
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 March 2018
Sec. Microbial Symbioses
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00404
 Xuchen Hu1†
Xuchen Hu1† Yuxia Wang2†
Yuxia Wang2† Li Gao1
Li Gao1 Wenxin Jiang1
Wenxin Jiang1 Wenzhen Lin1
Wenzhen Lin1 Chenguang Niu1
Chenguang Niu1 Keyong Yuan1
Keyong Yuan1 Rui Ma1*
Rui Ma1* Zhengwei Huang1*
Zhengwei Huang1*The luxS gene is present in a wide range of bacteria and is involved in many cellular processes. LuxS mutation can cause autoinducer(AI)-2 deficiency and methyl metabolism disorder. The objective of this study was to demonstrate that, in addition to AI-2-mediated quorum sensing (QS), methyl metabolism plays an important role in LuxS regulation in Streptococcus mutans. The sahH gene from Pseudomonas aeruginosa was amplified and introduced into the S. mutans luxS-null strain to complement the methyl metabolism disruption in a defective QS phenotype. The intracellular activated methyl cycle (AMC) metabolites [S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), homocysteine (HCY), and methionine] were quantified in wild-type S. mutans and its three derivatives to determine the metabolic effects of disrupting the AMC. Biofilm mass and structure, acid tolerance, acid production, exopolysaccharide synthesis of multispecies biofilms and the transcriptional level of related genes were determined. The results indicated that SAH and SAM were relatively higher in S. mutans luxS-null strain and S. mutans luxS null strain with plasmid pIB169 when cultured overnight, and HCY was significantly higher in S. mutans UA159. Consistent with the transcriptional profile, luxS deletion-mediated impairment of biofilm formation and acid tolerance was restored to wild-type levels using transgenic SahH. These results also suggest that methionine methyl metabolism contributes to LuxS regulation in S. mutans to a significant degree.
Streptococcus mutans is naturally present in the human oral microbiota and is considered a primary etiological agent of caries, which is the most prevalent oral disease (Loesche, 1986; Ghasempour et al., 2013). The coordination of communication and group behavior in S. mutans has a significant impact on the cariogenic ability of this bacterial species (Li et al., 2008). Quorum sensing (QS) is a well-known cell-to-cell communication mechanism defined as a process whereby cells collectively regulate gene expression by producing, secreting, and responding to the accumulation of small chemical signal molecules known as autoinducers (AIs) (Bassler, 1999). Among the potential QS signal molecules, AI-2 is produced by a broad range of bacteria and may participate in interspecies communication (Blehert et al., 2003; Merritt et al., 2003). AI-2 is a byproduct of the activated methyl cycle (AMC; Figure 1A), which generates activated methyl groups for the methylation of DNA, RNA, and proteins (Winzer et al., 2002; Sun et al., 2004; Parveen and Cornell, 2011; Redanz et al., 2012). In the AMC, S-adenosylmethionine (SAM), the main methyl donor, is catalyzed by the enzyme MetK to form S-adenosine homocysteine (SAH). Then SAH is converted into homocysteine (HCY) through a two-step process. SAH uses the enzyme Pfs to generate S-ribosyl-homocysteine, then converted into homocysteine (HCY) by the enzyme LuxS (Díaz et al., 2014). For bacteria without LuxS/Pfs pathway, including Pseudomonas aeruginosa, S-adenosyl-L-homocysteine hydrolase (SahH) encoded by sahH is used to complete the AMC (Cataldi et al., 2009; Fernandez-Sanchez et al., 2009). In contrast to the LuxS/Pfs-dependent two-step pathway, SahH directly catalyzes the toxic intermediate SAH into HCY without generating AI-2 (Wang et al., 2012).
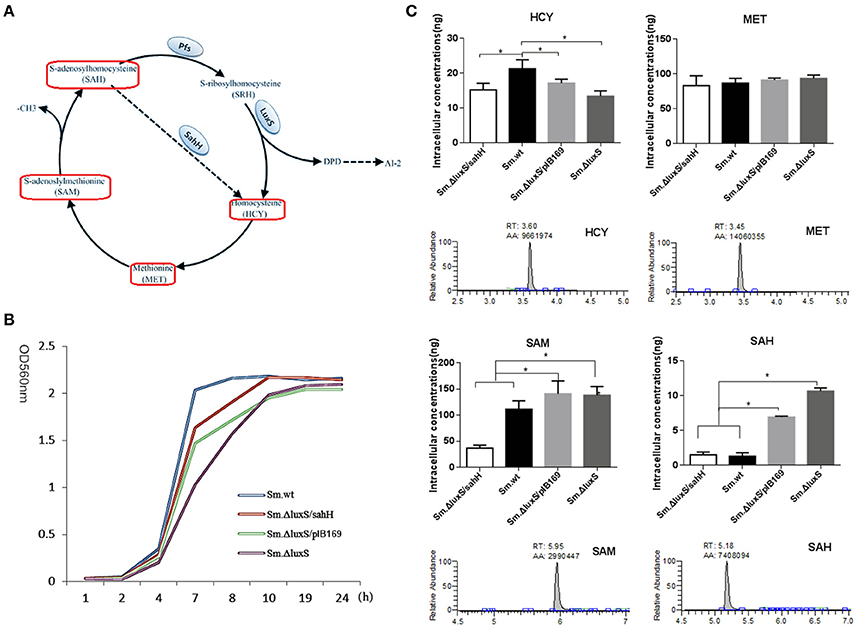
Figure 1. Intracellular concentration of four activated methyl cycle (AMC)-related metabolites in wild-type Sm.wt and derivative strains. (A) The AMC. The detected compounds are circled on the diagram (red box). (B) Bacterial growth curve of the four strains. (C) Intracellular concentrations of the four metabolites and their representative chromatograms. Cell extracts were analyzed with HPLC-MS/MS to determine the intracellular concentrations of methionine, homocysteine, S-adenosyl methionine, and S-adenosyl homocysteine when bacterial strains are cultured overnight and compared to their chromatographic standards. The data shown correspond to the mean calculated intracellular concentrations ± standard deviations of three independent cultures. *Significant differences at P < 0.05.
By knocking out luxS from the genome, many studies have shown that LuxS mutation impairs biofilm formation, acid tolerance, and acid production, and this impairment is the result of AI-2 mediated QS (Wen and Burne, 2004; Yoshida et al., 2005; Huang et al., 2009). However, in the AMC, deletion of luxS may cause metabolic disruption and AI-2 deficiency. Therefore, a debate about whether changes caused by luxS mutation should be attributed solely to QS disruption has arisen. Yoshida et al. (2005) observed that the LuxS defect could be rescued by using supernatants from Streptococcus gordoniae and Streptococcus sorbrinus, which produce AI-2. Similar results were obtained by Rickard et al. (2006) by using synthetic AI-2, suggesting a role of QS in the alterations. However, some studies reported a lack of effect of synthetic AI-2 on the changes induced by LuxS mutation. For example, in a study of S. mutans, 30% of the genes altered by luxS mutation, including genes related to biofilm formation and acid tolerance, were not restored by synthetic AI-2 supplementation (Sztajer et al., 2008). Similar results were observed in Salmonella enterica (De Keersmaecker et al., 2005) and Lactobacillus rhamnosus (Lebeer et al., 2007). Moreover, a genomic analysis by Rezzonico (Rezzonico and Duffy, 2008) revealed that the AI-2 receptor was restricted to Vibrionales, suggesting that luxS has a role other than QS in most bacteria.
Based on the contradictory evidence of the predominant role of QS in luxS mutation-induced changes and the evidence suggesting the absence of AI-2 receptors in most bacterial species (Blehert et al., 2003), we hypothesized that metabolism plays a critical role in LuxS-based changes in S. mutans. Since S. mutans does not have a sahH gene, to confirm this hypothesis, the exogenous sahH gene from P. aeruginosa was used to complete the disrupted AMC in an S. mutans luxS-null strain, and consequently to distinguish the contribution of metabolism from the original functions of LuxS. The hypothesis was verified by determining whether metabolism, phenotypes, and gene expression were restored. To the best of our knowledge, this experimental approach has not been used previously.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and P. aeruginosa were grown in Luria-Bertani (LB) medium, and chloramphenicol (20 μg/mL) was added when necessary. S. mutans UA159 (wild-type; Sm.wt) and its derivatives were routinely grown in Brain Heart Infusion (BHI) medium (Oxoid, Wesel, Germany) with or without erythromycin 10 μg/mL or chloramphenicol 20 μg/mL. For transformation, a S. mutans luxS-null strain (Sm.ΔluxS) was cultured in Todd-Hewitt broth (Becton, Dickinson and Company) with yeast extract (0.2% w/v), heat-inactivated horse serum (5% v/v), and erythromycin (10 μg/mL). The transformation products were grown on Mitis Salivarius (MS) agar (Becton, Dickinson and Company) with erythromycin (10 μg/mL), and chloramphenicol (20 μg/mL). For biofilm assay, semi-defined biofilm medium (BM) containing glucose (0.8% w/v), and sucrose (0.5% w/v) (BMGS) as the alternative carbohydrate source was used (Wen and Burne, 2004). The primers used in this study are listed in Table 2.
sahH were amplified from the genomic DNA of P. aeruginosa PAO1. The E. coli-streptococcal shuttle plasmid pIB169 was kindly provided by Indranil Biwas (University of Kansas Medical Center, Kansas, USA) (Biswas et al., 2008). After DNA digestion and ligation, sahH were cloned into pIB169, generating the plasmids pIB-sahH. After sequencing, pIB-sahH and pIB169 were individually transformed into Sm.ΔluxS using the competence-stimulating peptide (CSP), and generating two S. mutans-derived strains: Sm.ΔluxS/sahH (S. mutans luxS-null strain with pIB-sahH) and Sm.ΔluxS/pIB169 (S. mutans luxS-null strain with pIB169). CSP (amino acid sequence: SGSLSTFFRLFNRSFTQALGK) (Cvitkovitch, 2001) was synthesized by GL Biochem (Shanghai, China). The protocol for CSP transformation was described previously (Biswas et al., 2007). The transformants were selected on MS agar using erythromycin (10 μg/mL) and chloramphenicol (20 μg/mL). The growth curves of these four strains were determined in BHI medium under anaerobic conditions.
Ice-cold isotonic saline was used to wash the S. mutans cultures twice to remove the extracellular components present in the growth medium. After that, 1 mL of an ice-cold extracting solution (methyl alcohol:acetonitrile:water ratio of 4:4:2 v/v/v, containing 0.1% methane acid) and 0.7 g of acid-washed glass beads were mixed with the bacterial cells for the mechanical rupture of the cell walls. Moreover, an ice-cold extracting solution was used to quench and permeabilize the cells, inhibit any residual enzymatic activity by protein precipitation, and maximize the extraction of AMC metabolites. The mixture was centrifuged at 12,000 g for 20 min at 4°C, and the supernatant was collected. The procedure was repeated twice, and all the supernatants were pooled. Subsequently, the supernatants were treated by adding 20 μL of 1 mol/L dithiothretiol and incubated at 65°C for 30 min to ensure the complete reduction of homocysteine (Jiang et al., 2017). The obtained solution was freeze-dried and stored at −80°C. A spectrophotometric method for the determination of tryptophan (Edelhoch, 1967) was used to quantify the residual protein of bacterial sediments, maintain an identical biomass between individual samples, and improve the batch-to-batch consistency of the method.
Stock solutions of SAM, SAH, HCY, methionine (MET), and [13CD3] methionine ([13CD3]MET) ([13CD3]MET is not a hazardous substance or mixture according to Regulation (EC) No. 1272/2008) (Sigma-Aldrich, St. Louis, MO, USA) were prepared at 1 mg/mL in water. A standard solution was prepared by combining the stock solutions and diluting them with water immediately before use. The working standard solutions were 2, 10, 20, 50, 500, and 1,000 ng/mL. [13CD3]MET at a concentration of 200 ng/mL in water was used as the internal standard (IS) (Halliday et al., 2010). All stock solutions were stored at −80°C. Before analysis, the samples were redissolved in 1 mL of water. The samples and standards were centrifuged at 12,000 g for 5 min at 4°C. A 30-μL aliquot of each supernatant was transferred to HPLC vials. A triple quadrupole Quattro Ultima mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) and an ACQUITY UPLC BEH Amide Column (2.1 × 100 mm, 1.7 μm) (Waters, Milford, USA) were used to quantify the intracellular AMC metabolites. The column was maintained at 30°C and the mobile phase flow rate was set at 200 μL/min. The gradient elution mobile phases were methyl alcohol (solution A) and 0.1% methane acid in acetonitrile (solution B). The following ramped gradient was used: 0 min at 95% A; 0.5 min at 95% A; 8 min at 5% A; 10 min at 5% A; 11 min at 95% A; and 13 min at 95% A (Da Silva et al., 2016). The analytes were quantified using tandem electrospray mass spectrometry in the positive mode (+ESI). The pressure of the collision gas (argon) was set at 1.5 mTorr. The software Xcalibur version 2.1 was used for instrument control, data acquisition, and data analysis.
Crystal violet (CV)-staining was used to quantify biofilm formation by S. mutans (7, 35). Sm.wt and its derivatives were grown in BHI broth overnight at 37°C under anaerobic conditions. After initial dilutions to achieve a similar cell density (OD560 = 0.8), the cultures were diluted 1:10 with fresh BMGS medium. Two-hundred microliters of the cell dilutions were transferred to the 96-well flat-bottom microtiter plate (Corning, Corning, NY, USA). Wells containing uninoculated growth medium were used as negative controls. The plates were incubated at 37°C under anaerobic conditions for 16–24 h without agitation. The liquid medium was removed and the wells were gently rinsed three times with sterile distilled water to remove the planktonic or loosely bound cells. The plates were air-dried and stained with 0.1% (w/v) CV (50 μl per well) for 15 min at room temperature. Excess dye was removed and the plates were rinsed three times with distilled water. After air-drying, 200 μl of 99% ethanol was added to each experimental well and the plates were shaken for several minutes to induce dye release. The amount of biofilm was determined by spectrophotometric reading at OD570 using a Varioskan Flash plate reader (Thermo Fisher Scientific). Each assay was performed in triplicate.
Culture dilutions were prepared using the quantitative assay protocol described above. Alexa Fluor 647-labeled dextran conjugate (Ren et al., 2016) (1 M; Life Technologies, Carlsbad, CA, USA) was added to the BHIS medium before inoculation. One milliliter of each diluted culture was transferred to individual wells of a 24-well flat-bottom plate with a glass coverslip on the bottom of each well. The plate was incubated at 37°C overnight under aerobic conditions. The biofilm was stained with a MSYTO 9 green fluorescent nucleic acid stain (Life Technologies). After incubation at room temperature in the dark for 15 min, all samples were scanned using a Leica TCS SP2 confocal laser scanning microscope (CLSM, Leica, Wetzlar, Germany) at 1-μm intervals, and images were recorded from the moment the signal appeared until the signal disappeared. The images were randomly captured from each sample.
Sm.wt and its derivatives were cultured under anaerobic conditions in 24-well flat-bottom culture plates overnight at 37°C. After incubation, the culture medium was carefully removed and replaced with 2 mL of sterile PBS. The cells were resuspended and vortexed. The suspension was centrifuged at 6,000 g for 10 min at 4°C and the supernatant was collected. This procedure was repeated twice and all the supernatants containing the water-soluble extracellular polymeric substances (EPS) were pooled. Water-insoluble EPS was extracted from the samples using 1.0 M NaOH with agitation for 2 h at 37°C, as previously described (Ren et al., 2016). The concentration of alkali-soluble carbohydrate in the supernatant was determined using the anthrone-sulfuric method (Chen et al., 2017). Cell dry weight was measured to standardize the number of bacteria.
Acid tolerance was assessed by exposing the biofilms to an acidic environment. Biofilms were incubated in 96-well plates as described for the quantitative biofilm assay. After incubation at 37°C under aerobic conditions without agitation for 8 h, the supernatants were removed and the formed biofilm was rinsed with sterile double distilled water. Fresh BMGS at pH 5.8, 4.3, and 2.8 in series was added to the wells, and the plates were incubated for 24 h. BMGS medium (pH 7.4) was used as a standard control. The resultant biofilm was quantified using the quantitative assay protocol. The decrease in biofilm formation at lower pH-values (5.8, 4.3, and 2.8) compared with the respective neutral controls was calculated to determine the acid suppression effect.
The disks containing biofilms were rinsed with cysteine peptone water to remove loose bacteria. Each disk was placed in a new 24-well plate with 1.5 mL of buffered peptone water (BPW) supplemented with 0.2% sucrose. BPW medium was used to maintain the stability of the biofilms during the 3-h incubation. BPW is suitable for this purpose because its relatively high buffering capacity prevents the decrease in pH, and low pH hinders bacterial acid production. Disks with biofilms were incubated at 37°C for 3 h to allow the biofilms to produce acid. After 3 h, the BPW solutions were stored for later lactate analysis using an enzymatic (lactate dehydrogenase) method (Powers et al., 2007). A microplate reader (SpectraMax M5, Molecular Devices, Sunnyvale, CA, USA) was utilized to measure the absorbance of the collected BPW solutions at an OD of 340. Standard curves were prepared using a lactic acid standard (Sigma-Aldrich).
Quantitative reverse transcription PCR (qRT-PCR) was used to determine the effects of the compounds on the expression of mRNAs smu.238, gtfB, gtfC, gtfD, gbpA, gbpC, smu.44, smu.46, ciaH, ldH, and aguA. The S. mutans strains were cultured overnight. The isolation, purification, and reverse transcription of total bacterial RNA into cDNA was performed as described previously (Zuo et al., 2017). qRT-PCR was performed using the Bio-Rad CFX96 system (Bio-Rad, Marnes-la-Coquette, France). All primers used are listed in Table 2. The reaction mixture (20 μL) contained 1X SYBR green PCR master mix (Takara-Bio, Otsu, Japan), template cDNA, and forward and reverse primers (10 mM each). The thermal cycling conditions were as follows: initial denaturation at 95°C for 30 s followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. An additional step at 95°C for 15 s and 60°C for 1 min (0.05°C s−1) was used to establish a melting curve.
All experiments were performed at least in triplicate and reproduced three separate times. Statistical analysis was conducted with SPSS software version 21.0 using one-way analysis of variance and Tukey's multiple comparisons test. P < 0.05 was considered statistically significant.
Sm.ΔluxS/pIB169 and Sm.ΔluxS/sahH strains were successfully constructed and the transformation and expression of sahH gene in Sm.ΔluxS/sahH were verified by PCR and RT-PCR (Figure S1). A HPLC-MS/MS method was developed and the standard curves of the four metabolites were shown in Figure S2. By using the HPLC-MS/MS method, changes in the synthesis of metabolites in the four strains were investigated, and significant differences were observed among these strains. Under the culture conditions used (in the stationary phase, Figure 1B), the amount of SAM and MET was one to two orders of magnitude higher than the amount of SAH and HCY. Furthermore, the amount of SAH and SAM was significantly higher in Sm.ΔluxS and Sm.ΔluxS/pIB169 strains than in their counterparts, and HCY was higher in Sm.wt (Figure 1C, P < 0.05). The results suggested that complementation of SahH partially restored the AMC metabolism in S.mutans.
The biofilm formation of S. mutans plays an essential role in the etiology and pathogenesis of dental caries. In our study, quantification of biofilm was conducted to investigate whether SahH complementation could restore the luxS-deletion induced alteration of biofilm formation. The amount of biofilm was quantified by staining with 0.1% CV and absorbance reading at an OD of 570 nm. luxS deletion in Sm.ΔluxS caused a decrease in biofilm mass compared to Sm.wt (Figure 2A). The decrease was completely restored by the supplementation of transgenic SahH, and the amount of biofilm was similar between Sm.ΔluxS/sahH and Sm.wt. In contrast, the amount of biofilm was not restored in Sm.ΔluxS/pIB169 and was decreased compared to Sm.wt. Thus, a contributory effect of the functional AMC on biofilm formation was verified by quantification.
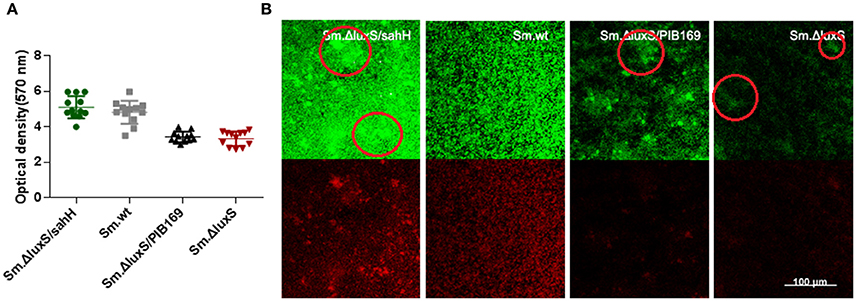
Figure 2. Quantification of biofilms by CV staining and CLSM of biofilms formed by S. mutans and derivative strains. (A) Quantification of biofilms. Two independent experiments were performed (n = 3 for each strain). The results are shown as mean ± SD, and the asterisks represent a significant difference (P < 0.05) compared to the Sm.ΔluxS strain. (B) The CLSM of biofilms. Double-labeling imaging of bacteria (green) and EPS (red) of biofilm formed on the surface of glass coverslips, showing the topographic features of biofilm. Images are representative of compressed biofilms in strains Sm.wt, Sm.ΔluxS, Sm.ΔluxS/sahH, Sm.ΔluxS/pIB169. The distinct bulges which were resulting from high-density cell aggregates in Sm.ΔluxS/sahH, Sm.ΔluxS, and Sm.ΔluxS/pIB169 were circled. Images were randomly captured from each sample.
To further investigate the restoration of biofilm formation by SahH, a structural analysis was made with CLSM. Compared with the dense, uniform biofilm with complete coverage of the slide surface by Sm.wt, the biofilm formed by Sm.ΔluxS was heterogeneous and organized, cell aggregates, and conspicuous empty surfaces were scattered throughout the matrix (the cell aggregates were circled in Figure 2B). Besides, no textural repair was observed in Sm.ΔluxS/pIB169 biofilms. However, for the Sm.ΔluxS/sahH biofilm, although pronounced bulges resulted from the aggregation of high density cells could still be found, a dense structure was restored and a smaller empty surface was present. This suggested that partial restoration of biofilm texture was obtained by Sm.ΔluxS/sahH due to complementation of SahH (Figure 2B).
For phenotypic confirmation of the transcriptional profiling, acid tolerance of the four S. mutans strains was evaluated by exposure to low pH (5.8, 4.3, and 2.8), and to neutral medium (pH 7.4) as a control. Decreased biofilm production at lower pH was a reflection of acid inhibition and thus reversed acid tolerance. The results indicated that a weakly acidic environment (pH 5.8) did not strongly suppress biofilm formation. However, at pH 4.3, biofilm formation was decreased, and this decrease was categorized into two groups: a higher-decrease group, including Sm.ΔluxS and Sm.ΔluxS/pIB169; and a lower-decrease group, including Sm.wt and Sm.ΔluxS/sahH (P < 0.05). This suggested better resistance to acid inhibition by Sm.wt and Sm.ΔluxS/sahH than Sm.ΔluxS and Sm.ΔluxS/pIB169. A similar profile was observed at pH 2.8 (Figure 3A). Hence, the greater sensitivity of S. mutans luxS null strain to acid inhibition that resulted from luxS deletion was compensated by SahH complementation.
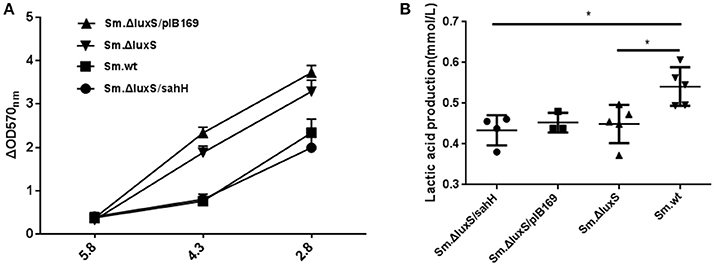
Figure 3. Acid tolerance and lactic acid production of four bacterial strains in the presence of biofilms. (A) Acid tolerance of four bacterial strains. ΔOD570 nm is the difference in optical density at 570 nm between the experimental groups (pH 4.3 or pH 2.8) and reflects the acid-inhibition effect. The results are shown as mean ± SEM. (B) Variability in acid production in biofilms from the four strains. Lactic acid production was higher in the Sm.wt strain than in Sm.ΔluxS and Sm.ΔluxS/sahH strains. *Significant differences at P < 0.05.
S. mutans in biofilms can metabolize carbohydrates to acids (mainly lactic acid), causing demineralization of the tooth structure and EPS are extremely important in the processes of biofilm formation and stabilization. Therefore, the discoveries of lactic acid production and EPS synthesis abilities have great significance. In the study, the ability of the four bacterial strains to produce lactic acid and extracellular polysaccharides were investigated using the lactate dehydrogenase and anthrone-sulfuric method. The results indicated that lactic acid production in Sm.wt was higher than that in Sm.ΔluxS and Sm.ΔluxS/sahH when cultured for 24 h (Figure 3B, P < 0.05). However, there was no significant difference between Sm.ΔluxS/sahH and Sm.ΔluxS. Thus, the lactic acid production ability was not restored by SahH supplementation. Besides, the anthrone-sulfuric results indicated that there was no significant difference in EPS (water-soluble and insoluble) between Sm.ΔluxS, Sm.wt, and Sm.ΔluxS/sahH (Figure S3, P > 0.05). The EPS of biofilm formed by S. mutans and derivatives were showed by CLSM. The results (Figure 2B, stained red) showed that with the changes in the number of bacteria in the biofilm, the amount of bacterial EPS also changes.
The transcriptional changes of 11 genes involved in biofilm formation, acid tolerance, lactic acid production, and EPS synthesis in S. mutans were quantified by qRT-PCR. Four genes involved in biofilm formation (smu.238, gtfD, gbpA, and gbpC), four genes related to acid tolerance (smu.44, smu.46, ciaH, and aguA), two genes involved in extracellular polysaccharide synthesis (gtfB and gtfC), and one gene related to lactic acid production in S. mutans (ldH) (Yamada and Carlsson, 1975; Koo et al., 2006; Sztajer et al., 2008; Wen et al., 2011) were chosen to evaluate gene transcription. Six genes (smu.44, smu.46, ciaH, smu.238, gtfD, and gbpA) were restored by SahH supplementation, and the most significant changes were observed in smu.46, with a maximal downregulation of 69% by luxS mutation and a maximal upregulation of 196% by transgenic SahH (Figure 4). Although luxS mutation did not induce transcriptional changes in the aguA gene (P > 0.05 between Sm.ΔluxS and Sm.wt), SahH supplementation caused a strong upregulation, with an increase of 200% relative to Sm.wt. However, the upregulation of gbpC gene by luxS mutation was not significantly restored by sahH supplementation, and the expression of gtfB, gtfC, and ldH was not compensated in Sm.ΔluxS/sahH.
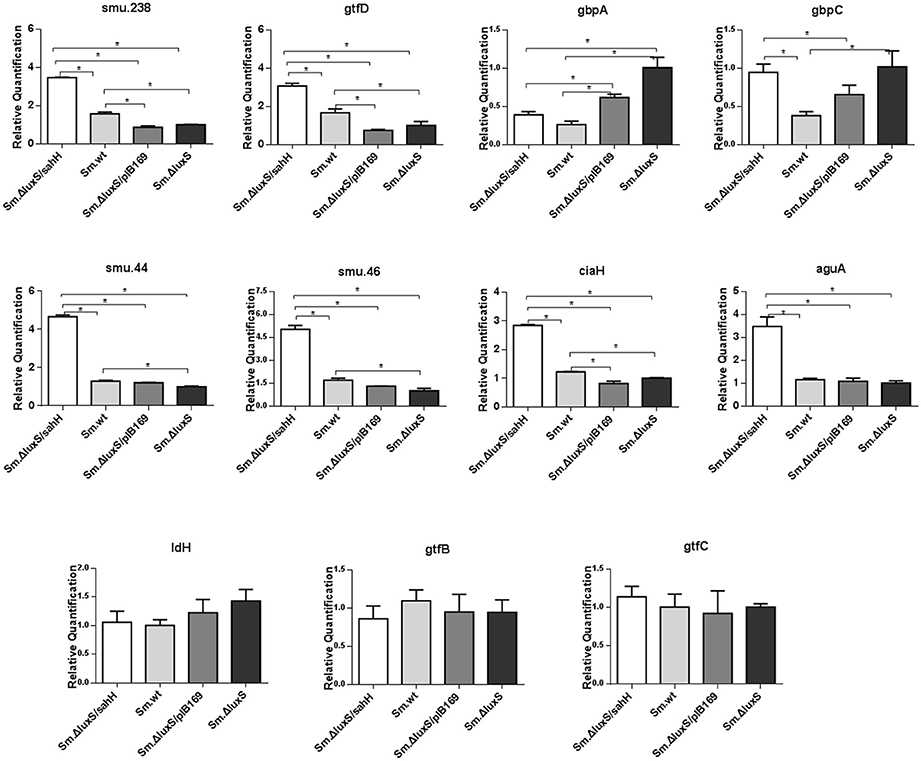
Figure 4. Transcriptional comparison of studied genes with qRT-PCR. The 11 target genes were relevant to biofilm formation (smu.238, gtfD, gbpA, and gbpC), aciduricity (smu.44, smu.46, ciaH, and aguA), acid generation (ldH), and synthesis of extracellular polymeric substances (gtfB, gtfC). RNAs were quantified with qRT-PCR and normalized to 16S rRNA transcripts. Results represent mean ± SD of relative quantification. *Significant differences at P < 0.05.
AI-2-dependent QS is considered a primary contributor to the impairment of biofilm formation and acid tolerance by luxS-deletion in S. mutans (Li et al., 2002; Merritt et al., 2003). However, evidence has suggested that many changes induced by luxS-mutation failed to respond to exogenous AI-2 (Sztajer et al., 2008). For this reason, the predominant role of QS in LuxS regulation was questioned. Moreover, luxS deletion caused AI-2-mediated QS disruption and metabolic disorders. Therefore, the latter could be a contributor to the disruptions caused by luxS mutation. In our study, the AMC defect in the luxS mutant strain with an AI-2 defect phenotype was restored using the transgenic SahH heterologously expressed in Sm.ΔluxS, and the sahH mRNA was exclusively expressed in Sm.ΔluxS/sahH (data not shown). The difference in the amount of AMC metabolites for four important cariogenic phenotypic effects (biofilm formation, acid tolerance, lactic acid production, EPS synthesis ability) and the relative gene transcription levels of the four strains were confirmed.
Changes in metabolite levels by luxS mutation and sahH supplementation were determined to elucidate the AMC in S. mutans. The AMC metabolites were extracted, identified, and quantified with HPLC-MS/MS. The results revealed that the concentration of SAH, SAM, and HCY were significantly different between the S. mutans strains. After culturing these strains overnight, the levels of SAM and SAH were significantly higher in Sm.ΔluxS/pIB169 and Sm.ΔluxS than in Sm.ΔluxS/sahH and Sm.wt. It can be speculated that, because of the SahH pathway which presumably converts SAH to HCY, the levels of SAM and SAH in the Sm.ΔluxS/sahH strain were lower than those in Sm.ΔluxS/pIB169 and Sm.ΔluxS but similar to those in Sm.wt, remained at a relatively lower level. In contrast, SAM and SAH were unable to normal metabolize in ΔluxS/plasmid and Sm.ΔluxS because mutations in luxS genes inhibited the LuxS enzyme. Consequently, the concentrations of SAH and SAM continued to increase in Sm.ΔluxS/pIB169 and Sm.ΔluxS. Therefore, the metabolism of SAH and SAM was complemented by SahH supplementation.
HCY was significantly lower in the Sm.ΔluxS, Sm.ΔluxS/pIB169 compared with Sm.wt. This difference may be because of the lack of recycling of interactive metabolites in these mutant strains and other changes in the SahH pathway in Sm.ΔluxS/sahH strain. HCY can be synthesized de novo from cystathionine using sulfate and oxaloacetate as precursors in a series of biochemical steps, and this synthesis served to replenish the AMC metabolites (Walters et al., 2006). Halliday et al. (2010) considered that the de novo synthesis mechanism explained why HCY was still detectable in LuxS-mutant strains. The significantly lower concentration of HCY in AMC mutants suggested that under the growth conditions used, the loss of the pool concentration of HCY was not being compensated for by increased de novo synthesis. Instead, the cycle was likely restored via the uptake of MET from the growth medium. The concentration of MET in bacterial cells was similar between the four evaluated strains. MET is a crucial amino acid, it has extensive cellular functions in addition to its role in the supply of activated methyl groups (Sekowska et al., 2000). The similar concentration of MET in bacterial cells between the four evaluated strains may reflect the need for the bacteria to maintain a metabolite pool with a relatively high and stable concentration of the metabolite. Thus, it can be speculated that part of the metabolic defect caused by mutations in luxS genes in S. mutans was compensated by the supplementation of SahH.
In this study, the biofilm mass was markedly decreased by luxS deletion, which agrees with the results of other studies (Merritt et al., 2003; Yoshida et al., 2005). It is of note that when the metabolic disruption was reversed by SahH supplementation, the decrease in biofilm mass in Sm.ΔluxS/sahH was reversed to a level similar to that of Sm.wt. Biofilm quantification supported the predominant role of methionine metabolism in biofilm formation in S. mutans. In addition, along with the changes described previously, the expression levels of genes smu.238, gtfD and gbpA were restored in Sm.ΔluxS/sahH. The gbpC gene was upregulated by luxS-deletion. GbpC is believed to promote dextran-dependent bacterial aggregation and biofilm formation (Sato et al., 2002) and was not restored by SahH supplementation. However, other unknown mechanisms may be involved in the transcriptional regulation of gbpC by luxS.
The biofilm structure of the four bacterial strains was evaluated with CLSM. Our results revealed that luxS-mutation impaired biofilm formation, and this result agrees with those of previous studies (Wen and Burne, 2004; Lebeer et al., 2007). In contrast to the complete restoration of biofilm formation by SahH, the parental structural phenotype of homogeneous biofilm was not fully rescued in Sm.ΔluxS/sahH. However, although the distinct bulges resulting from cell aggregates were found in Sm.ΔluxS/sahH biofilm, dense structure was regained and smaller fissures were present. Therefore, the methionine metabolism may partially contribute to the structure phenotype of biofilm.
Acid tolerance, acid production, and extracellular polysaccharides are crucial cariogenic traits of S. mutans (Huang et al., 2002; Wen and Burne, 2004). In this study, the aciduric assay revealed that at pH 4.3 and 2.8, Sm.ΔluxS/sahH and Sm.wt had a higher ability of acid resistance than Sm.ΔluxS and Sm.ΔluxS/pIB169, and the increased ability to restore genes smu.44, smu.46, and ciaH. This result indicates that the acid tolerance defect caused by luxS mutation in Sm.ΔluxS was repaired by transgenic SahH supplementation. The aguA gene encodes an enzyme that produces ammonia, and the enzyme can reduce the intracellular pH in streptococci that are less acid-tolerant (Griswold et al., 2006). Sztajer found that aguA transcription was strongly downregulated (72.97-fold) by luxS mutation (Sztajer et al., 2008). In contrast, the transcriptional level of aguA was non-significantly downregulated by luxS mutation in our study. However, transcription was markedly increased by SahH supplementation, which supports a potential regulatory effect of the functional AMC on gene expression.
Few studies to date have evaluated the variability in acid generation by knocking off the luxS gene in S. mutans. In our study, the ability of biofilms to produce lactic acid in Sm.wt was higher than that of the other three strains when cultured 24 h. Furthermore, no significant changes in the expression of the ldH gene were observed between the four strains. For this reason, acid production may not be responding to SahH supplementation. In this respect, a defect in LuxS may lead to a decline in the production of lactate dehydrogenase. With regard to the production of exopolysaccharides, Ye et al. (2008) found an increase in EPS production in Vibrio alginolyticus. However, the amount of EPS in Sm.wt, Sm.ΔluxS, and Sm.ΔluxS/sahH was similar in this study. We hypothesize that AI-2-mediated QS impairment or methionine methyl metabolism defect does not affect EPS synthesis in S. mutans, and the transcriptional levels of gtfB and gtfC confirmed this hypothesis.
However, apart from AMC metabolism, many intracellular activities are found to be involved in LuxS-related functions, including the AI-2-LsrK axis in Yersinia pestis (Fitts et al., 2016) and the LuxS/AI-2/Rbf regulatory cascade in Staphylococcus aureus (Ma et al., 2017). The potential AI-2 receptors may also have a role in the LuxS system, including LuxP (in V. harveyi) (Chen et al., 2002), LsrB (in Bacillus cereus, Escherichia coli, and S. enterica), and RbsB (in Haemophilus influenza) (Pereira et al., 2013; Ali et al., 2017). Moreover, environmental stress conditions may affect the LuxS/AI-2 system (Park et al., 2017). The relationship between these intracellular and extracellular processes and methyl metabolism is unclear and more studies are needed to elucidate these mechanisms.
Our results demonstrated the occurrence of changes in AMC metabolites, biofilm formation, acid tolerance, lactic acid production, EPS synthesis ability, and transcription levels of related genes in four S. mutans stains (Sm.ΔluxS, Sm.ΔluxS/pIB169, Sm.ΔluxS/sahH, and Sm.wt), and provided evidence of the crucial contribution of methyl metabolism to LuxS-regulated functions. In conclusion, the study verified the hypothesis that apart from quorum-sensing, methionine methyl metabolism also contributes to LuxS regulation in S. mutans to a significant degree. However, the growth and other factors, like metabolic differences, were not normalized in the cultivation of biofilm in the study, which may have a certain impact on the CV staining results. Besides, exogenous transformation of sahH gene has not completely restored the luxs-mutant strain's function defects, like the concentration of HCY and lactic acid production ability. Thus, further studies are needed to elucidate the mechanisms underlying the change caused by luxS mutations.
ZH and RM: conceived and designed the work; XH and YW: performed all the experiments, analyzed the results; CN, LG, XH and WJ: contributed reagents, materials, analysis tools; LG, WL, XH, KY and WJ: wrote the paper, ZH and RM: revised the manuscript.
This study was funded by the National Natural Science Foundation of China (NSFC) (Program No. 81570964/81371143/81070826) and Shanghai Committee of Science and Technology (Grant No. 17140903500).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Prof. Indranil Biswas for kindly providing plasmid pIB169, to Prof. Hu Zhou and his team for their support in the quantification of AMC metabolites by HPLC-MS/MS.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00404/full#supplementary-material
AMC, Activated methyl cycle; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; HCY, Homocysteine; MET, Methionine; [13CD3]MET, [13CD3] methionine; Sm.wt; S. mutans UA159; Sm.ΔluxS, S. mutans luxS-null strain; Sm.ΔluxS/sahH, S. mutans luxS-null strain with pIB-sahH; Sm.ΔluxS/pIB169, S. mutans luxS-null strain with pIB169.
Ali, L., Goraya, M. U., Arafat, Y., Ajmal, M., Chen, J. L., and Yu, D. (2017). Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 18:E960. doi: 10.3390/ijms18050960
Bassler, B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. doi: 10.1016/S1369-5274(99)00025-9
Biswas, I., Drake, L., Johnson, S., and Thielen, D. (2007). Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. BioTechniques 42, 487–490. doi: 10.2144/000112414
Biswas, I., Jha, J. K., and Fromm, N. (2008). Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154(Pt 8), 2275–2282. doi: 10.1099/mic.0.2008/019265-0
Blehert, D. S., Palmer, R. J. Jr., Xavier, J. B., Almeida, J. S., and Kolenbrander, P. E. (2003). Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185, 4851–4860. doi: 10.1128/JB.185.16.4851-4860.2003
Cataldi, T. R., Bianco, G., Abate, S., and Mattia, D. (2009). Analysis of S-adenosylmethionine and related sulfur metabolites in bacterial isolates of Pseudomonas aeruginosa (BAA-47) by liquid chromatography/electrospray ionization coupled to a hybrid linear quadrupole ion trap and Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 23, 3465–3477. doi: 10.1002/rcm.4274
Chen, J., Li, T., Zhou, X., Cheng, L., Huo, Y., Zou, J., et al. (2017). Characterization of the clustered regularly interspaced short palindromic repeats sites in Streptococcus mutans isolated from early childhood caries patients. Arch. Oral Biol. 83, 174–180. doi: 10.1016/j.archoralbio.2017.07.023
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Cvitkovitch, D. G. (2001). Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med 12, 217–243. doi: 10.1177/10454411010120030201
Da Silva, L., Collino, S., Cominetti, O., Martin, F. P., Montoliu, I., Moreno, S. O., et al. (2016). High-throughput method for the quantitation of metabolites and co-factors from homocysteine-methionine cycle for nutritional status assessment. Bioanalysis 8, 1937–1949. doi: 10.4155/bio-2016-0112
De Keersmaecker, S. C., Varszegi, C., van Boxel, N., Habel, L. W., Metzger, K., Daniels, R., et al. (2005). Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J. Biol. Chem. 280, 19563–19568. doi: 10.1074/jbc.M412660200
Díaz, A., Martínez, E., Puerta, L., Méndez, D., Rodríguez, E., Fang, L., et al. (2014). A CoMSIA study to design antagonist ligands for the LuxS protein. New J. Chem. 38:1235. doi: 10.1039/c3nj01162c
Edelhoch, H. (1967). Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954. doi: 10.1021/bi00859a010
Fernandez-Sanchez, M. E., Gonatopoulos-Pournatzis, T., Preston, G., Lawlor, M. A., and Cowling, V. H. (2009). S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol. Cell. Biol. 29, 6182–6191. doi: 10.1128/MCB.00973-09
Fitts, E. C., Andersson, J. A., Kirtley, M. L., Sha, J., Erova, T. E., Chauhan, S., et al. (2016). New insights into autoinducer-2 signaling as a virulence regulator in a mouse model of pneumonic plague. mSphere 1:e00342-16. doi: 10.1128/mSphere.00342-16
Ghasempour, M., Rajabnia, R., Irannejad, A., Hamzeh, M., Ferdosi, E., and Bagheri, M. (2013). Frequency, biofilm formation and acid susceptibility of Streptococcus mutans and Streptococcus sobrinus in saliva of preschool children with different levels of caries activity. Dent. Res. J. 10, 440–445.
Griswold, A. R., Jameson-Lee, M., and Burne, R. A. (2006). Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188, 834–841. doi: 10.1128/JB.188.3.834-841.2006
Halliday, N. M., Hardie, K. R., Williams, P., Winzer, K., and Barrett, D. A. (2010). Quantitative liquid chromatography-tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 403, 20–29. doi: 10.1016/j.ab.2010.04.021
Huang, Z., Meric, G., Liu, Z., Ma, R., Tang, Z., and Lejeune, P. (2009). luxS-based quorum-sensing signaling affects Biofilm formation in Streptococcus mutans. J. Mol. Microbiol. Biotechnol. 17, 12–19. doi: 10.1159/000159193
Huang, Z. W., Xiao, Y., Liu, T. J., Zhou, X. D., Li, J. Y., and Zhan, L. (2002). [The in vitro study on Polistes mandarinus' effects on cariogenic bacteria]. Shanghai Kou Qiang Yi Xue 11, 50–52. doi: 10.3969/j.issn.1006-7248.2002.01.018
Jiang, Y., Mistretta, B., Elsea, S., and Sun, Q. (2017). Simultaneous determination of plasma total homocysteine and methionine by liquid chromatography-tandem mass spectrometry. Clin. Chim. Acta 464, 93–97. doi: 10.1016/j.cca.2016.11.017
Koo, H., Sheng, J., Nguyen, P. T., and Marquis, R. E. (2006). Co-operative inhibition by fluoride and zinc of glucosyl transferase production and polysaccharide synthesis by mutans streptococci in suspension cultures and biofilms. FEMS Microbiol. Lett. 254, 134–140. doi: 10.1111/j.1574-6968.2005.00018.x
Lebeer, S., De Keersmaecker, S. C., Verhoeven, T. L., Fadda, A. A., Marchal, K., and Vanderleyden, J. (2007). Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189, 860–871. doi: 10.1128/JB.01394-06
Li, Y. H., Tang, N., Aspiras, M. B., Lau, P. C., Lee, J. H., Ellen, R. P., et al. (2002). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184, 2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002
Li, Y. H., Tian, X. L., Layton, G., Norgaard, C., and Sisson, G. (2008). Additive attenuation of virulence and cariogenic potential of Streptococcus mutans by simultaneous inactivation of the ComCDE quorum-sensing system and HK/RR11 two-component regulatory system. Microbiology 154(Pt 11), 3256–3265. doi: 10.1099/mic.0.2008/019455-0
Loesche, W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50, 353–380.
Ma, R., Qiu, S., Jiang, Q., Sun, H., Xue, T., Cai, G., et al. (2017). AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 307, 257–267. doi: 10.1016/j.ijmm.2017.03.003
Merritt, J., Qi, F., Goodman, S. D., Anderson, M. H., and Shi, W. (2003). Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71, 1972–1979. doi: 10.1128/IAI.71.4.1972-1979.2003
Park, H., Lee, K., Yeo, S., Shin, H., and Holzapfel, W. H. (2017). Autoinducer-2 quorum sensing influences viability of Escherichia coli O157:H7 under osmotic and in vitro gastrointestinal stress conditions. Front. Microbiol. 8:1077. doi: 10.3389/fmicb.2017.01077
Parveen, N., and Cornell, K. A. (2011). Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 79, 7–20. doi: 10.1111/j.1365-2958.2010.07455.x
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Powers, J. L., Kiesman, N. E., Tran, C. M., Brown, J. H., and Bevilacqua, V. L. (2007). Lactate dehydrogenase kinetics and inhibition using a microplate reader. Biochem. Mol. Biol. Educ. 35, 287–292. doi: 10.1002/bmb.74
Redanz, S., Standar, K., Podbielski, A., and Kreikemeyer, B. (2012). Heterologous expression of sahH reveals that biofilm formation is autoinducer-2-independent in Streptococcus sanguinis but is associated with an intact activated methionine cycle. J. Biol. Chem. 287, 36111–36122. doi: 10.1074/jbc.M112.379230
Ren, Z., Cui, T., Zeng, J., Chen, L., Zhang, W., Xu, X., et al. (2016). Molecule targeting glucosyltransferase inhibits Streptococcus mutans biofilm formation and virulence. Antimicrob. Agents Chemother. 60, 126–135. doi: 10.1128/AAC.00919-15
Rezzonico, F., and Duffy, B. (2008). Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. doi: 10.1186/1471-2180-8-154
Rickard, A. H., Palmer, R. J. Jr., Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G., et al. (2006). Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60, 1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x
Sato, Y., Senpuku, H., Okamoto, K., Hanada, N., and Kizaki, H. (2002). Streptococcus mutans binding to solid phase dextran mediated by the glucan-binding protein C. Oral Microbiol. Immunol. 17, 252–256. doi: 10.1034/j.1399-302X.2002.170408.x
Sekowska, A., Kung, H. F., and Danchin, A. (2000). Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2, 145–177.
Sun, J., Daniel, R., Wagner-Dobler, I., and Zeng, A. P. (2004). Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. doi: 10.1186/1471-2148-4-36
Sztajer, H., Lemme, A., Vilchez, R., Schulz, S., Geffers, R., Yip, C. Y., et al. (2008). Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190, 401–415. doi: 10.1128/JB.01086-07
Walters, M., Sircili, M. P., and Sperandio, V. (2006). AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188, 5668–5681. doi: 10.1128/JB.00648-06
Wang, Q., He, Z., Hu, Y., Jiang, Y., Ma, R., Tang, Z., et al. (2012). luxS mutant regulation: quorum sensing impairment or methylation disorder? Sensors 12, 6176–6185. doi: 10.3390/s120506176
Wen, Z. T., and Burne, R. A. (2004). LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186, 2682–2691. doi: 10.1128/JB.186.9.2682-2691.2004
Wen, Z. T., Nguyen, A. H., Bitoun, J. P., Abranches, J., Baker, H. V., and Burne, R. A. (2011). Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol. Oral Microbiol. 26, 2–18. doi: 10.1111/j.2041-1014.2010.00581.x
Winzer, K., Hardie, K. R., Burgess, N., Doherty, N., Kirke, D., Holden, M. T., et al. (2002). LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148(Pt 4), 909–922. doi: 10.1099/00221287-148-4-909
Yamada, T., and Carlsson, J. (1975). Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J. Bacteriol. 124, 55–61.
Ye, J., Ma, Y., Liu, Q., Zhao, D. L., Wang, Q. Y., and Zhang, Y. X. (2008). Regulation of Vibrio alginolyticus virulence by the LuxS quorum-sensing system. J. Fish. Dis. 31, 161–169. doi: 10.1111/j.1365-2761.2007.00882.x
Yoshida, A., Ansai, T., Takehara, T., and Kuramitsu, H. K. (2005). LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71, 2372–2380. doi: 10.1128/AEM.71.5.2372-2380.2005
Zuo, C., Zhang, W., Chen, Z., Chen, B., and Huang, Y. (2017). RNA sequencing reveals that endoplasmic reticulum stress and disruption of membrane integrity underlie dimethyl trisulfide toxicity against Fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 8:1365. doi: 10.3389/fmicb.2017.01365
Keywords: Streptococcus mutans, methionine metabolism, LuxS, SahH, high-performance liquid chromatography–tandem mass spectrometry
Citation: Hu X, Wang Y, Gao L, Jiang W, Lin W, Niu C, Yuan K, Ma R and Huang Z (2018) The Impairment of Methyl Metabolism From luxS Mutation of Streptococcus mutans. Front. Microbiol. 9:404. doi: 10.3389/fmicb.2018.00404
Received: 04 December 2017; Accepted: 21 February 2018;
Published: 12 March 2018.
Edited by:
Russell T. Hill, University of Maryland Center for Environmental Science, United StatesReviewed by:
Jing Yan, Princeton University, United StatesCopyright © 2018 Hu, Wang, Gao, Jiang, Lin, Niu, Yuan, Ma and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Ma, bWFydWkxNzIzQHNpbmEuY24=
Zhengwei Huang, aHVhbmd6aGVuZ3dlaUBzaHNtdS5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.