
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 May 2018
Sec. Microbiotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00864
This article is part of the Research TopicEffects of Plant-Microbiome Interactions on Phyto- and Bio-Remediation CapacityView all 12 articles
Although plants of the genus Pennisetum can accelerate the removal of atrazine from its rhizosphere, the roles played by this plant in adjusting the soil environment and soil microorganism properties that might contribute to pollutant removal are incompletely understood. We selected Pennisetum americanum (L.) K. Schum (P. americanum) as the test plant and investigated the interaction between P. americanum and atrazine-contaminated soil, focusing on the adjustment of the soil biochemical properties as well as bacterial functional and community diversity in the rhizosphere using Biolog EcoPlates and high-throughput sequencing of the 16S rRNA gene. The results demonstrate that the rhizosphere soil of P. americanum exhibited higher catalase activity, urease activity and water soluble organic carbon (WSOC) content, as well as a suitable pH for microorganisms after a 28-day incubation. The bacterial functional diversity indices (Shannon and McIntosh) for rhizosphere soil were 3.17 ± 0.04 and 6.43 ± 0.86 respectively, while these indices for non-rhizosphere soil were 2.95 ± 0.06 and 3.98 ± 0.27. Thus, bacteria in the P. americanum rhizosphere exhibited better carbon substrate utilization than non-rhizosphere bacteria. Though atrazine decreased the richness of the soil bacterial community, rhizosphere soil had higher bacterial community traits. For example, the Shannon diversity indices for rhizosphere and non-rhizosphere soil were 5.821 and 5.670 respectively. Meanwhile, some bacteria, such as those of the genera Paenibacillus, Rhizobium, Sphingobium, and Mycoplana, which facilitate soil nutrient cycling or organic pollutants degradation, were only found in rhizosphere soil after a 28-day remediation. Moreover, redundancy analysis suggests that the soil biochemical properties that were adjusted by the test plant exhibited correlations with the bacterial community composition and functional diversity. These results suggest that the soil environment and bacterial properties could be adjusted by P. americanum during phytoremediation of atrazine-contaminated soil.
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is one of the most widely used herbicides in agriculture. It is primarily applied to control broadleaf weeds in the crops such as maize, sorghum and sugar cane. Though atrazine has been proved highly persistent in the environment with the reported half-life ranges between 10 and 5824 days (Salazar-Ledesma et al., 2018), it also could be metabolized in environment according to microbiological degradation and some kinds of physicochemical process (Sun et al., 2010; Roustan et al., 2014). As a result, atrazine and its metabolites are the most commonly detected pesticide contaminants in groundwater and surface water due to their mobility in soil (Yang et al., 2014). In addition, atrazine has been classified as a priority pollutant since many researches proved it could affect the endocrine system of various kinds of organisms (Lasserre et al., 2009). In addition, there were also some other reports reveal that atrazine also could cause obvious toxic affection on the microorganisms in soil (Muñoz-Leoz et al., 2011; Imfeld and Vuilleumier, 2012). As a result, the soil nutrients cycling, as well as soil physical and chemical properties were also affected simultaneously. Consequently, the toxicity of atrazine has raised serious concerns and innovative strategies for remediating atrazine-contaminated soils are critically needed (Lima et al., 2009; Pandey et al., 2009).
In recent years, phytoremediation has aroused increasing concern in the field of contaminated soil remediation (Hamdi et al., 2012; Wang et al., 2012; Albright et al., 2013; Jagtap et al., 2014). Unlike the conventional physical or chemical technologies for soil remediation, which have the disadvantages of high economic costs, formulation of secondary contaminants and damage to soil organisms, phytoremediation has been considered a cost-effective, environmentally friendly strategy to solve soil contamination (Agnello et al., 2016; Gil-Díaz et al., 2016). Several literatures have been reported that phytoremediation can decrease the residual level of organic contaminates in soil by the interaction between plant roots and the specific microorganisms harbored in the rhizosphere (Cai et al., 2010). The plant-stimulated bioremediation of organic pollutants by rhizospheric microorganisms described above is also termed rhizoremediation (Hussain et al., 2018). Rhizoremediation is the major mechanism for phytoremediation of organic-polluted soil because it stimulates the population growth and activity of degrading microorganisms around roots through the rhizosphere effect (Truu et al., 2015). It is well known that the plant roots can create a nutrient-rich micro-environment for pollutants-degrading microbes, as well as that the microorganisms in rhizosphere can enhance plant growth by providing plant nutrients and protection against the stress caused by contaminants (Hussain et al., 2018; Lacalle et al., 2018). Therefore, the rhizosphere has long been considered as the most biologically active microsites in soil and the organic compounds are degraded here by the stimulated microbial biomass and the activity that is part of the rhizosphere effect (Liu et al., 2014).
Root exudates, which consist of low-molecular-weight carbohydrates, amino acids and organic polymers, can be used as an energy and carbon source by soil microbes during their metabolic processes (Martin et al., 2014). Therefore, root exudation is considered as a potential driving force for stimulated rhizoremediation and is the most important factor affecting microbial drift in the rhizosphere (Hussain et al., 2018). The distinct microorganism community shifts in contaminated soils mentioned above suggest the alteration of microbial catabolic activity and selection of specific microbial strains. Therefore, it is widely considered that root exudates play a distinctive role in shaping the rhizosphere microbiome in polluted soil (Thijs et al., 2016).
The selection of a suitable plant species is an indispensable component of phytoremediation success (Lacalle et al., 2018). It is widely supposed that a plant with the potential for organic pollution remediation may selectively enrich the specific pollutant degraders harbored in rhizosphere by releasing a variety of root exudates (Fang et al., 2001). In addition, the different plant select for certain microorganism or sharpen characteristics, and the types of microorganism that thrive on the rhizoplane mainly depend on the plant types (Chen et al., 2016). This is mainly because the constituents or the concentrations of the root exudates might different between the various species of plants (El Amrani et al., 2015). Therefore, further investigation of the distinct microorganism community or activity stimulating traits by the selected plant used for rhizoremediation is essential to illustrate the phytoremediation mechanism of contaminated soil.
The genus Pennisetum has been useful for remediation of soils contaminated with atrazine (Singh et al., 2004). However, little information about the interaction between the Pennisetum genus plant roots and the soil microorganisms, especially how this plant affects the soil biochemical properties or bacterial functional diversity and community structure, is available. The objectives of this study were (1) to assess the impact of the Pennisetum americanum (L.) K. Schum (P. americanum) rhizosphere on atrazine-contaminated soil biochemical properties, and (2) to investigate the variations in the composition, diversity, and functions of bacterial community across the P. americanum root-associated compartments, and (3) to examine which environmental factors or soil biochemical properties are important in shaping the structure and carbon substrates utilization diversity of soil bacterial community. All the results could help us better understanding the importance of the interaction between P. americanum roots and the soil microorganism during atrazine-polluted soil phytoremediation.
The soil used in this research was collected from a farmland in Harbin, Heilongjiang Province which is located in the black soil region of northeast China. The sampled soil was air dried, passed through a 2-mm sieve and detected to be no atrazine. Total organic carbon, ammonium nitrogen, rapidly available phosphorus, rapidly available potassium and pH which are the basic properties of the soil were 20.04 g kg-1, 89.33 mg kg-1, 70.15 mg kg-1, 501.00 mg kg-1 and 6.28, respectively.
To illustrate the regulatory role of the P. americanum rhizosphere on bacterial functional diversity and the bacterial community structure of atrazine-contaminated soil, three treatments were set up: (1) soil without any addition of atrazine or P. americanum, which served as the control treatment (CK); (2) soil without planting of P. americanum but with addition of 20 mg kg-1 atrazine, which served as pollution treatment (PT); (3) soil with planting of P. americanum and addition of 20 mg kg-1 atrazine as phytoremediation treatment (RT). To separate the non-rhizosphere soil (RN) from the rhizosphere soil (R) in the phytoremediation treatment (RT), a rhizobox was used (Figure 1). A 23 μm nylon mesh was used to divide the rhizobox (100 mm length × 100 mm width × 85 mm height) into three sections: the rhizosphere zone, which was in the center of the rhizobox (34 mm in width), and the two non-rhizosphere zones, which were located on the left and right sides of the rhizobox (each 33 mm in width). Four hundred twenty grams of air-dried soil was put in the rhizobox. In the phytoremediation treatment, P. americanum was only sown in the central zone. In addition, plastic pot of the same size containing identical amounts of soil were used in the CK and PT treatments, but without dividing these boxes into three parts and without sowing the seeds.
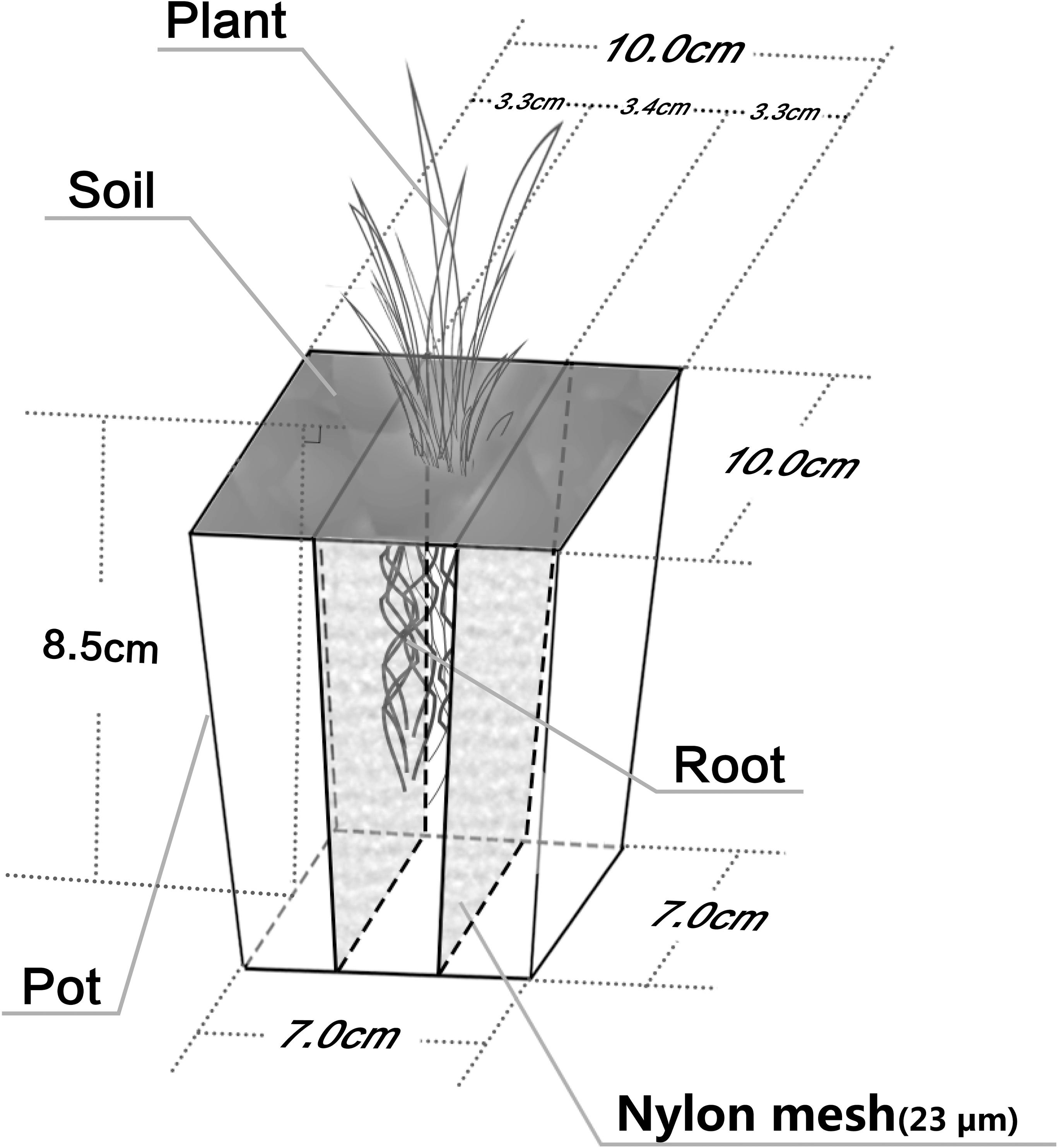
FIGURE 1. Rhizobox used in phytoremediation treatment (RT). The rhizobox was divided into three parts by a 23 μm nylon mesh. The test plant was only sowed in the middle part and this part represents rhizosphere zone. Additionally, the other two parts in the two side of the rhizobox without sowing test plant were designed as non-rhizosphere.
To prepare the soil used for the treatments of PT and RT, atrazine was dissolved in acetone, followed by being completely mixed with a small part of the soil, and then the spiked soil was put in the fume hood to make acetone vaporize thoroughly. Finally, the soil was mixed into a large amount of the soil homogeneously. The final atrazine concentration in the soil was 20 mg kg-1 (dry weight).
Seeds of P. americanum were soaked in distilled water for 5 h and then were surface-sterilized in 30% H2O2 solution for 10 min. Afterward, the sterilized seeds were rinsed several times with deionized water and were placed in a culture dish with moist filter paper for germination overnight at 28°C. The germinated seeds were sown in the rhizosphere zone of the rhizobox mentioned above. Pot experiments were performed in a greenhouse. The temperature of the greenhouse was kept at 27 ± 1°C during the day and 20 ± 1°C during the night. All the samples were watered with distilled water every 2 days to keep the plants at approximately 50% of the water holding capacity. Three replicates were conducted for each treatment mentioned above.
The day of sowing the tested plant seeds described in Section “Experimental Design and Compartmented System” was set as day 0. The soil of CK and PT was sampled on day 0 and the collected samples were named CK0 and PT0, respectively. Additionally, the soil of CK, PT, RN and R were sampled when a 28-day cultural period (as described above) was finished and the collected samples were marked as CK28, PT28, RN28 and R28, respectively. Each sample mentioned above was divided into two sets. One was stored at -20°C until soil microbial community structure and function assessment, and the other was stored at 4°C to measure atrazine concentration and other soil characteristics, such as pH, Eh, catalase activity and urease activity, water-soluble organic carbon and microbial biomass carbon.
Some typical physicochemical properties and the microbial biomass carbon of the soil samples mentioned above were detected by the methods described below. (1) Soil pH was determined in water (1:2.5, soil/water) with a pH meter (Rex PHS-3C, China). (2) Soil Eh was measured with an ORP electrode (Rex 501, China). (3) The water- soluble organic carbon (WSOC) was measured using a TOC analyzer (Shimadzu TOC-VCPN, Japan) according to the method reported by Nan et al. (2016). Soil samples (6 g) were shaken with distilled water (ratio of 1:5, w/v) for 1 h at 25°C and 180 r min-1 and centrifuged (4500 rpm for 5 min), and the supernatants were filtered through 0.45 μm filter membrane. The extract was analyzed. (4) Soil catalase activity was determined by measuring the hydrogen peroxide (H2O2)-catalyzing ability when soil was incubated in H2O2 solution. Two grams fresh soil was added to 40 mL hydrogen peroxide solution (0.03%, w/v) and cultured at 37°C, 150 r min-1 for 30 min. The enzymatic reaction was stopped by adding 5 mL of 3.0 M H2SO4. Then, 25 mL filtrate was titrated by 0.1 M KMnO4 and the soil catalase activity was calculated basing on the change in H2O2 concentration as reported by Cao et al. (2015). (5) Soil urease activity was determined by a sodium phenolate and sodium hypochlorite spectrophotometry. Five grams of soil (wet weight) was placed in tested tube and 1 mL toluene, 10 mL urea (10%, w/v) and 20 mL citrate buffer (pH = 6.7). The mixture was cultured at 37°C for 24 h, then the solution was filtered and measured by spectrophotometer (Shimadzu UV-1800, Japan) at the wavelength of 578 nm (Cao et al., 2015). (6) The soil microbial biomass carbon (MBC) was measured by the chloroform-fumigation-extraction method (Vance et al., 1987). The extracted organic C was determined using the TOC analyze and a KEC of 0.45 was used to convert the difference between the organic C extracted with 0.5 M K2SO4 from the chloroform fumigated and unfumigated soil samples.
Biolog EcoPlates (MicroPlate., BIOLOG Inc., Hayward, CA, United States) were employed to study the microbial physiological metabolic characteristics. Four grams of soil (wet weight) was added to 36 mL of sterilized 0.85% NaCl/water solution. Tenfold serial dilutions were made and the 10-3 dilution was added into the Biolog EcoPlates. Then, the plates were cultured at 25 ± 1°C in the incubator in the dark avoided light for 7 days. Color development in the plates was recorded with an automated microplate reader (Biotek Epoch, United States) every 24 h at 590 nm. Plate readings at 96 h of incubation were used to calculate the average well color development (AWCD), Shannon index (H′), Simpson index(D) and McIntosh index(U), since 96 corresponded to the time of maximal microbial growth that allowed the best resolution among the treatments.
DNA was extracted from the soil samples (0.4 g wet weight) with E.Z.N.A Soil DNA (OMEGA, United States) according to the manufacturer’s instructions. The V4 hypervariable region of bacterial 16S rRNA gene fragments were amplified in triplicate from each of the resulting DNA extracts using the primers 515F (5′-GTG CCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The amplification was carried out in 20 μL mixture 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase and 10 ng of template DNA. The amplification conditions involved an initial denaturing step at 95°C for 2 min followed by 25 cycles (95°C for 30 s, 56°C for 30 s, 72°C for 30 s) and a final extension at 72°C for 5 min.
Amplicons were purified using QIAquick PCR Purification Kit (Qiagen, China) and quantified using QuantiFluor-ST fluorometer (Promega, United States) according to the manufacturer’s instructions. Then the qualified libraries mentioned above were sequenced pair-end on the Illumina HiSeq System (Illumina, United States) by the sequencing strategy PE250.
The raw data were quality-filtered using QIIME (version 1.17) with the following criteria: (1) Sequence reads not having an average quality of 20 over a 25 bp sliding window based on the phred algorithm were truncated. Meanwhile, we trimmed and removed the reads with lengths less than 75% of their original length. (2) We removed reads contaminated by adapters (default parameter of 15 base overlapped by reads and adapter, as well as a maximal of 3 bases mismatch allowed). (3) We removed of reads with ambiguous base; (4) removal of low complexity reads that contain more than 10 of the same base consecutively. The filtered paired-end reads were combined to tags based on overlaps by FLASH (v1.2.11). The tags with 97% pairwise identity were binned into operational taxonomic units (OTU) by USEARCH (v7.0.1090). The abundance of each OTU was calculated according to the USEARCH_global method. The most abundant sequence of each OTU was selected as the representative OTU sequence. Taxonomic designation of OTUs was assigned by comparing the representative OTU sequence against the Greengenes database using RDP Classifier (v.2.2). The bacterial community structures diversity of the samples were further analyzed according to the OTU taxonomic richness and number.
The results of the soil typical physicochemical property for each experiment treatment were given as means and standard deviations of three replicates. Statistical significance between treatments was performed using SPSS 19.0 with two-way ANOVA and least significant difference (LSD) at p < 0.05.
Principal component analysis (PCA) was performed in Canoco for Windows 4.5 to compare the differences of the microbial physiological metabolic characteristics of the studied treatments based on Biolog EcoPlates data. Redundancy analysis (RDA) was carried out in Canoco for Windows 4.5 to determine which soil environmental variables best explained the changes in the frequency distributions of microbial metabolic functions under various treatments.
The bacterial community structure diversity indices were calculated using Mothur (v1.31.2). Principal coordinate analyses based on pairwise unweighted and weighted UniFrac distances were calculated in the “ade4” package of R software (v3.1.1). The information of common and unique OTUs among various treatments was plotted by “VennDiagram” package of R (v3.1.1). The log10-transformed relative abundance of genus-level OTUs was used to construct a heat map using the “gplots” package for R software (v3.1.1). A hierarchical cluster analysis was performed using BrayeCurtis distances. The relationship between the bacterial community structure and environmental factors was visualized according to redundancy analysis (RDA), which was performed with Canoco for Windows 4.5.
The typical physical and biochemical characteristics, such as pH, Eh, water-soluble organic carbon (WSOC), catalase and urease activity, and MBC of the soil samples collected during the experimental period are summarized in Table 1. CK0 and PT0 only differed in catalase activity (0.56 mg KMnO4 g-1 h-1 and 0.71 mg KMnO4 g-1 h-1, respectively). However, CK, PT, R and RN exhibited various soil physicochemical properties and MBC contents on day 28. Nearly all the indices mentioned above (except Eh and MBC) of R28 were significantly higher than those of CK28 and PT28 (P < 0.05). In addition, R28 presented higher pH, catalase activity, urease activity and WSOC than RN28, whereas Eh and MBC were significantly lower in R28 than RN28. Furthermore, CK28 presented higher catalase activity and lower Eh than CK0. On the other hand, PT28 and PT0 only differed in Eh.
The results of sampled soil microbial function assessment are shown in Table 2. The soil microbial functional diversity indices, such as AWCD, H′, D and U, were not significantly different between CK0 and PT0. In contrast, the soil on day 28 in the four treatment groups exhibited various microbial function diversities. Nearly all the indices mentioned above (except D) of R28 were significantly higher than those of CK28 and PT28 (P < 0.05). In addition, R28 presented higher AWCD, H′, and U than did RN28, whereas D was not significantly different between R28 and RN28. Furthermore, CK28 presented lower AWCD, H′, D and U than did CK0. On the other hand, PT28 presented lower AWCD and U than PT0 did.
PCA was performed to reduce the dimensionality of the Biolog EcoPlate data set, as well as to compare the differences in the microbial physiological metabolic characteristics of the researched treatments. The substrate utilization patterns of the researched treatments are shown in Figure 2A. The PCA of the substrate utilization patterns extracted two principle components, which explained 74.9% of the total variance together. In addition, the first principle component (PC1) exhibited great power of separation, as it explained 56.4% of the total variance. The PC1 axis showed that the carbon substrate utilization pattern of CK28 was significantly different from that of CK0. Similarly, PT28 exhibited a different carbon substrate utilization pattern from that of PT0. Moreover, PT28 and CK28 were located together, and RN28 was separated from PT28, by PC2. In addition, R28 was completely separated from RN28 and PT28.
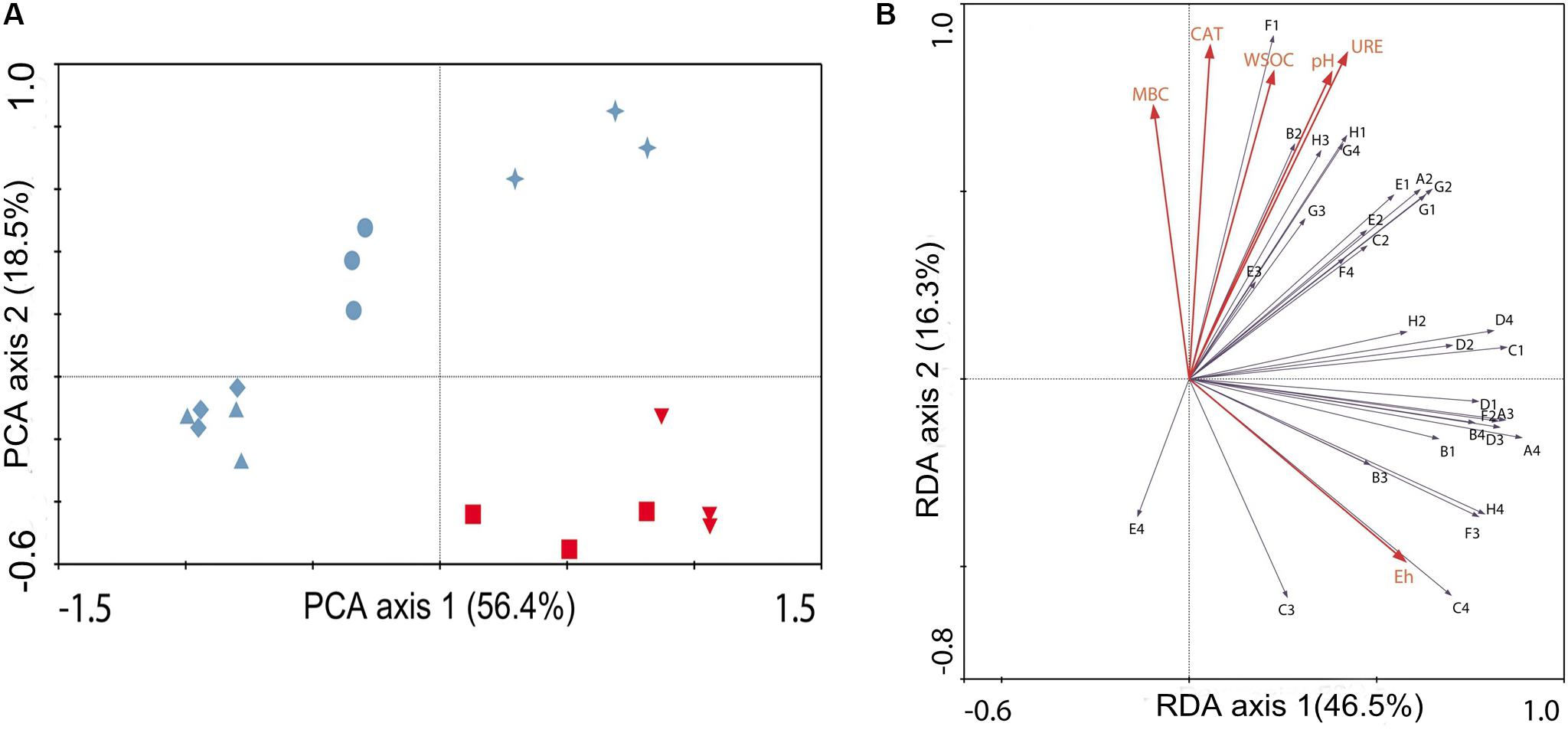
FIGURE 2. Analysis of functional diversity of bacterial communities in each researched treatment. (A) principle component analysis performed with the absorbance values of 31 carbon sources in Biolog ECO plates, measured at 96 h of incubation after adding serial dilutions of soil collected from the treatments. Red downturned triangle ( ) represents CK0, red square (
) represents CK0, red square ( ) represents PT0, blue upturned triangle (
) represents PT0, blue upturned triangle ( ) represents CK28, blue diamond (
) represents CK28, blue diamond ( ) represents PT28, blue circle (
) represents PT28, blue circle ( ) represents RN28 and blue star (
) represents RN28 and blue star ( ) represents R28. (B) RDA was performed with absorbance values mentioned above and the significant environmental parameters (red arrows, explanatory variables), such as pH, catalase activity (CAT), urease activity (URE), Eh, water soluble organic carbon (WSOC) and microbial biomass carbon (MBC). The arrows show the positions of functional microbial groups (metabolizing specific substrates): A2, β-Methyl-D-glucoside; A3, D-Galactonic acid γ-Lactone; A4, L-Arginine; B1, Pyruvic acid methyl ester; B2, D-Xylose; B3, D-Galacturonic acid; B4, L-Asparagine; C1, Tween 40; C2, I-Erythritol; C3, 2-Hydroxy benzoic acid; C4, L-Phenylalanine; D1, Tween 80; D2, D-Mannitol; D3, 4-Hydroxy benzoic acid; D4, L-Serine; E1, α-Cyclodextrin; E2, N-Acetyl-D-glucosamine; E3, γ-Hydroxybutyric acid; E4, L-Threonine; F1, Glycogen; F2, D-Glucosaminic acid; F3, Itaconic acid; F4, Glycyl-L-glutamic acid; G1, D-Cellobiose; G2, Glucose-1-phosphate; G3, α-Ketobutyric acid; G4, Phenylethylamine; H1, α-D-Lactose; H2, D,L-α-Glycerol phosphate; H3, D-Malic acid; and H4, Putrescine.
) represents R28. (B) RDA was performed with absorbance values mentioned above and the significant environmental parameters (red arrows, explanatory variables), such as pH, catalase activity (CAT), urease activity (URE), Eh, water soluble organic carbon (WSOC) and microbial biomass carbon (MBC). The arrows show the positions of functional microbial groups (metabolizing specific substrates): A2, β-Methyl-D-glucoside; A3, D-Galactonic acid γ-Lactone; A4, L-Arginine; B1, Pyruvic acid methyl ester; B2, D-Xylose; B3, D-Galacturonic acid; B4, L-Asparagine; C1, Tween 40; C2, I-Erythritol; C3, 2-Hydroxy benzoic acid; C4, L-Phenylalanine; D1, Tween 80; D2, D-Mannitol; D3, 4-Hydroxy benzoic acid; D4, L-Serine; E1, α-Cyclodextrin; E2, N-Acetyl-D-glucosamine; E3, γ-Hydroxybutyric acid; E4, L-Threonine; F1, Glycogen; F2, D-Glucosaminic acid; F3, Itaconic acid; F4, Glycyl-L-glutamic acid; G1, D-Cellobiose; G2, Glucose-1-phosphate; G3, α-Ketobutyric acid; G4, Phenylethylamine; H1, α-D-Lactose; H2, D,L-α-Glycerol phosphate; H3, D-Malic acid; and H4, Putrescine.
Figure 2B shows how the soil microbial community functional diversity varied with the potential explanatory variables. Four RDA axes were extracted, and the eigenvalues for these axes were 0.465, 0.163, 0.054, and 0.022, respectively. In addition, the variance in soil microbial functional data could be better explained by first RDA axis, while the soil microbial functional data exhibited a positive correlation with environmental data, with the correlation coefficient of 0.909. The results of the RDA also suggest that functional microbial groups among various treatments were significantly affected by the studied environmental variables, such as Eh, MBC, catalase activity and urease activity. These environmental variables, respectively, explained 12.3, 8, 5.9, and 4.5% of the total variance in the soil microbial functional data. In addition, the Eh of soil positively correlated with the use of L- phenylalanine (C4). Urease activity was strongly negatively correlated with the use of L-threonine (E4) and strongly positively correlated with the use of D-xylose (B2). Moreover, the use of the carbon substrates, such as D-malic acid (H3), α-D-lactose (H1) and phenylethylamine (G4), exhibited higher responses to urease activity. Furthermore, other carbon substrates, including L-threonine (E4) and 2-hydroxy benzoic acid (C3), exhibited a lower response in the proximity of the MBC and catalase activity.
A total of 167,992 high-quality 16S rRNA gene tags generated from all samples were clustered into 2686 OTUs. The relative abundances of the OTUs mentioned above at the phylum level are illustrated in Figure 3A. In total, 33 identified phyla were observed. Proteobacteria, Actinobacteria, and Acidobacteria were the three dominant phyla in all soil samples. The relative abundance of Proteobacteria phyla was significantly higher in PT0 (42.50%) than CK0 (37.70%). Furthermore, the relative abundances of Actinobacteria (19.90%), Verrucomicrobia (7.54%), Bacteroidetes (6.18%), and Cyanobacteria (0.33%) in R28 were significantly higher than those of other treatments. In contrast, the relative abundances of Proteobacteria and Acidobacteria were lower than other treatments. Moreover, Fibrobacteres phylum was only found in R28. In addition, the genera Kaistobacter, Candidatus_Nitrososphaera and Arthrobacter were the dominant genera of the present samples. Particularly, the relative abundances of Luteolibacter, Streptomyces Phycicoccus, and Chitinophaga in R28 were significantly higher than those in other samples, while there were lower relative abundances of Sporosarcina, Lactococcus, and Kaistobacter in R28 than other samples (Figure 3B).
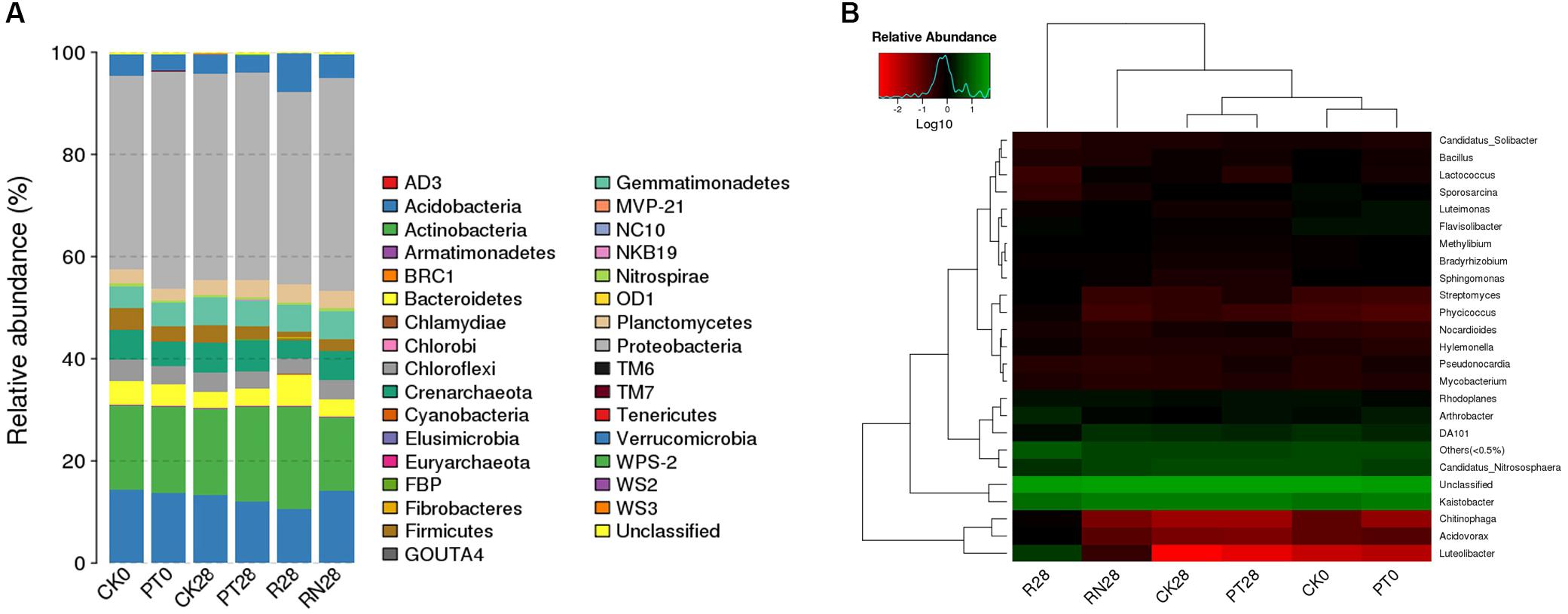
FIGURE 3. Comparison of the bacterial community structures of the researched treatments at (A) phylum level and (B) genus level. (A) showed the relative abundance of different bacterial phyla by bar plot and (B) exhibit the species of clustering heat map based on the relative abundance of different bacterial genus within the researched treatments, including CK, control treatment; PT, pollution treatment; RN, non-rhizosphere zone of the phytoremediation treatment; R, rhizosphere zone of phytoremediation treatment. The sampling point was day 0 (CK0 and PT0) and day 28 (CK28, PT28, R28 and RN28) respectively.
The alpha bacterial community diversity indices, such as observed species, including Chao, ACE, Shannon-Weaver and Simpson, are shown in Table 3. PT0 exhibited lower richness and diversity compared to the CK0 according to the four calculated indices in Table 3. Furthermore, significantly greater diversity was observed in R28 compared to CK28, PT28 and RN28 based on their Shannon indices, while the difference in bacterial richness and diversity among CK28, PT28 and RN28 was not obvious. The time period changed the bacterial richness of CK treatments slightly, as the Chao of CK0 and CK28 were 1800.860 and 1790.369, respectively, and the ACE indices were 1860.425 and 1851.134.
PCA was applied to identify the differences in bacterial community structure within all the researched treatments. The two principle components (PC1 and PC2) in Figure 4A explained 64.36% of the total variance. In addition, the six treatments of this study clustered into four groups. R28 was widely separated from the other five treatments. However, CK28 and PT28, as well as CK0 and PT0, were grouped together and clustered into two respective groups. RN28 itself was clustered into a new group, but it was located near CK28 and PT28.
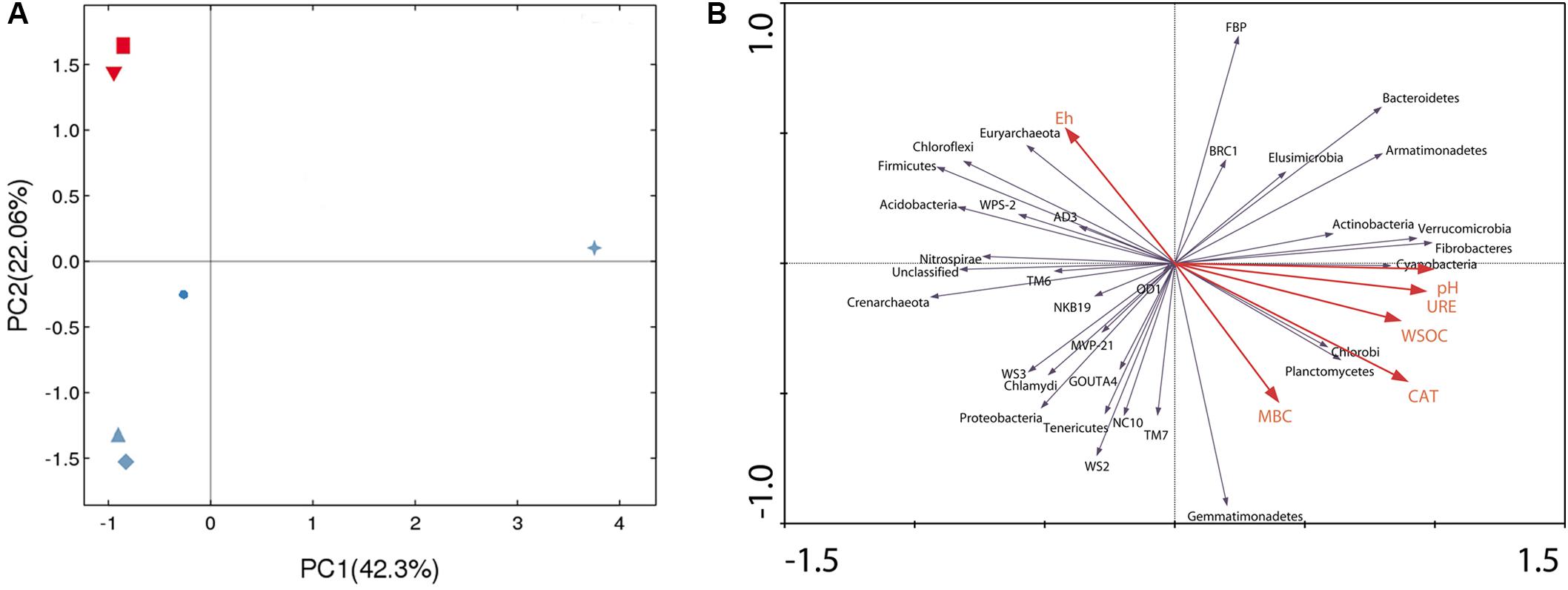
FIGURE 4. (A) Principle component analysis of soil bacterial community composition based on operational taxonomic units abundance of the different treatments, CK, control treatment; PT, pollution treatment; RN, non-rhizosphere zone of the phytoremediation treatment; R, rhizosphere zone of phytoremediation treatment. The sampling point was day 0 and day 28 respectively. Red downturned triangle ( ) represents CK0, red square (
) represents CK0, red square ( ) represents PT0, blue upturned triangle (
) represents PT0, blue upturned triangle ( ) represents CK28, blue diamond (
) represents CK28, blue diamond ( ) represents PT28, blue circle (
) represents PT28, blue circle ( ) represents RN28 and blue star (
) represents RN28 and blue star ( ) represents R28. (B) Ordination plots of the results from the redundancy analysis to identify the relationships among the bacterial populations (blue arrows), soil physico-chemical characteristics and microbial biomass carbon (red arrows). CAT, catalase activity; URE, urease activity; WSOC, water soluble organic carbon; MBC, microbial biomass carbon (MBC).
) represents R28. (B) Ordination plots of the results from the redundancy analysis to identify the relationships among the bacterial populations (blue arrows), soil physico-chemical characteristics and microbial biomass carbon (red arrows). CAT, catalase activity; URE, urease activity; WSOC, water soluble organic carbon; MBC, microbial biomass carbon (MBC).
RDA analysis was performed to show the effect of main soil physicochemical and biological characteristics on the bacterial communities (phylum level). Figure 4B shows that the first two axes explained 86.5% of the total variance, indicating that pH, urease activity, catalase activity, and WSOC were the most influential factors driving the changes in the composition and diversity of the bacterial communities. Specifically, the soil pH was strongly negatively correlated with Nitrospirae and Crenarchaeota, while it was strongly positively correlated with Cyanobacteria, Fibrobacteres and Verrucomicrobia. Urease activity was strongly negatively correlated with Acidobacteria. Catalase activity was strongly negatively correlated with Chloroflexi and Firmicutes but strongly positively correlated with Chlorobi and Planctomycetes. Acidobacteria and Firmicutes exhibited higher responses to WSOC.
The plants of the Pennisetum genus exhibits tolerance to herbicide atrazine and potential to decrease the atrazine residual level in the rhizosphere (Zhang et al., 2014; Jiang et al., 2016). A higher microbial biomass might be the main reason for the enhanced degradation of atrazine in the rhizosphere (Singh et al., 2004). Therefore, the rhizosphere is widely considered as a hot spot of pollutants rhizoremediation for its higher microbial activity (Velasco et al., 2013). Furthermore, it has been proved that the high biomass and diversity microbita in rhizosphere is mainly due to the interaction of plant and microorganism during the rhizoremediation period (Velasco et al., 2010; Velasco et al., 2013). However, there is little detailed information about the differences in the soil physicochemical properties, microbial metabolize and bacterial community diversitycharacteristics between the rhizosphere and non-rhizosphere soil of Pennisetum genus plants in remediation of contaminated soil. This paper is mainly intends to further illustrate the relationships among P. americanum, rhizosphere soil physicochemical properties and bacterial community traits during the phytoremediation of atrazine-contaminated soil.
Catalase in soil is responsible for removal of the hydrogen peroxide (H2O2) and alleviating the oxidative damage to microorganisms and plants. Table 1 shows that significantly greater catalase activity was found in PT0 than CK0. This phenomenon might be mainly because of the oxidative stress response of soil microorganisms to the addition of atrazine. This inference could be further supported by the well-known viewpoint that atrazine causes oxidative stress on various types of organisms (Zhang et al., 2012; Jiang et al., 2016). Catalase activity could also be used to evaluate the metabolic activity of soil microbial communities (Samuel et al., 2011), and urease activity exhibits a strong correlation with the organic nitrogen transfer ability by microbes. Therefore, the higher catalase and urease activities in R28 suggest that the P. americanum rhizosphere could accelerate the metabolic activity of soil microorganisms by releasing various types of nutrient substances (root exudates) to boost the functional microbial survival, or by changing the soil micro-environment to make it favorable to the microorganisms mentioned above. These possibilities are in line with the result that the concentration of water-soluble organic carbon (WSOC), which might be released by the roots of P. americanum, in R28 was higher than those of other treatments. They also could be supported by the result that the pH in the rhizosphere of P. americanum was much closer to the suitable pH range (6–8) for microorganisms.
The effect of the P. americanum rhizosphere on the bacterial catabolic ability was assessed by determining the community-level physiological profiles (CLPP) of soil bacteria using Biolog EcoPlates. Meanwhile, redundancy analysis (RDA) was employed to further investigate the interaction of environmental factors and the carbon-containing substrate utilization characteristics of the soil bacteria (Figure 3B). We found that the studied soil physical, chemical and biological properties, such as Eh, MBC, catalase activity and urease activity, exhibited strong positive or negative correlations with the utilization of some types of substrates in the Biolog EcoPlates. Combined with the results described above that the soil physical, chemical and biological properties were affected by the P. americanum rhizosphere, it is reasonable to infer that the bacterial catabolic ability of the P. americanum rhizosphere might be different from those of other treatments. CLPP-based principal component analysis (PCA) suggested that the substrate utilization pattern of R28 was completely different from other treatments, based on the two principal components in Figure 3A. Moreover, the higher bacterial functional diversity indices (AWCD, H′, and U) in R28 revealed that the soil in P. americanum rhizosphere exhibited much greater carbon substrate utilization ability, since the indices mentioned above are commonly proposed to measure the functional diversity or catabolic ability of bacterial community (Villeger et al., 2008). These results might partly illustrate why higher organic pollutants removal efficiency could be found in rhizosphere soil (Li et al., 2011; Blaud et al., 2015).
It has been thought that the soil microbial activity especially the carbon utilization ability could be affected by the species-specific root exudates released from various types of plant species, since the root exudates are the most important sources of readily available carbon for rhizosphere microorganisms (Martineau et al., 2014; Yuan et al., 2016). Because the microorganisms differ in their ability to metabolize and compete for different carbon sources, it is reasonable to consider that the structure of microorganism communities might change during the variation of microorganism functional diversity, as well as the soil physical and chemical properties (Berg and Smalla, 2009). In this paper, high throughput sequencing technology based on the Illumina HiSeq platform was selected to access the bacterial community information of the researched treatments. The redundancy analysis (RDA) based on the bacterial community information and the soil physical and chemical properties further showed that the detected soil properties, such as pH, urease activity, catalase activity, and WSOC, affected the bacterial composition of soil samples collected from various treatments. Since the typical soil physical and chemical properties of the P. americanum rhizosphere were different from that of non-rhizosphere treatments (Table 1), it also can be concluded that the test plant P. americanum could shape the rhizosphere environment, as well as the bacterial community. Additionally, the pincipal component analysis (PCA) of the bacterial community information extracted from the treatments in Figure 4A further show that the P. americanum rhizosphere exhibited a different bacterial community characteristics. Therefore, a strong evolutionary relationship between P. americanum and bacteria might exist in the rhizosphere. These observations are in agreement with results by that Lacalle et al. (2018) that Brassica napus plants not only increased the activity of microbial communities in contaminated soils, but also its functional diversity by creating suitable conditions for microbial growth in the rhizosphere.
This study also found that though R28 exhibit a higher bacterial diversity (Shannon indice was 5.821), the greatest bacterial species richness (Chao and ACE) was not found in R28 (Table 3). This phenomenon might have been due to the succession of the bacterial communities in the P. americanum rhizosphere which triggers an environmental filtering shift of bacteria community composition (Zappelini et al., 2015; Chen et al., 2016). This inference is in line with the data presented in this study that some genera of bacteria, such as such as Arthrobacter, Chitinophaga, Streptomyces, Sporosarcina and Phycicoccus, were very sensitive to the P. americanum rhizosphere environment, as the relative abundances of these genera in R28 were significantly different from those of other treatments. In addition, we found that there were 178 unique OTUs in R28 by comparing the bacterial community composition with those of CK28, PT28 and RN28 (Figure 5). Taxonomic analysis indicated that some of the unique OTUs mentioned above belonged to the phyla Chloroflexi and Cyanobacteria, the order Acidimicrobiales, as well as the genera Paenibacillus and Rhizobium, which can facilitate soil nutrient cycling (Bustamante et al., 2006; Rodrigues et al., 2015; Zimmermann et al., 2015; Redding et al., 2016). In addition, some other OTUs which represent the bacteria with the potential to degrade organic pollutants were found in the P. americanum rhizosphere. These OTUs consisted of the genus Sphingobium which could participate in the degradation of herbicide (Sun et al., 2008), the genus Mycoplana which can decompose 2,4-dichlorophenol (Manikandan et al., 2008), and the family Sphingomonadaceae, which degrades aromatic compounds (Lafortune et al., 2009). It is worth to note that the class Fibrobacteria and the order Rhizobiales, two types of bacteria frequently appearing around the rhizosphere that can decompose fiber and fix nitrogen, respectively, only were detected in R28. These results further suggest that the P. americanum also could effectively boost the potential of nutrient metabolism and pollutants degradation in rhizosphere by enhancing the kind or abundance of the bacteria with the corresponding ecology function. It could be greatly supported by the results described above that the abundance of the Arthrobacter genus bacteria was obviously enhanced in rhizosphere of P. americanum (Figure 2B), since much more atrazine-degrading strains has been identified as Arthrobacter genus (Zhang et al., 2011). This inference could be further supported by our previous published results that P. americanum could obviously accelerate the atrazine removal in soil than that of the treatment without any plant (Zhang et al., 2014). Indeed, further research might continually focus on the variety of microbial functional genes, which responsible for atrazine degradation and soil nutrient cycling, to further illustrate the phytoremediation mechanism of atrazine-contaminated soil by P. americanum.
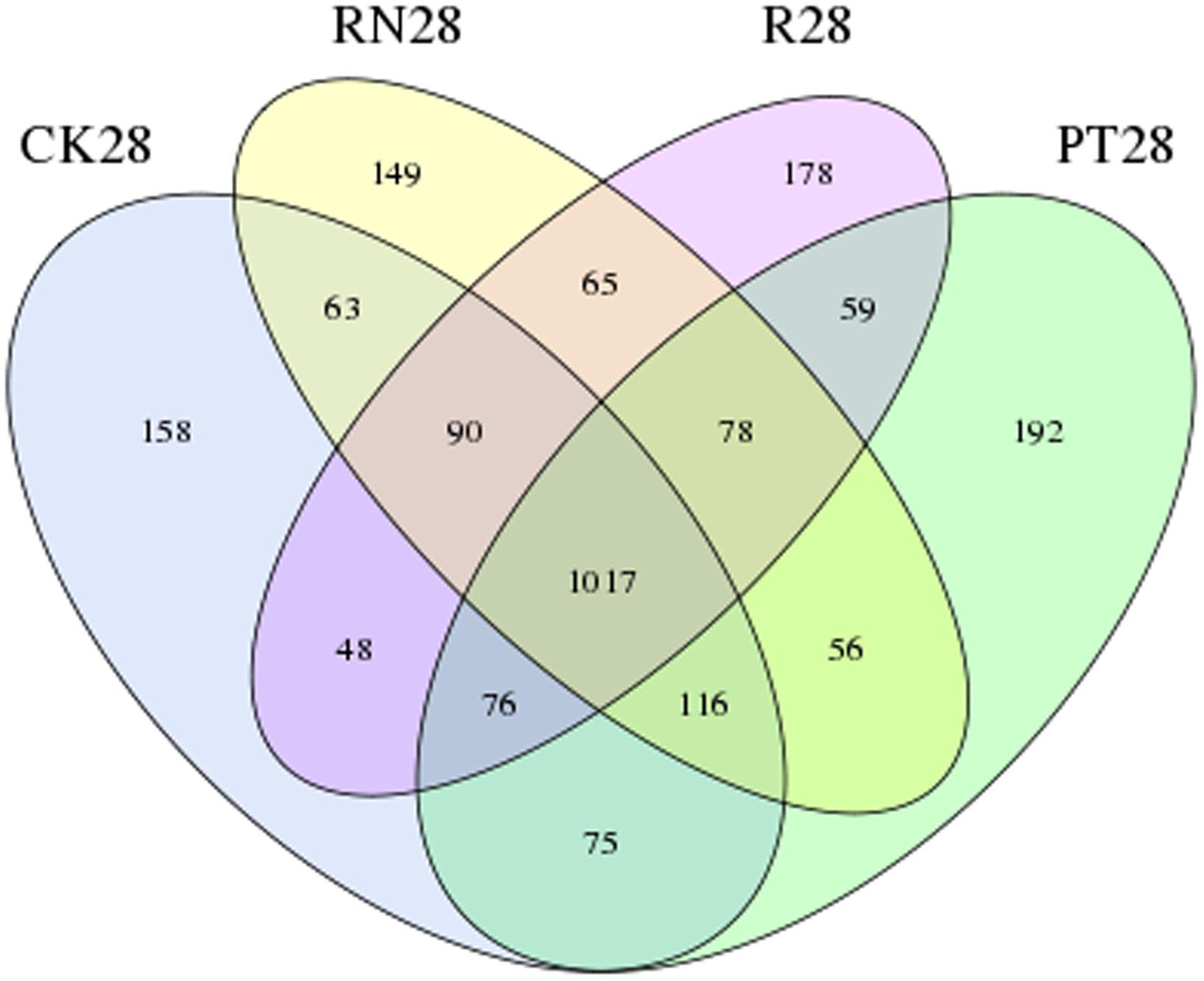
FIGURE 5. Venn diagram of operational taxonomic units (OTUs) distribution among the samples collected at day 28 of the researched treatments, including CK, control treatment; PT, pollution treatment; RN, non-rhizosphere zone of the phytoremediation treatment; R, rhizosphere zone of phytoremediation treatment. The number in the Venn diagram depicting the shared and unique OTUs present in the four treatments mentioned above.
Pennisetum americanum planted in atrazine-contaminated soil shaped the bacterial communities and enhanced the bacterial functional diversity of the rhizosphere by re-shaping the soil physicochemical properties, such as catalase activity, urease activity, WSOC and pH, to be more suitable to soil microorganisms. Additionally, some unique types of bacteria that could facilitate soil nutrient cycling or organic pollutant degradation were only found in the rhizosphere of P. americanum. This study provides insight into how that the interaction between the P. americanum, soil physicochemical environment as well as the soil bacterial properties (community and functional diversity) plays an important role during the phytoremediation process of atrazine-contaminated soil.
BC, as the first author of this manuscript, was mainly responsible for writing the whole manuscript and analyzing the results about the microorganism community diversity. YZ and ZJ designed the whole experiment together and calculated the data. ML mainly prepared the rhizobox that was used in this research, as well as did much work for soil microbial physiological metabolic characteristics analysis using Biology Eco plates. FY worked on the soil sample collection. ZW detected the soil physicochemical properties of the soil samples. DJ was responsible for the extraction of the soil microorganism DNA and the detection of the soil microbial biomass carbon. In addition, ZJ was also responsible for submitting the manuscript to the journal.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by the National Science Fund for Distinguished Young Scholars (41625002); Natural Science Foundation of Heilongjiang (C2016020); Training program for young creative talents of ordinary undergraduate colleges and universities in Heilongjiang Province (UNPYSCT-2016155); Backbone Project of Northeast Agricultural University (17XG07); National Natural Science Foundation of China (31300433); Agricultural Research Outstanding Talents and Innovation Team.
Agnello, A. C., Bagard, M., van Hullebusch, E. D., Esposito, G., and Huguenot, D. (2016). Comparative bioremediation of heavy metals and petroleum hydrocarbons co-contaminatedsoil by natural attenuation, phytoremediation, bioaugmentation and bioaugmentation-assisted phytoremediation. Sci. Total Environ. 563, 693–703. doi: 10.1016/j.scitotenv.2015.10.061
Albright, V. C., Murphy, I. J., Anderson, J. A., and Coats, J. R. (2013). Fate of atrazine in switchgrass-soil column system. Chemosphere 90, 1847–1853. doi: 10.1016/j.chemosphere.2012.09.097
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Blaud, A., Lerch, T. Z., Phoenix, G. K., and Osborn, A. M. (2015). Arctic soil microbial diversity in a changing world. Res. Microbiol. 166, 796–813. doi: 10.1016/j.resmic.2015.07.013
Bustamante, M. M. C., Medina, E., Asner, G. P., Nardoto, G. B., and Garcia-Montiel, D. C. (2006). Nitrogen cycling in tropical and temperate savannas. Biogeochemistry 79, 209–237. doi: 10.1007/s10533-006-9006-x
Cai, Z., Zhou, Q., Peng, S., and Li, K. (2010). Promoted biodegradation and microbiological effects of petroleum hydrocarbons by Impatiens balsamina L. with strong endurance. J. Hazard. Mater. 183, 731–737. doi: 10.1016/j.jhazmat.2010.07.087
Cao, J., Ji, D., and Wang, C. (2015). Interaction between earthworms and arbuscular mycorrhizal fungi on the degradation of oxytetracycline in soils. Soil Biol. Biochem. 90, 283–292. doi: 10.1016/j.scitotenv.2016.07.077
Chen, L., Brookes, P. C., Xu, J., Zhang, J., Zhang, C., Zhou, X., et al. (2016). Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biol. Biochem. 98, 1–10. doi: 10.1016/j.soilbio.2016.04.004
El Amrani, A., Dumas, A. S., Wick, L. Y., Yergeau, E., and Berthome, R. (2015). “Omics” insights into PAH degradation toward improved green remediation biotechnologies. Environ. Sci. Technol. 49, 11281–11291. doi: 10.1021/acs.est.5b01740
Fang, C., Radosevich, M., and Fuhrmann, J. J. (2001). Atrazine and phenanthrene degradation in grass rhizosphere soil. Soil Biol. Biochem. 33, 671–678. doi: 10.1016/S0038-0717(00)00216-9
Gil-Díaz, M., González, A., Alonso, J., and Lobo, M. C. (2016). Evaluation of the stability of ananoremediation strategy using barley plants. J. Environ. Manag. 165, 150–158. doi: 10.1016/j.jenvman.2015.09.032
Hamdi, H., Benzarti, S., Aoyama, I., and Jedidi, N. (2012). Rehabilitation of degraded soils containing aged PAHs based on phytoremediation with alfalfa (Medicago sativa L.). Int. Biodeterior. Biodegradation 67, 40–47. doi: 10.1016/j.ibiod.2011.10.009
Hussain, I., Puschenreiter, M., Gerhard, S., Schoftner, P., Yousaf, S., Wang, A. J., et al. (2018). Rhizoremediation of petroleum hydrocarbon-contaminated soils: improvement opportunities and field applications. Environ. Exp. Bot. 147, 202–219. doi: 10.1016/j.envexpbot.2017.12.016
Imfeld, G., and Vuilleumier, S. (2012). Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 49, 22–30. doi: 10.1016/j.ejsobi.2011.11.010
Jagtap, S. S., Woo, S. M., Kim, T.-S., Dhiman, S. S., Kim, D., and Lee, J.-K. (2014). Phytoremediation of diesel-contaminated soil and saccharification of the resulting biomass. Fuel 116, 292–298. doi: 10.1016/j.fuel.2013.08.017
Jiang, Z., Ma, B., Erinle, K. O., Cao, B., Liu, X., Ye, S., et al. (2016). Enzymatic antioxidant defense in resistant plant: Pennisetum americanum (L.) K. Schum during long-term atrazine exposure. Pestic. Biochem. Physiol. 133, 59–66. doi: 10.1016/j.pestbp.2016.03.003
Lacalle, R. G., Gomez-Sagasti, M. T., Artetxe, U., Garbisu, C., and Becerril, J. M. (2018). Brassica napushas a key role in the recovery of the health of soils contaminated with metals and diesel by rhizoremediation. Sci. Total Environ. 618, 347–356. doi: 10.1016/j.scitotenv.2017.10.334
Lafortune, I., Juteau, P., Deziel, E., Lepine, F., Beaudet, R., and Villemur, R. (2009). Bacterial diversity of a consortium degrading high-molecular-weight polycyclic aromatic hydrocarbons in a two-liquid phase biosystem. Microb. Ecol. 57, 455–468. doi: 10.1007/s00248-008-9417-4
Lasserre, J. P., Fack, F., Revets, D., Planchon, S., Renaut, J., Hoffmann, L., et al. (2009). Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J. Proteome Res. 8, 5485–5496. doi: 10.1021/pr900480f
Li, J., Jin, Z., and Gu, Q. (2011). Effect of plant species on the function and structure of the bacterial community in the rhizosphere of lead-zinc mine tailings in Zhejiang. China. Can. J. Microbiol. 57, 569–577. doi: 10.1139/W11-054
Lima, D., Viana, P., Andre, S., Chelinho, S., Costa, C., Ribeiro, R., et al. (2009). Evaluating a bioremediation tool for atrazine contaminated soils in open soil microcosms: the effectiveness of bioaugmentation and biostimulation approaches. Chemosphere 74, 187–192. doi: 10.1016/j.chemosphere.2008.09.083
Liu, R., Xiao, N., Wei, S., Zhao, L., and An, J. (2014). Rhizosphere effects of PAH-contaminated soil phytoremediation using a special plant named Fire Phoenix. Sci. Total Environ. 47, 350–358. doi: 10.1016/j.scitotenv.2013.12.027
Manikandan, R., Prabhu, H. J., and Sivashanmugam, P. (2008). Biodegradation of 2,4-dichlorophenol using Mycoplana dimorpha extracts and evaluation of kinetic parameters. Afr. J. Biotechnol. 7, 2038–2048. doi: 10.5897/AJB2008.000-5053
Martin, B. C., George, S. J., Price, C. A., Ryan, M. H., and Tibbett, M. (2014). The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbondegradation: current knowledge and future directions. Sci. Total Environ. 472, 642–653. doi: 10.1016/j.scitotenv.2013.11.050
Martineau, N., McLean, J. E., Dimkpa, C. O., Britt, D. W., and Anderson, A. J. (2014). Components from wheat roots modify the bioactivity of ZnO and CuO nanoparticles in a soil bacterium. Environ. Pollut. 187, 65–72. doi: 10.1016/j.envpol.2013.12.022
Muñoz-Leoz, B., Ruiz-Romera, E., Antigüedad, I., and Garbisu, C. (2011). Tebuconazole application decreases soil microbial biomass and activity. Soil Biol. Biochem. 43, 2176–2183. doi: 10.1016/j.soilbio.2011.07.001
Nan, W., Yue, S., Li, S., Huang, H., and Shen, Y. (2016). The factors related to carbon dioxide effluxes and production in the soil profiles of rain-fed maize fields. Agric. Ecosyst. Environ. 216, 177–187. doi: 10.1016/j.agee.2015.09.032
Pandey, J., Chauhan, A., and Jain, R. K. (2009). Integrative approaches for assessing the ecological sustainability of in situ bioremediation. FEMS Microbiol. Rev. 33, 324–375. doi: 10.1111/j.1574-6976.2008.00133.x
Redding, M. R., Shorten, P. R., Lewis, R., Pratt, C., Paungfoo-Lonhienne, C., and Hill, J. (2016). Soil N availability, rather than N deposition, controls indirect N2O emissions. Soil Biol. Biochem. 95, 288–298. doi: 10.1016/j.soilbio.2016.01.002
Rodrigues, R. R., Pineda, R. P., Barney, J. N., Nilsen, E. T., Barrett, J. E., and Williams, M. A. (2015). Plant invasions associated with change in root-zone microbial community structure and diversity. PLoS One 10:e0141424. doi: 10.1371/journal.pone.0141424
Roustan, A., Aye, M., De Meo, M., and Di Giorgio, C. (2014). Genotoxicity of mixtures of glyphosate and atrazine and their environmental transformation products before and after photoactivation. Chemosphere 108, 93–100. doi: 10.1016/j.chemosphere.2014.02.079
Salazar-Ledesma, M., Prado, B., Zamora, O., and Siebe, C. (2018). Mobility of atrazine in soils of a wastewater irrigated maize field. Agr. Ecosyst. Environ. 255, 73–83. doi: 10.1016/j.agee.2017.12.018
Samuel, A. D., Domuta, C., Ciobanu, G., Sandor, M., Ciobanu, C., and Brejea, R. (2011). Enzymological study of the evolution of the technogenic soil submitted to biological recultivation in the bauxite mine from Padurea Craiului (Romania). J. Environ. Prot. Ecol. 12, 535–542.
Singh, N., Megharaj, M., Kookana, R. S., Naidu, R., and Sethunathan, N. (2004). Atrazine and simazine degradation in Pennisetum rhizosphere. Chemosphere 56, 257–263. doi: 10.1016/j.chemosphere.2004.03.010
Sun, K., Gao, B., Zhang, Z. Y., Zhang, G. X., Zhao, Y., and Xing, B. S. (2010). Sorption of atrazine and phenanthrene by organic matter fractions in soil andsediment. Environ. Pollut. 158, 3520–3526. doi: 10.1016/j.envpol.2010.08.022
Sun, Y., Zhou, Q., and Diao, C. (2008). Effects of cadmium and arsenic on growth and metal accumulation of Cd-hyperaccumulator Solanum nigrum L. Bioresour. Technol. 99, 1103–1110. doi: 10.1016/j.biortech.2007.02.035
Thijs, S., Sillen, W., Rineau, F., Weyens, N., and Vangronsveld, J. (2016). Towards an enhanced understanding of plant–microbiome interactions to improve phytoremediation: engineering the metaorganism. Front. Microbiol. 7:341. doi: 10.3389/fmicb.2016.00341
Truu, J., Truu, M., Espenberg, M., Nõlvak, H., and Juhanson, J. (2015). Phytoremediation andplant-assisted bioremediation in soil and treatment wetlands: a review. Open. Biotechnol. J. 9, 85–92. doi: 10.2174/1874070701509010085
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Velasco, A. G. V., Kowalchuk, G. A., Manero, F. J. G., Ramos, B., Yergeau, E., and Lucas, J. A. (2013). Increased microbial activity and nitrogen mineralization coupled to changes in microbial community structure in the rhizosphere of Bt corn. Appl. Soil Ecol. 68, 46–56. doi: 10.1016/j.apsoil.2013.03.010
Velasco, A. G. V., Probanza, A., Manero, F. J. G., Solano, B. R., and Lucas, J. A. (2010). Characterization of the rhizosphere microbial community from different Arabidopsis thaliana genotypes using phospholipid fatty acids (PLFA) analysis. Plant Soil 329, 315–325. doi: 10.1007/s11104-009-0160-5
Villeger, S., Mason, N. W., and Mouillot, D. (2008). New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology 89, 2290–2301. doi: 10.1890/07-1206.1
Wang, M. C., Chen, Y. T., Chen, S. H., Chien, S. W. C., and Sunkara, S. V. (2012). Phytoremediation of pyrene contaminated soils amended with compost and planted with ryegrass and alfalfa. Chemosphere 87, 217–225. doi: 10.1016/j.chemosphere.2011.12.063
Yang, Y. X., Cao, H. B., Peng, P., and Bo, H. M. (2014). Degradation and transformation of atrazine under catalyzed ozonation process with TiO2 as catalyst. J. Hazard. Mater. 279, 444–451. doi: 10.1016/j.jhazmat.2014.07.035
Yuan, H., Zhu, Z., Liu, S., Ge, T., Jing, H., Li, B., et al. (2016). Microbial utilization of rice root exudates: 13C labeling and PLFA composition. Biol. Fert. Soils 52, 615–627. doi: 10.1007/s00374-016-1101-0
Zappelini, C., Karimi, B., Foulon, J., Lacercat-Didier, L., Maillard, F., Valot, B., et al. (2015). Diversity and complexity of microbial communities from a chlor-alkali tailings dump. Soil Biol. Biochem. 90, 101–110. doi: 10.1016/j.soilbio.2015.08.008
Zhang, Y., Ge, S., Jiang, M., Jiang, Z., Wang, Z., and Ma, B. (2014). Combined bioremediation of atrazine-contaminated soil by Pennisetum and Arthrobacter sp. strain DNS10. Environ. Sci. Pollut. Res. Int. 21, 6234–6238. doi: 10.1007/s11356-013-2410-6
Zhang, Y., Jiang, Z., Cao, B., Hu, M., Wang, Z., and Dong, X. (2011). Metabolic ability and gene characteristics of Arthrobacter sp. strain DNS10, the sole atrazine-degrading strain in a consortium isolated from black soil. Int. Biodeterior. Biodegradation 65, 1140–1144. doi: 10.1016/j.ibiod.2011.08.010
Zhang, Y., Meng, D., Wang, Z., Guo, H., Wang, Y., Wang, X., et al. (2012). Oxidative stress response in atrazine-degrading bacteria exposed to atrazine. J. Hazard. Mater. 22, 434–438. doi: 10.1016/j.jhazmat.2012.05.054
Keywords: rhizosphere, atrazine, Pennisetum, phytoremediation, microbial succession
Citation: Cao B, Zhang Y, Wang Z, Li M, Yang F, Jiang D and Jiang Z (2018) Insight Into the Variation of Bacterial Structure in Atrazine-Contaminated Soil Regulating by Potential Phytoremediator: Pennisetum americanum (L.) K. Schum. Front. Microbiol. 9:864. doi: 10.3389/fmicb.2018.00864
Received: 26 January 2018; Accepted: 13 April 2018;
Published: 04 May 2018.
Edited by:
Piotr Rozpądek, Jagiellonian University, PolandReviewed by:
M. Oves, King Abdulaziz University, Saudi ArabiaCopyright © 2018 Cao, Zhang, Wang, Li, Yang, Jiang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao Jiang, amlhbmd6aGFvXzA4MjhAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.