
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 May 2018
Sec. Microbial Symbioses
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00924
This article is part of the Research Topic Advances and New Perspectives in Beneficial Plant-Microbial Interactions View all 23 articles
 Osama A. A. Mohamad1,2*†
Osama A. A. Mohamad1,2*† Li Li1,3†
Li Li1,3† Jin-Biao Ma1
Jin-Biao Ma1 Shaimaa Hatab4
Shaimaa Hatab4 Lin Xu5
Lin Xu5 Jian-Wei Guo1,6
Jian-Wei Guo1,6 Bakhtiyor A. Rasulov1,7
Bakhtiyor A. Rasulov1,7 Yong-Hong Liu1
Yong-Hong Liu1 Brian P. Hedlund3
Brian P. Hedlund3 Wen-Jun Li1,8*
Wen-Jun Li1,8*Endophytic bacteria associated with medicinal plants possess unique strategies that enhance growth and suvival of host plants, many of which are mediated by distinctive secondary metabolites. These bacteria and their secondary metabolites are important subjects for both basic and applied research aimed at sustainable agriculture. In the present study, 114 endophytic strains isolated from the wild ethnomedicinal plant Glycyrrhiza uralensis (licorice) were screened for their in vitro antimicrobial activities against common fungal pathogens of tomato (Fusarium oxysporum f. sp., Fulvia fulva, Alternaria solani), cotton (Fusarium oxysporum f. sp. Vesinfectum, Verticillium dahliae), pomegranite (Ceratocystis fimbriata), Cymbidinium (Colletotrichum gloeosporioides), and Tsao-ko (Pestalotiopsis microspora and Fusarium graminearum) and the common bacteria Staphylococcus aureus, Bacillus cereus, Salmonella enteritidis, and Escherichia coli. Several Bacillus strains, particularly Bacillus atrophaeus and Bacillus mojavensis, had a broad spectrum of antifungal and antibacterial activity. A total of 16 strains, selected based on broad antimicrobial activity, were shown to contain at least one putative secondary metabolite-encoding gene (i.e., polyketide synthase or non-ribosomal peptide synthetase) and/or one lytic enzyme (i.e., protease, cellulase, lipase, chitinase), which may be important mediators of antagonistic activity against pathogens. Five strains, representing Bacillus atrophaeus and Bacillus mojavensis, were selected for plant growth chamber experiments based on strong in vitro antifungal activities. All five strains significantly reduced disease severity in Arabidopsis thaliana plants challenged with V. dahlia infection. Gas-chromatography/mass-spectrometry analysis of cell-free extracts of Bacillus atrophaeus strain XEGI50 showed that at least 13 compounds were produced only during co-cultivation with V. dahlia, including putative compounds known to have antimicrobial activity, such as 1,2-benzenedicarboxylic acid, bis (2-methylpropyl) ester; 9,12-octadecadienoic acid (Z,Z)-, methyl ester; 9-octadecenoic acid, methyl ester, (E)-; and decanedioic acid, bis(2-ethylhexyl) ester. To our knowledge, this study is the first to report that bacteria isolated from G. uralensis have biocontrol abilities. Our findings provide new insights into the antimicrobial activities of natural endophytes, particularly B. atrophaeus, and suggest this species may a promising candidate as a biocontrol agent to confer resistance to Verticillium wilt disease and other phytopathogens in cotton and other crops.
Fungal disease is the main threat to both crop yields and global food security (Fisher et al., 2012). Vascular wilts are devastating plant diseases that cause major losses to food crops and destroy natural ecosystems (Yadeta and Thomma, 2013). Two major genera of pathogenic fungi, Fusarium and Verticillium, enter their host plants through the roots or are transmitted by beetles and cause vascular wilts. Both are characterized by a wide host range (Juzwik et al., 2008; Harwood et al., 2011). The signs of Verticillium wilts are similar to those of Fusarium, starting with yellowing of the older leaves, followed by chlorosis and necrosis. As a result, vascular discoloration and stunting may be visible (Ting, 2014). In China, approximately 3 million hectares of cotton crops are affected by Verticillium wilt, accounting for an annual loss of yield of 10–30% (Bibi et al., 2013). In addition, no fungicides are registered for controlling Verticillium wilt disease in cotton (Göre et al., 2009). Xinjiang Province, located in the northwest of China, produces 11% of the global cotton fiber yield and is disproportionately harmed by Verticillium wilt disease (Peng et al., 2016).
Controlling vascular wilt pathogens can be challenging due to the fact that there are no efficient approaches to treat infected plants. Chemical fungicides are costly, inefficient, and have adverse impacts on the environment and human health (Qing et al., 2015). Increasing concern about the environmental and human health impacts of traditional fungicides has spawned intense interest in the development of safer alternatives. Biological control, an eco-friendly alternative, is mediated by microbial antagonists that possess unique traits that enable them to inhibit the growth of fungal pathogens (Walker et al., 1998; Chernin and Chet, 2002; Berg and Hallmann, 2006). Several mechanisms are responsible for these antagonistic activities, including inhibition of pathogen growth via antibiotics, toxins, surface-active compounds (antibiosis), and extracellular digestive enzymes such as proteases, cellulases and chitinases (Chernin and Chet, 2002; de Souza et al., 2003). However, several studies of biocontrol agents have reported dissimilarities between the antagonistic effects in vitro and the corresponding in situ efficacy (Berg et al., 2000). Therefore, additional research is needed to better understand the basis by which plant-associated bacteria suppress fungal diseases in vivo (Santhanam et al., 2014), not only for basic science purposes, but also for the development of improved biocontrol approaches to support sustainable eco-friendly crop production (Weller, 1988; Sturz et al., 2000).
Endophytic bacteria have the capability to systematically colonize plant tissues and establish a symbiotic relationship with the host, which makes them highly efficient biocontrol agents (Bakker et al., 2013). Several reports have investigated bacterial endophytes as possible biocontrol agents against diverse pathogenic fungi (Lacava et al., 2007; Erdogan and Benlioglu, 2010; Egamberdieva et al., 2017a,b). Recently, several studies have described bacterial endophytes with biological control activity on a number of crops as potential sources of antimicrobial metabolites. The host plants in these studies have included Solanum trilobatum (Bhuvaneswari et al., 2013), Nicotiana attenuata (Santhanam et al., 2014), Solanum trilobatum melongena, and Solanum torvum (Achari and Ramesh, 2014).
In view of the importance of endophytes to plant health, and increased focus on traditional herbal remedies as alternatives to synthetic pharmaseuticals, recent studies have begun to probe the importance of endophytic bacteria to medicinal plants, particularly those growing in unusual or stressed environments (Strobel and Daisy, 2003; Vieira et al., 2011; Liu Y.-H. et al., 2016; Egamberdieva et al., 2017a; Liu et al., 2017). Many of these studies have fingered the genus Bacillus, due to its widespread abundance in different plants, its broad-spectrum antimicrobial activities (Zheng et al., 2013; Jiang et al., 2015; Gao et al., 2017), and its ability to form endospores that are highly resistant to abiotic stresses such as UV light, desiccation, and extremes of pH, salinity, and temperature (Horikoshi, 2008).
Bacteria associated with medicinal plants have rarely been explored with regard to antagonistic activity against plant pathogens (Bakker et al., 2013; Bhuvaneswari et al., 2013; Egamberdieva et al., 2017a). Licorice (Glycyrrhiza uralensis) is a popular traditional Chinese medicine. Licorice contains bioactive compounds such as phenolics, flavonoids, triterpene saponins, and coumarins (Zhang and Ye, 2009). According to Chinese medicine theory, licorice has many important pharmacological activities, including antimicrobial and antiviral activity, histamine inhibition, anti-inflammatory activity, detoxification, and antioxidant and antitumor activities (Liao et al., 2012). Despite the economic interest and broad pharmacological effects of the popular medicinal usage of G. uralensis, very little research has been conducted on its endophytes or the potential for its endophytes to be used as biocontrol agents (Asl and Hosseinzadeh, 2008). Therefore, the objectives of the present study were: (1) to screen a diverse collection of endophytic bacteria (18 genera and 34 species) isolated from wild populations of G. uralensis for activity against a variety of pathogens in vitro; (2) to evaluate selected isolates for their biological control efficiency against the vascular wilt pathogen V. dahliae in vivo; and (3) to identify prevalent volatile organic compounds (VOCs) produced by endophytes only in the presence of V. dahliae, which are likely to be among the effectors of the antimicrobial properties.
A collection of 114 endophytes were previously isolated from wild populations of G. uralensis (Li et al., 2018) from three areas in Xinjiang province, representing 18 genera and 34 species. All isolates were submitted to NCBI GenBank under Accession Number (KY127308 – KY127422) and used in this study. Bacterial isolates were stored in 20% glycerol at the Key Laboratory of Biogeography and Bioresource in Arid Land, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences under -20°C. Bacteria were routinely cultured on ISP2 growth medium, and incubated at 28 ± 2°C for 48–72 h.
A modification of the agar disk diffusion method for detecting antagonism was used against the four common bacteria listed in Table 1 (Nie et al., 2012). The common bacteria and endophytes were each pre-cultured overnight, and 5 mL-1 of each culture was centrifuged at 604 × g for 5 min. The pellets were resuspended in sterile phosphate buffered saline (PBS) in a laminar air flow cabinet and density adjusted to 108 colony forming units (CFU) mL-1 by using Densicheck plus (BioMérieux, United States). A total of 200 μL of the common bacteria cell concentrate was inoculated and evenly spread by sterile cotton swaps onto the surface of the medium, and then four 5-mm-diameter pieces of sterile filter paper were placed on each corner of the petri dish. A total of 10 μL of each endophyte strain was then added dropwise to the filter paper. All plates were wrapped with parafilm, incubated at 37 ± 2°C for 24 h and observed for the inhibition of the common bacteria (Cho et al., 2007). Antibacterial activity was assessed by measuring the diameter of the clear zone of growth inhibition. An equivalent volume of sterile phosphate buffered saline (PBS) instead of the endophytic bacteria was used as a negative control.
The antifungal activity of each bacterial endophyte was screened for antagonistism against the pathogenic fungi in Table 2 by the plate diffusion method (Sun et al., 2017). Bacterial strains were grown in ISP2 medium at 28°C overnight. The fungal pathogens were grown on potato dextrose agar (PDA) plates. A 5-mm agar plug containing 6-day-old mycelial growth was placed at the center of a 9-cm modified culture PDA plate, which favors for the growth of both endophytes and the pathogen. A concentrate of each endophyte was placed onto the agar surface at 8 equidistant points, 2.5 cm from the plate periphery (Loqman et al., 2009). All plates were wrapped with parafilm and incubated at 28 ± 2°C for 3–5 days and observed for the inhibition of the pathogen. Plates with pathogenic fungi alone served as a control. The percentage of growth inhibition was calculated by measuring the diameter of the inhibition zone by using the following formula:
where Fcd is the fungal colony diameter on the control PDA base plate, Tcd is the fungal colony diameter on the experimental PDA base plate, and F0 is the diameter of the test fungus agar disks (5 mm) (Aeron et al., 2011). Each experiment was performed with three replicates, and the analysis was repeated to ensure consistency of the results.
A total of 16 strains were used for screening for natural product biosynthetic gene clusters by PCR. Three sets of degenerate primers targeting biosynthetic genes were used for PCR amplification: KSF (5′-GTSCCSGTSSCRTGSSHYTCSA-3′) and KSR (5′-CGCTCCATGGAYCCSCARCA-3′), targeting polyketide synthase (PKS)-I KS and methyl malonyl transferase domains (Kun et al., 2010); KSαF (5′-TSGCSTGCTTGGAYGCSATC-3′) and KSαR (5′-TGGAANCCGCCGAABCCGCT-3′), targeting PKS-II KSα genes (Metsä-Ketelä et al., 1999); and A3F (5′-GCSTACSYSATSTACACSTCSGG-3′) and A7R (5′-SASGTCVCCSGTSCGGTAS-3′), targeting non-ribosomal peptide synthetase (NRPS) genes. The reactions were performed in a (BIO-RAD C1000 Thermal Cycler) in a total volume 25 μl consisting of 50 ng of genomic DNA, 10 pmol of each primer, 2.5 mM of each deoxynucleotide triphosphates, 1X PCR buffer, and 1 U of Taq DNA polymerase. Gradient PCR was performed under the following conditions: initial denaturation step at 95°C for 5 min, followed by 32 cycles of denaturation at 96°C for 1 min, annealing at 56, 62.1, and 52.5°C for PKS-I, PKS-II, and NRPS genes, respectively, for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. The amplified PCR products were analyzed by electrophoresis on 1% agarose gels with TAE buffer. A negative control without DNA template was included with each PCR.
A total of 16 isolates were used for screening their ability to produce digestive enzymes. Cellulase activity was assayed with modified DSMZ1 medium 65 without CaCO3 and supplemented with carboxymethyl cellulose (5 g L-1; Sigma-Aldrich) in place of glucose. After incubation for 3–4 days at 28°C, plates were stained with a Congo red solution and destained with a NaCl solution (Li et al., 2018). A clear or lightly colored halo around the colonies indicated a positive reaction. Protease activity was assayed with YEM agar medium containing 5% (v/v) skim milk. After incubation for 3–4 days at 28°C, a clear halo around the bacterial colonies due to hydrolysis of milk indicated a positive reaction. Lipase enzyme activity was assayed with modified Sierra lipolysis agar supplemented with beef extract (3 g L-1) and ferrous citrate (0.2 g L-1). After autoclaving, 50 mL of Victoria Blue B solution (0.1 g 150 mL-1) and 10 mL of Tween-80 was added to the medium. After 5–6 days incubation at 28°C, white calcium precipitates around the bacterial colonies indicated a positive reaction (Li et al., 2018).
Colloidal chitin was prepared from commercial chitin by the method of Agrawal and Kotasthane, 2012. Chitin was hydrolyzed in concentrated HCl by stirring at 4°C overnight, followed by extraction of colloidal chitin in 200 mL of ice-cold 99% ethanol, neutralization at room temperature overnight, and centrifugation at 1677 × g for 10 min at 4°C. The pellet was washed with sterile distilled water by centrifugation at 2415 × g for 5 min at 4°C till the smell of alcohol was completely removed and the pH was 7. The colloidal chitin had a soft pasty consistency with 90% moisture and was stored at 4°C until further use. Chitinase detection medium composed of (L-1) 0.3 g of MgSO4⋅7H2O, 3.0 g of (NH4)2SO4, 2.0 g of KH2PO4, 1.0 g of citric acid monohydrate, 15 g of agar, 200 μL of Tween-80, 4.5 g of colloidal chitin, and 0.15 g of bromocresol purple and then autoclaved at 121°C for 15 min. To test for chitinase activity, inoculated plates were incubated at 25 ± 2°C and were observed for formation of a colored zone. For all the tests mentioned above, sterile nutrient agar was used as a control for bacterial growth. All experiments were performed twice with three replicates for each isolate.
Five strains, XEGI33, XEGI38, XEGI44, XEGI50, and XEGI78, were selected for greenhouse experiments based on antagonistic activity against V. dahliae in vitro and presence of at least one biosynthetic gene and at least one digestive enzyme. Arabidopsis thaliana was used as a model plant and V. dahliae was used as a model pathogen (Maldonado-González et al., 2015). A. thaliana seeds with uniform shape and size were surface-sterilized with 99% ethanol for 0.5 min, and then washed with sterile distilled water 5–6 times. The seeds were placed on MS plates at 25 ± 2°C for 3–5 days. Arabidopsis thaliana seedlings with true stage leaves were then transplanted singly into pots of 8 cm diameter containing 60 g of sterilized soil. After 2 days of transplantation, plants were divided into five groups and each group was given different treatments. V. dahliae was cultivated in advance in Czapek-Dox broth at 28°C for 4 days. After 72 h of transplantation, 10 mL of the fungal mycelia suspension (108 CFU mL-1) was spread on the soil surface above fine roots by using a sterile syringe, and then after another 48 h, 10 mL of an endophytic bacteria suspension (108 CFU ml-1) was added, as generally described by Chen et al., 2014. Two controls were used in this experiment: seedlings treated with sterile water (CK+), and seedlings inoculated with the pathogen alone (CK-). The pots were placed in a growth chamber with the following conditions: 25–30°C, 60% humidity, and 16 h of daylight alternating with 8 h of darkness (Jiang et al., 2015). Three replicates were done for each treatment. Plant phenotypes were observed after 10 days of pathogen inoculation, while the development of wilt disease signs was recorded after 40 days.
A disease index, based on yellowing and chlorosis of cotyledons and leaves after 40 days, was used to classify disease signs for each leaf into six grades (i.e., grade 0, 1, 2, 3, 4, and 5) (Zhang et al., 2012), and then the percentage of affected leaves was calculated and categorized (≤ 25, 25–35, 35–45, 45–55, and 55–80%). The final disease index (DI) was calculated according to the following formula: DI = [(Σ disease grades × number of infected)/(total checked plants × 5)] × 100 (Zhang et al., 2012). The DI represents a comprehensive and objective measure of plant health, with high DI values corresponding to serious infections.
Strain XEGI50 was inoculated into 500 mL-1 of ISP2 broth at 28°C for 10 days with agitation at 120 rpm and used as a control (1). V. dahliae was cultivated in 500 mL-1 in Czapek broth at 28°C for 10 days with agitation at 120 rpm and used as a control (2). The antibiosis experiment was carried out by co-cultivation of strain XEGI50 with V. dahliae in 500 mL-1 of broth medium at 28°C for 12 days with agitation at 120 rpm. All cells were collected by centrifugation at 5000 × g for 10 min. The cell-free supernatant was divided into equal volumes. After that, the supernatant was adjusted to pH 7 and pH 3 with 500 mL-1 of 1 N HCl and an equal volume (1:1) of ethyl acetate was added and mixed by vigorous shaking for 30 min and allowed to settle. The organic solvent phase was collected and evaporated at 40°C under vacuum, using a rotary evaporator model (IKA, HB10 basic). The ethyl acetate extract was dissolved in 5 mL of Tris–Cl buffer (0.02 M, pH 7.0) and used for gas-chromatography/mass-spectrometry (GC-MS).
The GC-MS analysis of the cell-free extracts was performed using a gas chromatograph (Model 7890A, Agilent, Palo Alto, CA, United States) equipped with a split-splitless injector, an Agilent model 7693 autosampler, and an Agilent HP-5MS fused silica column (5% phenyl-methylpolysiloxane, 30 m length, 0.25 mm I.D., film thickness 0.25 mm). Injecting volume was 1 μL, and the GC conditions included programmed heating from 50 to 300°C at 10°C/min, followed by 10 min at 300°C. The injector was maintained at 280°C. Helium was the carrier gas, at 1.0 mL min-1, and the split mode was 5:1. The GC was fitted with a quadrupole mass spectrometer with an Agilent model 5975 detector. The MS conditions were as follows: ionization energy, 70 eV; electronic impact ion source temperature, 230°C; quadrupole temperature, 150°C; scan rate, 3.2 scan/s; mass range, 50–1000 μ. The compounds were identified based on the match with their mass spectra and retention indices with the NIST/Wiley 275 library (Wiley, New York, NY, United States). Relative abundance of each feature was calculated from Total Ion Chromatogram (TIC) computationally.
The morphological response of XEGI50 to V. dahliae was observed under a laser microscope (Olympus SZX2-ILLT, Japan) at different magnifications. Bacteria were incubated in ISP2 medium. V. dahliae was grown on PDA medium. A 6-day-old mycelial disk (5 mm) was placed at the center of a 7 cm modified culture PDA plate. The bacteria were placed at four corners on the bacterial lawn at four equidistant points of 2.5 cm from the plate periphery. All plates were wrapped with parafilm, incubated at 28 ± 2°C for 6 days, and observed for the inhibition of the pathogen. Plates with pathogenic fungi alone served as control.
A total of 114 endophytes isolated from wild populations of G. uralensis, representing 18 genera and 34 species, were screened for their ability to inhibit four common bacteria, representing the Gram-positive phylum Firmicutes and the Gram-negative phylum Proteobacteria (Table 1). Of the 114 isolates examined, 56 (49.1%) displayed inhibitory activity, ranging from 8.6 to 11.8 mm against S. aureus, B. cereus, and S. enteritidis; in contrast, only 14 (12.3%) were antagonistic to E. coli, ranging from 7.5 to 10.3 mm (Supplementary Figures S1A,B). These 14 strains belonged to 6 different genera, namely Bacillus, Microbacterium, Brevibacterium, Phyllobacterium, Pantoea, and Stenotrophomonas. The genus Bacillus showed the highest antimicrobial activity against the selected Gram-positive and Gram-negative bacteria (Figure 1A).
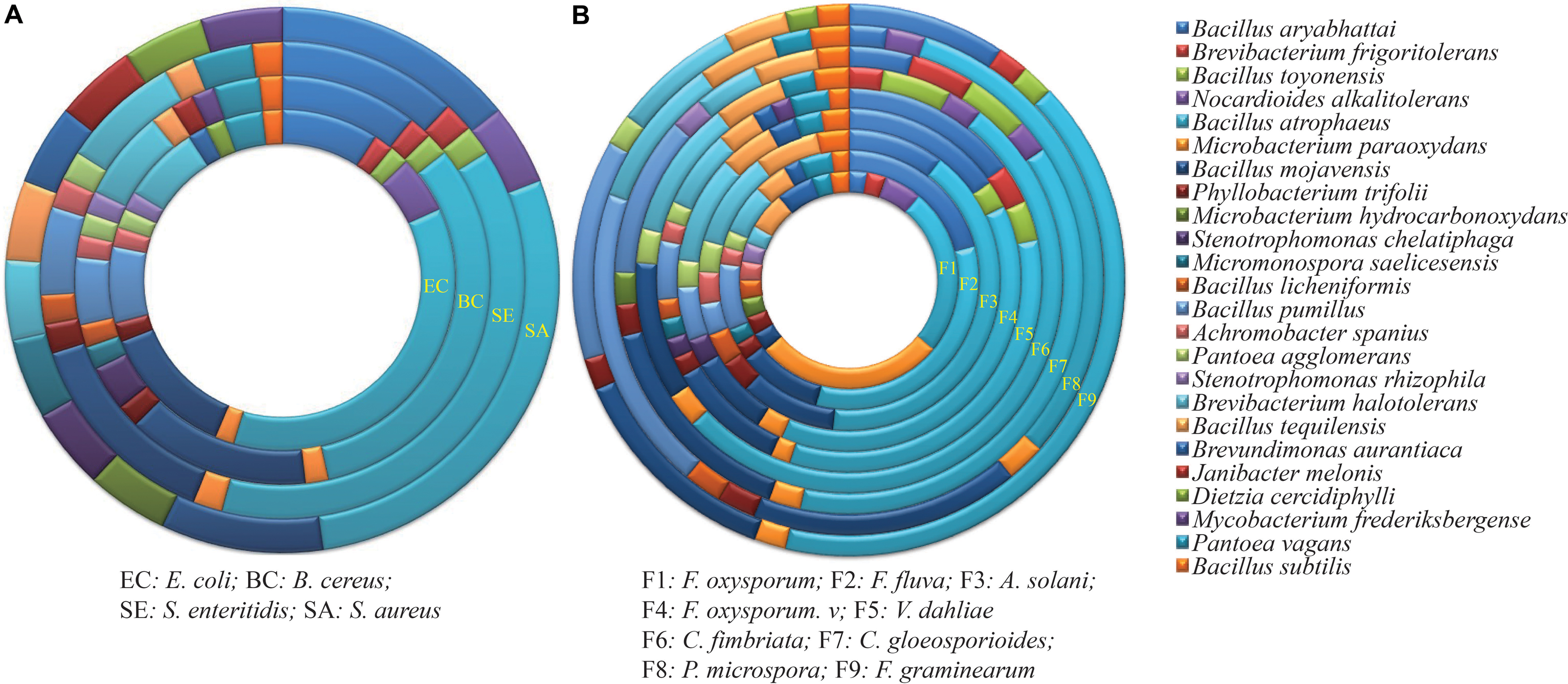
FIGURE 1. Antimicrobial activity of endophytes against (A) four common bacteria and (B) nine fungal pathogens. Each ring represents the total number of strains with activity against the pathogen, and is divided into colors based on the proportion of each genus to that total. Details on the common bacteria and fungal pathogens are shown in Tables 1, 2.
The inhibitory effect of all endophytic isolates was tested against the nine fungal phytopathogens listed in Table 2. The endophytes varied in their ability to inhibit the growth of the fungi, with the percentage of inhibition ranging from 12.3 to 75.3% (Supplementary Figures S1C,D). A total of 44, 75, and 34 strains (38.6, 65.8, and 29.8%) were antagonistic to the tomato pathogens F. oxysporum f. sp. (F1), F. fulva (F2), and A. solani (F3), respectively. For F. oxysporum f. sp., the largest inhibition zone (72.1%) was observed for strain XEGI74, belonging to Bacillus halotolerans. For F. fulva, the largest inhibition zone (72.0%), was observed for Bacillus atrophaeus strain XEGI51. And for A. solani, the largest inhibition zone (63.0%) was observed for strain XEGI15, belonging to Brevibacterium frigoritolerans.
For the pathogens causing cotton wilt diseases, the antibiosis assay demonstrated that 48 and 83 endophytic strains (42.1 and 72.8%) were antagonistic to F. oxysporum f. sp. Vesinfectum (F4) and V. dahliae (F5), respectively. For F. oxysporum f. sp. Vesinfectum, the largest inhibition zone (70.1%) was observed for strain XEGI9, belonging to Nocardioides alkalitolerans; for V. dahliae, the largest inhibition zone (75.5%) was observed for strain XEGI50, belonging to B. atrophaeus.
For the pathogens Ceratocystis fimbriata, Colletotrichum gloeosporioides, Fusarium graminearum, and the receently described pathogen Pestalotiopsis microspora (Guo J. W. et al., 2016), the antibiosis assay demonstrated that 60, 45, 57, and 40 strains (52.6, 39.5, 50.0, and 35.1%) were antagonistic to C. fimbriata (F6), C. gloeosporioides (F7), P. microspore (F8), and F. graminearum (F9), respectively. For C. fimbriata, the largest inhibition zone (55.4%) was observed for strain XEGI46, which belonged to Bacillus mojavensis. For C. gloeosporioides, the largest inhibition zone (48.0%) was observed for B. atrophaeus strain XEGI10. For P. microspore, the largest inhibition zone (47.9%) was observed for strain XEGI44, belonging to Bacillus mojavensis; and for F. graminearum, the largest inhibition zone (34.0%) was observed for strain XEGI39, which belonged to B. atrophaeus. Based on our investigation, members of the genus Bacillus had the highest antagonistic activity of any of the bacterial genera (Figure 1B), and V. dahliae was the most susceptible fungal pathogen.
The presence of putative biosynthetic genes encoding PKSs and NRPS of peptide antibiotics were investigated in the 16 strains exhibiting the strongest antimicrobial activities by using 3 sets of degenerate PCR primers. Among the 16 strains, 12 were positive for PKSs genes (Table 3); the translated amino acid sequences of these PKS-I genes shared moderate to high amino acid identity (54–99%) with those from members of the phylum Actinobacteria. A total of seven were positive for NRPS genes (Table 3). The NRPS sequences shared 40–70% amino acid sequence identities with peptide synthetase genes of the genus Bacillus, except the sequence from Microbacterium paraoxydans strain XEGI12, which was most closely related to enzymes of genus Microbacterium. Moreover, of the 16 isolates, 7 strains were positive for amplification of all 3 biosynthetic genes; 2 strains were positive for 2 genes; 4 strains were positive for only 1 gene; and only 3 strains were negative for all 3 PKS and NRPS genes (Table 3 and Supplementary Figure S2).
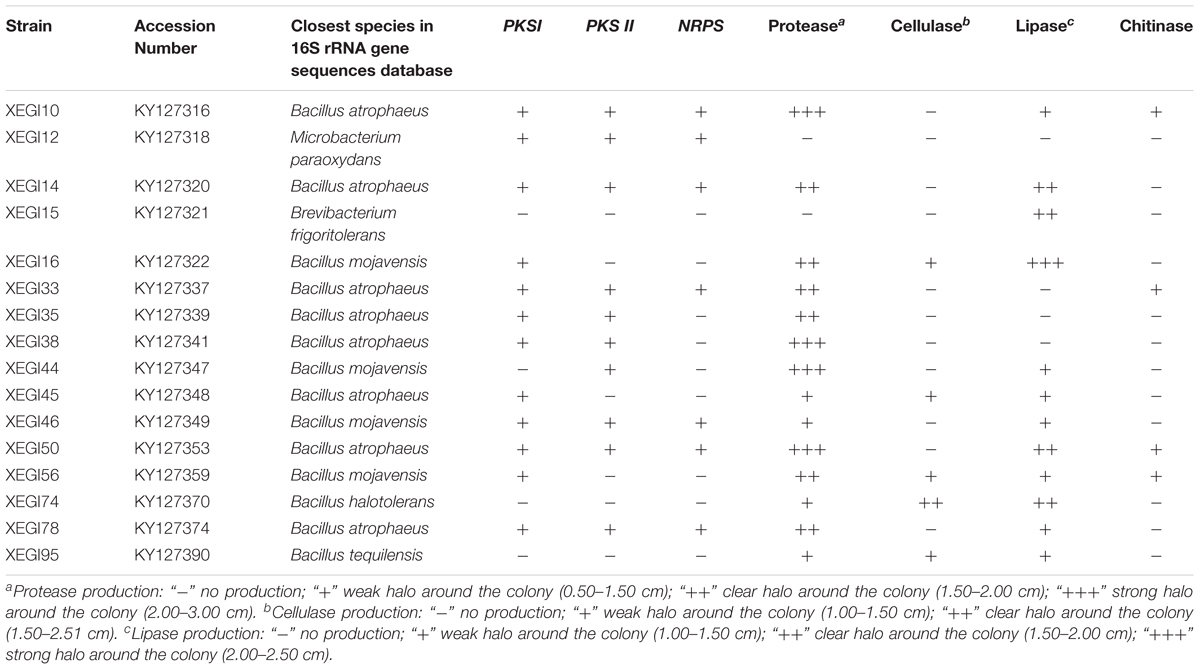
TABLE 3. Presence of biosynthetic genes (PKSI, PKSII, and NRPS) and digestive enzyme activity of the 16 most active strains.
The same 16 strains were assayed for protease, cellulase, lipase, and chitinase, which are potentially involved in lysis of phytopathogens or modulation of virulence factors. Some of the tested strains produced one or more lytic enzyme, as assessed by a change in pH in plates containing soluble chitin and bromocresol purple (chitinase) or by the diameter of the halo zone on media containing skim milk (protease), carboxymethyl cellulose (cellulase), or Sierra lipolysis agar (lipase) (Table 3 and Supplementary Figure S3). Proteases and lipases were the most common enzymes detected. The Bacillus strains were more active in these assays than the other two genera tested.
An infection time course of A. thaliana and V. dahliae was developed by evaluating plant disease signs over a 5-week period. The first signs developed within 7 days of inoculation, including yellowing of leaves, and leaf chlorosis, which began with older leaves and progressed to younger leaves. Severe signs were seen on the leaves of the plants challenged with V. dahliae in the absence of endophytes (Figure 2). After 5 weeks, the control plantlets under pathogen-challenged conditions showed severe signs, while those inoculated with endophytes showed mild signs of disease. Although all tested endophytes conferred some degree of Verticillium wilt resistance, the distribution of disease grades varied dramatically (Figure 3A). The disease severity index (DSI) in plantlets treated with strains XEGI33, XEGI38, XEGI44, XEGI50, and XEGI78 were each significantly reduced (44.5, 50.0, 44.5, 33.3, and 48.6%), compared to those grown in the absence of the biocontrol agent, which was 78.0% (Figure 3B). XEGI50 conferred the best protection (33.3% DSI) by suppressing yellowing other and wilt signs. This result suggested that XEGI50 could slow disease development, and the expression of signs were delayed compared with other treatments.
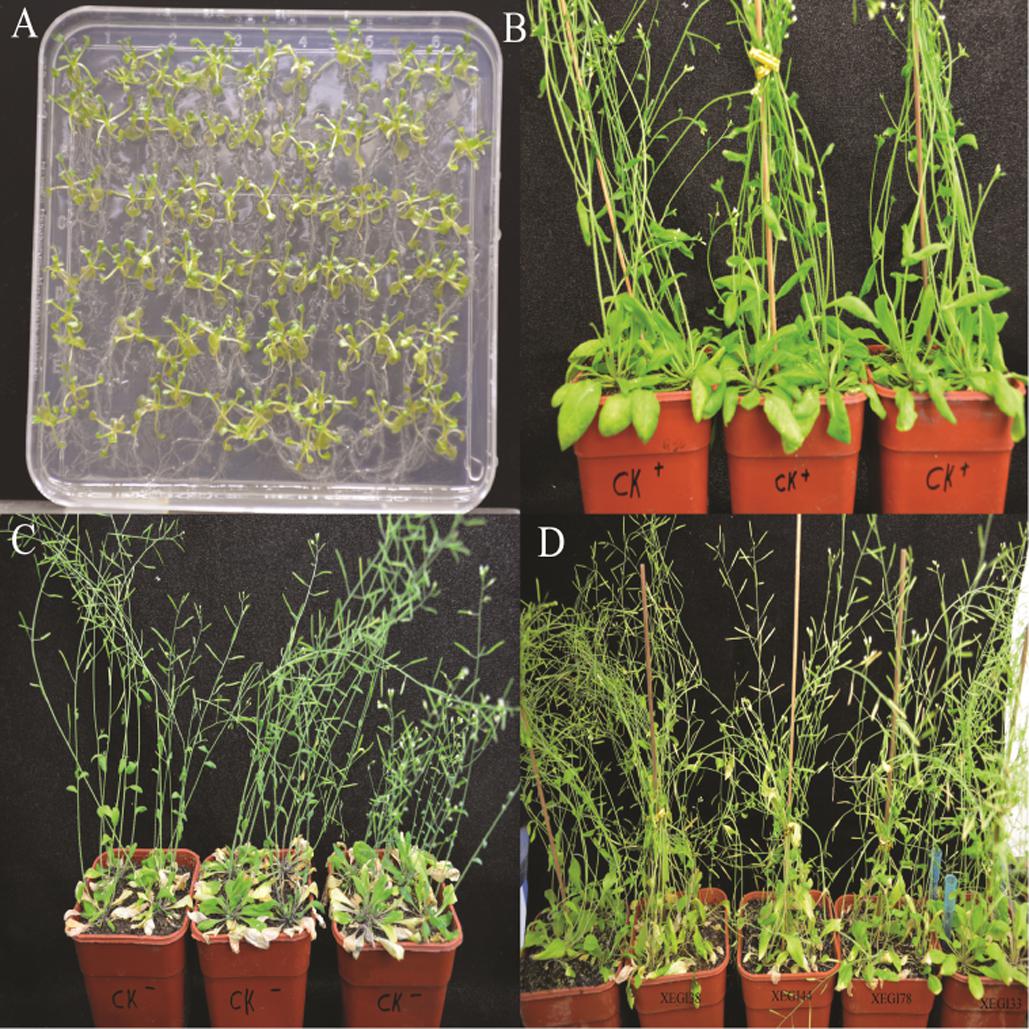
FIGURE 2. Defense response to V. dahliae in Arabidopsis thaliana plants (A) A. thaliana seedling; (B) A. thaliana without inoculation of V. dahliae; (C) Response of A. thaliana to V. dahliae; (D) Response of A. thaliana to V. dahliae inoculated with different endophytic strains after 5 weeks.
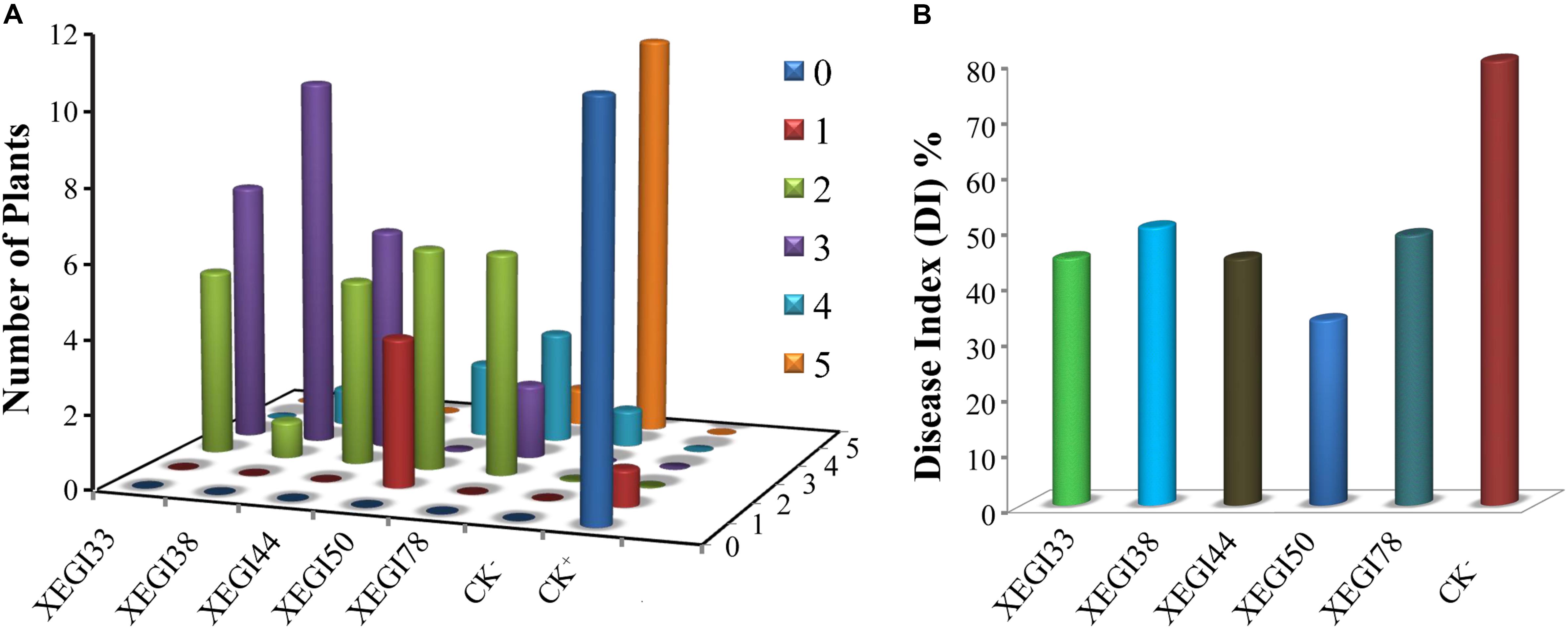
FIGURE 3. Effect of antagonistic isolates on disease grades and disease index of Arabidopsis thaliana plants to V. dahliae over 5 weeks after inoculation of the pathogen. (A) Signs were rated along a scale that assigned disease grades ranging from ‘0’ to ‘5’ (0: no signs, 1: ≤ 25, 2: 25–35, 3: 35–45, 4: 45–55, 5: 55–80%); (B) disease index of Arabidopsis thaliana.
To determine the prevalent organic compounds produced by the most bioactive strain, B. atrophaeus XEGI50, ethyl acetate extracts of cell supernatant buffered at pH 7 and pH 3 were concentrated from cultures of strain XEGI50, V. dahlia, and a co-culture. GC–MS analysis showed that different features (putative compounds) were produced at pH 7 and pH 3. Features were tentatively identified based on comparison of spectra avaiable through the National Institute of Standards and Technology (NIST) database, and biological activities were interpreted primarily based on Dr. Duke’s Phytochemical and Ethnobotanical Databases created by Dr. Jim Duke of the Agricultural Research Service/USDA.
The GC-MS analysis of crude extracts buffered at pH 7 from strain XEGI50 cultivated alone revealed at least 21 features (Supplementary Table S1) and 36 features at pH 7 (Supplementary Table S2). Nine distinctive features in the pH 7 extract, compared to pH 3, were present at RT 3.897, 4.171, 17.302, 19.849, 20.701, 21.511, 22.301, 22.374, and 24.331, suggesting the presence of benzene, 1,3-dimethyl-; o-xylene; dibutyl phthalate; heptadecane; eicosane; tetracosane; pentacosane; bis (2-ethylhexyl) phthalate; and decanedioic acid, bis (2-ethylhexyl) ester (Figure 4A). The pH 3 extract showed three major peaks that were different from the pH 7 extract at RT 3.908, 18.954, and 19.859, suggestive of p-xylene; heneicosane; and docosane (Figure 4B).
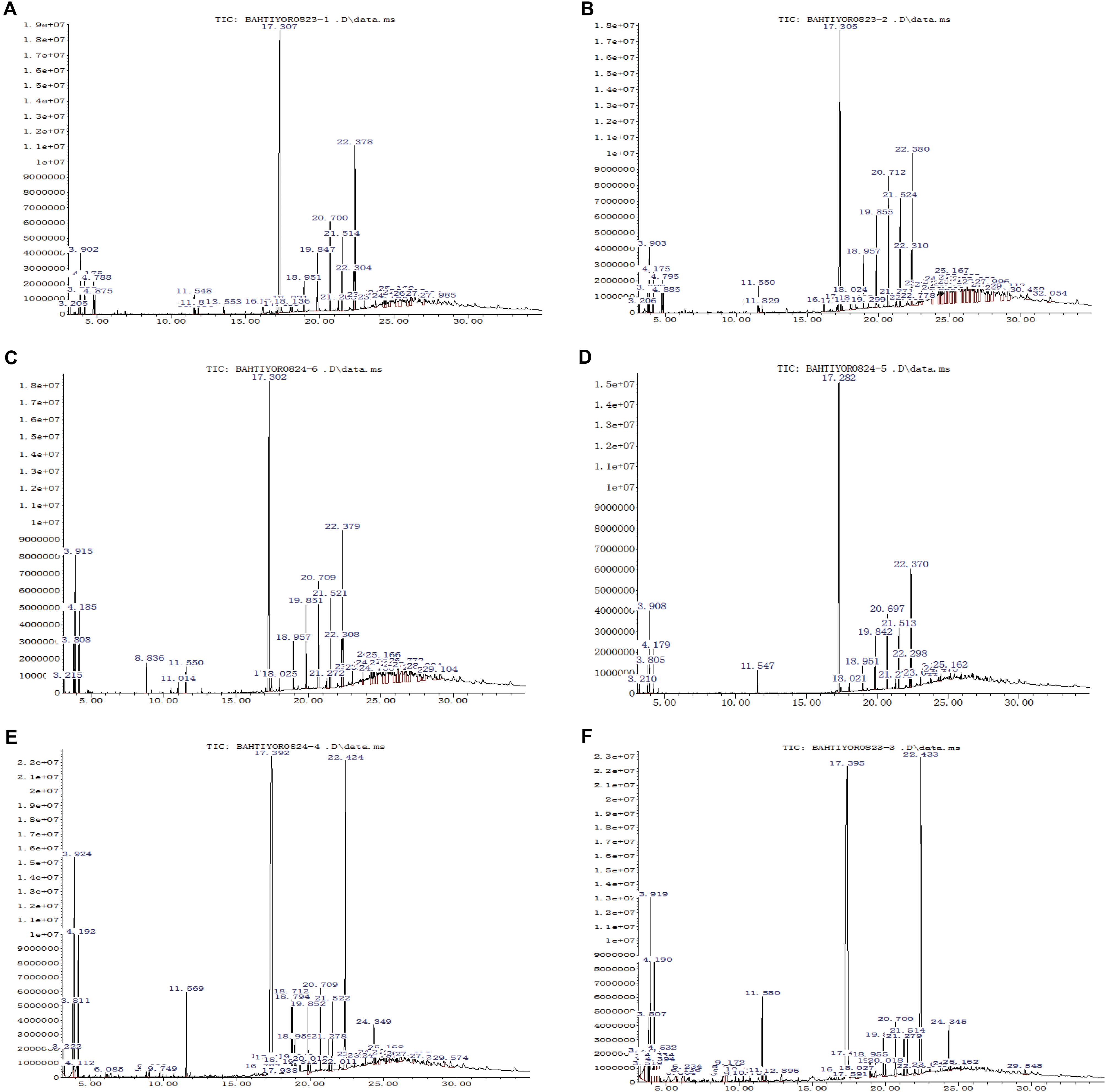
FIGURE 4. GC-MS analysis of bioactive compound of ethyl acetate extract sample. (A) The crude extract of XEGI50 at pH 7; (B) The crude extract of XEGI50 at pH 3; (C) The crude extract of V. dahliae at pH 7; (D) The crude extract of V. dahliae at pH 3; (E) Antibiosis crude extract of XEGI50 and V. dahliae mixture at pH 7; (F) Antibiosis crude extract of XEGI50 and V. dahliae mixture at pH 3.
The GC-MS analysis of crude extracts from V. dahliae cultivated alone showed 32 compounds in the pH 7 extract (Supplementary Table S3) and 17 compounds in the pH 3 extract (Supplementary Table S4). Six major features were obtained from the pH 7 extract at RT 3.802, 3.918, 4.181, 17.302, 20.711, and 22.384, suggestive of ethylbenzene; p-xylene; benzene 1,3-dimethyl-; dibutyl phthalate; heptadecane; and bis (2-ethylhexyl) phthalate (Figure 4C). The pH 3-buffered extract from V. dahliae showed one major peak are different from the crude extract at pH 7 at RT 22.374, suggestive of phthalic acid, di (2-propylpentyl) ester (Figure 4D).
The GC-MS resolved several features in the extracts of XEGI50 and V. dahliae mixture: 37 compounds from the pH 7 extract (Supplementary Table S5) and 32 compounds from pH 3 extract (Supplementary Table S6). A total of 12 major peaks were obtained from the pH 7 extract at RT 3.813, 3.929, 4.192, 11.568, 17.397, 18.712, 18.796, 19.848, 20.711, 21.521, 22.426, and 24.352, suggestive of ethylbenzene; p-xylene; dimethyl phthalate; 1,2-benzenedicarboxylic acid; bis (2-methyl propyl) ester; 9,12-octadecadienoic acid (Z,Z)- methyl ester; 9-octadecenoic acidmethyl ester, (E)-; eicosane; heptadecane; tetracosane; bis (2-ethylhexyl) phthalate; and decanedioic acid, bis (2-ethylhexyl) ester (Figure 4E). The pH 3 extract from the co-culture contained most of the same compounds, and one additional feature at RT 4.192, suggestive of o-xylene (Figure 4F).
Many compounds tentatively identified in the GC-MS analysis are known to have antimicrobial activity. Therefore, to determine the prevalent compounds that were induced by co-cultivation, putative compounds were compared between the three different datasets. At least 13 compounds were detected only in the co-culture, consisting of mainly fatty acid esters, phenols, alkanes, alkenes, and aromatic chemicals. Four of them showed high peaks at RT 17.397, 18.712, 18.796, and 24.35, suggestive of 1,2-benzenedicarboxylic acid butyl 2-ethylhexyl ester; 9,12-octadecadienoic acid (Z,Z)-, methyl ester; 9-octadecenoic acid, methyl ester, (E)-; and decanedioic acid, bis (2-ethylhexyl) ester (Figures 4E,F). In addition, several minor peaks were present, including butanoic acid, 2-hydroxy-3-methyl-; eicosane9-cyclohexyl-, heptadecanoic acid, 16-methyl-, methyl ester; hexadecanoic acid, methyl ester; hexanedioic acid, dioctyl ester; naphthalene, 1-methyl-; naphthalene, 2-methyl-; n-decanoic acid; pentanoic acid, 4-methyl-; and picolinamide (Supplementary Tables S5, S6).
The results showed that V. dahliae could not grow after 7 days of incubation with antagonistic strains (Figures 5Aa,b). The morphological response of XEGI50 to V. dahliae was observed under a laser microscope at different magnifications (1.25×, 2.5×, 4×). The microscopic characteristics of strains XEGI50 based on laser microscopy showed that at 1.25× magnification the endophytic strain XEGI50 was able to control the growth of the fungal mycelium after 5 days of cultivation (Figure 5B) and at 4×, a white powder appeared only on the side facing the pathogenic fungi, we hypothesize that strain XEGI50 may secrete some antifungal compound (Figure 5C). In addition, at the same magnification of 2.5×, strain XEGI50 looked intense at the bacteria/fungi interface, but expanded in the direction away from V. dahliae (Figure 5D).
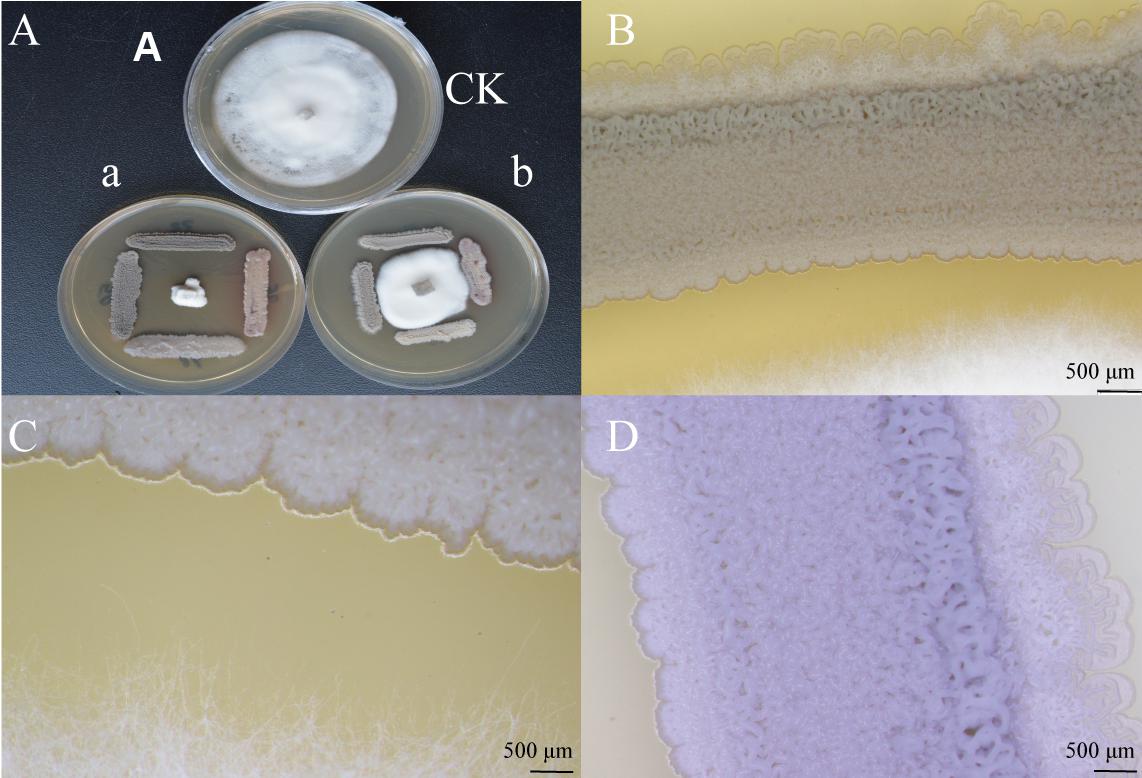
FIGURE 5. Intelligent live digital imaging of the response of endophytic strain XEGI50 to V. dahliae under a laser microscope. (A) In vitro evaluation of antagonistic activity of strain XEGI50; (a) V. dahliae could not grow after seven days of incubation with antagonistic strains, (b) V. dahliae grows after three days then we inoculate the antagonistic stains at four sides of the agar plate. (B) Response of strain XEGI50 to V. dahliae at 1.25× magnification; (C) Response of strain XEGI50 to V. dahliae at 4× magnification showed white powder appeared only on one the side which is facing the pathogenic fungi; (D) Different changes of the behavior of strain XEGI50 on the both sides due to the stress at 2.5× magnification.
Biological control of plant pathogens using microorganisms can be a safe, cost-effective, and efficient method for suppressing plant diseases. Endophytes accociated with medicinal plants are rich sources of secondary metabolites with antimicrobial activity, and they spend their whole life cycle within plant tissues without causing any infections or signs of disease (Bacon and White, 2000; Saikkonen et al., 2004). In addition, it has also been documented that endophytes associated with medicinal plants may produce the same metabolites in vitro and within host plant tissue (Kusari et al., 2013; Dos Santos et al., 2016). In the present study, we analyzed the antimicrobial activity of a diverse collection of endophytes previously isolated from wild populations of G. uralensis (Li et al., 2018), consisting of 18 genera and 34 species, with a goal of determining which strains offer the strongest antagonistic activities against pathogens, and to identify microbial products that may confer these antagonistic activities.
In vitro screens for antagonistic activity were conducted by co-cultivating the G. uralensis endophytes with common fungal pathogens of tomato (F. oxysporum f. sp., F. fulva, A. solani), cotton (F. oxysporum f. sp. Vesinfectum, V. dahliae), pomegranite (C. fimbriata), Cymbidinium (C. gloeosporioides), and Tsao-ko (P. microspora and F. graminearum), and the common bacteria S. aureus, B. cereus, S. enteritidis, and E. coli. In these assays, a significant fraction of the endophytic bacteria displayed antagonistic effects. The genus Bacillus (Figures 1A,B) was the dominant genus, with high antimicrobial activity against all indicator pathogens used in this study. Several studies have observed similar trends. For example, other studies have demonstrated that Bacillus strains associated with medicinal plants exhibit antibacterial activity against common bacteria such as S. aureus, Streptococcus pyogenes, Pseudomonas aeruginosa, and E. coli (Slepecky and He, 1992; Nejatzadeh-Barandozi, 2013; Egamberdieva et al., 2017a). Moreover, other studies have shown that Bacillus strains isolated from medicinal plants inhibited the mycelial growth of diverse vascular wilts caused by pathogenic fungi (Cho et al., 2002; Ebrahimi et al., 2010; Jiang et al., 2015; Akinsanya et al., 2015; Gao et al., 2017).
In our study, 16 strains showing antagonistic activity toward at least 7 pathogenic fungi and at least 1 common bacterium, were screened for the presence of PKS-I, PKS-II, and NRPS gene clusters to determine whether their genomes contain these genes. These 16 strains consisted of 1 strain each of Microbacterium paraoxydans and B. frigoritolerans, and 14 strains of Bacillus, belonging to Bacillus atrophaeus (8 strains), Bacillus mojavensis (four strains), Bacillus halotolerans (1 strain), and Bacillus tequilensis (1 strain). Only 12, 10, and 7 strains were positive for PCR amplification of fragments of PKS-I, PKS-II, and NRPS genes, respectively (Table 3). Several isolates had antimicrobial activity, but biosynthetic genes were not successfully amplified; they were B. frigoritolerans XEGI15, Bacillus halotolerans XEGI74, and Bacillus tequilensis XEGI95. The absence of amplification products may be due to the lack of natural product biosynthetic genes or because they contain divergent or novel genes that are not recognized by the degenerate primers used in this study (Courtois et al., 2003; Finking and Marahiel, 2004). Moreover, not all NRPS genes are involved in the biosynthesis of bioactive secondary metabolites, and there might be additional types of bioactive agents or mechanisms that may be involved in the generation of antimicrobial activities (Schneemann et al., 2010; Liu L. et al., 2016).
The 16 most antagonistic strains each produced at least 1 digestive enzyme, such as protease, cellulase, lipase, and chitinase (Table 3). Thus, these endophytes may protect the plant from fungi and insects by degrading the fungal cell wall or cell membrane, by degrading cell membrane proteins or extracellular virulence factors, or by stimulating systemic resistance in plants (Frankowski et al., 2001). In our previous investigation, endophytic bacteria isolated from the medicinal plants Ferula songorica, Hypericum perforatum, and Ferula sinkiangensis secreted similar digestive enzymes (Liu Y.-H. et al., 2016; Liu et al., 2017). Moreover, similar work done by Egamberdieva et al. (2017a) reported that endophytic bacteria associated with the medicinal plant Ziziphora capital were able to produce chitinolytic enzymes.
Verticillium dahliae, which infects cotton and several other plants, causes wilt diseases and crop losses of varying severity as well as natural ecosystems (Erdogan and Benlioglu, 2010). The signs of Verticillium wilt disease start with yellowing followed by chlorosis and necrosis of leaves (Ting, 2014). Arabidopsis thaliana is an excellent tool to identify traits involved in V. dahliae biocontrol by endophytic bacteria (Maldonado-González et al., 2015). In the current study, five endophytic bacterial strains, XEGI33, XEGI38, XEGI44, XEGI50, and XEGI78, representing B. atrophaeus and B. mojavensis, were selected for growth chamber experiments to suppress V. dahliae pathogenesis in A. thaliana. These five strains were chosen based on in vitro effects against a large number of pathogens. All five strains seemed to colonize A. thaliana and significantly reduced the DSI (Figure 3). In accordance with these results, several previous studies have shown that endophytic Bacillus species control fungal pathogens, including B. mojavensis, B. subtilis (Bacon and Hinton, 2002; Bacon et al., 2005; Cazorla et al., 2007), B. tequilensis, B. velezensis, B. amyloliquefaciens (Alvarez et al., 2012; Akinsanya et al., 2015; Gao et al., 2017), and B. megaterium, (Cho et al., 2002; Lin et al., 2013). In addition, Guo Y. et al. (2016) isolated and characterized B. atrophaeus strain OSY-7LA and showed that it exhibited a strong antagonistic activity against Listeria innocua, a food-borne pathogen that can survive at extreme pH, temperature, and high salt concentration.
Among the endophytes, B. atrophaeus XEGI50 was selected for exometabolomic studies by GC-MS, based on its ability to decrease the DSI and suppress the growth of V. dahliae. A total of 13 features that were expressed only in co-cultures of XEGI50 and V. dahliae were tentatively identified as compounds with known antimicrobial, antiphrastic, antitumor, and anticancer properties. Among these compounds, four of them were major peaks in cell-free extracts from the co-culture, suggesting they play an important role in antimicrobial activities; these compounds were 1,2-benzenedicarboxylic acid, butyl 2-ethylhexyl ester (Kavitha et al., 2010); 9,12-octadecadienoic acid (Z,Z)-, methyl ester (Sermakkani and Thangapandian, 2012); 9-octadecenoic acid, methyl ester, (E)-; and decanedioic acid, bis (2-ethylhexyl) ester (Tambekar et al., 2014). In addition, several putative antimicrobial compounds were identified as minor peaks: eicosane, 9-cyclohexyl (Hsouna et al., 2011); heptadecanoic acid, 16-methyl-, methyl ester (Kandasamy et al., 2012); hexadecanoic acid, methyl ester (Chandrasekaran et al., 2011); hexanedioic acid, dioctyl ester (Rodríguez-Meizoso et al., 2010); naphthalene, 1-methyl; naphthalene, 2-methyl- (Rokade and Sayyed, 2009); and pentanoic acid, 4-methyl- (Sharma et al., 2016). Since these compounds were not produced by pure cultures of XEGI50, they were likely induced by the presence of fungal pathogens such as V. dahliae, and very likely play a role in antagonism of pathogens.
The genus Bacillus is well known for the natural production of secondary metabolites with antibacterial and antifungal activities and has a strong potential to control plant diseases (Lodewyckx et al., 2002; Cavaglieri et al., 2005; Radhakrishnan et al., 2017). This study further illustrates its potential role as a biological agent for controlling phytopathogens. In recent years, the development of biological agents derived from Bacillus isolates, such as “Avogreen” (Korsten et al., 1997; Janisiewicz and Korsten, 2002) and “Shemer” (Droby, 2005), has been shown to be effective biocontrols against some plant diseases. In the present study, we provide insights about plant beneficial traits of culturable endophytic bacteria associated with the medicinal plant G. uralensis. Our results may provide a new biological control agent for controlling V. dahliae and improve our understanding of the biocontrol mechanism of natural endophytes belonging to the genus Bacillus. These results support the development of natural products that may minimize the need for the application of chemical fungicides, which would be an environmentally friendly approach and preserve biological resources in a sustainable agricultural system.
Our study revealed that natural endophytes of natural populations of the medicinal plant G. uralensis have a variety of antimicrobial activities in vitro and in vivo. The genus Bacillus, particularly B. atrophaeus and B. mojavensis, were the most effective biocontrol agents, with most strains exhibiting broad antibacterial and antifungal activities. Most of these bacteria contained genes for PKS and non-ribosomal proteins, both known to encode antimicrobial compounds, as well as extracellular digestive enzymes that may destroy or neutralize a variety of pathogens, including chitinases, cellulases, lipases, and proteases. Strain XEGI50, which belongs to Bacillus atrophaeus, was the most effective at reducing disease signs in A. thaliana in plant growth chambers. XEGI50 produces at least 13 compounds when co-cultivated with V. dahlia, many of which were putatively identified as compounds with known antimicrobial effects. To our knowledge, this is the first report establishing that B. atrophaeus produces bioactive compounds with antimicrobial activity. Future studies are needed to unequivocally identify these compounds, to establish their effects individually in model plant systems, and to better understand genetic and biochemical pathways for synthesis of these compounds.
OAAM, LL and W-JL participated in the design of the study, performed all the experiments and the interpretation of results, and wrote the manuscript. J-BM and SH participated in the antimicrobial experiments in vitro condition and provided the pathogenic bacteria, and also conducted the greenhouse experiments and data analysis. LX and J-WG conducted to antifungal activity in vitro condition and also provided some of the pathogenic fungi. BR did the GC-MS analyses and data analysis. Y-HL helped for preparing enzymes activity test, microscopic analysis, and PCR works. BH revised the revision and improve the language and structure of the manuscript. W-JL and OAAM revised the manuscript and supervised the hall experiments. All authors edited and critically revised the manuscript.
This research was supported by the National Key Research and Development Program of China (2017YFD0200503), National Natural Science Foundation of China (Grant No. 31650110479), Xinjiang Uygur Autonomous Region Regional Coordinated Innovation Project (Shanghai Cooperation Organization Science and Technology Partnership Program) (Grant No. 2017E01031), and West Light Foundation of the Chinese Academy of Sciences (Grant Nos. XBBS201305 and XBBS201201). W-JL was also supported by Guangdong Province Higher Vocational Colleges and Schools Pearl River Scholar Funded Scheme (2014). LL was supported by China Scholarship Council to study in the United States (Grant No. 201509655013). OAAM was supported by Available Position Talented Young Scientists Program of Ministry of Science and Technology of China (Grant No. P-EG-16-01), and also funded by Chinese Academy of Sciences President’s International Fellowship Initiative (Grant No. 2016PB024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00924/full#supplementary-material
FIGURE S1 |In vitro evaluation of antagonistic activity of endophytic bacterial isolates associated with G. uralensis. (A) Bacillus cereus (BC); (B) Salmonella enteritidis (SE); (C) Verticillium dahliae; (D) Fusarium oxysporum.
FIGURE S2 | Agarose gel showing PCR results for PKS-I, PKS-II, and NRPS from selected strains. (M: Marker 1,500 bp).
FIGURE S3 | Ability of endophytes strains to produce lytic enzymes. (A) breakdown of chitin into N-acetyl glucosamine causes an increase in pH and a change from yellow to purple zone around bacterial colony; (B) control; (C) clear zone formation in cellulose medium around bacterial colony; (D) Clear zone formation in protases medium around bacterial colony.
TABLE S1 | GC-MS identified components of the crude extract of XEGI50 at pH7 (Volatile compounds are listed in ascending order of Retention Time).
TABLE S2 | GC-MS identified components of the crude extract of XEGI50 at pH3 (Volatile compounds are listed in ascending order of Retention Time).
TABLE S3 | GC-MS identified components of the crude extract of V. dahliae at pH7 (Volatile compounds are listed in ascending order of Retention Time).
TABLE S4 | GC-MS identified components of the crude extract of V. dahliaeat pH3 (Volatile compounds are listed in ascending order of Retention Time).
TABLE S5 | GC-MS identified components of the antibiosis crude extract of XEGI50 and V. dahliae mixture at pH7 (Volatile compounds are listed in ascending order of Retention Time).
TABLE S6 | GC-MS identified components of the antibiosis crude extract of XEGI50 and V. dahliae mixture at pH3 (Volatile Compounds are listed in ascending order of Retention Time).
Achari, G. A., and Ramesh, R. (2014). Diversity, biocontrol, and plant growth promoting abilities of xylem residing bacteria from solanaceous crops. Int. J. Microbiol. 2014, 1–14. doi: 10.1155/2014/296521
Aeron, A., Dubey, R., Maheshwari, D., Pandey, P., Bajpai, V. K., and Kang, S. C. (2011). Multifarious activity of bioformulated Pseudomonas fluorescens PS1 and biocontrol of Sclerotinia sclerotiorum in Indian rapeseed (Brassica campestris L.). Eur. J. Plant Pathol. 131, 81–93. doi: 10.1007/s00248-009-9531-y
Agrawal, T., and Kotasthane, A. S. (2012). Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. Springerplus 1:73. doi: 10.1186/2193-1801-1-73
Akinsanya, M. A., Goh, J. K., Lim, S. P., and Ting, A. S. Y. (2015). Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol. Lett. 362:fnv184. doi: 10.1093/femsle/fnv184
Alvarez, F., Castro, M., Príncipe, A., Borioli, G., Fischer, S., Mori, G., et al. (2012). The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J. Appl. Bacteriol. 112, 159–174. doi: 10.1111/j.1365-2672.2011.05182.x
Asl, M. N., and Hosseinzadeh, H. (2008). Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 22, 709–724. doi: 10.1002/ptr.2362
Bacon, C., Hinton, D., and Snook, M. (2005). Tentative identification of Bacillus mojavensis antifungal inhibitor. Phytopathology 95:S5.
Bacon, C. W., and Hinton, D. M. (2002). Endophytic and biological control potential of Bacillus mojavensis and related species. Biol. Control 23, 274–284. doi: 10.1006/bcon.2001.1016
Bakker, P. A., Berendsen, R. L., Doornbos, R. F., Wintermans, P. C., and Pieterse, C. M. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 4:165. doi: 10.3389/fpls.2013.00165
Berg, G., and Hallmann, J. (2006). “Control of plant pathogenic fungi with bacterial endophytes,” in Microbial Root Endophytes, eds B. J. E. Schulz, C. J. C. Boyle, and T. N. Sieber (Berlin: Springer), 53–69.
Berg, G., Kurze, S., Buchner, A., Wellington, E. M., and Smalla, K. (2000). Successful strategy for the selection of new strawberry-associated rhizobacteria antagonistic to Verticillium wilt. Can. J. Microbiol. 46, 1128–1137. doi: 10.1139/w00-101
Bhuvaneswari, S., Madhavan, S., and Panneerselvam, A. (2013). Enumertion of endophytic bacteria from Solanum trilobatum L. World J. Pharm. Res. 3, 2270–2279.
Bibi, N., Zhang, G., Li, F., Fan, K., Yuan, S., and Wang, X. (2013). Utilization of Vd toxin for rapid screening of cotton germplasm against Verticillium dahliae. Pak. J. Bot. 45, 2157–2162.
Cavaglieri, L., Orlando, J., Rodriguez, M., Chulze, S., and Etcheverry, M. (2005). Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res. Microbiol. 156, 748–754. doi: 10.1016/j.resmic.2005.03.001
Cazorla, F., Romero, D., Pérez-García, A., Lugtenberg, B., Vicente, A. D., and Bloemberg, G. (2007). Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol 103, 1950–1959. doi: 10.1111/j.1365-2672.2007.03433.x
Chandrasekaran, M., Senthilkumar, A., and Venkatesalu, V. (2011). Antibacterial and antifungal efficacy of fatty acid methyl esters from the leaves of Sesuvium portulacastrum L. Eur. Rev. Med. Pharmacol. Sci. 15, 775–780.
Chen, Y., Gao, X., Chen, Y., Qin, H., Huang, L., and Han, Q. (2014). Inhibitory efficacy of endophytic Bacillus subtilis EDR4 against Sclerotinia sclerotiorum on rapeseed. Biol. Control 78, 67–76. doi: 10.1016/j.biocontrol.2014.07.012
Chernin, L., and Chet, I. (2002). Microbial Enzymes in Biocontrol of Plant Pathogens and Pests. Enzymes in the Environment: Activity, Ecology, and Applications. New York, NY: Marcel Dekker, 171–225.
Cho, K. M., Hong, S. Y., Lee, S. M., Kim, Y. H., Kahng, G. G., Lim, Y. P., et al. (2007). Endophytic bacterial communities in ginseng and their antifungal activity against pathogens. Microb. Ecol. 54, 341–351. doi: 10.1007/s00248-007-9208-3
Cho, S. J., Park, S. R., Kim, M. K., Lim, W. J., Ryu, S. K., An, C. L., et al. (2002). Endophytic Bacillus sp. isolated from the interior of balloon flower root. Biosci. Biotechnol. Biochem. 66, 1270–1275.
Courtois, S., Cappellano, C. M., Ball, M., Francou, F.-X., Normand, P., Helynck, G., et al. (2003). Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69, 49–55.
de Souza, J. T., de Boer, M., de Waard, P., van Beek, T. A., and Raaijmakers, J. M. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 69, 7161–7172.
Dos Santos, P. J. C., Savi, D. C., Gomes, R. R., Goulin, E. H., Senkiv, C. D. C., Tanaka, F. A. O., et al. (2016). Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 186, 153–160. doi: 10.1016/j.micres.2016.04.002
Droby, S. (2005). Improving quality and safety of fresh fruits and vegetables after harvest by the use of biocontrol agents and natural materials. Acta Hortic. 709, 45–52.
Ebrahimi, A., Asgharian, S., and Habibian, S. (2010). Antimicrobial activities of isolated endophytes from some Iranian native medicinal plants. Iran. J. Pharm. Sci. 6, 217–222. doi: 10.17795/zjrms-2482
Egamberdieva, D., Wirth, S., Behrendt, U., Ahmad, P., and Berg, G. (2017a). Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 8:199. doi: 10.3389/fmicb.2017.00199
Egamberdieva, D., Wirth, S. J., Shurigin, V. V., Hashem, A., and Abd_Allah, E. F. (2017b). Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under Salt Stress. Front. Microbiol. 8:1887. doi: 10.3389/fmicb.2017.01887
Erdogan, O., and Benlioglu, K. (2010). Biological control of Verticillium wilt on cotton by the use of fluorescent Pseudomonas spp. under field conditions. Biol. Control 53, 39–45. doi: 10.1016/j.biocontrol.2009.11.011
Finking, R., and Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58, 453–488. doi: 10.1146/annurev.micro.58.030603.123615
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Frankowski, J., Lorito, M., Scala, F., Schmid, R., Berg, G., and Bahl, H. (2001). Purification and properties of two chitinolytic enzymes of Serratia plymuthica HRO-C48. Arch. Microbiol. 176, 421–426. doi: 10.1007/s002030100347
Gao, Z., Zhang, B., Liu, H., Han, J., and Zhang, Y. (2017). Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control 105, 27–39. doi: 10.1016/j.biocontrol.2016.11.007
Göre, M. E., Caner,Ö. K., Altın, N., Aydın, M. H., Erdoǧan, O., Filizer, F., et al. (2009). Evaluation of cotton cultivars for resistance to pathotypes of Verticillium dahliae. Crop Prot. 28, 215–219. doi: 10.1016/j.cropro.2008.10.004
Guo, J. W., Yang, L. F., Liu, Y. H., Yang, J., Wang, H. F., Li, L., et al. (2016). First report of pseudostem black spot caused by Pestalotiopsis microspora on Tsao-ko in Yunnan, China. Plant Dis. 100, 1021–1021. doi: 10.1094/PDIS-08-15-0920-PDN
Guo, Y., Huang, E., Yang, X., Zhang, L., Yousef, A. E., and Zhong, J. (2016). Isolation and characterization of a Bacillus atrophaeus strain and its potential use in food preservation. Food Control 60, 511–518. doi: 10.1016/j.foodcont.2015.08.029
Harwood, T., Tomlinson, I., Potter, C., and Knight, J. (2011). Dutch elm disease revisited: past, present and future management in Great Britain. Plant Pathol. 60, 545–555. doi: 10.1111/j.1365-3059.2010.02391.x
Horikoshi, K. (2008). Past, present and future of extremophiles. Extremophiles 12, 1–2. doi: 10.1007/s00792-007-0127-5
Hsouna, A. B., Trigui, M., Mansour, R. B., Jarraya, R. M., Damak, M., and Jaoua, S. (2011). Chemical composition, cytotoxicity effect and antimicrobial activity of Ceratonia siliqua essential oil with preservative effects against Listeria inoculated in minced beef meat. Int. J. Food Microbiol. 148, 66–72. doi: 10.1016/j.ijfoodmicro.2011.04.028
Janisiewicz, W. J., and Korsten, L. (2002). Biological control of postharvest diseases of fruits. Annu. Rev. Phytopathol. 40, 411–441.
Jiang, C.-H., Wu, F., Yu, Z.-Y., Xie, P., Ke, H.-J., Li, H.-W., et al. (2015). Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol. Res. 170, 95–104. doi: 10.1016/j.micres.2014.08.009
Juzwik, J., Harrington, T. C., MacDonald, W. L., and Appel, D. N. (2008). The origin of Ceratocystis fagacearum, the oak wilt fungus. Annu. Rev. Phytopathol. 46, 13–26. doi: 10.1146/annurev.phyto.45.062806.094406
Kandasamy, S., Sahu, S. K., and Kandasamy, K. (2012). In Silico studies on fungal metabolite against skin cancer protein (4, 5-Diarylisoxazole HSP90 Chaperone). ISRN Dermatol. 2012, 626214. doi: 10.5402/2012/626214
Kavitha, A., Prabhakar, P., Narasimhulu, M., Vijayalakshmi, M., Venkateswarlu, Y., Rao, K. V., et al. (2010). Isolation, characterization and biological evaluation of bioactive metabolites from Nocardia levis MK-VL_113. Microbiol. Res. 165, 199–210. doi: 10.1016/j.micres.2009.05.002
Korsten, L., De Villiers, E., Wehner, F., and Kotzé, J. (1997). Field sprays of Bacillus subtilis and fungicides for control of preharvest fruit diseases of avocado in South Africa. Plant Dis. 81, 455–459. doi: 10.1094/PDIS.1997.81.5.455
Kun, L., Zhixiang, Z., and Xiaofei, L. (2010). Screening for PKS gene from soil metagenomics library and identification of the actives against root-knot nematodes. Plant Prot. 36, 38–42.
Kusari, S., Pandey, S. P., and Spiteller, M. (2013). Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 91, 81–87. doi: 10.1016/j.phytochem.2012.07.021
Lacava, P. T., Li, W., Arau´jo, W. L., Azevedo, J. L. C., and Hartung, J. S. (2007). The endophyte Curtobacterium flaccumfaciens reduces symptoms caused by Xylella fastidiosa in Catharanthus roseus. J. Microbiol. 45, 388–393.
Li, L., Mohamad, O. A. A., Ma, J., Friel, A. D., Su, Y., Wang, Y., et al. (2018). Synergistic plant–microbe interactions between endophytic bacterial communities and the medicinal plant Glycyrrhiza uralensis F. Antonie Van Leeuwenhoek 2018, 1–14. doi: 10.1007/s10482-018-1062-4
Liao, W. C., Lin, Y. H., Chang, T. M., and Huang, W. Y. (2012). Identification of two licorice species, Glycyrrhiza uralensis and Glycyrrhiza glabra, based on separation and identification of their bioactive components. Food Chem. 132, 2188–2193. doi: 10.1016/j.foodchem.2011.12.051
Lin, T., Zhao, L., Yang, Y., Guan, Q., and Gong, M. (2013). Potential of endophytic bacteria isolated from ‘Sophora alopecuroides’ nodule inbiological control against Verticillium wilt disease. Aust. J. Crop Sci. 7:139.
Liu, L., Salam, N., Jiao, J.-Y., Jiang, H.-C., Zhou, E.-M., Yin, Y.-R., et al. (2016). Diversity of culturable thermophilic Actinobacteria in hot springs in Tengchong, China and studies of their biosynthetic gene profiles. Microb. Ecol. 72, 150–162. doi: 10.1007/s00248-016-0756-2
Liu, Y.-H., Guo, J.-W., Salam, N., Li, L., Zhang, Y.-G., Han, J., et al. (2016). Culturable endophytic bacteria associated with medicinal plant Ferula songorica: molecular phylogeny, distribution and screening for industrially important traits. 3 Biotech 6:209. doi: 10.1007/s13205-016-0522-7
Liu, Y., Guo, J., Li, L., Asem, M. D., Zhang, Y., Mohamad, O. A., et al. (2017). Endophytic bacteria associated with endangered plant Ferula sinkiangensis KM Shen in an arid land: diversity and plant growth-promoting traits. J. Arid Land 9, 432–445. doi: 10.1007/s40333-017-0015-5
Lodewyckx, C., Vangronsveld, J., Porteous, F., Moore, E. R., Taghavi, S., Mezgeay, M., et al. (2002). Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 21, 583–606. doi: 10.1080/0735-260291044377
Loqman, S., Barka, E. A., Clément, C., and Ouhdouch, Y. (2009). Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J. Microbiol. Biotechnol. 25, 81–91. doi: 10.1007/s11274-008-9864-6
Maldonado-González, M. M., Bakker, P. A., Prieto, P., and Mercado-Blanco, J. (2015). Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front. Microbiol. 6:266. doi: 10.3389/fmicb.2015.00266
Metsä-Ketelä, M., Salo, V., Halo, L., Hautala, A., Hakala, J., Mäntsälä, P., et al. (1999). An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180, 1–6. doi: 10.1016/S0378-1097(99)00453-X
Nejatzadeh-Barandozi, F. (2013). Antibacterial activities and antioxidant capacity of Aloe vera. Org. Med. Chem. Lett. 3:5. doi: 10.1186/2191-2858-3-5
Nie, Y., Zeng, X.-C., Yang, Y., Luo, F., Luo, X., Wu, S., et al. (2012). A novel class of antimicrobial peptides from the scorpion Heterometrus spinifer. Peptides 38, 389–394. doi: 10.1016/j.peptides.2012.09.012
Peng, H., Thevs, N., Beckmann, V., and Abdusalih, N. (2016). Economic performance of cotton and fruit plantations in arid regions: observation from the Tarim River Basin, NW China. Asian J. Agric. Ext. Econ. Sociol. 8, 1–15. doi: 10.9734/AJAEES/2016/22254
Qing, W., Zuo, J.-H., Qian, W., Yang, N., and Gao, L.-P. (2015). Inhibitory effect of chitosan on growth of the fungal phytopathogen, Sclerotinia sclerotiorum, and sclerotinia rot of carrot. J. Integr. Agric 14, 691–697. doi: 10.1016/S2095-3119(14)60800-5
Radhakrishnan, R., Hashem, A., and Abd_Allah, E. F. (2017). Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 8:667. doi: 10.3389/fphys.2017.00667
Rodríguez-Meizoso, I., Jaime, L., Santoyo, S., Señoráns, F., Cifuentes, A., and Ibáñez, E. (2010). Subcritical water extraction and characterization of bioactive compounds from Haematococcus pluvialis microalga. J. Pharm. Biomed. Anal. 51, 456–463. doi: 10.1016/j.jpba.2009.03.014
Rokade, Y., and Sayyed, R. (2009). Naphthalene derivatives: a new range of antimicrobials with high therapeutic value. Rasayan J. Chem. 2, 972–980.
Saikkonen, K., Wäli, P., Helander, M., and Faeth, S. H. (2004). Evolution of endophyte–plant symbioses. Trends Plant Sci. 9, 275–280. doi: 10.1016/j.tplants.2004.04.005
Santhanam, R., Groten, K., Meldau, D. G., and Baldwin, I. T. (2014). Analysis of plant-bacteria interactions in their native habitat: bacterial communities associated with wild tobacco are independent of endogenous jasmonic acid levels and developmental stages. PLoS One 9:e94710. doi: 10.1371/journal.pone.0094710
Schneemann, I., Nagel, K., Kajahn, I., Labes, A., Wiese, J., and Imhoff, J. F. (2010). Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 76, 3702–3714. doi: 10.1128/AEM.00780-10
Sermakkani, M., and Thangapandian, V. (2012). GC-MS analysis of Cassia italica leaf methanol extract. Asian J. Pharm. Clin. Res. 5, 90–94.
Sharma, D., Pramanik, A., and Agrawal, P. K. (2016). Evaluation of bioactive secondary metabolites from endophytic fungus Pestalotiopsis neglecta BAB-5510 isolated from leaves of Cupressus torulosa D. Don. 3 Biotech 6:210. doi: 10.1007/s13205-016-0518-3
Slepecky, R., and He, H. (1992). “The genus Bacillus-Nonmedical,” in The Prokaryotes, eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (New York, NY: Springer-Verlag), 1663–1696.
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/MMBR.67.4.491-502.2003
Sturz, A. V., Christie, B. R., and Nowak, J. (2000). Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19, 1–30. doi: 10.1080/07352680091139169
Sun, G., Yao, T., Feng, C., Chen, L., Li, J., and Wang, L. (2017). Identification and biocontrol potential of antagonistic bacteria strains against Sclerotinia sclerotiorum and their growth-promoting effects on Brassica napus. Biol. Control. 104, 35–43. doi: 10.1016/j.biocontrol.2016.10.008
Tambekar, D., Tiwari, A., and Tambekar, S. (2014). Studies on production of antimicrobial substances from Bacillus species isolated from Lonar Lake. Indian J. Appl. Res. 4, 502–506. doi: 10.15373/2249555X/August2014/131
Ting, A. S. Y. (2014). “Biosourcing endophytes as biocontrol agents of wilt diseases,” in Advances in Endophytic Research, eds V. Verma and A. Gange (New Delhi: Springer), 283–300.
Vieira, M. L., Hughes, A. F., Gil, V. B., Vaz, A. B., Alves, T. M., Zani, C. L., et al. (2011). Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae). Can. J. Microbiol. 58, 54–66. doi: 10.1139/w11-105
Walker, R., Powell, A. A., and Seddon, B. (1998). Bacillus isolates from the spermosphere of peas and dwarf French beans with antifungal activity against Botrytis cinerea and Pythium species. J. Appl. Microbiol. 84, 791–801. doi: 10.1046/j.1365-2672.1998.00411.x
Weller, D. M. (1988). Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26, 379–407. doi: 10.1146/annurev.py.26.090188.002115
Yadeta, K. A., and Thomma, B. P. (2013). The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4:97. doi: 10.3389/fpls.2013.00097
Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, Q., and Ye, M. (2009). Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J. Chromatogr. A 1216, 1954–1969. doi: 10.1016/j.chroma.2008.07.072
Keywords: medicinal plants, endophytes, environmental microbiology, biological control, Verticillium dahliae, Bacillus atrophaeus, Licorice
Citation: Mohamad OAA, Li L, Ma J-B, Hatab S, Xu L, Guo J-W, Rasulov BA, Liu Y - H, Hedlund BP and Li W-J (2018) Evaluation of the Antimicrobial Activity of Endophytic Bacterial Populations From Chinese Traditional Medicinal Plant Licorice and Characterization of the Bioactive Secondary Metabolites Produced by Bacillus atrophaeus Against Verticillium dahliae. Front. Microbiol. 9:924. doi: 10.3389/fmicb.2018.00924
Received: 01 January 2018; Accepted: 20 April 2018;
Published: 09 May 2018.
Edited by:
Paula García-Fraile, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaCopyright © 2018 Mohamad, Li, Ma, Hatab, Xu, Guo, Rasulov, Liu, Hedlund and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Osama A. A. Mohamad, ZHIub3NhbWFhYnVzYXVkQGhvdG1haWwuY29t Wen-Jun Li, bGl3ZW5qdW4zQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.