kramer et al., 2009). In total, six genes were selected for knock-out in Z. tritici, including three potential brlA homologs due to the importance of this gene family in controlling asexual sporulation in A. nidulans (Table 2).
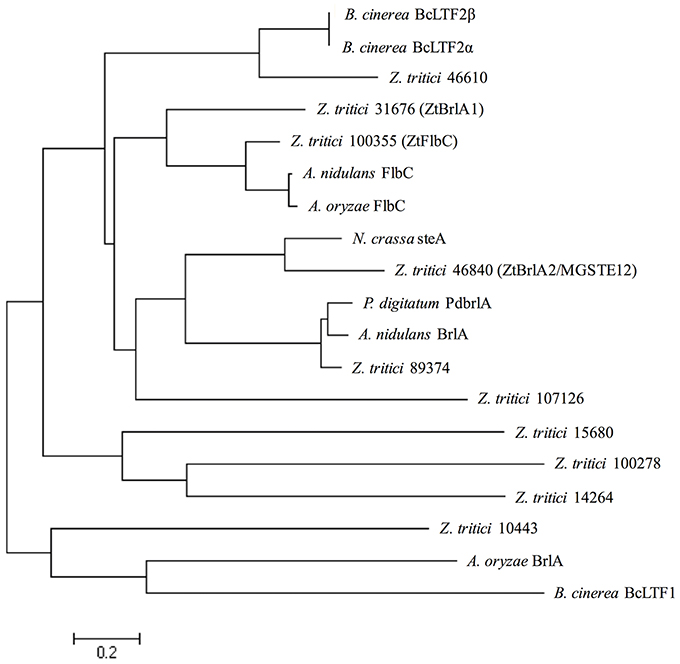
Figure 1. Zymoseptoria tritici IPO323 protein sequences in relation to BrlA and FlbC protein sequences from the ascomycete fungi Aspergillus nidulans, Aspergillus oryzae, Neurospora crassa, Penicillium digitatum, and Botrytis cinerea. Neighbour-joining phylogenetic trees were constructed using FASTA protein sequences obtained from the National Centre for Biotechnology Information (NCBI) database and the Z. tritici genome database (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html). Branch length represents 0.2 amino acid substitutions per site. The Z. tritici protein 100355 shares similarity with FlbC from A. nidulans. The Z. tritici proteins 31676 and 46840 share similarities with BrlA-like proteins from A. nidulans, P. digitatum and N. crassa.
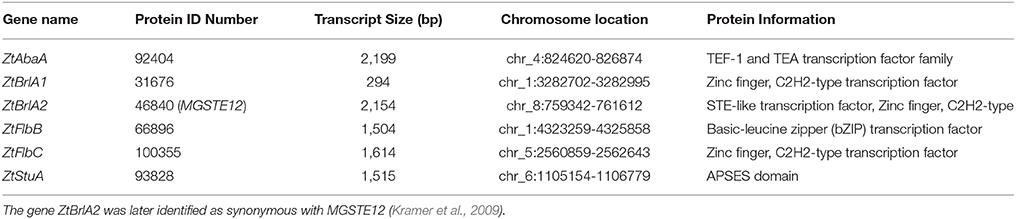
Table 2. The six sporulation genes selected for knock-out in Zymoseptoria tritici, listing each gene name, protein ID, transcript size, chromosome location and protein information obtained from the Z. tritici genome database.
Generation of Z. tritici Gene Deletion Mutants
Gene deletion mutants were generated in Z. tritici to assess the role of ZtAbaA, ZtBrlA1, ZtBrlA2, ZtFlbB, ZtFlbC, and ZtStuA in asexual sporulation. Knock-out plasmids were generated in a yeast-adapted version of pCAMBIA_0380 by yeast-based homologous recombination. The knock-out construct consisted of a Hygromycin-trpC resistance cassette (indirectly derived from pCB1003; Carroll et al., 1994) flanked by two 1.5 kb regions amplified from either side of the gene of interest, to allow efficient targeting by Agrobacterium-mediated transformation (Supplementary Table 1). The ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC mutants were successfully generated in the wild-type IPO323 background with 10–40% targeting efficiency. Despite several attempts, we were not able to isolate any ΔztstuA mutants in the wild-type background. However, these were successfully generated in the Δku70 background which had a higher targeting efficiency (Bowler et al., 2010).
Transformants were identified and purified using selective media containing Hygromycin B and confirmed by PCR (Supplementary Figure 1 and Supplementary Table 3). The first primer pair amplified across the flanking region to the IPO323 wild-type gene, and the second pair was used to amplify across the flanking region to the Hygromycin-trpC resistance cassette. Successful disruption was indicated by both loss of the wild-type amplicon and gain of the knockout amplicon. Throughout this study, three independent deletion strains and the corresponding parental IPO323 or Δku70 strain were analyzed for each of the genes selected.
The ΔZtbrlA1, ΔZtflbC and ΔZtstuA Mutants Have Defects in Vegetative Growth In vitro
Phenotypic differences between the IPO323 or Δku70 parental strain and the deletion mutants were assessed by growing the fungi in liquid PDB and on the solid media PDA, CDV8, and YPDA for 7 days. The ΔztabaA, ΔztbrlA2 and ΔztflbB knock-out mutants showed no difference in phenotype to the parental IPO323 strain when grown in liquid PDB or on PDA, CDV8 or YPDA solid media. By 7 days post-inoculation (d.p.i.), all liquid cultures were rosy-pink in color with some melanisation, and microscopy analyses showed all cultures had both yeast-like and filamentous hyphae. On solid media, the ΔztabaA, ΔztbrlA2, and ΔztflbB mutants showed no difference in vegetative growth compared to IPO323 (Figure 2). Although ΔztbrlA2 and ΔztflbB vegetative growth sometimes differed to IPO323 on PDA, this was not consistent and was therefore attributed to inherent variation in Z. tritici growth.
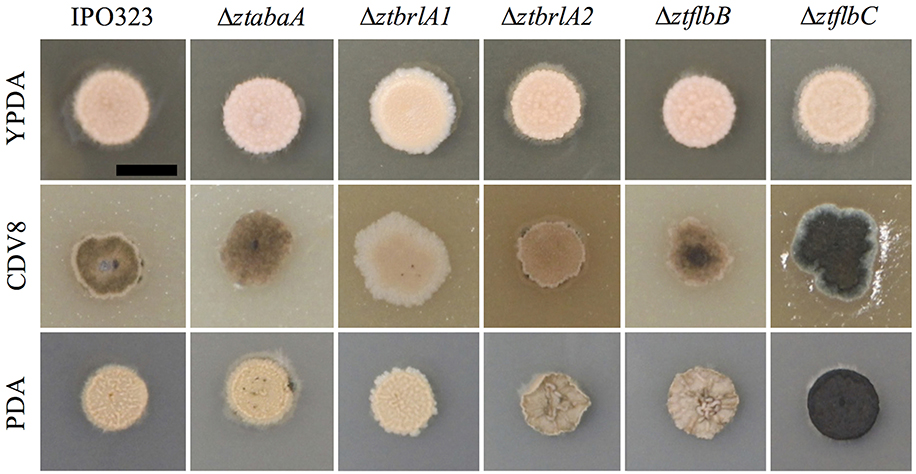
Figure 2. Zymoseptoria tritici wild-type (IPO323) and knockout mutants growing on YPDA, CDV8 agar, and PDA at 20°C, under white light (16:8 light:dark cycles), at 7 days post-inoculation (d.p.i.). Images are representative of three independent ΔztabaA, Δztbrla1, ΔztbrlA2, ΔztflbB, and Δztflbc strains tested for each mutant. Scale bar = 5 mm. The ΔztabaA, ΔztbrlA2, and ΔztflbB mutants are similar phenotypically to IPO323 on all three media types tested. However, the ΔztflbC mutant melanised faster than IPO323 on CDV8 and PDA. The ΔztbrlA1 strain melanised slower than the IPO323 strain on all three media types.
The ΔztflbC mutant had normal morphology in liquid PDB, however, on solid media PDA and CDV8, colonies of ΔztflbC melanised faster than IPO323 (Figure 2). Unlike the wild-type, the ΔztbrlA1 mutant liquid cultures stayed rosy-pink in color and showed no melanisation at seven d.p.i. In addition, the ΔztbrlA1 strain also took longer to melanise than IPO323 on all three solid media types tested. Microscopy of liquid culture showed that the ΔztbrlA1 vegetative cells were short, unbranching and yeast-like, with only a few starting to undergo the transition to hyphal growth.
The ΔztstuA mutant differed from the Δku70 parent when grown in liquid PDB or on solid media. In PDB, the mutant cultures were pale yellow to rosy-pink in color and did not melanise by 7 d.p.i. The ΔztstuA liquid culture also showed aggregation of the cells to make mycelial pellets. On observation under the microscope, the ΔztstuA mutant cells were not yeast-like, but instead formed a mass of long thread-like hyphae (Figure 3).
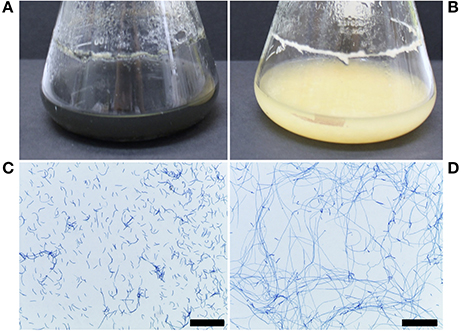
Figure 3. Z. tritici Δku70 and ΔztstuA growth in PDB at 20°C on a rotary shaker at 200 rpm, 7 and 10 days post-inoculation (d.p.i.). Images are representative of the three independent ΔztstuA strains tested. Scale bar = 200 μm.; (A) Δku70 liquid cultures at 10 d.p.i. are black and melanised (B) ΔztstuA liquid cultures at 10 d.p.i. remain pale yellow in color with no melanisation; (C) Δku70 cultures at 7 d.p.i. stained with lactophenol cotton blue are yeast-like with some branching hyphal cells; (D) ΔztstuA cultures at 7.d.p.i. stained with lactophenol cotton blue have minimal yeast-like growth and instead produce long thread-like hyphae.
When grown on solid media, the Δku70 strain grew as rosy-pink colonies of yeast-like cells which began to melanise by 7 d.p.i. However, the ΔztstuA mutant did not form yeast-like cells and did not melanise, instead colonies of this mutant produced white aerial hyphae on all three types of media tested with no melanisation even after 10 d.p.i (Figure 4).
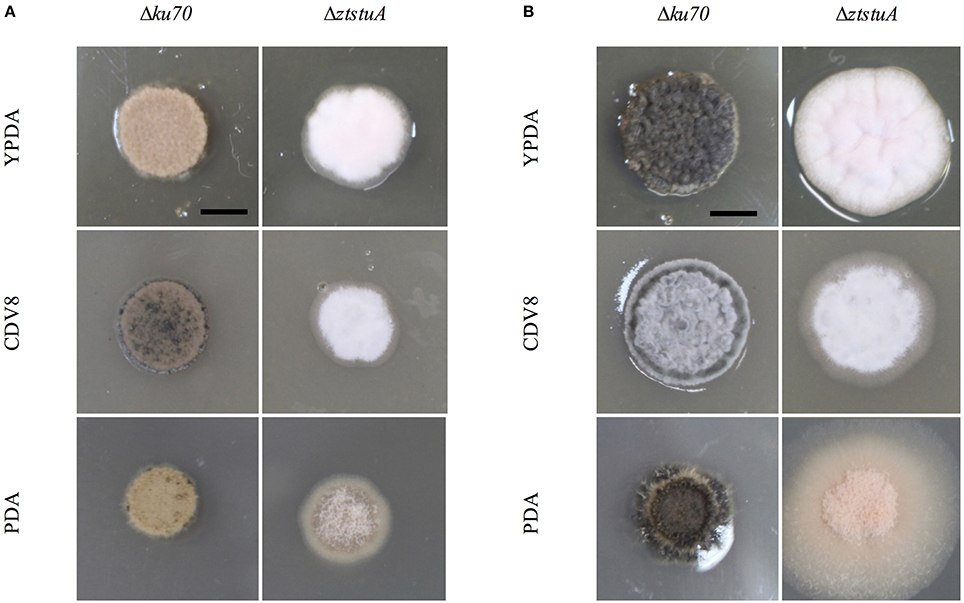
Figure 4. Zymoseptoria tritici Δku70 and ΔztstuA growth on YPDA agar, CDV8, and PDA at 20°C, under white light (16:8 light:dark cycles), at 7 days post-inoculation (d.p.i.) and 10 d.p.i. Scale bar = 5 mm; (A) at 7 d.p.i. the Δku70 strain grows as rosy-pink yeast-like cells with some melanisation on CDV8 and PDA media. The ΔztstuA mutant grows as a white hyphal colony with no signs of yeast-like growth or melanisation; (B) at 10 d.p.i. the Δku70 strain is melanised on all three types of media, but the ΔztstuA strain remains white or rosy-pink in color with aerial hyphae.
ΔztstuA Mutants Are Unable to Undergo Asexual Sporulation In vitro
Asexual reproduction was established in vitro by incubating the Z. tritici mutants and respective background strain on wheat leaf extract agar (WEA) under UV-A light for 28 days. This method was used to test whether the gene deletion mutants could produce pycnidia in vitro, as it enables the uncoupling of pathogenicity from asexual sporulation.
By 14 d.p.i. the IPO323 wild-type strain grew lateral hyphae radiating away from the point of inoculation both into and over the agar surface. Leading hyphae at the colony margins formed white hyphal knots by 21 d.p.i. These hyphal knots were 50–200 μm in diameter which formed dense clusters on the agar surface. By 28 d.p.i. the hyphal knots developed into dark brown, globose structures, each with an ostiole. These pycnidial structures were 50–100 μm in diameter and resembled the pycnidia typically observed in planta. The structures were present both within and on the surface of the agar, and some oozed a cloudy white liquid which was similar to cirrhus observed in planta.
The ΔztabaA, Δztbrla1, Δztbrla2, ΔztflbB and ΔztflbC mutants were all able to produce pycnidia in vitro similar to the IPO323 strain. In addition, the pycnidia produced by the mutant strains followed the same developmental pathway and timings as IPO323 (Figure 5).
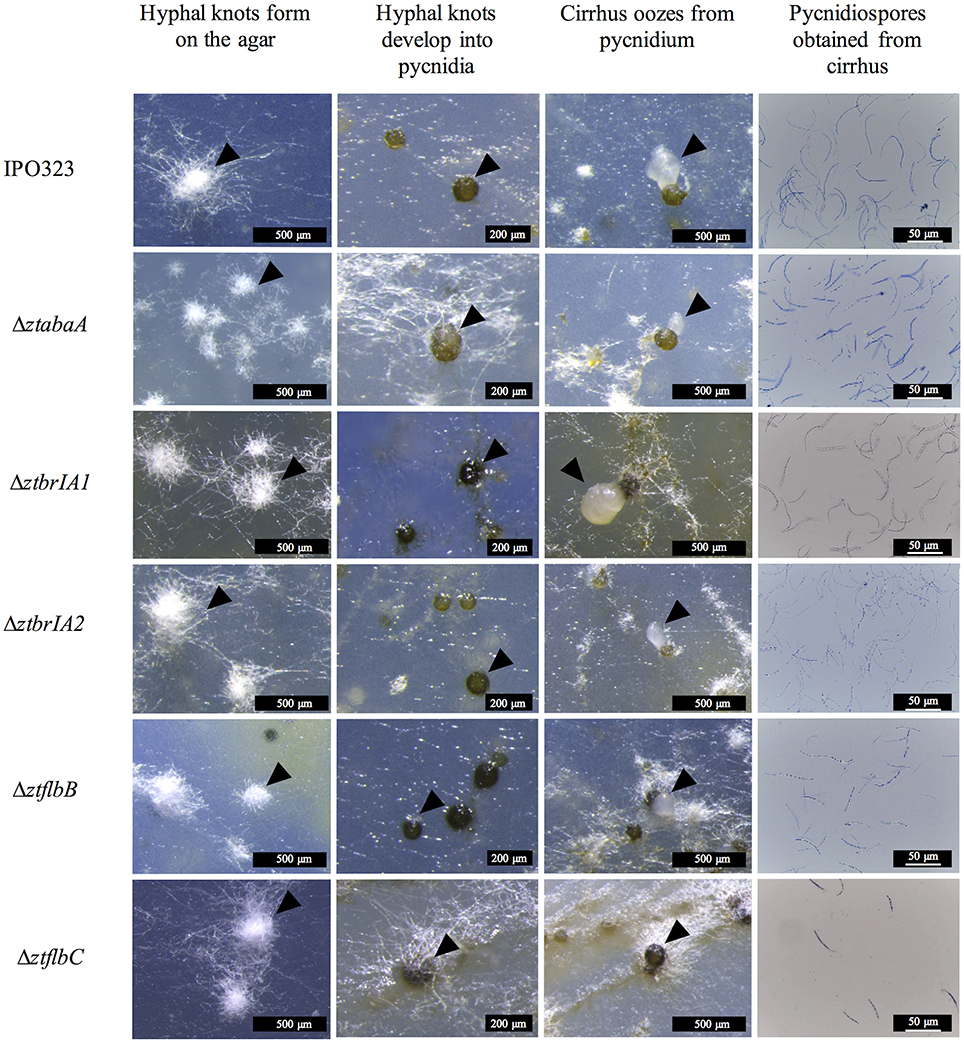
Figure 5. Pycnidia and pycnidiospore production on wheat extract agar (WEA) for IPO323 and selected knock-out mutants, grown at 20°C under UV-A light (16:8 light:dark cycles), 21 – 28 days post-inoculation (d.p.i.). Images are representative of the three independent ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC strains tested for each knockout. The different structures formed at each stage of pycnidia development are indicated by black arrows. At 21 d.p.i. IPO323 and ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC all develop white hyphal knots on the WEA. The hyphal knots progress to form round brown/black pycnidia by 28 d.p.i. The pycnidia produced by the strains oozed cirrhus, which was extracted and stained with lactophenol cotton blue. The cirrhus obtained from the IPO323 and ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC strains all contained curved unbranched pycnidiospores.
The pycnidia-like structures from IPO323, ΔztabaA, Δztbrla1, Δztbrla2, ΔztflbB, and ΔztflbC all exuded a cirrhus-like ooze. The cirrhus was collected, stained using lactophenol cotton blue, and observed under a microscope. The cirrhus obtained from the mutant strains contained pycnidiospores which were morphologically indistinguishable from IPO323 pycnidiospores (Figure 5).
In contrast, the ΔztstuA mutant did not form hyphal knots, and hence no fruiting bodies or spores under these conditions. Instead, the mutant produced a thin hyphal mat over the surface of the agar, but these did not aggregate into knots or form pycnidia (Figure 6).
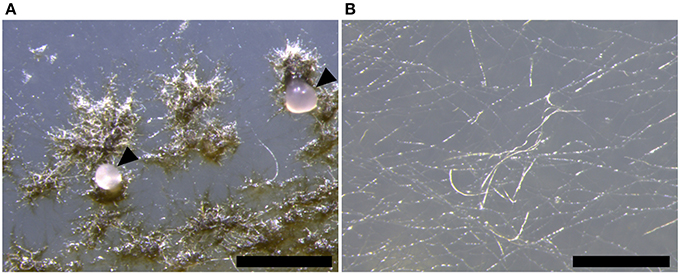
Figure 6. Zymoseptoria tritici Δku70 and ΔztstuA in vitro growth on wheat extract agar (WEA) at 20°C under continuous UV-A light at 28 days post-inoculation (d.p.i.). Images are representative of the three independent ΔztstuA strains tested. Scale bar = 500 μm.; (A) by 28 d.p.i., the Δku70 strain produces round dark brown pycnidia on the WEA. The pycnidia ooze cirrhus which contains pycnidiospores (indicated by arrows); (B) the ΔztstuA strain does not form pycnidia, even after 28 d.p.i. Instead, the ΔztstuA mutant grows as white hyphae across the surface of the WEA.
ΔZtbrlA2 and ΔZtstuA Mutants Have Reduced Virulence In planta
In order to assess whether the candidate genes knocked-out in Z. tritici have a role in pathogenicity or sporulation, susceptible wheat plants were infected with the mutant strains and IPO323 or Δku70 parental strain. The infection progression was recorded until 28 d.p.i., when the pycnidia were fully developed. Three independent deletion mutants were tested against the respective parental strain for each target gene, and experiments were performed on two independent occasions.
Wheat leaves infected with the ΔztabaA, ΔztbrlA1, ΔztflbB, and ΔztflbC mutants showed normal disease progression and normal symptoms. By 28 d.p.i., these lesions contained pycnidia which oozed pycnidiospores under conditions of high humidity (Figure 7). The ΔztbrlA2 mutant produced milder symptoms than IPO323, but was still able to make pycnidia and pycnidiospores by 28 d.p.i. To test whether the difference in symptom severity was due to an inability to establish initial infection, the yeast-like cells of the mutant were infiltrated into the leaves using a syringe and disease progression was measured as before. When this method was used, the ΔztbrlA2 mutant was able to produce symptoms. The time to chlorosis was the same as for IPO323 delivered by this route, however, the necrotic lesions produced were less severe than those made by IPO323, and these lesions were not as densely covered with pycnidia.
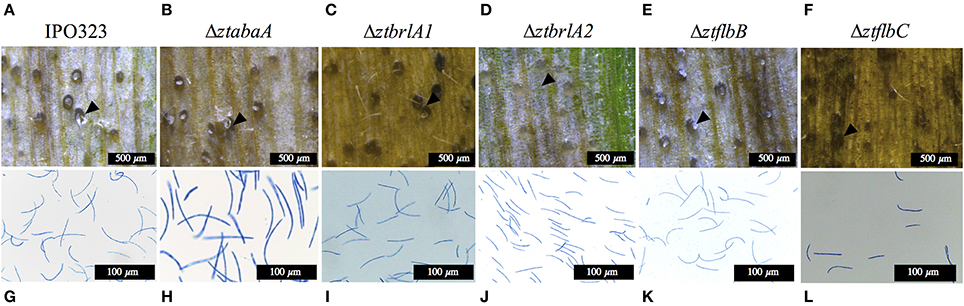
Figure 7. Images of pycnidia (indicated by arrows) and pycnidiospores from wheat leaves infected with 4 × 106 spores/ml from IPO323 and ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC mutants. (A,G) IPO323; (B,H) ΔztabaA; (C,I) Δztbrla1; (D,J) ΔztbrlA2; (E,K) ΔztflbB; (F,L) ΔztflbC at 28 days post-infection (d.p.i.). The knock-out mutant strains were all able to produce pycnidia and pycnidiospores, and these were morphologically similar to the IPO323 strain. The ΔztbrlA2 mutant produced fewer pycnidia, and these took longer to develop compared to the IPO323 strain. Fewer spores were harvested from the ΔztbrlA2 and ΔztflbC mutants.
The ΔztstuA mutant was unable to make yeast-like spores on solid culture so the conventional inoculation method was not possible. Therefore, to prepare inoculum, both the Δku70 and ΔztstuA strains were grown in PDB for 7 days. The cultures were then homogenized and the O.D.600 adjusted to the equivalent of 4 × 106 spores/ml. The cells were harvested by centrifugation and the pellet was resuspended in 0.1% Tween 20. This suspension was used to infect the wheat leaves using a cotton bud.
Whilst the Δku70 parent generated symptoms in the normal manner, the ΔztstuA mutant was unable to form productive necrotic lesions on the wheat leaves. Leaves infected with the ΔztstuA mutants did display some patchy chlorosis in the inoculated area and developed occasional small necrotic lesions. Careful inspection of these infected leaves with a dissection microscope showed no evidence of pycnidia within the substomatal cavities of the leaves (Figure 8).
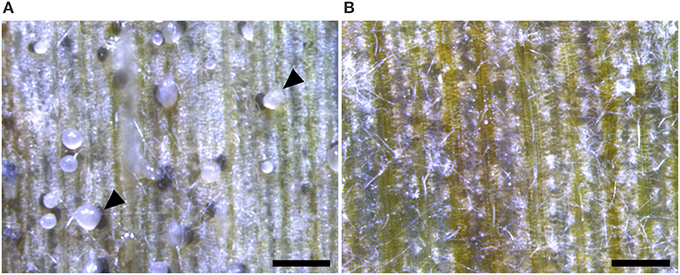
Figure 8. Wheat leaves infected with 4 × 106 spores/ml Zymoseptoria tritici Δku70 strain and the ΔztstuA mutant at 28 days post-infection (d.p.i.). Images representative of three independent ΔztstuA strains tested. Scale bar = 500 μm.; (A) the Δku70 strain produces necrotic lesions and pycnidia on the wheat leaf. The pycnidia ooze cloudy cirrhus which contains pycnidiospores (indicated by arrows); (B) leaves infected with the ΔztstuA mutant show signs of chlorosis but no pycnidia. Instead, the ΔztstuA mutant grows as white hyphae which traverse the leaf surface.
The ΔztbrlA2 and ΔZtflbC Mutants Have Reduced Pycnidiospore Production In planta
Pycnidia numbers and pycnidiospore production of the ΔztabaA, Δztbrla1, Δztbrla2, ΔztflbB, and ΔztflbC deletion mutants was compared to the IPO323 strain. There was no significant difference between the numbers of pycnidia or pycnidiospores produced by the ΔztabaA, ΔztbrlA1 or ΔztflbB mutant strains. Although ΔztflbC mutants had reduced numbers of pycnidia compared to IPO323, this was not found to be significant (p > 0.05). The ΔztbrlA2 strain produced significantly fewer pycnidia compared to IPO323 (p < 0.05) (Figure 9). In addition, both the ΔztbrlA2 and ΔztflbC mutant strains produced significantly fewer pycnidiospores compared to IPO323 (p < 0.05) (Figure 9).
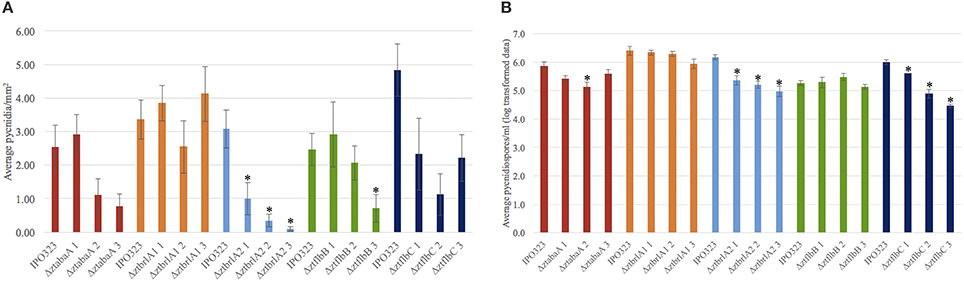
Figure 9. Average numbers of pycnidia and pycnidiospores produced in planta by IPO323 and the various mutant strains, assessed at 28 days post-infection (d.p.i.). Three independent knock-out strains were tested for each ΔztabaA, ΔztbrlA1, ΔztbrlA2, ΔztflbB, and ΔztflbC mutant. Bars represent mean SE across 5–10 technical repeats from a single experiment. Bars labeled with an asterisk (*) are knock-out strains with a significant difference in average pycnidia/mm2 compared to IPO323 (p < 0.05). The ΔztbrlA2 strains have significantly reduced numbers of pycnidia/mm2 compared to IPO323, however, there is no consistent significant difference between pycnidia/mm2 produced by the ΔztabaA, ΔztbrlA1, ΔztflbB or ΔztflbC strains and those made by IPO323. The ΔztbrlA2 and ΔztflbC strains have significantly reduced numbers of pycnidiospores/ml compared to IPO323.
Pycnidiospore viability was tested by harvesting the spores from leaves infected with the ΔztabaA, ΔztbrlA1, and ΔztflbB mutants and using them directly to infect fresh plants. These results showed that ΔztabaA, ΔztbrlA1, and ΔztflbB pycnidiospores were able to cause normal disease in planta (data not shown). Pycnidiospore viability was not tested for the ΔztflbC orΔztbrlA2 mutants due to low levels of production.
Discussion
Asexual sporulation by Z. tritici enables the pathogen to spread rapidly in the field (Eyal et al., 1987), therefore, targeting production of these pycnidiospores and the pycnidia which make them could be an effective way to reduce disease spread. Although Z. tritici makes use of a complex pycnidial fruiting body for asexual spore production, bioinformatic analyses showed that Z. tritici has potential homologs to key A. nidulans conidiation genes including abaA, brlA, flbB, flbC, and stuA. The presence of these indicates possible genetic conservation and perhaps the use of a similar pathway to regulate asexual sporulation
Results presented here suggest that the ZtAbaA and ZtFlbB genes do not have equivalent roles to abaA and flbB in A. nidulans, as deletion did not affect growth or sporulation in vitro or in planta. We note that RNA-seq data from the literature (Rudd et al., 2015) shows that ZtAbaA expression does not change significantly during infection, again perhaps making its involvement in sporulation less likely. ZtFlbB expression shows a decrease from 1 to 21 d.p.i. (Rudd et al., 2015), suggesting recruitment of this gene in a different process. Only one homolog to abaA was identified in Z. tritici, so the only likely candidate was deleted in the fungus. However, two other potential flbB homologs exist in the pathogen (Table 1). Therefore, genetic redundancy might explain why deletion of ZtFlbB did not affect development or pathogenicity in Z. tritici.
The ZtFlbC, ZtBrlA2, and ZtStuA genes were all shown to have roles in asexual sporulation in Z. tritici. In A. nidulans, deletion of flbC results in delayed and reduced rates of conidiation (Kwon et al., 2010). A similar outcome was seen here in Z. tritici, where the ΔztflbC strains had no significant difference in pycnidia production compared to IPO323. However, these mutants did have a significant reduction in pycnidiospore numbers. This could be as a result of a defects in pycnidia viability or ability to release the pycnidiospores. Further evidence for the importance of ZtFlbC has been documented in two separate RNA-seq studies, which show a significant change in the expression of this gene during infection, (Yang et al., 2013; Rudd et al., 2015). Therefore, this adds further evidence for the possible role of ZtFlbC in a process such as asexual sporulation in Z. tritici.
The brlA gene is a master regulator of asexual sporulation in A. nidulans and is essential for the switch from vegetative growth to asexual sporulation. Null mutants can form the conidiophore stalk, but this grows indeterminately and does not form a mature spore-producing structure. In contrast, over-expression of brlA can induce hyphal tips to produce conidiophores (Clutterbuck, 1969; Adams et al., 1988). Whilst the ΔztbrlA1 mutant displayed normal asexual sporulation, disruption of ZtBrlA2 clearly reduced numbers of both pycnidia and pycnidiospores, severely impacting on asexual reproduction.
The milder effects on fruiting body numbers and pycnidiospores seen in the ΔztflbC mutants compared to ΔztbrlA2 would agree with findings in A. nidulans where flbC null-mutants show similar but weaker phenotypes than brlA in terms of impacts on conidiation, consistent with the role of FlbC as one of the upstream activators for brlA (Kwon et al., 2010).
The deletion of ZtStuA shows that this gene is required for asexual sporulation in Z. tritici. The ΔztstuA mutants were completely impaired in their ability to make either pycnidia or pycnidiospores, suggesting that ZtStuA is essential for this process. The use of the WEA media shows that the lack of sporulation of ΔztstuA in planta is not simply a consequence of the reduced virulence of the fungus, as sporulation was prevented in vitro on inductive media. These mutant phenotypes are different to those observed in A. nidulans, where stuA mutants make abnormal conidiophores and some conidia. However, the results from these studies agree with work carried out on the wheat pathogen P. nodorum. The deletion of SnStuA from P. nodorum results in mutants which fail to produce pycnidia or pycnidiospores. In addition, like ΔztstuA mutants, the SnStuA mutants are non-pathogenic and produce thick white aerial hyphae on V8PDA media (IpCho et al., 2010). These similar mutant phenotypes suggest a conserved role of StuA in these two fungi. As P. nodorum and Z. tritici are both Dothidiomycete cereal pathogens, this could be attributed to their common lifestyle and evolutionary history. Therefore, it could be that both fungal species employ stuA-like genes as key regulators of asexual sporulation and pathogenicity, in addition to other un-characterized downstream genes.
Deletion of stuA homologs has been carried out in both Ascomycete and Basidiomycete fungi, demonstrating that this gene may have essential roles within the wider Dikarya. For example, deletion of the Magnaporthe oryzae stuA homolog, Mstu1, results in a reduction in asexual reproduction and attenuated pathogenicity due to defects in appressorium formation (Nishimura et al., 2014). Deletion of the stuA homolog, Ust1, in Ustilago maydis also impacts vegetative growth and pathogenicity (García-pedrajas et al., 2010). Therefore, stuA may be a core gene among both the Ascomycota and Basidiomycota with essential roles in vegetative growth, pathogenicity and sporulation.
From our initial work here, we propose a rudimentary genetic pathway controlling asexual sporulation in Z. tritici. In this pathway, the ZtStuA gene encodes an upstream positive regulator of asexual sporulation (ZtStuA). Downstream of ZtStuA are the ZtFlbC and ZtBrlA2 genes which encode ZtFlbC and ZtBrlA2 respectively, and these are required for pycnidiospore formation (Figure 10).
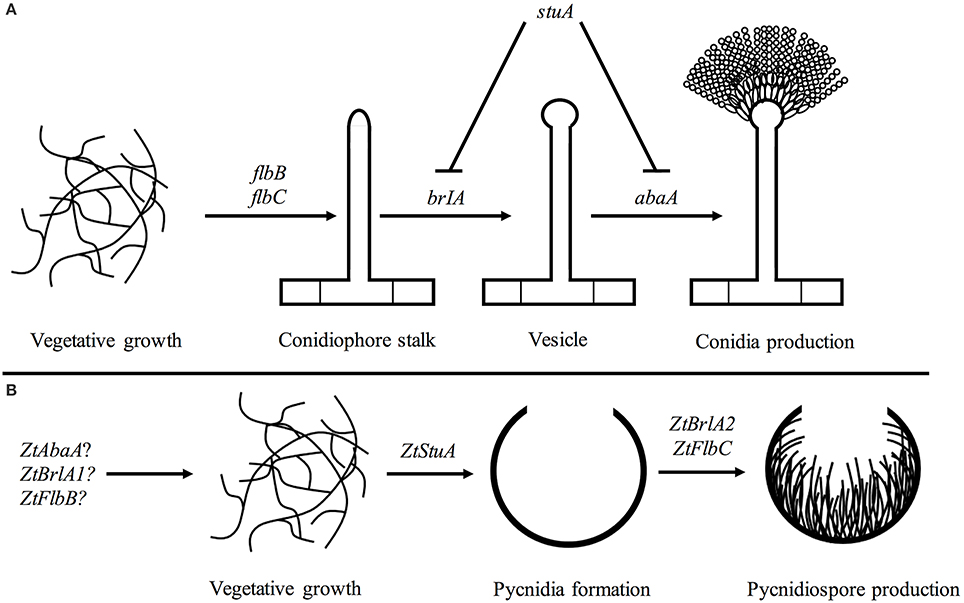
Figure 10. Pathways involved in asexual sporulation in A. nidulans, compared to the pathway proposed for Z. tritici; (A) in A. nidulans, asexual sporulation is controlled by the key regulatory genes including brlA and abaA. These are required for formation of the conidiophore stalk, vesicle and conidia. The stuA gene is required for spatial patterning of the conidiophore. The flbB and flbC genes are required for activation of asexual sporulation (reviewed in Adams et al., 1998); (B) in Z. tritici the ZtStuA gene may encode a key positive regulator of pycnidia production, with ZtFlbC and ZtBrlA2 acting downstream to induce pycnidiospore formation. Disruption of ZtAbaA, ZtBrlA1, and ZtFlbB had no impact on pathogenicity or asexual sporulation in Z. tritici, highlighting differences in regulation of sporulation between A. nidulans and Z. tritici, however these genes may be involved in other Z. tritici processes such as sexual sporulation.
It is likely that additional pathways involved in light signaling may also act upstream of ZtStuA, ZtFlbC and ZtBrlA2 and initiate asexual sporulation in Z. tritici, however, the interaction of these Z. tritici elements is yet to be determined.
Taken together, our findings identify some of the key genes involved in controlling asexual sporulation in this pathogen. We have shown that Z. tritici may be using a similar genetic pathway to control initiation of asexual sporulation as A. nidulans, however some components of the Z. tritici sporulation pathway may differ and could employ currently un-characterized genes. A comparison of the established functions of key A. nidulans asexual sporulation genes and the Z. tritici homologs examined in this study are summarized in Table 3.
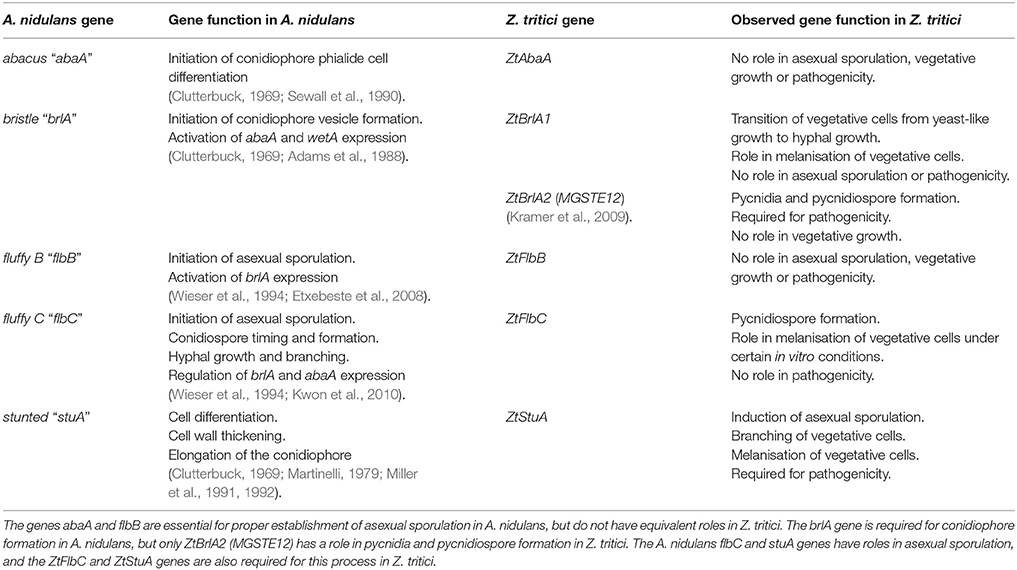
Table 3. Comparison of the functions of the key genes regulating asexual sporulation in Aspergillus nidulans, and the newly established functions of their homologs in Zymoseptoria tritici from the current study.
The difference in regulatory pathways (e.g., lack of involvement of ZtAbaA or ZtFlbB) used by Z. tritici and A. nidulans to control asexual sporulation may be explained by the divergent lifestyles and evolutionary histories of the two fungi. A. nidulans conidiophores develop as elongated stalks among the colony of exposed hyphae. In contrast, Z. tritici pycnidia form within the enclosed space of the plant sub-stomatal cavity. Although the upstream initiator of sporulation, stuA, may be shared by these two fungi, downstream regulators may differ because of the variation in fungal fruiting body structure.
From these results we suggest that the ZtStuA gene is a key regulator of asexual reproduction in Z. tritici which controls initiation of pycnidia development and is likely to be a key regulator in other related species. The ZtFlbC and ZtBrlA2 genes may be downstream regulators which regulate pycnidia and pycnidiospore production. The Z. tritici genes such as ZtStuA, ZtFlbC, and ZtBrlA2 could therefore be explored as targets for sporulation inhibitors. The findings from this project therefore help further understanding of an essential developmental process in this under-studied pathogen and open up promising avenues for research in Z. tritici.
Author Contributions
AB devised the project, and contributed to experimental design and analysis. AT designed and performed the experiments, analyzed the data, and wrote the manuscript. AB and GF supervised the project and provided feedback on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AT was supported by the BBSRC SWBio Doctoral Training Partnership. We thank Josie Miller for assistance in construction of the ΔztflbB mutant strains. This work was performed under DEFRA license number 51046-198767.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01859/full#supplementary-material
References
Adams, T. H., Boylan, M. T., and Timberlake, W. E. (1988). brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362. doi: 10.1016/0092-8674(88)90198-5
Adams, T. H., Wieser, J. K., and Yu, J.-H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54.
Andrianopoulos, A., and Timberlake, W. E. (1994). The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14, 2503–2515.
Bowler, J., Scott, E., Tailor, R., Scalliet, G., Ray, J., and Csukai, M. (2010). New capabilities for Mycosphaerella graminicola research. Mol. Plant Pathol. 11, 691–704. doi: 10.1111/j.1364-3703.2010.00629.x
Carroll, A. M., Sweigard, J. A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 41:22. doi: 10.4148/1941-4765.1367
Clutterbuck, A. J. (1969). A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63, 317–327.
Dancer, J., Daniels, A., Cooley, N., and Foster, S. (1999). “Septoria tritici and Stagonospora nodorum as model pathogens for fungicide discovery,” in Septoria on Cereals: A Study of Pathosystems, eds J. A. Lucas, P. Bowyer, and H. M. Anderson (Wallingford: CAB International), 316–331.
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Derbyshire, M. C., Michaelson, L., Parker, J., Kelly, S., Thacker, U., Powers, S. J., et al. (2015), Analysis of cytochrome b5 reductase-mediated metabolism in the phytopathogenic fungus Zymoseptoria tritici reveals novel functionalities implicated in virulence. Fung. Genet. Biol. 82, 69–84. doi: 10.1016/j.fgb.2015.05.008.
Duncan, K. E., and Howard, R. J. (2000). Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola. Mycol. Res. 104, 1074–1082. doi: 10.1017/S0953756299002294
Etxebeste, O., Garzia, A., Espeso, E. A., and Ugalde, U. (2010). Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18, 569–576. doi: 10.1016/j.tim.2010.09.007
Etxebeste, O., Ni, M., Garzia, A., Kwon, N. J., Fischer, R., Yu, J. H., et al. (2008). Basic-zipper-type transcription factor flbb controls asexual development in Aspergillus nidulans. Eukaryotic Cell 7, 38–48. doi: 10.1128/EC.00207-07
Eyal, Z., Scharen, A. L., Prescott, J. M., and van Ginkel, M. (1987). The Septoria Diseases of Wheat: Concepts and Methods of Disease Management, 1st Edn. CIMMYT.
García-pedrajas, M. D., Baeza-Montañez, L., and Gold, S. E. (2010). Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain. Mol. Plant Microbe Interact. 23, 211–222. doi: 10.1094/MPMI-23-2-0211
Hu, P., Wang, Y., Zhou, J., Pan, Y., and Liu, G. (2015). AcstuA, which encodes an APSES transcription regulator, is involved in conidiation, cephalosporin biosynthesis and cell wall integrity of Acremonium chrysogenum. Fungal Genet. Biol. 83, 26–40. doi: 10.1016/j.fgb.2015.08.003
IpCho, S. V., Tan, K. C., Koh, G., Gummer, J., Oliver, R. P., Trengove, R. D., et al. (2010). The transcription factor StuA regulates central carbon metabolism, mycotoxin production, and effector gene expression in the wheat pathogen Stagonospora nodorum. Eukaryot. Cell 9, 1100–1108. doi: 10.1128/EC.00064-10
Kema, G. H. J., Yu, D., Rijkenberg, F. H. J., Shaw, M. W., and Baayen, R. P. (1996). Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathol 86, 777–786.
Keon, J., Antoniw, J., Carzaniga, R., Deller, S., Ward, J. L., Baker, J. M., et al. (2007). Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 20, 178–193. doi: 10.1094/MPMI-20-2-0178
Kramer, B., Thines, E., and Foster, A. J. (2009). MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet. Biol. 46, 667–681.doi: 10.1016/j.fgb.2009.06.001
Kwon, N. J., Garzia, A., Espeso, E. A., Ugalde, U., and Yu, J. H. (2010). FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77, 1203–1219 doi: 10.1111/j.1365-2958.2010.07282.x
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Liu, D., Coloe, S., Baird, R., and Pederson, J. (2000). Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38:471.
Lovell, D. J., Parker, S. R., Hunter, T., Welham, S. J., and Nichols, A. R. (2004) Position of inoculum in the canopy affects the risk of Septoria tritici blotch epidemics in winter wheat. Plant Pathol. 53 11–21. doi: 10.1046/j.1365-3059.2003.00939.x
Marshall, M. A., and Timberlake, W. E. (1991). Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 11, 55–62.
Martinelli, S. D. (1979). Phenotypes of double conidiation mutants of Aspergillus nidulans. J. Gen. Microbiol. 114, 277–287.
Mehrabi, R., Ben M'Barek, S., van der Lee, T. A., Waalwijk, C., de Wit, P. J. G. M., and Kema, G. H. J. (2009). Gα and Gβ proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola. Eukaryot. Cell 8, 1001–1013.doi: 10.1128/EC.00258-08
Miller, K. Y., Toennis, T. M., Adams, T. H., and Miller, B. L. (1991). Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. 227, 285–292.
Miller, K. Y., Wu, J., and Miller, B. L. (1992). StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6, 1770–1782.
Mirabito, P. M., Adams, T. H., and Timberlake, W. E. (1989). Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57, 859–868.
Mirzadi Gohari, A., Mehrabi, R., Robert, O., Ince, I. A., Boeren, S., Schuster, M., et al. (2014). Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 15, 394–405.
Motteram, J., Küfner, I., Deller, S., Brunner, F., Hammond-Kosack, K. E., Nürnberger, T., et al. (2009). Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 22, 790–799. doi: 10.1094/MPMI-22-7-0790
Nishimura, M., Fukada, J., Moriwaki, A., Fujikawa, T., Ohashi, M., Hibi, T., et al. (2014). Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci. Biotechnol. Biochem. 73, 1779–1786. doi: 10.1271/bbb.90146
Ohara, T., and Tsuge, T. (2004). FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, fungal plant pathogen Fusarium oxysporum. Eukaryot. Cell 3, 1412–1422. doi: 10.1128/EC.3.6.1412-1422.2004
Palmer, C. L., and Skinner, W. (2002). Pathogen profile Mycosphaerella graminicola: latent infection, crop devastation and genomics. Mol. Plant Pathol. 3, 63–70. doi: 10.1046/j.1464-6722.2002.00100.x
Park, H., and Yu, J. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Ponomarenko, A., Goodwin, S. B., and Kema., G. H. J. (2011). Septoria Tritici Blotch (STB) of Wheat. Available online at: https://www.apsnet.org/edcenter/intropp/lessons/fungi/ascomycetes/Pages/Septoria.aspx [Accessed on 20th March, 2018]
Prade, R. A., and Timberlake, W. E. (1993). The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 12, 2439–2447.
Rudd, J. J., Kanyuka, K., Hassani-Pak, K., Derbyshire, M., Andongabo, A., Devonshire, J., et al. (2015). Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle. Plant Physiol. 167, 1158–1185. doi: 10.1104/pp.114.255927
Sewall, T. C., Mims, C. W., and Timberlake, W. E. (1990). abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 2, 731–739.
Sheppard, D. C., Doedt, T., Chiang, L. Y., Kim, H. S., Chen, D., Nierman, W. C., et al. (2005). The Aspergillus fumigatus StuA protein governs the up- regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16, 5866–5879. doi: 10.1091/mbc.e05-07-0617
Skinner, W. (2001). Non-Pathogenic Mutants of Mycosphaerella Graminicola. Doctoral thesis, Bristol: University of Bristol, UK.
Tong, X., Zhang, X., Plummer, K. M., Stowell, K. M., Sullivan, P. A., and Farley, P. C. (2007). GcSTUA, an APSES transcription factor, is required for generation of appressorial turgor pressure and full pathogenicity of Glomerella cingulata. Mol. Plant Microbe Interact. 20, 1102–1111. doi: 10.1111/mpp.12249
Torriani, S. F., Melichar, J. P., Mills, C., Pain, N., Sierotzki, H., and Courbot, M. (2015). Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet. Biol. 79, 8–12. doi: 10.1094/MPMI-20-9-1102
Tottman (1987). The decimal code for the growth stages of cereals, with illustrations. Ann. Appl. Biol. 110, 441–454.
Wieser, J., Lee, B. N. L., Fondon, J. W. III., and Adams, T. H. (1994). Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27, 62–69.
Keywords: Zymoseptoria tritici, Aspergillus nidulans, ascomycete, septoria tritici blotch, asexual sporulation, pycnidia, pycnidiospores
Citation: Tiley AMM, Foster GD and Bailey AM (2018) Exploring the Genetic Regulation of Asexual Sporulation in Zymoseptoria tritici. Front. Microbiol. 9:1859. doi: 10.3389/fmicb.2018.01859
Received: 11 May 2018; Accepted: 24 July 2018;
Published: 14 August 2018.
Edited by:
Mohamed Hijri, Université de Montréal, CanadaReviewed by:
Todd B. Reynolds, University of Tennessee, Knoxville, United StatesBirinchi Kumar Sarma, Banaras Hindu University, India
Copyright © 2018 Tiley, Foster and Bailey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna M. M. Tiley, anna.tiley@ucd.ie
Andy M. Bailey, andy.bailey@bristol.ac.uk
†Present Address: Anna M. M. Tiley, Molecular Plant Pathology group, School of Agriculture and Food Science, O'Brien Centre for Science, University College Dublin, Dublin, Ireland
 Anna M. M. Tiley
Anna M. M. Tiley Gary D. Foster
Gary D. Foster Andy M. Bailey
Andy M. Bailey