
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 June 2018
Sec. Extreme Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01318
This article is part of the Research TopicActinobacteria in Special and Extreme Habitats: Diversity, Function Roles and Environmental Adaptations, Second Edition View all 17 articles
Marine actinobacteria are potential producers of various secondary metabolites with diverse bioactivities. Among various bioactive compounds, anti-complement agents have received great interest for drug discovery to treat numerous diseases caused by inappropriate activation of the human complement system. However, marine streptomycetes producing anti-complement agents are still poorly explored. In this study, a marine-derived strain Streptomyces sp. DUT11 showing superior anti-complement activity was focused, and its genome sequence was analyzed. Gene clusters showing high similarities to that of tunicamycin and nonactin were identified, and their corresponding metabolites were also detected. Subsequently, tunicamycin I, V, and VII were isolated from Streptomyces sp. DUT11. Anti-complement assay showed that tunicamycin I, V, VII inhibited complement activation through the classic pathway, whereas no anti-complement activity of nonactin was detected. This is the first time that tunicamycins are reported to have such activity. In addition, genome analysis indicates that Streptomyces sp. DUT11 has the potential to produce novel lassopeptides and lantibiotics. These results suggest that marine Streptomyces are rich sources of anti-complement agents for drug discovery.
Marine environments cover more than 70% of the surface of the Earth, and are habitat of diverse microorganisms. Marine actinomycetes are rich sources for a myriad of bioactive natural products. Among various marine actinomycetes, marine streptomycetes have received the most attention due to their biosynthetic potential for producing novel bioactive compounds (Subramani and Aalbersberg, 2012; Agustina et al., 2016; Prietodavó et al., 2016).
Genome sequences of marine streptomycetes not only provide insights in mechanisms of marine environmental adaptation (Ian et al., 2014; Tian et al., 2016), but also benefit discovery of their biosynthetic potential (Zhao and Yang, 2011). Various useful metabolites were identified based on genome mining of Streptomyces (Xu et al., 2016; Ye et al., 2017; Yu et al., 2018). However, so far studies on genome mining of marine Streptomyces remain limited (Zhang et al., 2015; Chen et al., 2016; Jin et al., 2018).
Complement system is one of the important human immune defense systems, and it plays an important role in eliminating foreign microorganisms, clearance of damaged cells, adaptive immunity, inflammation, tissue regeneration, and tumor growth (Barnum, 2017). However, the improper activation of complement system may lead to a variety of diseases, such as rheumatoid arthritis and Alzheimer’s disease (Morgan and Harris, 2015). Chemical synthesis of anti-complementary agents has the limitations such as high cost, low selectivity, and unwanted side effects. Therefore, natural products with anti-complement activities have received increasing attention. Up to now, various anti-complementary compounds were isolated from algae and plants (Jin et al., 2015; Wen et al., 2017), whereas compounds with such activities have been poorly studied in microorganisms. The only known microbial-derived anti-complement compounds are complestatin and its analogs, which are produced by Streptomyces lavendulae SANK 60477 (Kaneko et al., 1989). Microbial production of anti-complement agents has the advantages to reduce production cost by rapid accumulation of metabolites and easy scale-up fermentation. In addition, novel compounds can also be obtained by genetic modifications of the microbial producers. Therefore, exploration of microbial strains with the potential to produce anti-complement compounds is of great interest, and the information of related genome sequences of the producer will promote rapid discovery of such active compounds.
During the screening of marine streptomycetes producing anti-complement compounds, we found that Streptomyces sp. DUT11 has superior anti-complement activities. In this study, the anti-complement compounds of Streptomyces sp. DUT11 were studied by genome mining of Streptomyces sp. DUT11, and its biosynthetic potential was also explored. Our current report indicates that marine Streptomyces strains are valuable sources of anti-complement compounds for drug discovery.
Strain Streptomyces sp. DUT11 was isolated from the marine sediment about 10 m below sea level in Xinghai Bay (39° 52′N, 121° 35′E), Dalian, China. This strain was deposited in the China General Microbiological, and the strain number was CGMCC 14581. The strain was maintained on ISP4 agar slants at 4°C and in 20% (v/v) glycerol stock at -80°C. For plate culture, strain DUT11 was grown at 28°C on Bennett’s agar. For seed culture, tryptic soy broth (TSB) medium was used.
To prepare samples for bioactivity assays, Streptomyces sp. DUT11 was inoculated into 20 mL TSB medium in 50 mL tubes to culture at 28°C for 2–3 days. Then, the seed culture was inoculated into the M33 fermentation media with an inoculum size of 2% (v/v) to culture at 28°C for 7 days. To prepare metabolites from agar culture for molecular networking analysis, 50 μL seed culture was spread over the 6 cm plate containing A1 agar medium to culture at 28°C for 7 days. The compositions of all the cultural media used in this study were provided in Supplementary Table S1.
The genome of Streptomyces sp. DUT11 was sequenced through Pacbio RS technology (English et al., 2012) in State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, and the acquired data were assembled by Canu v1.5 (Berlin et al., 2015). The open reading frames (ORFs) prediction and genome annotation were acquired by RAST (Rapid Annotation using Subsystem Technology) (Brettin et al., 2015). The Clusters of Orthologous Groups (COGs) of gene functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway prediction was performed through WebMGA (Altschul et al., 1990; Ogata et al., 1999).
Identification of potential biosynthetic gene clusters (BGCs) for secondary metabolites were proposed by bioinformatics tool website antiSMASH (Weber et al., 2015). Further gene function analysis was through manual BLAST on NCBI and viewed on visual software Artemis Release 16.0 (Carver et al., 2012). The alignment of the genomes was performed by the website Double ACT v21 and viewed through software Artemis Comparison Tool (ACT) (Carver et al., 2005). The genome sequence data have been deposited in the GenBank database and was assigned the accession number PRJNA351245 (CP025511).
16S rRNA PCR amplification was performed using the methods described previously (Lee et al., 2003; Li et al., 2007). The universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1429R (5′-AAGGAGGTGATCCAAGCCGCA-3′) were used to amplify the 16S rRNA sequence. The amplified 16S rRNA gene was sequenced by TaKaRa and uploaded to web-based EzTaxon-e program2 (Kim et al., 2012) for comparison. A 16S rRNA phylogenetic tree was created by software Geneious (Kearse et al., 2012) based on the EzTaxon-e database and partially on BLAST of some closely related strains.
The whole-genome based phylogenetic trees with peptide sequences or DNA sequences were generated with Composition Vector Tree v3 (CVtree v3) (Xu and Hao, 2009), and OrthoANI was used to generate OAT heat map (Ouk Kim et al., 2016). The genomes used for analysis were obtained from GenBank3. Nine closely related genomes were chosen for analysis. Due to the poor quality of the genome of S. bacillaris NBRC 13487, it was excluded in the genome alignments.
Seven media, namely TSB, TSBY, TYDM, M3, M9, M33, and A1 medium (Supplementary Table S1) were chosen as fermentation media to test the production of anti-complement agents. The fermentation broth was centrifuged at 6,000 rpm for 15 min to obtain supernatant and mycelia. Subsequently, the supernatant was extracted by EtOAc and the mycelia was extracted by MeOH. Then the extracts were dried and re-dissolved into 1 mL 5% dimethyl sulfoxide (DMSO) solution to acquire crude extracts for anti-complement test.
A hemolytic assay was used to determine the inhibition of complement activation in the classical pathway (CP) (Morgan and Harris, 2015) with minor modifications. 1 × BBS (barbital buffer solution) buffer was used for solution buffer. The fresh sheep red blood cell (SRBC) was diluted to 2% for the test. Heparin was used as a positive control, which was dissolved in 1 × BBS. After preliminary test, 1/160 serum was chosen to be submaximal lysis in the absence of complement inhibitors. Sensitized erythrocytes (EAs) were prepared by incubating 2% SRBS with equal volumes of 1:1000 hemolysin. The samples were dissolved in 1 mL 5% DMSO, which were diluted 10 times in 5% DMSO to test anti-complement activities. For the anti-complement activity test, every 100 μL diluted sample and 100 μL serum solution (SS) were mixed and incubated at 37°C for 10 min, after which the mixture was cooled down on ice. Subsequently, 200 μL of EAs was added and the volume was filled to 600 μL with 1 × BBS. The mixtures were then incubated at 37°C for 30 min. Control groups were incubated under the same conditions, which include: (1) standard control: 200 μL EAs and 100 μL serum in 300 μL 1 × BBS; (2) 100% lysis: 200 μL EAs in water (400 μL); (3) sample blank: 100 μL dilution of each sample in 500 μL 1 × BBS; (4) sample test: 200 μL SS with 200 μL EAs in 200 μL 1 × BBS (Zhu et al., 2008). After incubation, the reacted mixtures were centrifuged at 5,000 rpm, 4°C for 10 min immediately. Optical density of the supernatants (200 μL) was measured at 405 nm with a spectrophotometer (Multiskan GO 1510, Thermo Fisher Scientific, Finland). The inhibition percentage of each sample was calculated by excluding the controls and blanks and then divided by a standard value, and the anti-complementary activity (%) is calculated from the formula (Wang et al., 2008):
For analysis the secondary metabolites in strain DUT11, the cultured plates were cut into small pieces and extracted with 1:1 MeOH/H2O or EtOAc. The samples were dried and re-dissolved into MeOH to analyze by UPLC-MTQ MS (Agilent1290- Bruker MicroTOF-Q II) using the gradient of 5–100% ACN/H2O containing 0.1% formic acid for 9 min, and 100% ACN with 0.1% formic acid for 1 min with a 0.5 mL/min flow rate (Agilent Infinity 1290 HPLC, Supelco Discover C18 250 mm × 4.6 mm, 5 μm column and Bruker MTQ MS system). The acquired LC-MS data was uploaded to Global Natural Products Social Molecular Networking (GNPS) website (Wang et al., 2016) to generate the molecular networking map and the map is viewed by Cytoscape v3.4. The open accession of the GNPS data in this study via MassIVE is ftp://massive.ucsd.edu/MSV000082328.
To isolate active components from fermentation broth and mycelia, Streptomyces sp. DUT11 was cultured in modified M33 medium at 30°C with 200 rpm agitation for 7 days for extraction and purification of the active compounds. After 7 days cultivation, the fermentation broth (45 L) was centrifuged at 6,000 rpm, after which the mycelia were extracted by MeOH and the supernatant was extracted by EtOAc. The resultant organic extracts were concentrated in vacuo until dried. The crude extracts of supernatant and mycelia were separated by C18 column with H2O/MeOH (v/v, 100:0, 70:30, 50:50, 30:70, 0:100), to afford five fractions for each samples. Fractions 4 and 4′ (70% MeOH) with high anti-complement activity were purified by HPLC on the C18 column (XDB-C18, 4.6 mm × 250 mm, 5 μm at a flow rate of 0.4 mL/min using a gradient solvent from 5 to 95% CH3CN with UV detector set at 260 nm, and compound A was isolated. The active fractions 5 and 5′ (100% MeOH) were purified using the same method to isolate compound B and compound C.
LC/MS (Agilent1290-MS6230 Trap system) was used for further detection of the compounds. LC/MS analysis was performed on a XDB-C18 Column (Agilent, 150 mm × 4.6 mm, 5 μm column) at a flow rate of 0.4 mL/min using a gradient solvent from 5 to 95% ACN over 50 min.
A Waters ACQUITYTM series mass spectrometer equipped with an electrospray ionization source was used for UPLC-Q/TOF MS analysis of compounds. MS/MS detection mode was set as: mass range, from m/z 100 to 2,000 in positive mode; capillary, 3 kV; sample cone voltage, 35 V; desolvation gas temperature, 350°C; flow rate of desolvation gas, 600 L/h. The column temperature was maintained at 40°C. Mobile phase A (H2O, 0.1% formic acid) and B (ACN, 0.1% formic acid) were used; and the gradient program was as follows: 0–5 min 10% B, 30–31 min 40–10% B, 40 min 10% B; flow rate 0.35 mL/min; and injection volume was 1 μL.
Streptomyces sp. DUT11 was selected due to its superior anti-complement activity during our primary assay of the marine strain library. This strain grows well on A1, M33, and ISP4 agar, generating white spores (Figure 1A). DUT11 tolerates up to 10% NaCl in TSB media, and achieved the highest biomass with 3% NaCl (data not shown). The 16S rRNA sequence of Streptomyces sp. DUT11 is the same with that of the type strain S. bacillaris NBRC 13487 (100% similarity), and is the most closest to that of S. globisporus NBRC 12867 (Waksman and Lechevalier, 1953) (99.65% similarity). However, so far the genome sequences of these two type strains are still not available (Figure 1B).
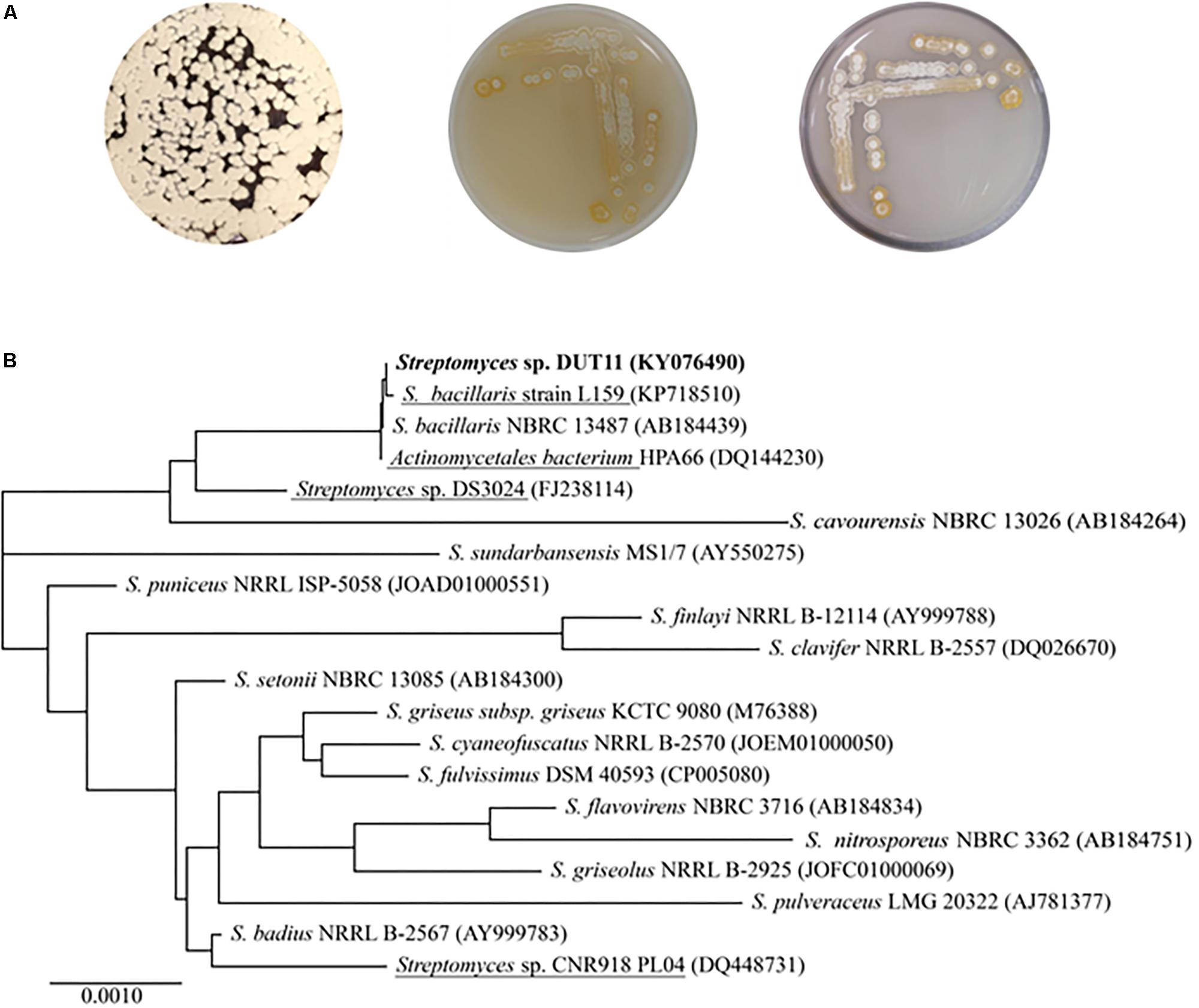
FIGURE 1. Morphological characteristics and phylogenetic analyses of Streptomyces sp. DUT11. (A) Phenotype of DUT11 cultured on A1 (left), M33 (middle) and ISP4 medium (right), respectively. (B) Neighbor-joining tree showing the phylogenetic relationships of DUT11 and related strains based on 16S rRNA sequences. The strains underlined were isolated from marine environment. The accession numbers of the 16S rRNA sequences were provided in the parentheses.
Due to the complicate procedures of anti-complement assay, we decided to use genome mining to facilitate the discovery of anti-complement agents from Streptomyces sp. DUT11. The genome of this strain was sequenced and analyzed. The genome size of Streptomyces sp. DUT11 is 8,027,164 bp with 71.83% GC content. There are 7,745 ORFs covering 6.76 Mb with 72.22% GC content. The coding percentage is 84.7% and the average length of ORFs is 872 bp. Totally 63 tRNA and 9 rRNA genes were also predicted. From the genome circle view, it was observed that the majority of the secondary metabolic BGCs were located near the ends of the genomes (Figure 2A). By blasting the COG database, two third of the annotated genes were classified, and there are 291 genes annotated to related with secondary metabolites (Supplementary Figure S1).
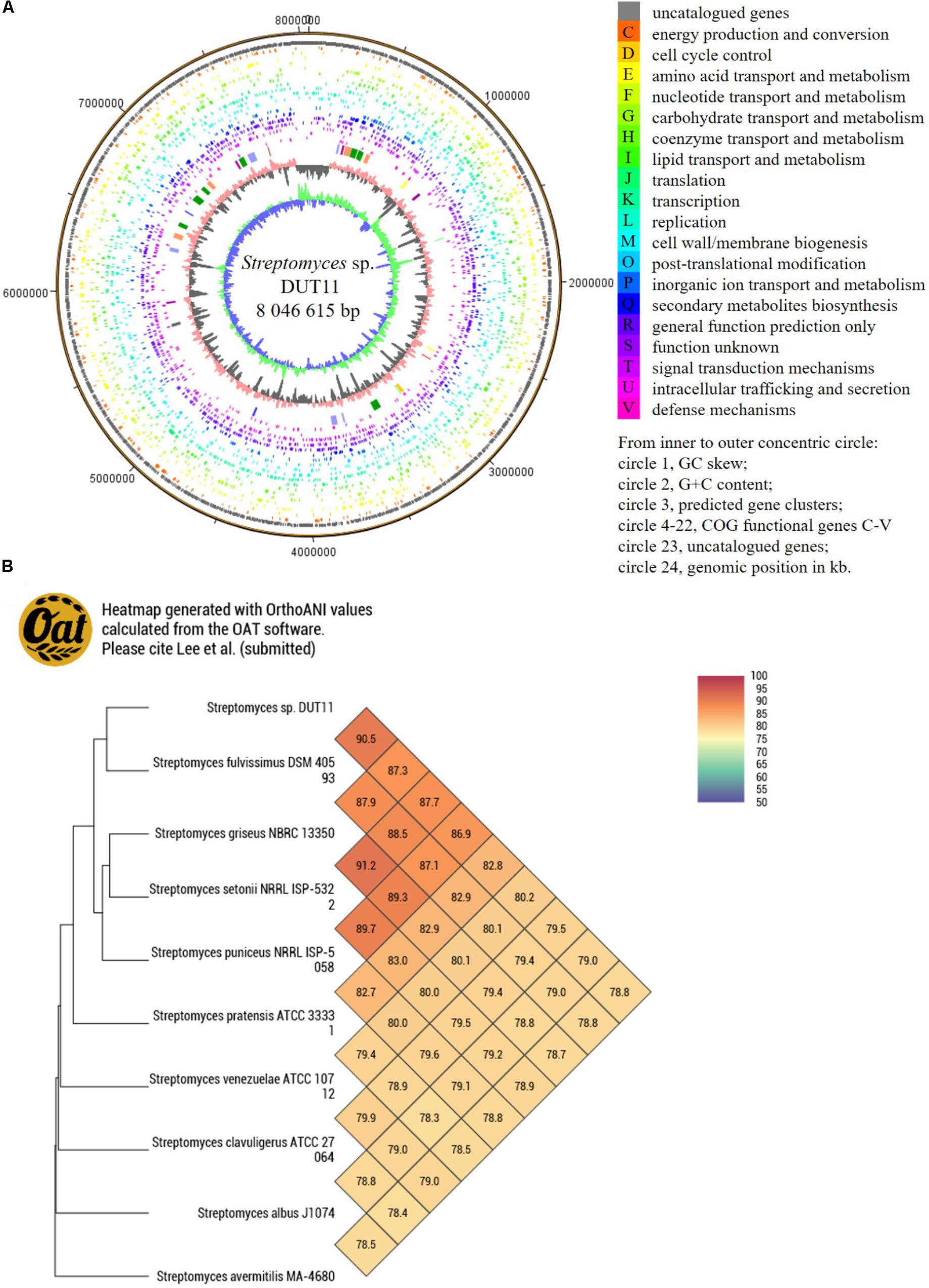
FIGURE 2. Circular map of Streptomyces sp. DUT11 genome and comparative genome analysis. (A) Circular map of the genome. (B) Heat map generated with OrthoANI values between strain DUT11 and nine closely related strains.
The genome sequence reported in this study thus provide basis to study related genomes. Among the closely related strains of DUT11, there are three marine actinomycetes, namely, Streptomyces sp. DS3024 (Hao et al., 2009), S. bacillaris strain L159 (Petříčková et al., 2015), and Actinomycetales bacterium HPA66 (Zhang et al., 2006). According the CVTree based on global protein or DNA sequences (Supplementary Figure S2), the most related strain with DUT11 is S. fulvissimus DSM 40593 (Myronovskyi et al., 2013), which shared a 16S rRNA similarity of 99.38% with DUT11. Furthermore, the OAT heat map (Figure 2B) also showed that S. fulvissimus DSM 40593 is the closest strain from genome scale, and the ANI (average nucleotide identity) score is 90.4% between the two strains. The second closest strain is S. griseus NBRC 13350 (87.9% similarity). The genomes of the rest strains are 78–83% similar to that of Streptomyces sp. DUT11.
There are 33 BGCs including at least seven non-ribosomal peptide synthases (NRPSs), five polyketide synthases (PKSs), six post-translationally modified peptides (PTMPs), and five terpene BGCs in the genome sequence. Furthermore, BGCs involve in the biosynthesis of ectoine, siderophore, bacteriocin, and butyrolactone which are common in most of the actinobacteria were also identified in DUT11 (Table 1).
Biosynthetic gene clusters potentially involved in biosynthesis of three known compounds, namely, tunicamycin, nonactin, and medermycin were focused. Among these BGCs, Cluster 18 was annotated as tunicamycin BGC by analyses with antiSMASH. Through further comparison with other tunicamycin BGCs in different strains, the cluster in DUT11 has the closest similarity to the gene cluster in S. chartreuis (Chen et al., 2010) (Figure 3A), and all the genes share around 94–98% identities with the corresponding genes in S. chartreuis (Supplementary Table S2). Strain DUT11 also lacks tunM and tunN which encodes methyltransferase and NUDIX hydrolase, respectively, but these genes are not essential for tunicamycin biosynthesis (Chen et al., 2010).
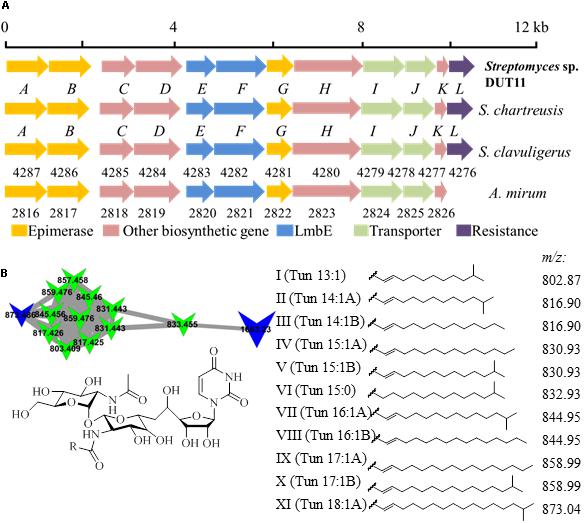
FIGURE 3. Organization of the tunicamycin BGC and identification of tunicamycins in Streptomyces sp. DUT11. (A) The tunicamycin BGC from strain DUT11. (B) Molecular networking map generated by GNPS and corresponding structures of the tunicamycin analogs.
We further analyzed tunicamycin production in both agar culture and liquid fermentation. Firstly, we identified a cluster of tunicamycin analogs in GNPS map which has been annotated by GNPS (Wang et al., 2016). Depending on the reported structures and molecular weight range, at least eleven tunicamycin analogs with different lengths of alkane tails ranging from 13 to 18 carbons were identified from the A1 agar culture of DUT11 (Figure 3B) by comparing the MS spectrum with m/z shifts (Supplementary Figure S3).
Anti-complement activities of the extracts from both mycelia and fermentation broth of Streptomyces sp. DUT11 were detected. As shown in Table 2, when different culture media were compared, the highest anti-complement activities were observed using the M33 medium. In addition, similar anti-complement activities were observed for the extracts of supernatant and mycelia grown in M33 fermentation medium.
We further isolated active fractions from the liquid culture using the M33 medium. The extracts of mycelia and supernatant were concentrated to yield 10 g and 8.5 g. The two organic extracts were separated by C18 column with H2O/MeOH (v/v, 100:0, 70:30, 50:50, 30:70, 0:100), to afford five fractions. The fractions 3, 4, and 5 from the crude extract of supernatant eluted using 50%, 70%, and 100% methanol showed good anti-complement activities, which were 58.9%, 77.1%, and 70.4%, respectively. Similarly, the fractions 3′, 4′, and 5′ eluted using 50%, 70%, and 100% methanol from the crude extracts of mycelia showed better anti-complement activity, which were 50.1%, 86.03%, and 58.6%, respectively. We deduced that the strain DUT11 can produce a variety of substances with anti-complement activity (Figures 4A,B).
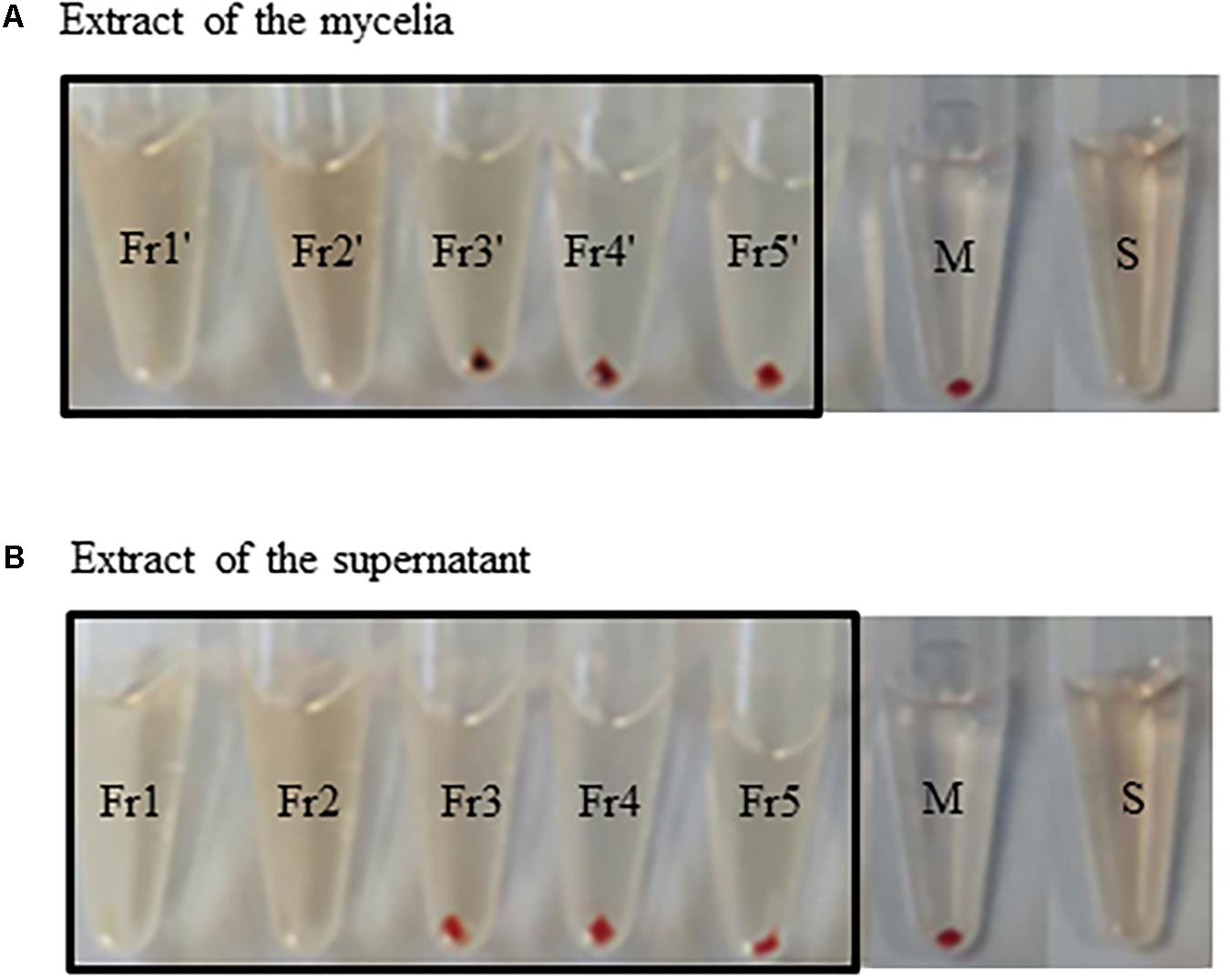
FIGURE 4. Anti-complement activity of the extracts of Streptomyces sp. DUT11. (A) The anti-complement activity of fractions from the extracts of mycelia obtained with different MeOH concentrations: Fr1′: H2O/MeOH (v/v, 100:0); Fr2′: H2O/MeOH (v/v, 70:30); Fr3′: H2O/MeOH (v/v, 50:50); Fr4′: H2O/MeOH (v/v, 30:70); Fr5′: H2O/MeOH (v/v, 0:100); S: full hemolysis; M: mechanical hemolysis. (B) The anti-complement activity of fractions from the extracts of supernatant. Fr1: H2O/MeOH (v/v, 100:0); Fr2: H2O/MeOH (v/v, 70:30); Fr3: H2O/MeOH (v/v, 50:50); Fr4: H2O/MeOH (v/v, 30:70); Fr5: H2O/MeOH (v/v, 0:100); S: full hemolysis control; M: mechanical hemolysis control.
The active fractions 4, 4′, 5, and 5′ were further analyzed by HPLC, and tunicamycins were detected. The profile of tunicamycin standard was presented in Figure 5A. Tunicamycin analogs compound A (retention time, Rt: 19.5 min), compound B (Rt: 21.9 min), and compound C (Rt: 23.5 min) were detected (Figures 5B,C) in the active fractions. In the tunicamycin standard we purchased, only two tunicamycin analogs, namely tunicamycin V and VII, which are corresponding to compounds B and C, were present (Figure 5A). MS/MS analysis of tunicamycin V and VII was shown in Figures 5D,E. We further purified the three tunicamycin analogs in the active fractions. Compound A gives [M + H]+/[M + Na]+ ions at m/z 803.3949/825.3766 (calcd. for C36H59N4O16: 803.3921, ppm: 3.49; C36H58N4O16Na: 825.3740, ppm: 3.15). Compound B gives [M + H]+/[M + Na]+ ions at m/z 831.4202/853.4023 (calcd. for C38H63N4O16: 831.4234, ppm: -3.84; C38H62N4O16Na: 853.4053, ppm: -3.52) and compound C gives [M + H]+/[M + Na]+ ions at m/z 845.4393/867.4218 (calcd. for C39H65N4O16: 845.4390, ppm: 0.35; C39H64N4O16Na: 867.4210, ppm: 0.92), respectively. The peaks generated by MS/MS analysis showed that the main fragment ions are at 600.3, 582.3, and 546.3; 628.3, 610.3, and 574.3; as well as 642.4, 624.3, and 588.3, respectively (Figures 6A–C). Fragmentation of molecular adduct ions give rise to [M + H-221]+ and [M + H-203]+ ions across the tunicamycin α β-1, 1′-glycosidic bonds to generate stable deglycosylated species, the relative masses of which are diagnostic of the attached N-acyl group (-R) (Figure 6D). Comparing with the structural spectroscopic data in the literature (Tsvetanova and Price, 2001; Chen et al., 2010) and the tunicamycin standard, we confirmed that compounds A, B, and C are tunicamycin I, V, and VII (Figures 6E–G), respectively. The other compounds in these active fractions are being investigated in our ongoing work.
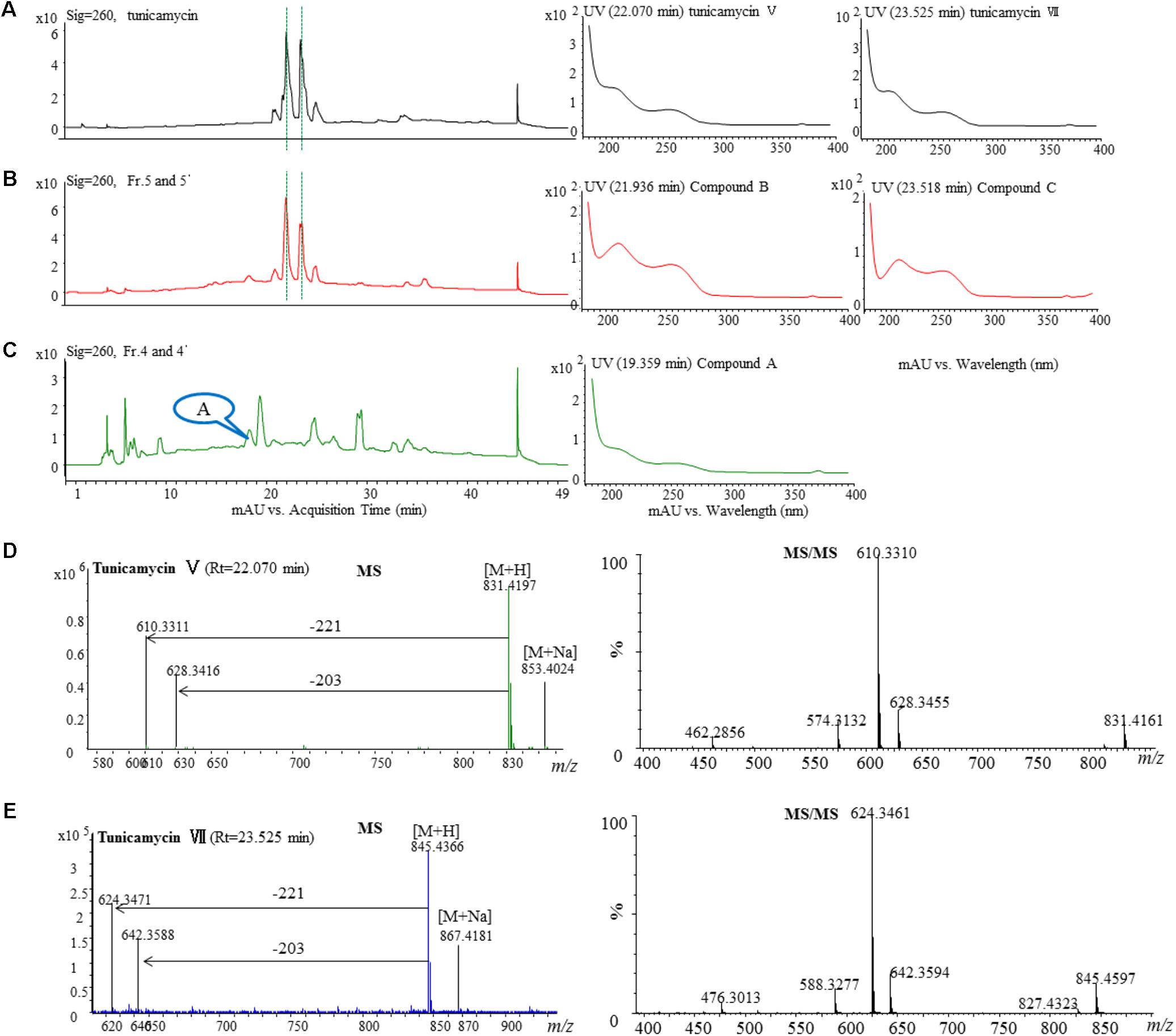
FIGURE 5. HPLC, MS, and MS/MS analysis of the tunicamycin standard and HPLC analysis of the active fractions. Each sample was purified by HPLC on the C18 column (XDB-C18, 4.6 mm × 250 mm, 5 μm at a flow rate of 0.4 mL/min using a gradient solvent from 5 to 95% CH3CN with UV detector set at 260 nm. (A) HPLC analysis of the tunicamycin standard under 260 nm and the UV-Vis spectra of the tunicamycin standard. (B) HPLC analysis of the active fractions 5 and 5′ under 260 nm and UV-Vis spectra of compounds B and C which were isolated from the fractions. The HPLC profiles of fraction 5 and 5’ are the same. (C) HPLC analysis of the active fractions 4 and 4′ under 260 nm and UV-Vis spectra of compound A which was isolated from the fractions. The HPLC profiles of fraction 5 and 5′ are the same. (D,E), MS-MS analysis of the standard tunicamycin V and VII, respectively.
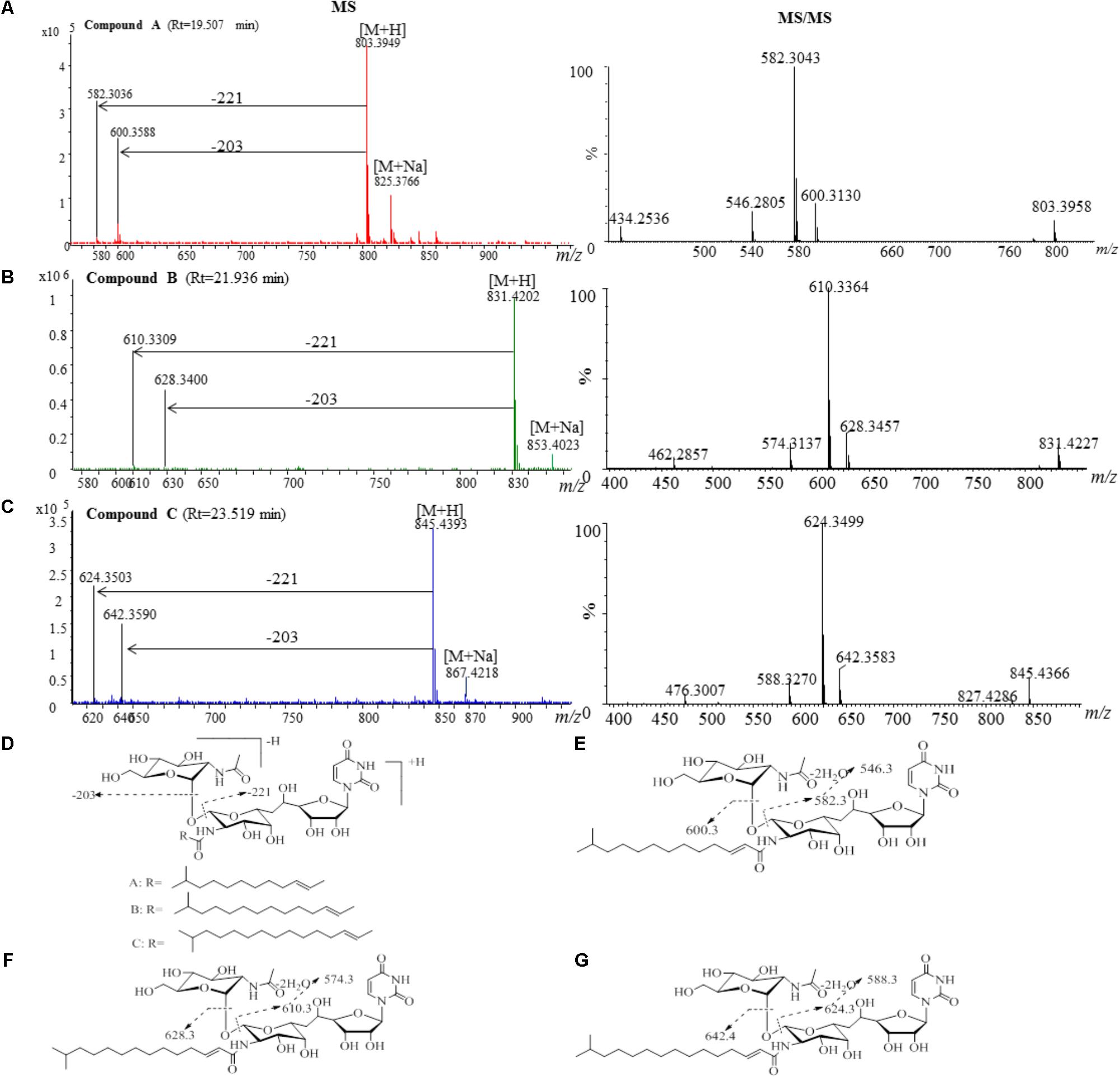
FIGURE 6. MS and MS/MS analysis of the purified compounds A, B and C. (A) The component of compound A generates a [M + H]+ ion at m/z 803.3926, which is fragmented into main ions of 600.3, 582.3, 546.3, etc. (B) The component of compound B generates a [M + H]+ ion at m/z 831.4141, which is fragmented into main ions of 628.3, 610.3, 574.3, etc. (C) The component of compound C generates a [M + H]+ ion at m/z 845.4366, which is fragmented into main ions of 642.4, 624.3, 588.3, etc. (D) Voltage gradient-induced ions at [M + H - 221]+ and [M + H - 203]+ indicate fragmentations across the compound A, B and C. MS/MS fragmentation pattern of the compound A (E), compound B (F), and compound C (G).
We further confirmed anti-complementary activities of the isolated tunicamycins produced by Streptomyces sp. DUT11. As shown in Figure 7, the concentrations that resulted in 50% hemolysis inhibition (CH50) of tunicamycin I, tunicamycin V, and tunicamycin VII were 0.071 ± 0.01 mg/mL (0.088 ± 0.012 mM), 0.060 ± 0.009 mg/mL (0.072 ± 0.011 mM), and 0.045 ± 0.009 mg/mL (0.053 ± 0.011 mM), respectively. The CH50 of the positive control heparin was 0.115 ± 0.05 mg/mL. The CH50 of tunicamycin standard V and VII were 0.059 ± 0.008 mg/mL (0.071 ± 0.009 mM), and 0.046 ± 0.009 mg/mL (0.054 ± 0.011 mM), respectively. This is the first time that tunicamycins are proved to have anti-complement activities.
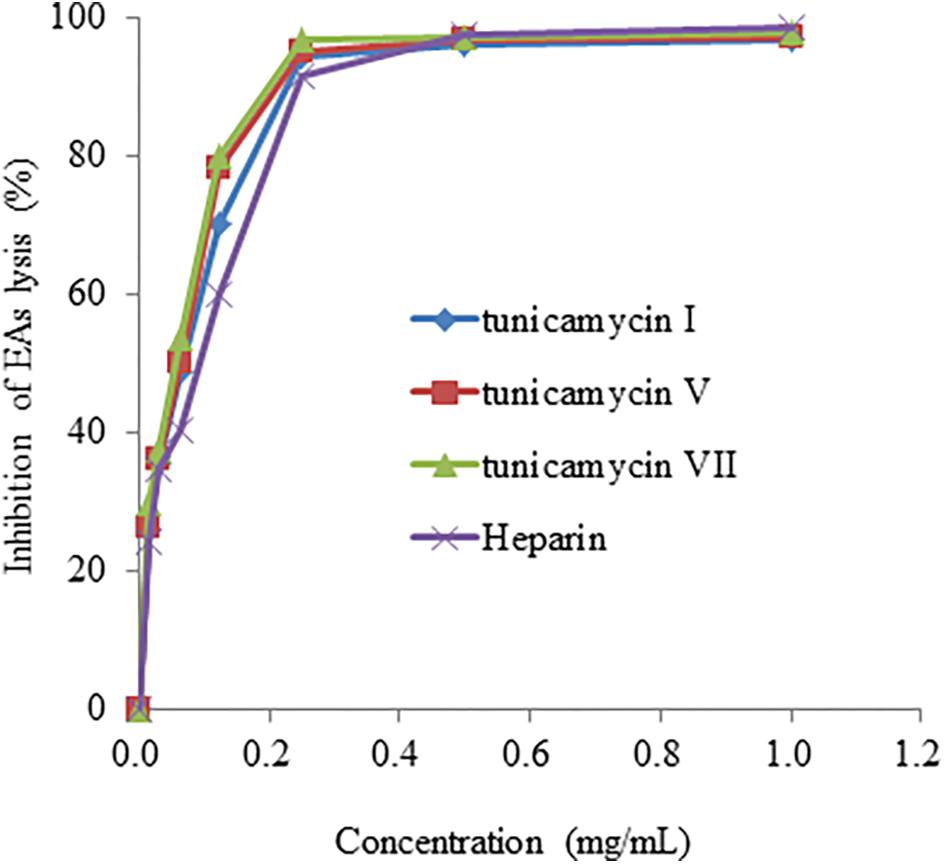
FIGURE 7. The anti-complement activity of tunicamycin I, V and VII. A hemolytic assay was used to determine the inhibition of complement activation in the classical pathway.
Cluster 26 was originally identified as type II PKS gene cluster by antiSMASH. With further manual comparison, we found that the gene cluster is close to that of nonactin gene cluster from S. griseus ETH A7796 (Supplementary Figure S4A) (Walczak et al., 2000). Most of the genes in Cluster 26 shared 85–90% identities with that in the nonactin BGC (Supplementary Table S3). However, there is no counterpart of nonI (AAD37458.1), which is a putative acetoacetate reductase in this gene cluster. In the primary search in the GNPS annotation, only monactin was found annotated in one molecular cluster. Subsequently, based on the information from GNPS and MS spectra comparison, seven cyclized nonactin analogs were found around the monactin node, including nonactin, dinactin, trinactin, and tetranactin (Smith et al., 2000) (Supplementary Figures S4B–D). Besides, several hydrolyzed liner nonactin analogs were also observed in the same molecular cluster. However, no anti-complement activity of nonactin was detected under our assay conditions in this study (data not shown).
Cluster 21 is another type II PKS BGC in DUT11, which has 66% similarity to that of the medermycin BGC from Streptomyces sp. K73 (Ichinose, 2003). The organization of Cluster 21 is also similar to the medermycin BGC. Nevertheless, the significant difference between the two BGC is that the two genes encoding phosphopantetheinyl transferase and putative carbohydrate kinase, respectively, are not present in Cluster 21. The other parts of the genes shares around 70–80% similarities between Cluster 21 and medermycin BGC (Supplementary Table S4). Due to the inability to get the medermycin standard sample, we did not test the anti-compliment activity of this compound.
Cluster 7 probably produces a lantibiotic peptide with four S-S cycles. The peptide sequence including leader peptide and core peptide is quite similar to the reported venezuelin peptide sequence (Kersten et al., 2011). However, five amino acid in the leader peptide and three in the core peptide are different (Figure 8A). The putative product of venL in the reference BGC includes both protein kinase C (PK C) like superfamily domain and lanC-like superfamily domain. Nevertheless, DNA sequences encoding such two domains are divided into two separate genes in Cluster 7. The domains are highly conserved with around 72% similarities although the intergenic region is quite different.
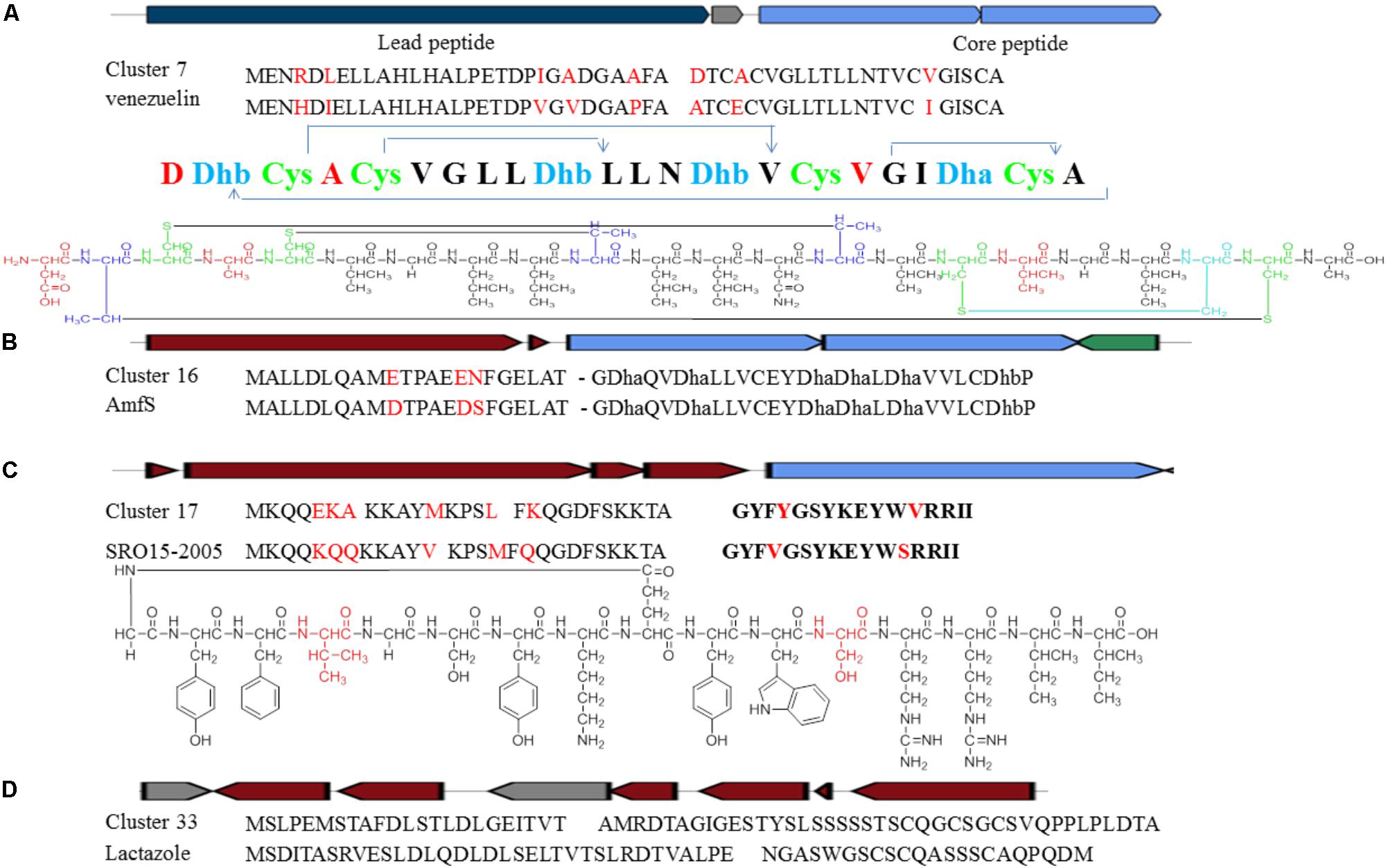
FIGURE 8. Identification of post-translationally modified peptide class BGCs in Streptomyces sp. DUT11. (A) Comparison of Cluster 7 with the venezuelin peptide gene cluster. (B) Comparison of Cluster 16 with the AmfS gene cluster. (C) Comparison of Cluster 17 with the lassopeptide SRO15-2005 gene cluster. (D) Comparison of Cluster 33 with the lactazole gene cluster.
Cluster 16 is a class III lantipeptide BGC and quite similar to AmfS BGC (Figure 8B), the product of which is a morphogen, a biological surfactant which positively regulates the formation of erect aerial mycelium (Ueda et al., 2002; Willey and Gaskell, 2011). The five genes involved in the biosynthesis share around 80% similarities with the reference ones. Compared to AmfS BGC, the core peptide encoded by related gene in this cluster is the same but the leader peptide contains three mutations.
Cluster 17 is a lassopeptide BGC and close to SRO15-2005 BGC (Maksimov et al., 2012) (Figure 8C). There are only two amino acid changes in the core peptide, but there are six amino acid differences in the leader peptide. The gene construction of Cluster 33 is partially close to lactazole BGC and might be a lassopeptide BGC hybrid with an additional NRPS and PKS (Hayashi et al., 2014) (Figure 8D). It is still not clear how the PKS and the NPRS affect the formation of the final products. The precursor peptides are quite different between the two BGCs, indicating that strain DUT11 has the potential to produce novel lassopeptides.
In addition, there are quite abundant BGCs of PTMPs in the DUT11 genome. Two of the tree lantibiotic BGCs and one lassopeptide found their close reference sequences which would help to identify the structures. Two more thiopeptide BGCs were identified as novel ones and the main backbone could be predicted.
Cluster 3 was supposed to be PKS I-NRPS hybrid BGC. There should be 7 PKS modules in this gene cluster and three A-domains of NRPS. This is the biggest PKS related BGC in the DUT11 genome. Through comparison and analysis, we found that there might be several key domains lost in the modules, and the backbone based on the function prediction was speculated. Another type III PKS BGC Cluster 6 might only include 2 genes and shared 71% similarities to naringenin BGC (Alvarez-Alvarez et al., 2015). The rest one was Cluster 29 and is a small type I PKS-NRPS hybrid gene cluster. This BGC only contains one PKS module including KS, AT, DH, KR, and ER domains and one A-domain responsible for ornithine. The BGC construction is quite close to the BGC class of polycyclic tetramate macrolactams including SGR PTMs BGC (Luo et al., 2013) and frontalamides BGC (Blodgett et al., 2010). Furthermore, eight proposed NRPS related BGCs were observed in the DUT11 genome, even though most of those only contained small NRPS genes. Cluster 4 and Cluster 5 were found to be quite close to BGC of griseobactin (Patzer and Braun, 2009) and coelichelin (Bentley et al., 2002), respectively. The predicted structures should be the same with reference compounds. Cluster 14 was found to be able to synthesize a four amino acid peptide backbone, namely, gly-ser-ser-ser. Cluster 24 was supposed to be a NRPS-PKS hybrid BGC which contains five A-domains coding leu-ser-dhb-cys-gly and one putative PKS sets. Further analysis of expression of these BGCs and their corresponding metabolites will provide functional insights of the Streptomyces sp. DUT11 genome.
Marine streptomycetes are rich sources of various secondary metabolites for drug discovery. In this study, we report the biosynthetic potential of the marine strain Streptomyces sp. DU11, which was revealed by analysis of its genome sequence. We found that genome sequences of many species having close phylogenetic relationship with stain DUT11 are still not available. Therefore, the available genome information reported in this study will assist the studies of these closely related species.
Complement system plays important roles in defense of invading microorganisms, clearance of damaged cells, adaptive immunity, and tissue regeneration (Hourcade et al., 2016). However, inappropriate and excessive activation of complement system will cause tissue damage diseases, such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), systemic inflammatory response syndrome (SIRS), and acute respiratory distress syndrome (ARDS) (Qu et al., 2009). Anti-complement compounds are potential drugs to cure disorder of complement system, but so far such agents are mainly from plants rather than microbial source (Kaneko et al., 1989). Tunicamycins are a family of nucleoside antibiotic consisting of uracil, N-acetylglucosamine, a unique 11-carbon 2-aminodiadose sugar (tunicamine), and an N-acyl chain with variable lengths (Tsvetanova and Price, 2001; Price and Tsvetanova, 2007). Tunicamycins target bacterial cell wall biosynthesis by inhibiting early stage of peptidoglycan biosynthesis (Winn et al., 2010). They are inhibitors of eukaryotic protein N-glycosylation to induce endoplasmic reticulum stress and consequently become drugs to study apoptotic cell death related diseases (Cóppola-Segovia et al., 2016; Guo et al., 2017). The antagonistic activity of tunicamycin against Bacillus subtilis was known (Chen et al., 2010). In the previous study, tunicamycin was proved to be able to block the glycosylation on the synthesis of pro-C4, C2 and factor B and inhibited the secretion of these proteins, which led to decrease of the activity of the complement system, and the study was performed using tissue culture of guinea-pig peritoneal macrophages (Matthews et al., 1982), where the authors incubated for 6 h in the experiment. However, in our current study, we added tunicamycins directly to combine complement proteins in vitro to inhibit the complement complex activity, and our assay is more rapid (less than 1 h). We found that tunicamycin I, V and VII have anti-complement activity comparable to, if not stronger, than other natural anti-complementary agents (Table 3). Although complestatins show better anti-complement activity, these compounds have very poor solubility. Therefore, searching for new microbial-derived anti-complement agents are of great interest. Our results suggest that other compounds with known structures may also be explored to their anti-complement activities.
Nonactin and related analogs are a central class of macrocyclic ionophores consisting of 32-membered ring (a cyclodotricontane) built with 24 carbon and 8 oxygen atoms (Martinez-Haya et al., 2017). These compounds show antibiotic, antitumor and anti-virus activities and are also widely used for the preparation of ion-selective electrodes and sensors (Zhan and Zheng, 2016). Although production of nonactin has been attempted through chemical synthesis and optimization of biosynthesis, the yields are not satisfied so far (Zhan and Zheng, 2016). Therefore, manipulation of strain DUT11 may provide alternative source of nonactin and its analogs.
Medermycin is featured with a fused three-ring structure composed of a benzene ring, a quinone and a stereospecific pyran ring, and possesses significant antitumor activities against many types of cancer cells as well as antibacterial activity (He et al., 2015; Lü et al., 2015). Our studies indicate that DUT11 is a promising producer of medermycin for drug discovery.
Genome mining and metabolite analysis indicates that strain DUT11 has great potential in secondary metabolite production. Besides the production of tunicamycin, nonactin and medermycin analogs with known structures, many other BGCs in strain DUT11 may produce novel metabolites. For example, there are quite abundant PTMP BGCs in the genome sequence of DUT11. PTMPs are ribosomal peptides with various activities including antimicrobial, antitumor and antivirus activities (Arnison et al., 2012). We have found that the extracts of mycelia and supernatant of strain DUT11 have good antifungal and antibacterial activities (Supplementary Figure S5), and the novel PTMPs would be further explored to produce such bioactive compounds. Furthermore, up to four NRPS BGCs, three PKS BGCs and four terpene BGCs are quite different from the known BGCs in the literature, and novel metabolites may be produced by further genome mining.
From the phylogenetic tree based on 16S rRNA sequences, strain DUT11 is close to S. bacillaris, and up to now, the only active compound was reported in S. bacillaris strain L159 (Hu and Macmillan, 2012), which revealed a novel peptide with autophagy inhibitory activity. There is no other report about the genome analysis and potential analysis of secondary metabolites produced by the closely related strains of DUT11. Therefore, exploration of the novel metabolites in DUT11 will provide more insights on the biosynthetic potential of marine microorganisms.
Streptomyces sp. DUT11 tolerates up to 10% NaCl and grows well in 3% NaCl in the liquid TSB medium (Supplementary Figure S6). Its fast growth and ability of producing various secondary metabolites make it suitable to be further explored to produce useful metabolites using sea water. Our studies here indicate that marine streptomycetes are valuable sources for developing anti-complement agents and other novel metabolites for biotechnological applications.
CS, F-WB, and X-QZ conceived the project. X-NX and L-YC performed the study. X-NX, L-YC, and CC analyzed the data and drafted the manuscript. X-QZ, CS, and Y-JT critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the Open Funding Project of State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, China; National Natural Science Foundation of China (Grant Nos. 31600038 and 31600004) and Fundamental Research Funds for the Central Universities (GK201703035), China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Dr. Pieter C. Dorrestein at University of California at San Diego, La Jolla, CA, United States for the kind support to identify metabolites through molecular networking analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01318/full#supplementary-material
Agustina, U., Fabrizio, B., Claverías, F. P., Myriam, G., Moore, E. R., Michael, S., et al. (2016). Exploring the diversity and antimicrobial potential of marine actinobacteria from the Comau fjord in Northern Patagonia, Chile. Front. Microbiol 7:1135. doi: 10.3389/fmicb.2016.01135
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarez-Alvarez, R., Botas, A., Albillos, S. M., Rumbero, A., Martin, J. F., and Liras, P. (2015). Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Fact. 14:178. doi: 10.1186/s12934-015-0373-7
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2012). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/c2np20085f
Barnum, S. R. (2017). Complement: a primer for the coming therapeutic revolution. Pharmacol. Ther. 172, 63–72. doi: 10.1016/j.pharmthera.2016.11.014
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Berlin, K., Koren, S., Chin, C., Drake, J. P., Landolin, J. M., and Phillippy, A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630. doi: 10.1038/nbt.3238
Blodgett, J. A., Oh, D. C., Cao, S., Currie, C. R., Kolter, R., and Clardy, J. (2010). Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 11692–11697. doi: 10.1073/pnas.1001513107
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365, doi: 10.1038/srep08365
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Carver, T. J., Rutherford, K. M., Berriman, M., Rajandream, M. A., Barrell, B. G., and Parkhill, J. (2005). ACT: the artemis comparison tool. Bioinformatics 21, 3422–3423. doi: 10.1093/bioinformatics/bti553
Chen, L., Lai, Y. M., Yang, Y. L., and Zhao, X. (2016). Genome mining reveals the biosynthetic potential of the marine-derived strain Streptomyces marokkonensis M10. Synth. Syst. Biotechnol. 1, 56–65. doi: 10.1016/j.synbio.2016.02.005
Chen, W., Qu, D., Zhai, L., Tao, M., Wang, Y., Lin, S., et al. (2010). Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1, 1093–1105. doi: 10.1007/s13238-010-0127-6
Chung, I. M., Song, H. K., Kim, S. J., and Moon, H. I. (2011). Anticomplement activity of polyacetylenes from leaves of Dendropanax morbifera Leveille. Phytother. Res. 25, 784–786. doi: 10.1002/ptr.3336
Cóppola-Segovia, V., Cavarsan, C., Maia, F. G., Ferraz, A. C., Nakao, L. S., Lima, M. M., et al. (2016). ER stress induced by tunicamycin triggers alpha-synuclein oligomerization, dopaminergic neurons death and locomotor impairment: a new model of Parkinson’s disease. Mol. Neurobiol. 54, 5798–5806. doi: 10.1007/s12035-016-0114-x
English, A. C., Richards, S., Han, Y., Wang, M., Vee, V., Qu, J., et al. (2012). Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7:e47768. doi: 10.1371/journal.pone.0047768
Guo, Q., Li, H., Liu, J., Xu, L., Yang, L., Sun, Z., et al. (2017). Tunicamycin aggravates endoplasmic reticulum stress and airway inflammation via PERK-ATF4-CHOP signaling in a murine model of neutrophilic asthma. J. Asthma. 54, 125–133. doi: 10.1080/02770903.2016.1205085
Hao, J. J., Meng, Q. X., Yin, J. F., and Kirk, W. W. (2009). Characterization of a new Streptomyces strain, DS3024, that causes potato common scab. Plant. Dis. 93, 1329–1334.
Hayashi, S., Ozaki, T., Asamizu, S., Ikeda, H., Ômura, S., Oku, N., et al. (2014). Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem. Biol. 21, 679–688. doi: 10.1016/j.chembiol.2014.03.008
He, Q., Li, L., Yang, T., Li, R., and Li, A. (2015). Functional characterization of a ketoreductase-encoding gene med-ORF12 involved in the formation of a stereospecific pyran ring during the biosynthesis of an antitumor antibiotic medermycin. PLoS One 10:e0132431. doi: 10.1371/journal.pone.0132431
Hourcade, D. E., Akk, A. M., Mitchell, L. M., Zhou, H. F., Hauhart, R., and Pham, C. T. (2016). Anti-complement activity of the Ixodes scapularis salivary protein Salp20. Mol. Immunol. 69, 62–69. doi: 10.1016/j.molimm.2015.11.008
Hu, Y., and Macmillan, J. B. (2012). A new peptide isolated from a marine derived Streptomyces bacillaris. Nat. Prod. Commun. 2, 211–214.
Ian, E., Malko, D. B., Sekurova, O. N., Bredholt, H., Rückert, C., Borisova, M. E., et al. (2014). Genomics of sponge-associated Streptomyces spp. closely related to Streptomyces albus J1074: insights into marine adaptation and secondary metabolite biosynthesis potential. PLoS One. 9:e96719. doi: 10.1371/journal.pone.0096719
Ichinose, K. (2003). Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 149, 1633–1645. doi: 10.1099/mic.0.26310-0
Jin, J., Yang, X., Liu, T., Xiao, H., Wang, G., Ma, M., et al. (2018). Fluostatins M-Q featuring a 6-5-6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs. 16:87. doi: 10.3390/md16030087
Jin, W., Zhang, W., Liang, H., and Zhang, Q. (2015). The structure-activity relationship between marine algae polysaccharides and anti-complement activity. Mar. Drugs. 14:3. doi: 10.3390/md14010003
Kaneko, I., Kamoshida, K., and Takahashi, S. (1989). Complestatin, a potent anti-complement substance produced by Streptomyces lavendulae. J. Antibiot. (Tokyo) 42, 236–241. doi: 10.7164/antibiotics.42.236
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kersten, R. D., Yang, Y. L., Xu, Y., Cimermancic, P., Nam, S. J., Fenical, W., et al. (2011). A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7, 794–802. doi: 10.1038/nchembio.684
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Micr. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Lee, Y. K., Kim, H. W., Liu, C. L., and Lee, H. K. (2003). A simple method for DNA extraction from marine bacteria that produce extracellular materials. J. Microbiol. Methods. 52, 245–250. doi: 10.1016/S0167-7012(02)00180-X
Li, W. J., Xu, P., Schumann, P., Zhang, Y. Q., Pukall, R., Xu, L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Micr. 57, 1424–1428. doi: 10.1099/ijs.0.64749-0
Lü, J., He, Q., Huang, L., Cai, X., Guo, W., He, J., et al. (2015). Accumulation of a bioactive benzoisochromanequinone compound kalafungin by a wild type antitumor-medermycin-producing streptomycete strain. PLoS One 10:e0117690. doi: 10.1371/journal.pone.0117690
Luo, Y., Huang, H., Liang, J., Wang, M., Lu, L., Shao, Z., et al. (2013). Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Comms. 4:2894. doi: 10.1038/ncomms3894
Maksimov, M. O., Pan, S. J., and James, L. A. (2012). Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 29, 996–1006. doi: 10.1039/c2np20070h
Martinez-Haya, B., Aviles-Moreno, J. R., Hamad, S., and Elguero, J. (2017). On the ionophoric selectivity of nonactin and related macrotetrolide derivatives. Phys. Chem. Chem. Phys. 19, 1288–1297. doi: 10.1039/c6cp05324f
Matthews, W. J. Jr., Goldberger, G., Marino, J. T. Jr., Einstein, L. P., Gash, D. J., and Colten, H. R. (1982). Complement proteins C2, C4 and factor b. effect of glycosylation on their secretion and catabolism. Biochem. J. 204, 839–846. doi: 10.1042/bj2040839
Morgan, B. P., and Harris, C. L. (2015). Complement, a target for therapy in inflammatory and degenerative diseases. Nat. Rev. Drug. 14, 857–877. doi: 10.1038/nrd4657
Mu, L. H., Zhao, J. Y., Zhang, J., and Liu, P. (2016). Cycloartane triterpenes from Beesia calthaefolia and their anticomplement structure-activity relationship study. J. Asian. Nat. Prod. Res. 18, 1101–1107. doi: 10.1080/10286020.2016.1174698
Myronovskyi, M., Tokovenko, B., Manderscheid, N., Petzke, L., and Luzhetskyy, A. (2013). Complete genome sequence of Streptomyces fulvissimus. J. Biotechnol. 168, 117–118. doi: 10.1016/j.jbiotec.2013.08.013
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 27, 29–34. doi: 10.1093/nar/28.1.27
Ouk Kim, Y., Chun, J., Lee, I., and Park, S. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Micr. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Patzer, S. I., and Braun, V. (2009). Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 192, 426–435. doi: 10.1128/JB.01250-09
Petříčková, K., Chroňáková, A., Zelenka, T., Chrudimský, T., Pospíšil, S., Petříček, M., et al. (2015). Evolution of cyclizing 5-aminolevulinate synthases in the biosynthesis of actinomycete secondary metabolites: outcomes for genetic screening techniques. Front. Microbiol. 6:814. doi: 10.3389/fmicb.2015.00814
Price, N. P., and Tsvetanova, B. (2007). Biosynthesis of the tunicamycins: a review. J. Antibiot. Antibiot. 60, 485–491. doi: 10.1038/ja.2007.62
Prietodavó, A., Dias, T., Gomes, S. E., Rodrigues, S., Pareravaladez, Y., Borralho, P. M., et al. (2016). The madeira archipelago as a significant source of marine-derived actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 7:1594. doi: 10.3389/fmicb.2016.01594
Qin, Y., Wen, Q., Cao, J., Yin, C., Chen, D., and Cheng, Z. (2015). Flavonol glycosides and other phenolic compounds from Viola tianshanica and their anti-complement activities. Pharm. Biol 54, 1140–1147. doi: 10.3109/13880209.2015.1055635
Qu, H., Ricklin, D., and Lambris, J. D. (2009). Recent developments in low molecular weight complement inhibitors. Mol. Immunol. 47, 185–195. doi: 10.1016/j.molimm.2009.08.032
Quan, G. H., Oh, S. R., Kim, J. H., Lee, H. K., Kinghorn, A. D., and Chin, Y. W. (2010). Xanthone constituents of the fruits of Garcinia mangostana with anticomplement activity. Phytother. Res. 24, 1575–1577. doi: 10.1002/ptr.3177
Smith, W., Xiang, L., and Shen, B. (2000). Genetic localization and molecular characterization of the nons gene required for macrotetrolide biosynthesis in streptomyces griseus DSM40695. Antimicrob. Agents. Chemother. 44, 1809–1817. doi: 10.1128/AAC.44.7.1809-1817.2000
Subramani, R., and Aalbersberg, W. (2012). Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol. Res. 167, 571–580. doi: 10.1016/j.micres.2012.06.005
Tian, X., Zhang, Z., Yang, T., Chen, M., Li, J., Chen, F., et al. (2016). Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front. Microbiol. 7:998. doi: 10.3389/fmicb.2016.00998
Tsvetanova, B. C., and Price, N. P. (2001). Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Anal. Biochem. 289, 147–156. doi: 10.1006/abio.2000.4952
Ueda, K., Oinuma, K., Ikeda, G., Hosono, K., Ohnishi, Y., Horinouchi, S., et al. (2002). AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J. Bacteriol. 184, 1488–1492. doi: 10.1128/JB.184.5.1488-1492.2002
Waksman, S. A., and Lechevalier, H. A. (1953). Guide to the Classification and Identification of the Actinomycetes and their Antibiotics. Baltimore, MD: The Williams & Wilkins Co Press.
Walczak, R. J., Woo, A. J., Strohl, W. R., and Priestley, N. D. (2000). Nonactin biosynthesis: the potential nonactin biosynthesis gene cluster contains type II polyketide synthase-like genes. FEMS. Microbiol. Lett. 183, 171–175. doi: 10.1111/j.1574-6968.2000.tb08953.x
Wang, H., Cong, W. L., Fu, Z. L., Chen, D. F., and Wang, Q. (2017). Anti-complementary constituents of Viola kunawarensis. Nat. Prod. Res. 31, 2312–2315. doi: 10.1080/14786419.2017.1297446
Wang, L., Zhang, H., Zhang, X., and Chen, Z. (2008). Purification and identification of a novel heteropolysaccharide RBPS2a with anti-complementary activity from defatted rice bran. Food. Chem. 110, 150–155. doi: 10.1016/j.foodchem.2008.01.041
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837. doi: 10.1038/nbt.3597
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic. Acids. Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Wen, Q., Lu, Y., Chao, Z., and Chen, D. F. (2017). Anticomplement triterpenoids from the roots of Ilex asprella. Bioorg. Med. Chem. Lett. 27, 880–886. doi: 10.1016/j.bmcl.2017.01.007
Willey, J. M., and Gaskell, A. A. (2011). Morphogenetic signaling molecules of the streptomycetes. Chem. Rev. 111, 174–187. doi: 10.1021/cr1000404
Winn, M., Goss, R. J., Kimura, K., and Bugg, T. D. (2010). Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat. Prod. Rep. 27, 279–304. doi: 10.1039/b816215h
Xu, M., Wang, Y., Zhao, Z., Gao, G., Huang, S. X., Kang, Q., et al. (2016). Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl. Environ. Microbiol. 82, 5795–5805. doi: 10.1128/AEM.01383-16
Xu, Z., and Hao, B. (2009). CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic. Acids. Res. 37, W174–W178. doi: 10.1093/nar/gkp278
Ye, S., Molloy, B., Braña, A. F., Zabala, D., Olano, C., Cortés, J., et al. (2017). Identification by genome mining of a type I polyketide gene cluster from Streptomyces argillaceus involved in the biosynthesis of pyridine and piperidine alkaloids argimycins P. Front. Microbiol 8:194. doi: 10.3389/fmicb.2017.00194
Yin, X., Lu, Y., Cheng, Z. H., and Chen, D. F. (2016). Anti-complementary components of Helicteres angustifolia. Molecules 21:1506. doi: 10.3390/molecules21111506
Yu, Y., Tang, B., Dai, R., Zhang, B., Chen, L., Yang, H., et al. (2018). Identification of the streptothricin and tunicamycin biosynthetic gene clusters by genome mining in Streptomyces sp. strain fd1-xmd. Appl. Microbiol. Biotechnol. 102, 2621–2633. doi: 10.1007/s00253-018-8748-4
Zhan, Y., and Zheng, S. (2016). Efficient production of nonactin by Streptomyces griseus subsp. griseus. Can. J. Microbiol. 62, 711–714. doi: 10.1139/cjm-2016-0248
Zhang, H., Lee, Y. K., Zhang, W., and Lee, H. K. (2006). Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Anton. van. Leeuw. 90, 159–169. doi: 10.1007/s10482-006-9070-1
Zhang, Y., Huang, H., Xu, S., Wang, B., Ju, J., Tan, H., et al. (2015). Activation and enhancement of fredericamycin a production in deepsea-derived Streptomyces somaliensis SCSIO ZH66 by using ribosome engineering and response surface methodology. Microb. Cell. Fact. 14:64. doi: 10.1186/s12934-015-0244-2
Zhao, X., and Yang, T. (2011). Draft genome sequence of the marine sediment-derived actinomycete Streptomyces xinghaiensis NRRL B24674T. J. Bacteriol. 193:5543. doi: 10.1128/JB.05689-11
Keywords: marine streptomycetes, genome mining, secondary metabolites, tunicamycin, anti-complement activity
Citation: Xu X-N, Chen L-Y, Chen C, Tang Y-J, Bai F-W, Su C and Zhao X-Q (2018) Genome Mining of the Marine Actinomycete Streptomyces sp. DUT11 and Discovery of Tunicamycins as Anti-complement Agents. Front. Microbiol. 9:1318. doi: 10.3389/fmicb.2018.01318
Received: 31 March 2018; Accepted: 30 May 2018;
Published: 20 June 2018.
Edited by:
Wen-Jun Li, Sun Yat-sen University, ChinaCopyright © 2018 Xu, Chen, Chen, Tang, Bai, Su and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Su, c3VjaHVuQHNubnUuZWR1LmNu Xin-Qing Zhao, eHF6aGFvQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.