- 1Graduate Program in Genetics, University of Iowa, Iowa City, IA, United States
- 2Immunity to Pulmonary Pathogens Section, Laboratory of Intracellular Parasites, Hamilton, MT, United States
- 3Genomics Core, Research Technologies Branch, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Hamilton, MT, United States
- 4Department of Microbiology, University of Iowa, Iowa City, IA, United States
Francisella tularensis is a highly infectious bacterial pathogen that causes the potentially fatal disease tularemia. The Live Vaccine Strain (LVS) of F. tularensis subsp. holarctica, while no longer licensed as a vaccine, is used as a model organism for identifying correlates of immunity and bacterial factors that mediate a productive immune response against F. tularensis. Recently, it was reported that two biovars of LVS differed in their virulence and vaccine efficacy. Genetic analysis showed that they differ in ferrous iron homeostasis; lower Fe2+ levels contributed to increased resistance to hydrogen peroxide in the vaccine efficacious LVS biovar. This also correlated with resistance to the bactericidal activity of interferon γ-stimulated murine bone marrow-derived macrophages. We have extended these findings further by showing that a mutant lacking bacterioferritin stimulates poor protection against Schu S4 challenge in a mouse model of tularemia. Together these results suggest that the efficacious biovar of LVS stimulates productive immunity by a mechanism that is dependent on its ability to limit the toxic effects of oxidative stress by maintaining optimally low levels of intracellular Fe2+.
Introduction
Francisella tularensis is a highly infectious bacterial pathogen that parasitizes the cytosol of host cells and causes the lethal disease tularemia in humans. The low infectious dose and high morbidity and mortality associated with tularemia have led the United States Centers for Disease Control and Prevention to designate F. tularensis a Tier 1 Select Agent. Fear of the intentional misuse of F. tularensis has spurred research into understanding the pathogenesis of the organism and the development of a vaccine. No licensed vaccine for F. tularensis infection is currently available for widespread use, although, the F. tularensis Live Vaccine Strain (LVS) has, in recent years, been used to vaccinate laboratory staff in the US and in other countries and large numbers of individuals have been vaccinated in the past (Eigelsbach and Downs, 1961). LVS is an attenuated derivative of F. tularensis subsp. holarctica that was generated in the Soviet Union in the mid- twentieth century but an incomplete understanding of the genetic changes, and therefore the underlying mechanisms, leading to the attenuation of the strain have led to its discontinuation as a licensed vaccine. Although not available as a human vaccine, LVS is routinely used as a model to understand the genetic requirements of a F. tularensis strain to stimulate host immunity and as a tool to discover and characterize correlates of immunity in the host. Each of these areas of understanding are critical in the rational design of a future vaccine against virulent F. tularensis species.
Despite significant differences in virulence, members of the Francisella genus are very similar at the genetic level; many of the observed differences are genomic rearrangements and single nucleotide polymorphisms (SNPs) (Rohmer et al., 2006, 2007). The genome similarities of the Francisella strains have made identification of specific factors that mediate the high level of virulence displayed by Type A and Type B strains difficult, although genome comparisons between F. tularensis subsp. tularensis Schu S4 and less virulent LVS have identified some of the attenuating mutations. These mutations include fusion of the fupA/B genes and deletion of pilA (Lindgren et al., 2009). Another noted difference between the highly virulent Type A strains and the intermediately virulent type B strains (including LVS) is the levels of intracellular iron, with virulence being inversely correlated with iron levels (lower intracellular iron correlates with higher virulence and higher intracellular iron correlates with lower virulence) (Lindgren et al., 2011). Iron is an essential micronutrient for bacteria, and numerous studies have shown that it is highly sought after by bacterial pathogens, as host sequestration of iron (a part of nutritional immunity) can restrict the growth of numerous pathogens (Cassat and Skaar, 2013; Parrow et al., 2013). Apparently, the virulence strategy of F. tularensis represents an exception to this paradigm as Lindgren et al., found that higher virulence subspecies had lower levels of intracellular iron (Lindgren et al., 2011).
Francisella species import iron via siderophore-bound ferric iron via the Fsl and Fup systems, and ferrous iron via FeoB and FupA/B; the latter system is unique to LVS (Ramakrishnan et al., 2008; Thomas-Charles et al., 2013; Perez and Ramakrishnan, 2014; Ramakrishnan and Sen, 2014). The feo operon typically consists of feoAB, and less frequently feoABC; the Francisella chromosome encodes feoA and feoB separately, and lacks feoC (Cartron et al., 2006; Perez and Ramakrishnan, 2014). FeoB is a large transmembrane protein that has an N-terminal G-protein domain and a multi-pass transmembrane C-terminal domain (Marlovits et al., 2002; Andrews et al., 2003; Hantke, 2003). In other organisms, the Fe2+ import activity of FeoB requires interaction with the small protein FeoA, though the details of how FeoA stimulates FeoB activity are unknown (Su et al., 2010; Kim et al., 2012; Lau et al., 2013; Weaver et al., 2013). The general importance of iron in bacterial pathogenesis is reflected by the numerous ways that bacterial pathogens have evolved to obtain iron in a host, and several research groups have demonstrated that feoB contributes to or is required for full pathogenesis of Salmonella, Campylobacter, Helicobacter, Legionella, and others (Velayudhan et al., 2000; Robey and Cianciotto, 2002; Naikare et al., 2006; Aranda et al., 2009; Cassat and Skaar, 2013; Nagy et al., 2014). Multiple studies have characterized the Fe2+ uptake function of the F. tularensis FeoB and have linked this to its pathogenesis (Thomas-Charles et al., 2013; Perez and Ramakrishnan, 2014; Lindgren et al., 2015). However, it is not clear from these studies if the modulation of growth among feoB mutants is a direct result of differential ability to use and take up iron or if there are indirect effects of excess iron that impact bacterial replication. For example, while iron is necessary, excess Fe2+ can contribute to toxicity for an organism as ferrous iron can participate in the Fenton reaction with H2O2, poisoning iron-sulfur cluster enzymes in essential metabolic pathways (Imlay, 2013).
Francisella tularensis virulence is connected to the ability of the organism to avoid the detrimental effects of reactive oxygen species (ROS), such as H2O2 (Bou-Abdallah et al., 2002; Lindgren et al., 2007; McCaffrey et al., 2010; Lindemann et al., 2011; Crane et al., 2014; Ma et al., 2014). The bacterium couples glutamate metabolic pathways to H2O2 neutralization, and also maintains optimally low levels of intracellular Fe2+ such that Fenton reaction-mediated damage appears to be minimized (Lindgren et al., 2011; Ramond et al., 2014). When this optimal iron threshold is surpassed, numerous antioxidant enzymes are employed to protect the organism from toxic H2O2; mutants lacking enzymes like the Dyp peroxidase, superoxide dismutase and catalase are more sensitive to H2O2 damage, fail to inhibit host inflammatory signaling, and are often attenuated in murine infection models (Bakshi et al., 2006; Melillo et al., 2009, 2010; Llewellyn et al., 2011; Binesse et al., 2015; Rabadi et al., 2015; Shakerley et al., 2015). A growing body of literature has shown that F. tularensis utilizes numerous strategies to avoid activation of ROS-dependent host signaling pathways and killing by host-generated reactive oxygen and reactive nitrogen species (Buchan et al., 2009; Melillo et al., 2009, 2010; McKenna et al., 2010; Langmead and Salzberg, 2012; Binesse et al., 2015; Griffin et al., 2015; Rabadi et al., 2015; Shakerley et al., 2015). Furthermore, at least one phenotypic difference between the virulent F. tularensis Schu S4 strain and the non-pathogenic F. novicida strain is that the latter is considerably more sensitive to H2O2; this sensitivity is associated with activation of the AIM2 inflammasome (Zubay et al., 1972).
In this work we describe how regulation of iron uptake by F. tularensis LVS has a critical impact on the mouse virulence of the organism and on the resulting ability of the strain to induce effective protection against challenge with virulent F. tularensis. We first demonstrate how genetic variability in FeoB-mediated Fe2+ uptake observed in low vs. highly efficacious strains of LVS contributes to H2O2 sensitivity of F. tularensis LVS. We show that a LVS biovar that exhibits greater virulence in mice and engenders strong protective immunity possesses a single nucleotide polymorphism in the feoB gene that confers significantly higher resistance to H2O2 (due to lower iron acquisition) than other LVS biovars. In addition, we confirm that the modulation of iron uptake and the resulting sensitivity to H2O2 is not exclusive to feoB, but can be extended to other iron homeostasis systems in the bacterium. We previously identified a F. tularensis gene involved in iron homeostasis from a mutagenesis screen to identify genes important for growth in human monocyte derived macrophages (MDMs) (Salomonsson et al., 2009). This earlier work identified the bacterioferritin (bfr) gene as important for F. tularensis Schu S4 growth in MDMs infected with pools of transposon mutants (Salomonsson et al., 2009). Specifically, we demonstrate that deletion of bacterioferritin (bfr) increases sensitivity of the bacterium to H2O2, decreases virulence in vivo, and renders LVS poorly protective against challenge with virulent F. tularensis. Together, our findings connect the nutrient acquisition of LVS with its ability to provoke strong vaccine induced immunity.
Materials and Methods
Mutant Construction
Deletion of feoB (FTL_0133) and bfr (FTL_0617) genes was achieved by homologous recombination using derivatives of the non-replicating plasmid pJC84. Primer sequences are shown in Table 1. Briefly, upstream and downstream flanking DNA was amplified via PCR, and amplicons were cloned into the multiple cloning site of pCR2.1 (Life Technologies). A spectinomycin resistance cassette was cloned into the AvrII site in the 3′ end of each upstream PCR fragment. The upstream-spectinomycin resistance fragment was removed by digestion with AscI, and cloned into the AscI site in the 5′ region of the downstream PCR plasmid. The entire upstream-spectinomycin resistance-downstream fragment was cloned into the BamHI site of the suicide plasmid pJC84. Finally, the spectinomycin resistance cassette was removed by digestion with AvrII and the plasmid was re-ligated. Plasmids were electroporated into LVS (2.5 kV, 25 μF, and 600), and the bacteria were plated onto MMH agar with 50 μg/mL kanamycin after 2–3 h of outgrowth. Organisms were then grown overnight in broth lacking kanamycin, and were plated onto MMH agar with 8% sucrose for sacB-mediated counter-selection. Kanamycin sensitive colonies were screened by colony PCR to detect deletion of the feoB or bfr gene. Primers were designed so that they flanked the coding sequence of each gene such that an amplicon would be produced regardless of genotype.
Bacterial Strains and Growth Conditions
Plasmids and bacterial strains used in this work at listed in Tables 2, 3, respectively. Three different biovars of F. tularensis subsp. holarctica LVS were used in this work. LVS obtained from Karen Elkins (FDA, Bethesda, MD) and routinely utilized at the University of Iowa, here called Iowa LVS (University of Iowa) and the RML LVS and ATCC LVS, (Rocky Mountain Laboratories) (Su et al., 2007). Bacteria were routinely cultured on modified Mueller-Hinton (supplemented with 1% glucose, 0.025% ferric pyrophosphate, and 2% IsoVitaleX) agar or in broth, with 50 μg/mL of kanamycin or spectinomycin, as needed. Mueller-Hinton agar was also prepared without ferric pyrophosphate supplementation to assay for growth in lower iron conditions. For some experiments, bacteria were cultured in Chamberlain's defined medium with either 35 μM FeSO4 or 350 nM FeSO4, supplemented with antibiotics as needed. Agar plates were incubated at 37°C with humidity and 5% CO2 while broth cultures were grown at 37°C, shaken at 200 rpm.
Murine Infection
Eight to Ten week old female C57BL/6 mice were maintained on corncob bedding for 1 week prior to infection. Mice were intranasally infected with 25 μL of various doses of F. tularensis LVS strains and mutant derivatives that had been resuspended in PBS. Inocula were calculated by measuring the OD600 value of mid- to late-log phase grown organisms, and were confirmed by serial dilution onto modified Mueller-Hinton agar and enumeration after 2–3 days growth at 37°C with humidity, 5% CO2. Moribund animals (defined as having lost 25% of the initial body weight) were sacrificed in accordance with the protocol approved by the University of Iowa Institutional Animal Care and Use Committee.
Genome Sequencing
Genomic DNA from RML LVS and ATCC LVS was prepared from overnight cultures using DNeasy tissue kit (Qiagen) according to the instructions of the manufacturer. DNA was sequenced using an Illumina HiSeq2500 platform by the Institute for Genomic Medicine Genomic Center (University of California, San Diego, San Diego, CA), resulting in ~7 million reads per sample. Reads were trimmed for adapter sequence and trimmed and filtered for low quality bases using the FASTX-Toolkit (Hannon Lab, CSHL). Resulting reads were mapped to the LVS genome (NC_007880) using Bowtie2 (Ashkenazy et al., 2010). Analysis of genetic modifications, e.g., SNPs and gene deletions was performed using GATK comparing both RML LVS and ATCC LVS genomes to each other and the annotated LVS genome (Landau et al., 2005).
Iron-Regulated β-Galactosidase Reporter Activity
To assess iron uptake-regulated gene expression, Miller assays were performed with Iowa LVS, RML LVS, and ATCC LVS carrying the fslA promoter-lacZ fusion on the pBB103 plasmid (Troxell and Hassan, 2013). Bacteria were grown in Chamberlain's defined medium (supplemented with 50 μg/mL spectinomycin) overnight, and β-galactosidase activity was assayed using the standard Miller assay (Sullivan et al., 2006).
FeoB Function in a Heterologous Reporter System
The coding sequence of feoB was PCR-amplified from both Iowa and RML LVS using the high fidelity Phusion polymerase (New England Biolabs). Sanger sequencing was performed to confirm that the SNP observed in the RML LVS background was present. The feoA coding sequence was cloned into the KpnI/SalI sites of a derivative of pTrc99a, immediately downstream of the groE promoter. The entire PgroE-feoA fragment was removed by digestion with BamHI and SalI, and ligated into the same sites in pBB103. Both this plasmid and the pWKS30 containing feoB were transformed into the E. coli H1771 strain (MC4100 aroB feoB7 fhuF::plac Mu). Control strains were also generated that carried only one of the plasmids. Miller assays were performed to measure FeoB Fe2+ import activity via the readout of Fur repression of fhuF::placZ. All strains were grown in LB supplemented with 50 μg/mL of spectinomycin and 100 μg/mL of ampicillin, as necessary.
Hydrogen Peroxide Sensitivity
To measure resistance to H2O2-mediated killing, bacteria were grown in modified Mueller-Hinton broth and mid- to late-log phase organisms were pelleted at 13,200 RPM for 5 min, washed in PBS, and ~106 CFU were resuspended in 200 μL PBS with or without 100 μM hydrogen peroxide (H2O2) in a 96-well dish. The samples were incubated at 37°C with humidity and 5% CO2 for 1 h. The culture from each well was then serially diluted in sterile PBS, plated onto modified Mueller-Hinton agar (with antibiotic when necessary) and the number of surviving organisms for each strain was enumerated after 2–3 days.
In Vitro Infections
LVS infections of bone marrow derived macrophages (BMM) were performed as previously described (Su et al., 2007). Briefly, BMM were differentiated from femurs of C57Bl/6 mice over the course of 7 days in complete DMEM (cDMEM, DMEM supplemented with 10% heat-inactivated fetal calf serum [FCS], 0.2 mM l-glutamine, 1 mM HEPES, and 0.1 mM non-essential amino acids [NEAA], all from Life Technologies) supplemented with M-CSF (Peprotech). Immediately prior to infection medium was removed and reserved for addition after infection. The indicated bacterial strains were added at a MOI of 50 and co-incubated with BMM for 90 min. Bacteria were then removed and fresh medium containing 50 μg/ml of gentamicin was added to kill extracellular organisms. After 45 min, BMM were washed extensively with D-PBS (Life Technologies) and the reserved medium was returned to the wells. IFN-γ (Peprotech) was then added to the indicated groups at a final concentration of 10 U/ml. At the indicated time points, medium was collected and assessed for cytokines as described below. BMM were washed 3 times with D-PBS followed by lysis with water. Lysates were serially diluted and plated on to MMH agar for enumeration of viable organisms.
Statistics
All statistics were calculated with Graphpad Prism software (San Diego, CA). To compare ß-galactosidase activity across multiple strains we used one-way ANOVA with Dunnett's multiple comparisons test. For comparisons of multiple strains across multiple conditions, we utilized two-way ANOVA with either Sidak's or Tukey's multiple comparisons tests. When applicable, an unpaired two-tailed t-test was used. The LD50 values were calculated using the method of Reed and Muench. To compare survival after F. tularensis SchuS4 challenge, a log-rank sum test was utilized. The error bars in the presented data represent the standard error of the mean. A p < 0.0001 is represented by ****p < 0.01 is represented by **p < 0.05 is represented by *, and “ns” indicates “not significant.”
Results
Genome Sequencing of F. tularensis LVS Strain
It was recently reported that there are differences in virulence and vaccine efficacy between LVS isolates (Deng et al., 2006; Su et al., 2007). Specifically, a low passage strain, termed RML LVS, had increased virulence in C57Bl/6 mice but also engendered complete protection against challenge with virulent F. tularensis subsp tularensis. In contrast, LVS strain ATCC29684 (ATCC LVS) had lower virulence in C57Bl/6 mice and failed to protect animals from lethality following F. tularensis challenge. The specific genetic changes that could account for the dramatic in vivo differences between these two strains were not identified. Therefore, in this work we have sequenced the genomes of these two LVS biovars to identify and characterize these genetic differences. Surprisingly, we detected very few differences in the sequences between RML LVS and ATCC LVS. We detected a 93 base pair deletion in the gene encoding a Dyp-type peroxidase that has been previously characterized (Binesse et al., 2015). Additionally, we detected a single nucleotide polymorphism in the feoB gene encoded by RML LVS. The C to A substitution leads to an aspartate to tyrosine mutation in the coding sequence of feoB. The D471Y mutation maps to a cytoplasmic loop of the FeoB protein, adjacent to a highly conserved glycine residue that is predicted to be functionally important by the ConSurf program (Kammler et al., 1993; Buchan et al., 2008). Although the genome of the lab stock of Iowa LVS from the Jones lab has not been completely sequenced, the strain was also included in the experiments described here. Sanger sequencing confirmed that the Iowa LVS feoB encodes an aspartate at residue 471, similar to the published LVS genome. FeoB is an inner membrane protein that imports ferrous iron into the bacterial cytoplasm and has previously been linked to virulence as one of two major iron uptake pathways in LVS (Schulert et al., 2009; Thomas-Charles et al., 2013; Perez and Ramakrishnan, 2014). The substitution of a large, hydrophobic tyrosine for an aspartate at residue 471 led us to hypothesize that the FeoB protein encoded by the RML LVS allele imports Fe2+ poorly, or not at all. We determined if differences in iron acquisition could account for the varied virulence and vaccine efficacy of RML LVS vs. ATCC LVS. Since iron plays an integral role in host-induced oxidative stress, it is likely that controlling intracellular iron levels is a significant component of the virulence strategy of F. tularensis. Furthermore, sensitivity to oxidative stress may contribute to the vaccine efficacy of LVS given that the ATCC biovar of LVS has an aspartate at residue 471 in FeoB and the 93 base pair dyp deletion, and is both relatively attenuated in murine infections and stimulates a weak immune response against a challenge with virulent F. tularensis Schu S4 (Griffin et al., 2015).
RML LVS Displays Phenotype Consistent With Lower Intracellular Fe2+ Than the ATCC LVS or Iowa LVS
Our first set of experiments was designed to test the hypothesis that RML LVS has less intracellular iron than either the Iowa LVS or the ATCC LVS. Strains were grown overnight in standard modified Mueller-Hinton (MMH) broth, serially diluted in PBS, and ten-fold dilutions of liquid bacterial cultures were spotted onto MMH agar with varying concentrations of iron (Figure 1A). Growth was identical among each strain when spotted onto the control agar containing the typical concentration of iron (0.0025% ferric pyrophosphate) routinely used for propagation of F. tularensis. When the iron concentration was reduced by 50%, both the Iowa LVS and ATCC LVS had growth patterns similar to that observed on the control plate, however, RML LVS exhibited growth restriction at this concentration of iron. Fewer isolated colonies were observed and lawn growth was less luxurious when compared to the Iowa and ATCC LVS. On MMH agar lacking added iron, all three strains exhibited growth restriction, however the reduced growth phenotype of RML LVS was exacerbated, with extremely poor growth even at the lowest dilution plated. Iowa and ATCC LVS both grew as lawns at these dilutions, indicating that they were able to scavenge sufficient iron from the agar plate environment, while RML LVS could not. The decrease in RML LVS growth was approximately two orders of magnitude greater than that observed for either the Iowa or ATCC LVS, consistent with the hypothesis that RML LVS has less intracellular iron under conditions of iron limitation. We interpreted these results to mean that RML LVS had inherently lower levels of intracellular iron and that when growing the strain on agar with less iron, the amount that RML LVS could acquire for growth was limiting, thus the strain grew poorly relative to the Iowa LVS or ATCC LVS.
Figure 1. RML LVS has relatively less intracellular iron than other LVS biovars. (A) Bacteria were grown overnight in standard MMH broth, serially diluted in sterile PBS and plated for enumeration on MMH agar with 0.025%, 0.0125%, or no added Fe3+PPi. Data from a representative experiment is shown. (B) Miller assay for each LVS carrying a plasmid with the Fur regulated PfslA-lacZ reporter. Bacteria were grown overnight in Chamberlain's defined medium with 35 μM (high iron) or 350 nM (low iron) FeSO4. Two-way ANOVA with Sidak's multiple comparisons was used to test for significance. Shown is a representative Miller assay from replicate experiments. (C) Strains overexpressing fur were grown in MMH broth overnight, serially diluted into sterile PBS and plated on standard MMH agar. Shown are representative data from two similarly designed experiments.
To confirm that the mutation carried in feoB by RML LVS had a direct consequence on iron levels, we performed experiments designed to give a semi-quantitative measure of intracellular iron levels. To avoid toxicity associated with the Fenton reaction that include lipid peroxidation, DNA damage, and poisoning of the iron-sulfur cluster enzymes of essential metabolic pathways, bacteria have evolved regulatory mechanisms to maintain iron homeostasis. One such example is the Fur system (Troxell and Hassan, 2013). The Fur transcriptional repressor is an allosteric regulator that uses Fe2+ as a co-repressor; when iron levels are sufficient, the Fur-Fe2+ complex binds to Fur Box sequence motifs in the promoters of iron uptake genes to mediate their repression. When iron is limiting, less Fe2+ is bound to Fur, decreasing its ability to bind DNA. This leads to de-repression of iron uptake genes. Since several studies have demonstrated that Francisella has a Fur regulatory system that functions similarly to other organisms (Deng et al., 2006; Sullivan et al., 2006; Buchan et al., 2008; Ramakrishnan et al., 2008), we were able to design experiments to indirectly measure F. tularensis LVS intracellular iron levels. A plasmid carrying a Fur-regulated PfslA-lacZ construct was introduced into the RML LVS, Iowa LVS, and ATCC LVS. Iron limitation is known to relieve Fur repression at the fslA promoter, so increased PfslA-lacZ reporter activity correlates with decreasing concentrations of intracellular Fe2+ (Buchan et al., 2009). Each LVS strain was grown in Chamberlain's defined medium (CDM) with 35 μM (high iron) or 350 nM (low iron) FeSO4, and β-galactosidase activity was measured after overnight growth (Figure 1B). As predicted, iron starvation induced high expression of PfslA-lacZ in all three strains; however, the lacZ reporter expression was approximately 2-fold higher in RML LVS than that observed for Iowa LVS or ATCC LVS. The higher activity of the PfslA-lacZ construct in the RML LVS is consistent with the hypothesis that this strain has lower levels of intracellular Fe2+ when iron is limiting in the growth medium.
To assess intracellular iron levels in RML LVS by another method, we reduced the ability of each strain to import iron by overexpression of Fur (which downregulates genes encoding iron uptake systems). The F. tularensis fur gene was placed under the control of the groE promoter, and the construct was introduced into each strain. Bacteria were grown in MMH broth with standard iron concentrations overnight (no differences in growth rate were observed between the two strains in iron replete conditions), serially diluted in PBS and plated onto standard MMH agar (Figure 1C). Iowa LVS and ATCC LVS, when overexpressing Fur, exhibited no significant difference in colony growth compared to vector controls, indicating that intracellular pools of iron were still sufficiently high to support normal growth, even with Fur overexpression. In contrast, RML LVS had much smaller colony size and poor lawn formation. We interpret these results to indicate that Fur overexpression in RML LVS had a greater impact on growth because Fur repression of iron uptake genes decreased the intracellular Fe2+ concentration to levels such that the strain could not grow normally, even though sufficient iron was present in the agar media. These results are consistent with the hypothesis that intracellular Fe2+ pools of RML LVS are lower than in Iowa LVS or ATCC LVS and can be manipulated to become limiting for growth in conditions that are not limiting for the other two strains.
FeoB D471Y Does Not Complement E. coli ΔfeoB fhuf::λplac
The data demonstrating that RML LVS has lower intracellular pools of iron than either Iowa LVS or ATCC LVS suggest that the single nucleotide change in the feoB gene of the RML strain encodes a protein with significantly reduced Fe2+ import activity. To assess the ability of the FeoB D471Y protein to transport iron, we made use of a well-characterized E. coli iron reporter strain that lacks feoB (Kammler et al., 1993; Weaver et al., 2013). This strain, E. coli H1771, has a chromosomal transcriptional lacZ reporter in the Fur-regulated gene fhuF which can be used to report the relative intracellular iron concentrations within the strain. When Fe2+ levels are high, expression of the fhuF::lacZ reporter is low and when Fe2+ levels are low, high β-galactosidase activity is observed. The feoB alleles from Iowa LVS and RML LVS (D471Y) were introduced into E. coli H1771 on the low copy number plasmid pWKS30, and Miller assays were performed (Figure 2). Initial experiments supplying only feoB (either allele) showed no repression of fhuF::lacZ, indicating that FeoB alone is not sufficient for Fe2+ import in this reporter system (data not shown). In most bacterial species encoding a feo system, the small feoA gene is encoded either in an operon with feoB or elsewhere on the chromosome and the encoded protein is required to stimulate FeoB Fe2+ import activity (Cartron et al., 2006; Lau et al., 2013; Weaver et al., 2013). When the Francisella feoA was supplied on pBB103 in concert with feoB (Iowa/ATCC allele) on pWKS30, significant repression of the fhuF-lacZ reporter was observed (~ 80% reduction in expression relative to the empty vector control), indicating that Fe2+ import was significantly increased. In contrast, complementation of H1771 with feoA and feoB D471Y (RML allele) failed to repress fhuF::lacZ. This result is consistent with the hypothesis that the FeoB D471Y protein from the RML LVS is significantly impaired in its ability to import Fe2+.
Figure 2. The RML LVS feoB allele is non-functional in a heterologous iron reporter system. The coding sequence of each feoB allele was cloned into the low copy plasmid pWKS30 and introduced to E. coli H1771. The coding sequence of feoA was fused to the groE promoter in a derivative of plasmid pBB103 and supplied to E. coli H1771 in tandem with the feoB expression plasmids, and Miller assays were performed as described in the Materials and Methods. A one-way ANOVA with Dunnett's multiple comparisons test was used to test for significance against the control pWKS30 plasmid. Significance was set at p < 0.05. **Indicates a significant difference between the labeled samples and pWKS30. Shown is a representative experiment from three replicates.
The feoB d471Y Allele Is Associated With Resistance to H2O2
Since we have provided several lines of evidence consistent with the hypothesis that RML LVS has less intracellular Fe2+ than either the Iowa or ATCC LVS strains and Lindgren et al. demonstrated that strains with lower intracellular iron concentrations are more resistant to H2O2, we hypothesized that RML LVS would be more resistant to killing by H2O2 than Iowa LVS or ATCC LVS. Bacteria were exposed to PBS alone or PBS with 100 μM H2O2 in a 96-well dish for 1 h at 37°C with 5% CO2 and humidity. Following incubation, the bacteria were serially diluted in PBS and plated onto MMH agar (Figure 3A). The RML LVS was typically ten-fold more resistant to H2O2 than the Iowa strain, and approximately 100-fold to 1,000-fold more resistant than ATCC strain. The genome of ATCC encodes a Dyp peroxidase with a 93-base pair deletion, and it has been reported that this deletion can contribute to increased sensitivity to H2O2; an Iowa LVS Δdyp mutant matches the H2O2 sensitivity of ATCC LVS (data not shown) and is in agreement with the data from Binesse et al. (2015).
Figure 3. RML LVS is more resistant to H2O2 as a result of the point mutation in its feoB. (A) Strains were grown overnight in MMH broth, resuspended to ~106 CFU in PBS and exposed to 100 μM H2O2 for 1 h at 37°C. Samples were serially diluted in sterile PBS and plated in triplicate. Shown is the CFU/ml in PBS alone and PBS + H2O2 from a representative experiment. A two-way ANOVA with Tukey's multiple comparisons test was used to test for significance. Shown is a representative of 3 replicates. (B) Each feoB allele was constitutively expressed from the groE promoter in a derivative of the multi-copy plasmid pBB103. These plasmids were introduced into the RML LVS and H2O2 sensitivity assays were performed as before, with the addition of a 30-min time point. Shown is the CFU/ml in PBS alone and PBS + H2O2 from a representative experiment. A two-way ANOVA with Sidak's multiple comparison test was used to test for significance. Shown is a representative of two replicates. (C) An Iowa LVS ΔfeoB mutant was complemented with each feoB allele in trans, and H2O2 sensitivity assays were performed as before. A two-way ANOVA with Tukey's multiple comparisons test was used to test for significance. Significance was set at p < 0.05. ****Indicates a significant difference between the two samples bracketed. Shown is a representative of two replicates.
Given the increased sensitivity of Iowa LVS to H2O2, and the functional complementation of an E. coli feoB mutant by the Iowa feoB allele, we next tested if the RML LVS could be sensitized to H2O2 by increasing the intracellular Fe2+ pool via overexpression of a functional feoB. This was achieved by transforming the strain with a plasmid carrying constructs Pgro-feoB (Iowa/ATCC allele–high Fe2+ transport) and Pgro-feoB (D471Y; RML allele–low Fe2+ transport) which both overexpress feoB. H2O2 sensitivity assays were performed as described previously, except that a 30 min H2O2 exposure time point was also included in case overexpression of the feoB genes led to substantially faster killing by hydrogen peroxide (Figure 3B). Nearly one log of killing was observed at 30 min for the RML + Pgro-feoB (Iowa/ATCC), while RML + Pgro- feoB (D471Y) only had a two-fold reduction in viability. The effect was significantly more pronounced at the 1 h time point, with no viable bacteria recovered at the level of detection (102 CFU) from RML + Pgro-feoB (Iowa/ATCC). In contrast, over 104 CFU were recovered at 1 h from the RML + Pgro- feoB (D471Y). These data demonstrate that the single nucleotide change in the feoB gene (D471Y0 encoded in the RML LVS genome results in significantly higher resistance to H2O2 compared to the feoB allele in either Iowa LVS or ATCC LVS.
To confirm that the H2O2 resistance phenotype conferred by feoB D471Y is not unique to the RML genetic background, we constructed a ΔfeoB mutant in the Iowa LVS background and complemented it with the Pgro-feoB constructs. The Iowa LVS strain background was chosen over the ATCC LVS background, as the latter strain was approximately 100-fold more sensitive to H2O2 than the RML LVS due to the 93 base pair deletion in the dyp peroxidase gene, while RML LVS and Iowa LVS both encode a full length dyp. H2O2 sensitivity assays were performed as described previously and, consistent with results in the RML strain background, overexpression of feoB D471Y mediated significant resistance to H2O2 in the Iowa LVS ΔfeoB mutant, relative to that when the Iowa/ATCC feoB was overexpressed (Figure 3C).
Bacterioferritin Protects Against Hydrogen Peroxide
To assess the role of the bfr gene in F. tularensis iron homeostasis and oxidative stress resistance, a Δbfr strain was constructed in the RML LVS and PfslA-lacZ assays, as well as H2O2 sensitivity assays were performed as described above. No growth differences were observed in the Δbfr mutant compared to the wild type strain. The RML LVS Δbfr strain exhibited a modest decrease (~20%) in PfslA-lacZ reporter activity relative to the parent strain (Figure 4A), while in the H2O2 sensitivity assay, as before, the RML LVS exhibited relatively high level resistance to H2O2. The Δbfr mutant had ~10-fold fewer viable organisms, similar to Iowa LVS (Figure 4B). A possible explanation for these results is that in the absence of the Bfr iron storage protein, intracellular ferrous iron levels are elevated relative to the parent RML LVS, which may lead to increased ferrous iron available for the Fenton reaction in the presence of H2O2 and the increased toxicity observed in the assay. Consistent with this, PfslA-lacZ activity was modestly reduced (~20%) in the RML LVS Δbfr mutant (Figure 4B). As a result, we conclude that bacterioferritin contributes to protection against oxidative stress, consistent with bacterioferritin function in Neisseria gonorrhea and Mycobacterium tuberculosis.
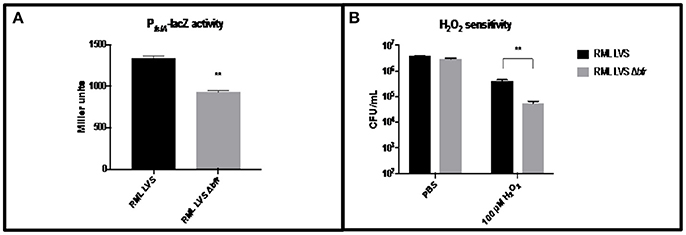
Figure 4. Iron related phenotypes of RML LVS Δbfr. (A) PfslA-lacZ reporter activity is decreased in the RML LVS relative to wild type. Welch's t-test was used to test for significance. Shown is a representative of five replicates. (B) The RML LVS Δbfr is more sensitive to H2O2 killing than the wild type. A two-way ANOVA with Sidak's multiple comparisons test was used to test for significance. Significance was set at p < 0.05. **Indicates a significant difference between the samples indicated. Shown is a representative of four replicates.
Expression of the Iowa/ATCC LVS feoB Allele Is Deleterious in IFN-γ-Stimulated Macrophages
One mechanisms by which IFN-γ can control intracellular bacteria is to provoke production of reactive nitrogen and reactive oxygen species. It has been reported that IFN-γ mediated control of Francisella replication can be independent of RNS and ROS (Edwards et al., 2010). However, this may be due to its ability to limit toxicity of ROS by restricting iron. Therefore, we hypothesized that strains of LVS that were unable to control iron acquisition similarly to wild type RML LVS would be more sensitive to host cell IFN-γ mediated killing. We first compared RML LVS, ATCC LVS, RML LVS ΔfeoB and RML LVS overexpressing RML feoB or Iowa/ATCC feoB. In agreement with our hypothesis RML LVS was the least sensitive to IFN-γ mediated killing among the strains tested (Figures 5A,B). Deletion of the feoB gene rendered the bacteria significantly more sensitive to IFN-γ mediated killing (Figures 5A,B). However, resistance to IFN-γ was partially restored in bacteria complemented with RML feoB. Complementation with ATCC feoB did not significantly change RML LVS sensitivity to IFN-γ mediated killing. We next tested the ability of RML LVSΔbfr to resist IFN-γ mediated killing. Similar to RML LVS ΔfeoB, RML LVS Δbfr was significantly more sensitive to IFN-γ mediated killing compared to wild type controls (Figures 5C,D). Together these data suggest that alterations in ferrous iron homeostasis confer sensitivity to IFN-γ mediated control of intracellular replication of LVS.
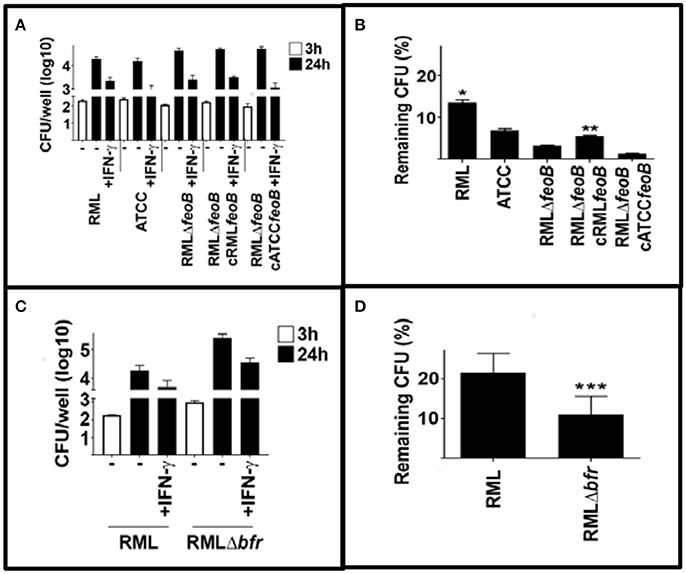
Figure 5. Mutations in iron acquisition enhance sensitivity of LVS to IFN-γ mediated killing. BMM were infected with the indicated strains of LVS and were treated with IFN-γ (10 U/ml) immediately after infection. Three and 24 h after infection intracellular bacteria were enumerated. (A,B) RML LVS is more resistant to IFN-γ mediated killing compared to all other strains tested. Deletion of the feoB gene rendered RML LVS more sensitive to IFN-γ mediated killing and complementation with homologous feoB, but not ATCC feoB partially restored resistance to IFN-γ. (C,D) Deletion of the bacterioferritin (bfr) also increased RML LVS sensitivity to IFN-γ mediated killing. Error bars represent SD. Significance was tested via two-way ANOVA followed by Tukey's multiple comparison of means. Significance was set at p < 0.05. *, significantly greater than all other samples. **, significantly greater than RMLΔfeoB. ***, significantly less than RML LVS. Data is representative of three experiments of similar design.
RML LVS Δbfr Is Attenuated in Murine Infection and Fails to Protect Against F. tularensis SchuS4
As strains with increased sensitivity to H2O2 are associated with IFNγ-mediated killing in BMMs, we sought to determine if strains with increased sensitivity to H2O2 in vitro might also be attenuated in a mouse model of respiratory tularemia. Disappointingly, several efforts to assess virulence of Iowa LVS, its isogenic ΔfeoB mutant and complemented derivatives gave inconclusive results, likely due to plasmid instability in the LVS strains in the absence of antibiotic selection in vivo. Thus, we pursued an alternative approach to assay for the role of altered ferrous iron homeostasis in vivo and the ability of RML LVS to provide protection against a challenge with F. tularensis SchuS4. To this end, we used the H2O2 sensitive RML LVS Δbfr mutant in murine intranasal infections. RML LVS was previously reported to have a LD50 of 174 CFU in C57BL6 mice (Griffin et al., 2015). We confirmed this value by performing a similar infection experiment and calculated a LD50 value for RML LVS in C57BL/6 mice of 195 CFU (Figure 6A). To test if bacterioferritin contributed to the virulence of the F. tularensis RML LVS, three groups of mice (n = 5 per group) were intranasally infected with 215 CFU, 2.15 × 103, or 2.15 × 104 CFU of the RML LVS Δbfr mutant, and their weight and health status was followed daily over the course of the experiment. All mice that received 215 CFU survived (average weight loss of ~12 percent) while forty percent of the mice survived infection with 2 × 103 CFU (average weight loss of ~22%); none survived 2 × 104 CFU. Importantly, it took >2,000 CFU of the RML LVS Δbfr strain to yield survival curves and weight losses in mice that approximated that observed for mice infected with only 133 CFU of RML LVS, a >15-fold difference in inoculum (Figures 6B,C). The estimated LD50 of the RML LVS Δbfr mutant was 1,464 CFU. These results demonstrate that the bacterioferritin protein contributes to the virulence of RML LVS in mice; however, future work is needed to determine if this is a consequence of the H2O2 sensitivity of this mutant.
Figure 6. Bacterioferritin is required for optimal RML LVS virulence and. (A) Groups of mice (n = 5) were intranasally infected with 25 μl inocula containing 133 or 1,330 CFU of RML LVS in PBS. Mice were weighed daily and humanely sacrificed after weight loss ≥ 25% of their initial weight. LD50 values were calculated via the method of Reed and Muench (1938). (B) Groups of mice (n = 5) were intranasally infected with 25 ul inocular containing 215, 2,150 or 21,500 CFU of RML LVS Δbrf in PBS. Mice were weighed daily and humanely sacrificed after weight loss ≥ 25% of their initial weight. LD50 values were calculated via the method of Reed and Muench (1938). (C) The average weight loss of mice infected with 102 or 103 of the RML LVS strain or 102 or 103 of the RML LVS Δbfr is represented by the bars in the graph. The significance of the weight loss of the mice infected with the RML LVS strain was compared to the weight loss of the mice infected with the RML LVS Δbfr strain Mice infected with RML LVS were closer to or at clinical endpoint at all doses, while mice infected with RML LVS Δbfr lost significantly less weight at the lowest infectious dose. Significance was set at p < 0.05. ****Indicates a significant difference between the two samples bracketed. Significance was tested via log-rank sum test.
Wild type RML LVS uniformly protects C57Bl/6 mice against challenge with virulent Francisella whereas ATCC LVS does not (Griffin et al., 2015). We have established that an important difference between RML LVS and ATCC LVS is their acquisition of iron and subsequent sensitivity to ROS that is inversely correlated with protective efficacy. Since mutation of bfr in RML LVS led to a decrease in PfslA-lacZ reporter activity, increased sensitivity to H2O2, and decreased virulence we hypothesized that vaccination with RML LVS Δbfr would provide poor protection against challenge with virulent F. tularensis SchuS4. Accordingly, 6 weeks after infection with the RML LVS Δbfr mutant, we challenged all animals that survived with F. tularensis SchuS4 (Figure 7). As a control, mice that had been intranasally infected with 121 CFU of RML LVS (n = 3) were also challenged with 25 CFU of F. tularensis SchuS4. As expected all mice that had previously been infected with RML LVS survived SchuS4 challenge. In contrast, none of the mice previously infected with RML LVS Δbfr strain were protected from the SchuS4 infection and all mice died within 8 days of challenge. Thus, while prior infection with RML LVS Δbfr did extend the mean time to death, it provoked incomplete protection against subsequent infection with fully virulent SchuS4. Together this suggests that Francisella iron homeostasis and its interplay with host elements that control bacterial replication are important factors that contribute to both survival of primary infection and induction of adaptive immune responses in vivo.
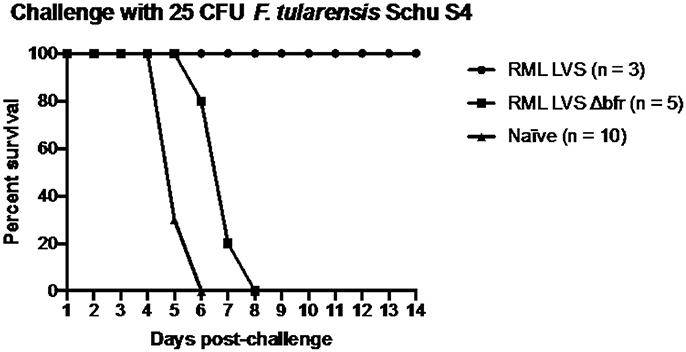
Figure 7. RML requires bacterioferritin to elicit protection against an intranasal challenge of 25 CFU F. tularensis Schu S4. Mice were infected with either 121 CFU of RML LVS or 215 CFU of the bacterioferritin mutant and allowed to recover for fix weeks before challenge. The challenge inoculum was suspended in 25 μl sterile PBS and administered intranasally. Health was monitored for the duration of infection and mice were humanely sacrificed if ≥25% of initial weight was lost. Data shown is from one replicate. Significance was tested via log-rank sum test.
Discussion
Francisella tularensis is a highly pathogenic bacterium for which no effective licensed vaccine exists. Due to concerns that the extreme pathogenicity of the organism might lead to its intentional release there is increased interest in developing an effective vaccine for F. tularensis infections. A complication with the LVS is that the genetic mutations that attenuated LVS are undefined, leaving open the possibility that the strain could revert to full virulence. While LVS is unlikely to be reapproved for human vaccination, it is used extensively as a standard by which other candidate Francisella vaccines can be measured in animal models, as well as a tool to discover correlates of immunity to F. tularensis. Indeed, in a previous report it was found that the RML LVS biovar stimulated protective immunity in a mouse model of vaccine against virulent F. tularensis Schu S4 by inducing higher numbers of effector T cells (Griffin et al., 2015). Building on this observation, we have sought to identify the genetic differences between biovars of LVS and explore how they may affect the outcomes of virulence and immunity in mouse models of infection. In this report, we have demonstrated that there is a significant difference between the RML LVS and the Iowa LVS or ATCC LVS biovars in ferrous iron homeostasis.
Iron is one of the most abundant metals on the planet but is usually found in the environment in an insoluble oxidized state, which has required bacteria to develop mechanisms to scavenge iron from their local environment (Crichton and Pierre, 2001). The need for bacterial pathogens to acquire iron within a host environment presents additional challenges to the microorganism as they must also contend with host responses, which include mounting an immune response and sequestering iron, both of which can severely limit the ability of the pathogen to grow within the host (Cassat and Skaar, 2013). As expected, F. tularensis requires iron for productive infection of host cells, as mutants in ferrous and ferric iron uptake systems in F. tularensis LVS and F. tularensis Schu S4 were incapable of intracellular growth in vitro and were attenuated for virulence in mice (Perez and Ramakrishnan, 2014; Perez et al., 2016). Other work has shown that F. tularensis significantly increases expression of host iron-related genes, including transferrin receptor, presumably to increase the labile iron pools in the cytosol of infected host cells (Pan et al., 2010). Although F. tularensis requires iron for growth in a host, it appears to be unusual among bacterial pathogens in that its virulence may depend, in part, on maintaining optimally low levels of iron within its cytoplasm, as the highly virulent subsp. tularensis strains had approximately 4- to 5-fold less intracellular iron than the moderately virulent subsp. holarctica strains (Lindgren et al., 2011).
Consistent with this model of Francisella virulence, we have shown that the more virulent RML LVS biovar encodes a non-synonymous mutation in the feoB gene that confers phenotypes consistent with lower ferrous iron levels. The strain grows similarly to the Iowa and ATCC LVS biovars in standard propagation media, yet is restricted for growth when the iron concentrations are significantly reduced. This indicates that when extracellular iron levels become limiting, the RML LVS may not be as proficient at scavenging iron (via the feoB system) as Iowa LVS and ATCC LVS. Numerous labs have reported that expression of the fsl iron uptake operon genes are tightly regulated (repressed in high iron, induced in low iron) by iron concentrations in the media, so we used a PfslA-lacZ reporter as an indirect measure of relative ferrous iron levels between the various LVS biovars (Deng et al., 2006; Ramakrishnan et al., 2008; Lindgren et al., 2009). As with growth phenotypes, there were no detectable differences in PfslA-lacZ expression in growth media with normal iron concentrations; however, the RML LVS had increased reporter activity (consistent with lower intracellular iron levels) relative to the Iowa LVS and ATCC LVS after growth in iron limiting media. We probed feoB function of the various LVS strains further by performing functional complementation experiments. Using a E. coli Δ feoB mutant with a Fur-repressed fhuF-lacZ reporter as the complementation strain, we observed that the RML LVS feoB allele did not mediate repression of the fhuF-lacZ reporter (low intracellular iron), while both the Iowa LVS and ATCC LVS alleles did (high intracellular iron concentrations). Collectively, these experimental results provide compelling genetic evidence that the RML LVS likely has lower basal levels of intracellular ferrous iron than the Iowa LVS and ATCC LVS biovars. To our knowledge this is the first direct genetic evidence that explains differences in iron content between F. tularensis strains, though it is likely unique to LVS biovars. A significant contribution of this work is that it helps to provide a rationale for why virulent F. tularensis strains may carefully control their levels of intracellular iron as it appears that they balance the requirement of iron for growth against the effects of the Fenton reaction, which may induce oxidant damage to the bacteria that could expose the organisms to the host immune response (Binesse et al., 2015).
The low ferrous iron phenotypes exhibited by subsp. tularensis have been reported to correlate with resistance to the lethal effects of H2O2, so we sought to characterize the relative sensitivity of each LVS biovar in vitro. Indeed, the RML LVS exhibited nearly ten-fold more resistance to hydrogen peroxide than the Iowa LVS, which encodes a functional FeoB, and 100-fold or more resistance than the ATCC LVS. The latter strain is significantly more sensitive to H2O2 as it is competent for Fe2+ uptake through its functional FeoB, and because it encodes a Dyp peroxidase lacking 31 amino acids near the C-terminal portion of the protein. Mutation of the dyp gene has been shown previously to increase H2O2 sensitivity (Binesse et al., 2015), and a clean deletion of dyp in the Iowa LVS recapitulates this phenotype (unpublished data). We found that the RML LVS became significantly more sensitive to H2O2 by introducing a functional feoB allele under constitutive expression on a plasmid; the H2O2 sensitivity was significantly lower when the nonfunctional RML allele was introduced into the strain. We further demonstrated that the increased H2O2 lethality associated with overexpression of a functional feoB was not unique to the RML LVS genetic background, as an Iowa LVS ΔfeoB mutant complemented with the mutant allele was more resistant to H2O2 than that complemented with the functional allele. These experiments are significant, as the strains only differed by a single nucleotide in feoB, yet exhibited nearly ten-fold differences in their sensitivities to H2O2, irrespective of genetic background. In sum, these experiments strongly suggest that RML LVS is better at avoiding Fe2+-associated toxicity in the context of the host environment virtue due to its lower functioning FeoB system.
In addition to characterizing the feoB D471Y mutation in RML LVS, we also characterized a mutant lacking the putative iron storage protein bacterioferritin. Bacterioferritin is known to oxidize ferrous iron and store the ferric oxide mineral in the hollow cavity formed by a 24-mer sphere in other organisms (Honarmand Ebrahimi et al., 2015). We reasoned that a F. tularensis RML LVS mutant lacking bacterioferritin may be unable to sequester free Fe2+ as effectively as the wild type strain, and that this would be reflected in iron-related gene expression. The PfslA-lacZ reporter indeed showed a modest decrease in expression in the Δbfr mutant, indicating that the strain may have more Fe2+ available to interact with Fur and repress expression of the reporter. This result is consistent with the hypothesis that F. tularensis LVS uses bacterioferritin to alter the pools of intracellular Fe2+ levels by sequestration. The PfslA-lacZ reporter expression decreased by only ~20% suggesting that the bacteria may utilize other iron storage mechanisms as well. As the iron-regulated reporter indicated that the Δbfr mutant may have increased levels of intracellular ferrous iron, we hypothesized that the strain would be more sensitive to H2O2. We found that the Δbfr mutant had approximately ~90% reduction in viability relative to the parental strain, indicating that bacterioferritin has a role in resistance to oxidative stress in F. tularensis LVS.
As the plasmid-based complementation strategy for the Iowa LVS ΔfeoB was unsuitable for in vivo studies, we utilized the RML LVS Δbfr mutant as an alternative for proof of principle, given the lower PfslA-lacZ reporter activity and the increased sensitivity to H2O2 of this mutant. Intranasal infections of mice confirmed that bacterioferritin was required for full virulence of the strain, with the LD50 of the mutant estimated to be ~1,500 CFU, nearly an eight-fold increase relative to the parental strain (<200 CFU). Additionally, the mice exhibited differences in the severity of disease, as mice infected with 133 CFU of RML LVS lost considerable weight during the infection, approximately twenty-three percent on average. In contrast, the mice infected with 215 CFU of the Δbfr mutant lost significantly less body weight during infection (~12%), even though the dose was almost two times higher. Only at a ten-fold higher dose of the Δbfr mutant did the mice lose similar amounts of weight as those infected with the lowest dose of the parent strain.
As the RML LVS has been shown to be more effective at stimulating a protective response to challenge with F. tularensis Schu S4 than the more H2O2 sensitive biovar of LVS (ATCC), we hypothesized that loss of H2O2 resistance would render it a poor vaccine strain. Consistent with our hypothesis, the RML LVS Δbfr did not stimulate robust protection in mice infected with this strain, as all succumbed from Schu S4 infection by day eight. These data suggest that H2O2 resistance may contribute to the efficacy of the RML LVS as a vaccine against tularemia in mice; however, it is possible that other interpretations of the data are valid. For example, the bacterioferritin protein is known to stimulate antibody production and T cell proliferation in LVS immunized mice, however it isn't clear that antibody responses are effective at controlling infection with virulent F. tularensis Schu S4 (Bakshi et al., 2008). In in vitro studies, F. tularensis Schu S4 is able to bind the active form of the host protease plasmin, which can cleave Francisella specific antibodies to avoid the ensuing bactericidal and inflammatory consequences of phagocytosis via antibody mediated opsonization (Crane et al., 2009). Furthermore, introduction of monoclonal antibodies against Francisella bacterioferritin into mice did not provide protection against lethal LVS challenge, even though other antibodies can be highly effective at protecting against this strain (Cole et al., 2009; Savitt et al., 2009). Taken together, these observations suggest that the lack of a bacterioferritin-specific antibody response is not likely to explain the poor efficacy of the RML LVS Δbfr at protecting against Schu S4.
This work is significant for several reasons. The observation that the RML LVS is better able to induce host immunity than other LVS strains and has phenotypes consistent with relatively lower levels of free ferrous iron indicates that there is an important inverse correlation between the two. This finding is of particular importance for research groups using F. tularensis LVS as a model organism for studying tularemia. Care should be taken as lab workers plate LVS cultures during routine work to not over-passage LVS and/or inadvertently select for isolates that carry mutations that lead to higher acquisition of iron. This may have the unintended consequence of working with LVS isolates that have reduced immunogenicity and/or virulence. In addition, our findings highlight further the virulence strategy of F. tularensis which appears to be optimizing its ability to grow while avoiding, as much as possible, its visibility to the host's immune surveillance systems. Presumably, this contributes significantly to the extreme virulence of this bacterial pathogen. Finally, we believe that this work provides new information that should help to guide efforts to design a rational vaccine for tularemia.
Ethics Statement
All experiments using recombinant DNA techniques or reagents with F. tularensis LVS were approved by the Institutional Biosafety Committee. All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies, and all animal work was approved by the University of Iowa Animal Care and Use committee (ACURF #1305086). Guidelines provided by the NIH were followed in all experimentation. The University of Iowa is PHS assured.
Author Contributions
JF designed and performed a majority of experiments and was responsible for writing the manuscript. DC and TW designed and performed experiments in which LVS strains were used to infect macrophages and stimulate cytokine production. They also participated in editing the manuscript. CM performed bioinformatics analysis of the genomic sequences of strains and was responsible for editing the manuscript. CB oversaw experiments in her lab, she is responsible for the first observation of virulence differences in LVS strains and she edited and helped to write the manuscript. BJ oversaw research work performed by JF in his lab, is responsible for the final state of the manuscript and is the corresponding author.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (the lab of CB and the lab of CM). Financial support for this work was provided to the lab BJ (NIH grants 2PO1 AI044642 and 2U54 AI057160). JF was supported by the Predoctoral Training Program in Genetics 5 T32 GM008629.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Aranda, J., Cortés, M. P., Garrido, M. E., Fittipaldi, N., Llagostera, M., Gottschalk, M., et al. (2009). Contribution of the FeoB transporter to Streptococcus suis virulence. Int. Microbiol. 12, 137–143. doi: 10.2436/20.1501.01.91
Ashkenazy, H., Erez, E., Martz, E., Pupko, T., and Ben-Tal, N. (2010). ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533. doi: 10.1093/nar/gkq399
Bakshi, C. S., Malik, M., Mahawar, M., Kirimanjeswara, G. S., Hazlett, K. R., Palmer, L. E., et al. (2008). An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine 26, 5276–5288. doi: 10.1016/j.vaccine.2008.07.051
Bakshi, C. S., Malik, M., Regan, K., Melendez, J. A., Metzger, D. W., Pavlov, V. M., et al. (2006). Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188, 6443–6448. doi: 10.1128/JB.00266-06
Binesse, J., Lindgren, H., Lindgren, L., Conlan, W., and Sjostedt, A. (2015). Roles of reactive oxygen species-degrading enzymes of Francisella tularensis SCHU S4. Infect. Immun. 83, 2255–2263. doi: 10.1128/IAI.02488-14
Bou-Abdallah, F., Lewin, A. C., Le Brun, N. E., Moore, G. R., and Chasteen, N. D. (2002). Iron detoxification properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J. Biol. Chem. 277, 37064–37069. doi: 10.1074/jbc.M205712200
Buchan, B. W., McCaffrey, R. L., Lindemann, S. R., Allen, L. A., and Jones, B. D. (2009). Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect. Immun. 77, 2517–2529. doi: 10.1128/IAI.00229-09
Buchan, B. W., McLendon, M. K., and Jones, B. D. (2008). Identification of differentially regulated francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 74, 2637–2645. doi: 10.1128/AEM.02882-07
Cartron, M. L., Maddocks, S., Gillingham, P., Craven, C. J., and Andrews, S. C. (2006). Feo–transport of ferrous iron into bacteria. Biometals 19, 143–157. doi: 10.1007/s10534-006-0003-2
Cassat, J. E., and Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. doi: 10.1016/j.chom.2013.04.010
Cole, L. E., Yang, Y., Elkins, K. L., Fernandez, E. T., Qureshi, N., Shlomchik, M. J., et al. (2009). Antigen-specific B-1a antibodies induced by Francisella tularensis LPS provide long-term protection against F. tularensis LVS challenge. Proc. Natl. Acad. Sci. U.S.A. 106, 4343–4348. doi: 10.1073/pnas.0813411106
Crane, D. D., Bauler, T. J., Wehrly, T. D., and Bosio, C. M. (2014). Mitochondrial ROS potentiates indirect activation of the AIM2 inflammasome. Front. Microbiol. 5:438. doi: 10.3389/fmicb.2014.00438
Crane, D. D., Warner, S. L., and Bosio, C. M. (2009). A novel role for plasmin-mediated degradation of opsonizing antibody in the evasion of host immunity by virulent, but not attenuated, Francisella tularensis. J. Immunol. 183, 4593–4600. doi: 10.4049/jimmunol.0901655
Crichton, R. R., and Pierre, J. L. (2001). Old iron, young copper: from Mars to Venus. Biometals 14, 99–112. doi: 10.1023/A:1016710810701
Deng, K., Blick, R. J., Liu, W., and Hansen, E. J. (2006). Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74, 4224–4236. doi: 10.1128/IAI.01975-05
Edwards, J. A., Rockx-Brouwer, D., Nair, V., and Celli, J. (2010). Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-gamma activation of macrophages. Microbiology 156, 327–339. doi: 10.1099/mic.0.031716-0
Eigelsbach, H. T., and Downs, C. M. (1961). Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87, 415–425.
Griffin, A. J., Crane, D. D., Wehrly, T. D., and Bosio, C. M. (2015). Successful protection against tularemia in C57BL/6 mice is correlated with expansion of Francisella tularensis-specific effector T cells. Clin. Vaccine Immunol. 22, 119–128. doi: 10.1128/CVI.00648-14
Hantke, K. (2003). Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 11, 192–195. doi: 10.1016/S0966-842X(03)00100-8
Honarmand Ebrahimi, K., Hagedoorn, P. L., and Hagen, W. R. (2015). Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 115, 295–326. doi: 10.1021/cr5004908
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol 11, 443–454. doi: 10.1038/nrmicro3032
Kammler, M., Schon, C., and Hantke, K. (1993). Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175, 6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993
Kim, H., Lee, H., and Shin, D. (2012). The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423, 733–738. doi: 10.1016/j.bbrc.2012.06.027
Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., et al. (2005). ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302. doi: 10.1093/nar/gki370
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lau, C. K., Ishida, H., Liu, Z., and Vogel, H. J. (2013). Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 195, 46–55. doi: 10.1128/JB.01121-12
Lindemann, S. R., Peng, K., Long, M. E., Hunt, J. R., Apicella, M. A., Monack, D. M., et al. (2011). Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 79, 581–594. doi: 10.1128/IAI.00863-10
Lindgren, H., Honn, M., Golovlev, I., Kadzhaev, K., Conlan, W., and Sjöstedt, A. (2009). The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect. Immun. 77, 4429–4436. doi: 10.1128/IAI.00702-09
Lindgren, H., Honn, M., Salomonsson, E., Kuoppa, K., Forsberg, Å., and Sjöstedt, A. (2011). Iron content differs between Francisella tularensis subspecies tularensis and subspecies holarctica strains and correlates to their susceptibility to H2O2-induced killing. Infect. Immun. 79, 1218–1224. doi: 10.1128/IAI.01116-10
Lindgren, H., Lindgren, L., Golovliov, I., and Sjostedt, A. (2015). Mechanisms of heme utilization by Francisella tularensis. PLoS ONE 10:e0119143. doi: 10.1371/journal.pone.0119143
Lindgren, H., Shen, H., Zingmark, C., Golovliov, I., Conlan, W., and Sjöstedt, A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309. doi: 10.1128/IAI.01717-06
Llewellyn, A. C., Jones, C. L., Napier, B. A., Bina, J. E., and Weiss, D. S. (2011). Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE 6:e24201. doi: 10.1371/journal.pone.0024201
Ma, Z., Banik, S., Rane, H., Mora, V. T., Rabadi, S. M., Doyle, C. R., et al. (2014). EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol. Microbiol. 91, 976–995. doi: 10.1111/mmi.12509
Marlovits, T. C., Haase, W., Herrmann, C., Aller, S. G., and Unger, V. M. (2002). The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16243–16248. doi: 10.1073/pnas.242338299
McCaffrey, R. L., Schwartz, J. T., Lindemann, S. R., Moreland, J. G., Buchan, B. W., Jones, B. D., et al. (2010). Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J. Leukoc. Biol. 88, 791–805. doi: 10.1189/jlb.1209811
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Melillo, A. A., Bakshi, C. S., and Melendez, J. A. (2010). Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J. Biol. Chem. 285, 27553–27560. doi: 10.1074/jbc.M110.144394
Melillo, A. A., Mahawar, M., Sellati, T. J., Malik, M., Metzger, D. W., Melendez, J. A., et al. (2009). Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J. Bacteriol. 191, 6447–6456. doi: 10.1128/JB.00534-09
Nagy, T. A., Moreland, S. M., and Detweiler, C. S. (2014). Salmonella acquires ferrous iron from haemophagocytic macrophages. Mol. Microbiol. 93, 1314–1326. doi: 10.1111/mmi.12739
Naikare, H., Palyada, K., Panciera, R., Marlow, D., and Stintzi, A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444. doi: 10.1128/IAI.00052-06
Pan, X., Tamilselvam, B., Hansen, E. J., and Daefler, S. (2010). Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 10:64. doi: 10.1186/1471-2180-10-64
Parrow, N. L., Fleming, R. E., and Minnick, M. F. (2013). Sequestration and scavenging of iron in infection. Infect. Immun. 81, 3503–3514. doi: 10.1128/IAI.00602-13
Perez, N., Johnson, R., Sen, B., and Ramakrishnan, G. (2016). Two parallel pathways for ferric and ferrous iron acquisition support growth and virulence of the intracellular pathogen Francisella tularensis Schu S4. Microbiologyopen 5, 453–468. doi: 10.1002/mbo3.342
Perez, N. M., and Ramakrishnan, G. (2014). The reduced genome of the Francisella tularensis live vaccine strain (LVS) encodes two iron acquisition systems essential for optimal growth and virulence. PLoS ONE 9:e93558. doi: 10.1371/journal.pone.0093558
Rabadi, S. M., Sanchez, B. C., Varanat, M., Ma, Z., Catlett, S. V., Melendez, J. A., et al. (2015). Antioxidant defenses of Francisella tularensis modulate macrophage function and production of proinflammatory cytokines. J. Biol. Chem. 291, 5009–5021. doi: 10.1074/jbc.M115.681478
Ramakrishnan, G., Meeker, A., and Dragulev, B. (2008). fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190, 5353–5361. doi: 10.1128/JB.00181-08
Ramakrishnan, G., and Sen, B. (2014). The FupA/B protein uniquely facilitates transport of ferrous iron and siderophore-associated ferric iron across the outer membrane of Francisella tularensis live vaccine strain. Microbiology 160, 446–457. doi: 10.1099/mic.0.072835-0
Ramond, E., Gesbert, G., Rigard, M., Dairou, J., Dupuis, M., Dubail, I., et al. (2014). Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog. 10:e1003893. doi: 10.1371/journal.ppat.1003893
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty percent endpoints. Am. J. Hygiene 27, 493–497.
Robey, M., and Cianciotto, N. P. (2002). Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70, 5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002
Rohmer, L., Fong, C., Abmayr, S., Wasnick, M., Larson Freeman, T. J., Radey, M., et al. (2007). Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. doi: 10.1186/gb-2007-8-6-r102
Rohmer, L., Brittnacher, M., Svensson, K., Buckley, D., Haugen, E., Zhou, Y., et al. (2006). Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 74, 6895–6906. doi: 10.1128/IAI.01006-06
Salomonsson, E., Kuoppa, K., Forslund, A. L., Zingmark, C., Golovliov, I., Sjöstedt, A., et al. (2009). Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 77, 3424–3431. doi: 10.1128/IAI.00196-09
Savitt, A. G., Mena-Taboada, P., Monsalve, G., and Benach, J. L. (2009). Francisella tularensis infection-derived monoclonal antibodies provide detection, protection, and therapy. Clin. Vaccine Immunol. 16, 414–422. doi: 10.1128/CVI.00362-08
Schulert, G. S., McCaffrey, R. L., Buchan, B. W., Lindemann, S. R., Hollenback, C., Jones, B. D., et al. (2009). Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77, 1324–1336. doi: 10.1128/IAI.01318-08
Shakerley, N. L., Chandrasekaran, A., Trebak, M., Miller, B. A., and Melendez, J. A. (2015). Francisella tularensis catalase restricts immune function by impairing TRPM2 channel activity. J Biol. Chem. 291, 3871–3881. doi: 10.1074/jbc.M115.706879
Su, J., Yang, J., Zhao, D., Kawula, T. H., Banas, J. A., and Zhang, J. R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101. doi: 10.1128/IAI.01865-06
Su, Y. C., Chin, K. H., Hung, H. C., Shen, G. H., Wang, A. H., and Chou, S. H. (2010). Structure of Stenotrophomonas maltophilia FeoA complexed with zinc: a unique prokaryotic SH3-domain protein that possibly acts as a bacterial ferrous iron-transport activating factor. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 66, 636–642. doi: 10.1107/S1744309110013941
Sullivan, J. T., Jeffery, E. F., Shannon, J. D., and Ramakrishnan, G. (2006). Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188, 3785–3795. doi: 10.1128/JB.00027-06
Thomas-Charles, C. A., Zheng, H., Palmer, L. E., Mena, P., Thanassi, D. G., Furie, M. B., et al. (2013). FeoB-mediated uptake of iron by Francisella tularensis. Infect. Immun. 81, 2828–2837. doi: 10.1128/IAI.00170-13
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Velayudhan, J., Hughes, N. J., McColm, A. A., Bagshaw, J., Clayton, C. L., Andrews, S. C., et al. (2000). Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286. doi: 10.1046/j.1365-2958.2000.01987.x
Weaver, E. A., Wyckoff, E. E., Mey, A. R., Morrison, R., and Payne, S. M. (2013). FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J. Bacteriol. 195, 4826–4835. doi: 10.1128/JB.00738-13
Wehrly, T. D., Chong, A., Virtaneva, K., Sturdevant, D. E., Child, R., Edwards, J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x
Keywords: Francisella tularensis, iron acquisition mechanism, microbial pathogenesis, inflammatory mechanisms, virulence mechanisms, intracellular pathogen
Citation: Fletcher JR, Crane DD, Wehrly TD, Martens CA, Bosio CM and Jones BD (2018) The Ability to Acquire Iron Is Inversely Related to Virulence and the Protective Efficacy of Francisella tularensis Live Vaccine Strain. Front. Microbiol. 9:607. doi: 10.3389/fmicb.2018.00607
Received: 08 December 2017; Accepted: 15 March 2018;
Published: 04 April 2018.
Edited by:
Leonard Peruski, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Anders Sjöstedt, Umeå University, SwedenNagendran Tharmalingam, Alpert Medical School, Brown University, United States
Copyright © 2018 Fletcher, Crane, Wehrly, Martens, Bosio and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley D. Jones, YnJhZGxleS1qb25lc0B1aW93YS5lZHU=
 Joshua R. Fletcher
Joshua R. Fletcher Deborah D. Crane2
Deborah D. Crane2 Catharine M. Bosio
Catharine M. Bosio Bradley D. Jones
Bradley D. Jones