
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01642
This article is part of the Research Topic Antimicrobial Resistance and the Food Chain: A One Health Perspective View all 14 articles
 Yinju Du1†
Yinju Du1† Chuanqing Wang2†
Chuanqing Wang2† Yulong Ye3†
Yulong Ye3† Yue Liu4†
Yue Liu4† Aimin Wang2
Aimin Wang2 Yong Li5
Yong Li5 Xiaoying Zhou6
Xiaoying Zhou6 Hanjian Pan7*
Hanjian Pan7* Jianmin Zhang8*
Jianmin Zhang8* Xuebin Xu9*
Xuebin Xu9*Emerging resistance to the antimicrobial agents of choice for treatment of thermophilic Campylobacter infections is becoming a serious threat to public health. In this study, 548 Campylobacter (372 C. jejuni and 176 C. coli) isolates from diarrheal patients and poultry meat were subjected for antibiotic susceptibility analysis to ciprofloxacin, tetracycline, gentamicin, erythromycin and clindamycin. Among them, 151 Campylobacter (32 C. jejuni and 119 C. coli) were identified as multidrug resistant isolates. PFGE analysis was performed on the 151 multidrug resistant isolates to determine their genetic relatedness, and 103 PFGE genotypes were determined. Some isolates from both human and chicken belonged to identical genotypes, indicating these clones might be able to spread between human and chicken. Antibiotic resistant genes of the 151 isolates were identified. The numbers of isolates carried tet (O), aadE, ermB, and aadE-sat4-aphA were 148 (98%), 89 (58.9%), 31 (20.5%), and 10 (6.6%), respectively. Almost all (n = 150, 99.3%) had gyrA mutation at codon 86. And the 23s rRNA A2075G point mutation was found in 56 (37.1%) isolates. Gene mutations at the cmeR-cmeABC intergenic region may lead to the activation of CmeABC multidrug efflux pump, and in this study novel sequence types of the intergenic region were identified in both C. jejuni and C. coli. This study determined the genetic prerequisites for antibiotic resistance of multidrug resistant Campylobacter isolates from diarrheal patients and poultry meat in Shanghai, China.
Campylobacter jejuni and Campylobacter coli are major causes of bacterial gastroenteritis in the world (Silva et al., 2011). Humans most often become infected by ingesting contaminated food, especially undercooked poultry. For example, a national-scale high-throughput molecular typing study in Scotland had demostrated that chickens were the dominant source of campylobacteriosis(Sheppard et al., 2009). And molecular epidemiology study of C. coli in New Zealand suggested poultry as a main source to the burden of human disease (Nohra et al., 2016).
Effective antimicrobial therapy is critical in the treatment of prolonged and severe campylobacteriosis (Silva et al., 2011). The most common antimicrobial agents used in clinic are erythromycin and ciprofloxacin. Tetracyclines have been suggested as an alternative choice but would rarely be used. Intravenous aminoglycosides are also considered in the treatment of serious bacteraemia and other systemic infections due to Campylobacter infection (Alfredson and Korolik, 2007). However, the emergence of antimicrobial resistance to a few common clinical drugs have been repeatedly reported in China (Zhang et al., 2010; Ma et al., 2017) and many other countries (Ge et al., 2013; Narvaez-Bravo et al., 2017). Significantly, multidrug resistant clones are emerging in East Asia in recent years. A study on tracking Campylobacter contamination along a broiler chicken production chain in China suggested 71.7% of C. jejuni and 98.0% of C. coli exhibited multidrug resistance (Ma et al., 2014). And 57.1% of C. jejuni and 70.9% of C. coli from retail chicken and duck were resistant to at least four antibiotics in South Korea (Wei et al., 2016). Multidrug resistance was detected in 99.0% of C. coli from diarrheal patients and food-producing animals (Zhang et al., 2014). Our previous study had identified several clones of multidrug resistant Campylobacter from infants younger than 2 years of age (Pan et al., 2015).
Several mechanisms responsible for resistance have been described, for example, fluoroquinolone resistance mainly due to mutations in the DNA gyrase gene (gyrA gene). Resistance to tetracyclines is conferred by the tet(O) gene. Macrolide resistance is the result of modification of the ribosome target binding site by mutation of the 23S rRNA, and activation of the CmeABC multidrug efflux pump leads to resistance to several antimicrobials (Luangtongkum et al., 2009; Iovine, 2013; Wieczorek and Osek, 2013). Interestingly, presences of antimicrobial resistant genes of Gram-positive origin, such as ermB, aadE and aadE-sat4-aphA are also associated with multidrug resistance in Campylobacter (Wang et al., 2014).
Antimicrobial resistance of some common clinical drugs is of particular concern. The objectives of the present study were to determine the genetic prerequisites for antibiotic resistance of multidrug resistant Campylobacter isolates from diarrheal patients and poultry meat in Shanghai, China.
A total of 548 isolates of Campylobacter were collected in Shanghai, China from 2009 to 2014, including 443 isolates from patients with acute watery diarrhea, 97 from chicken meat, 7 from duck meat and 1 from pigeon meat. Speciation of the Campylobacter isolates was determined by performing multiple PCR method using genes 16S rRNA (857 bp for Campylobacter genus, upper primer 5′ ATC TAA TGG CTT AAC CAT TAA AC 3′ and lower primer 5′ GGA CGG TAA CTA GTT TAG TAT T 3′), mapA (589 bp for jejuni species, upper primer 5′ CTA TTT TAT TTT TGA GTG CTT GTG 3′ and lower primer 5′ GCT TTA TTT GCC ATT TGT TTT ATT A 3′) and ceuE (462 bp for coli species, upper primer 5′ AAT TGA AAA TTG CTC CAA CTA TG 3′ and lower primer 5′ TGA TTT TAT TAT TTG TAG CAG CG 3′) (Denis et al., 1999). C. jejuni ATCC 33560 and C. coli ATCC 33559 were used as controls in antimicrobial susceptibility testing. The bacteria were incubated on Mueller-Hinton (MH) agar (Hopebiol, Qingdao, China) containing 5% (vol/vol) sheep blood at 42°C under a microaerophilic atmosphere (85% N2, 5% O2 and 10% CO2).
Minimum inhibitory concentrations (MICs) were determined using agar dilution method described by the National Antimicrobial Resistance Monitoring System (NARMS) report (CDC, 2012). Eight antimicrobials belonging to seven classes were selected for antimicrobial susceptibility testing based on their antimicrobial mechanisms and importance for human campylobacteriosis treatment. They included β-lactams (ampicillin, AMP), aminoglycosides (gentamicin, GEN), quinolones (nalidixic acid, NAL and ciprofloxacin, CIP), macrolides (erythromycin, ERY), lincosamides (clindamycin, CLI), tetracyclines (tetracycline, TET) and phenicols (chloramphenicol, CHL), MIC breakpoints of resistance were ≥32 μg/ml for CHL, ≥4 μg/ml for CIP, ≥8 μg/ml for CLI, ≥32 μg/ml for ERY, ≥4 μg/ml for GEN, ≥64 μg/ml for NAL, ≥16 μg/ml for TET. MIC breakpoint for AMP (≥32 μg/ml) was utilized according to MIC interpretive standards for Enterobacteriaceae (CLSI, 2012). The antimicrobials used in this study were purchased from Sigma (St. Louis, MO, United States). C. jejuni ATCC 33560 and C. coli ATCC 33559 were included in each batch of agar dilution tests for quality control. Multidrug resistance was defined as resistance to three or more antimicrobial groups.
Pulsed-field gel electrophoresis was performed on the multidrug-resistant Campylobacter identified in this study as previously described (Ribot et al., 2001). Briefly, plugs were prepared using cells removed from the surface of the culture plates. After lysis for 0.5 h, the plugs were washed four times at 54°C in a shaking water bath. A 2-mm-wide slice from each plug was cut and transferred to a tube containing restriction buffer solution (TaKaRa, Dalian, China). The plug slices were incubated in this restriction buffer at room temperature (23–25°C) for 5 min. Genomic DNA was digested with 40 U of SmaI (TaKaRa, Dalian, China) at room temperature for 2 h. Restriction fragments were separated by electrophoresis in 0.5 × Tris-Borate-EDTA buffer at 14°C for 18 h using a Chef Mapper electrophoresis system (Bio-Rad, Hercules, CA, United States) with pulse times of 6.75–38.35 s). Gel images were scanned and analyzed using BioNumerics v.6.6 (Applied Maths, Kortrijk, Belgium). Dendrograms were created from a matrix of band matching using the Jaccard coefficient and the unweighted pair group method with arithmetic mean analysis (UPGMA).
Putative antibiotic resistant genes gyrA, 23s rRNA, tet (O), aadE, ermB, aadE-sat4-aphA and the cmeR-cmeABC intergenic region were amplified and sequenced using primers listed in Table 2. Templates of the 151 multidrug resistant Campylobacter isolates and the negative control (Escherichia coli DH5α) were prepared by a boiling method. Briefly, an overnight broth culture (0.5 ml) was heated at 100°C for 10 min and centrifuged at 12,000 g for 5 min. The supernatant was used as DNA template. The cycling program was denaturation at 94°C for 1 min, annealing at a temperature specific to each primer pair for 1 min, and extension at 72°C for 2 min. The sequences were assembled using Laser-gene Seqman II software (DNAStar, Inc., Madison, WI, United States), and aligned using the CLUSTALWroutine of MegAlign software (v.6.1, DNA Star, Inc.). All PCR reactions were performed in duplicate.
The sequences of the cmeR-cmeABC intergenic regions have been deposited in GenBank, and their accession numbers were listed in Supplementary Table 1.
The surveillance and sampling protocols were approved by Shanghai Center for Disease Control and Prevention Review Board. This study did not involve any health-related patient interventions. A verbal informed consent was obtained from each subject; however, the patient identities have not been disclosed at any stage. The institutional board was informed of the specific needs with reference to the study setting and approved this mode of consent. A check box was included in the data form to document the consent-taking procedure.
The 548 Campylobacter isolates were identified as 372 C. jejuni and 176 C. coli. Table 1 provides details on the origins and sampling dates for all isolates. Among them, a total of 151 Campylobacter isolates (32 C. jejuni and 119 C. coli) were resistant to three or more antimicrobials, including 99 from diarrheal patients and 52 from poultry meat. Multidrug-resistance rates were much higher in C. coli than in C. jejuni (67.6%, n = 119 versus 8.6%, n = 32, respectively) (Table 2). A large proportion of the 151 isolates were resistant to tetracycline (99.3%), ciprofloxacin (97.4%), erythromycin (80.1%), gentamicin (76.2%) and clindamycin (71.5%). And the antimicrobial resistance rates in C. jejuni and C. coli were 100, 100, 65.6, 53.1, 37.5% and 99.2, 96.6, 84, 82.3, 80.7%, respectively (Table 3).
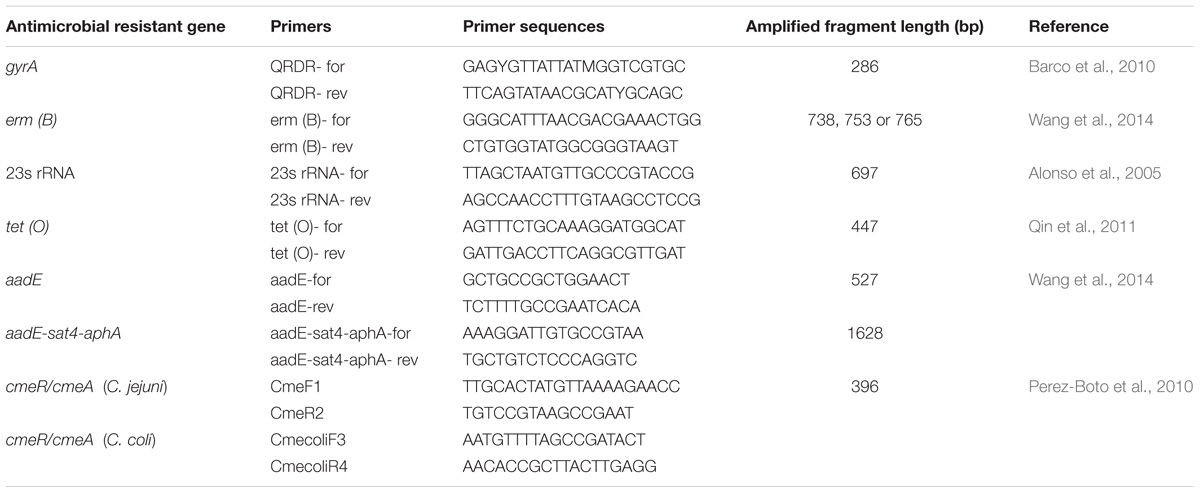
TABLE 2. Primers for the identification of antimicrobial resistance genes of the multidrug resistance Campylobacter jejuni and Campylobacter coli.
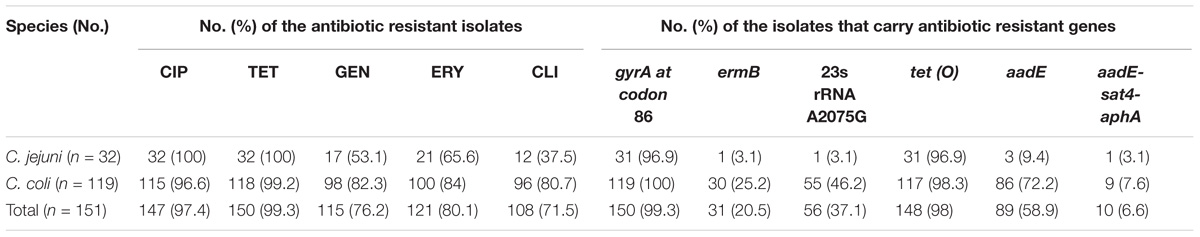
TABLE 3. The 151 multidrug-resistant Campylobacter isolates and the presence/mutant of the associated antibiotic resistant genes.
Genetic relatedness of the 151 multidrug-resistant Campylobacter isolates was analyzed by PFGE. Genetic diversity was significant among the 32 C. jejuni isolates, with 26 distinct PFGE genotypes (Figure 1). Interestingly, some human associated isolates shared identical PFGE genotype with those from chicken. For example, C. jejuni GFN-CN-JF-8S, SH11C09, SH11C429 isolated from patients were grouped together with SH12C138, which was isolated from chicken (Figure 1). In contrast, the 119 C. coli isolates represented 77 genotypes. It was also observed that isolates from different sources were grouped together. For example, in genotype B, C. coli SH11C353, GFN-CN-(3)-163 and GFN-CN-JF-130 that were isolated from human had identical genotype with the other three from chicken. And in genotype D, C. coli SH11C068 from human and the other 4 from chicken were grouped together (Figure 2).
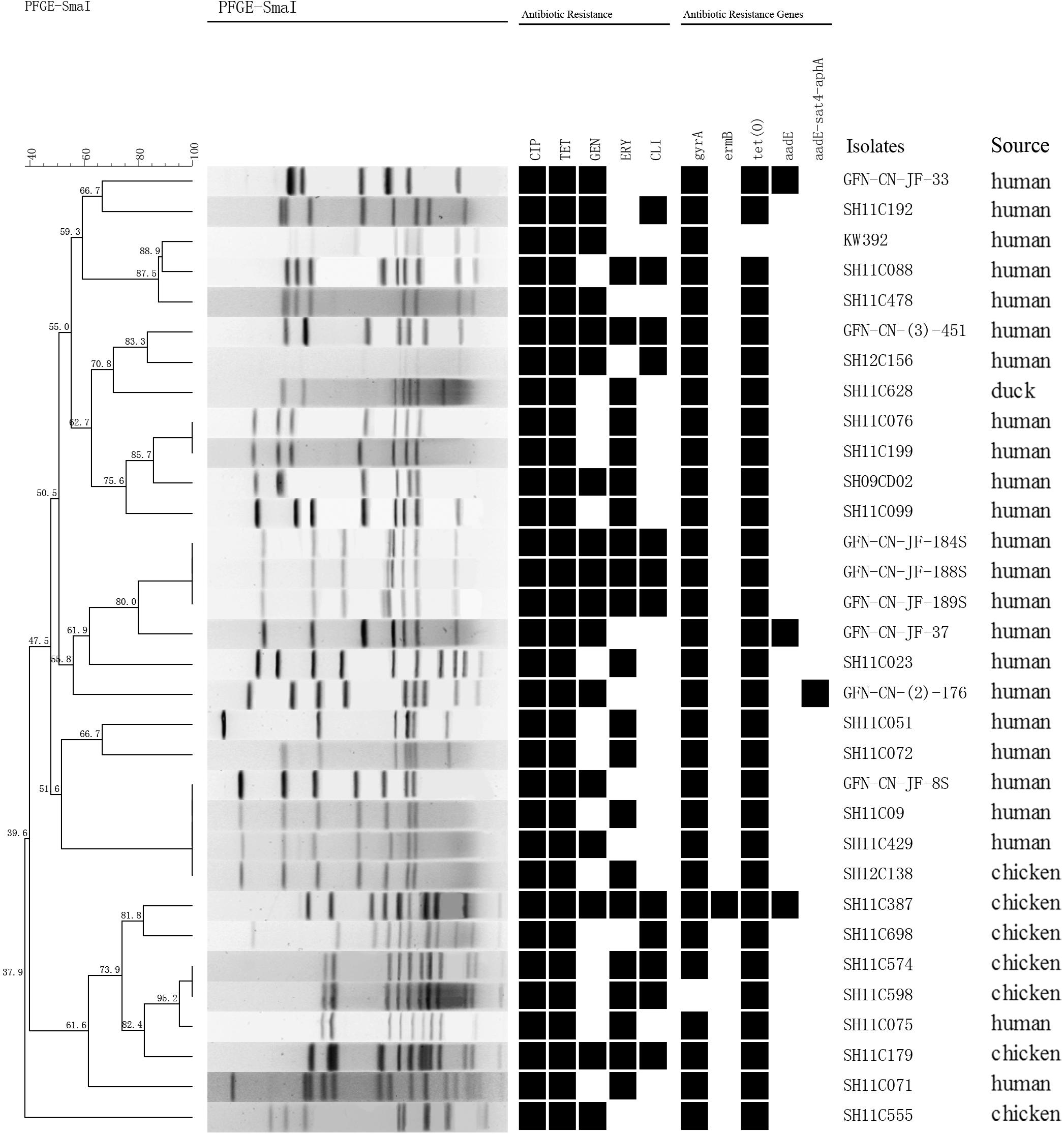
FIGURE 1. Dendrogram of PFGE profiles among the 32 multidrug-resistant Campylobacter jejuni. PFGE assay resulted into 26 different patterns. Corresponding antibiotic resistant patterns, antimicrobial resistant genes or mutants were listed for each isolate. CIP, ciprofloxacin; TET, tetracycline; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin.
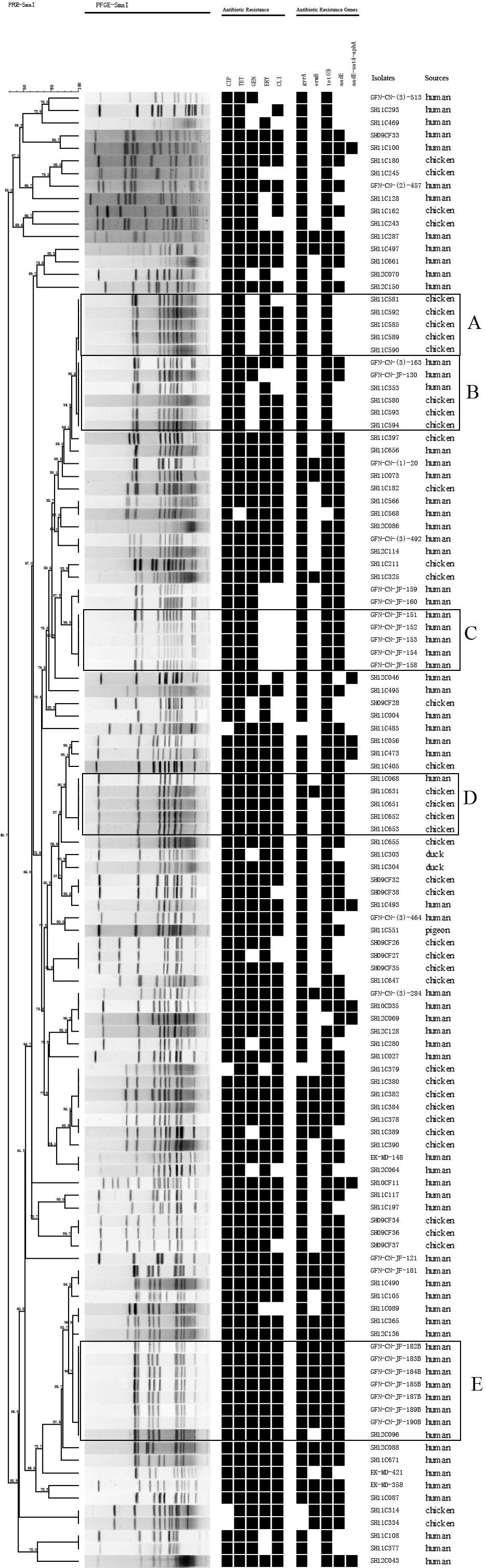
FIGURE 2. Dendrogram of PFGE profiles among the 119 multidrug-resistant Campylobacter coli. PFGE assay resulted into 77 different patterns. Corresponding antibiotic resistant patterns, antimicrobial resistant genes or mutants were listed for each isolate. Five prevalent genotypes (A–E) were identified. CIP, ciprofloxacin; TET, tetracycline; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin.
Among the 151 multidrug-resistant Campylobacter isolates, the numbers of isolates carried tet (O), aadE, ermB, and aadE-sat4-aphA were 148 (98%), 89 (58.9%), 31 (20.5%), and 10 (6.6%), respectively. Almost all (n = 150, 99.3%) had gyrA mutation at codon 86 that causes ciprofloxacin resistance. And the 23s rRNA A2075G point mutation, which is regarded to be associated with erythromycin resistance, was found in 56 (37.1%) isolates (Table 3). Isolates belonging to the same genotypes usually carry the same antimicrobial resistance genes and therefore exhibited similar antimicrobial resistance patterns. However, some exceptions were also observed. For example, C. coli SH11C631 carried a putative antibiotic resistance associated gene ermB, and GFN-CN-(3)-163 and GFN-CN-JF-130 carried aadE. We analyzed the relationship between PFGE genotype and the presence of antimicrobial resistance genes of Gram-positive origin ermB, aadE and aadE-sat4-aphA. No obvious links can be found.
Gene mutations were identified at the inverted repeat regions and flanking regions in the 32 C. jejuni and 119 C. coli multidrug-resistant isolates. As shown in Figure 3, the intergenic region of C. jejuni was 94 nucleotides, and the inverted repeat region was located from −46 to −31 upstream of the cmeABC (consensus sequence: TGTAATAAAAATTACA). While, the intergenic region of C. coli was 107 nucleotides, and the inverted repeat region was located from −50 to −35 upstream of the cmeABC (consensus sequence: TGTAATAAATATTACA). Nine and 15 different sequence types were determined in C. jejuni and C. coli isolates, respectively (Figure 3).
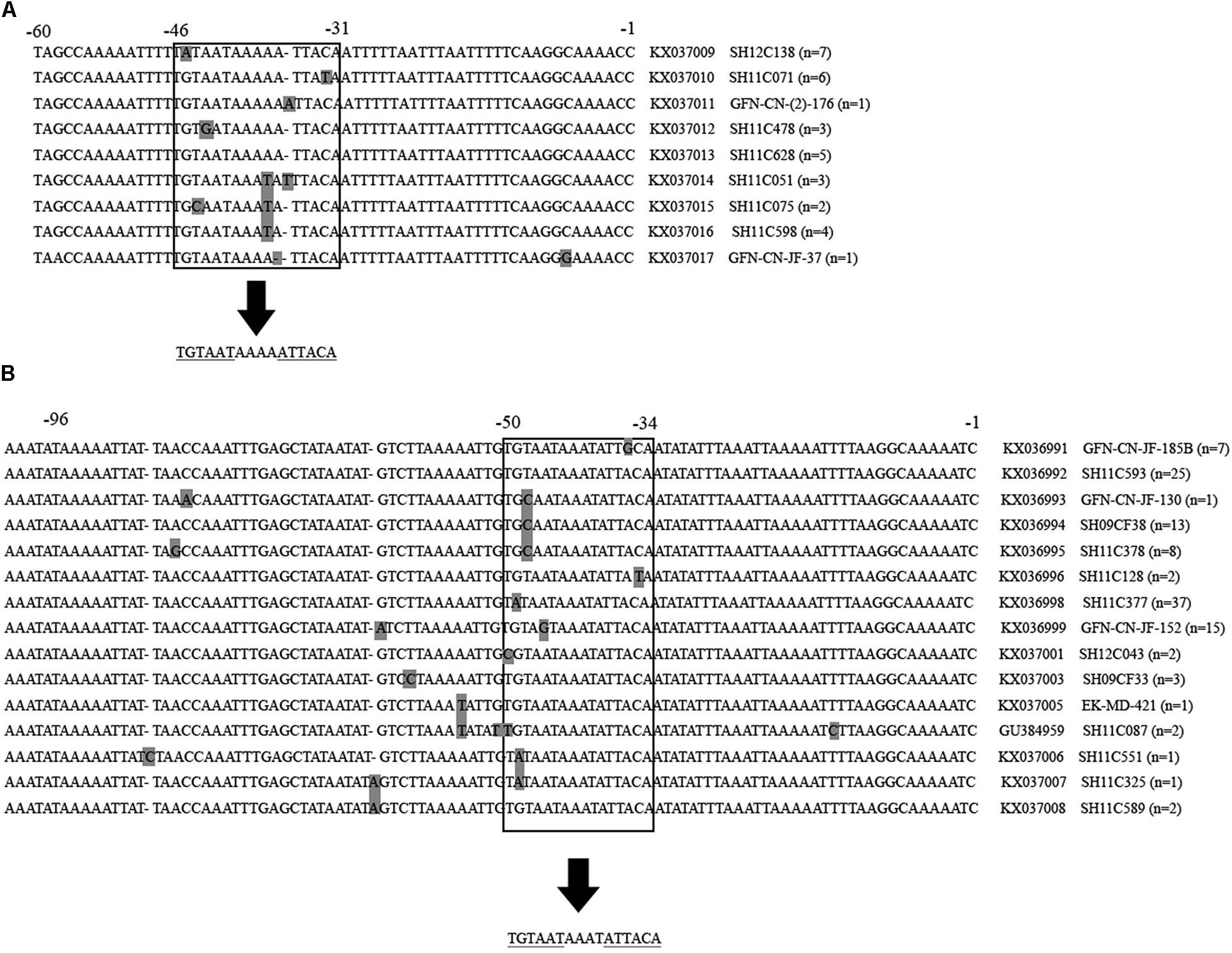
FIGURE 3. Partial multiple alignment of the intergenic regions between cmeR and cmeABC identified in multidrug-resistant Campylobacter jejuni and Campylobacter coli isolates. Polymorphisms are shown in gray squares. The inverted repeat regions are shown inside the open squares, and under the arrows the consensus sequence is shown. Repeated sequences are underlined. Positions are indicated on the top of the alignment. Partial sequences are shown. (A) Alignment of the 94 bp intergenic region of Campylobacter jejuni isolates. A total of 9 novel identified sequences are presented, followed by the accession number, a representative isolates of each sequence type and the number of isolates belonging to this sequence type. (B) Alignment of the 107 bp intergenic region of Campylobacter coli isolates. A total of 15 sequences were identified, including 14 novel ones, followed by the accession number, a representative isolates of each sequence type and the number of isolates belonging to this sequence type.
Antibiotic resistance is recognized as a global problem in human medicine. Several surveillance programs monitoring antimicrobial resistance of Campylobacter to ciprofloxacin, erythromycin, gentamicin, tetracycline, etc. have been well established in the developed world, such as the U.S. National Antimicrobial Resistance Monitoring Systems, the European Food Safety Authority, the European Centre for Disease Prevention and Control and national surveillance programs in several European countries (Ge et al., 2013). Compared with that of developed countries, knowledge on the resistance trends of Campylobacter in developing countries is more limited, which is largely hindered by the lack of an adequate surveillance system. In clinical settings, tetracyclines, fluoroquinolones and macrolides are widely used as the first choice to treat Campylobacter infections because of their low cost and ready availability. In this study, the resistance rates of tetracycline, ciprofloxacin and erythromycin were 99.3, 97.4, and 80.1%, which were much higher than those reported in Canada (Narvaez-Bravo et al., 2017), the United States and European Union (Ge et al., 2013).
As a zoonotic pathogen, Campylobacter has a broad animal reservoir. Antibiotic usage in both animal agriculture and human medicine can influence the development of antibiotic resistance (Luangtongkum et al., 2009). Unfortunately, multidrug resistant Campylobacter isolates are now creeping from food supply chain (Ma et al., 2014, 2017; Zhang et al., 2014, 2016; Wang et al., 2016; Wei et al., 2016; Zhong et al., 2016) into our community (Pan et al., 2015; Zhang et al., 2017). Multidrug resistant Campylobacter isolates are big risks to public health. Establishment of integrated surveillance networks that can monitering their transmission should be taken into consideration in China and other East Asia countries.
Molecular epidemiological studies have contributed to our understanding of transmission of foodborne bacteria. Multilocus sequence typing and PFGE have been developed as robust typing tools for this major pathogen. In this study, PFGE analysis was performed on the 151 multidrug resistant isolates, and a total of 103 PFGE genotypes were determined. Five prevalent genotypes (A–E) were identified in C. coli. It is noteworthy that 7 C. coli isolates in genotype E carried the same antibiotic resistance genes and resist to all the antibiotics (Figure 2). They were isolated from a public hospital in the same period of time (data not shown). In genotype B, isolates from human and chicken were grouped together, indicating this clone might be able to spread across species. We have added these clones into our high-risk clone database. Further studies on the spread of these clones should be carried out in order to rapidly control hospital-acquired infections and identify disease outbreaks. As the world’s third biggest city by population, Shanghai has 26 million people, including over 9 million migrant workers (Statistics of population and family planning, 2016). It is worthy of our concern that floating population may become a vehicle of the transmission of this major pathogen. Therefore, to effectively reduce Campylobacter infection in human, control and prevention strategies should be aimed at both food supply chain and susceptible population levels.
Bacterial resistance to a certain antibiotic is usually multifactorial. Some molecular mechanisms have been proven (Luangtongkum et al., 2009). By investigating the molecular mechanisms of multidrug resistant patterns, we found that antibiotic resistance phenotypes of the isolates correlated well with their antibiotic resistance genes. Almost all (150, 99.3%) had gyrA mutation at codon 86 that causes ciprofloxacin resistance; tet (O), the tetracycline resistance associated gene, was identified in 148 (98%) isolates; 23s rRNA A2075G point mutation, which is regarded to be associated with erythromycin, was found in 56 (37.1%) isolates; and 89 (58.9%) and 10 (6.6%) isolates carried aadE and aadE-sat4-aphA cluster respectively, and were regarded associated with both gentamicin and clindamycin resistance. More studies should be carried out on the horizontal spread of these Gram-positive origin genes by genome comparison (Florez-Cuadrado et al., 2017). In addition, the cmeR-cmeABC intergenic regions were investigated. Gene mutations were identified at the inverted repeat regions and flanking regions in both C. jejuni and C. coli isolates, and 9 and 15 differents sequence types were determined, respectively. These sequence types have potentials to be developed as tools for detecting multidrug resistant Campylobacter. Isolates belonging to the same genotype usually carry the same antimicrobial resistance genes and therefore exhibited similar antimicrobial resistance patterns. However, we did not find obvious links between PFGE genotype and the presence of antimicrobial resistance genes based on the limited data in this study.
This study identified 151 multidrug resistant Campylobacter isolates, and identified several clones that spread among human and poultry. Molecular assay of the antibiotic resistance genes revealed the mechanism of the antibiotic resistant phenotype. The results of this study may help better understand the situation of antibiotic resistance trends in China, and provide guidelines of antibiotic use for treatment of Campylobacter infection in clinic.
YD, JZ, and XX contributed to the conception of the study. CW and YY contributed significantly to analysis and manuscript preparation. HP performed data analysis and wrote the manuscript. YuL and AW helped perform analysis with constructive discussions. YoL and XZ collected samples and conducted the experiments.
This work was funded in part by the Key projects of the National Key R&D Program of China (2017YFC1600101, 2017YFC1600104).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01642/full#supplementary-material
Alfredson, D. A., and Korolik, V. (2007). Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277, 123–132. doi: 10.1111/j.1574-6968.2007.00935.x
Alonso, R., Mateo, E., Churruca, E., Martinez, I., Girbau, C., and Fernandez-Astorga, A. (2005). MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J. Microbiol. Methods 63, 99–103. doi: 10.1016/j.mimet.2005.03.013
Barco, L., Lettini, A. A., Dalla Pozza, M. C., Ramon, E., Fasolato, M., and Ricci, A. (2010). Fluoroquinolone resistance detection in Campylobacter coli and Campylobacter jejuni by Luminex xMAP technology. Foodborne Pathog. Dis. 7, 1039–1045. doi: 10.1089/fpd.2009.0505
CDC (2012). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2010. Atlanta: U.S. Department of Health and Human Services, CDC.
CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. Villanova, PA: CLSI, M100–M122.
Denis, M., Soumet, C., Rivoal, K., Ermel, G., Blivet, D., Salvat, G., et al. (1999). Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29, 406–410. doi: 10.1046/j.1472-765X.1999.00658.x
Florez-Cuadrado, D., Ugarte-Ruiz, M., Meric, G., Quesada, A., Porrero, M. C., Pascoe, B., et al. (2017). Genome comparison of erythromycin resistant campylobacter from turkeys identifies hosts and pathways for horizontal spread of erm(B) genes. Front. Microbiol. 8:2240. doi: 10.3389/fmicb.2017.02240
Ge, B., Wang, F., Sjolund-Karlsson, M., and McDermott, P. F. (2013). Antimicrobial resistance in campylobacter: susceptibility testing methods and resistance trends. J. Microbiol. Methods 95, 57–67. doi: 10.1016/j.mimet.2013.06.021
Iovine, N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. doi: 10.4161/viru.23753
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Ma, H., Su, Y., Ma, L., Ma, L., Li, P., Du, X., et al. (2017). Prevalence and characterization of Campylobacter jejuni isolated from retail chicken in Tianjin, China. J. Food Prot. 80, 1032–1040. doi: 10.4315/0362-028X.JFP-16-561
Ma, L., Wang, Y., Shen, J., Zhang, Q., and Wu, C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181, 77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023
Narvaez-Bravo, C., Taboada, E. N., Mutschall, S. K., and Aslam, M. (2017). Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int. J. Food Microbiol. 253, 43–47. doi: 10.1016/j.ijfoodmicro.2017.04.019
Nohra, A., Grinberg, A., Midwinter, A. C., Marshall, J. C., Collins-Emerson, J. M., and French, N. P. (2016). Molecular epidemiology of Campylobacter coli strains isolated from different sources in new Zealand between 2005 and 2014. Appl. Environ. Microbiol. 82, 4363–4370. doi: 10.1128/AEM.00934-16
Pan, H., Ge, Y., Xu, H., Zhang, J., Kuang, D., Yang, X., et al. (2015). Molecular characterization, antimicrobial resistance and Caco-2 cell invasion potential of Campylobacter Jejuni/coli from young children with diarrhea. Pediatr. Infect. Dis. J. 35, 330–334. doi: 10.1097/INF.0000000000001016
Perez-Boto, D., Lopez-Portoles, J. A., Simon, C., Valdezate, S., and Echeita, M. A. (2010). Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J. Antimicrob. Chemother. 65, 2083–2088. doi: 10.1093/jac/dkq268
Qin, S. S., Wu, C. M., Wang, Y., Jeon, B., Shen, Z. Q., Wang, Y., et al. (2011). Antimicrobial resistance in Campylobacter coli isolated from pigs in two provinces of China. Int. J. Food Microbiol. 146, 94–98. doi: 10.1016/j.ijfoodmicro.2011.01.035
Ribot, E. M., Fitzgerald, C., Kubota, K., Swaminathan, B., and Barrett, T. J. (2001). Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39, 1889–1894. doi: 10.1128/jcm.39.5.1889-1894.2001
Sheppard, S. K., Dallas, J. F., Strachan, N. J., MacRae, M., McCarthy, N. D., Wilson, D. J., et al. (2009). Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48, 1072–1078. doi: 10.1086/597402
Silva, J., Leite, D., Fernandes, M., Mena, C., Gibbs, P. A., and Teixeira, P. (2011). Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2:200. doi: 10.3389/fmicb.2011.00200
Statistics of population and family planning. (2016). Statistics of Population and Family Planning. Available at: http://www.frontiersin.org/Review/EnterReviewForum.aspx?activationno=c7418487-bafe-4c2a-8351-bea1b4c5432e
Wang, Y., Dong, Y., Deng, F., Liu, D., Yao, H., Zhang, Q., et al. (2016). Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008-14. J. Antimicrob. Chemother. 71, 666–669. doi: 10.1093/jac/dkv382
Wang, Y., Zhang, M., Deng, F., Shen, Z., Wu, C., Zhang, J., et al. (2014). Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 58, 5405–5412. doi: 10.1128/aac.03039-14
Wei, B., Cha, S.-Y., Yoon, R.-H., Kang, M., Roh, J.-H., Seo, H.-S., et al. (2016). Prevalence and antimicrobial resistance of Campylobacter spp. isolated from retail chicken and duck meat in South Korea. Food Control 62, 63–68. doi: 10.1016/j.foodcont.2015.10.013
Wieczorek, K., and Osek, J. (2013). Antimicrobial resistance mechanisms among Campylobacter. Biomed Res. Int. 2013:340605. doi: 10.1155/2013/340605
Zhang, M., Gu, Y., He, L., Ran, L., Xia, S., Han, X., et al. (2010). Molecular typing and antimicrobial susceptibility profiles of Campylobacter jejuni isolates from north China. J. Med. Microbiol. 59(Pt 10), 1171–1177. doi: 10.1099/jmm.0.022418-0
Zhang, M., Liu, X., Xu, X., Gu, Y., Tao, X., Yang, X., et al. (2014). Molecular subtyping and antimicrobial susceptibilities of Campylobacter coli isolates from diarrheal patients and food-producing animals in China. Foodborne Pathog. Dis. 11, 610–619. doi: 10.1089/fpd.2013.1721
Zhang, T., Dong, J., Cheng, Y., Lu, Q., Luo, Q., Wen, G., et al. (2017). Genotypic diversity, antimicrobial resistance and biofilm-forming abilities of Campylobacter isolated from chicken in Central China. Gut Pathog. 9:62. doi: 10.1186/s13099-017-0209-6
Zhang, T., Luo, Q., Chen, Y., Li, T., Wen, G., Zhang, R., et al. (2016). Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 8:48. doi: 10.1186/s13099-016-0132-2
Keywords: Campylobacter, PFGE, antimicrobial susceptibility, multidrug resistance, efflux pump
Citation: Du Y, Wang C, Ye Y, Liu Y, Wang A, Li Y, Zhou X, Pan H, Zhang J and Xu X (2018) Molecular Identification of Multidrug-Resistant Campylobacter Species From Diarrheal Patients and Poultry Meat in Shanghai, China. Front. Microbiol. 9:1642. doi: 10.3389/fmicb.2018.01642
Received: 09 February 2018; Accepted: 02 July 2018;
Published: 31 July 2018.
Edited by:
Lucas Dominguez, Complutense University of Madrid, SpainReviewed by:
Ben Pascoe, University of Bath, United KingdomCopyright © 2018 Du, Wang, Ye, Liu, Wang, Li, Zhou, Pan, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanjian Pan, b3VjaGFwcHlAMTYzLmNvbQ== Jianmin Zhang, anVuZmVuZy12QDE2My5jb20= Xuebin Xu, eHV4dWViaW5Ac2NkYy5zaC5jbg==
†These authors have contributed equally to the work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.