
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 01 June 2018
Sec. Extreme Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01191
This article is part of the Research Topic Recent Advances in Bioremediation/biodegradation by Extreme Microorganisms View all 13 articles
Metals and radionuclides (M&Rs) are a worldwide concern claiming for resilient, efficient, and sustainable clean-up measures aligned with environmental protection goals and global change constraints. The unique defense mechanisms of extremophilic bacteria and archaea have been proving usefulness towards M&Rs bioremediation. Hence, extremophiles can be viewed as microfactories capable of providing specific and controlled services (i.e., genetic/metabolic mechanisms) and/or products (e.g., biomolecules) for that purpose. However, the natural physiological plasticity of such extremophilic microfactories can be further explored to nourish different hallmarks of M&R bioremediation, which are scantly approached in the literature and were never integrated. Therefore, this review not only briefly describes major valuable extremophilic pathways for M&R bioremediation, as it highlights the advances, challenges and gaps from the interplay of ‘omics’ and biological engineering to improve extremophilic microfactories performance for M&R clean-up. Microfactories’ potentialities are also envisaged to close the M&R bioremediation processes and shift the classical idea of never ‘getting rid’ of M&Rs into making them ‘the belle of the ball’ through bio-recycling and bio-recovering techniques.
Metals and radionuclides (M&Rs) are problematic pollutants sourced from different industrial sectors, nuclear power plants, electronic waste and, especially, from mining activities. Undoubtedly, mining industry nurtures a plethora of human needs (Carvalho, 2017), but past and ongoing mining activities represent a huge footprint of environmental (Marques et al., 2014) and health injuries (Pereira et al., 2014; Venkateswarlu et al., 2016). Still, future societal demands will force industrial growth, thus making M&R pollution a never-ending threat, mainly due to their persistence and non-biodegradability (Gupta and Diwan, 2017).
Bioremediation, though being often viewed as an ‘old fashion’ and slow-acting technology, it has been thriving for M&R reclamation. Its sustainability, low-cost and efficiency totally rival conventional remediation techniques (Li and Zhang, 2012). In particular, microbially-based bioremediation relies on microbes abilities, biogenic products and/or components to scavange, transform or immobilize M&Rs (Lloyd and Lovley, 2001; Voica et al., 2016). Despite the interesting nature-based self-healing character of this approach, the tolerance of microbes to high M&R levels, extreme physical (e.g., radiation), chemical (e.g., acidic pH) and climate changing conditions may constrain (in situ) bioremediation efficiencies (Ramos et al., 2011). Extremophilic microbes, though, have attractive skills as bioremediation tiny factories (microfactories), which performance can even be improved and customized for M&R clean-up. This review will hence focus: (1) the peculiar mechanisms of extremophilic bacteria and archaea exploitable as ‘microfactories’ for M&R bioremediation, (2) the groundbreaking opportunities leveraged from nature and science (‘omics’ plus biological engineering) interplay to enhance extremophilic-microfactories-based bioremediation, (3) M&R recovery and recycling conducted by extremophilic microfactories as a sustainable and value-added management of M&R-bioremediation-resulting wastes. Overall, this is an innovative overview on the potentialities of extremophilic bacteria/archaea in three M&R bioremediation hallmarks: application, optimization, end-waste management (Figure 1).
Extremophilic bacteria and archaea evolved specialized mechanisms to endure physical and/or geochemical extreme environments (Rothschild and Mancinelli, 2001). Acidophilic and/or metalophilic microorganisms are especially appealing agents for bioremediation, given their defense mechanisms against M&Rs and acidity. They synthesize extremophilic enzymes (extremozymes) (Elleuche et al., 2014) and biomolecules (Raddadi et al., 2015) that keep active and/or stable under harsh conditions. The stability of thermophilic enzymes, like the esterase EstATII (Mohamed et al., 2013), helps facing acidity and metal stress, thanks to stiff folds sustained by ion-pair networks, to compact hydrophobic cores and aminoacid arrangements/packing (Elleuche et al., 2014). Additionally, extremophiles evolved sharpened metal detoxification pathways (Voica et al., 2016) due to fast-adapting transcriptional and translational mechanisms that activate and/or inhibit many anti-oxidative stress, metal-binding, metal-transport, and membrane-permeability responses (Mukherjee et al., 2012; Dekker et al., 2016). Their membranes exhibit a specific structure (Singh and Singh, 2017), composition and a positively-charged inner layer that promote metal-transporters functioning (Dekker et al., 2016) and minimize metals and protons entrance, thereby controlling acidity and M&R toxicity (Slonczewski et al., 2009; Krulwich et al., 2011; Navarro et al., 2013; Zhang et al., 2016). Notwithstanding, since extremophiles can express defense mechanisms active against multiple extremes simultaneously (Rothschild and Mancinelli, 2001), thermophilic, halophilic, radiophilic, and polyextremophilic bacteria/archaea are additional M&R clean-up agents (Amoozegar et al., 2012; Wheaton et al., 2015). Either as whole-cells or providers of economically-valuable bio-services and/or biomolecules (Figure 2), extremophiles can be explored as tiny factories (microfactories) for M&R remediation.
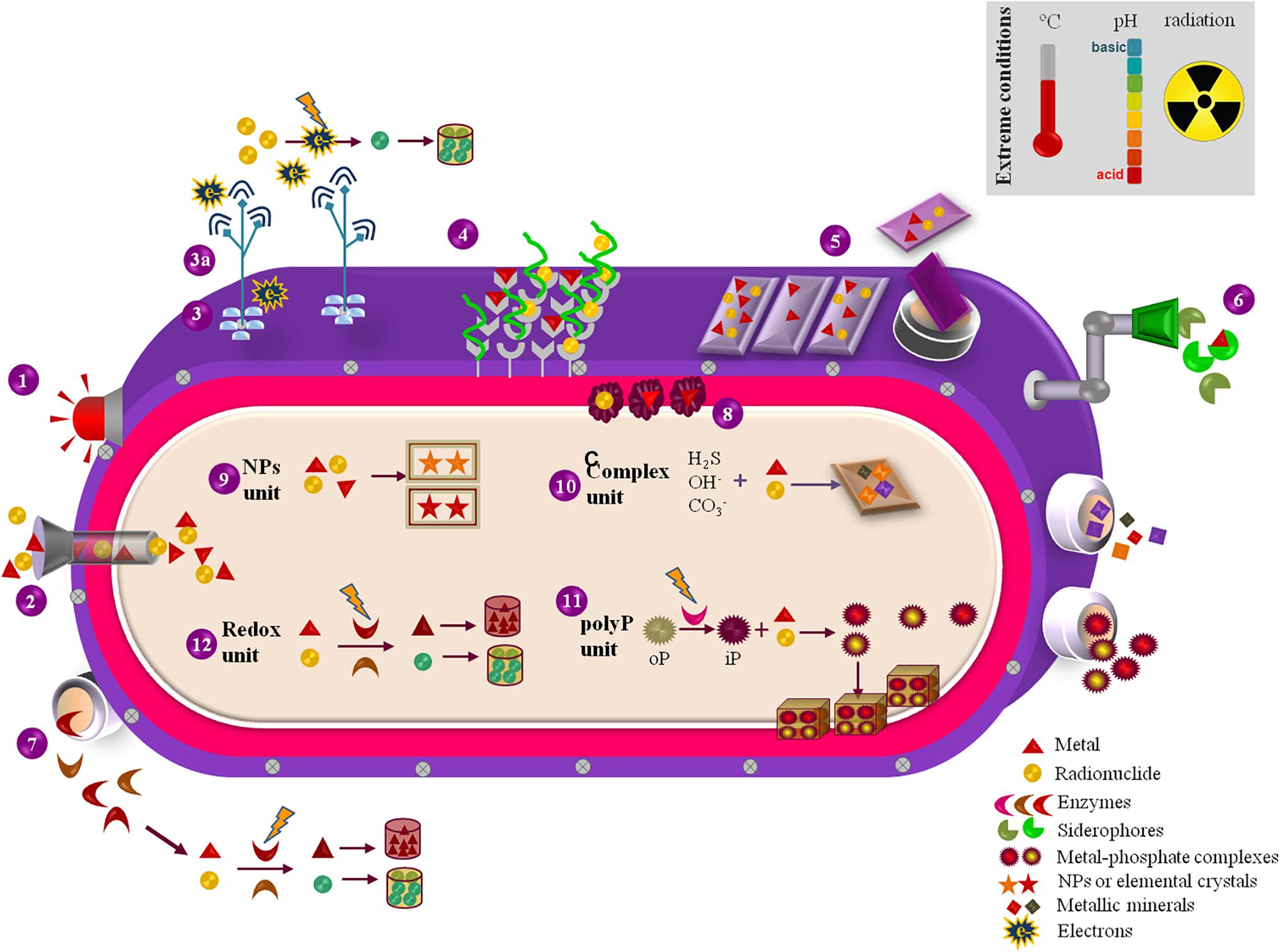
FIGURE 2. Representation of the exploitable biosynthetic and metabolic features of extremophilic microfactories with relevance for M&R bioremediation purposes. (1) M&R sensing by regulatory proteins that trigger downstream genetic and metabolic pathways coping with M&R exposure. (2) M&R uptake through energy-dependant transporters as resistance nodulation cell division carriers. (3) Surface c-cytochromes that promote electron transference to pili (3a), which electron-conductive ability promotes the extracellular reduction and precipitation of M&Rs. (4) M&R binding sites at the cell-surface cationic and anionic functional groups and S-layer proteins. (5) Extracellular polymeric substances (EPS) that function as panels for M&R capture and sorption, either attached to the cell surface or released to the extracellular environment. (6) Biosynthetic siderophores extracellularly excreted for M&R sorption. (7) Redox enzymes liberated for catalyzing the reduction/oxidation of M&Rs that, if reduced, may precipitate and accumulate in the outer environment. (8) Metallic chaperones located in the periplasm space that promote M&Rs quenching and mobilization. (9) M&R-based nanoparticles (NPs) or nanocrystals formed and accumulated in the cytoplasm. (10) M&R biotransformation through complexation with hydroxides, carbonates, sulfides, hence forming M&R-oxides, M&R-carbonates and M&R-sulfides complexes either intracellularly accumulated or released to the exterior environment. (11) Enzymatic hydrolysis of organic poly-phosphates into inorganic phosphates for M&R precipitation and accumulation as metal-phosphate granules in the inner space or expelled to the exterior. (12) Reduction/oxidation of M&Rs in the cytoplasm driven by biosynthesized redox enzymes.
Sensing M&Rs presence is the alarm button to trigger the activation/repression of specific regulatory proteins and downstream cascades that control the expression of resistance genes/operons encoding enzymes and biomolecules for M&R detoxification (Das et al., 2016). Along this, the uptake/efflux of M&Rs across extremophiles cell wall and membranes may occur through energy-dependent P-type ATPases (copA1Af, copA2Af, and copBAf), resistance nodulation cell division carriers (cusAAf, cusBAf, and cusCAf) and/or chaperones (cusFAf and copCAf) as in the copper-resistant Acidithiobacillus ferrooxidans (Navarro et al., 2009). A comprehensive description of the genetic characteristics and functioning of regulatory and M&R uptake/efflux systems was performed namely for thermoacidophilic, acidophilic, and metalophilic bacteria and/or archaea (Nies, 2000; Orell et al., 2010, 2013; Navarro et al., 2013; Wheaton et al., 2015).
Extremophiles-derived cell wall/capsule, S-layer proteins, extracellular polymer substances (EPS) and siderophores present unusual structural and functional properties for M&Rs biosorption under severe stress. Llorens et al. (2012) suggested uranium sorption by S-layer proteins on the metalophilic Cupriavidus metallidurans at pH1; while acidophilic (Masaki et al., 2015) and moderate halophilic bacteria/archaea adsorbed M&Rs onto cell surface (Amoozegar et al., 2007, 2008, 2012). EPS are the most relevant structures for M&R biosorption with proved efficiency namely on psychrotolerant (Pseudoalteromonas sp.; Qin et al., 2007) and acidophilic (A. ferroxidans; Sand and Gehrke, 2006) bacteria. Either attached on the cell surface or extracellularly released, extremophilic EPS promote cell adhesion, biofilm formation, and M&Rs biosorption (Poli et al., 2011) due to a greater abundance of negatively-charged metal-binding sites of its components (e.g., polysaccharides, proteins, nucleic acids, and lipids) (Nies, 2000; Navarro et al., 2013; Shukla et al., 2017). An improved structure/composition favors enzyme stabilization and protein anchoring, hence boosting bacteria/archaea proliferation (Qin et al., 2007). Some halophilic archae can synthesize EPS with specialized jellifying properties due to abundant glucuronic acids and sulfates that stabilize EPS matrix and enhances M&R sorption (Singh and Singh, 2017). Siderophores are small biomolecules released to the environment for metal scavaging. De Serrano et al. (2016) reviewed regulatory mechanisms and chemical features of siderophores synthesized by extremophiles, evidencing their potential as bioremediation agents for metal chelation.
Extremophiles mediate the chemical transformation of M&R through the biosynthesis of enzymes that intracellularly or extracellularly catalyze M&Rs oxidation (solubilization) or reduction (precipitation) (Das et al., 2016). Overall, the redox reactions may occur through direct or indirect activity of extremophiles (e.g., sulfur-/iron-oxidizing or sulfate-reducing), under aerobiosis and/or anaerobiosis (Zhang et al., 2016; Shukla et al., 2017). Normally, M&R reduction leads to the bioprecipitation/bioaccumulation of organic- and mineral-M&R complexes, and M&R-based nanocrystals. Halophilic (Amoozegar et al., 2012; Srivastava et al., 2013), thermo-tolerant (Özdemir et al., 2017a,b), thermophilic (Kashefi and Lovley, 2000), metalophilic (Lovley et al., 1991), radiophilic (Appukuttan et al., 2011) and acidophilic bacteria and archaea (Orell et al., 2012; Masaki et al., 2015), proved to precipitate tellurite, uranium, iron, silver, chromium, or copper as elemental nanocrystals, carbonate complexes, magnetite and metal-sulfides. Inorganic polyphosphate granules enzymatically-transformed by acidophilic and metalophilic bacteria (e.g., Caulobacter crescentus, Acidithiobacillus sp.) and archaea (e.g., Sulfolobus metallicus) can intracellularly and extracellularly accumulate M&Rs (Hu et al., 2005; Navarro et al., 2009; Orell et al., 2012). M&R bioaccumulation/bioprecipitation was indeed favored by highly reductive pili (Cologgi et al., 2011) and c-cytochromes (Marshall et al., 2006; Shelobolina et al., 2007) that increase electron transport, which attached to Geobacter sp. or Shewanella oneidensis outer membranes, promote the extracellular reduction of U(VI) into the precipitable U(IV) along with cell viability and growth. These strategies are excellent towards in situ bioremediation, as already applied for uranium clean-up in groundwater (Coates and Anderson, 2000).
Despite the great biotechnological potential of extremophilic microfactories, remediation efficiency, biomass productivity, and economic profitability may be challenging, especially for in situ applications due to extreme optima, interspecies competition and interaction/communication inhibiting bioremediation, and limited proliferation (Ramos et al., 2011; Johnson et al., 2015). Consequently, extremophiles-based M&R bioremediation may become less attractive considering mining industry demands, hence requiring function-directed improvements.
‘Omics’ and biological engineering are leading the achievement of more proficient extremophilic microfactories for M&R bioremediation to keep pace with current economical, technological, social, and climate change defiance.
A fundamental step is to untie the genomic and metabolic complexity governing extremophiles-M&R interactions. Therefore, genomic approaches involving amplicon and whole-genome sequencing, comparative genomics and in silico genome mining have been employed to target essential genes encoding regulatory proteins, promoters, metal-binding proteins, enzymes, and biomolecules involved in radionuclides (e.g., Makarova et al., 2001; Methé et al., 2003; Zivanovic et al., 2009) and metals (e.g., Nies, 2000; Osorio et al., 2008; Cárdenas et al., 2010; Guo et al., 2014; Kernan et al., 2017) detoxification in extremophilic bacteria (e.g., Geobacter sulfurreducens, Acidithiobacillus spp., Deinococcus radiodurans, Sulfobacillus thermosulfidooxidans) and archaea (e.g., Thermococcus gammatolerans, Metallosphaera sedula).
Despite the relevance of this knowledge, the study of M&R-constrained gene expression and regulation through transcriptomics assumes a pivotal role for bioremediation purposes. DNA microarrays applied to the As-resistant Thiomonas spp. evidenced genomic rearrangements and horizontal gene transfer (Arsène-Ploetze et al., 2010), which can be exploited to transfer M&R resistance. Other studies observed overexpression of metallochaperones and enzymes mediating metal-phosphate granules formation in acidophilic bacteria (A. ferooxidans) (Navarro et al., 2009) and (thermo)acidophilic archaea (Sulfolobus metallicus, Metallosphaera sedula) challenged with M&Rs (Orell et al., 2013; Wheaton et al., 2016).
Nevertheless, many transcripts keep with unassigned functions or activities. Proteomics, through 2-DE gel coupled with mass spectroscopy (e.g., LC–MS, MALDI-ToF, and ICPL) tools and protein libraries, is thereby elucidating the type and abundance of proteins synthesized under M&R stress (Desai et al., 2010). Among them, those associated with EPS and biofilm production, oxidative stress responses, regulation of RND-type Cus system and metal uptake/efflux pumps (Zivanovic et al., 2009; Almárcegui et al., 2014; Martínez-Bussenius et al., 2016) are hugely relevant.
Additionally, metabolomics enlightens the metabolic fingerprinting of M&R-induced responses on extremophiles (Mosier et al., 2013). A diverse toolbox of chromatographic (GC and HPLC) and spectroscopic (FTIR and NMR) techniques have hence been employed to harness the genetic, transcriptional and translational processes sustaining the cellular biomolecules/metabolite fluxes (i.e., fluxomics) (Hold et al., 2009; Desai et al., 2010) leading to M&R sequestration. In silico reconstruction of unknown metabolic networks relevant for M&R bioremediation based on sequenced genomes (Sun et al., 2009), robust bioinformatics and libraries, is becoming a powerful fine-tuning approach, though requiring a cautious analysis of metal-microbe interactions to prevent misinterpretations (Johnson et al., 2015). In this context, community-level meta-omics (metagenomics, metatranscriptomics, and metaproteomics) offer great avenues by avoiding extremophiles culturing hurdles, while uncovering pools of novel biomolecules/enzymes for M&R bioremediation without previous sequence data (Mohamed et al., 2013), and providing ecologically-relevant analysis of community shifts and functions (Cowan et al., 2015; Garris et al., 2016; Mirete et al., 2016) along the bioremediation processes. Multi-‘omics’ integration, either at individual or community levels, constitutes another robust approach to unravel new M&R-bioremediation agents or monitoring bio-devices. Indeed, Wilkins et al. (2011) used a proteogenomic analysis to develop a biomarker of G. sulfurreducens activity during uranium bioremediation.
The advent of DNA recombinant techniques for tailoring existent abilities or to create synthetic ones, in natural or newly designed extremophiles, is a doorway to boost M&R bioremediation.
The peculiar mechanisms of extremophiles can be customized through the insertion of genes or gene clusters, as to enhance their skills and robustness for M&R attenuation (Cárdenas et al., 2010). Metal-resistant (Cupriavidus metallidurans; Rojas et al., 2011), radiophilic (D. radiodurans; Brim et al., 2000) and thermoacidophilic (D. geothermalis; Brim et al., 2003) bacteria were transformed with genes/clusters encoding enzymes involved in individual- or multi-M&Rs reduction/oxidation. This is biotechnologically worthy given the mixture of M&Rs in mine/nuclear wastes. In order to enhance M&R bioprecipitation, D. radiodurans was indeed manipulated to express a Cd(II)-binding synthetic phytochelatin (EC20) and metallothionein (smtA) (Chaturvedi and Archana, 2014), and a periplasmic acid phosphatase (PhoN), which hydrolysed organic into inorganic phosphates to precipitate uranium (Appukuttan et al., 2006, 2011). However, some extremophiles are more resistant to transformation due to their robust protective mechanisms (Chen et al., 2011; Liu et al., 2011), hence forcing the construction of adapted vectors (Meng et al., 2013).
Recently, the innovative project NANOBINDERS (PTDC/AAG-REC/3004/2014) is enrolled in the creation of biogenic nanopolymers functionalized to bind M&Rs (the NANOBINDERS) from uranium mine effluents. These M&R-binding nanopolymers are self-assembled inside a host transformed with new constructs harboring nanopolymer- and metal-binding-peptides-encoding genes, originally obtained from microbes isolated in a uranium mine. Such functional nanobeads are a revolutionary option to engineered microbes, which in situ release still raises regulatory concerns, besides genetic instability and limited efficiency under real scenarios (de Lorenzo, 2009). Alternatively, manipulated extremophiles may house suicidal genes (Paul et al., 2005) activated upon undetectable M&R levels, for future in situ application.
Native microbial communities have been used to rehabilitate M&R-contaminated areas by natural attenuation (Shukla et al., 2017), which can be engineered through biostimulation (i.e., supplementation of compounds/substrates enhancing microbial proliferation and activities) and/or bioaugmentation measures (i.e., addition of microbes endowed with particular functional mechanisms). In mesocosms, Ñancucheo and Johnson (2011) reduced acid mine drainage production when consortia of metabolically-cooperating microalgae, acidophilic and/or acid-tolerant bacteria were added to promote iron- and/or sulfate-reduction of tailings. The biostimulation of Geobacter sp. activity by acetate enrichment had enhanced uranium reduction, thereby proving successful in situ bioremediation (Anderson et al., 2003). However, metabolic fluxes modeling (Hold et al., 2009) should be applied in the future to design novel extremophilic consortia with tailored co-activities for M&R clean-up.
Synthetic biology (SynBio) moves beyond the simple transformation of genes or operons to the insertion of newly designed and constructed synthetic biological parts and circuits to create robust, profitable, programmable and customized microfactories (Martínez-Garcia and de Lorenzo, 2016). The genetic instability and limited performance of engineered microbes for M&R bioremediation, can be overcome by DNA de novo synthesis and genome editing tools [e.g., Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs)- and protein Cas; Mougiakos et al., 2016, 2017]. Hence, extremophiles can be engineered as novel platforms (or chassis) housing stable and multiple designed functions together with the transcriptional and translational machinery to be active from laboratory- to field-scale extreme scenarios (Adams, 2016). Likewise, Gerber et al. (2015) took advantage of Deinococcus genetic plasticity and robustness to construct a chassis towards different applications. Deinococcus spp. was also successfully engineered with metal-resistance genes, demonstrating their genetic flexibility for heterologous and co-expression of different metal-resistance determinants under extreme temperature and radiation (Brim et al., 2000, 2003). Thus, constructing extremophilic-microfactories chassis is an appealing new wave for enhanced M&R bioremediation, especially if universal genetic toolkits could be created (Adams, 2016).
The joint endeavor of SynBio and metabolic engineering of communities is indeed enabling the design and building of synthetic consortia (Shong et al., 2012) for M&R remediation. Developing synthetic consortia demands the engineering of cell–cell communication, i.e., quorum-sensing (QS) systems (Brenner et al., 2008). QS modulates behavioral, metabolic and structural dynamics in microbial communities through signaling molecules [e.g., acyl homoserine lactones family (AHL)] (Shong et al., 2012). QS is pivotal namely in EPS synthesis and biofilm formation (Farah et al., 2005; McDougald et al., 2012), as well as AHLs can mediate copper resistance in A. ferrooxidans (Wenbin et al., 2011). Thus, putting effort on the modulation or creation of new QS systems and signaling molecules biosynthesis can fine-tune the pool of relevant functions in extremophilic consortia for M&R removal (e.g., biomass production and EPS synthesis) (Brune and Bayer, 2012; Shong et al., 2012).
A discouraging issue in M&R bioremediation is the disposal of final wastes. Nevertheless, extremophilic microfactories can be successfully explored for M&Rs recycling and/or recovery after being remediated, thereby sustainably closing M&R bioremediation processes.
Biomining is usually conducted by extremophilic bacteria and archaea for the recovery of M&R from ores and mine/metal-rich/nuclear wastes, through biooxidation (minerals oxidation for metal release) and/or bioleaching (metal solubilization) (Brune and Bayer, 2012; Johnson, 2014). These bioprocesses take advantage of the anaerobic metabolism, redox pathways, intercellular communication, biofilm formation (Vera et al., 2013), and resistance to heat, acid and metals of (thermo)acidophiles consortia (e.g., Leptospirillum spp., Acidithiobacillus spp., Sulfobacillus spp., Ferroplasma spp., Acidiplasma spp.) (Orell et al., 2010, 2013). Insoluble metal sulfides are converted into soluble metal sulfates by iron- and sulfur-oxidizing extremophiles, thereby facilitating the recovery of economically-relevant metals (e.g., Cu, Fe, Au, Ni, and Zn) (Watling et al., 2015; Wheaton et al., 2015).
Microbial fuel cells (MFCs) can be a valuable recovery method since it uses the catalytic activity of microbes to generate electrical energy from the oxidation of organic substrates (Mathuriya and Yakhmi, 2014). They are composed by an anode that captures electrons from substrate oxidation, a cathode and an intermediate cation-specific membrane. Since M&Rs have a high redox potential, they can be reduced and precipitated by receiving electrons from the cathode. Geobacter sulfurreducens biofilms possessing electron-conductive nanowires (pili) have been exploited for increased electricity generation into MFCs (Reguera et al., 2006). The generated electrons can be further used to reduce U(VI) into removable U(IV), hence synergizing Geobacter sp. skills for bioremediation-derived-waste recycling. An anoxic sludge containing Geobacter spp. recovered the noble metal silver, while generating electrical energy in a MFC (Ho et al., 2017). Other extremophiles have indeed been used in MFCs (Shrestha et al., 2018), especially towards metal recovery (Dopson et al., 2015).
Extremophiles biosynthesize economically-lucrative inorganic NPs through intracellular or extracellular transformation/preci-pitation of M&Rs (Ulloa et al., 2016), hence providing another profitable M&R recycling strategy. The metal-tolerant Cupriavidus metallidurans CH34 mineralized Au and precipitated it as nanoparticulate Au0 in the periplasm (Reith et al., 2009), whilst the alkalotolerant Rhodococcus sp. accumulated Au-NPs in the cytoplasm and cell wall (Ahmad et al., 2003). Spherical Ag-NPs were also extracellularly synthesized at 80°C by the thermophilic Ureibacillus thermosphaericus (Juibari et al., 2011). CdS-NPs with enhanced stability under acidic conditions were produced by Acidithiobacillus spp., being a gainful option for Cd turnover (Ulloa et al., 2016). Uraninite, which is a worthy biogenic NP for uranium in situ remediation (Bargar et al., 2008), was synthesized by Geobacter metallireducens upon extracellular U(VI) reduction (Gorby and Lovley, 1992). The precious metal Pd(II) used in many industrial sectors was precipitated in G. sulfurreducens biofilms as Pd(0)-NPs and easily extracted by centrifugation (Yates et al., 2013).
Extremophiles enclose a pool of genetic and metabolic opportunities that can be harnessed as microfactories for multiple M&R-bioremediation hallmarks. Nevertheless, the complexity of extremophiles’ cellular mechanisms, interspecies relationships and M&R interplay under real scenarios of co-occurring extreme conditions may rise some challenges. Multi-‘omics’ applied side-by-side with genetic engineering techniques are covering knowledge gaps, allowing gene expression and metabolic pathways customization for improved bioremediation. Synthetic biology, however, through an iterative design-build-test-analyze framework is further revolutionizing M&R bioremediation and opening new perspectives on the creation of robust extremophilic recombinants or chassis with de novo-designed biologically-, energetically-, and economically-viable traits for specific M&R removal processes. Future trends will hence target adaptation or construction of multi-skilled extremophilic microfactories to sustainably empower M&R bioremediation and end-waste recycling, while keeping up with global changes, natural resources availability, and cost-efficiency requirements.
CM was responsible for doing all the tasks concerning the development of the work: conceptualized the idea and goals of the mini-review, performed literature search and revision, and wrote the manuscript.
This work was funded by the Portuguese Foundation for Science and Technology (FCT) and by the European Regional Development Fund (ERDF) through the Portugal 2020 Partnership Agreement between Portugal and the European Union, the Competitiveness and Internationalization Operational Program (COMPETE 2020) and the Regional Operational Program Lisboa (POR-Lisboa), under the project NANOBINDERS (PTDC/AAG-REC/3004/2014). CM was granted as an Invited Scientist within the project NANOBINDERS (ref. BCC/UI88/5626/2016). Thanks are due for the financial support to CESAM (UID/AMB/50017 – POCI-01-0145-FEDER-007638), to FCT/MCTES through national funds (PIDDAC), and the co-funding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adams, B. L. (2016). The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth. Biol. 5, 1328–1330. doi: 10.1021/acssynbio.6b00256
Ahmad, A., Senapati, S., Khan, M. I., Kumar, R., Ramani, R., Srinivas, V., et al. (2003). Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnology 14, 824–828. doi: 10.1088/0957-4484/14/7/323
Almárcegui, R. J., Navarro, C. A., Paradela, A., Albar, J. P., von Bernath, D., and Jere, C. A. (2014). New copper resistance determinants in the extremophile Acidithiobacillus ferrooxidans: a quantitative proteomic analysis. J. Proteome Res. 13, 946–960. doi: 10.1021/pr4009833
Amoozegar, M. A., Ashengroph, M., Malekzadeh, F., Reza, R. M., Naddaf, S., and Kabiri, M. (2008). Isolation and initial characterization of the tellurite reducing moderately halophilic bacterium, Salinicoccus sp. strain QW6. Microbiol. Res. 163, 456–465. doi: 10.1016/j.micres.2006.07.010
Amoozegar, M. A., Ghasemi, A., Razavi, M. R., and Naddaf, S. (2007). Evaluation of hexavalent chromium reduction by chromate-resistant moderately halophile, Nesterenkonia sp. strain MF2. Process. Biochem. 42, 1475–1479. doi: 10.1016/j.procbio.2007.07.001
Amoozegar, M. A., Khoshnoodi, M., Didari, M., Hamedi, J., Ventosa, A., and Baldwin, S. A. (2012). Tellurite removal by a tellurium-tolerant halophilic bacterial strain, Thermoactinomyces sp. QS-2006. Ann. Microbiol. 62, 1031–1037. doi: 10.1007/s13213-011-0343-1
Anderson, R. T., Vrionis, H. A., Ortiz-Bernad, I., Resch, C. T., Long, P. E., Dayvault, R., et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003
Appukuttan, D., Rao, A. S., and Apte, S. K. (2006). Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from diluted nuclear waste. Appl. Environ. Microbiol. 72, 7873–7878. doi: 10.1128/AEM.01362-06
Appukuttan, D., Seetharam, C., Padma, N., Rao, A. S., and Apte, S. K. (2011). PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation. J. Biotechnol. 154, 285–290. doi: 10.1016/j.jbiotec.2011.05.002
Arsène-Ploetze, F., Koechler, S., Marchal, M., Coppée, J. Y., Chandler, M., Bonnefoy, V., et al. (2010). Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet. 6:e1000859. doi: 10.1371/journal.pgen.1000859
Bargar, J. R., Bernier-Latmani, R., Giammar, D. E., and Tebo, B. M. (2008). Biogenic uraninite nanoparticles and their importance for uranium remediation. Elements 4, 407–412. doi: 10.2113/gselements.4.6.407
Brenner, K., You, L., and Arnold, F. H. (2008). Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 26, 483–489. doi: 10.1016/j.tibtech.2008.05.004
Brim, H., McFarlan, S. C., Fredrickson, J. K., Minton, K. W., Zhai, M., Wackett, L. P., et al. (2000). Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18, 85–90. doi: 10.1038/71986
Brim, H., Venkateswaran, A., Kostandarithes, H. M., Fredrickson, J. K., and Daly, M. J. (2003). Engineering Deinococcus geothermalis for bioremediation of high-temperature radioactive waste environments. Appl. Environ. Microbiol. 69, 4575–4582. doi: 10.1128/AEM.69.8.4575-4582.2003
Brune, K. D., and Bayer, T. S. (2012). Engineering microbial consortia to enhance biomining and bioremediation. Front. Microbiol. 3:203. doi: 10.3389/fmicb.2012.00203
Cárdenas, J. P., Valdés, J., Quatrini, R., Duarte, F., and Holmes, D. S. (2010). Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl. Microbiol. Biotechnol. 88, 605–620. doi: 10.1007/s00253-010-2795-9
Carvalho, F. P. (2017). Mining industry and sustainable development time for change. Food Energy Sec. 6, 61–77. doi: 10.1002/fes3.109
Chaturvedi, R., and Archana, G. (2014). Cytosolic expression of synthetic phytochelatin and bacterial metallothionein genes in Deinococcus radiodurans R1 for enhanced tolerance and bioaccumulation of cadmium. Biometals 27, 471–482. doi: 10.1007/s10534-014-9721-z
Chen, D., Lin, J., Che, Y., Liu, X., and Lin, J. (2011). Construction of recombinant mercury resistant Acidithiobacillus caldus. Microbiol. Res. 166, 515–520. doi: 10.1016/j.micres.2010.10.003
Coates, J. D., and Anderson, R. T. (2000). Emerging techniques for anaerobic bioremediation of contaminated environments. Trends Biotechnol. 18, 408–412. doi: 10.1016/S0167-7799(00)01478-5
Cologgi, D. L., Lampa-Pastirk, S., Speers, A. M., Kelly, S. D., and Reguera, G. (2011). Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc. Natl. Acad. Sci. U.S.A. 108, 15248–15252. doi: 10.1073/pnas.1108616108
Cowan, D. A., Ramond, J. B., Makhalanyane, T. P., and De Maayer, P. (2015). Metagenomics of extreme environments. Curr. Opin. Microbiol. 25, 97–102. doi: 10.1016/j.mib.2015.05.005
Das, S., Dash, H. R., and Chakraborty, J. (2016). Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 100, 2967–2984. doi: 10.1007/s00253-016-7364-4
de Lorenzo, V. (2009). Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin. Microbiol. Infect. 1, 63–65. doi: 10.1111/j.1469-0691.2008.02683.x
De Serrano, L. O., Camper, A. K., and Richards, A. M. (2016). An overview of siderophores for iron acquisition in microorganisms living in the extreme. Biometals 29, 551–571. doi: 10.1007/s10534-016-9949-x
Dekker, L., Arsène-Ploetze, F., and Santini, J. M. (2016). Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res. Microbiol. 167, 234–239. doi: 10.1016/j.resmic.2016.01.007
Desai, C., Pathak, H., and Madamwar, D. (2010). Advances in molecular and “-omics” to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour. Technol. 101, 1558–1569. doi: 10.1016/j.biortech.2009.10.080
Dopson, M., Ni, G., and Sleutels, T. H. (2015). Possibilities for extremophilic microorganisms in microbial electrochemical systems. FEMS Microbiol. Rev. 40, 164–181. doi: 10.1093/femsre/fuv044
Elleuche, S., Schröder, C., Sahm, K., and Antranikian, G. (2014). Extremozymes — biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 29, 116–123. doi: 10.1016/j.copbio.2014.04.003
Farah, C., Vera, M., Morin, D., Haras, D., Jerez, C. A., and Guiliani, N. (2005). Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 71, 7033–7040. doi: 10.1128/AEM.71.11.7033-7040.2005
Garris, H. W., Baldwin, S. A., van Hamme, J. D., Gardner, W. C., and Fraser, L. H. (2016). Genomics to assist mine reclamation: a review. Restor. Ecol. 24, 165–173. doi: 10.1111/rec.12322
Gerber, E., Bernard, R., Castang, S., Chabot, N., Coze, F., Dreux-Zigha, A., et al. (2015). Deinococcus as new chassis for industrial biotechnology: biology, physiology and tools. J. Appl. Microbiol. 119, 1–10. doi: 10.1111/jam.12808
Gorby, Y. A., and Lovley, D. R. (1992). Enzymatic uranium precipitation. Environ. Sci. Technol. 26, 205–207. doi: 10.1021/es00025a026
Guo, X., Yin, H., Liang, Y., Hu, Q., Zhou, X., Xiao, Y., et al. (2014). Comparative genome analysis reveals metabolic versatility and environmental adaptations of Sulfobacillus thermosulfidooxidans strain ST. PLoS One 9:e99417. doi: 10.1371/journal.pone.0099417
Gupta, P., and Diwan, B. (2017). Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 13, 58–71. doi: 10.1016/j.btre.2016.12.006
Ho, N. A. D., Babel, S., and Kurisu, F. (2017). Bio-electrochemical reactors using AMI-7001S and CMI-7000S membranes as separators for silver recovery and power generation. Bioresour. Technol. 244, 1006–1014. doi: 10.1016/j.biortech.2017.08.086
Hold, C., Andrews, B. A., and Asenjo, J. A. (2009). A stoichiometric model of Acidithiobacillus ferrooxidans ATCC 23270 for metabolic flux analysis. Biotechnol. Bioeng. 102, 1448–1459. doi: 10.1002/bit.22183
Hu, P., Brodie, E. L., Suzuki, Y., McAdams, H. H., and Andersen, G. L. (2005). Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187, 8437–8449. doi: 10.1128/JB.187.24.8437-8449.2005
Johnson, D. B. (2014). Biomining-biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 30, 24–31. doi: 10.1016/j.copbio.2014.04.008
Johnson, D. R., Helbling, D. E., Men, Y., and Fenner, K. (2015). Can meta-omics help to establish causality between contaminant biotransformations and genes or gene products? Environ. Sci. 1, 272–278. doi: 10.1039/C5EW00016E
Juibari, M. M., Abbasalizadeh, S., Jouzani, G. S., and Noruzi, M. (2011). Intensified biosynthesis of silver nanoparticles using a native extremophilic Ureibacillus thermosphaericus strain. Mater. Lett. 65, 1014–1017. doi: 10.1016/j.matlet.2010.12.056
Kashefi, K., and Lovley, D. R. (2000). Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66, 1050–1056. doi: 10.1128/AEM.66.3.1050-1056.2000
Kernan, T., West, A. C., and Banta, S. (2017). Characterization of endogenous promoters for control of recombinant gene expression in Acidithiobacillus ferrooxidans. Biotechnol. Appl. Biochem. 64, 793–802. doi: 10.1002/bab.1546
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Li, J., and Zhang, Y. (2012). Remediation technology for the uranium contaminated environment: a review. Procedia Environ. Sci. 13, 1609–1615. doi: 10.1016/j.proenv.2012.01.153
Liu, W., Lin, J., Pang, X., Cui, S., Mi, S., and Lin, J. (2011). Overexpression of rusticyanin in Acidithiobacillus ferrooxidans ATCC19859 increased Fe(II) oxidation activity. Curr. Microbiol. 62, 320–324. doi: 10.1007/s00284-010-9708-0
Llorens, I., Untereiner, G., Jaillard, D., Gouget, B., Chapon, V., and Carriere, M. (2012). Uranium interaction with two multi-resistant environmental bacteria: Cupriavidus metallidurans CH34 and Rhodopseudomonas palustris. PLoS One 7:e51783. doi: 10.1371/journal.pone.0051783
Lloyd, J. R., and Lovley, D. R. (2001). Microbial detoxification of metals and radionuclides. Curr. Opin. Biotechnol. 12, 248–253. doi: 10.1016/S0958-1669(00)00207-X
Lovley, D. R., Phillips, E. J. P., Gorby, Y. A., and Landa, E. R. (1991). Microbial reduction of uranium. Nature 350, 413–416. doi: 10.1038/350413a0
Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., et al. (2001). Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79. doi: 10.1128/MMBR.65.1.44-79.2001
Marques, C. R., Caetano, A. L., Haller, A., Goncalves, F., Pereira, R., and Rombke, J. (2014). Toxicity screening of soils from different mine areas - A contribution to track the sensitivity and variability of Arthrobacter globiformis assay. J. Hazard. Mater. 274, 331–341. doi: 10.1016/j.jhazmat.2014.03.066
Marshall, M. J., Beliaev, A. S., Dohnalkova, A. C., Kennedy, D. W., Shi, L., and Wang, Z. (2006). c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268. doi: 10.1371/journal.pbio.0040268
Martínez-Bussenius, C., Navarro, C. A., Orellana, L., Paradela, A., and Jerez, C. A. (2016). Global response of Acidithiobacillus ferrooxidans ATCC 53993 to high concentrations of copper: a quantitative proteomics approach. J. Proteomics 145, 37–45. doi: 10.1016/j.jprot.2016.03.039
Martínez-Garcia, E., and de Lorenzo, V. (2016). The quest for the minimal bacterial genome. Curr. Opin. Biotechnol. 42, 216–224. doi: 10.1016/j.copbio.2016.09.001
Masaki, Y., Hirajima, T., Sasaki, K., and Okibe, N. (2015). Bioreduction and immobilization of hexavalent chromium by the extremely acidophilic Fe(III)-reducing bacterium Acidocella aromatica strain PFBC. Extremophiles 19, 495–503. doi: 10.1007/s00792-015-0733-6
Mathuriya, A. S., and Yakhmi, J. V. (2014). Microbial fuel cells to recover heavy metals. Environ. Chem. Lett. 12, 483–494. doi: 10.1007/s10311-014-0474-2
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D., and Kjelleberg, S. (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Meng, J., Wang, H., Liu, X., Lin, J., Pang, X., and Lin, J. (2013). Construction of small plasmid vectors for use in genetic improvement of the extremely acidophilic Acidithiobacillus caldus. Microbiol. Res. 168, 469–476. doi: 10.1016/j.micres.2013.04.003
Methé, B. A., Nelson, K. E., Eisen, J. A., Paulsen, I. T., Nelson, W., Heidelberg, J. F., et al. (2003). Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302, 1967–1969. doi: 10.1126/science.1088727
Mirete, S., Morgante, V., and González-Pastor, J. E. (2016). Functional metagenomics of extreme environments. Curr. Opin. Biotechnol. 38, 143–149. doi: 10.1016/j.copbio.2016.01.017
Mohamed, Y. M., Ghazy, M. A., Sayed, A., Ouf, A., El-Dorry, H., and Siam, R. (2013). Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea brine pool. Sci. Rep. 3:3358. doi: 10.1038/srep03358
Mosier, A. C., Justice, N. B., Bowen, B. P., Baran, R., Thomas, B. C., Northen, T. R., et al. (2013). Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics. mBio 4:e00484-12. doi: 10.1128/mBio.00484-12
Mougiakos, I., Bosma, E. F., de Vos, W. M., van Kranenburg, R., and van der Oost, J. (2016). Next generation prokaryotic engineering: the crispr-cas toolkit. Trends Biotechnol. 34, 575–587. doi: 10.1016/j.tibtech.2016.02.004
Mougiakos, I., Mohanraju, P., Bosma, E. F., Vrouwe, V., Finger Bou, M., Naduthodi, M. I. S., et al. (2017). Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 8:1647. doi: 10.1038/s41467-017-01591-4
Mukherjee, A., Wheaton, G. H., Blum, P. H., and Kelly, R. M. (2012). Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proc. Natl. Acad. Sci. U.S.A. 109, 16702–16707. doi: 10.1073/pnas.1210904109
Ñancucheo, I., and Johnson, D. B. (2011). Significance of microbial communities and interactions in safeguarding reactive mine tailings by ecological engineering. Appl. Environ Microbiol. 77, 8201–8208. doi: 10.1128/AEM.06155-11
Navarro, C. A., Orellana, L. H., Mauriaca, C., and Jerez, C. A. (2009). Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Appl. Environ. Microbiol. 75, 6102–6109. doi: 10.1128/AEM.00308-09
Navarro, C. A., von Bernath, D., and Jerez, C. A. (2013). Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol. Res. 46, 363–371. doi: 10.4067/S0716-97602013000400008
Nies, D. H. (2000). Significance of microbial communities and interactions in safeguarding reactive mine tailings by ecological engineering. Appl. Environ. Microbiol. 77, 8201–8208. doi: 10.1128/AEM.06155-11
Orell, A., Navarro, C. A., Arancibia, R., Mobarec, J. C., and Jerez, C. A. (2010). Life in blue: copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 28, 839–848. doi: 10.1016/j.biotechadv.2010.07.003
Orell, A., Navarro, C. A., Rivero, M., Aguilar, J. S., and Jerez, C. A. (2012). Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 16, 573–583. doi: 10.1007/s00792-012-0457-9
Orell, A., Remonsellez, F., Arancibia, R., and Jerez, C. A. (2013). Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea 2013:289236. doi: 10.1155/2013/289236
Osorio, H., Martínez, V., Nieto, P. A., Holmes, D. S., and Quatrini, R. (2008). Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol. 8:203. doi: 10.1186/1471-2180-8-203
Özdemir, S., Oduncu, M. K., Kilinc, E., and Soylak, M. (2017a). Resistance, bioaccumulation and solid phase extraction of uranium (VI) by Bacillus vallismortis and its UV-vis spectrophotometric determination. J. Environ. Radioact. 171, 217–225. doi: 10.1016/j.jenvrad.2017.02.021
Özdemir, S., Oduncu, M. K., Kilinc, E., and Soylak, M. (2017b). Tolerance and bioaccumulation of U(VI) by Bacillus mojavensis and its solid phase preconcentration by Bacillus mojavensis immobilized multiwalled carbon nanotube. J. Environ. Manage. 187, 490–496. doi: 10.1016/j.jenvman.2016.11.004
Paul, D., Pandey, G., Pandey, J., and Jain, R. K. (2005). Accessing microbial diversity for bioremediation and environmental restoration. Trends Biotechnol. 23, 135–142. doi: 10.1016/j.tibtech.2005.01.001
Pereira, R., Barbosa, S., and Carvalho, F. P. (2014). Uranium mining in Portugal: a review of the environmental legacies of the largest mines and environmental and human health impacts. Environ. Geochem. Health 36, 285–301. doi: 10.1007/s10653-013-9563-6
Poli, A., Di Donato, P., Abbamondi, G. R., and Nicolaus, B. (2011). Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by archaea. Archaea 2011:693253. doi: 10.1155/2011/693253
Qin, G., Zhu, L., Chen, X., Wang, P. G., and Zhang, Y. (2007). Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology 153, 1566–1572. doi: 10.1099/mic.0.2006/003327-0
Raddadi, N., Cherif, A., Daffonchio, D., Neifar, M., and Fava, F. (2015). Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 99, 7907–7913. doi: 10.1007/s00253-015-6874-9
Ramos, J. L., Marqués, S., van Dillewijn, P., Espinosa-Urgel, M., Segura, A., Duque, E., et al. (2011). Laboratory research aimed at closing the gaps in microbial bioremediation. Trends Biotechnol. 29, 641–647. doi: 10.1016/j.tibtech.2011.06.007
Reguera, G., Nevin, K. P., Nicoll, J. S., Covalla, S. F., Woodard, T. L., and Lovley, D. R. (2006). Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72, 7345–7348. doi: 10.1128/AEM.01444-06
Reith, F., Etschmann, B., Grosse, C., Moors, H., Benotmane, M. A., Monsieurs, P., et al. (2009). Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U.S.A. 106, 17757–17762. doi: 10.1073/pnas.0904583106
Rojas, L. A., Yáñez, C., González, M., Lobos, S., Smalla, K., and Seeger, M. (2011). Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS One 6:e17555. doi: 10.1371/journal.pone.0017555
Rothschild, L. J., and Mancinelli, R. L. (2001). Life in extreme environments. Nature 409, 1092–1101. doi: 10.1038/35059215
Sand, W., and Gehrke, T. (2006). Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 157, 49–56. doi: 10.1016/j.resmic.2005.07.012
Shelobolina, E. S., Coppi, M. V., Korenevsky, A. A., DiDonato, L. N., Sullivan, S. A., Konishi, H., et al. (2007). Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. doi: 10.1186/1471-2180-7-16
Shong, J., Diaz, M. R. J., and Collins, C. H. (2012). Towards synthetic microbial consortia for bioprocessing. Curr. Opin. Biotechnol. 23, 798–802. doi: 10.1016/j.copbio.2012.02.001
Shrestha, N., Chilkoor, G., Vemuri, B., Rathinam, N., Sani, R. K., and Gadhamshetty, V. (2018). Extremophiles for microbial-electrochemistry applications: a critical review. Bioresour. Technol. 255, 318–330. doi: 10.1016/j.biortech.2018.01.151
Shukla, A., Parmar, P., and Saraf, M. (2017). Radiation, radionuclides and bacteria: an in-perspective review. J. Environ. Radioact. 180, 27–35. doi: 10.1016/j.jenvrad.2017.09.013
Singh, A., and Singh, A. K. (2017). Haloarchaea: worth exploring for their biotechnological potential. Biotechnol. Lett. 39, 1793–1800. doi: 10.1007/s10529-017-2434-y
Slonczewski, J. L., Fujisawa, M., Dopson, M., and Krulwich, T. A. (2009). Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 55, 1–79. doi: 10.1016/S0065-2911(09)05501-5
Srivastava, P., Bragança, J., Ramanan, S. R., and Kowshik, M. (2013). Synthesis of silver nanoparticles using haloarchaeal isolate Halococcus salifodinae BK3. Extremophiles 17, 821–831. doi: 10.1007/s00792-013-0563-3
Sun, J., Sayyar, B., Butler, J. E., Pharkya, P., Fahland, T. R., Famili, I., et al. (2009). Genome-scale constraint-based modeling of Geobacter metallireducens. BMC Syst. Biol. 3:15. doi: 10.1186/1752-0509-3-15
Ulloa, G., Collao, B., Araneda, M., Escobar, B., Álvarez, S., and Bravo, D. (2016). Use of acidophilic bacteria of the genus Acidithiobacillus to biosynthesize CdS fluorescent nanoparticles (quantum dots) with high tolerance to acidic pH. Enzyme Microb. Technol. 95, 217–224. doi: 10.1016/j.enzmictec.2016.09.005
Venkateswarlu, K., Nirola, R., Kuppusamy, S., Thavamani, P., Naidu, R., and Megharaj, M. (2016). Abandoned metalliferous mines: ecological impacts and potential approaches for reclamation. Rev. Environ. Sci. Biotechnol. 15, 327–354. doi: 10.1007/s11157-016-9398-6
Vera, M., Krok, B., Bellenberg, S., Sand, W., and Poetsch, A. (2013). Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics 13, 1133–1144. doi: 10.1002/pmic.201200386
Voica, D. M., Bartha, L., Banciu, H. L., and Oren, A. (2016). Heavy metal resistance in halophilic Bacteria and Archaea. FEMS Microbiol. Lett. 363, 1–9. doi: 10.1093/femsle/fnw146
Watling, H. R., Shiers, D. W., and Collinson, D. M. (2015). Extremophiles in mineral sulphide heaps: some bacterial responses to variable temperature, acidity and solution composition. Microorganisms 3, 364–390. doi: 10.3390/microorganisms3030364
Wenbin, N., Dejuan, Z., Feifan, L., Lei, Y., Peng, C., Xiaoxuan, Y., et al. (2011). Quorum-sensing system in Acidithiobacillus ferrooxidans involved in its resistance to Cu2+. Lett. Appl. Microbiol. 53, 84–91. doi: 10.1111/j.1472-765X.2011.03066.x
Wheaton, G., Counts, J., Mukherjee, A., Kruh, J., and Kelly, R. (2015). The confluence of heavy metal biooxidation and heavy metal resistance: implications for bioleaching by extreme thermoacidophiles. Minerals 5, 397–451. doi: 10.3390/min5030397
Wheaton, G. H., Mukherjee, A., and Kelly, R. M. (2016). Transcriptomes of the extremely thermoacidophilic archaeon Metallosphaera sedula exposed to metal “shock” reveal generic and specific metal responses. Appl. Environ. Microbiol. 82, 4613–4627. doi: 10.1128/AEM.01176-16
Wilkins, M. J., Callister, S. J., Miletto, M., Williams, K. H., Nicora, C. D., Lovley, D. R., et al. (2011). Development of a biomarker for Geobacter activity and strain composition; proteogenomic analysis of the citrate synthase protein during bioremediation of U(VI). Microb. Biotechnol. 4, 55–63. doi: 10.1111/j.1751-7915.2010.00194.x
Yates, M. D., Cusick, R. D., and Logan, B. E. (2013). Extracellular palladium nanoparticle production using Geobacter sulfurreducens. ACS Sustainable Chem. Eng. 1, 1165–1171. doi: 10.1021/sc4000785
Zhang, X., Liu, X., Liang, Y., Fan, F., Zhang, X., and Yin, H. (2016). Metabolic diversity and adaptive mechanisms of iron- and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environ. Microbiol. Rep. 8, 738–751. doi: 10.1111/1758-2229.12435
Keywords: extremophilic bacteria and archaea, meta-‘omics’, genetic engineering, synthetic biology, mine wastes, metal-radionuclide recycling/recovering
Citation: Marques CR (2018) Extremophilic Microfactories: Applications in Metal and Radionuclide Bioremediation. Front. Microbiol. 9:1191. doi: 10.3389/fmicb.2018.01191
Received: 28 January 2018; Accepted: 16 May 2018;
Published: 01 June 2018.
Edited by:
Kian Mau Goh, University of Technology, MalaysiaReviewed by:
M. Oves, King Abdulaziz University, Saudi ArabiaCopyright © 2018 Marques. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catarina R. Marques, Y3JtYXJxdWVzQHVhLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.