
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 June 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01217
 Patricia Alba1
Patricia Alba1 Pimlapas Leekitcharoenphon2
Pimlapas Leekitcharoenphon2 Alessia Franco1
Alessia Franco1 Fabiola Feltrin1
Fabiola Feltrin1 Angela Ianzano1
Angela Ianzano1 Andrea Caprioli1
Andrea Caprioli1 Fiorentino Stravino1
Fiorentino Stravino1 Rene S. Hendriksen2
Rene S. Hendriksen2 Valeria Bortolaia2
Valeria Bortolaia2 Antonio Battisti1*
Antonio Battisti1*Colistin resistance by mobilisable mcr genes has been described in bacteria of food-animal origin worldwide, which has raised public health concerns about its potential foodborne transmission to human pathogenic bacteria. Here we provide baseline information on the molecular epidemiology of colistin-resistant, mcr-positive Escherichia coli and Salmonella isolates in food-producing animals in Italy in 2014-2015. A total 678, 861 and 236 indicator E. coli, Extended Spectrum Beta-Lactamase (ESBL)/AmpC-producing E. coli, and Salmonella isolates, respectively, were tested for colistin susceptibility. These isolates were collected according to the EU harmonized antimicrobial resistance monitoring program and are representative of at least 90 and 80% of the Italian poultry (broiler chickens and turkeys) and livestock (pigs and bovines < 12 months) production, respectively. Whole genome sequencing by Illumina technology and bioinformatics (Center for Genomic Epidemiology pipeline) were used to type 42 mcr-positive isolates by PCR. Colistin resistance was mainly observed in the ESBL/AmpC E. coli population, and was present in 25.9, 5.3, 6.5, and 3.9% of such isolates in turkeys, broilers, pigs, and bovines, respectively. Most colistin-resistant isolates (141/161, 87.5%) harbored genes of the mcr-1 group. mcr-1 was also detected in a small proportion of Salmonella isolates (3/146, 2.0%) in turkeys. Additional mcr types were mcr-3 in four ESBL-producing E. coli from bovines, and two mcr-4 in ESBL (n = 1) and indicator E. coli (n = 1) from pigs and bovines. We describe notable diversity of mcr variants with predominance of mcr-1.1 and mcr-1.2 on conjugative IncX4 plasmids in E. coli and in Salmonella serovars Typhimurium, Newport, Blockley from turkey. A new variant, mcr-1.13 was detected in the chromosome in E. coli in turkey and pig isolates. Additionally, we describe mcr-3.2 and mcr-4.3 in E. coli from bovines, and mcr-4.2 in E. coli from pigs. These findings elucidate the epidemiology of colistin resistance in food-producing animals in Italy along with its genetic background, and highlight the likelihood of mcr horizontal transfer between commensal bacteria and major food-borne pathogens (Salmonella) within the same type of productions. Thorough action and strategies are needed in order to mitigate the risk of mcr transfer to humans, in a “One Health” perspective.
Colistin is a polymyxin classified among the Highest Priority Critically Important Antimicrobials for human medicine by the World Health Organization (WHO), and it is considered a last resort antimicrobial for the treatment of infections by carbapenem-resistant Enterobacteriaceae in humans (Poirel et al., 2017).
In 2015, colistin resistance mediated by mcr-1, a phosphoethanolamine transferase gene located on a transferable plasmid was first reported in Escherichia coli from animals, food and patients from China (Liu et al., 2016). Since then, at least 32 countries from the five continents have found mcr-1 in E. coli isolates from different sources including humans, animals and foods (Xavier et al., 2016).
In Europe, the presence of mcr-1 was first detected in E. coli from poultry meat and humans in Denmark (Hasman et al., 2015). Subsequently, this gene was found in Enterobacteriaceae from different sources in almost all European countries, including Italy (Battisti, 2016; Cannatelli et al., 2016). In 2016, a new mcr gene, mcr-2, was described in E. coli in calves and piglets in Belgium (Xavier et al., 2016), followed by the description of three additional mobile colistin resistance genes in 2017, namely mcr-3 (Yin et al., 2017) in E. coli isolated from pig in China, mcr-4 (Carattoli et al., 2017) in Salmonella and E. coli from pigs in Italy, Spain and Belgium, and mcr-5 (Borowiak et al., 2017) in Salmonella Paratyphi B from poultry and environmental sources from Germany.
At present, eleven mcr-1 variants (KP347127, KX236309, KU934208, KY041856, KY283125, KY352406, KY488488, KY683842, KY964067, KY853650, and LC337668), one mcr-2 variant (LT598652), six mcr-3 variants (KY924928, NPZH01000177, FLXA01000011, MF463699, MG214533, and MG489958), three mcr-4 variants (MF543359, MG581979, and ERS1801979) and mcr-5 (KY807921) have been described in Enterobacteriaceae according to GenBank records (last accessed 21st February 2018).
It is clear that the epidemiology of transferable mcr-mediated colistin resistance is evolving rapidly and timely information on prevalence and molecular epidemiology of mcr-positive isolates is needed to enhance surveillance, and implement measures to prevention and to control further spread of colistin resistance. In the European Union (EU), the harmonized antimicrobial resistance (AMR) monitoring and reporting in poultry and livestock, which includes colistin susceptibility testing in E. coli and Salmonella, ensures that prevalence of colistin-resistant bacteria in a representative proportion of the food-animal population is reported from each Member State (MS) (Decision 2013/652/EU). However, the lack of molecular data limits the epidemiologic monitoring of colistin resistance (Schrijver et al., 2017).
The aim of this study is to determine the prevalence of colistin resistance, and the molecular epidemiology of mcr-mediated colistin resistance genes and their genetic environment in commensal E. coli, Extended Spectrum Beta-Lactamase (ESBL)/AmpC-producing E. coli, and Salmonella spp. in food-animals in Italy in 2014-2015.
Study design and sampling were performed according to Decision 2013/652/EU1, which mandates each EU Member State (MS) to collect caecal content samples from different epidemiological units of poultry flocks (broiler chickens, fattening turkeys), fattening pigs and bovines < 12 months at slaughter.
Samples were collected from broiler chickens (n = 300) and fattening turkeys (n = 300) in 2014, and from fattening pigs (n = 304) and bovines < 12 months (n = 223) in 2015 (Table 1). The regional stratification of samples represented at least 90 and 80% of the Italian poultry (broiler chickens and turkeys) and livestock (pigs and bovines < 12 months) production, respectively.
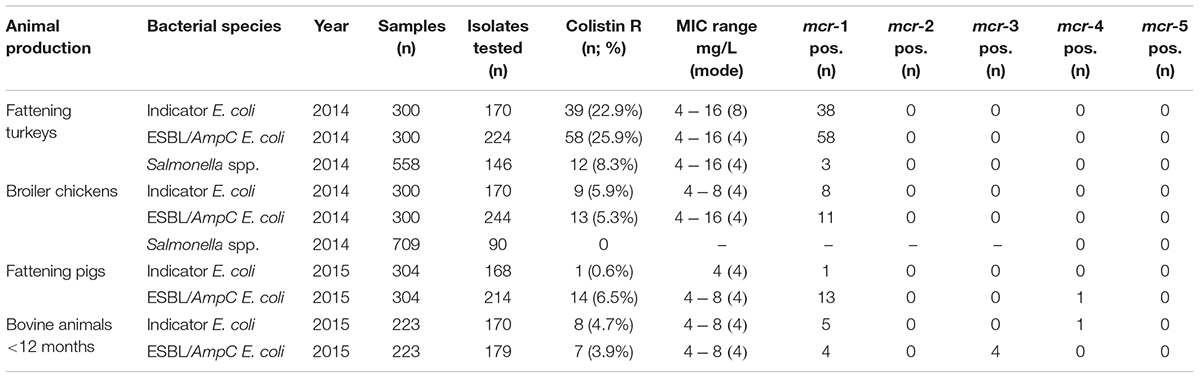
TABLE 1. Colistin-resistance and mcr genes in Escherichia coli and Salmonella sp. from caecal samples in animal primary productions, Italy, 2014–2015.
In addition, 558 and 709 samples from fattening turkey and broiler chicken flocks, respectively, were collected within the voluntary national Salmonella monitoring framework (Decision 2013/652/EU) in 2014 (Table 1). In compliance with Decision 2013/652/EU, E. coli isolation and identification, were performed according to the EURL-AR protocols2, whereas Salmonella spp., isolation, identification and serotyping were performed according to the ISO 6579:2002/Amd 1:2017 protocols.
Antimicrobial susceptibility was tested by minimum inhibitory concentration (MIC) determination using the broth microdilution method, and consensus 96-well microtiter plates (TREK Diagnostic Systems, Westlake, OH, United States). Antimicrobials tested, dilution ranges and interpretation of MIC values were in accordance with Decision 2013/652/EU.
The presence of mcr was investigated in all isolates displaying colistin MIC above the epidemiological cut-off (i.e., > 2 mg/L). A multiple PCR was used to detect mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 (Rebelo et al., 2018).
ESBL/AmpC-producing E. coli positive for mcr were further screened for blaCTX-M, blaSHV, blaTEM, blaOXA, blaCMY-1, and blaCMY -2 using primers and PCR conditions previously described (Donati et al., 2014; Franco et al., 2015). Obtained amplicons were Sanger sequenced and analyzed as previously described (Donati et al., 2014; Franco et al., 2015).
Four representative E. coli and three Salmonella sp. isolates from turkeys were selected as donors. Conjugation experiments were performed as previously described (Franco et al., 2015), with the only modification regarding the MacConkey agar plates selective for transconjugants which contained 2 mg/L colistin sulfate and 100 mg/L rifampicin in this study.
A total 42 isolates which tested mcr-positive by PCR, (28 E. coli and three Salmonella enterica isolates from turkeys, five E. coli from pigs and six E. coli from cattle), and the seven E. coli K-12 isolates result of the conjugation experiments were Whole Genome Sequenced. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Libraries were prepared for Illumina pair-end sequencing using the Illumina (Illumina, Inc., San Diego, CA, United States) NexteraXT® Guide 150319425031942. Sequencing was performed using an Illumina platform (MiSeq or HiSeq2000). Raw sequence data were submitted to the European Nucleotide Archive3 under study accession no.: PRJEB23728, PRJEB23778, PRJEB21546, and PRJEB26479.
Raw reads were assembled and analyzed using the pipeline from the Center for Genomic Epidemiology (CGE4, Thomsen et al., 2016), with default settings. This pipeline performed de novo assembly (Velvet based), species identification (KmerFinder 2.1), Multilocus Sequence Typing (MLST 1.6), identification of virulence (VirulenceFinder 1.2) and antimicrobial resistance genes (ResFinder 2.1), identification of plasmid incompatibility groups (PlasmidFinder 1.2) and plasmid MLST (pMLST 1.4). When identity values for mcr were less than 100% in the ResFinder output, the sequence was submitted to online BLAST5 (Zhang et al., 2000) to identify the exact mcr variant.
Manual annotation of the contigs containing selected mcr variants was performed using RAST (Aziz et al., 2008), BLAST (Zhang et al., 2000) and ISfinder (Siguier et al., 2006). The contigs harboring the new mcr variants were compared with reference sequences using BLAST and EasyFig (Sullivan et al., 2011). The references sequences used for comparison were CP016034, KP347127 and KY924928.
In 2014, colistin resistance was detected in 25.9% (58/224) and 5.3% (13/244) of the ESBL/AmpC-producing E. coli from fattening turkeys and broilers, respectively. In turkey flocks, all but two colistin-resistant E. coli were multidrug-resistant (MDR) isolates, (i.e., resistant to three antimicrobial classes). In MDR ESBL/AmpC-producing E. coli population of turkey flocks, colistin resistance was associated with concurrent fluoroquinolone microbiological resistance (ciprofloxacin MIC >0.064 mg/L) in 51 of 58 isolates (87.9%), with 35/58 (60.3%) displaying fluoroquinolone clinical resistance (MIC >1 mg/L, mode 8 mg/L) (Supplementary Table 1).
A similar prevalence was observed among the indicator commensal E. coli, with colistin resistance occurring in 22.9% (39/170) and 5.9% (9/170) of isolates from turkeys and broilers, respectively (Table 1). Nearly all colistin-resistant E. coli from turkey and broilers tested PCR-positive for mcr-1 independent of colistin MIC (Table 1).
In 2014, colistin resistance was detected in 8.3% (12/146) and 0% (0/90) of Salmonella spp. isolates from fattening turkeys and broiler chickens, respectively. mcr-1 was detected only in three of twelve isolates, which displayed colistin MIC = 8 mg/L. The remaining isolates (n = 9) did not yield any mcr amplicon (Table 1).
In 2015, colistin resistance occurred in 6.5% (14/214) and 3.9% (7/179) of ESBL/AmpC-producing E. coli from fattening pigs and bovines <12 months, respectively. Among the indicator E. coli population, colistin resistance occurred in 0.6% (1/168) and 4.7% (8/170) of isolates from fattening pigs and bovines <12 months, respectively (Table 1). All colistin-resistant E. coli from pigs tested positive for mcr genes, 14 out of 15 harbored mcr-1 and one ESBL-producing isolate tested positive for mcr-4 (Table 1). Only five out of eight colistin-resistant indicator E. coli from bovines yielded mcr-1 and one out of eight presented mcr-4 (Table 1). All seven ESBL/AmpC-producing E. coli from bovines yielded mcr genes (Table 1). Thus, mcr-1 was detected in three isolates, mcr-3 in three isolates and both mcr-1 and mcr-3 in one isolate (Supplementary Table 1).
High diversity of Sequences Types (STs) was evident when studying the genome of the 28 E. coli and three Salmonella enterica isolates from turkeys, five E. coli from pigs and six E. coli from bovine <12 months selected for WGS. In E. coli from turkeys, 20 STs were identified. The most represented STs were ST-155 and ST-156 with four isolates each, followed by ST-744 with three isolates and ST-101 with two isolates (Supplementary Table 1). The five E. coli from pigs and six E. coli from cattle isolates presented different ST. The serotype and the STs of the three S. enterica were also different: S. Typhimurium ST-3515, S. Blockley ST-52 and S. Newport ST-45 (Supplementary Table 1).
mcr-1.1 (NG_050417.1) was present in 24 isolates: 18 E. coli and two Salmonella isolates from turkeys, three E. coli from pigs and one from bovines. mcr-1.2 (KX236309.1) was detected in eight E. coli and one Salmonella from turkeys. In addition, one E. coli isolate from turkey presented a silent mutation, C801T, and two E. coli isolates, one from turkey and one from fattening pigs showed a new variant (hereafter termed mcr-1.13) of mcr-1.1 with two non-synonymous mutations: M2V and S14G. Four E. coli from bovines <12 months presented a mcr-3 variant, identified as mcr-3.2 (NG_055523.1), and one of them presented both mcr-1.1 and mcr-3.2 (Supplementary Table 1). Also, one ESBL(CTX-M-1)-producing E. coli isolate from pigs and one indicator E. coli from bovines presented mcr-4.2 and mcr-4.3, respectively.
The two E. coli isolates harboring mcr-1.13 allele, ID:14077295 from turkey and ID:15056414 from fattening pig, were genetically different, as shown by MLST (ST-69; ST-5995) and different resistance gene content (Supplementary Table 1). The virulence gene gad was detected in both isolates, but it was the only virulence-associated gene found in the pig isolate (Supplementary Table 1). Both isolates harbored the IncF replicon plasmid, but with different plasmid MLST: IncF [F1:A1:B20] and IncF [F46:A-:B42], for the turkey and the pig isolate, respectively (Supplementary Table 1).
All the nine isolates presenting mcr-1.2 (IDs:14087995, 14069546, 14062120, 14044802, 14091902, 14045775, 14047606, 14083136, and 14085183) were isolated from turkey. These isolates were one S. Blockley, and eight E. coli isolates with different STs (Supplementary Table 1). The resistance and virulence genes were variable among those isolates (Supplementary Table 1). For example, one of them (ID:14044802) was only resistant to colistin and only presented the accessory gene mcr-1.2, while all other isolates were MDR and ESC-R, and presented ESBL genes such as blaCTX-M-1 or blaTEM-52 or other different beta-lactamase genes (Supplementary Table 1). A variety of plasmid incompatibility groups was observed including the plasmids IncFII, IncFI, IncHI2, IncI, IncN in different proportions, but all the nine isolates presented the plasmid IncX4 (Supplementary Table 1). In the isolate ID:14065450, the contig containing mcr-1.2 also contained the replicon plasmid IncX4 (contig 35).
The four WGS sequenced isolates harboring mcr-3.2 (IDs:15054212, 15038100, 15056874, and 15078696), all of them E. coli from bovines <12 months, had different STs and serotype (Supplementary Table 1). All four were MDR and presented the ESBL gene blaCTX-M-55, although always found in contigs other than those containing mcr-3.2. The four isolates also shared other resistance accessory genes as sul3, aac(3)-Iid, aadA2, or floR. A number of different plasmids were detected in all four isolates, with only IncF (F46:A-:B20), IncX1 and IncR detected in all of them. The contigs harboring mcr-3.2 did not contain any plasmid replicon type (Supplementary Table 1).
One of the mcr-3.2-positive E. coli (ID:15054212), presented also mcr-1.1. This isolate was ST-744 and contained the following plasmids: IncFIB, IncX4, IncFIC(FII), IncX1, Col156, IncR. mcr-1.1 was located in a 32,823 bp contig that included also the replicon sequence of IncX4 (Supplementary Table 1).
The mcr-4.2 variant was isolated from an E. coli from fattening pig (ID: 15057173-5). This isolate, ST-410, presented the following accessory resistance genes: blaCTX-M-1, tet(B)-like, sul1,sul2, aadA1, aadB, aph(3′)-Ic-like, strA, strB, mph(A), floR-like and qnrB42-like (Supplementary Table 1). mcr-4.2 and ColE10 replicon sequence were located in the same contig (contig 105; length 7699 bp). In addition to the ColE10 plasmid replicon, this isolate presented other plasmids (Supplementary Table 1).
The mcr-4.3 variant was harbored in a E. coli (ST-399) isolated from a bovine <12 months (ID: 15050011-1) that also presented other accessory resistance genes as blaTEM-1B, tet(B), sul1-like, sul3, dfrA1, aac(3)-IIa, aadA1, aph(3′)-Ic-like, strA, strB and catA1-like (Supplementary Table 1). A variety of plasmids was found, including IncFIA, IncHI1A, IncHI1B(R27), ColE10, IncQ1, ColRNAI, p0111, and Col(MG828), but none in the same contig containing mcr-4.3.
mcr-1.13 was identified in E. coli strain 14077295 (contig 564; 10,310 bp lenght) from turkeys and in E. coli strain 15056414 (contig 27; 50,840 bp lenght) from pigs. The BLAST alignment of the two contigs showed 99% identity across the entire length of the shortest contig (10,310 bp). No plasmid replicon was detected in any of the two contigs. mcr-1.13 (from nt 33,836 to nt 35,462 in contig 27,) was upstream to a PAP2 superfamily hypothetical protein (from nt 35,509 to nt 36,256). Insertion sequences (IS) were detected flanking the mcr cassette: upstream mcr-1.13 a truncated IS66 (from nt 33,335 to 33,413 bp and from 33,455 to 33,651 bp) and downstream a small fragment of IS66 (from nt 36,261 to nt 36,392) and IS110 (from nt 36,449 to nt 36,816). Upstream the cassette, there was the gene coding for 50S ribosome-binding GTPase family protein (from nt 15,712 to nt 16,627) and downsteam the cassette, there was the coding gene of the subunit YeeA of the methylase (from nt 37,032 to nt 39,849) (Figure 1). In BLAST results, contig sequences showed 93–99% identity to E. coli chromosome sequence CP016034 except for the region flanked by ISs and containing mcr-1.13 and PAP2 superfamily coding genes that had 99 and 100% identity with the same region of the mcr plasmid pHNSHP45(KP347127), for the mcr gene and the PAP2 superfamily coding gene, respectively (Figure 1).
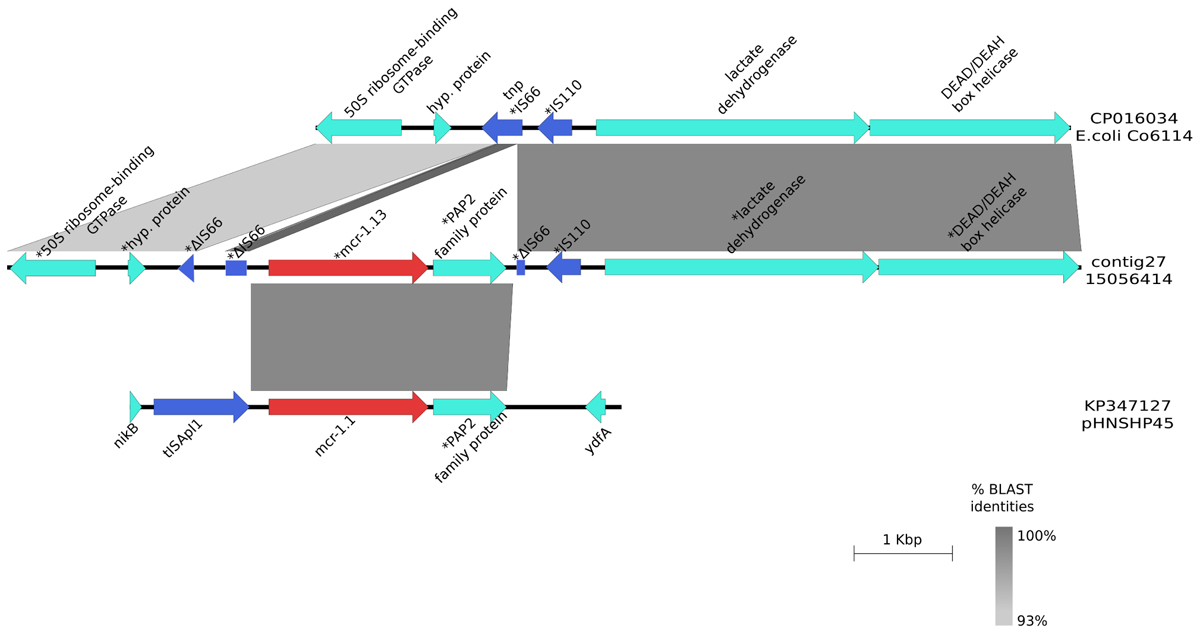
FIGURE 1. Graphical representation of the mcr-1.13 contig. Graphical representation of the BLAST analysis between the contig harboring mcr-1.13 from the pig-origin E. coli isolate (contig27; 15056414) and the mcr plasmid pHNSHP45 (KP347127; region 21000bp- 26000bp) and genomic DNA of E. coli Co6114 (CP016034; region 91844bp – 99527bp). ∗Indicated that the gene was manually annotated. The gray area represents the blast identities, the percentage of identity is indicated in the legend. Gene colors: red, mcr-1; blue, transposases or IS elements.
mcr-3.2 was detected in E. coli isolates 15054212 (contig 77), 15038100 (contig 128), 15056874 (contig 52), and 15078696 (contig 197) measuring 3,346, 9,285, 5,921, and 5.279 bp, respectively. All contigs were 100% identical to the shortest one. Upstream and downstream mcr-3.2 (from 766 to 2,392 bp in contig 77) a diacylglycerol kinase gene (from 268 to 249 bp) and a gene coding for a NimC/NimA putative family protein (from 2,436 to 2,696 bp) were detected, respectively. These three genes were flanked by IS3 (from 1 to 127 bp) and Tn3 (from 2,943 to 3,346 bp) (Figure 2). The shortest contig harboring these genes (contig number 77) showed a 99% identity with the pWJ1 plasmid containing mcr-3.1 (KY924928) region from 160,180 to 163,525 bp. The genes surrounding the cassette in our strains were not found in the pWJ1 plasmid (KY924928).
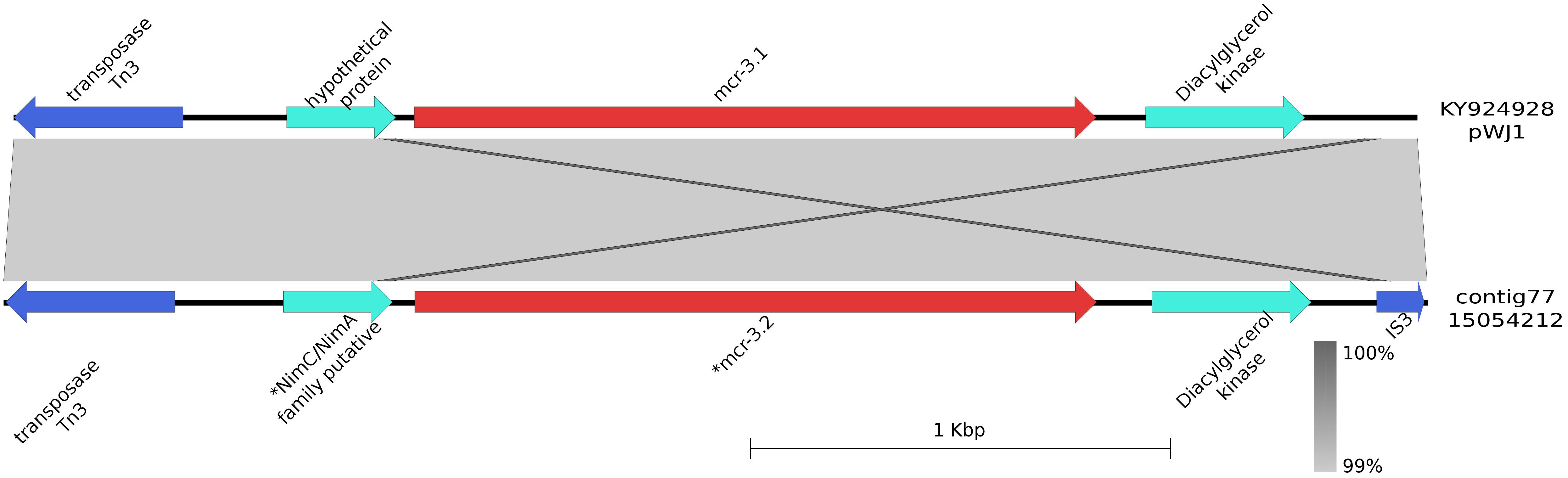
FIGURE 2. Graphical representation of the mcr-3.2 contig. Graphical representation of the BLAST analysis between the contig harboring mcr-3.2 in the bovine-origin E. coli (contig77; 15054212) and the mcr plasmid pWJ1 (KY924928; region 160180 to 163525 bp). ∗Indicated that the gene was manually annotated. The gray area represents the blast identities, the percentage of identity is indicated in the legend. Gene colors: red, mcr-1; blue, transposases or IS elements.
E. coli harboring mcr-1.1 (IDs:14043377 and 14065450), mcr-1.2 (ID:14047606) and mcr-1.13 (ID:14077295), S. Typhimurium harboring mcr-1.1 (ID:14043372), S. Newport harboring mcr-1.1 (ID:14038647) and S. Blockley harboring mcr-1.2 (ID:14085183) were used as donors for conjugation experiments (Table 2 and Supplementary Table 1). All donors, except the mcr-1.13-harboring E. coli, transferred mcr by conjugation to the E. coli K-12 recipient strain. mcr-1.1 and mcr-1.2 were carried by IncX4 plasmids that transferred either alone or in combination with additional plasmids (Table 2 and Supplementary Table 1). One isolate (ID: 14065450) displayed in the same contig both mcr-1.1 and the replicon of the IncX4 plasmid (contig 68).
The high diversity of transferable colistin resistance mediated by mcr genes and alleles, quickly spreading across pathogenic enterobacteria globally (Kluytmans, 2017), is an emerging challenge for treatment of Gram-negative infections due to increased occurrence of Healthcare-Associated Extremely Drug Resistant bacterial pathogens (EARS-Net, 2015).
In the present study, we found high prevalence (∼25%) of colistin resistance in both indicator commensal E. coli and ESBL/AmpC-producing E. coli in turkeys in Italy. In other Italian primary productions such as broilers, fattening pigs and bovines <12 months, colistin-resistant E. coli occurred at relatively lower, though still relevant levels.
From a (Veterinary) Public Health perspective, the spread of colistin resistance across production types is of major concern also considering that it was often associated with resistance to multiple drugs and notably also to fluoroquinolones. For instance, MDR E. coli exhibiting co-resistance to three Critically Important Antimicrobials for humans were detected in approximately 23% of turkey flocks investigated.
By using WGS, the vast diversity of mcr types and variants was evident within and across the major production types. At least three mcr-1 alleles were identified in turkeys, pigs and bovines <12 months in a 2-year-period, one of which has not been previously described. The majority of isolates harbored mcr-1.1, but mcr-1.2, mcr-1.13, mcr-3.2, mcr-4.2 and mcr-4.3 were also detected.
The core genomes of the E. coli and Salmonella isolates harboring mcr-1 were quite variable as shown by diversity of MLST types, in agreement with what has been observed in different contexts previously (Hasman et al., 2015; Bernasconi et al., 2016; Cannatelli et al., 2016; Zhao et al., 2017).
Conjugation experiments and WGS data analysis confirmed that mcr-1.1 and mcr-1.2 were located on IncX4 plasmids in E. coli and Salmonella spp. from turkey. In Europe, IncX4 plasmids have been described in association with mcr-1 in isolates of animal origin in Belgium (Xavier et al., 2016; Garcia-Graells et al., 2017) and from humans in Italy (Di Pilato et al., 2016).
The mcr-1.2 gene was first described on an IncX4 plasmid in a Klebsiella pneumoniae isolated from a human sample in the Tuscany Region (Italy) (Di Pilato et al., 2016). In our study, this variant was found on IncX4 plasmids in intestinal Salmonella and E. coli spread throughout the Italian fattening turkey production. Our observations suggest the transmission of mcr-positive IncX4 plasmids between different bacterial species, with the possibility of transmission from animals to humans, or vice versa.
The mcr-3 gene was first described on an IncHI plasmid (Yin et al., 2017), whereas the mcr-3.2 variant has been described from the whole genome shotgun sequence of a Shigella sonnei strain (NG_055523; NPZH01000177) isolated in the United States, but to our understanding, no formal description has been published to date. In our study, four ESBL/AmpC-producing E. coli isolates from bovines <12 months presented mcr-3.2. The WGS data analysis performed on these isolates did not allow us to determine the genomic location of mcr-3.2. However, only one of these isolates harbored an IncHI plasmid. A ST heterogeneity of E. coli isolates harboring the mcr-3.2 gene was evident. Further conjugation or transformation experiments are needed in order to elucidate if the mcr-3.2 found in E. coli in Italian cattle is plasmid-borne and transferable.
One E. coli from cattle was simultaneously positive for mcr-3.2 and mcr-1.1. In this isolate, mcr-1.1, was located on an IncX4 plasmid. An E. coli carrying both mcr-1.1 and mcr-3.2 and also isolated from cattle has been recently described in Spain (Hernández et al., 2017). Interestingly, all four mcr-3.2-positive E. coli isolated detected in bovines <12 months in Italy carried the ESBL blaCTX-M-55 gene, apparently an emerging variant in veal calves in Italy. The same observation has been made by other authors (Hernández et al., 2017; Roer et al., 2017). Taken together, these features could be suggestive of a genetic linkage between the two genes. Our study, however, provides preliminary data that the mcr-3.2 and the blaCTX-M-55 genes may not be linked on the same genetic element, and that this aspect needs further investigation.
The mcr-4.2 gene was described recently from two S. enterica Typhimurium (monophasic variant) isolated from human samples in Italy (Carretto et al., 2018). So far, this variant has not been associated to any plasmid. In our study, the mcr-4.2 gene identified in a ESBL(CTX-M-1)-producing E. coli isolated from a fattening pig, was located on a ColE plasmid, exactly the same type of plasmid in which mcr-4.1 was described by Carattoli et al., 2017. The mcr-4.3 variant was first described from a Salmonella Kedougou isolated from pigs in Spain (Rebelo et al., 2018). In our study, mcr-4.3 was harbored by an indicator E. coli from a bovine caecal sample. All the mcr-4 variants described so far have been found in Salmonella or E. coli from animals and from humans, despite their localisation on non-conjugative plasmids, similarly to what has been found with mcr-4.1 by Carattoli et al. (2017).
The mcr-1.13 gene was located on a mobile genetic element inserted in the chromosome. The mcr cassette found is different from the chromosomal mcr cassettes described so far, because of the presence of ISs other than ISplA (Snesrud et al., 2016; Li et al., 2017). By considering the high rate of self-transferability of the IncX plasmids (Sun et al., 2017) and that IncX4 is dominant in the Italian primary productions surveyed, this finding suggests that the mcr cassette may have been acquired from an IncX4 plasmid which lacks ISplA insertion sequence (Bernasconi et al., 2016; Matamoros et al., 2017; Sun et al., 2017). It is likely that the insertion occurred in different moments, since the two E. coli isolates derived from two different animal productions and presented different genome content. The ability of the mcr cassette to jump into several types of plasmids (IncI2, IncX4, IncHI2, and IncP) or into the chromosome is highly concerning also in view of the possibility of insertion into plasmids already present in MDR isolates, or even of the creation of a new mega-plasmid as occurred in Salmonella Infantis (Franco et al., 2015). This would imply that colistin resistance could be co-selected by the use of others antimicrobials. Indeed, multidrug resistance in colistin-resistant, mcr-positive isolates is a constant feature we have observed in these population-based studies. These findings highlight how rapidly mobile genetic elements can be acquired by Enterobacteriaceae and how genes can mutate in environments with high selective pressure as that occurring in the intensive farming systems. In this respect, the selection pressure exerted by the use of colistin and the complex co-selection mechanisms triggered by the overall high use of several antibiotic classes in the meat-producing industry is likely to have played a major role. For instance, the overall exposure to antibiotics in food-producing animals in Italy in 2014 was estimated around 360 mg/Population Correction Unit (PCU, i.e., mg per kilogram of biomass of farmed animals), and the exposure to colistin only was estimated around 29 mg/PCU (ESVAC, 2016). The mean, median and range of total sales in Europe (29 countries) were 108, 66, and 3 – 419 mg/PCU, respectively, while 22 out of 29 countries reported sales of colistin ≤5 mg/PCU (range of the 29 EU countries: 0.06–36.10 mg/PCU). It is well known that the intensively farmed, meat-producing animals (turkeys, broilers, pigs, bovines <12 months) are the population categories at risk of exposure to higher amounts of colistin in modern farming systems, and the results of this study strongly support the general concept that the spread of mcr-mediated colistin resistance has been favored by the semi-continuous and high exposure to colistin in these production chains.
Whether animals are an important source for human extraintestinal pathogenic E. coli (ExPEC) infections in humans is still a matter of debate (Bélanger et al., 2011). Few studies provide information and comparison on STs and plasmids harboring ESBLs or transferable AmpCs (Leverstein-van Hall et al., 2011) for both human and food-producing animal isolates. Although conclusive epidemiological evidence is still lacking, it has been proposed that some human ExPEC infections could arise from poultry and pig ExPEC reservoirs (Jakobsen et al., 2010). That being said, it is interesting to notice that at least one-fourth of E. coli isolates described in our study belong to the same STs as isolates associated with human ExPEC infections in Europe. In some cases (Leverstein-van Hall et al., 2011; Mavroidi et al., 2012; Brolund et al., 2014) they also share the same ESBL (e.g., ST69 and CTX-M-14, ST410 and CTX-M-1). As for colistin-resistant E. coli, one of the mcr-1.1 positive isolates from turkeys here described belongs to ST131, although it lacks the virulence gene markers and ESBL generally associated with the globally spread human clinical clone. Indeed, a mcr-1-positive, ESBL-negative ST-131 E. coli was also described in human bloodstream infections in Italy in 2017 (Corbella et al., 2017). Overall, these observations should be taken very cautiously, since methods for genome analysis and parameters for assessing relatedness among both core genomes and accessory genomes are quickly evolving, which implies that it may be necessary to re-evaluate any earlier conclusions on relatedness or source attribution based on partial molecular characteristics, as previously shown (De Been et al., 2014).
In conclusion, harmonized cross sectional studies at slaughter like the ones implemented by the EU represent a very important tool for a deep insight into trends and emergence of antimicrobial resistance traits and patterns in major food-borne pathogens and commensal opportunistic bacteria. Especially when occurring at high prevalence, the spread of transferable colistin resistance in E. coli (both indicator commensal and ESBL/AmpC-producing isolates) is to be considered a concern per se. Additionally, as a general principle, the high spread of resistance increases the probability of transfer of specific resistance traits also to major zoonotic pathogens, such as Salmonella spp. The hypothesis that horizontal transfer, so far, has played a major role in spread of colistin resistance among bacteria in Italian meat-producing animals is supported by the observed heterogeneity of mcr-positive E. coli. Indeed, at least in the Italian turkey productions, we demonstrated that the same transferable determinant of colistin-resistance is being carried on the same conjugative plasmid in both E. coli and major Salmonella serotypes detected in the same intensive-farming industry.
For the above reasons, quick and thorough action should be taken by the farming industry and by the Competent Authorities to drastically reduce the use of colistin in food-producing animals, especially in turkeys, following the recommendations of the European Medicines Agency (≤5 mg/PCU). EU Member States were encouraged to set stricter national targets, ideally below 1 mg/PCU colistin. We also strongly recommend reducing the overall use of all other classes of antibiotics at primary production level, in order to mitigate the effects of the complex mechanisms behind co-selection and multidrug resistance toward Critically Important Antimicrobials, in a “Consumer Protection” and a “One Health” perspective.
PA, PL, AF, RH, VB, and AB conceived and designed the experiments. PA, PL, FF, AI, and FS performed the experiments. PA, AF, FF, AI, RH, VB, and AB analyzed the data. PA, PL, AF, AC, RH, VB, and AB contributed reagents, materials, and analysis tools. PA, AF, RH, VB, and AB wrote the paper.
This work was supported by the project ‘Establishing Next Generation sequencing Ability for Genomic analysis in Europe’ (ENGAGE) co-funded by the European Food Safety Authority (EFSA, GP/EFSA/AFSCO/2015/01/CT1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The conclusions, findings and opinions expressed in this scientific paper reflect only the view of the authors and not the official position of the European Food Safety Authority.
The authors wish to thank Beatriz Guerra for the fruitful discussion on the manuscript, and Serena Lorenzetti, Roberta Amoruso, Carmela Buccella, Luigi Sorbara, and Roberta Onorati for outstanding technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01217/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Battisti, A. (2016). Antibiotic Resistance - Italy: Colistin, mcr-1, E. coli, Turkeys, 2014. Available at: http://www.promedmail.org/
Bélanger, L., Garenaux, A., Harel, J., Boulianne, M., Nadeau, E., and Dozois, C. M. (2011). Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 62, 1–10. doi: 10.1111/j.1574-695X.2011.00797.x
Bernasconi, O. J., Kuenzli, E., Pires, J., Tinguely, R., Carattoli, A., Hatz, C., et al. (2016). Travelers can import colistin-resistant Enterobacteriaceae, including those possessing the plasmid-mediated mcr-1 gene. Antimicrob. Agents Chemother. 60, 5080–5084. doi: 10.1128/AAC.00731-16
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Brolund, A., Edquist, P. J., Mäkitalo, B., Olsson-Liljequist, B., Söderblom, T., Wisell, K. T., et al. (2014). Epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Sweden 2007-2011. Clin. Microbiol. Infect. 20, O344–O352. doi: 10.1111/1469-0691.12413
Cannatelli, A., Giani, T., Antonelli, A., Principe, L., Luzzaro, F., and Rossolini, G. M. (2016). First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob. Agents Chemother. 60, 3257–3258. doi: 10.1128/AAC.00246-16
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589
Carretto, E., Brovarone, F., Nardini, P., Russello, G., Barbarini, D., Pongolini, S., et al. (2018). Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 23:821. doi: 10.2807/1560-7917.ES.2018.23.2.17-00821
Corbella, M., Mariani, B., Ferrari, C., Comandatore, F., Scaltriti, E., Marone, P., et al. (2017). Three cases of mcr-1-positive colistin-resistant Escherichia coli bloodstream infections in Italy, August 2016 to January 2017. Euro Surveill. 22:30517. doi: 10.2807/1560-7917.ES.2017.22.16.30517
De Been, M., Lanza, V. F., de Toro, M., Scharringa, J., Dohmen, W., Du, Y., et al. (2014). Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776. doi: 10.1371/journal.pgen.1004776
Di Pilato, V., Arena, F., Tascini, C., Cannatelli, A., Henrici De Angelis, L., Fortunato, S., et al. (2016). mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob. Agents Chemother. 60, 5612–5615. doi: 10.1128/AAC.01075-16
Donati, V., Feltrin, F., Hendriksen, R. S., Svendsen, C. A., Cordaro, G., García-Fernández, A., et al. (2014). Extended-spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistance in Klebsiella spp. from companion animals in Italy. PLoS One 9:e90564. doi: 10.1371/journal.pone.0090564
EARS-Net. (2015). “European centre for disease prevention and control. Antimicrobial resistance surveillance in Europe 2015,” in Proceedings of the Annual Report of the European Antimicrobial Resistance Surveillance Network (Stockholm: ECDC).
ESVAC (2016). European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption, ‘Sales of Veterinary Antimicrobial Agents in 29 European Countries in 2014’. (EMA/61769/2016). Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Report/2016/10/WC500214217.pdf
Franco, A., Leekitcharoenphon, P., Feltrin, F., Alba, P., Cordaro, G., Iurescia, M., et al. (2015). Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10:e0144802. doi: 10.1371/journal.pone.0144802
Garcia-Graells, C., De Keersmaecker, S. C. J., Vanneste, K., Pochet, B., Vermeersch, K., Roosens, N., et al. (2017). Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog. Dis. 15, 114–117. doi: 10.1089/fpd.2017.2329
Hasman, H., Hammerum, A. M., Hansen, F., Hendriksen, R. S., Olesen, B., Agersø, Y., et al. (2015). Detection of mcr-1 encoding plasmid-mediated colistin-resistant isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20, 1–5. doi: 10.2807/1560-7917.ES.2015.20.49.30085
Hernández, M., Iglesias, M. R., Rodríguez-Lázaro, D., Gallardo, A., Quijada, N., Miguela-Villoldo, P., et al. (2017). Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 22:30586. doi: 10.2807/1560-7917.ES.2017.22.31.30586
Jakobsen, L., Spangholm, D. J., Pedersen, K., Jensen, L. B., Emborg, H. D., Agersø, Y., et al. (2010). Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int. J. Food Microbiol. 142, 264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025
Kluytmans, J. (2017). Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveill. 22:30588. doi: 10.2807/1560-7917.ES.2017.22.31.30588
Leverstein-van Hall, M. A., Dierikx, C. M., Cohen Stuart, J., Voets, G. M., van den Munckhof, M. P., van Essen-Zandbergen, A., et al. (2011). National ESBL surveillance group. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17, 873–880. doi: 10.1111/j.1469-0691.2011.03497.x
Li, R., Yu, H., Xie, M., Chen, K., Dong, N., Lin, D., et al. (2017). Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 51, 578–585. doi: 10.1016/j.ijantimicag.2017.11.015
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Matamoros, S., van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Mavroidi, A., Miriagou, V., Malli, E., Stefos, A., Dalekos, G. N., Tzouvelekis, L. S., et al. (2012). Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents 39, 247–250. doi: 10.1016/j.ijantimicag.2011.11.003
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Roer, L., Hansen, F., Stegger, M., Sönksen, U. W., Hasman, H., and Hammerum, A. M. (2017). Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 22:22846. doi: 10.2807/1560-7917.ES.2017.22.31.30584
Schrijver, R., Stijntjes, M., Rodríguez-Baño, J., Tacconelli, E., Babu Rajendran, N., and Voss, A. (2017). Review of antimicrobial resistance surveillance programmes in livestock and their meat in Europe, with a focus on antimicrobial resistance patterns in humans. Clin. Microbiol. Infect. 24, 577–590. doi: 10.1016/j.cmi.2017.09.013
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., McGann, P., et al. (2016). Model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 6973–6976. doi: 10.1128/AAC.01457-16
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Fang, L. X., Wu, Z., Deng, H., Yang, R. S., Li, X. P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7:424. doi: 10.1038/s41598-017-00095-x
Thomsen, M. C., Ahrenfeldt, J., Cisneros, J. L., Jurtz, V., Larsen, M. V., Hasman, H., et al. (2016). A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17
Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. doi: 10.1089/10665270050081478
Keywords: epidemiology, colistin resistance, mcr genes, whole genome sequencing, food-producing animals, Italy, E. coli, Salmonella
Citation: Alba P, Leekitcharoenphon P, Franco A, Feltrin F, Ianzano A, Caprioli A, Stravino F, Hendriksen RS, Bortolaia V and Battisti A (2018) Molecular Epidemiology of mcr-Encoded Colistin Resistance in Enterobacteriaceae From Food-Producing Animals in Italy Revealed Through the EU Harmonized Antimicrobial Resistance Monitoring. Front. Microbiol. 9:1217. doi: 10.3389/fmicb.2018.01217
Received: 21 March 2018; Accepted: 18 May 2018;
Published: 12 June 2018.
Edited by:
Axel Cloeckaert, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Isabelle Kempf, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceCopyright © 2018 Alba, Leekitcharoenphon, Franco, Feltrin, Ianzano, Caprioli, Stravino, Hendriksen, Bortolaia and Battisti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Battisti, YW50b25pby5iYXR0aXN0aUBpenNsdC5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.