- State Key Laboratory of Animal Nutrition, College of Animal Science and Technology, China Agricultural University, Beijing, China
Our previous reports suggested that Dietary L-arginine supplementation attenuated gut injury of broiler chickens infected with Clostridium perfringens by enhancing intestinal immune responses, absorption and barrier function, but its effect on the gut microbiome of broiler chickens remains unclear. This experiment aimed at evaluating the effects of Dietary L-arginine supplementation on the gut bacterial community composition and function of broiler chickens challenged with C. perfringens. In total, 105 1-day-old male Arbor Acres broiler chickens were assigned to three groups: Control (CTL), C. perfringens-challenged (CP), and C. perfringens-challenged and fed diet supplemented with 0.3% L-arginine (ARGCP) groups. The challenge led to macroscopic and histomorphological gut lesions, decreased villus height and increased the number of Observed species, Shannon, Chao1 and ACE indices of ileal microbiota, whereas L-arginine addition reversed these changes. Moreover, the three treatments harbored distinct microbial communities (ANOSIM, P < 0.05). At the genus level, 24 taxa (e.g., Nitrosomonas spp., Coxiella spp., Ruegeria spp., and Thauera spp.) were significantly more abundant in CP group than in CTL group (P < 0.05), whereas the levels of 23 genera of them were significantly decreased by L-arginine supplementation (P < 0.05). The abundances of only 3 genera were different between CTL and ARGCP groups (P < 0.05). At the species level, the challenge promoted the relative abundance of Nitrospira sp. enrichment culture clone M1-9, Bradyrhizobium elkanii, Nitrospira bacterium SG8-3, and Pseudomonas veronii, which was reversed by L-arginine supplementation (P < 0.05). Furthermore, the challenge decreased the levels of Lactobacillus gasseri (P < 0.05). Predictive functional profiling of microbial communities by PICRUSt showed that compared with CP group, ARGCP group had enriched pathways relating to membrane transport, replication and repair, translation and nucleotide metabolism and suppressed functions corresponding to amino acid and lipid metabolisms (P < 0.05). The relative abundances of KEGG pathways in L-arginine-fed broilers were almost equal to those of the controls. In conclusion, L-arginine alleviated the gut injury and normalized the ileal microbiota of C. perfringens-challenged chickens to resemble that of unchallenged controls in terms of microbial composition and functionality.
Introduction
Necrotic enteritis (NE) is a universal enteric bacterial disease in poultry and is related to extensive production losses worldwide (Wade and Keyburn, 2015). The etiologic agent, Clostridium perfringens is a spore-forming, gram-positive anaerobe and is a normal inhabitant of the intestinal tract in healthy birds (Cooper and Songer, 2009). However, under certain pre-disposing conditions, this bacterium can proliferate to high numbers and meanwhile produce extracellular toxins, causing an outbreak of NE (Van Immerseel et al., 2009). NE can present as clinical and subclinical conditions. Clinical symptoms of this disease are characterized by severe necrosis of intestine and high flock morbidity and mortality, whereas the subclinical infection causes intestinal mucosal damage with no mortality, thus led to poor nutrient digestion and absorption, and further impaired growth performance (Van Immerseel et al., 2009; Timbermont et al., 2011).
Emerging evidence has shown that NE is strongly associated with intestinal microbiota in broiler chickens. There are strikingly varied microbial compositions in the onset and progression of NE (Stanley et al., 2012; Antonissen et al., 2016). For instance, C. perfringens challenge significantly reduces the level of Weissella confusa but significantly increases the abundance of uncultured Mollicutes related to human problems in chickens (Stanley et al., 2012). Reduction of Lachnospiraceae and Ruminococcaceae levels and the promotion of Gammaproteobacteria by NE have also been reported (Li et al., 2017). However, whether these shifts in microbiota communities are the results of other pre-disposing factors or whether they are more a consequence of C. perfringens multiplication and necrosis is still unclear.
A shift in diet can quickly affect the composition of the gut microbial community (David et al., 2014). Studies have suggested that dietary administration can affect the population of C. perfringens in the intestine (Liu et al., 2010) and influence the occurrence of this disease (Timbermont et al., 2011). With the removal of growth-promoting antibiotics, nutritional strategies are becoming a potential intervention for NE.
l-arginine is an essential amino acid for chickens and can be metabolized to produce important molecules such as nitric oxide, polyamines and creatine. L-arginine has presented great benefits in protecting intestinal health in numerous disease models. Tan et al. (2014a) reported that arginine-enriched diets alleviated intestinal villus damage, crypt dilation and goblet cell depletion induced by coccidia in broiler chickens. Moreover, these diets played a protective role in mice subjected to exertional hyperthermia by decreasing intestinal permeability and bacterial translocation (Costa et al., 2014). Furthermore, our results have recently shown that Dietary L-arginine addition protects the gut mucosa by improving innate immune responses, intestinal absorption and barrier function and by suppressing the colonization of C. perfringens in necrotic enteritis-challenged broiler chickens (Zhang et al., 2017).
Recent findings suggest that L-arginine regulates microbial composition and metabolism. Zheng et al. (2017) demonstrated that dentifrice supplemented with arginine favorably modulated the oral microbiome of individuals with dental caries, which was characterized by increasing the abundance of alkali-generating bacteria and reducing the content of acidogenic bacteria. With L-arginine addition, net utilization of alanine, glycine, threonine, lysine, leucine and isoleucine by the small-intestine bacteria of pig was reduced in vitro (Dai et al., 2012). However, to our knowledge, no studies have reported the function of L-arginine in the homeostasis of ileal microbial communities in broiler chickens. This study was undertaken to investigate the effects of arginine-supplemented diets on ileal microflora composition and function in broiler chickens challenged with C. perfringens by using 16S rRNA gene sequencing.
Materials and Methods
Experimental Animals, Diets, and Treatments
This study was approved by the China Agricultural University Animal Care and Use Committee (statement no. CAU20170601-2). A total of 105 1-day-old male Arbor Acres broiler chickens were randomly divided into three groups: Control (CTL), C. perfringens-challenged (CP), and C. perfringens-challenged and fed diet supplemented with 0.3% L-arginine (ARGCP) groups. Each group, involving 35 birds, was housed in a separate rearing isolator (160 × 70 × 70 cm) in a common room with controlled temperature. The experimental period lasted 21 days. All chickens were provided with feed and water ad libitum and received a 23 h light-1 h dark program every day. In addition, the chickens were vaccinated according to a routine immunization program. Corn-soybean meal diets were formulated according to the nutrient requirements for broilers as recommended by National Research Council (1994). The diet composition and nutrient levels are shown in Table S1. All diets were pelleted and crumbled. Dietary amino acid contents were determined by HPLC.
C. perfringens Challenge
Avian C. perfringens type A field strain (CVCC2030), received from the China Veterinary Culture Collection Center (Beijing, China), has been successfully used to establish NE model in broilers in our lab (Li et al., 2017). Briefly, chickens in the CP and ARGCP groups were infected with 1.0 mL of actively growing culture of C. perfringens at 2~3 × 108 CFU/mL by oral gavage each day from day 14 to day 20. Birds in the CTL group received an equal volume of sterile meat medium.
Sample Collection
On d 21, seven chickens per group were randomly selected and intravenously injected into pentobarbital sodium in a dose of 30 mg/kg body weight. Then these chickens were sacrificed by jugular exsanguination. The mid regions of the jejunum (~1 cm) were cut off and the digesta inside were gently rinsed with ice-cold saline. Then, the jejunal rings were immediately fixed in 4% paraformaldehyde for further intestinal morphological examination. ~2 g of digesta from the midpoint of the ileum was aseptically collected into sterile tubes, snap-frozen in liquid nitrogen, and stored at −80°C for DNA extraction.
Intestinal Lesion Score
The small intestine of each chicken was cut longitudinally and scored blindly as described by Dahiya et al. (2005) with some modifications. Lesions were evaluated with a scoring system from 0 to 4.0 = no gross lesions; 0.5 = severely congested serosa and mesenteric hyperemia; 1 = thin-walled and brittle intestines with small hemorrhagic spots (>5); 2 = small amounts of gas production and focal necrotic lesions 3 = gas-filled intestine and necrotic plaques (1–2 cm long); and 4 = large amounts of gas in the intestine and diffused necrosis.
Intestinal Morphological Analyses and Observation
The jejunum segments fixed in 4% paraformaldehyde were embedded in paraffin. Tissue rings were cut to a thickness of 5 μm and stained by hematoxylin and eosin. The slides were photographed by a Leica microscope (Wetzlar, Germany, Model DMi8). Villus height and crypt depth were measured from eight villi and crypts per slide by a blinded viewer using the Image-Pro Plus (version 6.0) software and averaged. Villus height was defined as the distance from the villus tip to the villus-crypt junction, and the crypt depth was measured from the villus-crypt junction to the base of the crypt. The means of villus height and crypt depth were calculated to obtain the villus height-to-crypt depth ratio (VCR). Morphological analyses and observation were conducted at magnifications of 50 × for each slide.
DNA Extraction and High-Throughput Sequencing
Bacterial DNA was extracted from ileal digesta with a QIAamp DNA Stool Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. The concentrations of DNA extracts were measured on a NanoDrop 2000 spectrophotometer (Thermo Scientific, MA, USA). The V4 region of the bacterial 16S rRNA gene was amplified with the barcoded primer pair 515F/806R (515F: 5′-GTG CCA GCM GCC GCG GTA A-3′, 806R: 5′-GGA CTA CHV GGG TWT CTA AT-3′) according to previously described methods (Wang et al., 2017). After amplification, PCR products run on a 2% agarose gel and were purified using a QIAquick Gel Extraction Kit (Qiagen, Germany). Pyrosequencing of 16S rDNA was performed on an Illumina HiSeq2500 PE250 platform (Illumina, San Diego, USA) at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China).
Sequence Processing and Bioinformatics Analysis
Raw tags were generated by merging paired-end reads using FLASH software (v1.2.7) (Magoc and Salzberg, 2011). High-quality clean tags were obtained by QIIME (v1.7.0) analysis (Caporaso et al., 2010), and chimera sequences were removed to obtain effective tags by using the UCHIME algorithm (Edgar et al., 2011). Sequences were analyzed by UPARSE software (v7.0.1001) and clustered into operational taxonomic units (OTUs) at a similarity level of 97% (Edgar, 2013). Each OTU was annotated with the Greengenes database (DeSantis et al., 2006). Rarefaction curve and Venn diagram were created using R software (v2.15.3). Analysis of microbial alpha diversity was conducted using QIIME software (Caporaso et al., 2010) with Python scripts. Beta diversity was evaluated by principal component analysis (PCA) to show the differences of bacterial community structures, and the significance of separation was tested via ANOSIM using R (v2.15.3). PICRUSt analysis was used to predict the functional potential of bacteria communities (Langille et al., 2013). OTUs were normalized by copy number, and metagenome prediction was further categorized into Kyoto Encyclopedia of Genes and Genomes (KEGG) at levels 2 and 3 (Kanehisa et al., 2012).
Statistical Analysis
Results are shown as the mean and SEM. Differences in the intestinal lesion score, jejunal morphology, alpha diversity indices, the relative abundance of top-10 phyla, and the bacterial functional pathways were analyzed by one-way ANOVA, followed by the Duncan multiple comparison tests (SPSS, version 18.0, Chicago, IL, USA). Comparisons of the relative abundances of genera and species and the Firmicutes/Bacteroidetes ratio between 2 groups were performed by unpaired Student's t-test. Significant difference was declared when P < 0.05.
Accession Number
The 16S rRNA gene amplicon sequencing results were submitted to the Sequence Read Archive of the NCBI (accession number: SRP134059).
Results
Intestinal Lesion Score
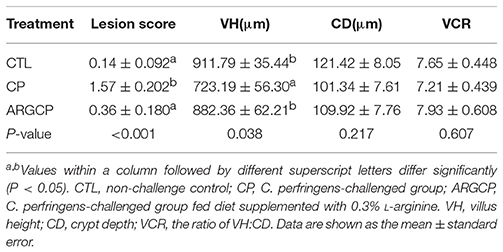
In our study, no intestinal lesions occurred in CTL birds, except that slight congestion appeared in the intestine of 2 birds. In the CP group, all birds exhibited macroscopic gut lesions such as thin-walled intestines, focal hemorrhagic lesions and small amounts of gas production. The average lesion score was significantly lower in the challenged birds fed L-arginine-enriched diets than in the challenged birds (P < 0.05, Table 1). No mortality was observed in the unchallenged chickens, but C. perfringens challenge caused one bird's death in each of the CP and ARGCP groups.
Intestinal Morphology
As shown in Table 1, the villus height in the jejunum in the CP group was significantly lower than that in the CTL and ARGCP groups (P < 0.05). There was no significant difference between the ARGCP group and the CTL group villus heights (P > 0.05). Neither C. perfringens challenge nor L-arginine addition affected the crypt depth and villus height/crypt depth ratio in the jejunum (P > 0.05). As depicted in Figure 1, control birds showed the normal appearance of the intestinal villus, whereas C. perfringens challenge led to severe pathological changes with the disappearance of the normal villus architecture. The ARGCP group exhibited mild pathological changes with defects of a small number of epithelial cells.
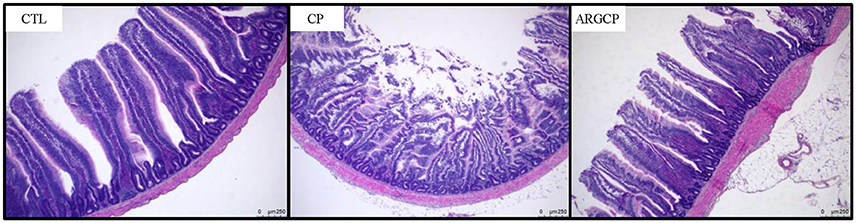
Figure 1. Representative photomicrographs of the jejunal cross section in chicken. Original magnification is 50 ×. CTL demonstrates the physiological features of the intestinal villus, whereas CP shows severe pathological changes affecting the normal villus architecture. ARGCP illustrates mild pathological changes with defects in a small number of epithelial cells. CTL, non-challenge control; CP, C. perfringens-challenged group; ARGCP, C. perfringens-challenged group fed diet supplemented with 0.3% L-arginine.
The Quality of Sequencing Data
After OTUs were assigned and chimeras were removed, sequencing of 21 samples generated 1,538,607 effective sequences with an average of 73,267 ± 2,356 (mean ± standard error) sequences per sample (Table S2). The median read length was 253 base pairs (bp), with a range from 253 to 255 bp (Table S2). A total of 3,597 OTUs were identified based on >97% sequencing similarity. Wherein 536 OTUs were common in three groups, and 96, 1,438, and 123 OTUs were exclusive in CTL group, CP group and ARGCP group, respectively (Figure S1). The Good's coverage estimators (Table S2) and the rarefaction curves (Figure S2) indicated that sufficient sequencing coverage was achieved.
The Alpha and Beta Diversity of Gut Microbiota
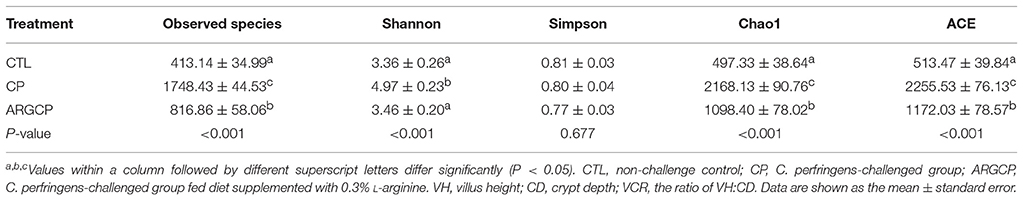
The alpha diversity of ileal microbiota was shown in Table 2. Compared with CTL group, both richness indices (observed species, Chao1 and ACE) and diversity indices (Shannon) significantly increased in the CP group (P < 0.001). However, these indices significantly decreased in ARGCP group compared with CP group (P < 0.001). Beta diversity analysis was illustrated via PCA in Figure 2, showing notable differentiation of the microbial community structure among the three groups. The composition of the microbiota in the ARGCP group was more similar to that in the CTL group than that in the CP group. ANOSIM results (P < 0.05) indicated that these significant differences in the microbial community structure depended on the three different treatments (Table 3).
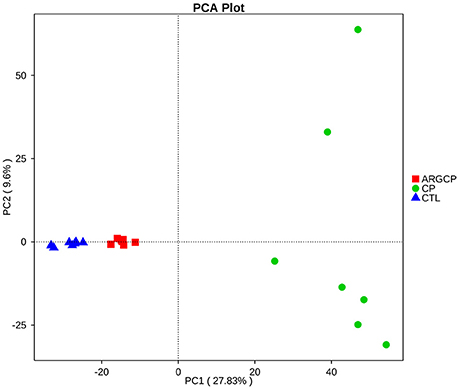
Figure 2. Comparison of the compositions of the ileal microbiota by principal component analysis (PCA). CTL, non-challenge control; CP, C. perfringens-challenged group; ARGCP, C. perfringens-challenged group fed diet supplemented with 0.3% L-arginine.
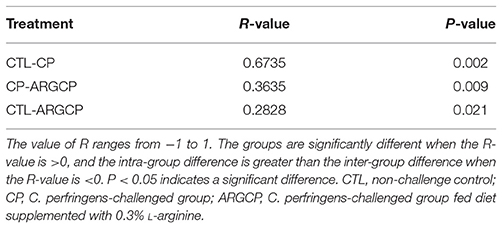
Table 3. Comparison of similarities in microbiota composition between the three treatments by ANOSIM analysis.
Relative Abundances of the Dominant Phyla
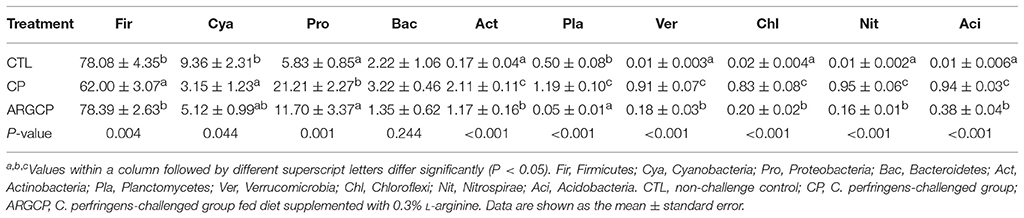
The most abundant (top 10) phyla of bacteria are presented in Table 4 and Figure S3. At the phylum level, the ileal microbiota was dominated by Firmicutes (53~91%), Cyanobacteria (1~34%), Proteobacteria (3~33%) and Bacteroidetes (0.1~8%), together making up over 88% of the total sequences. The C. perfringens-challenged birds (CP group) had markedly lower Firmicutes and Cyanobacteria abundances than the control chickens (P < 0.05). However, the relative abundance of Firmicutes was enhanced in the ARGCP group compared with that in the CP group. Moreover, C. perfringens challenge resulted in significantly greater abundances of Proteobacteria, Actinobacteria, Planctomycetes, Verrucomicrobia, Chloroflexi, Nitrospirae, and Acidobacteria in the CP group than in unchallenged birds (P ≤ 0.001); however, L-arginine addition significantly reversed these changes found in the CP group (P ≤ 0.001). Little difference was observed in the relative abundances of Firmicutes, Cyanobacteria and Proteobacteria in the ARGCP and CTL groups (P > 0.05), and no differences were shown in the relative abundances of Bacteroidetes among the three treatments (P > 0.05). Moreover, the Firmicutes/Bacteroidetes ratio tended to be greater in the CP group than in the CTL group (P = 0.083), but L-arginine addition remarkably increased this ratio (P < 0.05, Figure S4).
Differentially Abundant Gut Bacterial Genera
T-test analysis was performed to evaluate the differentially abundant genera (relative abundance >0.1%) in Figure 3. A total of 24 taxa were significantly more abundant in the CP group than in the CTL group; among these taxa, 14 genera belonged to Proteobacteria (e.g., Nitrosomonas spp., Coxiella spp., Ruegeria spp., and Thauera spp., P < 0.05), 4 to Planctomycetes (e.g., Planctomyces spp. and Blastopirellula spp., P < 0.05), 3 to Bacteroidetes (Phaeodactylibacter spp., Lutimonas spp., and Terrimonas spp., P < 0.05), 1 to Nitrospirae, 1 to Actinobacteria, and 1 to Acidobacteria (Figure 3A and Table S3). However, compared with the CP group, ARGCP group had significantly less of most of these genera (Figure 3B, P < 0.05, except for Caulobacter spp.). In addition, the relative abundance of Pseudomonas spp. in the ARGCP group was significantly lower than that in the CP group (Figure 3B, P < 0.05). As shown in Figure 3C, the relative abundances of only three genera were significantly different between the CTL and ARGCP groups (P ≤ 0.001). The relative abundance of Candidatus jettenia spp. in the CTL group was significantly higher than that in the ARGCP group (P = 0.001), whereas Thauera spp. and Gaiella spp. were more abundant in the ARGCP group than in the control group (P < 0.001).
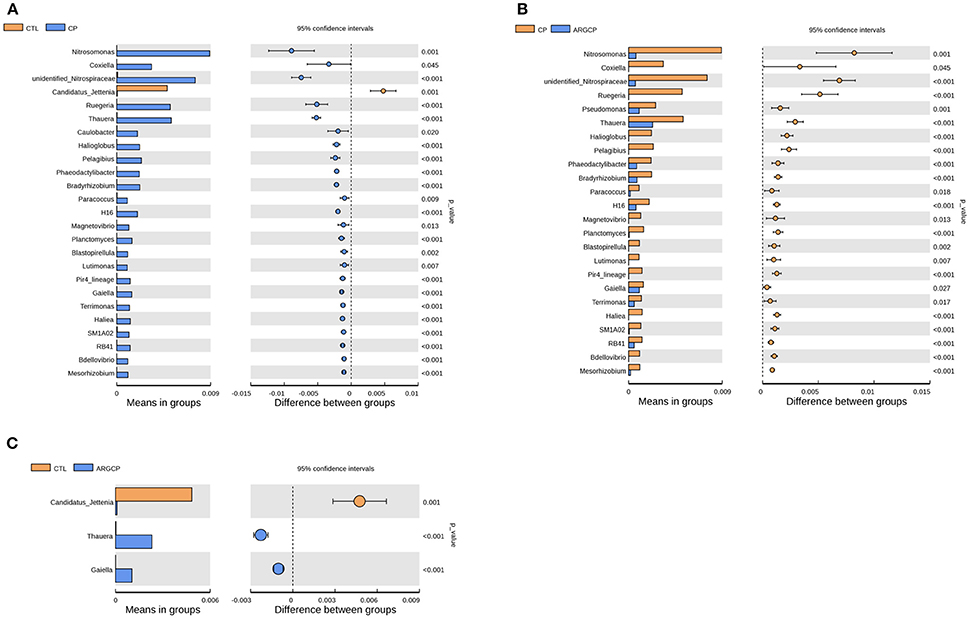
Figure 3. The genera differentially abundant between three treatments in the ileum by t-test analysis. These figures illustrate the differences in the microbiota in terms of the relative abundances of genera between groups CTL and CP (A), groups CP and ARGCP (B), and groups CTL and ARGCP (C). Only data whose differences with P-values lower than 0.05 and the relative abundances higher than 0.1% in either of the pairs are shown. CTL, non-challenge control; CP, C. perfringens-challenged group; ARGCP, C. perfringens-challenged group fed diet supplemented with 0.3% L-arginine.
Differentially Abundant Gut Bacterial Species
At the species level, the relative abundance lower than 0.1% in all groups was filtered. The relative abundance of Lactobacillus gasseri dramatically decreased in CP group when compared to CTL group (Figure 4A, P < 0.05). Furthermore, the relative abundance of four bacterial species including Nitrospira sp. enrichment culture clone M1-9, Bradyrhizobium elkanii, Nitrospira bacterium SG8-3, and Pseudomonas veronii was significantly increased by the C. perfringens challenge (Figure 4A, P ≤ 0.001). However, the increases of the relative abundance of the above four bacterial species were significantly reduced by L-arginine supplementation (Figure 4B, P ≤ 0.001). There was no differentially abundant species between CTL group and ARGCP group (P > 0.05).
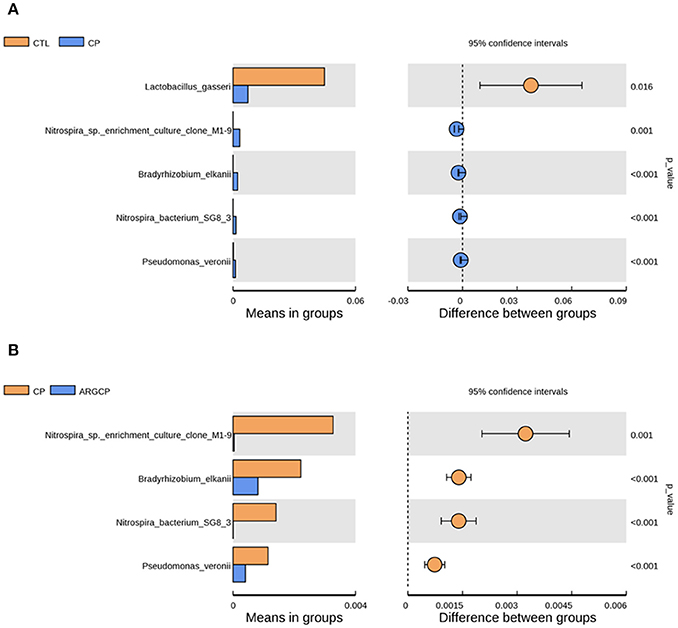
Figure 4. The species differentially abundant between three treatments in the ileum by t-test analysis. These figures illustrate the differences in the microbiota in terms of the relative abundances of species between groups CTL and CP (A), and groups CP and ARGCP (B). Only data whose differences with P-values lower than 0.05 and the relative abundances higher than 0.1% in either of the pairs are shown. CTL, non-challenge control; CP, C. perfringens-challenged group; ARGCP, C. perfringens-challenged group fed diet supplemented with 0.3% L-arginine. There was no differentially abundant species between CTL group and ARGCP group.
Predicted Functional Changes in Gut Microbial Communities
Alterations in the presumptive functions of the ileal microbiota of broilers were evaluated using PICRUSt analysis. Figure 5 shows the top 10 predicted microbial functions at level 2 of the KEGG pathways. Compared with CTL group, the CP group had significantly less abundance of KEGG pathways affiliated with membrane transport, replication and repair, translation and nucleotide metabolism (P < 0.05) and remarkably larger abundances of KEGG pathways belonging to amino acid metabolism and lipid metabolism (P < 0.05). L-arginine significantly reversed the changes in the abundances of these pathways (P < 0.05). A predicted microbial function comparison at level 3 of the KEGG pathways is shown in Table 5. Compared with the CTL group, the CP group had significantly lower abundance of 19 functional pathways, including two pathways in membrane transport (“Transporters” and “Phosphotransferase system”), five pathways in carbohydrate metabolism (e.g., “Amino sugar and nucleotide sugar metabolism” and “Glycolysis/Gluconeogenesis”), five pathways in replication and repair (e.g., “DNA repair and recombination proteins” and “Chromosome”), two pathways in energy metabolism (“Photosynthesis proteins” and “Photosynthesis”), three pathways in translation (e.g., “Ribosome” and “Ribosome Biogenesis”) and two pathways in nucleotide metabolism (“Purine metabolism” and “Pyrimidine metabolism”), while most of these changes were reversed by L-arginine supplementation (P < 0.05, except “Photosynthesis proteins” and “Photosynthesis”). Three pathways in carbohydrate metabolism (e.g., “Pyruvate metabolism” and “Butanoate metabolism”), two pathways in amino acid metabolism (“Arginine and proline metabolism” and “Glycine, serine and threonine metabolism”) and two pathways in energy metabolism (“Oxidative phosphorylation” “Carbon fixation pathways in prokaryotes”) were represented more in the CP group than in the CTL group (P < 0.05), whereas the ARGCP group had significantly less abundance of these pathways than the CP group (P < 0.05). Table 5 illustrated that in L-arginine-fed broilers, the relative abundances of KEGG pathways at level 3 were almost the same as those of the controls (except for 8 microbial functions whose abundances differed between the CTL and ARGCP groups).
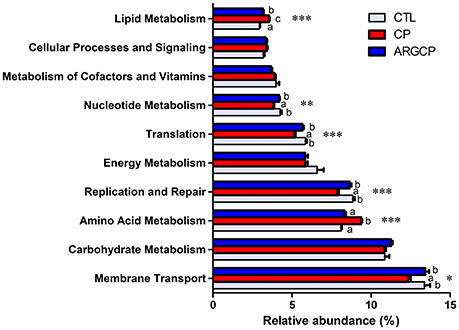
Figure 5. The 10 most abundant microbial pathways grouped into level-2 functional categories using PICRUSt. CTL, non-challenge control; CP, C. perfringens-challenged group; ARGCP, C. perfringens-challenged group fed diet supplemented with 0.3% L-arginine. Different lowercase letters at each column indicate significant differences. Values are means with their standard errors. *P < 0.05, **P < 0.01, ***P < 0.001.
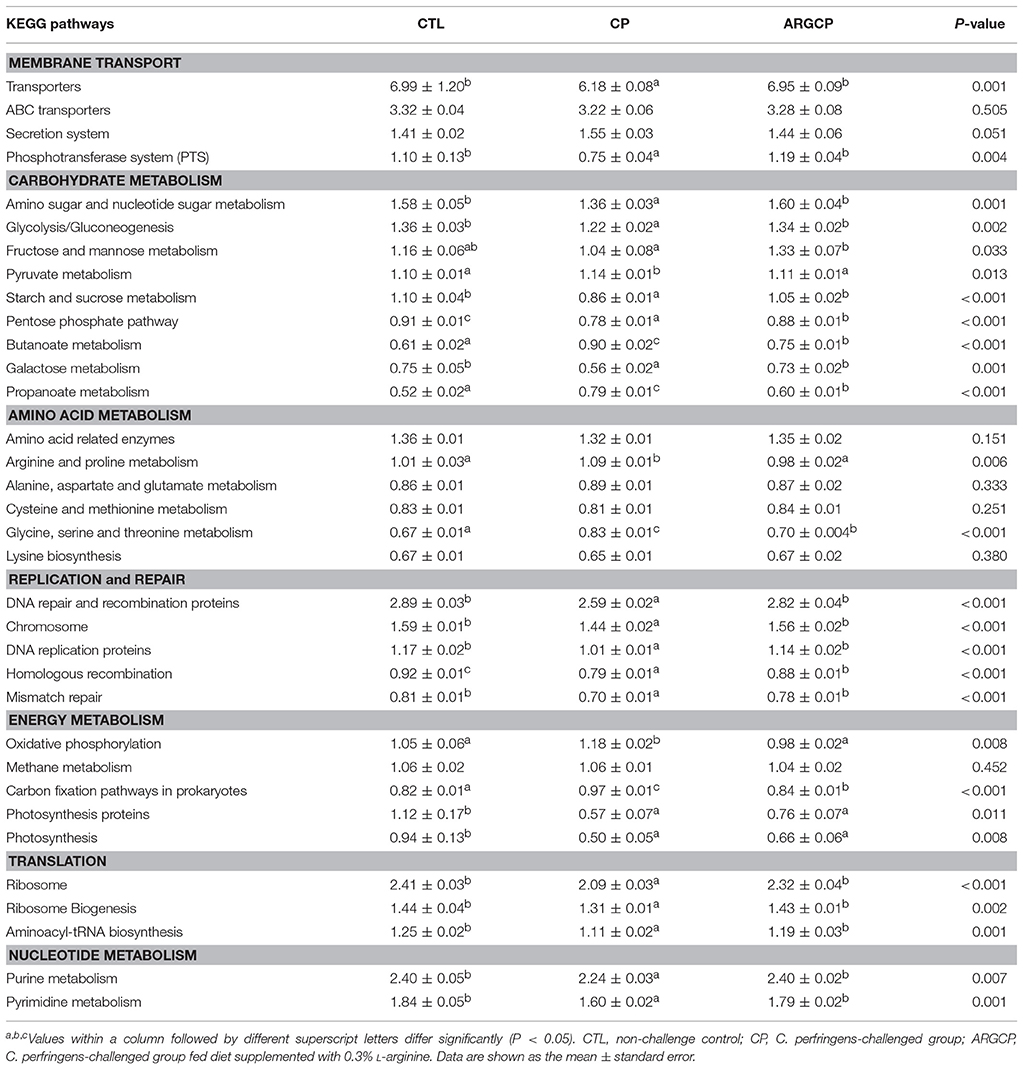
Table 5. The selected main microbial pathways grouped into level-3 functional categories using PICRUSt.
Discussion
A surge of information regarding the gut microbiota and its impacts on health has been reported over the past decade. The gut microbial community structure is strongly affected by many factors such as diet, age, immunological response, host genotype, antibiotic addition, and pathogen infection (Luo et al., 2017). Previous researches have demonstrated that diet has a great influence (estimated at 57%, compared with 12% for genetic factors) on the gut microbial community structure (Tomasello et al., 2014). The community structure of microbiota can be quickly altered by the changes of dietary components (David et al., 2014). L-arginine possesses potential benefits for both humans and animals against numerous diseases, such as alleviating necrotizing enterocolitis of premature neonates and coccidiosis and infectious bursal disease of broiler chickens (Amin et al., 2002; Tan et al., 2014a, 2015). In our previous studies, Dietary L-arginine supplementation protected the gut mucosa by improving innate immune responses, intestinal absorption and barrier function and by suppressing the colonization of C. perfringens in necrotic enteritis-challenged broiler chickens (Zhang et al., 2017). However, little is known regarding how L-arginine influences the intestinal microbiota in broiler chickens.
In this experiment, all of the C. perfringens-challenged birds appeared obvious pathological changes in the small intestines such as hyperemia, small red petechiae, thinner walls and gas production in the intestinal tract, which has been documented by other researchers (Van Immerseel et al., 2004, 2009; Dahiya et al., 2005; Liu et al., 2010, 2012). Hematoxylin-eosin staining showed that C. perfringens challenge led to pathological changes and the disappearance of normal villus architecture (Figure 1). Also, previous studies of our lab reported that C. perfringens challenge impaired growth performance and led to mucosal atrophy and increased plasma endotoxin levels (Liu et al., 2012; Guo et al., 2014). All these data proved that the experimental NE model was successfully established. Moreover, L-arginine supplementation played a beneficial role in C. perfringens-challenged birds, as characterized by decreased gut lesion scores, enhanced villus height and preserved gut morphology.
The community structure of intestinal flora is relatively stable and similar in healthy hosts while intestinal microbiota disturbance can cause many diseases (Karlsson et al., 2013). The result of alpha diversity in our study showed that C. perfringens challenge significantly increased the richness and diversity of ileal microbiota (Table 2), which was also proven by Xu et al. (2018). Nevertheless, Li et al. (2017) reported that C. perfringens challenge dramatically reduced the observed species, Chao1 and ACE indices of the ileal microbiota. The discrepancy may be attributed to the different sections of ileum in which the digesta was collected. Here, the C. perfringens challenge-induced increases of microbial richness and diversity were reversed by L-arginine supplementation (Table 2), which reflected that L-arginine could prevent the intestinal flora disorder.
PCA and ANOSIM tests showed that bacterial communities varied across the different treatments (Figure 2 and Table 3). PCA indicated that the composition of the microbiota in the ARGCP group was more similar to that in the CTL group than that in the CP group (Figure 2), which might be due to the regulation of L-arginine. Firmicutes, Cyanobacteria, Proteobacteria and Bacteroidetes were the four pre-dominant bacterial taxa at the phylum level in the ileal microbiota (Table 4), which is similar to previous studies of broilers (Li et al., 2017; Wang et al., 2017). It has been reported that Firmicutes improved the utilization of energy in the diet and the ratio of Firmicutes to Bacteroides was often positively associated with weight gain (Ley et al., 2006; Bervoets et al., 2013). In the current study, the relative abundance of Firmicutes and the ratio of Firmicutes to Bacteroides were remarkably higher in the ARGCP group than in the CP group (Table 4 and Figure S4), which may be correlated with the effect of L-arginine promoting broiler growth performance (Tan et al., 2014a,b). In this research, Proteobacteria was more abundant in the CP group than in the CTL group (Table 4). Gut bacteria belonging to Proteobacteria include a wide variety of pathogens such as Rickettsia spp. (Ferla et al., 2013), Brucella spp. (de Figueiredo et al., 2015) and Neisseria spp. (Criss and Seifert, 2012). However, significantly less Proteobacteria was observed in the ARGCP group than in the CP group (Table 4), which was consistent with reports that L-arginine treatment reduced the number of harmful bacteria belonging to the phylum Proteobacteria, such as Escherichia coli (Liu et al., 2017) and Helicobacter pylori (Chaturvedi et al., 2007). Moreover, C. perfringens challenge resulted in significantly higher relative abundances of Actinobacteria, Planctomycetes, Verrucomicrobia, and Chloroflexi in the CP group than in unchallenged birds (Table 4). In some pathological cases such as cardiovascular diseases, non-alcoholic fatty liver disease and ulcerative colitis, the relative abundance of Actinobacteria was higher in patients than in healthy individuals (Lepage et al., 2011; Dinakaran et al., 2014; Del Chierico et al., 2017). In addition, Verrucomicrobia and Chloroflexi, despite usually being present in low abundance (a few percent or less), are consistent components in most individuals and animals, and their levels become elevated in some diseases (Abusleme et al., 2013; Campbell et al., 2014). Planctomycetes is phylogenetically closely related to Verrucomicrobia and Chlamydiae and widely distributed in vast anoxic zones of the ocean (Fuerst and Sagulenko, 2011). Planctomycetes spp. are major participants in the global nitrogen cycle since 50% of the nitrogen molecules in the atmosphere are generated by this genus (Fuerst and Sagulenko, 2011). However, the role that Planctomycetes plays in animals is unclear. The alterations of the gut microbiota community by L-arginine treatment in our research were associated with recovery from intestinal injury, suggesting that L-arginine-mediated reduction of the Proteobacteria, Actinobacteria, Verrucomicrobia, Chloroflexi, and Planctomycetes abundances played a benign role in the development of necrotic enteritis.
In the t-test analysis at the genus level, more than half of the genera differentially abundant between the three treatments belonged to the phylum Proteobacteria (Figure 3 and Table S3). In this phylum, the genera of Nitrosomonas, Coxiella, Ruegeria, and Thauera were more dominant in the CP group than in other groups (Figures 3A,B), and Nitrosomonas spp. were the most abundant species. Nitrification is conducted by two different types of bacteria. The first type are the ammonia-oxidizing bacteria, which oxidize ammonia to nitrite. The other type, the nitrite-oxidizing bacteria, convert nitrite to nitrate (Chauret et al., 2008). Nitrosomonas, a major genus of the ammonia-oxidizing bacteria, utilizes ammonia as its sole energy source (Lek Noophan et al., 2009). L-arginine addition inhibits the nitrite formation and protein synthesis of Nitrosomonas europaea in vitro (Clark and Schmidt, 1967). In our study, L-arginine treatment decreased the population of ammonia oxidizers, which might be due to different C and N availability, intrinsic amino acid characteristics and interactions with the host and heterotrophic microorganisms (Tzanakakis and Paranychianakis, 2017). Consistent with the changes in Nitrosomonas spp., the abundance of Unidentified_Nitrospiraceae, a genus of nitrite-oxidizing bacteria, was also markedly lower in the ARGCP group than in the CP group (Figure 3B). This result may reflect the synergistic efforts of ammonia-oxidizing bacteria and nitrite-oxidizing bacteria in nitrification. Coxiella spp. include different species of bacteria that use ticks as effective vectors to spread a range of disease (Khoo et al., 2016; Machado-Ferreira et al., 2016). A typical example is Coxiella burnetii, which is the causative agent for Q fever, an infectious disease with a global distribution commonly affecting animals and human (Khoo et al., 2016). Members of Ruegeria were linked to yellow band disease in Fungiidae corals (Apprill et al., 2013). The bacterium Ruegeria pomeroyi is more abundant in corals with white patch syndrome or skeletal growth anomalies than in healthy corals (Sere et al., 2013; Li et al., 2015). The genus Thauera, of the class Betaproteobacteria, has been detected in sediments, sludge, soils, and wastewater treatment plants (Liu et al., 2013), and it has been recently reported in research on human (Smith et al., 2016; Xu et al., 2017). Xu et al. (2017) observed that Thauera was significantly more abundant in individuals with substance use disorders than in healthy individuals. Similar to the result of Xu et al. (2017), Thauera was represented more in the CP group than in the CTL group in our study, indicating that this genus may comprise opportunistic pathogens. However, determining the specific functions of these bacteria requires further testing and verification. Furthermore, the abundances of only 3 genera differed between the CTL group and the ARGCP group, and the abundances of most genera were not significantly different between these pairs (Figure 3C), which indicated that the diet supplemented with L-arginine normalized the ileal microbiota of broiler chickens challenged with C. perfringens, returning its microbial structure to resemble that of healthy controls, and that the beneficial effects of L-arginine on intestinal injury may be largely mediated by the composition of the gut microbiota.
Lactobacillus gasseri is one kind of the well-established probiotic microorganisms. In humans, Lactobacillus gasseri was shown to competitively inhibit the growth of harmful bacteria, produce bacteriocin and modulate the innate and adaptive systems, eliciting many health benefits (Selle and Klaenhammer, 2013). On the species level, Lactobacillus gasseri was less abundant in CP group than in CTL group (Figure 4A), which reflected that C. perfringens challenge inhibited the growth of some probiotic bacteria. In our study, Nitrospira sp. enrichment culture clone M1-9 and Nitrospira bacterium SG8-3, belonged to the genus of unidentified_Nitrospiraceae, were more abundant in CP group than other groups (Figures 4A,B). This was in accordance with our observation on the changes of the relative abundance of unidentified_Nitrospiraceae spp. Pseudomonas veronii has been found in natural mineral waters, soils and animal intestines. This bacterium was reported to be the aetiological agent of human intestinal inflammatory pseudotumour (Cheuk et al., 2000). Here, L-arginine supplementation reversed the increased relative abundance of Pseudomonas veronii caused by C. perfringens challenge. This suggested that L-arginine supplementation could inhibit this potential opportunistic pathogen.
According to the predictive functional profiles of microbial communities determined by PICRUSt analysis, the most abundant functions were membrane transport, carbohydrate metabolism and amino acid metabolism (Figure 5), and clear differences were observed in the level 3 KEGG pathways between the three treatments (Table 5). Membrane transport pathways, such as those involving transporters and ABC transporters, are essential to cell viability and growth and therefore crucial for the survival of bacteria in the gut ecosystem (Lyons et al., 2017). Odamaki et al. (2016) demonstrated that such predicted transporter functions were connected with nutrient-associated changes in gut microbiota composition. In our study, L-arginine supplementation significantly increased the proportion of transporters, suggesting that L-arginine might play an important role in affecting the gut microbiota composition of birds challenged with C. perfringens. A phosphotransferase system (PTS) is used by bacteria for sugar uptake and uses phosphoenolpyruvate, a key intermediate in glycolysis, as the source of energy (Erni, 2013). PTS pathways are more abundant in the ARGCP group than in the CP group, which may be closely associated with the promotion of various sugar metabolism pathways (for instance, glycolysis/gluconeogenesis, fructose, mannose, sucrose, and galactose metabolisms) (Table 5). The results suggested that L-arginine could play an active role in the progress that intestinal microbiota sensed and utilized sugars as resources for the production of energy and synthesis of cellular components.
Our data showed the amino acid and lipid metabolisms were overrepresented in the ileal microbiota of challenged chickens (Figure 5). Similarly, according to the research of Davenport et al. (2014), bacterial functional pathways were notably altered in the inflamed tissue of ulcerative colitis patients, with decreased carbohydrate metabolism and promoted lipid and amino acid metabolisms. The author inferred that the supply of carbohydrates was reduced and that bacteria that metabolize amino acids and lipids dominate in the inflamed gut. However, in our study, carbohydrate metabolism was not affected by the three treatments. This result may be related to differences in the hosts and disease models used in the studies. The small intestine is rich in monosaccharides, disaccharides and amino acids, which supports the growth of certain bacteria, particularly Proteobacteria and Lactobacillales (Kamada et al., 2013). In our data, the reduction in amino acid metabolism may be closely related to the low abundance of Proteobacteria in the ARGCP group.
In this study, the C. perfringens challenge enriched pathways related to arginine and proline metabolism and glycine, serine and threonine metabolism (Table 5). As summarized by previous reviews, arginine can be used as an energy source by various pathogens such as Pseudomonas aeruginosa, Helicobacter pylori and Salmonella typhimurium (Gogoi et al., 2016; Xiong et al., 2016). The bacteria degrade arginine and produce urea and ammonia, which can diminish the availability of arginine substrate for host-cell inducible nitric oxide synthase (iNOS), thereby reducing nitric oxide production and facilitating evasion from its antibacterial effects (Gobert et al., 2001). In this experiment, the acceleration of arginine metabolism by the gut microbiota in challenged birds may cause arginine deprivation in the host, leading to compromised T-cell function and eventually resulting in increased susceptibility to infection (Popovic et al., 2007). L-arginine supplementation might normalize arginine metabolism, which may result in that L-arginine significantly enhancing T-cell proliferation and function (Ochoa et al., 2001; Taheri et al., 2001; Tan et al., 2015) and thus inhibiting pathogens catabolizing L-arginine (Stadelmann et al., 2013). In the current experiment, L-arginine treatment depleted the pathways related to glycine, serine and threonine metabolism (Table 5). This finding was consistent with the results of Dai et al. (2012) and demonstrated that the net utilization of glycine, serine and threonine by mixed ileal bacteria was reduced with arginine addition in vitro. Recent research showed that the pathogenic bacteria Staphylococcus aureus utilizes serine and threonine for producing hemolysin, which contributes to the virulence of the bacteria (Burnside et al., 2010).
Conclusion
In summary, we found that Dietary L-arginine supplementation promoted the intestinal health and modulated the ileal microbiota of broiler chickens challenged with C. perfringens to result in a consortium similar to that of healthy controls, characterized by enriched helpful bacteria and suppressed harmful species.
Author Contributions
YG and BZ designed the research. BZ conducted the animal feeding. BZ, ZPL, and GL performed sample collection and detection. BZ, ZL, and WW analyzed the data. BZ and YG wrote the manuscript. All authors approved the final manuscript.
Funding
This research was funded by the China Agricultural Research System (CARS-41-G11).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01716/full#supplementary-material
References
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Amin, H. J., Zamora, S. A., McMillan, D. D., Fick, G. H., Butzner, J. D., Parsons, H. G., et al. (2002). Arginine supplementation prevents necrotizing enterocolitis in the premature infant. J. Pediatr. 140, 425–431. doi: 10.1067/mpd.2002.123289
Antonissen, G., Eeckhaut, V., Van Driessche, K., Onrust, L., Haesebrouck, F., Ducatelle, R., et al. (2016). Microbial shifts associated with necrotic enteritis. Avian Pathol. 45, 308–312. doi: 10.1080/03079457.2016.1152625
Apprill, A., Hughen, K., and Mincer, T. (2013). Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ. Microbiol. 15, 2063–2072. doi: 10.1111/1462-2920.12107
Bervoets, L., Van Hoorenbeeck, K., Kortleven, I., Van Noten, C., Hens, N., Vael, C., et al. (2013). Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathog. 5:10. doi: 10.1186/1757-4749-5-10
Burnside, K., Lembo, A., de Los Reyes, M., Iliuk, A., Binhtran, N. T., Connelly, J. E., et al. (2010). Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE 5:e11071. doi: 10.1371/journal.pone.0011071
Campbell, A. G., Schwientek, P., Vishnivetskaya, T., Woyke, T., Levy, S., Beall, C. J., et al. (2014). Diversity and genomic insights into the uncultured Chloroflexi from the human microbiota. Environ. Microbiol. 16, 2635–2643. doi: 10.1111/1462-2920.12461
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chaturvedi, R., Asim, M., Lewis, N. D., Algood, H. M., Cover, T. L., Kim, P. Y., et al. (2007). L-arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect. Immun. 75, 4305–4315. doi: 10.1128/IAI.00578-07
Chauret, C., Smith, C., and Baribeau, H. (2008). Inactivation of Nitrosomonas europaea and pathogenic Escherichia coli by chlorine and monochloramine. J. Water Health 6, 315–322. doi: 10.2166/wh.2008.052
Cheuk, W., Woo, P. C., Yuen, K. Y., Yu, P. H., and Chan, J. K. (2000). Intestinal inflammatory pseudotumour with regional lymph node involvement: identification of a new bacterium as the aetiological agent. J. Pathol. 192, 289–292. doi: 10.1002/1096-9896(2000)9999:9999 < ::AID-PATH767>3.0.CO;2-F
Clark, C., and Schmidt, E. L. (1967). Growth response of Nitrosomonas europaea to amino acids. J. Bacteriol. 93, 1302–1308.
Cooper, K. K., and Songer, J. G. (2009). Necrotic enteritis in chickens: a paradigm of enteric infection by Clostridium perfringens type A. Anaerobe 15, 55–60. doi: 10.1016/j.anaerobe.2009.01.006
Costa, K. A., Soares, A. D., Wanner, S. P., Santos, R., Fernandes, S. O., Martins Fdos, S., et al. (2014). L-arginine supplementation prevents increases in intestinal permeability and bacterial translocation in male Swiss mice subjected to physical exercise under environmental heat stress. J. Nutr. 144, 218–223. doi: 10.3945/jn.113.183186
Criss, A. K., and Seifert, H. S. (2012). A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat. Rev. Microbiol. 10, 178–190. doi: 10.1038/nrmicro2713
Dahiya, J. P., Hoehler, D., Wilkie, D. C., Van Kessel, A. G., and Drew, M. D. (2005). Dietary glycine concentration affects intestinal Clostridium perfringens and lactobacilli populations in broiler chickens. Poult. Sci. 84, 1875–1885. doi: 10.1093/ps/84.12.1875
Dai, Z. L., Li, X. L., Xi, P. B., Zhang, J., Wu, G., and Zhu, W. Y. (2012). Regulatory role for L-arginine in the utilization of amino acids by pig small-intestinal bacteria. Amino Acids 43, 233–244. doi: 10.1007/s00726-011-1067-z
Davenport, M., Poles, J., Leung, J. M., Wolff, M. J., Abidi, W. M., Ullman, T., et al. (2014). Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 20, 723–731. doi: 10.1097/mib.0000000000000011
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
de Figueiredo, P., Ficht, T. A., Rice-Ficht, A., Rossetti, C. A., and Adams, L. G. (2015). Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am. J. Pathol. 185, 1505–1517. doi: 10.1016/j.ajpath.2015.03.003
Del Chierico, F., Nobili, V., Vernocchi, P., Russo, A., Stefanis, C., Gnani, D., et al. (2017). Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65, 451–464. doi: 10.1002/hep.28572
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/aem.03006-05
Dinakaran, V., Rathinavel, A., Pushpanathan, M., Sivakumar, R., Gunasekaran, P., and Rajendhran, J. (2014). Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS ONE 9:e105221. doi: 10.1371/journal.pone.0105221
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Erni, B. (2013). The bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS): an interface between energy and signal transduction. J. Iran. Chem. Soc. 10, 593–630. doi: 10.1007/s13738-012-0185-1
Ferla, M. P., Thrash, J. C., Giovannoni, S. J., and Patrick, W. M. (2013). New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE 8:e83383. doi: 10.1371/journal.pone.0083383
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413. doi: 10.1038/nrmicro2578
Gobert, A. P., McGee, D. J., Akhtar, M., Mendz, G. L., Newton, J. C., Cheng, Y., et al. (2001). Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. U.S.A. 98, 13844–13849. doi: 10.1073/pnas.241443798
Gogoi, M., Datey, A., Wilson, K. T., and Chakravortty, D. (2016). Dual role of arginine metabolism in establishing pathogenesis. Curr. Opin. Microbiol. 29, 43–48. doi: 10.1016/j.mib.2015.10.005
Guo, S., Liu, D., Zhao, X., Li, C., and Guo, Y. (2014). Xylanase supplementation of a wheat-based diet improved nutrient digestion and mRNA expression of intestinal nutrient transporters in broiler chickens infected with Clostridium perfringens. Poult. Sci. 93, 94–103. doi: 10.3382/ps.2013-03188
Kamada, N., Chen, G. Y., Inohara, N., and Nunez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. doi: 10.1093/nar/gkr988
Karlsson, F. H., Tremaroli, V., Nookaew, I., Bergstrom, G., Behre, C. J., Fagerberg, B., et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103. doi: 10.1038/nature12198
Khoo, J. J., Lim, F. S., Chen, F., Phoon, W. H., Khor, C. S., Pike, B. L., et al. (2016). Coxiella detection in ticks from wildlife and livestock in Malaysia. Vector Borne Zoonotic Dis. 16, 744–751. doi: 10.1089/vbz.2016.1959
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lek Noophan, P., Sripiboon, S., Damrongsri, M., and Munakata-Marr, J. (2009). Anaerobic ammonium oxidation by Nitrosomonas spp. and anammox bacteria in a sequencing batch reactor. J. Environ. Manage. 90, 967–972. doi: 10.1016/j.jenvman.2008.03.003
Lepage, P., Hasler, R., Spehlmann, M. E., Rehman, A., Zvirbliene, A., Begun, A., et al. (2011). Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236. doi: 10.1053/j.gastro.2011.04.011
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, C., Guo, S., Gao, J., Guo, Y., Du, E., Lv, Z., et al. (2015). Maternal high-zinc diet attenuates intestinal inflammation by reducing DNA methylation and elevating H3K9 acetylation in the A20 promoter of offspring chicks. J. Nutr. Biochem. 26, 173–183. doi: 10.1016/j.jnutbio.2014.10.005
Li, Z., Wang, W., Liu, D., and Guo, Y. (2017). Effects of Lactobacillus acidophilus on gut microbiota composition in broilers challenged with Clostridium perfringens. PLoS ONE 12:e0188634. doi: 10.1371/journal.pone.0188634
Liu, B., Mao, Y., Bergaust, L., Bakken, L. R., and Frostegard, A. (2013). Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ. Microbiol. 15, 2816–2828. doi: 10.1111/1462-2920.12142
Liu, D., Guo, S., and Guo, Y. (2012). Xylanase supplementation to a wheat-based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens. Avian Pathol. 41, 291–298. doi: 10.1080/03079457.2012.684089
Liu, D., Guo, Y., Wang, Z., and Yuan, J. (2010). Exogenous lysozyme influences Clostridium perfringens colonization and intestinal barrier function in broiler chickens. Avian Pathol. 39, 17–24. doi: 10.1080/03079450903447404
Liu, G., Ren, W., Fang, J., Hu, C. A., Guan, G., Al-Dhabi, N. A., et al. (2017). L-Glutamine and L-arginine protect against enterotoxigenic Escherichia coli infection via intestinal innate immunity in mice. Amino Acids 49, 1945–1954. doi: 10.1007/s00726-017-2410-9
Luo, Y., Zhang, L., Li, H., Smidt, H., Wright, A. G., Zhang, K., et al. (2017). Different types of dietary fibers trigger specific alterations in composition and predicted functions of colonic bacterial communities in BALB/c Mice. Front. Microbiol. 8:966. doi: 10.3389/fmicb.2017.00966
Lyons, P. P., Turnbull, J. F., Dawson, K. A., and Crumlish, M. (2017). Phylogenetic and functional characterization of the distal intestinal microbiome of rainbow trout Oncorhynchus mykiss from both farm and aquarium settings. J. Appl. Microbiol. 122, 347–363. doi: 10.1111/jam.13347
Machado-Ferreira, E., Vizzoni, V. F., Balsemao-Pires, E., Moerbeck, L., Gazeta, G. S., Piesman, J., et al. (2016). Coxiella symbionts are widespread into hard ticks. Parasitol. Res. 115, 4691–4699. doi: 10.1007/s00436-016-5230-z
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
National Research Council (1994). Nutrient Requirements of Poultry, 9th Edn. Washington, DC: National Academies Press.
Ochoa, J. B., Strange, J., Kearney, P., Gellin, G., Endean, E., and Fitzpatrick, E. (2001). Effects of L-arginine on the proliferation of T lymphocyte subpopulations. JPEN J. Parenter. Enteral Nutr. 25, 23–29. doi: 10.1177/014860710102500123
Odamaki, T., Kato, K., Sugahara, H., Hashikura, N., Takahashi, S., Xiao, J. Z., et al. (2016). Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16:90. doi: 10.1186/s12866-016-0708-5
Popovic, P. J., Zeh, H. J. III., and Ochoa, J. B. (2007). Arginine and immunity. J. Nutr. 137, 1681S−1686S. doi: 10.1093/jn/137.6.1681S
Selle, K., and Klaenhammer, T. R. (2013). Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol. Rev. 37, 915–935. doi: 10.1111/1574-6976.12021
Sere, M. G., Tortosa, P., Chabanet, P., Turquet, J., Quod, J. P., and Schleyer, M. H. (2013). Bacterial communities associated with Porites white patch syndrome (PWPS) on three western Indian Ocean (WIO) coral reefs. PLoS ONE 8:e83746. doi: 10.1371/journal.pone.0083746
Smith, A. D., Zhang, Y., Barber, R. C., Minshall, C. T., Huebinger, R. M., and Allen, M. S. (2016). Common lung microbiome identified among mechanically ventilated surgical patients. PLoS ONE 11:e0166313. doi: 10.1371/journal.pone.0166313
Stadelmann, B., Hanevik, K., Andersson, M. K., Bruserud, O., and Svard, S. G. (2013). The role of arginine and arginine-metabolizing enzymes during Giardia-host cell interactions in vitro. BMC Microbiol. 13:256. doi: 10.1186/1471-2180-13-256
Stanley, D., Keyburn, A. L., Denman, S. E., and Moore, R. J. (2012). Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet. Microbiol. 159, 155–162. doi: 10.1016/j.vetmic.2012.03.032
Taheri, F., Ochoa, J. B., Faghiri, Z., Culotta, K., Park, H. J., Lan, M. S., et al. (2001). L-arginine regulates the expression of the T-cell receptor zeta chain (CD3zeta) in Jurkat cells. Clin Cancer Res 7(Suppl. 3), 958s−965s.
Tan, J., Applegate, T. J., Liu, S., Guo, Y., and Eicher, S. D. (2014a). Supplemental dietary L-arginine attenuates intestinal mucosal disruption during a coccidial vaccine challenge in broiler chickens. Br. J. Nutr. 112, 1098–1109. doi: 10.1017/s0007114514001846
Tan, J., Liu, S., Guo, Y., Applegate, T. J., and Eicher, S. D. (2014b). Dietary L-arginine supplementation attenuates lipopolysaccharide-induced inflammatory response in broiler chickens. Br. J. Nutr. 111, 1394–1404. doi: 10.1017/s0007114513003863
Tan, J. Z., Guo, Y. M., Applegate, T. J., Du, E. C., and Zhao, X. (2015). Dietary L-arginine modulates immunosuppression in broilers inoculated with an intermediate strain of infectious bursa disease virus. J. Sci. Food Agric. 95, 126–135. doi: 10.1002/jsfa.6692
Timbermont, L., Haesebrouck, F., Ducatelle, R., and Van Immerseel, F. (2011). Necrotic enteritis in broilers: an updated review on the pathogenesis. Avian Pathol. 40, 341–347. doi: 10.1080/03079457.2011.590967
Tomasello, G., Tralongo, P., Damiani, P., Sinagra, E., Di Trapani, B., Zeenny, M. N., et al. (2014). Dismicrobism in inflammatory bowel disease and colorectal cancer: changes in response of colocytes. World J. Gastroenterol. 20, 18121–18130. doi: 10.3748/wjg.v20.i48.18121
Tzanakakis, V. A., and Paranychianakis, N. V. (2017). Divergent response of ammonia oxidizers to various amino acids. Appl. Soil Ecol. 114, 45–51. doi: 10.1016/j.apsoil.2017.02.019
Van Immerseel, F., De Buck, J., Pasmans, F., Huyghebaert, G., Haesebrouck, F., and Ducatelle, R. (2004). Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 33, 537–549. doi: 10.1080/03079450400013162
Van Immerseel, F., Rood, J. I., Moore, R. J., and Titball, R. W. (2009). Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 17, 32–36. doi: 10.1016/j.tim.2008.09.005
Wang, W., Li, Z., Lv, Z., Zhang, B., Lv, H., and Guo, Y. (2017). Effects of Kluyveromyces marxianus supplementation on immune responses, intestinal structure and microbiota in broiler chickens. PLoS ONE 12:e0180884. doi: 10.1371/journal.pone.0180884
Xiong, L., Teng, J. L., Botelho, M. G., Lo, R. C., Lau, S. K., and Woo, P. C. (2016). Arginine metabolism in bacterial pathogenesis and cancer therapy. Int. J. Mol. Sci. 17, 363. doi: 10.3390/ijms17030363
Xu, S., Lin, Y., Zeng, D., Zhou, M., Zeng, Y., Wang, H., et al. (2018). Bacillus licheniformis normalize the ileum microbiota of chickens infected with necrotic enteritis. Sci. Rep. 8:1744. doi: 10.1038/s41598-018-20059-z
Xu, Y., Xie, Z., Wang, H., Shen, Z., Guo, Y., Gao, Y., et al. (2017). Bacterial diversity of intestinal microbiota in patients with substance use disorders revealed by 16S rRNA gene deep sequencing. Sci. Rep. 7:3628. doi: 10.1038/s41598-017-03706-9
Zhang, B., Lv, Z., Li, H., Guo, S., Liu, D., and Guo, Y. (2017). Dietary L-arginine inhibits intestinal Clostridium perfringens colonisation and attenuates intestinal mucosal injury in broiler chickens. Br. J. Nutr. 118, 321–332. doi: 10.1017/s0007114517002094
Keywords: L-arginine, gut microbiota, Clostridium perfringens, PICRUSt predicted functions, 16S rRNA high-throughput sequencing, broiler chicken
Citation: Zhang B, Lv Z, Li Z, Wang W, Li G and Guo Y (2018) Dietary L-arginine Supplementation Alleviates the Intestinal Injury and Modulates the Gut Microbiota in Broiler Chickens Challenged by Clostridium perfringens. Front. Microbiol. 9:1716. doi: 10.3389/fmicb.2018.01716
Received: 08 March 2018; Accepted: 10 July 2018;
Published: 31 July 2018.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Muhammad Zubair Shabbir, University of Veterinary and Animal Sciences, PakistanAggeliki Saridaki, Technical University of Crete, Greece
Copyright © 2018 Zhang, Lv, Li, Wang, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuming Guo, Z3VveXVtQGNhdS5lZHUuY24=
 Beibei Zhang
Beibei Zhang Zengpeng Lv
Zengpeng Lv Weiwei Wang
Weiwei Wang