- 1Teagasc Food Research Centre, Cork, Ireland
- 2School of Microbiology, University College Cork, Cork, Ireland
- 3APC Microbiome Ireland, Cork, Ireland
Anaerobic sporeformers, specifically spoilage and pathogenic members of the genus Clostridium, are a concern for producers of dairy products, and of powdered dairy products in particular. As an alternative to testing for individual species, the traditional, and still current, approach to detecting these sporeformers, including non-spoilage/non-pathogenic species, in dairy products has involved testing for a sulphite reducing phenotype [Sulphite reducing Clostridia (SRCs)] under anaerobic conditions. This phenotype is conserved throughout the Order Clostridia. Unfortunately, however, this phenotype is exhibited by other sulphite reducing bacteria (SRBs) also, potentially leading to potential for false positives. Here, this risk was borne out through the identification of several SRBs from industry samples that were identified as Proteus mirabilis and various Bacillus/Paenibacillus sp. Genome wide comparison of a number of representative SRCs and SRBs was employed to determine phylogenetic relationships, especially among SRCs, and to characterize the genes responsible for the sulphite reducing phenotype. This screen identified two associated operons, i.e., asrABC in SRCs, and cysJI in Bacillus/Paenibacillus spp. and P. mirabilis. This screen identified spp. belonging to sensu stricto, Lachnospiraceae and Cluster XIV of the Clostridia all producing the SRC phenotype. This study highlights the inaccuracy of the industry standard SRC test but highlights the potential to generate an equivalent molecular test designed to detect the genes responsible for this phenotype in clostridia.
Introduction
Raw milk is populated by a variety of metabolically and taxonomically diverse bacteria, the majority of which are inactivated by commercial pasteurization (Wells-Bennik et al., 2016). While this process reduces the overall bacterial load and diversity of the milk, it selects for thermoduric and, in particular, sporeforming, bacteria. This is notable as sporeforming bacteria, including many anaerobic sporeformers, are present in niches throughout the dairy chain, extending from farm to factory (Wells-Bennik et al., 2016) and are a significant concern for the dairy industry (Doyle et al., 2015). The majority of strictly anaerobic sporeformers of concern to the dairy industry belong to the Clostridium genus, specifically to Cluster I and Cluster II, and are also known as the Clostridium sensu stricto (McAuley et al., 2014; Doyle et al., 2015). From a spoilage perspective, some of these Clostridium spp. can cause late-blowing defects in cheese due to butyric acid production (Bassi et al., 2015). Clostridium tyrobutyricum is most commonly associated with this defect but Clostridium sporogenes, Clostridium butyricum, Clostridium beijerickii and, to a lesser extent, Clostridium tertium may also cause or contribute to this defect include (Bermúdez et al., 2016). From a public health perspective, Clostridium perfringens, Clostridium botulinum, and Clostridium tetani are of greatest concern due to their toxigenic potential. C. perfringens is the most prevalent of these species from a foodborne illness perspective, and causes in excess of 1 million incidences of foodborne illness in the United States per annum (Scallan et al., 2011). Although, between 1998 and 2008 only one case of foodborne illness in the United States was attributed to a dairy related vector (Bennett et al., 2013), C. perfringens has recently been isolated throughout the dairy farm environment in Australia, including in raw milk (McAuley et al., 2014), has been detected in defective cheese in Italy (Bassi et al., 2015) and its presence in powdered infant formula (PIF) has been reported (Barash et al., 2010). In the case of C. botulinum, while the presence of the pathogen in PIF has been associated with two incidences of infant botulism previously, one in the United States and one in the United Kingdom in 2001, the links were not conclusively established (Barash et al., 2010). Regardless, this species remains a concern for dairy producers, particularly for those that produce products for infant consumption, as the infectious dose for botulinum spores in infant botulism is thought to be extremely low (ICMSF, 2014) and the reputational damage associated with an outbreak would likely be great. Indeed, the inaccurate reporting of the presence of C. botulinum in PIF originating from New Zealand has previously resulted in a significant recall (Doyle and Glass, 2013). To our knowledge C. tetani has not been associated with any incidences of foodborne illness associated with the consumption of dairy product, nor has it been reported to have been detected in dairy products. Nonetheless it remains of concern to producers because of its ability to produce a neurotoxin.
Because of the toxigenicity of some members of the Clostridia, coupled with the potential of some members of the sensu stricto to cause spoilage in dairy products, it is routine to test dairy products for the presence of these sporeformers. The test employed most frequently, primarily for historical reasons, involves the enumeration of sulphite reducing Clostridia (SRC) and relies on the ability of the majority of Clostridium spp. of concern to the dairy industry to reduce sulphite to sulfide (Weenk et al., 1995; Doyle et al., 2015), most frequently through use the asrABC operon involved in dissimilatory sulphite reduction (Czyzewski and Wang, 2012). The asrABC operon has previously been described in Clostridium spp. and Salmonella enteria (Huang and Barrett, 1991). However, other bacteria referred to as sulphite reducing bacteria (SRBs) may have other genes (cysJI) that produce the same phenotype (Standards, 2003) and result in false positives (Weenk et al., 1991; Doyle et al., 2015). Indeed, aerobic sporeformers and even Gram negative bacteria have caused such false positive results in the past (Sugiyama, 1951; Fischer et al., 2012). Ultimately, the distribution of the SRC phenotype throughout the heterogeneous Clostridium genus, including many species that were previously considered Clostridium, (Ludwig et al., 2009) is not well understood, making the relevance of the SRC assay unclear.
The objectives of this study were to determine the identity of SRCs, and SRBs, isolated from a variety of dairy sources, and to employ comparative genomics to identify genetic features common among SRCs with a view to the identification of conserved loci that could be used for alternative, DNA-based, diagnostic approaches.
Materials and Methods
Isolation and Identification of Sulphite Reducing Isolates
Anaerobic sulphite reducing bacteria were isolated from dairy powders, cheese and raw bulk tank milk using standard protocols (Standards, 2004). This method includes a heat inactivation step (80°C for 10 min) that is intended to eliminate non-sporeforming bacteria. Black colonies were then aseptically picked and grown in pure culture in reinforced Clostridium media before DNA was extracted using the Mericon Bacteria plus kit (Qiagen). The 16S rRNA gene was amplified from each isolate using the CO1 and CO2 primers (Simpson et al., 2003). This PCR was conducted using the following parameters; 94°C for 5 min, followed by 30 amplification cycles, each consisting of three 1 min stages at 94°C, 60°C, and 72°C, with a final extension of 5 min at 72°C. Amplified DNA was then purified using the GenElute PCR cleanup kit (Sigma Aldrich, Wexford, Ireland) before Sanger sequencing was carried out (Source Bioscience, Waterford, Ireland). The resulting sequences were than subjected to BLAST analysis (Altschul et al., 1997) against the NCBI database with a view to determining their identity.
Whole Genome Sequencing
Genomic DNA, which had been extracted as described above, was cleaned up using the Powerclean kit (Mo Bio, Carlsbad, CA). Genomic DNA was then quantified using the Qubit high sensitivity kit (Bioscience, Dublin, Ireland), prepared for sequencing using the Nextera XT library preparation kit (Illumina) and sequenced on the Illumina Miseq platform using paired-end 2 × 250 base pair reads at the Teagasc Sequencing Centre, Teagasc Food Research Centre, Moorepark. Raw reads were processed and filtered based on quality and quantity and trimmed to 200 bp with a combination of Picardtools (https://github.com/broadinstitute/picard) and SAMtools (Li et al., 2009). Quality was visualized using FastQC (Andrews, 2010). Sequences were assembled using IDBA-UD (Peng et al., 2012), removing all contigs smaller than 500 bp.
Annotation, Phylogenetic Comparison and Analysis of Core Genes of Clostridium Genus
Assembled contigs from sequenced isolates and genome scaffolds from the NCBI genome repository were annotated using Prokka (Seemann, 2014). Global alignment of amino acid sequences was carried out using Phylophlan (Segata et al., 2013). A phylogenetic tree was created from this alignment using FastTree (Price et al., 2009). The phylogenetic tree was then visualized using Graphlan (Asnicar et al., 2015). Using the .gff files from Prokka, Roary (Page et al., 2015) was used to compare the annotated genes from all SRBs using a BLASTp threshold of 50. In addition, core genes within SRC were also identified using Roary (Page et al., 2015) setting a BLASTp threshold of 50% for both comparisons. Sequences for each species can be found on ENA under accession numbers ERS1887784, ERS1887785, ERS1887786, ERS1887787, ERS1887788, and ERS1887789.
In silico Screening for Sulphite Reducing Genes Among SRBs
A protein database was created containing all the annotated genomes of the SRBs listed in Table S1. For the SRC phenotype, query amino acid sequences for the A, B, and C subunits of the asr gene cluster from the type C. butyricum strain, DSM 10702, were BLASTed against this database (Altschul et al., 1997). For the non-SRC SRB blastp query searches, the amino acid sequences for the assimilatory sulphite reducing genes cysI and cysJ from B. licheniformis were selected as this was the most frequently isolated Bacillus SRB in the surveillance.
Analysis of Amino Acid Sequence Homology in asrABC and cysIJ
The sample sequences for BLASTp hit for each gene were retrieved from the BLASTp searches and converted into fasta format and aligned using MUSCLE (Edgar, 2004) for visual inspection of conservation. Aligned sequences from each gene were visualized using Jalview (Waterhouse et al., 2009). The amino acid sequences of the A, B and C subunits of the asr operon were examined for the presence of conserved functional domains. Furthermore, the cysI and cysJ genes were also analyzed for conserved amino acid domains. The structure of these proteins was also modeled using Phyre2 (Kelley et al., 2015).
Results and Discussion
Identification of SRBs in Dairy Products
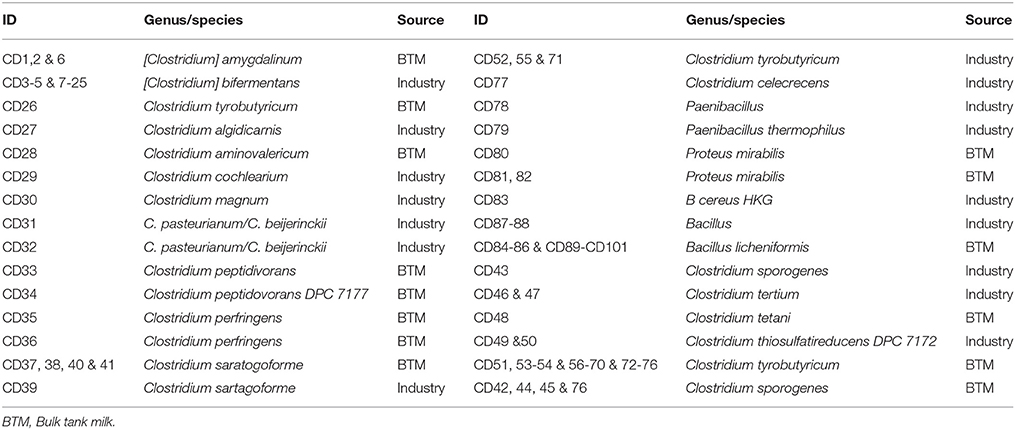
In order to better understand the prevalence and identity of SRBs in the Irish dairy chain, 101 positive SRB isolates were identified by Sanger sequencing of the corresponding 16S rRNA gene. 77 isolates were identified as clostridia (SRCs), 19 were Bacillus sp., 3 isolates were Proteus mirabilis and 2 Paenibacillus sp. (Table 1). It was thus apparent that the SRBs present in the dairy chain were relatively heterogeneous, with the proportion of non-clostridia being particularly notable in light of the purpose of the assay i.e., to detect SRCs. The basis for positive phenotypes was anticipated to reflect the presence of asrABC operons [i.e., those associated with Clostridium spp. (Czyzewski and Wang, 2012)], and cysJI operons [i.e., those previously found in P. mirabilis, Bacillus/Paenibacillus and other genera (Guillouard et al., 2002; Turnbull and Surette, 2008)].
Among the 101 isolates, the pathogens detected were C. perfringens and C. tetani. Although not a pathogen, the presence of C. sporogenes is notable in that it can be difficult to distinguish between C. sporogenes and C. botulinum because of the significant genomic synteny shared between the two species. C. sporogenes may also contribute to gas defects in continental style cheeses (Bermúdez et al., 2016). The presence of C. tyrobutyricum, C. beijerinckii and C. tertium is notable as these species have previously been associated with late blowing defects in cheese (Cocolin et al., 2004; Bermúdez et al., 2016). Other clostridia detected were C. amygdalinum, C. bifermentans, C. algidcarnis, C. aminovelerium, C. peptidoveorans, C. sartagoforme, C. thiosulfatireducens, C. cochlearium, and C. celecrescens. Of these, C. bifermentans has been associated with a pediatric infection previously (Brook, 1995) and both it and C. cochlearium have previously been isolated from powdered infant formula (Barash et al., 2010) as well as dairy farm effluent (Gupta and Brightwell, 2017), the latter observation potentially highlighting a source of these microbes in the dairy chain. To our knowledge, the presence of C. amygdalinum, C. algidcarnis, C. aminovelerium, C. peptidoveorans, C. sartagoforme, C. thiosulfatireducens, and C. celecrescens has not previously been reported in dairy sources.
Among the non-clostridia were 16 Bacillus licheniformis and 1 B. cereus strains. These are spoilage and pathogenic species, respectively, that have been associated with dairy foods (McHugh et al., 2017), bulk tank milk (BTM) (Miller et al., 2015; Sadiq et al., 2016) and dairy farm effluent (Gupta and Brightwell, 2017). Finally, three SRB isolates were identified as P. mirabilis. The detection of this Gram negative bacterium was unusual as it would be expected that Proteus would be inactivated by the heat-treatment step within the assay. Regardless, it is notable that P. mirabilis (Kawabata, 1980) and B. licheniformis (Harmon et al., 1971; Weenk et al., 1995; Fischer et al., 2012), though not B. cereus, have previously been found to cause false positive results in a SRC assay. Two Paenibacillus spp.(including Paenibacillus thermophilus) were isolated in this screen, this species has frequently been isolated from raw milk and processed dairy products previously (Ivy et al., 2012).
SRB In-silico Genome Characterization
In-silico genome characterization was utilized to further investigate SRB taxonomy, and associated sulphite reducing genes. This analysis included genome sequences that were representative of the species detected in the dairy products and were already available on the NCBI database, as well as sequences corresponding to other cluster 1 Clostridium, including C. botulinum group and other known sulphite reducing Clostridium spp. and members of the sensu stricto, Paenibacillus lactis (due to the non-availability of a P. thermophilus genome sequence) and Salmonella enterica typhimurium LT2, as this species has both an asrABC gene cluster for dissimilatory sulphite reduction and a cysIJ operon for assimilatory sulphite reduction. Six additional Clostridium strains isolated from this study, i.e., Clostridium aminovelericum DPC 7173, Clostridium thiosulfatireducens DPC 7172, Clostridium cochlerium DPC 7174, Clostridium tertium DPC 7175, Clostridium amygdalinum DPC 7176 and Clostridium peptidovorans DPC 7177, were selected for whole genome sequencing due to the absence of existing whole genome sequences at the time of analysis. A list of all the genomes used for this analysis and a summary of the assembly statistics can be found in Table S1.
After sequence assembly and annotation, global genome alignment was carried out using Phylophlan and Roary. Phylophlan uses 300 marker genes common to all bacteria, while Roary uses all the annotated genes of each genome. The Phylophlan tree (Figure 1) highlights the phylogenetic diversity which exists across bacteria with the SRB phenotype. The division between species that use the cysJI operon to reduce sulphite to sulfide (Bacillus spp., P. lactis, P. mirabilis and S. enterica) and those that use the asr operon is apparent. A similar functional separation, i.e., consistent with the presence or absence of asrABC, is observed in the gene presence absence Multidimensional scaling (MDS) plot generated from the Roary results (Figure 2). As noted, S. enterica has both asrABC and cysJI operons. In the Phylophlan tree, the newly sequenced C. amygdalinum and C. aminovelericum genomes cluster closely with that of C. celecrescens. In addition, C. thiosulfatireducens shows relatedness to P. bifermentans and C. difficile. These six species form a distinct branch which is distant from the rest of the Clostridium spp. Indeed, some of species falling within this subgroup have been recently reclassified, for instance the bacterium formally known as Clostridium difficile is now designated as Clostridioides difficile (Lawson et al., 2016). The separation of C. amygdalinum and C. aminovelericum from the sensu stricto can be seen in Figure 2. This is expected as they are members of Clostridium Clusters XIV and III respectively. It is also evident that the genomes of C. tunisiense and C. sulfidigenes form a distinct clade separate from the rest of the sensu stricto Clostridium spp. Neither of these bacteria were isolated during the present study but were included in the analysis as they are known to reduce sulphite (Thabet et al., 2004; Sallam and Steinbüchel, 2009). The distinct clustering of these strains was not anticipated and warrants future investigation. This separation of these two species in Figure 1 suggests that they belong to a distinct subcluster within the sensu stricto.
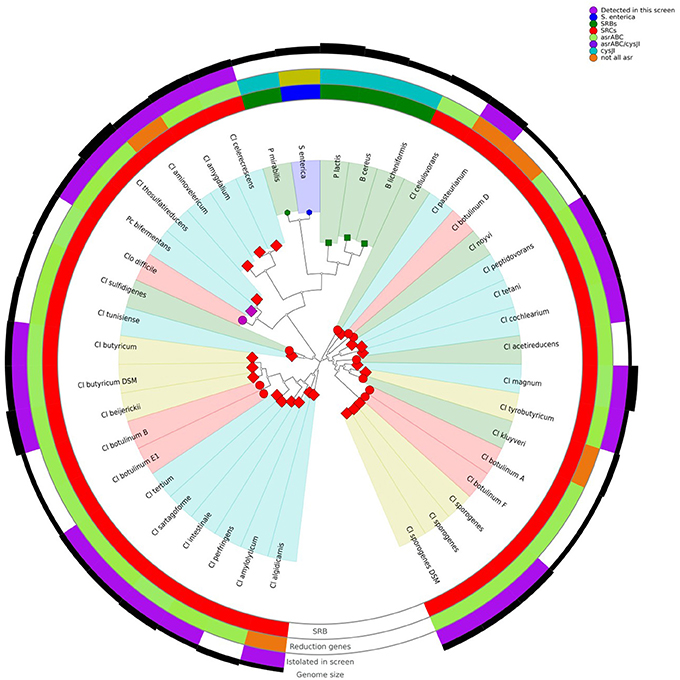
Figure 1. Phylophlan tree of SRBs, including those isolated during this screen, and presence/absence of specific sulphite reducing genes.
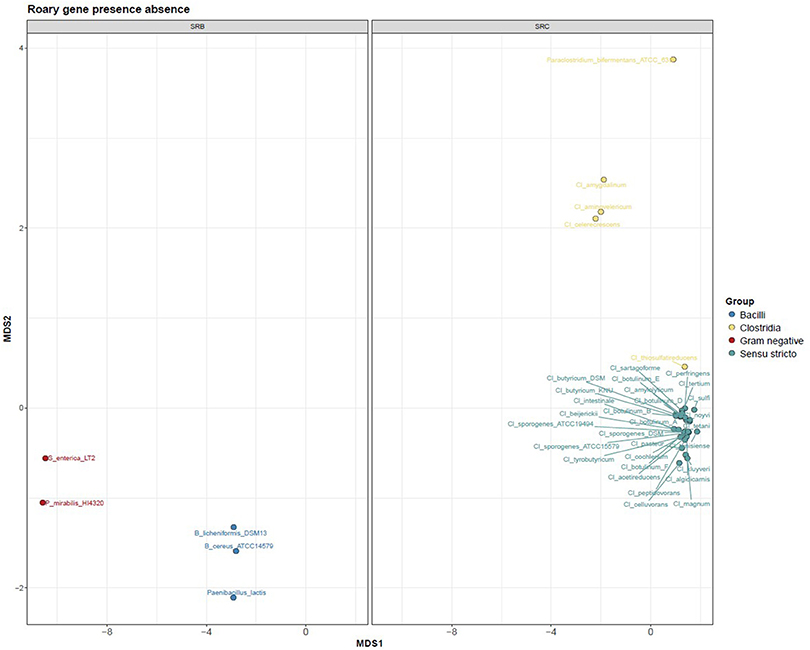
Figure 2. Bray Curtis PCoA profile depicting the dissimilarity of SRB genomes (faceted based on phylogeny).
Both the Phylophlan tree and the MDS plot highlight the diversity of sulphite reducing microbes of interest to the dairy industry. While a great number of bacteria can reduce sulphite to sulphite via different pathways (Dahl et al., 2008), it would appear from these analyses that it is only bacteria which utilize the asrABC or the cysIJ operons which give a positive test for the SRC assay employed by dairy producers. More specifically, the clostridia that utilize the asrABC sulphite reduction pathway are of most concern as they include pathogenic and spoilage-associated bacteria belonging to the genus Clostridium. These results highlight the heterogeneity that exists within the Clostridia. While this has already been shown from the context of the 16S rRNA gene sequence (Wiegel et al., 2006), whole genome-wide heterogeneity has until now been examined for this Order of bacteria. Although many Clostridium spp. have been reclassified and placed with new or existing genera (Lawson et al., 2016), there is still an issue with Clostridium nomenclature. For instance, C. aminovelericum and C. celecrescens belong to Cluster III of the clostridia (Wiegel et al., 2006), while C. amygdalinum belongs to clostridia Cluster XIV based on its phylogeny and high GC content (Parshina et al., 2003). The SRC phenotype is distributed across this heterogeneous group of bacteria.
Sulphite Reducing Protein Homology in Dairy-Associated SRBs
While the previous section examined the phylogeny of the SRC and SRB groups, this section details the homology associated with the proteins responsible for these phenotypes. Annotated complete sulphite reducing gene clusters for all the SRBs in the database can be seen in Figure S1B. Figure 3A depicts the BLASTp bit-score results for asrABC queries for all of the genomes in the constructed SRB database; the bit score is used to highlight proteins that are similar. The asrA protein sequence is present at a high degree of homology in the majority of Clostridium spp. and at a lower degree of homology in C. celecresens. Furthermore, there were no BLASTp hits for the asrA query for C. acetireducens, C. algicarnis, C. aminovelericum, C. botulinum D, C. kluyveri, C. noyvi, and C. pasteurianum. The asrB protein sequence was present in the all of the Clostridium genomes in the database (Figure S2). However, levels of homology found in C. aetireducens, C. algicarnis, C. aminovelericum, C. botulinum D, C. kluyveri, C. noyvi, and C. pasteurianum for this query were much lower than that within other Clostridium genomes. Similarly, for the asrC protein, the bit-scores for this query were again low for C. algicarnis, C. aminovelericum, C. botulinum D, C. kluyveri, and C. noyvi and no corresponding gene was found in the C. acetireducens and C. pasteurianum genomes. Furthermore, this sulphite reducing operon is present at a high degree of homology in sensu stricto Clostridium spp. Interestingly, C. acetireducens, C. kluyveri, C. algicarnis, C. noyvi, and C. pasteurianum, which are members of the sensu stricto, did not have the full operon based on these results and highlights that not all asrABC genes are necessary to confer this phenotype.
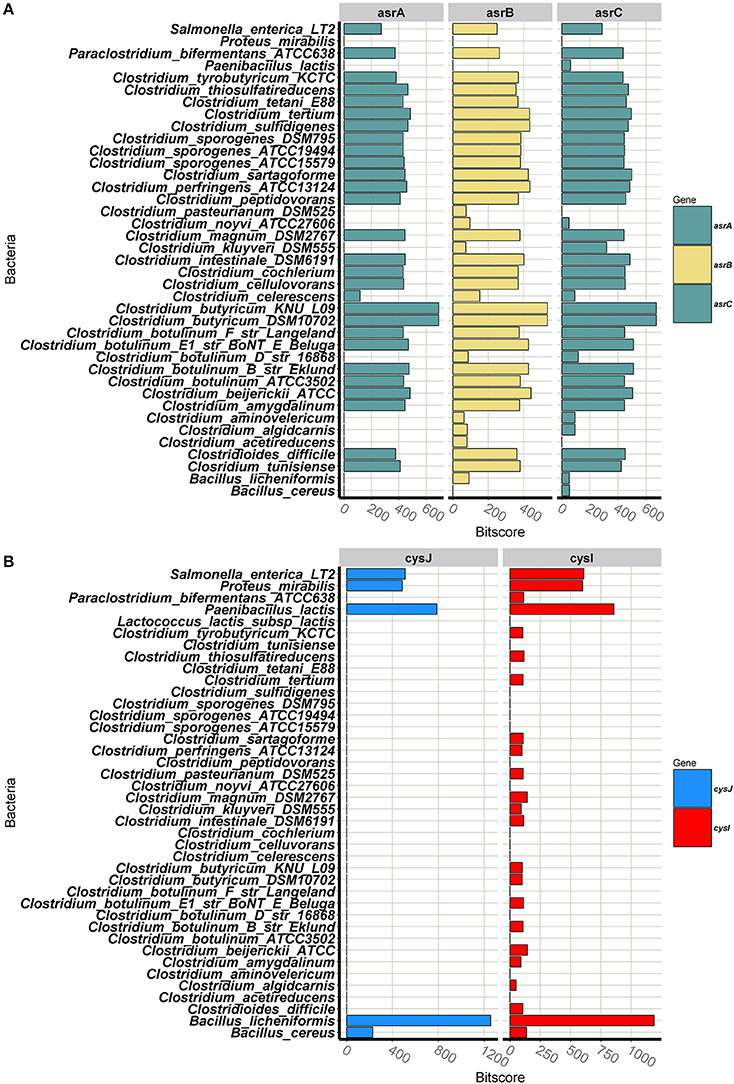
Figure 3. (A) Bar plot depicting bit-scores of BlastP query hits for asrABC from C. butyricum, and fnt from C. perfringes. (B) Bar plot depicting bit-scores of BlastP query hits for cysJI from B. licheniformis.
The bar plot in Figure 3B shows the BLASTp results for the cysJI queries for all genomes in the constructed SRB database. For the dissimilatory sulphite reducing pathway involving cysJI, the BLASTp bit-scores indicate that the cysJ gene is only present in B. licheniformis, B. cereus, P. lactis, P. mirabilis, and S. enterica and not in any of the other SRB genomes in the constructed database (Figure 3B). For the BLASTp with the cysI query, again the highest homology is shared with B. licheniformis, P. lactis, P. mirabilis, S. enteria and to a lesser extent B. cereus. The presence of this dissimilatory sulphite reductase gene cluster in these species is consistent with what is reported in the literature (Huang and Barrett, 1991).
The results from the BLASTp queries of the sulphite reducing genes of Clostridium prompted further examination of conserved amino acid domains with each amino acid sequence in this cluster. Conserved domains could act as targets for a nucleic acid-based detection assay for SRCs as an alternative to the non-specific agar-based approach. It was observed that asrA and C genes contain areas with conserved cysteine motifs. These 4Fe-4S clusters have been observed in asrA and C in Salmonella previously (Huang and Barrett, 1991). They have 4 conserved cysteine residues, with a proline toward the C terminus end of the domain. Amino acid sequence alignments can be seen in (Figure S3). The asrC protein also contains a siroheme binding site which is annotated in indigo (Figure S5). The asrB gene is involved in nucleotide binding (Ostrowski et al., 1989) (Figure S4). These alignments show the conservation in the functional regions of these genes. While similar functional domains might exist in other sulphite reducing bacteria using alternative pathways to the asrABC mediated reduction, the conserved proline appears to be a unique feature in the asr 4Fe-4S clusters. To verify that these conserved domains do not exist on other dairy associated SRBs, we examined the dissimilatory sulphite reducing genes in other SRBs from this analysis. The cysJI operon was also examined for conserved functional domains. The alignment for the alpha-subunit cysJ is shown in (Figure S6). The beta-subunit cysI contains a similar 4Fe-4S cluster to that in asrA and C (Figure S7). It is interesting to note that this sulphite binding cluster does not contain the conserved proline which is a feature of asr 4Fe-4S clusters. This shows that differences exist not only in the proteins used by these SRBs to reduce sulphite but also in the functional domains within this proteins compared to SRCs.
Conclusion
Here, the extent to which the agar-based SRC assay fails to distinguish between SRCs and SRBs that are facultative anaerobes was the focus of an extensive investigation. It is apparent that there is a need for a more rapid assay with increased discriminatory power to distinguish between SRCs and the wider group of SRB. The failure of the conventionally applied culture based method to differentiate between Clostridia and other SRBs such as Bacillus and Proteus could potentially be overcome by applying a PCR based assay which targets the genetic basis for this phenotype in different microbes. Our genome-wide phylogenetic comparison of the dairy-associated SRB phenotype has shown the diversity that exists within this group of microbes. In addition to the noted distribution of this phenotype across Gram positive and negative bacteria, this phenotype is observed throughout the Order Clostridia, with isolates from the sensu stricto, Lachnospiraceae and Cluster XIV of the Clostridia all producing this phenotype. Furthermore, we have carried out a genomic characterization of the SRBs of interest to the dairy industry, with specific focus on Clostridia. This has highlighted the heterogeneity that exists within the SRC phenotype. The wider SRB phenotype can be divided into two further phenotypes based on each isolate's phylogeny and the pathway (asrABC or cysJI) they utilize to produce the sulphite reducing phenotype. While asrABC mediated sulphite reduction has been studied in S. enterica and C. difficile, it has not been previously examined in the context of SRC phenotype in the dairy industry. Here, we have carried out an in-silico screen for the genes of this operon in dairy-associated SRBs and have provided more clarity to what defines a SRC is on the basis of the presence or absence of the asrABC operon.
Author Contributions
PC and PO developed the concept with subsequent input from CD. CD carried out the research with input from PO and PC. CD drafted and edited the manuscript. Revised and edited by PO and PC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Dr. Fiona Crispie and Ms. Laura Finnegan of Teagasc DNA Sequencing Platform for their contributions to DNA sequencing. Thanks also to Dr. Raul Cabrera-Rubio and Mr. Aaron Walsh of the Food Bioscience Department at Teagasc Food Research Centre, Moorepark, and Mr. Hugh Harris of the School of Microbiology, University College Cork for discussions relating to whole genome sequencing and analysis. The project was funded through a Teagasc Walsh Fellowship (2013030) to CD and internal Teagasc funding (RMIS6364) to PC.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01507/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C., and Segata, N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029
Barash, J. R., Hsia, J. K., and Arnon, S. S. (2010). Presence of soil-dwelling clostridia in commercial powdered infant formulas. J. Pediatr. 156, 402–408. doi: 10.1016/j.jpeds.2009.09.072
Bassi, D., Puglisi, E., and Cocconcelli, P. S. (2015). Understanding the bacterial communities of hard cheese with blowing defect. Food Microbiol. 52, 106–118. doi: 10.1016/j.fm.2015.07.004
Bennett, S. D., Walsh, K. A., and Gould, L. H. (2013). Foodborne disease outbreaks caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus—United States, 1998–2008. Clin. Infect. Dis. 57, 425–433. doi: 10.1093/cid/cit244
Bermúdez, J., González, M. J., Olivera, J. A., Burgueño, J. A., Juliano, P., Fox, E. M., et al. (2016). Seasonal occurrence and molecular diversity of clostridia species spores along cheesemaking streams of 5 commercial dairy plants. J. Dairy Sci. 99, 3358–3366. doi: 10.3168/jds.2015-10079
Brook, I. (1995). Clostridial infection in children. J. Med. Microbiol. 42, 78–82. doi: 10.1099/00222615-42-2-78
Cocolin, L., Innocente, N., Biasutti, M., and Comi, G. (2004). The late blowing in cheese: a new molecular approach based on PCR and DGGE to study the microbial ecology of the alteration process. Int. J. Food Microbiol. 90, 83–91. doi: 10.1016/S0168-1605(03)00296-4
Czyzewski, B. K., and Wang, D. N. (2012). Identification and characterization of a bacterial hydrosulphide ion channel. Nature 483, 494–497. doi: 10.1038/nature10881
Doyle, C. J., Gleeson, D., Jordan, K., Beresford, T. P., Ross, R. P., Fitzgerald, G. F., et al. (2015). Anaerobic sporeformers and their significance with respect to milk and dairy products. Int. J. Food Microbiol. 197, 77–87. doi: 10.1016/j.ijfoodmicro.2014.12.022
Doyle, M. E., and Glass, K. (2013). Spores of Clostridium botulinum in Dried Dairy Products. Madison, WI: Food Research Institute.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fischer, M., Zhu, S., and de Ree, E. (2012). Culture media for the detection and enumeration of clostridia in food. Handb. Cult. Media Food Water Microbiol. 66–89. doi: 10.1039/9781847551450-00066
Guillouard, I., Auger, S., Hullo, M. F., Chetouani, F., Danchin, A., and Martin-Verstraete, I. (2002). Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184, 4681–4689. doi: 10.1128/JB.184.17.4681-4689.2002
Gupta, T. B., and Brightwell, G. (2017). Farm level survey of spore-forming bacteria on four dairy farms in the Waikato region of New Zealand. Microbiologyopen 6:e00457. doi: 10.1002/mbo3.457
Harmon, S. M., Kautter, D. A., and Peeler, J. T. (1971). Improved medium for enumeration of Clostridium perfringens. Appl. Microbiol. 22, 688–692.
Huang, C. J., and Barrett, E. L. (1991). Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J. Bacteriol. 173, 1544–1553. doi: 10.1128/jb.173.4.1544-1553.1991
ICMSF (2014). Usefulness of Testing for Clostridium botulinum in Powdered Infant Formula and Dairy-based Ingredients for Infant Formula. ICMSF.
Ivy, R. A., Ranieri, M. L., Martin, N. H., den Bakker, H. C., Xavier, B. M., Wiedmann, M., et al. (2012). Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl. Environ. Microbiol. 78, 1853–1864. doi: 10.1128/AEM.06536-11
Kawabata, N. (1980). Studies on the sulfite reduction test for clostridia. Microbiol. Immunol. 24, 271–279. doi: 10.1111/j.1348-0421.1980.tb02830.x
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Ludwig, W., Schleifer, K. H., and Whitman, W. B. (2009). “Revised road map to the phylumFirmicutes,” in Bergey's Manual® of Systematic Bacteriology, eds P. Vos, G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, K.-H. Schleifer, and W. Whitman (New York, NY: Springer), 1–13.
McAuley, C. M., McMillan, K., Moore, S. C., Fegan, N., and Fox, E. M. (2014). Prevalence and characterization of foodborne pathogens from Australian dairy farm environments. J. Dairy Sci. 97, 402–412. doi: 10.3168/jds.2014-8735
McHugh, A. J., Feehily, C., Hill, C., and Cotter, P. D. (2017). Detection and enumeration of spore-forming bacteria in powdered dairy products. Front. Microbiol. 8:109. doi: 10.3389/fmicb.2017.00109
Miller, R. A., Kent, D. J., Watterson, M. J., Boor, K. J., Martin, N. H., and Wiedmann, M. (2015). Spore populations among bulk tank raw milk and dairy powders are significantly different. J. Dairy Sci. 98, 8492–8504. doi: 10.3168/jds.2015-9943
Ostrowski, J., Barber, M. J., Rueger, D. E., Miller, B. E., Siegel, L. M., and Kredich, N. M. (1989). Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli. Physicochemical and catalytic properties, amino acid sequence deduced from DNA sequence of cysJ, and comparison with NADPH-cytochrome P-450 reductase. J. Biol. Chem. 264, 15796–15808.
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Parshina, S. N., Kleerebezem, R., Sanz, J. L., Lettinga, G., Nozhevnikova, A. N., Kostrikina, N. A., et al. (2003). Soehngenia saccharolytica gen. nov., sp. nov. and Clostridium amygdalinum DPC 7176 sp. nov., two novel anaerobic, benzaldehyde-converting bacteria. Int. J. Syst. Evol. Microbiol. 53, 1791–1799. doi: 10.1099/ijs.0.02668-0
Peng, Y., Leung, H. C., Yiu, S. M., and Chin, F. Y. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Sadiq, F. A., Li, Y., Liu, T., Flint, S., Zhang, G., Yuan, L., et al. (2016). The heat resistance and spoilage potential of aerobic mesophilic and thermophilic spore forming bacteria isolated from Chinese milk powders. Int. J. Food Microbiol. 238, 193–201. doi: 10.1016/j.ijfoodmicro.2016.09.009
Sallam, A., and Steinbüchel, A. (2009). Clostridium sulfidigenes sp. nov., a mesophilic, proteolytic, thiosulfate-and sulfur-reducing bacterium isolated from pond sediment. Int. J. Syst. Evol. Microbiol. 59, 1661–1665. doi: 10.1099/ijs.0.004986-0
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Segata, N., Börnigen, D., Morgan, X. C., and Huttenhower, C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/ncomms3304
Simpson, P. J., Stanton, C., Fitzgerald, G. F., and Ross, R. P. (2003). Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185, 2571–2581. doi: 10.1128/JB.185.8.2571-2581.2003
Standards, I. O. O. (2003). Microbiology of Food and Animal Feeding Stuffs - Horizontal Method for the Enumeration of Sulfite-Reducing Bacteria Growing Under Anaerobic Conditions. ISO 2003.
Standards, I. O. O. (2004). Microbiology of Food And Animal Feeding Stuffs – Horizontal Method for the Enumeration of Clostridium perfringens – Colony-count Technique. ISO 2004.
Sugiyama, H. (1951). Studies on factors affecting the heat resistance of spores of Clostridium botulinum. J. Bacteriol. 62, 81–96.
Thabet, O. B. D., Fardeau, M.-L., Joulian, C., Thomas, P., Hamdi, M., Garcia, J.-L., et al. (2004). Clostridium tunisiense sp. nov., a new proteolytic, sulfur-reducing bacterium isolated from an olive mill wastewater contaminated by phosphogypse. Anaerobe 10, 185–190. doi: 10.1016/j.anaerobe.2004.04.002
Turnbull, A. L., and Surette, M. G. (2008). L-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar typhimurium. Microbiology 154, 3410–3419. doi: 10.1099/mic.0.2008/020347-0
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Weenk, G., Fitzmaurice, E., and Mossel, D. (1991). Selective enumeration of spores of Clostridium species in dried foods. J. Appl. Microbiol. 70, 135–143.
Weenk, G., Van den Brink, J., Struijk, C., and Mossel, D. (1995). Modified methods for the enumeration of spores of mesophilic Clostridium species in dried foods. Int. J. Food Microbiol. 27, 185–200. doi: 10.1016/0168-1605(94)00164-2
Keywords: sulphite reducing bacteria, SRCs, sulphite reducing clostridia, genomics, food microbes
Citation: Doyle CJ, O'Toole PW and Cotter PD (2018) Genomic Characterization of Sulphite Reducing Bacteria Isolated From the Dairy Production Chain. Front. Microbiol. 9:1507. doi: 10.3389/fmicb.2018.01507
Received: 28 February 2018; Accepted: 18 June 2018;
Published: 05 July 2018.
Edited by:
Rosalba Lanciotti, Università degli Studi di Bologna, ItalyReviewed by:
Jorge Reinheimer, National University of the Littoral, ArgentinaMaria Schirone, Università di Teramo, Italy
Copyright © 2018 Doyle, O'Toole and Cotter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul D. Cotter, cGF1bC5jb3R0ZXJAdGVhZ2FzYy5pZQ==
 Conor J. Doyle
Conor J. Doyle Paul W. O'Toole2,3
Paul W. O'Toole2,3 Paul D. Cotter
Paul D. Cotter