
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 03 August 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01746
This article is part of the Research Topic Microbial Biotechnology Providing Bio-based Components for the Food Industry View all 14 articles
In the 1950s, the idea of a single harmonized list of food additives for the European Union arose. Already in 1962, the E-classification system, a robust food safety system intended to protect consumers from possible food-related risks, was introduced. Initially, it was restricted to colorants, but at later stages also preservatives, antioxidants, emulsifiers, stabilizers, thickeners, gelling agents, sweeteners, and flavorings were included. Currently, the list of substances authorized by the European Food Safety Authority (EFSA) (referred to as “E numbers”) comprises 316 natural or artificial substances including small organic molecules, metals, salts, but also more complex compounds such as plant extracts and polymers. Low overall concentrations of such compounds in natural producers due to inherent regulation mechanisms or production processes based on non-regenerative carbon sources led to an increasing interest in establishing more reliable and sustainable production platforms. In this context, microorganisms have received significant attention as alternative sources providing access to these compounds. Scientific advancements in the fields of molecular biology and genetic engineering opened the door toward using engineered microorganisms for overproduction of metabolites of their carbon metabolism such as carboxylic acids and amino acids. In addition, entire pathways, e.g., of plant origin, were functionally introduced into microorganisms, which holds the promise to get access to an even broader range of accessible products. The aim of this review article is to give a systematic overview on current efforts during construction and application of microbial cell factories for the production of food additives listed in the EU “E numbers” catalog. The review is focused on metabolic engineering strategies of industrially relevant production hosts also discussing current bottlenecks in the underlying metabolic pathways and how they can be addressed in the future.
In industrial food production, consistent quality of foodstuff, and protection against contamination by harmful microorganisms must be guaranteed. For this purpose, a broad range of natural or artificial food additives are used in today's food applications, which not only serve for increasing the shelf life of foodstuff, but also for modifying its color, taste, odor, or texture. For preventing an arbitrary use of potentially harmful substances, there was strong desire toward establishing obligatory guidelines for the use of food additives. The additives sector has played a very important role in the approximation of food law in the European Union (EU). The first regulation, on which the six member states of the European Economic Community (EEC) (Belgium, Germany, France, Italy, Luxembourg, and the Netherlands) were able to agree, was the “Directive on food colorants” in 1962 (Directive 62/2645/EEC), which marks the beginning of a comprehensive harmonization effort. With this directive the E-classification system, a robust food safety system intended to protect consumers from possible food-related risks, was introduced. This classification system lists food additives approved by the EU (or its predecessor organizations) in form of “E numbers” allowing an unambiguous identification of a single compound (which might have several designations, e.g., trivial names), a set of chemically similar compounds or a plant extract.
In 1965, the regulation was extended to preservative substances which may be used in foodstuff (Directive 65/66/EEC). In a similar manner, other classes of food additives were included, e.g., anti-oxidants, emulsifiers, stabilizers, thickeners, and gelling agents in 1978 (Directives 78/663/EEC and 78/664/EEC) and sweeteners in 1994 (Directive 94/35/EC). In parallel, a number of additional intermediate steps in the form of European directives, e.g., for monitoring of purity criteria and for maintenance of national bans on the use of certain additives, was adopted. In 2008, the Framework Regulation (EC) No. 1333/2008 on food additives, and thus directly applicable law, was finally adopted and revised in 2010 in the Directive (EU) No. 257/2010. The principles described therein include health safety, technological necessity and prevention of misleading of food additives. In its current form the list of food additives approved by the EU includes 316 compounds, which are classified according to their major application as colors (E100–E199), preservatives (E200–E299), antioxidants and acidity regulators (E300–E399), thickeners, stabilizers, and emulsifiers (E400–E499), anti-caking agents (E500–E599), flavor enhancers (E600–E699), antibiotics (E700–E799), glazing agents, gases, and sweeteners (E900–E999) and additional additives (E1000–1599). Not all E numbers have been assigned and some E numbers were removed from the list.
The compounds listed in the E numbers catalog comprise small organic molecules, metals, inorganic salts, but also polymers and complex compounds derived from plants or animals (e.g., beeswax, E901). There are in principle three sources of the approved compounds: extraction from the natural producer (e.g., plant material), chemical production or production in (engineered) microorganisms. Not every source is suitable for the production of each of the above-mentioned type of compounds, e.g., metals, inorganic salts, and synthetic colors cannot be produced by microorganisms, whereas complex plant-derived substances (e.g., Gummi arabicum, E414) can often not be produced by chemical synthesis. For this reason, this review article focuses on the production of natural small organic molecules, which are accessible through metabolic engineering of microorganisms either because they represent natural intermediates of their carbon metabolism or because they can be produced after functional introduction of heterologous pathways (Table 1).
In many cases, heterologous enzymes from plants were functionally introduced in microorganisms allowing for the production of natural compounds, which can otherwise only be obtained from plants (Marienhagen and Bott, 2013). A generalized strategy for extraction of natural products from plant material is in most cases not applicable as plants produce such compounds in low amounts and typically harbor complex mixtures of chemically very similar compounds. In addition, the concentrations are subject to regional and seasonal differences and several compounds are only produced under special environmental conditions (e.g., after infections). Although a few plant compounds can nowadays be obtained by direct extraction from plant material (Dai and Mumper, 2010), the above-mentioned drawbacks serve as a strong motivation for reconstructing plant pathways in a controllable and reliable metabolic environment in microbial cell factories (Milke et al., 2018).
The E numbers catalog also includes small organic molecules such as carboxylic acids and aromatic compounds, which are typically produced by the chemical industry. Here, in particular environmental concerns for the production from non-regenerative raw material and the use of toxic chemicals are forces toward establishing alternative production platforms. The fact that microorganisms are naturally capable of producing carboxylic acids and aromatic compounds was recognized as an excellent starting point for metabolic engineering work into this direction in the recent decades (Abbott et al., 2009; Gosset, 2009).
The E numbers catalog classifies food additives according to their major application in foodstuff. However, also distinct and metabolically rather unrelated classes of small organic molecules can have similar application ranges, e.g., aliphatic terpenoids and aromatic phenols are both used as anti-oxidants (Table 1). For this reason, compounds mentioned in the following sections are subdivided based on similar biosynthetic pathways or by use of the same precursor metabolites for production.
The underlying metabolic pathways relevant for the production of food additives have significant influence on the choice of the microorganism used for production and on the metabolic engineering strategies followed for estabilishing and improving production. For some products microorganisms already naturally overproducing the desired compound are tested. In contrast, for products which naturally do not occur in microorganisms it is reasonable to use microbial hosts, which can be easily genetically manipulated. General requirements for microbial production strains include fast growth, cultivation in cheap, and defined growth media to high biomass concentrations, non-pathogenicity, and in particular important when food additives are supposed to be produced the obtained product should have GRAS (“generally recognized as safe”) status. In this review article the categories are chosen based on related biosynthetic pathways as shown in Figure 1 and in each of the following sections the products are sorted by their respective E numbers.
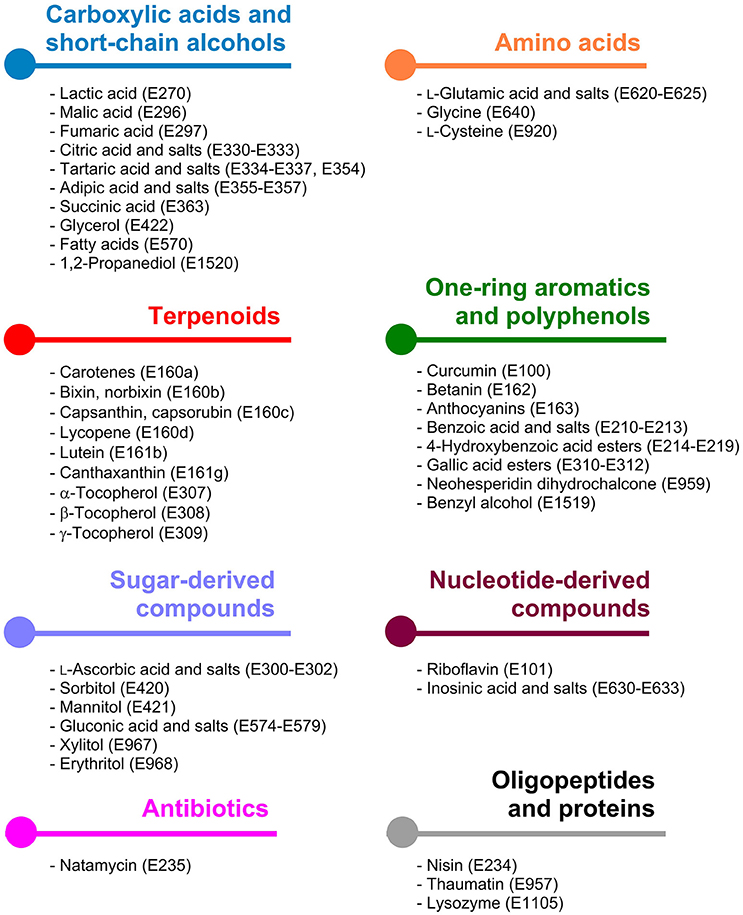
Figure 1. Classification of microbially accessible food additives according to the metabolic pathways being involved in their biosynthesis. Colors indicate the same compound class and are consistently used in Figures 2–8.
Aliphatic carboxylic acids are for the most part used as preservatives or acidity regulators. Most of the carboxylic acids listed in the E numbers catalog can be derived from glycolysis or the tricarboxylic acid (TCA) cycle (Figure 2). These comprise lactic acid, citric acid, succinic acid, fumaric acid, and malic acid.
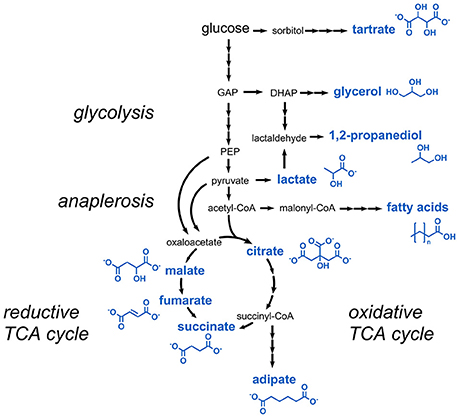
Figure 2. Metabolic pathways involved in the production of carboxylic acids and short-chain alcohols. Three arrows indicate several reaction steps which are not shown in detail. DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate.
Production of lactic acid (E270) was established in several microorganisms, e.g., lactic acid bacteria (Lactobacillus sp. and Lactococcus sp.), Bacillus subtilis, Escherichia coli, Corynebacterium glutamicum, yeasts, and microalgae (Abdel-Rahman et al., 2013). In addition to glucose also lignocellulosic biomass, agro-industrial, and food waste or glycerol were used as raw materials. Typically, yields of 0.85–0.98 g lactic acid per g carbon source were obtained. A maximal titer of 225 g/L lactic acid with a yield of 0.99 g/g was e.g., obtained from glucose during fed-batch cultivation of a non-engineered alkaliphilic Bacillus species (Meng et al., 2012). In addition to the use as food additive lactic acid is also used in large amounts for the production of the biodegradable polyester polylactic acid (Auras et al., 2011).
The TCA cycle intermediate malic acid (E296) is typically produced in Aspergillus species, e.g., A. flavus, A. niger, and A. oryzae (Battat et al., 1991; West, 2011). The latter was further engineered for increased production of malic acid from glycolysis-derived pyruvate by the reductive TCA cycle (Figure 2). An engineered strain overexpressing the genes coding for pyruvate carboxylase, malate dehydrogenase, and a C4-dicarboxylate transporter was capable of producing 154 g/L malic acid with a yield of 1.02 g/g glucose corresponding to 69% of the maximal theoretical yield (Brown et al., 2013). Similar metabolic engineering strategies were also followed in S. cerevisiae, which produced 59 g/L malic acid with a yield of 0.31 g malic acid per g glucose (Zelle et al., 2008). Malic acid production from glycerol was achieved in the plant pathogen Ustilago trichophora by combining adaptive laboratory evolution and metabolic engineering approaches (Zambanini et al., 2017). The highest malic acid titer obtained with the optimized strain was 134 g/L. In E. coli, the introduction of a mutation into the fumarate reductase gene along with the deletion of genes coding for two malic enzymes and lactate dehydrogenase led to the production of 34 g/L malic acid with a product yield of 1.06 g/g glucose (Zhang et al., 2011b). In very recent studies, additional strategies toward malic acid production in E. coli were tested and production from alternative carbon sources such as xylose was demonstrated (Li et al., 2018; Martinez et al., 2018).
For the production of fumaric acid (E297), which is directly formed from malic acid by fumarase activity, strains of Rhizopus nigricans or R. oryzae are applied as these fungi naturally overproduce fumaric acid. A titer of 85 g/L and yield of 0.85 g fumaric acid per g glucose were obtained during cultivation of R. oryzae self-immobilized on plastic discs of a rotary biofilm contactor (Cao et al., 1996). A succinate-producing E. coli mutant was turned into a fumarate producer by deletion of three fumarase-encoding genes (Li et al., 2014b). Additional strain engineering toward reducing the production of acetate led to an optimized strain producing 42 g/L fumaric acid from glycerol with 70% of the maximum theoretical yield.
Production of citric acid (E330) at industrial scale is typically conducted based on wild-type A. niger (Papagianni, 2007). This yeast naturally overproduces citric acid while the molecular mechanism for its accumulation and secretion is still not entirely understood (Karaffa and Kubicek, 2003). In a very recent study, an industrially used A. niger strain obtained from random mutagenesis, which is capable to produce 160 g/L citric acid, was analyzed by transcriptome analysis in comparison to strains producing lower amounts of citric acid (Yin et al., 2017). Significantly regulated genes coding for enzymes of glycolysis and the TCA cycle as well as for putative citrate transporters are the basis for a detailed analysis of the mechanism for citric acid accumulation and thus for future engineering work. Yarrowia lipolytica was recognized as an alternative microorganism for citric acid production from inexpensive carbon sources. Under nitrogen-limiting conditions production of 82 g/L citric acid was shown from glycerol-containing biodiesel waste (Kamzolova et al., 2015).
The dihydroxylated C4 dicarboxylic acid tartaric acid (E334) is structurally closely related to the TCA cycle intermediates succinic acid and malic acid, and can be converted to oxaloacetic acid in one single dehydration reaction (Hurlbert and Jakoby, 1965). Astonishingly, tartaric acid is not derived from a precursor of the TCA cycle but from a completely unrelated metabolic pathway. In higher plants, it was shown that tartaric acid is produced from L-ascorbic acid via 5-keto D-gluconic acid (DeBolt et al., 2006) (Figure 2). The latter is a natural product obtained from sugar oxidation in the acetic acid bacterium Gluconobacter oxydans subsp. suboxydans. It was shown that tartaric acid was spontaneously formed from 5-keto D-gluconic acid in culture medium for G. oxydans containing vanadium salts (Klasen et al., 1992). A titer of 2.1 g/L tartaric acid from 20 g/L of sorbitol was obtained with this organism (Chandrashekar et al., 1999). However, for commercial applications, large amounts of tartaric acid are obtained today as a by-product from wine industry or by chemical conversion of maleic acid or fumaric acid (Church and Blumberg, 1951; Zhang et al., 2011a).
The C6 dicarboxylic acid adipic acid (E355) serves as a major building block for the synthesis of nylon polymers, but is also used in foodstuff as acidulant. Adipic acid can be microbially accessed by several pathways, e.g., by the fatty acid synthesis pathway, by degradation of aromatic compounds, or starting from the TCA cycle intermediates succinyl-CoA or 2-oxoglutarate (Polen et al., 2013; Kallscheuer et al., 2017a) (Figure 2). Current efforts for establishing a bio-based production of adipic acid in microorganisms are summarized in a number of recent review articles (Deng et al., 2016; Kallscheuer et al., 2017b; Kruyer and Peralta-Yahya, 2017). The highest published titer for adipic acid of 2.5 g/L was obtained in E. coli (Cheong et al., 2016). The constructed strain harbors a heterologous catabolic pathway for adipic acid ultimately leading to succinyl-CoA, which was exploited in the reverse direction for adipic acid biosynthesis. The same pathway is present in the thermophilic bacterium Thermobifida fusca in which the overexpression of a gene for an endogenous adipyl-CoA dehydrogenase enabled the production of 2.2 g/L adipic acid without further strain engineering (Deng and Mao, 2015).
The TCA cycle intermediate succinic acid (E363) serves as a flavor enhancer for desserts, dry soups, and drink powder due to its mildly acidic and at the same time slightly salty flavor (Glassner and Datta, 1992). Succinic acid is typically produced via the reductive TCA cycle (Figure 2) under microaerobic or anaerobic conditions for achieving sufficient NADH supply. Several organisms were engineered for its production, e.g., Actinobacillus succinogenes, Basfia succiniciproducens, Mannheimia succiniciproducens, E. coli, C. glutamicum, and S. cerevisiae (Yan et al., 2014; Choi et al., 2016; Meng et al., 2016; Salvachúa et al., 2016; Lange et al., 2017; Mao et al., 2018). The tested strategies during strain engineering included increasing the flux into the pentose phosphate pathway for increasing NADH supply, elimination of competing pathways leading to acetate or lactate formation and overexpression of genes coding for succinate exporter proteins. With these strategies titers in the range of 30–90 g/L and yields of 0.6–1.3 g product per g carbon source were obtained (Ahn et al., 2016).
Nowadays, the triol glycerol (E422) is recognized as a cheap carbon source rather than as a product. This is due to its availability in large quantities as a by-product of biodiesel production (Quispe et al., 2013). However, in the 1990s in particular yeast was extensively engineered for production of glycerol and a maximal titer of 130 g/L and a yield of 0.63 g/g glucose was obtained in the osmotolerant yeast Candida glycerinogenes (Wang et al., 2001). In the recent years, there are still several studies reporting on glycerol production, e.g., in the model cyanobacterium Synechocystis sp. PCC6803 (Savakis et al., 2015), and also production in yeast strains is still further optimized (Tilloy et al., 2014; Yu et al., 2014; Murashchenko et al., 2016; Semkiv et al., 2017).
The E number E570 lists fatty acids and refers in particular to the long-chain fatty acid stearic acid, which is used as anti-caking agent or as plasticizer in chewing gum. For the production of fatty acids typically oleaginous yeasts such as Y. lipolytica, Trichosporon dermatis, or Rhodosporidium toruloides are employed, which are capable of accumulating long-chain fatty acids with a typical length of 16 or 18 carbon atoms. During fed-batch fermentations lipid contents of 25 to 68% (w/w) with a different composition of long-chain fatty acids were obtained (Papanikolaou et al., 2002; Li et al., 2007; Huang et al., 2012; Qiao et al., 2015). The key enzyme for further improving fatty acid synthesis is the acetyl-CoA carboxylase, which catalyzes the ATP-dependent carboxylation of acetyl-CoA to malonyl-CoA. This enzyme catalyzes the first committed step in the fatty acid synthesis pathways and is strictly regulated by different regulation mechanisms for avoiding fatty acid overproduction (Brownsey et al., 2006; Wei et al., 2016).
The short-chain alcohol 1,2-propanediol (1,2-PD, E1520, also known as propylene glycol) is mainly used as a carrier in the production of flavors for foodstuffs. Several microorganisms were found to be capable of fermenting sugars to 1,2-PD and two metabolic routes for its biosynthesis were identified (Saxena et al., 2010). 1,2-PD can either be derived from the glycolysis intermediate dihydroxyacetone phosphate, which is first dephosphorylated to methylglyoxal and subsequently reduced to lactaldehyde and finally to 1,2-PD (Figure 2). Alternatively, in the second pathway lactic acid is reduced to lactaldehyde, which is then further reduced to 1,2-PD. Already in the 1980s, non-engineered Clostridium thermosaccharolyticum strains were found to produce 7.9 g/L 1,2-PD with a yield of 0.27 g/g glucose via methylglyoxal (Cameron and Cooney, 1986). Functional introduction of this pathway in an E. coli strain optimized for glycerol assimilation allowed for the production of 5.6 g/L 1,2-PD from glycerol (Clomburg and Gonzalez, 2011). The second pathway for 1,2-PD production by degradation of lactic acid was identified in Lactobacillus buchneri (Elferink et al., 2001). In E. coli, lactaldehyde was identified as a side product from cleavage of L-fucose (Cocks et al., 1974). Lactaldehyde-derived production of 84 mg/L 1,2-PD was reported in a mutant strain of E. coli constitutively expressing a gene coding for a propanediol dehydrogenase during growth on L-fucose (Cocks et al., 1974). It turned out that the strain was also capable of utilizing 1,2-PD as sole carbon and energy source (Sridhara et al., 1969). Thus, a functional reversal of the underlying catabolic pathway (e.g., by using anaerobic cultivation conditions) might be the basis for establishing 1,2-PD production via lactic acid in E. coli.
Three proteinogenic amino acids are approved by the EU as flavor enhancers, namely L-glutamic acid (E620), glycine (E640), and L-cysteine (E920). L-Glutamic acid and in particular its monosodium salt (E621) are the major compounds responsible for the “umami” taste of foodstuff. L-Cysteine is also used as flour treatment agent to improve baking functionality. All three amino acids can in principle be isolated from hydrolyzed protein or can be produced by engineered microorganisms. In case of glycine, both strategies are not followed for industrial production as this amino acid can be more easily produced by chemical synthesis (Zeng et al., 2016). Especially the expensive step for purification of the L-form from a racemic mixture of chemically synthesized amino acids is not required for glycine as it lacks stereogenic centers.
The L-glutamic acid-overproducing C. glutamicum was isolated in Japan in 1956 during a screening campaign for identifying glutamate-producing bacteria (Kinoshita et al., 1957). In the following decades the mechanism for glutamate secretion was investigated in detail and C. glutamicum was extensively engineering toward increased product titers (Hirasawa and Wachi, 2016). Combined activity during strain engineering and optimization of process conditions led to a strain capable of producing 100 g/L L-glutamic acid with a yield of 0.6 g per g glucose (Ault, 2004). Metabolic engineering efforts for increasing L-glutamic acid production in C. glutamicum are summarized in several books and review articles (Kimura, 2003; Eggeling and Bott, 2005; Sano, 2009). In recent years, alternative organisms such as Pantoea ananatis were also found to be “talented” producers of L-glutamic acid (Katashkina et al., 2009).
L-Cysteine is nowadays for the most part obtained from protein hydrolysis obtained from animal material, e.g., poultry feathers. The natural biosynthesis pathway starting from L-serine and acetyl-CoA was used for engineered L-cysteine production in E. coli and C. glutamicum (Wada and Takagi, 2006) (Figure 3). The rate-limiting reaction in the pathway is the initial step catalyzed the serine O-acetyltransferase (CysE), which is strictly feedback-inhibited by L-cysteine with enzyme inhibition constants in the micromolar range (Denk and Böck, 1987). Metabolic engineering efforts focused on introducing mutations in the respective gene cysE for obtaining less feedback-sensitive enzymes (Kai et al., 2006). By functional introduction of heterologous feedback-insensitive CysE isoenzymes from Arabidopsis thaliana into E. coli an L-cysteine titer of 1.7 g/L was obtained (Takagi et al., 1999). In a subsequent study, deletion of the gene yciW, which encodes an oxidoreductase putatively involved in L-cysteine metabolism in E. coli, led to an increased production of L-cysteine, however the titer of 1.7 g/L already obtained in 1999 was not exceeded (Kawano et al., 2015; Takagi and Ohtsu, 2016). Very recently, additional overexpression of the genes coding for SerA (3-phosphoglycerate dehydrogenase), SerB (phosphoserine phosphatase), and SerC (phosphoserine aminotransferase) involved in the synthesis of the precursor L-serine and deletion of genes coding for enzymes involved in the degradation of L-serine and L-cysteine was tested (Liu et al., 2018). The optimized strain produced 5.1 g/L L-cysteine during fed-batch fermentations.
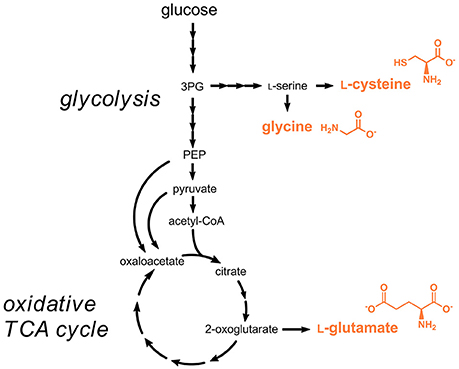
Figure 3. Production pathways for amino acids approved as food additives by the EU. Three arrows indicate several reaction steps which are not shown in detail. 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate.
Terpenoids (also referred to as isoprenoids) are mostly aliphatic compounds derived from units of isoprene (2-methyl-1,3-butadiene). Most of the terpenoids listed in the E numbers catalog are used as colors and antioxidants. Based on the number of condensed isoprene units (isoprene: C5) the products are designated monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), or tetraterpenes (C40). The isoprene units are naturally supplied in form of the two isomers dimethylallyl pyrophosphate (DMAPP) or isopentenyl pyrophosphate (IPP). Two independent pathways responsible for DMAPP/IPP formation were identified: the mevalonate pathway and the methyl-D-erythritol 4-phosphate (MEP) pathway (non-mevalonate pathway) (Goldstein and Brown, 1990; Eisenreich et al., 2004) (Figure 4). The mevalonate pathway produces DMAPP/IPP from three molecules of acetyl-CoA, whereas the MEP pathway requires pyruvate and glyceraldehyde 3-phosphate. The linear, non-cyclic precursor compounds, from which all terpenoids with the corresponding length derive, are geranyl pyrophosphate (C10), farnesyl pyrophosphate (C15), geranylgeranyl pyrophosphate (C20), and phytoene (C40). These precursors are cyclized by downstream enzymes and are often further modified, e.g., by hydroxylation reactions.
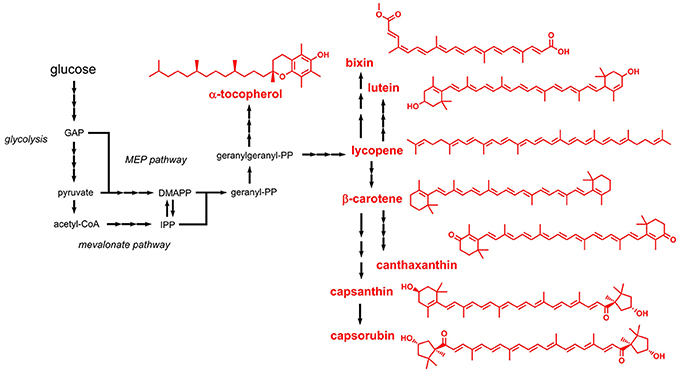
Figure 4. Biosynthetic pathways leading to terpenoids approved as food additives by the EU. Three arrows indicate several reaction steps which are not shown in detail. GAP, glyceraldehyde 3-phosphate; MEP, methyl erythritol phosphate; DMAPP, dimethylallylpyrophosphate; IPP, isopentenylpyrophosphate.
Lycopene (E160d) is a central tetraterpene, which represents the precursor for biosynthesis of β-carotene (E160a), capsanthin and capsorubin (both E160c), lutein (E161b), and canthaxanthin (E161g) (Figure 4). Before starting metabolic engineering toward lycopene production in E. coli, the DMAPP/IPP-forming mevalonate pathway was reconstituted in vitro and the steady-state kinetic and biochemical parameters were analyzed (Zhu et al., 2015). Subsequent coupling of the optimized pathway with downstream enzymes required for converting DMAPP/IPP to lycopene led to an E. coli strain producing 1.4 g/L lycopene during fed-batch fermentation. Similar titers in the range of 0.9 - 1.1 g/L lycopene were also obtained in related studies performed with E. coli as production host (Zhang et al., 2015b; Xu et al., 2018). Alternative hosts for lycopene production include e.g., Blakeslea trispora and Y. lipolytica (Xu et al., 2007; Matthäus et al., 2014), which are also subject of a recent review article on engineered lycopene production (Hernández-Almanza et al., 2016).
Production of lycopene-derived β-carotene was shown in engineered yeasts such as S. cerevisiae and Rhodotorula glutinis (Bhosale and Gadre, 2001; Li et al., 2013). In S. cerevisiae, genes from the carotenoid-producing yeast Xanthophyllomyces dendrorhous were expressed, which led to the production of 5.9 mg β-carotene per g dry weight (Verwaal et al., 2007). One of the introduced genes codes for a 3-hydroxy-3-methylglutaryl-CoA reductase which catalyzes the rate-limiting step in the mevalonate pathway supplying the precursors DMAPP/IPP (Chappell et al., 1995). The entire mevalonate pathway and the β-carotene biosynthetic pathway were also functionally introduced into E. coli (Yoon et al., 2009). In this study, the best-performing strain with plasmid-borne expression of 10 heterologous genes produced 0.47 g/L β-carotene in complex medium with 2% glycerol.
Apocarotenoids such as bixin and norbixin (E160b) are obtained by oxidative cleavage of lycopene (Figure 4). Both compounds were produced by functional introduction of the lycopene cleavage dioxygenase from the plant achiote (Bixa orellana) together with an aldehyde dehydrogenase and a carboxyl methyltransferase into an engineered lycopene-producing E. coli strain (Bouvier et al., 2003). A production level of 5 mg bixin/g dry weight was obtained.
Capsanthin and capsorubin (E160c) belong to the class of xanthophylls (oxygen-containing carotenoids) and are the major carotenoids in red pepper fruits (Lefebvre et al., 1998). Both compounds can be produced from lycopene by six reactions steps including two cyclization and two hydroxylation steps and subsequent epoxidation and de-epoxidation. Production of capsanthin and capsorubin has not been achieved in microorganisms until today, but strategies for production in E. coli were proposed (Misawa, 2013).
Lutein (E161b), which is also a plant-derived xanthophyll, is produced from lycopene by two cyclization and two hydroxylation reactions (Kim and DellaPenna, 2006). Biotechnological lutein production focused exclusively on the use of microalgae, which naturally produce lutein (Fernández-Sevilla et al., 2010). It turned out that lutein production capabilities strongly depend on environmental and operating factors. A third xanthophyll, canthaxanthin (E161g), is naturally produced from β-carotene by two reaction steps, which are both catalyzed by the β-carotene ketolase CrtW. Several microorganisms naturally producing canthaxanthin were identified, e.g., Haloferax alexandrinus, Gordonia jacobaea, and Dietzia natronolimnaea (de Miguel et al., 2000; Asker and Ohta, 2002; Gharibzahedi et al., 2012). With mutated strains and by choosing cultivation conditions promoting canthaxanthin production, product titers in the range of 2–8 mg/L were obtained (Gharibzahedi et al., 2012; Rostami et al., 2014). The β-carotene ketolase gene crtW from Agrobacterium aurantiacum was already functionally expressed in an E. coli strain engineered for β-carotene production, which led to the production of detectable amounts of canthaxanthin in this organism (Misawa et al., 1995).
Some carotenoid biosynthetic pathways lead to the production of aromatic compounds, e.g., okenone and isorenieratene (these compounds are not listed in the E numbers catalog). In such cases the aliphatic rings, e.g., in β-carotene, are oxidized to aromatic (benzene) rings. Tocopherols (E307, E308, and E309) also contain an aromatic ring but represent an exception to the above-mentioned biosynthesis strategy. Tocopherols can be classified as terpenoids because they are derived from geranylgeranyl pyrophosphate (Figure 4). However, the aromatic ring present in tocopherol is not formed during terpenoid biosynthesis but is obtained from homogentisate (2,5-dihydroxyphenylacetate), which in turn is produced from the aromatic amino acid L-tyrosine (Arias-Barrau et al., 2004). Thus, microbial production of tocopherol requires engineering of two unrelated pathways, namely the terpenoid biosynthetic pathway and the L-tyrosine-forming shikimate pathway. This might be a reason why tocopherol production focused on natural producers, namely photosynthetic microorganisms such as Dunaliella tertiolecta and Euglena gracilis (Tani and Tsumura, 1989; Carballo-Cárdenas et al., 2003). In E. gracilis, a titer of 144 mg/L α-tocopherol was achieved.
Aromatic compounds approved as food additives by the EU can be classified into two large groups: one-ring aromatics and polyphenols. One-ring aromatics are typically benzoic acid derivatives, while polyphenols comprise a large group of structurally diverse and more complex secondary metabolites in plants (Quideau et al., 2011). All compounds of these two classes are derived from the aromatic amino acid-forming shikimate pathway, which requires the pentose phosphate pathway-derived erythrose 4-phosphate and glycolysis-derived phosphoenolpyruvate as precursors (Figure 5). One-ring aromatics are typically produced from intermediates of the shikimate pathway (e.g., shikimate or chorismate), whereas all polyphenols are derived from phenylpropanoids, which in turn are produced by deamination of the aromatic amino acids L-phenylalanine or L-tyrosine (Vogt, 2010) (Figure 5).
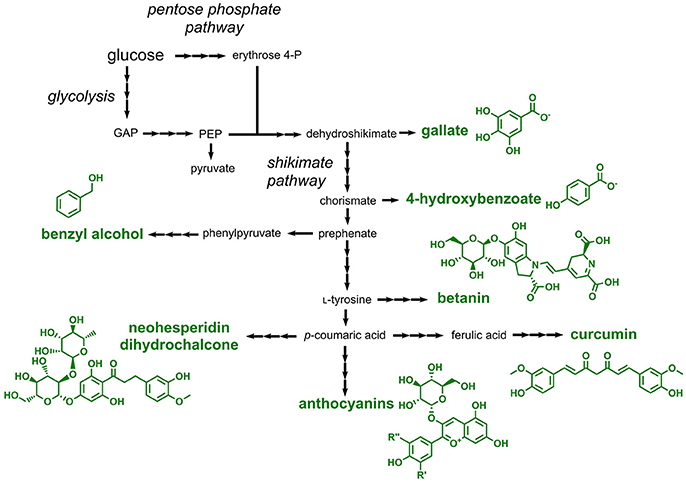
Figure 5. Pathways involved in the biosynthesis or aromatic food additives approved by the EU. Three arrows indicate several reaction steps which are not shown in detail. GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate.
Four different one-ring aromatics, which can be accessed with microorganisms, are listed as E numbers: benzoic acid and different salts derived thereof (E210–E213), 4-hydroxybenzoic acid esters (E214–E219), gallic acid esters (E310–E312), and benzyl alcohol (E1519). Although a natural pathway for the production of benzoic acid by chain-shortening of the phenylpropanoid cinnamic acid was identified in plants (Moerkercke et al., 2009), this compound was not produced in engineered microorganisms until today. This is probably due to the fact that benzoic acid can be easily obtained from petroleum-derived toluene (Kaeding et al., 1965). In contrast, several microorganisms were engineered for the production of 4-hydroxybenzoic acid and gallic acid (3,4,5-trihydroxybenzoic acid), which are direct precursors for the corresponding methyl-, ethyl- and propyl esters approved as food additives. 4-hydroxybenzoic acid is microbially produced by cleavage of the shikimate pathway intermediate chorismate (Siebert et al., 1994) (Figure 5). In addition to the functional introduction of the required chorismate pyruvate lyase the feedback-regulation at the initial step of the shikimate pathway needs to be abolished. For this purpose, feedback-resistant enzymes catalyzing this initial step are applied (Weaver and Herrmann, 1990; Fukuda et al., 1991; Jossek et al., 2001). By following this strategy, 4-hydroxybenzoic acid was produced e.g., in E. coli, C. glutamicum, S. cerevisiae, and Pseudomonas putida (Meijnen et al., 2011; Noda et al., 2016; Averesch et al., 2017; Kallscheuer and Marienhagen, 2018). Very recently, a titer of 36.6 g/L and a yield of 0.31 g/g glucose was obtained in engineered C. glutamicum during fed-batch fermentation (Kitade et al., 2018). An E. coli strain overproducing the shikimate pathway intermediate 3-dehydroshikimate was capable of producing 20 g/L gallic acid, when an endogenous gene coding for a dehydroshikimate dehydrogenase was overexpressed (Kambourakis et al., 2000). 114 mg/L benzyl alcohol was produced from glucose in E. coli using a non-natural pathway starting from the shikimate pathway intermediate phenylpyruvate (Pugh et al., 2015).
Plant-derived polyphenols present in fruits and vegetables are part of our daily diet. Thus, it is surprising that only four polyphenols are approved as food additives by the EU. These include curcumin (E100, a yellow colorant), betanin (E162, a red colorant), anthocyanins (E163, a large class of plant colorants), and neohesperidin dihydrochalcone (E959, a natural sweetener).
Curcumin is produced starting from the phenylpropanoid p-coumaric acid, which in turn is obtained from the non-oxidative deamination of L-tyrosine (Katsuyama et al., 2009). p-Coumaric acid is first converted to the phenylpropanoid ferulic acid by hydroxylation and subsequent O-methylation. Ferulic acid is then CoA-activated by the activity of a CoA-ligase. The key enzyme curcumin synthase subsequently condenses the CoA-thioesters of ferulic acid and of a diketide derived from ferulic acid yielding curcumin. Curcumin production in the range of 60 mg/L from supplemented ferulic acid was demonstrated in E. coli by expression of heterologous genes coding for the CoA ligase and the curcumin synthase (Katsuyama et al., 2008). In a more recent study, curcumin production with a titer of 0.6 mg/L was achieved in E. coli starting from L-tyrosine (Wang et al., 2015).
Betanin (E162) is a glycosylated betacyanine, which is a major compound present in beetroot. This natural red dye is produced starting from L-tyrosine, which is first hydroxylated yielding L-3,4-dihydroxyphenylalanine (also known as Levodopa or L-DOPA). By the activity of two independent enzymes one molecule of L-DOPA is converted to cyclo-DOPA (precursor 1) and a second one to betalamic acid (precursor 2). Precursor 1 and 2 are subsequently ligated yielding betanidin, which is subsequently 5-O-glycosylated to betanin (Tanaka et al., 2008). The entire pathway was recently reconstructed in S. cerevisiae by functional expression of genes from various plants (Grewal et al., 2018). The constructed strains were not only capable of producing 17 mg/L betanin, but also converted alternative aromatic amines (instead of cyclo-DOPA) to the corresponding betacyanine dyes.
In addition to betanin, also plant-derived anthocyanins (E163) find an application as colorants in foodstuff. For the synthesis of anthocyanins the L-tyrosine-derived and CoA-activated phenylpropanoid p-coumaroyl-CoA acid is first condensed with three molecules of malonyl-CoA yielding a tetraketide intermediate, which is cyclized to the compound naringenin chalcone. All steps during this reaction, the malonyl-CoA-dependent chain elongation as well as the cyclization step, are catalyzed by chalcone synthases (Ferrer et al., 1999). By the activity of chalcone isomerases, naringenin chalcone is subsequently isomerized to the flavanone naringenin. Naringenin, the first compound in the pathway harboring the typical flavonoid core structure, is then further converted by plant dioxygenases and reductases giving rise to the anthocyanidin pelargonidin, which is further stabilized by O-glycosylation at C3 (the glycosylated anthocyanidin is then designated anthocyanin). Taken together, the overall pathway from p-coumaric acid includes seven reaction steps (1: CoA activation, 2: chain elongation and cyclization, 3: isomerization, 4: hydroxylation, 5: reduction, 6: oxidation, 7: glycosylation) (Figure 5). The resulting pelargonidin-3-O-glucoside can be further converted to related anthocyanins by ring hydroxylation or O-methylation reactions. For achieving microbial anthocyanin production in particular E. coli strains were constructed (Yan et al., 2005, 2008; Lim et al., 2015). Recently, the production of 9.5 mg/L pelargonidin-3-O-glucoside (callistephin) from glucose was achieved by a co-culture of four different E. coli strains together expressing 15 heterologous genes (Jones et al., 2017).
Neohesperidin dihydrochalcone (E959) is a rutinosylated chalcone, i.e., it is decorated with rutinose (rhamnose-α-1,6-glucose) (Figure 5). This chalcone is present in citrus fruits and is applied as a natural sweetener in beverages, yogurt and ice cream. Neohesperidin dihydrochalcone was not produced in engineered microbes so far, but a very similar compound, naringin dihydrochalcone, which differs from neohesperidin dihydrochalcone only by one O-methyl-group, was obtained in engineered S. cerevisiae (Eichenberger et al., 2017). The constructed strain heterologously expressed nine genes and produced 12 mg/L naringin dihydrochalcone from glucose. Based on these results a microbial production of neohesperidin dihydrochalcone can be achieved by including an additional O-methyltransferase capable of converting naringin dihydrochalcone to neohesperidin dihydrochalcone.
Subject of this section are food additives listed in the E numbers catalog, which are more or less directly derived from the sugar metabolism. Many different compounds can in principle be “derived from sugar”; hence only compounds derived from glycolysis and the pentose phosphate pathway are included here (Figure 6).
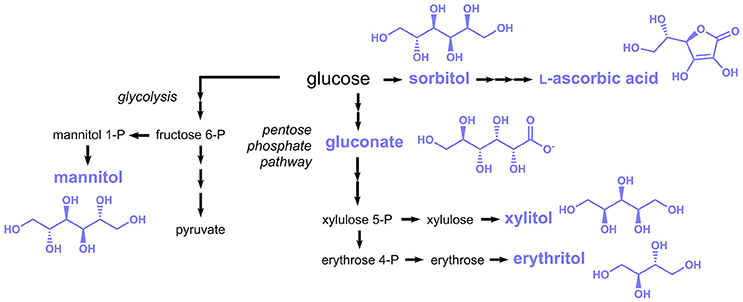
Figure 6. Biosynthesis of sugar-derived compounds approved as food additives by the EU. Three arrows indicate several reaction steps which are not shown in detail.
L-Ascorbic acid (E300, also known as vitamin C) is a vitamin traditionally consumed with fruits and vegetables, which is today produced at the scale of more than 100,000 tons per year (Pappenberger and Hohmann, 2013). L-Ascorbic acid is also extensively added to foods, beverages and pharmaceuticals. The established industrial production process of L-ascorbic acid is based on D-sorbitol or sorbose, which are first oxidized to 2-keto-L-gulonic acid by Gluconobacter oxydans or Ketogulonicigenium vulgare, repectively and then chemically converted to L-ascorbic acid (Pappenberger and Hohmann, 2013). G. oxydans was capable to produce 130 g/L 2-keto-L-gulonic acid from 150 g/L sorbitol (Saito et al., 1997), while a titer of 60 g/L was achieved from 70 g/L sorbose in K. vulgare (Ning et al., 1988).
Sorbitol (E420), the precursor for industrial L-ascorbic acid production, is also a food additive approved by the EU and is mainly used as natural sweetener, e.g., in chewing gum. Although sorbitol is traditionally synthesized by catalytic hydrogenation of glucose (Kusserow et al., 2003), it was also produced in engineered microorganisms from glucose in a single reduction step. In Zymomonas mobilis the enzyme glucose-fructose oxidoreductase was shown to catalyze the reduction of glucose to sorbitol by simultaneously oxidizing fructose to gluconolactone (Zachariou and Scopes, 1986). Not surprisingly, this organism was exploited for the production of sorbitol, in particular in the 1990s (Silveira and Jonas, 2002). In more recent studies also alternative hosts were applied, e.g., the cyanobacterium Synechocystis sp. and the lactic acid bacterium Lactobacillus plantarum (Jan et al., 2017; Chin et al., 2018).
Additional polyols with applications similar to sorbitol comprise mannitol (E421), xylitol (E967), and erythritol (E968) (Figure 6). These are also produced industrially by catalytic hydrogenation (Schiweck et al., 2000), but several microorganisms were engineered toward their production. Mannitol is typically produced in lactic acid bacteria, e.g., Lactococcus lactis or Lactobacillus reuteri via the glycolysis intermediate fructose-6-phosphate, which is reduced to mannitol-1-phosphate and subsequently dephosphorylated. Yields of 0.5–0.6 g mannitol per g glucose or fructose were obtained (Song and Vieille, 2009). A constructed C. glutamicum strain expressing genes coding for a mannitol dehydrogenase from Leuconostoc pseudomesenteroides and a glucose/fructose transporter from Z. mobilis was capable to produce 87 g/L mannitol from 94 g/L fructose (Bäumchen and Bringer-Meyer, 2007). The xylose reductase of Pichia stipitis was introduced into S. cerevisiae, which led to a xylose conversion rate of 95% in the recombinant strain (Hallborn et al., 1991). In a more recent study, the sugar import of a Kluyveromyces marxianus strain harboring a xylose reductase from Neurospora crassa was engineered and a final titer of 312 g/L xylitol was obtained during fed-batch fermentation (Zhang et al., 2015a).
For the production of erythritol, erythrose 4-phosphate (E4P) derived from the pentose phosphate pathway is first dephosphorylated to erythrose and subsequently reduced to erythritol (Figure 6). In some microorganisms, e.g., lactic acid bacteria, E4P is first reduced to erythritol 4-phosphate and then dephosphorylated. Yields of 0.3 to 0.4 g/g glucose and product titers in the range of 120–250 g/L were obtained in microorganisms engineered toward erythritol production (Moon et al., 2010).
Oxidation of glucose at the C1-atom leads to gluconic acid (E574) (Figure 6), which is used in food applications as an acidity regulator. The enzymatically-driven oxidation reaction can either be coupled to the reduction of a cofactor, e.g., NAD(P)+, FAD or a quinone (glucose 1-dehydrogenase) or alternatively molecular oxygen can serve as oxidant (glucose oxidase). The latter reaction also yields H2O2 as byproduct, which needs to be rapidly detoxificated by a catalase. Production of gluconic acid was achieved in an A. niger strain with high activity of glucose oxidase and catalase (Znad et al., 2004). After 60 h of cultivation a nearly complete conversion of 150 g/L glucose to 150 g/L gluconic acid was observed. Further optimization of the process conditions reduced the production time to 15 h and improved the yield and final titer to 1.05 g/g and 311 g/L respectively (Lu et al., 2015).
Riboflavin (E101, vitamin B2), a yellow colorant, and inosinic acid (E630), a flavor enhancer, are compounds derived from the nucleotide metabolism. Biosynthesis of riboflavin requires two precursors, namely the pentose phosphate pathway intermediate ribulose 5-phosphate and the nucleotide guanosine 5'-triphosphate (GTP), which is derived from the purine metabolism (Figure 7). Inosinic acid, which is also designated inosine 5'-monophosphate (IMP), is a pathway intermediate in the purine metabolism leading to biosynthesis of GTP. Thus, it is not surprising that similar metabolic engineering strategies were followed for establishing production of riboflavin and inosinic acid.
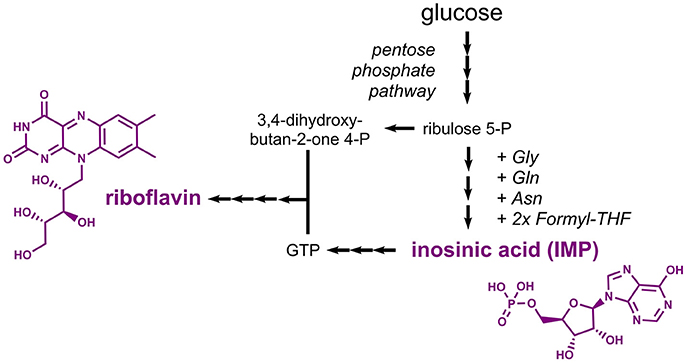
Figure 7. Pathways leading to production of the nucleotide-derived compounds inosinic acid and riboflavin. Three arrows indicate several reaction steps which are not shown in detail. Gly, glycine; Gln, glutamine; Asn, asparagine; THF, tetrahydrofolate; IMP, inosine monophosphate; GTP, guanosine triphosphate.
Microbially produced riboflavin is typically obtained from the filamentous fungus Ashbya gossypii or from the yeast Candida famata as both are natural riboflavin overproducers (Stahmann et al., 2000). A. gossypii was extensively engineered toward increased riboflavin titers in the recent years. To this end, rational approaches were followed to increase the carbon fluxes into the purine and riboflavin biosynthetic pathways. It turned out that reduced expression of the adenylosuccinate synthase gene ADE12 led to an increased flux from IMP into the guanosine monophosphate (GMP) branch and reduced the competing flux into the adenosine monophosphate (AMP) branch. The overexpression of five genes coding for enzymes of the riboflavin synthesis pathway in a strain with reduced ADE12 expression allowed for the production of 0.52 g/L riboflavin (Ledesma-Amaro et al., 2015). Very recently, the riboflavin production capabilities were further improved based on a 13C metabolic network analysis, which improved the final titer by 45% (the actual titer is not mentioned) (Schwechheimer et al., 2018). Metabolic engineering work in E. coli focused on overexpression of the native riboflavin biosynthesis genes ribABDEC along with additional modifications leading to reduced by-product formation and an increased flux into the pentose phosphate pathway (Lin et al., 2014). The best strain produced 2.7 g/L riboflavin with a yield of 0.14 g/g glucose. In engineered B. subtilis strains, riboflavin titers of 4.9 g/L were obtained (Wang et al., 2014).
Already in the 1960s, it was found that an adenine-auxotrophic Micrococcus glutamicus strain accumulated inosinic acid to a concentration of 0.75 g/L (IMP Na2 x 7.5 H2O) (Nakayama et al., 1964). Production was also achieved in Corynebacterium ammoniagenes strains obtained after random mutagenesis experiments, which were capable of accumulating 7.5 g/L inosinic acid (Tomita et al., 1991). Production of inosine, which can be obtained from inosinic acid by one dephosporylation step, was demonstrated in B. subtilis (Asahara et al., 2010). The bacterium was engineered by initially deleting the genes coding for enzymes involved in the conversion of inosinic acid to GMP and AMP. Subsequently, expression of purine biosynthetic genes organized in the 12-gene pur cluster was deregulated by deletion of the gene encoding the regulator PurR and by removing a riboswitch. The resulting mutant strain was shown to produce 6 g/L inosine from 30 g/L glucose (Asahara et al., 2010). Enzymes suitable for the required conversion of inosine into inosinic acid were characterized in vitro (Liu et al., 2012) and were more recently also applied in a whole-cell biotransformation (Yuan et al., 2016). As an alternative strategy, inosinic acid can also be produced by deamination of AMP (Li et al., 2017).
The E numbers catalog also includes oligopeptides and proteins, which serve as preservatives, sweeteners or antibiotics. Nisin (E234), an antibiotic oligopeptide consisting of 34 amino acids, is naturally produced by L. lactis and shows activity against Gram-positive bacteria (Severina et al., 1998). Nisin is encoded by the gene nisA and thus produced by ribosomes and not by nonribosomal peptide synthetases as many other peptide antibiotics (Kaletta and Entian, 1989). Strains of the natural producer L. lactis produced nisin by fermentation of milk or whey. It turned out that utilization non-filtrated milk whey allows for much higher nisin titers compared to filtrated milk whey. With different L. lactis strains nisin titers of 11.1 g/L were obtained (de Arauz et al., 2008). Metabolic engineering efforts toward increased production of nisin in L. lactis have been reviewed very recently (Özel et al., 2018).
Thaumatin (E957) is a mixture of six proteins (thaumatin I, II, III, a, b, and c, all consisting of 207 amino acids). These sweet-tasting proteins are produced in berries of the plant katamfe (Thaumatococcus daniellii) (Wel and Loeve, 1972), which grows in western and central Africa (Adansi and Holloway, 1975). Already in 1982, one of the thaumatin genes was functionally expressed in E. coli (Edens et al., 1982). Later, thaumatin II was produced in Aspergillus awamori strains as well as in B. subtilis by expression of a codon-optimized thaumatin II gene and subsequent protein secretion (Illingworth et al., 1988; Moralejo et al., 1999). Thaumatin production in A. awamori was improved by deleting a gene encoding the protease aspergillopepsin B and by simultaneous overexpression of bipA encoding an endoplasmic reticulum chaperone. With this strategy, a titer of 13 mg/L properly folded thaumatin II was obtained (Moralejo et al., 2002; Lombraña et al., 2004).
Due to its antimicrobial activity lysozyme (E1105) is used as a preservative in foodstuff. Lysozyme cleaves 1,4-β-bonds in peptidoglycan, which disturbs the integrity of the cell wall and leads to lysis of bacterial cells (Mir, 1977). Currently, commercially available lysozyme is obtained from egg white, which has several drawbacks, e.g., it requires laborious purification and can cause immunological problems in humans. As an alternative to extraction of lysozyme from egg white, recombinant A. niger was shown to be capable of producing this enzyme with a titer of 0.21 g/L (Gheshlaghi et al., 2005). In the recent years, there was increased interest in establishing microbial production of human lysozyme with higher activity compared to hen egg lysozyme, e.g., using Pichia pastoris, E. coli, S. cerevisiae, and Kluyveromyces lactis (Ercan and Demirci, 2016). Titers in the range of 0.03–0.13 g/L human lysozyme were obtained by expressing the human gene in microbial host organisms.
Natamycin (E235), a macrolide antibiotic from Streptomyces natalensis, has a similar mechanism of action as nisin (formation of pores in cell membranes), but acts exclusively against fungi, e.g. Candida, Aspergillus and Penicillium species (Pedersen, 1992). Natamycin is approved as a preservative for the surface treatment of hard or semi-hard cheese. It is also used for dried and salted sausages. Although nisin and natamycin share a similar mechanism of action, both compounds are structurally and metabolically unrelated. Production of natamycin is initiated by a polyketide synthase which uses methylmalonyl-CoA and 12 molecules of malonyl-CoA as substrates (Liu et al., 2015) (Figure 8). The resulting compound, referred to as natamycinolide, is further modified by carboxylation, glycosylation, and epoxidation, which give rise to natamycin. The biosynthesis pathway for natamycin is highly complex and (amongst others) involves the large polyketide synthases PimS1 and PimS2 with a length of 6,797 and 9,507 amino acids, respectively. Thus, it is not surprising that production of natamycin focused on natural producers such as S. natalensis and closely related species. The natamycin production capabilities of wild-type S. natalensis were optimized by testing different sources of carbon, nitrogen and phosphate. With glucose, potassium dihydrogen phosphate and a mixture of beef extract and yeast extract production of 1.5 g/L natamycin was observed (Farid et al., 2000). In a related study, it was tested whether the supplementation of short-chain acids or alcohols has a positive impact on natamycin production. As a result, it turned out that 1-propanol supplementation increased the natamycin titer to 10.4 g/L, which was 17% higher than the control strain without supplementation (Li et al., 2014a). The same effect was also observed for the supplementation of acetic acid and propionic acid (Elsayed et al., 2013). In this study a final titer of 4.0 g/L natamycin was achieved. Natamycin production in the natural producer Streptomyces gilvosporeus was improved by coupling the expression of the natamycin biosynthetic gene cluster to the expression of a kanamycin resistance gene (Wang et al., 2016). After seven iterative rounds of mutagenesis and selection for increased kanamycin resistance a strain producing 14.4 g/L natamycin was obtained.
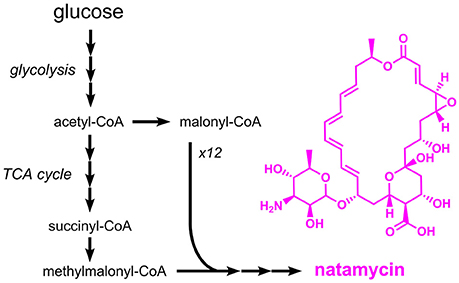
Figure 8. Overview on the biosynthetic pathway for the macrolide antibiotic natamycin. Three arrows indicate several reaction steps which are not shown in detail.
Forty-two of the 316 compounds currently approved as food additives by the EU can in principle be produced in microorganisms until today (Table 1). It is impressive that a broad range of different compounds ranging from small organic acids to more complex secondary metabolites or polymers such as oligopeptides can now be accessed by tailor-made microbial cell factories (Table 2). Engineering efforts toward metabolite overproduction follow some basic strategies or principles, which generally apply for all tested microorganisms. These include increasing the activity of enzymes catalyzing rate-limiting reactions, the abolishment of natural regulatory circuits, e.g., transcriptional control of gene expression or allosteric feedback inhibition mechanisms, and the elimination of complex networks of competing pathways for preventing side product formation. Strain modification also targets increased carbon source uptake or product export or an increased resistance of the host against the product or a pathway intermediate. In addition to increasing product titers there is often also room for reduction of production costs. This can e.g., be achieved by stable integration of heterologous genes into the host genome and expression from constitutive promoters, which allows for avoiding the use of antibiotics (for plasmid maintenance) and inducer compounds. In several cases, changing the expression rate of endogenous genes can be superior to gene deletion. This is especially true when deletions lead to auxotrophic strains, which can be circumvented by downregulating the gene instead of deleting it.
Interestingly, the range of host organisms exploited for production of food additives is not restricted to commonly used organisms such as E. coli and S. cerevisiae, but comprises a broad spectrum of bacteria, fungi, and microalgae (Table 2). Although it is reasonable to use established platform organisms, for which a wealth of metabolic engineering tools is available, it is also important to sample the natural diversity of microorganisms (Pei and Schmidt, 2018). This is in particular true for those organisms already naturally overproducing compounds of interest. Here, the development of tools for strain modification not only serves for increasing the overall product titers. It also contributes to extend our knowledge of the microbial physiology and its diversity and might enable identification of novel compounds or pathway intermediates. This information might also enable the use of alternative (or synthetic) pathways for production.
Currently, production approaches for approved food additives are at three different stages reflecting economical viability of large-scale production, which are designated stage I, II, and III here. The titers required for an economically viable production (corresponds to stage III) strongly depend on the complexity and value of the product, thus only a rough classification for each stage can be given here. For several products microbial production is already economically viable and industrial processes are well-established, e.g., for L-glutamate, riboflavin, L-ascorbic acid, succinic acid, and lactic acid, which can be classified as stage III. For these compounds product titers of 100 g/L or more are typically obtained and also methods and processes for product purification are established. Stage II comprises products, for which titers in the range of 10–100 g/L are obtained. For such products titers obtained with microorganisms are close to economical viability and it can be expected that industrial production is in reach. Examples for products at stage II are adipic acid and one-ring aromatics such as 4-hydroxybenzoic acid. As one example, the company Verdezyne opened a pilot plant for bio-based adipic acid production with yeast in 2011 (Tetzlaf, 2011). Current challenges during microbial overproduction of stage III and II compounds are metabolic imbalances in the producer strain (e.g., with regard to the availability of reducing equivalents or ATP), or a depletion of metabolites required for sufficient biomass formation, but also product toxicity and insufficent product export. These challenges can be addressed by additional strain engineering and by choosing suitable process conditions (e.g., using two stage cultivation strategies with a production phase decoupled from biomass formation).
Products with titers in the mg/L range belong for the most part to stage I. In particular for more complex plant-derived polyphenols and terpenoids titers obtained in engineered microorganism are often still too low for establishing production at larger scale and today most of these compounds are still obtained by extraction from plants containing the desired compounds in larger amounts. Major challenges during production of plant natural products with microorganisms are often directly associated to their bioactivity as many of them have anti-microbial activities or tend to react with oxygen or radicals (anti-oxidants). For these products the optimization of cultivation conditions should focus on preventing oxidation and on reducing toxicity for the production host.
As only naturally-occurring metabolic pathways were exploited for food additive production in engineered microorganisms all compounds discussed in the main section of this review article are natural compounds. This is somehow self-evident when taking into consideration that synthetic compounds can typically not be produced using natural metabolic pathways. On the contrary, this does not mean that only pathways evolved by nature must be followed for production. One recent example is the plant-derived stilbene resveratrol, which is naturally produced from the phenylpropanoid p-coumaric acid. For circumventing a bottleneck reaction at the stage of aromatic amino acids the functional reversal of a bacterial catabolic pathway was exploited as a novel route toward resveratrol production (Kallscheuer et al., 2016). In another study, the relaxed substrate specificity of enzymes was exploited for producing small organic compounds, which cannot be obtained by pathways evolved by nature (Cheong et al., 2016).
In the recent years there was an increasing interest in replacing synthetic food additives by their natural counterparts. This is due to that fact that many synthetic food additives were shown to have negative effects on human health. Scientists have been pointing out for years that synthetic dyes may be involved in the development of attention deficit hyperactivity syndrome (ADHD) (Stevens et al., 2013). Since July 2010, the EU has made the warning label “May affect the activity and attention of children” mandatory for all manufacturers using the controversial substances. The new regulation applies to the dyes tartrazine (E102), quinoline yellow (E104), yellow-orange S (E110), azorubine (E122), cochineal red (E124), and allurred (E129). Also negative effects on health of artificial sweeteners are currently discussed. Cyclamic acid is banned in the US and UK due its potential links to cancer, but it is still approved as food additive in the EU as E952. Also the role of artificial sweeteners in the prevention of obesity is controversially discussed (Suez et al., 2014).
In contrast, natural products approved by the EU are generally recognized as safe, well-accepted by customers and some even show health-promoting effects (Vauzour et al., 2010). It can be expected that future efforts for increasing microbial production of such natural compounds will profit from novel methods such as CRISPR/Cas9 and from combination of rational strain engineering, adaptive laboratory evolution and high-throughput screening approaches (Schallmey et al., 2014; Eggeling et al., 2015; Shalem et al., 2015). One promising example are transcription factor-based biosensors for screening of strain libraries, e.g., for production of L-lysine, succinic acid, adipic acid, and also for plant polyphenols such as naringenin or quercetin (Eggeling et al., 2015). It is highly likely that such novel synthetic biology approaches will render microbial production especially of many plant natural compounds economically viable within the next 10–15 years.
The author confirms being the sole contributor of this work and approved it for publication.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to thank Dr. Jan Marienhagen for critical reading of the manuscript.
Abbott, D. A., Zelle, R. M., Pronk, J. T., and Van Maris, A. J. (2009). Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 9, 1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x
Abdel-Rahman, M. A., Tashiro, Y., and Sonomoto, K. (2013). Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 31, 877–902. doi: 10.1016/j.biotechadv.2013.04.002
Adansi, M., and Holloway, H. (1975). “The cultivation of Katamfe (Thaumatococcus daniellii, Benth.),” in IV Africa Symposium on Horticultural Crops, Vol. 53 (Kumasi) 403–406.
Ahn, J. H., Jang, Y.-S., and Lee, S. Y. (2016). Production of succinic acid by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 42, 54–66. doi: 10.1016/j.copbio.2016.02.034
Arias-Barrau, E., Olivera, E. R., Luengo, J. M., Fernández, C., Galán, B., García, J. L., et al. (2004). The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186, 5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004
Asahara, T., Mori, Y., Zakataeva, N. P., Livshits, V. A., Yoshida, K.-I., and Matsuno, K. (2010). Accumulation of gene-targeted Bacillus subtilis mutations that enhance fermentative inosine production. Appl. Microbiol. Biotechnol. 87, 2195–2207. doi: 10.1007/s00253-010-2646-8
Asker, D., and Ohta, Y. (2002). Production of canthaxanthin by Haloferax alexandrinus under non-aseptic conditions and a simple, rapid method for its extraction. Appl. Microbiol. Biotechnol. 58, 743–750. doi: 10.1007/s00253-002-0967-y
Ault, A. (2004). The monosodium glutamate story: the commercial production of MSG and other amino acids. J. Chem. Educ. 81:347. doi: 10.1021/ed081p347
Auras, R. A., Lim, L.-T., Selke, S. E., and Tsuji, H. (2011). Poly (Lactic Acid): Synthesis, Structures, Properties, Processing, and Applications. Hoboken, NJ: John Wiley and Sons.
Averesch, N. J., Prima, A., and Krömer, J. O. (2017). Enhanced production of para-hydroxybenzoic acid by genetically engineered Saccharomyces cerevisiae. Bioprocess. Biosyst. Eng. 40, 1283–1289. doi: 10.1007/s00449-017-1785-z
Battat, E., Peleg, Y., Bercovitz, A., Rokem, J. S., and Goldberg, I. (1991). Optimization of L-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol. Bioeng. 37, 1108–1116. doi: 10.1002/bit.260371117
Bäumchen, C., and Bringer-Meyer, S. (2007). Expression of glf Zm increases D-mannitol formation in whole cell biotransformation with resting cells of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76, 545–552. doi: 10.1007/s00253-007-0987-8
Bhosale, P., and Gadre, R. (2001). β-carotene production in sugarcane molasses by a Rhodotorula glutinis mutant. J. Ind. Microbiol. Biotechnol. 26, 327–332. doi: 10.1038/sj.jim.7000138
Bouvier, F., Dogbo, O., and Camara, B. (2003). Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300, 2089–2091. doi: 10.1126/science.1085162
Bringer-Meyer, S., and Sahm, H. (1991). Process for Obtaining Sorbitol and Gluconic Acid by Fermentation, and Cell Material Suitable for This Purpose. US patent US5017485A. Alexandria, VA: United States Patent and Trademark Office.
Brown, S. H., Bashkirova, L., Berka, R., Chandler, T., Doty, T., McCall, K., et al. (2013). Metabolic engineering of Aspergillus oryzae NRRL 3488 for increased production of L-malic acid. Appl. Microbiol. Biotechnol. 97, 8903–8912. doi: 10.1007/s00253-013-5132-2
Brownsey, R., Boone, A., Elliott, J., Kulpa, J., and Lee, W. (2006). Regulation of Acetyl-CoA Carboxylase. London: Portland Press Limited.
Cameron, D. C., and Cooney, C. L. (1986). A Novel fermentation: the production of R (–)−1,2–propanediol and acetol by Clostridium thermosaccharolyticum. Nat. Biotechnol. 4, 651–654. doi: 10.1038/nbt0786-651
Cao, N., Du, J., Gong, C., and Tsao, G. (1996). Simultaneous production and recovery of fumaric acid from immobilized Rhizopus oryzae with a rotary biofilm contactor and an adsorption column. Appl. Environ. Microbiol. 62, 2926–2931.
Carballo-Cárdenas, E. C., Tuan, P. M., Janssen, M., and Wijffels, R. H. (2003). Vitamin E (α-tocopherol) production by the marine microalgae Dunaliella tertiolecta and Tetraselmis suecica in batch cultivation. Biomol. Eng. 20, 139–147. doi: 10.1016/S1389-0344(03)00040-6
Chandrashekar, K., Felse, P. A., and Panda, T. (1999). Optimization of temperature and initial pH and kinetic analysis of tartaric acid production by Gluconobacter suboxydans. Bioprocess. Eng. 20, 203–207. doi: 10.1007/PL00009044
Chappell, J., Wolf, F., Proulx, J., Cuellar, R., and Saunders, C. (1995). Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 109, 1337–1343. doi: 10.1104/pp.109.4.1337
Cheong, S., Clomburg, J. M., and Gonzalez, R. (2016). Energy-and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 34, 556–561. doi: 10.1038/nbt.3505
Chin, T., Okuda, Y., and Ikeuchi, M. (2018). Sorbitol production and optimization of photosynthetic supply in the cyanobacterium Synechocystis PCC 6803. J. Biotechnol. 276, 25–33. doi: 10.1016/j.jbiotec.2018.04.004
Choi, S., Song, H., Lim, S. W., Kim, T. Y., Ahn, J. H., Lee, J. W., et al. (2016). Highly selective production of succinic acid by metabolically engineered Mannheimia succiniciproducens and its efficient purification. Biotechnol. Bioeng. 113, 2168–2177. doi: 10.1002/bit.25988
Church, J. M., and Blumberg, R. (1951). Synthesis of tartaric acid. Ind. Eng. Chem. Res. 43, 1780–1786. doi: 10.1021/ie50500a030
Clomburg, J. M., and Gonzalez, R. (2011). Metabolic engineering of Escherichia coli for the production of 1, 2-propanediol from glycerol. Biotechnol. Bioeng. 108, 867–879. doi: 10.1002/bit.22993
Cocks, G., Aguilar, J., and Lin, E. (1974). Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J. Bacteriol. 118, 83–88.
Dai, J., and Mumper, R. J. (2010). Plant phenolics: extraction, analysis and their antioxidant and anticancer properties. Molecules 15, 7313–7352. doi: 10.3390/molecules15107313
de Arauz, L. J., Jozala, A. F., Pinheiro, G. S., Mazzola, P. G., Junior, A. P., and Vessoni Penna, T. C. (2008). Nisin expression production from Lactococcus lactis in milk whey medium. J. Chem. Technol. Biotechnol. 83, 325–328. doi: 10.1002/jctb.1813
de Miguel, T., Sieiro, C., Poza, M., and Villa, T. G. (2000). Isolation and taxonomic study of a new canthaxanthin-containing bacterium, Gordonia jacobaea MV-1 sp. nov. Int. Microbiol. 3, 107–111.
DeBolt, S., Cook, D. R., and Ford, C. M. (2006). L-Tartaric acid synthesis from vitamin C in higher plants. Proc. Natl. Acad. Sci. U.S.A. 103, 5608–5613. doi: 10.1073/pnas.0510864103
Del Campo, J. A., Moreno, J., Rodriguez, H., Vargas, M. A., Rivas, J., and Guerrero, M. G. (2000). Carotenoid content of chlorophycean microalgae: factors determining lutein accumulation in Muriellopsis sp. (Chlorophyta). J. Biotechnol. 76, 51–59. doi: 10.1016/S0168-1656(99)00178-9
Deng, Y., and Mao, Y. (2015). Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J. Appl. Microbiol. 119, 1057–1063. doi: 10.1111/jam.12905
Deng, Y., Ma, L., and Mao, Y. (2016). Biological production of adipic acid from renewable substrates: Current and future methods. Biochem. Eng. J. 105, 16–26. doi: 10.1016/j.bej.2015.08.015
Denk, D., and Böck, A. (1987). L-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. Microbiology 133, 515–525. doi: 10.1099/00221287-133-3-515
Edens, L., Heslinga, L., Klok, R., Ledeboer, A. M., Maat, J., Toonen, M. Y., et al. (1982). Cloning of cDNA encoding the sweet-tasting plant protein thaumatin and its expression in Escherichia coli. Gene 18, 1–12. doi: 10.1016/0378-1119(82)90050-6
Eggeling, L., and Bott, M. (2005). Handbook of Corynebacterium glutamicum. Boca Raton, FL: CRC Press.
Eggeling, L., Bott, M., and Marienhagen, J. (2015). Novel screening methods—biosensors. Curr. Opin. Biotechnol. 35, 30–36. doi: 10.1016/j.copbio.2014.12.021
Eichenberger, M., Lehka, B. J., Folly, C., Fischer, D., Martens, S., Simón, E., et al. (2017). Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab. Eng. 39, 80–89. doi: 10.1016/j.ymben.2016.10.019
Eisenreich, W., Bacher, A., Arigoni, D., and Rohdich, F. (2004). Biosynthesis of isoprenoids via the non-mevalonate pathway?. Cell. Mol. Life Sci. 61, 1401–1426. doi: 10.1007/s00018-004-3381-z
Elferink, S. J. O., Krooneman, J., Gottschal, J. C., Spoelstra, S. F., Faber, F., and Driehuis, F. (2001). Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 67, 125–132. doi: 10.1128/AEM.67.1.125-132.2001
Elsayed, E. A., Farid, M. A. F., and El Enshasy, H. A. (2013). Improvement in natamycin production by Streptomyces natalensis with the addition of short-chain carboxylic acids. Process Biochem. 48, 1831–1838. doi: 10.1016/j.procbio.2013.09.009
Ercan, D., and Demirci, A. (2016). Recent advances for the production and recovery methods of lysozyme. Crit. Rev. Biotechnol. 36, 1078–1088. doi: 10.3109/07388551.2015.1084263
Farid, M. A., El-Enshasy, H. A., El-Diwany, A. I., and El-Sayed, E. S. A. (2000). Optimization of the cultivation medium for natamycin production by Streptomyces natalensis. J. Basic Microbiol. 40, 157–166. doi: 10.1002/1521-4028(200007)40:3<157::AID-JOBM157>3.0.CO;2-1
Fernández-Sevilla, J. M., Fernández, F. A., and Grima, E. M. (2010). Biotechnological production of lutein and its applications. Appl. Microbiol. Biotechnol. 86, 27–40. doi: 10.1007/s00253-009-2420-y
Ferrer, J.-L., Jez, J. M., Bowman, M. E., Dixon, R. A., and Noel, J. P. (1999). Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Mol. Biol. 6, 775–784. doi: 10.1038/11553
Fukuda, K., Watanabe, M., Asano, K., Ouchi, K., and Takasawa, S. (1991). A mutated ARO4 gene for feedback-resistant DAHP synthase which causes both o-fluoro-DL-phenylalamine resistance and β-phenethyl-alcohol overproduction in Saccharomyces cerevisiae. Curr. Genet. 20, 453–456. doi: 10.1007/BF00334771
Gharibzahedi, S. M. T., Razavi, S. H., Mousavi, S. M., and Moayedi, V. (2012). High efficiency canthaxanthin production by a novel mutant isolated from Dietzia natronolimnaea HS-1 using central composite design analysis. Ind. Crops Prod. 40, 345–354. doi: 10.1016/j.indcrop.2012.03.030
Gheshlaghi, R., Scharer, J., Moo-Young, M., and Douglas, P. (2005). Medium optimization for hen egg white lysozyme production by recombinant Aspergillus niger using statistical methods. Biotechnol. Bioeng. 90, 754–760. doi: 10.1002/bit.20474
Glassner, D. A., and Datta, R. (1992). Process for the Production and Purification of Succinic Acid. US patent US5143834A. Alexandria, VA: United States Patent and Trademark Office.
Goldstein, J. L., and Brown, M. S. (1990). Regulation of the mevalonate pathway. Nature 343, 425–430. doi: 10.1038/343425a0
Gosset, G. (2009). Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 20, 651–658. doi: 10.1016/j.copbio.2009.09.012
Grabar, T., Gong, W, and Yocum, R. R. (2014). Metabolic Evolution of Escherichia coli Strains that Produce Organic Acids. US patent US8871489B2. Alexandria, VA: United States Patent and Trademark Office.
Grewal, P. S., Modavi, C., Russ, Z. N., Harris, N. C., and Dueber, J. E. (2018). Bioproduction of a betalain color palette in Saccharomyces cerevisiae. Metab. Eng. 45, 180–188. doi: 10.1016/j.ymben.2017.12.008
Hallborn, J., Walfridsson, M., Airaksinen, U., Ojamo, H., Hahn-Hägerdal, B., Penttilä, M., et al. (1991). Xylitol production by recombinant Saccharomyces cerevisiae. Nat. Biotechnol. 9:1090. doi: 10.1038/nbt1191-1090
Hernández-Almanza, A., Montañez, J., Martínez, G., Aguilar-Jiménez, A., Contreras-Esquivel, J. C., and Aguilar, C. N. (2016). Lycopene: progress in microbial production. Trends Food Sci. Technol. 56, 142–148. doi: 10.1016/j.tifs.2016.08.013
Hirasawa, T., and Wachi, M. (2016). “Glutamate fermentation-2: mechanism of L-glutamate overproduction in Corynebacterium glutamicum,” in Amino Acid Fermentation, eds A. Yokota, and M. Ikeda (Berlin: Springer), 57–72. doi: 10.1007/10_2016_26
Huang, C., Chen, X.-F., Xiong, L., and Ma, L.-L. (2012). Oil production by the yeast Trichosporon dermatis cultured in enzymatic hydrolysates of corncobs. Bioresour. Technol. 110, 711–714. doi: 10.1016/j.biortech.2012.01.077
Hurlbert, R. E., and Jakoby, W. B. (1965). Tartaric acid metabolism I. Subunits of L (+)-tartaric acid dehydrase. J. Biol. Chem. 240, 2772–2777.
Illingworth, C., Larson, G., and Hellekant, G. (1988). Secretion of the sweet-tasting plant protein thaumatin by Bacillus subtilis. Biotechnol. Lett. 10, 587–592. doi: 10.1007/BF01027135
Jan, K. N., Tripathi, A. D., Singh, S., Surya, D., and Singh, S. P. (2017). Enhanced Sorbitol Production under Submerged Fermentation using Lactobacillus plantarum. Appl. Food Biotechnol. 4, 85–92. doi: 10.22037/afb.v4i2.13514
Jeya, M., Lee, K.-M., Tiwari, M. K., Kim, J.-S., Gunasekaran, P., Kim, S.-Y., et al. (2009). Isolation of a novel high erythritol-producing Pseudozyma tsukubaensis and scale-up of erythritol fermentation to industrial level. Appl. Microbiol. Biotechnol. 83, 225–231. doi: 10.1007/s00253-009-1871-5
Jones, J. A., Vernacchio, V. R., Collins, S. M., Shirke, A. N., Xiu, Y., Englaender, J. A., et al. (2017). Complete biosynthesis of anthocyanins using E. coli polycultures. MBio 8, e00621–e00617. doi: 10.1128/mBio.00621-17
Jossek, R., Bongaerts, J., and Sprenger, G. A. (2001). Characterization of a new feedback-resistant 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol. Lett. 202, 145–148. doi: 10.1016/S0378-1097(01)00311-1
Kaeding, W. W., Lindblom, R., Temple, R., and Mahon, H. (1965). Oxidation of toluene and other alkylated aromatic hydrocarbons to benzoic acids and phenols. Ind. Eng. Chem. Process Des. Dev. 4, 97–101. doi: 10.1021/i260013a022
Kai, Y., Kashiwagi, T., Ishikawa, K., Ziyatdinov, M., Redkina, E., Kiriukhin, M., et al. (2006). Engineering of Escherichia coli L-serine O-acetyltransferase on the basis of crystal structure: desensitization to feedback inhibition by L-cysteine. Protein Eng. Des. Sel. 19, 163–167. doi: 10.1093/protein/gzj015
Kaletta, C., and Entian, K.-D. (1989). Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J. Bacteriol. 171, 1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989
Kallscheuer, N., and Marienhagen, J. (2018). Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb. Cell Fact. 17:70. doi: 10.1186/s12934-018-0923-x
Kallscheuer, N., Gätgens, J., Lübcke, M., Pietruszka, J., Bott, M., and Polen, T. (2017a). Improved production of adipate with Escherichia coli by reversal of β-oxidation. Appl. Microbiol. Biotechnol. 101, 2371–2382. doi: 10.1007/s00253-016-8033-3
Kallscheuer, N., Polen, T., Bott, M., and Marienhagen, J. (2017b). Reversal of β-oxidative pathways for the microbial production of chemicals and polymer building blocks. Metab. Eng. 42, 33–42. doi: 10.1016/j.ymben.2017.05.004
Kallscheuer, N., Vogt, M., and Marienhagen, J. (2016). A novel synthetic pathway enables microbial production of polyphenols independent from the endogenous aromatic amino acid metabolism. ACS Synth. Biol. 6, 410–415. doi: 10.1021/acssynbio.6b00291
Kambourakis, S., Draths, K., and Frost, J. (2000). Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis. J. Am. Chem. Soc. 122, 9042–9043. doi: 10.1021/ja000853r
Kamzolova, S. V., Vinokurova, N. G., Lunina, J. N., Zelenkova, N. F., and Morgunov, I. G. (2015). Production of technical-grade sodium citrate from glycerol-containing biodiesel waste by Yarrowia lipolytica. Bioresour. Technol. 193, 250–255. doi: 10.1016/j.biortech.2015.06.092
Karaffa, L., and Kubicek, C. P. (2003). Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl. Microbiol. Biotechnol. 61, 189–196. doi: 10.1007/s00253-002-1201-7
Katashkina, J. I., Hara, Y., Golubeva, L. I., Andreeva, I. G., Kuvaeva, T. M., and Mashko, S. V. (2009). Use of the λ red-recombineering method for genetic engineering of Pantoea ananatis. BMC Mol. Biol. 10:34. doi: 10.1186/1471-2199-10-34
Katsuyama, Y., Kita, T., Funa, N., and Horinouchi, S. (2009). Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J. Biol. Chem. 284, 11160–11170. doi: 10.1074/jbc.M900070200
Katsuyama, Y., Matsuzawa, M., Funa, N., and Horinouchi, S. (2008). Production of curcuminoids by Escherichia coli carrying an artificial biosynthesis pathway. Microbiology 154, 2620–2628. doi: 10.1099/mic.0.2008/018721-0
Kawano, Y., Ohtsu, I., Takumi, K., Tamakoshi, A., Nonaka, G., Funahashi, E., et al. (2015). Enhancement of L-cysteine production by disruption of yciW in Escherichia coli. J. Biosci. Bioeng. 119, 176–179. doi: 10.1016/j.jbiosc.2014.07.006
Kim, J., and DellaPenna, D. (2006). Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. U.S.A. 103, 3474–3479. doi: 10.1073/pnas.0511207103
Kimura, E. (2003). “Metabolic engineering of glutamate production,” in Microbial Production of l-Amino Acids, eds R. Faurie and J. Thommel (Berlin: Springer), 37–57.
Kinoshita, S., Udaka, S., and Shimono, M. (1957). Studies on the amino acid fermentation. J. Gen. Appl. Microbiol. 3, 193–205. doi: 10.2323/jgam.3.193
Kitade, Y., Hashimoto, R., Suda, M., Hiraga, K., and Inui, M. (2018). Production of 4-hydroxybenzoic acid by an aerobic growth-arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl. Environ. Microbiol. 84, e02587–17. doi: 10.1128/AEM.02587-17
Klasen, R., Bringer-Meyer, S., and Sahm, H. (1992). Incapability of Gluconobacter oxydans to produce tartaric acid. Biotechnol. Bioeng. 40, 183–186. doi: 10.1002/bit.260400126
Kruyer, N. S., and Peralta-Yahya, P. (2017). Metabolic engineering strategies to bio-adipic acid production. Curr. Opin. Biotechnol. 45, 136–143. doi: 10.1016/j.copbio.2017.03.006
Kusserow, B., Schimpf, S., and Claus, P. (2003). Hydrogenation of glucose to sorbitol over nickel and ruthenium catalysts. Adv. Synth. Catal. 345, 289–299. doi: 10.1002/adsc.200390024
Lange, A., Becker, J., Schulze, D., Cahoreau, E., Portais, J.-C., Haefner, S., et al. (2017). Bio-based succinate from sucrose: high-resolution 13C metabolic flux analysis and metabolic engineering of the rumen bacterium Basfia succiniciproducens. Metab. Eng. 44, 198–212. doi: 10.1016/j.ymben.2017.10.003
Ledesma-Amaro, R., Serrano-Amatriain, C., Jiménez, A., and Revuelta, J. L. (2015). Metabolic engineering of riboflavin production in Ashbya gossypii through pathway optimization. Microb. Cell Fact. 14:163. doi: 10.1186/s12934-015-0354-x
Lefebvre, V., Kuntz, M., Camara, B., and Palloix, A. (1998). The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol. Biol. 36, 785–789. doi: 10.1023/A:1005966313415
Li, M., Chen, S., Li, J., and Ji, Z. (2014a). Propanol addition improves natamycin biosynthesis of Streptomyces natalensis. Appl. Biochem. Biotechnol. 172, 3424–3432. doi: 10.1007/s12010-014-0766-9
Li, N., Zhang, B., Wang, Z., Tang, Y.-J., Chen, T., and Zhao, X. (2014b). Engineering Escherichia coli for fumaric acid production from glycerol. Bioresour. Technol. 174, 81–87. doi: 10.1016/j.biortech.2014.09.147
Li, Q., Sun, Z., Li, J., and Zhang, Y. (2013). Enhancing beta-carotene production in Saccharomyces cerevisiae by metabolic engineering. FEMS Microbiol. Lett. 345, 94–101. doi: 10.1111/1574-6968.12187
Li, S., Chen, L., Hu, Y., Fang, G., Zhao, M., Guo, Y., et al. (2017). Enzymatic production of 5′-inosinic acid by AMP deaminase from a newly isolated Aspergillus oryzae. Food Chem. 216, 275–281. doi: 10.1016/j.foodchem.2016.07.171
Li, Y., Zhao, Z. K., and Bai, F. (2007). High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb. Technol. 41, 312–317. doi: 10.1016/j.enzmictec.2007.02.008
Li, Z.-J., Hong, P.-H., Da, Y.-Y., Li, L.-K., and Stephanopoulos, G. (2018). Metabolic engineering of Escherichia coli for the production of L-malate from xylose. Metab. Eng. 48, 25–32. doi: 10.1016/j.ymben.2018.05.010
Lim, C. G., Wong, L., Bhan, N., Dvora, H., Xu, P., Venkiteswaran, S., et al. (2015). Development of a recombinant Escherichia coli strain for overproduction of the plant pigment anthocyanin. Appl. Environ. Microbiol. 81, 6276–6284. doi: 10.1128/AEM.01448-15
Lin, Z., Xu, Z., Li, Y., Wang, Z., Chen, T., and Zhao, X. (2014). Metabolic engineering of Escherichia coli for the production of riboflavin. Microb. Cell Fact. 13, 104. doi: 10.1186/s12934-014-0104-5
Liu, H., Fang, G., Wu, H., Li, Z., and Ye, Q. (2018). L-Cysteine production in Escherichia coli based on rational metabolic engineering and modular strategy. Biotechnol. J. 13:1700695. doi: 10.1002/biot.201700695
Liu, S.-P., Yuan, P.-H., Wang, Y.-Y., Liu, X.-F., Zhou, Z.-X., Bu, Q.-T., et al. (2015). Generation of the natamycin analogs by gene engineering of natamycin biosynthetic genes in Streptomyces chattanoogensis L10. Microbiol. Res. 173, 25–33. doi: 10.1016/j.micres.2015.01.013
Liu, Z.-Q., Zhang, L., Sun, L.-H., Li, X.-J., Wan, N.-W., and Zheng, Y.-G. (2012). Enzymatic production of 5′-inosinic acid by a newly synthesised acid phosphatase/phosphotransferase. Food Chem. 134, 948–956. doi: 10.1016/j.foodchem.2012.02.213
Lombraña, M., Moralejo, F. J., Pinto, R., and Martín, J. F. (2004). Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 70, 5145–5152. doi: 10.1128/AEM.70.9.5145-5152.2004
Lu, F., Ping, K., Wen, L., Zhao, W., Wang, Z., Chu, J., et al. (2015). Enhancing gluconic acid production by controlling the morphology of Aspergillus niger in submerged fermentation. Process Biochem. 50, 1342–1348. doi: 10.1016/j.procbio.2015.04.010
Mao, Y., Li, G., Chang, Z., Tao, R., Cui, Z., Wang, Z., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for efficient production of succinate from lignocellulosic hydrolysate. Biotechnol. Biofuels 11:95. doi: 10.1186/s13068-018-1094-z
Marienhagen, J., and Bott, M. (2013). Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 163, 166–178. doi: 10.1016/j.jbiotec.2012.06.001
Martinez, I., Gao, H., Bennett, G. N., and San, K.-Y. (2018). High yield production of four-carbon dicarboxylic acids by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 45, 53–60. doi: 10.1007/s10295-017-1991-3
Matthäus, F., Ketelhot, M., Gatter, M., and Barth, G. (2014). Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 80, 1660–1669. doi: 10.1128/AEM.03167-13
Meijnen, J.-P., Verhoef, S., Briedjlal, A. A., de Winde, J. H., and Ruijssenaars, H. J. (2011). Improved p-hydroxybenzoate production by engineered Pseudomonas putida S12 by using a mixed-substrate feeding strategy. Appl. Microbiol. Biotechnol. 90, 885–893. doi: 10.1007/s00253-011-3089-6
Meng, J., Wang, B., Liu, D., Chen, T., Wang, Z., and Zhao, X. (2016). High-yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli. Microb. Cell Fact. 15:141. doi: 10.1186/s12934-016-0536-1
Meng, Y., Xue, Y., Yu, B., Gao, C., and Ma, Y. (2012). Efficient production of L-lactic acid with high optical purity by alkaliphilic Bacillus sp. WL-S20. Bioresour. Technol. 116, 334–339. doi: 10.1016/j.biortech.2012.03.103
Milke, L., Aschenbrenner, J., Marienhagen, J., and Kallscheuer, N. (2018). Production of plant-derived polyphenols in microorganisms: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 1575–1585. doi: 10.1007/s00253-018-8747-5
Mir, M. A. (1977). Lysozyme: a brief review. Postgrad. Med. J. 53, 257–257. doi: 10.1136/pgmj.53.619.257
Misawa, N. (2013). Construction of an Efficient System for the Production of Red Pepper Carotenoids and New Derivatives. Research Grant, Noda Institute for Scientific Research, Japan.
Misawa, N., Kajiwara, S., Kondo, K., Yokoyama, A., Satomi, Y., Saito, T., et al. (1995). Canthaxanthin biosynthesis by the conversion of methylene to keto groups in a hydrocarbon β-carotene by a single gene. Biochem. Biophys. Res. Commun. 209, 867–876. doi: 10.1006/bbrc.1995.1579
Moerkercke, A. V., Schauvinhold, I., Pichersky, E., Haring, M. A., and Schuurink, R. C. (2009). A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 60, 292–302. doi: 10.1111/j.1365-313X.2009.03953.x
Moon, H.-J., Jeya, M., Kim, I.-W., and Lee, J.-K. (2010). Biotechnological production of erythritol and its applications. Appl. Microbiol. Biotechnol. 86, 1017–1025. doi: 10.1007/s00253-010-2496-4
Moralejo, F. J., Cardoza, R. E., Gutierrez, S., Lombraña, M., Fierro, F., and Martín, J. F. (2002). Silencing of the aspergillopepsin B (pepB) gene of Aspergillus awamori by antisense RNA expression or protease removal by gene disruption results in a large increase in thaumatin production. Appl. Environ. Microbiol. 68, 3550–3559. doi: 10.1128/AEM.68.7.3550-3559.2002
Moralejo, F.-J., Cardoza, R.-E., Gutierrez, S., and Martin, J. F. (1999). Thaumatin production in Aspergillus awamori by use of expression cassettes with strong fungal promoters and high gene dosage. Appl. Environ. Microbiol. 65, 1168–1174.
Murashchenko, L., Abbas, C., Dmytruk, K., and Sibirny, A. (2016). Overexpression of the truncated version of ILV2 enhances glycerol production in Saccharomyces cerevisiae. Yeast 33, 463–469. doi: 10.1002/yea.3161
Nakayama, K., Suzuki, T., Sato, Z., and Kinoshita, S. (1964). Production of nucleic acid-related substances by fermentative processes. V. Accumulation of inosinic acid by an adenine-auxotroph of Micrococcus glutamicus. J. Gen. Appl. Microbiol. 10, 133–142. doi: 10.2323/jgam.10.133
Ning, W., Tao, Z., Wang, C., Wang, S., Yan, Z., and Yin, G. (1988). Fermentation Process for Producing 2-keto-L-gulonic Acid. European patent EP0278447A2. Munich: European Patent Office.
Noda, S., Shirai, T., Oyama, S., and Kondo, A. (2016). Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 33, 119–129. doi: 10.1016/j.ymben.2015.11.007
Özel, B., Simşek, Ö., Akçelik, M., and Saris, P. E. (2018). Innovative approaches to nisin production. Appl. Microbiol. Biotechnol. 102, 6299–6307. doi: 10.1007/s00253-018-9098-y
Papagianni, M. (2007). Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol. Adv. 25, 244–263. doi: 10.1016/j.biotechadv.2007.01.002
Papanikolaou, S., Chevalot, I., Komaitis, M., Marc, I., and Aggelis, G. (2002). Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl. Microbiol. Biotechnol. 58, 308–312. doi: 10.1007/s00253-001-0897-0
Pappenberger, G., and Hohmann, H.-P. (2013). “Industrial production of L-ascorbic acid (Vitamin C) and D-isoascorbic acid,” in Biotechnology of Food and Feed Additives, eds H. Zorn and P. Czermak (Berlin: Springer), 143–188.
Pedersen, J. C. (1992). Natamycin as a fungicide in agar media. Appl. Environ. Microbiol. 58, 1064–1066.
Pei, L., and Schmidt, M. (2018). Fast-growing engineered microbes: new concerns for gain-of-function research?. Front. Genet. 9:207. doi: 10.3389/fgene.2018.00207
Polen, T., Spelberg, M., and Bott, M. (2013). Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 167, 75–84. doi: 10.1016/j.jbiotec.2012.07.008
Pugh, S., McKenna, R., Halloum, I., and Nielsen, D. R. (2015). Engineering Escherichia coli for renewable benzyl alcohol production. Met. Eng. Commun. 2, 39–45. doi: 10.1016/j.meteno.2015.06.002
Qiao, K., Abidi, S. H. I., Liu, H., Zhang, H., Chakraborty, S., Watson, N., et al. (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 29, 56–65. doi: 10.1016/j.ymben.2015.02.005
Quideau, S., Deffieux, D., Douat-Casassus, C., and Pouysegu, L. (2011). Plant polyphenols: chemical properties, biological activities, and synthesis. Angew. Chem. Int. Ed Engl. 50, 586–621. doi: 10.1002/anie.201000044
Quispe, C. A., Coronado, C. J., and Carvalho, Jr, J. A. (2013). Glycerol: production, consumption, prices, characterization and new trends in combustion. Renew. Sust. Energ. Rev. 27, 475–493. doi: 10.1016/j.rser.2013.06.017
Rostami, F., Razavi, S. H., Sepahi, A. A., and Gharibzahedi, S. M. T. (2014). Canthaxanthin biosynthesis by Dietzia natronolimnaea HS-1: effects of inoculation and aeration rate. Braz. J. Microbiol. 45, 447–456. doi: 10.1590/S1517-83822014005000046
Saito, Y., Ishii, Y., Hayashi, H., Imao, Y., Akashi, T., Yoshikawa, K., et al. (1997). Cloning of genes coding for L-sorbose and L-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-L-gulonate, a precursor of L-ascorbic acid, in a recombinant G. oxydans strain. Appl. Environ. Microbiol. 63, 454–460.
Salvachúa, D., Mohagheghi, A., Smith, H., Bradfield, M. F., Nicol, W., Black, B. A., et al. (2016). Succinic acid production on xylose-enriched biorefinery streams by Actinobacillus succinogenes in batch fermentation. Biotechnol. Biofuels 9:28. doi: 10.1186/s13068-016-0425-1
Sano, C. (2009). History of glutamate production. Am. J. Clin. Nutr. 90, 728S−732S. doi: 10.3945/ajcn.2009.27462F
Savakis, P., Tan, X., Du, W., dos Santos, F. B., Lu, X., and Hellingwerf, K. J. (2015). Photosynthetic production of glycerol by a recombinant cyanobacterium. J. Biotechnol. 195, 46–51. doi: 10.1016/j.jbiotec.2014.12.015
Saxena, R., Anand, P., Saran, S., Isar, J., and Agarwal, L. (2010). Microbial production and applications of 1, 2-propanediol. Indian J. Microbiol. 50, 2–11. doi: 10.1007/s12088-010-0017-x
Schallmey, M., Frunzke, J., Eggeling, L., and Marienhagen, J. (2014). Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors. Curr. Opin. Biotechnol. 26, 148–154. doi: 10.1016/j.copbio.2014.01.005
Schiweck, H., Bär, A., Vogel, R., Schwarz, E., and Kunz, M. (2000). Sugar Alcohols, Hoboken, NJ: Wiley Online Library; John Wiley and Sons.
Schwechheimer, S. K., Becker, J., Peyriga, L., Portais, J.-C., Sauer, D., Müller, R., et al. (2018). Improved riboflavin production with Ashbya gossypii from vegetable oil based on 13C metabolic network analysis with combined labeling analysis by GC/MS, LC/MS, 1D, and 2D NMR. Metab. Eng. 47, 357–373. doi: 10.1016/j.ymben.2018.04.005
Semkiv, M. V., Dmytruk, K. V., Abbas, C. A., and Sibirny, A. A. (2017). Metabolic engineering for high glycerol production by the anaerobic cultures of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 101, 4403–4416. doi: 10.1007/s00253-017-8202-z
Severina, E., Severin, A., and Tomasz, A. (1998). Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J. Antimicrob. Chemother. 41, 341–347. doi: 10.1093/jac/41.3.341
Shalem, O., Sanjana, N. E., and Zhang, F. (2015). High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 16, 299. doi: 10.1038/nrg3899
Siebert, M., Severin, K., and Heide, L. (1994). Formation of 4-hydroxybenzoate in Escherichia coli: characterization of the ubiC gene and its encoded enzyme chorismate pyruvate-lyase. Microbiology 140, 897–904. doi: 10.1099/00221287-140-4-897
Silveira, M., and Jonas, R. (2002). The biotechnological production of sorbitol. Appl. Microbiol. Biotechnol. 59, 400–408. doi: 10.1007/s00253-002-1046-0
Song, S. H., and Vieille, C. (2009). Recent advances in the biological production of mannitol. Appl. Microbiol. Biotechnol. 84, 55–62. doi: 10.1007/s00253-009-2086-5
Sridhara, S., Wu, T., Chused, T., and Lin, E. (1969). Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1,2-propanediol. J. Bacteriol. 98, 87–95.
Stahmann, K.-P., Revuelta, J., and Seulberger, H. (2000). Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53, 509–516. doi: 10.1007/s002530051649
Stevens, L. J., Kuczek, T., Burgess, J. R., Stochelski, M. A., Arnold, L. E., and Galland, L. (2013). Mechanisms of behavioral, atopic, and other reactions to artificial food colors in children. Nutr. Rev. 71, 268–281. doi: 10.1111/nure.12023
Suez, J., Korem, T., Zeevi, D., Zilberman-Schapira, G., Thaiss, C. A., Maza, O., et al. (2014). Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186. doi: 10.1038/nature13793
Takagi, H., and Ohtsu, I. (2016). “L-Cysteine metabolism and fermentation in microorganisms,” in Amino Acid Fermentation, eds A. Yokota and M. Ikeda (Berlin: Springer), 129–151.
Takagi, H., Awano, N., Kobayashi, S.-I., Noji, M., Saito, K., and Nakamori, S. (1999). Overproduction of L-cysteine and L-cystine by expression of genes for feedback inhibition-insensitive serine acetyltransferase from Arabidopsis thaliana in Escherichia coli. FEMS Microbiol. Lett. 179, 453–459. doi: 10.1016/S0378-1097(99)00429-2
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Tani, Y., and Tsumura, H. (1989). Screening for tocopherol-producing microorganisms and α-tocopherol production by Euglena gracilis Z. Agric. Biol. Chem. 53, 305–312. doi: 10.1271/bbb1961.53.305
Tetzlaf, D. (2011). Verdezyne Opens Pilot Plant to Produce Bio Based Adipic Acid for Renewable ‘Green’ Nylon, Available online at: https://verdezyne.com/uncategorized/verdezyne-opens-pilot-plant-to-produce-bio-based-adipic-acid-for-renewable-green-nylon/
Tilloy, V., Ortiz-Julien, A., and Dequin, S. (2014). Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl. Environ. Microbiol. 80, 2623–2632. doi: 10.1128/AEM.03710-13
Tomita, K., Nakanishi, T., and Kuratsu, Y. (1991). Stimulation by L-proline of 5′-inosinic acid production by mutants of Corynebacterium ammoniagenes. Agric. Biol. Chem. 55, 2221–2225. doi: 10.1271/bbb1961.55.2221
Vauzour, D., Rodriguez-Mateos, A., Corona, G., Oruna-Concha, M. J., and Spencer, J. P. (2010). Polyphenols and human health: prevention of disease and mechanisms of action. Nutrients 2, 1106–1131. doi: 10.3390/nu2111106
Verwaal, R., Wang, J., Meijnen, J.-P., Visser, H., Sandmann, G., van den Berg, J. A., et al. (2007). High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 73, 4342–4350. doi: 10.1128/AEM.02759-06
Wada, M., and Takagi, H. (2006). Metabolic pathways and biotechnological production of L-cysteine. Appl. Microbiol. Biotechnol. 73, 48–54. doi: 10.1007/s00253-006-0587-z
Wang, G., Bai, L., Wang, Z., Shi, T., Chen, T., and Zhao, X. (2014). Enhancement of riboflavin production by deregulating gluconeogenesis in Bacillus subtilis. World J. Microbiol. Biotechnol. 30, 1893–1900. doi: 10.1007/s11274-014-1611-6
Wang, S., Zhang, S., Xiao, A., Rasmussen, M., Skidmore, C., and Zhan, J. (2015). Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab. Eng. 29, 153–159. doi: 10.1016/j.ymben.2015.03.011
Wang, Y., Tao, Z., Zheng, H., Zhang, F., Long, Q., Deng, Z., et al. (2016). Iteratively improving natamycin production in Streptomyces gilvosporeus by a large operon-reporter based strategy. Metab. Eng. 38, 418–426. doi: 10.1016/j.ymben.2016.10.005
Wang, Z., Zhuge, J., Fang, H., and Prior, B. A. (2001). Glycerol production by microbial fermentation: a review. Biotechnol. Adv. 19, 201–223. doi: 10.1016/S0734-9750(01)00060-X
Weaver, L. M., and Herrmann, K. M. (1990). Cloning of an aroF allele encoding a tyrosine-insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J. Bacteriol. 172, 6581–6584. doi: 10.1128/jb.172.11.6581-6584.1990
Wei, J., Zhang, Y., Yu, T.-Y., Sadre-Bazzaz, K., Rudolph, M. J., Amodeo, G. A., et al. (2016). A unified molecular mechanism for the regulation of acetyl-CoA carboxylase by phosphorylation. Cell Discov. 2:16044. doi: 10.1038/celldisc.2016.44
Wel, H., and Loeve, K. (1972). Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. FEBS J. 31, 221–225. doi: 10.1111/j.1432-1033.1972.tb02522.x
West, T. P. (2011). Malic acid production from thin stillage by Aspergillus species. Biotechnol. Lett. 33, 2463–2467. doi: 10.1007/s10529-011-0720-7
Xu, F., Yuan, Q.-P., and Zhu, Y. (2007). Improved production of lycopene and β-carotene by Blakeslea trispora with oxygen-vectors. Process Biochem. 42, 289–293. doi: 10.1016/j.procbio.2006.08.007
Xu, J., Xu, X., Xu, Q., Zhang, Z., Jiang, L., and Huang, H. (2018). Efficient production of lycopene by engineered E. coli strains harboring different types of plasmids. Bioprocess. Biosyst. Eng. 41, 489–499. doi: 10.1007/s00449-017-1883-y
Yan, D., Wang, C., Zhou, J., Liu, Y., Yang, M., and Xing, J. (2014). Construction of reductive pathway in Saccharomyces cerevisiae for effective succinic acid fermentation at low pH value. Bioresour. Technol. 156, 232–239. doi: 10.1016/j.biortech.2014.01.053
Yan, Y., Chemler, J., Huang, L., Martens, S., and Koffas, M. A. (2005). Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71, 3617–3623. doi: 10.1128/AEM.71.7.3617-3623.2005
Yan, Y., Li, Z., and Koffas, M. A. (2008). High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 100, 126–140. doi: 10.1002/bit.21721
Yin, X., Shin, H. -d., Li, J., Du, G., Liu, L., and Chen, J. (2017). Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci. Rep. 7:41040. doi: 10.1038/srep41040
Yoon, S.-H., Lee, S.-H., Das, A., Ryu, H.-K., Jang, H.-J., Kim, J.-Y., et al. (2009). Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J. Biotechnol. 140, 218–226. doi: 10.1016/j.jbiotec.2009.01.008
Yu, J., Dong, J., Zhang, C., Li, J., and Xiao, D. (2014). “Effect of GPD1 and GPD2 Deletion on the Production of Glycerol and Ethanol in the Yeast Saccharomyces cerevisiae, in Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012), (Berlin: Springer), 171–179.
Yuan, H., Jia, Z.-F., He, J.-H., Fan, X.-G., and Chen, N. (2016). “Production of 5′-Inosinic acid by whole-cell biocatalyst expressing a mutated acid phosphatase/phosphotransferase,” in International Conference on Applied Biotechnology (Berlin: Springer), 605–614.
Zachariou, M., and Scopes, R. K. (1986). Glucose-fructose oxidoreductase, a new enzyme isolated from Zymomonas mobilis that is responsible for sorbitol production. J. Bacteriol. 167, 863–869. doi: 10.1128/jb.167.3.863-869.1986
Zambanini, T., Tehrani, H. H., Geiser, E., Sonntag, C. K., Buescher, J. M., Meurer, G., et al. (2017). Metabolic engineering of Ustilago trichophora TZ1 for improved malic acid production. Metab. Eng. Commun. 4, 12–21. doi: 10.1016/j.meteno.2017.01.002
Zelle, R. M., de Hulster, E., van Winden, W. A., de Waard, P., Dijkema, C., Winkler, A. A., et al. (2008). Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 74, 2766–2777. doi: 10.1128/AEM.02591-07
Zeng, Y., Li, Z., and Demopoulos, G. P. (2016). Process for glycine production by antisolvent crystallization using its phase equilibria in the ethylene glycol–NH4Cl–water system. Ind. Eng. Chem. Res. 55, 2426–2437. doi: 10.1021/acs.iecr.5b04144
Zhang, J., Zhang, B., Wang, D., Gao, X., and Hong, J. (2015a). Improving xylitol production at elevated temperature with engineered Kluyveromyces marxianus through over-expressing transporters. Bioresour. Technol. 175, 642–645. doi: 10.1016/j.biortech.2014.10.150
Zhang, K., Wang, M., and Gao, C. (2011a). Tartaric acid production by ion exchange resin-filling electrometathesis and its process economics. J. Membr. Sci. 366, 266–271. doi: 10.1016/j.memsci.2010.10.013
Zhang, T.-C., Li, W, Luo, X.-G., Feng, C.-X., Zhang, M.-H., Du, W., et al. (2015b). “Increase of the lycopene production in the recombinant strains of Escherichia coli by supplementing with fructose,” in Advances in Applied Biotechnology, eds T.-C. Zhang and M. Nakajima (Berlin: Springer), 29–35.
Zhang, X., Wang, X., Shanmugam, K., and Ingram, L. (2011b). L-malate production by metabolically engineered Escherichia coli. Appl. Environ. Microbiol. 77, 427–434. doi: 10.1128/AEM.01971-10
Zhu, F., Lu, L., Fu, S., Zhong, X., Hu, M., Deng, Z., et al. (2015). Targeted engineering and scale up of lycopene overproduction in Escherichia coli. Process Biochem. 50, 341–346. doi: 10.1016/j.procbio.2014.12.008
Zhuge, J., and Fang, H.-Y. (1995). A Novel Strain and its Breeding Method for Glycerol Production by Aerobic Fermentation. Chinese Patent CN 1082608A. Beijing: Chinese State Intellectual Property Office.
Keywords: food additives, E numbers, European Union, microbial production, metabolic engineering, plant natural products
Citation: Kallscheuer N (2018) Engineered Microorganisms for the Production of Food Additives Approved by the European Union—A Systematic Analysis. Front. Microbiol. 9:1746. doi: 10.3389/fmicb.2018.01746
Received: 20 June 2018; Accepted: 12 July 2018;
Published: 03 August 2018.
Edited by:
Laurent Dufossé, Université de la Réunion, FranceReviewed by:
Francesco Grieco, Istituto di Scienze delle Produzioni Alimentari (ISPA), ItalyCopyright © 2018 Kallscheuer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolai Kallscheuer, bi5rYWxsc2NoZXVlckBmei1qdWVsaWNoLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.