
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 July 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01436
This article is part of the Research Topic New Insights & Updates on The Molecular Epidemiology and Antimicrobial Resistance of MRSA in Humans in the Whole-Genome Sequencing Era View all 10 articles
 Stefan Monecke1,2,3*
Stefan Monecke1,2,3* Peter Slickers1,2
Peter Slickers1,2 Darius Gawlik1,2
Darius Gawlik1,2 Elke Müller1,2
Elke Müller1,2 Annett Reissig1,2
Annett Reissig1,2 Antje Ruppelt-Lorz3
Antje Ruppelt-Lorz3 Patrick E. Akpaka4
Patrick E. Akpaka4 Dirk Bandt5
Dirk Bandt5 Michele Bes6
Michele Bes6 Samar S. Boswihi7
Samar S. Boswihi7 David C. Coleman8
David C. Coleman8 Geoffrey W. Coombs9
Geoffrey W. Coombs9 Olivia S. Dorneanu10
Olivia S. Dorneanu10 Vladimir V. Gostev11
Vladimir V. Gostev11 Margaret Ip12
Margaret Ip12 Bushra Jamil13,14
Bushra Jamil13,14 Lutz Jatzwauk15
Lutz Jatzwauk15 Marco Narvaez15
Marco Narvaez15 Rashida Roberts4
Rashida Roberts4 Abiola Senok16
Abiola Senok16 Anna C. Shore8
Anna C. Shore8 Sergey V. Sidorenko12
Sergey V. Sidorenko12 Leila Skakni17
Leila Skakni17 Ali M. Somily18
Ali M. Somily18 Muhammad Ali Syed19
Muhammad Ali Syed19 Alexander Thürmer3
Alexander Thürmer3 Edet E. Udo8
Edet E. Udo8 Teodora Vremerǎ10
Teodora Vremerǎ10 Jeannete Zurita20,21
Jeannete Zurita20,21 Ralf Ehricht1,2
Ralf Ehricht1,2ST239-MRSA-III is probably the oldest truly pandemic MRSA strain, circulating in many countries since the 1970s. It is still frequently isolated in some parts of the world although it has been replaced by other MRSA strains in, e.g., most of Europe. Previous genotyping work (Harris et al., 2010; Castillo-Ramírez et al., 2012) suggested a split in geographically defined clades. In the present study, a collection of 184 ST239-MRSA-III isolates, mainly from countries not covered by the previous studies were characterized using two DNA microarrays (i) targeting an extensive range of typing markers, virulence and resistance genes and (ii) a SCCmec subtyping array. Thirty additional isolates underwent whole-genome sequencing (WGS) and, together with published WGS data for 215 ST239-MRSA-III isolates, were analyzed using in-silico analysis for comparison with the microarray data and with special regard to variation within SCCmec elements. This permitted the assignment of isolates and sequences to 39 different SCCmec III subtypes, and to three major and several minor clades. One clade, characterized by the integration of a transposon into nsaB and by the loss of fnbB and splE was detected among isolates from Turkey, Romania and other Eastern European countries, Russia, Pakistan, and (mainly Northern) China. Another clade, harboring sasX/sesI is widespread in South-East Asia including China/Hong Kong, and surprisingly also in Trinidad & Tobago. A third, related, but sasX/sesI-negative clade occurs not only in Latin America but also in Russia and in the Middle East from where it apparently originated and from where it also was transferred to Ireland. Minor clades exist or existed in Western Europe and Greece, in Portugal, in Australia and New Zealand as well as in the Middle East. Isolates from countries where this strain is not epidemic (such as Germany) frequently are associated with foreign travel and/or hospitalization abroad. The wide dissemination of this strain and the fact that it was able to cause a hospital-borne pandemic that lasted nearly 50 years emphasizes the need for stringent infection prevention and control and admission screening.
Staphylococcus aureus is a bacterial species that colonizes the skin and mucous membranes of a high percentage of the human population (van Belkum et al., 2009) and several animal species. It can cause localized infections, such as skin and soft tissue infections (SSTIs), bone, joint and implant infections or pneumonia as well as sepsis and toxicoses including toxic shock syndrome. Resistance toward antibiotics in S. aureus is a highly relevant issue. Methicillin resistance and resistance to most beta-lactams is due to the production of modified penicillin-binding proteins encoded by mec genes. The mecA/mecC genes are located on large, complex and potentially mobile staphylococcal cassette chromosome (SCCmec) elements (while mecB was observed on a plasmid; Becker et al., 2018). SCCmec elements additionally encode regulatory elements and, variably, genes encoding resistance to other antimicrobials, such as aminoglycosides, macrolides, tetracyclines, fusidic acid and to heavy metals (Oliveira et al., 2000; Ito et al., 2001). The comparatively older SCCmec types I, II, and III are typically restricted to MRSA strains involved in healthcare-associated infections (HCA-MRSA).
The HCA-MRSA, sequence type (ST) 239 MRSA, as designated by multilocus sequence typing (MLST), is of special interest. As ST240 and ST241 are single locus variants of ST239 (which differ only by mutations in MLST marker genes pta or yqil), these STs are here discussed together as clonal complex (CC) 239. CC239 harboring SCCmec type III have been designated various names in different geographic regions including “Wiener Epidemiestamm” (Vienna Epidemic Strain), the Hungarian Clone, UK-EMRSA−1,−4,−7,−9, or−11, Irish Phenotype III, Irish AR01,−09,−44, and−23, the Brazilian Clone, Australian Epidemic MRSA−2 and−3 as well as Canadian MRSA−3 or−6. CC239-MRSA-III is probably the oldest truly pandemic MRSA strain. In contrast to other early MRSA strains, it is still common and widespread, at least in some parts of the world.
CC239-MRSA-III has been reported in many European countries including the UK (Edgeworth et al., 2007), Ireland (Humphreys et al., 1990; Shore et al., 2005), Spain (Cuevas et al., 2007), Portugal (Smyth et al., 2010), Italy (Campanile et al., 2009), Malta (Scicluna et al., 2010), Croatia (Budimir et al., 2009), Germany (Witte et al., 1997; Albrecht et al., 2011), Austria (Krziwanek et al., 2008), the Czech Republic (Melter et al., 2003), Hungary (Conceicao et al., 2007), and Greece (Aires de Sousa et al., 2003a). It is also frequently observed in Romania (Cirlan et al., 2005; Chen et al., 2010; Monecke et al., 2014a) and Russia (Afanas'ev et al., 2010; Baranovich et al., 2010; Yamamoto et al., 2012; Gostev et al., 2017). CC239-MRSA-III is common to abundant in Mediterranean and Middle Eastern countries such as Greece (Aires de Sousa et al., 2003a), Turkey (Alp et al., 2009; Tekeli et al., 2016), Egypt (El-baz et al., 2017), Morocco, Tunisia, Algeria (Abdulgader et al., 2015), Iran (Fatholahzadeh et al., 2009), Saudi Arabia (Cirlan et al., 2005; Monecke et al., 2012c; Senok et al., 2016), Abu Dhabi (Weber et al., 2010), and Kuwait (Boswihi et al., 2016). The strain's presence has been reported in China (Chen et al., 2014a,b) including Hong Kong (Ip et al., 2005), Taiwan (Aires de Sousa et al., 2003b; Takano et al., 2007), Singapore (Hsu et al., 2007), Malaysia (Ghaznavi-Rad et al., 2010), Mongolia (Orth et al., 2006), Pakistan (Shabir et al., 2010; Zafar et al., 2011; Arfat, 2013; Jamil et al., 2017), India (D'Souza et al., 2010; Neetu and Murugan, 2016), South Korea (Cha et al., 2005; Peck et al., 2009), Laos (Yeap et al., 2017), and Thailand (Smyth et al., 2010). There are reports from several African countries including Ghana, Kenya, Niger, Nigeria, Senegal, and South Africa (Jansen van Rensburg et al., 2011; Abdulgader et al., 2015). In Eastern Australia and New Zealand it was a common cause of HCA infection, and it has been reported in association with large MRSA outbreaks (Coombs et al., 2004; Howden et al., 2010). In the western hemisphere, CC239-MRSA-III has been reported from the United States (Schaefler et al., 1981), Brazil (Vivoni et al., 2006), Paraguay (Mayor et al., 2007), Ecuador (Zurita et al., 2016), and Trinidad & Tobago where it recently was still the predominant MRSA strain (Akpaka et al., 2007; Monecke et al., 2012b, 2014b).
In terms of genetic structure, ST239 is an interesting clone, with six of the seven MLST housekeeping genes being identical to ST8 (Robinson and Enright, 2004a,b). However ST239 as well as ST240 and ST241 harbor a very different arcC-allele (arcC-2, rather than arcC-3). The difference in the arcC-allele and the presence of some other features that differentiate ST239 from canonical ST8 (including the presence of the collagen adhesion gene cna, capsule type 5 and RIDOM spa types t030 or t037), indicate the integration of a large fragment of genomic DNA of clonal complex (CC) 30 origin into a CC8 chromosome (Robinson and Enright, 2004b; Holden et al., 2009). The fragment consists of ~635,000 base pairs, which is ~20% of a S. aureus genome. The mechanism of integration is yet not known.
Another interesting recent observation has been the discovery of sasX/sesI in CC239-MRSA, a virulence factor thought to have a key role in nasal colonization, pathogenesis of lung disease, and abscess formation (Li et al., 2012). The sasX gene is located on a 127 kb lysogenic prophage phiSPbeta (Li et al., 2012) and it encodes the surface-anchored protein X, an LPxTG motif surface-anchored protein, and does not have orthologues in any of the other sequenced S. aureus genomes. A highly similar gene, sesI, is present in the S. epidermidis phiSPbeta region and has also been identified in other coagulase-negative staphylococci such as S. capitis (GenBank JGYJ) and S. cohnii (GenBank LATU and LATV).
With the rise of Next Generation Sequencing (NGS) technologies, Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012) sequenced a large collection of CC239-MRSA-III from very diverse geographic origins. They suggested a phylogenetic framework in which isolates of CC239-MRSA-III clustered in several major “clades” and a couple of isolated branches. The clades are largely associated with geographic background and thus were referred to as “European”, “Latin American”, “Turkish”, and “Asian” clades.
In the present study, a collection of CC239-MRSA-III isolates was characterized, primarily using previously published DNA microarray technology targeting typing markers, virulence and resistance genes and SCCmec subtypes (Monecke et al., 2008b, 2011, 2016). Published whole-genome sequence data, in particular those by Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012), were re-analyzed with regard to the presence or absence of the marker genes as used experimentally for array hybridization and with regard to variation within SCCmec elements. A comparison between our strain collection, mainly from countries that were not covered by the Harris and Castillo-Ramírez' work, to published genome sequences was then performed in order to see if and how they fit into their proposed phylogenetic framework and to look for epidemiological connections and suitable marker genes.
In total, 214 clinical or screening isolates were included in the present study. These isolates originated from hospitals in Ireland, Germany, Romania, Kuwait, Saudi Arabia, Russia, Pakistan, China/Hong Kong, Australia, Trinidad&Tobago, and Ecuador as well as some reference strains (see Supplemental Table 1 and below). Some of the isolates were a convenience sample from previous studies (Monecke et al., 2008a, 2011, 2012b,c, 2014a,b, 2016; Albrecht et al., 2011; Boswihi et al., 2016; Senok et al., 2016; Zurita et al., 2016; Gostev et al., 2017; Jamil et al., 2017). Others came from ongoing routine diagnostics, outbreak investigations or typing tasks performed by the authors and have not been published previously. Only one isolate per patient was included. Isolates were stored frozen using cryobank tubes (Microbank, Pro-Lab Diagnostics, Richmond Hill, Canada) at −80°C. Isolates were routinely cultured on Columbia blood agar plates, and DNA preparation was performed as previously described (Monecke et al., 2008b, 2011).
Two microarrays were used in the study, and were applied to 184 of the isolates investigated herein (another 30 were directly subjected to sequencing; see below). Both arrays have previously been described including probe and primer sequences, details of DNA extraction, labeling, amplification, hybridization protocols as well as data analysis and interpretation. The first microarray (Monecke et al., 2008b, 2011) detects genes associated with antibiotic resistance and virulence as well as a multitude of genes that can be used for typing purposes, such as genes related to agr or capsule types, set/ssl genes and genes coding for adhesion factors. In addition to the detection of individual genes, the array also allows the assignment of isolates to MLST CCs, to known epidemic strains and to SCCmec types. The second microarray (Monecke et al., 2016) was designed to subtype SCCmec elements. It also includes probes for sasX/sesI and for heavy metal resistance genes. Furthermore, it included a set of probes termed “SCCterm” followed by a number (see Table 1) which were designed to recognize intergenic regions alternative to dcs, between orfX and the first codon on the SCCmec element.
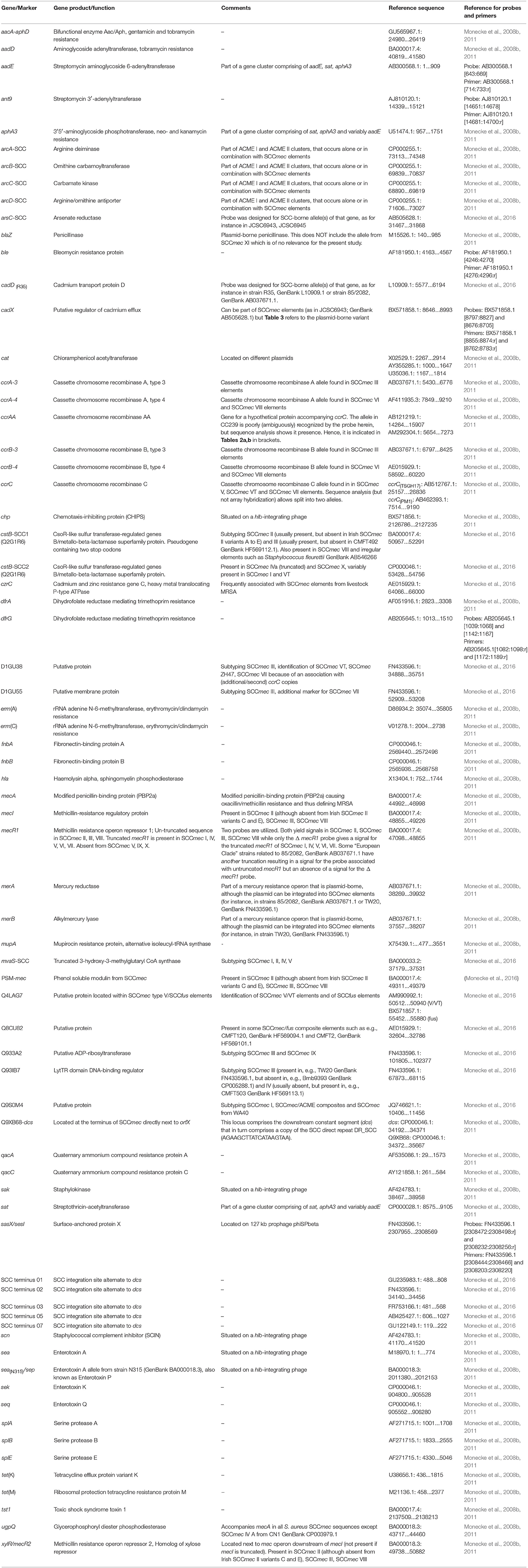
Table 1. SCCmec-associated genes and other relevant markers that were targeted by probes used for this study.
All markers of relevance for SCCmec subtyping are listed in the Supplemental Table 1. Markers that were detected among isolates in the present study are listed, and presented in more detail, in Table 1.
A third assay was used to identify and categorize alleles and variants of the mecA/C gene as previously described (Monecke et al., 2012a). While there are a multitude of mecA variants (also named mecA1) in staphylococcal species other than S. aureus, only four different mecA/C alleles are of relevance in S. aureus/MRSA that also differ in amino acid sequences encoded, i.e., (named with respect to representative GenBank entries), mecA(CP000046) (as in the CC8-MRSA-I strain COL), mecA(BA000018) (as in the CC5-MRSA-II strain N315), mecA(GQ902038) (as in the CC398-MRSA-VT strain UMCG-M4) and mecC (GenBank NG_047955.1). This array was applied to 30 isolates representing at least one isolate per SCCmec subtype.
The guidelines of the International Working Group (IWG-SCC, 2009) were used for the assignment of Roman numerals to SCCmec types defined by the class of mec gene complex and type of cassette chromosome recombinase (ccr) genes. As proposed by Shore and Coleman (2013) we named elements lacking ccr recombinase genes “pseudoSCCmec elements.” Composite elements are indicated by listing relevant components in square brackets. Heavy metal resistance genotypes were described by adding chemical symbols rather than individual gene designations e.g., SCC [mec III+Cd/Hg+ccrC]).
Genes aadD, aacA-aphD, ant9, ble, and erm(A) as well as tet genes were not included in the analysis of the SCCmec III subtypes. Although these genes may be situated on SCCmec elements, they also may be found on plasmids or other mobile genetic elements at various locations [see below for erm(A) and ant9]. Neither array hybridization nor those NGS technologies that yield a high number of short contigs can provide reliable information on the actual localizations of genes, or whether a plasmid [such as pT181/tet(K)] was free or integrated into the genome. Consequently, we did not differentiate between SCCmec III and IIIA (Vandenesch et al., 2003).
In contrast, the mercury resistance operon was included into the analysis of the SCCmec III subtypes. In CC239 this operon is part of a composite SCC element, although it can indeed be found outside of SCC elements (e.g., GenBank: AB179623.1).
All previously sequenced variants were tagged with the designation of one reference strain in which they have been sequenced [e.g., the particular variant of SCCmec III from the strain TW20 is indicated SCC [mec III+Cd/Hg+ccrC] (TW20)]. If we were not able to identify a reference sequence to a given SCC hybridization pattern, we added “unknown” followed by the clonal complex(es) and, if there were several similar such elements, by chronologically assigned numbers [as in SCC [mec III+Cd+ccrC] (Unknown, ST239−3)].
The genomes of 30 CC239 isolates from Perth/Australia have been sequenced with Illumina MiSEQ. Sequencing libraries were prepared with the Nextera kit (Illumina).
Sequencing reads of 30 CC239 isolates from Perth/Australia as well as several read sets downloaded from NCBI Short read Archive (https://www.ncbi.nlm.nih.gov/sra/; see Table 4 and Supplemental Table 1) were assembled with SPAdes version 3.10.1 (Bankevich et al., 2012). No attempts were made to close gaps between contigs. Contigs shorter than 500 nt were excluded from further analysis.
To date, several thousand either partially or fully assembled genomes of S. aureus isolates are available in NCBI GenBank. Fully assembled genomes comprise one or several sequences representing complete replicons (the bacterial chromosome and a variable number of plasmids). Partially assembled sequences consist of a set of contigs. The contigs usually end in repeats and the sequencing reads do not comprise enough information to link contigs unambiguously. The number of contigs varies between about 10 and several hundred depending on the sequencing method, read length, fragment size, coverage depth and assembling strategy and settings. Partially assembled contigs are available in NCBI Genbank (http://www.ncbi.nlm.nih.gov/Traces/wgs/) with special accession numbers assigned which start with four letters followed by eight digits. The entire set of contigs is referred to by a accession number which has all digits set to zero (e.g., AICH00000000.1). For the sake of conciseness, we will refer to these four-letter codes as an unambiguous identifier of a specific genome here. To genomes which we have assembled from raw sequencing reads obtained from the Short Read Archive (https://www.ncbi.nlm.nih.gov/sra/), we refer to henceforth by the BioSample accession number (e.g., SAMEA1029552).
A total of 215 genome sequences of CC239 available in the NCBI database (Table 4 and Supplemental Table 1) as well as genome sequences of 30 previously unpublished Australian study isolates were subjected to an in silico analysis or “virtual hybridization” that allowed a direct comparison to array hybridization experiments (Tables 2a,b, 3 and Supplemental Table 1). Hybridization patterns were generated from complete or from partially assembled genomic sequences.
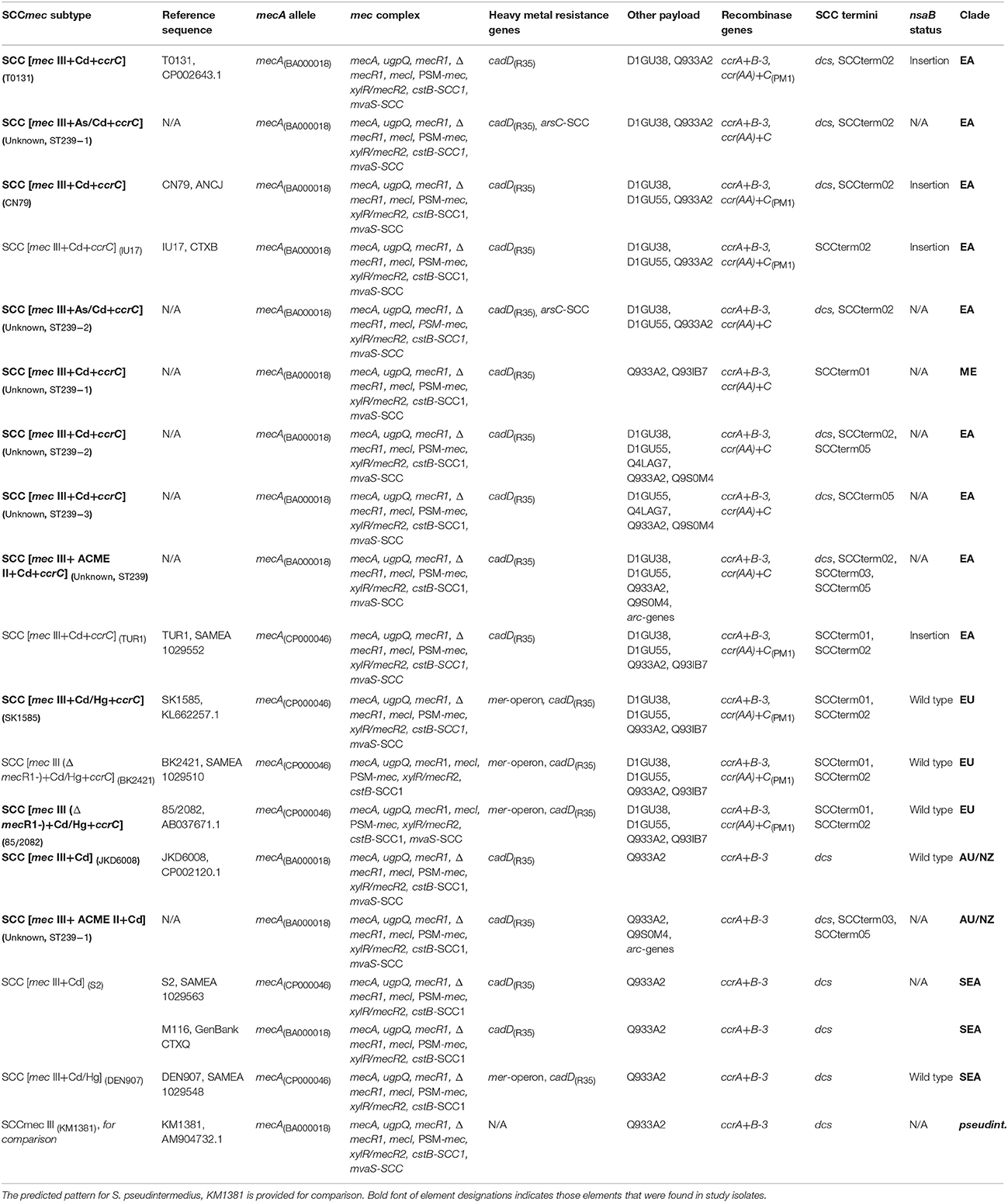
Table 2a. SCCmec-associated patterns observed in, or derived from sequences of, CC239-MRSA assigned to the “Eurasian” (EA), “European” (EU), “Australian/NZ” (AU/NZ) Clades, to the “unassigned Middle Eastern Strain“ (ME), or to the atypical cluster of “South-East Asian Clade” isolates (S2, DEN907; SEA).
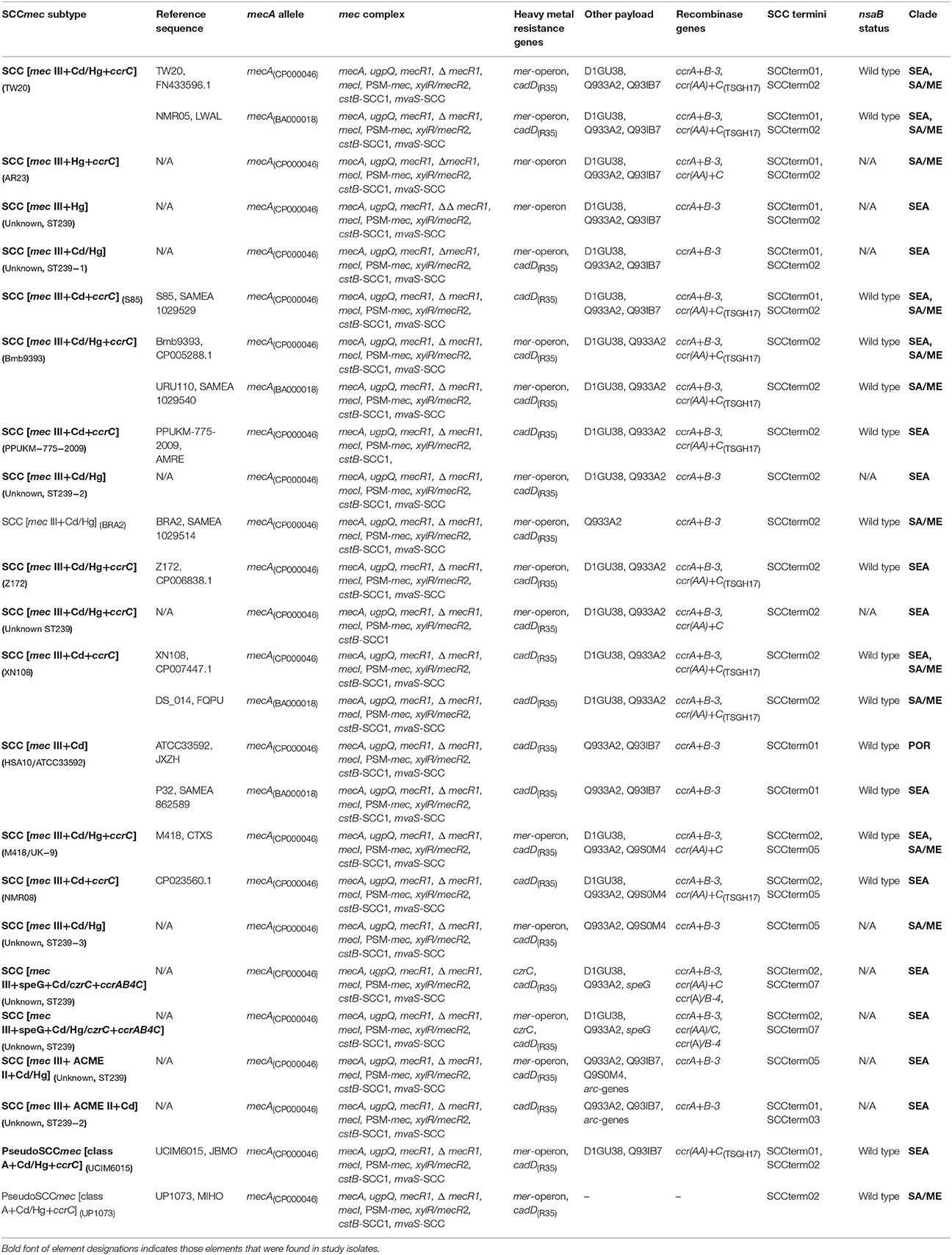
Table 2b. SCCmec-associated patterns observed in, or derived from sequences of, CC239-MRSA assigned to the “South-East Asian” (SEA), “South American/Middle Eastern” (SA/ME) and “Portuguese” (POR) clades.
Probe sequences were mapped on contigs using the program blastn (Camacho et al., 2009) from the NCBI blast+ suite and all sites were identified that matched the probe sequences with less than four mismatches. A signal value between 0 and 1 was assigned to each probe based on the actual number of mismatches derived from, and mimicking the normalized signals from a real hybridization experiment. A probe without mismatches was assigned signal intensity of 0.9; with 1 mismatch, a signal of 0.6; with two mismatches, a signal of 0.3; with three mismatches, a signal of 0.1. Probes with four—or more—mismatches were set as 0. These numerical values were then analyzed exactly as data from real hybridization experiments (Monecke et al., 2008b, 2011).
This approach has been developed and optimized based on real experiments performed with fully assembled strains (such as MSSA476, GenBank BX571857; N315, GenBank BA000018; COL, GenBank CP000046; MRSA252, GenBank BX571856; see also Monecke et al., 2016). For three strains (ATCC33592, UK-EMRSA-4, isolate Russia_0085) full genome sequences were available and we have done real as well as virtual hybridization experiments that were analyzed in parallel.
Some CC239 genomes comprise a site-specific insertion of a mobile element into the chromosomale genes nsaB (locus tag SATW20_27600) or yeeE (locus tag SAT0131_RS10920). Two query sequences with a size of 80 nt were used to evaluate assembled genomes for the presents of uninterrupted nasB and yeeE genes. The two query sequences were choosen to span the insertion sites (for nasB, FN433596.1[2933542:2933621:r] and for yeeE, CP002643.1[2151962:2152041:r]). These query sequences were mapped on all full genome sequences with blastn. If they did not match for their full length, we assumed that the target gene was interrupted.
Thirty-nine different variants of SCCmec III or SCCmec III-derived composite SCC elements or pseudoSCC-elements were observed in the 425 CC239-MRSA-III isolates and sequences. A description of these variants is provided in Tables 2a,b. Full profiles for individual isolates and sequences are shown in Supplemental Table 1.
Alleles of mecA in CC239-MRSA-III were assigned to two alleles matching the CC8-MRSA-I strain COL, CP000046 (among study isolates tested or sequenced, n = 21) and the CC5-MRSA-II strain N315, BA000018 (among study isolates, n = 38; one sequence not unambiguously assigned). When analyzing the binding sites of the probes used, the difference between the alleles is an “A” or, respectively, a “G” in position 737 (of the TW20 mecA sequence). Among the sequences and isolates investigated, mecA alleles largely correlate with the SCCmec subtypes and strains within CC239, i.e., a single mecA allele was found in association with each SCCmec subtype. However, there were five exceptions, i.e., SCCmec subtypes in which both mecA alleles were detected. This included the more common subtypes and strains (SCC [mec III+Cd/Hg+ccrC] (TW20), SCC [mec III+Cd/Hg+ccrC] (Bmb9393), SCC [mec III+Cd] (S2), SCC [mec III+Cd+ccrC] (XN108), SCC [mec III+Cd] (HSA10/ATCC33592)). This might suggest that the mecA sequence, and its allele assignment, is not a reliable phylogenetic marker but subject to random mutation (or to sequencing errors).
All SCCmec III elements from CC239 include a cadmium resistance operon for which cadD(R35) was used as a marker.
Fifteen SCCmec III elements in CC239 (“Eurasian” strains with ccrC being located elsewhere not included) were composite elements that additionally harbor the recombinase gene ccrC. Sequence analysis identified two different ccrC alleles (Tables 1, 2a,b) that could not be differentiated with the current set of probes. Genes accompanying ccrC are ccrAA (although the present allele yields usually only ambiguous signals with the probes used herein) and D1GU38. Nineteen SCCmec III elements in CC239 also include the mercury resistance operon; this is often but not always linked to ccrC.
Composite elements that include ACME II (that is, arc genes present but opp genes absent), an arsenic resistance operon, genes speG and czrC (zinc/cadmium resistance) were occasionally found (see Tables 2a,b). No isolates were identified that harbored composite elements involving ACME I (arc and opp genes), ACME III (opp genes only) or SCCfus (fusidic acid resistance, fusC).
The presence of dcs and a SCC terminus sequence or multiple SCC terminus sequences suggests the presence of composite elements. Sequence analyses has shown that SCC terminus sequences are not necessarily situated “terminally” toward orfX but can be found within a composite element demarking it components. For example in TW20, SCCterm02 (GenBank FN433596.1; positions 34,140 to 34,456) is situated toward orfX (positions 33,660 to 34,139) while SCCterm01 can be found between the region including the mercury resistance operon and the SCCmec element (positions 67,191 to 67,511). As described below, SCCterm02 can also be associated with integration of a transposon into the nsaB gene, i.e., at a distant position from orfX.
Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012) divided CC239-MRSA-III into several clades based on SNP analysis. We re-analyzed published sequences from this project, other previously published sequences as well as our own sequences and array hybridization patterns regarding presence of SCCmec III subtypes (see above), the gene sasX/sesI (see below), the enterotoxin genes sek and seq (representative for S. aureus pathogenicity island 3), certain resistance markers and regarding other conspicuous features such as spa types or deletions of individual genes.
Individual results as well as clade/strain assignments for each isolate and sequence are listed in the Supplemental Table 1. A list of strains and clades as well as of their genotypic features is provided in Table 3. Strain definitions are mainly based on SCCmec subtypes plus other notable features.

Table 3. Clades and Strains and their genotypic characterization, as identified in this study or when analyzing published genome sequences.
Harris and Castillo-Ramírez describe a distinct and rather homogenous “Turkish Clade” (Harris et al., 2010; Castillo-Ramírez et al., 2012). We found that this clade was not restricted to Turkey. It also included isolates and sequences (including T0131, CN79, 16K, 3HK and others) that originate from Eastern Europe and the Balkans, Russia, Pakistan as well as from China (mainly Northern China, but also Hong Kong). Thus we suggest renaming this clade “Eurasian Clade.”
Isolates and sequences assigned to this clade are characterized by the presence of mecA(BA000018) and dcs. Harris' strains TUR1 and TUR9 differ in this regard being an intermediate to the “European Clade” or a product of a horizontal gene transfer of another variant of a SCCmec III element.
Furthermore, “Eurasian Clade” strains are characterized by integration of a IS431 based transposon—carrying several genes that appear to origin from the SCCmec III element—into the nsaB gene which is located 140,000 nt away from the SCC element (T0131, GenBank CP002643: position 2,779,896 to 2,803,726). This transposon consists of ccrC, ccrAA, SCCterm02 and additional genes such as erm(A), ant9, hsdR2-WIS, D1GU60, A9UFT0, Q93IA1, A5INT3, Q9KX75, Q0P7G0, Q93IE0, Q3T2M7, Q4LAG3, D2N370, D1GU38, Q2FKL3, transposase genes and IS431 sequences as well as aacA-aphD (absent from T0131, but present in 16K; Yamamoto et al., 2012). This disruption of the nsaB gene was described first in isolates from Romania (Chen et al., 2010) and Russia (Yamamoto et al., 2012) but it can be detected in all “Eurasian Clade” sequences. A insertion into nsaB is also present in TUR1, SAMEA1029552 and TUR9, SAMEA985415 but due to fragmentation of the sequences into a high number of contigs, the gene content of their insertion cannot be reliably determined.
All other sequences and clades of CC239-MRSA-III present with an un-truncated, wildtype nsaB gene. This includes, despite their similarity to the “Eurasian Clade”, JKD6008 and related strains from Australia/New Zealand (see below). However, erm(A) and ant9 are not restricted to the insertion into nsaB. These two genes can frequently be found in CC239-MRSA strains without the nsaB insertion, where they are present in different localizations. In the previously published genome sequence Bmb9393, a transposon carrying these two genes disrupts radC (SABB_05268) while in TW20, this transposon is present twice, once integrated into radC and once co-localized with the SCCmec III element. In JKD6008, there are also two copies of this transposon, one within radC (SAA6008_01621) and one disrupting ywqG (SAA6008_00825). Therefore the detection of erm(A) and ant9 cannot be used as a surrogate marker for the identification of the “Eurasian Clade” but the insertion into nsaB can.
Other features of the “Eurasian Clade” include a predominance of RIDOM spa type t030 (with all isolates previously assigned to spa type t030 belonging to this clade; Gostev et al., 2017) or t632 (Moscow and Saint Petersburg), the uniform absence of the mercury resistance operon, the adhesion factor gene fnbB and the protease gene splE while splA and splB are present.
The basal “European Clade” (Harris et al., 2010) consists of isolates and sequences that share the SCC [mec III+Cd/Hg+ccrC] (SK1585) element and variants thereof from which some genes are fully (mvaS) or partially (mecR1) deleted. This particular SCCmec element is a composite SCCmec III/heavy metal resistance element that was first observed in a strain isolated in Australia as early as in 1973 (see Nimmo et al., 2015 and section Discussions).
The “European Clade” includes a cluster of homogenous sequences and isolates from, or with epidemiological connection to, Greece. These include Harris' Greek sequences (Harris et al., 2010), isolates from Saxony/Germany epidemiologically linked to Greece (see below and Albrecht et al., 2011) and the Greek reference strain from the Harmony collection (Greece 1_3680). It also includes two isolates from Morocco.
Some “European Clade” strains have a characteristic deletion of 166 nt (in 85/2082, GenBank AB037671.1; corresponding the region in TW20 of FN433596.1 [79030 to 79195]) in the mecR1 gene that results in the paradoxical observation that the probe associated with mecR1 yields a signal while the one for ΔmecR1 does not. These include genome sequences from Australia and the US (ANS46, LHH1, BK2421) as well as epidemic strains British UK-MRSA-01 and Irish AR01.
The sequences assigned by Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012) to the (South-East) “Asian Clade” all contain the sasX/sesI gene which was absent from all sequences not assigned to this clade. Consequently, sasX/sesI was used in the present study as an identifying marker for this clade. In all sasX/sesI-positive CC239 sequences analyzed, the sasX/sesI prophage is localized in the same position within the genome, splitting yeeE (FN433596.1; positions 2180899 to 2181684 downstream of the phage insertion, and 2308889 to 2309177 upstream). This, and the observation that the entire cluster appeared in previous sequencing studies to be monophyletic (Harris et al., 2010; Castillo-Ramírez et al., 2012) suggest that the “South-East Asian Clade” is one distinct lineage resulting from one single acquisition of the sasX/sesI prophage. Differences affecting mobile genetic elements (including SCCmec) and the presence of the alpha haemolysin gene hla (which is absent from several sequences, mainly from the Middle East and Thailand) could then be considered secondary.
SCCmec elements in the “South-East Asian Clade” are complex composite elements consisting of SCCmec III [usually, but not always, with mecA(CP000046)], ccrC, cadD as well as of SCCterm01 and/or 02 sequences. The mercury resistance operon is nearly always present, and the few exceptions might be regarded as secondary deletions.
Isolates and sequences originate from South-East Asia, including Hong Kong and Southern China, India, Australia, the Middle East, Western Europe and, surprisingly, Trinidad & Tobago. The clade comprises also TW20 (NCTC13626), AUS-EMRSA-3 and Harmony Collection Finland E24_98541.
Furthermore, there is a large clade encompassing a wide variety of isolates and sequences and that show identical or very similar SCCmec types as the “South-East Asian Clade” (Table 2b) but that are sasX/sesI-negative. This includes the “South American Clade” sequences (Harris et al., 2010; Castillo-Ramírez et al., 2012) with three different SCCmec subtypes (SCC [mec III+Cd/Hg+ccrC] (Bmb9393), SCC [mec III+Cd/Hg] (BRA2) without ccrC and associated genes and pseudoSCCmec [class A+Cd/Hg] (UP1073) without any ccr genes). However, there are also isolates and sequences that originate mainly from the Middle East, but also from Europe and Russia, and that have identical or similar SCCmec types. Hence, we referred to this clade herein as the “South American/Middle Eastern Clade.” This clade includes Bmb9393, ATCC BAA-39, NCTC13131, UK-EMRSA-4, UK-EMRSA-7, UK-EMRSA-9 and UK-EMRSA-11, Irish AR09 and AR23 and the unique tst1-positive strain from Krasnoyarsk, Russia.
Furthermore, there are additional geographically restricted clades and strains that do not fit into the larger clades as defined by Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012).
One group of sequences and isolates could be named the “Australian/New Zealand Clade” consisting of a number of isolates from Australia and New Zealand with JKD6009 and JKD6008 being representative genome sequences. Isolates lacked sasX/sesI, the mer operon and ccrC. They carried mecA(BA000018). Similar to the “Eurasian Clade” strains, they harbored dcs but they differed in the absence of the secondary SCC-like gene cluster inserted into nsaB.
Another clade comprises a very homogenous cluster of Portuguese genome sequences from Harris' work and a similar strain, ATCC 33592, from New York City. The isolates and sequences lack sasX/sesI, dcs, ccrC, the mercury resistance operon and an integration into nsaB, but harbor SCCterm01.
We also observed a cluster of isolates from the Middle East, Libya and Russia that did not match any published sequences. These isolates carried mecA(BA000018) and ccrC but lacked the mercury resistance operon, the nsaB integration, sasX/sesI and hla.
An overview on geographic origins by clade and strain is provided in Table 4. In the following paragraph a short summary by countries and different sampling sites is given.
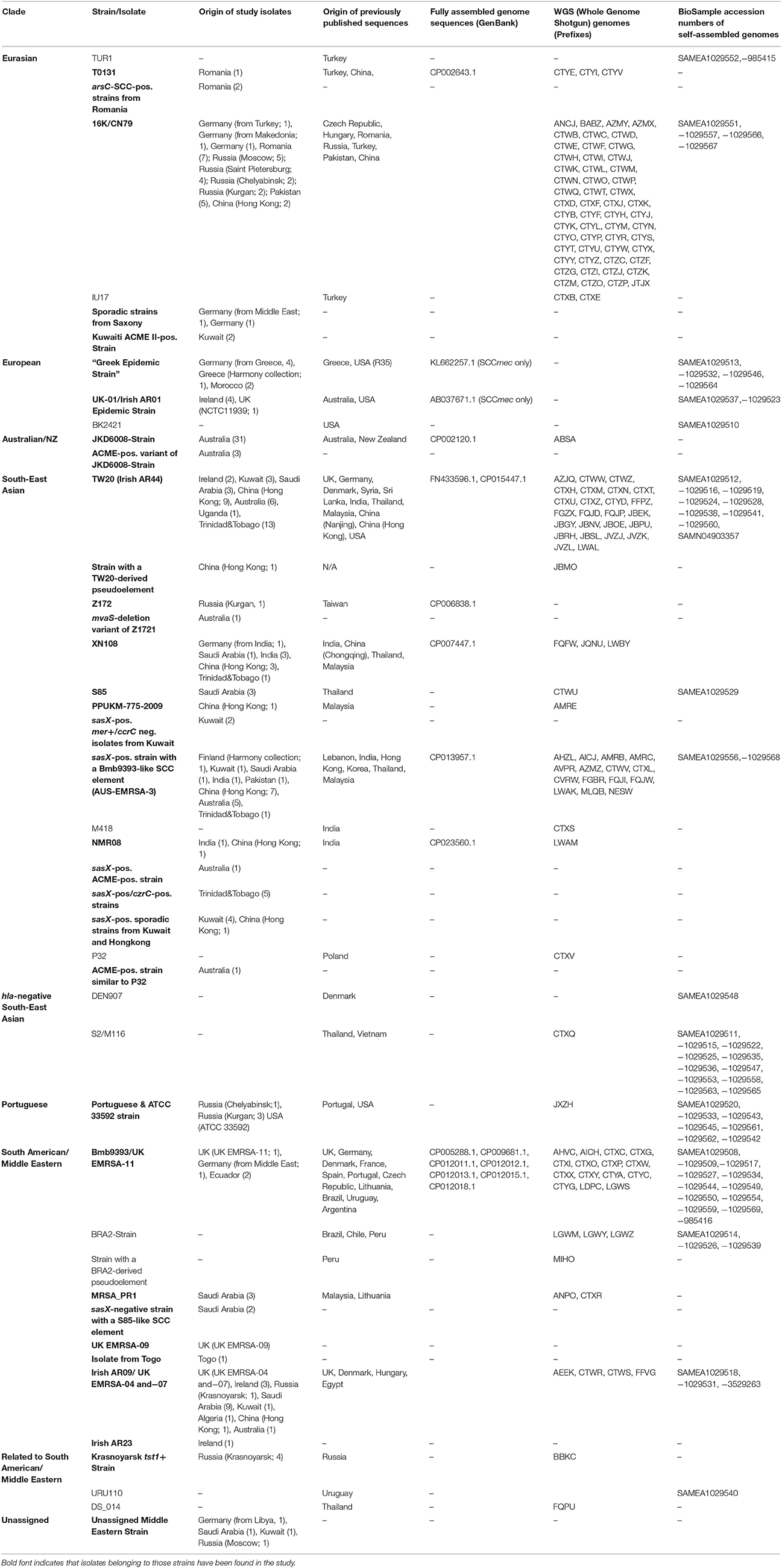
Table 4. Clades and strains of CC239-MRSA-III, geographic origin of isolates and sequences, GenBank and BioSample accession numbers.
Although CC239-MRSA-III has been observed in several African contries (Jansen van Rensburg et al., 2011; Abdulgader et al., 2015), there are insufficient data on epidemiological trends or molecular epidemiology.
Six isolates from five different African countries were included in the study. One isolate from Algeria was a “South American/Middle Eastern Clade” strain carrying SCCmec III+Cd/Hg+ccrC (TW20), and was identical to Irish AR09. One isolate from a Libyan patient (who was brought to North-Eastern Germany for humanitarian aid) belonged to the unassigned “Middle Eastern” strain. Two isolates from Morocco matched the “European Clade”, being most similar to the “Greek Strain” but differed from the other isolates in the presence of sek/seq and the absence of cadX, erm(C) and tet(M). One isolate from Togo matched the “South American/Middle Eastern Clade” but harbored an unique SCCmec subtype [designated SCCmec III+Cd/Hg (Unknown, ST239−3) in Table 2b]. This isolate might be a derivative of UK EMRSA-9 that has lost ccrC and accompanying genes. One isolate from Uganda (Monecke et al., 2013a) was sasX/sesI-positive and belonged to the “South-East Asian Clade.”
CC239-MRSA-III have been present in Australia for decades (Dubin et al., 1991; Coombs et al., 2004; Howden et al., 2010; Nimmo et al., 2015) with distinct variants (Aus-2 EMRSA and Aus-3 EMRSA) being distinguished based on mercury susceptibility or resistance, respectively (Coombs et al., 2006).
Eighteen Australian isolates were genotyped by microarray, and Ilumina NGS sequences of an additional 30 isolates were analyzed. The majority of isolates (n = 31) were very similar to JKD6008, GenBank CP002120.1, and JKD6009, GenBank ABSA from New Zealand and Australia, respectively, forming a distinct “Australian/New Zealand (NZ) Clade” (correlating to the mercury susceptible “Aus-2 EMRSA”). Three isolates harbored an ACME II cluster as well as Q9S0M4 and yielded additional signals for SCCterm03 and 05. The isolates might be regarded as direct derivatives of the JKD6008/6009 strain.
Thirteen isolates belonged to the “South-East Asian Clade” possibly indicating foreign importation which, given the geographic links between Australia and Asia, seems likely. Interestingly, two of these isolates also harbored the ACME II cluster. One isolate matched the Middle Eastern/Irish AR09 strain. One previously published Australian CC239 genome sequence, ANS46 (SAMEA1029537) belonged to the “European Clade,” UK-01/AR01.
The SCCmec element of the “Greek Strain,” SCC [mec III+Cd/Hg+ccrC] (SK1585), was also previously observed in Australia, in the chimeric strain, ST2249-MRSA-III, SK1585 (GenBank AYLT and KL662257.1) (Nimmo et al., 2015).
CC239-MRSA-III has been epidemic in China for decades. In Hong Kong, a presence of the “South-East Asian Clade” was reported while in Northern China, “Eurasian Clade” strains emerge and spread (Ip et al., 2005; Chen et al., 2010, 2014a; Wang et al., 2014).
Twenty-seven isolates originated from Hong Kong. The majority (n = 23) belonged to the “South-East Asian Clade” with the most common strains carrying SCC [mec III+Cd/Hg+ccrC] (TW20) or SCC [mec III+Cd/Hg+ccrC] (Bmb9393) (nine and seven isolates, respectively). Another isolate had a SCC [mec III+Cd/Hg+ccrC] (TW20)-derived pseudoSCCmec element. Two isolates belonged to the “Eurasian Clade”, likely to indicate influx from mainland China (Wang et al., 2014) and two isolates were assigned to the “South American/Middle Eastern Clade.”
In Ecuador, 2005-2013, CC239-MRSA-III was the second most common MRSA strain (Zurita et al., 2016) and contrarily to other Latin American countries it is still common there (Arias et al., 2017).
Two isolates were assigned to the “South American/Middle Eastern Clade” being sasX/sesI negative while carrying SCC [mec III+Cd/Hg+ccrC] (Bmb9393).
CC239-MRSA-III is not an epidemic strain in the German state of Saxony, or at least it has not been since 2000 and many cases are related to foreign travel (Albrecht et al., 2011).
Eleven isolates were included into the study. Nine of them were obtained from patients with known travel history (including admission to foreign healthcare facilities), with nosocomial contact to travelers, or from immigrant patients.
Five isolates were assigned to the “Eurasian Clade.” Two of them were obtained from Macedonian and Turkish nationals, respectively, the latter with history of hospitalization in Turkey after trauma. A third patient with an “Eurasian Clade” isolate appeared to have a Middle Eastern background while for two remaining cases, no history of immigration or travel was known. The “Greek Strain” CC239-MRSA-[III+Cd/Hg+ccrC] (SK1585) was found in four outbreak isolates, with an index patient who was repatriated from Greece after trauma and emergency care (Albrecht et al., 2011). One patient with a “South American/Middle Eastern Clade” strain had a Middle Eastern background indeed. Finally, one isolate from a patient of Indian background belonged to the “South-East Asian Clade” and matched the SCCmec element of XN108.
CC239-MRSA-III appear to be common and widespread in India; and although other strains emerged meanwhile, it is, at least regionally, still a dominant MRSA strain (D'Souza et al., 2010; Abimanyu et al., 2012; Neetu and Murugan, 2016).
In addition to one isolate from an Indian patient in Saxony (see above), five isolates with an Indian background were tested. All belonged to the “South-East Asian Clade”; three matched genome sequence of XN108 and its SCCmec subtype, and one the Indian genome sequences of NMR07/08. A fifth isolate was sasX/sesI positive but had a Bmb-9393-like SCCmec element.
CC239-MRSA-III predominated in Irish hospitals in the mid-to-late 1980s [locally known as phenotype III and antibiogram-resistogram (AR) types 01 and 09] but has since only been recovered sporadically or as part of localized outbreaks, represented by AR15 and AR23 isolates recovered in 1992/93 and AR44 recovered in 2002 (Carroll et al., 1989; Rossney et al., 1994; Shore et al., 2005). The 10 Irish ST239-MRSA-III isolates investigated clustered into three clades and four strains.
Firstly, AR01/AR15 isolates with a distinct truncation of mecR1 matched Harris' “Basal/European Clade,” being identical to sequences of Ans46 and LHH1 (from the US and Australia) as well as to UK-EMRSA-1.
Secondly, AR09/Phenotype III isolates harbored SCC [mec III+Cd/Hg+ccrC] (TW20), lacked sasX/sesI and matched Middle Eastern isolates. This fits to the observation that this strain was first brought to Ireland with an oil worker who was repatriated from Iraq in 1985 with a subsequent major outbreak (Humphreys et al., 1990).
AR23 could be considered a variant of the AR09 strain that was cadD-negative.
Thirdly, AR44 harbored SCC [mec III+Cd/Hg+ccrC] (TW20) and were sasX/sesI-positive. It has been suggested previously that this strain was imported from Singapore (Rossney, 2003), and indeed that this variant predominates in South-East Asia. This outbreak was contained and did not spread beyond one unit (Shore et al., 2005).
In Kuwait, CC239-MRSA-III accounted for more than 50% of typed MRSA isolates collected in a period from 1992 to 2010 (Boswihi et al., 2016). Currently, it is still present although it appears to be replaced by community-acquired MRSA strains (Udo and Al-Sweih, 2017).
Fourteen Kuwaiti isolates belonged to nine different strains and were assigned to the “Eurasian,” “South American/Middle Eastern,” and the “South-East Asian” clades. Two isolates of the “Eurasian Clade” harbored ACME II elements thus differing from all other isolates of that clade. One was identical to the Irish AR09 outbreak strain that was reported to originate from the Middle East (see above and Humphreys et al., 1990). Three “South-East Asian Clade” isolates were essentially identical to TW20 and one had a Bmb9393-like SCCmec element. Six others represented sporadic variants that were characterized by a loss of ccrC although the usually accompanying D1GU38 was present. One isolate belonged to an unassigned strain that was found mainly in the Middle East.
There are few studies on genotyping of MRSA from Pakistan indicating a presence of CC239-MRSA-III in hospitals (Shabir et al., 2010; Zafar et al., 2011; Arfat, 2013; Jamil et al., 2017) but its absence in the community.
Five Pakistani isolates from Rawalpindi (as well as the one previously published genome sequence from Pakistan, NCTR #32S, GenBank JTJX) belonged to the “Eurasian Clade”, CN79/16K-Strain. One isolate was assigned to “South-East Asian Clade” strain with a BMB939-like SCCmec element.
CC239-MRSA-III matching the “Eurasian Clade” has been reported to be common in Romania after 2000 (Cirlan et al., 2005; Chen et al., 2010; Monecke et al., 2014a).
All 10 Romanian isolates included, as well as previously published sequences, clustered into the “Eurasian Clade”, lacking sasX/sesI but carrying mecA(BA000018), dcs as well as SCCterm02 (indicating the secondary SCC-like gene cluster inserted into nsaB). Seven matched genome sequences 16K and CN79, and one T0131. Two isolates had unsequenced composite SCCmec elements including the arsenic resistance gene arsC that might be regarded as variants of 16K/CN79- and T0131-like elements, respectively.
After 2000, CC239-MRSA-III was found to be common in different regions across Russia, not only in hospitals but also in the community (Afanas'ev et al., 2010; Baranovich et al., 2010; Yamamoto et al., 2012; Khokhlova et al., 2015; Gostev et al., 2017). Previous reports (Afanas'ev et al., 2010; Baranovich et al., 2010; Yamamoto et al., 2012; Gostev et al., 2017) and sequences (16 K) indicated a presence of the “Eurasian Clade” as well as of other variants (Gostev et al., 2017). Besides, there was a notable emergence of a distinct tst1-positive variant in the city of Krasnoyarsk (Khokhlova et al., 2015).
The majority of Russian isolates (13/24), from Moscow, Saint Petersburg, Kurgan and Chelyabinsk, matched the “Eurasian Clade” and genome sequences CN79 and 16 K. However, three isolates from Saint Petersburg differed from the others in the presence of the sea(N315)/sep allele. Four isolates from Krasnoyarsk represent a local epidemic strain harboring tst1 and SCC [mec III+Cd/Hg+ccrC] (Bmb9393). Isolates were identical (although one isolate lacked presumably plasmid-borne cat, encoding chloramphenicol resistance) to the genome sequence of MRSA-OC3, GenBank BBKC, SAMD00019145 which also originated from this town. The four isolates from Kurgan and Chelyabinsk were essentially identical to ATCC 33592 (representing a clade previously known from Portugal and the USA). One isolate from Kurgan was identical to the Taiwanese genome sequence Z172 (SAMN02370325). One isolate from Krasnoyarsk belonged to the “South American/Middle Eastern Clade” and one isolate from Moscow matched the unassigned strain that was otherwise found in the Middle East.
Although CC239-MRSA-III is known to be present in the Middle Eastern/Gulf region for decades (Humphreys et al., 1990), molecular data confirming a presence of CC239-MRSA-III in the Kingdom of Saudi Arabia have been published only in recent years (Cirlan et al., 2005; Al-Obeid et al., 2010; Monecke et al., 2012c; Senok et al., 2016), and differences in carriage of ccrC, merA/B and aminoglycoside resistance genes indicated a simultaneous existence of different variants of this strain (Monecke et al., 2012c).
Twenty-three isolates from two different hospitals in Riyadh were characterized. Fourteen were assigned to the “South American/Middle Eastern Clade”, nine had SCC [mec III+Cd/Hg+ccrC] (TW20) thus matching Irish AR09 (see above and Humphreys et al., 1990). However, the 14 contemporary Saudi Arabian isolates lacked splE. As isolates were obtained from two hospitals in one city this might indicate a recent outbreak situation. The differences in SCCmec subtypes (absence of the mer operon resulting in SCC [mec III+Cd+ccrC] (S85) and of mer, SCCterm01 and Q93IB7 resulting in SCC [mec III+Cd+ccrC] (XN108)) would then only be secondary to the loss of splE.
Another eight isolates belonged to the “South-East Asian Clade” (being also all splE-negative) and one belonged to the unassigned “Middle Eastern Strain.”
CC239-MRSA-III has been the dominant MRSA clone in Trinidad & Tobago at least from 2000/01 (Akpaka et al., 2007) until 2012/2013 (Monecke et al., 2012b, 2014b).
Twenty-one isolates from Trinidad & Tobago where characterized. All were sasX/sesI-positive. This observation places CC239 from Trinidad & Tobago into the “South-East Asian Clade,” rather than any other Latin American strains. The majority (n = 14) carried SCC [mec III+Cd/Hg+ccrC] (TW20). Four isolates harbored composite SCC elements that included czrC, speG and additional recombinase genes ccrA/B-4. Based on the overall patterns, the isolates could be regarded as derivatives of SCC [mec III+Cd/Hg+ccrC] (Bmb9393).
CC239 is closely linked to SCCmec III. Only a few other lineages carry this type of SCCmec element. These include CC5 (Kwon et al., 2006; Shittu et al., 2009; Monecke et al., 2013b), CC398 (Nemati et al., 2008) and Staphylococcus pseudintermedius (KM1381, GenBank AM904732.1; KM241, GenBank AM904731). Virtually all CC239-MRSA isolates harbor one of the many different subtypes and composite elements of SCCmec III.
A relatively simple SCCmec III element, i.e., harboring ccrAB3 and a class A mec complex without additional ccr genes, heavy metal resistance markers, integrated transposons etc. has only been observed in S. pseudintermedius (KM1381, GenBank AM904732.1). However, it cannot safely be assumed that SCCmec III was initially transmitted from S. intermedius/pseudintermedius as, to the best of our knowledge, the earliest observation of methicillin resistance in “S. intermedius” was reported in 1984 (Roy et al., 1984). The most similar SCCmec III element in S. aureus can be found in the Sanger sequenced “Eurasian Clade” strain T0131 (CP002643) where it is only supplemented by the integration of a cadmium resistance operon (for which cadD(R35) is used as marker herein). The secondary set of SCC markers—ccrC, ccrAA, SCCterm02, D1GU38, erm(A) and ant9—are integrated elsewhere in the genomes of this lineage, distant from SCCmec and orfX. This is not only an interesting oddity, but raises the very practical question whether other SCC and SCCmec elements exist at alternative chromosomal sites away from orfX. If they do, this would have major consequences for rapid molecular MRSA tests as these assays target the integration of SCCmec into orfX.
Another relatively simple SCCmec element can be found in JKD6008 (CP002120.1) that, however, harbors additional resistance genes cadD(R35), aadD and ble. Hybridization patterns consistent with this element were observed in most “Australian/NZ Clade” isolates and in a livestock-associated CC5-MRSA-III isolate (see Monecke et al., 2013b and Supplemental Table 1). KM1381, JKD6008 as well as all “Australian/NZ Clade” and “Eurasian Clade” isolates and sequences (except TUR1 and TUR9; Harris et al., 2010) have dcs rather than other SCC terminal sequences and mecA(BA000018).
TUR1/TUR9 and strains of the “European” (85/2082, AB037671.1), “South-East Asian” (TW20, FN433596.1; XN108, CP007447; Z172, CP006838.1) and “South American/Middle Eastern” (Bmb9393, CP005288.1) clades harbor more complex SCCmec elements that include the cadmium resistance operon, ccrC, ccrAA and D1GU38 as well as (often, but not always) erm(A) and ant9, tet(K) and the mercury resistance operon. These isolates do not have dcs and a vast majority of isolates carry mecA(CP000046). “European Clade” sequences share the ccrC allele (ccrC (PM1)) with the “Eurasian Clade” while “South-East Asian” and “South American/Middle Eastern” clades have, if present, a different ccrC allele [ccrC (TSGH17)].
The presence of the mer genes raises the question for the benefit of mercury resistance in S. aureus/MRSA. One possible explanation could be the past medical use of mercury (e.g., for the treatment of syphilis, in topical agents such as merbromin, or in dental restorative materials such as amalgam) that could pose a selective pressure also on staphylococci colonizing the patients in question, regardless of whether they belonged to S. aureus or to other, coagulase-negative staphylococcal species. This could mean that SCCmer elements predated SCCmec in the same way as mercury use predated the clinical use of antibiotics. If SCCmec elements evolved indeed already after the introduction of penicillin (Harkins et al., 2017), there may have been a couple of decades of time for the evolution and selection of composite SCCmec/SCCmer elements.
The composite SCC [mec III+Cd/Hg+ccrC] (SK1585) element (SK1585, KL662257.1) existed at least already in the very early 1970s (as it was found in a strain epidemic in Australia from 1973 on; see below) and it appears to be ancestral to many SCC elements in CC239-MRSA. All SCC elements in “European,” “South-East Asian,” and “South American/Middle Eastern” clades could easily be described as variants of this particular element that have either acquired additional genes (ACME II, speG, czrC, ccrA/B4) or lost some or several genes. These latter genes have either no known function, are redundant (in case of the transposon with erm(A) and ant9 as a second copy is present elsewhere in the genome) or may no longer be of major advantage anymore because the compounds that provide a selective pressure are no longer frequently used (as in the case of mercury).
When mapping the presence of subtypes of SCCmec III on the phylogenetic trees as proposed by Harris and Castillo-Ramírez (Harris et al., 2010; Castillo-Ramírez et al., 2012), it becomes clear that identical subtypes can be observed in different clades (e.g., SCC [mec III+Cd/Hg+ccrC] (TW20), SCC [mec III+Cd/Hg+ccrC] (Bmb939)). There might be two different, mutually non-exclusive explanations. Firstly, these elements are subject to horizontal transfer so that SCCmec elements may be lost, acquired and exchanged after differentiation into different clades. Secondly, many of the SCCmec III subtypes differ in losses or acquisitions of accessory, purposeless or redundant genes (see above), and such events may have occurred several times. For instance, SCC [mec III+Cd/Hg+ccrC] (TW20) and SCC [mec III+Cd/Hg+ccrC] (Bmb9393) differ only in absence of the redundant transposon carrying erm(A) and ant9, Q93IB7 and SCCterm01 from the latter. It seems to be possible that this loss (or other similar losses) may have happened multiple times, independently from each other, to different lineages harboring SCC [mec III+Cd/Hg+ccrC] (TW20). Among a cluster of “South American/Middle Eastern Clade” isolates from Riyadh with a characteristic splE deletion, we observed three SCCmec subtypes suggesting that the losses of the mercury operon, SCCterm01 and Q93IB7 are secondary only to the loss of this gene and that they may spontaneously change SCC [mec III+Cd/Hg+ccrC] (TW20) to SCC [mec III+Cd+ccrC] (S85) and SCC [mec III+Cd+ccrC] (XN108).
An example for multiple acquisitions of one gene cluster is the observation of ACME II in dissimilar SCC elements of rather unrelated Australian and Kuwaiti strains. Likewise, repeated and independent acquisitions of arc genes have already been observed in Singapore (Hsu et al., 2015).
Based on accumulation of SNPs and mutation rates, previous work (Harris et al., 2010; Castillo-Ramírez et al., 2012) estimated the emergence of CC239 to have occurred in the mid- or late 1960s. The preservation of CC239-MRSA isolated in 1971 in a Norwegian strain collection (Smyth et al., 2010) also hints to an emergence and early spread of this strain in the late 1960s to 1970.
When analyzing gene content, one needs to assume two major recombination events to have occurred. One event was a horizontal gene transfer of a large segment of CC30 DNA into a CC8 genome (Robinson and Enright, 2004b; Holden et al., 2009). The other was a transfer of a SCCmec III element either before that “CC8/CC30 hybridization” into the CC30 ancestral strain, or afterwards, into the CC239 chimeric strain. It is not clear which gene transfer happened first. We are not aware of a CC30-MRSA-III strain that may have posed as a donor for the CC30 core genomic DNA and the SCCmec III element. CC239-MSSA strains have been identified (Strain 21178, GenBank AGRN and Luedicke et al., 2010), but they might be secondary deletion variants that lost SCCmec III rather than methicillin-susceptible ancestors to CC239-MRSA-III.
Our own observations also indicate that at least the “Greek Strain” and its SCC [mec III+Cd/Hg+ccrC] (SK1585) must have existed already in the early 1970s. There are reports of CC239 from Australia from this time and another strain, ST2249-MRSA-III was present in Melbourne, Australia, from 1973-1979 (predating the oldest Australian isolates of CC239 by 3 years). ST2249-MRSA-III is a chimeric strain (Nimmo et al., 2015) that combined features of CC45, CC30, and CC8 parental strains. The CC30- and CC8-like parts of its genome can be seen as one continuous segment originating from a CC239 parental strain, also including the SCC [mec III+Cd/Hg+ccrC] (SK1585) element that is characteristic for the “Greek Strain”. This allows two assumptions. Firstly, an importation of the “Greek Strain” of CC239 (or of the ST2249 chimera after the hybridization event) from Greece to Melbourne appears not improbable given a large Greek community in this city. Secondly, if the recombination that gave raise to ST2249-MRSA-III happened in 1973 or earlier, the “Greek Strain” CC239 and its SCCmec III element must have existed already some time before allowing for its emergence and spread as far as Australia. As discussed above, the SCC [mec III+Cd/Hg+ccrC] (SK1585) element could conveniently be regarded ancestral to many SCCmec elements in “European,” “South-East Asian,” and “South American/Middle Eastern” clades assuming that these elements emerged by serial or multiple deletions, and, occasionally, by acquisitions of genes. The other “European Clade” strains characterized by a distinct mecR1 deletion may have evolved in the early 1980s and spread in a rather limited way, i.e., in Ireland, UK, Australia, New Zealand and the USA during that decade (Ito et al., 2001; Shore et al., 2005; Harris et al., 2010). “European Clade” strains have been in recent years still of some relevance in Greece (and in travelers returning from there), but otherwise they have been replaced by other MRSA strains.
Then there is the “Eurasian Clade” (or Harris' and Castillo-Ramírez' “Turkish Clade”; Harris et al., 2010; Castillo-Ramírez et al., 2012). A comparatively low number of distinct strains within this clade might indicate a rather recent emergence, and earliest sequences identified (Harris et al., 2010) originate from Eastern Europe and Turkey, from the mid-/late 1990s. Genotyping data indicate relatedness to the “European Clade” (Harris et al., 2010; Castillo-Ramírez et al., 2012) but the “Eurasian Clade” and the “Australian/NZ Clade” differ from others by harboring less complex composite SCC elements with dcs and mecA(BA000018). Whether this indicates an independent, second acquisition of SCCmec III cannot yet be determined. The absence of splE and fnbB from all isolates and sequences indicate a monophyletic, clonal origin of the entire clade. TUR1 and TUR9 differ from other strains suggesting yet another horizontal gene transfer (possibly of a SCCmec III element from a “European” strain into an “Eurasian” strain with interrupted nsaB). The “Eurasian Clade” can be found in Turkey where it is frequently isolated and widespread (Tekeli et al., 2016). Furthermore it occurs in Eastern Europe including Macedonia (from where one study patient came from, see paragraph on Saxony/Germany) and, especially, Romania. In Hungary, it was common in the 1990s but it is declining since then, being replaced by other strains (Conceicao et al., 2007). It is also present in Russia, Pakistan and China. Recent reports from China indicate an emergence of the “Eurasian Clade” (with spa t030) at the expense of other CC239 strains (that is, of the “South-East Asian Clade”) following a North South gradient. Its distribution within China suggests import from Central Asia and/or a spill-over across the Russian border (Chen et al., 2014b). It appears to replicate faster than the “South-East Asian Clade” strains (Shang et al., 2016) and this advantage appears to outweigh in direct competition whatever advantage the presence of sasX/sesI may confer to the “South-East Asian” strains.
As mentioned, we found isolates matching Harris' and Castillo-Ramírez' “South American Clade” also in Russia and the Middle East. This raises the question where it emerged and to where it spread secondarily. Castillo-Ramírez estimated “the introduction into South America to have occurred approximately…in 1992 (late 1989, 1993)” (Castillo-Ramírez et al., 2012). Since the Irish AR09/Phenotype III outbreak strain was brought to Ireland from Iraq in 1985, and since it was described to be similar to a strain sampled in Baghdad as early as 1984 (Humphreys et al., 1990) we assume that this clade evolved earlier, possibly in the Middle East from where it may have spread to India, Russia and Europe. Again, travel from India to the Middle East and back as well as from the Middle East to Europe might have played a role. Strains of this clade may have come to Latin America from Europe or directly from the Middle East, and it became common and widespread in several Latin American countries (Harris et al., 2010; Castillo-Ramírez et al., 2012). Recent evidence, however, shows that this clade is declining or disappearing, except possibly in Ecuador and Peru (Arias et al., 2017). Many sequences of the “South American/Middle Eastern Clade” originated from Brazil (Harris et al., 2010; Castillo-Ramírez et al., 2012). While it is tempting to assume a link between Portugal and Brazil, a majority of Portuguese sequences clearly belong to a separate, geographically restricted, clade; and the few sequences that match the “South American/Middle Eastern Clade” might be re-imported by travelers (Harris et al., 2010; Castillo-Ramírez et al., 2012). While it is receding in Latin America and in India (D'Souza et al., 2010) and while it largely disappeared from Ireland, this clade still appears to be endemic in the Middle East and in Russia. One notable strain carrying tst1 has been endemic in the Russian town of Krasnoyarsk for several years (first observed in 2008; Iwao et al., 2012). The presence of the tst1 gene in CC239 is rather unique although this or a similar strain has also been described from Iran (Havaei et al., 2013).
The “South-East Asian Clade” most likely evolved from a “South American/Middle Eastern Clade” strain (or from a common ancestor of both clades) by acquisition of a prophage carrying sasX/sesI. Providing that this gene was acquired only once, which is the most parsimonious assumption, it might be assumed that this happened between the split of the related “South-East Asian,” “Portuguese,” and “South American/Middle Eastern” lineages and the proliferation of different strains within “South-East Asian Clade”, i.e., between ca. 1969 and 1985 based on Castillo-Ramírez' data (Castillo-Ramírez et al., 2012). The oldest published genome sequences originate from 1997 (CUHK_HK1997), 1998 (CHI59), 2001 (DEN907), and 2003 (TW20). The “South-East Asian Clade” spread in South-East Asia, including India, Thailand, Malaysia, Singapore, and China. Although it is still present in Hong Kong and in Southern Mainland China, in Northern China “Eurasian Clade” strains predominate nowadays (see above and Chen et al., 2014b; Shang et al., 2016). The “South-East Asian Clade” was also occasionally introduced to Europe, most likely by travelers (DEN907, TW20, AR44, P32, Finland E24_98541 from the Harmony collection), without becoming endemic there, and it also has been identified in Canada and the USA. The presence of the “South-East Asian Clade” and particularly of strains that appear to originate from India and South-East Asia in Kuwait and Saudi Arabia may easily be attributed to the large number of Indian and South-East Asian workers in the Gulf States (Birks et al., 1988). An interesting observation is the presence of this clade, rather than of the “South American” one, on the Caribbean islands of Trinidad & Tobago. A possible explanation is the Indian/South Asian descent of a high proportion of inhabitants of Trinidad & Tobago (ca. 38% of the total population, or 1.4 million people; http://www.tt.undp.org/content/dam/trinidad_tobago/docs/DemocraticGovernance/Publications/TandT_Demographic_Report_2011.pdf). Another 35% are of African descent, but no sufficient subtyping data for African CC239-MRSA are available. Possibly, an importation of MRSA by visits to ancestral lands might have played a greater role in the case of Trinidad & Tobago than just the mere geographic proximity to Latin America.
Some of Harris' strains from Thailand, one from Vietnam and one from Denmark are placed into the “South-East Asian Clade” by presence of sasX/sesI and by sequence analysis (Harris et al., 2010; Castillo-Ramírez et al., 2012). However, they differ from other strains of that clade in lacking hla and in harboring dcs instead of other SCC terminal sequences. The latter could indicate that their SCCmec elements rather originated from a horizontal gene transfer, maybe from the Australian/NZ lineage (see Table 2a).
Finally, there are some isolates and sequences that do not fit into the major clades. This includes the “Portuguese Clade” and the “Australian/New Zealand Clade.” The former is, according to sequence analysis (Harris et al., 2010; Castillo-Ramírez et al., 2012), related to the “South-East Asian” and “South American/Middle Eastern” clades. The latter was not represented by SNP-based studies and its SCCmec element might be more related to the one in non-CC239 strains (including S. pseudintermedius KM1381) than to the SCCmec elements in other CC239. We identified a cluster of Middle Eastern isolates (including one from Libya and one from Russia) that might constitute yet another clade. Finally there are strains such as DS_014, UR110 and P32 that could be assigned to the major clades but that still differ from them in particular features (such as mecA alleles). They may have evolved by further horizontal gene transfers. They also could be representatives of separate lineages or clades of CC239 that may be restricted to certain geographic regions poorly, or not at all, covered by previous typing and sequencing work. It might be expected that there are even more such unrecognized clades because CC239 was common in Western Europe and the USA before modern typing and sequencing technologies emerged, and because it is now common in countries were such technologies are not extensively applied.
Regarding typing technologies, NGS methods and DNA array hybridization profiling allow assignment to clades and strains. Arrays are currently cheaper and more convenient in a clinical setting. NGS can achieve a higher resolution although the definition of a “breakpoint for identity or non-identity” (i.e., how many differences between related isolates rule out direct transmission) still poses a challenge. This is quite a relevant issue for practical purposes. Traditionally, a “group of isolates that can be distinguished from other isolates of the same genus and species by phenotypic characteristics or genotypic characteristics or both” were regarded as a strain or clone (Tenover et al., 1995; Dijkshoorn et al., 2000). However, recent typing technologies achieve a level of resolution that is sufficiently informative to differentiate dozens of variants within one “strain” such as CC239-MRSA-III (as seen in the tables herein). Therefore, defining “strains” may still be useful for epidemiological purposes, but it is somewhat awkward and prone to subjectivity. Both approaches, short read NGS methods and microarray hybridization profiling, have difficulties with gene duplications and translocations if potentially mobile genes are flanked by repetitive and multi-copy sequences. Practically, this means that both technologies are useful for typing, but for the reconstruction of phylogenetic relationships, conventional sequencing still is unsurpassed.
On a very practical level, the definition of clades or variants can be useful for infection control purposes. For this pandemic strain it was possible to define such clades and to link molecular identifiers to geographic origins. Analyses of markers discussed herein, regardless whether by array hybridization, multiplex PCR, or by genome sequencing, can help assigning clinical isolates to these clades or variants and thus help to identify the provenance of an isolate and to discern imported from locally acquired cases. This is relevant as this strain was able to cause large hospital-born outbreaks upon importation with travelers or repatriated patients, as for instance the Irish experience (Humphreys et al., 1990; Shore et al., 2005), the TW20 outbreak in London (Holden et al., 2009), our own observations of the “Greek Strain” in Saxony or the spread of the “South American/Middle Eastern” clade in Latin America or of the “Eurasian Clade” in China showed. Based on European and North American experience, it is tempting to assume that CC239-MRSA-III has been side-lined by other clones or has even become extinct. Given the increasing scale of global travel and migration, there is still a possibility of re-importation and secondary spread. One should keep in mind that this strain still frequently detected in hospitals serving literally more than half of the world's population, i.e., China, India, South-East Asia, Turkey and the Middle East, Romania, Russia and parts of Latin America.
In conclusion, CC239-MRSA-III is a truly pandemic strain that, for nearly half a century, traveled around the world, infecting and even killing thousands of patients. This pandemic does not originate from elusive animals hosts in jungles and savannahs but from professionals working in the cleanest and most hygienic environments possible, that is, hospitals and operating theaters. Typing techniques allow following these movements, and even pinpointing individual index patients from whom this strain was brought into certain countries. However, understanding of a pandemic does not automatically results in an ability to prevent it. The very fact that an exclusively hospital-borne pandemic can spread that far and can last that long emphasizes an urgent need for improved hand hygiene, mandatory screening of staff and admitted patients, and decolonization procedures, a prudent use of antimicrobial agents and in general far more effective infection prevention and control measures.
SM designed the study, supervised and analyzed experiments, and wrote the manuscript. PS designed the primers and probes for the arrays used herein and analyzed genome sequences as well as experimental data. DG, EM, AR, AR-L, and RR performed experiments. SB and VG performed experiments and obtained isolates. PA, DB, MB, OD, MI, BJ, LJ, MN, AS, SS, LS, AMS, MS, AT, EU, TV, and JZ obtained isolates and provided clinical/epidemiological data. DC, GC, and ACS obtained isolates, provided clinical/epidemiological data and revised the manuscript. RE designed the study, supervised experiments, and revised the manuscript.
The collection of Romanian isolates was done as part of project PNII–IDEI, code ID_1586/2008 supported by CNCSIS–UEFISCSU. Collection and preliminary typing of isolates from Russia was supported by The Russian Science Foundation (research project no. 15-15-00185).
SM, PS, DG, EM, AR, and RE are employees of Abbott (Alere Technologies GmbH), the company that manufactures the microarrays used herein.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the clinical and laboratory staff at their respective institutions for collecting, identifying, and preserving isolates. During preparation of this manuscript we were sorry to hear that our esteemed colleague LS died. We had the privilege to work together with her for several years and will always remember her.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01436/full#supplementary-material
Supplementary Table 1. Full hybridization profiles for isolates characterized by microarray, and predicted hybridization profiles for analyzed sequences.
Abdulgader, S. M., Shittu, A. O., Nicol, M. P., and Kaba, M. (2015). Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front. Microbiol. 6:348. doi: 10.3389/fmicb.2015.00348
Abimanyu, N., Murugesan, S., and Krishnan, P. (2012). Emergence of methicillin-resistant Staphylococcus aureus ST239 with high-level mupirocin and inducible clindamycin resistance in a Tertiary Care Center in Chennai, South India. J. Clin. Microbiol. 50, 3412–3413. doi: 10.1128/JCM.01663-12
Afanas'ev, M. V., Karakashev, S. V., Il'ina, E. N., Salem Al-Salami, A. M., Sidorenko, S. V., and Govorun, V.M. (2010). [Molecular characteristic of methicillin-resistant Staphylococcus aureus strains isolated in Moscow clinics]. Mol. Gen. Mikrobiol. Virusol. 2, 20–24.
Aires de Sousa, M., Bartzavali, C., Spiliopoulou, I., Sanches, I. S., Crisóstomo, M. I., and de Lencastre, H. (2003a). Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41, 2027–2032. doi: 10.1128/JCM.41.5.2027-2032.2003
Aires de Sousa, M., Crisostomo, M. I., Sanches, I. S., Wu, J. S., Fuzhong, J., Tomasz, A., et al. (2003b). Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41, 159–163. doi: 10.1128/JCM.41.1.159-163.2003
Akpaka, P. E., Kissoon, S., Rutherford, C., Swanston, W. H., and Jayaratne, P. (2007). Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates from regional hospitals in Trinidad and Tobago. Int. J. Infect. Dis. 11, 544–548. doi: 10.1016/j.ijid.2007.03.004
Albrecht, N., Jatzwauk, L., Slickers, P., Ehricht, R., and Monecke, S. (2011). Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS ONE 6:e28189. doi: 10.1371/journal.pone.0028189
Al-Obeid, S., Haddad, Q., Cherkaoui, A., Schrenzel, J., and François, P. (2010). First detection of an invasive Staphylococcus aureus STRAIN (D958) with reduced susceptibility to glycopeptides in Saudi Arabia. J. Clin. Microbiol. 48, 2199–2204. doi: 10.1128/JCM.00954-09
Alp, E., Klaassen, C. H., Doganay, M., Altoparlak, U., Aydin, K., Engin, A., et al. (2009). MRSA genotypes in Turkey: persistence over 10 years of a single clone of ST239. J. Infect. 58, 433–438. doi: 10.1016/j.jinf.2009.04.006
Arfat, Y. (2013). Genotyping of Methicillin Resistant Staphylococcus aureus (MRSA) from Local Hospital of Rawalpindi/Islamabad, Pakistan. Ph.D., Quaid-i-Azam University, Islamabad.
Arias, C. A., Reyes, J., Carvajal, L. P., Rincon, S., Diaz, L., Panesso, D., et al. (2017). A prospective cohort multicenter study of molecular epidemiology and phylogenomics of Staphylococcus aureus bacteremia in nine Latin American countries. Antimicrob. Agents Chemother. 61:e00816-17. doi: 10.1128/AAC.00816-17
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baranovich, T., Zaraket, H., Shabana, I. I., Nevzorova, V., Turcutyuicov, V., and Suzuki, H. (2010). Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. 16, 575–582. doi: 10.1111/j.1469-0691.2009.02891.x
Becker, K., van Alen, S., Idelevich, E. A., Schleimer, N., Seggewiß, J., Mellmann, A., et al. (2018). Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerging Infect. Dis. 24, 242–248. doi: 10.3201/eid2402.171074
Birks, J. S., Seccombe, I. J., and Sinclair, C. A. (1988). Labour migration in the Arab Gulf states: patterns, trends and prospects. Int. Migr. 26, 267–286. doi: 10.1111/j.1468-2435.1988.tb00649.x
Boswihi, S. S., Udo, E. E., and Al-Sweih, N. (2016). Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait Hospitals: 1992-2010. PLoS ONE 11:e0162744. doi: 10.1371/journal.pone.0162744
Budimir, A., Deurenberg, R. H., Bosnjak, Z., Stobberingh, E. E., Cetkovic, H., and Kalenic, S. (2009). A variant of the Southern German clone of methicillin-resistant Staphylococcus aureus is predominant in Croatia. Clin. Microbiol. Infect. 16, 1077–1083. doi: 10.1111/j.1469-0691.2009.03042.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Campanile, F., Bongiorno, D., Borbone, S., and Stefani, S. (2009). Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Ann. Clin. Microbiol. Antimicrob. 8:22. doi: 10.1186/1476-0711-8-22
Carroll, J. D., Pomeroy, H. M., Russell, R. J., Arbuthnott, J. P., Keane, C. T., McCormick, O. M., et al. (1989). A new methicillin- and gentamicin-resistant Staphylococcus aureus in Dublin: molecular genetic analysis. J. Med. Microbiol. 28, 15–23. doi: 10.1099/00222615-28-1-15
Castillo-Ramírez, S., Corander, J., Marttinen, P., Aldeljawi, M., Hanage, W. P., Westh, H., et al. (2012). Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus. Genome Biol. 13:R126. doi: 10.1186/gb-2012-13-12-r126
Cha, H. Y., Moon, D. C., Choi, C. H., Oh, J. Y., Jeong, Y. S., Lee, Y. C., et al. (2005). Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean Hospital. J. Clin. Microbiol. 43, 3610–3614. doi: 10.1128/JCM.43.8.3610-3614.2005
Chen, H., Yang, X., Wang, Q., Zhao, C., Li, H., He, W., et al. (2014a). Insights on evolution of virulence and resistance from the whole-genome analysis of a predominant methicillin-resistant Staphylococcus aureus clone sequence type 239 in China. Chin. Sci. Bull. 59, 1104–1112. doi: 10.1007/s11434-014-0149-1
Chen, L., Mediavilla, J. R., Smyth, D. S., Chavda, K. D., Ionescu, R., Roberts, R. B., et al. (2010). Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob. Agents Chemother. 54, 3347–3354. doi: 10.1128/AAC.00001-10
Chen, Y., Liu, Z., Duo, L., Xiong, J., Gong, Y., Yang, J., et al. (2014b). Characterization of Staphylococcus aureus from distinct geographic locations in china: an increasing prevalence of spa-t030 and SCCmec type III. PLoS ONE 9:e96255. doi: 10.1371/journal.pone.0096255
Cirlan, M., Saad, M., Coman, G., Bilal, N. E., Elbashier, A. M., Kreft, D., et al. (2005). International spread of major clones of methicillin resistant Staphylococcus aureus: nosocomial endemicity of multi locus sequence type 239 in Saudi Arabia and Romania. Infect. Genet. Evol. 5, 335–339. doi: 10.1016/j.meegid.2004.09.005
Conceicao, T., Aires-de-Sousa, M., Fuzi, M., Toth, A., Paszti, J., Ungvari, E., et al. (2007). Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13, 971–979. doi: 10.1111/j.1469-0691.2007.01794.x
Coombs, G. W., Nimmo, G. R., Bell, J. M., Huygens, F., O'Brien, F. G., Malkowski, M. J., et al. (2004). Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42, 4735–4743. doi: 10.1128/JCM.42.10.4735-4743.2004
Coombs, G. W., Pearson, J. C., O'Brien, F. G., Murray, R. J., Grubb, W. B., and Christiansen, K. J. (2006). Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis. 12, 241–247. doi: 10.3201/eid1202.050454
Cuevas, O., Cercenado, E., Bouza, E., Castellares, C., Trincado, P., Cabrera, R., et al. (2007). Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Spain: a multicentre prevalence study (2002). Clin. Microbiol. Infect. 13, 250–256. doi: 10.1111/j.1469-0691.2006.01672.x
Dijkshoorn, L., Ursing, B. M., and Ursing, J. B. (2000). Strain, clone and species: comments on three basic concepts of bacteriology. J. Med. Microbiol. 49, 397–401. doi: 10.1099/0022-1317-49-5-397
D'Souza, N., Rodrigues, C., and Mehta, A. (2010). Molecular characterization of Methicillin resistant Staphylococcus aureus (MRSA) with emergence of epidemic clones ST 22 and ST 772, in Mumbai, India. J. Clin. Microbiol. 48, 1806–1811. doi: 10.1128/JCM.01867-09
Dubin, D. T., Matthews, P. R., Chikramane, S. G., and Stewart, P. R. (1991). Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrobial Agents Chemother. 35, 1661–1665.
Edgeworth, J. D., Yadegarfar, G., Pathak, S., Batra, R., Cockfield, J. D., Wyncoll, D., et al. (2007). An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin. Infect. Dis. 44, 493–501. doi: 10.1086/511034
El-baz, R., Rizk, D. E., Barwa, R., and Hassan, R. (2017). Virulence characteristics and molecular relatedness of methicillin resistant Staphylococcus aureus harboring different staphylococcal cassette chromosome mec. Microb. Pathog. 113, 385–395. doi: 10.1016/j.micpath.2017.11.021
Fatholahzadeh, B., Emaneini, M., Aligholi, M., Gilbert, G., Taherikalani, M., Jonaidi, N., et al. (2009). Molecular characterization of methicillin-resistant Staphylococcus aureus clones from a teaching hospital in Tehran. Jpn. J. Infect. Dis. 62, 309–311. Available online at: http://www0.nih.go.jp/JJID/62/309.html
Ghaznavi-Rad, E., Nor Shamsudin, M., Sekawi, Z., Khoon, L. Y., Aziz, M. N., Hamat, R. A., et al. (2010). Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J. Clin. Microbiol. 48, 867–872. doi: 10.1128/JCM.01112-09
Gostev, V., Kruglov, A., Kalinogorskaya, O., Dmitrenko, O., Khokhlova, O., Yamamoto, T., et al. (2017). Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation. Infect. Genet. Evol. 53, 189–194. doi: 10.1016/j.meegid.2017.06.006
Harkins, C. P., Pichon, B., Doumith, M., Parkhill, J., Westh, H., Tomasz, A., et al. (2017). Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 18:130. doi: 10.1186/s13059-017-1252-9
Harris, S. R., Feil, E. J., Holden, M. T., Quail, M. A., Nickerson, E. K., Chantratita, N., et al. (2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474. doi: 10.1126/science.1182395
Havaei, S. A., Azimian, A., Fazeli, H., Naderi, M., Ghazvini, K., Samiee, S. M., et al. (2013). Isolation of Asian endemic and livestock associated clones of methicillin resistant Staphylococcus aureus from ocular samples in Northeastern Iran. Iran. J. Microbiol. 5, 227–232.
Holden, M. T. G., Lindsay, J. A., Corton, C., Quail, M. A., Cockfield, J. D., Pathak, S., et al. (2009). Genome sequence of a recently emerged highly-transmissible, multi-antibiotic and antiseptic resistant, variant of methicillin-resistant Staphylococcus aureus (MRSA) sequence-type 239 (TW). J. Bacteriol. 192, 888–892. doi: 10.1128/JB.01255-09
Howden, B. P., Seemann, T., Harrison, P. F., McEvoy, C. R., Stanton, J. A., Rand, C. J., et al. (2010). Complete genome sequence of Staphylococcus aureus strain JKD6008, a ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate level vancomycin resistance. J. Bacteriol. 192, 5848–5849. doi: 10.1128/JB.00951-10
Hsu, L.-Y., Harris, S. R., Chlebowicz, M. A., Lindsay, J. A., Koh, T.-H., Krishnan, P., et al. (2015). Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 16:81. doi: 10.1186/s13059-015-0643-z
Hsu, L.-Y., Loomba-Chlebicka, N., Koh, Y.-L., Tan, T.-Y., Krishnan, P., Lin, R. T.-P., et al. (2007). Evolving EMRSA-15 epidemic in Singapore hospitals. J. Med. Microbiol. 56, 376–379. doi: 10.1099/jmm.0.46950-0
Humphreys, H., Carroll, J. D., Keane, C. T., Cafferkey, M. T., Pomeroy, H. M., and Coleman, D. C. (1990). Importation of methicillin-resistant Staphylococcus aureus from Baghdad to Dublin and subsequent nosocomial spread. J. Hosp. Infect. 15, 127–135. doi: 10.1016/0195-6701(90)90121-4
Ip, M., Yung, R. W. H., Ng, T. K., Luk, W. K., Tse, C., Hung, P., et al. (2005). Contemporary methicillin-resistant Staphylococcus aureus clones in Hong Kong. J. Clin. Microbiol. 43, 5069–5073. doi: 10.1128/JCM.43.10.5069-5073.2005
Ito, T., Katayama, Y., Asada, K., Mori, N., Tsutsumimoto, K., Tiensasitorn, C., et al. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001
Iwao, Y., Khokhlova, O. E., Takano, T., Hung, W. C., Isobe, H., Peryanova, O. V., et al. (2012). Fatal pneumonia in HIV-infected patients from a novel ST239 methicillin-resistant Staphylococcus aureus carrying the toxic shock syndrome toxin-1 gene in Krasnoyarsk, Siberian Russia. Jpn. J. Infect. Dis. 65, 184–186. Available online at: http://www.nih.go.jp/niid/images/JJID/65-2/184.pdf
IWG-SCC (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. doi: 10.1128/AAC.00579-09
Jamil, B., Gawlik, D., Syed, M. A., Shah, A. A., Abbasi, S. A., Müller, E., et al. (2017). Hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) from Pakistan: molecular characterisation by microarray technology. Eur. J. Clin. Microbiol. Infect. Dis. 37, 691–700. doi: 10.1007/s10096-017-3161-y
Jansen van Rensburg, M. J., Eliya Madikane, V., Whitelaw, A., Chachage, M., Haffejee, S., and Gay Elisha, B. (2011). The dominant methicillin-resistant Staphylococcus aureus clone from hospitals in Cape Town has an unusual genotype: ST612. Clin. Microbiol. Infect. 17, 785–792. doi: 10.1111/j.1469-0691.2010.03373.x
Khokhlova, O. E., Hung, W.-C., Wan, T.-W., Iwao, Y., Takano, T., Higuchi, W., et al. (2015). Healthcare- and community-associated methicillin-resistant Staphylococcus aureus (MRSA) and fatal pneumonia with pediatric deaths in Krasnoyarsk, Siberian Russia: unique MRSA's multiple virulence factors, genome, and stepwise evolution. PLoS ONE 10:e0128017. doi: 10.1371/journal.pone.0128017
Krziwanek, K., Luger, C., Sammer, B., Stumvoll, S., Stammler, M., Sagel, U., et al. (2008). MRSA in Austria-an overview. Clin. Microbiol. Infect. 14, 250–259. doi: 10.1111/j.1469-0691.2007.01896.x
Kwon, N. H., Park, K. T., Jung, W. K., Youn, H. Y., Lee, Y., Kim, S. H., et al. (2006). Characteristics of methicillin resistant Staphylococcus aureus isolated from chicken meat and hospitalized dogs in Korea and their epidemiological relatedness. Vet. Microbiol. 117, 304–312. doi: 10.1016/j.vetmic.2006.05.006
Li, M., Du, X., Villaruz, A. E., Diep, B. A., Wang, D., Song, Y., et al. (2012). MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18, 816–819. doi: 10.1038/nm.2692
Luedicke, C., Slickers, P., Ehricht, R., and Monecke, S. (2010). Molecular fingerprinting of Staphylococcus aureus from bone and joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 29, 457–463. doi: 10.1007/s10096-010-0884-4
Mayor, L., Ortellado, J., Menacho, C., Lird, G., Courtier, C., Gardon, C., et al. (2007). Molecular characterization of methicillin-resistant Staphylococcus aureus isolates collected in Asuncion, Paraguay. J. Clin. Microbiol. 45, 2298–2300. doi: 10.1128/JCM.00040-07
Melter, O., Aires de Sousa, M., Urbášková, P., Jakubu, V., Žemličková, H., and de Lencastre, H. (2003). Update on the major clonal types of methicillin-resistant Staphylococcus aureus in the Czech Republic. J. Clin. Microbiol. 41, 4998–5005. doi: 10.1128/JCM.41.11.4998-5005.2003
Monecke, S., Coombs, G., Shore, A. C., Coleman, D. C., Akpaka, P., Borg, M., et al. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e17936. doi: 10.1371/journal.pone.0017936
Monecke, S., Jatzwauk, L., Muller, E., Nitschke, H., Pfohl, K., Slickers, P., et al. (2016). Diversity of SCCmec elements in Staphylococcus aureus as observed in South-Eastern Germany. PLoS ONE 11:e0162654. doi: 10.1371/journal.pone.0162654
Monecke, S., Jatzwauk, L., Weber, S., Slickers, P., and Ehricht, R. (2008a). DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 14, 534–545. doi: 10.1111/j.1469-0691.2008.01986.x
Monecke, S., Muller, E., Buechler, J., Rejman, J., Stieber, B., Akpaka, P. E., et al. (2013a). Rapid detection of panton-valentine leukocidin in Staphylococcus aureus cultures by use of a lateral flow assay based on monoclonal antibodies. J. Clin. Microbiol. 51, 487–495. doi: 10.1128/JCM.02285-12
Monecke, S., Müller, E., Dorneanu, O. S., Vremeră, T., and Ehricht, R. (2014a). Molecular typing of MRSA and of clinical Staphylococcus aureus isolates from Iaşi, Romania. PLoS ONE 9:e97833. doi: 10.1371/journal.pone.0097833
Monecke, S., Muller, E., Schwarz, S., Hotzel, H., and Ehricht, R. (2012a). Rapid microarray based identification of different mecA alleles in Staphylococci. Antimicrob. Agents Chemother. 56, 5547–5554. doi: 10.1128/AAC.00574-12
Monecke, S., Nitschke, H., Slickers, P., Ehricht, R., Swanston, W., Manjunath, M., et al. (2012b). Molecular epidemiology and characterisation of MRSA isolates from Trinidad and Tobago. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1497–1500. doi: 10.1007/s10096-011-1469-6
Monecke, S., Ruppelt, A., Wendlandt, S., Schwarz, S., Slickers, P., Ehricht, R., et al. (2013b). Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet. Microbiol. 162, 806–812. doi: 10.1016/j.vetmic.2012.10.018
Monecke, S., Skakni, L., Hasan, R., Ruppelt, A., Ghazal, S. S., Hakawi, A., et al. (2012c). Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 12:146. doi: 10.1186/1471-2180-12-146
Monecke, S., Slickers, P., and Ehricht, R. (2008b). Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53, 237–251. doi: 10.1111/j.1574-695X.2008.00426.x
Monecke, S., Stieber, B., Roberts, R., Akpaka, P. E., Slickers, P., and Ehricht, R. (2014b). Population structure of Staphylococcus aureus from Trinidad & Tobago. PLoS ONE 9:e89120. doi: 10.1371/journal.pone.0089120
Neetu, T. J., and Murugan, S. (2016). Genotyping of methicillin resistant Staphylococcus aureus from Tertiary Care Hospitals in Coimbatore, South India. J. Glob. Infect. Dis. 8, 68–74. doi: 10.4103/0974-777X.182119
Nemati, M., Hermans, K., Lipinska, U., Denis, O., Deplano, A., Struelens, M., et al. (2008). Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 52, 3817–3819. doi: 10.1128/AAC.00613-08
Nimmo, G. R., Steen, J. A., Monecke, S., Ehricht, R., Slickers, P., Thomas, J. C., et al. (2015). ST2249-MRSA-III: a second major recombinant methicillin-resistant Staphylococcus aureus clone causing healthcare infection in the 1970s. Clin. Microbiol. Infect. 21, 444–450. doi: 10.1016/j.cmi.2014.12.018
Oliveira, D. C., Wu, S. W., and de Lencastre, H. (2000). Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphisms of this region. Antimicrob. Agents Chemother. 44, 1906–1910. doi: 10.1128/AAC.44.7.1906-1910.2000
Orth, D., Grif, K., Erdenechimeg, L., Battogtokh, C., Hosbayar, T., Strommenger, B., et al. (2006). Characterization of methicillin-resistant Staphylococcus aureus from Ulaanbaatar, Mongolia. Eur. J. Clin. Microbiol. Infect. Dis. 25, 104–107. doi: 10.1007/s10096-006-0102-6
Peck, K. R., Baek, J. Y., Song, J. H., and Ko, K. S. (2009). Comparison of genotypes and enterotoxin genes between Staphylococcus aureus isolates from blood and nasal colonizers in a Korean hospital. J. Korean Med. Sci. 24, 585–591. doi: 10.3346/jkms.2009.24.4.585
Robinson, D., and Enright, M. (2004a). Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 10, 92–97. doi: 10.1111/j.1469-0691.2004.00768.x
Robinson, D. A., and Enright, M. C. (2004b). Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186, 1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004
Rossney, A. S. (2003). A New Strain of MRSA Imported from Singapore. EPI-Insight, Disease Surveillance Report of NDSC, Ireland.
Rossney, A. S., Coleman, D. C., and Keane, C. T. (1994). Evaluation of an antibiogram-resistogram typing scheme for methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 41, 441–447. doi: 10.1099/00222615-41-6-441
Roy, A. F., Newman, S. S., Cox, H. U., and Hoskins, J. D. (1984). Effect of beta-lactamase of Staphylococcus intermedius on disk agar diffusion susceptibility tests. Cornell Vet. 74, 354–360.
Schaefler, S., Jones, D., Perry, W., Ruvinskaya, L., Baradet, T., Mayr, E., et al. (1981). Emergence of gentamicin- and methicillin-resistant Staphylococcus aureus strains in New York City hospitals. J. Clin. Microbiol. 13, 754–759.
Scicluna, E., Shore, A., Thuermer, A., Ehricht, R., Slickers, P., Borg, M., et al. (2010). Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur. J. Clin. Microbiol. Infect. Dis. 29, 163–170. doi: 10.1007/s10096-009-0834-1
Senok, A., Ehricht, R., Monecke, S., Al-Saedan, R., and Somily, A. (2016). Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect 14, 13–18. doi: 10.1016/j.nmni.2016.07.009
Shabir, S., Hardy, K. J., Abbasi, W. S., McMurray, C. L., Malik, S. A., Wattal, C., et al. (2010). Epidemiological typing of meticillin-resistant Staphylococcus aureus isolates from Pakistan and India. J. Med. Microbiol. 59, 330–337. doi: 10.1099/jmm.0.014910-0
Shang, W., Hu, Q., Yuan, W., Cheng, H., Yang, J., Hu, Z., et al. (2016). Comparative fitness and determinants for the characteristic drug resistance of ST239-MRSA-III-t030 and ST239-MRSA-III-t037 strains isolated in China. Microb. Drug Resist. 22, 185–192. doi: 10.1089/mdr.2015.0226
Shittu, A., Nubel, U., Udo, E., Lin, J., and Gaogakwe, S. (2009). Characterization of meticillin-resistant Staphylococcus aureus isolates from hospitals in KwaZulu-Natal province, Republic of South Africa. J. Med. Microbiol. 58, 1219–1226. doi: 10.1099/jmm.0.011452-0
Shore, A., Rossney, A. S., Keane, C. T., Enright, M. C., and Coleman, D. C. (2005). Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49, 2070–2083. doi: 10.1128/AAC.49.5.2070-2083.2005
Shore, A. C., and Coleman, D. C. (2013). Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359. doi: 10.1016/j.ijmm.2013.02.002
Smyth, D. S., McDougal, L. K., Gran, F. W., Manoharan, A., Enright, M. C., Song, J. H., et al. (2010). Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS ONE 5:e8582. doi: 10.1371/journal.pone.0008582
Takano, T., Saito, K., Teng, L. J., and Yamamoto, T. (2007). Spread of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in Taipei, Taiwan in 2005, and comparison of its drug resistance with previous hospital-acquired MRSA. Microbiol. Immunol. 51, 627–632. doi: 10.1111/j.1348-0421.2007.tb03949.x
Tekeli, A., Ocal, D. N., Ozmen, B. B., Karahan, Z. C., and Dolapci, I. (2016). Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Bloodstream Isolates in a Turkish University Hospital Between 2002 and 2012. Microb. Drug Resist. 22, 564–569. doi: 10.1089/mdr.2015.0116
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Udo, E. E., and Al-Sweih, N. (2017). Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS ONE 12:e0179563. doi: 10.1371/journal.pone.0179563
van Belkum, A., Melles, D. C., Nouwen, J., van Leeuwen, W. B., van Wamel, W., Vos, M. C., et al. (2009). Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 9, 32–47. doi: 10.1016/j.meegid.2008.09.012
Vandenesch, F., Naimi, T., Enright, M., Lina, G., Nimmo, G., Heffernan, H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerging Infect. Dis. 9, 978–984. doi: 10.3201/eid0908.030089
Vivoni, A. M., Diep, B. A., de Gouveia Magalhaes, A. C., Santos, K. R., Riley, L. W., Sensabaugh, G. F., et al. (2006). Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J. Clin. Microbiol. 44, 1686–1691. doi: 10.1128/JCM.44.5.1686-1691.2006
Wang, Z., Zhou, H., Wang, H., Chen, H., Leung, K. K., Tsui, S., et al. (2014). Comparative genomics of methicillin-resistant Staphylococcus aureus ST239: distinct geographical variants in Beijing and Hong Kong. BMC Genomics 15:529. doi: 10.1186/1471-2164-15-529
Weber, S., Ehricht, R., Slickers, P., Abdel-Wareth, L., Donnelly, G., Pitout, M., et al. (2010). “Genetic fingerprinting of MRSA from Abu Dhabi,” in ECCMID 2010 (Vienna).
Witte, W., Kresken, M., Braulke, C., and Cuny, C. (1997). Increasing incidence and widespread dissemination of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in central Europe, with special reference to German hospitals. Clin. Microbiol. Infect. 3, 414–422. doi: 10.1111/j.1469-0691.1997.tb00277.x
Yamamoto, T., Takano, T., Higuchi, W., Iwao, Y., Singur, O., Reva, I., et al. (2012). Comparative genomics and drug resistance of a geographic variant of ST239 methicillin-resistant Staphylococcus aureus emerged in Russia. PLoS ONE 7:e29187. doi: 10.1371/annotation/05a851e1-8627-4ea4-b12a-cfdba09f1e61
Yeap, A. D., Woods, K., Dance, D. A. B., Pichon, B., Rattanavong, S., Davong, V., et al. (2017). Molecular epidemiology of Staphylococcus aureus skin and soft tissue infections in the Lao People's Democratic Republic. Am. J. Trop. Med. Hyg. 97, 423–428. doi: 10.4269/ajtmh.16-0746
Zafar, A., Stone, M., Ibrahim, S., Parveen, Z., Hasan, Z., Khan, E., et al. (2011). Prevalent genotypes of meticillin-resistant Staphylococcus aureus: report from Pakistan. J. Med. Microbiol. 60, 56–62. doi: 10.1099/jmm.0.022707-0
Keywords: Staphylococcus aureus, MRSA, ST239-MRSA-III, SCCmec element, molecular epidemiology
Citation: Monecke S, Slickers P, Gawlik D, Müller E, Reissig A, Ruppelt-Lorz A, Akpaka PE, Bandt D, Bes M, Boswihi SS, Coleman DC, Coombs GW, Dorneanu OS, Gostev VV, Ip M, Jamil B, Jatzwauk L, Narvaez M, Roberts R, Senok A, Shore AC, Sidorenko SV, Skakni L, Somily AM, Syed MA, Thürmer A, Udo EE, Vremerǎ T, Zurita J and Ehricht R (2018) Molecular Typing of ST239-MRSA-III From Diverse Geographic Locations and the Evolution of the SCCmec III Element During Its Intercontinental Spread. Front. Microbiol. 9:1436. doi: 10.3389/fmicb.2018.01436
Received: 27 March 2018; Accepted: 11 June 2018;
Published: 06 July 2018.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Balaji Veeraraghavan, Christian Medical College & Hospital, IndiaCopyright © 2018 Monecke, Slickers, Gawlik, Müller, Reissig, Ruppelt-Lorz, Akpaka, Bandt, Bes, Boswihi, Coleman, Coombs, Dorneanu, Gostev, Ip, Jamil, Jatzwauk, Narvaez, Roberts, Senok, Shore, Sidorenko, Skakni, Somily, Syed, Thürmer, Udo, Vremerǎ, Zurita and Ehricht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Monecke, bW9uZWNrZUByb2NrZXRtYWlsLmNvbQ==; c3RlZmFuLm1vbmVja2VAYWxlcmUuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.