
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 July 2018
Sec. Terrestrial Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01692
Microbial co-inoculation is considered to be an innovative approach and had been applied worldwide. However, the underlying mechanisms of microbial co-inoculants constructions, especially the trait-based combination of distinctly different microbial species remains poorly understood. In this study, we constructed two microbial co-inoculants with the same three strains with emphasis on the microbial, soil and plant traits. Microbial co-inoculants 1 (M1) were constructed according to soil fertility, microbial activity and cucumber nutrient requirement with a 2:1:2 ratio (Ensifer sp. NYM3, Acinetobacter sp. P16 and Flavobacterium sp. KYM3), while microbial co-inoculants 2 (M2) were constructed according to soil fertility and cucumber nutrient requirement with a 1:10:1 ratio without considering the difference in the nutrient supply capability of microbial species. The results showed that M1 and M2 both obviously increased cucumber yields. The M1 had significant highest pH value, total nitrogen (TN) and invertase activity (IA). The M2 had significant highest available phosphate (AP), NO3-N, urea activity (UA), and alkaline phosphatase activity (APA). Gammaproteobacteria, Acidobacteria, Nitrospirae, and Armatimonadetes were significantly increased, while Actinobacteria and Firmicutes were significantly decreased by microbial co-inoculations (M1 and M2). The bacterial lineages enriched in M1 were Gammaproteobacteria and TM7. Acidobacteria, Bacteroidetes, and Deltaproteobacteria were enriched in M2. Principal coordinate analysis (PCoA) analysis showed that the bacterial communities were strongly separated by the different microbial inoculation treatments. The functional groups of intracellular_parasites were highest in M1. The functional groups of phototrophy, photoautotrophy, nitrification, fermentation, cyanobacteria, oxygenic_photoautotrophy, chitinolysis and animal_parasites_or_symbionts were highest in M2. Based on correlation analysis, it inferred that the M1 and M2 might promote cucumber yields by mediating bacterial community structure and function about nitrogen fixing and urea-N hydrolysis, respectively. Collectively, these results revealed that microbial co-inoculants had positive effects on cucumber yields. Trait-based integration of different microbial species had significant effects on soil properties and bacterial communities. It indicated that microbial activity should be considered in the construction of microbial co-inoculants. This will expand our knowledge in bacteria interaction, deepen understanding of microbial inoculants in improving plant performance, and will guide microbial fertilizer formulation and application in future.
Microbial inoculants, especially plant growth-promoting bacteria (PGPB) inoculants, are an alternative method of increasing crop productivity that can reduce the use of pesticides and/or chemical fertilizers (Adesemoye et al., 2009; Beneduzi et al., 2012; Baris et al., 2014; Pereg and McMillan, 2015; Baez-Rogelio et al., 2017). PGPB can promote plant growth through providing nutrient, producing phyto-hormone, defending against pathogens, alleviating stress and so on (Saleem et al., 2007; Bhattacharyya and Jha, 2012; Ambrosini et al., 2016). In order to enhance the reliability and efficacy of microbial inoculants in agriculture, combining these PGPB by co-inoculation had been applied worldwide for more than a decade. As co-inoculation would mimic the natural situation more closely and allow the combination of various mechanisms without the need for genetic engineering (Madhaiyan et al., 2010; Rodrigues et al., 2013). Microbial co-inoculants construction includes species selection and species integration. Microbial co-inoculants were usually composed of two or more compatible strains with different functions. The common selections were nitrogen fixing bacteria (Azospirillum sp. or Rhizobia) and biocontrol agents (Pseudomonas sp. or Bacillus sp.) (Figueiredo et al., 2008; Atieno et al., 2012b). Species integrations were essentially the ratios of microbial species in co-inoculants. The ratios of microbial species in co-inoculants were mostly 1:1 (Maheshwari et al., 2010; Yegorenkova et al., 2016). Saleem et al. (2016) revealed that species richness had positive effects on efficiency and stabilize of microbial co-inoculants and different ratios of a specie had different effects on co-inoculants efficiency. However, Paredes et al. (2018) demonstrated that the effects of microbial co-inoculants on plant physiology were mostly additive and independent of bacterial abundances. Therefore, multi-species with different ratios in inoculants should be paid more attentions.
After releasing in field, microbial inoculants efficiency is not only associated with its beneficial features, but also with the complex network of interactions occurring in the soil ecosystem (Johnke et al., 2014; Ambrosini et al., 2016; Shi et al., 2016). The knowledge of the effects of microbial inoculants on microbial community has been of great interest since changes in the structure of the indigenous microbial communities might affect the function of microbes plays in soil (Berendsen et al., 2012; Mueller and Sachs, 2015). Schwieger and Tebbe (2000) firstly reported the effect of field inoculation with Sinorhizobium meliloti L33 on bacterial communities in rhizospheres of Medicago sativa by SSCP (single-strand conformation polymorphism). The results showed that Sinorhizobium meliloti L33 inoculation decreased γ-proteobacteria and increased α-proteobacteria. Then many investigates indicated that application of microbial inoculants, such as rhizobia, Azospirillum, AMF, biocontrol agents and co-inoculation, could influence the resident microbial communities and commonly increased soil microbial diversity by various culture-independent methods, such as DGGE (denaturing-gradient gel electrophoresis), PLFA (phospholipid fatty acid) and the latest high-throughput sequencing (Barriuso et al., 2008; Felici et al., 2008; Trabelsi and Mhamdi, 2013; Pang et al., 2017; Rodriguez-Caballero et al., 2017). The positive effects of microbial inoculants on microbial diversity had also been demonstrated in many ecosystems (Xi et al., 2015; Zhang et al., 2017). However, the effect of different ratios of multi-species in inoculants on soil indigenous microbial community is rarely explored.
In this study, we constructed two microbial co-inoculants according to soil fertility, microbial activity and cucumber nutrient requirement. The two microbial co-inoculants had the same three strains but with different ratios. The present study aims to determine whether the two different ratios (2:1:2 vs. 1:10:1) of the integrated multi-species, comprising of three selected bio-inoculants, viz. Ensifer sp. NYM3 (Alphaproteobacteria), Acinetobacter sp. P16 (Gammaproteobacteria), and Flavobacterium sp. KYM3 (Bacteroidetes), have the same effects on cucumber yield, soil properties, especially on soil indigenous bacterial community in the field experiment. The ultimate goal is to choose the better construction of microbial co-inoculants. This work will provide valuable suggestion for microbial fertilizer formulation and application in future.
The experiment was performed in a cucumber (Jing Pin Xin Xiu, Dalian Shuangfeng seeds Co. Ltd., China) field in Anshan, Liaoning Province, China (41°00′ N, 123°00′ E). This region has a warm temperate continental monsoon climate with an average annual temperature of 7.5°C and a mean annual precipitation of 707 mm. The soil type is classified as brown earth. The soil had the following physio-chemical properties: pH 6.8, organic carbon 23.5 g/kg, total nitrogen (TN) 1.42 g/kg, total phosphorus 0.107 g/kg, total potassium 3.1 g/kg, available nitrogen 147 mg/kg, available phosphorus 6 mg/kg, available potassium 376 mg/kg.
The Ensifer sp. NYM3, the other name is Sinorhizobium sp. NYM3 (KY203668), Acinetobacter sp. P16 (KY203666), and Flavobacterium sp. KYM3 (KY203667) were previously isolated from Mount Huang in Anhui Province, China by Ashby culture medium, inorganic phosphate culture medium and K-releasing culture medium, respectively (Xu et al., 2015; Yin et al., 2015). The nitrogen-fixing activity of Ensifer sp. NYM3 was 921 nmol C2H2/h⋅mg protein. The phosphate solubilizing ability of Acinetobacter sp. P16 was 120 mg/L. The potassium solubilizing ability of Flavobacterium sp. KYM3 was 68 mg/L. Previous pot experiment showed that all the three strains had positive effects on cucumber. Previous antagonistic experiments showed that there was no antagonism among the three strains.
Microbial co-inoculants 2 (M2) were constructed according to soil fertility and cucumber nutrient requirement without considering the difference in the nutrient supply capability of microbial species. Soil nitrogen and potassium were about 10 times as much as soil phosphorus. The amount of nitrogen, phosphorus and potassium absorbed by 100 kg cucumber yields was about 0.4 kg, 0.4 kg, and 0.5 kg, respectively. Therefore, the ratio of Ensifer sp. NYM3, Acinetobacter sp. P16 and Flavobacterium sp. KYM3 in M2 was set as 1:10:1.
Microbial co-inoculants 1 (M1) were constructed according to soil fertility, microbial activity and cucumber nutrient requirement. We calculated the ratio of Acinetobacter sp. P16 and Flavobacterium sp. KYM3 as follows:
Where AP is the available phosphorus, TP is the total phosphorus, AP/TP is the soil phosphate solubilizing ability, 120 is the phosphate solubilizing ability of Acinetobacter sp. P16, X is the quantity of Acinetobacter sp. P16, AK is the available potassium, TK is the total potassium, AK/TK is the soil potassium solubilizing ability, 68 is the potassium solubilizing ability of Flavobacterium sp. KYM3, X is the quantity of Flavobacterium sp. KYM3, 0.4:0.5 was the ratio of phosphorus and potassium absorbed by 100 kg cucumber yields. Through this formula, we get the ratio of Acinetobacter sp. P16 and Flavobacterium sp. KYM3 to about 1:2. Because nitrogen is the largest nutrient element of plant demand, we set the ratio of Ensifer sp. NYM3, Acinetobacter sp. P16 and Flavobacterium sp. KYM3 in M1 as 2:1:2.
The fertilization experiments were established in 2014 and were designed in a randomized complete block with three replicates, each measuring 40 m2. The treatments were the non-inoculated (CK), microbial co-inoculants 1 (M1), microbial co-inoculants 2 (M2). Each plot received 4.08 kg organic and chemical compound fertilizer [lignite 70%, KH2PO4 5.51%, (NH4)2PO4 24.49%] at three different growth stages (base fertilizer, flowering period, and fruiting period), respectively. Microbial co-inoculants (1.5 × 1014cfu/ha) were applied with organic and chemical compound fertilizer and were only added in plots treated with M1 and M2.
The yields of cucumber from the entire plot were weighed and recorded during harvest period. Surface soils (0–20 cm) near cucumber roots were randomly sampled from 10 points in each treatment plot with a 2.5 cm diameter auger after cucumber harvest on July 2014, and thoroughly mixed into one composite sample. A total of nine composite samples (3 treatments × 3 replicates = 9 samples) were collected. Each soil sample was pooled in a sterile plastic bag, and transported to the laboratory on ice. The samples were sieved (2 mm) and then stored at 4°C (for soil characterization) or −80°C (for soil DNA isolation) until analysis.
The soil pH, organic matter (OM), total nitrogen (TN), total phosphorus (TP), total potassium (TK), available nitrogen (AN), available phosphorus (AP), available potassium (AK), nitrate nitrogen (NO3-N), invertase activity (IA), urease activity (UA), alkaline phosphatase activity (APA), and catalase activity (CA) were determined using commercial chemical assay kits (Suzhou Comin Biotechnology Co., Ltd., China), respectively, following the manufacturer’s instructions.
To analyze the bacterial community richness and composition, soil DNA was isolated from the three treatments by the PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA, United States). The genomic DNA were quantified with a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States) and stored at −80°C until amplification.
Briefly, the bacterial hypervariable domain V4 was amplified with region-specific primers (515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′) that included the Illumina flowcell adapter sequences (Caporaso et al., 2012). The reverse amplification primers contained a 10-base barcode sequence. The purified PCR products were normalized in equimolar amounts in a single tube for MiSeq 2000 sequence runs (Illumina, San Diego, CA, United States), producing bidirectional reads.
The 16S rRNA gene sequences were processed and analyzed by QIIME 1.9 (Caporaso et al., 2012). Initially, sequences shorter than 200 bp, containing unresolved nucleotides, exhibiting an average quality score lower than 25, harbor mismatches longer than 3 bp in the forward primer, or possessing homopolymers longer than 6 bp were removed with split_libraries.py. Chimeric sequences were removed using UCHIME with Ribosomal Database Project (RDP) as reference dataset (trainset10_082014_rmdup.fasta). Operational taxonomic unit (OTU) determination was performed at a genetic divergence of 3% (species level) with pick_open_reference_otus.py. Singletons, chloroplasts, unclassified OTUs and extrinsic domain OTUs were removed by employing filter_otu_table.py. We used a randomly selected subset of 6,500 sequences per sample to calculate the diversities and distances between samples using core_diversity_analyses.py. Differentially abundant OTUs between treatment groups were identified using linear discriminant analysis (LDA) effect size (LEfSe) analysis (Segata et al., 2011). Microbial community function were predicted by Functional Annotation of Prokaryotic Taxa (FAPROTAX) using python collapse_table.py (Louca et al., 2016).
The 16S rRNA gene sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRP127639.
All statistical analyses were performed using R (version 3.1.1) and IBM SPSS Statistics 19 (SPSS Inc.). The effects of the different treatments on the cucumber yields, soil physicochemical properties, soil enzymes activity, α-diversity indices and relative abundance of abundant taxa were tested using one-way ANOVA followed by Tukey’s post hoc test. Principal coordinate analysis (PCoA) and analysis of similarity (ANOSIM) based on weighted UniFrac distance were performed to evaluate the overall differences in the bacterial community. Permutational multivariate analysis of variance (PERMANOVA) with ADONIS function was conducted to quantitatively evaluate the contribution of different treatments to the variations of the bacterial community (Xiong et al., 2015). These analyses were performed in R with the ‘vegan’ package. Spearman’s rank correlations between abundant taxa, cucumber yield, and soil properties were calculated using IBM SPSS Statistics 19 (SPSS Inc.). A heatmap that illustrates the Functional Annotation of Prokaryotic Taxa (FAPROTAX) data underlying the clustering patterns was generated using a software package of HemI (Heatmap Illustrator, version 1.0.1) (Deng et al., 2014).
Microbial co-inoculation treatments (M1 and M2) obviously increased cucumber yields compared to the non-inoculated treatment (CK) (Figure 1). The M1 increased 44.23% while M2 increased 36.32% of cucumber yields. There was no obvious difference in cucumber yields between the microbial co-inoculation treatments. The M1 treatment resulted in the higher yield of cucumber (39.16 t/ha) that that of M2 (37.02 t/ha).
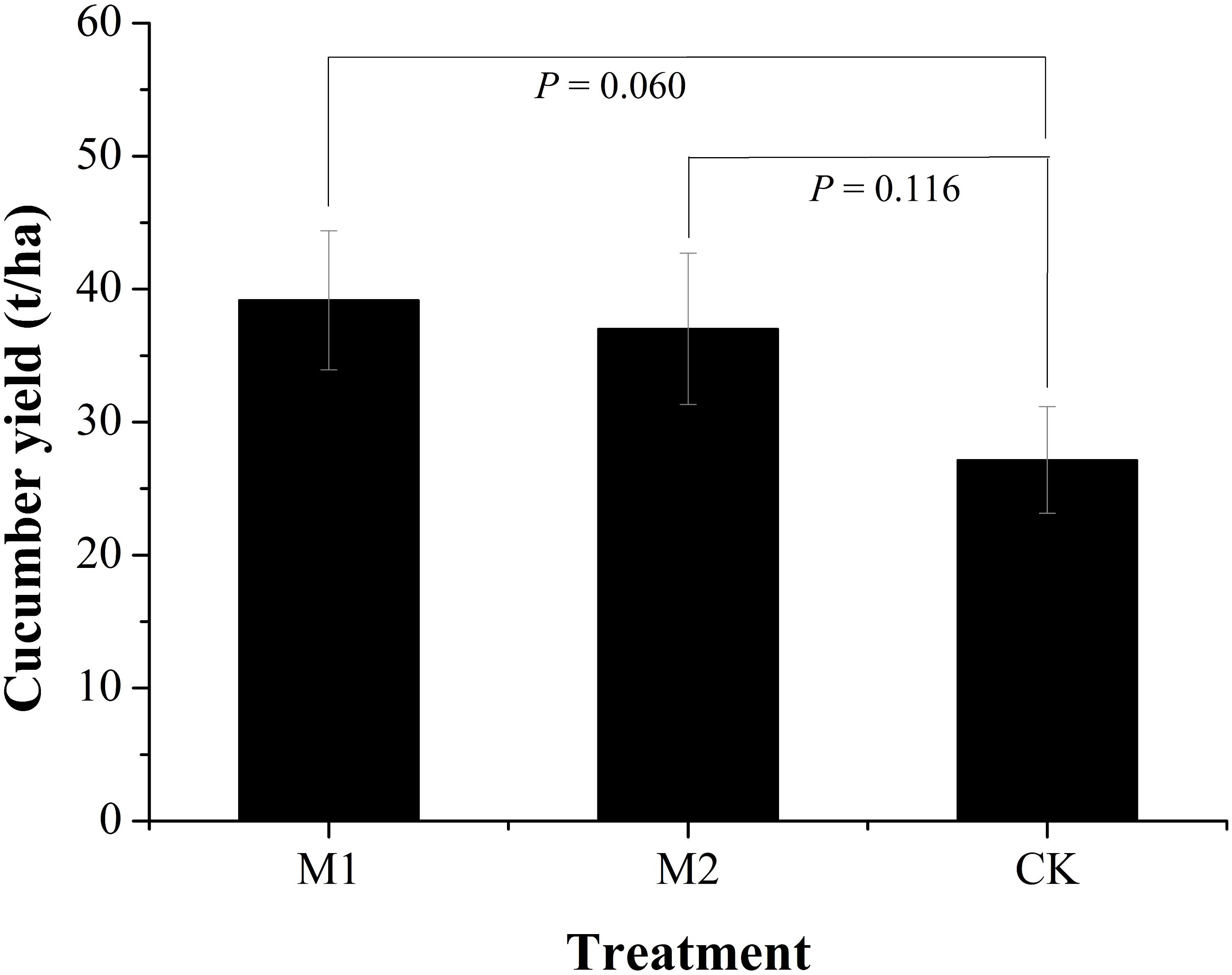
FIGURE 1. Cucumber yields under different treatments. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2; CK, non-inoculated.
The soil pH, TN, AK, NO3-N, IA, UA and APA contents differed significantly between the treatments (P < 0.05) (Table 1). The soil pH, TN and IA were significantly higher in M1 than that in other treatments. The UA and PA was significantly higher in M2 than in other treatments. M1 had significantly higher TN than M2, and M2 had significantly higher TN than CK. M2 had significantly higher UA than M1, and M1 had significantly higher UA than CK. M2 had significantly higher AK and NO3-N than that in M1.
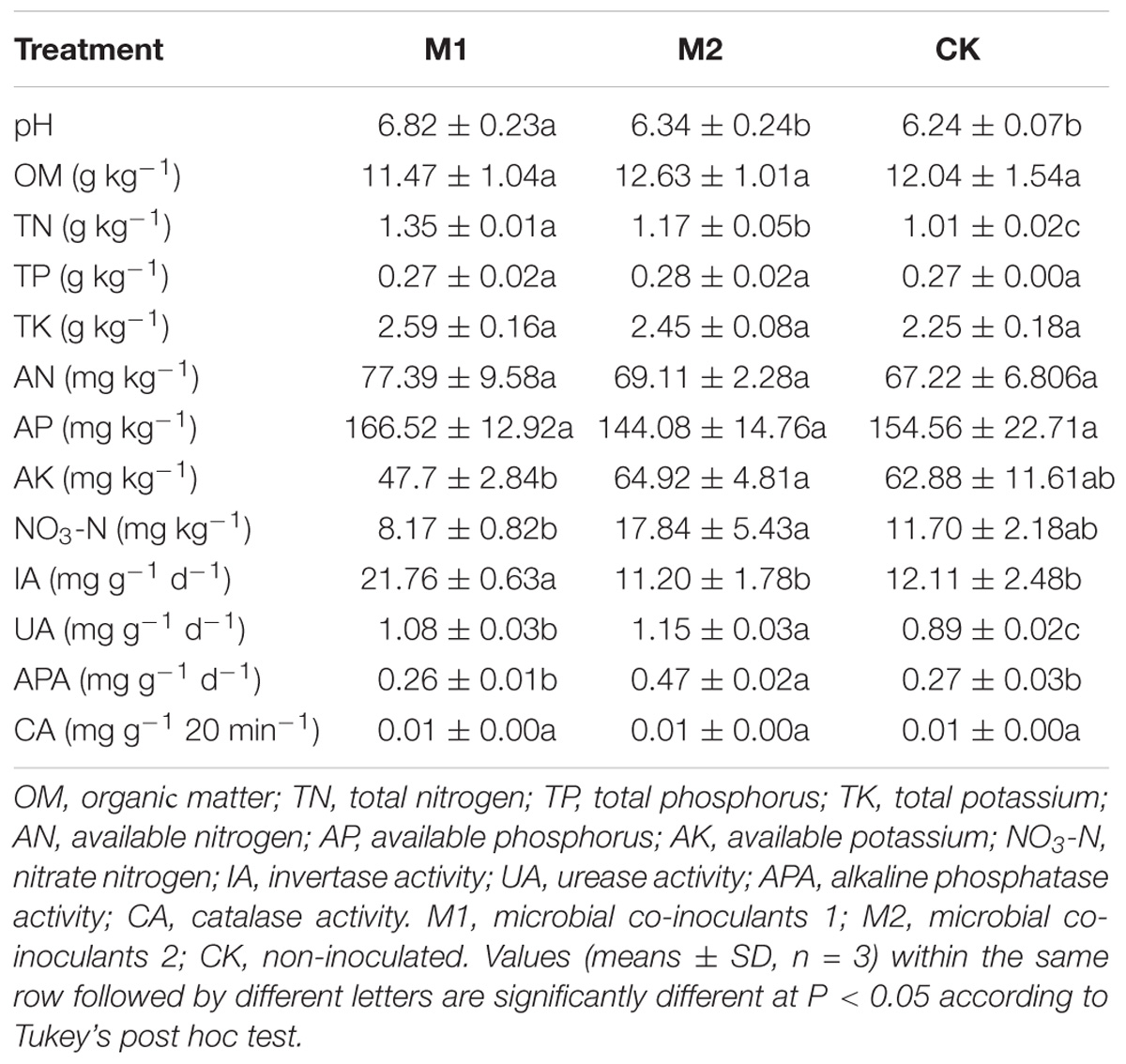
TABLE 1. Physicochemical and biological characteristics of cucumber soil under different treatments.
Across all samples, we obtained a total of 133,394 high-quality sequences and 6534–31354 sequences per sample (mean = 14,821). These high-quality sequences were clustered into 13, 226 OTUs at 97% sequence similarity, with 1521–2571 OTUs per sample. After rarefied to 6,500 sequences per sample, Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, Acidobacteria, Chloroflexi, Betaproteobacteria, TM7, Deltaproteobacteria, Gemmatimonadetes, Firmicutes, Planctomycetes, Verrucomicrobia, Nitrospirae, and Armatimonadetes were the dominant phyla (proteobacterial classes) (>1%) across all treatments. These dominant phyla were accounting for more than 97% of the bacterial sequences from each soil sample (Figure 2). The phyla (proteobacterial classes) Actinobacteria, Gammaproteobacteria, Bacteroidetes, Acidobacteria, Chloroflexi, Betaproteobacteria, TM7, Deltaproteobacteria, Gemmatimonadetes, Firmicutes, Verrucomicrobia, Nitrospirae, and Armatimonadetes varied significantly (P < 0.05) between the different treatments (Table 2). Gammaproteobacteria, Acidobacteria, Nitrospirae, and Armatimonadetes were significantly increased by microbial co-inoculation (M1 and M2) (P < 0.05). Conversely, Actinobacteria and Firmicutes showed the opposite pattern. M1 had highest TM7 and lowest Chloroflexi (P < 0.05). M2 had highest Gemmatimonadetes and Verrucomicrobia (P < 0.05). M2 had significant higher Betaproteobacteria than CK (P < 0.05). Bacteroidetes and Deltaproteobacteria were significant highest in M2 and lowest in M1 (P < 0.05). Additionally, no significant difference in Alphaproteobacteria and Planctomycetes was observed between different treatments.
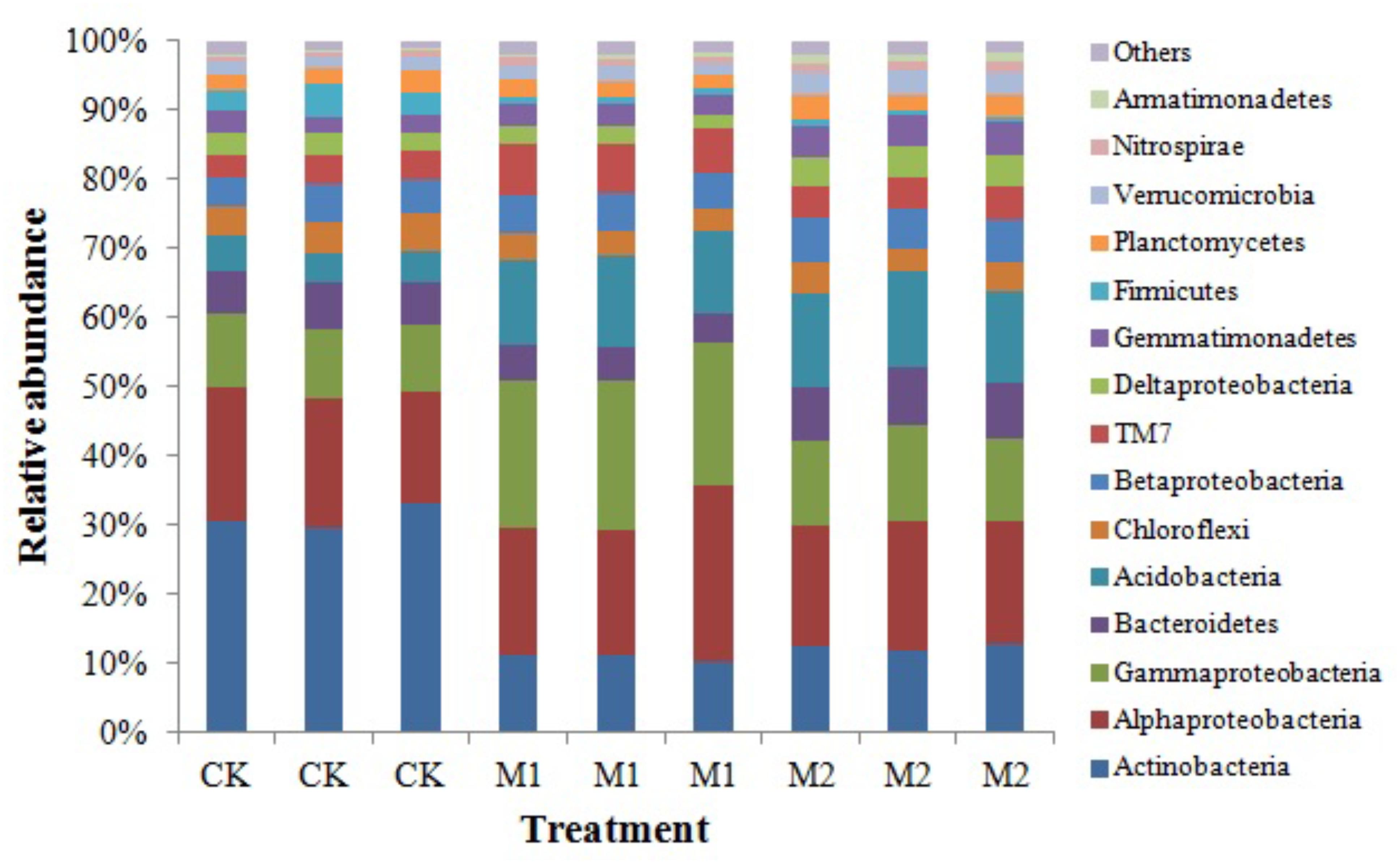
FIGURE 2. Relative abundances of the dominant bacterial phyla (proteobacterial classes) under different treatments. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2; CK, non-inoculated.
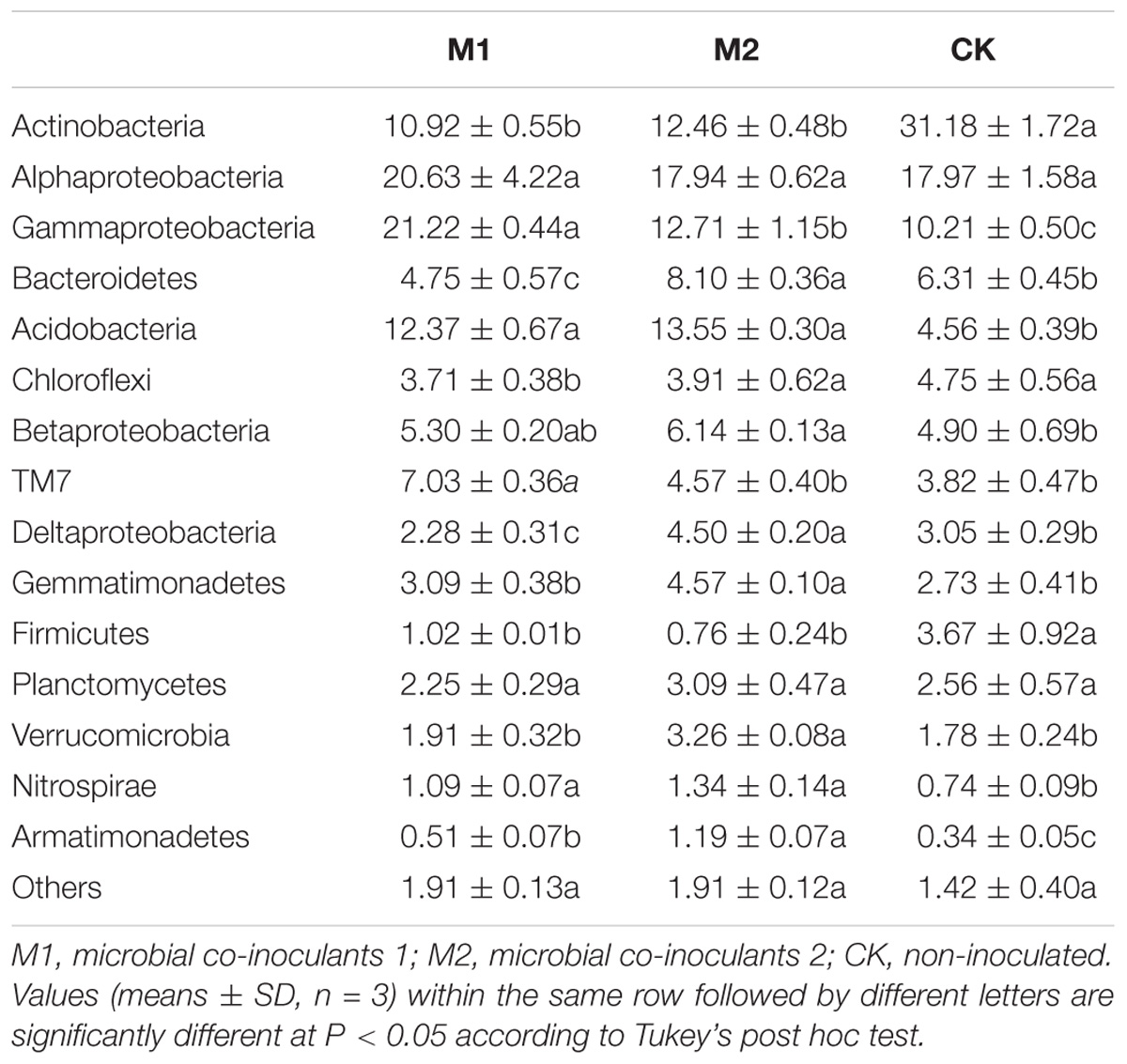
TABLE 2. Statistically significant differences in the dominant bacterial phyla (proteobacterial classes) under different treatments.
The high throughout sequencing results also showed that the relative abundances of Flavobacterium was significant higher than that of Rhizobiaceae_other and Acinetobacter sp. in M1. The ratio of Rhizobiaceae_other, Acinetobacter sp. and Flavobacterium sp. in M1 was about 2:1:14. The relative abundances of Flavobacterium sp. was significant higher than that of Rhizobiaceae_other in M2. The relative abundances of Rhizobiaceae_other was significant higher than that of Acinetobacter sp. in M2. The ratio of Rhizobiaceae_other, Acinetobacter sp. and Flavobacterium sp. in M2 was about 12:1:24. The relative abundances of Rhizobiaceae_other, Acinetobacter sp. and Flavobacterium sp. had no significant difference in CK. The ratio of Rhizobiaceae_other, Acinetobacter sp. and Flavobacterium sp. in CK was about 14: 2: 10 (Supplementary Figure S1).
The bacterial α-diversity in the individual samples under the different treatments was calculated. The number of OTUs, phylogenetic diversity, and the Shannon–Weaver index had no significant changes among the three different treatments (Supplementary Figure S2). It was obvious that the bacterial α-diversity (the number of OTUs, phylogenetic diversity, and the Shannon–Weaver index) in M2 were generally higher than the other treatments.
The PCoA revealed that the bacterial communities were obviously separated by the three treatments (Figure 3). ANOSIM analysis showed that microbial co-inoculation treatments (global R = 1, P = 0.003) were significant factors in shaping the bacterial community composition. Accordingly, ADONIS analysis showed that microbial co-incubation treatment contributed 82.3% (P = 0.01) of community variance. Collectively, these results demonstrated that the composition and structure of microbial communities were predominantly affected by the different microbial co-inoculation treatments.
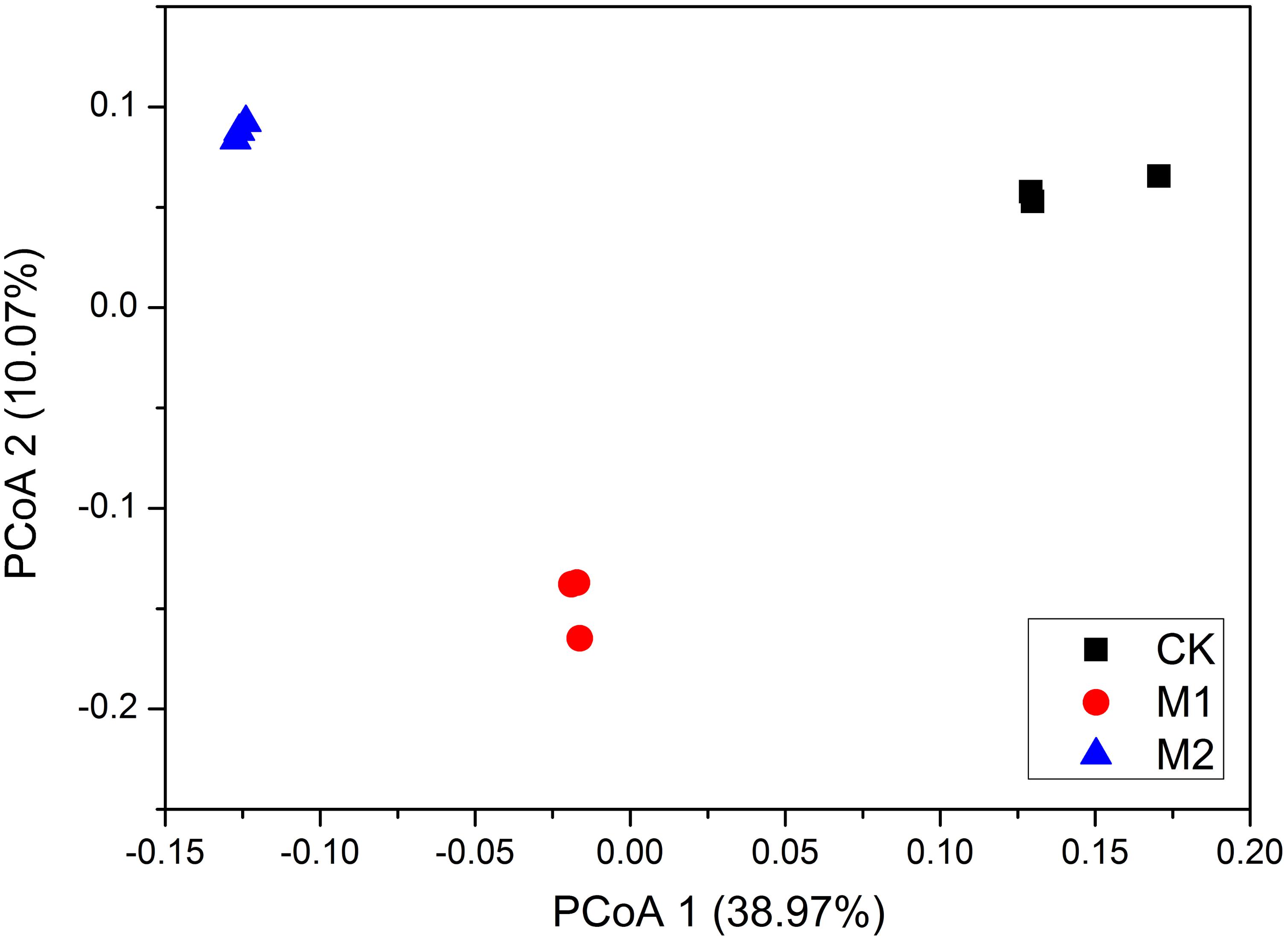
FIGURE 3. Principal coordinate analysis (PCoA) of weighted UniFrac distances of the bacterial communities under different treatments. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2; CK, non-inoculated.
To further investigate which bacterial taxa were distinct among the groups, LEfSe analysis was applied. A total of 153 bacterial groups were distinct to at least one treatment using the default logarithmic (LDA) value of 2 (Supplementary Figure S3). Cladograms show taxa with LDA value of 4 for clarity (Figure 4). The bacterial lineages enriched in M1 were Proteobacteria (the phylum and its class Gammaproteobacteria, and its orders Xanthomonadales, and genus of Sphingomonas) and TM7 (from phylum to class). There were three groups of bacteria enriched in M2, namely, Acidobacteria (the phylum and its class Acidobacteria_6, and its order iii1_15), Bacteroidetes, and Deltaproteobacteria. The Actinobacteria (from phylum to its class actinobacteria, and its orders actinomycetales and solirubrobacterales, and families of micrococcaceae and nocardioidaceae) and Firmicutes (from phylum to its class Bacilli, and to its order Bacillales) were enriched in CK (Figure 4).

FIGURE 4. Cladogram (A) and LDA score (B) of LEfSe analysis of bacterial community among CK (Red), M1 (green) and M2 (blue) treatments. Only the taxa with meeting a significant LDA threshold value of >4 were shown. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2; CK, non-inoculated.
FAPROTAX analysis showed that 30.26% (213 out of 704) records were assigned to at least one group. Dominant function (>1%) across all samples were chemoheterotrophy, aerobic_chemoheterotrophy, nitrate_reduction, phototrophy, photoautotrophy, nitrate_respiration, nitrogen_respiration, photoheterotrophy, anoxygenic_photoautotrophy_S_oxidizing, anoxygenic_photoautotrophy, nitrate_denitrification, nitrite_denitrification, nitrous_oxide_denitrification, denitrification, nitrite_respiration, nitrification, aerobic_nitrite_oxidation and fermentation (Figure 5). Microbial co-inoculation (M1 and M2) significantly increased functional groups of aerobic_nitrite_oxidation, and decreased functional groups of chloroplasts, cellulolysis, ureolysis, manganese_oxidation, human_gut, and mammal_gut (P < 0.05). The functional groups of intracellular_parasites were higher in M1 than in other treatments. The functional groups of phototrophy, photoautotrophy, nitrification, fermentation, cyanobacteria, oxygenic_photoautotrophy, chitinolysis and animal_parasites_or_symbionts were higher in M2 than in other treatments. The functional groups of methylotrophy, methanol_oxidation and aerobic_ammonia_oxidation were higher in CK than in other treatments (Table 3).
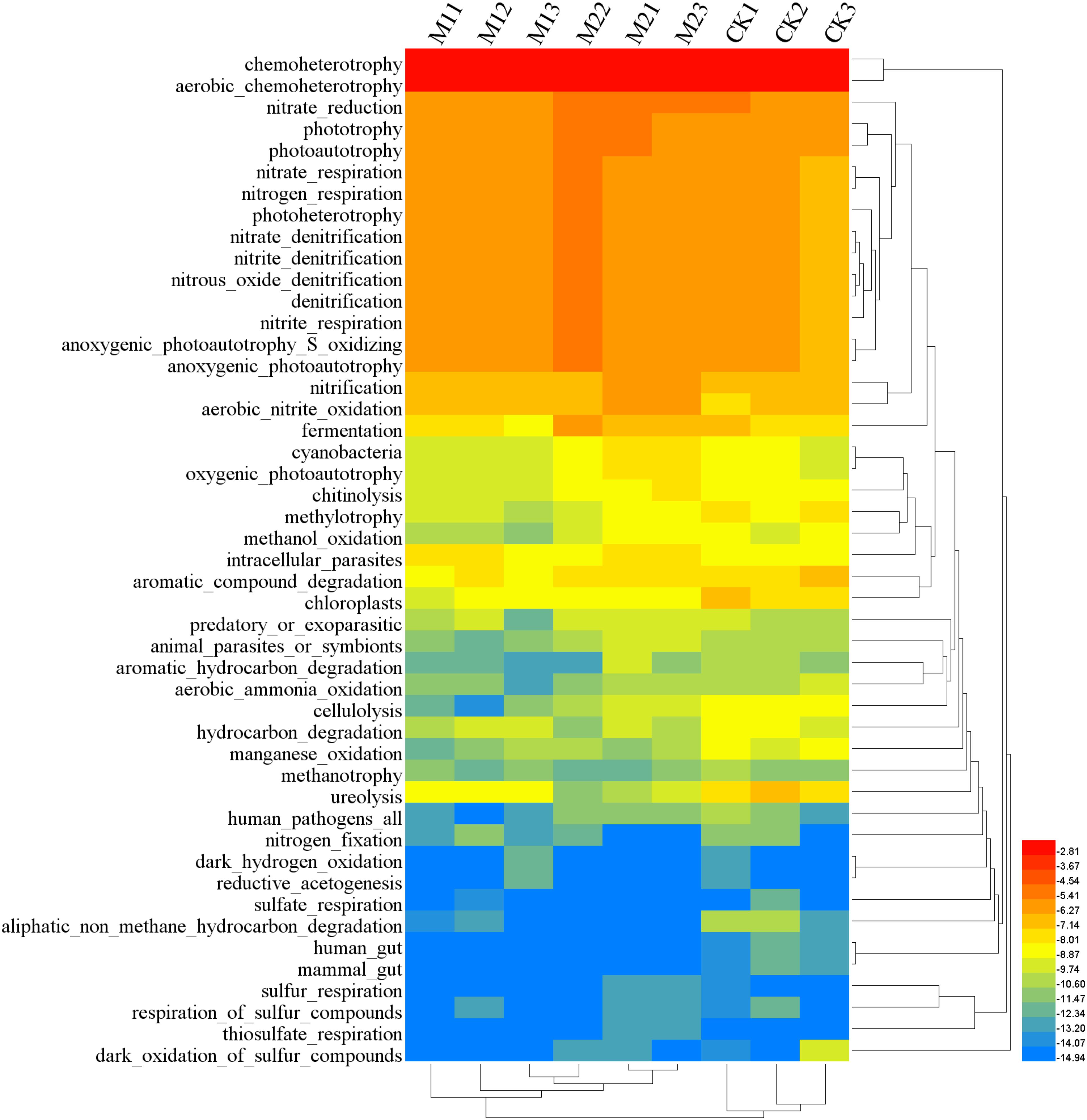
FIGURE 5. Heatmap of functional community profiles under different treatments. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2; CK, non-inoculated.
The PCoA revealed that the bacterial community function were obviously separated by the different microbial co-inoculation treatments (Supplementary Figure S4). ANOSIM analysis showed that microbial co-inoculation treatments (global R = 0.6049, P = 0.012) were significant factors in shaping the bacterial community function. Accordingly, ADONIS analysis showed that microbial co-incubation treatment contributed 58.7% (P = 0.01) of community variance. Collectively, these results revealed that the microbial community function were predominantly affected by the microbial co-inoculation.
Pearson’s rank correlation coefficient was used to evaluate the relationships among cucumber yield, soil characteristics and specific bacterial taxa obtained by LEfSe (Supplementary Tables S1, S2). Cucumber yield was significantly positively correlated with TN and UE. Four specific bacterial taxa of M1 (TM7-1, Legionellales, Sinobacteraceae, PRR-10) were significantly positively correlated with cucumber yield. Most of the specific bacterial taxa of M1 were significantly positively correlated with TN and IA. Some of the specific bacterial taxa of M1 were significantly positively correlated with TK, AN, UE, and significantly negatively correlated with AK, NO3-N, and CA (Supplementary Table S1). Ten specific bacterial taxa of M2 (Acidobacteria-6, RB25, OM190, iii1-15, Bacteroidales, N1423WL, mb2424, SJA-101, JG37-AG-70) were significantly positively correlated with cucumber yield. Most of the specific bacterial taxa of M2 were significantly positively correlated with UE and AKP. Some of the specific bacterial taxa of M2 were significantly positively correlated with APA, UE, CA, NO3-N, TK, AK, TN and significantly negatively correlated with AI (Supplementary Table S2).
Microbial co-inoculation is considered to be an innovative approach and had been applied worldwide (Baris et al., 2014; Baez-Rogelio et al., 2017). However, the underlying mechanisms of microbial co-inoculants constructions, especially the trait-based combination of distinctly different microbial species remains poorly understood. Microbial co-inoculants construction includes species selection and species integration. Most researches selected two or more compatible strains with different functions to construct co-inoculants. The common selections were nitrogen fixing bacteria (Azospirillum sp or Rhizobia) and biocontrol agents (Pseudomonas sp. or Bacillus sp.) (Figueiredo et al., 2008; Atieno et al., 2012b). In this study we selected three compatible strains evolving functions about nitrogen fixing, phosphate solubilizing and potassium solubilizing. The species selection was based on the consideration of three major nutrients (nitrogen, phosphorus, and potassium) in plant needs. Species integrations are essentially the ratios of microbial species in co-inoculants. The ratios of microbial species in co-inoculants were mostly 1:1 (Maheshwari et al., 2010; Yegorenkova et al., 2016). In this study, we set two ratios according to soil fertility, microbial activity and cucumber nutrient requirement. Although the influences of various factors in soil on the activity of bacteria were not considered and the formulas were not very scientific rigor, it provided a conception for rational construction of microbial co-inoculants.
The positive effects of microbial co-inoculation on plant yield, soil physiochemical properties, soil enzymes activity and soil microbial community had been extensively investigated (Atieno et al., 2012a; García de Salamone et al., 2012; Prasanna et al., 2012; Korir et al., 2017). However, multi-species, especially more than two species, with different ratios in inoculants on these factors had not been explored much. In this study, we illustrated the effect of the two ratios of multi-species in inoculants on cucumber yield, soil properties, and bacterial community in the field experiment. The ultimate goal is to choose the better construction of microbial co-inoculants.
Co-inoculation has been proved to increase colonization (Maheshwari et al., 2010; Figueredo et al., 2017). Our study revealed that the ratio of microbial species in inoculants did have some effects on colonization (Supplementary Figure S1). The ratio of 2:1:2 (M1) significantly increased Flavobacterium sp. proportions while decreased Rhizobiaceae_other and Acinetobacter proportions compared to CK. The ratio of 1:10:1 (M2) significantly increased Flavobacterium sp. and Rhizobiaceae_other proportions while decreased Acinetobacter proportions compared to CK. The ratio of Flavobacterium sp. was significant highest in M1. The ratio of Acinetobacter sp. was significant lowest in M2. Since Ensifer sp. did not detect in the high throughout sequencing results, Rhizobiaceae_other was used to represent Ensifer sp. NYM3. The colonization of microbial co-inoculants was not direct related to the inoculation quantity. This indicated that Flavobacterium sp. might have higher colonization activity while Acinetobacter sp. might have lower colonization activity. The colonization activity of microbial inoculants was related to strain characteristics, microbial interaction and environmental factors (van Veen et al., 1997). For more accurate evaluation of colonization, the absolute abundance of inoculated species will be future studied. Ensifer sp. could effectively establish the nitrogen-fixing symbiosis with leguminous crop plants (Miethling et al., 2000). The colonization activity of Acinetobacter sp. and Flavobacterium sp. was rarely reported.
A few works had been focus on the effects of microbial co-inoculation on soil microbial community (Trabelsi et al., 2011). However, the effect of the ratio of microbial species in inoculation was rarely described. Our results revealed that the ratio of microbial species in inoculants had significant effects on bacterial community diversity and function. This might due to the different interactions in microbial inoculants and with indigenous bacterial community. Statistics analysis showed that Gammaproteobacteria, Acidobacteria, Nitrospirae, and Armatimonadetes were significantly increased, while Actinobacteria and Firmicutes were significantly decreased by microbial co-inoculation (M1 and M2). Both the LEfSe analysis and statistics analysis revealed that the M1 had highest Gammaproteobacteria and TM7 while The M2 had highest Bacteroidetes, Deltaproteobacteria, Gemmatimonadetes, Armatimonadetes, and Verrucomicrobia. Some works showed that Bacteroidetes might result in a healthier development for plants as animals (Perez-Jaramillo et al., 2017). This was consistence with our results that higher Bacteroidetes with higher cucumber yields. Most species of Bacteroidetes in our study were Chitinophagaceae (54∼63%) in family level. Chitinophagaceae has been proposed for their potential role in protection against soil-borne pathogens (Yin et al., 2013; Perez-Jaramillo et al., 2017). This indicated that our microbial inoculants had potential biocontrol function. Effect of inoculation with Ensifer sp. (Sinorhizobium sp.) on soil bacterial community had been deeply investigated. Field inoculation with Sinorhizobium meliloti L33 decreased γ-proteobacteria but increased α-proteobacteria in rhizospheres of Medicago sativa (Schwieger and Tebbe, 2000). Cocktail of Ensifer strains had no effect on the bacterial community in the rhizosphere of Acacia senegal mature trees (Herrmann et al., 2012). Acinetobacter sp. had been widely applied to removal of organic and metal pollution from contaminated system (Bhattacharya et al., 2014; Rojas-Tapias et al., 2014). Flavobacterium sp. were well known identified as phosphate solubilizer and bioinoculants by many researchers (Rathi et al., 2014). The effects of Acinetobacter sp. or Flavobacterium sp. inoculation on soil bacterial community had not been reported.
There was few report about the effect of microbial co-inoculation on soil bacterial community function. Only Babic et al. (2008) demonstrated that two indigenous Sinorhizobium meliloti strains inoculation affected genes abundance involved in nitrogen turnover using qPCR. The differences in bacterial diversity may result in the differences in the bacterial community function. Although the coverage of FAPROTAX was not enough, it did give some information and could be used for comparison between different treatments (Louca et al., 2016). Statistics analysis showed that functional groups of aerobic_nitrite_oxidation were higher in microbial co-inoculation (M1 and M2) treatments. This was due to the higher Nitrospirales in these microbial co-inoculation (M1 and M2) treatments. The functional groups of intracellular_parasites were higher in M1 than in other treatments. This was due to the higher Chlamydia, Rickettsiales, and Legionellales in M1. The functional groups of phototrophy, photoautotrophy, nitrification, fermentation, cyanobacteria, oxygenic_photoautotrophy, chitinolysis, and animal_parasites_or_symbionts were higher in M2 than in other treatments. This was due to the higher Cyanobacteria, Rhodoplanes, Rhodobacter, Ectothiorhodospiraceae, Nitrospirales, Nitrosovibrio, Actinomycetales, Bacteroides, Bacilli, Clostridiales, Skermanella, Erwinia, Vibrionaceae, Opitutus, Lysobacter, Prevotella, Coprococcus, Francisellaceae, Acinetobacter, Stenotrophomonas, and Chthoniobacteraceae in M2. It is need further study on how microbial inoculants mediated the bacterial community structure and function in future.
The positive effect of microbial co-inoculants on soil properties had been reported by many works. Bacteria and cyanobacteria co-inoculation could improve soil nitrogen fixing potential, organic carbon, microbial biomass carbon, and enzymes activities (Prasanna et al., 2012). Co-inoculation of M. oryzae CBMB20 with A. brasilense CW903 or B. pyrrocinia CBPB-HOD could increase the activity of nitrogenase, urease and phosphatase enzymes in soil (Madhaiyan et al., 2010). However, the effect of different ratios of microbial species in inoculants on soil properties was few reported. This study showed that M1 had significant higher TN, IA, and pH (P < 0.05), M2 had significant higher urease activity and phosphatase activity (P < 0.05) than other treatments. This indicated that (i) microbial co-inoculants with different ratios of microbial species both had positive effect on soil properties; (ii) the ratio of microbial species in inoculants had significant effect on soil properties. The M1 and M2 increased different properties of soil. This might due to different bacterial community structure, function and activity mediated by different microbial inoculants. The ratio of Flavobacterium sp. was significant higher in M1 and M2 than that in CK (Supplementary Figure S1). However, the available potassium (AK) in M1 and M2 was not significant higher than CK. This might indicate that the K-solubilizing activity of Flavobacterium sp. was different in soil ecological systems. Our previous work showed that Flavobacterium sp. KYM3 could growth on Ashby medium plates (data not shown). This indicated that Flavobacterium sp. KYM3 had the nitrogen fixing ability. The higher TN in microbial co-inoculation treatments (M1 and M2) might be relate with the higher ratio of Flavobacterium sp. in microbial co-inoculants (M1 and M2) (Supplementary Figure S1).
The positive effect of microbial co-inoculants on plant had been widely investigated. Bacillus sp. CHEP5 were reported improved Bradyrhizobium sp. SEMIA6144 root surface colonization and increased the yield of peanut seeds from 2.15% to 16.69% (Figueredo et al., 2017). Application of Sinorhizobium meliloti RMP1 and Pseudomonas aeruginosa GRC(2) could significantly increased biomass and yield of B. juncea as compared to control in field trials (Maheshwari et al., 2010). Co-inoculation of Thiobacillus sp. with Rhizobium under field condition resulted in significantly higher nodule number, nodule dry weight, plant biomass and pod yield (Anandham et al., 2007). Our study firstly revealed that co-inoculation of Ensifer sp. NYM3, Acinetobacter sp. P16 and Flavobacterium sp. KYM3 with different ratios under field condition could promote cucumber yields. Our results also showed that the different ratios of the microbial species in inoculants (M1 and M2) had few effect on cucumber yields. This will expand our knowledge in bacteria interaction and will guide the construction and manufacture of microbial co-inoculation or fertilizers.
Microbial co-inoculants might promote cucumber yield by both the direct and indirect effects. The direct effects of microbial inoculants promote plant growth had been extensively described (Colla et al., 2015; Wang et al., 2017). Nowadays, some works had been suggested that regulation of soil bacterial community structure is one of the plant growth-promoting mechanisms of microbial inoculants (Kang et al., 2013; Rodriguez-Caballero et al., 2017). Our results did support this opinion. The results showed that M1 significantly increased TN, IA, Proteobacteria, TM7, aerobic_nitrite_oxidation and intracellular_parasites. Invertase catalyzes the hydrolysis of disaccharides to monosaccharides. Invertase plays a critical role in releasing low molecular weight sugars that are important as energy sources of microorganisms (Song et al., 2014). Cucumber yield was significantly positively correlated with TM7-1, Legionellales, Sinobacteraceae, PRR-10, TN, and UA. Most of the specific taxa of M1 was significant positive related with TN. It indicated that M1 might promote cucumber yields by mediating bacterial community structure and function about nitrogen fixing (Figure 6A). The results showed that M2 significantly increased Bacteroidetes, Deltaproteobacteria, Gemmatimonadetes, Armatimonadetes, Verrucomicrobia, phototrophy, photoautotrophy, nitrification, fermentation, cyanobacteria, oxygenic_photoautotrophy, chitinolysis, animal_parasites_or_symbionts, urease activity (UA) and phosphatase activity (APA). The enzyme urease catalyzes urea-N hydrolysis to NH3 and is produced by plants and many soil microorganisms (Fisher et al., 2017). Cucumber yield was significantly positively correlated with Acidobacteria-6, RB25, OM190, iii1-15, Bacteroidales, N1423WL, mb2424, SJA-101, JG37-AG-70, TN and UA. Most of the specific taxa of M2 was significant positive related with UA. It inferred that M2 might promote cucumber yields by mediating bacterial community structure and function about urea-N hydrolysis (Figure 6B). Due to the complex soil-microbe-plant ecosystem, the plant growth-promoting mechanism of microbial co-inoculants in field is still not well documented and needs to be further investigated. We will specially pay more attentions on plant nutrient contents, which could be more related to differences in soil properties as well as to bacterial community structures in future.
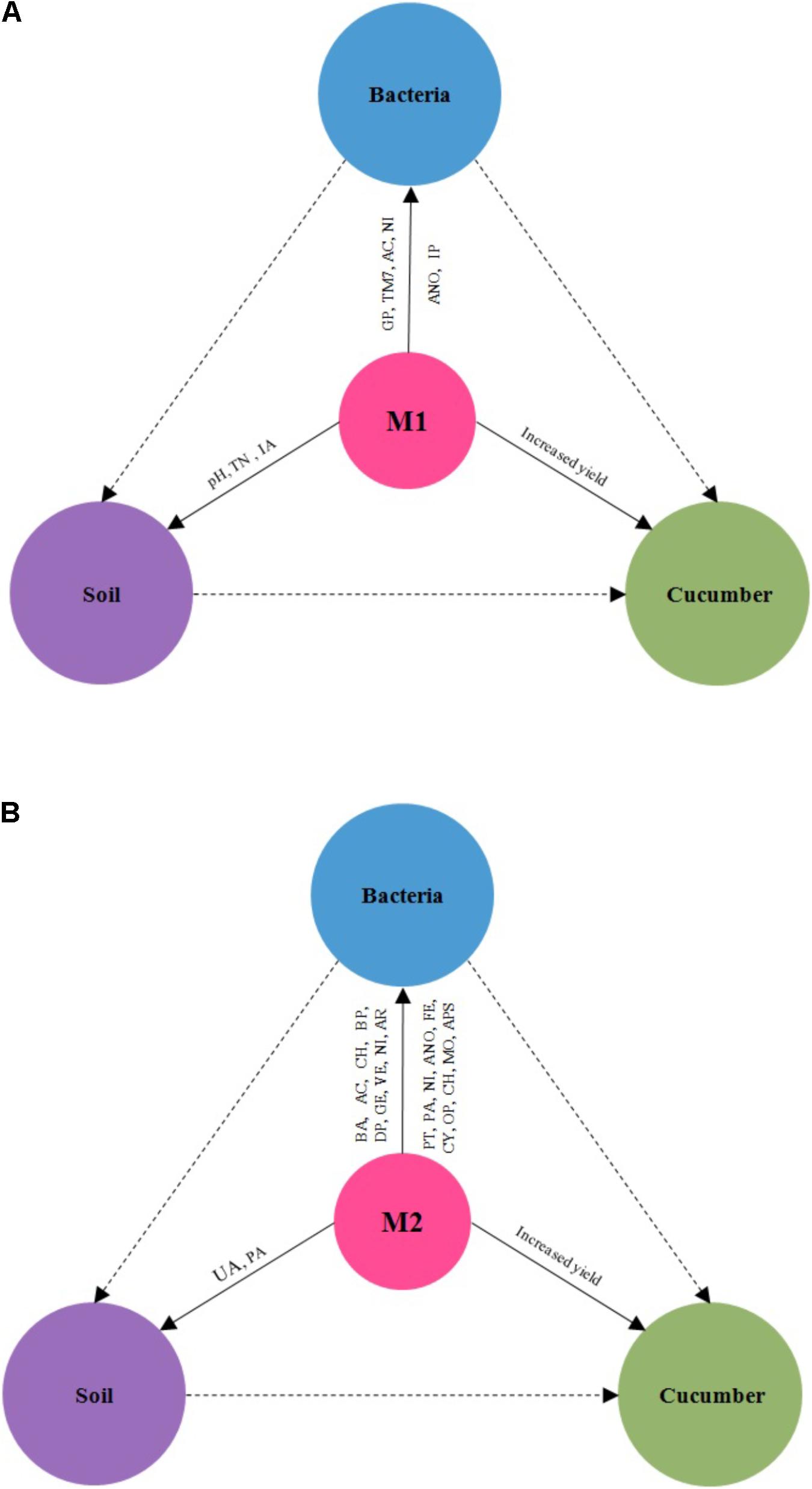
FIGURE 6. The effects of microbial co-inoculants 1 (A) and microbial co-inoculants 2 (B) on bacterial community, soil properties, and cucumber yield. M1, microbial co-inoculants 1; M2, microbial co-inoculants 2.
This study described the effect of multi-species with two different ratios in inoculants on cucumber yield, soil physiochemical properties, soil enzyme activity, and bacterial community. Inoculants with different ratios of microbial species both had positive effects on cucumber yields. The ratio of microbial species in inoculants had significant effect on soil properties and bacterial community. Inoculants with different ratios of microbial species might increase cucumber yields by different ways of mediating bacterial community structure and function. This revealed that in the process of microbial co-inoculants construction, trait-based integration of different microbial species had significant effects on soil bacterial communities. It also indicated that microbial activity should be considered in the construction of microbial co-inoculants. This will expand our knowledge in bacteria interaction and plant growth promoting mechanism of bacteria and will guide microbial fertilizer formulation and application in future.
ZH, JW, and CS: conception of the study. JW, SX, QL, CS, and ZH: designed the experiments. JW, QL, WZ, SX, and YL: performed the experiments. JW and WZ: interpretation of the results. JW, CS, and ZH: wrote the manuscript.
This study was supported by National Natural Science Foundation of China (No. 31500424) and Open Research Fund of Key Laboratory of Western China’s Mineral Resources of Gansu Province (WCRMGS-2014-02) and Key Research Program of the Chinese Academy of Sciences (NO. KFZD-SW-219).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Associate Editor Zhongjun Jia and the reviewers for their insightful comments and advice on this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01692/full#supplementary-material
Adesemoye, A. O., Torbert, H. A., and Kloepper, J. W. (2009). Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb. Ecol. 58, 921–929. doi: 10.1007/s00248-009-9531-y
Ambrosini, A., de Souza, R., and Passaglia, L. M. P. (2016). Ecological role of bacterial inoculants and their potential impact on soil microbial diversity. Plant Soil 400, 193–207. doi: 10.1007/s11104-015-2727-7
Anandham, R., Sridar, R., Nalayini, P., Poonguzhali, S., Madhaiyan, M., and Sa, T. (2007). Potential for plant growth promotion in groundnut (Arachis hypogaea L.) cv. ALR-2 by co-inoculation of sulfur-oxidizing bacteria and Rhizobium. Microbiol. Res. 162, 139–153. doi: 10.1016/j.micres.2006.02.005
Atieno, M., Herrmann, L., Okalebo, R., and Lesueur, D. (2012a). Efficiency of different formulations of Bradyrhizobium japonicum and effect of co-inoculation of Bacillus subtilis with two different strains of Bradyrhizobium japonicum. World J. Microbiol. Biotechnol. 28, 2541–2550. doi: 10.1007/s11274-012-1062-x
Atieno, M., Herrmann, L., Okalebo, R., and Lesueur, D. (2012b). Efficiency of different formulations of Bradyrhizobium japonicum and effect of co-inoculation of Bacillus subtilis with two different strains of Bradyrhizobium japonicum. World J. Microbiol. Biotechnol. 28, 2541–2550. doi: 10.1007/s11274-012-1062-x
Babic, K. H., Schauss, K., Hai, B., Sikora, S., Redzepovic, S., Radl, V., et al. (2008). Influence of different Sinorhizobium meliloti inocula on abundance of genes involved in nitrogen transformations in the rhizosphere of alfalfa (Medicago sativa L.). Environ. Microbiol. 10, 2922–2930. doi: 10.1111/j.1462-2920.2008.01762.x
Baez-Rogelio, A., Morales-Garcia, Y. E., Quintero-Hernandez, V., and Munoz-Rojas, J. (2017). Next generation of microbial inoculants for agriculture and bioremediation. Microb. Biotechnol. 10, 19–21. doi: 10.1111/1751-7915.12448
Baris, O., Sahin, F., Turan, M., Orhani, F., and Gulluce, M. (2014). Use of plant growth promoting rhizobacteria (PGPR) seed inoculation as an alternative fertilizer inputs in wheat and barley production. Commun. Soil Sci. Plant Anal. 45, 2457–2467. doi: 10.1080/00103624.2014.912296
Barriuso, J., Ramos Solano, B., Santamaria, C., Daza, A., and Gutierrez Manero, F. J. (2008). Effect of inoculation with putative plant growth-promoting rhizobacteria isolated from Pinus spp. on Pinus pinea growth, mycorrhization and rhizosphere microbial communities. J. Appl. Microbiol. 105, 1298–1309. doi: 10.1111/j.1365-2672.2008.03862.x
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35(Suppl. 4), 1044–1051. doi: 10.1590/S1415-47572012000600020
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhattacharya, A., Gupta, A., Kaur, A., and Malik, D. (2014). Efficacy of Acinetobacter sp B9 for simultaneous removal of phenol and hexavalent chromium from co-contaminated system. Appl. Microbiol. Biotechnol. 98, 9829–9841. doi: 10.1007/s00253-014-5910-5
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME. J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Colla, G., Rouphael, Y., Di Mattia, E., El-Nakhel, C., and Cardarelli, M. (2015). Co-inoculation of Glomus intraradices and Trichoderma atroviride acts as a biostimulant to promote growth, yield and nutrient uptake of vegetable crops. J. Sci. Food Agric. 95, 1706–1715. doi: 10.1002/jsfa.6875
Deng, W. K., Wang, Y. B., Liu, Z. X., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988.
Felici, C., Vettori, L., Giraldi, E., Forino, L. M. C., Toffanin, A., Tagliasacchi, A. M., et al. (2008). Single and co-inoculation of Bacillus subtilis and Azospirillum brasilense on Lycopersicon esculentum: effects on plant growth and rhizosphere microbial community. Appl. Soil Ecol. 40, 260–270. doi: 10.1016/j.apsoil.2008.05.002
Figueiredo, M. V. B., Burity, H. A., Martinez, C. R., and Chanway, C. P. (2008). Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil Ecol. 40, 182–188. doi: 10.1016/j.apsoil.2008.04.005
Figueredo, M. S., Tonelli, M. L., Ibanez, F., Morla, F., Cerioni, G., Tordable, M. D., et al. (2017). Induced systemic resistance and symbiotic performance of peanut plants challenged with fungal pathogens and co-inoculated with the biocontrol agent Bacillus sp CHEP5 and Bradyrhizobium sp SEMIA6144. Microbiol. Res. 197, 65–73. doi: 10.1016/j.micres.2017.01.002
Fisher, K. A., Yarwood, S. A., and James, B. R. (2017). Soil urease activity and bacterial ureC gene copy numbers: effect of pH. Geoderma 285, 1–8. doi: 10.1016/j.geoderma.2016.09.012
García de Salamone, I. E., Funes, J. M., Di Salvo, L. P., Escobar-Ortega, J. S., D’Auria, F., Ferrando, L., et al. (2012). Inoculation of paddy rice with Azospirillum brasilense and Pseudomonas fluorescens: impact of plant genotypes on rhizosphere microbial communities and field crop production. Appl. Soil Ecol. 61(Suppl. C), 196–204. doi: 10.1016/j.apsoil.2011.12.012
Herrmann, L., Sanon, K., Zoubeirou, A. M., Dianda, M., Sall, S., Thuita, M., et al. (2012). Seasonal changes of bacterial communities in the rhizosphere of Acacia senegal mature trees inoculated with ensifer strains in burkina faso and niger. Agric. Ecosyst. Environ. 157, 47–53. doi: 10.1016/j.agee.2011.12.014
Johnke, J., Cohen, Y., de Leeuw, M., Kushmaro, A., Jurkevitch, E., and Chatzinotas, A. (2014). Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 27, 185–190. doi: 10.1016/j.copbio.2014.02.003
Kang, Y., Shen, M., Wang, H., and Zhao, Q. (2013). A possible mechanism of action of plant growth-promoting rhizobacteria (PGPR) strain Bacillus pumilus WP8 via regulation of soil bacterial community structure. J. Gen. Appl. Microbiol. 59, 267–277. doi: 10.2323/jgam.59.267
Korir, H., Mungai, N. W., Thuita, M., Hamba, Y., and Masso, C. (2017). Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 8:141. doi: 10.3389/fpls.2017.00141
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Madhaiyan, M., Poonguzhali, S., Kang, B. G., Lee, Y. J., Chung, J. B., and Sa, T. M. (2010). Effect of co-inoculation of methylotrophic Methylobacterium oryzae with Azospirillum brasilense and Burkholderia pyrrocinia on the growth and nutrient uptake of tomato, red pepper and rice. Plant Soil 328, 71–82. doi: 10.1007/s11104-009-0083-1
Maheshwari, D. K., Kumar, S., Kumar, B., and Pandey, P. (2010). Co-inoculation of urea and DAP tolerant Sinorhizobium meliloti and Pseudomonas aeruginosa as integrated approach for growth enhancement of Brassica juncea. Ind. J. Microbiol. 50, 425–431. doi: 10.1007/s12088-011-0085-6
Miethling, R., Wieland, G., Backhaus, H., and Tebbe, C. C. (2000). Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 40, 43–56. doi: 10.1007/s002480000021
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Pang, G., Cai, F., Li, R. X., Zhao, Z., Li, R., Gu, X. L., et al. (2017). Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil 416, 181–192. doi: 10.1007/s11104-017-3178-0
Paredes, S. H., Gao, T. X., Law, T. F., Finkel, O. M., Mucyn, T., Teixeira, P. J., et al. (2018). Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 16:e2003962. doi: 10.1371/journal.pbio.2003962
Pereg, L., and McMillan, M. (2015). Scoping the potential uses of beneficial microorganisms for increasing productivity in cotton cropping systems. Soil Biol. Biochem. 80, 349–358. doi: 10.1016/j.soilbio.2014.10.020
Perez-Jaramillo, J. E., Carrion, V. J., Bosse, M., Ferrao, L. F. V., de Hollander, M., Garcia, A. A. F., et al. (2017). Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 11, 2244–2257. doi: 10.1038/ismej.2017.85
Prasanna, R., Joshi, M., Rana, A., Shivay, Y. S., and Nain, L. (2012). Influence of co-inoculation of bacteria-cyanobacteria on crop yield and C-N sequestration in soil under rice crop. World J. Microbiol. Biotechnol. 28, 1223–1235. doi: 10.1007/s11274-011-0926-9
Rathi, M. S., Paul, S., and Thakur, J. K. (2014). Response of wheat to inoculation with mycorrhizae alone and combined with selected rhizobacteria including Flavobacterium Sp as a potential bioinoculant. J. Plant Nutr. 37, 76–86. doi: 10.1080/01904167.2013.848889
Rodrigues, A. C., Antunes, J. E., da Costa, A. F., de Paula Oliveira, J., and do Vale Barreto Figueiredo, M. (2013). Interrelationship of Bradyrhizobium sp. and plant growth-promoting bacteria in cowpea: survival and symbiotic performance. J. Microbiol. 51, 49–55. doi: 10.1007/s12275-013-2335-2
Rodriguez-Caballero, G., Caravaca, F., Fernandez-Gonzalez, A. J., Alguacil, M. M., Fernandez-Lopez, M., and Roldan, A. (2017). Arbuscular mycorrhizal fungi inoculation mediated changes in rhizosphere bacterial community structure while promoting revegetation in a semiarid ecosystem. Sci. Total Environ. 584, 838–848. doi: 10.1016/j.scitotenv.2017.01.128
Rojas-Tapias, D. F., Bonilla, R., and Dussan, J. (2014). Effect of inoculation and co-inoculation of Acinetobacter sp RG30 and Pseudomonas putida GN04 on growth, fitness, and copper accumulation of maize (Zea mays). Water Air Soil Poll. 225:2232. doi: 10.1007/s11270-014-2232-2
Saleem, M., Arshad, M., Hussain, S., and Bhatti, A. S. (2007). Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 34, 635–648. doi: 10.1007/s10295-007-0240-6
Saleem, M., Fetzer, I., Harms, H., and Chatzinotas, A. (2016). Trophic complexity in aqueous systems: bacterial species richness and protistan predation regulate dissolved organic carbon and dissolved total nitrogen removal. Proc. Roy. Soc. B-Biol. Sci. 283:20152724. doi: 10.1098/rspb.2015.2724
Schwieger, F., and Tebbe, C. C. (2000). Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album) – Linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol. 66, 3556–3565. doi: 10.1128/AEM.66.8.3556-3565.2000
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shi, S., Nuccio, E. E., Shi, Z. J., He, Z., Zhou, J., and Firestone, M. K. (2016). The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936. doi: 10.1111/ele.12630
Song, Y. Y., Song, C. C., Tao, B. X., Wang, J. Y., Zhu, X. Y., and Wang, X. W. (2014). Short-term responses of soil enzyme activities and carbon mineralization to added nitrogen and litter in a freshwater marsh of Northeast China. Eur. J. Soil Biol. 61, 72–79. doi: 10.1016/j.ejsobi.2014.02.001
Trabelsi, D., and Mhamdi, R. (2013). Microbial inoculants and their impact on soil microbial communities: a review. Biomed Res. Int. 2013:863240. doi: 10.1155/2013/863240
Trabelsi, D., Mengoni, A., Ben Ammar, H., and Mhamdi, R. (2011). Effect of on-field inoculation of Phaseolus vulgaris with rhizobia on soil bacterial communities. FEMS Microbiol. Ecol. 77, 211–222. doi: 10.1111/j.1574-6941.2011.01102.x
van Veen, J. A., van Overbeek, L. S., and van Elsas, J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135.
Wang, J. J., Wang, H., Yin, T. T., Xu, S., Zhao, W., Wang, J., et al. (2017). The persistence and performance of phosphate-solubilizing Gluconacetobacter liquefaciens qzr14 in a cucumber soil. 3 Biotech 7:294.
Xi, B. D., He, X. S., Dang, Q. L., Yang, T. X., Li, M. X., Wang, X. W., et al. (2015). Effect of multi-stage inoculation on the bacterial and fungal community structure during organic municipal solid wastes composting. Bioresour. Technol. 196, 399–405. doi: 10.1016/j.biortech.2015.07.069
Xiong, J., Chen, H., Hu, C., Ye, X., Kong, D., and Zhang, D. (2015). Evidence of bacterioplankton community adaptation in response to long-term mariculture disturbance. Sci. Rep. 5:15274. doi: 10.1038/srep15274
Xu, S., Liu, Y., Wang, J., Yin, T., Han, Y., Wang, X., et al. (2015). Isolation and potential of Ochrobactrum sp. NW-3 to increase the growth of cucumber. Int. J. Agric. Policy Res. 3, 341–350.
Yegorenkova, I. V., Tregubova, K. V., Burygin, G. L., Matora, L. Y., and Ignatov, V. V. (2016). Assessing the efficacy of co-inoculation of wheat seedlings with the associative bacteria Paenibacillus polymyxa 1465 and Azospirillum brasilense Sp245. Can. J. Microbiol. 62, 279–285. doi: 10.1139/cjm-2015-0647
Yin, C. T., Hulbert, S. H., Schroeder, K. L., Mavrodi, O., Mavrodi, D., Dhingra, A., et al. (2013). Role of bacterial communities in the natural suppression of rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 79, 7428–7438. doi: 10.1128/AEM.01610-13
Yin, T., Wang, J., Liu, Y., Liang, Y., Wang, X., Han, Y., et al. (2015). Isolation of efficient phosphate-solubilizing bacteria and the primary study about its mechanism of plant-growth-promoting. Biotechnol. Bull. (China)31, 1–6.
Keywords: microbial co-inoculants, integration, bacterial community structure and function, soil properties, soil enzyme, cucumber
Citation: Wang J, Li Q, Xu S, Zhao W, Lei Y, Song C and Huang Z (2018) Traits-Based Integration of Multi-Species Inoculants Facilitates Shifts of Indigenous Soil Bacterial Community. Front. Microbiol. 9:1692. doi: 10.3389/fmicb.2018.01692
Received: 09 February 2018; Accepted: 06 July 2018;
Published: 26 July 2018.
Edited by:
Zhongjun Jia, Institute of Soil Science (CAS), ChinaReviewed by:
Muhammad Saleem, University of Kentucky, United StatesCopyright © 2018 Wang, Li, Xu, Zhao, Lei, Song and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Wang, d2FuZ19qakB0aWIuY2FzLmNu; Chunhui Song, c29uZ2NoaEBsenUuZWR1LmNu; Zhiyong Huang, aHVhbmdfenlAdGliLmNhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.