Abstract
Sulfate-reducing bacteria are a group of microorganisms that use sulfate as an electron acceptor. These bacteria are useful in the bioremediation of heavy metal pollution since they can reduce/precipitate metals. Previously, we identified the Alishewanella strain WH16-1 from soil of a copper and iron mine and determined that it can reduce sulfate and chromate and that it was tolerant to many heavy metals. In this study, we investigated the chromate reduction mechanism of strain WH16-1 through Tn5 transposon mutagenesis. A cytochrome bd (cytbd) Tn5 mutant was generated (Δcytbd), and a detail analysis showed that the following: (1) gene cydE (coding for a GbsR-type regulator) was co-transcribed with the two subunits coding genes of the Cytochrome bd complex (Cytbd), namely, cydA and cydB, based on RT-PCR analysis, and similar gene arrangements were also found in other Alteromonadaceae family strains; (2) the chromate resistance level was dramatically decreased and chromate reduction efficiency also decreased in strain Δcytbd compared to the wild-type and a complemented strain (Δcytbd-C); (3) Cytbd could catalyze the decomposition of H2O2 according to the analyses of H2O2 decomposition ability, cellular H2O2 contents, H2O2 inhibition zone, and H2O2 sensitivity tests; (4) surprisingly, chromate was not an inducer of the expression of Cytbd, but sulfate induced expression of Cytbd, and sulfate/sulfide resistance levels were also decreased in the Δcytbd strain; (5) the addition of sulfate enhanced the chromate resistance level and reduction efficiency; (6) Cytbd expression was repressed by CydE and derepressed by sulfate based on an in vivo bacterial one hybrid system and in vitro EMSA tests; and (7) DNA footprinting and short-fragment EMSA tests revealed two binding sites of CydE in its promoter region. All these results showed that Cytbd is negatively regulated by CydE and derepressed by sulfate. In addition, Cytbd contributes to the resistance of sulfate and sulfide, and sulfide could be used as a reductant to reduce chromate. Moreover, Cytbd is essential to decompose H2O2 to decrease cellular oxidative stress. Thus, the regulation and function of Cytbd may explain why sulfate could enhance chromate reduction.
Introduction
Sulfate-reducing bacteria (SRB) are a diverse group of prokaryotes that use sulfate as the terminal electron acceptor and produce H2S (Muyzer and Stams, 2008). They are widely distributed and play a key role in the environment (Muyzer and Stams, 2008; Barton and Fauque, 2009). Various SRB have exhibited great potential for environmental bioremediation applications, such as participating in the precipitation of heavy metals to produce metal sulfides (Xia et al., 2016; Zhou et al., 2016), reduction of toxic metals (Barton and Fauque, 2009), degradation of azo dyes (Pandey et al., 2007), and nitroaromatic compound respiration (Barton and Fauque, 2009).
Chromate [Cr(VI)] is highly soluble and can easily cross cellular membranes. Once inside the cell, chromate exhibits a variety of toxic, mutagenic, and carcinogenic effects since it induces reactive oxidative species and affects both DNA and protein functions (O’Brien, 2003; Sobol and Schiestl, 2012). However, its reduction product, Cr(III), is insoluble and has low toxicity (Dhal et al., 2013; Viti et al., 2014). In addition to Cr(VI) reduction, other bacterial Cr(VI) detoxification mechanisms have been found, such as efflux (Viti et al., 2014), reduction of cellular oxidative stress (Ramírez-Díaz et al., 2007; Branco et al., 2008), and DNA repair (Ramírez-Díaz et al., 2007; Viti et al., 2014). In addition, sulfur metabolism is found to be relevant to Cr(VI) detoxification in many bacteria (Cheung and Gu, 2007; Ramírez-Díaz et al., 2007; Thatoi et al., 2014; Viti et al., 2014; Joutey et al., 2015).
Chromate is chemically analogous to sulfate and enters cells mediated by the sulfate ABC transporter CysPUWA in various bacteria (Viti et al., 2014). Some SRB are also chromate-reducing bacteria (CRB) (Viti et al., 2014; Xia et al., 2016). Accordingly, Cr(VI) induces the expression of the sulfate transporter and competes with sulfate in some bacteria (Viti et al., 2014). Moreover, the products of sulfur assimilation are involved in Cr(VI) detoxification. H2S, cysteine, and glutathione (GSH) are capable of directly reducing Cr(VI) (Cheung and Gu, 2007; Thatoi et al., 2014; Joutey et al., 2015). In addition, GSH also plays an important role in maintaining cellular sulfhydryl groups in their reduced form when exposed to oxidative stress induced by Cr(VI) (Presnell et al., 2013; Viti et al., 2014). However, many details concerning the effects of sulfur metabolism on Cr(VI) detoxification remain unclear.
Cytbd is a terminal respiratory oxidase found in many prokaryotes and is composed of two subunits, CydA and CydB (Giuffre et al., 2014). CydC and CydD are also needed for the assembly of Cytbd in Escherichia coli (Borisov et al., 2011), while CydX is also essential for the activity of Cytbd in some bacteria (Sun et al., 2012; VanOrsdel et al., 2013; Chen H. et al., 2015). Cytbd is involved in energy supply, bacterial virulence, and resistance to oxidative and nitrosative stresses (Borisov et al., 2013; Giuffre et al., 2014; Roop et al., 2015). Recently, Cytbd was also found to be associated with sulfide resistance in E. coli, since sulfide could inactivate heme–copper family respiratory oxygen reductases (cytochrome bo3) but not the copper-free Cytbd (Forte et al., 2016; Korshunov et al., 2016). In addition, the expression of Cytbd is regulated by the transcriptional regulators Arc, Fnr, and CydR, depending on environmental oxygen concentrations in E. coli and Azotobacter vinelandii (Wu et al., 1997; Borisov et al., 2011). Potential regulator genes containing helix-turn-helix (HTH) conserved domain sequences were observed adjacent to the cytbd operon in some bacteria (Degli Esposti et al., 2015). However, these potential regulators, such as the GbsR type (Nau-Wagner et al., 2012; Lee et al., 2013), have not been reported to regulate the expression of Cytbd. Furthermore, no study concerning the relevance between Cytbd and chromate resistance has been reported thus far.
Alishewanella sp. WH16-1 (=CCTCC M201507) was isolated from soil of a copper and iron mine. It possesses great potential in metal bioremediation since it reduces sulfate and chromate or generates CdS/PbS precipitation (Xia et al., 2016, 2018; Zhou et al., 2016). Strain WH16-1 also showed a high tolerance to Cr(VI) (MIC of 45 mM) (Zhou et al., 2016). However, its Cr(VI) resistance and reduction mechanisms remain to be explored. A first step of this study was to investigate the Cr(VI) detoxification mechanism of strain WH16-1 through Tn5 transposon mutagenesis. Later, we found that Cytbd was relevant to chromate, sulfate, and sulfide resistance. Interestingly, the transcription of cytbd was repressed by a GbsR-type regulator (named CydE) and depressed by sulfate.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1, and the primers are listed in Supplementary Table S2. Alishewanella sp. WH16-1, E. coli, and their derivative strains were cultured at 37°C in Luria-Bertani (LB) medium unless otherwise noted. Stock solutions of rifampin (Rif, 50 mg mL−1), kanamycin (Km, 50 mg mL−1), chloramphenicol (Cm, 25 mg mL−1), tetracycline (Tet, 5 mg mL−1), K2CrO4 (1 M), Na2SO4 (1 M), and Na2S (0.1 M) were added when required.
Transposon Mutagenesis and Construction of a Complemented Strain
To identify the molecular mechanism of Cr(VI) detoxification of strain WH16-1, Tn5 transposon mutagenesis was used for screening Cr(VI) resistance genes. Transposon insertion mutants were generated with a suicide plasmid pRL27 (Larsen et al., 2002) transferred from the donor strain E. coli S17-1 to the recipient strain WH16-1 using the filter mating method (Smith and Guild, 1980). After conjugation, the Tn5 (Kmr) transposon was randomly inserted into the chromosome DNA of strain WH16-1, generating a library of insertion mutants. Selection was done on LB plates with Rif (50 μg/mL) and Km (50 μg/mL) to obtain strains in which transposition had occurred. The transconjugants were then plated on two LB plates with or without 20 mM K2CrO4, and the colonies that were unable to grow in the presence of K2CrO4 were reserved and subjected to further analyses. Cloning of genes neighboring the Tn5 transposon was performed according to the plasmid rescue method described before (Chen F. et al., 2015). The resulting neighboring sequences were searched against the whole genome of strain WH16-1 (Xia et al., 2016) using the NCBI BLAST server.
To identify the function of Cytbd, a complemented strain was constructed. The whole cytbd operon was cloned into the pCT-Zori plasmid using SacI and HindIII restriction enzyme sites. The generated plasmid was transferred into the mutant strain Δcytbd by conjugation from E. coli S17-1 to obtain a complemented strain, Δcytbd-C.
Analysis of cytbd Operon and Co-transcription
For analysis of cytbd conservation, homologous operon sequences from members of the Alteromonadaceae were selected from their genomes. They were Alteromonas macleodii HOT1A3T (NZ_CP012202), Alteromonas marina AD001T (NZ_JWLW01000010), Alteromonas sp. Mex14 (CP018023), Alteromonas sp. Nap 26 (LSMP01000036), Alteromonas australica H17T (NZ_CP008849), Salinimonas chungwhensis DSM 16280T (NZ_KB899391), Glaciecola pallidula DSM 14239T (NZ_AUAV01000023), Alishewanella agri BL06T (AKKU01000001), Alishewanella jeotgali KCTC 22429T (AHTH01000001), Paraglaciecola arctica BSs20135T (NZ_BAEO01000055), and Lacimicrobium alkaliphilum YelD216T (NZ_CP013650). Phylogenetic analysis was carried out based on the cytbd operon (GbsR family regulator CydE, CydA, and CydB) amino acid sequences. The analysis was performed by MEGA 6.0 (Tamura et al., 2013) with a neighbor joining algorithm, and 1,000 bootstrap repetitions were computed to estimate the reliability of the tree. In addition, the operon arrangement in these strains was also analyzed.
For co-transcription analysis, strain WH16-1 was incubated to an OD600 of approximately 0.3 in 100 mL LB broth, followed by incubation with 1 mM K2CrO4 for 3 h. Total RNA was extracted by Trizol reagent (Invitrogen), and DNA was removed by digestion with DNase I (Takara). Reverse transcription was conducted with a RevertAid First Strand cDNA Synthesis Kit (Thermo) with 300 ng total RNA for each sample. The resulting cDNA was used as a template to amplify the fragments between genes in the cytbd operon. Genomic DNA was used as a positive control. The total RNAs of strain WH16-1 and ddH2O were used as negative controls. Primers are shown in Supplementary Table S2.
Reporter Gene Construction
The putative promoter and promoter-cydE regions (Supplementary Figure S1A) were each PCR amplified from genomic DNA of strain WH16-1. Each DNA fragment was then cloned into plasmid pLSP-kt2lacZ using EcoRI–BamHI restriction enzyme sites. The resulting constructs were designated as pLSP-promoter-lacZ (Supplementary Figure S1B) and pLSP-promoter-cydE-lacZ (Supplementary Figure S1C). E. coli DH5α containing pLSP-promoter-lacZ or pLSP-promoter-cydE-lacZ was incubated in LB medium. Overnight cultures were diluted 100 times with fresh medium and incubated for approximately 4 h (OD600 approximately 0.3). Next, Na2SO4 (0, 5, 25, and 50 mM) and K2CrO4 (0, 1, and 5 mM) were added to the cultures. The cultures were then distributed into tubes after a 6-h incubation. β-Galactosidase enzymatic assays were performed using the method described by Li et al. (2015).
Chromate/Sulfate/Sulfide Sensitivity and Chromate Reduction Assay
Strains WH16-1, Δcytbd, and Δcytbd-C were each inoculated into 5 mL LB and incubated at 37°C with shaking at 150 rpm. When the OD600 reached approximately 0.8–1.0, the strains were each inoculated into 100 mL LB with the presence of 500 mM Na2SO4, 200 μM Na2S, 3 mM K2CrO4, or no addition. Na2SO4 powder was added to LB medium before sterilization, while K2CrO4 and Na2S were added from stock solutions. In addition, LB plates with 0 or 3 mM K2CrO4 were used for Cr(VI) sensitivity analysis. For observing Cr(VI) reduction, strains were incubated in LB broth with 1 mM K2CrO4. To maintain consistent growth conditions, K2CrO4 was added when OD600 reached 0.6. At designated times, culture samples were taken for measuring OD600 and chromate amounts by spectrophotometry (DU800, Beckman) and atomic absorption spectrometry (AAS; 986A, Beijing Puxi General Instrument Co., Beijing, China), respectively.
Effects of Cytbd on Cellular Oxidative Stress
To analyze the effects of Cytbd on oxidative stress, membrane proteins of strains WH16-1, Δcytbd, and Δcytbd-C were extracted to react with H2O2 and chromate. The membrane protein extraction was performed as described previously by Das et al. (2005), and the protein concentration was determined by the Lowry method (Lowry et al., 1951). Then, 10 mg/L of membrane proteins was reacted with 10 mM hydroquinone and 10 mM H2O2 or K2CrO4 in Tris–HCl (pH 8.5) buffer for 30 min under anoxic conditions in a N2 chamber. The residual H2O2 was measured by the Amplex red/horseradish peroxidase assay (Mishin et al., 2010), and chromate concentrations were determined as mentioned above.
To gain more insight into the effects of Cytbd on cellular oxidative stress, cellular H2O2 contents of the WH16-1, Δcytbd, and Δcytbd-C strains were determined. The strains were incubated to an OD600 of approximately 0.3 in 100 mL LB. K2CrO4 was then added to the cultures until the final concentrations reached 1 mM. Cells were centrifuged and washed twice with potassium phosphate buffer (50 mM, pH 7.7). The pellets were lysed via sonication on ice for 3 min and centrifuged for 5 min at 12,000 rpm to remove particulate materials. H2O2 amounts were measured as mentioned above.
Moreover, inhibition zone and H2O2 sensitivity tests were performed. For the inhibition zone test, cultures of each strain (OD600 approximately 0.8–1.0) were added to LB agar medium, and 200 μL of 3% H2O2 was added to the Oxford cup (Wang et al., 2009). For the H2O2 sensitivity assay, 5 μL overnight cultures of WH16-1, Δcytbd, and Δcytbd-C (OD600 0.8–1.0) were dropwise added onto LB agar media containing various amounts of H2O2 (0, 0.05, 0.1, 0.5, and 1 mM).
Effects of Sulfate on Chromate Reduction and Resistance
Chemically defined medium (CDM) was selected to test the effects of sulfate on Cr(VI) reduction and resistance in strain WH16-1. The components of the CDM medium were the same as previously described (Weeger et al., 1999), except for replacing sodium lactate, magnesium sulfate and sodium sulfate with maltose, magnesium, and sodium chloride, respectively. This medium contained 0.12 mM SO4−2 and no other forms of sulfur. Strain WH16-1 was incubated with or without 100 μM Cr(VI) and additional 0, 5, or 10 mM sulfate in CDM medium. The remaining Cr(VI) in the medium was determined as mentioned above.
Bacterial One-Hybrid System Assay
The DNA binding activity of CydE was tested in vivo with a bacterial one-hybrid system (Guo et al., 2009). The cydE coding sequence was amplified and cloned into the pTRG vector using BamHI–EcoRI restriction enzyme sites to obtain a plasmid pTRG-cydE. The promoter sequence of the cydE (Supplementary Figure S1A) was amplified and inserted directly into XcmI site of pBXcmT, yielding the pBX-promoter plasmid. The next steps were followed as previously described (Guo et al., 2009; Shi et al., 2017). The pTRG-cydE and pBX-promoter plasmids were co-transformed into E. coli XL1-Blue and grew on selective screening medium plates (Guo et al., 2009). In addition, E. coli XL1-Blue containing the pBX-MthspXp and pTRG-Rv3133c plasmids served as the positive controls, while E. coli XL1-Blue containing the empty vectors pBXcmT or pTRG was used as negative controls (Guo et al., 2009; Shi et al., 2017).
Cloning, Expression, and Purification of CydE
The CydE coding sequence was also amplified from DNA of strain WH16-1 using specific primers (Supplementary Table S2) that were designed to contain the restriction sites for BamHI and HindIII. The PCR product was digested with these enzymes and cloned into pET28a generating plasmid pET28a-cydE. After DNA sequencing confirmation, the plasmid was introduced into E. coli BL21 (DE3) cells. CydE was overexpressed by adding 0.1 mM IPTG to cells at an OD600 of 0.3–0.4 that were further cultured for 4 h at 28°C. The cells were then harvested by centrifugation (8,000 rpm for 10 min at 4°C). After washing twice with 50 mM Tris–HCl (pH 8.0), the pellets were lysed via French Press at 120 MPa. Next, the soluble supernatant was mixed with 1 mL pre-equilibrated Ni-NTA His Bind Resin (7sea Biotech) and gently agitated at 4°C for 1 h. The resin was transferred into a 10-mL gravity-flow column and washed with 4 mL Tris–HCl with 200 mM imidazole to elute the miscellaneous proteins. The His-tagged CydE protein was eluted in 1 mL Tris–HCl with 500 mM imidazole, and the eluted fractions were analyzed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The quality and quantity of the proteins were assessed with spectrophotometry (NanoDrop 2000, Thermo) and SDS–PAGE.
Electrophoretic Mobility Shift Assay (EMSA)
The DNA probe of cytbd promoter sequence (Supplementary Figure S1) was generated using the primer pair PromoterF/PrompterR (Supplementary Table S2). The PrompterR primer was labeled by fluorophore FAM when needed. In general, DNA binding assay was performed in a 20 μL reaction volume containing 2 μL 10× binding buffer (10 mM Tris, pH 7.5, 10 mM EDTA, 1 mM KCl, 1 mM DTT, 50% glycerol, and 0.1 mg/mL BSA), 100 ng FAM-labeled probe, and different concentrations (0, 0.1, 0.2, and 0.4 μg) of the purified CydE. For competition assay, 0.2, 1, and 2 μg unlabeled probes were added to reaction mixtures containing 0.4 μg CydE and the 100-ng labeled probe. All reaction mixtures were incubated at 37°C for 30 min before being loaded onto an 8% native polyacrylamide gel (Shi et al., 2017). After 1 h of electrophoresis at 120 V in 0.5× TGE buffer (6 mM Tris, 47.5 mM glycine, 0.25 mM EDTA, pH 8.0), gels were exposed to a phosphor imaging system (Fujifilm FLA-5100). For derepression analysis, 0.4 μg CydE was incubated with different concentrations of Na2SO4 (0, 1, 10, and 100 mM) and K2CrO4 (1, 10, and 100 mM) for 15 min, and then, 100 ng FAM-labeled probe and other components were added. Gel analysis was carried out as described above.
DNA Footprinting
One hundred nanograms of FAM-labeled DNA probe was incubated with 0, 0.2, and 0.4 μg CydE, respectively (the reaction system was the same as in EMSA), then digested by 6 × 10−4 U/μL DNase I (New England Biolabs) for 10 min at room temperature. Next, the reaction was stopped by addition of 50 mM EDTA and incubation in a water bath at 65°C for 10 min. The digested DNA fragments were purified with a PCR clean-up Gel extraction kit (Macherey-Nagel). Samples were analyzed in a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, United States), and the electropherograms were aligned with GeneMapper v3.5 (Applied Biosystems, Foster City, CA, United States). For verification of the binding sites, a short-fragment EMSA test was used. The DNA sequences of the two binding sites identified by DNA footprinting were synthesized by Tsingke (Biological Technology Company, Beijing, China). The process of short-fragment EMSA was performed as described above. The final gel was stained by ethidium bromide.
Results
Characterization of the Chromate-Sensitive Mutants by Transposon Mutagenesis
Random mutants were generated by mobilization of the suicide plasmid pRL27 from the donor strain E. coli S17-1 into the recipient strain WH16-1. Approximately 8,000 Km- and Rif-resistant clones were randomly chosen and initially tested for their ability to grow on LB plates containing 20 mM K2CrO4. After 48 h of incubation, 40 Cr(VI)-sensitive mutants were obtained. Mutation sites were identified in 13 mutants with decreased Cr(VI) resistance. These mutant genes encoded CydB, ChrB, ferredoxin, iron transporter, DNA repair proteins (UvrC, UvrD, RecA, RecB, RecC, and YebC), and three other proteins (ComEC, ScpA, and a hypothetical protein). The cydB mutant (Δcytbd) was selected for this study since it may reveal potentially novel Cr(VI) detoxification mechanisms. The BLAST results showed that the Tn5 was inserted in the middle of the cydB (AAY72_09260) gene.
The cytbd Operon in Strain WH16-1
The cytbd operon sequence is conserved in the Alteromonadaceae strains based on phylogenetic analysis (Figure 1A). Moreover, the gene arrangement is also similar in these strains (Figure 1A). The genes coding for CydE (AAY72_09270) and CydA (AAY72_09265) were identified adjacent to cydB. The operon was located in contig 1 of the genome sequence. To gain more insight, RT-PCR was carried out. The forward and inverse primers used for RT-PCR were designed to overlap each two adjacent genes. The results of RT-PCR showed that DNA fragments between the three genes (cydE/cydA and cydA/cydB) were amplified with DNA and cDNA templates. It implied that cydE, cydA, and cydB were co-transcribed in an operon (Figure 1B). To verify the function of Cytbd, a complementation experiment was carried out. The complete cytbd operon including cydE, cydA, and cydB was introduced into the mutant strain Δcytbd and confirmed by PCR using primers cydBF/cydBR (Figure 1C) and DNA sequencing. This generated the complemented strain Δcytbd-C.
FIGURE 1

Analysis of cytbd gene operon, cydB mutation, and complementation. (A) The amino acid-based phylogenetic relationship of the cytbd gene cluster and its adjacent gene arrangement in Alteromonadaceae family members. (B)cydE, cydA, and cydB were co-transcribed. (C) Physical evidence of cydB mutation and complementation. Lanes 1–3 are the PCR-amplified products of wild-type, Δcytbd, and Δcytbd-C by primers cydBF/cydBR, respectively. Lane 4 is a negative control. The Tn5 insert fragment is also approximately 1.8 kb. Consequently, the PCR product of the mutant strain is approximately 1.8 kb longer than PCR products of the wild-type and complemented strains.
The Expression of Cytbd Was Induced by Sulfate
Escherichia coli DH5α-pLSP-promoter-lacZ and E. coli DH5α- pLSP-promoter-cydE-lacZ were constructed (Supplementary Figure S1) to test the expression of Cytbd protein. When cells were incubated in LB medium for 4 h without sulfate, the β-galactosidase activity was higher without CydE, indicating that CydE repressed the activity of the cydE promoter (Figure 2A). Furthermore, the β-galactosidase activity of E. coli DH5α-pLSP-promoter-cydE-lacZ was upregulated when sulfate was added (Figure 2A). However, Cytbd was constitutively expressed when chromate or sulfide were added (data not shown).
FIGURE 2

Cytbd is induced by sulfate and contributes to sulfate and sulfide resistance. (A) Sulfate effects on Cytbd expression. β-Galactosidase activities were increased after adding sulfate or removing cydE behind the promoter. The growth of WH16-1 (wild type), Δcytbd (mutant strain), and Δcytbd-C (complemented strain) with 0 (B), 500 mM Na2SO4(C), or 200 μM Na2S (D) in LB medium. The values represent averages and standard deviations of three replicates.
Cytbd Contributes to Sulfide and Sulfate Resistance
The wild-type, mutant, and complemented strains were used in sulfate- and sulfide-sensitivity tests. Cultures containing corresponding strains without sulfate and sulfide were used as controls (Figure 2B). The results showed that with the addition of 500 mM Na2SO4 or 200 μM Na2S, the wild-type strain grew almost as well as the ones without the addition of Na2SO4 or Na2S (Figures 2C vs. B). However, the growth of strain Δcytbd was partially inhibited with the addition of sulfate (Figure 2C) and completely inhibited with the addition of sulfide (Figure 2D), and the complemented strains were partially recovered to the wild-type levels. These results indicated that Cytbd was weakly associated with sulfate resistance, but it was very essential for sulfide resistance in strain WH16-1. Sulfate appeared not to be very toxic to strain WH16-1 since the addition of 500 mM sulfate had almost no effect on its growth.
Cytbd Contributes to Chromate Resistance and Reduction
A chromate sensitivity test was also performed. The chromate sensitivity test was performed on LB plates and in LB medium. The results showed that the chromate resistance of the mutant strain was noticeably weaker than in the wild-type and the complemented strain (Figures 3A,B). The chromate minimal inhibition concentration (MIC) of the mutant strain was 3 mM, while for the wild type, it was 45 mM (Xia et al., 2016). In addition, the Cr(VI) reduction ability of the mutant strain was also somewhat weaker than the wild-type and complemented strain (Figure 3D) under similar growth conditions (Figure 3C).
FIGURE 3

Effects of Cytbd on chromate resistance and reduction. (A) The growth of WH16-1 (wild type), Δcytbd (mutant strain), and Δcytbd-C (complemented strain) with 0 or 3 mM K2CrO4 on an LB medium plate. (B) The growth curve of WH16-1, Δcytbd, and Δcytbd-C with 3 mM K2CrO4 in LB medium. K2CrO4 was added at the beginning (A and B). The growth (C) and K2CrO4 reduction (D) curves of WH16-1, Δcytbd, and Δcytbd-C with 1 mM K2CrO4 in LB medium. To maintain similar growth conditions, K2CrO4 was added until OD600 reached 0.6 (C and D). Every sample was prepared in triplicate, and the results are presented as the mean values.
Cytbd Protects Against Cellular Oxidative Stress
To achieve a better understanding of how Cytbd contributes to chromate resistance, a series of experiments were carried out. First, the membrane protein of the wild-type, mutant, and complemented strain was extracted and reacted with H2O2 and chromate. The H2O2 decomposition activity of the Δcytbd membrane protein was noticeably lower than the wild type and Δcytbd-C (Supplementary Figure S2A), while chromate reduction showed no significant difference (data not shown). Furthermore, the cytoplasmic H2O2 contents were measured to reflect the cellular oxidative stress. Without K2CrO4, there were no significant differences in H2O2 content among strains WH16-1, Δcytbd, and Δcytbd-C (Supplementary Figure S2B). When exposed to 1 mM K2CrO4, the H2O2 contents of all three strains were increased (Supplementary Figure S2B). However, the H2O2 content in the mutant strain was higher than that in the wild-type and the complemented strain with K2CrO4 (Supplementary Figure S2B). A similar result was obtained based on the inhibition zone test for H2O2 sensitivity. The diameter of the inhibition zone of the mutant strain was visibly larger than for the other two strains (Supplementary Figure S2C). This finding means that the mutant strain was more sensitive to H2O2. In addition, the MIC to H2O2 of the mutant strain was 0.5 mM, which was lower than that of the wild-type and the complemented strain (Supplementary Figure S2D).
Interestingly, the mutant strain lost partial capability for H2O2 decomposition. Hence, it was more sensitive to H2O2 compared to the wild-type and complemented strain. As a result, we inferred that the Cytbd catalyzes the reduction of cytoplasmic oxidative stress to enhance Cr(VI) resistance in strain WH16-1, but it does not directly catalyze chromate reduction.
Sulfate Enhances Chromate Resistance Level and Reduction Efficiency
The effects of sulfate on chromate resistance and reduction were examined since sulfur metabolism is relevant to chromate metabolism in many bacteria (Viti et al., 2014), and the above results showed that sulfate and chromate resistance are both associated with Cytbd. The growth of strain WH16-1 showed no significant difference with or without additional Na2SO4 in the absence of added K2CrO4 (data not shown) but was affected after adding K2CrO4 (Figure 4A). However, growth was much better with the addition of Na2SO4 (Figure 4A). Additionally, the Cr(VI) reduction ability of strain WH16-1 increased with increasing concentrations of Na2SO4 (Figure 4B).
FIGURE 4

Effects of sulfate on chromate resistance and reduction. (A) The growth curve of strain WH16-1 with various amounts of additional sulfate in CDM medium. (B) The chromate reduction curve of strain WH16-1 with various additional Na2SO4. Data are shown as the mean of three biological replicates ± SD.
Interaction Between Regulator CydE and the Promoter Region of cytbd Operon
CydE is homologous to the GbsR-type regulator based on the results of BLASTP in NCBI. It shares 18.8% and 19.4% similarities with GbsR and OpcR, respectively. Next, we aligned CydE with the two reported GbsR regulators (Figure 5A). The results showed that CydE harbored the same conserved amino acids as the GbsR-type regulators. These conserved amino acid residues may be involved in DNA binding. GbsR-type regulators usually act as repressors of gene expression (Nau-Wagner et al., 2012; Lee et al., 2013).
FIGURE 5
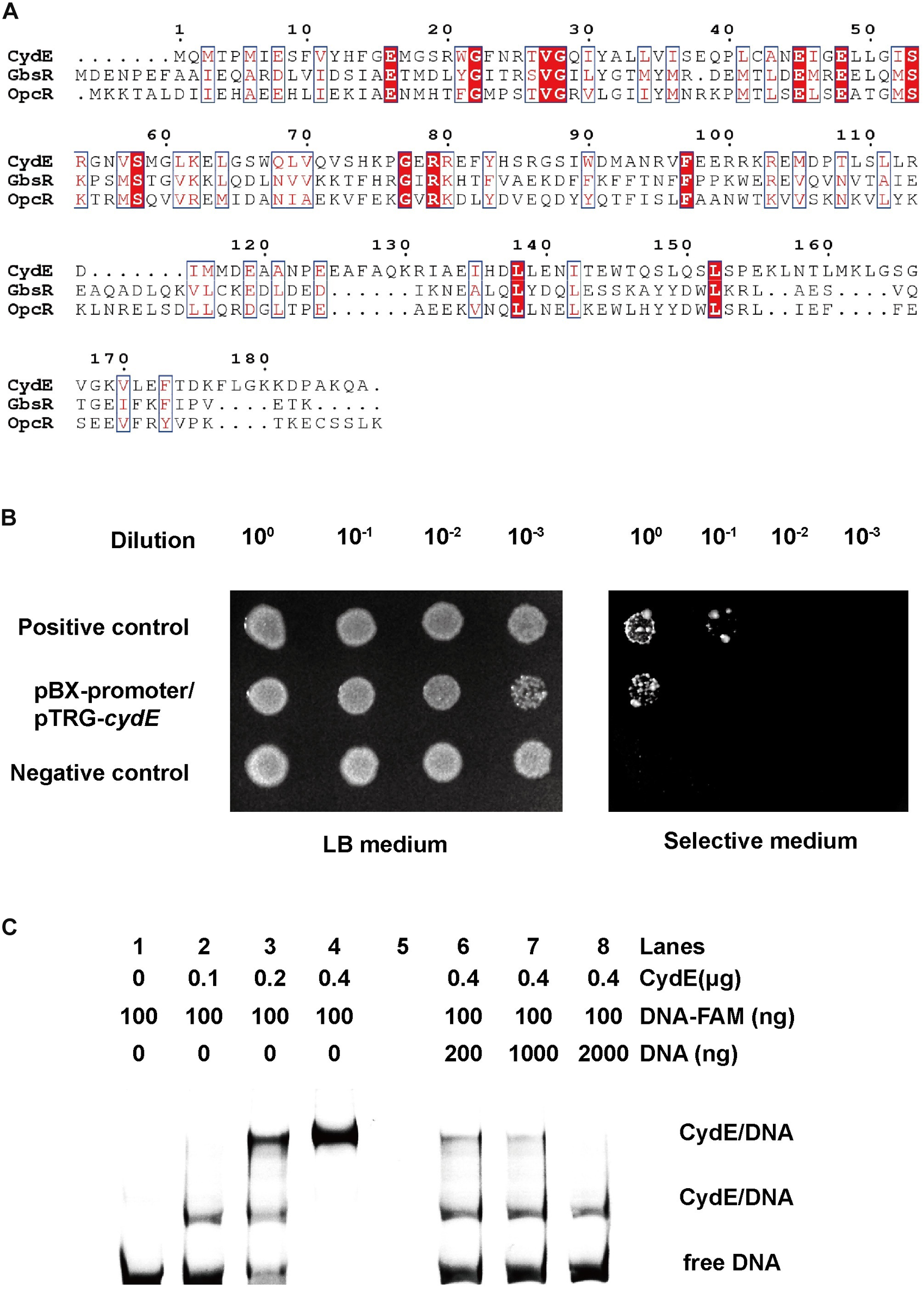
Sequence alignment of CydE and its interaction with the cydE promoter region. (A) Multiple sequence alignment was performed using ClustalW and Espript 3.0. The identical and elevated level of similarity residues are shown in red backgrounds and red boxes, respectively. The amino acid sequences of Bacillus subtilis GbsR (P71015) and Bacillus subtilis_OpcR (O34709) were selected from the UniProt database. (B) Bacterial one-hybrid assay. Co-transformants containing pBX-Mt2031p/pTRG-Rv3133c (Rv3133c protein can integrate with Mt2031p promoter) and empty vector pBXcmT/pTRG (without protein and promoter DNA) were used as positive and negative controls, respectively. Cells of positive and negative controls and the reporter strain containing plasmids pBX-promoter (promoter sequence of cydE) and pTRG-cydE (coding for CydE protein) were grown to an OD600 of 1.0, and 2 μL of each was spotted onto His-selective medium (+3AT, +Strr) and LB plates (–3AT, –Strr). (C) EMSA assay. Lanes 1–4, band shifts were enhanced by increased CydE. Lanes 6–8, FAM-labeled DNA was competed by unlabeled DNA. The result showed two migration bands. It indicated that there are two binding sites for the CydE in the cydE promoter region.
To examine the regulation function of CydE, we first used a bacterial one-hybrid system to test the protein–DNA interaction based on the transcriptional activation of HIS3 (imidazoleglycerol-phosphate dehydratase gene involved in histidine biosynthesis) and aadA (streptomycin resistance gene) (Guo et al., 2009). The promoter of cydE (Supplementary Figure S1A) was cloned into upstream of HIS3–aadA in the reporter vector pBXcmT, while the CydE coding region (Supplementary Figure S1A) was introduced into the pTRG vector. Both constructed vectors were then transferred into a histidine synthesis defective and streptomycin (Str) sensitive strain. The generated strain and positive control strain grew well on the screening plate (Guo et al., 2009) containing 3-amino-1,2,4-triazole (3-AT) and Str, while the negative control strain did not grow. The results demonstrated that CydE could interact with the promoter of the cytbd operon in vivo (Figure 5B).
Next, the purified His-tag CydE (Supplementary Figure S3) and cydE promoter DNA (Supplementary Figure S1A) were used to test the interaction in vitro using EMSA. With increasing amounts of CydE, the free DNA substrates gradually disappeared, while the intensity of the shifted DNA band increased (Figure 5C). Moreover, the unlabeled DNA substrate could competitively inhibit the binding of CydE to the labeled DNA substrate (Figure 5C).
To identify the binding site of CydE, DNA footprinting was carried out. With increasing amounts of CydE, decreases in two sites of the peaks were observed (Figure 6A), indicating that there are two binding sites. This is consistent with the EMSA results. The sequences of the binding sites were TATTTCAGAAATTTCTGAAAGTTCA and GGGATGCGCATATGCAAAT (Figure 6B). The two binding site sequences were synthesized and then incubated with CydE. The EMSA result showed that both binding sites could interact with CydE (Figure 6C).
FIGURE 6

CydE binding sites and derepression analysis. (A) DNA footprinting. FAM-labeled DNA was the same as used in the EMSA and it was incubated with CydE in various amounts. Electropherograms indicated the protection pattern of DNA in different concentrations of CydE. (B) The promoter sequence of cydE. The red and purple highlighted sequences are binding sites 1 and 2, respectively. (C) EMSA for verification of the binding sites of CydE in the promoter region of cydE. The DNA of binding sites 1 and 2 sequences was synthesized and then incubated with CydE protein. The result showed that both the binding sites were shifted after adding CydE. (D) Derepression analysis. The free DNA band is shown with the addition of Na2SO4, while no similar phenomenon was observed with K2CrO4.
CydE Is a Repressor and Can Be Derepressed by Sulfate
To investigate the repression ability of CydE, an EMSA derepression experiment was performed. The results showed that the free DNA increased when more sulfate was added (Figure 6D). However, the phenomenon was not observed when chromate was added (Figure 6D). These results are coincided with those of the lacZ reporter assay (Figure 2A). All these results demonstrated that CydE can repress the expression of the cytbd operon and sulfate addition results in derepression.
Discussion
Alishewanella sp. WH16-1 is a sulfate- and chromate-reducing bacterium. According to our previous study, this strain can produce H2S during cultivation and has complete sulfate assimilation reduction pathway genes (cysCDNHIJ) (Xia et al., 2016; Zhou et al., 2016). It possesses high chromate resistance and reduction ability (Xia et al., 2016, 2018). In this study, we found that Cytbd was involved in sulfide and chromate resistance in Alishewanella sp. WH16-1. The function of Cytbd in sulfide resistance was previously reported in E. coli (Forte et al., 2016; Korshunov et al., 2016), where sulfide can inactivate heme–copper family cytochrome oxidase but not Cytbd (Forte et al., 2016; Korshunov et al., 2016). Under sulfide stress, Cytbd may play a key role in cell respiration. To our knowledge, this is the first report showing that Cytbd is associated with chromate resistance and reduction.
In strain WH16-1, Cytbd is induced by sulfate and is essential for decomposing H2O2 to reduce cellular oxidative stress. In addition, Cytbd contributed to sulfide resistance, and sulfide can be used as a reductant to reduce chromate. These findings explain why Cytbd is important in coupling with chromate stress and the chromate resistance mechanism of strain WH16-1 appears to be indirect (Figure 7). Cytbd also plays an important role in resistance to other environmental stresses such as low oxygen, nitrosative, and oxidative stresses, since Cytbd can use O2, NO, and H2O2 as electron acceptors (Borisov et al., 2013; Giuffre et al., 2014; Roop et al., 2015). The electron transformation models between Cytbd and O2/NO have been clarified (Giuffre et al., 2014). Under low oxygen conditions, Cytbd is regulated by Arc, Fnr, or CydR in E. coli (Borisov et al., 2011) and A. vinelandii (Wu et al., 1997).
FIGURE 7

The proposed mechanism by which Cytbd contributes to sulfide and chromate resistance in Alishewanella sp. WH16-1. CydE, GbsR family regulator; CydA, Cytochrome bd complex subunit A; CydB, Cytochrome bd complex subunit B; ChrA, chromate efflux protein; CysPUWA, sulfate/chromate transporter; CysND, sulfate adenylyltransferases; CysC, adenylylsulphate kinase; CysH phosphoadenylylsulphate reductase; CysIJ, sulfite reductase.
Another important achievement of this study is the finding of a novel regulation mechanism of Cytbd transcription. Previously, a GbsR-family protein was reported as an intracellular choline sensor (Nau-Wagner et al., 2012; Lee et al., 2013). In this study, we identified a GbsR-family protein, CydE, which is Cytbd’s repressor and it is inactivated by high sulfate concentration. In this way, high amount of sulfate can stimulate Cytbd transcription. Thus, sulfate could enhance chromate resistance in Alishewanella sp. WH16-1 (Figure 7). We speculate that numerous factors including the following may cause such enhancement. (i) Sulfate assimilation products such as S2-, Cys, and GSH can directly reduce chromate (Thatoi et al., 2014; Joutey et al., 2015; Qian et al., 2016). (ii) S2- can be used for Fe–S cluster synthesis. A potential Cr(VI) reductase (4Fe–4S ferredoxin, AAY72_06850) was also identified by the Tn5 transposon mutagenesis in this study. Previously, ferredoxin and hydrogenase, which contain the Fe–S cluster as the active group, were reported to be associated with chromate reduction (Chardin et al., 2003). In addition, proteins associated with Fe–S cluster biogenesis, such as IscRs, are involved in multiple stress responses (Liu et al., 2015; Romsang et al., 2015). (iii) Sulfate induces the expression of Cytbd, and Cytbd is essential for chromate resistance and reduction.
On the other hand, sulfate was reported to have no effect on Cr(VI) reduction in some bacteria (Shen and Wang, 1994; Campos et al., 1995; Liu et al., 2006) or even inhibited Cr(VI) reduction in some cases (Wang, 2000; Çetin et al., 2008). The different phenomena reflect various Cr(VI) reduction mechanisms of bacteria. Some bacteria cannot reduce sulfate to produce H2S and do not use sulfate and chromate as terminal electron acceptors (Liu et al., 2006). Accordingly, sulfate has no noticeable effect on chromate reduction in these bacteria. Other bacteria use chromate as a terminal electron acceptor under anaerobic conditions (Wang, 2000; Liu et al., 2006). In some cases, sulfate could inhibit the activity of chromate reductase competitively (Park et al., 2000) and consequently inhibit chromate reduction in these microorganisms. These reports and our results suggest that the chromate detoxification mechanisms of selected bacteria are quite varied.
Conclusion
We showed that Cytbd contributes to chromate resistance, which can be explained by the ability of Cytbd to catalyze the decomposition of H2O2 to protect against H2O2-related oxidative stresses. Furthermore, Cytbd contributes to the resistance of sulfide, and sulfide could act as a reductant to reduce chromate. In addition, Cytbd’s expression is negatively regulated by the GbsR family regulator CydE and derepressed by sulfate. Hence, sulfate could enhance chromate resistance and reduction in Alishewanella sp. WH16-1.
Statements
Author contributions
XX designed, analyzed and interpreted the experiments, and prepared the manuscript. SW participated in the lacZ report gene and sulfate-/sulfide-sensitive experiments. LL and BX participated in the Tn5 transposon test. GW designed the study and revised the draft manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (2016YFD0800702) and National Natural Science Foundation of China (31470226).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01849/full#supplementary-material
References
1
BartonL.FauqueG. (2009). Biochemistry, physiology and biotechnology of sulfate-reducing bacteria.Adv. Appl. Microbiol.6841–98. 10.1016/S0065-2164(09)01202-7
2
BorisovV. B.ForteE.DavletshinA.MastronicolaD.SartiP.GiuffreA. (2013). Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress.FEBS Lett.5872214–2218. 10.1016/j.febslet.2013.05.047
3
BorisovV. B.GennisR. B.HempJ.VerkhovskyM. I. (2011). The cytochrome bd respiratory oxygen reductases.Biochim. Biophys. Acta18071398–1413. 10.1016/j.bbabio.2011.06.016
4
BrancoR.ChungA. P.JohnstonT.GurelV.MoraisP.ZhitkovichA. (2008). The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium(VI) and superoxide.J. Bacteriol.1906996–7003. 10.1128/JB.00289-08
5
CamposJ.Martinez-PachecoM.CervantesC. (1995). Hexavalent-chromium reduction by a chromate-resistant Bacillus sp. strain.Antonie Van Leeuwenhoek68203–208. 10.1007/BF00871816
6
ÇetinD.DönmezS.DönmezG. (2008). The treatment of textile wastewater including chromium (VI) and reactive dye by sulfate-reducing bacterial enrichment.J. Environ. Manage.8876–82. 10.1016/j.jenvman.2007.01.019
7
ChardinB.Giudici-OrticoniM. T.De LucaG.GuigliarelliB.BruschiM. (2003). Hydrogenases in sulfate-reducing bacteria function as chromium reductase.Appl. Microbiol. Biotechnol.63315–321. 10.1007/s00253-003-1390-8
8
ChenF.CaoY.WeiS.LiY.LiX.WangQ.et al (2015). Regulation of arsenite oxidation by the phosphate two-component system PhoBR in Halomonas sp. HAL1.Front. Microbiol.6:923. 10.3389/fmicb.2015.00923
9
ChenH.LuoQ.YinJ.GaoT.GaoH. (2015). Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis.Biochim. Biophys. Acta1850318–328. 10.1016/j.bbagen.2014.10.005
10
CheungK. H.GuJ. D. (2007). Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: a review.Int. Biodeterior. Biodegrad.598–15. 10.1016/j.ibiod.2006.05.002
11
DasA.Silaghi-DumitrescuR.LjungdahlL. G.KurtzD. M.Jr. (2005). Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica.J. Bacteriol.1872020–2029. 10.1128/JB.187.6.2020-2029.2005
12
Degli EspostiM.Rosas-PerezT.Servin-GarciduenasL. E.BolanosL. M.RosenbluethM.Martinez-RomeroE. (2015). Molecular evolution of cytochrome bd oxidases across proteobacterial genomes.Genome Biol. Evol.7801–820. 10.1093/gbe/evv032
13
DhalB.ThatoiH. N.DasN. N.PandeyB. D. (2013). Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: a review.J. Hazard. Mater.25272–291. 10.1016/j.jhazmat.2013.01.048
14
ForteE.BorisovV. B.FalabellaM.ColacoH. G.Tinajero-TrejoM.PooleR. K.et al (2016). The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth.Sci. Rep.6:23788. 10.1038/srep23788
15
GiuffreA.BorisovV. B.AreseM.SartiP.ForteE. (2014). Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress.Biochim. Biophys. Acta18371178–1187. 10.1016/j.bbabio.2014.01.016
16
GuoM.FengH.ZhangJ.WangW.WangY.LiY.et al (2009). Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system.Genome Res.191301–1308. 10.1101/gr.086595.108
17
JouteyN. T.SayelH.BahafidW.El GhachtouliN. (2015). Mechanisms of hexavalent chromium resistance and removal by microorganisms.Rev. Environ. Contam. Toxicol.23345–69. 10.1007/978-3-319-10479-9_2
18
KorshunovS.ImlayK. R.ImlayJ. A. (2016). The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide.Mol. Microbiol.10162–77. 10.1111/mmi.13372
19
LarsenR. A.WilsonM. M.GussA. M.MetcalfW. W. (2002). Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria.Arch. Microbiol.178193–201. 10.1007/s00203-002-0442-2
20
LeeC. H.WuT. Y.ShawG. C. (2013). Involvement of OpcR, a GbsR-type transcriptional regulator, in negative regulation of two evolutionarily closely related choline uptake genes in Bacillus subtilis.Microbiology159(Pt 10), 2087–2096. 10.1099/mic.0.067074-0
21
LiJ.WangQ.LiM.YangB.ShiM.GuoW.et al (2015). Proteomics and genetics for identification of a bacterial antimonite oxidase in Agrobacterium tumefaciens.Environ. Sci. Technol.495980–5989. 10.1021/es506318b
22
LiuH.ZhuangW.ZhangS.RensingC.HuangJ.LiJ.et al (2015). Global regulator IscR positively contributes to antimonite resistance and oxidation in Comamonas testosteroni S44.Front. Mol. Biosci.2:70. 10.3389/fmolb.2015.00070
23
LiuY. G.XuW. H.ZengG. M.LiX.GaoH. (2006). Cr(VI) reduction by Bacillus sp. isolated from chromium landfill.Process Biochem.411981–1986. 10.1016/j.procbio.2006.04.020
24
LowryO. H.RosebroughN. J.FarrA. L.RandallR. J. (1951). Protein measurement with the Folin phenol reagent.J. Biol. Chem.193265–275.
25
MishinV.GrayJ. P.HeckD. E.LaskinD. L.LaskinJ. D. (2010). Application of the amplex red/horseradish peroxidase assay to measure hydrogen peroxide generation by recombinant microsomal enzymes.Free Radic Biol. Med.481485–1491. 10.1016/j.freeradbiomed.2010.02.030
26
MuyzerG.StamsA. J. (2008). The ecology and biotechnology of sulphate-reducing bacteria.Nat. Rev. Microbiol.6441–454. 10.1038/nrmicro1892
27
Nau-WagnerG.OpperD.RolbetzkiA.BochJ.KempfB.HoffmannT.et al (2012). Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor.J. Bacteriol.1942703–2714. 10.1128/JB.06642-11
28
O’BrienT. (2003). Complexities of chromium carcinogenesis: role of cellular response, repair and recovery mechanisms.Mutat. Res.5333–36. 10.1016/j.mrfmmm.2003.09.006
29
PandeyA.SinghP.IyengarL. (2007). Bacterial decolorization and degradation of azo dyes.Int. Biodeterior. Biodegrad.5973–84. 10.1016/j.ibiod.2006.08.006
30
ParkC. H.KeyhanM.WielingaB.FendorfS.MatinA. (2000). Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase.Appl. Environ. Microbiol.661788–1795. 10.1128/AEM.66.5.1788-1795.2000
31
PresnellC. E.BhattiG.NumanL. S.LercheM.AlkhateebS. K.GhalibM.et al (2013). Computational insights into the role of glutathione in oxidative stress.Curr. Neurovasc. Res.10185–194. 10.2174/1567202611310020011
32
QianJ.WeiL.LiuR.JiangF.HaoX.ChenG. H. (2016). An exploratory study on the pathways of Cr (VI) reduction in sulfate-reducing up-flow anaerobic sludge bed (UASB) reactor.Sci. Rep.6:23694. 10.1038/srep23694
33
Ramírez-DíazM. I.Díaz-PérezC.VargasE.Riveros-RosasH.Campos-GarcíaJ.CervantesC. (2007). Mechanisms of bacterial resistance to chromium compounds.Biometals21321–332. 10.1007/s10534-007-9121-8
34
RomsangA.Duang-NkernJ.WirathornW.VattanaviboonP.MongkolsukS. (2015). Pseudomonas aeruginosa IscR-regulated ferredoxin NADP(+) reductase gene (fprB) functions in iron-sulfur cluster biogenesis and multiple stress response.PLoS One10:e0134374. 10.1371/journal.pone.0134374
35
RoopR. M.LeclercJ.RosenfeldE.TraininiM.MartinB.MeuricV.et al (2015). The cytochrome bd oxidase of Porphyromonas gingivalis contributes to oxidative stress resistance and dioxygen tolerance.PLoS One10:e0143808. 10.1371/journal.pone.0143808
36
ShenH.WangY.-T. (1994). Biological reduction of chromium by E. coli.J. Environ. Eng.120560–572. 10.1061/(ASCE)0733-9372(1994)120:3(560)
37
ShiK.FanX.QiaoZ.HanY.McDermottT. R.WangQ.et al (2017). Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4.Sci. Rep.7:43252. 10.1038/srep43252
38
SmithM. D.GuildW. R. (1980). Improved method for conjugative transfer by filter mating of Streptococcus pneumoniae.J. Bacteriol.144457–459.
39
SobolZ.SchiestlR. H. (2012). Intracellular and extracellular factors influencing Cr(VI) and Cr(III) genotoxicity.Environ. Mol. Mutagen.5394–100. 10.1002/em.20679
40
SunY. H.de JongM. F.den HartighA. B.RouxC. M.RolanH. G.TsolisR. M. (2012). The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus.Front. Cell. Infect. Microbiol.2:47. 10.3389/fcimb.2012.00047
41
TamuraK.StecherG.PetersonD.FilipskiA.KumarS. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0.Mol. Biol. Evol.302725–2729. 10.1093/molbev/mst197
42
ThatoiH.DasS.MishraJ.RathB. P.DasN. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review.J. Environ. Manage.146383–399. 10.1016/j.jenvman.2014.07.014
43
VanOrsdelC. E.BhattS.AllenR. J.BrennerE. P.HobsonJ. J.JamilA.et al (2013). The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity.J. Bacteriol.1953640–3650. 10.1128/JB.00324-13
44
VitiC.MarchiE.DecorosiF.GiovannettiL. (2014). Molecular mechanisms of Cr(VI) resistance in bacteria and fungi.FEMS Microbiol. Rev.38633–659. 10.1111/1574-6976.12051
45
WangY.LuZ.WuH.LvF. (2009). Study on the antibiotic activity of microcapsule curcumin against foodborne pathogens.Int. J. Food Microbiol.13671–74. 10.1016/j.ijfoodmicro.2009.09.001
46
WangY. T. (2000). “Microbial reduction of chromate,” inEnvironmental Microbe-Metal Interactions, ed.LovleyD. R. (Washington, DC: American Society of Microbiology Press), 225–235. 10.1128/9781555818098.ch10
47
WeegerW.LievremontD.PerretM.LagardeF.HubertJ. C.LeroyM.et al (1999). Oxidation of arsenite to arsenate by a bacterium isolated from an aquatic environment.Biometals12966–844. 10.1023/A:1009255012328
48
WuG.HillS.KellyM. J.SawersG.PooleR. K. (1997). The cydR gene product, required for regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii, is an Fnr-like protein.Microbiology143(Pt 7), 2197–2207. 10.1099/00221287-143-7-2197
49
XiaX.LiJ.LiaoS.ZhouG.WangH.LiL.et al (2016). Draft genomic sequence of a chromate- and sulfate-reducing Alishewanella strain with the ability to bioremediate Cr and Cd contamination.Stand. Genomic Sci.11:48. 10.1186/s40793-016-0169-3
50
XiaX.WuS.LiN.WangD.ZhengS.WangG. (2018). Novel bacterial selenite reductase CsrF responsible for Se(IV) and Cr(VI) reduction that produces nanoparticles in Alishewanella sp. WH16-1.J. Hazard. Mater.342499–509. 10.1016/j.jhazmat.2017.08.051
51
ZhouG.XiaX.WangH.LiL.WangG.ZhengS.et al (2016). Immobilization of lead by Alishewanella sp. WH16-1 in pot experiments of Pb-contaminated paddy soil.Water Air Soil Pollut.227:399. 10.1007/s11270-016-3040-7
Summary
Keywords
chromate resistance, sulfate reducing, cytochrome bd, Alishewanella, CydE
Citation
Xia X, Wu S, Li L, Xu B and Wang G (2018) The Cytochrome bd Complex Is Essential for Chromate and Sulfide Resistance and Is Regulated by a GbsR-Type Regulator, CydE, in Alishewanella Sp. WH16-1. Front. Microbiol. 9:1849. doi: 10.3389/fmicb.2018.01849
Received
07 October 2017
Accepted
24 July 2018
Published
10 August 2018
Volume
9 - 2018
Edited by
Dennis A. Bazylinski, University of Nevada, Las Vegas, United States
Reviewed by
Toivo Kallas, University of Wisconsin–Oshkosh, United States; A. Andrew Pacheco, University of Wisconsin–Milwaukee, United States
Updates
Copyright
© 2018 Xia, Wu, Li, Xu and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gejiao Wang, gejiao@mail.hzau.edu.cn
This article was submitted to Microbiological Chemistry and Geomicrobiology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.