
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 May 2018
Sec. Terrestrial Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00885
Nitrification is the two-step aerobic oxidation of ammonia to nitrate via nitrite in the nitrogen-cycle on earth. However, very limited information is available on how fertilizer regimes affect the distribution of nitrite oxidizers, which are involved in the second step of nitrification, across aggregate size classes in soil. In this study, the community compositions of nitrite oxidizers (Nitrobacter and Nitrospira) were characterized from a red soil amended with four types of fertilizer regimes over a 26-year fertilization experiment, including control without fertilizer (CK), swine manure (M), chemical fertilization (NPK), and chemical/organic combined fertilization (MNPK). Our results showed that the addition of M and NPK significantly decreased Nitrobacter Shannon and Chao1 index, while M and MNPK remarkably increased Nitrospira Shannon and Chao1 index, and NPK considerably decreased Nitrospira Shannon and Chao1 index, with the greatest diversity achieved in soils amended with MNPK. However, the soil aggregate fractions had no impact on that alpha-diversity of Nitrobacter and Nitrospira under the fertilizer treatment. Soil carbon, nitrogen and phosphorus in the soil had a significant correlation with Nitrospira Shannon and Chao1 diversity index, while total potassium only had a significant correlation with Nitrospira Shannon diversity index. However, all of them had no significant correlation with Nitrobacter Shannon and Chao1 diversity index. The resistance indices for alpha-diversity indexes (Shannon and Chao1) of Nitrobacter were higher than those of Nitrospira in response to the fertilization regimes. Manure fertilizer is important in enhancing the Nitrospira Shannon and Chao1 index resistance. Principal co-ordinate analysis revealed that Nitrobacter- and Nitrospira-like NOB communities under four fertilizer regimes were differentiated from each other, but soil aggregate fractions had less effect on the nitrite oxidizers community. Redundancy analysis and Mantel test indicated that soil nitrogen, carbon, phosphorus, and available potassium content were important environmental attributes that control the Nitrobacter- and Nitrospira-like NOB community structure across different fertilization treatments under aggregate levels in the red soil. In general, nitrite-oxidizing bacteria community composition and alpha-diversity are depending on fertilizer regimes, but independent of the soil aggregate.
Nitrification is of great importance in the nitrogen (N) cycle of agricultural ecosystems, including the oxidation processes of ammonia and nitrite. Ammonia oxidation is the first and rate-limiting step in nitrification, which is a critical aerobic process, resulting in the formation of nitrite by ammonia-oxidizing bacteria (AOB) and archaea (AOA) (Kowalchuk and Stephen, 2001; Leininger et al., 2006; Gubry-Rangin et al., 2010; Hayatsu et al., 2017), as well as some taxa called comammox belonging to the Nitrospira lineage II (Daims et al., 2015; van Kessel et al., 2015). Nitrite-oxidation is the second step in nitrification; catalyzed by nitrite oxidizing bacteria (NOB) which can convert nitrite to nitrate (Prosser, 1989). Compared with the study on the community structure of AOB and AOA (Kowalchuk and Stephen, 2001; Webster et al., 2005; He et al., 2007; Le Roux et al., 2008; Zhang et al., 2012), intensive investigation of the ecology of the nitrite oxidizing community has received little attention (Attard et al., 2010; Ward et al., 2011; Daims et al., 2016; Han et al., 2017).
Nitrite-oxidizers mainly comprise six bacterial genera: Nitrobacter, Nitrospira, Nitrotoga, Nitrococcus, Nitrospina, and Nitrolancetus, which are affiliated with Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria, as well as the phyla Chloroflexi (Nitrolancetus) and Nitrospirae (Nitrospira defluvii), respectively (Gould and Lees, 1960; Watson and Waterbury, 1971; Daims et al., 2001; Alawi et al., 2007; Sorokin et al., 2012). However, only Nitrobacter- and Nitrospira-like NOB are believed to play important functional roles in terrestrial ecosystems (Bartosch et al., 2002; Kim and Kim, 2006; Ke et al., 2013). Nitrobacter-like NOB are r-strategists, which prefer high substrate concentrations and have lower substrate affinity, while Nitrospira-like NOB are K-strategists with affinity for lower nitrite and oxygen concentrations (Daims et al., 2001; Blackburne et al., 2006; Nowka et al., 2015).
Nitrobacter-like NOB are one of the key functional players within the NOB community in tilled soils with high N availability and a high nitrite oxidation activity (Attard et al., 2010). In addition, the selection of a dominant population of Nitrobacter was observed under no-tillage soils. The application of sulfadiazine-contaminated pig manure decreased the diversity of Nitrobacter-like NOB community in the root-rhizosphere under Merzenhausen agricultural soil (Ollivier et al., 2013). In paddy pot experiment soils, Nitrobacter-like NOB were found to be more active in the rice rhizosphere than in the bulk soil (Ke et al., 2013). An opposite trend was also observed for Nitrospira-like NOB, which suggests a different habitat preference of Nitrobacter-like NOB and Nitrospira-like NOB in paddy soils. In this case, nitrite oxidation in surface soil was dominated by Nitrospira spp. (Ke et al., 2013). They also found that soil depth (related to O2 content) and sampling site (related to rice rhizosphere) affect Nitrobacter activities in paddy soils. Long-term straw applications selected several lineages II Nitrospira, which were probably affected by the increment of available soil potassium, NH4+ and NO3-, as well as decrements of available phosphorus in paddy-upland soil (Luo et al., 2017). Our previous study also showed that Nitrospira-like NOB community composition is more sensitive than Nitrobacter to land management in acid and straw application soils of a rapeseed-rice rotation field trial, and the abundance of Nitrospira was always greater than Nitrobacter; Nitrospira was affected by the straw application, while Nitrobacter was not (Han et al., 2017). It is worth mentioning that previously detected Nitrospira lineage II may have been comammox (Pester et al., 2014; Bertagnolli et al., 2016; Han et al., 2017). The nxrB from comammox Nitrospira does not form an independent phylogenetic clade from nitrite-oxidizing Nitrospira (Daims et al., 2015). Therefore, depicting the diversity and community composition of comammox Nitrospira using the nxrB gene remains a great challenge.
Soil aggregates include organic matter, minerals and microbes in the structured particles (Tisdall and Oades, 1982; Jastrow, 1996). Soil aggregates provide spatially heterogeneous habitats for a wide range of microorganisms characterized by different organic matter, predation pressure, oxygen concentrations and water contents (Six et al., 2000; Sessitsch et al., 2001; Davinic et al., 2012). Previous studies have suggested that fertilization was the major factor affecting soil AOB community structure, and aggregate fractions exhibited the secondary effect (Jiang et al., 2014). AOB community structure seemed to be more sensitive to different soil fractions and fertilizer treatments (Zhang et al., 2017). The absorption of ammonia to soil particles was found to limit its toxicity to NOB (Venterea et al., 2015). These studies suggested soil aggregates putatively shape a functional microbial community. However, little information is available regarding the effects of soil aggregation on different functional microbial communities.
To investigate the impact of both long-term fertilization treatments and soil aggregate fractions on nitrite-oxidizing bacterial community diversity, resistance and populations, a study using high-throughput sequencing of the marker genes associated with Nitrobacter- and Nitrospira-like NOB was conducted from an experimental field of red soil in Hunan, China. We aimed at identifying the main environmental factors that affect the diversity and composition of Nitrobacter- and Nitrospira-like NOB communities in this red soil. We hypothesized the following: (i) a higher influence on the NOB community putatively came from the long-term fertilization treatments than from the aggregate fraction sizes; (ii) the resistance of the NOB alpha-diversity index in response to environmental changes is both related to the strength of disturbances (associated with the fertilization regimes) and their inhabiting microenvironments (diverged by different soil particle size classes); (iii) Nitrospira-like NOB and Nitrobacter-like NOB communities were shaped by different soil properties.
The experimental site was located at Qiyang Red Soil Experimental Station (26°45′N, 111°52′E), Hunan Province, China. The experiment was started in 1990 with a winter wheat (Triticum aestivum L.) and summer maize (Zea mays L.) rotation system, including four treatments with three replicate plots for each treatment-type in a randomized plot design. Wheat was planted after fertilization in October and maize was planted after fertilization in April. The treatments included the following: control without fertilizers (CK, pH = 6.87); swine manure (M, pH = 6.52); chemical fertilization (nitrogen, phosphate, and potassium fertilizers, NPK, pH = 4.83); or chemical/organic combined fertilization (nitrogen, phosphate, potassium and swine manure fertilizers, MNPK, pH = 5.90). The nitrogen fertilizer was provided as urea or swine manure at 300 kg N ha-1, phosphate (P) as a single application of superphosphate [Ca(H2PO4)2] at 53 kg P ha-1, and potassium (K) as potassium chloride (KCl) at 100 kg K ha-1.
Soil samples were collected in November 2016 at a depth of 0–20 cm. Six soil cores (approximately 5 cm in diameter) were taken from each plot and mixed to form one composite sample. Samples were placed in a sterile plastic bag for transport to the laboratory within 24 h after collection. Each soil sample was divided into portions. One portion was used for DNA extraction and stored at -80°C and another portion was air-dried at room temperature for analysis of soil chemical properties.
Field fresh soils were gently broken apart along natural planes of weakness and then used for aggregate fractionation (Elliot, 1986). Three water-stable aggregate-size classes were manually fractionated through sieving 100 g of fresh soil on a series of three sieves (2000–250, 250–53, and <53 μm) as follows: large macroaggregates (2000–250 μm, LA), microaggregates (250–53 μm, MA) and silt+clay (<53 μm, SA). Fresh soil was processed using the wet-sieving method as follows: 50 g of soil was submerged in deionized water for 5 min at room temperature on top of a 2000 μm sieve; the sieve was manually moved up and down 3 cm, 50 times over a 2-min period. The fraction remaining on the 2000 μm sieve was collected in a plastic cup. Water plus the filtered soil was poured through a 250 μm sieve and the sieving procedure was repeated. Water plus the 250 μm fraction of soil was poured through a 53 μm sieve, and the sieving procedure repeated; the remaining 50 g of soil was repeated with the above procedure and mixed with the above obtained aggregates and then freeze-dried and weighed to determine the proportion of the entire soil weight.
Soil aggregate total carbon (TC) and total nitrogen (TN) were measured using a Vario Max element analyzer (Elementar, Hanau, Germany). Total soil phosphorus (TP) and potassium (TK) were digested by HClO4 and determined by molybdenum-blue colorimetry and flame photometry, respectively (Jackson, 1958). Soil total organic carbon (SOC) was measured using a potassium dichromate oxidation spectrophotometric method. The total available phosphorus (AP) was determined according to the methods described by Olsen et al. (1954). The available potassium (AK) was extracted for 30 min with 1 M NH4OAc (w/v, 1:10) and analyzed by atomic absorption spectrophotometry (Lanyon and Heald, 1982). Soil exchangeable ammonium (NH4+-N) contents were determined on a FIAstar 5000 Analyzer after extraction of soil aggregate with 2 M KCl (w/v, 1:5).
Soil DNA was extracted from 0.5 g samples by a bead mill homogenization procedure, using a FastDNA SPIN kit for soil and FastPrep®-24 (MP Biomedicals, Santa Ana, CA, United States) according to the manufacturer’s protocol. The quality and concentration of DNA were determined using a Nanodrop-2000 spectrophotometer (Nano Drop Technologies, Wilmington, DE, United States). All extracted DNA samples were then stored under -20°C for future molecular analysis.
Quantitative PCR assays were conducted using an ABI7500 FAST Real-time PCR system with nxrA (for Nitrobacter-like NOB) primers (Attard et al., 2010) and nxrB (for Nitrospira-like NOB) primers (Pester et al., 2014). The 20 μl PCR reaction mixtures contained 10 μl SYBR Premix Ex Taq II (2x) (Takara Bio Inc., Shiga, Japan), 0.9 μl of a 10 mM solution of each primer, 6.2 μl of DEPC-treated water, 2.0 μl of soil extract (diluted 10-fold) or a 2.0 μl standard plasmid. A standard curve was generated using 10-fold serial dilutions of a plasmid containing a copy of the target gene. All of the qPCR was performed in triplicate and gel electrophoresis analyses were conducted to confirm the amplification specificity. To avoid inhibitory effects on qPCR, the samples were diluted 10-fold based on a pre-experiment.
PCR amplification of nxrA and nxrB genes was performed using an Illumina MiSeq platform with the nxrA primers F1norA and R2norA (Attard et al., 2010) for Nitrobacter-like NOB and nxrB gene primers nxrB169f and nxrB638r (Pester et al., 2014) for Nitrospira-like NOB. To distinguish amplicons originating from the different soil samples, barcode oligonucleotides of 7 bp in length were ligated to each side of the purified PCR products and an equal amount of PCR products for each sample were combined in a single tube to run on the sequencing platform. Illumina reads were split based on the barcodes, which were obtained using QIIME (Caporaso et al., 2010), and the sequences of low quality (quality score < 25, length < 200 bp or 350 bp for nxrA and nxrB genes) were removed. The remaining sequences were further screened for frame shifts using the tool FrameBot from RDP’s FunGene Pipeline1. The remaining quality-screened sequences were clustered into operational taxonomic units (OTU) based on 95% sequence identity. In addition, values of the OTU abundance lower than 0.001% of the total sample sequence were removed and then OTU table is used for subsequent analysis.
Two-way analysis of variance (ANOVA) was used to analyze effects of soil aggregates and fertilization treatments on the soil variables and alpha-diversity index (Shannon and Chao1 index) and abundances by SPSS 19.0 statistical software (IBM Co., Armonk, NY, United States). Spearman’s correlation was used to determine whether the correlation between each soil variables and alpha-diversity was significant. Redundancy analysis (RDA) was carried out using the Canoco 4.5. Monte Carlo permutation test with 999 unrestricted permutations were performed to determine the extent of the environmental parameter(s) that were used to explain the nitrifier community. The two-way permutational multivariate analysis of variance (PerMANOVA) was conducted to analyze effects of the fertilization treatment and soil aggregate fractions on the nitrifier community by using PC-ORD 5.0 (Bruce McCune and MJM Software). Principal co-ordinates analysis (PCoA) was performed2 in the R using the “ape” package. Multi-response permutation procedures (MRPP) were performed based on the community data by using R ‘vegan’ package to test significant differences in the community composition. Aggregated boosted trees (ABT) analysis, a statistical learning method that aimed to attain both accurate prediction and explanation, was carried out using the ‘gbmplus’ package.
The stabilities of α-diversity index (Shannon and Chao1 index), in terms of their resistance to fertilization treatments under soil aggregate levels, were quantified by resistance indices as described by Orwin and Wardle (2004) and Delgado-Baquerizo et al. (2014) with the following equation:
where RS, the value of the resistance index of alpha-diversity index (Shannon or Chao1 index) in response to a fertilization regime. C0, the value of alpha-diversity index from the control; S0, the value of alpha-diversity index from a treated plot. For example, values from the CK and fertilization treatments under soil aggregate levels were used to calculate the resistance index for the fertilization treatments. The value of the resistance index (Rs) is bounded between -1 and 1. If the index value reaches 1, this indicates that the treatment did not cause any change in the response variable. If the index value is 0 or negative, this means that there is a 100% change or greater than 100% change in the response variable in the treatment compared with that in the control (Orwin et al., 2006).
The results of the soil aggregate chemical properties are shown in Table 1. The content of soil TC and TN is the highest in macroaggregates, followed by microaggregates and silt+clay. The soil SOC, P, K, and NH4+ content was higher in macroaggregates and microaggregates and lowest in silt+clay. However, less influence on TN, TP, TK, and NH4+ across different soil aggregates was observed. Fertilizations significantly (p < 0.05) increased the content of all the measured soil geochemical factors, except soil TK and NH4+ content. The soil TC, TN, and SOC content are highest in the M treatment, followed by MNPK and NPK plots. The AK content was highest in the MNPK, followed by the M and NPK plots. Two-way ANOVA indicated that the fertilization treatments and soil aggregate fractions (except NH4+) had significant impacts on those measured properties (Supplementary Table S1).
As shown in Figure 1, the abundances of the Nitrobacter- and Nitrospira-like NOB, represented by the copy numbers of the nxrA gene and nxrB gene, ranged from 0.78 ± 0.42 × 104 to 19.0 ± 5.2 × 104 g-1 dry soil, and from 0.40 ± 0.24 × 106 to 6.2 ± 1.1 × 104 g-1 dry soil, respectively. The Nitrobacter and Nitrospira abundance was significantly (p < 0.05) higher in the M and MNPK treatment than CK (Figures 1A,B), indicated that manure fertilizer could promote the abundances of NOB. No significant difference of Nitrobacter and Nitrospira abundance was observed among various soil aggregates sizes in CK and M plot. Two-way ANOVA data indicated that the Nitrobacter and Nitrospira abundance was significantly affected by fertilization treatments (F = 24.2, F = 67.9, respectively, p < 0.01).
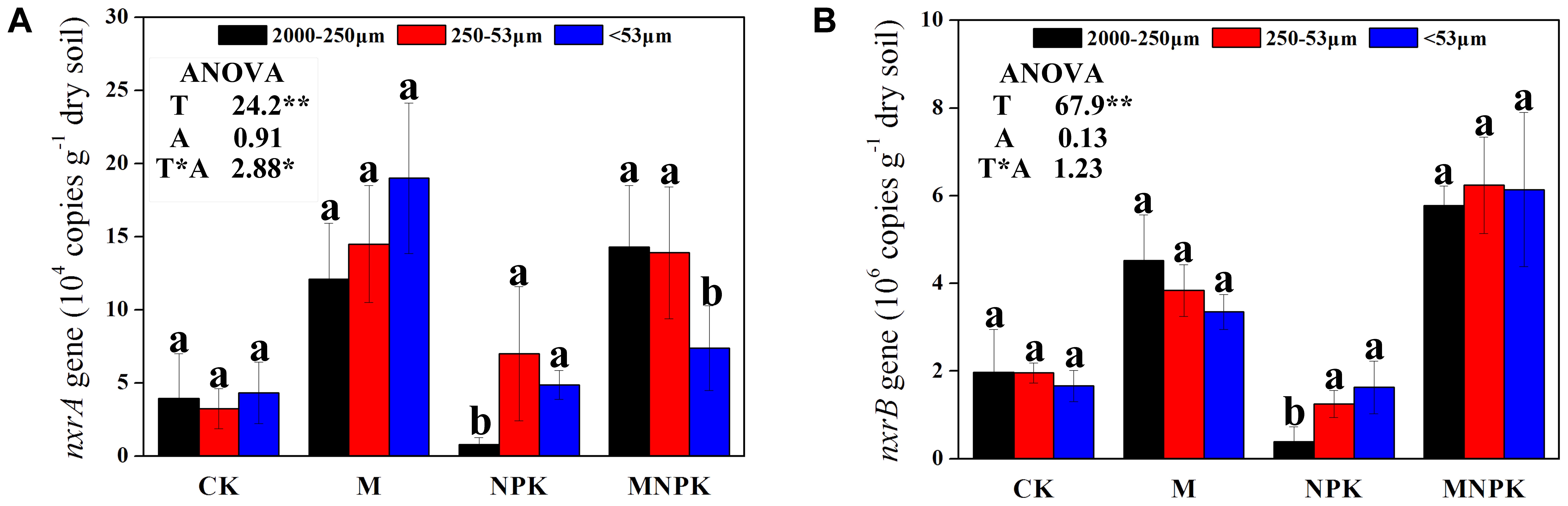
FIGURE 1. Abundances of Nitrobacter- (A) and Nitrospira-like NOB (B) of three soil fractions under different fertilizer treatments. Error bars represent standard error (n = 3) and are followed by a lowercase letter indicating a significant difference among three factions within each fertilizer according to the Tukey test (p < 0.05). T, fertilization treatment; A, soil aggregate size. ∗, indicate a significant difference at p < 0.05, ∗∗, indicate a significant difference at p < 0.01.
The community structures of Nitrobacter- and Nitrospira-like NOB were characterized by sequencing analysis of the nxrA and nxrB genes, respectively. Sequencing yielded an average of 26635 and 15812 high-quality sequences per sample for nxrA and nxrB, respectively. The constructed unique operational taxonomic units (OTUs, based on a 95% cut off) dataset was utilized in the following analysis after being normalized.
For Nitrobacter-like NOB, the Shannon index was remarkably decreased in M and NPK treatment plots and increased in MNPK compared to the CK plots (Figure 2A). In addition, this index was similar in different size of soil aggregate fractions under the same fertilization. For Nitrospira-like NOB, the Shannon index was significantly increased in M and MNPK treatments, but decreased in the NPK plot (Figure 2C). The index was also similar in different sizes of soil aggregate fractions under the fertilization. The trend of Chao1 index is similar to that of Shannon index for Nitrobacter and Nitrospira (Figures 2B,D). Two-way ANOVA showed that fertilization treatments have a strong impact (p < 0.01) on the Shannon and Chao1 indexes of Nitrobacter- and Nitrospira-like NOB, but soil aggregate sizes do not (Supplementary Table S1). Spearman’s rank coefficient correlation analysis showed that soil carbon, nitrogen and phosphorus in the soil had a significant correlation with Nitrospira Shannon and Chao1 diversity index, while total potassium only had a significant correlation with Nitrospira Shannon diversity index (Supplementary Table S2). However, this correlation was not detected in the Nitrobacter-like NOB.
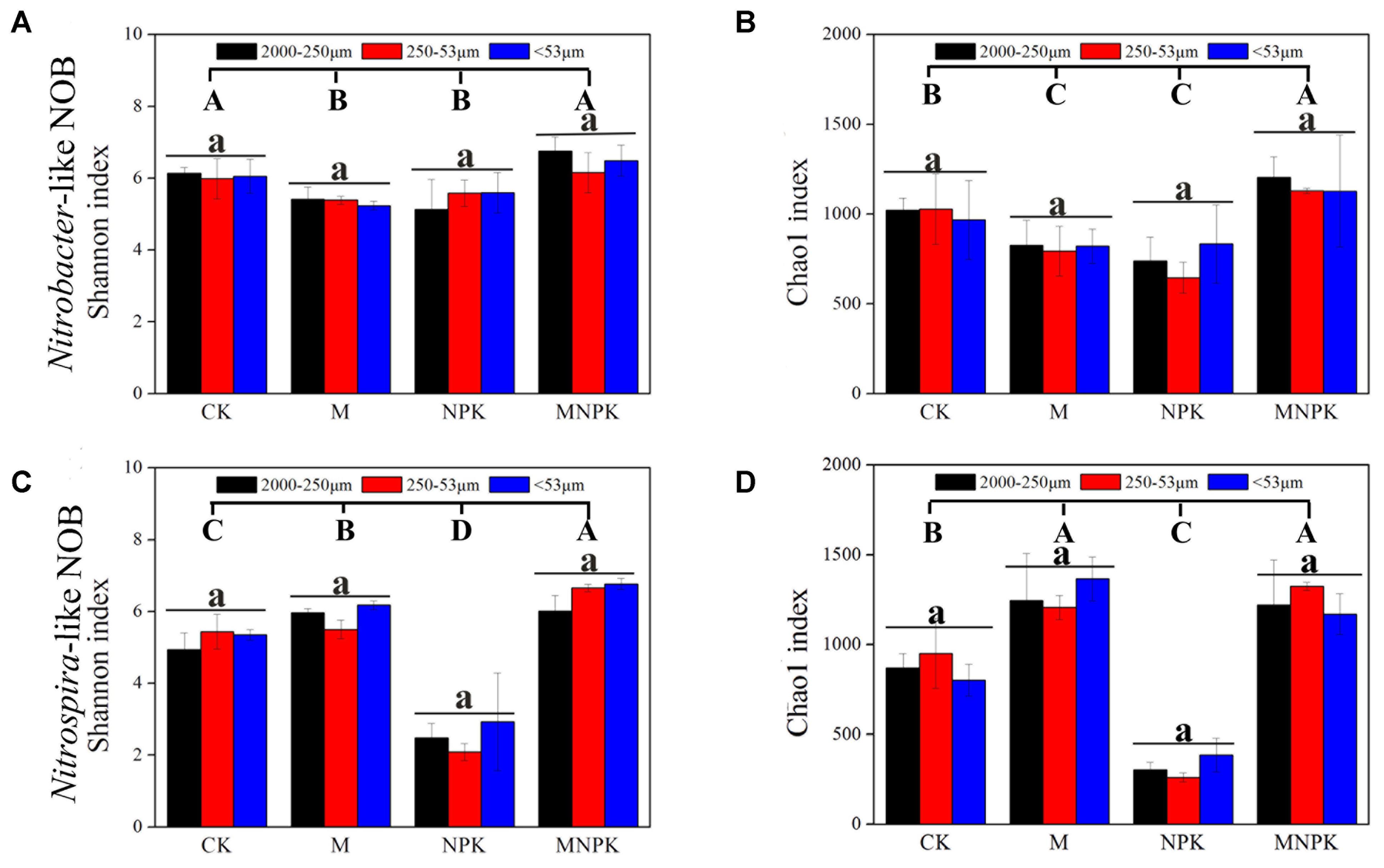
FIGURE 2. The Shannon and Chao1 diversity index of Nitrobacter (A,B) and Nitrospira (C,D). Error bars represent standard error (n = 3) and are followed by a lowercase letter indicating a significant difference among three factions within each fertilizer and the capital letter indicating a significant difference among four fertilization treatments under a same soil aggregates size according to Tukey’s test (p < 0.05).
Aggregated boosted trees analysis indicated that soil variables were scaled differently to the alpha-diversity (Shannon and Chao1 index) of Nitrobacter- and Nitrospira-like NOB (Figure 3). In particular, soil AP, TK, NH4+, and AK were the most important driving factors for Nitrobacter-like NOB Shannon and Chao1 diversity index (Figures 3A,B). However, soil NH4+, TP, AK, and AP have shown a strong effect on Nitrospira-like NOB Shannon and Chao1 diversity index (Figures 3C,D). Furthermore, soil NH4+ content independently accounted for more variance in Nitrospira (34.4%) than in Nitrobacter (17.4%) in both of diversity indexes; conversely, the explanatory variables of soil AP to Nitrobacter (26.7%) were greater than Nitrospira (14.2%).
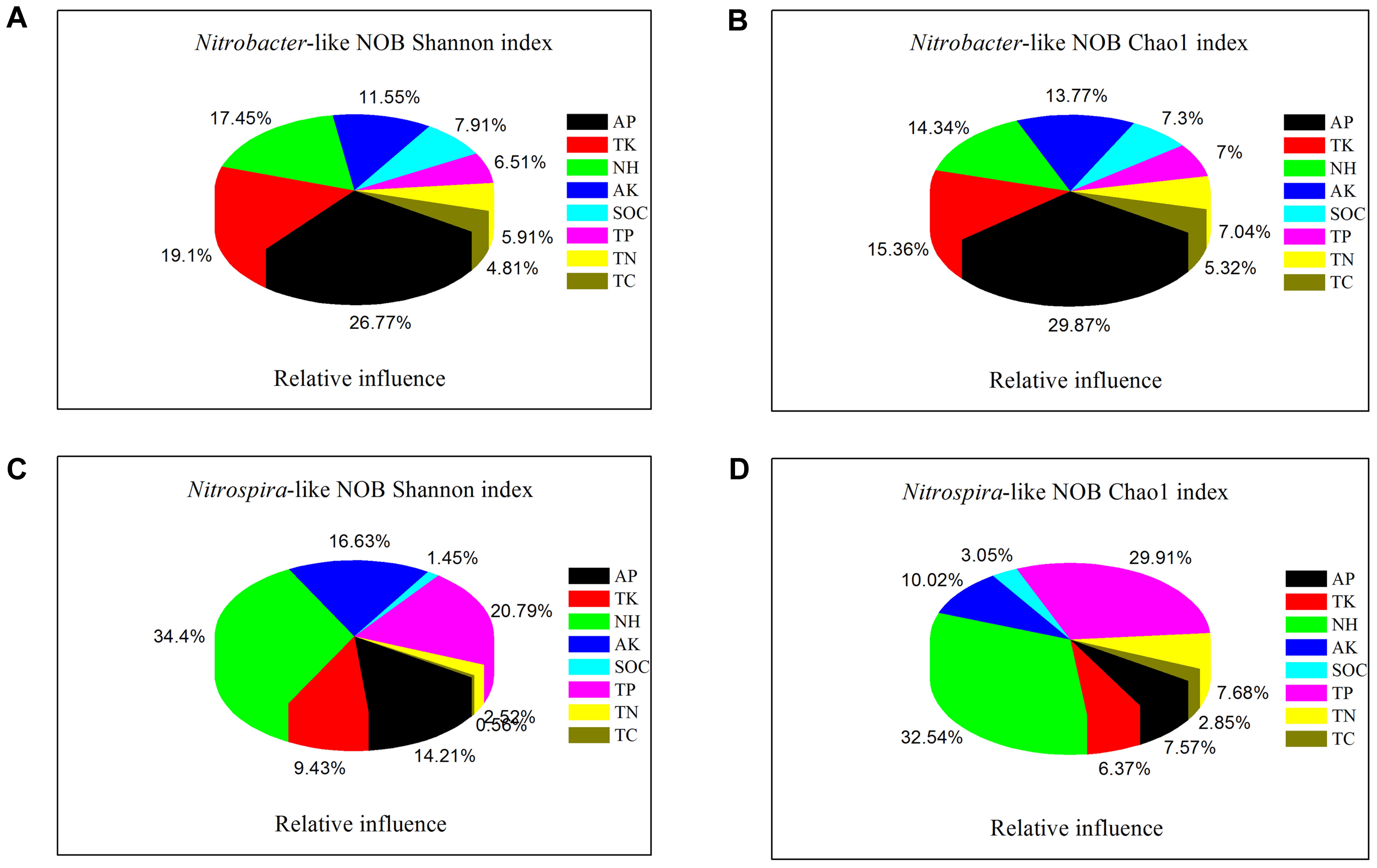
FIGURE 3. Relative influence of the driving factors for the Shannon and Chao1 diversity index of Nitrobacter (A,B) and Nitrospira (C,D) across fertilization and soil aggregate fractions by aggregated boosted trees (ABT) analysis. TC, total carbon; TN, total nitrogen; TP, total phosphorus; TK, total potassium; SOC, soil organic carbon; AP, available phosphorus; AK, available potassium; NH4+-N, ammonium nitrogen.
To monitor the resistance of alpha-diversity of nitrifiers, the resistance of the Shannon and Chao1 index was calculated at three fertilization treatments under the soil aggregate levels (Figure 4). A higher resistance (Rs > 0.7) of Shannon index was observed in Nitrobacter-like NOB (Figure 4A). However, different fertilization treatments and soil aggregate fractions have less effect on both of Shannon and Chao1 index resistance (Figures 4A,B). For Nitrospira-like NOB Shannon index resistance, the M plot has a higher resistance, followed by MNPK, and NPK has a significant (p < 0.05) and the lowest resistance compared to that of the M and MNPK treatments (Figure 4C); this outcome means that additional M and MNPK fertilizer has the capability of improving the resistance of Nitrospira. The trend of Chao1 index resistance of Nitrospira is similar to that of Shannon index (Figure 4D). In addition, soil aggregate sizes also have a small impact on both of Shannon and Chao1 index resistance of Nitrospira. Spearman’s rank coefficient correlation analysis showed that the Shannon index resistance of Nitrospira-like NOB was significantly associated with soil TN (p < 0.01), TP (p < 0.01), AK (p < 0.01), and NH4+ (p < 0.01) content, but this correlation was not detected in the Nitrobacter-like NOB (Supplementary Table S2).
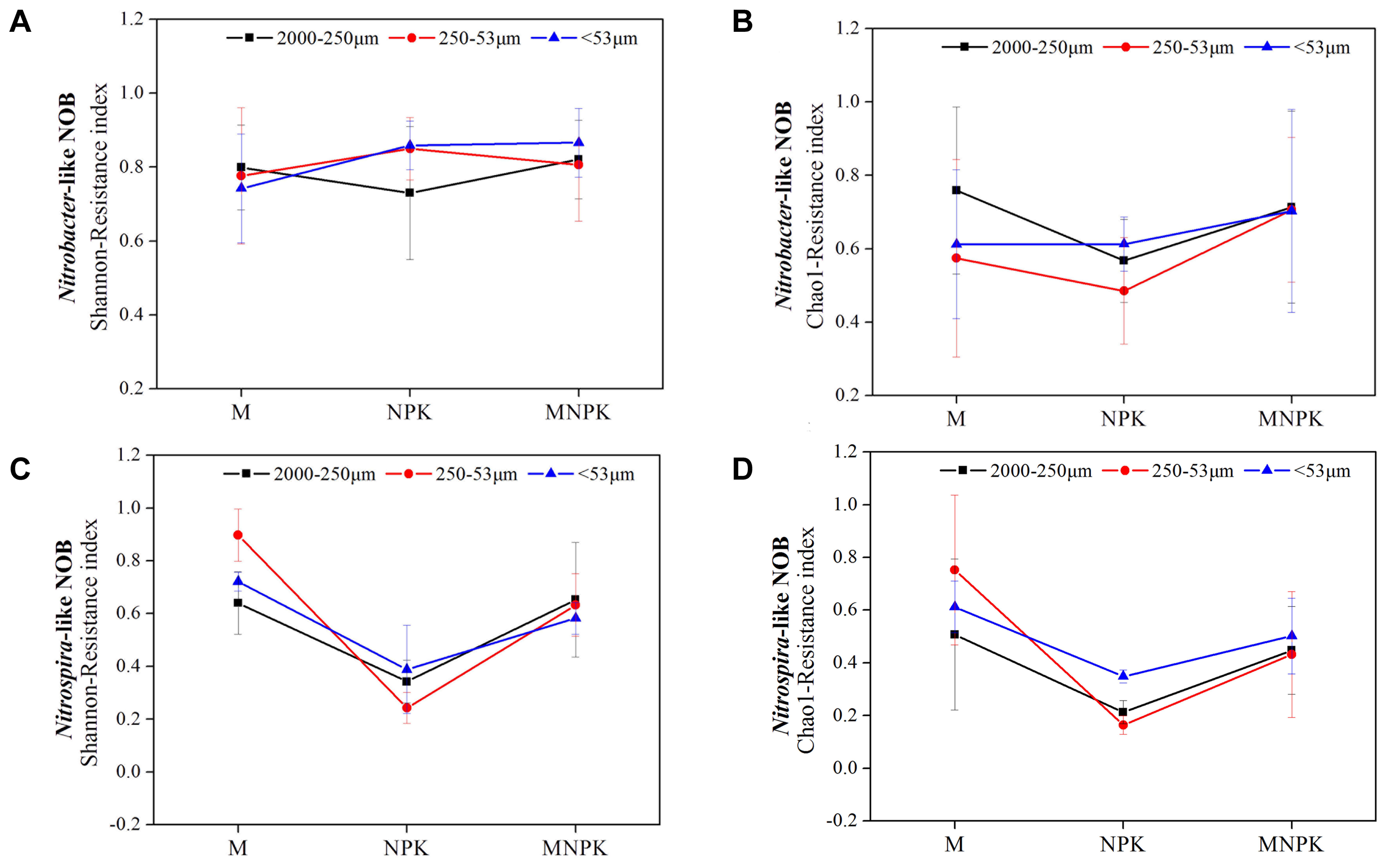
FIGURE 4. The resistance indices of Nitrobacter (A,B) and Nitrospira (C,D) Shannon and Chao1 diversity index. Error bars represent standard error (n = 3).
Principal co-ordinates analysis revealed that the Nitrobacter-like NOB community under four fertilizer regimes was well differentiated from each other along axis 1 (Figure 5). The Nitrobacter-like NOB community of CK was well separated from those of M, NPK, and MNPK along axis 2 (Figure 5A). In addition, a similar trend was also observed for the Nitrospira-like NOB community (Figure 5B). However, the soil aggregate fractions had no effect on the Nitrobacter- and Nitrospira-like NOB community composition (Figures 5C,D). MRPP analysis showed that the compositions of both Nitrobacter- and Nitrospira-like NOB community were distinctly different (p < 0.01) among fertilizer treatments across all soil aggregate sizes classes (Supplementary Table S3). However, significant differences among various soil aggregate classes were not detectable in all types of the fertilizer plots. Those results suggested that the long-term fertilization regimes were the main drivers in regulating NOB community structure. PerMANOVA results also indicated that both of Nitrobacter- and Nitrospira-like NOB community structure were significantly (p < 0.001) altered by soil fertilization treatments, and soil aggregate fractions exhibits less effects (Supplementary Table S4). Those analyses revealed that Nitrobacter- and Nitrospira-like NOB community structures were closely associated with fertilization treatments rather than soil aggregate fractions.
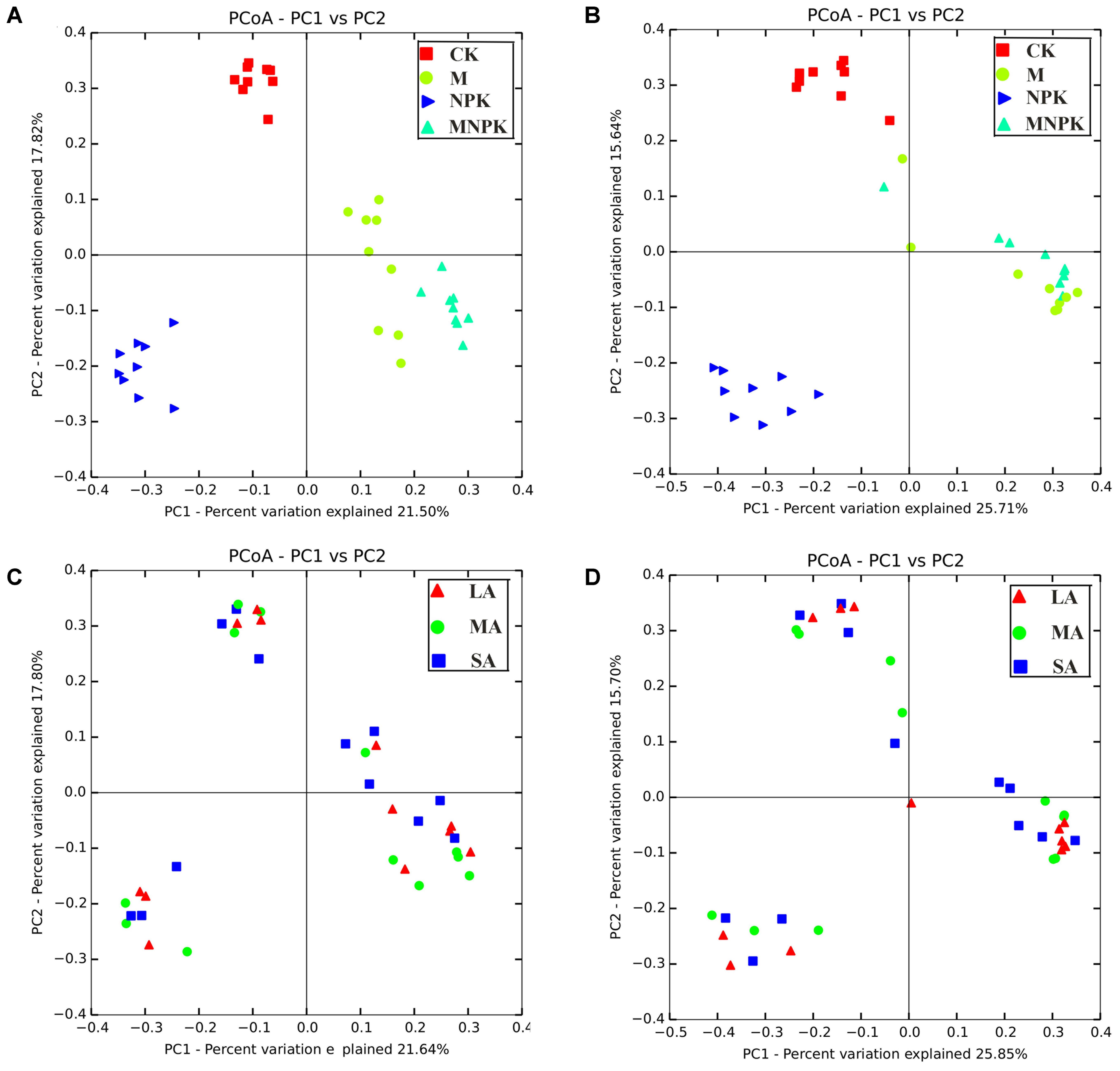
FIGURE 5. Principal co-ordinates analysis (PCoA) based on the unweighted unifrac distance showing the changes in Nitrobacter (A,C) and Nitrospira (B,D) community composition. LA, large macroaggregates (2000–250 μm), MA, microaggregates (250–53 μm), SA, silt+clay (<53 μm).
Both RDA analysis and Mantel test were performed to show the relationship between Nitrobacter- and Nitrospira-like NOB community structure and the environmental factors (Figure 6 and Table 2). The Monte Carlo test and Mantel results indicated that soil TC (p < 0.01), TN (p < 0.01), TP (p < 0.01), AP (p < 0.01), AK (p < 0.01), SOC (p < 0.01), and NH4+ (p < 0.01) content were important environmental attributes that control the Nitrobacter- and Nitrospira-like NOB community structure across different fertilization treatments in the red soil (Table 2 and Supplementary Table S5). RDA first two components explained approximately 21% of the total variation in both of the Nitrobacter- and Nitrospira-like NOB communities (Figures 6A,B). The soil Nitrobacter- and Nitrospira-like NOB communities formed distinct clusters by fertilization treatments on the RDA plots, respectively. In addition, the soil Nitrobacter- and Nitrospira-like NOB community was weakly grouped into clusters by soil aggregate fractions under the same fertilization treatments.
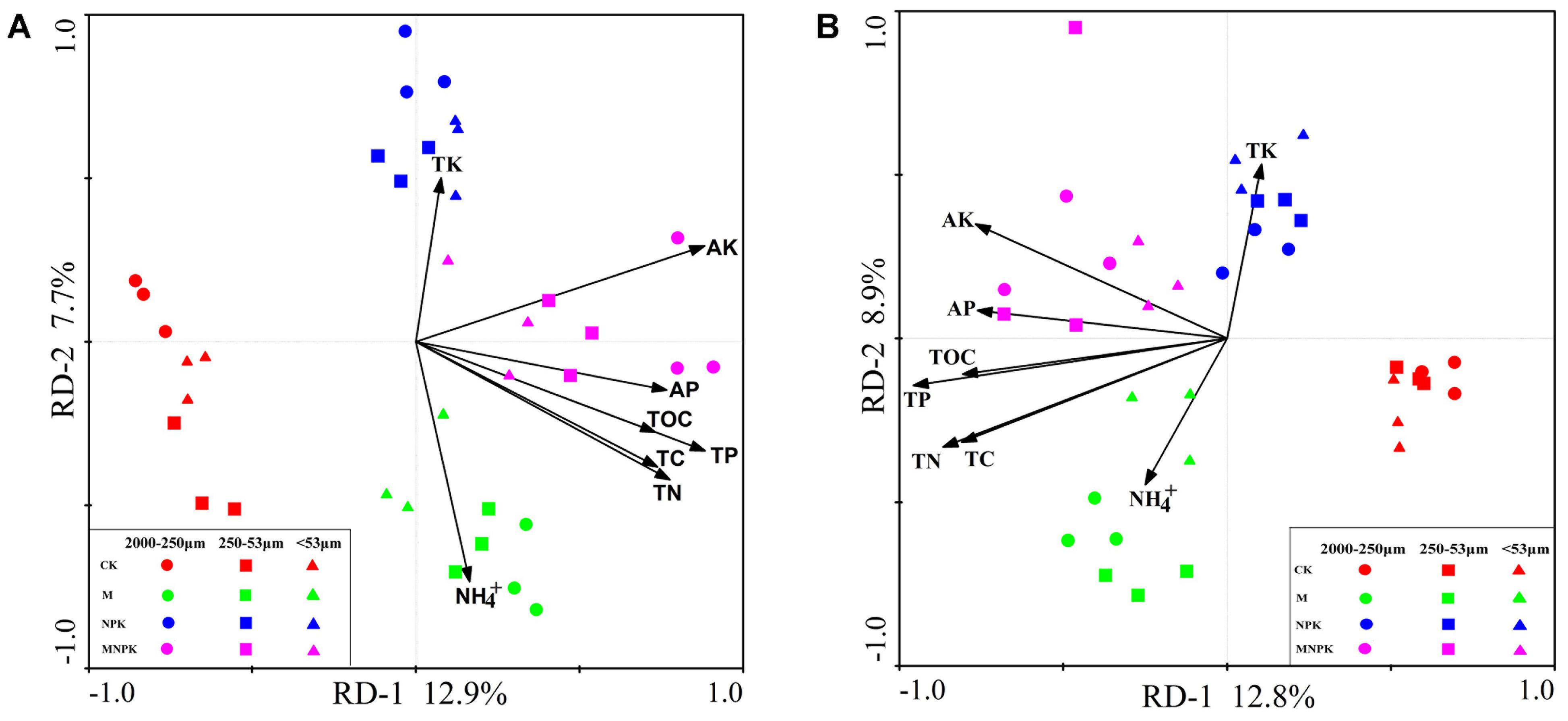
FIGURE 6. Redundancy analyses (RDA) of the correlations between soil properties and microbial Nitrobacter (A) and Nitrospira (B) community composition. TC, total carbon; TN, total nitrogen; TP, total phosphorus; TK, total potassium; SOC, soil organic carbon; AP, available phosphorus; AK, available potassium; NH4+-N, ammonium nitrogen.
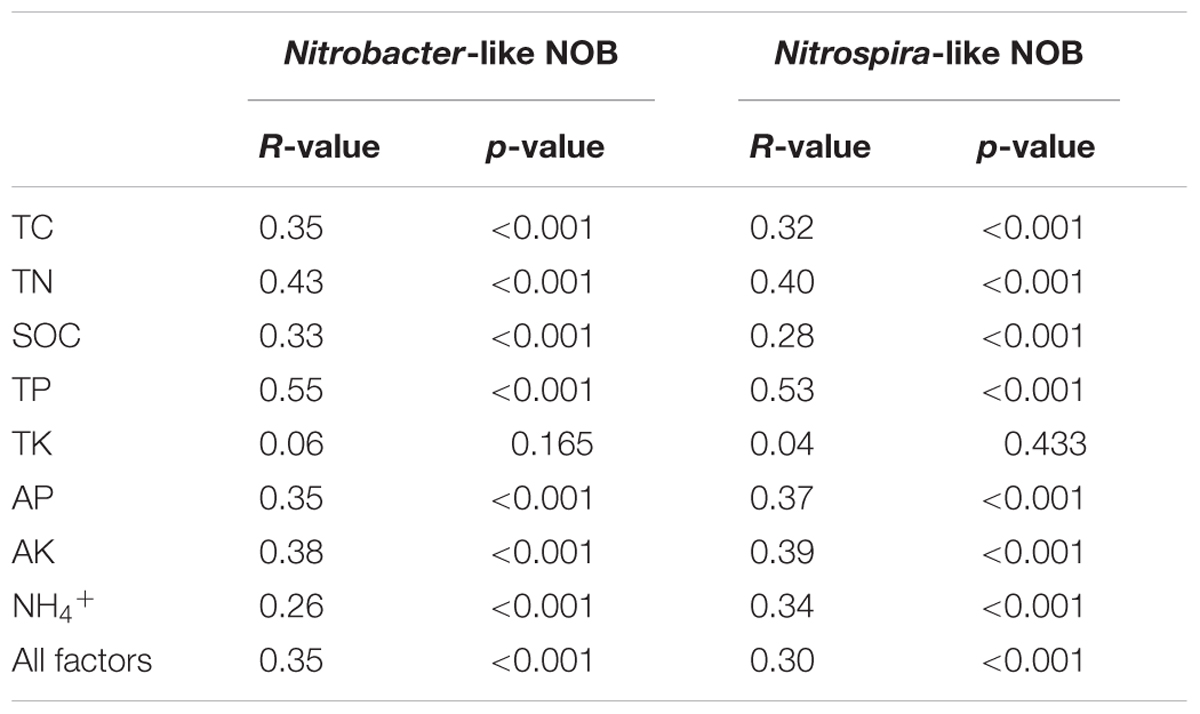
TABLE 2. Mantel test results to discern correlation between the composition of Nitrobacter- and Nitrospira-like NOB community and soil geochemical variables.
Different soil aggregate fractions represent different physical and chemical properties, which reflect the differentials of environmental factors and maintenance of distinct microbial assemblages within aggregate fraction (Davinic et al., 2012; Tiemann et al., 2015). Study on the soil aggregate levels potentially gives us a different point of view because different fractions represent different nutrient pools, which reflect the differentials of environmental factors. The main focus of this study was to construct a linkage between the Nitrobacter- and Nitrospira-like NOB community, soil aggregate fractions and fertilization treatments. Understanding these relationships further could be crucial for nitrogen biogeochemical cycling and sustainable agriculture.
Estimating the species diversity of microorganisms is of great importance in predicting, maintaining and managing microbial communities (Hubbell, 2001). We have highlighted the significant effect of environmental variation and spatial variation on the estimation of soil nitrite-oxidizing bacterial species diversity (Figure 2). Our results showed that, among soil aggregate sizes, the diversity of Nitrobacter- and Nitrospira-like NOB varies to a lesser extent, while the diversity among fertilizer regimes were clearly different from the CK plot. For Nitrobacter, the diversity was highest in the CK and MNPK treatments, with similar values in the M and NPK treatment, which indicated that M and NPK have inhibitory effects on the diversity under aggregate levels. In particular, mixed fertilizer (chemical and organic) may recovery microbial diversity for unknown reasons. In contrast, for Nitrospira, the diversity was highest in the MNPK plot, followed by M and CK, and NPK treatments was the lowest. The use of organic fertilizer may offset the loss of diversity caused by chemical fertilizers. Thus, we revealed that the addition of organic fertilizer had a greater positive impact on Nitrospira diversity and can result in increased Nitrospira diversity compared to chemical fertilization under aggregate levels. In addition, genomic data showed that comammox Nitrospira spp. potently assimilate short-chain amides (Palomo et al., 2016). It is reasonable to hypothesize that the abundance of some comammox Nitrospira spp. probably increased in the soil particles from the M and MNPK fertilizer plots. However, whether manure fertilizer can really enrich comammox in soil remains an opening question.
Freitag et al. (2005) has reported the first evidence for the influence of agricultural N management regimes on the diversity of nitrite-oxidizing bacteria but failed to note the NOBs diversity regulated by which environmental factors. In this study, ABT analysis was used to better understand how the diversity could be explained by soil variables and the specific contribution of each soil variable. Soil AP, K, and NH4+ were the most important driving factors for Nitrobacter-like NOB community diversity. However, soil NH4+, P, and AK have shown a strong effect on Nitrospira-like NOB. Furthermore, ABT analysis also indicated that the alpha-diversity of Nitrospira was more sensitive to the nitrogen variables than Nitrobacter (Figure 3). In contrast, the alpha-diversity of Nitrobacter was more sensitive to the phosphorus variables than Nitrospira. The distinct responses of the alpha-diversity of Nitrobacter- and Nitrospira-like NOB to shift in soil factors suggests different mechanisms in controlling the composition of Nitrobacter- and Nitrospira-like NOB, which are subordinate to different phylogenetic classification and life strategies.
The responses of target variables to disturbances can be quantified by a resistance index to evaluate the amount of change in response to variables caused by a disturbance as proposed by Orwin and Wardle (2004). Upon closer inspection, the alpha-diversity of Nitrobacter was more resistant to fertilizer regimes compared with the Nitrospira but was not under soil aggregate level (Figure 4). This may have attributed to the stronger environmental adaptability and higher resource utilization efficiency of the Nitrobacter community, as indicated by the lack of correlations among the diversity of Nitrobacter, the resistance index of Nitrobacter and environmental factors (Supplementary Table S2). In contrast, Nitrospira diversity was more disturbed in different fertilizer systems. However, manure fertilizer plays an important role enhancing Nitrospira diversity resistance. The distinct environmental preferences between Nitrobacter and Nitrospira may result in different responses of these functional microbial groups to fertilization treatments. We argue that knowledge on what controls soil Nitrobacter- and Nitrospira-like NOB community stability is pivotal for predicting the impacts of fertilization treatments and soil aggregate fractions on soil nitrifies community and the nitrification processes that they drive.
Organic and chemical fertilization, common agricultural practice, can significantly influence the soil aggregate size distribution, soil properties and the composition of the microbial community in the soil (Aoyama et al., 1999; Kong et al., 2005; Zhang et al., 2014, 2015). Due to the spatial and physico-chemical aspects of heterogeneous soil aggregates, soil microbes have different distribution patterns at the aggregate level (Zhang et al., 2014). Microbial community shifts were also observed with fertilizer management across soil particle-size fractions (Mummey and Stahl, 2004; Davinic et al., 2012; Lazcano et al., 2013; Zhang et al., 2017).
In this study, we found physico-chemical heterogeneity between different sizes of soil aggregates (Table 1). The characterization of the NOB community structure in soil aggregate level was well clustered together according to fertilization types (Figure 5), which indicated that fertilization treatments rather than soil aggregate sizes influence the NOB community structure in this red soil. We also observed that the fertilizer regime is a driving force of NOB community compositions. Distinctly differences in NOB community were found among fertilizer plots across the variation of soil aggregate sizes (Supplementary Table S3). Several different types of fertilizer (chemical and organic) have been added to soils to increase its nutrient content, and particularly to improve soil microenvironment fertility. Under the condition of applying organic fertilizer (M and MNPK plots), fungi can convert various organic materials into bioavailable forms in a soil ecosystem (Hoorman, 2011). It is possible that some NOB tended to use the simple organic carbon from the organic fertilizer or the organic products decomposed by fungi. Previous studies have shown that facultative/mixotrophic NOB were found to assimilate a narrow range of simple organic carbon compounds (e.g., acetate, D-lactate, pyruvate and glycerol) (Bock, 1976; Starkenburg et al., 2008). Fertilizer-induced shifts in nitrifier community composition may be explained by the direct/indirect effects of selective feeding on certain nutrient substance types as a result of the general effect of fertilizer on environmental conditions. The RDA and Mantel test indicated that soil SOC content (p < 0.01) was the important contributor to nitrifier community variation, which can partially support this explanation (Figure 6 and Table 2). Thus, we observed a greater shift in Nitrobacter- and Nitrospira-like NOB community composition in soils amended with organic materials than those only treated with chemical fertilizers and no fertilizers.
A striking result of the present study is that we found significant relationships between all the measured soil environmental factors (except TK content) and nitrifier community compositions. Interestingly, the effects of soil variables, which were significantly (p < 0.01) enhanced by fertilization (except NH4+ content), on the Nitrobacter- and Nitrospira-like NOB community responded in a uniform way (Figure 6 and Table 2). However, these measured environmental factors have low explanatory variables (20.6 and 21.7%, respectively) for Nitrobacter- and Nitrospira-like NOB communities; this outcome suggests that other uncharacterized biotic and abiotic factors affect the community structure of Nitrobacter and Nitrospira in soil aggregate size fractions, as one would expect for complicated microenvironment in red soil. Maixner et al. (2006) have shown that the nitrite concentration influences the structure of a Nitrospira-like bacterial community; however, soil aggregate nitrite is leached well-below the detection limit. Water content and oxygen availability, which are necessary for the nitrite oxidizers activity, are also the probable environmental factors that impact the nitrite-oxidizer community structure (Van Cleemput and Samater, 1996; Liu and Wang, 2013). Han et al. (2017) showed that the Nitrospira community changes were significantly shaped by the soil pH, whereas that of the Nitrobacter community was not in a rapeseed-rice rotation field trial. Also, distinct differences in Nitrospira community were found between the straw fertilizer and no straw. However, no obvious differences in the Nitrobacter were detectable. Because the water-stable soil aggregates are separated by wet-sieving method, the soil aggregate pH was not able to be accurately measured and thus not determined in this study. Therefore, in addition to the abiotic factors, the synergistic or competitive relationship between nitrite oxidizers and other microorganisms (e.g., anaerobic ammonium oxidizing bacteria) would also contribute to modulating the NOB community structure.
We examined the effect of fertilizer regimes and soil aggregate fractions on soil nitrite oxidizers microbial community structure, diversity and resistance by focusing on functional genes. High throughput sequencing data indicated that the addition of M and NPK significantly decreased Nitrobacter diversity, while M and MNPK remarkably increased Nitrospira diversity, and NPK considerably decreased Nitrospira diversity, with the greatest diversity achieved in soils amended with MNPK. However, the soil aggregate fractions had no impact on the diversity of Nitrobacter and Nitrospira under the fertilizer treatment. Moreover, changes in diversity were largely attributed to soil phosphorus, potassium and NH4+ content. Fertilization treatments have more and less effect on the Nitrobacter- and Nitrospira-like NOB community diversity resistance index, respectively, but not soil aggregate fractions. Importantly, fertilizer regimes had significant influences on the composition and structure of soil nitrite oxidizers community; conversely, soil aggregate fractions had less effect. In addition, soil Nitrobacter- and Nitrospira-like NOB community structure were driven by nitrogen, carbon, phosphorus and available potassium content across different fertilization treatments under aggregate levels in the red soil. This work represents an important step forward in understanding nitrite oxidizers in soil aggregate levels under long-term fertilization.
WC and QH designed the study. SH and XiL conducted the experiment and analyzed the data. WC and SW collected the soil samples. SW conducted the field experiments. SH and XuL wrote the initial draft of the manuscript. WC, QH, SH, XiL, XuL, and SW contributed to revising the manuscript.
This work was supported by the National Basic Research Program of China (973, Grant No. 2015CB150504), and the Fundamental Research Funds for the Central Universities (Project Nos. 2662015PY016 and 2662015PY116). Sequencing service was provided by Personal Biotechnology Co., Ltd., Shanghai, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00885/full#supplementary-material
Alawi, M., Lipski, A., Sanders, T., Pfeiffer, E. M., and Spieck, E. (2007). Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 1, 256–264. doi: 10.1038/ismej.2007.34
Aoyama, M., Angers, D. A., and N’dayegamiye, A. (1999). Particulate and mineral-associated organic matter in water-stable aggregates as affected by mineral fertilizer and manure applications. Can. J. Soil Sci. 79, 295–302. doi: 10.4141/S98-049
Attard, E., Poly, F., Commeaux, C., Laurent, F., Terada, A., Smets, B. F., et al. (2010). Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326. doi: 10.1111/j.1462-2920.2009.02070.x
Bartosch, S., Hartwig, C., Spieck, E., and Bock, E. (2002). Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43, 26–33. doi: 10.1007/s00248-001-0037-5
Bertagnolli, A. D., McCalmont, D., Meinhardt, K. A., Fransen, S. C., Strand, S., Brown, S., et al. (2016). Agricultural land usage transforms nitrifier population ecology. Environ. Microbiol. 6, 1918–1929. doi: 10.1111/1462-2920.13114
Blackburne, R., Vadivelu, V. M., Yuan, Z., and Keller, J. (2006). Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res. 41, 3033–3042. doi: 10.1016/j.watres.2007.01.043
Bock, E. (1976). Growth of Nitrobacter. Presence of organic matter.2. Chemo-organotrophic growth of Nitrobacter-Agilis. Arch. Microbiol. 108, 305–312. doi: 10.1007/BF00454857
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K. H., and Wagner, M. (2001). In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001
Davinic, M., Fultz, L. M., Acosta-Martinez, V., Calderón, F. J., Cox, S. B., Dowd, S. E., et al. (2012). Pyrosequencing and mid-infrared spectroscopy reveal distinct aggregate stratification of soil bacterial communities and organic matter composition. Soil Biol. Biochem. 46, 63–72. doi: 10.1016/j.soilbio.2011.11.012
Delgado-Baquerizo, M., Maestre, F. T., Escolar, C., Gallardo, A., Ochoa, V., Gozalo, B., et al. (2014). Direct and indirect impacts of climate change on microbial and biocrust communities alter the resistance of the N cycle in a semiarid grassland. J. Ecol. 102, 1592–1605. doi: 10.1111/1365-2745.12303
Elliot, E. T. (1986). Aggregate structure and carbon, nitrogen and phosphorus in native and cultivated soils. Soil Sci. Soc. Am. J. 50, 518–524.
Freitag, T. E., Chang, L., Clegg, C. D., and Prosser, J. I. (2005). Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl. Environ. Microbiol. 71, 8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005
Gould, G. W., and Lees, H. (1960). The isolation and culture of the nitrifying organisms: part I. Nitrobacter. Can. J. microbiol. 6, 299–307. doi: 10.1139/m60-034
Gubry-Rangin, C., Nicol, G. W., and Prosser, J. I. (2010). Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 74, 566–574. doi: 10.1111/j.1574-6941.2010.00971.x
Han, S., Luo, X. S., Liao, H., Nie, H. L., Huang, Q. Y., and Chen, W. L. (2017). Nitrospira are more sensitive than Nitrobacter to land management in acid, fertilized soils of a rapeseed-rice rotation field trial. Sci. Total Environ. 599, 135–144. doi: 10.1016/j.scitotenv.2017.04.086
Hayatsu, M., Tago, K., Uchiyama, I., Toyoda, A., Wang, Y., Shimomura, Y., et al. (2017). An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 11, 1130–1141. doi: 10.1038/ismej.2016.191
He, J. Z., Shen, J. P., Zhang, L. M., Zhu, Y. G., Zheng, Y. M., Xu, M. G., et al. (2007). Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and archaea of a Chinese upland red soil under long term fertilization practices. Environ. Microbiol. 9, 2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x
Hoorman, J. J. (2011). The Role of Soil Fungus. Ohio State University Extension, Agriculture and Natural Resources SAG. Columbus, OH: Ohio State University.
Hubbell, S. P. (2001). A Unified Neutral Theory of Biodiversity and Biogeography. Princeton, NJ: Princeton University Press.
Jastrow, J. D. (1996). Soil aggregate formation and the accrual of particulate and mineral associated organic matter. Soil Biol. Biochem. 28, 665–676. doi: 10.1016/0038-0717(95)00159-X
Jiang, Y., Jin, C., and Sun, B. (2014). Soil aggregate stratification of nematodes and ammonia oxidizers affects nitrification in an acid soil. Environ. Microbiol. 16, 3083–3094. doi: 10.1111/1462-2920.12339
Ke, X., Angel, R., Lu, Y., and Conrad, R. (2013). Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ. Microbiol. 15, 2275–2292. doi: 10.1111/1462-2920.12098
Kim, D. J., and Kim, S. H. (2006). Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res. 40, 887–894. doi: 10.1016/j.watres.2005.12.023
Kong, A. Y. Y., Six, J., Bryant, D. C., Denison, R. F., and van Kessel, C. (2005). The relationship between carbon input, aggregation, and soil organic carbon stabilization in sustainable cropping systems. Soil Sci. Soc. Am. J. 69, 1078–1085. doi: 10.2136/sssaj2004.0215
Kowalchuk, G. A., and Stephen, J. R. (2001). Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55, 485–529. doi: 10.1146/annurev.micro.55.1.485
Lanyon, L. E., and Heald, W. R. (1982). “Magnesium, calcium, strontium and barium,” in Methods of Soil Analysis, 22nd Edn, eds A. L. Page, R. H. Miller, and D. R. Keeney (Madison WI: American Society of Agronomy), 247–262.
Lazcano, C., Gómez-Brandón, M., Revilla, P., and Domínguez, J. (2013). Short-term effects of organic and inorganic fertilizers on soil microbial community structure and function. Biol. Fert. Soils 49, 723–733. doi: 10.1007/s00374-012-0761-7
Le Roux, X., Poly, F., Currey, P., Commeaux, C., Hai, B., Nicol, G. W., et al. (2008). Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J. 2, 221–231. doi: 10.1038/ismej.2007.109
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Liu, G., and Wang, J. (2013). Long-term low DO enriches and shifts nitrifier community in activated sludge. Environ. Sci. Technol. 47, 5109–5117. doi: 10.1021/es304647y
Luo, X. S., Han, S., Lai, S. S., Huang, Q. Y., and Chen, W. L. (2017). Long-term straw returning affects Nitrospira-like nitrite oxidizing bacterial community in a rapeseed-rice rotation soil. J. Basic Microbiol. 57, 309–315. doi: 10.1002/jobm.201600400
Maixner, F., Noguera, D. R., Anneser, B., Stoecker, K., Wegl, G., Wagner, M., et al. (2006). Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ. Microbiol. 8, 1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x
Mummey, D. L., and Stahl, P. D. (2004). Analysis of soil whole-and inner-microaggregate bacterial communities. Microb. Ecol. 48, 41–50. doi: 10.1007/s00248-003-1000-4
Nowka, B., Daims, H., and Spieck, E. (2015). Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753. doi: 10.1128/AEM.02734-14
Ollivier, J., Schacht, D., Kindler, R., Groeneweg, J., Engel, M., Wilke, B. M., et al. (2013). Effects of repeated application of sulfadiazine-contaminated pig manure on the abundance and diversity of ammonia and nitrite oxidizers in the root-rhizosphere complex of pasture plants under field conditions. Front. Microbiol. 4:22. doi: 10.3389/fmicb.2013.00022
Olsen, S. R., Cole, C. V., Watanabe, F. S., and Dean, L. A. (1954). Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate. Washington, DC: US Department of Agriculture, 19.
Orwin, K. H., and Wardle, D. A. (2004). New indices for quantifying the resistance and resilience of soil biota to exogenous disturbances. Soil Biol. Biochem. 36, 1907–1912. doi: 10.1016/j.soilbio.2004.04.036
Orwin, K. H., Wardle, D. A., and Greenfield, L. G. (2006). Context-dependent changes in the resistance and resilience of soil microbes to an experimental disturbance for three primary plant chronosequences. Oikos 112, 196–208. doi: 10.1111/j.0030-1299.2006.13813.x
Palomo, A., Fowler, S. J., Gülay, A., Rasmussen, S., Sicheritz-Ponten, T., and Smets, B. F. (2016). Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 10, 2569–2580. doi: 10.1038/ismej.2016.63
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lucker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071. doi: 10.1111/1462-2920.12300
Prosser, J. I. (1989). Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30, 125–181. doi: 10.1016/S0065-2911(08)60112-5
Sessitsch, A., Weilharter, A., Gerzabek, M. H., Kirchmann, H., and Kandeler, E. (2001). Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67, 4215–4224. doi: 10.1128/AEM.67.9.4215-4224.2001
Six, J., Paustian, K., Elliott, E. T., and Combrink, C. (2000). Soil structure and organic matter. I. Distribution of aggregate-size classes and aggregate-associated carbon. Soil Sci. Soc. Am. J. 64, 681–689. doi: 10.2136/sssaj2000.642681x
Sorokin, D. Y., Lücker, S., Vejmelkova, D., Kostrikina, N. A., Kleerebezem, R., Rijpstra, W. I. C., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256. doi: 10.1038/ismej.2012.70
Starkenburg, S. R., Arp, D. J., and Bottomley, P. J. (2008). D-Lactate metabolism and the obligate requirement for CO2 during growth on nitrite by the facultative lithoautotrop Nitrobacter hamburgensis. Microbiology 8, 2473–2481. doi: 10.1099/mic.0.2008/018085-0
Tiemann, L. K., Grandy, A. S., Atkinson, E. E., Marin-Spiotta, E., and McDaniel, M. D. (2015). Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecol. Lett. 18, 761–771. doi: 10.1111/ele.12453
Tisdall, J. M., and Oades, J. (1982). Organic matter and water-stable aggregates in soils. J. Soil Sci. 33, 141–163. doi: 10.1111/j.1365-2389.1982.tb01755.x
Van Cleemput, O., and Samater, A. H. (1996). Nitrite in soils: accumulation and role in the formation of gaseous N compounds. Fert. Res. 45, 81–89. doi: 10.1007/BF00749884
van Kessel, M. A., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Venterea, R. T., Clough, T. J., Coulter, J. A., and Breuillin-Sessoms, F. (2015). Ammonium sorption and ammonia inhibition of nitrite-oxidizing bacteria explain contrasting soil N2O production. Sci. Rep. 5:12153. doi: 10.1038/srep20720
Ward, B. B., Arp, D. J., and Klotz, M. G. (2011). Nitrification. Washington, DC: American Society for Microbiology Press.
Watson, S. W., and Waterbury, J. B. (1971). Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen.nov. sp. and Nitrococcus mobilis nov. gen.nov.sp. Arch. Microbiol. 77, 203–230. doi: 10.1007/BF00408114
Webster, G., Embley, T. M., Freitag, T. E., Smith, Z., and Prosser, J. (2005). Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ. Microbiol. 7, 676–684. doi: 10.1111/j.1462-2920.2005.00740.x
Zhang, H., Ding, W., He, X., Yu, H., Fan, J., and Liu, D. (2014). Influence of 20-year organic and inorganic fertilization on organic carbon accumulation and microbial community structure of aggregates in an intensively cultivated sandy loam soil. PLoS One 9:e92733. doi: 10.1371/journal.pone.0092733
Zhang, L. M., Hu, H. W., Shen, J. P., and He, J. Z. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045. doi: 10.1038/ismej.2011.168
Zhang, Q., Liang, G., Myrold, D. D., and Zhou, W. (2017). Variable responses of ammonia oxidizers across soil particle-size fractions affect nitrification in a long-term fertilizer experiment. Soil Biol. Biochem. 105, 25–36. doi: 10.1016/j.soilbio.2016.11.005
Keywords: nitrite oxidizers, Nitrobacter, Nitrospira, fertilizer regimes, soil aggregate fractions
Citation: Han S, Li X, Luo X, Wen S, Chen W and Huang Q (2018) Nitrite-Oxidizing Bacteria Community Composition and Diversity Are Influenced by Fertilizer Regimes, but Are Independent of the Soil Aggregate in Acidic Subtropical Red Soil. Front. Microbiol. 9:885. doi: 10.3389/fmicb.2018.00885
Received: 14 January 2018; Accepted: 17 April 2018;
Published: 08 May 2018.
Edited by:
Jeanette M. Norton, Utah State University, United StatesReviewed by:
Brett Mellbye, Oregon State University, United StatesCopyright © 2018 Han, Li, Luo, Wen, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenli Chen, d2xjaGVuQG1haWwuaHphdS5lZHUuY24= Qiaoyun Huang, cXlodWFuZ0BtYWlsLmh6YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.