- 1Guangdong Province Key Laboratory of Microbial Signals and Disease Control, South China Agricultural University, Guangzhou, China
- 2Guangdong Province Key Laboratory of New Technology in Rice Breeding, Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 3Integrative Microbiology Research Centre, South China Agricultural University, Guangzhou, China
Mitogen-activated protein kinase (MAPK) pathways are ubiquitous and evolutionarily conserved signal transduction modules directing cellular respond to a diverse array of stimuli, in the eukaryotic organisms. In this study, PlMAPK10 was identified to encode a MAPK in Peronophythora litchii, the oomycete pathogen causing litchi downy blight disease. PlMAPK10, containing a specific and highly conserved dual phosphorylation lip sequence SEY (Serine-Glutamic-Tyrosine), represents a novel group of MAPKs as previously reported. Transcriptional profiling showed that PlMAPK10 expression was up-regulated in zoospore and cyst stages. To elucidate its function, the PlMAPK10 gene was silenced by stable transformation. PlMAPK10 silence did not impair oospore production, sporangium germination, zoospore encyst, or cyst germination but hindered hyphal growth, sporulation, pathogenicity, likely due to altering laccase activity. Over all, our results indicated that a MAPK encoded by PlMAPK10 gene in P. litchii is important for pathogenic development.
Introduction
Litchi (Litchi chinensis Sonn.) is native to China with the hugest cultivated area in the world and production due to its lovely shape, beautiful color, delicious taste, and medicinal value (Jiang et al., 2006). However, litchi fruit does not reach maximum export volume from China, owing to suffering from serious plant diseases and insect pests, in which, litchi downy blight is the most serious disease (Wang et al., 2009). Litchi downy blight is caused by the oomycete pathogen Peronophythora litchii, with a broad range of infection stages and tissues including tender leaf, flower, and mature fruit period, and even extends to post-harvest and transport (Xu et al., 2013). Oomycetes are evolutionarily related to marine algae and has a far relationship from fungi although it grows filamentous mycelia and includes various of plant and animal pathogens responsible for many economically important diseases (Ye et al., 2016).
Mitogen-activated protein kinase (MAPK), with a conserved serine/threonine domain, is the center of cell signal transduction network. A variety of external or internal signals were shown to be transduced by MAPK pathway and regulate cell growth, development, stress response, and pathogenicity, etc. (Chen and Thorner, 2007). In the model fungus, Saccharomyces cerevisiae, five important cellular development events, namely sporulation, cell wall integrity, osmoregulation, sexual mating, and filamentous (pseudohyphal) growth, have been shown to be regulated respectively by five established MAPK pathways (Chen and Thorner, 2007; Vallejo and Mayinger, 2015). Specifically, S. cerevisiae Hog1 is integral to the osmoregulatory signal transduction cascade (Nancy et al., 2015), Fus3/Kss1 are essential for sexual reproduction and regulate chronological life span (Maneesha et al., 2017), and MPK1 is involved in cell wall integrity (Kim et al., 2018). In addition, a specialized type of MAPKs in fungi and mammalian was termed as stress-activated protein kinases (SAPKs), as it is responsible for stress response, including heat shock, hyperosmolarity, ultraviolet (UV) light irradiation, and oxidative stress (Brown et al., 2014). Examples of SAPKs from fungi include Magnaporthe oryzae YPD1 regulating penetration and conidiation (Mohanan et al., 2017), Mycosphaerella graminicola Hog1 involved in osmosensitivity and dimorphic transition in fungal pathogenicity (Mehrabi et al., 2006), and Bipolaris oryzae SRM1 for tolerance to hyperosmolarity, hydrogen peroxide (H2O2), and UV exposure (Moriwaki et al., 2006). On the other hand, mammalian p38s and JNKs are involved in the inflammatory and stress responses (Kyriakis et al., 2004; Chadee and Kyriakis, 2010). In the plant pathogenic oomycetes, three MAPKs/SAPKs in Phytophthora sojae, namely PsMPK1, PsMPK3 (PsSAK1) and PsMPK7, have been demonstrated essential for spore development, osmotic and oxidative stresses responses, and pathogenicity (Li et al., 2010, 2014; Gao et al., 2015).
In the long-term interaction between plant and microbial pathogens, plant has evolved defense responses, one of which is rapidly elevating level of reactive oxygen species (ROS) at the infection site (Park et al., 2018). As the first parclose for host plant against pathogens, ROS is further utilized by peroxidase and combines with polyphenols, such as callose deposition, lignin, and polymeric polyphenols synthesized (Torres and Dangl, 2005; de Leon and Montesano, 2017). In addition, ROS triggers the pathogen-associated molecular pattern-triggered immunity (PTI) by acting as a secondary messenger (Neill et al., 2002; Nürnberger et al., 2004; Zhang et al., 2009; Sang and Macho, 2017). Meanwhile, plant pathogens evolved ability to detoxify plant-derived ROS either by enzymatic (Staerck et al., 2017) or non-enzymatic mechanism, to ensure successful infection (Garg and Chandel, 2015).
In this study, we firstly functionally study PlMAPK10, a P. litchii ortholog to P. sojae MAPK10 (Ye et al., 2015), by generation and characterization of PlMAPK10-silenced mutants, and examining the expression pattern during pathogenic development. Our results showed that PlMAPK10 was highly expressed in infection-related structures, including zoospores and cysts, compared to vegetative mycelia. Silence of PlMAPK10 gene led to reduced mycelial growth rate, reduced sporulation, weakened pathogenicity and altered laccase activity, indicating an important function of MAPK signal pathway in oomycete pathogenicity.
Materials and Methods
Bioinformatics Analysis
Genome sequence data of P. litchii were obtained from NCBI (BioProject ID: PRJNA290406) (Ye et al., 2016). JGI1 is used for prediction of P. sojae and Phytophthora ramorum genomic DNA and protein sequence. Genomic DNA and protein sequences of Phytophthora infestans were obtained from the Broad Institute2. The five STKc_MAPKs in the alignment of the indicated MAP kinases were predicted by SMART3 and CDD4 in NCBI. The phylogenetic dendrograms were constructed with Neighbor-Joining algorithm (with setting of 1000 bootstrap replications), by the MEGA 7.0 program5.
P. litchii Strain and Culture Conditions
Peronophythora litchii strain was isolated from a litchi fruit with litchi downy blight disease in Guangdong province, and purified by single-spore isolation using MSM400 Dissection Microscope (Singer Instruments) (Jiang et al., 2017). The strain was cultured on carrot juice agar (CJA) media (juice from 200 g carrot topped up to 1 L, 15 g/L agar for solid media) at 25°C in the dark. The P. litchii transformants of PlMAPK10-silencing were maintained on CJA media containing 50 μg/L geneticin (G418) at 25°C in the dark and then transferred to be cultured on another CJA media plate without G418 at the same condition as wild type strain. Sporangia were harvested by flooding the mycelia, which had been cultured on CJA media for 5 days, with sterile distilled water, then filtering the subsequent suspension through a 100 μm mesh size of strainer. The suspension was incubated with sterile distilled water at 16°C for 0.5 and 2 h, respectively, for zoospores release. 10 μL sporangium suspension was sampled to calculate the release rate of zoospores under microscope.
RNA Extraction and Gene Expression Analysis
Total RNA was extracted from different developmental stages and different time-points post-mycelial mat-inoculation on tender litchi leaves (Ye et al., 2011), using the Total RNA Kit (Omega, Cat. No.: R6834-01). RNA integrity was examined by agarose gel electrophoresis. For reverse transcription, 1 μg total RNA was used to synthesize the first-strand cDNA by oligo(dT) priming with the M-MLV reverse transcriptase kit (Invitrogen, Cat. No.: S28025-014). Transcription of PlMAPK10 was analyzed with qRT-PCR assay using primers MAPK10-QRT-F: GCTCCACTTTAAGCCGAATG and MAPK10-QRT-R: ATTCGTCACCTTGCAGCTCT. P. litchii actin gene (using primers PlAct-F: TCACGCTATTGTTCGTCTGG and PlAct-R: TCATCTCCTGGTCGAAGTCC) was used as loading control and the relative fold change was calculated using the 2-ΔΔCT method.
To analyze transcription of PlMAPK10 gene in response to oxidative stress, WT hyphae were cultured at 25°C in the dark for 3 days using liquid carrot juice medium followed by washed with liquid Plich medium (0.5 g/L KH2PO4, 0.25 g/L MgSO4⋅7H2O, 1 g/L asparagine, 1 mg/L thiamine, 0.5 g/L yeast extract, 10 mg/L β-sitosterol, 5 g/L glucose, 15 g/L bacto-agar for solid media), and subsequently incubation in 20 mL liquid Plich medium (van West et al., 1999). After 0, 45, or 55 min respectively, appropriate H2O2 was added to the liquid Plich medium to reach a final concentration of 5 mM. At 60 min post-inoculation in liquid Plich medium, all samples were harvested and subject to examination of PlMAPK10 expressional level, with a set of mycelial culture in liquid Plich medium for 60 min, untreated by H2O2, as control.
Transformation of P. litchii
PlMAPK10-silenced mutants were generated by a polyethylene glycol (PEG)-mediated protoplast transformation strategy. The full-length open reading frame of PlMAPK10 was amplified with primers MAPK10-FL-F: AAACGTACGATGACGAGCACGTCGTTG and MAPK10-FL-R:s AAAATCGATTTACTCGGCGATAGTCTCCTGT by PCR. The PCR fragment was ligated into the pTOR vector6 digested with ClaI and BsiWI in antisense orientation and sequenced for confirmation. 30 μg of the resulting construct pTOR::PlMAPK10 was transformed, a selection construct carrying the geneticin gene nptII, into protoplasts of P. litchii wild type strain.
Preliminary transformants were identified according to their ability to grow on CJA media containing 50 μg/mL geneticin. Genome PCR analysis were then performed to confirm plasmid integration in these transformants, with primers ham34-F: GCTTTTGCGTCCTACCATCCG and MAPK10-F: AAACGTACGATGACGAGCACGTCGTTG. These transformants were then subjected to qRT-PCR assay for analysis of PlMAPK10 expression level and gene-silencing efficiency.
Pathogenicity Assays on Litchi Leaves
Pathogenicity on litchi was assayed using zoospore suspensions of P. litchii with two inoculation methods: (1) 10 μL zoospore suspension (104 zoospores/L) was directly inoculated onto reverse side of a litchi tender leaf. Each strain was tested on 10 litchi leaves. The proportion of leaves displaying disease phenotype was calculated; (2) 30 mL sporangium suspension (104 zoospores/L) was sprayed to three tender litchi branches, each of which contained approximately 30 leaves. According to the proportions of leaves with different disease levels, a disease severity value was calculated. Disease severity = [Σ (number of symptomatic leaves in certain index) × disease index]/[(total number of leaves investigated) × (the highest disease index)] × 100. The leaves were maintained at 80% humidity in the dark at 25°C. The symptoms were examined at 48 h post-inoculation. This experiment was repeated three times independently, with three biological replications per repeat.
Extracellular Enzyme Activity Assays
Detection of peroxidase secretion was performed following the reported procedure (Sheng et al., 2015), based on laccase activity detection. At least three independent repeats, each with three biological replica, were performed for each instance.
Identification, Selection, and Quantification of Putative Extracellular Laccase and Peroxidase Genes
We firstly searched the predicted extracellular laccase and peroxidase encoding genes in P. litchii genome according to their orthologs that annotated in the P. sojae genome and their conserved Cu-oxidase domains (IPR001117, IPR011706, and IPR011707). All the identified genes were quantified by qRT-PCR and analyzed by Multi-experiment Viewer program to select for those with significant fold change, during the life cycle in the wild type. All the selected genes were used to compare expression with wild type and CK strain by qRT-PCR. This experiment was repeated three independent times, and for each repeat with three biological replications.
Hypersensitivity Toward Oxidative Stress
The sensitivity of PlMAPK10-silenced transformants to hydrogen peroxide (H2O2) was evaluated on modified Plich medium plates following established procedure (Sheng et al., 2015). At least three independent repeats, which contained three replicates, were performed for each instance. Growth inhibition rate was calculated, and the inhibition rate = (growth rate on plates without H2O2-growth rate on plates with H2O2)/growth rate on plates with H2O2.
Results
PlMAPK10 Encodes a Typical and Conserved Mitogen-Activated Protein Kinase With a Dual Phosphorylation Lip
The available genome data of P. litchii was searched against using full-length sequence of the P. sojae MAPK10 protein (PsMAPK10) through the program TBLASTN integrated in SeqHunter2.0 software (Ye et al., 2010), with an expected (E) value of 1.0E-20. A single gene, designated PlMAPK10, was identified in the search which encoded a protein of 85% amino acid identity with PsMAPK10 and the case spanning more than 90% of the length of the sequence. The corrected PlMAPK10 gene model is 3249 bp in length with no intron, and encodes a protein of 1082 amino acids. We predicted a typically conservative serine/threonine protein kinase catalytic (STKc) MAPK domain (E = 8.14e-143; amino acids 748–1083) in PlMAPK10, using the CD-Search program on NCBI website. Sequence conservation between full-length PlMAPK10 protein and its orthologs in P. sojae, P. infestans, and P. parasitica is 85, 90, and 90%, respectively. We also noticed that a same predicted dual phosphorylation lip sequence SEY present in both P. litchii (amino acids 894–896) and several other oomycetes (Figure 1).

FIGURE 1. Amino acid sequences arrangement and phylogenetic analysis with PlMAPK10 protein and its orthologs. Amino acid sequences alignment of PlMAPK10 and its orthologs from Phytophthora infestans (Phi), Phytophthora sojae (Phs), Hyaloperonospora arabidopsidis (Hya), and Peronospora tabacina (Pet). The red boxes and three red triangles represent STKc_MAPK domains and predicted dual phosphorylation lip sequences, respectively.
Besides the 10 oomycete MAPKs, we search the orthologous sequence of PlMAPK10 by BLASTP using its full length sequence, in the pathogenic fungi including genus of Ustilago (taxid: 5269), Sporisorium (taxid: 63265), Fusarium (taxid: 5506), Botrytis (taxid: 33196), and in the rice-blast fungus Magnaporthe oryzae 70–15 (taxid: 242507). The top 3–10 hits (>40% amino acid identity, E value ≤ 1.0E-20) from each search were retrieved for sequence alignment and phylogenetic analysis. We also searched the similar MAPKs from plant Arabidopsis thaliana, and from the yeasts S. cerevisiae and Schizosaccharomyces pombe by BLink, and included them for phylogenetic analysis. The phylogenetic analysis showed that the 10 oomycete MAPK10 orthologs were in a clade that seperated from the other kinases (Figure 2), and they were mostly conserved in the serine/threonine protein kinase catalytic (STKc) MAPK domain. Therefore we conclude that PlMAPK10 was conserved with its orthologs of the indicated two Phytophthora species and three downy mildew species in full length protein sequence.
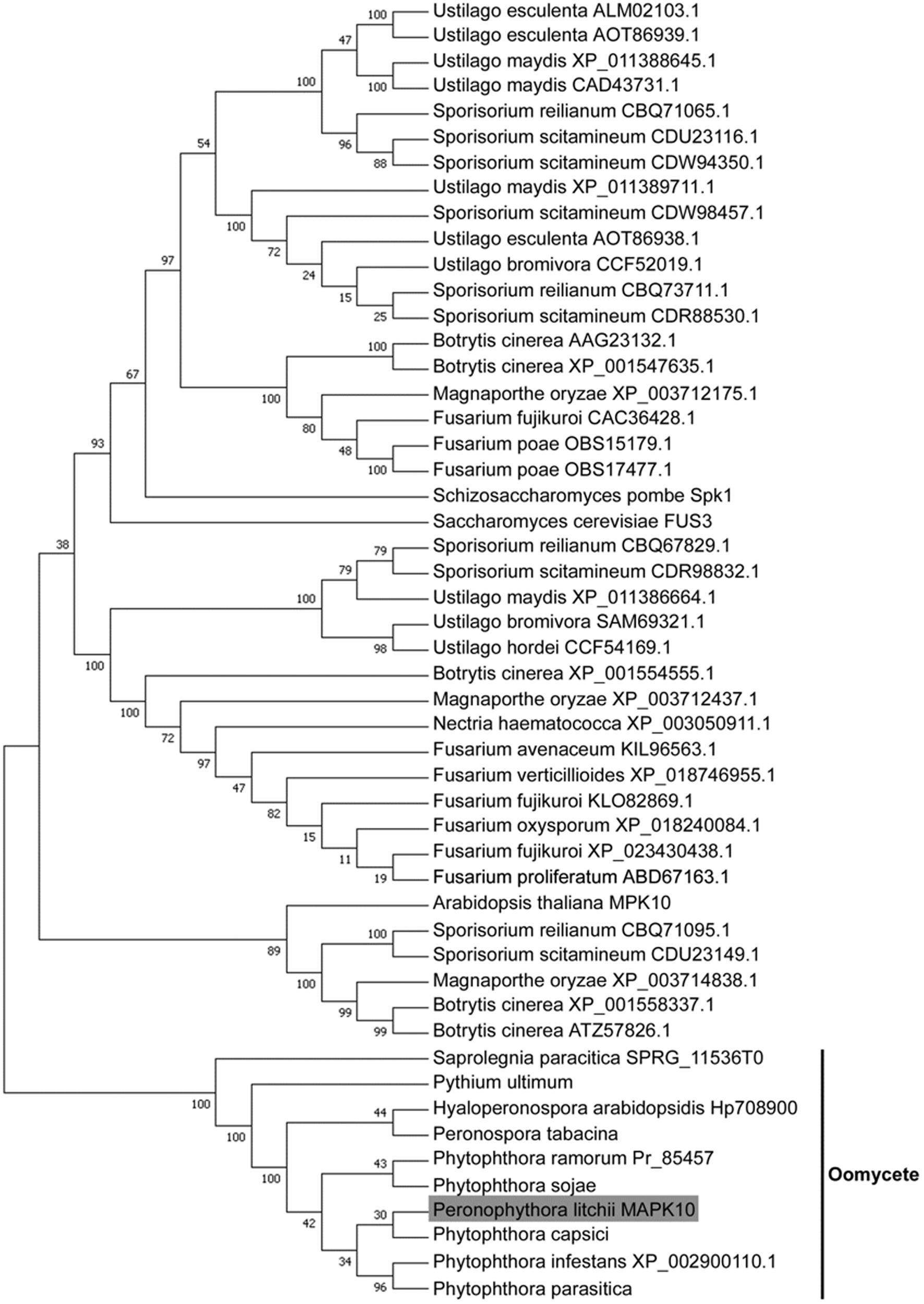
FIGURE 2. Phylogenetic analysis of MAPK10 proteins in P. litchii and in other oomycetes, pathogenic fungi, yeasts, and plant. Each sequence was labeled with its taxonomic name and accession number. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 4.63768252 is shown. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The analysis involved 51 amino acid sequences. All positions with less than 50% site coverage were eliminated. That is, fewer than 50% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 371 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
Generation of PlMAPK10-Silenced Transformants
Next we examine the transcriptional patterns of PlMAPK10 in mycelia, sporangia, zoospores, cysts, germination cysts and infection stages, by quantitative real-time PCR (qRT-PCR). PlMAPK10 Transcripts in zoospores and cysts were estimated as twofold of that in mycelium, while not obviously changed in sporangia, germination cysts and during the infection process (Figure 3A). Thus, we infer that PlMAPK10 may play a crucial role in zoospores and cysts.
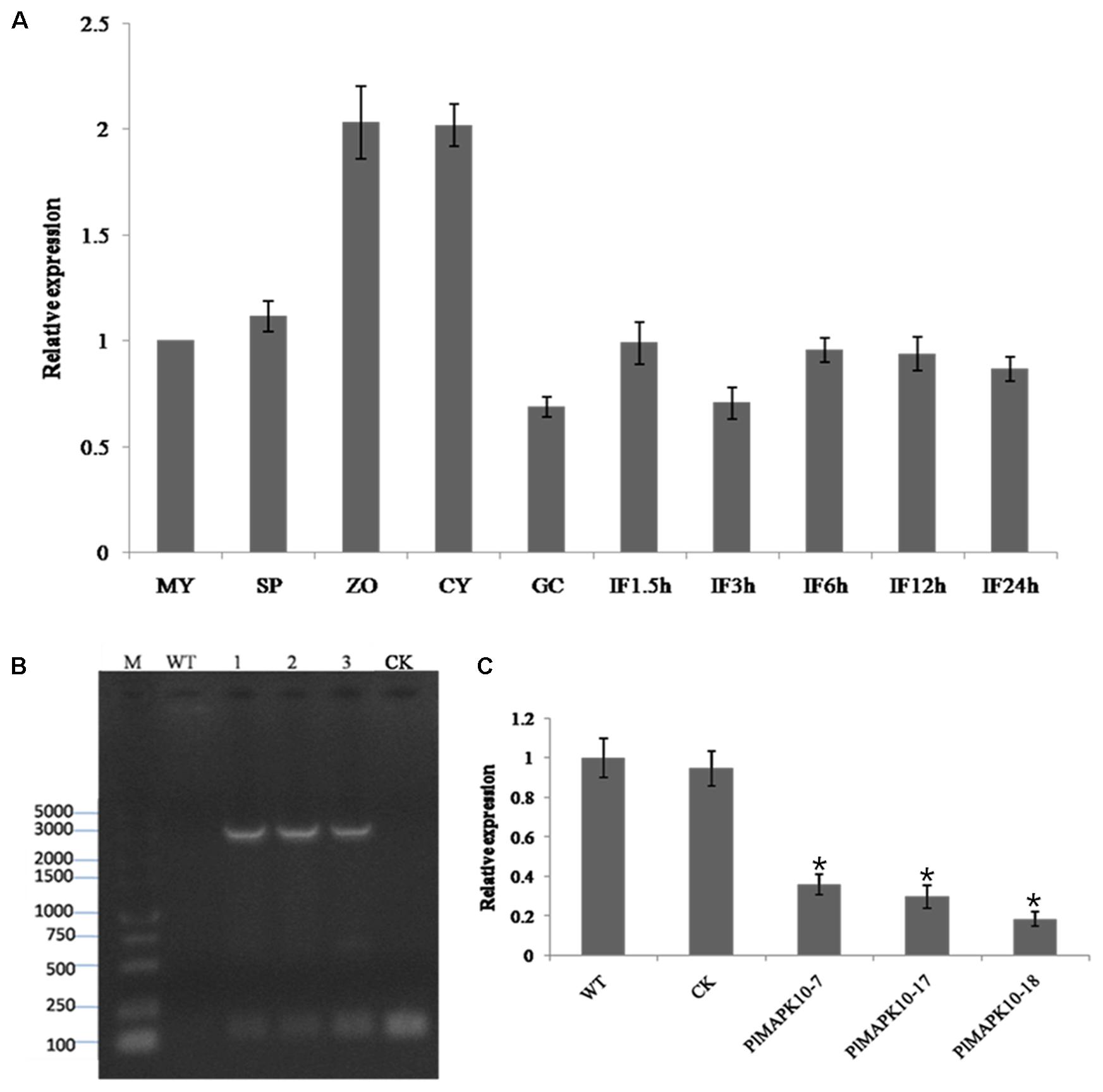
FIGURE 3. Transcriptional profile of PlMAPK10 gene in the life cycle and infection stages, and identification and selection of PlMAPK10-silenced transformants. (A) Relative expression levels of PlMAPK10 were determined by qRT-PCR with total RNA extracted from specific-stages of life cycle: MY, mycelia; SP, sporangia; ZO, zoospores; CY, cysts; GC, germinating cysts. IF1.5h–IF24h, infection stage of 1.5, 3, 6, 12, and 24 h post-inoculation, using mycelial mats on the tender litchi leaves. Expression levels were normalized using the MY values as ‘1.’ P. litchii Actin gene was used as endogenous gene. The experiment was repeated two times with independent sampling. (B) Genome PCR identification of three transformants, CK strain and WT strain. Marker: DL 5000. Lane 1, lane 2, and lane 3 were the three PlMAPK10-silenced transformants, named PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18, respectively. (C) Relative gene expression levels of PlMAPK10 in PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18 and the WT and CK strains. PlMAPK10 expression was normalized to that of WT value (set as “1.0”). P. litchii Actin gene serves as a loading control. Asterisks represent significant difference (P < 0.05) based on statistics analysis using SPSS (version 19.0). These experiments were repeated three times independent, and for each repeat with three biological replications.
To explore the biological functions of PlMAPK10 in P. litchii development, stress response, and pathogenicity, we generated an antisense direction of PlMAPK10 coding region, driven by the oomycete constitutive pHAM34 promoter in a pTOR vector (Whisson et al., 2007) and transformed it into WT P. litchii protoplast cells following a well-established protocol (Jiang et al., 2017). Transformation with pTOR vector served as blank control and named as CK strain. In total, we obtained 132 transformants by antibiotic resistance screening. Among them, 49 transformants with stable integration of silence cassette into P. litchii genome, were identified by genome PCR amplification (Figure 3B). Finally, using qRT-PCR we identified 3 (T7, T17, and T18), out of 49 PlMAPK10-integrated transformants, showing significant (P < 0.05) lower transcriptional level of PlMAPK10 than that in the wild type or CK control (Figure 3C). In the following we characterized mycelium growth, sporulation, and zoospore release of these three PlMAPK10-silenced strains and compared to WT and CK strains.
PlMAPK10-Silenced Transformants Showed Slower Mycelium Growth and Less Sporulation But No Influence in Zoospore Release
We first measured colony diameters of the three silenced transformants as well as WT and CK strains. As shown in Figure 4A, the colony size of the PlMAPK10-silenced strains was obviously smaller than that of WT or CK strains. We further calculated the mycelial radial growth rate, and found that the rates of three PlMAPK10-silenced transformants were ranged from 6.36 to 7.00 mm/d, and were significantly (P < 0.05) lower than that of wild type (7.74 mm/d) and CK strains (7.71 mm/d) (Figure 4B).
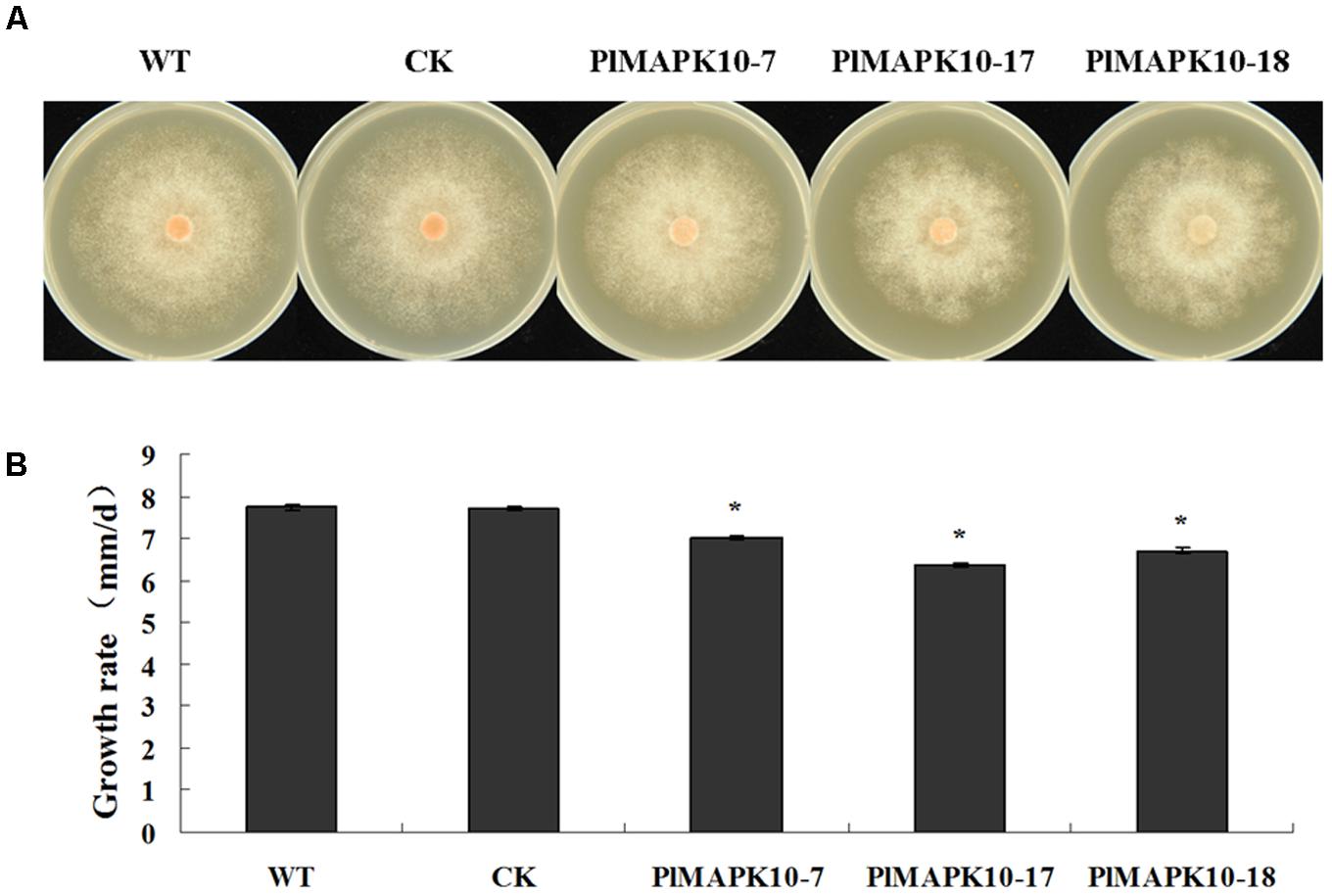
FIGURE 4. Measurement of colony diameter of WT, CK, and the PlMAPK10-silenced transformants, PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18. (A) Photographs of colony of WT, CK, and the PlMAPK10-silenced transformants, PlMAPK10-7, PlMAPK10-17 and PlMAPK10-18, were taken at 7 days. (B) Measurement of colony growth rate of WT, CK, and the three PlMAPK10-silenced transformants. With “∗” on the top of each bar represents significant difference between PlMAPK10-silenced transformants and two controls based on the statistics analysis of Student’s t-test (∗P < 0.05). This experiment was repeated three times independent, and for each repeat with three biological replications.
Next we assessed production of sporangia in the silenced strains. The average numbers in per μL sporangium suspension of the three PlMAPK10-silenced transformants were 21 in PlMAPK10-7, 17 in PlMAPK10-17, and 20 in PlMAPK10-18 respectively, significantly (P < 0.05) less than 32 in wild type and 31 in CK control (Figure 5A). However, zoospore release from an equal number of sporangia was not affected by PlMAPK10 silencing. Zoospore release was induced in sporangium suspensions of three silenced transformants and two controls, by incubation at 16°C for 0.5 and 2 h, respectively. As shown in Figure 5B, after 0.5 h of incubation, an average of 63.15, 64.76, and 64.10% of the sporangia of PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18, respectively, released zoospores, which was comparable to that in the controls (66.72% for WT and 64.16% for CK strain). After 2 h of incubation, an average of 95.77, 98.12, and 96.94% of the sporangia of PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18, respectively, released zoospores, compared with 95.96 and 95.59% in the controls (WT and CK, respectively).
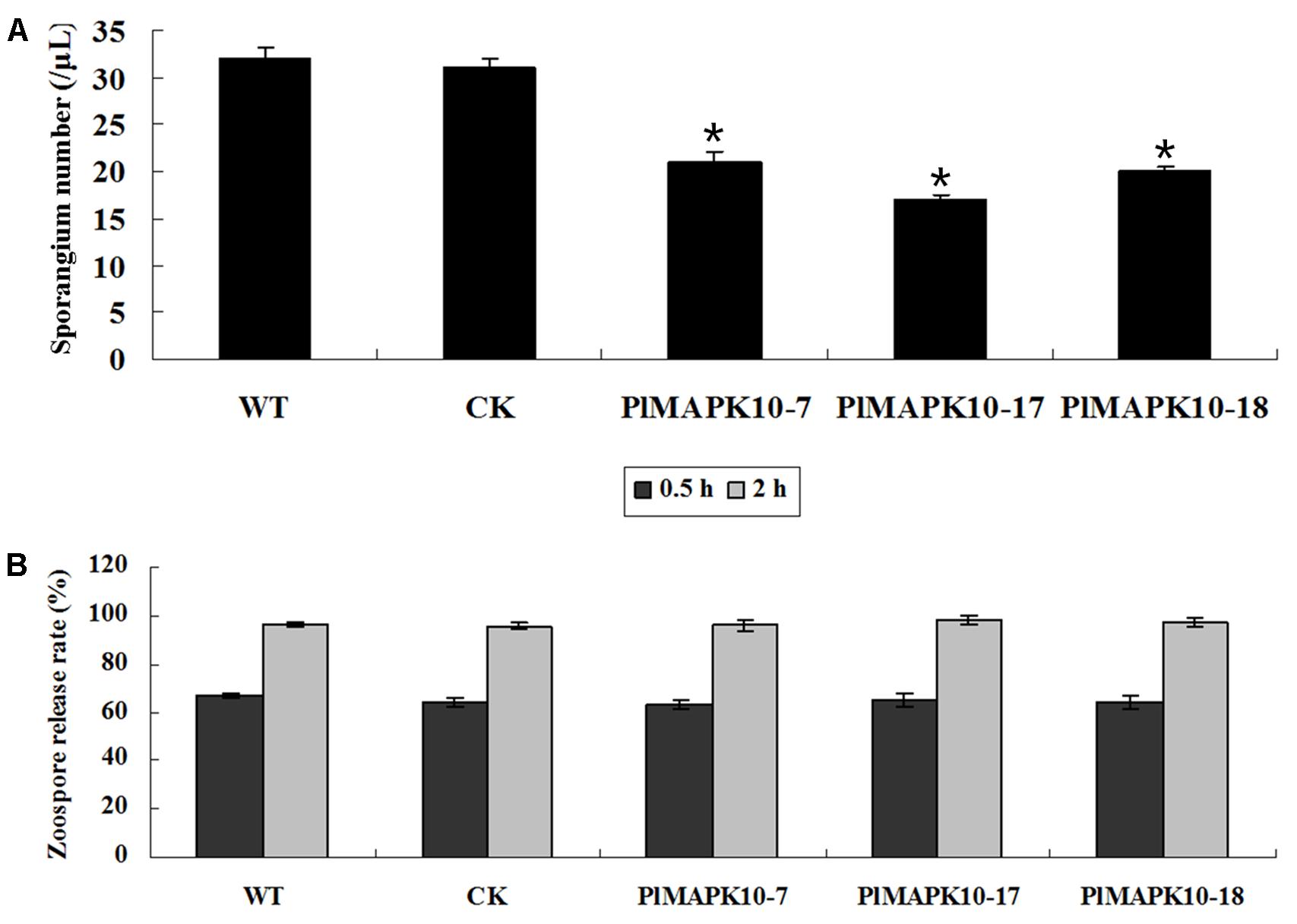
FIGURE 5. Measurement of sporangia number and zoospores release of PlMAPK10-silenced transformants, wild type and CK strain. (A) Mean ± SE sporangia number in 1 μl sporangia suspension. (B) Mean ± SE zoospore release rate. With “∗” on the top of each bar represents significant difference between PlMAPK10-silenced transformants and two controls (P < 0.05), based on the statistics analysis using SPSS (version 19.0) with statistical analysis of two-tailed t-test. This experiment was repeated three times independent, and for each repeat with three biological replications.
PlMAPK10 Is Required for Pathogenicity
To explore the roles of PlMAPK10 in P. litchii pathogenicity we performed infection assay with tender litchi leave. Sporangium and zoospore suspensions of the three silenced transformants and the two control strains were respectively inoculated onto the leave surface and leave vein. For leave surface infection, zoospore suspensions were inoculated on the reverse side of tender litchi leaves and symptoms were examined on 2 days post-inoculation (dpi). We observed that the leaves inoculated with control zoospores showed typical disease symptoms and water-soaked lesions at 2 dpi (Figure 6A), while the lesion caused by the three PlMAPK10-silenced transformants was smaller (Figure 6A). To mimic a more realistic pathogenic process, sporangia suspensions were sprayed to tender litchi branches. According to the proportions of leaves of different disease levels, a disease severity value was calculated (see section “Materials and Methods”). As shown in Figure 6B, after 2 days of inoculation, an average disease severity value of 70.19, 58.89, and 56.30 of PlMAPK10-7, PlMAPK10-17, and PlMAPK10-18 sporangia, respectively, significantly (P < 0.05) lower when compared with 98.89 and 97.78 in the controls (WT and CK, respectively). Therefore, PlMAPK10 might be critical for full pathogenicity of P. litchii.
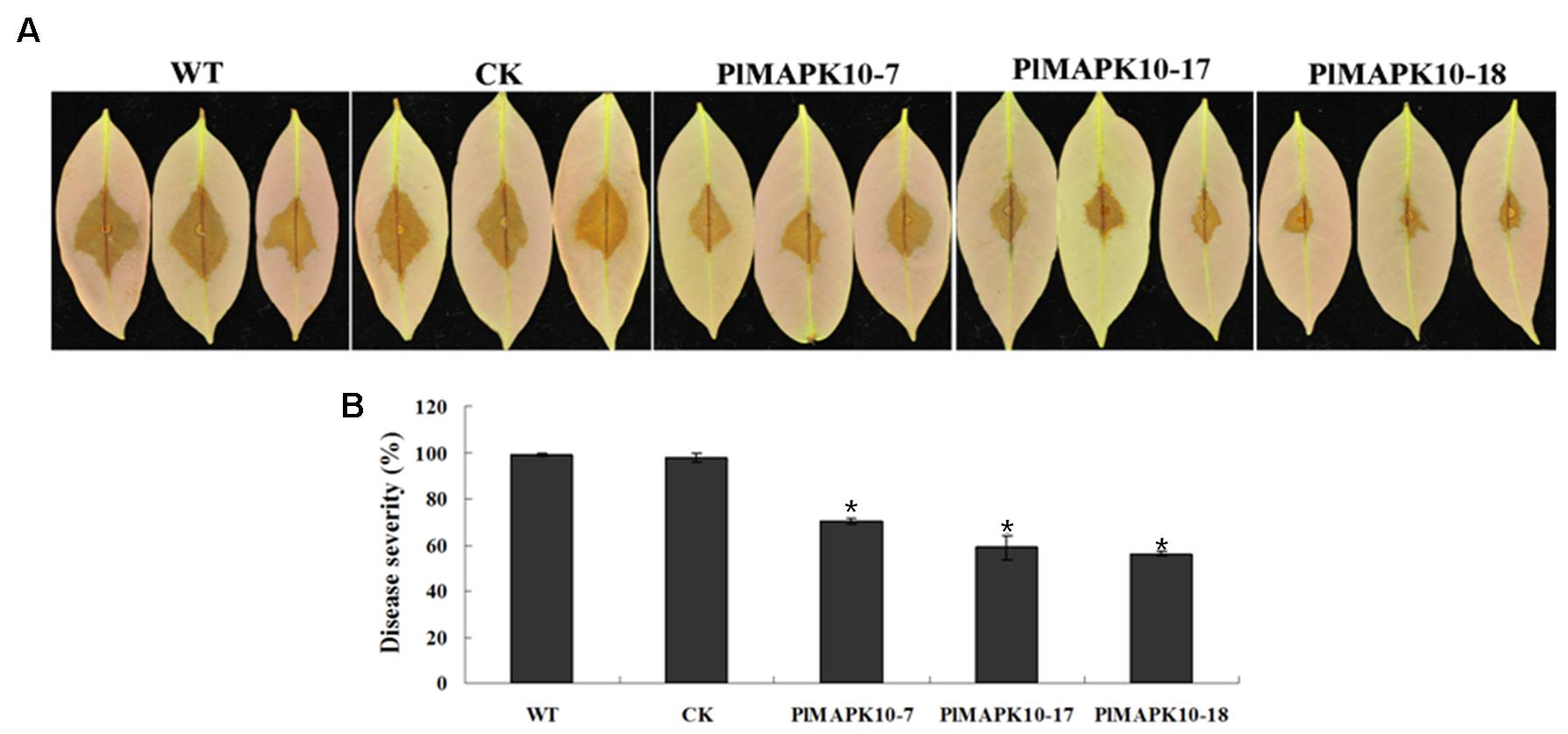
FIGURE 6. Pathogenicity test of Peronophythora litchii PlMAPK10-silenced transformants, WT and CK. (A) Zoospore suspension was inoculated on tender litchi leaves (n = 30 for each strain) for 48 h at 25°C in the dark. Images are three representatives for each instance. (B) Quantification of pathogenicity by disease severity values. Zoospore suspension was sprayed to tender litchi branches (containing approximately 30 leaves), for three independent repeats. With “∗” on the top of each bar represents significant difference between PlMAPK10-silenced transformants and two controls (P < 0.05), based on the statistics analysis using SPSS (version 19.0) with statistical analysis of two-tailed t-test. This experiment was repeated three times independent, and for each repeat with three biological replications.
Silencing of PlMAPK10 Decreases Extracellular Laccase Activity Rather Than Peroxidase Activity Through Regulating the Transcription of Relative Gene
Extracellular laccase or peroxidase activity has been shown to contribute to fungal or oomycete pathogenicity, therefore we further investigate any alteration in such extracellular enzymes activities in the PlMAPK10-silenced transformants, which may account for their reduced pathogenicity. We first assessed peroxidase activity based on Congo Red (CR) degradation. As shown in Figure 7A (Upper panel), diameters of the halo caused by the three PlMAPK10-silenced transformants were comparable to that of controls. Subsequently, we examined laccase activity based on oxidation assay of ABTS [2,2′-azino-bis (3-ethylbenzothiazoline-6 sulfonic acid)]. Three PlMAPK10-silenced transformants showed significantly decreased accumulation of ABTS, visualized by dark purple staining around the mycelial mat compared with wild type and CK control (Figure 7A, Lower panel). These results indicated that PlMAPK10 may regulate extracellular laccase activity to ensure successful host infection.
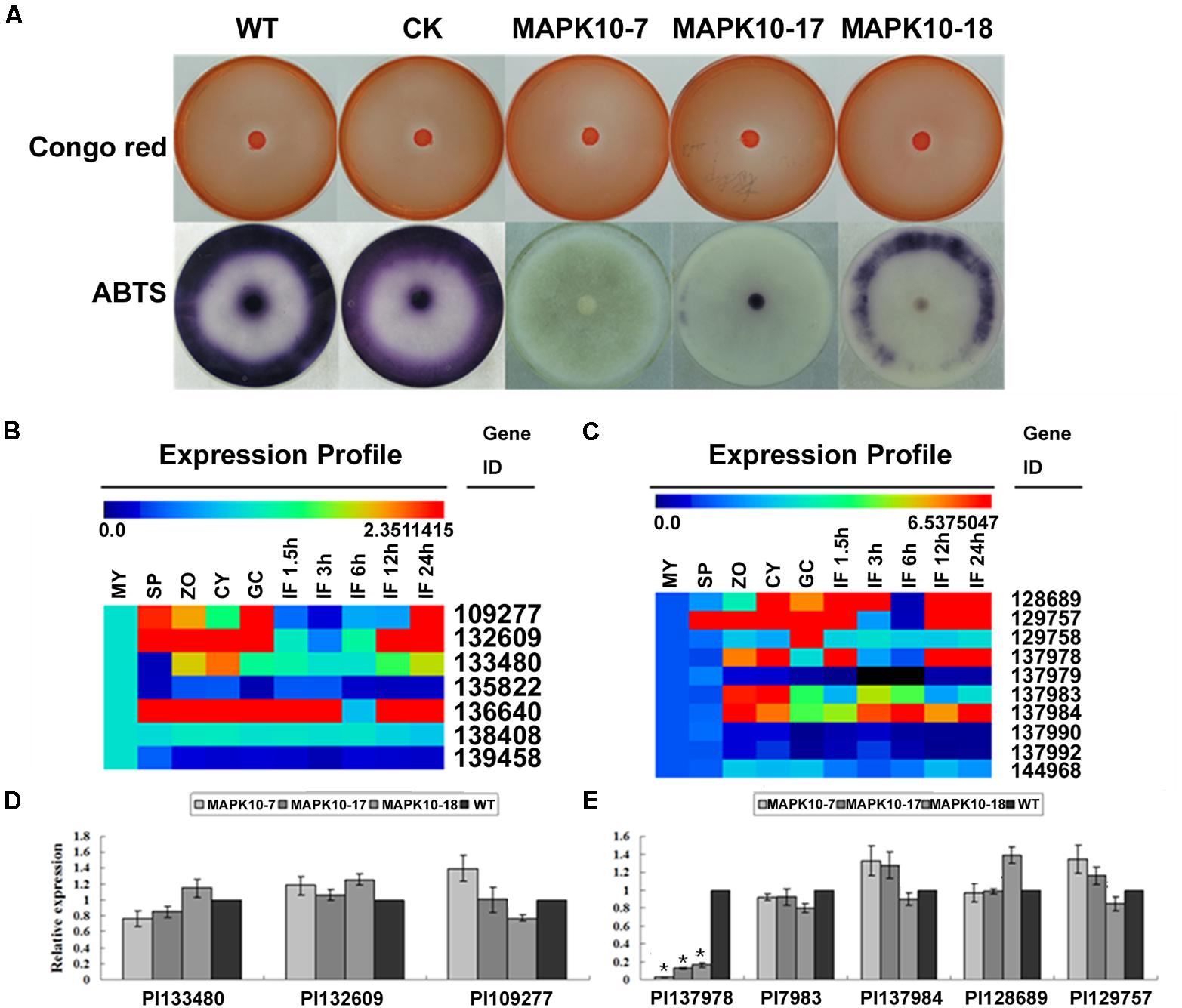
FIGURE 7. Detection of extracellular peroxidase and laccase activity, transcriptional patterns and relative expression of their putative genes in the PlMAPK10-silenced transformants. (A) Upper panel: peroxidase activity assay. Mycelial mats of WT, CK, and three PlMAPK10-silenced transformants were inoculated on solid Plich medium containing Congo Red at a final concentration of 500 μg/ml, as indicator. The strains were incubated for 24 h before Congo Red discoloration was assessed. Lower panel: laccase activity assay. Mycelial mats of the aforementioned strains were inoculated on LBA media containing 0.2 mM ABTS for 10 days, and the oxidized ABTS (dark purple) was measured as an index of laccase activity. (B,C) Transcriptional levels of selected peroxidase-encoding genes and laccase-encoding genes were respectively treated with Multi-experiment Viewer according to the genome of P. litchii and homology genes in P. sojae and their conserved domains. (D,E) Quantitative real-time PCR analysis of P. litchii putative peroxidase-encoding genes and laccase-encoding genes in three PlMAPK10-silenced transformants and wild type strain, respectively. This experiment was repeated three times independent, and for each repeat with three biological replications. Asterisks represent significant difference (p < 0.05) based on statistics analysis using SPSS (version 19.0).
To further find out the reason for the reduced laccase activity, we examined the transcription of putative peroxidase or laccase encoding genes. These genes were selected from a thorough search in P. litchii genome, using their orthologs annotated in the P. sojae genome and their conserved domains. As shown in Figures 7B,C, seven predicted peroxidase encoding genes (Pl_109277, Pl_132609, Pl_133480, Pl_135822, Pl_136640, Pl_138408, Pl_139458) and 10 predicted laccase encoding genes (Pl_128689, Pl_129757, Pl_129758, Pl_137978, Pl_137979, Pl_137983, Pl_137984, Pl_137990, Pl_137992, Pl_144968) existed in P. litchii genome. Among them, three of putative peroxidase genes (Pl_109277, Pl_132609, Pl_133480) and five of laccase genes (Pl_128689, Pl_129757, Pl_137978, Pl_137983, Pl_137984) were indicated to express differentially during infection stages based on Multi-experiment Viewer program analysis7, and were selected for further confirmation by qRT-PCR. Compared with wild type and CK, the transcript levels of three predicted peroxidase genes were not obviously changed in three PlMAPK10-silenced transformants (Figure 7D). In contrast, one laccase encoding gene (Pl_137978) out of the five predicted laccase genes, showed significant decrease in transcript level in three PlMAPK10-silenced transformants (73–97%) compared to that of WT and CK (Figure 7E). Since silencing of PlMAPK10 affects the transcript level of a predicted laccase gene, we infer that PlMAPK10 act as a regulator in P. litchii pathogenicity through direct or indirect regulation on the transcription of laccase gene expression, likely serving a function in detoxification of raised ROS level caused by plant defense response.
Expression of PlMAPK10 Gene Is Not Regulated by Hydrogen Peroxide, While Silencing of PlMAPK10 Has No Effect on Sensitivity to H2O2
Given that PlMAPK10 is required for transcriptional induction of laccase gene Pl_137978, we would like to investigate whether PlMAPK10 is essential for oxidative stress response. A sensitivity assay was performed with the three silenced strains and two control strains. The three PlMAPK10-silenced transformants displayed comparable H2O2 sensitivity with that of the control strains, as the diameters of all the examined strains were similar (Supplementary Figure S1A). As shown in Supplementary Figure S1B, inhibition rate of three PlMAPK10-silenced transformants and wild type were approximate in both 2 mM H2O2 and 5 mM H2O2. Based on this, we further detected the relative expression of PlMAPK10 gene in mycelia treated with 5 mM H2O2 for different time. qRT-PCR assay showed that transcription of PlMAPK10 gene did not change with H2O2 for different time (Supplementary Figure S1C). Thus, we inferred that expression of PlMAPK10 gene is not induced by oxidative stress.
Discussion
Mitogen-activated protein kinase is protein kinase highly conserved in eukaryotic organisms as signal transducers (Román et al., 2007). In this study, we identified a P. litchii ortholog of MAPK10 gene, that is highly conserved in sequenced Phytophthora spp. and in the Hyaloperonospora arabidopsidis genome. Particularly, a typical STKc_MAPKs domains with the dual phosphorylation lip sequence SEY is present in all the oomycete MAPK10s.
The transcription of PlMAPK10 gene was constitutive throughout the P. litchii life cycle, while up-regulated specifically in the zoospores and cysts. Given that signal transduction through MAPK pathways depends on a protein phosphorylation/dephosphorylation cascade, we could not ruled out that PlMAPK10 may also play a role in infection process, transcriptional levels did not changed obviously in the time-course examination. Silencing of PlMAPK10 expression in P. litchii reduced mycelial growth rate and sporangium sporulation, and without difference on zoospore release. PlMAPK10-silenced transformants lost pathogenicity to tender litchi leaves, which is consistant with the well-established function of MAPKs fungal development and pathogenesis (Xu, 2000; Zhao et al., 2005; Román et al., 2007). For example, knocking out PMK1 and CHK1 in Magnaporthe grisea and Cochliobolus heterostrophus, affects on appressoria and conidia formation respectively (Zhao et al., 2005; Eliahu et al., 2007). FMK1 in Fusarium oxysporum, GPMK1 in Fusarium graminearum, BMP1 in Botrytis cinerea, BMK1 in Bipolaris oryzae, and MgFUS3 in Mycosphaerella graminicola were demonstrated to be essential for pathogenicity (Zheng et al., 2000; Jenczmionka et al., 2003; Moriwaki et al., 2006; Kramer et al., 2009; Rispail and Di, 2009). In addition, deletion of Botrytis cinerea SAK1 resulted in interruptived conidia formation, increased sclerotial development, and lost penetration to unwounded plant tissue (Segmüller et al., 2007), and knocking out of Mycosphaerella graminicola Hog1 also affects on filamentous growth and pathogenicity (Mehrabi et al., 2006). Although functionally conserved, the amino acid sequences of oomycete MAPK10s were distance from a variety of fungal MAPKs, providing an addition evidence for the independent evolutionary lineage between phytopathogenic oomycetes and fungi.
During a long term of plant–microbe interaction, plants evolve a series of defense responses to protect themselves from the pathogens. During the process of coevolution with their hosts, pathogenic fungi or oomycetes developed a set of mechanisms to escape or overcome host plant cellular environment, the first parclose (Kim et al., 2009). We showed that the extracellular laccase activity of PlMAPK10-silenced transformants were much lower, and one putative laccase encoding gene was down-regulated due to the silence of PlMAPK10 gene, suggesting a possible reason why the PlMAPK10 is required for full pathogenicity. Silencing of PsMPK7 in P. sojae leads to a similar result of reduced pathogenicity and extracellular laccase activity in the transformants compared with wild type and CK strains (Gao et al., 2015). Given that laccase or peroxidase has been widely identified in fungi as an important virulence factor (Zhu and Williamson, 2004; Molina and Kahmann, 2007; Howlett et al., 2009; Staerck et al., 2017), we infer that reduced pathogenicity of PlMAPK10-silenced transformants may be due to a severely compromise in degradation of callose deposition, lignin accumulation, and/or other stress tolerance capabilities during infection. However, it remains unclear whether PlMAPK10 regulates the secretion pathway of extracellular enzymes.
Overall, our research suggests that PlMAPK10 is required for oomycete pathogenicity, likely via regulation of expression of gene encoding extracellular enzymes, which is consistant with several plant pathogenic fungi as reported. However, the regulatory mechanisms related to development and pathogenicity of P. litchii awaits elucidation. The functional characterization of P. litchii MAPK10 will add to our knowledge of oomycete morphogenesis, pathgenesis, extracellular enzymes synthesis and secretion pathway, and provide a potential target of disease control.
Author Contributions
This study was conceived and designed by ZJ, PX, and LJ. All experiments in this study were performed by LJ, JS, LW, and YC. Data was analyzed by LJ and DX. The guidance for this study was provided by ZJ, PX, and YD. This manuscript was written by LJ, ZJ, PX, and YD. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This research was supported by the earmarked fund for China Agriculture Research System (CARS-33-07) and the Natural Science Foundation of Guangdong Province, China (2016A030313408).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Yuanchao Wang for providing pTOR vector and post-transcriptional interference strategy on P. sojae as a reference and, to Dr. Wenwu Ye for critical reading and useful suggestions for the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00426/full#supplementary-material
Footnotes
- ^ http://www.jgi.doe.gov/
- ^ http://www.broad.mit.edu/
- ^ http://smart.embl-heidelberg.de
- ^ https://www.ncbi.nlm.nih.gov/cdd/
- ^ http://megasoftware.net
- ^ https://www.ncbi.nlm.nih.gov/nuccore/EU257520.1
- ^ http://www.tm4.org
References
Brown, A. J., Budge, S., Kaloriti, D., Tillmann, A., Jacobsen, M. D., Yin, Z., et al. (2014). Stress adaptation in a pathogenic fungus. J. Exp. Biol. 217, 144–155. doi: 10.1242/jeb.088930
Chadee, D. N., and Kyriakis, J. M. (2010). Activation of SAPK/JNKs in vitro. Methods Mol. Biol. 661, 59–73. doi: 10.1007/978-1-60761-795-2_3
Chen, R. E., and Thorner, J. (2007). Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773, 1311–1340. doi: 10.1016/j.bbamcr.2007.05.003
de Leon, I. P., and Montesano, M. (2017). Adaptation mechanisms in the evolution of moss defenses to microbes. Front. Plant Sci. 8:366. doi: 10.3389/fpls.2017.00366
Eliahu, N., Igbaria, A., Rose, M. S., Horwitz, B. A., and Lev, S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 6, 421–429. doi: 10.1128/EC.00264-06
Gao, J., Cao, M., Ye, W., Li, H., Kong, L., Zheng, X., et al. (2015). PsMPK7, a stress-associated mitogen-activated protein kinase (MAPK) in Phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxification, cyst germination, sexual reproduction and infection of soybean. Mol. Plant Pathol. 16, 61–70. doi: 10.1111/mpp.12163
Garg, N., and Chandel, S. (2015). Role of arbuscular mycorrhiza in arresting reactive oxygen species (ROS) and strengthening antioxidant defense in Cajanus cajan (L.) Millsp. nodules under salinity (NaCl) and cadmium (Cd) stress. Plant Growth Regul. 75, 521–534. doi: 10.1007/s10725-014-0016-8
Howlett, B. J., Chi, M. H., Park, S. Y., Kim, S., and Lee, Y. H. (2009). A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5:e1000401. doi: 10.1371/journal.ppat.1000401
Jenczmionka, N. J., Maier, F. J., Lösch, A. P., and Schäfer, W. (2003). Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43, 87–95. doi: 10.1007/s00294-003-0379-2
Jiang, L. Q., Ye, W., Situ, J., Chen, Y., Yang, X., Kong, G., et al. (2017). A Puf RNA-binding protein encoding gene PlM90 regulates the sexual and asexual life stages of the litchi downy blight pathogen Peronophythora litchii. Fungal Genet. Biol. 98, 39–45. doi: 10.1016/j.fgb.2016.12.002
Jiang, Y. M., Wang, Y., Song, L., Liu, H., Lichter, A., Kerdchoechuen, O., et al. (2006). Postharvest characteristics and handling of litchi fruit-an overview. Aust. J. Exp. Agric. 46, 1541–1556. doi: 10.1071/EA05108
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282.
Kim, H., Thak, E. J., Yeon, J. Y., Sohn, M. J., Choo, J. H., Kim, J. Y., et al. (2018). Functional analysis of Mpk1-mediated cell wall integrity signaling pathway in the thermotolerant methylotrophic yeast Hansunula polymorpha. J. Microbiol. 56, 72–82. doi: 10.1007/s12275-018-7508-6
Kim, S., Park, S. Y., Kim, K. S., Rho, H. S., Chi, M. H., Choi, J., et al. (2009). Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5:e1000757. doi: 10.1371/journal.pgen.1000757
Kramer, B., Thines, E., and Foster, A. J. (2009). MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet. Biol. 46, 667–681. doi: 10.1016/j.fgb.2009.06.001
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kyriakis, J. M., Liu, H., and Chadee, D. N. (2004). Activation of SAPKs/JNKs and p38s in vitro. Methods Mol. Biol. 250, 61–68.
Li, A. N., Wang, Y. L., Tao, K., Dong, S. M., Huang, Q., Dai, T. T., et al. (2010). PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant Microbe Interact. 23, 1022–1031. doi: 10.1094/MPMI-23-8-1022
Li, A. N., Zhang, M., Wang, Y., Li, D., Liu, X., Tao, K., et al. (2014). PsMPK1, an SLT2-type mitogen-activated protein kinase, is required for hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity in Phytophthora sojae. Fungal Genet. Biol. 65, 14–24. doi: 10.1016/j.fgb.2014.01.003
Maneesha, A., Tori, M., Anne-Kathryn, L. V., Shilpa, C., and Matthew, T. (2017). Mitogen-activated protein kinase, Fus3 and Kss1, regulate chronological lifespan in yeast. Aging 9, 2587–2609. doi: 10.18632/aging.101350
Mehrabi, R., Zwiers, L. H., de Waard, M. A., and Kema, G. H. (2006). MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19, 1262–1269.
Mohanan, V. C., Chandarana, P. M., Chattoo, B. B., Patkar, R. N., and Manjrekar, J. (2017). Fungal histidine phosphotransferase plays a crucial role in photomorphogenesis and pathogenesis in Magnaporthe oryzae. Front. Chem. 5:31. doi: 10.3389/fchem.2017.00031
Molina, L., and Kahmann, R. (2007). An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19, 2293–2309.
Moriwaki, A., Kubo, E., Arase, S., and Kihara, J. (2006). Disruption of SRM1, a mitogen-activated protein kinase gene, affects sensitivity to osmotic and ultraviolet stressors in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 257, 253–261.
Nancy, V. Z., Miriam, R. G., Rocio, N. O., Laura, O. L., Laura, K., Francisco, T. O., et al. (2015). Ineffective phosphorylation of mitogen-activated protein kinase Hog1p in response to high osmotic stress in the yeast Kluyveromyces lactis. Eukaryot. Cell 14, 922–930. doi: 10.1128/EC.00048-15
Neill, S., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5, 388–395. doi: 10.1016/S1369-5266(02)00282-0
Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Park, E., Nedo, A., Caplan, J. L., and Dinesh-Kumar, S. P. (2018). Plant-microbe interactions: organelles and the cytoskeleton in action. New Phytol. 217, 1012–1028. doi: 10.1111/nph.14959
Rispail, N., and Di, P. A. (2009). Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant Microbe Intreact. 22, 830–839. doi: 10.1094/MPMI-22-7-0830
Román, E., Arana, D. M., Nombela, C., Alonso-Monge, R., and Pla, J. (2007). MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15, 181–190. doi: 10.1016/j.tim.2007.02.001
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sang, Y. Y., and Macho, A. P. (2017). Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol. Biol. 1578, 143–153. doi: 10.1007/978-1-4939-6859-6_11
Segmüller, N., Ellendorf, U., Tudzynski, B., and Tudzynski, A. P. (2007). BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 6, 211–221. doi: 10.1128/EC.00153-06
Sheng, Y. T., Wang, Y., Meijer, H. J., Yang, X., Hua, C., Ye, W., et al. (2015). The heat shock transcription factor PsHSF1 of Phytophthora sojae is required for oxidative stress tolerance and detoxifying the plant oxidative burst. Environ. Microbiol. 17, 1351–1364. doi: 10.1111/1462-2920.12609
Staerck, C., Gastebois, A., Vandeputte, P., Calenda, A., Larcher, G., Gillmann, L., et al. (2017). Microbial antioxidant defense enzymes. Microb. Pathog. 110, 56–65. doi: 10.1016/j.micpath.2017.06.015
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Vallejo, M. C., and Mayinger, P. (2015). Delayed turnover of unphosphorylated Ssk1 during carbon stress activates the yeast Hog1 MAP kinase pathway. PLoS One 10:e0137199. doi: 10.1371/journal.pone.0137199
van West, P., Kamoun, S., van’t Klooster, J., and Govers, F. (1999). Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3, 681–692. doi: 10.1016/S1097-2765(00)80461-X
Wang, H. C., Sun, H. Y., Stammler, G., Ma, J. X., and Zhou, M. G. (2009). Baseline and differential sensitivity of Peronophythora litchii (lychee downy blight) to three carboxylic acid amide fungicides. Plant Pathol. 58, 571–576. doi: 10.1111/j.1365-3059.2008.01990.x
Whisson, S. C., Boevink, P. C., Moleleki, L., Avrova, A. O., Morales, J. G., Gilroy, E. M., et al. (2007). A translocation signal for delivery of oomycete effector protein into host plant cells. Nature 450, 115–118. doi: 10.1038/nature06203
Xu, J. R. (2000). Map kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. doi: 10.1006/fgbi.2000.1237
Xu, L. X., Xue, J., Wu, P., Wang, D., Lin, L., Jiang, Y., et al. (2013). Antifungal activity of hypothemycin against Peronophythora litchii in vitro and in vivo. J. Agric. Food Chem. 61, 10091–10095. doi: 10.1021/jf4030882
Ye, W. W., Dou, D., and Wang, Y. (2010). SeqHunter: a bioinformatics toolbox for local blast and sequence analysis. Chin. J. Bioinform. 8, 364–377.
Ye, W. W., Li, A., Wang, X., and Wang, Y. (2015). Genome-wide identification and transcriptional analysis of the MAPK genes in Phytophthora sojae. Acta Phytopathol. Sin. 46, 338–346. doi: 10.13926/j.cnki.apps.2016.03.007
Ye, W. W., Wang, X., Tao, K., Lu, Y., Dai, T., Dong, S., et al. (2011). Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant Microbe Interact. 24, 1530–1539. doi: 10.1094/MPMI-05-11-0106
Ye, W. W., Wang, Y., Shen, D., Li, D., Pu, T., Jiang, Z., et al. (2016). Sequencing of the litchi downy blight pathogen reveals it is a Phytophthora species with downy mildew-like characteristics. Mol. Plant Microbe Interact. 29, 573–583. doi: 10.1094/MPMI-03-16-0056-R
Zhang, H. J., Fang, Q., Zhang, Z. G., Wang, Y. C., and Zheng, X. B. (2009). The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J. Exp. Bot. 60, 3109–3122. doi: 10.1093/jxb/erp146
Zhao, X., Kim, Y., and Park, G. J. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17, 1317–1329. doi: 10.1105/tpc.104.029116
Zheng, L., Campbell, M., Murphy, J., Lam, S., and Xu, J. R. (2000). The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 13, 724–732. doi: 10.1094/MPMI.2000.13.7.724
Keywords: Peronophythora litchii, mitogen-activated protein kinase, gene silencing, pathogenicity, laccase activity
Citation: Jiang L, Situ J, Deng YZ, Wan L, Xu D, Chen Y, Xi P and Jiang Z (2018) PlMAPK10, a Mitogen-Activated Protein Kinase (MAPK) in Peronophythora litchii, Is Required for Mycelial Growth, Sporulation, Laccase Activity, and Plant Infection. Front. Microbiol. 9:426. doi: 10.3389/fmicb.2018.00426
Received: 17 December 2017; Accepted: 22 February 2018;
Published: 08 March 2018.
Edited by:
Helio K. Takahashi, Federal University of São Paulo, BrazilReviewed by:
Jing Si, Beijing Forestry University, ChinaGuotian Li, University of California, Davis, United States
Copyright © 2018 Jiang, Situ, Deng, Wan, Xu, Chen, Xi and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zide Jiang, emRqaWFuZ0BzY2F1LmVkdS5jbg== Pinggen Xi, eHBnQHNjYXUuZWR1LmNu
 Liqun Jiang1,2
Liqun Jiang1,2 Zide Jiang
Zide Jiang