- 1Laboratoire Chrono-Environnement, UMR CNRS 6249, Université Bourgogne Franche-Comté, Montbéliard, France
- 2Département d’Informatique des Systèmes Complexes, Institut FEMTO-ST (UMR 6174 CNRS), Université Bourgogne Franche-Comté, Besançon, France
- 3Faculté des Sciences et Technologies, Université de Lorraine, Nancy, France
The successful restoration of well-engineered tailings storage facilities is needed to avoid mine tailings problems. This study characterized the bacterial communities from vegetated and non-vegetated soils from a red gypsum landfill resulting from the industrial extraction of titanium. A set of 275 bacteria was isolated from vegetated soil and non-vegetated soil areas and taxonomically characterized using BOX-PCR. The study also evaluated the ability of a subset of 88 isolated bacteria on their ability to produce plant growth promoting (PGP) traits [indoleacetic acid (IAA) production, phosphate solubilization, and siderophore production] and their tolerance to potentially toxic elements (PTEs). Twenty strains were chosen for further analysis to produce inoculum for birch-challenging experiments. Principal component analysis (PCA) showed that the set of pedological parameters (pH, granulometry, carbon, organic matter, and Mg content) alone explained approximately 40% of the differences between the two soils. The highest density of total culturable bacteria was found in the vegetated soil, and it was much higher than that in the non-vegetated soil. The Actinobacteria phyla dominated the culturable soil community (70% in vegetated soil and 95% in non-vegetated soil), while the phyla Firmicutes (including the genus Bacillus) and Bacteroides (including the genera Pedobacter and Olivibacter) were found only in the vegetated soil fraction. Additional genera (Rhizobium, Variovorax, and Ensifer) were found solely in the vegetated soil. The vegetated soil bacteria harbored the most beneficial PGP bacteria with 12% of the isolates showing three or more PGP traits. The strains with higher metal tolerances in our study were Phyllobacterium sp. WR140 (RO1.15), Phyllobacterium sp. WR140 (R01.34), and Streptomyces sp. (R04.15), all isolated from the vegetated soil. Among the isolates tested in challenging experiments, Phyllobacterium (R01.34) and Streptomyces sp. (R05.33) have the greatest potential to act as PGP rhizobacteria and therefore to be used in the biological restoration of tailings dumps.
Introduction
Mining operations produce significant volumes of waste substrates that originated from the physical or chemical treatment of waste rocks from which ores have been extracted (Kumaresan et al., 2017). Such extractive (anthropogenic) activities produce potentially toxic elements (PTEs) that may contaminate the environment and induce human health problems. Artificial substrates generated by these extractive activities have not been subjected to weathering (Cross et al., 2017) and represent substantial volumes of wastes that are vulnerable to water and wind erosion, and thus potentially transported to long distances (Honeker et al., 2017). They are frequently characterized by a poor physico-hydrological structure, resulting in an unstable geochemical nature, and the presence of potentially toxic chemicals (Wang et al., 2017). Factually, management of tailings plans have often been concentrated on their confinement and containment and little attention has been paid to the long-term alteration of the tailings materials, including changes of their biological, chemical or physical, properties (Santini and Banning, 2016). In addition, they contain abundant by-products, which could be potentially used as amendment in land farming, although they obviously encompass substantial abiotic constraints for the survival of plant and microorganisms.
Revegetation is often encouraged on these tailings since it can efficiently control the erosion of tailings particles by wind and water and may advance the landscape of these waste areas (Mendez and Maier, 2008). The successful restoration of tailings storage amenities may be indeed the best technique to limit mine tailings tragedies (Cross et al., 2017). Salisbury et al. (2017) demonstrated the effectiveness of a vegetative cover to retain some PTE in the upper soil horizons for some decades, and thus playing a considerable function in reclaiming contaminated land. Plant may indeed tolerate PTE contamination through various mechanisms (exclusion, hyperaccumulation traits, and microbe functions) that allow growth and reproduction in such severe environments. Nonetheless, the in situ remediation of tailings is likely to necessitate the addition of amendments that may accelerate substrate weathering (Santini and Banning, 2016). The addition of a topsoil to the rooting soil area represents an efficient method to alleviate the abiotic constraints existing in original tailings and is likely to hasten the reappearance of microbial functions (Huang et al., 2012). Choosing plant species that are endemic to the tailings areas is recognized as a suitable choice for a successful revegetation (Wang et al., 2008; Jana et al., 2012; Wanat et al., 2014). Among plant species that readily colonize tailings, Betula species have a recognized ability to quickly colonize bare areas and are characterized by their poor affinity for any specific soil category and their capacity to grow in nutrient poor substrates (Atkinson, 1992; Jana et al., 2012).
There is abundant literature on the characterization of microbial communities from forest or agricultural soils contaminated by PTE or polycyclic aromatic hydrocarbon (PAH) (Tardy et al., 2015; Yergeau et al., 2015; Foulon et al., 2016a,b). Microorganisms occurring in mine tailings have also drew significant interest in the past decade, especially in acid mine drainage dumps (Méndez-García et al., 2015; Bruneel et al., 2017; Gupta et al., 2017; Mesa et al., 2017). However, in other environments with different soil characteristics (bauxite and red gypsum), there are considerably fewer studies. Microorganisms are both relevant indicators of ecological functions and facilitators of the soil metabolic activities that are required for further aboveground-plant reestablishment. However, the mechanisms involved on how microorganisms facilitate restoration of degraded lands such as post-mining lands remain poorly understood. Wubs et al. (2016) reported that the addition of microbial inoculum could foster ecosystem restoration, while emphasizing that the origin of the inoculum was a major factor to promote the establishment of plant communities. Therefore, efforts to characterize endogenous microbial communities from these soils are urgently needed to achieve optimal plant recovery.
Among soil bacteria, Actinobacteria constitute a group of microorganisms found in high amounts in soils and play key roles in the recycling of natural compounds or xenobiotic such as pesticides and PTE, due to their metabolic capacity (Kieser et al., 2000). Currently, Actinobacteria are considered among the most prosperous colonizers in most extreme environments, in contrast to being conventionally considered as endogenous soil and freshwater microorganisms (Álvarez et al., 2017). Actinobacteria can directly promote plant growth by supplying the plant with bacterial-synthesized compounds or by facilitating soil nutrient uptake by the plant (Barka et al., 2016). Actinobacteria may also prevent infection by deleterious microorganisms, which is achieved through biocontrol or antagonism toward soil plant pathogens. Despite these recognized traits, the plant growth promoting (PGP) rhizospheric potential of Streptomyces has been poorly studied, although there are widely recognized as efficient root colonizers and able to cope with unfavorable growth conditions by forming spores. Actinomycetes strains were isolated from birch rhizospheric soils, birch being one of the few native tree able to succeed on a coal mine dump (Ostash et al., 2013).
The present work focused on plant root microbe interactions occurring in a titanium tailings dump that has been naturally recolonized by birch trees, to increase our understanding on how these interactions may be favorable to plant redeployment on such stressful environment. The primary objectives of the present study were: (i) to isolate indigenous bacteria from birch (Betula spp.) based on physiological and morphological traits as well as using 16S rRNA gene sequencing, (ii) to test PGP functional traits from isolated bacteria, such as indoleacetic acid (IAA) production, siderophore production, phosphate solubilization and metal tolerance, and (iii) to study the PGP potential of bacterial isolates under controlled conditions.
Materials and Methods
Study Site Location
The study site belongs to an 80 ha titanium industry effluent treatment unit located at Thann in northeastern France in the southern part of the Alsace plain (47°47′47.7″N 7°08′18.5″E). The study was carried out in a tailing dump consisting of an embankment, where byproducts produced during the neutralization of titanium dioxide extraction effluents have been stored since the 1930s. The dump surface studied here has not been used since the early 2000s, which has allowed natural revegetation with flora that is not very abundant and is distributed in a heterogeneous way. We may thus observe heavily vegetated areas and, in contrast, areas completely bare of vegetation. The flora at this dumpsite is almost exclusively dominated by the woody species Betula sp.
Sampling
The samples were collected on October 27, 2015. They consisted of samples from two areas, a vegetated and a non-vegetated areas (Figure 1). Five birches distributed over the vegetated area were harvested, and the soil fraction adhering to the root system was collected (vegetated soil or VS). For the non-vegetated area, five samples (non-vegetated soil or NVS) were also sampled using an auger at a depth approximately close to that of the root system for the vegetated area. The whole samples were packed on site in plastic bags and transported to the laboratory at a temperature approaching 4°C.
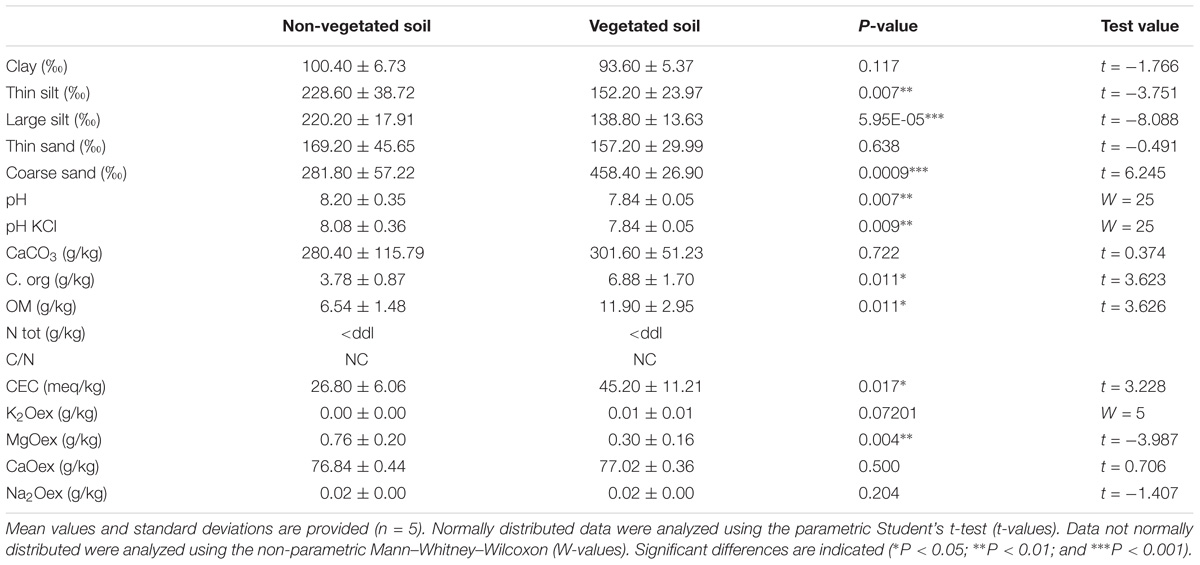
FIGURE 1. (A) Diagram of the sampling site with soils taken in a vegetated area under birch trees (VS, top-left photo, green dotted line) or soil taken in a non-vegetated area (NVS, bottom-left photo). (B) PCA carried out with soil data and soil element concentrations discriminating between the two sampling areas, n = 5.
Pedological Characterization
The soils were dried at 40°C and then ground by hand at 2 mm. The soil analyses were carried out by a service provider in accordance with the following French standards for grinding (NF ISO 11464), residual humidity (NF ISO 11465), granulometry (5 fractions – NFX 31-107), pH water + KCl (NF ISO 10390), total organic carbon and organic matter (NF ISO 14235), total nitrogen (NF ISO 13878), CEC Metson (NFX 31-130), bore soluble boiling water (NFX 31-122), oligoelements, K2O, MgO, CaO, Na2O (French Norm X 31-108), and total available phosphorus (Joret Hebert method French Norm X 31-161).
In addition, pseudo-total concentrations in the soils were measured using inductively coupled plasma atomic emission spectrometry (ICP-AES, Thermo Fischer Scientific, Inc., Pittsburgh, PA, United States) analysis after the acid digestion of 500 mg of a sample in a microwave digestion system (Mars Xpress, CEM, Saclay, France), using a mix of 2 mL of 67% nitric acid, 6 mL of 34% hydrochloric acid, and 2 mL of 48% hydrofluoric acid. To assess the analytical quality, a standard reference material (loamy clay soil) was used. To determine the extractable fractions of PTE, 5 g of 2 mm sieved soil was dried at 60°C for 48 h (or air-dried) and incubated with 50 mL of 10 mM CaCl2 under agitation (40 rpm) for 2 h at room temperature. The mixture was first filtered with ash-free filters, passed hrough a 0.45 μm mesh, and acidified at 2% (v/v) with HNO3 prior to ICP-AES analysis.
Microbial Characterization
The vegetated and non-vegetated soil fractions were homogenized in 45 ml of 10 mM MgSO4 and stirred at 100 rpm for 15 min at room temperature. One milliliter was used to perform serial dilutions in 10-fold series, and 100 μL was plated onto a 284-agar medium (Becerra-Castro et al., 2011b) in duplicate dilutions and kept for 7 days at 25°C. The 284 medium contains (per liter): 6.06 g Tris–HCl, 4.68 g NaCl, 1.49 g KCl, 1.07 g NH4Cl, 0.43 g Na2SO4, 0.2 g MgCl2.6H2O, 0.03 g CaCl2.2H2O, 0.04 g Na2HPO4.2H2O, and 10 mL Fe(III)NH4 citrate solution (containing 48 mg/100 mL) plus micronutrients (1.5 mg FeSO4.7H2O, 0.3 mg H3BO4, 0.19 mg CoCl2.H2O, 0.1 mg MnCl2.4H2O, 0.08 mg ZnSO4.7H2O, 0.02 mg CuSO4.5H2O, and 0.036 mg Na2MoO4.2H2O) adjusted to a pH of 7. The medium was supplemented with a mixture of different carbon sources: lactate (0.7 g/L), glucose (0.5 g/L), gluconate (0.7 g/L), fructose (0.5 g/L), and succinate (0.8 g/L). Culturable bacterial densities were calculated and expressed as CFU per gram dry soil. Single morphotypes were isolated by plating them twice onto 284 medium-agar plates. The isolates were further stored in cryotubes in a brain heart infusion broth (Roth, D) with 15% glycerol glucosate at −80°C.
Genotypic Characterization
DNA Extraction and BOX-PCR
For DNA preparation, the isolates were grown in the 284 liquid medium at 25°C for 7 days at 250 rpm (Gallenkamp Orbital Incubator). After centrifugation, DNA was extracted from the pellets using an EZNA bacterial DNA isolation kit (Omega Bio-Tek, Inc., Norcross, GA, United States) according to the manufacturer’s instructions. The BOX-PCR fingerprinting method was used to group genotypic profiles at a similarity level of 90% as previously described (Becerra-Castro et al., 2011a). BOX reactions were performed in a reaction volume of 25 μL containing 12.5 μL of Ready Mix PCR Master Mix (Thermo Fisher Scientific, Carlsbad, CA, United States), 2 μM BOX A1R primer (5′- CTACGGCAAGGCGACGCTGACG-3′, Eurofins Genomics, Paris, France), and 5 μL of bacterial DNA. DNA amplification was carried out in a thermocycler (Mastercycler gradient, Eppendorf, Hamburg, Germany) under the following conditions: 1 cycle of 5 min at 95°C, 40 cycles of 25 s at 95°C, 35 s at 55°C, and 1.05 min at 72°C with an additional 5 min cycle at 72°C. The amplicons obtained were separated by electrophoresis on a 1.8% agarose gel at 45 V for 3 h. The gel images were analyzed with the software Gel.J (Heras et al., 2015) using the Pearson correlation coefficient and a UPGMA clustering algorithm. VS and NVS bacteria were treated separately.
16S Taxonomic Assignment
PCR was performed on one representative of each BOX group using the following conditions: a volume of 50 μL containing 25 μL AccuStartTM II PCR ToughMix® (2×) (Quantas), 5 μM 27f (Escherichia coli positions 8–27, 5′-AGAGTTTGAT CCTGGCTCAG-3′) and 1492r (E. coli positions 1,492–1,510, 5′-ACGGTTACC TTGTTACGACTT-3′), which were used to amplify nearly full-length 16S rRNA genes (Mark Ibekwe et al., 2007), and 5 μL of cell lysate. The thermocycling conditions were as follows: 1 cycle of 94°C for 3 min, 40 cycles of 25 s at 94°C, 25 s at 49.4°C, and 1.30 min at 72°C. Alignments were performed using the SILVA website1.
Functional Traits
Characterization of PGP Traits
Isolates were screened for their ability to solubilize inorganic phosphate and the production of siderophores, organic acids, and IAA. All of these analyses were conducted for one representative of each BOX group. The ability to solubilize inorganic phosphate was assessed in a modified NBRIP agar medium (1.8%) supplied with 5 g/L of hydroxyapatite and incubated at 28°C for 5 days [10.0 g glucose, 5.0 g MgCl2.6H2O, 0.25 g MgSO4.7H2O, 0.2 g KCl, 0.1 g (NH4)2SO4, and 0.1 g yeast extract in 1 L deionized water adjusted to a pH of 7.0 modified from Nautiyal, 1999]. A clear halo around the bacterial colony indicated the solubilization of mineral phosphate. Siderophore production was detected in a modified 284 liquid medium (without Fe) using the Chrome Azurol S (CAS) method described by Schwyn and Neilands (1987). All glassware used in this assay was previously cleaned with 30% HNO3 followed by washing in distilled water (Cox, 1994).
The ability to produce organic acids was tested on an agar medium containing 0.002% bromocresol purple (per liter medium): 10.0 g glucose, 1.0 g tryptone, 0.5 g yeast extract, 0.5 g NaCl, and 0.03 g CaCl2.2H2O. Colonies forming a yellow halo after 1 day of growth at 28°C indicated a pH change in the medium, and they were considered acid producers. IAA production was evaluated in liquid medium [5.0 g glucose, 1.0 g (NH4)2SO4, 2.0 g K2HPO4, 0.5 g CaCO3, 0.5 g MgSO4.7H2O, 0.1 g NaCl, and 0.1 g yeast extract adjusted to a pH of 7 modified from Sheng et al. (2008); supplemented with 0.5 mg/mL tryptophan]. After 5 days incubation at 28°C, the cultures were centrifuged, and the supernatant was incubated with the Salkowski reagent for 25 min. The production of IAA was identified by the presence of red coloring, and isolates were considered IAA producers when the concentration of IAA determined was more than 4 mg/L culture.
Metal Tolerance
Metal tolerance was tested for Cr, Mn, and Zn using 284 agar medium (see above) supplemented with increasing concentrations of Cr [0.1, 0.25, 0.5, 1.0, 2.5, and 5.0 mM; added as Cr(NO3)3.9H2O], Zn [1.0, 2.5, 5.0, 10.0, and 25.0 mM added as Zn(SO4)2.7H2O] and Mn [5.0, 10.0, and 25.0 mM added as MnSO4.H2O] and incubated at 28°C for 7 days. The maximal tolerable concentration (MTC) of each metal was recorded for one selected isolate of each BOX-group.
Plant Inoculation Experimental Setup and Post-harvest Analysis
Birch seeds were germinated in a commercial potting mixture. Three-month-old birch seedlings were transplanted into pots containing 200 g of soil collected from the study site. After 1 week of plant adaptation, bacterial inoculation was carried out. Fresh cultures of bacterial strains were grown in an 869 liquid medium (Mergeay et al., 1985) for 24 h, harvested by centrifugation (6,000 rpm, 15 min) and re-suspended in 10 mM MgSO4 to a dry mass weight of 0.5 mg/L. Each pot was inoculated with 10 mL of bacterial suspension. The same amount of sterile 10 mM MgSO4 was added to the non-inoculated pots. Six replicates of each plant species were prepared for each inoculation strain. Plants were watered regularly to maintain soil moisture and incubated in a growth chamber in the following climatic conditions: daylight for 16 h (250–300 μmol m−2 s−1), day temperature of 22°C, night temperature of 18°C, and day and night humidities of 30%.
After a 3 months growth period, the plants were harvested and the shoot and root dry weight (DW) yields were determined. The plant material was washed in deionized water, oven-dried at 45°C, weighed and ground. The oven-dried plant material was digested in a 2:1 HNO3:HCl mixture, and the concentrations of P, K, Ca, Mg, Fe, Cd, Pb, and Zn were measured by ICP-AES.
Statistical Analyses
All statistical analyses were performed using R software v. 3.0.2 (R Core Team, 2013). Normality was tested with using Shapiro-Wilk (all data sets), and homoscedasticity was tested with Bartlett’s (abiotic dataset) and Levene (biomass dataset, PGP, and metal tolerance traits) tests using R. Data that were normally distributed were analyzed using a parametric test (Student’s t-test) in R. Data that were not normally distributed were analyzed using a non-parametric Mann–Whitney–Wilcoxon (soil data) or a Kruskal–Wallis (inoculation) test using R. The principal component analysis (PCA) was performed using the R ade4 package. Data expressed as % (PGP and metal tolerance traits) were analyzed using a chi-squared test in R.
Results and Discussion
Pedological Characterization of the Two Areas
The sampling zone where the vegetation was found (Betula pendula) is separated by approximately 60 m from the non-vegetated area (Figure 1A). Investigations carried out at the physico-chemical level show that the VS fraction differed significantly from the NVS fraction in several pedological parameters (Table 1) and elements (Table 2). Physico-chemical analysis revealed that the VS contained significantly less silt and more sand (Table 1) and was slightly more acidic than the NVS. It also contained more C and OM. Significant differences between the NVS and VS samples were found for the following parameters: Ti (+25.40% in VS), Mn (+60.04% in NVS), K (+27.66% in NVS), Sb (+28.88% in NVS), As (+32.43% in NVS), and B (+33.31% in NVS). In the CaCl2 extractable fraction, only B, Cr, Fe, K, Mg, Mn, Na, P, S, Si, Sr, Ti, and Zn were detected in significant amounts in this fraction (>0.01% from the total) (Table 2). However, for Fe, Mn, and Ti, the extractable fraction accounted for less than 0.01%. Conversely, for Cr, K, Mg, Si, S, and Sr, the extractable fraction accounted for approximately 2–10%. Only the total concentrations of As, B, Mg, Sr, Sb, and Ti differed significantly between the VS and NVS samples, while only the CaCl2 extractable fraction of Mg differed between the two soils.
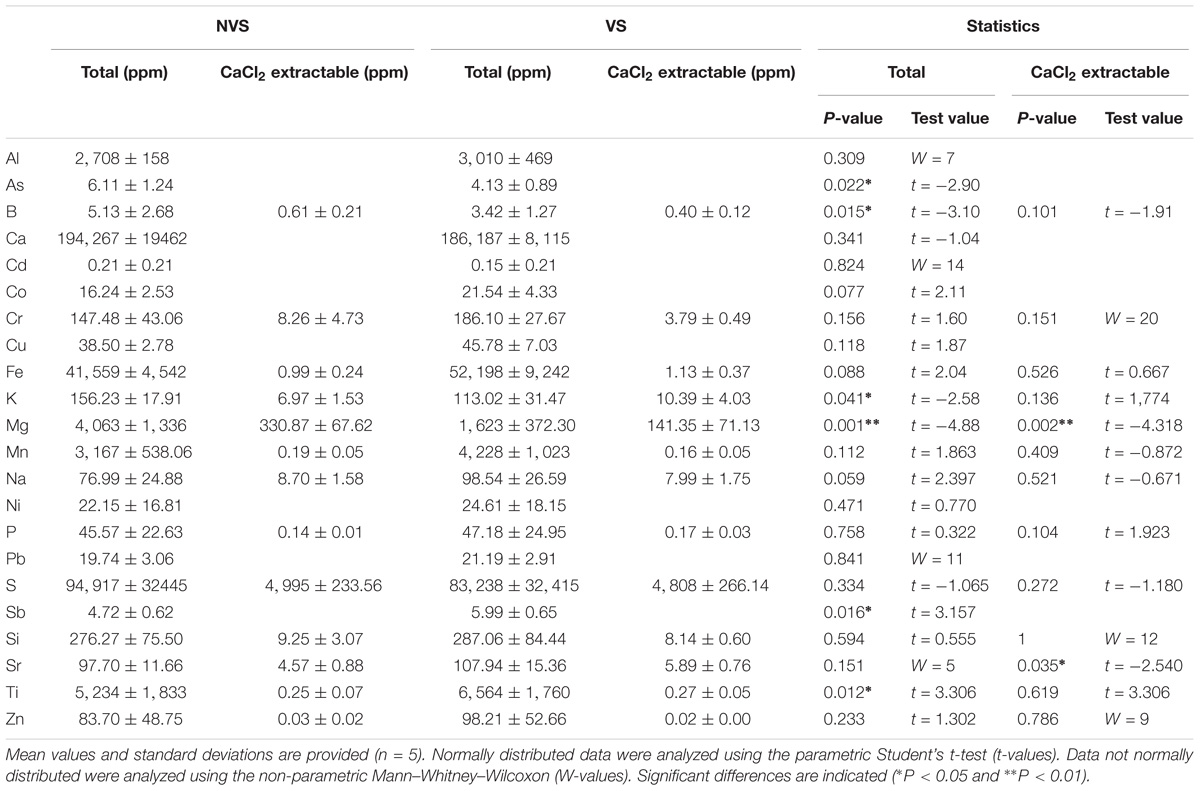
TABLE 2. Total and CaCl2 extractable element concentrations in vegetated soils (VS) and non-vegetated soils (NVS).
This set of data indicates that the soil of the tailings dumps is not suitable for revegetation due to its low nutrient content, very low N content (below the detection limit) and slightly alkaline pH. These extreme conditions have been shown to suppress tree root growth and to induce leaf chlorosis and decrease biomass production (Wang et al., 2017). The large amount of Fe and Mn, mostly in oxide forms (Carbonell, unpublished data), may also limit the availability of nutrients at this whole area. Previous studies were carried out on the interactions between plant richness and the physicochemical properties of tailings dumps and have identified pH, metal concentration and bioavailability as the major factors that may limit plant establishment on these sites (Santos et al., 2017).
Disparities between the VS and NVS emerged, as illustrated by the PCA (Figure 1B). PCA showed that the set of pedological parameters alone explains approximately 40% of the differences between the two soils. The VS also showed a significantly lower pH and higher CEC (Table 1). The differences between the VS and NVS could be due to the presence of the Betula trees. The lower pH in the vicinity of the birch roots, which could lead to an increase in the CEC in this area, is probably due to the root metabolic activity. Birch trees are known to exude several acids in the millimolar range, especially as monocarboxylic acids (Sandnes et al., 2005). The birch litter may also slightly contribute to the observed enrichment of the vegetated soil in C and OM.
Microbial Characteristics
The VS fraction had the highest density of culturable bacteria density, which was much higher than in the NVS fraction. In the latter samples, the CFUs were five times lower than the CFUs found in the VS (Figure 2). A total of 170 (VS) and 105 (NVS) bacteria were isolated in the present study. Based on their BOX-PCR profiles, the isolates were further placed into 53 (VS) and 43 (NVS) distinct groups and were further identified through comparative sequencing of their 16S rDNA. Isolates were recognized to include a total of 16 different bacterial genera, all of which belonging to four bacterial phyla. The Shannon diversity (H′) index was calculated based on the genera, and similar values were found for the VS (H′ = 1.16) and the NVS (H′ = 1.17).
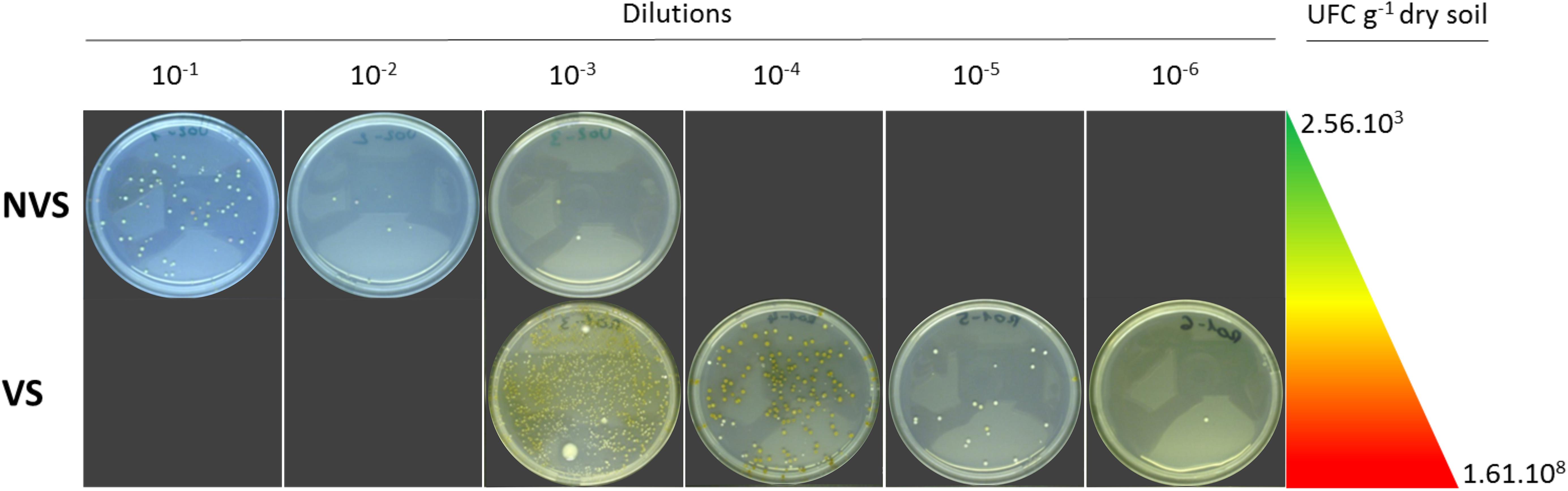
FIGURE 2. CFU culturable bacterial density expressed as UFC g−1 dry soil. The dilution series used to determine the density is represented by the Petri dishes after incubation for the non-vegetated soil (NVS) and the vegetated soil (VS).
The phyla Actinobacteria and Proteobacteria were represented in the two soils, while the Firmicutes (including the genus Bacillus) and Bacteroides (including the genera Pedobacter and Olivibacter) representatives were found only in the VS fraction. The Actinobacteria accounted for more than 95% in the NVS fraction. Within that phylum, the abundance of each genus differed between the two bacterial populations (Figure 3). In both soils, Streptomyces dominated and accounted for approximately 70% of the total isolates. Additional Actinobacteria were found in the NVS (Amycolatopsis, Nocardia, Nocardioides, and Paenarthrobacter) but were absent from the VS. The Rhodococcus isolates were found only in the VS. Within the Actinobacteria phylum, the two soils shared only Pseudoarthrobacter and Arthrobacter (Figure 3). The two soils also shared Proteobacteria members, although they were represented to a lower degree in the NVS. However, the two soils shared only Pseudomonas and Phyllobacterium isolates. Rhizobium, Variovorax, and Ensifer isolates were detected only in the VS fraction.
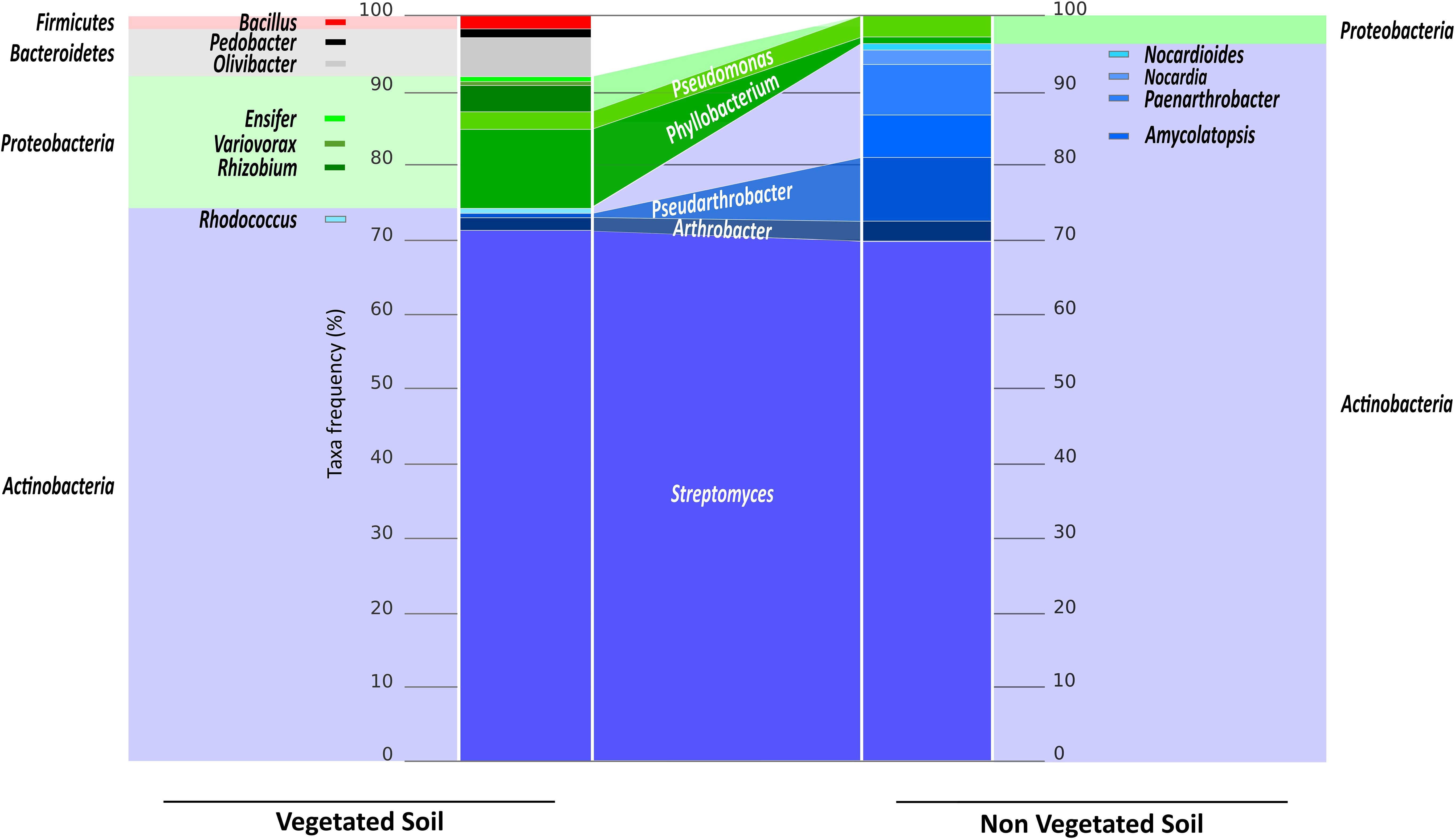
FIGURE 3. The cultivable bacterial communities from the VS and NVS fractions. The light-colored histograms at the edge represent the abundance of bacterial phyla expressed as a percentage of the total number of bacteria for the VS (Left) and the NVS (Right). The dark-colored central histogram represents the abundance of bacterial genera expressed as a percentage of the total number of bacteria for the VS (Left) and the NVS (Right). The names in white in the center indicate the shared bacteria between the two habitats.
The Actinobacteria phyla dominated the culturable soil community (70% in the VS and 95% in the NVS). These results agree with other data found in the literature. For example, the rhizospheric soil of PTE-hyperaccumulating plant Thlaspi caerulescens hosted bacterial communities that were analyzed and compared with that of contaminated bulk soil (Gremion et al., 2003). The sequences belonging to Actinobacteria dominated both the bulk and the rhizosphere soils in that study. Isolates from our soils were restricted to the genera Streptomyces, Arthrobacter, and Rhodococcus, as previously described (Álvarez-López et al., 2015). Streptomyces was also isolated from Mn-contaminated soils (Mo et al., 2017) and from the birch rhizosphere collected on the coal mining dump (Ostash et al., 2013). The rhizosphere from the Ni-hyperaccumulating plant Alyssum serpyllifolium hosts Ni-resistant bacteria that were predominantly belonging to the Arthrobacter and Streptomyces genera (Becerra-Castro et al., 2011b). Álvarez et al. (2017) isolated, using culture-dependent methods, and characterized more than 35 Actinobacteria genera that were shown to be PTE-tolerant. Actinobacteria members tolerant to PTE have been shown to be dominant in PTE-contaminated sites by Margesin et al. (2011), in addition to Proteobacteria members, and by Oliveira and Pampulha (2006). Most of these studies agreed on the fact that contaminated soils usually exhibited quantitatively lower culturable bacteria, although Actinobacteria were usually less affected by the PTE present in the soil than other culturable heterotrophic bacteria or nitrogen fixers. In addition to Streptomyces, Arthrobacter was the second most key bacterial genus concerning its PTE-tolerance and thereof it has a real potential for use in bioremediation. Alkaline environments are commonly encountered in soils contaminated with PTE, for instance with Cr. Due to its capacity to tolerate alkaline conditions, Elangovan et al. (2010) suggested the use of both intact Arthrobacter cells and cell-free extracts for the bioremediation of alkaline soils contaminated with chromate.
The Streptomyces genus, among the order Actinomycetales is notably the richest source of natural compounds, including antimetabolites, antibiotics, and antitumor compounds (Bérdy, 2005; Olano et al., 2009; Aigle et al., 2014). For instance, the Streptomyces genus produces around 80% of secondary metabolites known to be microbial bioactive compounds (Bérdy, 2005). Actinobacteria are able to grow under various life styles such as saprophytes in aquatic environments and soils, or plant commensals such as nitrogen-fixing symbionts. These key features render the Actinobacteria well-suited for research-based bioremediation technology.
Functional Traits of the Bacterial Isolates
The PGP properties of the bacterial collection comprising 53 (R) and 43 (S) BOX-PCR groups were tested in vitro for PGP traits such as IAA production, nutrient uptake, and the metabolism of bacterial compounds regulating plant growth (Figure 4). The bacteria isolated from the vegetated soil harbored the PGP bacteria with the higher beneficial potential, with 12% of the isolates exhibiting three or more PGP traits (Supplementary Table S1). The ability to produce IAA was detected in both the isolates from the VS and the NVS fractions, although they were statistically more abundant in the VS. Conversely, the siderophore-producing capacity was higher in the isolates from the NVS. Organic acid-producers and rare phosphate solubilizers were present in both populations and were not significantly different. The production of siderophores by Streptomyces isolates has already been shown by Ostash et al. (2013). The genetically and enzymatically based siderophore biosynthesis and transport are well-described in Streptomyces (Cruz-Morales et al., 2017). More generally, the Actinomycetes group got substantial prominence as PGP microorganism because of its recognized and strong antimicrobial potential and saprophytic behavior dominating numerous soils (Franco-Correa et al., 2010).
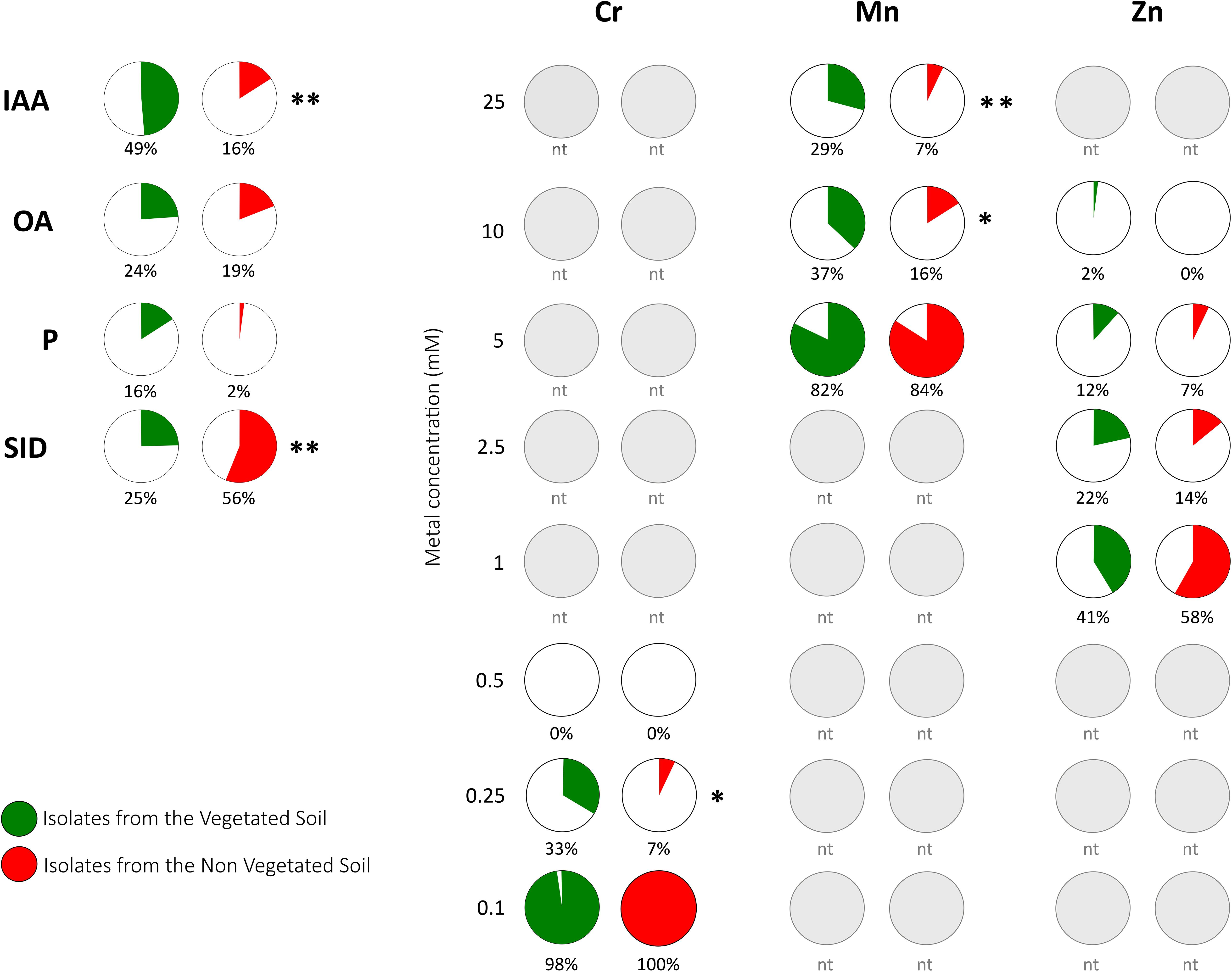
FIGURE 4. Functional traits of bacterial isolates from the Thann site. Plots representing the percentage of isolates that harbor the positive PGP trait or show metal tolerance. PGP traits were indoleacetic acid (IAA) production, organic acid production (OA), phosphorus solubilization (P), and siderophore production (SID). Metal tolerance was tested for Cr, Mn, and Zn at concentrations ranging from 0.1 to 25 mM. Gray plots indicate non-tested concentrations (nt) for a given metal. Significant differences are indicated (∗P < 0.05; ∗∗P < 0.01).
In this study, the tolerance to the three metals Zn, Cr, and Mn was investigated (Table 2) using the 284 growth medium, and thus our data may only be considered as relative. As indicated in Figure 4, the relative order of bacterial toxicity of the three metals was determined as follows: Mn > Zn > Cr. Tolerance to Cr and Zn was considered to be reached at concentrations higher than 0.5 and 1 mM, respectively (Navarro-Noya et al., 2012). In our samples, isolates tolerated Cr concentrations below that threshold that were much lower than that measured for Cupriavidus metallidurans (2.5 mM) (Zhao et al., 2012). Conversely, our isolates were more resistant to Zn with an MTC up to 10 mM for some VS isolates. Two percent of our isolates showed an MTC of 10 mM, which is on the same order of magnitude as that determined for the metal-resistant Cupriavidus metallidurans (Zhao et al., 2012). Zn-resistant rhizospheric as well as endophytic bacterial isolates of Zn-accumulating Salix trees were characterized by Kuffner et al. (2008).
Comparing isolates both from the VS and the NVS samples, the Mn and Cr tolerances were higher for the VS bacteria at the highest concentrations. The strains with the higher metal tolerance in our study were the Phyllobacterium sp. WR140 (RO1.15), Phyllobacterium sp. WR140 (R01.34) and Streptomyces sp. (R04.15), all of which were isolated from the VS fraction. The least tolerant species were isolated from the NVS fraction. The primary tolerant soil bacterium was Streptomyces flavovirens (U04.24). To the best of our knowledge, the Mn tolerance in either Phyllobacterium or Streptomyces has been rarely studied. The Proteobacteria, although they were a less abundant species in our isolation experiment, appeared to be the most metal tolerant bacteria (4 over 5 more tolerant). There is abundant literature on Mn oxidation by bacteria (Adams and Ghiorse, 1985; Wang et al., 2009). For instance, the Mn-oxidizing bacterium Brachybacterium strain isolated from the deep-sea was able to grow in liquid media supplemented with up to 55 mM MnCl (Wang et al., 2009). Bacterial cells have mechanisms to sense excess metals (Chandrangsu et al., 2017). Generally, the most efficient physiological mechanism that bacteria are exploiting to tolerate excess PTE is efflux. Some of our isolated bacteria (i.e., Phyllobacterium sp. WR140) exhibited a Mn MIC higher than that of Cupriavidus metallidurans (6 mM) (Zhao et al., 2012). In a previous study, the proportions of metal-tolerant bacterial isolates were primarily represented by Gram-negatives, and the Proteobacteria (Pseudomonas and Variovorax species) dominated (Piotrowska-Seget et al., 2005).
Abbes and Edwerds (1990) evaluated the toxicity of various PTE including Cd, Co, Cu, Cr, Hg, Ni, Zn, and Mn on 34 Streptomyces species representative of various taxonomic clusters. Another study described the isolation of several Streptomyces strains with resistances to different PTE from contaminated areas, and some exhibited multiple tolerances against different PTE (Álvarez et al., 2013). Due to the extreme abundance of the genus Streptomyces in our study, the further characterization of tolerant Streptomyces from this red gypsum dump might lead to a better assessment/new discovery of the physiological mechanisms involved in the metal tolerance in this genus and therefore to ecological applications.
However, we examined the tolerance to the Mn ionic form, while the Mn in the Thann soil was primarily in the form of Mn oxides (Zapata, unpublished results). A recent review also pointed out the use of bacteria in Mn biomining processes, describing distinct, taxonomically distant bacteria that have been described to reduce Mn either by enzymatic or non-enzymatic based mechanisms or Das et al. (2011). Mn is usually reduced to fulfill a nutritional need for soluble forms of Mn and Mn-reducing bacteria belong to either aerobes or facultative anaerobes.
Inoculation Tests
A set of 20 bacterial strains isolated from the VS was chosen to carry out an inoculation pot experiment. The bacterial strains were selected from our previous experiments (section “Functional Traits of the Bacterial Isolates”) and those with the best functional traits (PGP traits, siderophore production and metal resistance) were retained, as detailed in Supplementary Table S1. The selected bacterial genera were also representative of the most important phyla in our study and, when possibly, previously tested in the literature. After 3 months of growth, the biomass of the birch plants either inoculated or non-inoculated was analyzed (Figure 5). Among the 20 isolates, five increased the total biomass production significantly (P < 0.01), and an additional set of six isolates increased the total biomass production significantly (P < 0.05) compared with uninoculated control. This was primarily due to an increase in the root biomass, while the shoot biomass was only slightly affected (Figure 5). We also measured the elements in birch leaves and found no significant effect of the inoculated bacteria (data not shown).
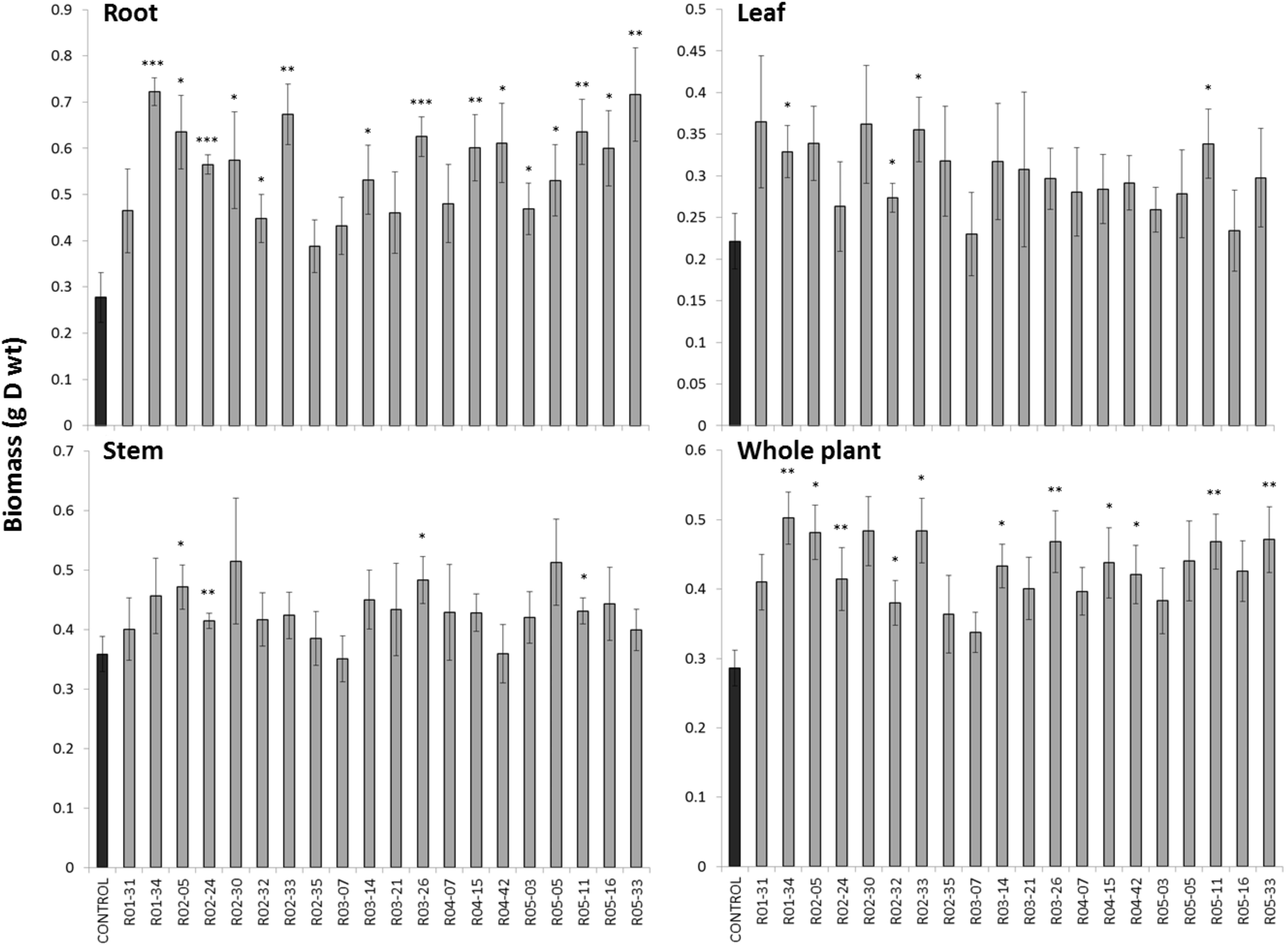
FIGURE 5. Impact of bacterial inoculation on birch growth. Birch trees were grown for 3 months on the Thann soil inoculated with 20 bacterial isolates collected from the VS fraction. Significant differences are indicated (∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001).
Among the isolates tested, the Phyllobacterium sp. isolate (R01.34) that showed the greatest performance on birch also exhibited multiple PGP traits, including the production of IAA and the solubilization of P. PGP rhizobacteria and bacterial endophytes have huge potential to increase the bioremediation of PTE-contaminated sites (Chen et al., 2010; Luo et al., 2011; Xinxian et al., 2011; Burges et al., 2017). Ma et al. (2013) demonstrated the similar performance of Phyllobacterium myrsinacearum on Sedum plumbizincicola growth, although this strain increased metal transfer to the shoot, which was not observed in our study. However, another Phyllobacterium strain examined in our study did not demonstrate any phytoremediation relevant features, which indicates intragenera variability. A Pseudomonas isolate also significantly increased birch biomass production. Similarly, Huang et al. (2016) isolated a Cd-resistant P. aeruginosa from a Cd-contaminated oil field, and the inoculations of Cd-polluted soil with that strain significantly elevated the shoot and root biomass. The Variovorax isolate (R05-11) also exhibited PGP traits in our study. Similarly, the inoculation of hyperaccumulating plants by Variovorax paradoxus isolated from plant rhizosphere significantly increased root biomass (Durand et al., 2018). However, growth promotion was not always linked to functional traits in our study. For instance, Streptomyces (R05-33) showed significant growth promotion effects without possessing any significant functional traits. In general, we found that Streptomyces strains did not show a great potential to promote plant growth. Other potential traits, such as the release of volatile organic compounds (VOCs), which are very well known in the genus Streptomyces (Dias et al., 2017), could be responsible for this beneficial effect. The Olivibacter soli (R04-07) isolate exhibited all functional traits while having no growth promoting effects. In greenhouse studies (Alekhya and Gopalakrishnan, 2017), Cicer arietinum plant growth was increased by Streptomyces due to root length and weight promotion, increase production of nodules, increase production of shoot biomass, pods, and pod weight as compared with the non-inoculated control, demonstrating the colonizing capability of bacteria belonging to this genera.
In addition to the Arthrobacter and Rhodococcus genera, Streptomyces bacteria have received considerable consideration for being used in efficient biotechnological method to clean up contaminated ecosystems. In addition to their physiological diversity, Streptomyces isolates may be appropriate for being used as soil inoculants since they are able to rapidly grow as mycelium in semi-selective substrates and their capacity to be genetically transformed (Álvarez et al., 2017). However, most of the studies described in this review paper concerned pesticide-degrading Actinobacteria. In the study by Ali et al. (2017), bioremediation was performed on PTE-contaminated mining soils by inoculation using Streptomyces pactum. Metal extraction amount data established that the strain Act12 stimulated the PTE uptake and transfer in Brassica juncea above ground tissues.
Conclusion
This study demonstrates that vegetated soils from red gypsum tailings dumps exhibited a higher bacterial diversity compared with non-vegetated soils, based on a culture-dependent method. The number of bacterial isolates having the capacity to produce IAA was higher for bacteria from the vegetated soil, while siderophore production was higher in the bacteria from the non-vegetated soil. Mn and Cr tolerance was also higher for the bacteria isolated from the VS samples. The potential of some bacterial isolates to promote birch growth was observed, although it was not always linked to PGP traits. However, the results of the inoculation tests and the dominance of some Streptomyces (within the VS Actinobacteria population) and Phyllobacterium (within the VS Proteobacteria population) associated with Betula growing on this soil suggests that those bacteria are involved in the early establishment of woody species in the dump. They appear to be good candidates to identify new approaches for the management of tailings dumps. The Phyllobacterium sp. isolate (R01-34) and Streptomyces (R05-33) appeared to be promising alternatives for improved inocula and their application at field levels.
Author Contributions
MC, CZ, VA-L, and NC planned and designed the research. VA-L and CZ performed the experiments and conducted the fieldwork. MC, VA-L, CZ, NC, and CG analyzed the data and wrote the manuscript.
Funding
This work was supported by the French National Research Agency (PHYTOCHEM ANR-13-CDII-0005-01), the French Environment and Energy Management Agency (PROLIPHYT ADEME-1172C0053), the Région Franche-Comté (Environnement-Homme-Territoire 2014-069) and the Pays de Montbéliard Agglomération (13/070-203-2015). VA-L received a post doc grant from the Région Franche-Comté. CZ received a Ph.D. grant from the French Ministry of Higher Education and Research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Dr. Nadia Morin-Crini and Caroline Amiot for the ICP-AES analyses. We thank Jean Michel Colin (CRISTAL Co., France) for providing us with access to the Thann site.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01772/full#supplementary-material
Footnotes
References
Abbes, A., and Edwerds, C. (1990). Effects of metals on Streptomyces coelicolor growth and actinorhodin production ALA. Appl. Environ. Microbiol. 56, 675–680.
Adams, L. F., and Ghiorse, W. C. (1985). Influence of maganese on growth of a sheathless strain of Leptothrix discophora. Appl. Environ. Microbiol. 49, 556–562.
Aigle, B., Lautru, S., Spiteller, D., Dickschat, J. S., Challis, G. L., Leblond, P., et al. (2014). Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 41, 251–263. doi: 10.1007/s10295-013-1379-y
Alekhya, G., and Gopalakrishnan, S. (2017). Biological control and plant growth-promotion traits of Streptomyces species under greenhouse and field conditions in chickpea. Agric. Res. 6, 410–420. doi: 10.1007/s40003-017-0278-2
Ali, A., Guo, D., Mahar, A., Wang, Z., Muhammad, D., Li, R., et al. (2017). Role of Streptomyces pactum in phytoremediation of trace elements by Brassica juncea in mine polluted soils. Ecotoxicol. Environ. Saf. 144, 387–395. doi: 10.1016/j.ecoenv.2017.06.046
Álvarez, A., Catalano, S. A., and Amoroso, M. J. (2013). Heavy metal resistant strains are widespread along Streptomyces phylogeny. Mol. Phylogenet. Evol. 66, 1083–1088. doi: 10.1016/j.ympev.2012.11.025
Álvarez, A., Saez, J. M., Davila Costa, J. S., Colin, V. L., Fuentes, M. S., Cuozzo, S. A., et al. (2017). Actinobacteria: current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 166, 41–62. doi: 10.1016/j.chemosphere.2016.09.070
Álvarez-López, V., Prieto-Fernández, Á, Becerra-Castro, C., Monterroso, C., and Kidd, P. S. (2015). Rhizobacterial communities associated with the flora of three serpentine outcrops of the Iberian Peninsula. Plant Soil 403, 1–20. doi: 10.1007/s11104-015-2632-0
Atkinson, M. (1992). Betula pendula Roth (B. verrucosa Ehrh.) and B. pubescens Ehrh. J. Ecol. 80, 837–870. doi: 10.2307/2260870
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Becerra-Castro, C., Kidd, P. S., Prieto-Fernández, Á, Weyens, N., Acea, M. J., and Vangronsveld, J. (2011a). Endophytic and rhizoplane bacteria associated with Cytisus striatus growing on hexachlorocyclohexane-contaminated soil: isolation and characterisation. Plant Soil 340, 413–433. doi: 10.1007/s11104-010-0613-x
Becerra-Castro, C., Prieto-Fernández, Á,Álvarez-López, V., Monterroso, C., Cabello-Conejo, M. I., Acea, M. J., et al. (2011b). Nickel solubilizing capacity and characterization of rhizobacteria isolated from hyperaccumulating and non-hyperaccumulating subspecies of Alyssum serpyllifolium. Int. J. Phytorem. 13(Suppl. 1), 229–244. doi: 10.1080/15226514.2011.568545
Bruneel, O., Mghazli, N., Hakkou, R., Dahmani, I., Filali Maltouf, A., and Sbabou, L. (2017). In-depth characterization of bacterial and archaeal communities present in the abandoned Kettara pyrrhotite mine tailings (Morocco). Extremophiles 21, 671–685. doi: 10.1007/s00792-017-0933-3
Burges, A., Epelde, L., Blanco, F., Becerril, J. M., and Garbisu, C. (2017). Ecosystem services and plant physiological status during endophyte-assisted phytoremediation of metal contaminated soil. Sci. Total Environ. 584–585, 329–338. doi: 10.1016/j.scitotenv.2016.12.146
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Chen, L., Luo, S., Xiao, X., Guo, H., Chen, J., Wan, Y., et al. (2010). Application of plant growth-promoting endophytes (PGPE) isolated from Solanum nigrum L. for phytoextraction of Cd-polluted soils. Appl. Soil Ecol. 46, 383–389. doi: 10.1016/j.apsoil.2010.10.003
Cox, C. D. (1994). Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 235, 315–329.
Cross, A. T., Stevens, J. C., and Dixon, K. W. (2017). One giant leap for mankind: can ecopoiesis avert mine tailings disasters? Plant Soil 421, 1–5. doi: 10.1007/s11104-017-3410-y
Cruz-Morales, P., Ramos-Aboites, H. E., Licona-Cassani, C., Selem-Mójica, N., Mejía-Ponce, P. M., Souza-Saldívar, V., et al. (2017). Actinobacteria phylogenomics, selective isolation from an iron oligotrophic environment and siderophore functional characterization, unveil new desferrioxamine traits. FEMS Microbiol. Ecol. 93:fix086. doi: 10.1093/femsec/fix086
Das, A. P., Sukla, L. B., Pradhan, N., and Nayak, S. (2011). Manganese biomining: a review. Bioresour. Technol. 102, 7381–7387. doi: 10.1016/j.biortech.2011.05.018
Dias, M. P., Bastos, M. S., Xavier, V. B., Cassel, E., Astarita, L. V., and Santarém, E. R. (2017). Plant growth and resistance promoted by Streptomyces spp. in tomato. Plant Physiol. Biochem. 118, 479–493. doi: 10.1016/j.plaphy.2017.07.017
Durand, A., Maillard, F., Alvarez-Lopez, V., Guinchard, S., Bertheau, C., Valot, B., et al. (2018). Bacterial diversity associated with poplar trees grown on a Hg-contaminated site: community characterization and isolation of Hg-resistant plant growth-promoting bacteria. Sci. Total Environ. 622–623, 1165–1177. doi: 10.1016/j.scitotenv.2017.12.069
Elangovan, R., Philip, L., and Chandraraj, K. (2010). Hexavalent chromium reduction by free and immobilized cell-free extract of arthrobacter rhombi-RE. Appl. Biochem. Biotechnol. 160, 81–97. doi: 10.1007/s12010-008-8515-6
Foulon, J., Zappelini, C., Durand, A., Valot, B., Blaudez, D., and Chalot, M. (2016a). Impact of poplar-based phytomanagement on soil properties and microbial communities in a metal-contaminated site. FEMS Microbiol. Ecol. 92:fiw163. doi: 10.1093/femsec/fiw163
Foulon, J., Zappelini, C., Durand, A., Valot, B., Girardclos, O., Blaudez, D., et al. (2016b). Environmental metabarcoding reveals contrasting microbial communities at two poplar phytomanagement sites. Sci. Total Environ. 571, 1230–1240. doi: 10.1016/j.scitotenv.2016.07.151
Franco-Correa, M., Quintana, A., Duque, C., Suarez, C., Rodríguez, M. X., and Barea, J. M. (2010). Evaluation of actinomycete strains for key traits related with plant growth promotion and mycorrhiza helping activities. Appl. Soil Ecol. 45, 209–217. doi: 10.1016/j.apsoil.2010.04.007
Gremion, F., Chatzinotas, A., and Harms, H. (2003). Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ. Microbiol. 5, 896–907. doi: 10.1046/j.1462-2920.2003.00484.x
Gupta, A., Dutta, A., Sarkar, J., Paul, D., Panigrahi, M. K., and Sar, P. (2017). Metagenomic exploration of microbial community in mine tailings of Malanjkhand copper project, India. Genomics Data 12, 11–13. doi: 10.1016/j.gdata.2017.02.004
Heras, J., Domínguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). GelJ – a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16:270. doi: 10.1186/s12859-015-0703-0
Honeker, L. K., Neilson, J. W., Root, R. A., Gil-Loaiza, J., Chorover, J., and Maier, R. M. (2017). Bacterial rhizoplane colonization patterns of buchloe dactyloides growing in metalliferous mine tailings reflect plant status and biogeochemical conditions. Microb. Ecol. 74, 853–867. doi: 10.1007/s00248-017-0998-7
Huang, J., Liu, Z., Li, S., Xu, B., Gong, Y., Yang, Y., et al. (2016). Isolation and engineering of plant growth promoting rhizobacteria Pseudomonas aeruginosa for enhanced cadmium bioremediation. J. Gen. Appl. Microbiol. 62, 258–265. doi: 10.2323/jgam.2016.04.007
Huang, L., Baumgartl, T., and Mulligan, D. (2012). Is rhizosphere remediation sufficient for sustainable revegetation of mine tailings? Ann. Bot. 110, 223–238. doi: 10.1093/aob/mcs115
Jana, U., Chassany, V., Bertrand, G., Castrec-Rouelle, M., Aubry, E., Boudsocq, S., et al. (2012). Analysis of arsenic and antimony distribution within plants growing at an old mine site in Ouche (Cantal, France) and identification of species suitable for site revegetation. J. Environ. Manage. 110, 188–193. doi: 10.1016/j.jenvman.2012.06.007
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norvich: John Innes Cent. Ltd, doi: 10.4016/28481.01
Kuffner, M., Puschenreiter, M., Wieshammer, G., Gorfer, M., and Sessitsch, A. (2008). Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil 304, 35–44. doi: 10.1007/s11104-007-9517-9
Kumaresan, D., Cross, A. T., Moreira-Grez, B., Kariman, K., Nevill, P., Stevens, J., et al. (2017). Microbial functional capacity is preserved within engineered soil formulations used in mine site restoration. Sci. Rep. 7:564.. doi: 10.1038/s41598-017-00650-6
Luo, S., Wan, Y., Xiao, X., Guo, H., Chen, L., Xi, Q., et al. (2011). Isolation and characterization of endophytic bacterium LRE07 from cadmium hyperaccumulator Solanum nigrum L. and its potential for remediation. Appl. Microbiol. Biotechnol. 89, 1637–1644. doi: 10.1007/s00253-010-2927-2
Ma, Y., Rajkumar, M., Luo, Y., and Freitas, H. (2013). Phytoextraction of heavy metal polluted soils using Sedum plumbizincicola inoculated with metal mobilizing Phyllobacterium myrsinacearum RC6b. Chemosphere 93, 1386–1392. doi: 10.1016/j.chemosphere.2013.06.077
Margesin, R., Płaza, G. A., and Kasenbacher, S. (2011). Characterization of bacterial communities at heavy-metal-contaminated sites. Chemosphere 82, 1583–1588. doi: 10.1016/j.chemosphere.2010.11.056
Mark Ibekwe, A., Kennedy, A. C., Halvorson, J. J., and Yang, C. H. (2007). Characterization of developing microbial communities in Mount St. Helens pyroclastic substrate. Soil Biol. Biochem. 39, 2496–2507. doi: 10.1016/j.soilbio.2007.05.010
Mendez, M., and Maier, R. M. (2008). Phytoremediation of mine tailings in temperate and arid environments. Rev. Environ. Sci. Biotechnol. 7, 47–59. doi: 10.1007/s11157-007-9125-4
Méndez-García, C., Peláez, A. I., Mesa, V., Sánchez, J., Golyshina, O. V., and Ferrer, M. (2015). Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 6:475. doi: 10.3389/fmicb.2015.00475
Mergeay, M., Nies, D. H., Schlegel, H. G., Gerits, J., Charles, P., and Gijsegem, F. V. (1985). Alcaligenes eutrophus CH34 ia a facultative Chemolithotrophy with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328.
Mesa, V., Gallego, J. L. R., González-Gil, R., Lauga, B., Sánchez, J., Méndez-García, C., et al. (2017). Bacterial, archaeal, and eukaryotic diversity across distinct microhabitats in an acid mine drainage. Front. Microbiol. 8:1756. doi: 10.3389/fmicb.2017.01756
Mo, P., Yu, Y. Z., Zhao, J. R., and Gao, J. (2017). Streptomyces xiangtanensis sp. nov., isolated from a manganese-contaminated soil. Antonie Van Leeuwenhoek 110, 297–304. doi: 10.1007/s10482-016-0797-z
Nautiyal, C. S. (1999). An eficiente microbiological growth médium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Navarro-Noya, Y. E., Hernández-Mendoza, E., Morales-Jiménez, J., Jan-Roblero, J., Martínez-Romero, E., and Hernández-Rodríguez, C. (2012). Isolation and characterization of nitrogen fixing heterotrophic bacteria from the rhizosphere of pioneer plants growing on mine tailings. Appl. Soil Ecol. 62, 52–60. doi: 10.1016/j.apsoil.2012.07.011
Olano, C., Méndez, C., and Salas, J. A. (2009). Antitumor compounds from marine actinomycetes. Mar. Drugs 7, 210–248. doi: 10.3390/md7020210
Oliveira, A., and Pampulha, M. E. (2006). Effects of long-term heavy metal contamination on soil microbial characteristics. J. Biosci. Bioeng. 102, 157–161. doi: 10.1263/jbb.102.157
Ostash, B., Gren, T., Hrubskyy, Y., Tistechok, S., Beshley, S., Baranov, V., et al. (2013). Cultivable actinomycetes from rhizosphere of birch ( Betula pendula ) growing on a coal mine dump in Silets. Ukraine. J. Basic Microbiol. 54, 851–857. doi: 10.1002/jobm.201200551
Piotrowska-Seget, Z., Cycoñ, M., and Kozdrój, J. (2005). Metal-tolerant bacteria occurring in heavily polluted soil and mine spoil. Appl. Soil Ecol. 28, 237–246. doi: 10.1016/j.apsoil.2004.08.001
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Salisbury, A. B., Reinfelder, J. R., Gallagher, F. J., and Grabosky, J. C. (2017). Long-term stability of trace element concentrations in a spontaneously vegetated urban brownfield with anthropogenic soils. Soil Sci. 182, 69–81. doi: 10.1097/SS.0000000000000193
Sandnes, A., Eldhuset, T. D., and Wollebæk, G. (2005). Organic acids in root exudates and soil solution of Norway spruce and silver birch. Soil Biol. Biochem. 37, 259–269. doi: 10.1016/j.soilbio.2004.07.036
Santini, T. C., and Banning, N. C. (2016). Alkaline tailings as novel soil forming substrates: reframing perspectives on mining and refining wastes. Hydrometallurgy 164, 38–47. doi: 10.1016/j.hydromet.2016.04.011
Santos, A. E., Cruz-Ortega, R., Meza-Figueroa, D., Romero, F. M., Sanchez-Escalante, J. J., Maier, R. M., et al. (2017). Plants from the abandoned Nacozari mine tailings: evaluation of their phytostabilization potential. PeerJ 5:e3280. doi: 10.7717/peerj.3280
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sheng, X.-F., Xia, J.-J., Jiang, C.-Y., He, L.-Y., and Qian, M. (2008). Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 156, 1164–1170. doi: 10.1016/j.envpol.2008.04.007
Tardy, V., Spor, A., Mathieu, O., Lévèque, J., Terrat, S., Plassart, P., et al. (2015). Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 90, 204–213. doi: 10.1016/j.soilbio.2015.08.010
Wanat, N., Joussein, E., Soubrand, M., and Lenain, J. F. (2014). Arsenic (As), antimony (Sb), and lead (Pb) availability from Au-mine Technosols: a case study of transfer to natural vegetation cover in temperate climates. Environ. Geochem. Health 36, 783–795. doi: 10.1007/s10653-014-9596-5
Wang, L., Ji, B., Hu, Y., Liu, R., and Sun, W. (2017). A review on in situ phytoremediation of mine tailings. Chemosphere 184, 594–600. doi: 10.1016/j.chemosphere.2017.06.025
Wang, W., Shao, Z., Liu, Y., and Wang, G. (2009). Removal of multi-heavy metals using biogenic manganese oxides generated by a deep-sea sedimentary bacterium - Brachybacterium sp. strain Mn32. Microbiology 155, 1989–1996. doi: 10.1099/mic.0.024141-0
Wang, X., Liu, Y., Zeng, G., Chai, L., Xiao, X., Song, X., et al. (2008). Pedological characteristics of Mn mine tailings and metal accumulation by native plants. Chemosphere 72, 1260–1266. doi: 10.1016/j.chemosphere.2008.05.001
Wubs, E. R. J., Van Der Putten, W. H., Bosch, M., and Bezemer, T. M. (2016). Soil inoculation steers restoration of terrestrial Ecosystems. Nat. Plants 2, 1–5. doi: 10.1038/NPLANTS.2016.107
Xinxian, L., Xuemei, C., Yagang, C., Woon-Chung, W. J., Zebin, W., and Qitang, W. (2011). Isolation and characterization endophytic bacteria from hyperaccumulator Sedum alfredii Hance and their potential to promote phytoextraction of zinc polluted soil. World J. Microbiol. Biotechnol. 27, 1197–1207. doi: 10.1007/s11274-010-0568-3
Yergeau, E., Bell, T. H., Champagne, J., Maynard, C., Tardif, S., Tremblay, J., et al. (2015). Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front. Microbiol. 6:1436. doi: 10.3389/fmicb.2015.01436
Zhao, X. Q., Wang, R. C., Lu, X. C., Lu, J. J., Li, J., and Hu, H. (2012). Tolerance and biosorption of heavy metals by Cupriavidus metallidurans strain XXKD-1 isolated from a subsurface laneway in the Qixiashan Pb-Zn sulfide minery in Eastern China. Geomicrobiol. J. 29, 274–286. doi: 10.1080/01490451.2011.619637
Keywords: Actinobacteria, red gypsum landfill, birch, plant growth promoting traits, biological restoration
Citation: Zappelini C, Alvarez-Lopez V, Capelli N, Guyeux C and Chalot M (2018) Streptomyces Dominate the Soil Under Betula Trees That Have Naturally Colonized a Red Gypsum Landfill. Front. Microbiol. 9:1772. doi: 10.3389/fmicb.2018.01772
Received: 02 May 2018; Accepted: 16 July 2018;
Published: 03 August 2018.
Edited by:
Piotr Rozpądek, Jagiellonian University, PolandReviewed by:
Wenli Chen, Huazhong Agricultural University, ChinaMariusz Cycoñ, Medical University of Silesia, Poland
Copyright © 2018 Zappelini, Alvarez-Lopez, Capelli, Guyeux and Chalot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michel Chalot, bWljaGVsLmNoYWxvdEB1bml2LWZjb210ZS5mcg==
 Cyril Zappelini
Cyril Zappelini Vanessa Alvarez-Lopez
Vanessa Alvarez-Lopez Nicolas Capelli1
Nicolas Capelli1 Christophe Guyeux
Christophe Guyeux Michel Chalot
Michel Chalot