- 1Anhui Provincial Key Laboratory of Microbial Pest Control, Anhui Agricultural University, Hefei, China
- 2School of Plant Protection, Anhui Agricultural University, Hefei, China
The Metarhizium genus of filamentous entomopathogenic fungi plays a pivotal role in regulating insect populations. Metalloproteases (MEPs) are a widely distributed and diverse family of hydrolytic enzymes that are important toxicity factors in the interactions between fungi and their hosts. Herein, we characterized two MEPs, Mrmep1 and Mrmep2, in Metarhizium robertsii using gene deletion. Growth rates of the resulting ΔMrmep1 and ΔMrmep2 mutants decreased by 16.2 and 16.5%, respectively, relative to the wild-type (WT) strain. Both mutants were less sensitive to cell wall-perturbing agents, sodium dodecyl sulfate and Congo red than the WT strain, whereas did not show any obvious changes in fungal sensitivity to ultraviolet B irradiation or heat stress. The conidial yield of ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 mutants decreased by 56.0, 23, and 53%, respectively. Insect bioassay revealed that median lethal time values against Galleria mellonella increased by 25.5% (ΔMrmep1), 19% (ΔMrmep2), and 28.8% (ΔMrmep1ΔMrmep2) compared with the WT strain at a concentration of 1 × 107 conidia mL-1, suggesting attenuated fungal virulence in the ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 strains. During fungal infection, transcription levels of Mrmep1 was 1.6-fold higher than Mrmep2 at 36 h post inoculation. Additionally, transcription levels of gallerimycin gene were 1.2-fold, 2.18-fold, and 2.5-fold higher in insects infected with the ΔMrmep1, ΔMrmep2, or ΔMrmep1ΔMrmep2 mutant than those infected with the WT strain, respectively. Our findings suggest that Mrmep1 and Mrmep2 are differentially contributed to the growth, sporulation, cell wall integrity, and virulence of M. robertsii.
Introduction
Metarhizium robertsii is an entomopathogenic fungus that is widely used in the biological control of a variety of insects that cause significant economic losses in agriculture (Frazzon et al., 2000; Lord, 2005; Faria and Wraight, 2007). Metarhizium spp. utilize their abundant hydrolytic enzymes, including chitinases, proteases, lipases, and esterases, to penetrate insect cuticles, which consist of proteins, chitins, and lipids (Adams, 2003; Leger and Wang, 2010; Wang and Feng, 2014). Proteases from pathogenic fungi not only degrade the insect body wall but also activate the insect immune system (Gillespie et al., 2000; Fernandes et al., 2012). Of these proteins, metalloproteinases are a type of protease that rely on metal ions for activation (Tallant et al., 2006). Zinc metalloproteases (MEPs) depend on zinc ions and have a small number of common HEXXH sequences. Depending on the nature of the protein and the position of the third ligand in the zinc ion, zinc MEPs are divided into three groups, metzincins, aspzincins, and gluzincins (Laursen et al., 2002; Glerup et al., 2005; Tallant et al., 2006).
Metalloproteases of pathogens have been linked to virulence (Zhang et al., 1999; Dow et al., 2000; Jia et al., 2000). In the rice blast fungus Magnaporthe oryzae, the zinc MEPs Avr-Pita triggers a signaling transduction cascade recognized by a cytoplasmic receptor Pita (Jia et al., 2000). The secreted fungalysin MEPs gene cgfl is strongly upregulated during the early stages of infection in Colletotrichum graminicola, suggesting a role in fungus-plant interaction (Vargas et al., 2012; Sanzmartín et al., 2015). ZrMEP1, the first reported MEPs from an entomopathogenic fungus Zoophthora radicans, is associated with pathogenesis but not a major host specific determinant (Xu et al., 2006).
The role of entomopathogenic fungal MEPs in growth, germination, stress tolerance, and virulence has not yet been characterized. In our previous study, two MEP genes, Mrmep1 (EFY97549) and Mrmep2 (EFY97706), were significantly upregulated in heat-treated conidia (Wang et al., 2014). Here, we characterized the two MEPs by generating gene-disruption mutants. Our results demonstrated that the two MEPs are involved in growth, sporulation, cell wall integrity, and virulence, but their contributions to fungal virulence are different.
Materials and Methods
Fungal Strains, Host Insects, and Culture Conditions
The fungus M. robertsii strain ARSEF 23 was kindly gifted by Dr. Chengshu Wang (Gao et al., 2011). The fungal strains were cultured on potato dextrose agar [PDA, 20% potato, 2% dextrose and 2% agar (w/v)] medium at 25°C for 14 days in the dark. Collected conidia were dispersed in sterile 0.05% Tween-80 solution and filtered through non-woven fabric to remove mycelia. The conidial suspension was inoculated into Sabouraud dextrose agar yeast extract culture medium (SDAY; 4% dextrose, 1% peptone, 1% yeast extract, and 1.5% agar) and incubated at 25°C for 3 days and hyphae/cultures were harvested by scraping from the cellophane. Escherichia coli DH5α were cultured at 37°C in Luria Bertani broth (LB; 1% tryptone, 0.5% yeast extract, and 1% NaCl [w/v]). The Agrobacterium tumefaciens AGL-1 stain containing target plasmid used as a T-DNA donor for fungal transformation and was incubated in yeast extract beef broth (YEB; 0.5% sucrose, 0.1% yeast extract, 1% peptone, and 0.05% MgSO4⋅7H2O) at 28°C for 16–20 h. For fungal virulence bioassay, larvae of the great wax moth Galleria mellonella were obtained from RuiQing Bait Co., Ltd. (Shanghai, China) and used for the bioassay.
Cloning, Bioinformatics, and Phylogenetic Analysis of MEPs
The sequences of genes encoding MrMEP1 (EFY97549) and MrMEP2 (EFY97706) were obtained from the NCBI database. Primers were designed to amplify the complete cDNA, including the 5′ untranslated region (UTR) and 3′ UTR, using a SMART RACE cDNA Amplification Kit (Clontech, Mountain View, CA, United States after which the products were cloned and sequenced. Domain analysis was performed using the conserved domain database (CDD1). Protein parameters were calculated using the ProtParam tool in ExPASy2 and signal peptide prediction was carried out using the SignalP 4.0 server3.
Homologous MEP sequences from different fungal species were retrieved from the NCBI database4. For phylogenetic analysis, MEP sequences were aligned using ClustalX and then a neighbor-joining (NJ) tree was generated using MEGA 7.0 software (Wang et al., 2017).
Generation of Gene Deletion and Complementation Mutants
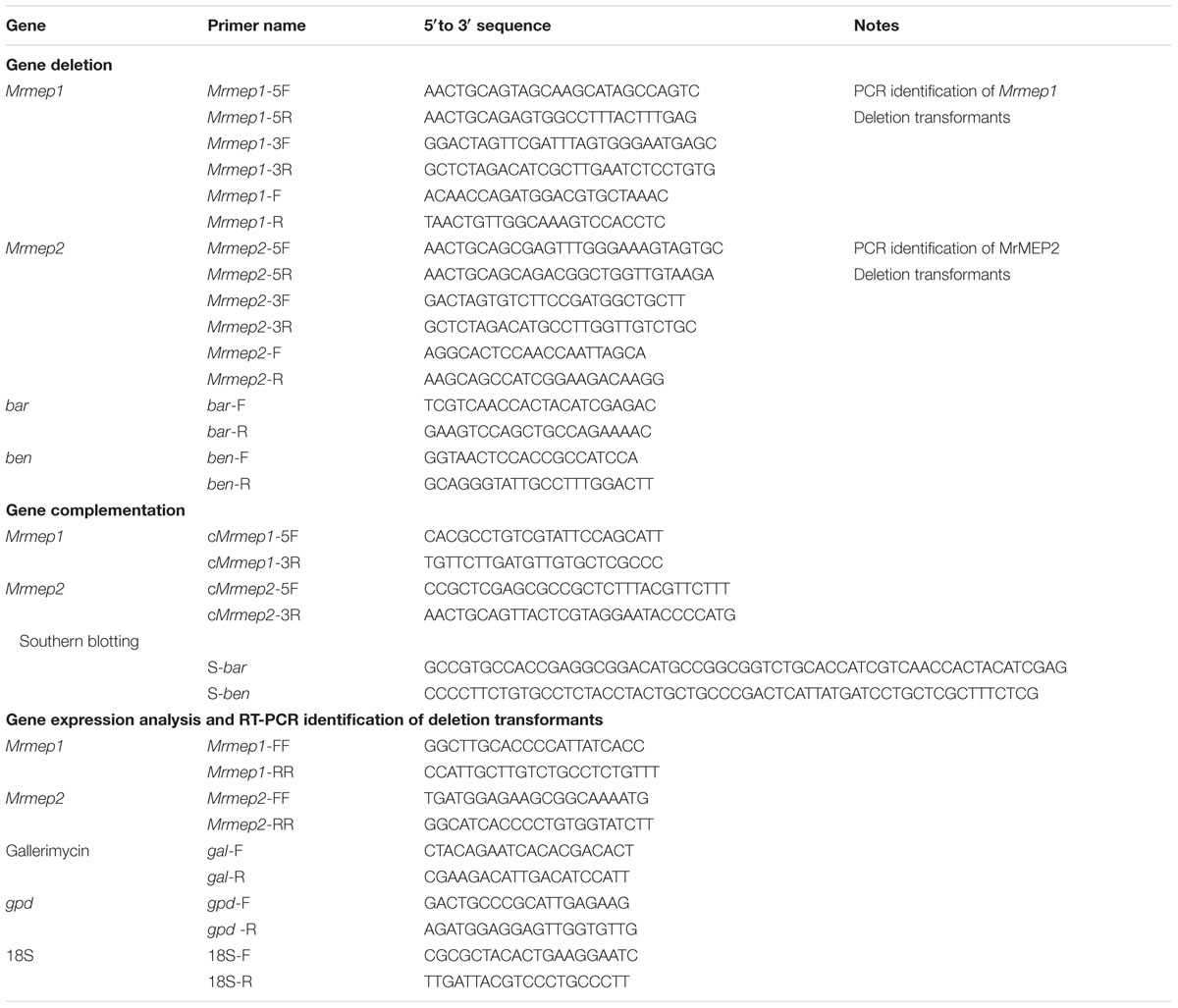
Gene deletion and complementation tests were conducted using a method described previously (Fang et al., 2006). All primers are listed in Table 1.
For gene deletion, fragments of the 5′- and 3′-end-flanking regions of Mrmep1 (1200 and 1000 bp, respectively) were amplified using genomic DNA as the PCR template and Dream Taq DNA Polymerase (Fermentas, Burlington, ON, Canada) with the primer pairs Mrmep1-5F/Mrmep1-5R and Mrmep1-3F/Mrmep1-3R. The purified 5′- and 3′-end-flanking fragments were subsequently cloned into the PstI and SpeI/XbaI restriction enzyme sites in the binary vector pDHt-SK-bar (conferring resistance against ammonium glufosinate) to produce the binary vector pbar-Mrmep1 for Agrobacterium-mediated fungal transformation where a 1993-bp fragment was replaced by the bar cassette. The Mrmep2 deletion mutant was constructed as described above but with the binary vector pDHt-SK-ben. The length of the 5′- and 3′-end flanking regions as well as the replaced fragment in the ΔMrmep2 mutant were 641, 550, and 581 bp, respectively.
For gene complementation, the complementation vector C-pben-Mrmep1 was constructed by inserting the entire Mrmep1 gene plus 1000 bp of the upstream sequence and 200 bp of the downstream sequence into the binary vector pDHt-SK-ben for fungal transformation. Construction of the Mrmep2 complementation vector and fungal transformation were the same as that for Mrmep1, except that the binary vector pDHt-SK-bar was used instead.
For double gene disruption, Mrmep2 was disrupted in a ΔMrmep1 background. For confirmation analysis, all mutants were verified by PCR and reverse transcription PCR (RT-PCR) (Table 1). For ΔMrmep1 deletion and complementation strains, PCR using genomic DNA and two primer sets (Mrmep1-F and Mrmep1-R; Mrmep1-F and bar-R) was performed for detecting the two fragments of interest (1000-bp partial Mrmep1 fragment and 2000-bp partial Mrmep1 plus partial bar fragment) in different strains; the bar gene was also detected using the primer set bar-F and bar-R in the same strains. RT-PCR analysis was conducted to detect the present partial Mrmep1 fragment in ΔMrmep1 using cDNA as the template; the glyceraldehyde 3-phosphate dehydrogenase (gpd) gene (MAA_07675) was used as an internal control. For the other single and double mutants, the same method as described above was used but with different primer sets.
For Southern blotting, 30 μg samples of genomic DNA extracted from the SDAY colonies of wild-type (WT) and each mutant were digested with BamHI and PstI. After separation on 0.7% agarose gels, the DNA fragments were transferred to a nitrocellulose membrane and probed with two bar and ben gene fragments labeled with the PCR DIG Labeling Mix Kit (Roche, Basel, Switzerland. Probe preparation, membrane hybridization, and visualization were performed according to manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit II; Roche).
Mutant Phenotype Assays
Phenotype assays were performed as described previously (Meng et al., 2017). The conidial suspension (1 μL; concentration, 1 × 107 conidia mL-1) of each strain was spotted onto various media unless otherwise noted.
For the growth assay, conidial suspensions of WT and mutant strains were spotted in the center of PDA plates and radial growth (colony diameter) of the vegetative mycelia at 25°C was measured daily.
For the sporulation assay, 30 μL conidia suspension at a concentration of 1 × 107 conidia mL-1 was spread onto PDA plates and incubated at 25°C for 14 days to evaluate sporulation capacity.
For chemical stress tolerance assays, conidial suspensions of WT and mutants were spotted in the center of PDA plates containing different chemical reagents, including the cell wall-disturbingcompounds Congo red (3 mg mL-1) or sodium dodecyl sulfate (SDS)(2.5 μg mL-1; Ying and Feng, 2011). PDA plates without reagents were used as controls. The rate of growth inhibition (RGI%) was calculated as (C - S)/C × 100, where C is the growth rate of the control and S is the growth rate under stress conditions (Wang et al., 2017).
For heat stress tolerance assays, we placed 1-mL conidial suspension aliquots of WT and each mutant strain in 1.5-mL Eppendorf tubes and incubated in a water bath at 40°C for 90 min. One hundred microliter conidial suspension from each tube were inoculated on PDA plates. Conidial germination was observed under the microscope after 24 h of incubation. Conidia with visible germ tubes were considered germinated.
To examine ultraviolet B (UV-B) stress tolerance, 30-μL aliquots of conidial suspensions were centrally smeared onto petri dishes and exposed to UV-B irradiation with a wavelength of 312 nm (280–320 nm) at 100 μJ cm-2 using a UV crosslinker (HL-2000 Hybrilinker; UVP, Upland, CA, United States) (Yao et al., 2010). After exposure, conidial germination on each plate was observed as described for the heat stress tolerance assay.
Bioassays were conducted using G. mellonella larvae as described previously (Wang et al., 2017). Insects were immerged in conidial suspensions for 90 s. All immersed larvae were maintained in large Petri dishes for 10 days at 25°C and examined every 12 h for mortality records. The SPSS statistical package was used to determine the median lethal time (LT50) values. All bioassays were repeated three times in triplicate with 15 insects per replicate.
Quantitative RT PCR (qRT-PCR)
First strand cDNA, to be used as the qPCR template, was synthesized from 1 μg total RNA extracted as described previously (Meng et al., 2017) with reverse transcriptase (TaKaRa, Dalian, China). qRT-PCR was carried out on a Real-Time PCR System (CFX Manager Software; Bio-Rad, Hercules, CA, United States) using a SYBR Green kit (SYBR Premix Ex Taq II; TaKaRa Dalian, China) according to manufacturer’s instructions. Amplification conditions for qPCR were 95°C for 5 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s; all reactions were run in triplicate. The threshold cycle (Ct) was determined using default threshold settings. We used the ΔΔCt method to calculate relative gene expression levels (Livak and Schmittgen, 2001) using gpd (GenBank accession number MAA_07675) as an internal control for each sample (Fang and Bidochka, 2006).
To investigate the expression patterns of Mrmep1 and Mrmep2 during infection, total RNA was extracted from mixed fungi and insect samples by TRIzol reagent (Invitrogen, Camarillo, CA, United States) and used for qRT-PCR q(with gpd as internal control) at different time points of 6, 12, 24, 36, and 48 h after inoculation. The gallerimycin gene is an antifungal peptide in the Lepidoptera insect that can be induced by a variety of bacteria, fungi, and even MEPs in the corpus adiposum. This peptide plays an important role in insect humoral immunity (Feng et al., 2015; Cen et al., 2017). Therefore, we chose the 36 h post inoculation (hpi) time point to examine how the insect responds to the fungus, relative to the 6 h expression levels of the host gallerimycin gene. Additionally, 18S rRNA (GenBank accession number AF286298) was used as the reference in qRT-PCR.
Statistical Analysis
All data were analyzed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, United States). Data were expressed as the mean ± standard error (SE) of at least three independent experiments. Student’s t-test was used to compare the differences between two means. For multiple comparisons, Tukey’s multiple comparison test was used for significance analysis. p-values equal to or less than 0.05 were considered significant.
Results
Characteristics of Mrmep1 and Mrmep2 in M. robertsii
The two MEP sequences were amplified using rapid-amplification of cDNA ends (RACE). Mrmep1 (EFY97549) has a 93-bp 5′ UTR and a 105-bp 3′ UTR and encodes a polypeptide of 611 amino acids. Mrmep2 (EFY97706) includes 5′ and 3′ UTRs of 74 and 101 bp, respectively, and encodes a 255-amino acid polypeptide.
The predicted molecular weight and theoretical isoelectric point (pI) of the deduced MrMEP1 and MrMEP2 proteins were 68.66 and 27.89 kDa, and 6.17 and 5.66, respectively. Both MrMEP1 and MrMEP2 were found to contain predicted signal peptide sequences, suggesting that they are extracellular proteins.
Bioinformatics analysis indicated that a ZnMC domain was present at residues spanning 118-273 in MrMEP1 and 88-245 in MrMEP2 (Figure 1A). Furthermore, the active site motifs HEVGHWLGLLHPHE (MrMEP1), HEAGHWLGLLHTFE (MrMEP2), the Met-Turn motifs HNYMTY (MrMEP1) and HNYMGY (MrMEP2) were identified by analyzing the amino acid sequences at the domain sites of MrMEP1 and MrMEP2. These data confirmed that both proteins are Zn2+ MEPs.
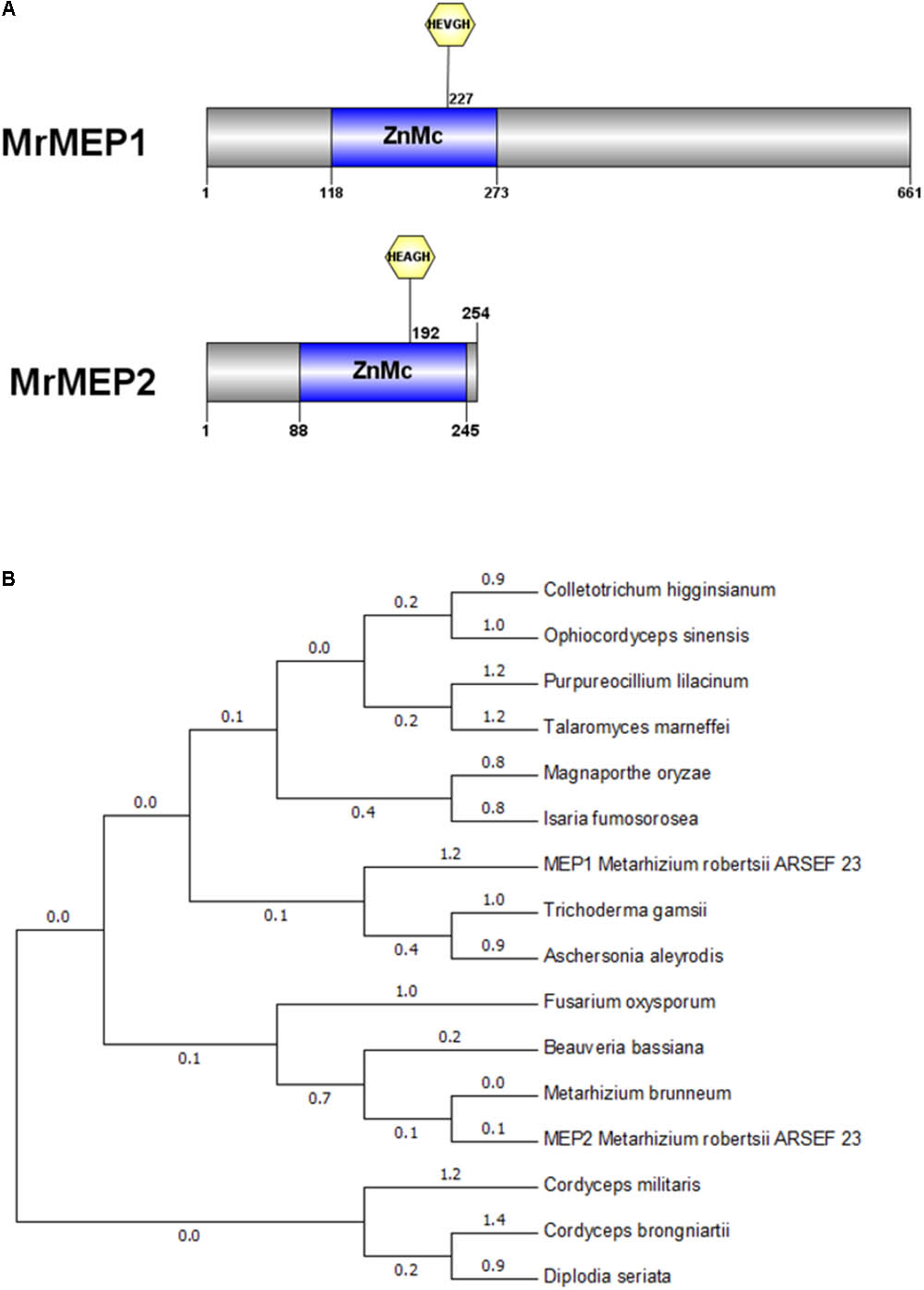
FIGURE 1. Bioinformatic analysis of MrMEP1 and MrMEP2 proteins. (A) The structural domain sites in MrMEP1 and MrMEP2 proteins. (B) Phylogenetic analysis of the fungal MEP proteins. Phylogenetic trees constructed with fungal MEP proteins. The protein accession numbers as follows, EWZ30531.1 hypothetical protein FOZG_16010 [Fusarium oxysporum Fo47]; PNP41669.1 hypothetical protein TGAMA5MH_06456 [Trichoderma gamsii]; XP_018155334.1 metalloprotease [Colletotrichum higginsianum IMI 349063]; XP_018179452.1 metalloprotease [Purpureocillium lilacinum]; OAA39196.1 metalloprotease 1 [Cordyceps brongniartii RCEF 3172]; XP_003714993.1 metalloprotease 1 [Magnaporthe oryzae 70-15]; XP_014540809.1 metalloprotease 1, partial [Metarhizium brunneum ARSEF 3297]; KZZ90284.1 metalloprotease MEP1 [Aschersonia aleyrodis RCEF 2490]; XP_008593388.1 metalloprotease MEP1 [Beauveria bassiana ARSEF 2860]; XP_002149130.1 metalloprotease MEP1 [Talaromyces marneffei ATCC 18224]; XP_002149130.1 metalloprotease MEP1 [Talaromyces marneffei ATCC 18224]; XP_018708221.1 peptidase M43, pregnancy-associated plasma-A [Isaria fumosorosea ARSEF 2679]; XP_006674200.1 peptidase M43B, pregnancy-associated plasma-A [Cordyceps militaris CM01]; EQL01370.1 protein related to metalloprotease MEP1 [Ophiocordyceps sinensis CO18]; KKY13372.1 putative metalloprotease 1 [Diplodia seriata]; EFY97549.1 metalloprotease MEP1-like protein [Metarhizium robertsii ARSEF 23]; EFY97706.1 metalloprotease 1 [Metarhizium robertsii ARSEF 23].
A phylogenetic tree of MEPs from Metarhizium spp. and related fungal species was constructed using Cordyceps militaris as the outgroup, revealing that MrMEP1 is most closely related to Trichoderma gamsii and Isaria fumosorosea MEPs, whereas MrMEP2 is more closely related to Metarhizium brunneum MEPs (Figure 1B).
Knockout and Complementation of Mrmep1 and Mrmep2
To investigate the roles of these two MEPs in M. robertsii, five gene replacement or complementation strains were generated, which include ΔMrmep1 (Mrmep1 disruption mutant), ΔMrmep2 (Mrmep2 disruption mutant), cpΔMrmep1 (Mrmep1 complementation strain), cpΔMrmep2 (Mrmep2 complementation strain), and ΔMrmep1ΔMrmep2 (Mrmep1 and Mrmep2 double disruption mutant) (Supplementary Figure S1). All mutant strains were confirmed by PCR using genomic DNA or cDNA as a template.
PCR analysis showed that an 853-bp Mrmep1 fragment was present in WT and cpΔMrmep1, but not in the ΔMrmep1 and ΔMrmep1ΔMrmep2 strains (Figure 2A). In addition, a 413-bp Mrmep2 fragment was detected in WT and cpΔMrmep2, but not in the ΔMrmep2 and ΔMrmep1ΔMrmep2 strains (Figure 2B). A partial fragment (434 bp) corresponding to the bar gene was present in ΔMrmep1 and ΔMrmep1ΔMrmep2 (Figure 2C) and a partial fragment (301 bp) corresponding to the ben gene was present in ΔMrmep2 and ΔMrmep1ΔMrmep2 (Figure 2D). Furthermore, PCR analysis showed that a 2084-bp fragment and a 1104-bp fragment were detected by the primer sets Mrmep1-F/bar-R and Mrmep2-F/ben-R in the ΔMrmep1 and ΔMrmep2 strains, respectively (Figures 2E,F). Moreover, RT-PCR analysis confirmed loss or regain of the target gene transcription in the gene disruption mutants and reverse complement strains (Figure 2G).
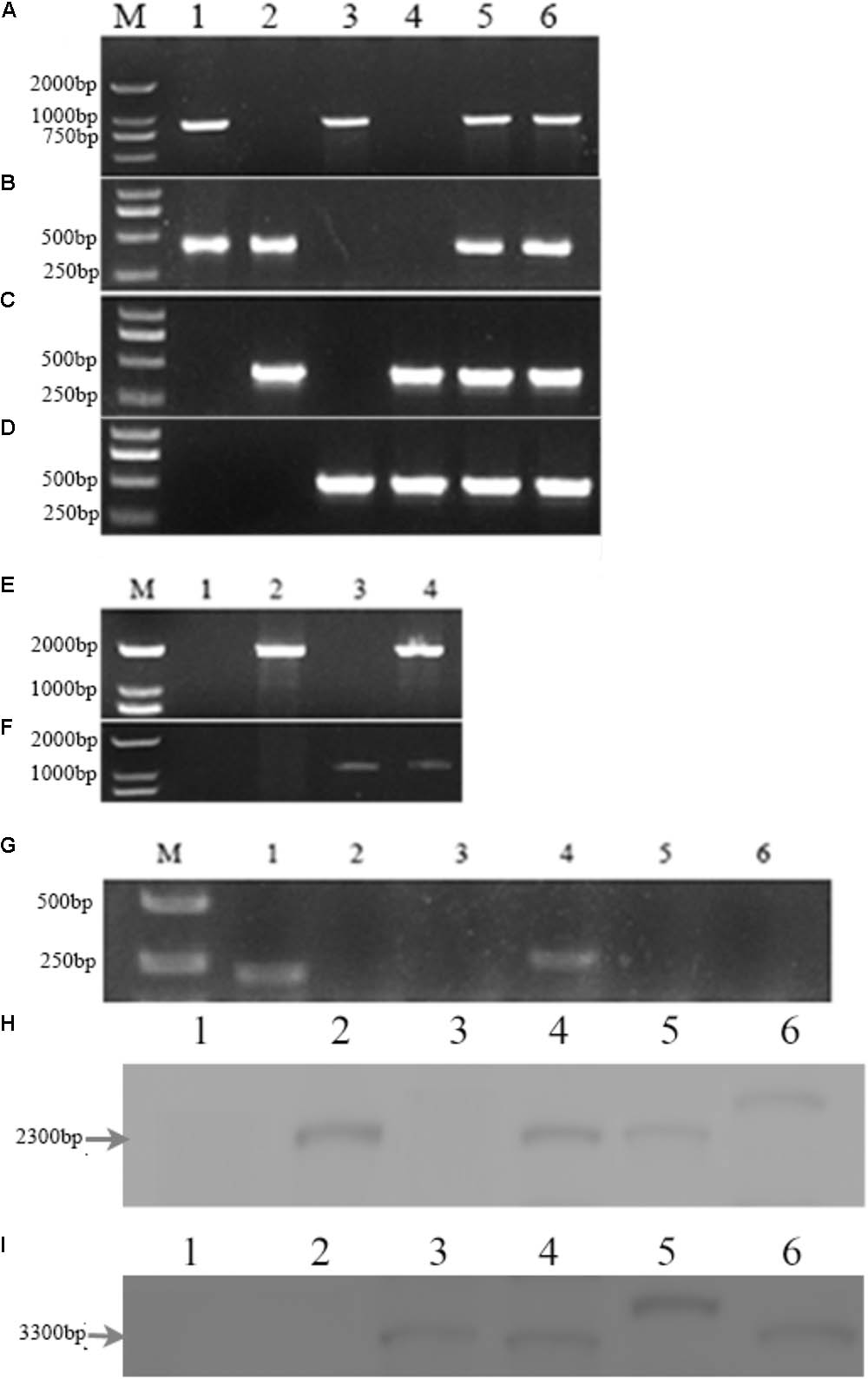
FIGURE 2. Deletion of the Mrmep1 and Mrmep2 gene in M. robertsii. (A–D) PCR analysis of the deleted genes using the primers Mrmep1-F and Mrmep1-R (A), the primers Mrmep2-F and Mrmep2-R (B), the primers bar-F and bar-R (C), and the primers ben-F and ben-R (D). M, Marker; 1, WT; 2, ΔMrmep1; 3, ΔMrmep2; 4, ΔMrmep1 ΔMrmep2; 5, cpΔMrmep1; 6, cpΔMrmep2. (E,F) PCR analysis for the bar (E) or ben (F) genes. M, Marker; 1, WT; 2, ΔMrmep1; 3, ΔMrmep2; 4, ΔMrmep1ΔMrmep2. (G) RT-PCR analysis for the deleted genes. M, Marker; 1 and 4, WT; 2, ΔMrmep1; 3 and 6, ΔMrmep1ΔMrmep2; 5, ΔMrmep2. (H,I) Southern blotting hybridization with bar or ben gene probes. 1, WT; 2, ΔMrmep1; 3, ΔMrmep2; 4, ΔMrmep1ΔMrmep2; 5, cpΔMrmep1; 6, cpΔMrmep2. Further information on primers can be found in Table 1.
Southern blotting further revealed the single copy insertion events in the mutants. The bar gene fragment probe detected a 2.3-kb BamHI band in the ΔMrmep1, ΔMrmep1ΔMrmep2, cpΔMrmep1, and cpΔMrmep2 strains, but not in the WT and ΔMrmep2 strains (Figure 2H). The ben gene fragment probe detected a 3.3-kb BamHI band in the ΔMrmep2, ΔMrmep1ΔMrmep2, cpΔMrmep1, and cpΔMrmep2 strains, but not in the WT and ΔMrmep1 strains (Figure 2I).
Mrmep1 and Mrmep2 Are Involved in Growth and Development
The effect on vegetative growth due to each MEP mutation was investigated. The growth rate of the ΔMrmep1 and ΔMrmep2 mutants were markedly reduced with a 16.2% (p < 0.05) and 16.5% (p < 0.05) decrease as compared to the WT strain, respectively (Figure 3A).
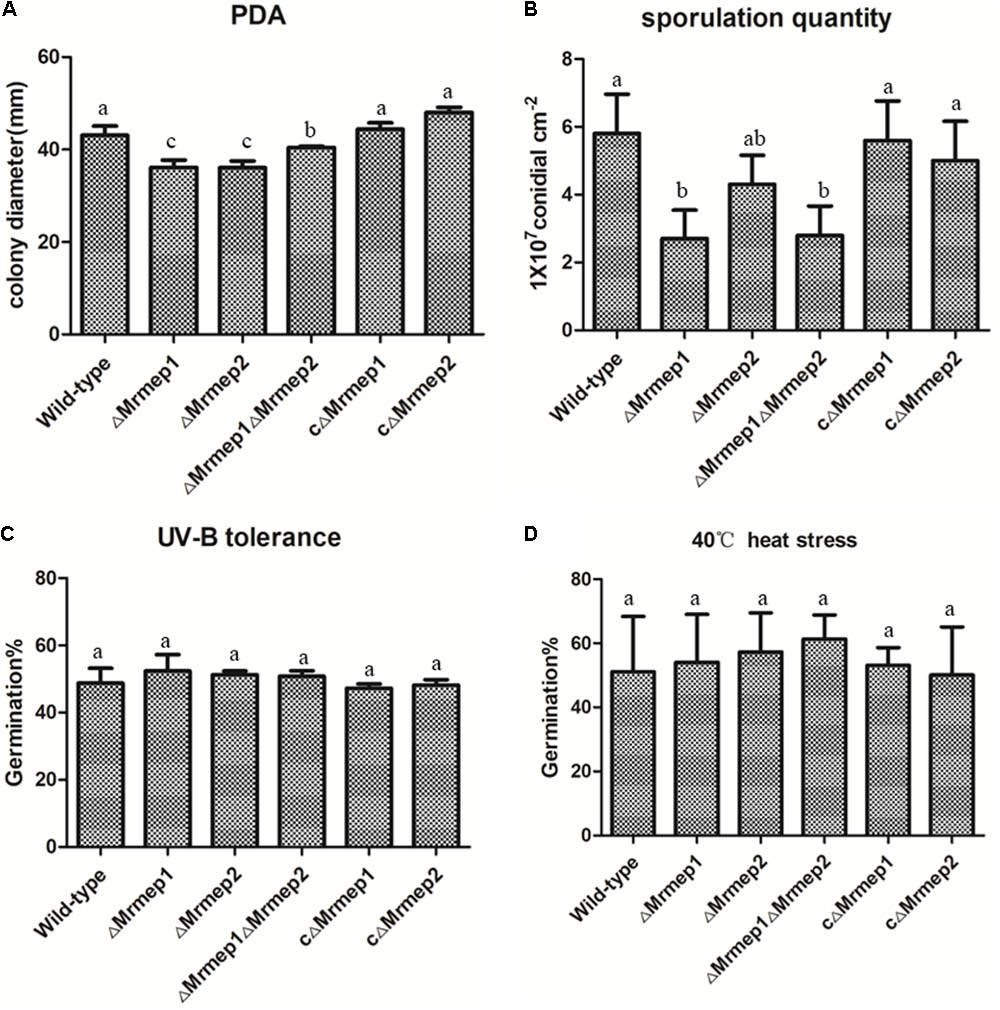
FIGURE 3. Wild-type (WT) and mutant strain growth rates and germination in PDA as well as heat and UV-B stress tolerance. (A) WT and mutant strains were spot inoculated onto PDA plates. (B) Sporulation quantity of the strains at 14 days. (C) WT and mutant strains were cultivated at 40°C under heat stress for 60 min. (D) WT and mutant strains were cultivated under UV-B irradiation for 90 s.
We next assessed the effect of disrupting MEPs on conidial yield. The results indicated that conidial yield in the ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 mutants was reduced by 56.0, 23, and 53%, respectively, compared with the WT. The ΔMrmep1 and ΔMrmep1ΔMrmep2 mutants exhibited a considerably larger decrease, suggesting that Mrmep1 has a more significant role than Mrmep2 in conidiation (Figure 3B).
Mrmep1 and Mrmep2 Are Involved in Cell Wall Integrity
To examine the role of Mrmep1 and Mrmep2 in cell wall integrity, we measured mycelial growth on PDA containing SDS and CR. The SDS RGI values for WT, ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 were 5.67, 1.53, 1.29, and 0.83%, respectively. Meanwhile, the CR RGI values for WT, ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 were 48.79, 13.24, 28.38, and 22.78%, respectively. Thus, the sensitivity of the mutants to SDS and CR decreased significantly compared to that of the WT strain, suggesting roles of Mrmep1 and Mrmep2 in cell wall integrity (Figure 4).
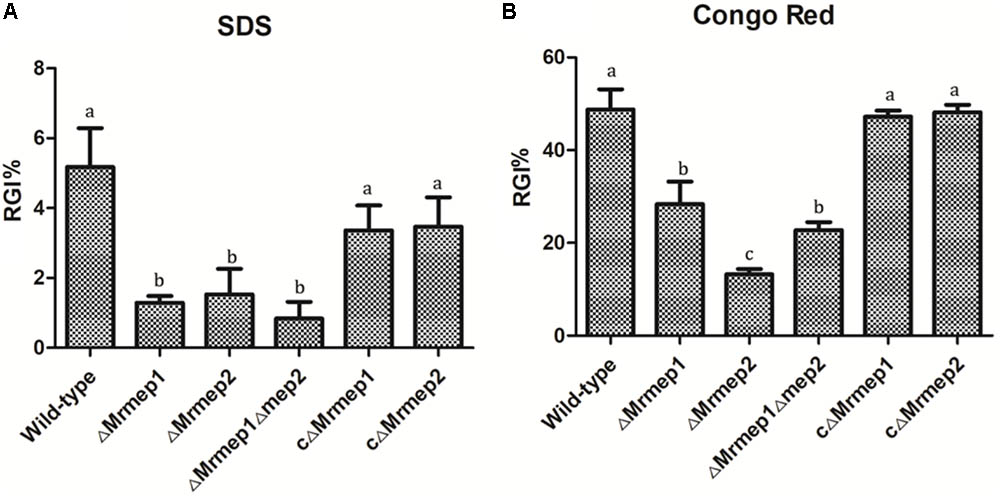
FIGURE 4. The rate of growth inhibition (RGI%) of the mutants in cell wall-disrupting compounds. (A) Mutant strains were spot inoculated onto PDA plates with 0.025% SDS. (B) Mutant strains were spot inoculated onto PDA plates with 0.3 mg mL-1 Congo red.
To investigate whether the mutants exhibited defects under different stress, we measured mycelial growth by the crossing method but the results indicated that the Mrmep mutations did not affect sensitivity to oxidative or osmotic stress (data not shown).
Furthermore, sensitivity of the gene mutations to UV irradiation and thermal stress was examined by measuring conidial germination. We found that conidial germination rates following UV-B irradiation were approximately 51.6, 53.9, and 54.0% for ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2, respectively, compared with 47.1% for the WT strain (Figure 3C). Similar results were obtained for conidial tolerance to high temperature (40°C); 51.0% of WT conidia germinated, compared with 54.0, 57.2, and 61.3% of conidia for ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2, respectively (Figure 3D). Through statistical analysis, there was no significant difference in conidial germination between WT and mutant strains after treatment with either UV or heat. Thus, it appears that Mrmep1 and Mrmep2 do not play a role in conidial tolerance to either UV irradiation or thermal stress.
Mrmep1 and Mrmep2 Are Required for Full Virulence
Insect bioassays using the larvae of G. mellonella were employed to assess the consequences of Mrmep1 and Mrmep2 loss on fungal virulence. Insects were infected topically—representing the natural route of infection—and mortality was monitored daily over a 12-day period. We found that insects infected with ΔMrmep1, ΔMrmep2, or ΔMrmep1ΔMrmep2 displayed an LT50 of 6.2, 5.8, and 6.3 days, respectively, while LT50 values for WT, cΔMrmep1, and cΔMrmep2 were 5.0, 4.9, and 4.3 days, respectively. Thus, ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 displayed a significant attenuation of virulence (p < 0.05) against G. mellonella with increased LT50 values by 25.5, 19, and 28.8%, respectively, compared with WT (Figure 5).
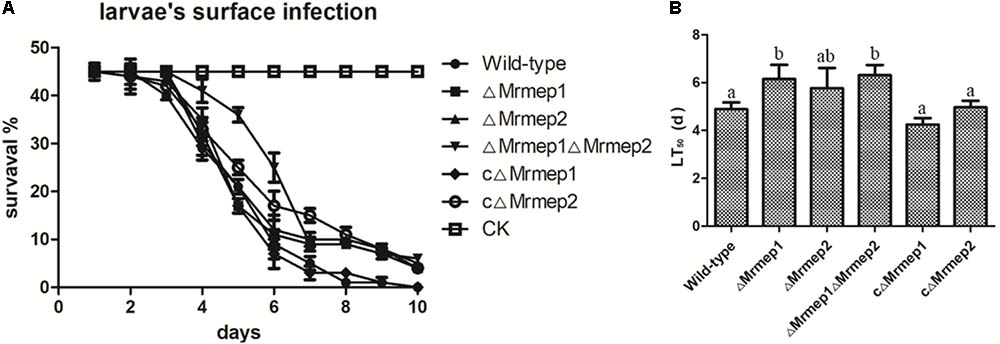
FIGURE 5. Insect bioassays to assess the loss of Mrmep1 and Mrmep2 using the larvae of G. mellonella as hosts. (A) Survival curves of G. mellonella larvae after injection with fungal conidia of WT, ΔMrmep1, ΔMrmep2, ΔMrmep1ΔMrmep2, cΔMrmep1, or cΔMrmep2 strains. Conidial suspensions (3 μL) at 1 × 107 conidia mL-1 were injected into each larva. (B) LT50 of the different mutant strains.
qRT-PCR analysis indicated that expression levels of Mrmep1 and Mrmep2 reached a peak at 36 hpi and decreased at 48 hpi, compared with their levels at 6 hpi in the WT Mr23 strain. Furthermore, we also found that the expression levels of Mrmep1 are always higher than those of Mrmep2 at each tested time point (Figure 6A). To further investigate how insects respond to the fungus, G. mellonella individuals were infected with WT, ΔMrmep1, or ΔMrmep2 strains and expression levels of the gallerimycin gene were measured. The results showed that GAL mRNA levels were higher in the Mrmep mutants than in the WT as they increased by 1.2- and 2.18-fold in the ΔMrmep1 and ΔMrmep2 mutants, respectively, and 2.5-fold in the ΔMrmep1ΔMrmep2 double mutant (Figure 6B).
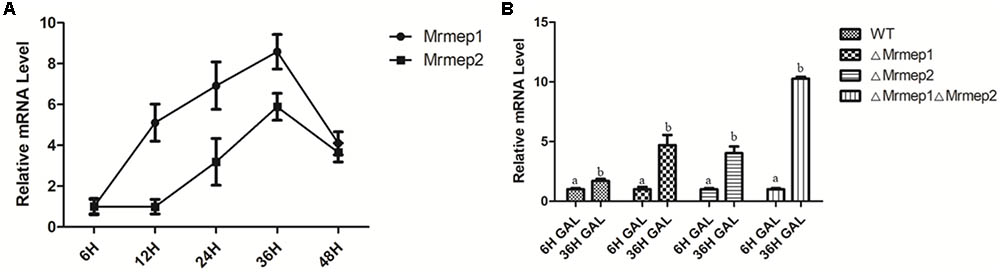
FIGURE 6. qRT-PCR analysis of the expression levels of different genes in the infection process. (A) During G. mellonella infection with the WT Mr23, we quantified the relative expression of Mrmep1 and Mrmep2 transcript levels in total RNA at 6, 12, 24, 36, and 48 hpi. (B) During the infection of G. mellonella at 36 hpi, the relative expression levels of the gal gene were used to quantify the transcript levels of GAL in the ΔMrmep1, ΔMrmep2, and ΔMrmep1ΔMrmep2 strains.
Discussion
In recent years, a large number of virulence factors have been identified (Wang and Wang, 2017). However, there have been few reports on the biological functions of MEPs in entomopathogenic fungi. Herein, we investigated two Zinc MEPS that are potential virulence factors in M. robertsii. Our results suggest that Mrmep1 and Mrmep2 function in growth, sporulation, cell wall integrity, and virulence and that Mrmep1 is more integral than Mrmep2 in sporulation.
We found that MrMEP1 and MrMEP2 belong to the ZnMc superfamily based on CDD analysis of their structural domains. Further analysis revealed that MrMEP1 can be subdivided into the ZnMc pappalysin-like family (M43 in the MEROPS database; Laursen et al., 2002) because MrMEP1 includes the characteristic HEVGHWLGLLH motif (where L is a bulky hydrophobic amino acid), coordinates with the zinc ion, and contains a catalytic Glu residue. ZnMc pappalysin-like family has also been linked to pregnancy in humans and other animals (Vallee and Auld, 1990; Tallant et al., 2006). However, it is unclear whether the protein play a similar role in the entomopathogenic fungus. Here, we found that the function of MrMEP1 is related to asexual sporulation in M. robertsii.
Disruption of either Mrmep1 or Mrmep2 caused an increase in tolerance to CR. However, the double mutant ΔMrmep1ΔMrmep2 did not display increased Congo red tolerance in comparison with the single deletion strains. A similar phenomenon was also observed in conidial production. We speculated that the pathway for conidial production or cell wall integrity involving MEPs may have been completely blocked while another pathway was activated for complementation in the ΔMrmep1ΔMrmep2 double mutant.
To validate the role of the two MEPs during pathogenesis, we developed M. robertsii null mutants lacking the Mrmep1 or Mrmep2 genes as well as a double mutant to test their ability in infecting G. mellonella larvae and verified the results by qRT-PCR. We found decreased sporulation on the insect cuticle for ΔMrmep1 than for ΔMrmep2, which is consistent with the conidial yield of the two mutants.
Moreover, as is well known, gallerimycin is an immune-related gene that confers resistance to fungal pathogens (Altincicek et al., 2007; Huang et al., 2015). Upregulation of gallerimycin at the transcriptional level will increase host defense by the innate immune response (Langen et al., 2006). In our study, the transcription levels of gallerimycin in the single and double mutants were upregulated, suggesting that it was easier for the mutants than for the WT to activate the insect innate immune response.
It has been reported that microbial metalloproteinases mediate the sensing of invading pathogens and activate innate immune responses, including the upregulation of gallerimycin, in G. mellonella (Altincicek et al., 2007). Therefore, we speculated that fungal metalloproteinases in Metarhizium may also digest hemolymph proteins, which leads to the formation of small peptide fragments that bind to danger-sensing receptors. The engagement of these receptors triggers Toll signaling pathways, resulting in the expression of antimicrobial peptides (AMPs) such as gallerimycin.
Furthermore, our previous research showed that Mrmep1 and Mrmep2 expression levels were significantly upregulated in conidia following heat treatment (Wang et al., 2014). Therefore, we speculated that these MEPs may be important in the response to heat stress. However, no apparent phenotypic differences between WT and mutants were observed in the heat stress experiments. This discrepancy between the phenotypic heat stress results and RNA-Seq data may be due to post-transcriptional or post-translational modifications in fungi.
In summary, MrMEP1 and MrMEP2 are Zinc MEPs that function in growth, sporulation, cell wall integrity, and virulence; however, the molecular mechanisms underpinning these functional differences remain unclear. Future studies on downstream signaling pathways could provide further insights.
Author Contributions
BH, XZ, and ZW conceived and designed the study. RZ wrote the manuscript, conducted the experiments, and analyzed the data. AF performed some of the experiments. BH and ZW edited the manuscript. BH supervised the project. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (2017YFD0200400) and the National Natural Science Foundation of China (Grant Nos. 31772226, 31471821, and 31572060).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01528/full#supplementary-material
FIGURE S1 | Deletion of genes encoding metalloproteases in M. robertsii. (A) The disruption and complementation plasmids of Mrmep1. (B) The disruption and complementation plasmids of Mrmep2.
Footnotes
- ^https://www.ncbi.nlm.nih.gov/cdd/
- ^http://web.expasy.org/compute_pi/
- ^http://www.cbs.dtu.dk/services/SignalP/
- ^https://www.ncbi.nlm.nih.gov/
References
Adams, J. (2003). The proteasome: structure, function, and role in the cell. Cancer Treat. Rev. 29, 3–9. doi: 10.1016/S0305-7372(03)00081-1
Altincicek, B., Linder, M. D., Preissner, K. T., and Vilcinskas, A. (2007). Microbial metalloproteinases mediate sensing of invading pathogens and activate innate immune responses in the lepidopteran model host Galleria mellonella. Infect. Immun. 75, 175–183. doi: 10.1128/IAI.01385-06
Cen, K., Li, B., Lu, Y., Zhang, S., and Wang, C. (2017). Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 13:e1006604. doi: 10.1371/journal.ppat.1006604
Dow, M., Newman, M. A., and Von, R. E. (2000). The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. doi: 10.1146/annurev.phyto.38.1.241
Fang, W., and Bidochka, M. J. (2006). Expression of genes involved in germination, conidiogenesis and pathogenesis in Metarhizium anisopliae using quantitative real-time RT-PCR. Mycol. Res. 110, 1165–1171. doi: 10.1016/j.mycres.2006.04.014
Fang, W., Pei, Y., and Bidochka, M. J. (2006). Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 52, 623–626. doi: 10.1139/w06-014
Faria, M. R. D., and Wraight, S. P. (2007). Mycoinsecticides and Mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43, 237–256. doi: 10.1016/j.biocontrol.2007.08.001
Feng, P., Shang, Y., Cen, K., and Wang, C. (2015). Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. U.S.A. 112, 11365–11370. doi: 10.1073/pnas.1503200112
Fernandes,É. K., Bittencourt, V. R. E., and Roberts, D. W. (2012). Perspectives on the potential of entomopathogenic fungi in biological control of ticks. Exp. Parasitol. 130, 300–305. doi: 10.1016/j.exppara.2011.11.004
Frazzon, A. P., Da, S. V. J., Masuda, A., Schrank, A., and Vainstein, M. H. (2000). In vitro assessment of Metarhizium anisopliae isolates to control the cattle tick Boophilus microplus. Vet. Parasitol. 94, 117–125. doi: 10.1016/S0304-4017(00)00368-X
Gao, Q., Jin, K., Ying, S. H., Zhang, Y., Xiao, G., Shang, Y., et al. (2011). Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. doi: 10.1371/journal.pgen.1001264
Gillespie, J. P., Bailey, A. M., Cobb, B., and Vilcinskas, A. (2000). Fungi as elicitors of insect immune responses. Arch. Insect. Biochem. Physiol. 44, 49–68. doi: 10.1002/1520-6327(200006)44:2<49::AID-ARCH1>3.0.CO;2-F
Glerup, S., Boldt, H. B., Overgaard, M. T., Sottrup-Jensen, L., Giudice, L. C., and Oxvig, C. (2005). Proteinase inhibition by proform of eosinophil major basic protein (pro-MBP) is a multistep process of intra- and intermolecular disulfide rearrangements. J. Biol. Chem. 280, 9823–9832. doi: 10.1074/jbc.M413228200
Huang, W., Shang, Y., Chen, P., Gao, Q., and Wang, C. (2015). MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ. Microbiol. 17, 994–1008. doi: 10.1111/1462-2920.12451
Jia, Y., Mcadams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Langen, G., Imani, J., Altincicek, B., Kieseritzky, G., Kogel, K. H., and Vilcinskas, A. (2006). Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth Galleria mellonella, confers resistance against pathogenic fungi in tobacco. Biol. Chem. 387, 549–557. doi: 10.1515/BC.2006.071
Laursen, L. S., Overgaard, M. T., Nielsen, C. G., Boldt, H. B., Hopmann, K. H., Conover, C. A., et al. (2002). Substrate specificity of the metalloproteinase pregnancy-associated plasma protein-A (PAPP-A) assessed by mutagenesis and analysis of synthetic peptides: substrate residues distant from the scissile bond are critical for proteolysis. Biochem. J. 367, 31–40. doi: 10.1042/bj20020831
Leger, R. J. S., and Wang, C. (2010). Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl. Microbiol. Biotechnol. 85, 901–907. doi: 10.1007/s00253-009-2306-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lord, J. C. (2005). From Metchnikoff to Monsanto and beyond: the path of microbial control. J. Invertebr. Pathol. 89, 19–29. doi: 10.1016/j.jip.2005.04.006
Meng, H., Wang, Z., Wang, Y., Zhu, H., and Huang, B. (2017). Identification and functional analysis of Dicer and Argonaute genes involved in RNA interference in the entomopathogenic fungus Metarhizium robertsii. Appl. Environ. Microbiol. 83:e03230-16. doi: 10.1128/AEM.03230-16
Sanzmartín, J. M., Pachecoarjona, J. R., Bellorico, V., Vargas, W. A., Monod, M., Díazmínguez, J. M., et al. (2015). A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Pathol. 17, 1048–1062. doi: 10.1111/mpp.12347
Tallant, C., Garcã-Castellanos, R., Seco, J., Baumann, U., and Gomis-Rüth, F. X. (2006). Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases. J. Biol. Chem. 281, 17920–17928. doi: 10.1074/jbc.M600907200
Vallee, B. L., and Auld, D. S. (1990). Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29, 5647–5659. doi: 10.1021/bi00476a001
Vargas, W. A., Martín, J. M. S., Rech, G. E., Rivera, L. P., Benito, E. P., Díazmínguez, J. M., et al. (2012). Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 158, 1342. doi: 10.1104/pp.111.190397
Wang, C., and Feng, M. G. (2014). Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol. Control 68, 129–135. doi: 10.1016/j.biocontrol.2013.06.017
Wang, C., and Wang, S. (2017). Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 62, 73–90. doi: 10.1146/annurev-ento-031616-035509
Wang, Y., Wang, T., Qiao, L., Zhu, J., Fan, J., Zhang, T., et al. (2017). DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 101, 1–12. doi: 10.1007/s00253-017-8197-5
Wang, Z. X., Zhou, X. Z., Meng, H. M., Liu, Y. J., Zhou, Q., and Huang, B. (2014). Comparative transcriptomic analysis of the heat stress response in the filamentous fungus Metarhizium anisopliae using RNA-Seq. Appl. Microbiol. Biotechnol. 98, 5589–5597. doi: 10.1007/s00253-014-5763-y
Xu, J., Baldwin, D., Kindrachuk, C., and Hegedus, D. D. (2006). Serine proteases and metalloproteases associated with pathogenesis but not host specificity in the Entomophthoralean fungus Zoophthora radicans. Can. J. Microbiol. 52, 550–559. doi: 10.1139/w06-004
Yao, S. L., Ying, S. H., Feng, M. G., and Hatting, J. L. (2010). In vitro and in vivo responses of fungal biocontrol agents to gradient doses of UV-B and UV-A irradiation. Biocontrol 55, 413–422. doi: 10.1007/s10526-009-9265-2
Ying, S. H., and Feng, M. G. (2011). A conidial protein (CP15) of Beauveria bassiana contributes to the conidial tolerance of the entomopathogenic fungus to thermal and oxidative stresses. Appl. Microbiol. Biotechnol 90, 1711–1720. doi: 10.1007/s00253-011-3205-7
Keywords: Metarhizium, metalloprotease, virulence, sporulation, cell wall integrity
Citation: Zhou R, Zhou X, Fan A, Wang Z and Huang B (2018) Differential Functions of Two Metalloproteases, Mrmep1 and Mrmep2, in Growth, Sporulation, Cell Wall Integrity, and Virulence in the Filamentous Fungus Metarhizium robertsii. Front. Microbiol. 9:1528. doi: 10.3389/fmicb.2018.01528
Received: 07 March 2018; Accepted: 19 June 2018;
Published: 06 July 2018.
Edited by:
Weiguo Fang, Zhejiang University, ChinaReviewed by:
Chengshu Wang, Shanghai Institutes for Biological Sciences (CAS), ChinaYongjun Zhang, Southwest University, China
Copyright © 2018 Zhou, Zhou, Fan, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Huang, Ymh1YW5nQGFoYXUuZWR1LmNu
 Rong Zhou1
Rong Zhou1 Bo Huang
Bo Huang