
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 May 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00986
Pseudomonas aeruginosa is a major pathogenic bacterium in chronic infections and is a model organism for studying biofilms. P. aeruginosa is considered an aerobic bacterium, but in the presence of nitrate, it also grows in anaerobic conditions. Oxygen diffusion through the biofilm generates metabolic and genetic diversity in P. aeruginosa growth, such as in ribonucleotide reductase activity. These essential enzymes are necessary for DNA synthesis and repair. Oxygen availability determines the activity of the three-ribonucleotide reductase (RNR) classes. Class II and III RNRs are active in the absence of oxygen; however, class II RNRs, which are important in P. aeruginosa biofilm growth, require a vitamin B12 cofactor for their enzymatic activity. In this work, we elucidated the conditions in which class II RNRs are active due to vitamin B12 concentration constraints (biosynthesis or environmental availability). We demonstrated that increased vitamin B12 levels during aerobic, stationary and biofilm growth activate class II RNR activity. We also established that the cobN gene is essentially responsible for B12 biosynthesis under planktonic and biofilm growth. Our results unravel the mechanisms of dNTP synthesis by P. aeruginosa during biofilm growth, which appear to depend on the bacterial strain (laboratory-type or clinical isolate).
Pseudomonas aeruginosa is an opportunistic pathogen that causes severe chronic infections in immunocompromised patients and other risk groups, such as cystic fibrosis (CF) or chronic obstructive pulmonary disease (COPD) patients. The key to P. aeruginosa survival in environments that range from soil to various living host organisms is its metabolic versatility. It subsists on various carbon sources for energy, uses nitrogen as a terminal electron acceptor under anaerobic conditions, requires minimal nutrients, and grows at temperatures up to 42°C. P. aeruginosa uses anaerobic metabolism to reduce nitrogen (N2) via the denitrification process (Schobert and Jahn, 2010; Arat et al., 2015), as an essential metabolic condition during chronic infection and biofilm growth (Yoon et al., 2006; Hassett et al., 2009; Crespo et al., 2016).
During P. aeruginosa infections, bacteria must multiply inside the infected organisms (plant, animal, insect, etc.), requiring active DNA synthesis for bacterial cell division. Ribonucleotide reductase (RNR) enzymes provide all living organisms with deoxyribonucleotide triphosphates (dNTP) supplying the monomers for DNA synthesis. Three different RNR classes exist (class I, subdivided into Ia, Ib, and Ic; class II and class III) that differ in their overall protein structure and cofactor requirements, but all possess allosteric regulation and use organic radicals to initiate catalysis through free radical chemistry (Jordan and Reichard, 1998; Cotruvo Jr and Stubbe, 2011; Hofer et al., 2012; Torrents, 2014). P. aeruginosa is one of the few organisms that encode three different RNR classes; the oxygen-dependent class Ia (encoded by the nrdAB genes), the oxygen-independent class II (encoded by the nrdJab genes) and the oxygen-sensitive class III (encoded by the nrdDG genes). Specifically, class II RNR activity depends on an external cofactor, adenosylcobalamin (AdoCob) or vitamin B12, to generate its radical independently of oxygen to reduce the different ribonucleotides to their corresponding deoxyribonucleotides.
Vitamin B12 is one of the most structurally complex cofactors synthesized by bacteria (Warren et al., 2002); however, not all microorganisms encode for the ~25 genes needed for the complete biosynthetic pathway. In nature, two vitamin B12 biosynthesis pathways exist: the aerobic, or late cobalt insertion pathway and the anaerobic, or early cobalt insertion pathway (Warren et al., 2002). One of the genes involved in the aerobic pathway that participates in cobalt insertion is the cobN gene described extensively in Pseudomonas denitrificans (Warren et al., 2002). The most studied anaerobic biosynthetic pathway involved in early cobalt insertion was described in Salmonella typhymurium (Roth et al., 1993).
Pseudomonas synthesizes vitamin B12 for different metabolic reactions, such as methionine synthesis, cobalamin biosynthesis, and RNR enzymes. One essential reaction is ribonucleotide synthesis by RNR. P. aeruginosa PAO1 has been demonstrated to grow in a filament cell morphology due to cellular stress by RNR activity depletion, such as the low expression levels of class III RNR under anaerobic conditions (Lee et al., 2012; Crespo et al., 2017) or the high nitric oxide levels in the denitrification process, which interacts with a cobalamin precursor of the vitamin B12 pathway (Broderick et al., 2005; Yoon et al., 2011; Sullivan et al., 2013). Therefore, this cell filamentation results from DNA replication impairment that affects P. aeruginosa PAO1 cell division, thus affecting infection (Sjöberg and Torrents, 2011; Crespo et al., 2017), anaerobic growth (Torrents et al., 2005; Torrents, 2014) and biofilm growth (Crespo et al., 2016). Class II and III RNR enzymes reduce ribonucleotides under these conditions. Thus, their activity is essential for cell division (Crespo et al., 2016, 2017).
Class II RNR (NrdJab) activity is oxygen independent, but it strictly depends on vitamin B12 availability. To date, the link between internal vitamin B12 biosynthesis or availability from the environment and the real class II RNR activity is unknown. Therefore, in this work, we analyzed P. aeruginosa vitamin B12 biosynthesis during aerobic growth, anaerobic growth and biofilm formation. We also determined the relationship between vitamin B12 biosynthesis and class II RNR activity under different growing conditions.
Pseudomonas aeruginosa and Escherichia coli strains, listed in Table 1, were grown in Luria-Bertani broth (LB) or minimum medium (MM) (Kjaergaard et al., 2000) at 37°C. MM containing 1% KNO3 (MMN) was used for anaerobic liquid growth in screw-cap tubes (Hungate tubes) (Garriga et al., 1996; Crespo et al., 2016) or in anaerobic plates using the GENbag system (bioMérieux) according to the manufacturer's instructions.
The medium was supplemented, when necessary, with the following antibiotics: 100 μg/ml or 50 μg/mL gentamicin, 300 μg/ml carbenicillin and 40 μg/ml tetracycline for P. aeruginosa, and 10 μg/ml gentamicin and 50 μg/ml ampicillin for E. coli.
Pseudomonas aeruginosa PAO1 with a mutation in the cobN gene (ETS126; ΔcobN) was constructed by inserting the gentamicin-resistance gene (aacC1) into the cobN gene by homologous recombination using the pEX18Tc vector, as previously described (Quenee et al., 2005; Sjöberg and Torrents, 2011). Briefly, two 400-bp areas surrounding the P. aeruginosa PAO1 cobN gene were amplified by PCR using the High-Fidelity PCR Enzyme Mix (Thermo Scientific) with the primer pairs, CobN1HIII-up/CobN2BI-low and CobN3BI-up/CobN4SI-low, listed in Table 2. The two amplicons were cloned separately into the pJET1.2 vector (Thermo Scientific). A plasmid containing both fragments was generated by BamHI/SacI digestion. The gentamicin resistance gene aacC1 was obtained using BamHI digestion of pUCGmlox, and the corresponding cassette was ligated to the two fragments. The construct was cloned into the sacB gene-based counter-selection pEX18Tc vector and transferred into the S17.1λpir strain for P. aeruginosa PAO1 conjugation as previously described (Crespo et al., 2016). Transconjugants were selected by plating them with tetracycline, gentamicin and sucrose (5%), used for sacB-mediated plasmid counter selection. aacC1 insertion was screened and verified by PCR with the primer pair CobN1HIII-up/CobN5-low and later confirmed by DNA sequencing.
Transcripts of RNR genes (nrdA, nrdJ and nrdD) were quantified using quantitative real-time PCR (qRT-PCR). P. aeruginosa was grown in planktonic conditions at the mid-exponential growth phase in which samples were treated with RNAprotect Bacterial Reagent (Qiagen). The RNeasy Mini Kit (Qiagen) was used to isolate and purify total RNA, and extra DNA was removed using DNase I (Turbo DNA-free, Applied Biosystems) per the manufacturer's instructions. DNA contaminations were verified by PCR. cDNA was synthetized using 0.5 μg of RNA with SuperScript III Reverse Transcriptase (Thermo Scientific). The primers used are listed in Table 2 (Crespo et al., 2016). RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). The gapA gene was used to normalize the transcript gene levels.
GFP fluorescence expressed in plasmids, pETS134 (PnrdA), pETS180 (PnrdJ) and pETS136 (PnrdD), was measured to determine each RNR gene's promoter activity. P. aeruginosa containing the nrd promoter fusion was grown to exponential phase, and three independent 1-ml samples were analyzed. Cells were fixed with 1 ml of freshly prepared PBS 1x solution containing 2% formaldehyde (Sigma) and stored in the dark at 4°C. GFP fluorescence was measured in a 96-well plate (Costar® 96-Well Black Polystyrene Plate, Corning) on an Infinite 200 Pro Fluorescence Microplate Reader (Tecan), as previously described (Crespo et al., 2015). Three measurements were performed per independent sample.
Continuous-flow cell biofilms were grown in MM + 0.2% glucose and performed as previously described (Baelo et al., 2015; Crespo et al., 2016). These in vitro formed biofilms are a more natural, mature biofilm with clear oxygen concentration stratification (Stewart and Franklin, 2008). Briefly, biofilms were grown in a three-channel flow cell with a constant flow rate of 42 μl per minute for each channel using an Ismatec ISM 943 pump (Ismatec). After 5 days of growth, biofilms were stained with the LIVE/DEAD BacLight Bacterial Viability Kit (Thermo Scientific) and visualized using a Zeiss LSM 800 confocal laser scanning microscope (CSLM). Images were generated using ImageJ Fiji software, and COMSTAT 2 software was used to quantify biomass and biofilm thickness (Heydorn et al., 2000).
Pseudomonas aeruginosa PAO1, PAET1, and PA14 strains were grown in MM or MMN medium for 20 h in aerobic or anaerobic conditions or for 5 days in a continuous-flow cell biofilm growth system. Cells were lysed using lysozyme (50 mg/ml) (Sigma) and sonicated five times on ice using a 6-mm sonication probe at 32% power for 20 s (Digital Sonifier, Branson). After centrifugation (4,000 × g at 4°C), the supernatants were filtrated with a 10-kDa Centricon column (Millipore). Samples were manipulated in the dark to avoid vitamin B12 degradation. Finally, 1% ammonium formate was added to each sample before HPLC-MS quantification. Samples of 10 μl were injected into the Luna 5-μm C18 100 Å (150 × 2 mm) column for HPLC-MS [4000 QTRAP (AB SCIEX) in an Aligent 1,200 Series] at the Separation Techniques platform of the Scientific Center Services of the Scientific Park of Barcelona (PCB). A calibration curve was constructed for vitamin B12 (Sigma) measured in the range of 0.1–100 ng/ml. Values were normalized, and the protein concentration was measured using a Bradford assay (Bio-Rad).
Pseudomonas aeruginosa strain cultures were stained using the LIVE/DEAD BacLight viability kit (Thermo Scientific) for 15 min at room temperature in the dark. Fluorescent bacteria were visualized with a Nikon ECLIPSE Ti-S/L 100 inverted fluorescence microscope (Nikon) coupled with a Nikon DS-Qi2 camera. Live cells were visualized in green (SYTO 9 dye), and dead cells were visualized in red (propidium iodide dye). ImageJ software was used for image analysis.
Vitamin B12, or adenosylcobalamin (AdoCob), acts as a radical generator for class II RNR enzyme activity, but the link between vitamin B12 biosynthesis and class II RNR (NrdJ) activity in P. aeruginosa is poorly understood, and further investigations are required, especially during bacterial biofilm growth.
We first analyzed the essentiality and role of the class II RNR enzyme under aerobic and anaerobic conditions depending on vitamin B12 availability. We used diverse P. aeruginosa PAO1 strains deficient for different RNR classes (ETS102, ΔnrdJ; ETS103, ΔnrdD, and ETS125, ΔnrdJΔnrdD) (see Table 1). We also used a specific mutant strain for the vitamin B12 biosynthesis pathway involved in cobalt insertion under aerobic conditions (ETS126, ΔcobN). As the nrdA mutation is unviable (Sjöberg and Torrents, 2011), we added 30 mM hydroxyurea (HU) to mimic an nrdA mutant strain. Hydroxyurea interferes with P. aeruginosa PAO1 growth, arresting DNA replication by inhibiting NrdA activity (Gale et al., 1964; Sjöberg and Torrents, 2011; Lee et al., 2012; Julian et al., 2015).
Aerobically, class Ia RNR inhibition by HU decreased P. aeruginosa PAO1 wild-type growth in minimal medium, as previously described (Jordan et al., 1999; Torrents et al., 2005), but after 48 h of aerobic incubation, some growth was observed (see the undiluted sample 0 with HU in P. aeruginosa wild-type and ΔnrdD) (Figure 1). However, any P. aeruginosa with either a class II RNR or a vitamin B12 biosynthesis gene mutation (ETS102ΔnrdJ, ETS125ΔnrdJΔnrdD, and ETS126ΔcobN) treated with HU showed no growth after 48 h (Figure 1) or even after 72 h (data not shown). This result indicates that after 48 h of HU treatment, class II RNR remains active and allows Pseudomonas growth. Adding vitamin B12 into the minimal medium containing 30 mM HU, re-establishes the optimal aerobic growth in the strains encoding an active class II RNR (NrdJ) enzyme (P. aeruginosa PAO1 wild-type; ETS103, ΔnrdD and ETS126, ΔcobN strains). Therefore, vitamin B12 availability (from biosynthesis or the environment) supports class II RNR activity and rescues class Ia RNR deficiency by HU inhibition under aerobic conditions. Thus, in this work, we demonstrated that a cobN gene mutation disrupted vitamin B12 biosynthesis and completely abolished class II RNR activity, inhibiting aerobic bacterial growth (ETS126, ΔcobN).
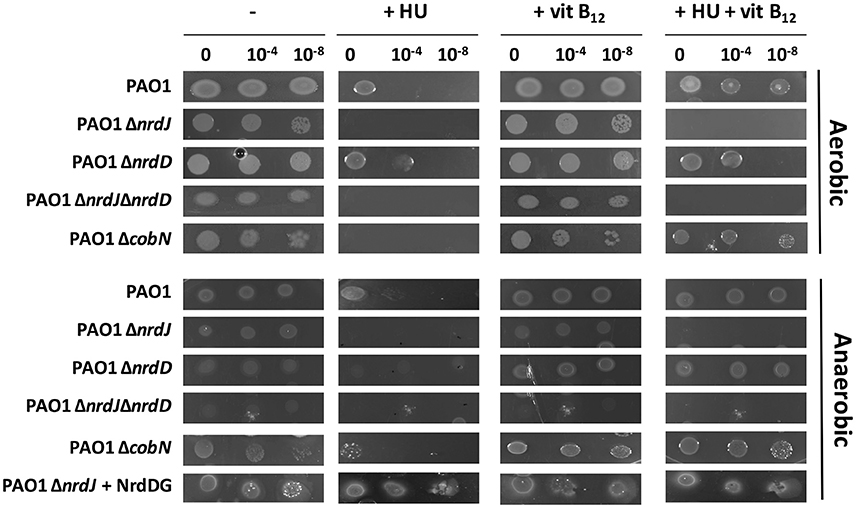
Figure 1. Effect of hydroxyurea and vitamin B12 on P. aeruginosa PAO1 wild-type, ΔnrdJ, ΔnrdD, ΔnrdJΔnrdD, and ΔcobN strain growth. Five-microliter drops were plated from a 0, 10−4, and 10−8 dilution into a solid medium containing 30 mM hydroxyurea (HU) and 1 μg/ml vitamin B12 (vit B12) for 48 h. Pictures represent three independent experiments.
However, under anaerobic conditions, class II and III RNR mutants (ΔnrdJ, ΔnrdD, and ΔnrdJΔnrdD) grew slightly less than the P. aeruginosa PAO1 wild-type and ΔcobN deficient strains (Figure 1). ΔcobN mutant strain growth was unaffected anaerobically (undiluted sample, 0). This result suggests that the cobN gene was uninvolved in P. aeruginosa vitamin B12 biosynthesis under anaerobic conditions. Another Pseudomonas strain, P. denitrificans, was shown to only synthesize vitamin B12 aerobically (Roth et al., 1996). Thus, P. aeruginosa PAO1 cannot sustain proper growth anaerobically unless RNR activity is increased by externally adding vitamin B12 to the medium (1 μg/ml) (this work) or by increasing class III RNR gene copy numbers by complementing extra external NrdDG copies [ETS103 (ΔnrdD)+pETS60 (+NrdDG)] as previously described (Crespo et al., 2017). Nevertheless, the vitamin B12 anaerobic internalization pathway remains unknown, and more experiments are required.
Therefore, we demonstrated that class II RNR (NrdJ) is active in both aerobic and anaerobic conditions if vitamin B12 is available in the medium. However, class Ia and III RNR enzymes preferentially supply the dNTPs required for aerobic (Sjöberg and Torrents, 2011) and anaerobic (Crespo et al., 2017) bacterial DNA replication, respectively. Lack of class Ia and III RNR activity in planktonic culture, due to class Ia RNR activity inhibition by HU or by low nrdD expression levels, causes cell filamentation growth in P. aeruginosa PAO1 (Sjöberg and Torrents, 2011; Crespo et al., 2017), thus increasing its nrd expression (Figure 2). Adding vitamin B12 returns its cellular morphology to rod-shaped by restoring the DNA replication impairment by activating class II RNR (Crespo et al., 2017) and slightly decreasing expression of the three nrd genes (Figure 2), independently of B12-riboswitch regulation (Vitreschak et al., 2003). Other vitamin B12-dependent enzymes (methionine, cobalamin biosynthesis and some ribonucleotide reductases from other microorganisms) are usually regulated by a B12-riboswitch on their promoter regions (Vitreschak et al., 2003; Borovok et al., 2006).
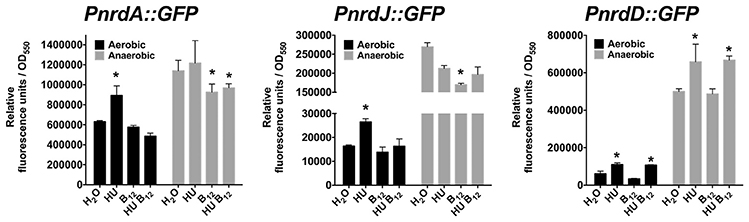
Figure 2. Expression analysis of the different P. aeruginosa PAO1 RNR classes under HU and vitamin B12 treatment under aerobic and anaerobic conditions. Cultures were treated in the presence of HU (30 mM) and vitamin B12 (1 μg/ml) for 20 min prior to measure the relative fluorescence units of PnrdA (pETS134), PnrdJ (pETS180) and PnrdD (pETS136). The results are the mean of three independent experiments ± standard deviation. Asterisks over bars (*) indicate statistical differences compared to those without treatment (H2O) (p < 0.05 in pairwise t-test calculated with GraphPad 6.0).
In addition, P. aeruginosa PAO1 planktonic cells treated with HU for 2 h in minimal medium under aerobic conditions cause filamentous morphology (Figure 3); however, at 24 h post-HU treatment (late stationary phase), P. aeruginosa PAO1 cells return to their rod-shaped morphology without adding external vitamin B12 (Figure 3), indicating that DNA synthesis was restored only by class II RNR activity. Nevertheless, disrupting vitamin B12 biosynthesis using the P. aeruginosa PAO1 ΔcobN mutant strain causes filamentous cells even after 24 h of HU treatment. These results highlight an active vitamin B12 biosynthesis in P. aeruginosa PAO1 that specifically requires the cobN gene under aerobic stationary growing conditions for class II RNR activity and thus for DNA biosynthesis. However, vitamin B12 levels are insufficient during the initial hours of P. aeruginosa PAO1 growth (2 h) and likely reach optimal physiological levels of vitamin B12 after 24 h. Vitamin B12 biosynthesis pathway regulation requires further investigation.
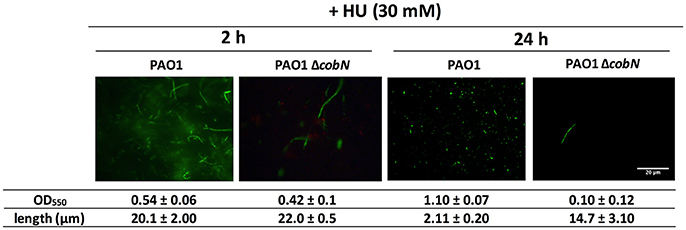
Figure 3. Cell morphology after hydroxyurea treatment. Fluorescence microscopy images from aerobic P. aeruginosa PAO1 wild-type and ΔcobN growth visualized after 2 and 24 h of HU treatment. Cells were stained with the LIVE/DEAD BacLight Bacterial Viability Kit, and the bacterial length was measured using ImageJ software. The images represent at least three different experiments. Scale bars, 20 μm.
Pseudomonas aeruginosa cell morphology under anaerobic conditions was filamentous due to the low class III RNR activity (Crespo et al., 2017). We also observed that the P. aeruginosa PAO1 ΔcobN mutant strain cell morphology was similar to the P. aeruginosa PAO1 strain, suggesting no vitamin B12 biosynthesis during anaerobic growth, even after 16 h (Figure 4). Hence, in anaerobic conditions, the P. aeruginosa PAO1 and ΔcobN strains growth needed external vitamin B12 supplementation for optimal class II RNR activity. This was demonstrated previously in the anaerobic P. aeruginosa PAO1 growth that was restored with an extra copy of nrdDG genes or by adding vitamin B12, enhancing RNR activity (Crespo et al., 2017).
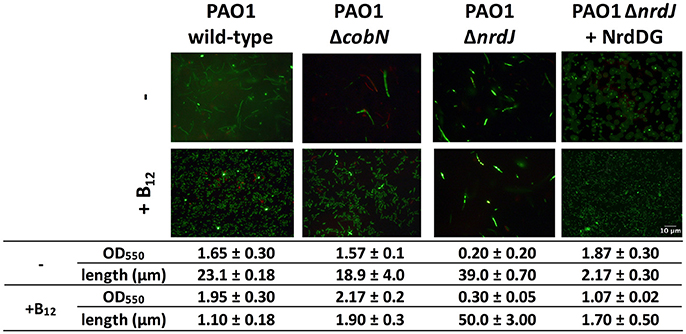
Figure 4. CobN is not involved in P. aeruginosa PAO1 anaerobic growth. Fluorescence microscopy pictures of P. aeruginosa PAO1 wild-type, ΔcobN, ΔnrdJ, and ΔnrdJ + NrdDG cells stained with the LIVE/DEAD BacLight Bacterial Viability Kit at 16 h of anaerobic growth in MM ± vitamin B12. The length was measured using ImageJ software. Scale bars, 10 μm.
Class II and III RNR enzymes are necessary for biofilm formation when class II RNR is highly expressed (Crespo et al., 2016). Currently, it is unknown whether vitamin B12 is synthesized and influences class II RNR activity under biofilm conditions. Thus, we analyzed different P. aeruginosa strains (wild-type and isogenic mutant strains for nrdJ, nrdD and cobN genes) grown in a continuous-flow cell biofilm system. Figure 5A shows that aerobic biofilm formation in minimal media, measured as total biofilm biomass and average thickness, decreased when class II and III RNR were mutated. We previously reported a similar result for biofilm cells grown in LB-rich media (Crespo et al., 2016). The P. aeruginosa ΔcobN mutant strain (vitamin B12 deficient) decreased in biofilm formation compared to the P. aeruginosa PAO1 strain, similar to the produced levels in any P. aeruginosa deficient for class II RNR (ΔnrdJ and ΔnrdJΔnrdD) (Figure 5A). Furthermore, the biomass and thickness levels in the P. aeruginosa PAO1 ΔnrdJ and ΔnrdJΔnrdD mutant strains did not reach the wild-type strain levels even when vitamin B12 was added. However, the P. aeruginosa PAO1 ΔnrdD and ΔcobN mutant strain biofilm thickness increased considerably when vitamin B12 was added, indicating active class II RNR activity.
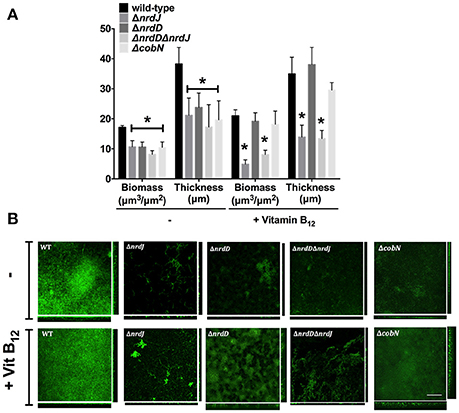
Figure 5. Vitamin B12 enables P. aeruginosa biofilm formation though class II RNR activity. (A) The continuous-flow cell biofilm of P. aeruginosa PAO1 wild-type, ΔnrdJ, ΔnrdD, ΔnrdJΔnrdD, and ΔcobN strains was grown for 5 days in MM ± vitamin B12 (1 μg/ml). Biomass and thickness were calculated using COMSTAT software. Data are the average of three independent experiments. Asterisks over bars (*) indicate statistical differences (p < 0.005 in pairwise t-test calculated with GraphPad 6.0). (B) Pictures of the sum and the orthogonal views of biofilms stained with the LIVE/DEAD BacLight Bacterial Viability Kit before visualization by a confocal microscope. Images represent three independent experiments. Bars represent 200 μm.
These results suggest an active vitamin B12 biosynthesis in the P. aeruginosa biofilm growth via the cobN gene. Moreover, supplying vitamin B12 enabled optimum P. aeruginosa PAO1 biofilm growth in the biofilm layers without active vitamin B12 biosynthesis due to oxygen concentration strengths, activating class II RNR. As expected, cell filamentation morphology, in the ΔnrdJ mutants, was restored by adding vitamin B12 to the continuous-flow biofilm (Figure 5B).
We described that P. aeruginosa needs vitamin B12 availability during planktonic and biofilm growth, essential for class II RNR enzymatic activity. Thus, we elucidated for the first time the amount of vitamin B12 available for P. aeruginosa growth under different conditions (planktonic aerobic or anaerobic and biofilm) in different P. aeruginosa strains.
Quantifying vitamin B12 by HPLC-MS showed this molecule only in cells that were grown aerobically and in the stationary phase (Table 3). However, at exponential growth and in anaerobic conditions, vitamin B12 was detected below the technique detection limit, corroborating previous results under these conditions. Surprisingly, under 5-day-old continuous-flow biofilm P. aeruginosa PAO1 growth, cells produced a 10-fold increase in vitamin B12 compared to aerobic growth, indicating this biosynthetic pathway is activated under this circumstance. We suggested that vitamin B12 biosynthesis in biofilm is only produced in the upper-aerobic biofilm layer because we detected no vitamin B12 levels in cells grown anaerobically (Table 3). Some studies suggested that vitamin B12 (cob) aerobic synthesis genes are expressed more during biofilm growth (Anderson et al., 2008) in the mucoid phenotype (Rao et al., 2008) and the stationary phase (Fung et al., 2010), with downregulated anoxic conditions (Alvarez-Ortega and Harwood, 2007).
Previous studies showed different RNR activity in other P. aeruginosa strains under aerobic and anaerobic conditions (Crespo et al., 2017). Therefore, we analyzed vitamin B12 levels in strains more recently isolated, such as the P. aeruginosa PA14 and PAET1 strains, and we observed different vitamin B12 levels between strains. In P. aeruginosa PA14 and PAET1 strains, we identified increased vitamin B12 levels under aerobic conditions (1.6 and 2.1 times, respectively) (Table 3) and lower vitamin B12 levels under biofilm growth conditions compared to the P. aeruginosa PAO1 strain. These different vitamin B12 levels may affect RNR activity and expression (Figure 2), but further experiments are required to validate this hypothesis. It may be due to an active class III RNR detected in the most recently isolated strains compared to the P. aeruginosa PAO1 strain (Crespo et al., 2017).
Increased vitamin B12 availability in the aerobic growth of P. aeruginosa PA14 and the clinical isolate PAET1 strains suggests higher class II RNR activity under this growing condition. We evaluated strain growth in cells with class Ia RNR inhibited by adding 30 mM HU, which were only growing with an active class II RNR. The results showed that any P. aeruginosa strain could grow when vitamin B12 was added (Figure 6A). In contrast to P. aeruginosa PAO1, the absence of external vitamin B12 in P. aeruginosa PA14 and the clinical isolate PAET1 strain did not affect their growth aerobically. Therefore, vitamin B12 is more available in these strains than in the P. aeruginosa PAO1 strain for class II RNR activity.
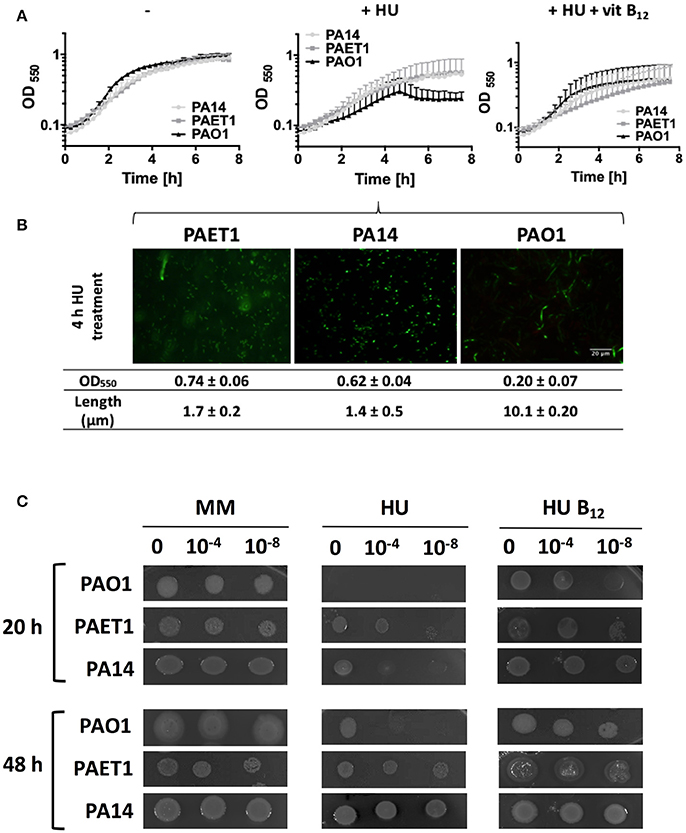
Figure 6. P. aeruginosa PAET1 and PA14 strains are resistant to HU. (A) Growth curve of P. aeruginosa PAO1, PAET1 and PA14 strains in MM containing 30 mM hydroxyurea (HU) and 1 μg/ml vitamin B12. (B) Cells were visualized 4 h after HU treatment. Images and values are representative of three independent experiments ± standard deviation. Bars represent 20 μm. (C) Aerobic P. aeruginosa PAO1, PAET1 and PA14 strain cell viability with HU and vitamin B12 at 20 or 48 h of incubation. Pictures represent three independent experiments. Cultures were diluted to 0, −4, and −8 for the growth.
Additionally, the cell morphology shown in the P. aeruginosa PAO1 strain after 4 h of HU treatment was filamentous (~10 μm). However, HU treatment of P. aeruginosa PA14 and PAET1 yielded rod-shaped cell morphology (1.4 and 1.7 μm), suggesting that their DNA replication was unimpaired (Figure 6B) after 4 h of treatment. However, nrd gene expression in P. aeruginosa clinical isolates strains was increased, suggesting that HU inhibited class I RNR as in the P. aeruginosa PAO1 strain (Table 4). This result was corroborated by analyzing their cell viability in a solid medium under HU treatment with and without vitamin B12. P. aeruginosa PA14 and PAET1 strains grew in as little as 20 h in the presence of HU (Figure 6C) compared with 48 h for P. aeruginosa PAO1.
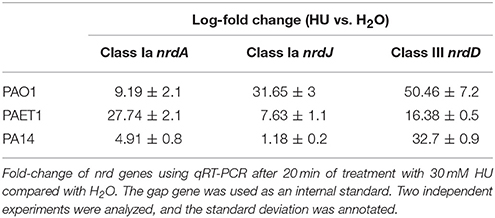
Table 4. Expression of nrdA, nrdJ, and nrdD genes of P. aeruginosa PAO1 and clinical isolates (PAET1 and PA14) strains with HU.
We demonstrated that vitamin B12 synthesis occurs under P. aeruginosa aerobic planktonic growth conditions with an active class Ia RNR that supplies dNTPs required for DNA replication (Figure 7A). Vitamin B12 cannot be synthesized anaerobically when P. aeruginosa cells are grown with class III RNR (Figure 7A; Crespo et al., 2017). Class II RNR is enzymatically active when vitamin B12 is available through internal biosynthesis or from the environment.
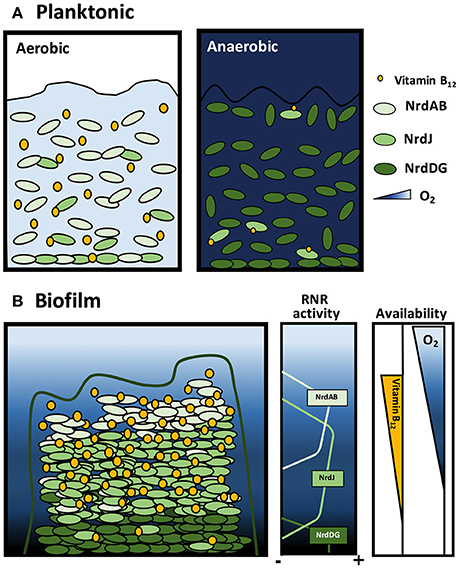
Figure 7. Model of ribonucleotide reductase activity and vitamin B12 availability during P. aeruginosa planktonic and biofilm growth. Scheme of P. aeruginosa growing in (A) aerobic and anaerobic planktonic cultures and (B) biofilm. Orange circles represent the vitamin B12 gradient, blue indicates oxygen concentration gradients and green indicates class Ia, II and III RNR activity.
Pseudomonas aeruginosa growing in biofilm differs and requires a more in-depth analysis. Oxygen diffusion through the complex biofilm structure generates an oxygen concentration gradient with apparent cell distribution with different RNR class activity (Figure 7B; Crespo et al., 2016). We suggest that in the superficial biofilm layers, aerobic cells express class Ia RNR, whereas in the internal layers, anaerobic conditions require cells to express class II or III RNR (Crespo et al., 2016). However, class II RNR is highly expressed during biofilm formation and in aerobic environments (Sjöberg and Torrents, 2011; Crespo et al., 2016) but is oxygen-independent and vitamin B12-dependent (aerobically synthetized). This leads us to ask under which conditions this RNR class enzymatically is active.
We suggest that external cells in a biofilm, which are in contact with aerobic environments, can synthesize vitamin B12, and it can diffuse through the biofilm structure creating a vitamin B12 concentration gradient along this structure. In this sense, class II RNR can be active in areas with microaerophilic conditions where class Ia or class III RNR are inactive (Figure 7B). Consequently, these results bring us closer to understanding the P. aeruginosa cell division mechanism through dNTP synthesis in planktonic and biofilm conditions.
AC, NB-C, and ET: designed the study; AC and NB-C: performed the experiments. All authors analyzed the data, wrote the paper, and read and approved the final version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported in part by grants to ET from the Spanish Ministerio de Economia y Competitividad (MINECO/FEDER) (BIO2015-63557-R), Generalitat de Catalunya (2014 SGR01260 and CERCA programme), the EIT Health Program, the Catalan Fibrosis Association and La Caixa Foundation.
Alvarez-Ortega, C., and Harwood, C. S. (2007). Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165. doi: 10.1111/j.1365-2958.2007.05772.x
Anderson, G. G., Moreau-Marquis, S., Stanton, B. A., and O'toole, G. A. (2008). In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76, 1423–1433. doi: 10.1128/IAI.01373-07
Arat, S., Bullerjahn, G. S., and Laubenbacher, R. (2015). A network biology approach to denitrification in Pseudomonas aeruginosa. PLoS ONE 10:e0118235. doi: 10.1371/journal.pone.0118235
Baelo, A., Levato, R., Julian, E., Crespo, A., Astola, J., Gavalda, J., et al. (2015). Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Control. Release 209, 150–158. doi: 10.1016/j.jconrel.2015.04.028
Borovok, I., Gorovitz, B., Schreiber, R., Aharonowitz, Y., and Cohen, G. (2006). Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J. Bacteriol. 188, 2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006
Broderick, K. E., Singh, V., Zhuang, S., Kambo, A., Chen, J. C., Sharma, V. S., et al. (2005). Nitric oxide scavenging by the cobalamin precursor cobinamide. J. Biol. Chem. 280, 8678–8685. doi: 10.1074/jbc.M410498200
Cotruvo, J. A. Jr, and Stubbe, J. (2011). Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767. doi: 10.1146/annurev-biochem-061408-095817
Crespo, A., Gavalda, J., Julian, E., and Torrents, E. (2017). A single point mutation in class III ribonucleotide reductase promoter renders Pseudomonas aeruginosa PAO1 inefficient for anaerobic growth and infection. Sci. Rep. 7:13350. doi: 10.1038/s41598-017-14051-2
Crespo, A., Pedraz, L., Astola, J., and Torrents, E. (2016). Pseudomonas aeruginosa exhibits deficient biofilm formation in the absence of Class, I. i., and iii ribonucleotide reductases due to hindered anaerobic growth. Front. Microbiol. 7:688. doi: 10.3389/fmicb.2016.00688
Crespo, A., Pedraz, L., and Torrents, E. (2015). Function of the Pseudomonas aeruginosa NrdR transcription factor: global transcriptomic analysis and its role on ribonucleotide reductase gene expression. PLoS ONE 10:e0123571. doi: 10.1371/journal.pone.0123571
de Lorenzo, V., Cases, I., Herrero, M., and Timmis, K. N. (1993). Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 175, 6902–6907. doi: 10.1128/jb.175.21.6902-6907.1993
Fung, C., Naughton, S., Turnbull, L., Tingpej, P., Rose, B., Arthur, J., et al. (2010). Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 59, 1089–1100. doi: 10.1099/jmm.0.019984-0
Gale, G. R., Kendall, S. M., Mclain, H. H., and Dubois, S. (1964). Effect of Hydroxyurea on Pseudomonas aeruginosa. Cancer Res. 24, 1012–1020.
Garriga, X., Eliasson, R., Torrents, E., Jordan, A., Barbe, J., Gibert, I., et al. (1996). nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229, 189–192. doi: 10.1006/bbrc.1996.1778
Hassett, D. J., Sutton, M. D., Schurr, M. J., Herr, A. B., Caldwell, C. C., and Matu, J. O. (2009). Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 17, 130–138. doi: 10.1016/j.tim.2008.12.003
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146 (Pt 10), 2395–2407. doi: 10.1099/00221287-146-10-2395
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hofer, A., Crona, M., Logan, D. T., and Sjöberg, B. M. (2012). DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63. doi: 10.3109/10409238.2011.630372
Jordan, A., and Reichard, P. (1998). Ribonucleotide reductases. Annu. Rev. Biochem. 67, 71–98. doi: 10.1146/annurev.biochem.67.1.71
Jordan, A., Torrents, E., Sala, I., Hellman, U., Gibert, I., and Reichard, P. (1999). Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181, 3974–3980.
Julian, E., Baelo, A., Gavalda, J., and Torrents, E. (2015). Methyl-hydroxylamine as an efficacious antibacterial agent that targets the ribonucleotide reductase enzyme. PLoS ONE 10:e0122049. doi: 10.1371/journal.pone.0122049
Kjaergaard, K., Schembri, M. A., Ramos, C., Molin, S., and Klemm, P. (2000). Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2, 695–702. doi: 10.1046/j.1462-2920.2000.00152.x
Lee, K. M., Go, J., Yoon, M. Y., Park, Y., Kim, S. C., Yong, D. E., et al. (2012). Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect. Immun. 80, 1639–1649. doi: 10.1128/IAI.06161-11
Quenee, L., Lamotte, D., and Polack, B. (2005). Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. BioTechniques 38, 63–67. doi: 10.2144/05381ST01
Rao, J., Digiandomenico, A., Unger, J., Bao, Y., Polanowska-Grabowska, R. K., and Goldberg, J. B. (2008). A novel oxidized low-density lipoprotein-binding protein from Pseudomonas aeruginosa. Microbiology 154, 654–665. doi: 10.1099/mic.0.2007/011429-0
Roth, J. R., Lawrence, J. G., and Bobik, T. A. (1996). Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181. doi: 10.1146/annurev.micro.50.1.137
Roth, J. R., Lawrence, J. G., Rubenfield, M., Kieffer-Higgins, S., and Church, G. M. (1993). Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175, 3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993
Schobert, M., and Jahn, D. (2010). Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 549–556. doi: 10.1016/j.ijmm.2010.08.007
Sjöberg, B. M., and Torrents, E. (2011). Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during infection. Infect. Immun. 79, 2663–2669. doi: 10.1128/IAI.01212-10
Stewart, P. S., and Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Sullivan, M. J., Gates, A. J., Appia-Ayme, C., Rowley, G., and Richardson, D. J. (2013). Copper control of bacterial nitrous oxide emission and its impact on vitamin B12-dependent metabolism. Proc. Natl. Acad. Sci. U.S.A. 110, 19926–19931. doi: 10.1073/pnas.1314529110
Torrents, E. (2014). Ribonucleotide reductases: essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 4:52. doi: 10.3389/fcimb.2014.00052
Torrents, E., Poplawski, A., and Sjöberg, B. M. (2005). Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: expression and transcriptional analysis of the aerobic enzymes. J. Biol. Chem. 280, 16571–16578. doi: 10.1074/jbc.M501322200
Vitreschak, A. G., Rodionov, D. A., Mironov, A. A., and Gelfand, M. S. (2003). Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9, 1084–1097. doi: 10.1261/rna.5710303
Warren, M. J., Raux, E., Schubert, H. L., and Escalante-Semerena, J. C. (2002). The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19, 390–412. doi: 10.1039/b108967f
Yoon, M. Y., Lee, K. M., Park, Y., and Yoon, S. S. (2011). Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 6:e16105. doi: 10.1371/journal.pone.0016105
Keywords: Vitamin B12, adenosylcobalamin, ribonucleotide reductases, Pseudomonas aeruginosa, NrdJ, bacterial growth, biofilm, anaerobiosis
Citation: Crespo A, Blanco-Cabra N and Torrents E (2018) Aerobic Vitamin B12 Biosynthesis Is Essential for Pseudomonas aeruginosa Class II Ribonucleotide Reductase Activity During Planktonic and Biofilm Growth. Front. Microbiol. 9:986. doi: 10.3389/fmicb.2018.00986
Received: 09 January 2018; Accepted: 26 April 2018;
Published: 15 May 2018.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
Paraskevi Papaioannidou, Aristotle University of Thessaloniki, GreeceCopyright © 2018 Crespo, Blanco-Cabra and Torrents. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduard Torrents, ZXRvcnJlbnRzQGliZWNiYXJjZWxvbmEuZXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.