- State Key Laboratories for Agrobiotechnology, Key Laboratory of Animal Epidemiology of the Ministry of Agriculture, National Animal Transmissible Spongiform Encephalopathy Laboratory, College of Veterinary Medicine, China Agricultural University, Beijing, China
Tuberculosis (TB) is one of the most fatal infectious diseases and a leading cause of mortality, with 95% of these deaths occurring in developing countries. The causative agent, Mycobacterium tuberculosis (Mtb), has a well-established ability to circumvent the host’s immune system for its intracellular survival. microRNAs (miRNAs) are small, non-coding RNAs having an important function at the post-transcriptional level and are involved in shaping immunity by regulating the repertoire of genes expressed in immune cells. It has been established in recent studies that the innate immune response against TB is significantly regulated by miRNAs. Moreover, differential expression of miRNA in Mtb infection can reflect the disease progression and may help distinguish between active and latent TB infection (LTBI). These findings encouraged the application of miRNAs as potential biomarkers. Similarly, active participation of miRNAs in modulation of autophagy and apoptosis responses against Mtb opens an exciting avenue for the exploitation of miRNAs as host directed therapy (HDT) against TB. Nanoparticles mediated delivery of miRNAs to treat various diseases has been reported and this technology has a great potential to be used in TB. In reality, this exploitation of miRNAs as biomarkers and in HDT is still in its infancy stage, and more studies using animal models mimicking human TB are advocated to assess the role of miRNAs as biomarkers and therapeutic targets. In this review, we attempt to summarize the recent advancements in the role of miRNAs in TB as immune modulator, miRNAs’ capability to distinguish between active and latent TB and, finally, usage of miRNAs as therapeutic targets against TB.
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) is one of the most fatal infectious diseases (Dye and Williams, 2010). Current estimations show that 1/3 of the world population is latently infected with Mtb. However, only 5–10% of infected people develop active TB in their lifetime (Bhatt and Salgame, 2007). It is the leading infectious cause of deaths, exceeding HIV/AID, resulting in approximately 1.3 million human deaths in 2016 (WHO, 2017). Mtb is an intracellular pathogen and its ability to survive inside host macrophages and tubercle granulomas constitutes the major attribute of its virulence. Macrophages are an important component of the host innate immune response and are able to generate a variety of antimicrobial molecular responses, such as antibacterial peptides, hydrolases and toxic reactive oxygen and nitrogen intermediates (Fenton and Vermeulen, 1996). Research on how Mtb is capable of surviving in this antibacterial environment remains an area of interest for many research groups. Accumulating evidence suggests that Mtb is capable of modulating cellular processes such as cytokine production, autophagy, apoptosis, MHC class II expression and phagolysosome maturation in macrophages and dendritic cells (Ahmad, 2011). Currently, many studies have reported that most of the above mentioned cellular mediated immune responses of eukaryotic cells are under the control of miRNAs (Bartel, 2004; Giraldez et al., 2005) and it is now established that modulation of miRNAs expression associated with these biological processes is one of the important strategies implemented by bacterial pathogens to survive inside host immune cells (Das et al., 2016).
miRNAs are small, non-coding RNAs having an important function at the post-transcriptional level to regulate gene expression (O’Connell et al., 2010; Watanabe and Kanai, 2011). Binding of miRNAs to their complementary sequences in the 3′ untranslated region (3′ UTR) of their respective protein coding mRNA targets results in transcript degradation or translational inhibition (Maute et al., 2014). The human genome may encode more than two thousands potentially functional miRNAs (Kozomara and Griffiths-Jones, 2014; Hammond, 2015) and it is estimated that one-half of all protein-coding transcripts are subjected to miRNA regulation (Bartel, 2009; Krol et al., 2010; Watanabe and Kanai, 2011; Vera et al., 2013). Each miRNA may suppress multiple genes and one mRNA can be targeted by multiple miRNAs. Therefore, disease-associated miRNAs represent a new class of diagnostic markers or therapeutic targets (Mayr et al., 2007; Pillai et al., 2007; Subramanian and Steer, 2010; Jackson and Levin, 2012; Mendell and Olson, 2012; Nana-Sinkam and Croce, 2013). Moreover, some bioinformatics studies hint at the existence of tens of thousands of short non-coding RNAs similar to miRNAs with the potential to regulate expression of most or all human genes, although it is unclear whether all of the small RNAs are functional or they are just fragments of larger RNAs (Brameier and Wiuf, 2007; Dieci et al., 2007; Bentwich, 2008). Previously, it was believed that transcription factors (TFs) are responsible for the regulation of gene expression for various cell phenotypes and responses of cells to environmental stimuli. But with the discovery of miRNAs, it is speculated that miRNAs, in combination with TFs, control the expression of thousands of mammalian genes, a process described as ‘tuning’ the transcriptional network (Zhou et al., 2007; Fabian et al., 2010). Some miRNAs, like miR-16, are widely expressed in body tissues while others are tissue or developmental stage specific (Poy et al., 2004). The tissue-specific expression of miRNAs is suggestive of their role in cell differentiation and function (Shi et al., 2010; Pauli et al., 2011).
It has been established that both adaptive and innate immune responses are regulated by miRNAs. For example, in the adaptive immune response, the differentiation of B cells, antibody generation, and T cell development and function are controlled by miRNAs (Singh et al., 2013). A number of recent studies described the regulation of mammalian miRNAs in response to bacterial infection (De Flora and Bonanni, 2011). Innate immune cell activation requires miRNAs, including miR-155, miR-146a, miR-21, and miR-9 (Belver et al., 2011). Components of inflammatory and immune pathways are regulated by miRNAs under the challenge of mycobacterial infections (Liu et al., 2011; Chatterjee et al., 2011). For example, TNF biosynthesis is inhibited by miR-125b in human alveolar macrophages during Mtb infection (Rajaram et al., 2011). Furthermore, many research groups have reported the differential expression of miRNAs in host cells challenged with Mtb, indicating the important role of these miRNAs in regulating the immune response in TB and proposing the miRNAs as potential biomarkers (Wang et al., 2011; Qi et al., 2012; Wu et al., 2012; Zhang et al., 2013). Many of these studies also provide an insight into the role of these differentially expressed miRNAs in TB, but a detailed understanding of the role of these miRNAs in the anti-mycobacterial response is a matter of highest interest for future investigations. These studies also provide a good foundation for the development of reliable biomarkers and therapeutic targets for TB.
Autophagy plays a pivotal role in controlling the bacterial load during TB (Deretic et al., 2013; Richetta and Faure, 2013). However, Mtb has evolved strategies to survive in macrophages by evading delivery to the lysosomes (Hmama et al., 2015) and like other intracellular pathogens Mtb also has the ability to exploit host degradative processes to breakdown cellular macromolecules into simple nutrients for its survival and multiplication (Steele et al., 2015). Similarly, cell apoptosis is also one of the important host defense mechanisms through which macrophages control Mtb infection (Weiss and Schaible, 2015). Mtb is able to inhibit phagolysosome biogenesis, inhibition of apoptosis as well as autophagy due to presence of lipoarabinomannan in its cell wall which is a major immunomodulatory lipoglycan (Vergne et al., 2015). The emerging roles of miRNAs in regulating Mtb–induced autophagy and apoptosis have attracted increased attention in recent years. Appreciating the potential to be exploited in host-directed therapies designed to control autophagy and apoptosis, many groups have focused their research on miRNAs (Ouimet et al., 2016; Zhang et al., 2016; Guo et al., 2017). These recent studies have opened a new avenue to exploit miRNAs in HDT. In the current review, we will summarize recent advances in the understanding of differential expression of miRNAs and their role in TB, and their potential to be used as biomarkers and therapeutic targets for diagnosis and treatment of this deadly disease.
Differential Expression of miRNAs in Different Cell Types Following Mtb Infection
Several studies have addressed the differential expression of miRNAs as reflecting disease prediction and progression in vivo and in vitro. For example, differentially expressed miRNAs have a close relationship with disease progression to hepatocellular carcinoma in both hepatitis B and C (Ura et al., 2009). Recently, attempts have been made to determine the effects of Mtb infection on the expression of miRNAs in the host (Ghorpade et al., 2012; Vegh et al., 2015; Zheng L. et al., 2015). In a recent study, mouse bone marrow derived macrophages (BMDMs) infected with Mtb showed up-regulation of miR-155 (Kumar et al., 2015) while infection of peripheral blood mononuclear cells (PBMCs) derived macrophages with the same pathogen led to down-regulation of miR-155 expression (Rajaram et al., 2011). This means different cell types may respond differently upon infection with Mtb. Sharbati et al. (2011) reported overexpression of let-7e, miR-29a and miR-886-5p in human monocyte derived macrophages (MDMs) in response to mycobacterial infection. Integrated analysis of microRNA and mRNA expression as well as target prediction revealed caspases 3 and 7 as potential targets of let-7e, and miR-29a, respectively. Liu et al. (2012) reported that differentially expressed hsa-mir-21 inhibits expression of two genes encoding vitamin D–dependent antimicrobial peptides, CAMP and DEFB4A, in human monocytes derived from leprosy patients. This inhibition is carried out by direct downregulation of Toll-like receptor (TLR) 2/1 heterodimer (TLR2/1)-induced CYP27B1 and IL1B expression as well as indirect upregulation of IL-10. Furci et al. (2013) studied Mtb-induced miRNA expression profile in primary human macrophages infected with virulent Mtb H37Rv and avirulent M. bovis BCG and showed that macrophages differentially expressed miRNAs, including miR-155, miR-146a, miR-145, miR-222∗, miR-27a, and miR-27b. In this study, miR-222∗, miR-27a, and miR-27b, which have been reported to control inflammatory response and lipid metabolism (McGregor and Choi, 2011; Graff et al., 2012) were significantly downregulated. miR-145, which has been reported to induce apoptosis (Spizzo et al., 2010), was also downregulated, in line with a reduced capacity of virulent Mtb strain to induce apoptosis (Keane et al., 2000; Arcila et al., 2007; Starczynowski et al., 2010; O’Neill et al., 2011). Downregulation of miR-145 results in overexpression of its targets and inhibition of apoptosis (Starczynowski et al., 2010).
Das et al. (2013) analyzed global changes in the miRNA expression profile using microarrays and found that nine miRNA genes (miR-30a, miR-30e, miR-155, miR-1275, miR-3665, miR-3178, miR-4484, miR-4668-5p, and miR-4497) were differentially expressed in THP-1 cells infected with Mtb H37Rv or Mtb H37Ra strains. These differentially expressed miRNAs perform various important functions. miR-30e is activated by β-catenin (Schepeler et al., 2012), while miR-30a inhibits the epithelial to mesenchymal transition (Kumarswamy et al., 2012). miR-1275 is associated with liver metastases of cancer (Kahlert et al., 2011) and miR-155 binds a negative regulator associated with TNF-α production (Rajaram et al., 2011). Spinelli et al. (2013) examined PBMCs and pleural fluid mononuclear cells (PFMCs), and reported that miRNAs expression is associated with IL-6 levels, a cytokine playing a substantial role in TB immunopathology. Lin et al. (2015) showed that the Beijing/W TB strains repressed a number of miRNAs in human macrophages as compared to the non-Beijing/W TB strains, which might reflect their virulence characteristics in altering the host response. Two other research groups identified a series of miRNA differentially expressed in PBMCs of Chinese patients with pulmonary TB by using miRNA expression profiling. Liu et al. (2011) showed that the expression of several miRNAs was significantly altered in patients with active TB, with miR-144∗ being mainly expressed in T cells. Functional analysis showed that miR-144∗ inhibits the secretion of two important cytokines, INF-γ and TNF-α, and also reduces T cell proliferation. Wang et al. (2011) demonstrated that miR-424 and miR-365 levels were significantly raised in patients with active TB compared to healthy controls. In recent years, many other studies have reported the differential expression of miRNAs in different cell types in response to mycobacterial infection (Ni et al., 2014; Zhang et al., 2015). The detailed role of these differentially expressed miRNAs in anti-mycobacterial response is a matter of high interest for many research groups. Furthermore, these studies also provide a good foundation for the development of reliable biomarkers for TB diagnosis.
miRNAs as Potential TB Biomarkers
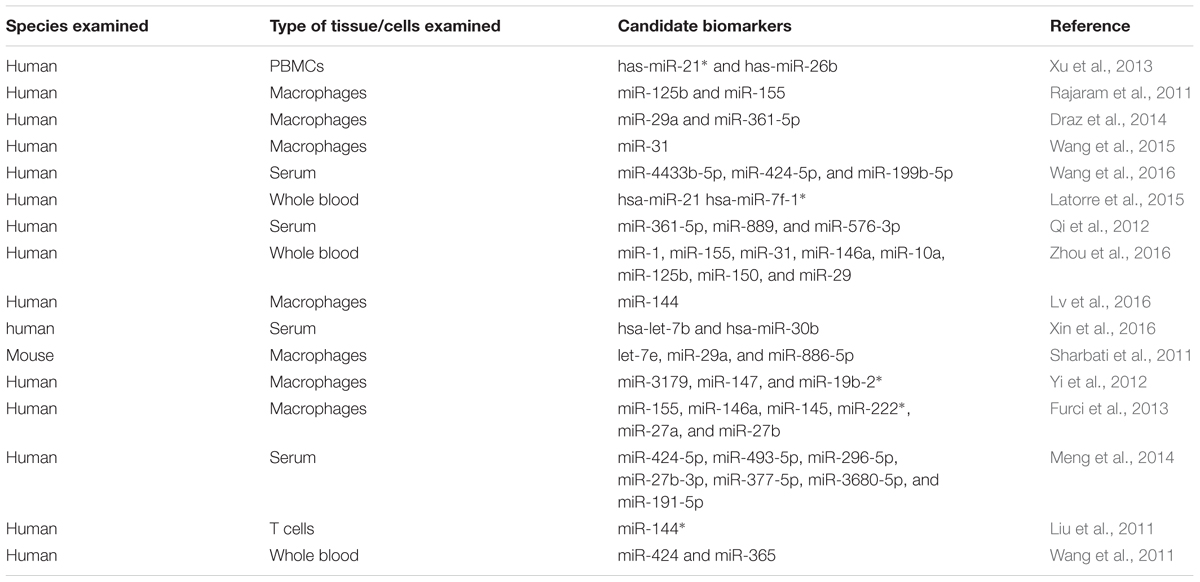
An essential method to effectively control the spread of TB is to diagnose it at an early stage. Currently used test systems are insufficient and unable in practice to discriminate between active TB and latent TB infection (LTBI). Altered miRNA expression profiles may help differentiate between active TB and LTBI and can also act as reliable biomarkers for the diagnosis of the disease. This potential of miRNAs has received much attention in recent years, with several in vivo and in vitro studies proposing miRNAs as potential biomarkers (Miotto et al., 2013; Weiner et al., 2013) although suitable miRNA biomarkers have not been established yet (Walz et al., 2011) (Table 1). In a study using human PBMCs, the authors suggested that differentially expressed miRNAs combined with predicted differentially expressed mRNAs from the same whole genome transcriptional profiling may be used as a new way to differentiate among active TB, LTBI and healthy controls (Xu et al., 2013). Maertzdorf et al. (2012) identified a cluster of differentially expressed miRNAs in patients having active TB and sarcoidosis. Interestingly, strong correlations between miRNAs and gene expression were found. Similarly, lipomannan from virulent Mtb stimulated the expression of miR-125b in human macrophages while lipomannan from avirulent M. smegmatis -resulted in increased expression of miR-155 (Rajaram et al., 2011). These reports revealed that components from related bacterial species, with different virulence, lead to differential expression of miRNAs and consequently modulated the immune response.
In a very recent study, co-regulatory networks consisting of transcription factors and miRNAs as well as their target genes were analyzed from whole blood of TB patients. This TF-gene network showed that SPI1, CEBPB, STAT1, STAT2, STAT3, STAT4, and STAT5A directly regulate 22, 12, 11, 1, 10, 1, and 3 genes, respectively. The results suggested that TF-miRNA gene co-regulatory networks may help provide a way to discover future biomarker and therapeutic targets (Lin et al., 2017). Wu et al. (2014) uncovered several miRNA-gene interactions differentiating among active TB, LTBI and healthy subjects. Wang et al. (2015) discovered that expression of miRNA-31 in pediatric TB patients was significantly lower compared with that in normal children. Furthermore, miRNA-31 expression was negatively correlated with serum levels of IL-6, TNF-α, NF-κ0, and IFN-overed several m- analysis study reported that the current data do not support any association between miR-146a/499 polymorphisms and genetic susceptibility of humans to TB (Lu et al., 2016). Latorre et al. (2015) reported nine differentially expressed miRNAs in active TB patients with respect to those with LTBI and healthy controls. Qi et al. (2012) suggested that altered levels of serum miRNAs have great potential to serve as biomarkers for early detection of pulmonary TB. Similarly, Zhou et al. (2016) identified the expression profile of circulating miRNAs and demonstrated that miRNAs may act as effective biomarkers for the early diagnosis of childhood TB.
Barry et al. (2015) examined the appropriateness of 12 miRNAs and RNU6B to normalize circulating plasma miRNA levels in individuals with active TB. These data identify miR-93 as a suitable miRNA that can be used for normalizing miRNA levels in TB patients. Ren et al. (2015) acquired miRNA expression data from sensitive Mtb and MDR Mtb strains by using next generation sequencing (NGS) and revealed that 142 miRNAs were differentially expressed in the MDR Mtb strains but not in sensitive TB strains. This suggests that miRNAs may have a role in the development of drug-resistance in Mtb strains. Miotto et al. (2013) identified 15 serum miRNAs as signature in pulmonary TB with a diagnostic accuracy of 82%. These studies have contributed substantially to present the differentially expressed miRNAs as potential biomarker candidates for diagnosis of TB, but there is no established miRNA biomarker so far. Several factors may contribute to this slow discovery, including heterogeneous study designs having small study group sizes, noticeable inter-individual variability of miRNA expression and inadequate statistical evaluation for candidate selection (Ueberberg et al., 2014). Moreover, many other pathological conditions may also induce similar miRNAs expression profiles. Keeping in view these limitations, additional investigations are a prerequisite for the selection and use of miRNAs as biomarkers for diagnosis of TB.
miRNAs as Regulators of TB Immunity
Regulation of Inflammatory and Immuno-Modulatory Cytokines
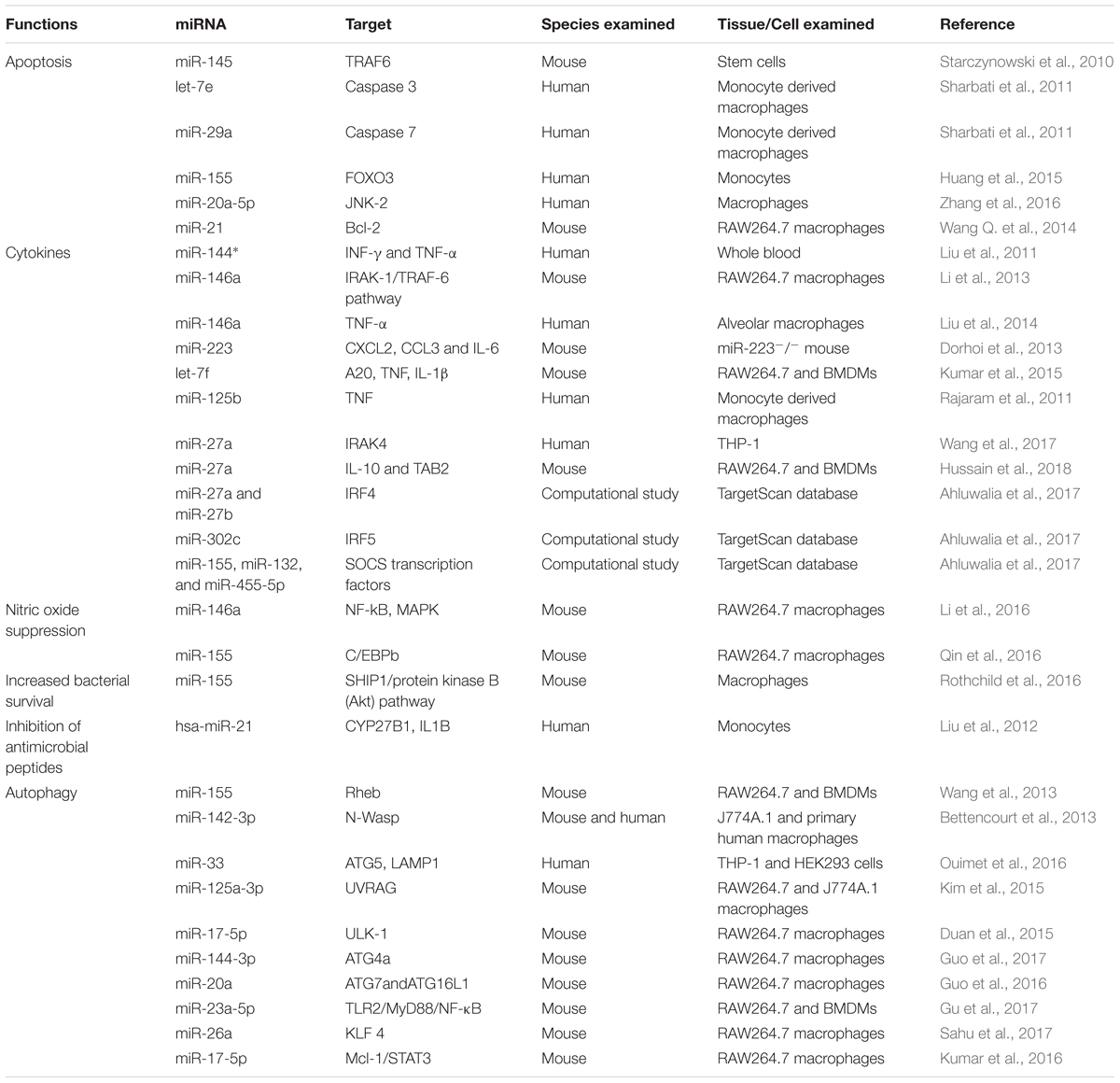
Several miRNAs have been demonstrated to regulate the inflammatory and immune response signaling pathways (Table 2) in response to challenge with Mtb (Guo et al., 2010; Chatterjee et al., 2011; Fu et al., 2011; Huang et al., 2011; Li et al., 2011; Liu et al., 2011). Innate immune cell activation is regulated by miR-155, miR-146a, miR-21, and miR-9 (Belver et al., 2011). Similarly, miR-155 is a positive regulator of TLR signaling, and is induced upon stimulation of murine macrophages with interferon beta (IFN-β) or TLR ligands (Gantier, 2010; Liston et al., 2010). TNF biosynthesis is inhibited by miR-125b in Mtb-infected human alveolar macrophages (Rajaram et al., 2011). miR-29 helps control innate and adaptive immune responses against Mtb by targeting interferon-γ and it is suggested as a biomarker for pulmonary tuberculosis because it is related to the clinical manifestation of the disease (Ma et al., 2011).
Role of miR-146a and miR-155 in Host Immune Response Regulation
With respect to TB, miR-146a and miR-155 are the most vastly studied miRNAs influencing the host–pathogen interaction. miR-146a expression is driven by the transcription factor NF-κB, predicted to base-pair with sequences in the 3′-UTRs of the TNF receptor-associated factor 6 and IL-1 receptor-associated kinase 1 genes (Taganov et al., 2006). Mice deficient in miR-146a showed fatal IFN-γ-dependent autoimmune disease, possibly involving the STAT1 signaling pathway (Lu et al., 2016). miR-146a also represses mycobacteria-mediated inflammatory response and facilitates bacterial proliferation in RAW264.7 macrophages via the IRAK-1/TRAF-6 pathway (Li et al., 2013). In addition, it is overexpressed in macrophages in response to M. bovis BCG while downregulated in alveolar macrophages of pulmonary TB patients and negatively regulating TNF-α (Liu et al., 2014). More recently, it has been described that miR-146a promotes mycobacterial survival in RAW264.7 macrophages through suppression of nitric oxide (NO) production (Li et al., 2016).
miR-155 also has been extensively studied and plays an important role in various physiological and pathological processes (Hacker and Karin, 2006; Faraoni et al., 2009; Kutty et al., 2010; Wang et al., 2010; Savan, 2014; Forster et al., 2015). It is induced upon infection of murine macrophages with mycobacteria and known to be a positive regulator of TLR signaling (Wang J. et al., 2014). Pharmacological inhibition of the Janus kinase (JNK) pathway blocked the induction of miR-155 in response to either polyriboinosinic: polyribocytidylic acid or TNF-α, suggesting that miR-155 signals through the JNK pathway (O’Connell et al., 2007). Iwai et al. (2015) unveiled that miRNA-155 knockout mice were susceptible to M. tuberculosis infection and died significantly earlier with significantly higher numbers of CFU in lungs, as compared to wild-type mice. These studies demonstrate the protective role of miRNA-155 against mycobacterial infection. On the other hand, miR-155 augments the survival of macrophages thereby providing a niche for the replication of Mtb, it promotes the survival of Mtb-specific T cells enabling an effective adaptive immune response. These effects of miR-155 on innate and adaptive immunity are associated with the observations that miR-155-deficient mice better control early Mtb infection, but are compromised in their ability to control late infection (Rothchild et al., 2016). Furthermore, miR-155 reportedly inhibits apoptosis in monocytes (Huang et al., 2015). Zhang et al. (2015) discovered that there was an inverse relationship between serum miR-155 abundance and NK cell cytotoxicity. Serum miR-155 levels were shown to be negatively associated with the TB-suppressing activity of NK cells. Similarly, Qin et al. (2016) reported that miR-155 expression is significantly increased in macrophages after M. marinum infection, resulting in decreased NO synthesis and increased mycobacterium load. These findings support the proposed theory that a single miRNA can target multiple genes at the same time and emphasizing the regulatory role of miRNAs (Mayr et al., 2007; Pillai et al., 2007).
Role of Other miRNAs in Host Immune Response Regulation
Besides these two extensively studied miRNAs, many other miRNAs have been reported to play a role in Mtb infection. miR-223 has a critical role in the control of TB and potentially other chronic inflammatory diseases. Chemoattractants like CXCL2, CCL3, and IL-6 have been identified as targets of miR-223 in myeloid cells (Dorhoi et al., 2013). Similarly, a transgenic mouse with endogenous blockage of miR-29 showed increased resistance against Mtb infection (Ma et al., 2011). A recent study has identified miR-27a as a restrainer of immune response in Mtb infection by targeting IRAK4. Expression levels of IFN-γ, IL-β, IL-6, and TNF-α were significantly decreased following transfection of miR-27a mimics (Wang et al., 2017). miRNAs expression and transcriptional regulation of target genes potentially affect the regulation of multiple immunological responses. These studies enhance our understanding of the role of miRNAs in host-pathogen interactions and offer a new way to improve our diagnostic tools and treatments regimes against TB.
miRNAs as Regulators of Autophagy
Autophagy is an intracellular process involving self-digestion or self-eating, whereby cytoplasmic constituents are transported to and degraded by lysosomes (Lamb et al., 2013). Besides being critical for other cell functions, autophagy plays a key role in immune responses against invading viral and bacterial pathogens (Huang and Brumell, 2014). Similarly, apoptosis is also one of the important host defense mechanisms through which the host cells limit the extent of damage caused by the infection. However, the capacity of Mtb to survive and replicate in host macrophages is central to Mtb pathogenesis and is often associated with its degree of virulence (Weiss and Schaible, 2015). It is important to explore how Mtb hijacks host immunomodulatory signaling pathways to survive and replicate in macrophages for appropriate control and treatment of the disease. It is well established that Mtb has developed several schemes to avoid the antimicrobial effects of macrophages to be able to survive intracellularly (Ahmad, 2011). One of these significant strategies is its ability to block phagosome maturation and deploy other countermeasures to impede autophagy, and evade the hostile environment of phagolysosomes (Vergne et al., 2004; Russell, 2011; Espert et al., 2015). Moreover, phagosome maturation and mycobacterial killing can be reinstated through exogenous induction of autophagy in infected macrophages (Deretic et al., 2013).
The pathways associated with autophagy are tightly regulated at the post-transcriptional level and have been well described, but the contribution of miRNAs in activation or inhibition of autophagy during Mtb infection had been largely unknown. However, in recent years many research groups have successfully unveiled the role of several miRNAs in autophagy regulation during Mtb infection. Ouimet et al. (2016) reported that miR-33 induction in THP-1 and HEK293 cells inhibits the integrated pathways involved in autophagy and also reprograms the host lipid metabolism for intracellular survival and persistence of Mtb. Guo et al. (2017) revealed that M. bovis BCG infection of macrophages leads to increased expression of miR-144-3p, which induces autophagy-related gene 4a (ATG4a) to inhibit autophagy. In another study, overexpression of miR-23a-5p dramatically prevented Mtb-induced activation of autophagy in macrophages by modulating TLR2/MyD88/NF-κB signaling (Gu et al., 2017). Similarly, miRNA-20a targets ATG7 and ATG16L1 and is able to inhibit autophagy (Guo et al., 2016).
In contrast to the inhibitory role of miRNAs in autophagy induction reported by the above studies, other studies have identified several miRNAs involved in enhancing autophagy during Mtb infection. Sahu et al. (2017) reported that miR-26a mimic attenuates Mtb survival in macrophages by targeting the transcription factor KLF4. This transcription factor is able to prevent trafficking of Mtb to lysosomes. miR-155 expression accelerates the autophagy-mediated anti-mycobacterial response by targeting Ras homolog enriched in brain (Rheb), a negative regulator of autophagy (Wang et al., 2013). Similarly, Kumar et al. (2016) demonstrated that a miR-17/PKCδ/STAT3 axis is involved in regulating autophagy during Mtb infection. Neural Wiskott-Aldrich syndrome protein (N-Wasp) is an actin-binding protein involved in phagocytosis during microbial challenge. Mtb induced expression of miR-142-3p targets N-Wasp leading to reduced phagocytosis (Bettencourt et al., 2013).
miRNAs as Regulators of Apoptosis
MiRNAs also play a role in regulation of apoptosis in Mtb infection. According to recent findings, miR-20a-5p functions as a negative regulator of mycobacterial-triggered apoptosis and inhibition of miR-20a-5p results in more efficient Mtb clearance (Zhang et al., 2016). MPT64 is one of the important secreted proteins by Mtb and it could inhibit apoptosis of RAW264.7 cells through the NF-kB-miRNA21-Bcl-2 pathway (Wang Q. et al., 2014). Huang et al. (2015) demonstrated that miR-155 targets FOXO3 and regulates apoptosis. Other studies have also reported the role of miRNAs in apoptosis in Mtb infection (Starczynowski et al., 2010; Sharbati et al., 2011). Collectively, these findings highlight the role of miRNAs in regulation of autophagy and apoptosis in TB (Figure 1). These findings also offer an opportunity for exogenous manipulation of miRNAs to restrain the multiplication and enhance the clearance of Mtb from the host cells.
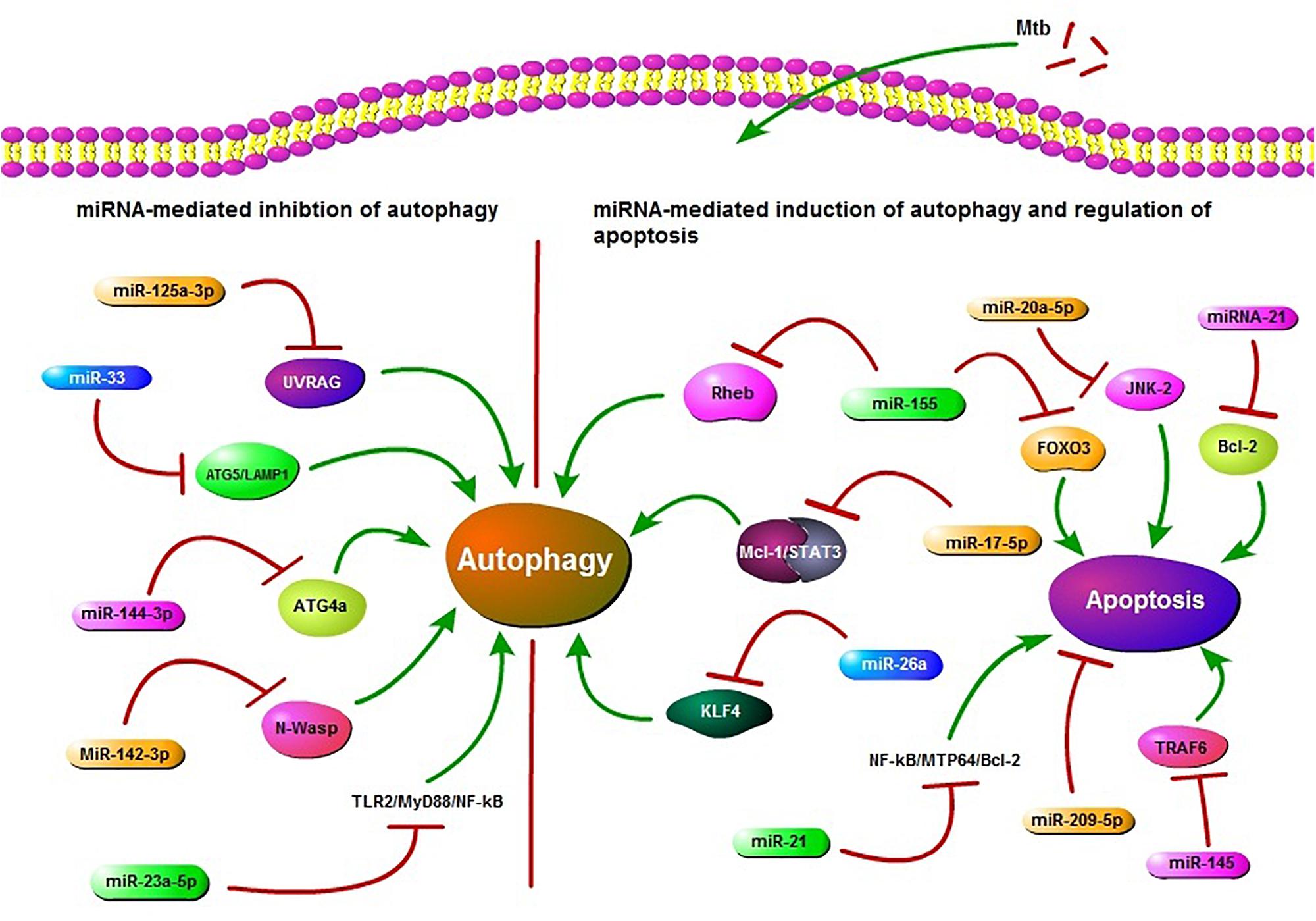
FIGURE 1. Schematic presentation of miRNA regulation of autophagy and apoptosis. miR-125a-3p, miR-33, miR-144-3p, miR-23a-5p, and miR-142-3p are potential inhibitors of autophagy in Mycobacterium tuberculosis (Mtb) infection. miR-125a-3p inhibits autophagy through targeting UV radiation resistance-associated gene (UVRAG), miR-33 targets ATG5/LAMP1, miR-144-3p targets autophagy-related gene 4a (ATG4a), miR-23a-5p inhibits the TLR2/MyD88/NF-κB leading to reduced autophagy and miR-33 also plays an inhibitory role via targeting some unknown factors. miR-142-3p targets Neural Wiskott-Aldrich syndrome protein (N-Wasp) leading to reduced phagocytosis. While miR-155, miR-17-5p, and miR-26a target Ras homolog enriched in brain (Rheb), Mcl-1/STAT3, KLF4, respectively, and play a positive role in autophagy regulation during Mtb infection. miR-155 also inhibits apoptosis by targeting FOXO3. miR-21 targets NF-kB-MTP64-Bcl-2 to down regulate the apoptosis. miR-20a-5p, miR-21, miR-209-5p, and miR-145 also regulate apoptosis. All of these miRNAs may act as potential targets in HDT.
miRNAs as Therapeutic Targets
Application of miRNAs as a novel class of drug targets for treatment of various diseases is an emerging area of research. For example, miRNAs have been used as therapeutic targets in hepatitis C virus infection, cancer and cardiovascular diseases (Lanford et al., 2010; Takeshita et al., 2010; Kotsinas et al., 2015). Exogenous manipulation in pathologically imbalanced miRNAs is able to transform the phenotype of cancer cells (Merhautova et al., 2016). Moreover, antitumor activity of miRNAs has been demonstrated and safety of use has been established in hepatic carcinoma (Callegari et al., 2015). The expression of miRNAs can be manipulated for therapeutic purposes either through positive or negative regulation. Novel intervention strategies exploit miRNA-mimics to restore optimal miRNA expression or anti-miRNAs to block abnormally produced miRNAs (Stenvang et al., 2012). For example, miRNAs can be targeted therapeutically to inhibit their maturation (Lee et al., 2006). Similarly, it is possible to increase the activity of downregulated anti-mycobacterial miRNAs by using synthetic oligos and reduce the effects of overexpressed pro-mycobacterial miRNAs through antisense oligonucleotides or anti-miRNA complementary to mature miRNA (Meister et al., 2004; Grimm et al., 2006; Baumann and Winkler, 2014). Another alternative strategy is to replace specific miRNA by gene therapy using viral vectors (Uprichard, 2005).
In Vivo Manipulation of miRNAs Expression
Besides these anti-miRNA techniques, substantial advances have been made concerning delivery of miRNAs into lungs and encouraging results have described modulation of TGF-β1 expression in a mouse model of TB (Rosas-Taraco et al., 2011). In vivo delivery of miRNAs has been performed by lentiviral vectors, lipid conjugates, or small exosome-like vesicles. Antigen-specific exosome-like nanovesicles have been transfected with mimics or anti-miRNA to restore miRNA expression in target cells (Bryniarski et al., 2013). In a similar study, primary B cells transfected with a specific anti-miRNA have been shown to deliver the molecule successfully to antigen-activated T cells (Almanza et al., 2013). Zheng D. et al. (2015) successfully silenced in vivo miR-195 expression by using anti-miR-195. Similarly, in vivo inhibition of miR-328 by intra-tracheal inoculation of anti-328 resulted in 4-fold enhanced non-typeable Haemophilus influenzae (NTHi) clearance from the lungs as compared to controls (Tay et al., 2015). Thus, miRNAs can be targeted in the lung to enhance host immunity against microbial infections and offer a potential new anti-mycobacterial approach for the treatment of TB. Further studies are required to define the most appropriate route of miRNA delivery and pharmaco-vigilance problems, providing a valid foundation for future miRNA-based HDTs of TB.
Nanoparticle-Mediated Delivery of miRNAs in TB
Technological developments at the micro and nanoscale can be used to modulate the immune response (Dacoba et al., 2017). Multiple materials and techniques have been developed to deliver drugs and a broad array of cargo to cells, including nanostructured materials (Vázquez-Hernández et al., 2017), synthetic nanoparticles (Ayer and Harm-Anton, 2017), proactive biomimetic delivery systems (Parodi et al., 2017), functional polymers (Jiang et al., 2017) or cell-penetrating peptides (Tashima, 2016), among others. Several materials have been proposed to help treat immune diseases with nanoparticles, including silica, iron oxide, gold, liposomes, or polylactic-co-glycolic acid (PLGA) (Prosperi et al., 2017). Of particular interest in infectious diseases are techniques that can encapsulate miRNAs and deliver them to the cells that are at the forefront of the immune response, i.e., macrophages or dendritic cells, taking advantage of the innate ability of these cells to internalize foreign bodies. The idea of miRNA delivery fits into the general concept of nanoparticle-based induction of immune responses by targeting phagocytes (i.e., macrophages), in contrast to the alternative method of targeting adaptive lymphocytes (i.e., T cells). MiRNA delivery has been explored for many applications related to inflammation (Zhou et al., 2013) and cancer (Fernandez-Piñeiro et al., 2017), and its potential use demonstrated in infectious diseases (Pegtel et al., 2010). MiRNAs can be incorporated into both natural –i.e., chitosan- and synthetic – i.e., (PLGA)- particle-forming materials. For example, nanoparticle- and liposome-loaded miRNAs have been delivered to macrophages (Duan et al., 2015; Moore et al., 2016). At the time of writing, however, very few reports are available on the implementation of this technique in TB. This implementation can be discussed by either describing current general uses of nanoparticles in immunology or broadly mentioning the utilization of nanoparticles with miRNAs. We will briefly combine these two views and describe nanoparticle methods used elsewhere that can be adopted in miRNA delivery in TB.
Direct Delivery of miRNAs to Modulate Host Immune Response
To help eliminate or substantially reduce mycobacterial activity by targeting macrophages and dendritic cells, several approaches can be implemented. An obvious approach is to directly deliver miRNAs that improve host immune responses against TB, or more generally that facilitate increased expression of anti-inflammatory molecules like IL-10. Among the candidates for this approach we can cite miR-155, miR-146a, miR-145, miR-99b, miR-19b-2∗, miR-27a, or miR-27b. Conversely, a second approach is to deliver siRNAs that help decrease expression of pro-inflammatory cytokines such as TNF-α that are induced by miRNAs. Candidates for this approach are those miRNAs that are overexpressed during TB infection, including let-7e, miR-29a, miR-886-5p, miR-147, or miR-3179. In addition to siRNAs, this approach could include the delivery of small molecules (drugs) that can selectively affect immune responses including dexamethasone or other small molecules which modify the host’s ability to regulate miRNA expression.
Direct miRNA delivery could knock-down, overexpress or downregulate appropriate host molecular targets against TB. An early example of this approach of direct delivery is the nanoparticle release of miR-223 in macrophages to facilitate phenotype transitions (Tran et al., 2016). To implement the direct delivery, the critical aspect is the selection of the miRNAs. When innate immune cell activation is desired, direct delivery of miR-155 may be an option, because of its role as a positive regulator of TLR signaling (Wang J. et al., 2014) and its promotion of autophagy and mycobacterial clearance (Wang et al., 2013). Similarly to miR-155, nanoparticle delivery of miR-125b during Mtb infection can be useful in inhibiting TNF biosynthesis. Delivery of these miRNAs have been demonstrated previously in pancreatic cancer cells (Su et al., 2016) or in lung cancer (Talekar et al., 2016), but not in infectious diseases. In general, this approach of direct delivery should be implemented for those miRNAs that are downregulated during TB infection, including miR-155, miR-146a, miR-145, miR-222∗, miR-27a, or miR-27b (Spizzo et al., 2010; Belver et al., 2011; McGregor and Choi, 2011; Graff et al., 2012). Only a few reports are available describing nanoparticle uses with mi-R155, miR-223, miR-146, or miR-125b, among the many miRNAs involved in TB cited in our manuscript.
Use of siRNAs to Downregulate the miRNAs
In the second approach mentioned –releasing siRNAs to downregulate miRNAs expression-, a clear candidate may be miR-146a, because it facilitates bacterial proliferation via the IRAK-1/TRAF-6 pathway, negatively regulates TNF-α and promotes mycobacterial survival in macrophages through suppression of NO production. Interestingly, nanoparticles loaded with miR-146 have been used to inhibit inflammation in keratinocytes (Urgard et al., 2016) and human dental pulp cells (Liu et al., 2016) and to inhibit kidney fibrosis (Morishita et al., 2015). This suggests that both methodologies –direct delivery and secondary molecular induction- can be complementary approaches in nanoparticle-mediated delivery of miRNAs. Delivery of molecular compounds that facilitate upregulation or downregulation of host’s expression of miRNAs is part of this second approach. For this to work, it would be necessary to know what class of molecules (or particular molecule) can directly regulate the host expression of the targeted miRNAs. In summary, there appears to be great potential for the broad implementation of nanoparticle-mediated release of miRNAs important during TB infection. Additional studies will be necessary to clarify the role of these differentially expressed miRNAs in mycobacterial infection and observe differential effects of direct miRNA delivery or indirect regulation to each particular set of cells.
Recent Studies Illustrating miRNAs as Potential Therapeutic Targets
In recent years, several research groups have pointed out many miRNAs that can be targeted to treat TB. For instance, miR-155 is negatively associated with the TB-suppressing activity of NK cells (Zhang et al., 2015). The same miRNA also leads to autophagy-mediated mycobacterial elimination by targeting Rheb (Wang et al., 2013). Ouimet et al. (2016) revealed that silencing of miR-33 and miR-33∗ by genetic or pharmacological means promotes autophagy flux through depression of key autophagy effectors and AMPK-dependent activation of the transcription factors FOXO3 and TFEB. In a similar study, miR-23a-5p modulated the host innate immune response by promoting Mtb survival and inhibiting autophagy induction through TLR2/MyD88/NF-κB pathway (Gu et al., 2017). Cytokines are important mediators of inflammatory and immune responses. miR-99b is able to inhibit the secretion of pro-inflammatory cytokines in Mtb infection, suggesting a new approach for designing miRNA-based therapies and control of TB (Singh et al., 2013). miR-206 has been suggested to function as an inflammatory regulator leading to the expression of MMP9 by targeting TIMP3 in Mtb infection (Fu et al., 2016). Similarly, Lou et al. (2017) demonstrated that miR-20b can alleviate the inflammatory response in TB mice by targeting the NLRP3/caspase-1/IL-1β pathway. Zhang et al. (2017) showed that the induction of miR-32-5p strongly increases the survival rate of Mtb by directly targeting Follistatin-like protein 1 (FSTL1) through the TLR-4/miRNA-32-5p/FSTL1 pathway. However, a possible limitation is that most of the miRNAs are not entirely gene-specific. One miRNA may target multiple mRNAs, suggesting that exogenous miRNA administration might exhibit off-target effects. In reality, the exploitation of miRNAs in HDT is still in its infancy stage but it has opened an exciting avenue for the control and treatment of TB.
Conclusion and Future Prospects
Tuberculosis (TB) is one of the world’s most deadly communicable diseases and Mtb is difficult to eradicate, due to its capability to persist within macrophages. Macrophages play a pivotal role in the host immune response against Mtb, a response tightly regulated by multiple factors, including miRNAs. The emerging role of miRNAs in regulating both adaptive and innate immune responses against Mtb has attracted increasing attention from many research groups in recent years. Many studies have revealed that differential expression of miRNA could reflect disease progression and may distinguish between active and LTBI. These findings provide insights into the potential utility of miRNAs as biomarkers for the diagnosis of TB. However, a possible limitation in the use of miRNAs as biomarkers is that most of the miRNAs are not completely gene-specific. Active participation of miRNAs in modulation of autophagy and apoptosis against Mtb, and the emerging role of nanotechnology in medicine opens an exciting avenue for the exploitation of nanoparticles-mediated delivery of miRNAs in host directed therapy (HDT) against TB.
These findings provide valuable information and a firm foundation for the development of HDT based on miRNAs. Moreover, these advancements in miRNA research have reduced our current limitations related to diagnostics, multidrug resistance and HDT for better control and treatment of TB. In this review, we present a comprehensive survey of the published literature on the differential expression and role of miRNAs, as well as recent advances in the development of miRNA-based biomarkers and therapeutics for TB. Further studies are required to elucidate the full potential of miRNAs as novel TB biomarkers, as well as their manipulation as adjunctive treatment for TB.
Author Contributions
NS collected the data and wrote the manuscript. TH and SS helped for figure and table compilation. AP assisted in writing the manuscript and provided the critical comments. XZ gave the idea behind the manuscript compilation. DZ reviewed the article before final submission. All authors read and approved the manuscript prior to submission.
Funding
This work was supported by China Agriculture Research System (No. CARS-36), the National Key Research and Development Program (No. 2017YFD0500901), the National Natural Science Foundation of China (No. 31572487); the National Science and Technology Pillar Program during the twelfth 5-year plan period (No. 2015BAI09B01), and the MoSTRCUK International Cooperation Project (Project No. 2013DFG32500); and the High-End Foreign Experts Recruitment Program (Project No. GDW20161100071).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahluwalia, P. K., Pandey, R. K., Sehajpal, P. K., and Prajapati, V. K. (2017). Perturbed microRNA expression by Mycobacterium tuberculosis promotes macrophage polarization leading to pro-survival foam cell. Front. Immunol. 8:107. doi: 10.3389/fimmu.2017.00107
Ahmad, S. (2011). Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin. Dev. Immunol. 2011:814943. doi: 10.1155/2011/814943
Almanza, G., Anufreichik, V., Rodvold, J. J., Chiu, K. T., DeLaney, A., Akerset, J. C., et al. (2013). Synthesis and delivery of short, noncoding RNA by B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 20182–20187. doi: 10.1073/pnas.1311145110
Arcila, M. L., Sanchez, M. D., Ortiz, B., Barrera, L. F., Garcia, L. F., and Rojas, M. (2007). Activation of apoptosis, but not necrosis, during Mycobacterium tuberculosis infection correlated with decreased bacterial growth: role of TNF-alpha, IL-10, caspases and phospholipase A2. Cell. Immunol. 249, 80–93. doi: 10.1016/j.cellimm.2007.11.006
Ayer, M., and Harm-Anton, K. (2017). Cell-mediated delivery of synthetic nano- and microparticles. J. Control. Release 259, 92–104. doi: 10.1016/j.jconrel.2017.01.048
Barry, S. E., Chan, B., Ellis, M., Yang, Y. R., Plit, M. L., Guan, G., et al. (2015). Identification of miR-93 as a suitable miR for normalizing miRNA in plasma of tuberculosis patients. J. Cell. Mol. Med. 19, 1606–1613. doi: 10.1111/jcmm.12535
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Baumann, V., and Winkler, J. (2014). MiRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 17, 1967–1984. doi: 10.4155/fmc.14.116
Belver, L., Papavasiliou, F. N., and Ramiro, A. R. (2011). MicroRNA control of lymphocyte differentiation and function. Curr. Opin. Immunol. 23, 368–373. doi: 10.1016/j.coi.2011.02.001
Bentwich, I. (2008). Identifying human microRNAs. Curr. Top. Microbiol. Immunol. 320, 257–269. doi: 10.1007/978-3-540-75157-1_12
Bettencourt, P., Marion, S., Pires, D., Santos, L. F., Lastrucci, C., Carmo, N., et al. (2013). Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front. Cell. Infect. Microbiol. 3:19. doi: 10.3389/fcimb.2013.00019
Bhatt, K., and Salgame, P. (2007). Host innate immune response to Mycobacterium tuberculosis. J. Clin. Immunol. 27, 347–362. doi: 10.1007/s10875-007-9084-0
Brameier, M., and Wiuf, C. (2007). Ab initio identification of human microRNAs based on structure motifs. BMC Bioinformatics 8:478. doi: 10.1186/1471-2105-8-478
Bryniarski, K., Ptak, W., Jayakumar, A., Püllmann, K., Caplan, M. J., Chairoungdua, A., et al. (2013). Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J. Allergy Clin. Immunol. 132, 170–181. doi: 10.1016/j.jaci.2013.04.048
Callegari, E., Gramantieri, L., Domenicali, M., D’Abundo, L., Sabbioni, S., and Negrini, M. (2015). MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 22, 46–57. doi: 10.1038/cdd.2014.136
Chatterjee, S., Dwivedi, V. P., Singh, Y., Siddiqui, I., Sharma, P., Van Kaer, L., et al. (2011). Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes protective T helper 17 cell responses in a Toll-Like receptor-2-dependent manner. PLoS Pathog. 7:e1002378. doi: 10.1371/journal.ppat.1002378
Dacoba, T. G., Olivera, A., Torres, D., Crecente-Campo, J., and Alonso, M. J. (2017). Modulating the immune system through nanotechnology. Semin. Immunol. 34, 78–102. doi: 10.1016/j.smim.2017.09.007
Das, K., Garnica, O., and Dhandayuthapani, S. (2016). Modulation of host miRNAs by intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 6:79. doi: 10.3389/fcimb.2016.00079
Das, K., Saikolappan, S., and Dhandayuthapani, S. (2013). Differential expression of miRNAs by macrophages infected with virulent and avirulent Mycobacterium tuberculosis. Tuberculosis 93, S47–S50. doi: 10.1016/S1472-9792(13)70010-6
De Flora, S., and Bonanni, P. (2011). The prevention of infection associated cancers. Carcinogenesis 32, 787–795. doi: 10.1093/carcin/bgr054
Deretic, V., Saitoh, T., and Akira, S. (2013). Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737. doi: 10.1038/nri3532
Dieci, G., Fiorino, G., Castelnuovo, M., Teichmann, M., and Pagano, A. (2007). The expanding RNA polymerase III transcriptome. Trends Genet. 23, 614–622. doi: 10.1016/j.tig.2007.09.001
Dorhoi, A., Iannaccone, M., Farinacci, M., Faé, K. C., Schreiber, J., Moura-Alves, P., et al. (2013). MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 123, 4836–4848. doi: 10.1172/JCI67604
Draz, N. I., el Hady, S. A., Elsayed, M. S., Korraa, E. E. A., and el Magd, N. M. (2014). Serum microRNA-29a and microRNA-361-5p as potential diagnostic biomarkers for active pulmonary tuberculosis. Egypt. J. Med. Microbiol. 23.
Duan, W., Yang, T., Zhou, S. F., Wang, Z. L., Zhou, Z. W., and He, Z. X. (2015). Novel targeting of PEGylated liposomes for codelivery of TGF-beta1 siRNA and four antitubercular drugs to human macrophages for the treatment of mycobacterial infection: a quantitative proteomic study. Drug Des. Dev. Ther. 9, 4441–4470. doi: 10.2147/DDDT.S79369
Dye, C., and Williams, B. G. (2010). The population dynamics and control of tuberculosis. Science 328, 856–861. doi: 10.1126/science.1185449
Espert, L., Beaumelle, B., and Vergne, I. (2015). Autophagy in Mycobacterium tuberculosis and HIV infections. Front. Cell. Infect. Microbiol. 5:49. doi: 10.3389/fcimb.2015.00049
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Faraoni, I., Antonetti, F. R., Cardone, J., and Bonmassar, E. (2009). MiR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta 1792, 497–505. doi: 10.1016/j.bbadis.2009.02.013
Fenton, M. J., and Vermeulen, M. W. (1996). Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect. Immun. 64, 683–690.
Fernandez-Piñeiro, I., Badiola, I., and Sanchez, A. (2017). Nanocarriers for microRNA delivery in cancer medicine. Biotechnol. Adv. 35, 350–360. doi: 10.1016/j.biotechadv.2017.03.002
Forster, S. C., Michelle, D. T., and Hertzog, P. J. (2015). MicroRNA as type i interferon-regulated transcripts and modulators of the innate immune response. Front. Immunol. 6:334. doi: 10.3389/fimmu.2015.00334
Fu, X. D., Zeng, L., Liu, Z., Ke, X., Lei, L., and Li, G. (2016). MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in Mycobacterium tuberculosis–infected THP-1 human macrophages. Biochem. Biophys. Res. Commun. 477, 167–173. doi: 10.1016/j.bbrc.2016.06.038
Fu, Y., Yi, Z., Wu, X., Wu, X., Li, J., Xu, F., et al. (2011). Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 49, 4246–4251. doi: 10.1128/JCM.05459-11
Furci, L., Schena, E., Miotto, P., and Daniela, M. C. (2013). Alteration of human macrophages microRNA expression profile upon infection with Mycobacterium tuberculosis. Int. J. Mycobacteriol. 2, 128–134. doi: 10.1016/j.ijmyco.2013.04.006
Gantier, M. P. (2010). New perspectives in MicroRNA regulation of innate immunity. J. Interferon Cytokine Res. 30, 283–289. doi: 10.1089/jir.2010.0037
Ghorpade, D. S., Leyland, R., Kurowska-Stolarska, M., Patil, S. A., and Balajia, K. N. (2012). MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol. Cell. Biol. 12, 2239–2253. doi: 10.1128/MCB.06597-11
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838. doi: 10.1126/science.1109020
Graff, J. W., Dickson, A. M., Clay, G., McCaffrey, A. P., and Wilson, M. E. (2012). Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 287, 21816–21825. doi: 10.1074/jbc.M111.327031
Grimm, D., Streetz, K. L., Jopling, C. L., Storm, T. A., Pandey, K., Davis, C. R., et al. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 7092, 537–541. doi: 10.1038/nature04791
Gu, X., Gao, Y., Mu, D. G., and Fu, E. Q. (2017). MiR-23a-5p modulates mycobacterial survival and autophagy during Mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp. Cell Res. 354, 71–77. doi: 10.1016/j.yexcr.2017.03.039
Guo, L., Zhao, J., Qu, Y., Yin, R., Gao, Q., Ding, S., et al. (2016). MicroRNA-20a inhibits autophagic process by targeting ATG7 and ATG16L1 and favors mycobacterial survival in macrophage cells. Front. Cell. Infect. Microbiol. 6:134. doi: 10.3389/fcimb.2016.00134
Guo, L., Zhou, L., Gao, Q., Zhang, A., Wei, J., Hong, D., et al. (2017). MicroRNA-144-3p inhibits autophagy activation and enhances Bacillus Calmette-Guerin infection by targeting ATG4a in RAW264.7 macrophage cells. PLoS One 12:e0179772. doi: 10.1371/journal.pone.0179772
Guo, W., Li, J. T., Pan, X., Wei, L., and Wu, J. Y. (2010). Candidate Mycobacterium tuberculosis genes targeted by human microRNAs. Protein Cell 1, 419–421. doi: 10.1007/s13238-010-0056-4
Hacker, H., and Karin, M. (2006). Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006:re13. doi: 10.1126/stke.3572006re13
Hammond, S. M. (2015). An overview of microRNAs. Adv. Drug Deliv. Rev. 87, 3–14. doi: 10.1016/j.addr.2015.05.001
Hmama, Z., Pena-Diaz, S., Joseph, S., and Av-Gay, Y. (2015). Immuno-evasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 264, 220–232. doi: 10.1111/imr.12268
Huang, J., and Brumell, J. H. (2014). Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol. 12, 101–114. doi: 10.1038/nrmicro3160
Huang, J., Junhua, J., Xu, W., Zhao, H., Zhang, C., Shi, Y., et al. (2015). MiR-155 is up-regulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol. Med. Rep. 12, 7102–7108. doi: 10.3892/mmr.2015.4250
Huang, Y., Shen, X. J., Zou, Q., Shen, X. J., Zou, Q., Wang, S. P., et al. (2011). Biological functions of microRNAs: a review. J. Physiol. Biochem. 67, 129–139. doi: 10.1007/s13105-010-0050-6
Hussain, T., Zhao, D., Shah, S. Z. A., Wang, J., Yue, R., Liao, Y., et al. (2018). MicroRNA 27a-3p regulates antimicrobial responses of murine macrophages infected by Mycobacterium avium subspecies paratuberculosis by targeting interleukin-10 and TGF-β-activated protein kinase 1 binding protein 2. Front. Immunol. 8:1915. doi: 10.3389/fimmu.2017.01915
Iwai, H., Funatogawa, K., Matsumura, K., Kato-Miyazawa, M., Kirikae, F., Kiga, K., et al. (2015). MicroRNA-155 knockout mice are susceptible to Mycobacterium tuberculosis infection. Tuberculosis 95, 246–250. doi: 10.1016/j.tube.2015.03.006
Jackson, A. L., and Levin, A. A. (2012). Developing microRNA therapeutics: approaching the unique complexities. Nucleic Acid Ther. 22, 213–225. doi: 10.1089/nat.2012.0356
Jiang, B., Yang, J., Rahoui, N., Taloub, N., and Huang, Y. D. (2017). Functional polymer materials affecting cell attachment. Adv. Colloid Interface Sci. 250, 185–194. doi: 10.1016/j.cis.2017.09.002
Kahlert, C., Klupp, F., Brand, K., Lasitschka, F., Diederichs, S., Kirchberg, J., et al. (2011). Invasion front specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci. 102, 1799–1807. doi: 10.1111/j.1349-7006.2011.02023.x
Keane, J., Remold, H. G., and Kornfeld, H. (2000). Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164, 2016–2020. doi: 10.4049/jimmunol.164.4.2016
Kim, J. K., Yuk, J. M., Kim, S. Y., Kim, T. S., Jin, H. S., and Yang, C. S. (2015). MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J. Immunol. 194, 5355–5365. doi: 10.4049/jimmunol.1402557
Kotsinas, A., Sigala, F., Garbis, S. D., Galyfos, G., Filis, K., Vougas, K., et al. (2015). MicroRNAs determining inflammation as novel biomarkers and potential therapeutic targets. Curr. Med. Chem. 22, 2666–2679. doi: 10.2174/0929867322666150716113304
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kumar, M., Sahu, S. K., Kumar, R., Subuddhi, A., Maji, R. K., Jana, K., et al. (2015). MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kB pathway. Cell Host Microbe 11, 345–356. doi: 10.1016/j.chom.2015.01.007
Kumar, R., Sahu, S. K., Kumar, M., Jana, K., Gupta, P., Gupta, U. D., et al. (2016). MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 18, 679–691. doi: 10.1111/cmi.12540
Kumarswamy, R., Mudduluru, G., Ceppi, P., Muppala, S., Kozlowski, M., Niklinski, J., et al. (2012). MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int. J. Cancer 130, 2044–2053. doi: 10.1002/ijc.26218
Kutty, R. K., Nagineni, C. N., Samuel, W., Vijayasarathy, C., Hooks, J. J., and Redmond, T. M. (2010). Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem. Biophys. Res. Commun. 402, 390–395. doi: 10.1016/j.bbrc.2010.10.042
Lamb, C. A., Yoshimori, T., and Tooze, S. A. (2013). The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774. doi: 10.1038/nrm3696
Lanford, R. E., Hildebrandt-Eriksen, E. S., Petri, A., Persson, R., Lindow, M., Munk, M. E., et al. (2010). Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201. doi: 10.1126/science.1178178
Latorre, I., Leidinger, P., Backes, C., Domínguez, J., de Souza-Galvão, M. L., Maldonado, J., et al. (2015). A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur. Respir. J. 45, 1173–1176. doi: 10.1183/09031936.00221514
Lee, Y., Han, J., Yeom, K. H., Jin, H., and Kim, V. N. (2006). Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol. 71, 51–57. doi: 10.1101/sqb.2006.71.041
Li, D., Wang, T., Song, X., Qucuo, M., Yang, B., Zhang, J., et al. (2011). Genetic study of two single nucleotide polymorphisms within corresponding microRNAs and susceptibility to tuberculosis in a Chinese Tibetan and Han population. Hum. Immunol. 72, 598–602. doi: 10.1016/j.humimm.2011.03.004
Li, M., Wang, J., Fang, Y., Gong, S., Li, M., Wu, M., et al. (2016). MicroRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci. Rep. 6:23351. doi: 10.1038/srep23351
Li, S., Yue, Y., Xu, W., and Xiong, S. (2013). MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS One 12:e81438. doi: 10.1371/journal.pone.0081438
Lin, Y., Duan, Z., Xu, F., Zhang, J., Shulgina, M. V., and Li, F. (2017). Construction and analysis of the transcription factor-microRNA co-regulatory network response to Mycobacterium tuberculosis: a view from the blood. Am. J. Transl. Res. 9, 1962–1976.
Lin, Z., Leung, E., Lee, N., Lui, G., To, K., and Chan, R. C. Y. (2015). Differential MicroRNA expression in human macrophages with Mycobacterium tuberculosis infection of Beijing/W and non-Beijing/W strain types. PLoS One 10:e0126018. doi: 10.1371/journal.pone.0126018
Liston, A., Linterman, M., and Lu, L. F. (2010). MicroRNA in the adaptive immune system, in sickness and in health. J. Clin. Immunol. 30, 339–346. doi: 10.1007/s10875-010-9378-5
Liu, L., Shu, S., Cheung, G. S., and Wei, X. (2016). Effect of miR-146a/bFGF/PEG-PEI nanoparticles on inflammation response and tissue regeneration of human dental pulp cells. Biomed Res. Int. 2016:3892685. doi: 10.1155/2016/3892685
Liu, P. T., Wheelwright, M., Teles, R., Komisopoulou, E., Edfeldt, K., Ferguson, B., et al. (2012). MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 18, 267–273. doi: 10.1038/nm.2584
Liu, Y. H., Wang, X. J., Jiang, J., Cao, Z. H., Yang, B. F., and Cheng, X. (2011). Modulation of T cell cytokine production by miR-144∗ with elevated expression in patients with pulmonary tuberculosis. Mol. Immunol. 48, 1084–1090.
Liu, Z., Zhou, G., Deng, X., Yuc, Q., Hu, Y., Sun, H., et al. (2014). Analysis of miRNA expression profiling in human macrophages responding to Mycobacterium infection: induction of the immune regulator miR-146a. J. Infect. 68, 553–561. doi: 10.1016/j.jinf.2013.12.017
Lou, J., Wang, Y., Zhang, Z., and Qiu, W. (2017). MiR-20b inhibits Mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp. Cell Res. 358, 120–128. doi: 10.1016/j.yexcr.2017.06.007
Lu, Y. J., Shen, N., and Wang, X. (2016). Genetic associations between miR-146a/499 polymorphisms and tuberculosis: a meta-analysis. Int. J. Clin. Exp. Med. 9, 6445–6452.
Lv, Y., Guo, S., Li, X., Chi, J., Qu, Y., and Zhong, H. (2016). Sputum and serum microRNA-144 levels in patients with tuberculosis before and after treatment. Int. J. Infect. Dis. 43, 68–73. doi: 10.1016/j.ijid.2015.12.014
Ma, F., Xu, S., Liu, X., Zhang, Q., Xu, X., Liu, M., et al. (2011). The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 12, 861–869. doi: 10.1038/ni.2073
Maertzdorf, J., Weiner, J., Mollenkopf, H. J., Bornot, T. B., Bauer, T., Prasse, A., et al. (2012). Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. U.S.A. 109, 7853–7858. doi: 10.1073/pnas.1121072109
Maute, R. L., Dalla-Favera, R., and Basso, K. (2014). RNAs with multiple personalities. Wiley Interdiscip. Rev. RNA 5, 1–13. doi: 10.1002/wrna.1193
Mayr, C., Hemann, M. T., and Bartel, D. P. (2007). Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579. doi: 10.1126/science.1137999
McGregor, R. A., and Choi, M. S. (2011). MicroRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 11, 304–316. doi: 10.2174/156652411795677990
Meister, G., Landthaler, M., Dorsett, Y., and Tuschl, T. (2004). Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10, 544–550. doi: 10.1261/rna.5235104
Mendell, J. T., and Olson, E. N. (2012). MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187. doi: 10.1016/j.cell.2012.02.005
Meng, Q. L., Liu, F., Yang, X., Liu, X., Zhang, X., Zhang, C., et al. (2014). Identification of latent tuberculosis infection related microRNAs in human U937 macrophages expressing Mycobacterium tuberculosis Hsp16.3. BMC Microbiol. 14:37. doi: 10.1186/1471-2180-14-37
Merhautova, J., Demlova, R., and Slaby, O. (2016). MicroRNA-based therapy in animal models of selected gastrointestinal cancers. Front. Pharmacol. 7:329. doi: 10.3389/fphar.2016.00329
Miotto, P., Mwangoka, G., Valente, I. C., Norbis, L., Sotgiu, G., Bosu, R., et al. (2013). MiRNA signatures in sera of patients with active pulmonary tuberculosis. PLoS One 8:e80149. doi: 10.1371/journal.pone.0080149
Moore, L. B., Sawyer, A. J., Saucier-Sawyer, J., Saltzman, W. M., and Kyriakides, T. R. (2016). Nanoparticle delivery of miR-223 to attenuate macrophage fusion. Biomaterials 89, 127–135. doi: 10.1016/j.biomaterials.2016.02.036
Morishita, Y., Imai, T., Yoshizawa, H., Watanabe, M., Ishibashi, K., Muto, S., et al. (2015). Delivery of microRNA-146a with polyethylenimine nanoparticles inhibits renal fibrosis in vivo. Int. J. Nanomedicine 11, 3475–3488. doi: 10.2147/IJN.S82587
Nana-Sinkam, S. P., and Croce, C. M. (2013). Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 93, 98–104. doi: 10.1038/clpt.2012.192
Ni, B., Rajaram, M. V. S., Lafuse, W. P., Landes, M. B., and Schlesinger, L. S. (2014). Mycobacterium tuberculosis decreases human macrophage IFN-g responsiveness through miR-132 and miR-26a. J. Immunol. 193, 4537–4547. doi: 10.4049/jimmunol.1400124
O’Connell, R. M., Rao, D. S., Chaudhuri, A. A., and Baltimore, D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122. doi: 10.1038/nri2708
O’Connell, R. M., Taganov, K. D., Boldin, M. P., Cheng, G., and Baltimore, D. (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 5, 1604–1609. doi: 10.1073/pnas.0610731104
O’Neill, L. A., Sheedy, F. J., and McCoy, C. E. (2011). MicroRNAs: the fine tuners of Toll-like receptor signaling. Nat. Rev. Immunol. 11, 163–175. doi: 10.1038/nri2957
Ouimet, M., Koster, S., Sakowski, E., Ramkhelawon, B., van Solingen, C., Oldebeken, S., et al. (2016). Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 17, 677–686. doi: 10.1038/ni.3434
Parodi, A., Molinaro, R., Sushnitha, M., Evangelopoulos, M., Martinez, J. O., Arrighetti, N., et al. (2017). Bio-inspired engineering of cell- and virus-like nanoparticles for drug delivery. Biomaterials 147, 155–168. doi: 10.1016/j.biomaterials.2017.09.020
Pauli, A., Rinn, J. L., and Schier, A. F. (2011). Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. doi: 10.1038/nrg2904
Pegtel, D. M., Cosmopoulos, K., Thorley-Lawson, D. A., van Eijndhoven, M. A., Hopmans, E. S., Lindenberg, J. L., et al. (2010). Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333. doi: 10.1073/pnas.0914843107
Pillai, R. S., Bhattacharyya, S. N., and Filipowicz, W. (2007). Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 17, 118–126. doi: 10.1016/j.tcb.2006.12.007
Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P. E., et al. (2004). A pancreatic islet specific microRNA regulates insulin secretion. Nature 432, 226–230. doi: 10.1038/nature03076
Prosperi, D., Colombo, M., Zanonia, I., and Granucci, F. (2017). Drug nanocarriers to treat autoimmunity and chronic inflammatory diseases. Semin. Immunol. 34, 61–67. doi: 10.1016/j.smim.2017.08.010
Qi, Y., Cui, L., Ge, Y., Shi, Z., Zhao, K., Guo, X., et al. (2012). Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect. Dis. 12:384. doi: 10.1186/1471-2334-12-384
Qin, Y., Wang, Q., Zhou, Y., Duan, Y., and Gao, Q. (2016). Inhibition of IFN-γ-induced nitric oxide dependent antimycobacterial activity by miR-155 and C/EBPβ. Int. J. Mol. Sci. 17:535. doi: 10.3390/ijms17040535
Rajaram, M. V., Ni, B., Morris, J. D., Brooks, M. N., Carlson, T. K., Bak-thavachalu, B., et al. (2011). Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2(MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. U.S.A. 108, 17408–17413. doi: 10.1073/pnas.1112660108
Ren, N., Gao, G., Sun, Y., Zhang, L., Wang, H., Hua, W., et al. (2015). MicroRNA signatures from multidrug-resistant Mycobacterium tuberculosis. Mol. Med. Rep. 12, 6561–6567. doi: 10.3892/mmr.2015.4262
Richetta, C., and Faure, M. (2013). Autophagy in antiviral innate immunity. Cell. Microbiol. 15, 368–376. doi: 10.1111/cmi.12043
Rosas-Taraco, A. G., Higgins, D. M., Sanchez-Campillo, J., Lee, E. J., Orme, I. M., and González-Juarrero, M. (2011). Local pulmonary immunotherapy with siRNA targeting TGFbeta1 enhances antimicrobial capacity in Mycobacterium tuberculosis infected mice. Tuberculosis 91, 98–106. doi: 10.1016/j.tube.2010.11.004
Rothchild, A. C., Sissonsa, J. R., Shafiani, S., Plaisier, C., Mina, D., Maia, D., et al. (2016). MiR-155–regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 113, E6172–E6181.
Russell, D. G. (2011). Mycobacterium tuberculosis and the intimated is course of a chronic infection. Immunol. Rev. 240, 252–268. doi: 10.1111/j.1600-065X.2010.00984.x
Sahu, S. K., Kumar, M., Chakraborty, S., Banerjee, S. K., Kumar, R., Gupta, P., et al. (2017). MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 13:e1006410. doi: 10.1371/journal.ppat.1006410
Savan, R. (2014). Post-transcriptional regulation of interferons and their signaling pathways. J. Interferon Cytokine Res. 5, 318–329. doi: 10.1089/jir.2013.0117
Schepeler, T., Holm, A., Halvey, P., Nordentoft, I., Lamy, P., Riising, E. M., et al. (2012). Attenuation of the beta-catenin/TCF4 complex in colorectal cancer cells induces several growth-suppressive microRNAs that target cancer promoting genes. Oncogene 31, 50–60. doi: 10.1038/onc.2011.453
Sharbati, J., Lewin, A., Kutz-Lohroff, B., Kamal, E., Einspanier, R., and Sharbati, S. (2011). Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One 6:e20258. doi: 10.1371/journal.pone.0020258
Shi, Y., Zhao, X., Hsieh, J., Wichterle, H., Impey, S., Banerjee, S., et al. (2010). MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 30, 14931–14936. doi: 10.1523/JNEUROSCI.4280-10
Singh, Y., Kau, V., Mehra, A., Chatterjee, S., Tousif, S., Dwivedi, V. P., et al. (2013). Mycobacterium tuberculosis controls MicroRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J. Biol. Chem. 288, 5056–5061. doi: 10.1074/jbc.C112.439778
Spinelli, S. V., Diaza, A., D’Attilio, L., Marchesinib, M. M., Bogueb, C., Baya, M. L., et al. (2013). Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol. Immunol. 53, 265–269. doi: 10.1016/j.molimm.2012.08.008
Spizzo, R., Nicoloso, M. S., Lupini, L., Lu, Y., Fogarty, J., Rossi, S., et al. (2010). MiR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 17, 246–254. doi: 10.1038/cdd.2009.117
Starczynowski, D. T., Kuchenbauer, F., Argiropoulos, B., Sung, S., Morin, R., Muranyi, A., et al. (2010). Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 16, 49–58. doi: 10.1038/nm.2054
Steele, S., Brunton, J., and Kawula, T. (2015). The role of autophagy in intracellular pathogen nutrient acquisition. Front. Cell. Infect. Microbiol. 5:51. doi: 10.3389/fcimb.2015.00051
Stenvang, J., Petri, A., Lindow, M., Obad, S., and Kauppinen, S. (2012). Inhibition of microRNA function by antimiR oligonucleotides. Silence 3:1. doi: 10.1186/1758-907X-3-1
Su, M. J., Aldawsari, H., and Amiji, M. (2016). Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of MicroRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci. Rep. 22:30110. doi: 10.1038/srep30110
Subramanian, S., and Steer, C. J. (2010). MicroRNAs as gatekeepers of apoptosis. J. Cell. Physiol. 223, 289–298. doi: 10.1002/jcp.22066
Taganov, D., Boldin, M. P., Chang, K., and Baltimore, D. (2006). NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses Konstantin. Proc. Natl. Acad. Sci. U.S.A. 33, 12481–12486. doi: 10.1073/pnas.0605298103
Takeshita, F., Patrawala, L., Osaki, M., Takahashi, R. U., Yamamoto, Y., Kosaka, N., et al. (2010). Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via down regulation of multiple cell-cycle genes. Mol. Ther. 18, 181–187. doi: 10.1038/mt.2009.207
Talekar, M., Trivedi, M., Shah, P., Ouyang, Q., Oka, A., Gandham, S., et al. (2016). Combination wt-p53 and MicroRNA-125b transfection in a genetically engineered lung cancer model using dual CD44/EGFR-targeting nanoparticles. Mol. Ther. 24, 759–769. doi: 10.1038/mt.2015.225
Tashima, T. (2016). Intelligent substance delivery into cells using cell-penetrating peptides. Bioorg. Med. Chem. Lett. 27, 121–130. doi: 10.1016/j.bmcl.2016.11.083
Tay, H. L., Kaiko, G. E., Plank, M., Li, J., Maltby, S., Essilfie, A. T., et al. (2015). Antagonism of miR-328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of nontypeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 11:e1004549. doi: 10.1371/journal.ppat.1004549
Tran, T., Krishnan, S., and Amiji, M. M. (2016). MicroRNA-223 induced repolarization of peritoneal macrophages using CD44 targeting hyaluronic acid nanoparticles for anti-inflammatory effects. PLoS One 11:e152024. doi: 10.1371/journal.pone.0152024
Ueberberg, B., Kohns, M., Mayatepek, E., and Jacobsen, M. (2014). Are microRNAs suitable biomarkers of immunity to tuberculosis? Mol. Cell. Pediatr. 1:8. doi: 10.1186/s40348-014-0008-9
Uprichard, S. L. (2005). The therapeutic potential of RNA interference. FEBS Lett. 579, 5996–6007. doi: 10.1016/j.febslet.2005.08.004
Ura, S., Honda, M., Yamashita, T., Ueda, T., Takatori, H., Nishino, R., et al. (2009). Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49, 1098–1112. doi: 10.1002/hep.22749
Urgard, E., Lorents, A., Klaas, M., Padari, K., Viil, J., Runnel, T., et al. (2016). Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J. Control. Release 235, 195–204. doi: 10.1016/j.jconrel.2016.06.006
Vázquez-Hernández, F., Granada-Ramírez, D. A., Arias-Cerón, J. S., Rodriguez-Fragoso, P., Mendoza-Álvarez, J. G. E., Ramón-Gallegos, A., et al. (2017). “Use of nanostructured materials in drug delivery,” Nanobiomaterials, ed. R. Narayan (Sawston: Woodhead Publishing), 503–549.
Vegh, P., Magee, D. A., Nalpas, N. C., Bryan, K., McCabe, M. S., John, A., et al. (2015). MicroRNA profiling of the bovine alveolar macrophage response to Mycobacterium bovis infection suggests pathogen survival is enhanced by microRNA regulation of endocytosis and lysosome trafficking. Tuberculosis 95, 60–67. doi: 10.1016/j.tube.2014.10.011
Vera, J., Lai, X., Schmitz, U., and Wolkenhauer, O. (2013). MicroRNA-regulated networks: the perfect storm for classical molecular biology, the ideal scenario for systems biology. Adv. Exp. Med. Biol. 774, 55–76. doi: 10.1007/978-94-007-5590-1_4
Vergne, I., Chua, J., Singh, S. B., and Deretic, V. (2004). Cell biology of Mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 20, 367–394. doi: 10.1146/annurev.cellbio.20.010403.114015
Vergne, I., Gilleron, M., and Nigou, J. (2015). Manipulation of the endocytic pathway and phagocyte functions by Mycobacterium tuberculosis lipoarabinomannan. Front. Cell. Infect. Microbiol. 4:187. doi: 10.3389/fcimb.2014.00187
Walz, G., Ronacher, K., Hanekom, W., Scriba, T. J., and Zumla, A. (2011). Immunological biomarkers of tuberculosis. Nat. Rev. Immunol. 11, 343–354. doi: 10.1038/nri2960
Wang, C., Liu, C., Wei, L., Shi, L., Pan, Z., Mao, L., et al. (2016). A group of novel serum diagnostic biomarkers for multidrug-resistant tuberculosis by iTRAQ-2D LC-MS/MS and Solexa sequencing. Int. J. Biol. Sci. 12, 246–256. doi: 10.7150/ijbs.13805
Wang, C., Yang, S., Sun, G., Tang, X., Lu, S., Neyrolles, O., et al. (2011). Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS One 6:e25832. doi: 10.1371/journal.pone.0025832
Wang, J., Wu, M., Wen, J., Yang, K., Li, M., Zhan, X., et al. (2014). MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP. Mol. Immunol. 62, 29–36. doi: 10.1016/j.molimm.2014.05.012
Wang, J., Yang, K., Zhou, L., Minhaowu, Wu, Y., Zhu, M., et al. (2013). MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 9:e1003697. doi: 10.1371/journal.ppat.1003697
Wang, J. X., Xu, J., Han, Y. F., Zhu, Y. B., and Zhang, W. J. (2015). Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genet. Mol. Res. 14, 17235–17243. doi: 10.4238/2015.December.16.23
Wang, J. Y., Jia, Z., Wei, B., Zhou, Y., Niu, C., Bai, S., et al. (2017). MicroRNA-27a restrains the immune response to Mycobacterium tuberculosis infection by targeting IRAK4, a promoter of the NF-κB pathway. Int. J. Clin. Exp. Pathol. 10, 9894–9901.
Wang, P., Hou, J., Lin, L., Wang, C., Liu, X., Li, D., et al. (2010). Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 185, 6226–6233. doi: 10.4049/jimmunol.1000491
Wang, Q., Liu, S., Tang, Y., Liu, Q., and Yao, Y. (2014). MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One 9:e100949. doi: 10.1371/journal.pone.0100949
Watanabe, Y., and Kanai, A. (2011). Systems biology reveals microRNA-mediated gene regulation. Front. Genet. 2:29. doi: 10.3389/fgene.2011.00029
Weiner, J., Maertzdorf, J., and Kaufmann, S. H. (2013). The dual role of biomarkers for understanding basic principles and devising novel intervention strategies in tuberculosis. Ann. N. Y. Acad. Sci. 1283, 22–29. doi: 10.1111/j.1749-6632.2012.06802.x
Weiss, G., and Schaible, U. E. (2015). Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203. doi: 10.1111/imr.12266
WHO (2017). Global Tuberculosis Report. Available at: http://apps.who.int/iris/bitstream/10665/259366/1/9789241565516-eng.pdf?ua=1
Wu, J., Lu, C., Diao, N., Zhang, S., Wang, S., Wang, F., et al. (2012). Analysis of microRNA expression profiling identifies miR-155 and miR-155∗ as potential diagnostic markers for active tuberculosis: a preliminary study. Hum. Immunol. 73, 31–37. doi: 10.1016/j.humimm.2011.10.003
Wu, S., Lee, S., Huang, K., Lee, T., Hsu, P. W., and Weng, J. T. (2014). Systematic expression profiling analysis identifies specific MicroRNA-gene interactions that may differentiate between active and latent tuberculosis infection. Biomed Res. Int. 2014:895179. doi: 10.1155/2014/895179
Xin, H., Yang, Y., Liu, J., Li, X., Li, M., Feng, B., et al. (2016). Association between tuberculosis and circulating microRNA hsa-let-7b and hsa-miR-30b: a pilot study in a Chinese population. Tuberculosis 99, 63–69. doi: 10.1016/j.tube.2016.04.005
Xu, Y., Ren, W., Liu, Y., Zhang, X., Li, C., and Sun, Z. (2013). Tuberculosis-related miRNAs have potential as disease biomarkers. J. Tuberc. Res. 1, 17–27. doi: 10.4236/jtr.2013.12005
Yi, Z., Fu, Y., Ji, R., Li, R., and Guan, Z. (2012). Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One 7:e43184. doi: 10.1371/journal.pone.0043184
Zhang, C., Xi, X., Wang, Q., Jiao, J., Zhang, L., and Zhao, H. (2015). The association between serum miR-155 and natural killer cells from tuberculosis patients. Int. J. Clin. Exp. Med. 8, 9168–9172.
Zhang, G., Liu, X., Wang, W., Cai, Y., Li, S., and Chen, Q. (2016). Down-regulation of miR-20a-5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle 15, 2527–2538. doi: 10.1080/15384101.2016.1215386
Zhang, X., Guo, J., Fan, S., Li, Y., Wei, L., Yang, X., et al. (2013). Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One 12:e81076. doi: 10.1371/journal.pone.0081076
Zhang, Z. M., Zhang, A. R., Xu, M., Lou, J., and Qiu, W. Q. (2017). TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages. Exp. Cell Res. 352, 313–321. doi: 10.1016/j.yexcr.2017.02.025
Zheng, D., Yu, Y., Li, M., Wang, G., Chen, R., Fan, G., et al. (2015). Inhibition of microRNA 195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J. Infect. Dis. 213, 1661–1670. doi: 10.1093/infdis/jiv760
Zheng, L., Leung, E., Lee, N., Lui, G., To, K., Chan, R. C. Y., et al. (2015). Differential MicroRNA expression in human macrophages with Mycobacterium tuberculosis infection of Beijing/W and non-Beijing/W strain types. PLoS One 10:e0126018. doi: 10.1371/journal.pone.0126018
Zhou, F., Ferguson, J., and Chang, J. T. (2007). Inter- and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genomics 8:396. doi: 10.1186/1471-2164-8-396
Zhou, M., Yu, G., Yang, X., Zhu, C., Zhang, Z., and Zhan, X. (2016). Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol. Med. Rep. 13, 4620–4626. doi: 10.3892/mmr.2016.5097
Keywords: Mycobacterium tuberculosis, miRNA expression, immune regulation, autophagy, apoptosis, biomarker, nanoparticles, host directed therapy
Citation: Sabir N, Hussain T, Shah SZA, Peramo A, Zhao D and Zhou X (2018) miRNAs in Tuberculosis: New Avenues for Diagnosis and Host-Directed Therapy. Front. Microbiol. 9:602. doi: 10.3389/fmicb.2018.00602
Received: 07 January 2018; Accepted: 15 March 2018;
Published: 29 March 2018.
Edited by:
Petros Karakousis, Johns Hopkins University School of Medicine, United StatesCopyright © 2018 Sabir, Hussain, Shah, Peramo, Zhao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangmei Zhou, emhvdXhtQGNhdS5lZHUuY24=
 Naveed Sabir
Naveed Sabir Tariq Hussain
Tariq Hussain Deming Zhao
Deming Zhao Xiangmei Zhou
Xiangmei Zhou