- School of Life Sciences, East China Normal University, Shanghai, China
Short-wave ultraviolet (UV-C) treatment represents a potent, clean and safe substitute to chemical sanitizers for fresh fruit preservation. However, the dosage requirement for microbial disinfection may have negative effects on fruit quality. In this study, UV-C was found to be more efficient in killing spores of Botrytis cinerea in dark and red light conditions when compared to white and blue light. Loss of the blue light receptor gene Bcwcl1, a homolog of wc-1 in Neurospora crassa, led to hypersensitivity to UV-C in all light conditions tested. The expression of Bcuve1 and Bcphr1, which encode UV-damage endonuclease and photolyase, respectively, were strongly induced by white and blue light in a Bcwcl1-dependent manner. Gene mutation analyses of Bcuve1 and Bcphr1 indicated that they synergistically contribute to survival after UV-C treatment. In vivo assays showed that UV-C (1.0 kJ/m2) abolished decay in drop-inoculated fruit only if the UV-C treatment was followed by a dark period or red light, while in contrast, typical decay appeared on UV-C irradiated fruits exposed to white or blue light. In summary, blue light enhances UV-C resistance in B. cinerea by inducing expression of the UV damage repair-related enzymes, while the efficiency of UV-C application for fruit surface disinfection can be enhanced in dark or red light conditions; these principles seem to be well conserved among postharvest fungal pathogens.
Introduction
Fresh fruits and vegetables are rich in moisture and nutrients, and thus susceptible to postharvest decays caused by microbial contamination and proliferation, especially pathogenic fungi (Sperber et al., 2009). Chemical sanitizers are commonly used for disinfection of the harvested crops. However, the long-term use of chemical fungicides frequently poses the risk of fungicide resistance in pathogens. More importantly, pesticide residue in fresh crops is an increasing health concern among consumers. To address these issues, developing alternative methods to synthetic fungicides for disease management purpose is in urgent need (Romanazzi et al., 2012, 2016).
Ultraviolet-C (UV-C, 200–280 nm) offers interesting possibilities for postharvest disease management as a safe alternative to conventional chemical fungicides. Although the UV-C portion of the cosmic rays is almost completely absorbed by the outer space atmosphere and is hardly observed in nature on the earth’s surface, UV-C radiation can be created by artificial lamps, and usually causes two distinct effects on fresh fruits and vegetables: one is the elicitation of disease resistance and quality improvement in fresh crops, while the other is reduction of microbial population due to its direct germicidal effect (Urban et al., 2016). The former effect on host crops is often defined as hormesis, that is, stimulation of favorable responses in plants exposed to low or sublethal doses of an agent such as a physical stressor (Luckey, 1982). It has been recognized that UV-C light at low hormetic doses reduced the postharvest decay of a wide range of crops (Luckey, 1982), although these beneficial effects depend on the dose and timing of UV-C exposure, the fruit or vegetable species and cultivars, and the exposed area (Allende and Artés, 2003; Vicente et al., 2005; Costa et al., 2006; Pombo et al., 2011; Topcu et al., 2015). UV-C can cause DNA damage, and is thus used as a sterilizing agent for air, water and food (Bintsis et al., 2000). However, the host hormesis-inducing and microbe-disinfecting effects of UV-C on fresh crops are somewhat incompatible: the disease resistance elicitation effect can be achieved only when the UV-C dosage is restricted to certain sublethal dosages, while the microbe-disinfecting effect can be produced by increasing UV-C dosage. Accordingly, UV-C treatment of fruits and vegetables needs to be optimized tactically to obtain a desirable balance between the beneficial changes in host plants along with efficient disinfection against pathogens (Urban et al., 2016). To address this issue, it is important to understand the regulation mechanisms of UV-C resistance in fungal pathogens, which still remains to be elucidated.
It is known that UV-C inhibits microbial growth mainly by inducing the formation of pyrimidine dimers that alter the DNA helix and block microbial cell replication (Bintsis et al., 2000). However, microorganisms can protect themselves against UV radiation by repairing damaged DNA (Sinha and Hader, 2002). Proteins such as DNA photolyases have been found in a variety of species and can restore the UV-damaged bases back to their original undamaged states (Bluhm and Dunkle, 2008; Brettel and Byrdin, 2010). Additionally, the UV-damage endonuclease (UVDE) can directly recognize and cleave damaged DNA, which is followed by lesion removal, gap-filling, and ligation reactions (Bowman et al., 1994; Freyer et al., 1995; Yajima et al., 1995). Therefore, the UV-C dosages for fungicidal purposes must be relatively high, usually ranging from 0.5 to 20 kJ/m2 (Bintsis et al., 2000).
Fungi can also sense visible light to promote tolerance against harmful UV radiation (Fuller et al., 2015). This has been validated in several fungi by functional studies on the orthologs of White collar complex (WCC), the blue light receptor of Neurospora crassa. These proteins can act both as photosensors as well as transcription factors to regulate the expression of light responsive genes (Ballario et al., 1996; Crosthwaite et al., 1997; Liu et al., 2003). DNA repair enzymes, including photolyases and UVDE, were shown to be induced by light via the conserved WCC signaling pathway in several fungal species, including Cryptococcus neoformans, Phycomyces blakesleeanus, and Ustilago maydis (Verma and Idnurm, 2013; Tagua et al., 2015; Brych et al., 2016). The light regulation of UV-C resistance in fungi implies that ultraviolet disinfection efficiency can be adjusted by orchestrating photic conditions.
Botrytis cinerea is the gray mold pathogen that causes enormous economic damage to fruits and vegetables, both in field and during postharvest procedures (Fillinger and Elad, 2016). The infection cycle of this pathogen usually starts with the attachment of conidia to the plant surface, followed by infection and rapid hyphal spreading inside the plant tissue leading to host collapse (Fillinger and Elad, 2016). B. cinerea shows varied developmental responses to different wavelengths of the light spectrum. The conserved WCC homologs of B. cinerea mediate transcriptional responses to the blue light spectrum and inhibit its conidiation. Furthermore, WCC is required for coping with excessive light, oxidative stress, and to achieve full virulence to host plants (Canessa et al., 2013). Recently, cryptochrome/photolyase homologs, BcCRY1 and BcCRY2, were characterized in B. cinerea, revealing that BcCRY1 acts as the major photolyase in photoprotection, whereas BcCRY2 acts as a cryptochrome with signaling function in regulating repression of conidiation (Cohrs and Schumacher, 2017). However, the mechanism of photoselective regulation of UV resistance in B. cinerea has not been fully elucidated yet.
The present study aims to reveal the mechanism of regulation of UV-C resistance in the fungal fruit spoilage agents. Using B. cinerea as a representative model, we find that blue light and Bcwcl1 are required for activating the expression of the UV-damage endonuclease and photolyase genes, Bcuve1 and Bcphr1 (or Bccry1 in an earlier report, Cohrs and Schumacher, 2017), respectively. Gene mutation analysis revealed that Bcuve1 and Bcphr1 are synergistically responsible for coping with UV-C induced damage in B. cinerea. More importantly, since blue light is the specific spectrum that supports DNA-damage repair activities in fungi, UV-C treatment followed by dark or red-light conditions was thus found to enhance microbe-killing efficiency thereby facilitating fruit spoilage management.
Materials and Methods
Fungal Strains and Culture
The reference strain B05.10 of Botrytis cinerea was designated as wild type for genetic modification. The other pathogenic fungi were originally isolated from fruits and vegetables (Table 1). Potato dextrose agar (PDA) was used to maintain the fungal cultures at their indicated optimum temperatures. Conidia of each fungal species were collected by flooding the sporulated colonies with sterilized water, followed by filtration through four layers of cheesecloth and centrifugation at 4500 rpm. The concentration of the resulting conidial suspension was measured by a hematocytometer.
Visible and UV-C Light Treatment
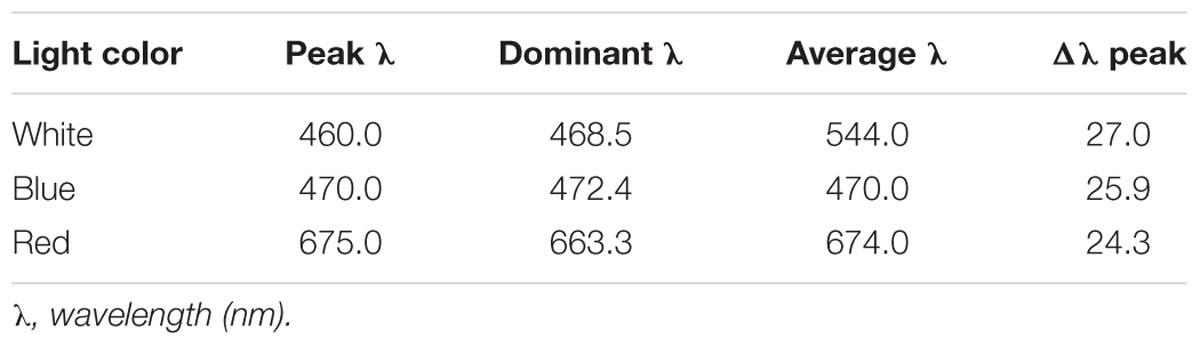
White, blue, and red visible light spectra were produced by a light-emitting diode (LED, Qiding Photo Electronic, Shanghai, China). The parameters of each light spectrum are listed in Table 2. Light intensities were fixed at 20 μmol m-2s-1, as measured by Quantum Light Meter (Spectrum Technologies, United States), and achieved by manually adjusting the power control switch of the LED devices. For dark treatment, samples were kept in a light-proof plastic box and incubated at a temperature similar to the light-treated ones. HL-2000 cross-linker lamps (UVP, United States) were used for UV-C radiation (254 nm) treatment. The UV-C dosage was recorded as either μJ/cm2 or kJ/m2.
Generation of Gene Deletion Mutant
Protoplast transformation mediated homologous recombination strategy was adopted to generate knock out mutants of target genes according to the previous method (Chung and Lee, 2015). 1 kb of the 5′ and 3′ untranslated regions (UTRs) of target genes were amplified from the DNA of the wild type strain, and selective marker genes, hygromycin (hyg) or nourseothricin resistant (nat) cassette, were PCR amplified from the plasmid pCAMBI1300 or pNR2, respectively. Overlap PCR was then performed to fuse the 5′- and 3′-UTRs with the selective marker genes, resulting in 5′ UTR-hyg (or nat) -3′ UTR constructs for protoplast transformation. Diagnostic PCR was performed to identify bonafide targeted disruption mutants among the emerged transformants as indicated in Supplementary Figure S1. The gene disruption mutants were further verified by Southern blot hybridization according to the protocol recommended in the DIG high prime DNA labeling and detection starter Kit II (Roche, Mannheim, Germany).
Construction of Bcuve1-GFP Fusion Expression Strain and Fluorescent Microscopy
The expression vector pNDN-OGG (Schumacher, 2012) carrying nourseothricin resistance (NAT) and GFP expression cassettes was digested with NcoI. The wild type Bcuve1 was amplified without the stop codon using the primers P45/46 (Supplementary Table S1) that are equipped with 22-bp overlaps corresponding to the sequences in the destination vector for the Gibson assembly-based cloning using the Hieff CloneTM Plus Multi One Step Cloning Kit (YEASEN, China). The resulted clones were identified by PCR diagnosis, and the positive ones were further confirmed for correctness by sequencing. The correct vector named pNDN-OGG-Bcuve1 carrying Bcuve1 upstream of the GFP gene was linearized by SacII digestion and transformed into Δbcuve1. The fungal transformation was selected by nourseothricin (50 μg/ml). The positive resistant transformants were purified by series of single spore culture, and the Δbcuve1-Bcuve1-GFP strain was obtained for UV-C sensitivity and microscopy analysis. Fluorescence and light microscopy was performed with a Zeiss Axio Imager Z2 microscope. Differential interference microscopy (DIC) was used for bright field images. GFP fluorescence was examined using excitation BP 470/40 and emission BP 525/50, DAPI staining with the excitation G 365 and emission BP 445/50. DIC, GFP, and DAPI images were merged via ImageJ soft ware.
Gene Expression Analysis
Aliquots (200 μl) of conidial suspensions (106 conidia/ml) were inoculated on cellophane-overlaid PDA and incubated at 25°C in dark for 24 h. Samples were subsequently divided into four groups each and placed under different light conditions (white, blue, red light, and darkness). After incubating for 1 h, mycelium samples (about 0.1 g) from each of the groups were harvested using cell scrapers in the dark, transferred into 2 ml Eppendorf tubes, and immediately frozen in liquid nitrogen. For total RNA extraction, each sample was submerged in 1 ml Trizol reagent (Invitrogen, Carlsbad, CA, United States), and homogenized by shaking along with four steel balls (2 mm diameter) at 70 Hz and 4°C for 3 min on a Tissuelyser (Jingxin Industrial, Shanghai, China). The resulting suspensions were extracted with chloroform according to the manufacturer’s instructions supplied with Trizol. One microgram of each RNA sample was used as a template for reverse transcription using the Prime ScriptTM RT reagent Kit (Perfect Real Time) (TakaRa Biotechnology, Co., Dalian, China). Real-time PCR amplifications were conducted in a CFX96TM Real-Time System (BIO-BAD, Inc., United States) using TakaRa SYBR Premix Ex Taq (TakaRa Biotechnology). Relative quantifications of the real-time PCR amplifications were performed with the following parameters, initial preheating at 95°C for 30 s followed by 39 cycles at 95°C for 5 s and 60°C for 30 s. The β-tubulin gene was analyzed as an internal reference. Experiments were repeated three times for each sample. The primers used in this study are listed in Supplementary Table S1. The gene expression levels were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). All experiments were repeated three times.
UV-C Sensitivity Assays
To evaluate fungal UV-C sensitivity, 200 μl of conidial suspensions (5 × 103 conidia/ml) were evenly inoculated on individual PDA surfaces using a cell spreader and subjected to UV-C irradiation, with dosages ranging from 0.6 to 1.2 kJ/m2. The samples were immediately incubated at 25°C for 2 h under white, blue, red light and dark conditions. Subsequently, all samples were continuously incubated in the dark for 2 days. Samples that were not subjected to UV-C treatment were used as reference controls. Fungal colonies arising on the plate were counted and survival rates were calculated by dividing the colony numbers on UV-C treated plates with those on non-UV-C treated ones.
To visualize the effect of visible light on fungal UV-C sensitivity, 5 μl of conidial suspension (106 conidia/ml) was dropped on cellophane overlaid with water agar. After UV-C radiation, the samples were similarly treated for 2 h under different light and dark conditions, and then incubated in the dark for another 22 h. Conidial germination of each sample was finally examined under a light microscope. Five replicates were conducted for all experiments.
Fruit Inoculation Assay
Wild type spores of B. cinerea were suspended in sterilized 1% sugar solution, and the concentration was adjusted to 106 conidia/ml. Table grapes were purchased from the local super market. Healthy berries with uniform size and maturity stage were selected for this assay. Before inoculation, the fruits were submerged in 0.5% sodium hypochlorite solution for 3 min to eliminate possible contaminating microorganisms on the surface of grape berries, followed by rinsing thrice with sterilized water. The fruits were then artificially wound-inoculated with 10 μl spore suspension at a site on the equatorial line of each grape berry. The fruits were then exposed to 1 kJ/m2 UV-C, and divided into four groups, each including 30 berries, and transferred to dark, white, blue, and red lights to incubate at 25°C for 2 h. Subsequently, all samples were placed in continuous dark, and 4 days later the disease symptoms were photographed, and the decay areas were measured via ImageJ software.
Statistical Analysis
The experiments in this study were repeated three times. The data obtained were analyzed by ANOVA followed by Duncan’s multiple range tests (p < 0.01) for means comparison with the use of SPSS 17.0.
Results
Blue Light Is Specifically Required for Inducing UV-C Resistance in a Bcwc1-Dependent Manner
In the UV-C sensitivity assay, wild type spores of B. cinerea were completely killed by 0.8 kJ/m2 UV-C in dark and 2 h red light-treated groups, while the spores exposed to white and blue light survived by more than 40%. Even when the UV-C dose reached 1.2 kJ/m2, the wild type spores illuminated in white and blue light maintained an approximately 10% survival rate (Figure 1A). Eventually, we confirmed that the blue light spectrum (but not the red light) is specifically effective in enhancing B. cinerea spore survivability after UV-C irradiation.
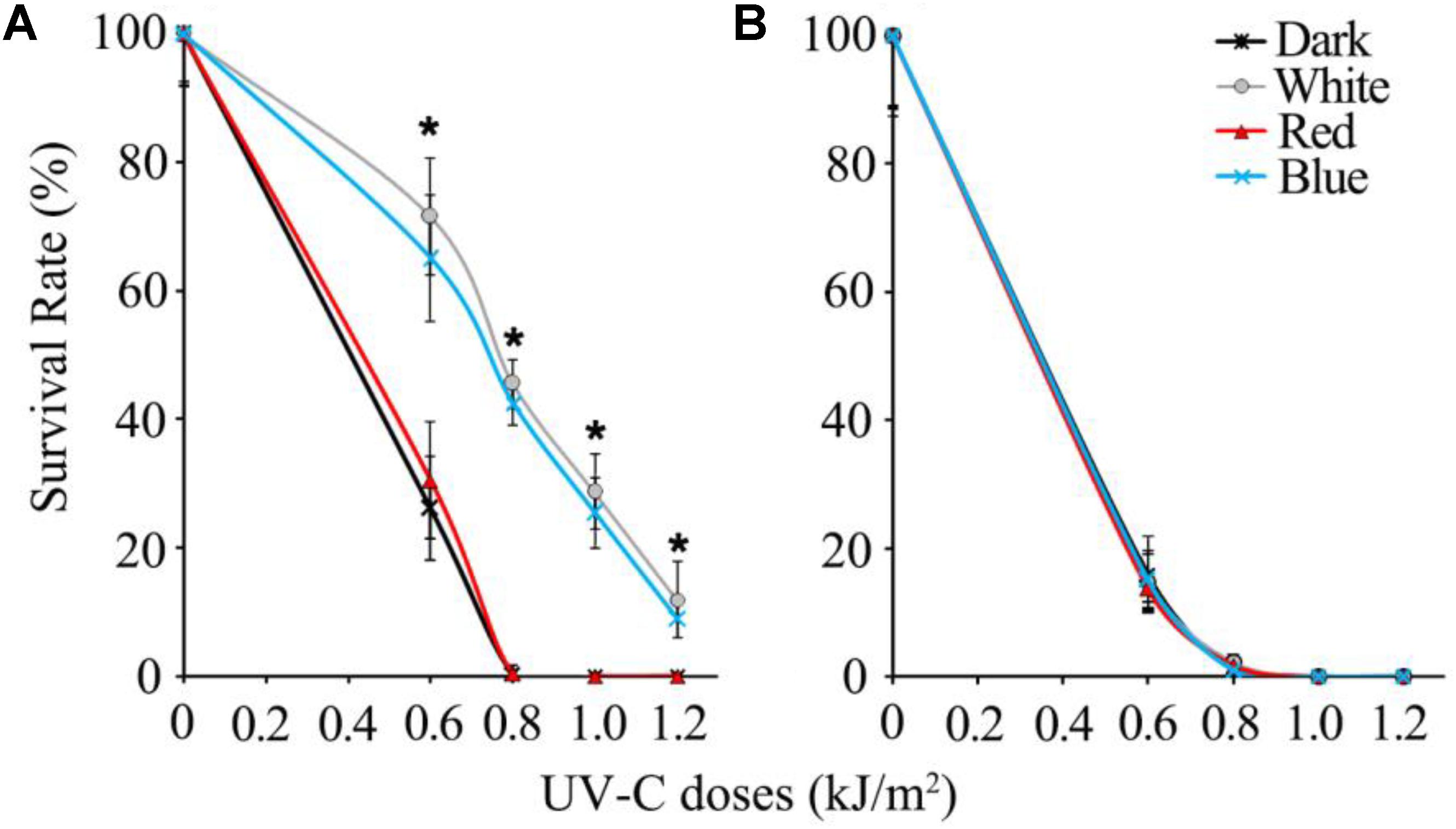
FIGURE 1. Survival rates of UV-C treated spores of wild type (A) and Δbcwcl1 (B) strains of B. cinerea. Error bars represent standard deviation. Asterisks (∗) indicate that survival of the spores illuminated by blue and white light after UV-C stress was significantly higher than those treated by red light and dark (P < 0.01). Error bars represent SD and n = 5.
Since blue light is known to be sensed by fungi via the WCC photoreceptors, the WC-1 homolog gene in B. cinerea, Bcwcl1 (Canessa et al., 2013), was disrupted by replacing the open reading frame with the hygromycin resistance cassette via homologous recombination. Δbcwcl1 mutant strains were confirmed by genomic PCR, and showed enhanced melanization and sporulation in contrast to the wild type strain (data not shown). These phenotypes were in agreement with a previous report in which WC-1 was shown to negatively regulate spore formation and melanin biosynthesis (Canessa et al., 2013). One representative Δbcwcl1 strain was thus used for the UV-C sensitivity assay. The results showed that neither blue nor white light could increase survivability of the mutant after UV-C radiation. The survival rates of the Δbcwc1 mutant dropped down to almost 0% in all the treatment groups when the UV-C dosage was above 0.8 kJ/m2 (Figure 1B). Taken together, we conclude that photoinduction of UV-C resistance in B. cinerea is specifically caused by the blue light spectrum via signaling through the light receptor encoded by Bcwcl1.
Expression of DNA Damage Repair Related Genes Is Regulated by Light via Bcwcl1
Since white collar 1 can function as both blue light receptor and transcription factor (Canessa et al., 2013), it is assumed that certain downstream genes regulated by this protein could be responsible for photo responsive phenotypes. Based on the transcriptomic data (Schumacher et al., 2014), two genes expected to contribute to DNA damage repair in B. cinerea were obtained: Bcuve1 (Bcin01g08960) encoding the protein homologous to the UV damage endonuclease Uve1 of Schizosaccharomyces pombe (GenBank: CAA19577.1), and Bcphr1 (Bcin05g08060, or Bccry1 in Cohrs and Schumacher, 2017) encoding the homolog of photolyase/cryptochrome of Neurospora crassa (GenBank: KHE81232.1). The deduced protein domains of these two gene products are presented in Figure 2A. The expression of Bcuve1 and Bcphr1 was analyzed via quantitative RT-PCR. The results showed that white and blue light treatments strongly induced the expression of Bcuve1 and Bcphr1 in the wild type, but not in the Δbcwcl1 mutant strain. However, red light exposure did not change the expression of these two genes in both WT and Δbcwcl1 strains (Figure 2B). Taken together, these results indicate that blue light signaling via the WC-1 homolog activates both endonuclease excision and photolyase pathways in B. cinerea.
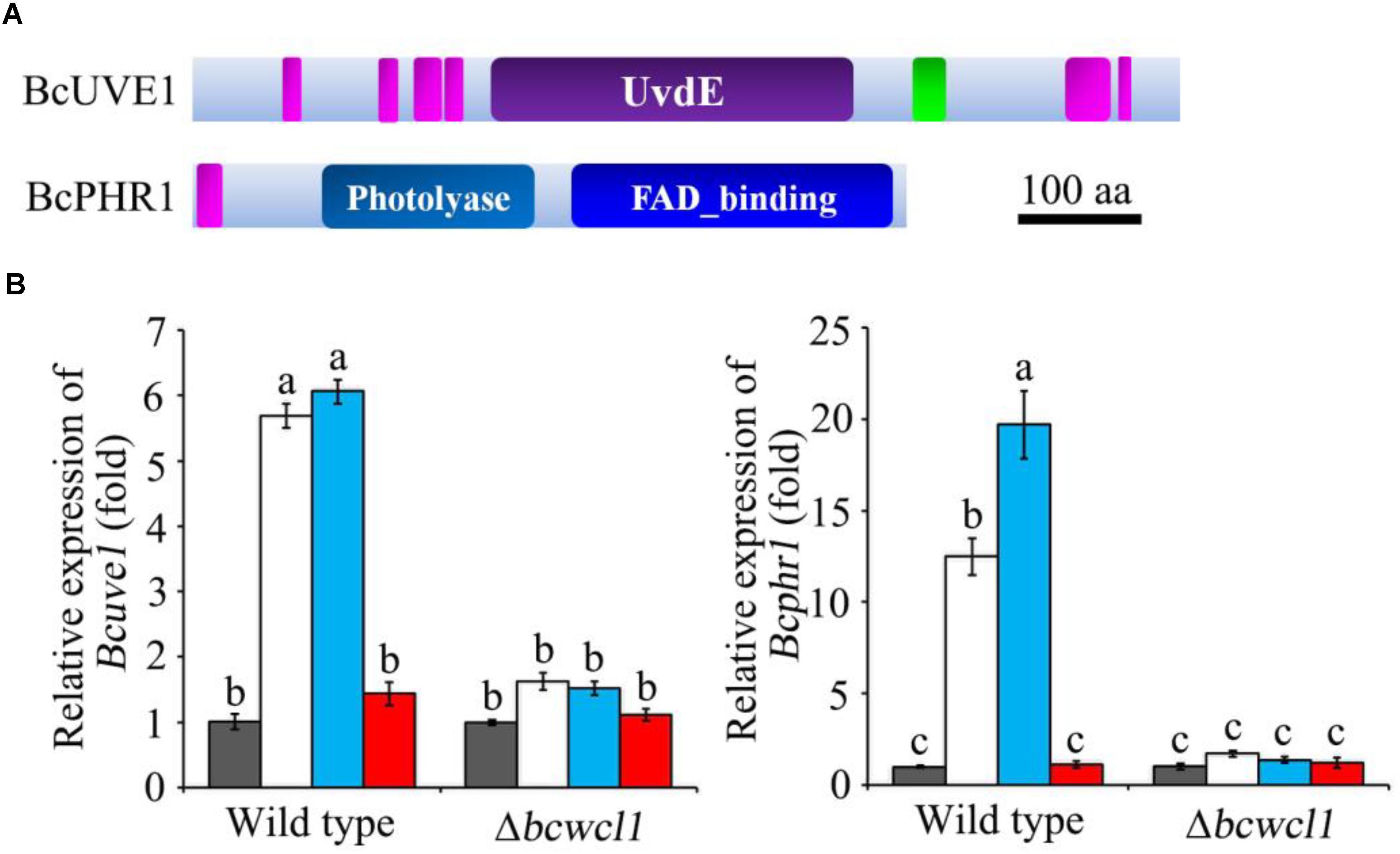
FIGURE 2. Expression analysis of DNA damage repair related genes. (A) The Ensembl Fungi IDs of Bcuve1 and Bcphr1 are Bcin01g08960.1 and Bcin05g08060.1, respectively. According to SMART analysis, the BcUVE1 protein contains a UV damage endonuclease domain (UvdE), and BcPHR1 contains a DNA_photolyase domain and a FAD_binding domain. The pink rectangles represent regions of low complexity, and the green one represents the coiled-coil region. The scale bar indicates a length of 100 amino acids. (B) Comparison of Bcuve1 and Bcphr1 expression levels between the wild type and Δbcwcl1 strains under different light conditions. Columns represent dark, white, blue, and red light exposure from left to right. Histograms marked with different characters are statistically different (p < 0.01). Error bars represent SD and n = 3.
Bcuve1 and Bcphr1 Synergistically Contribute to UV-C Resistance in B. cinerea
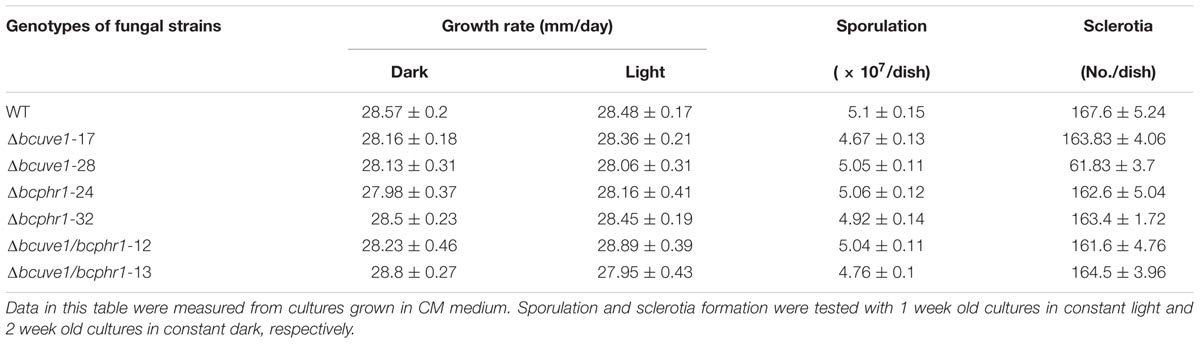
To confirm the roles of Bcuve1 and Bcphr1 in B. cinerea, single and double mutant strains were created via protoplast transformation and homologous recombination mediated gene replacement. Genomic PCR analysis and Southern blot confirmed mutation of the targeted locus in the colonies recovered (Supplementary Figure S1). The resulting mutants, Δbcuve1, Δbcphr1, and ΔΔbcuve1/bcphr1, showed growth rates, sporulation, sclerotial development (Figure 3 and Table 3), and virulence equivalent to the wild type strain when tested on grape berries (Figure 4), indicating that neither Bcuve1 nor Bcphr1 is involved in regulating the vegetative growth, development, and host-invasion processes.
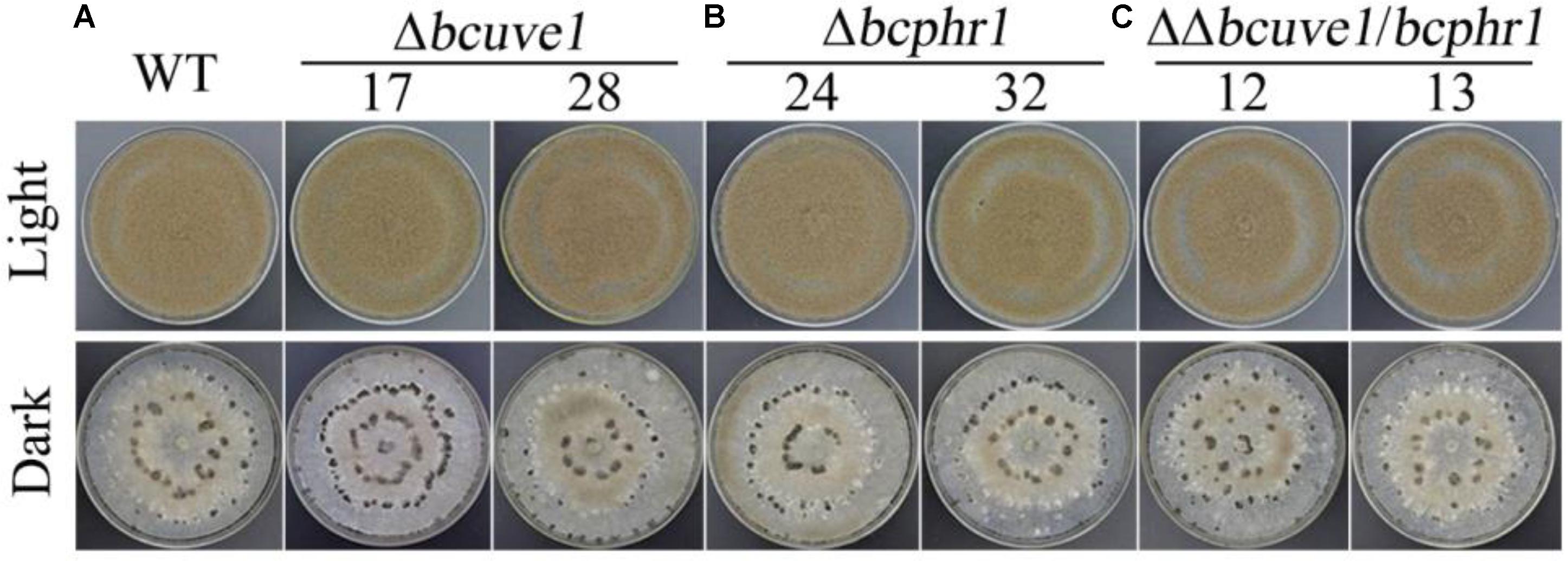
FIGURE 3. The three mutant strains, (A) Δbcuve1, (B) Δbcphr1, and (C) ΔΔbcuve1/bcphr1, formed similar sporulation colonies (1 week in light) and sclerotia (2 weeks in dark) on PDA.
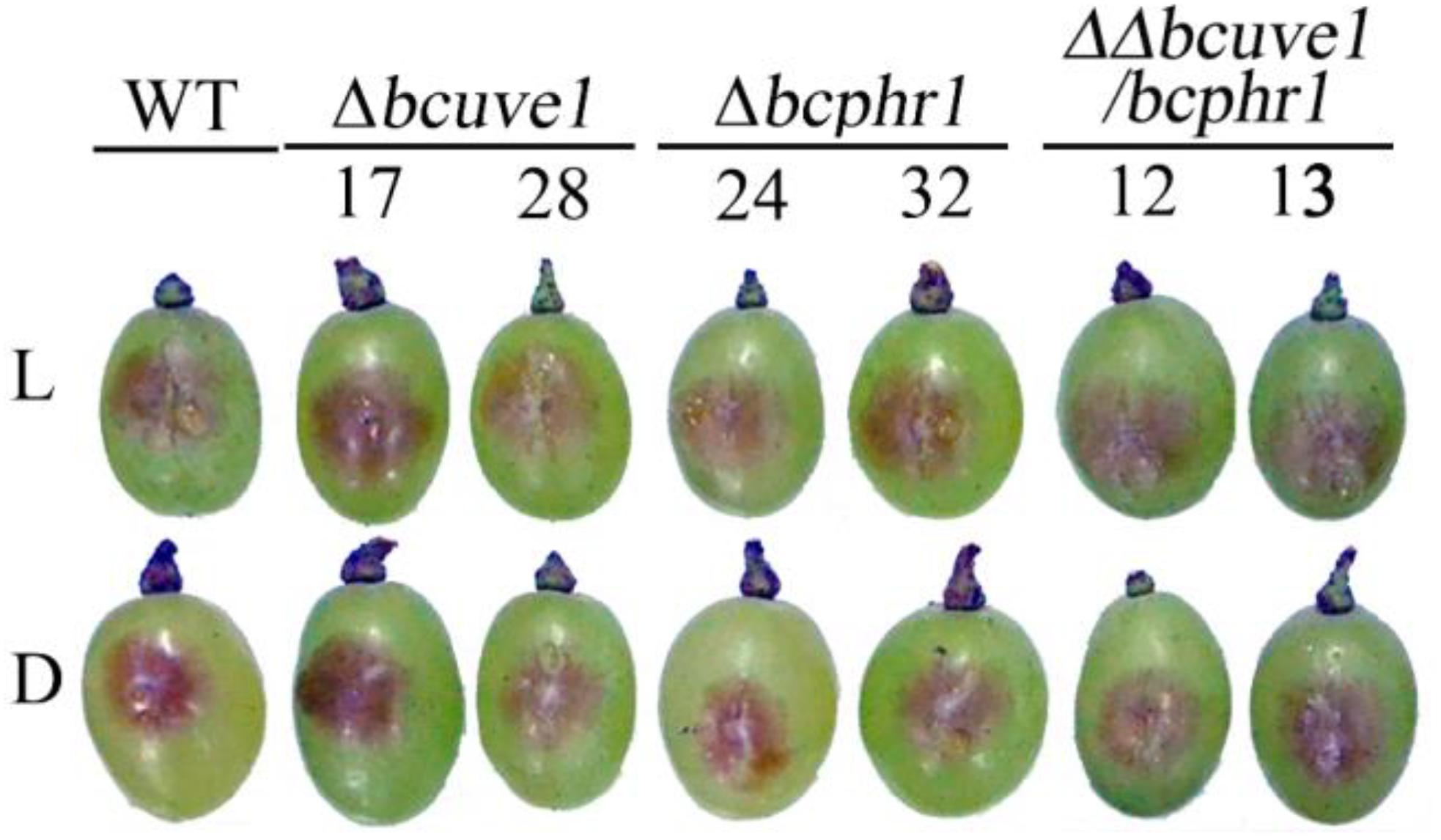
FIGURE 4. Bcuve1 and Bcphr1 are not required for virulence. Grape berries were inoculated with spore suspensions of each strain, and disease development was checked after 3 days of incubation at 22°C in light (L) or dark (D).
In the UV-C sensitivity assay, the Δbcuve1 mutant showed significantly reduced survival rates when compared to the wild type strain. However, blue and white lights were still capable of enhancing UV-C tolerance of the Δbcuve1 mutant when the dosage was 0.6 kJ/m2 (Figure 5). On the other hand, the Δbcphr1 mutant was relatively more tolerant to UV-C than Δbcuve1, although Δbcphr1 still showed significantly reduced survival rate under UV-C stress when compared to the wild type. Visible light treatment after UV-C radiation did not alter the survivability of Δbcphr1 (Figure 5). Moreover, the double mutant, ΔΔbcuve1/bcphr1, combined the patterns of the two single mutants, showing similar sensitivity to UV-C as the Δbcuve1 mutant in dark, and no change in survival rate as the Δbcphr1 mutant when treated with white light after UV-C treatment (Figure 5). These data together indicate that BcUVE1 and BcPHR1 play synergistic roles in UV-C damage repair in B. cinerea.
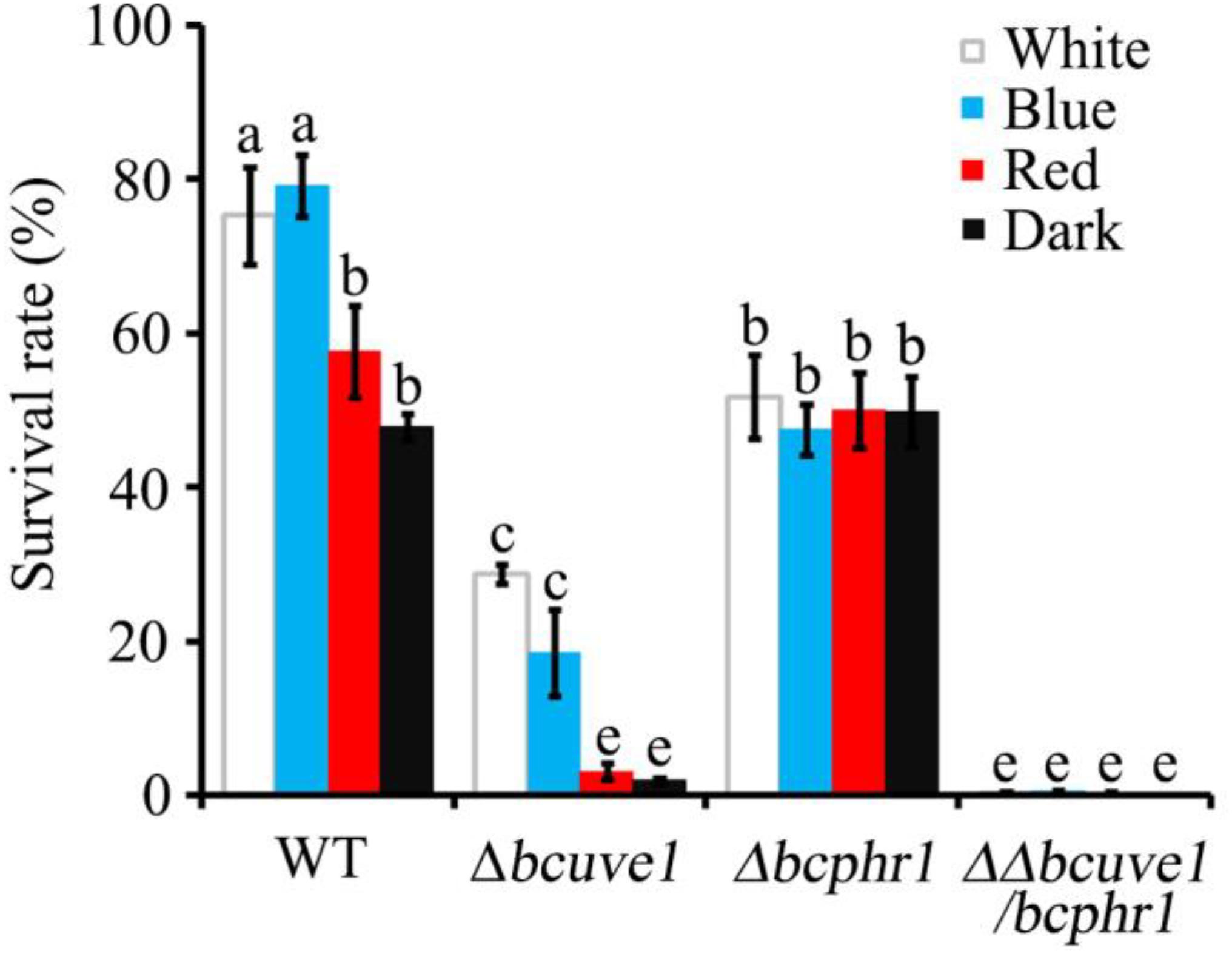
FIGURE 5. The three mutant strains, Δbcuve1, Δbcphr1, and ΔΔbcuve1/bcphr1, showed differentially altered sensitivities to UV-C (0.6 kJ/m2) when influenced with visible light qualities. Bars marked with non-identical characters are significantly different (P < 0.01). Error bars represent SD and n = 5.
Since UV radiation can stimulate organisms to generate reactive oxygen species (ROS), which can also cause DNA damage, we additionally tested the sensitivity of each strain to ROS stress upon treatment with hydrogen peroxide (H2O2). The results demonstrated that all the strains, i.e., Δbcuve1, Δbcphr1, ΔΔbcuve1/bcphr1 and WT, showed decreased survivability with increasing H2O2 concentration in the medium, however, no significant difference in susceptibility to H2O2 was observed between the mutants and wild type (Figure 6).
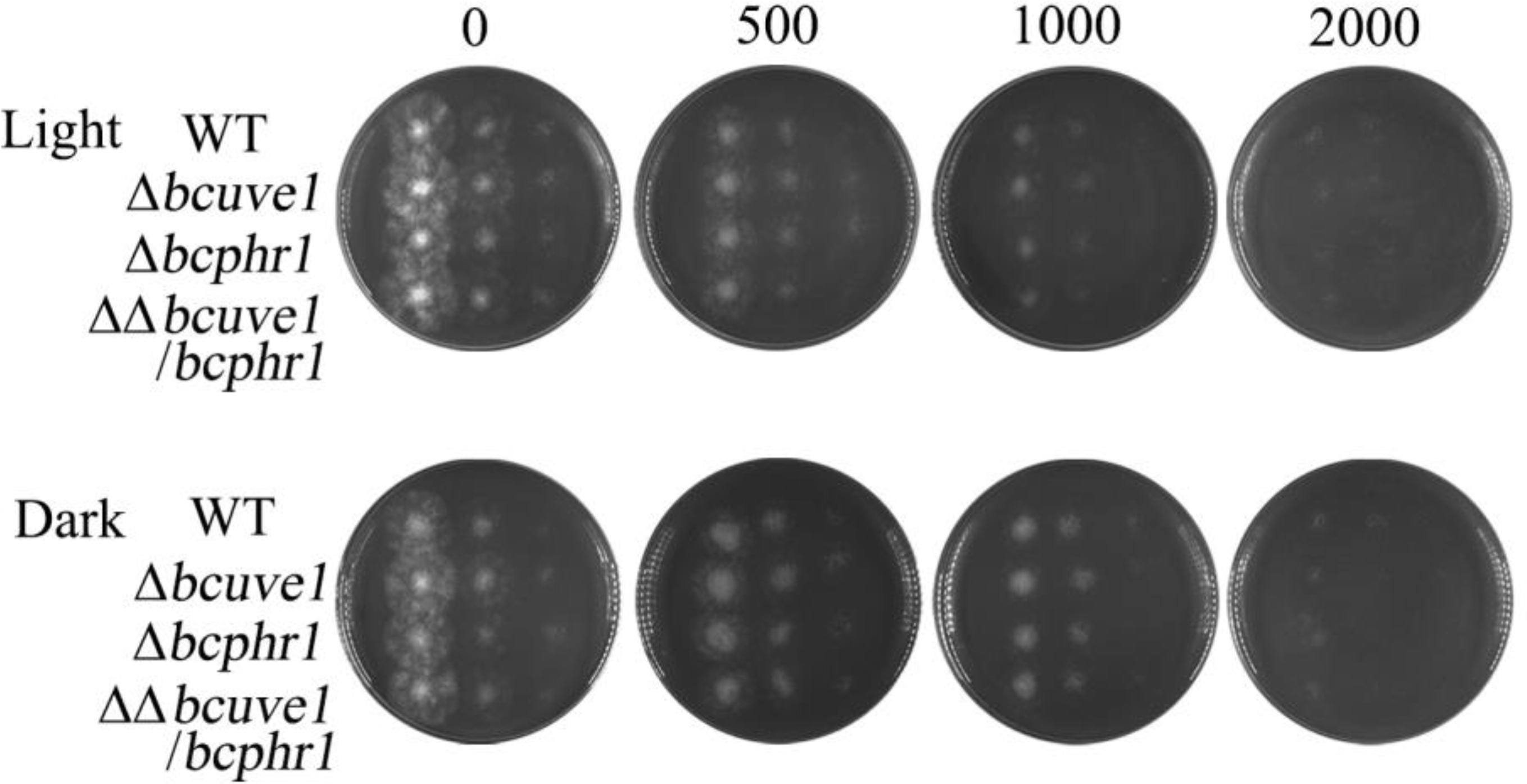
FIGURE 6. Sensitivity to hydrogen peroxide (H2O2). Spore suspensions of each tested strain were serially diluted to 106, 105, and 104 spores per ml, and 10 μl of each suspension was spotted on PDA medium (vertical array of 4 spots for each diluted suspension). The H2O2 contents (0–2000 mM) in the media are indicated for each column. The plates were photographed after 2 days of incubation at 22°C in light (L) or dark (D).
Role of BcUVE1 in UV-Damage Repair Is Confirmed by Genetic Complementation and Subcellular Localization
In order to confirm that the UV-tolerance deficiency of Δbcuve1 mutant is due to disruption of the Bcuve1 gene, wild type Bcuve1 was tagged with GFP at the 3′ end and transformed into Δbcuve1 to produce the complementation mutant strain Δbcuve1-Bcuve1-GFP. UV sensitivity assays showed that survival of Δbcuve1-Bcuve1-GFP was similar to the WT (Figure 7A). Since the UV-endonuclease is supposed to be involved in DNA damage repair, we determined the subcellular localization patterns of BcUVE1 by tracking the constitutively expressing GFP fusion proteins in the Δbcuve1-Bcuve1-GFP strain. As expected, the fused protein BcUVE1-GFP was found in the nuclei (Figure 7B).

FIGURE 7. Mycelial development of test fungal strains exposed to UV-C. (A) Bcuve1-GFP complementation to detect UV-sensitivity of the Δbcuve1 mutant. (B) Location of the fused marker protein in the nuclei of the Δbcuve1-Bcuve1-GFP mutant as shown in the light (upper) and fluorescent (lower) microscopes.
Additionally, yeast two hybrid experiments indicated that BcUVE1 and BcPHR1 do not interact with each other (Figure 8), even though BcPHR1 (or named as BcCRY1 earlier) is also localized in nuclei (Cohrs and Schumacher, 2017), and both BcUVE1 and BcPHR1 are involved in UV-damage repair.
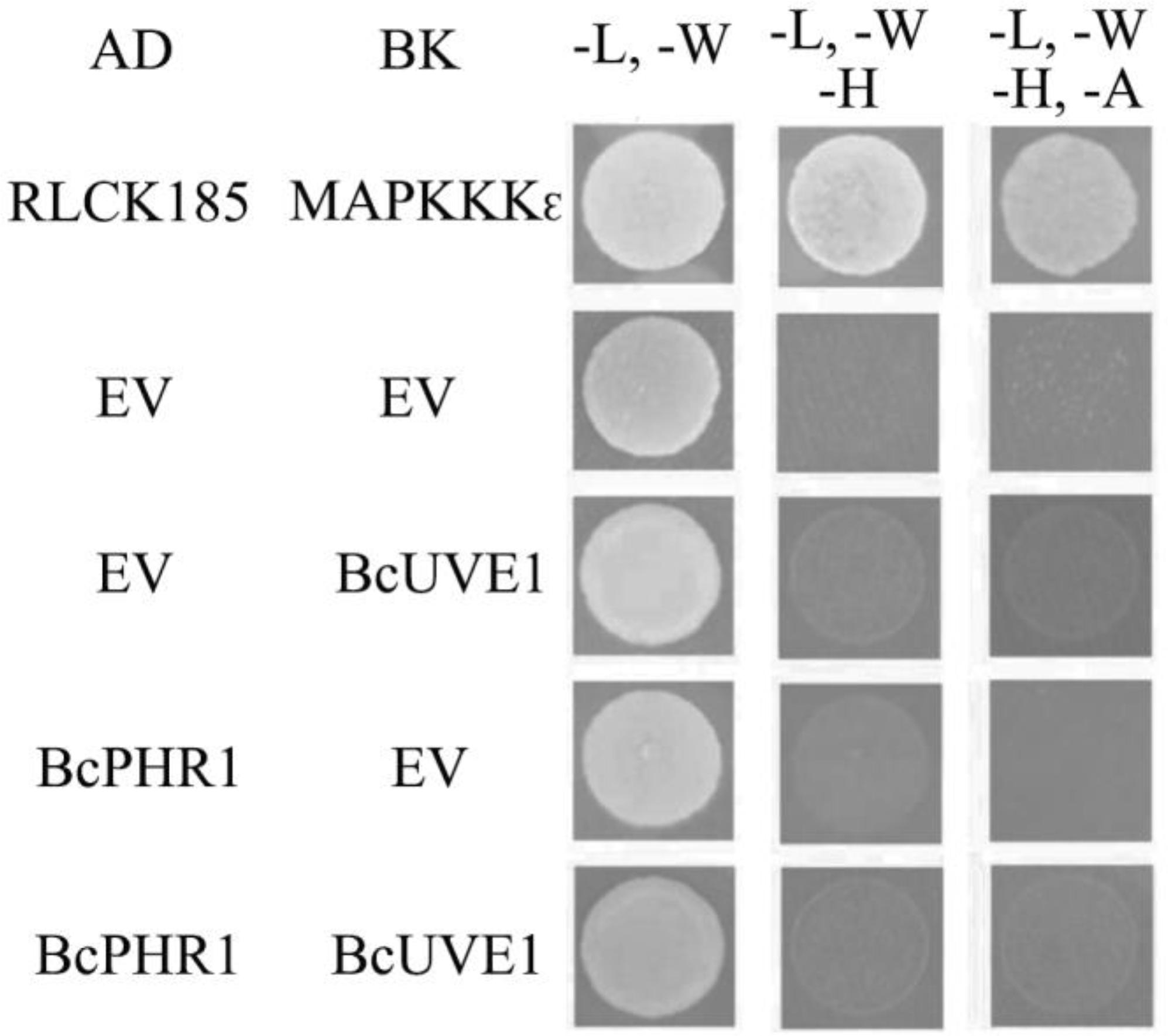
FIGURE 8. Yeast two hybrid assays: BcUVE1 and BcPHR1 do not interact with each. The cDNAs of either gene were cloned adjacent to the activation (AD) or DNA binding (BK) domains of S. cerevisiae Gal4. Constructs were transformed into the S. cerevisiae reporter strain AH109. Growth on medium without leucine (L), tryptophan (W), histidine (H), and adenine (A) indicate interactions between tested alleles to reconstitute the Gal4 protein. AD-RLCK185 and BK-MAPKKK𝜖 are positive controls that have been proved to physically interact with each other (Wang et al., 2017).
Fungicidal Efficiency of UV-C on Fruits Is Enhanced in Red Light and Dark Conditions but Not in Blue or White Light Conditions
The above study demonstrated that B. cinerea is more susceptible to UV-C stress in dark and red light than in blue and white light. These findings enabled us to make an association between the process of light-regulated UV damage repair mechanism and UV-C application for plant disease control, especially at the postharvest stage. The present assay indicated that artificial inoculation of wild type B. cinerea spores on grape berries would fail to cause decay symptoms if the UV-C (1 kJ/m2) treatment was followed by dark or red light (Figure 9). In contrast, exposure to blue and white light caused the UV-C treated samples to finally develop typical soft decay (Figure 9). Consequently, the in vitro and in vivo assays suggest that more satisfactory results of UV-C application for postharvest disease management can be expected if the UV-C damage repair activities of the fungal pathogens are suppressed.
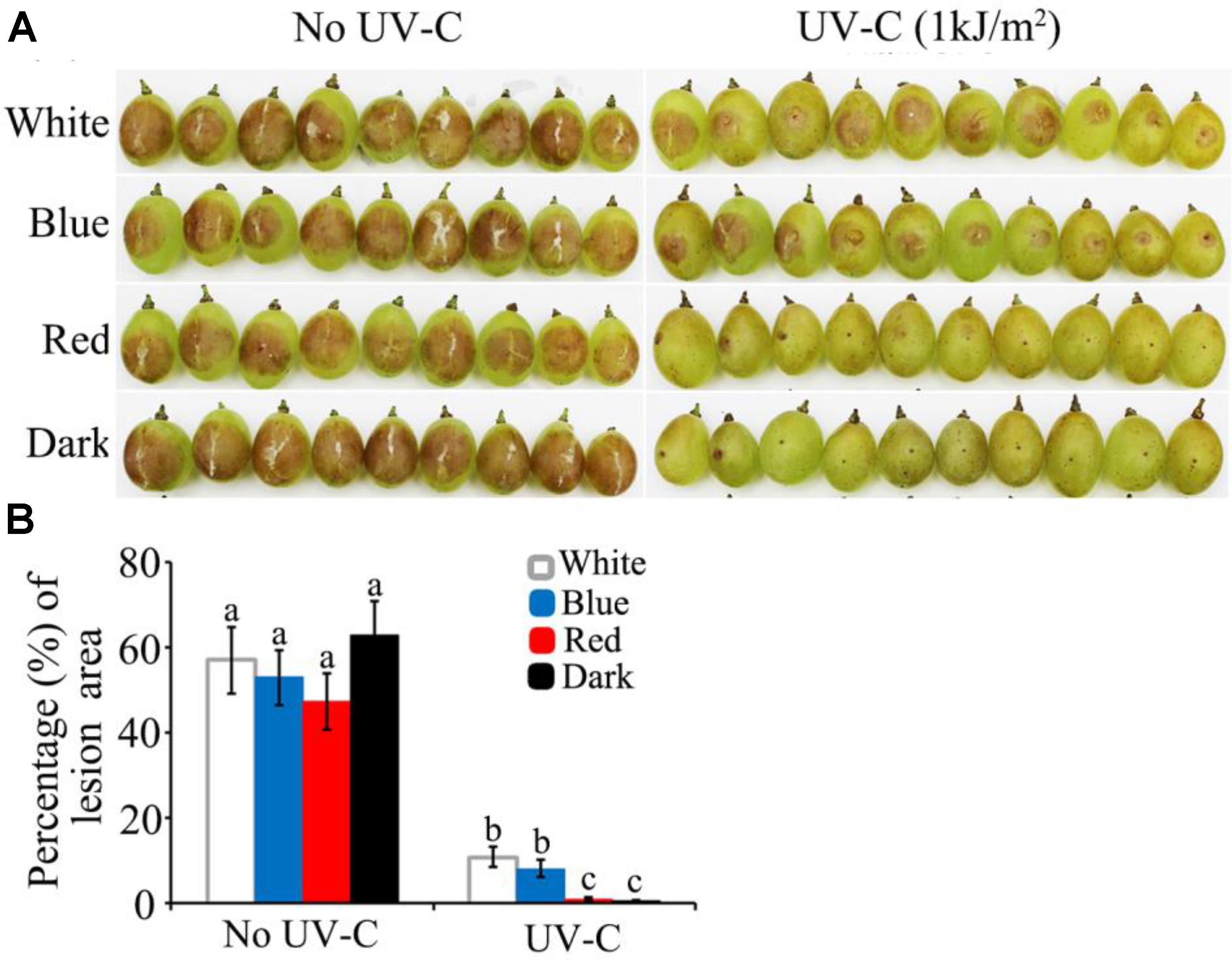
FIGURE 9. UV-C radiation achieved more efficient restriction of grape decay in red light and dark than in blue and white light conditions. (A) Photograph records of the infected grape berries on 4th day after inoculation. (B) Data analysis for the percentage of decayed area on each grape berry. White, blue, red, and dark represent light quality treatment. The data followed by different characters are statistically different (p < 0.01).
Visible Light Qualities Show Similar Effects on UV-C Sensitivity of Common Postharvest Fungal Pathogens
The regulatory mechanisms of UV-C resistance uncovered here are expected to be valid even in other pathogenic fungi. In this study, we additionally tested the UV-C sensitivity of other important postharvest fungal pathogens: Alternaria alternata, Penicillium digitatum, and Colletotrichum gloeosporioides. The results suggest that all of the fungi tested were killed much more easily by relatively lower dosages (less than 1 kJ/m2) of UV-C in red light and dark conditions, while the spores exposed to blue and white light could survive from higher UV-C dosages (Figure 10).
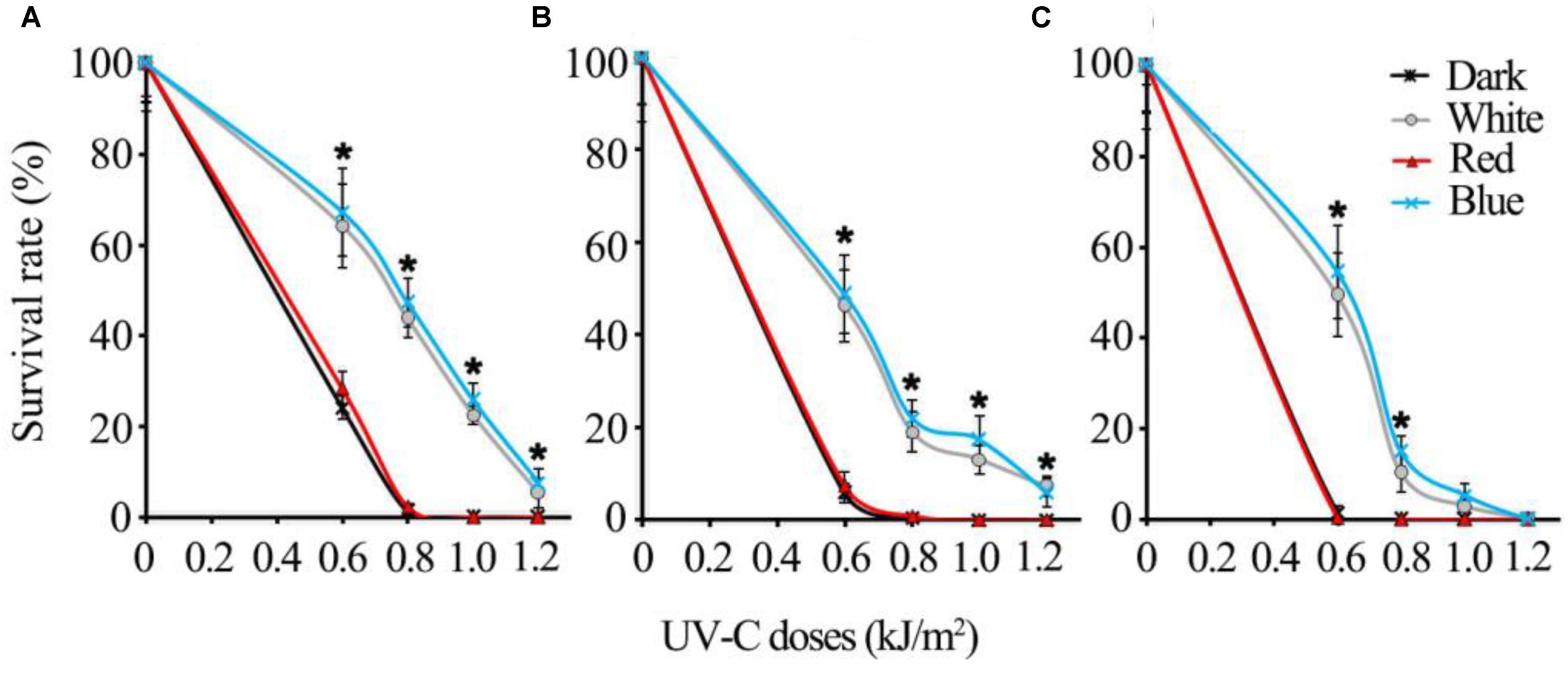
FIGURE 10. The UV-C sensitivity of three common postharvest fungal pathogens is regulated similarly by different visible light qualities (A) Alternaria alternata; (B) Penicillium digitatum; (C) Colletotrichum gloeosporioides. Asterisks (∗) indicate that survival of spores illuminated by blue and white light after UV-C stress was significantly higher than those treated by red light and dark (P < 0.01). Error bars represent SD and n = 5.
Discussion
Spoilage decay due to contamination by pathogenic fungi is one of the main causes of abundant postharvest losses of fresh fruits and vegetables. UV-C is an alternative to fungicides for control of postharvest diseases, due to its dual roles of inducing defense in plants and causing surface disinfection of the pathogenic microbes. Induced resistance to postharvest pathogens by UV-C was shown in a wide range of crops (Ben-Yehoshua et al., 1992; Mercier et al., 1993; Charles et al., 2008a,b). However, decontamination of the fruit surface by UV-C could still be interesting from a practical standpoint, as the irradiated tissues would be subject to less inoculum pressure in addition to being more disease resistant. Thus, the present work has been focused on the pathogen rather than the host.
From an evolutionary perspective, the presence of light may signal the upcoming threat of genotoxic ultraviolet radiation to microbes in the natural environment and thus activate UV-damage repair activities (Fuller et al., 2015). Therefore, our study attempted to address the knowledge gap of light-regulation mechanisms of UV-C tolerance in phytopathogenic fungi, and lay the foundation to optimize UV-C treatment parameters for better disinfection efficiency on fresh crop surfaces.
Through quantitative UV-C sensitivity assays with the model species B. cinerea, we found that the spores of this fungus incubated in red light and dark are more sensitive to UV-C than those incubated in white and blue light conditions. This phenomenon implies that the blue light spectrum is capable of inducing UV-C resistance in B. cinerea. Since the WCC proteins are known to be conserved blue light receptors in the fungal kingdom, we further investigated the role of the key component of the WCC, BcWCL1 of B. cinerea, in UV-C resistance. A Bcwcl1 deletion mutant showed substantially reduced UV-C resistance under any light conditions, confirming that the blue light receptor system of B. cinerea indeed regulates UV-C resistance. This is in accordance with the reports that WC-1 homologs are pivotal for environmental UV stress tolerance in several other fungal species (Idnurm and Heitman, 2005; Ruiz-Roldan et al., 2008; Kim et al., 2014).
The photoreceptor WCC can serve both signal input (LOV domain) and output (Zn-finger transcription factor domain) functions. Photo induction of DNA repair enzymes represents one of the downstream signaling targets of WCC (Fuller et al., 2015). Photoreactivating enzymes such as photolyases are induced by light via WCC homologues in the ascomycetes Neurospora crassa, Aspergillus fumigatus (Fuller et al., 2013), Aspergillus nidulans (Ruger-Herreros et al., 2011), Fusarium oxysporum, (Ruiz-Roldan et al., 2008) and Cercospora zeae-maydis (Bluhm and Dunkle, 2008; Kim et al., 2011), as well in as the basidiomycete Ustilago maydis (Brych et al., 2016). In these fungi, photolyases are recognized as the major enzymes responsible for UV damage repair. Visible light likely plays dual roles in enhancing photolyase-dependent UV resistance in fungi, one being the induction of expression of photolyase genes via WCC signaling, while the other being energy provision to support photoreactivation activity of the photolyases (Fuller et al., 2015). In B. cinerea, there are two cryptochome/photolyase homologs, BcCRY1 and BcCRY2, but only BcCRY1 was found to act as the major photolyase in photoprotection (Cohrs and Schumacher, 2017), which we therefore re-named as BcPHR1. We confirmed that the expression of Bcphr1 was induced by white and blue light in a Bcwcl1-dependent manner, and the deletion mutant Δbcphr1 showed increased UV-sensitivity when compared with WT as measured by quantitative spore survivability assay. However, Δbcphr1 was found to be relatively more resistant to UV-C than Δbcwcl1, implying that Bcphr1 is not the only member of the WCC downstream targets responsible for UV-damage repair.
Actually, as shown in the light-induced transcriptome data (Schumacher et al., 2014), Bcuve1 represents another candidate for repairing UV- induced damages. This gene encodes a protein that is homologous to UVDE in fission yeast Schizosaccharomyces pombe, in which UVDE is essential for excision repair of UV induced DNA damage (Bowman et al., 1994; Freyer et al., 1995; Yajima et al., 1995). Additionally, Uve1, the UVDE homolog in the basidiomycetes C. neoformans, is a direct target of WCC signaling and required for UV resistance (Verma and Idnurm, 2013). As shown in our study, expression of Bcuve1 is also enhanced by blue and white light in a Bcwcl1-dependent manner, and the deletion mutant Δbcuve1 is more sensitive to UV-C than WT. However, white and blue light still moderately elevated survival rate of Δbcuve1 spores after UV-C treatment, which is most probably due to the presence of functional Bcphr1 in this mutant. This hypothesis was confirmed by analysis of the double mutant ΔΔbcuve1/bcphr1, which showed almost similar deficiency of UV-C tolerance as the Δbcwcl1 mutant to any kind of light conditions. Taken together, WCC mediated blue light signaling in B. cinerea can activate both UV-endonuclease and photolyase to synergistically repair damages caused by UV-C radiation.
The major damage caused by UV-C to organisms is DNA lesions. Thus, subcellular localization of each DNA damage repair enzyme is indicative of its functional preference on either the nuclear or cytoplasmic (the mitochondrion) genomes. BcCRY1 (or BcPHR1 in this paper) was shown to solely localize in the nuclei (Cohrs and Schumacher, 2017). Interestingly, this study found that the UV-endonuclease BcUVE1 also accumulated in the nuclei as shown by analysis of the GFP tagged allele. Thus, these two DNA damage repairing enzymes are regulated by blue light signaling in B. cinerea, and are presumably responsible for removing UV-induced lesions in the nuclear genome. However, yeast-two-hybrid assays demonstrated that BcPHR1 and BcUVE1 did not interact with each other, further implying that these two enzymes mediated two independent DNA repair pathways. In addition, the spores of the Δbcuve1 strain were more sensitive to UV-C than those of Δbcphr1. This phenotype could possibly be explained by the different DNA damage precursors they repair. It is well known that the major DNA lesions induced by UV-C are cyclobutane pyrimidine dimers (CPD) (Watanabe et al., 2006), and other minor lesions are pyrimidine pyrimidone photoproducts (6-4PP) and some diverse rare DNA photoproducts (Stapleton, 1992). BcPHR1 (or BcCRY1) is phylogentically recognized as belonging to the CPD photolyase group (Cohrs and Schumacher, 2017), and therefore its target precursors may be limited to CPD lesions. On the other hand, UV-endonuclease was originally discovered in S. pombe to be able to recognize both CPD and 6-4PP and initiate their excision repair (Bowman et al., 1994; Freyer et al., 1995; Yajima et al., 1995), even though CPD and 6-4PP differ significantly with respect to the structural distortions that they induce in the DNA duplex. The homolog of UV-endonuclease in B. cinerea, BcUVE1, is probably more versatile than the photolyase BcPHR1 in its DNA damage repair capability.
White collar complex of B. cinerea were found to be involved in tolerance to ROS (Canessa et al., 2013), which can also cause DNA damage and affect virulence. However, the test of sensitivity against hydrogen peroxide demonstrated that neither Bcuve1 nor Bcphr1 was involved in detoxification of ROS stress. Besides, the deletion mutants did not show any notable defects in vegetative growth, sporulation, sclerotial formation, or virulence, suggesting that BcUVE1 and BcPHR1 specifically cope with UV stress in B. cinerea.
As discussed earlier, UV-C could be used as a potential agent for sanitization of fresh fruit and vegetable surfaces (Nigro et al., 1998). However, the efficacy of UV-C is dependent on the resistance of target mircroorganisms against UV-C light (Syamaladevi et al., 2013). Based on our study, it can be deduced that the germicidal effect of UV-C on fungal pathogens can be attenuated by exposure to visible light, especially the blue light spectrum, largely due to induction of DNA damage repair enzymes (BcUVE1 and BcPHR1) by light. So, these findings may theoretically verify the rationality of an earlier practical study reporting that dark period following UV-C treatment enhances killing of Botrytis cinerea conidia and controls gray mold of strawberries better in green houses (Janisiewicz et al., 2016). Furthermore, we expanded this photoselective enhancement of UV-C disinfection into postharvest disease management. Consequently, UV-C application for postharvest disease management can be more effective if the UV-C damage repair activities of the fungal pathogens are suppressed by either dark or red light conditions. Additionally, the UV-C sensitivities of several common fungal pathogens behaved similarly under different light conditions. These phenomena can be explained by the fact that the WCC homologs are widely conserved blue light receptors in the fungal kingdom (Fuller et al., 2015), with the UV damage repair systems being one of their common regulation targets (Idnurm and Heitman, 2005; Verma and Idnurm, 2013; Schumacher et al., 2014; Wu et al., 2014; Brych et al., 2016). As a result, common postharvest pathogenic fungi can be efficiently killed by relatively less amounts of UV-C by following the principles stated in this study. Based on the phenomena and their underlying mechanisms discovered in this study, a shelf device equipped with UV-C inside and a red monochromatic filter on the screen is proposed to be beneficial for better disease management of fresh postharvest crops (Supplementary Figure S2).
Although UV-C is directly germicidal to microbial agents of postharvest diseases, its application for conservation purpose of fresh fruit and vegetables is also largely influenced by its effect on physiological modifications of the commodities. The possibility of injuries to crops by higher UV-C doses could even cause an increase in the susceptibility of fruits to postharvest decays (Stevens et al., 1996). We achieved enhancement of UV-C disinfection efficiency on the pathogen with limited dosages that are significantly less than those being commonly used to irradiate fresh crops. Subsequently, future efforts should be focused on selecting proper UV-C parameters to obtain beneficial effects without causing detrimental changes on quality attributes.
Author Contributions
PZ and LX designed the experiments. QL, SA, YQ, YW, and PZ performed the experiments. QL, SA, YQ, PZ, YJ, and LX analyzed and interpreted the data. QL, SA, PZ, and LX wrote the paper with insight from all the authors.
Funding
This work was financially supported by the grant of National Key Research and Development Program of China (2016YFD0400105), National Natural Science Foundation of China (Nos. 31571902 and 31501536), and Science and Technology Commission of Shanghai Municipality (15YF1403300).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZZ and handling Editor declared their shared affiliation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01141/full#supplementary-material
FIGURE S1 | Strategies for deletion of Bcuve1 and Bcphr1. (A) Schematic illustration of the homologous recombination strategy to replace the 5′ end of the target gene with hygromycin B (hph) or nourseothricin (Nat) resistance cassette as selective markers. (B) Diagnostic PCR analysis for integration of the replacement fragment with genomic DNA. As demonstrated in (A), primer pairs PTF-PTR and PMF-PMR were used to test the presence or absence of target genes (Bcuve1 or Bcphr1) and selection markers (hph or Nat), respectively; the primer pairs PT5F-PMR′ and PMF′-PT3R were used to verify the correctness of integration sites at the 5′-UTR and 3′-UTR regions respectively. (C) Southern blot analysis of the WT, Δbcuve1 and Δbcphr1 strains. Genomic DNAs were digested with HindIII; the probes targeting the selection markers are indicated in (A). A single band of expected size in each mutant verified authentic homologous recombination events, and ruled out the possibility of multicopy insertion of the selection markers. (D) Reverse transcript-PCR confirmed the absence of expression of Bcuve1 and Bcphr1 in the respective mutants. The constitutively expressed gene β-tubulin was used as a reference.
FIGURE S2 | Shelf device equipped with UV-C inside and a monochromatic filter on the screen for fresh crop preservation and display.
TABLE S1 | Primers used in this study.
References
Allende, A., and Artés, F. (2003). UV-C radiation as a novel technique for keeping quality of fresh processed ‘Lollo Rosso’ lettuce. Food Res. Int. 36, 739–746. doi: 10.1016/S0963-9969(03)00054-1
Ballario, P., Vittorioso, P., Magrelli, A., Talora, C., Cabibbo, A., and Macino, G. (1996). White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15, 1650–1657.
Ben-Yehoshua, S., Rodov, V., Kim, J. J., and Carmeli, S. (1992). Preformed and induced antifungal materials of citrus fruits in relation to the enhancement of decay resistance by heat and ultraviolet treatments. J. Agr. Food Chem. 40, 1217–1221. doi: 10.1021/jf00019a029
Bintsis, T., Litopoulou-Tzanetaki, E., and Robinson, R. K. (2000). Existing and potential applications of ultraviolet light in the food industry – a critical review. J. Sci. Food Agr. 80, 637–645. doi: 10.1002/(SICI)1097-0010(20000501)80:6
Bluhm, B. H., and Dunkle, L. D. (2008). PHL1 of Cercospora zeae-maydis encodes a member of the photolyase/cryptochrome family involved in UV protection and fungal development. Fungal Genet. Biol. 45, 1364–1372. doi: 10.1016/j.fgb.2008.07.005
Bowman, K. K., Sidik, K., Smith, C. A., Taylor, J. S., Doetsch, P. W., and Freyer, G. A. (1994). A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 22, 3026–3032. doi: 10.1093/nar/22.15.3026
Brettel, K., and Byrdin, M. (2010). Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 20, 693–701. doi: 10.1016/j.sbi.2010.07.003
Brych, A., Mascarenhas, J., Jaeger, E., Charkiewicz, E., Pokorny, R., Bolker, M., et al. (2016). White collar 1-induced photolyase expression contributes to UV-tolerance of Ustilago maydis. Microbiologyopen 5, 224–243. doi: 10.1002/mbo3.322
Canessa, P., Schumacher, J., Hevia, M. A., Tudzynski, P., and Larrondo, L. F. (2013). Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the White Collar Complex. PLoS One 8:e84223. doi: 10.1371/journal.pone.0084223
Charles, M. T., Goulet, A., and Arul, J. (2008a). Physiological basis of UV-C induced resistance to Botrytis cinerea in tomato fruit: IV. Biochemical modification of structural barriers. Postharvest Biol. Tec. 47, 41–53. doi: 10.1016/j.postharvbio.2007.05.019
Charles, M. T., Mercier, J., Makhlouf, J., and Arul, J. (2008b). Physiological basis of UV-C-induced resistance to Botrytis cinerea in tomato fruit: I. Role of pre- and post-challenge accumulation of the phytoalexin-rishitin. Postharvest Biol. Tec. 47, 10–20. doi: 10.1016/j.postharvbio.2007.05.013
Chung, K. R., and Lee, M. H. (2015). “Split-marker-mediated transformation and targeted gene disruption in filamentous fungi,” in Genetic Transformation Systems in Fungi, Fungal Biology, Vol. 2, eds M. van den Berg, and K. Maruthachalam (Cham: Springer).
Cohrs, K. C., and Schumacher, J. (2017). The two cryptochrome/photolyase family proteins fulfill distinct roles in DNA photorepair and regulation of conidiation in the gray mold fungus Botrytis cinerea. Appl. Environ. Microb. 83, e812–e817. doi: 10.1128/AEM.00812-17
Costa, L., Vicente, A. R., Civello, P. M., Chaves, A. R., and Martínez, G. A. (2006). UV-C treatment delays postharvest senescence in broccoli florets. Postharvest Biol. Tec. 39, 204–210. doi: 10.1016/j.postharvbio.2005.10.012
Crosthwaite, S. K., Dunlap, J. C., and Loros, J. J. (1997). Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276, 763–769. doi: 10.1126/science.276.5313.763
Fillinger, S., and Elad, Y. (2016). Botrytis – the Fungus, the Pathogen and its Management in Agricultural Systems. New York, NY: Springer International Publishing. doi: 10.1007/978-3-319-23371-0
Freyer, G. A., Davey, S., Ferrer, J. V., Martin, A. M., Beach, D., and Doetsch, P. W. (1995). An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol. 15, 4572–4577. doi: 10.1128/MCB.15.8.4572
Fuller, K. K., Loros, J. J., and Dunlap, J. C. (2015). Fungal photobiology: visible light as a signal for stress, space and time. Curr. Genet. 61, 275–288. doi: 10.1007/s00294-014-0451-0
Fuller, K. K., Ringelberg, C. S., Loros, J. J., and Dunlap, J. C. (2013). The fungal pathogen aspergillus fumigatus regulates growth, metabolism, and stress resistance in response to light. mBio 4, e142–e113. doi: 10.1128/mBio.00142-13
Idnurm, A., and Heitman, J. (2005). Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3:e95. doi: 10.1371/journal.pbio.0030095
Janisiewicz, W. J., Takeda, F., Glenn, D. M., Camp, M. J., and Jurick, W. N. (2016). Dark period following UV-C treatment enhances killing of Botrytis cinerea conidia and controls gray mold of strawberries. Phytopathology 106, 386–394. doi: 10.1094/PHYTO-09-15-0240-R
Kim, H., Ridenour, J. B., Dunkle, L. D., and Bluhm, B. H. (2011). Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog 7:e1002113. doi: 10.1371/journal.ppat.1002113
Kim, H., Son, H., and Lee, Y. W. (2014). Effects of light on secondary metabolism and fungal development of Fusarium graminearum. J. Appl. Microbiol. 116, 380–389. doi: 10.1111/jam.12381
Liu, Y., He, Q., and Cheng, P. (2003). Photoreception in Neurospora: a tale of two White Collar proteins. Cell. Mol. Life Sci. 60, 2131–2138. doi: 10.1007/s00018-003-3109-5
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mercier, J., Arul, J., and Julien, C. (1993). Effect of UV-C on phytoalexin accumulation and resistance to Botrytis cinerea in stored carrots. J. Phytopathol. 139, 17–25.
Nigro, F., Ippolito, A., and Lima, G. (1998). Use of UV-C light to reduce Botrytis storage rot of table grapes. Postharvest Biol. Tec. 13, 171–181. doi: 10.1016/S0925-5214(98)00009-X
Pombo, M. A., Rosli, H. G., Martínez, G. A., and Civello, P. M. (2011). UV-C treatment affects the expression and activity of defense genes in strawberry fruit (Fragaria × ananassa. Duch.). Postharvest Biol. Tec. 59, 94–102. doi: 10.1016/j.postharvbio.2010.08.003
Romanazzi, G., Lichter, A., Gabler, F. M., and Smilanick, J. L. (2012). Recent advances on the use of natural and safe alternatives to conventional methods to control postharvest gray mold of table grapes. Postharvest Biol. Tec. 63, 141–147. doi: 10.1016/j.postharvbio.2011.06.013
Romanazzi, G., Smilanick, J. L., Feliziani, E., and Droby, S. (2016). Integrated management of postharvest gray mold on fruit crops. Postharvest Biol. Tec. 113, 69–76. doi: 10.1016/j.postharvbio.2015.11.003
Ruger-Herreros, C., Rodríguez-Romero, J., Fernández-Barranco, R., Olmedo, M., Fischer, R., Corrochano, L. M., et al. (2011). Regulation of conidiation by light in Aspergillus nidulans. Genetics 188, 809–822. doi: 10.1534/genetics.111.130096
Ruiz-Roldan, M. C., Garre, V., Guarro, J., Marine, M., and Roncero, M. I. (2008). Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot. Cell 7, 1227–1230. doi: 10.1128/EC.00072-08
Schumacher, J. (2012). Tools for Botrytis cinerea: new expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet. Biol. 49, 483–497. doi: 10.1016/j.fgb.2012.03.005
Schumacher, J., Simon, A., Cohrs, K. C., Viaud, M., and Tudzynski, P. (2014). The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 10:e1004040. doi: 10.1371/journal.pgen.1004040
Sinha, R. P., and Hader, D. P. (2002). UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 1, 225–236. doi: 10.1039/b201230h
Sperber, W. H., Doyle, M. P., Barth, M., Hankinson, T. R., Zhuang, H., and Breidt, F. (2009). Microbiological Spoilage of Fruits and Vegetables. eds W. H. Sperber, and M. P. Doyle (New York: Springer), 135–183.
Stapleton, A. E. (1992). Ultraviolet radiation and plants: burning questions. Plant Cell 4, 1353–1358. doi: 10.1105/tpc.4.11.1353
Stevens, C., Wilson, C. L., Lu, J. Y., Khan, V. A., Chalutz, E., Droby, S., et al. (1996). Plant hormesis induced by ultraviolet light-C for controlling postharvest diseases of tree fruits. Crop Prot. 15, 129–134. doi: 10.1016/0261-2194(95)00082-8
Syamaladevi, R. M., Lu, X., Sablani, S. S., Insan, S. K., Adhikari, A., Killinger, K., et al. (2013). Inactivation of Escherichia coli population on fruit surfaces using Ultraviolet-C light: influence of fruit surface characteristics. Food Bioprocess Tech. 6, 2959–2973. doi: 10.1007/s11947-012-0989-0
Tagua, V. G., Pausch, M., Eckel, M., Gutierrez, G., Miralles-Duran, A., Sanz, C., et al. (2015). Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 15130–15135. doi: 10.1073/pnas.1514637112
Topcu, Y., Dogan, A., Kasimoglu, Z., Sahin-Nadeem, H., Polat, E., and Erkan, M. (2015). The effects of UV radiation during the vegetative period on antioxidant compounds and postharvest quality of broccoli (Brassica oleracea L.). Plant Physiol. Bioch. 93, 56–65. doi: 10.1016/j.plaphy.2015.02.016
Urban, L., Charles, F., de Miranda, M. R., and Aarrouf, J. (2016). Understanding the physiological effects of UV-C light and exploiting its agronomic potential before and after harvest. Plant Physiol. Biochem. 105, 1–11. doi: 10.1016/j.plaphy.2016.04.004
Verma, S., and Idnurm, A. (2013). The Uve1 endonuclease is regulated by the white collar complex to protect Cryptococcus neoformans from UV damage. PLoS Genet. 9:e1003769. doi: 10.1371/journal.pgen.1003769
Vicente, A. R., Pineda, C., Lemoine, L., Civello, P. M., Martinez, G. A., and Chaves, A. R. (2005). UV-C treatments reduce decay, retain quality and alleviate chilling injury in pepper. Postharvest Biol. Tec. 35, 69–78. doi: 10.1016/j.postharvbio.2004.06.001
Wang, C., Wang, G., Zhang, C., Zhu, P., Dai, H., Yu, N., et al. (2017). OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant 10, 619–633. doi: 10.1016/j.molp.2017.01.006
Watanabe, K., Yamada, N., and Takeuchi, Y. (2006). Oxidative DNA damage in cucumber cotyledons irradiated with ultraviolet light. J. Plant Res. 119, 239–246. doi: 10.1007/s10265-006-0266-2
Wu, C., Yang, F., Smith, K. M., Peterson, M., Dekhang, R., Zhang, Y., et al. (2014). Genome-wide characterization of light-regulated genes in Neurospora crassa. G3 (Bethesda) 4, 1731–1745. doi: 10.1534/g3.114.012617
Keywords: ultraviolet-C, fungal pathogen, photoreceptor, postharvest decay, Botrytis cinerea
Citation: Zhu P, Li Q, Azad SM, Qi Y, Wang Y, Jiang Y and Xu L (2018) Fungal Gene Mutation Analysis Elucidating Photoselective Enhancement of UV-C Disinfection Efficiency Toward Spoilage Agents on Fruit Surface. Front. Microbiol. 9:1141. doi: 10.3389/fmicb.2018.01141
Received: 30 March 2018; Accepted: 14 May 2018;
Published: 12 June 2018.
Edited by:
Boqiang Li, Institute of Botany (CAS), ChinaReviewed by:
Zhanquan Zhang, Chinese Academy of Sciences, ChinaErnesto P. Benito, Universidad de Salamanca, Spain
Copyright © 2018 Zhu, Li, Azad, Qi, Wang, Jiang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pinkuan Zhu, cGt6aHVAYmlvLmVjbnUuZWR1LmNu Ling Xu, bHh1QGJpby5lY251LmVkdS5jbg==
 Pinkuan Zhu
Pinkuan Zhu Qianwen Li
Qianwen Li Yina Jiang
Yina Jiang Ling Xu
Ling Xu