
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 March 2018
Sec. Infectious Agents and Disease
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00342
This article is part of the Research Topic Bacterial Colonization: Bystander Microbiota or the Initial Step of Chronic Disease? View all 15 articles
Two-component systems (TCS) are modular signal transduction pathways that allow cells to adapt to prevailing environmental conditions by modifying cellular physiology. Staphylococcus aureus has 16 TCSs to adapt to the diverse microenvironments encountered during its life cycle, including host tissues and implanted medical devices. S. aureus is particularly prone to cause infections associated to medical devices, whose surfaces coated by serum proteins constitute a particular environment. Identification of the TCSs involved in the adaptation of S. aureus to colonize and survive on the surface of implanted devices remains largely unexplored. Here, using an in vivo catheter infection model and a collection of mutants in each non-essential TCS of S. aureus, we investigated the requirement of each TCS for colonizing the implanted catheter. Among the 15 mutants in non-essential TCSs, the arl mutant exhibited the strongest deficiency in the capacity to colonize implanted catheters. Moreover, the arl mutant was the only one presenting a major deficit in PNAG production, the main exopolysaccharide of the S. aureus biofilm matrix whose synthesis is mediated by the icaADBC locus. Regulation of PNAG synthesis by ArlRS occurred through repression of IcaR, a transcriptional repressor of icaADBC operon expression. Deficiency in catheter colonization was restored when the arl mutant was complemented with the icaADBC operon. MgrA, a global transcriptional regulator downstream ArlRS that accounts for a large part of the arlRS regulon, was unable to restore PNAG expression and catheter colonization deficiency of the arlRS mutant. These findings indicate that ArlRS is the key TCS to biofilm formation on the surface of implanted catheters and that activation of PNAG exopolysaccharide production is, among the many traits controlled by the ArlRS system, a major contributor to catheter colonization.
The extensive use of indwelling devices such as prosthetic joints, heart valves, and intravascular catheters in hospitalized patients has increased the incidence of device related infections (DRI) (Hogan et al., 2015). DRI constitute a growing healthcare concern because they are associated with a high morbidity and mortality and also with increased costs in health care systems. Staphylococcus aureus is a frequent etiological agent of DRI, especially those related with intravascular catheters, prosthetic joints, vascular grafts and pacemakers (Kluytmans et al., 1997; Lowy, 1998; Ellis et al., 2014).
The success of S. aureus as a pathogen depends on the combined action of multiple factors. On one hand, S. aureus produces a wide array of cell surface and secreted virulence factors such as exoenzymes (coagulase, lipases, and proteases), toxins (cytolytic toxins, enterotoxins, exfoliative toxins) and immune evasion mechanisms (protein A) (Foster et al., 2014). Cell surface virulence factors allow the organism to colonize the host through adhesion to mucosal surfaces and to interfere with normal immune system functions. Secreted factors, including exoenzymes and exotoxins, allow the organism to spread into surrounding tissues and access nutrients through cell damage. On the other hand, S. aureus exhibits a great capacity to adapt to many different environments including almost every organ of the human body (lung, heart, blood, bone, skin, muscles, eye, joints, and intestinal tract). For that, S. aureus possesses efficient signal transduction systems that facilitate the integration of environmental stimuli and adjust the cellular physiology in response (Novick, 2003; Cheung et al., 2004; Haag and Bagnoli, 2016). The sensory machinery of S. aureus is composed of the two-component systems (TCS) network, a Ser/Thr protein kinase, two Ser/Thr phosphatases and a c-di-AMP cyclase (Dac) and a phosphodiesterase (GpdP) involved in c-di-AMP metabolism. As regards the TCS network, most S. aureus strains contain 16 TCSs, with one of them being essential for bacterial viability (Kuroda et al., 2001). In a canonical TCS, extracellular stimuli induce the autophosphorylation of a histidine kinase (HK). The activated HK transfers the phosphoryl group to a conserved aspartate residue present on the response regulator (RR). The phosphorylation of the RR activates an output domain, which can then effect changes in cellular physiology often by regulating gene expression (Stock et al., 2000).
When S. aureus reaches the surface of an implanted medical device, attachment to the surface occurs through interactions between bacterial adhesins and plasma proteins deposited on the implant’s surface. Once attached, S. aureus needs to adjust bacterial physiology to the sessile lifestyle. For that, S. aureus produces an extracellular matrix mainly composed of exopolysaccharides, proteins and extracellular DNA (O’Gara, 2007; Rice et al., 2007). The exopolysaccharide PNAG, also named PIA (polysaccharide intercellular adhesin) is a major component of the staphylococcal biofilm matrix. PNAG, β-1,6-linked N-acetylglucosamine, is synthetized by the enzymes encoded in the icaADBC operon whose expression is tightly regulated by the transcriptional repressor IcaR. Some strains of S. aureus make use of extracellular proteins to build the biofilm matrix by interacting with eDNA and polysaccharides or in some cases, through polysaccharide-independent mechanisms (Cucarella et al., 2001; Corrigan et al., 2007; O’Neill et al., 2008; Vergara-Irigaray et al., 2009; Taglialegna et al., 2016). Although the molecular determinants underlying the choice of either a polysaccharide or protein-based biofilm matrix are not well understood, it is assumed that environmental signals determine the composition of the biofilm matrix (Vergara-Irigaray et al., 2009). Attempts to identify the specific TCSs involved in the biofilm formation process of S. aureus in vitro have been previously performed (Toledo-Arana et al., 2005). However, identification of TCSs important for colonization of implanted medical devices in vivo remains to be elucidated.
In this study, we used a collection of single mutants in each non-essential TCS of S. aureus and a murine subcutaneous catheter infection model to identify the TCSs controlling the production and composition of the biofilm matrix on the surface of the implanted device. Our results revealed that arlRS, and in a lower extent agr and srrAB mutants, have a significant decreased capacity in colonize the surface of implanted catheters. In the arl mutant background, expression of the icaADBC operon restores the capacity to colonize the catheter, indicating that the PNAG exopolysaccharide is required for efficient colonization of implanted catheters. This result is central to the development of novel therapeutic approaches to specifically target biofilm related infections.
Bacterial strains, plasmids and oligonucleotides (Stabvida) used in this study are listed in Supplementary Tables S1, S2. Escherichia coli strains were grown in LB broth (Conda-Pronadisa). S. aureus strains were grown in trypticase soy broth (TSB) (Conda-Pronadisa), TSB supplemented with glucose (TSBg) in strain ISP479r or NaCl 3% (TSB-NaCl) in the case of strain 132. When required for selection, medium was supplemented with appropriate antibiotics at the following concentrations: erythromycin (Em), 1.5 and 10 μg ml-1; ampicillin (Amp), 100 μg ml-1.
General DNA manipulations were performed using standard procedures. Plasmids were purified using the NucleoSpin Plasmid miniprep kit (Macherey-Nagel) according to the manufacturer’s protocol. FastDigest restriction enzymes and Rapid DNA ligation kit (Thermo Scientific) were used according to the manufacturer’s instructions. Plasmids were transformed into E. coli XL1-Blue strain (Stratagene) and S. aureus by electroporation, using previously described protocols (Cucarella et al., 2001). Staphylococcal electrocompetent cells were generated as previously described (Schenk and Laddaga, 1992).
We used a collection of single mutants in each TCS constructed in S. aureus MW2 (listed in Supplementary Table S1) by Villanueva et al. (2018) as follows. Fragments of at least 500 bp that flanked the left (primers A and B) and right sequences (primers C and D) of the region targeted for deletion were amplified by PCR. The PCR products AB and CD were used to obtain an overlapping PCR product named AD, which was cloned into the shuttle vector pMAD (Arnaud et al., 2004) or pMAD_lic. A LIC-modified pMAD vector was constructed in order to enable efficient directional cloning without restriction enzyme digestion or ligation reactions (Alanidis and Dejong, 1990). To create the pMAD_lic vector we oligomerized the primers pMAD_lic (EcoRI) and pMAD_lic (BamHI) listed in Supplementary Table S2, and cloned the double DNA strand dimer into pMAD using EcoRI and BamHI restriction enzymes. To produce specific non-complementary overhangs in the pMAD_lic vector, the ApaI linearized plasmid was treated with T4 DNA Polymerase (Novagen) in the presence of dTTP (Novagen) for 30 min at 22°C. PCR products with complementary overhangs were created using Phusion enzyme (Thermo Scientific) by building appropriate 5′ extensions into the primers. The PCR products were purified and then treated with T4 DNA Polymerase in the presence of dATP (Novagen) at 22°C. After 30 min the enzyme was inactivated. To anneal the insert into the pMAD_lic vector, the mix of vector and insert was incubated for 5 min at 22°C and then, EDTA (6.25 mM) was added and a further incubation of 5 min at 22°C was applied. To express the ica operon and mgrA gene under de cadmium promoter, we amplified the cadmium inducible promoter from pCN51 plasmid (Pcd) (Charpentier et al., 2004) and two fragments of at least 500 bp that flanked the left (primers A and B) and right sequences (primers C and D) of the start of the mRNA of both genes. Three fragments were fused by overlapped PCR. The purified fragment was cloned into the pMAD_lic plasmid. Allelic exchange in the absence of a selection marker was performed as previously described (Valle et al., 2003). White colonies, which no longer contained the pMAD plasmid, were tested to confirm the replacement by PCR using primers E and F (Supplementary Table S2). Note that cadmium was not used for the expression of Pcd since the leakage of expression of the Pcd promoter was sufficient to express PNAG and MgrA.
Overnight cultures of the strains tested were diluted 1:40 in the appropriate medium, and 2 ml of this cell suspension was used to inoculate sterile 24-well polystyrene microtiter plates (Sarstedt). After 16 h cell-surface PNAG production was detected as described previously (Valle et al., 2003). 5 μl of the extract or 5 μl of a dilution of the purified extract were spotted on a nitrocellulose filter using a Bio-Dot microfiltration apparatus (Bio-Rad), blocked overnight with 5% skim milk in phosphate-buffered saline with 0.1% Tween 20, and incubated for 2 h with an anti-PNAG antibody diluted 1:10,000 (Maira-Litrán et al., 2002). Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G anti-bodies (Invitrogen) 1:5,000.
To obtain an ica and mgrA transcriptional fusions, we amplified the ica promoter using primers AU59 and AU76 and the mgrA promoter using primers PmgrA-fw and Pmgra-rv (Supplementary Table S2) and cloned in pCN52 plasmid (Charpentier et al., 2004), giving pCN52-Pica(BS)-gfp and pCN52-PmgrA-gfp plasmids, respectively. Plasmids were transformed in S. aureus wild type strains and arl mutants. To analyze ica and mgrA expression, total protein extracts were recovered and analyzed by SDS-PAGE and Western blot Gfp was detected using anti-GFP [Living Color® A.v. Monoclonal Antibody (JL-8), Clontech] antibodies diluted 1:2,500 in 0.1% PBS-Tween 5% skim-milk. Peroxidase-conjugated goat anti-mouse Immunoglobulin G (Invitrogen) diluted 1:5000 in 0.1% PBS-Tween 5% skim-milk were used as secondary antibodies.
Staphylococcus aureus 132 and arl strains were grown in 10 ml of TSB-NaCl at 37°C under static conditions for 5 h. RNA extraction was performed as described previously (Lasa et al., 2011). Northern blots were performed as described (Ruiz de Los Mozos et al., 2013). Briefly, 8 μg of total RNA was separated in precast agarose gels (Sigma). RNAs were blotted onto Nytran membranes (0.2-μm pore size) (Amersham Biosciences), UV cross-linked, prehybridized in ULTRAhyb solution (Ambion) at 65°C, and labeled with strand-specific riboprobes specific for icaA, icaC, and icaR (Supplementary Table S2). Membranes were washed and autoradiography images were registered at different exposition times according to each gene.
Transfer of the 3xFlag sequence into IcaC was achieved by recombination using plasmid pMADicaC-3xFlag. To construct pMADicaC-3xFlag the C-terminal region of icaC was amplified using primers CFlag1 and CFlag2 (Supplementary Table S2). The CFlag2 primer contains 66 nt that code for the 3xFlag sequence. Strains containing IcaC-3xFlag were grown in TSB-NaCl or TSBg at 37°C under static conditions for 5 h. Then, cells were centrifuged and the pellets were resuspended in PBS buffer containing lysostaphin (12.5 μg/ml; Sigma) and DNase I for 2 h. Samples were subjected to electrophoresis on SDS-PAGE 12% CriterionTM TGX Stain-FreeTM Precast Gels (Bio-Rad). Proteins were transferred onto nitrocellulose membranes (Hybond Amersham Biosciences) and 3xFLAG fusion proteins were immunodetected by the use of anti-FLAG M2 mAbs conjugated with peroxidase (Sigma).
For detection of the levels of Ebh protein overnight cultures of the different strains were diluted 1:100 in 50 ml of TSB-glu medium and grown at 37°C to a mid-log exponential phase (OD = 0.8). Surface-associated proteins were extracted as previously described (Taglialegna et al., 2016). Proteins were subjected to electrophoresis on SDS-PAGE 7.5% TGX Stain-FreeTM FastCastTM Acrylamide Kit (Bio-Rad). For immunoblotting, proteins were transferred onto Hybond-ECL nitrocellulose membranes (Amersham Biosciences). Membranes were blocked for 2 h at room temperature with 5% skimmed milk in PBS with 0.1% Tween 20 and incubated overnight at 4°C with specific antibodies for the carboxy-terminal region of Ebh diluted 1:1,000. Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G anti-bodies (Invitrogen) 1:5,000.
To examine the role of the staphylococcal TCSs in implant colonization, an in vivo murine model of catheter-associated biofilm formation described by Cucarella et al. (2001) was used with some modifications. S. aureus strains grown overnight at 37°C on TSA plates were resuspended in PBS to an optical density of OD650 nm = 0.2 (108 CFU/mL). Groups of 5, 5 weeks old ICR female mice (Charles River Laboratories) were anesthetized with isoflurane (B. Braun). Two 19 mm intravenous catheters (24G; B. Braun) were aseptically implanted into the subcutaneous interscapular of each mouse and inoculated with 100 μl (107 CFU) of S. aureus strains. After 5 days, animals were opportunely euthanatized by isoflurane inhalation followed by cervical dislocation. Catheters were aseptically removed, placed in a sterile microcentrifuge tube containing 1 ml of PBS, and vortexed at high speed for 3 min. The number of bacteria was determined by plate count.
All animal studies were reviewed and approved by the Comité de Ética para la Experimentación Animal (CEEA) of the Universidad de Navarra (approved protocol 032-17). Work was carried out at the Centro de Investigación Médica Aplicada building (ES312010000132) under the principles and guidelines described in European Directive 2010/63/EU for the protection of animals used for experimental purposes.
Statistical analyses were performed with the GraphPad Prism 5.01 program. The multiple comparison Tukey’s test was used in mice infection assays to find means that are significantly different from each other.
Since the TCS network is essential for S. aureus to adapt to new environments, we hypothesized that one or several TCSs might be involved in environmental sensing and activation of genes that code for biofilm-related factors needed for implant colonization. To address this question, we proceeded to systematically analyze the involvement of each TCS in the colonization of subcutaneous foreign catheters (Cucarella et al., 2001; Vergara-Irigaray et al., 2009) (Figure 1) by comparing the levels of catheter-associated biofilm formation of the methicillin-resistant S. aureus wild type MW2 strain to those of a collection of single MW2 mutants in each TCS (Villanueva et al., 2018). Because differences in growth rates could affect the capacity to colonize the catheters, we first analyzed the growth rate of each mutant under the laboratory growth conditions. The results revealed no significant differences in growth rates between the collection of mutants deficient in TCS and the wild type strain (Supplementary Figure S1). Results showed that 3 TCS single mutants, namely arl, srr, and agr, showed significantly decreased colonization (Figure 2). Interestingly, the arl mutant showed the greatest deficit in colonization of implanted catheters amongst all single TCS mutants. These results indicated that ArlRS, SrrAB, and Agr TCSs are concomitantly needed for biofilm formation in this DRI model, with ArlRS playing a major role in the process.
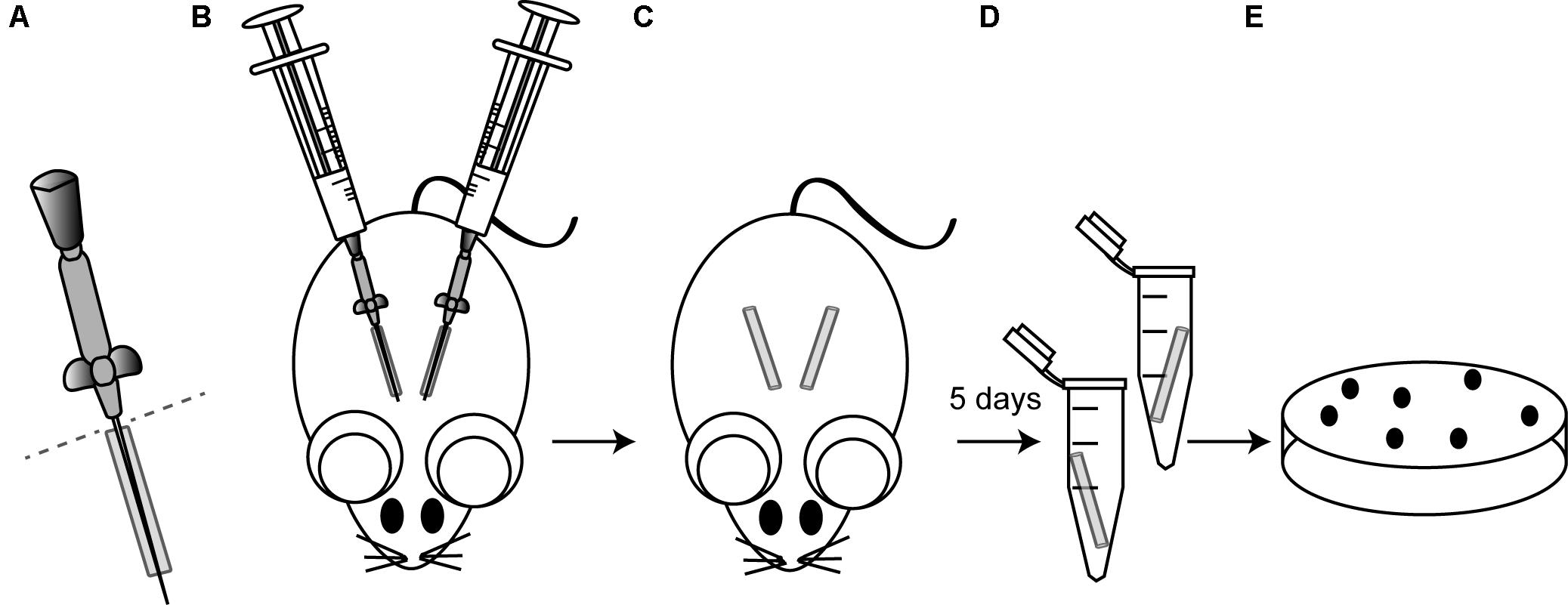
FIGURE 1. Schematic illustrating the murine model of catheter-associated biofilm formation. (A) ICR female mice were anesthetized by isoflurane and the skin was cleansed with alcohol prior to surgery. The bottom of 19 mm catheters (Introcan Safety 24G; B. Braun) was cut right before insertion (dotted line). (B) Then, two catheters were inserted into the subcutaneous interscapular space of each mice and 100 μl of the bacterial suspension containing 107 CFU of the strain under study were injected through the Introcan Devices into the catheters. In all the experiments, five mice were used for each strain under study, so that a total of ten catheters were inoculated with each strain. (C) Introcan Devices were carefully pulled out from mice and wounds were closured with the tissue adhesive Histoacryl® (B. Braun) so that catheters remained inside for 5 days. Note that although closuring the wounds, some catheters were naturally pulled out from mice during the course of experiments. (D) Mice were euthanized, catheters were aseptically removed, placed in a sterile microcentrifuge tube containing 1 ml of PBS and vigorously vortexed for 3 min to remove adherent bacteria. (E) Bacteria were enumerated by viable plate counts.
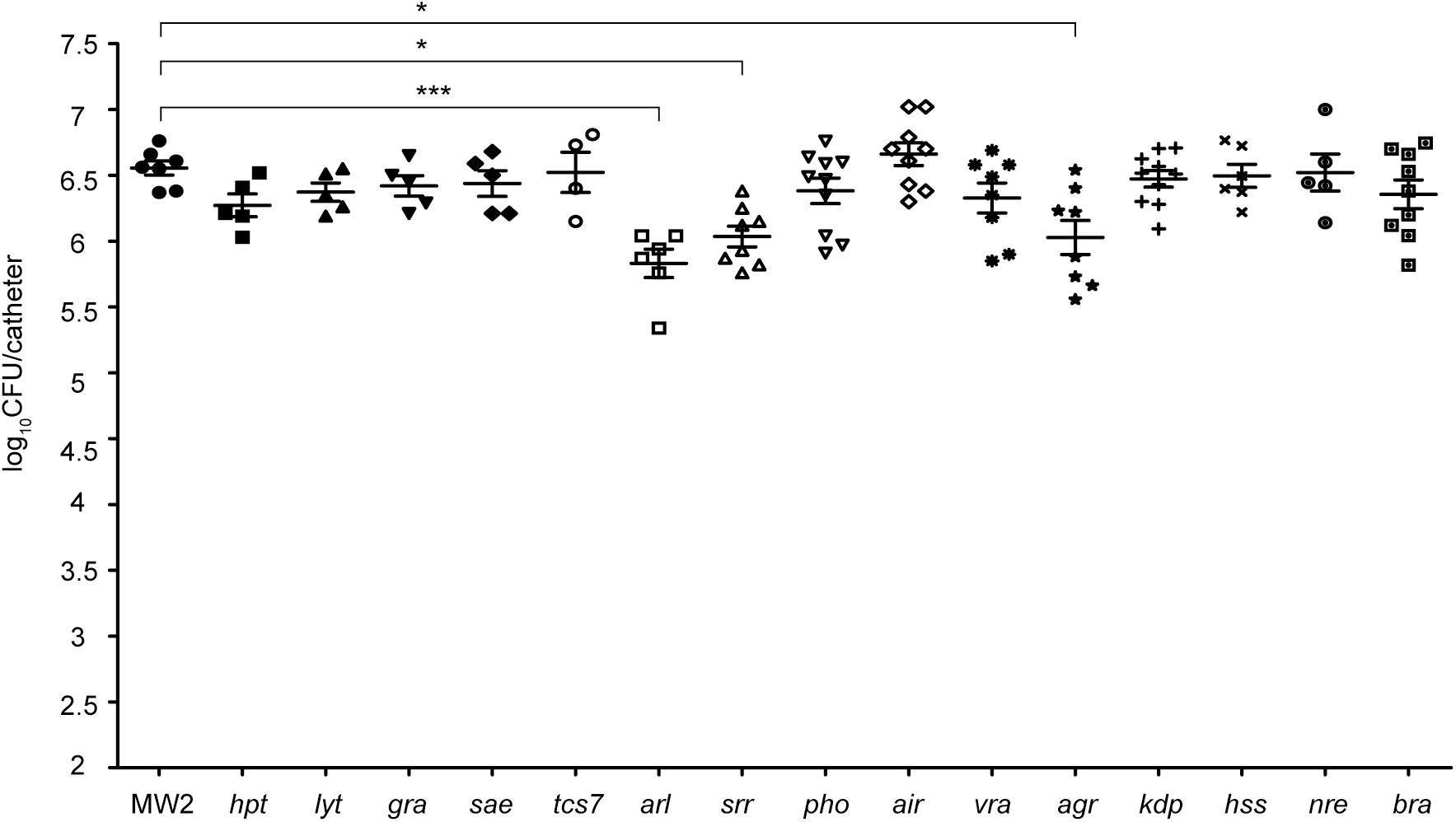
FIGURE 2. Systematic analysis of the contribution of the TCS signaling system to catheter colonization. Comparison of catheter colonization capacity of the wild type strain (MW2) and single mutants in each non-essential TCS. Bacteria were not detectable in control catheters that had been inoculated with PBS (detection limit 100 CFU/catheter). Note that although a total of ten catheters were inoculated with each strain, a variable number of catheters were recovered in each group due to natural catheter expulsion from mice during the course of the experiment. The plots display values obtained from individual catheters and the mean is represented by horizontal bars. Statistical significance was determined with one-way ANOVA followed by Tukey’s multiple comparison test comparing to the WT strain. ∗P < 0.05, ∗∗∗P < 0.001.
Since the exopolysaccharide PNAG is one of the main components of the S. aureus biofilm matrix, we next investigated whether the deficit in catheter colonization shown by the arl mutant strain correlated with an altered capacity to produce PNAG. To do so, we determined PNAG production in arl by dot-blot, using antibodies anti-PNAG, and compared it to that of the wild type strain and the rest of single MW2 mutants in each TCS. Interestingly, the arl mutant strain showed a null capacity to produce PNAG. In contrast, the rest of the mutants displayed a similar, slightly higher or slightly lower PNAG production capacity than the wild type strain (Figure 3A). To extend the observed PNAG deficient phenotype of the arl mutant to other strain backgrounds, we deleted the arlRS genes in the genetically unrelated S. aureus strains 132 (methicillin-resistant) and ISP479r (methicillin-susceptible). Deficiency in PNAG production was confirmed in both arl mutant strains (Figure 3B). Moreover, complementation of the S. aureus MW2 arl mutant with the pCN51 plasmid expressing the arl sensor and response regulator genes under the expression of the PCad promoter (parlRS) led to complete restoration of the capacity to produce PNAG (Figure 3C). Such restoration was completely dependent on the presence of icaADBC genes, since complementation of the arl mutant with parlRS in a ΔicaADBC background did not result in PNAG production (Figure 3C). Altogether, these results showed that the ArlRS TCS is required for PNAG production in S. aureus and suggested a link between the incapacity of arl to synthesize PNAG and the deficiency shown by this mutant in the in vivo model of subcutaneous catheter colonization.
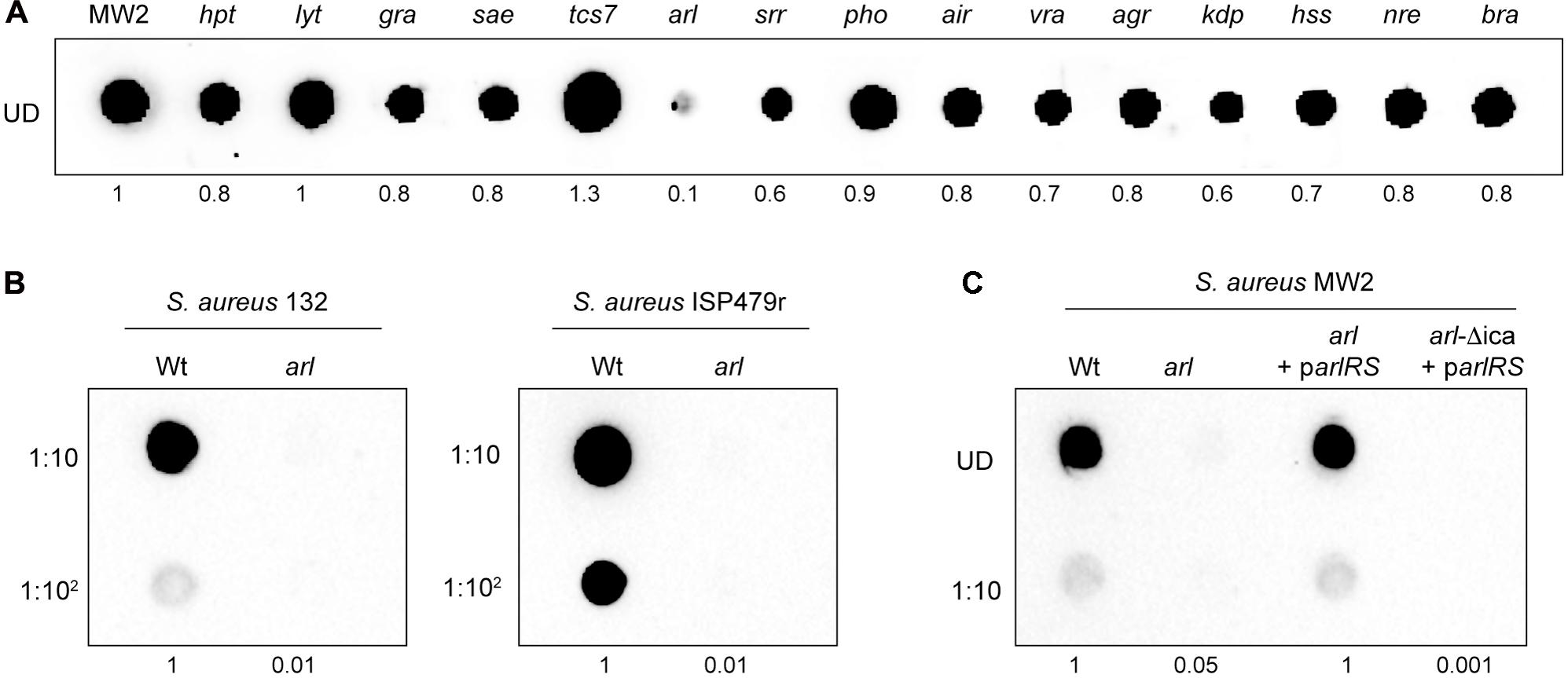
FIGURE 3. Staphylococcus aureus arlRS mutants do not synthesize PNAG. (A) Dot blot analysis of the PNAG exopolysaccharide synthesized by the wild type MW2 strain and the collection of single mutants in each non-essential TCS. (B) Dot blot analysis of PNAG synthesized by the wild type 132 and ISP479r strains and their corresponding arl mutants. (C) Dot blot analysis of PNAG produced by S. aureus MW2, a single arl and double arl-Δica mutant strains complemented with plasmid parlRS, which overexpresses the arlRS genes under the expression of their own promoter. In all cases, samples were analyzed after 16 h of static incubation, at 37°C, in TSB (MW2 and derivatives), TSB NaCl (132) and TSBg (ISP479r). Serial dilutions (1/10) of the samples were spotted onto nitrocellulose membranes and PNAG production was detected with specific anti PIA/PNAG antibodies. UD; undiluted sample. Numbers below the image show relative dot quantification according to densitometry analysis performed with ImageJ (http://rsbweb.nih.gov/ij/).
To investigate the possibility that catheter colonization deficiency of the arl mutant strain might be due to its failure to produce PNAG, we firstly analyzed catheter colonization capacity of an arl mutant in which the icaADBC operon was expressed from the chromosome, under the Pcd promoter (arl-Pcd_ica strain). Restoration of PNAG production in this strain (Figure 4A) resulted in colonization levels similar to the wild type strain (Figure 4B), indicating that the absence of PNAG in arl strain is in fact the reason for its defect in biofilm formation on subcutaneous catheters. Accordingly, a MW2 ΔicaADBC mutant, incapable of synthesizing PNAG (Figure 4A), showed a significantly reduced capacity to colonize catheters compared with the wild type strain (Figure 4B). Additionally, we investigated catheter colonization of S. aureus MW2 derivative mutants in surface proteins whose expression is known to be regulated by ArlRS and that are involved in multicellular behavior (Merino et al., 2009; Christner et al., 2010; Crosby et al., 2016). Results showed that neither protein played a role in biofilm formation on subcutaneous catheters (Supplementary Figure S2). Overall, these findings indicated that the ArlRS TCS governs the process of S. aureus catheter colonization through activation of the synthesis of the PNAG exopolysaccharide, which is fundamental to the formation of a biofilm on the surface of the subcutaneous implanted device.
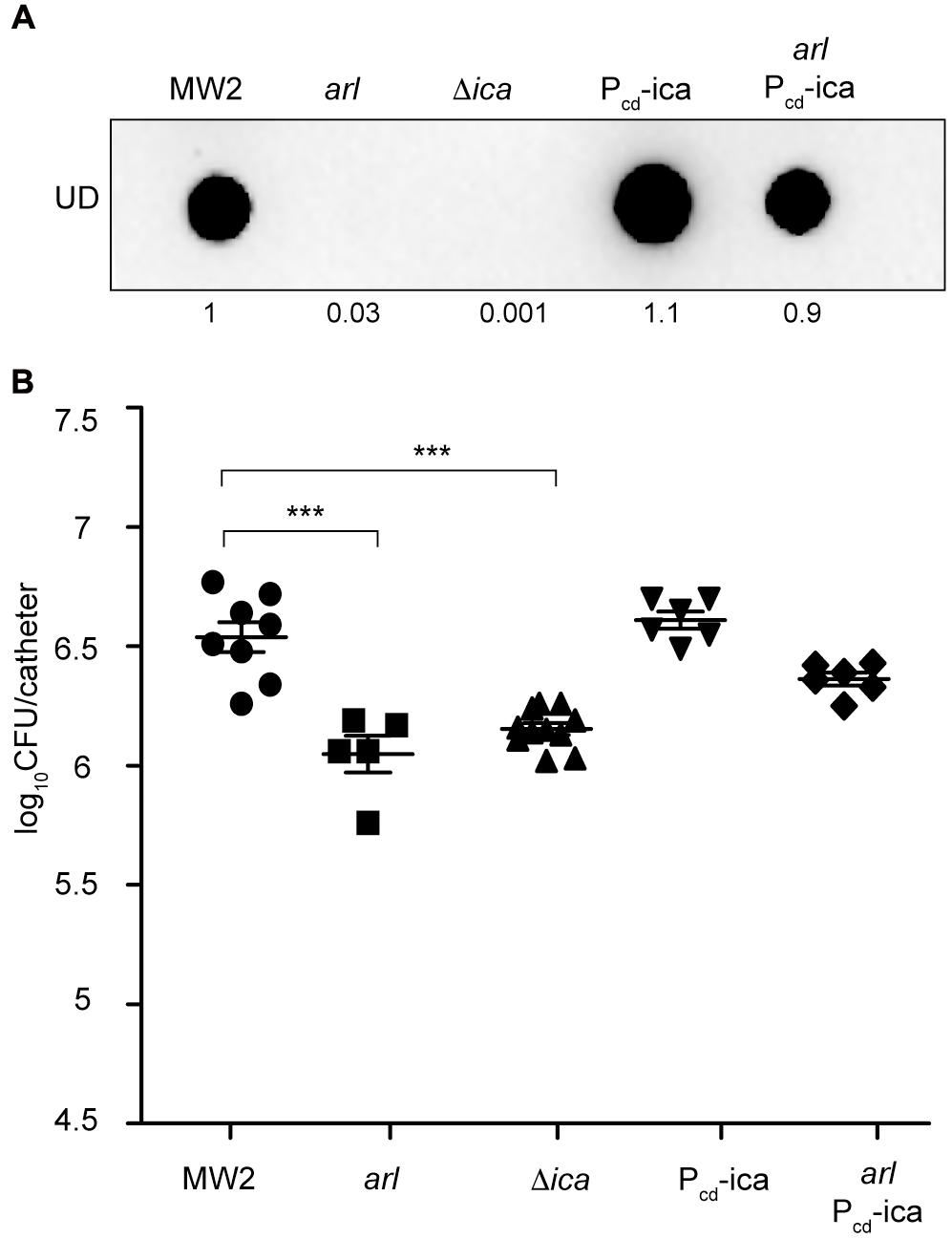
FIGURE 4. Overexpression of the icaADBC operon in an arl mutant restores catheter colonization. (A) Dot blot analysis of the PNAG exopolysaccharide synthesized by the wild type MW2, arl, Δica and the wild type and arl mutant that overproduce PNAG through the chromosomal expression of the icaADBC operon under the Pcad promoter (Pcd_ica and arl Pcd_ica, respectively). Samples were analyzed after 16 h of static incubation, at 37°C, in TSB media. Samples were spotted onto nitrocellulose membranes and PNAG production was detected with specific anti PIA/PNAG antibodies. Numbers below the image show relative dot quantification according to densitometry analysis performed with ImageJ (http://rsbweb.nih.gov/ij/). UD; undiluted sample. (B) Comparison of catheter colonization capacity of the strains shown in (A). Bacteria were not detectable in control catheters that had been inoculated with PBS (detection limit 100 CFU/catheter). Note that although a total of ten catheters were inoculated with each strain, a variable number of catheters were recovered in each group due to natural catheter expulsion from mice during the course of the experiment. The plots display values obtained from individual catheters and the mean is represented by horizontal bars. Statistical significance was determined with one-way ANOVA followed by Tukey’s multiple comparison test comparing to the WT strain. ∗∗∗P < 0.001.
It has previously been shown that ArlRS regulates S. epidermidis biofilm formation by activating icaADBC operon expression, probably through repression of IcaR (Wu et al., 2012). To analyse if this is the case in S. aureus, a sequence upstream of the ica operon, including the promoter region of icaADBC and the IcaR binding site at the 5′UTR of ica genes (-147 to +30), was amplified and cloned into the pCN52 vector (Charpentier et al., 2004), which contains a promoterless gfpmut2 gene, generating plasmid Pica(BS)-gfp. This plasmid was then introduced into S. aureus 132 and ISP479r wild type strains and their corresponding arl mutants, and expression of the Gfp reporter protein was determined by western-blot. Note that for this analysis, we chose to use such strains instead of S. aureus MW2 since as it is shown in Figure 3, levels of PNAG production in strains 132 and ISP479r are significantly higher than in strain MW2. Results showed that arl mutants expressed much lower levels of Gfp compared to wild type strains (Figure 5A), indicating that ArlRS activates the expression of the icaADBC promoter. Next, we examined, by Northern Blot, whether regulation of ica promoter expression by ArlRS leads to altered icaADBC mRNA levels, using two different riboprobes specific for icaA or icaC mRNA. Results showed a strong reduction in the levels of icaA and icaC mRNA in the arl mutant compared with the wild type strain (Figure 5B). Also, we investigated whether the decrease in the amount of icaADBC mRNA in the arl mutant correlated with lower levels of Ica proteins. To do so, we tagged the chromosomal copy of the icaC gene with the 3XFlag sequence in the wild type 132 and ISP479r strains, arl mutants and arl mutants complemented with a plasmid expressing the arlRS genes (arl parlRS), and then, we analyzed IcaC production by Western Blot. As expected, IcaC production was abolished in arl mutant strains and restored to wild type levels in arl parlRS strains (Figure 5C). Finally, considering that in S. epidermidis, Wu et al. (2012) observed an increase in icaR expression in an arlRS mutant, we wondered whether in S. aureus, the ArlRS TCS activates icaADBC operon expression through repression of IcaR. Thus, we determined the levels of icaR mRNA in the wild type 132 strain and compared them to the levels occurring in the arl mutant and the arl mutant complemented with a plasmid expressing the arlRS genes. As it is shown in Figure 5D, mutation of arl led to a significant increase in icaR mRNA levels that were lowered to approximately wild type levels when the arl mutant was complemented with the arlRS genes. Collectively, these findings indicated that in S. aureus, ArlRS regulates icaADBC expression at the transcriptional level, at least through downregulation of icaR expression, resulting in the activation of PNAG production.
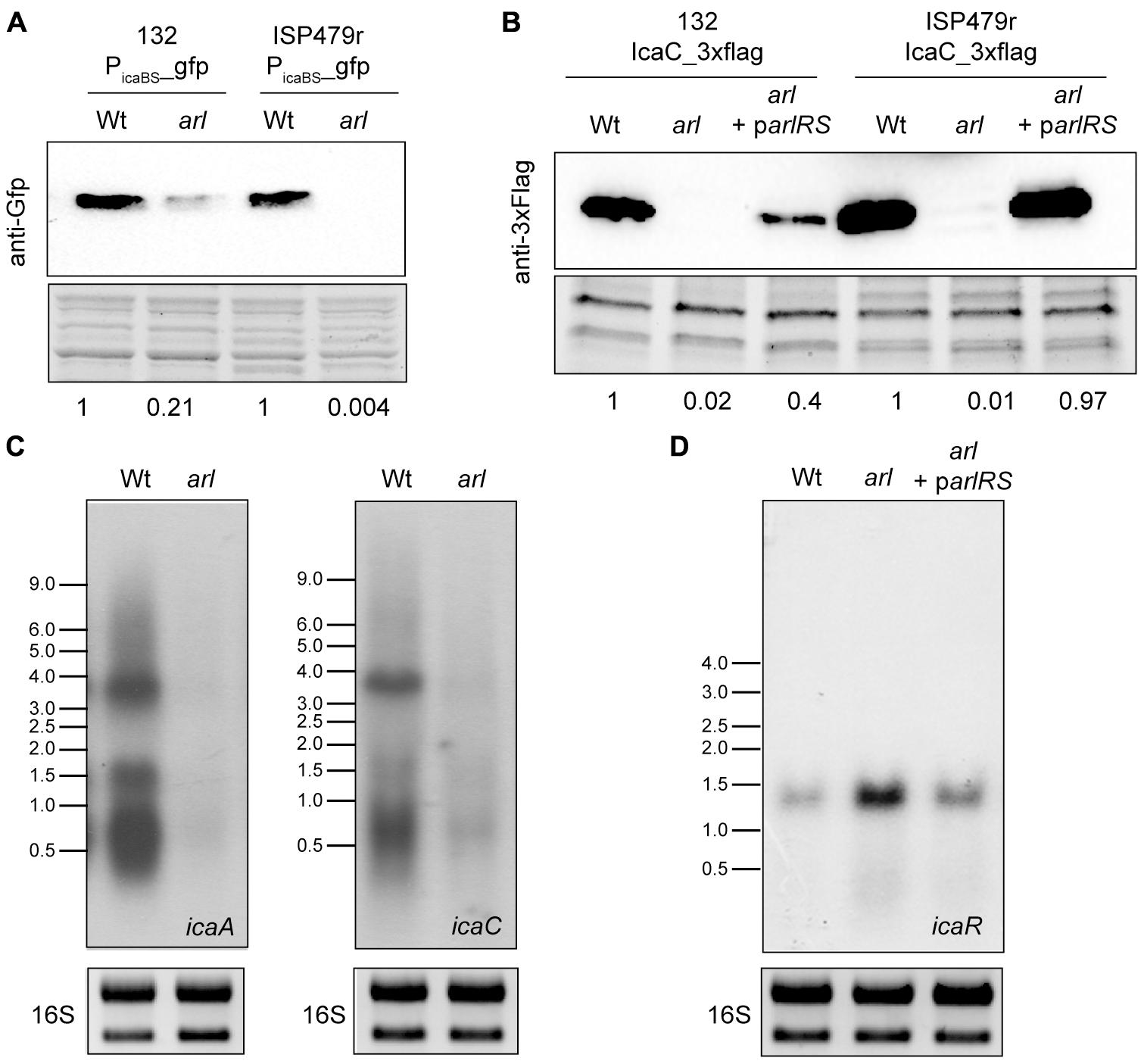
FIGURE 5. ArlRS transcriptionally activates icaADBC operon expression in S. aureus. (A) A representative Western blot showing GFP protein levels expressed from S. aureus 132 and ISP479r wild type strains and their corresponding arl mutants harboring plasmid Pica(BS)-gfp. The GFP protein was detected with commercial anti-GFP antibodies. A stain-free gel portion is shown as a loading control. Numbers below the image show relative band quantification according to densitometry analysis performed with ImageJ (http://rsbweb.nih.gov/ij/). (B) Representative Northern blots showing icaA and icaC mRNA of S. aureus 132 wild type and arl mutant strains grown at 37°C until exponential phase (OD600 nm = 0.8). Lower panels show 16S ribosome band stained with RedSafe Nucleic Acid Staining Solution as loading control. (C) A representative Western blot showing IcaC protein levels of the wild type 132 and ISP479r strains, arl mutants and arl mutants complemented with a plasmid expressing the arlRS genes (arl parlRS). The 3XFlag tagged IcaC protein was detected with commercial anti-3XFlag antibodies. A stain-free gel portion is shown as a loading control. Numbers below the image show relative band quantification according to densitometry analysis performed with ImageJ (http://rsbweb.nih.gov/ij/). (D) A representative Northern blot showing icaR mRNA of S. aureus 132 wild type strain, arl mutant and arl mutant complemented with a plasmid expressing the arlRS genes (arl parlRS) grown at 37°C until exponential phase (OD600 nm = 0.8). Lower panels show 16S ribosome band stained with RedSafe Nucleic Acid Staining Solution as loading control.
It is known that ArlRS and the global regulator MgrA form a regulatory cascade (Supplementary Figure S3A) in which MgrA acts downstream of ArlRS to control expression of a number of genes important for virulence, including those for several large surface proteins (Crosby et al., 2016). Also, MgrA seems to regulate PNAG synthesis, since a mutant in mgrA has been shown to produce lower levels of PNAG than its corresponding wild type strain (Trotonda et al., 2008). Taken these results into account, we decided to investigate the contribution of MgrA to catheter colonization mediated by the ArlRS TCS. To do so, we firstly constructed a mgrA mutant in the MW2 strain and analyzed its capacity to synthesize PNAG by dot blot, using anti PNAG antibodies. The mgrA mutant lacked all capacity to produce PNAG (Figure 6A). We then compared catheter colonization of the arl mutant with that of the mgrA mutant and verified that both strains showed a similar deficiency in implant colonization. Importantly, catheter colonization capacity of the mgrA mutant was restored when the icaADBC operon was expressed from the chromosome, under the Pcad promoter (mgrA-Pcd_ica strain) (Figure 6B), confirming the requirement of PNAG production for implant colonization. Then, we performed epistatic experiments by overexpressing MgrA in the arlRS mutant (Supplementary Figure S3B) and analyzing PNAG production and colonization capacity of the resulting strain (arl Pcd_mgrA). Results showed that MgrA overexpression in the arlRS background restored neither PNAG production nor implant colonization capacity, indicating that MgrA activity cannot counteract ArlRS absence.
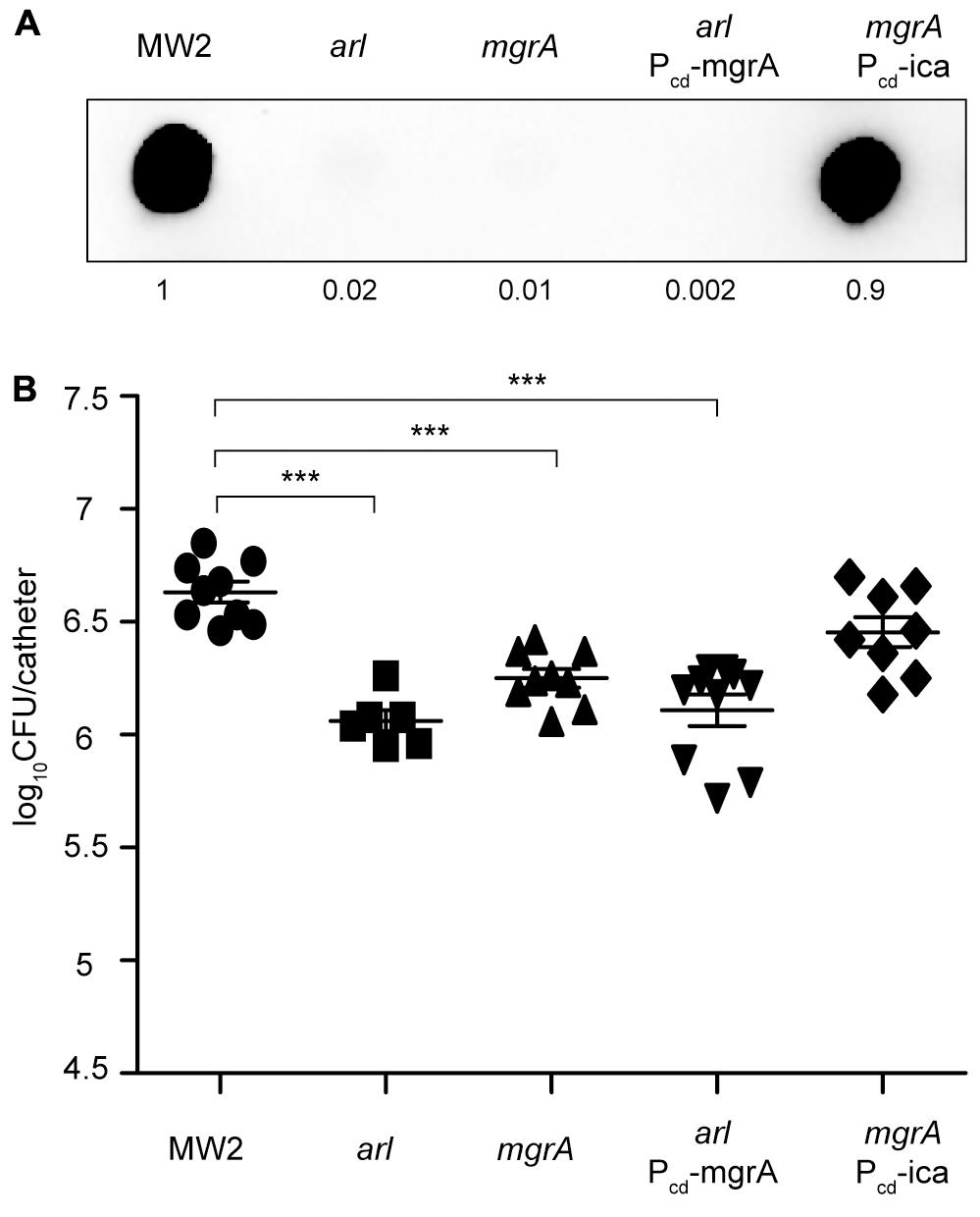
FIGURE 6. MgrA is unable to restore catheter colonization of an arl mutant. (A) Dot blot analysis of the PNAG exopolysaccharide synthesized by the wild type MW2, arl, mgrA, arl that overproduces MgrA through the chromosomal expression of the mgrA gene under the Pcad promoter (arl Pcd_mgrA), and mgrA that overproduces PNAG through the chromosomal expression of the icaADBC operon under the Pcad promoter (mgrA Pcd_ica). Samples were analyzed after 16 h of static incubation, at 37°C, in TSB media. Samples were spotted onto nitrocellulose membranes and PNAG production was detected with specific anti PIA/PNAG antibodies. Numbers below the image show relative dot quantification according to densitometry analysis performed with ImageJ (http://rsbweb.nih.gov/ij/). UD; undiluted sample. (B) Comparison of catheter colonization capacity of S. aureus strains shown in (A). Bacteria were not detectable in control catheters that had been inoculated with PBS (detection limit 100 CFU/catheter). Note that although a total of 10 catheters were inoculated with each strain, a variable number of catheters were recovered in each group due to natural catheter expulsion from mice during the course of the experiment. The plots display values obtained from individual catheters and the mean is represented by horizontal bars. Statistical significance was determined with one-way ANOVA followed by Tukey’s multiple comparison test comparing to the WT strain. ∗∗∗P < 0.001.
Many loss-of-function studies have been performed in several pathogenic bacteria to identify the TCSs required for virulence using in vivo infection models (Lasaro et al., 2014; Reboul et al., 2014; Cheng et al., 2015; Thomassin et al., 2017). However, to our knowledge, this is the first systematic study conducted to determine the role of each TCS in colonization and survival of S. aureus on the surface of an implanted medical device. We used a collection of single mutants in each non-essential TCS of S. aureus MW2 and a murine subcutaneous catheter infection model to identify TCSs controlling colonization of the implanted device. It is important to note that none of the single mutants in each TCS show a defect in growth rate when compared to the wild type strain, at least under the laboratory growth conditions tested (Supplementary Figure S1). The strongest phenotype in our screening was observed when the arlRS TCS was mutated, leading to a greater than 10-fold reduction in the colonization of the catheter. This result was somehow unexpected since other studies have shown that arlRS mutants display an increased ability to form a biofilm in vitro, due to the release of eDNA (Fournier and Hooper, 2000; Walker et al., 2013) and to the overexpression of SasG, a large surface protein implicated in biofilm formation (Crosby et al., 2016). Furthermore, Toledo-Arana et al. (2005) also showed that mutation of arlRS in S. aureus 15981 strain, which is naturally defective in agr, promoted biofilm formation in a chemically defined media (HHWm) through overexpression of the protein A surface protein. In contrast, mutation of arlRS in the closely related S. epidermidis reduces biofilm formation capacity both in vitro and in vivo (Wu et al., 2012, 2014). The molecular mechanism by which ArlRS regulates biofilm formation in S. epidermidis has been related with the capacity of ArlR to bind to the promoter region between icaR and icaADBC and to repress icaR expression, which indirectly results in an activation of icaADBC expression. In agreement with these results, we showed that ArlRS is involved in the regulation of icaADBC expression in S. aureus. Deletion of arlRS in three genetically unrelated S. aureus strains led to a deficiency in PNAG production in TSB media. Transcriptional fusions performed between the icaADBC promoter and the gfp reporter gene showed that the GFP levels were reduced significantly (about 30%) (Figure 5A) in the absence of ArlRS. In addition, icaC mRNA levels and consequently, IcaC protein levels, were reduced in the arl strain. Thus, we concluded that ArlRS regulates icaADBC operon expression at the level of transcription. The control that ArlRS exerts over icaADBC transcription might occur directly or through another regulatory protein(s). Similarly to what has been demonstrated in S. epidermidis, we observed that IcaR expression is upregulated in the absence of ArlRS, strongly suggesting that in S. aureus, regulation of icaADBC transcription by ArlR is mediated through upregulation of IcaR. Further studies are necessary to determine the direct interaction of ArlR with the promoter of the icaR gene.
It is believed that the properties provided by the biofilm lifestyle depend on the composition of the biofilm matrix, which can vary not only between strains but also within the same bacterial strain depending on the environmental conditions. S. aureus can form biofilms either made of the PNAG exopolysaccharide or composed by surface proteins such as fibronectin binding proteins, FnBPs, protein A, SasG or Bap (Cucarella et al., 2001; Corrigan et al., 2007; Merino et al., 2009; Vergara-Irigaray et al., 2009). Despite the mutation of arlRS leads to an increased expression of several surface proteins, including some involved in multicellular behavior, such as SasG, Ebh, and Spa (Fournier and Hooper, 2000; Walker et al., 2013; Crosby et al., 2016), in the present study we have shown that none of these proteins seem to play a role in the colonization of implanted catheters. On the contrary, S. aureus MW2 required PNAG to form a biofilm on the surface of the subcutaneous catheter. This result was again unexpected since S. aureus MW2 is a weak PNAG exopolysaccharide producer under laboratory conditions (Trotonda et al., 2008). This fact warns against the value of in vitro PNAG production analysis in order to infer PNAG relevance during infection.
In the present study, we used commercial polyurethane (PU) intravenous catheters. PU is one of the most commonly used material for catheter building due to its high excellent physical properties and good biocompatibility. Analysis of the molecular forces involved in PNAG-mediated adhesion using single-cell force spectroscopy revealed that cationic PNAG binds to anionic surfaces via multivalent electrostatic interactions (Formosa-Dague et al., 2016). During cell-to-cell interactions, it has been proposed that PNAG connects the cells together by electrostatic interaction between its positively charged groups and negatively charged molecules of wall teichoic acids on opposing cells (Gross et al., 2001; Vergara-Irigaray et al., 2008). In the case of cell-implant interaction it seems plausible that PNAG also mediates surface adhesion via electrostatic interactions. On the other hand, adhesion to biological surfaces is often mediated by hydrophobic interactions that are usually the strongest of all long-range non-covalent forces (Briandet et al., 2001; Cerca et al., 2005). It is known that hydrophobic materials favor bacterial adherence more than hydrophilic ones and consequently modification of the bacterial surface hydrophobicity can also affect the capacity to colonize the catheter surface (Pavithra and Doble, 2008). We have not addressed whether surface hydrophobicity is modified in the absence of ArlRS and future studies to clarify this possibility are needed. On the other hand, though bacteria can encounter the naked implant surface, we also need to consider that immediately following the implantation of the catheter, a layer of host proteins rapidly adsorbs on its surface, altering the properties of the polymer which may affect bacterial adhesion and biofilm formation. In our experimental model, catheters were infected immediately after implantation without allowing the catheter to be coated with plasma proteins. This model mimics those infection processes that occur during catheter implantation but we cannot exclude that colonization of a catheter surface coated by plasma may involve additional factors.
ArlR activates expression of one of the promoters of the transcriptional regulator MgrA, also called Rat or NorR (Fournier et al., 2000; Ingavale et al., 2003; Crosby et al., 2016). This global regulator modulates the expression of 5–10% of the S. aureus genome, and a large part of the ArlRS regulon depends on the expression of MgrA (Luong et al., 2003; Gupta et al., 2015; Crosby et al., 2016). Based on results shown by Trotonda et al. (2008) that indicate that although S. aureus mutants in mgrA have increased in vitro biofilm formation capacity, they produce lower levels of the PNAG exopolysaccharide, we hypothesized that MgrA might be related with the arlRS mutant deficiency in catheter colonization. Our results confirmed that an mgrA mutant has a decreased PNAG production capacity and also showed that in our catheter infection model, the mgrA mutant presents a colonization deficit similar to that of the arlRS mutant. Importantly, complementation of both arlRS and mgrA mutants with the icaADBC operon restored implant colonization capacity. However, epistatic experiments revealed that overexpression of MgrA in the arl mutant restored neither PNAG production nor implant colonization capacity. These results confirmed the impact that the PNAG exopolysaccharide has for catheter colonization and also indicated that as regards PNAG expression, MgrA activity cannot compensate for ArlRS absence. One plausible explanation for this might be that both regulators regulate icaADBC expression at a different level. According to Wu et al. (2012) and to our results, ArlRS regulates icaADBC expression at the transcriptional level through the icaR repressor. Instead, MgrA seems to regulate icaADBC expression at the posttranscriptional level (Trotonda et al., 2008). These last observations may explain why the ica genes have never been identified in studies carried out to define the MgrA regulon (Luong et al., 2003; Crosby et al., 2016).
Our results also showed that deletion of Agr and SrrAB TCSs cause a slight reduction in the capacity to colonize subcutaneous catheters. Agr controls biofilm architecture and cell dispersion through the regulation of phenol soluble modulins (PSMs) in a ica-independent manner (Otto, 2012). PSMs can aggregate and form amyloid fibrils that contribute to stability of the biofilm (Schwartz et al., 2012, 2014). Because the levels of the PNAG exopolysaccharide produced in vitro were similar between the wild type and agr mutant strains, it is likely that PNAG is not involved in the decreased colonization capacity of the agr mutant in vivo. On the other hand, SrrAB has been shown to induce biofilm formation under anaerobic conditions through activation of icaADBC gene transcription (Ulrich et al., 2007; Wu et al., 2015) and release of extracellular DNA (Mashruwala et al., 2017). Since implanted catheters are thought to comprise anaerobic microenvironments, it might be possible that the srrA mutant has a catheter colonization deficiency because of a defect linked to oxygen sensing.
In summary, we have shown that ArlRS plays a significant role in S. aureus implant colonization by affecting the expression of the icaADBC operon by directly modifying icaR transcription. Although different studies using different genetic strains coincide in showing that ArlRS plays an important role in biofilm mediated infections, it is also noticeable the strong strain and growth media dependency in the observed phenotypes. Thus, additional studies are necessary to determine the precise mechanisms of regulation and the signals to which this TCS responds.
SB, CS, IL, and JV designed the work and revised the manuscript. SB, CG, and JV completed all the experiments. SB and CG performed the statistically analysis and made the figures. JV, SB, IL, and CS wrote the manuscript.
JV was supported by SAF2015-74267-JIN. This research was supported by grants BIO2014-53530-R, BIO2017-83035-R and RTC-2015-3184-1 from the Spanish Ministry of Economy and Competitivity.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Gerald B. Pier for providing the anti-S. aureus PNAG antiserum.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00342/full#supplementary-material
Alanidis, C., and Dejong, P. J. (1990). Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074. doi: 10.1093/nar/18.20.6069
Arnaud, M., Chastanet, A., and Débarbouillé, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004
Briandet, R., Herry, J. M., and Bellon-Fontaine, M. N. (2001). Determination of the van der Waals, electron donor and electron acceptor surface tension components of static Gram-positive microbial biofilms. Colloids Surf. B Biointerfaces 21, 299–310. doi: 10.1016/S0927-7765(00)00213-7
Cerca, N., Pier, G. B., Vilanova, M., Oliveira, R., and Azeredo, J. (2005). Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res. Microbiol. 156, 506–514. doi: 10.1016/j.resmic.2005.01.007
Charpentier, E., Anton, A. I., Barry, P., Alfonso, B., Fang, Y., and Novick, R. P. (2004). Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70, 6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004
Cheng, A. T., Ottemann, K. M., and Yildiz, F. H. (2015). Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog. 11:e1004933. doi: 10.1371/journal.ppat.1004933
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H., and Xiong, Y.-Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40, 1–9.
Christner, M., Franke, G. C., Schommer, N. N., Wendt, U., Wegert, K., Pehle, P., et al. (2010). The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75, 187–207. doi: 10.1111/j.1365-2958.2009.06981.x
Corrigan, R. M., Rigby, D., Handley, P., and Foster, T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. doi: 10.1099/mic.0.2007/006676-0
Crosby, H. A., Schlievert, P. M., Merriman, J. A., King, J. M., Salgado-Pabón, W., and Horswill, A. R. (2016). The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 12:e1005604. doi: 10.1371/journal.ppat.1005604
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penadés, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Ellis, M. W., Schlett, C. D., Millar, E. V., Crawford, K. B., Cui, T., Lanier, J. B., et al. (2014). Prevalence of nasal colonization and strain concordance in patients with community-associated Staphylococcus aureus skin and soft-tissue infections. Infect. Control Hosp. Epidemiol. 35, 1251–1256. doi: 10.1086/678060
Formosa-Dague, C., Speziale, P., Foster, T. J., Geoghegan, J. A., and Dufrêne, Y. F. (2016). Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. U.S.A. 113, 410–415. doi: 10.1073/pnas.1519265113
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Fournier, B., Aras, R., and Hooper, D. C. (2000). Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182, 664–671. doi: 10.1128/JB.182.3.664-671.2000
Fournier, B., and Hooper, D. C. (2000). A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182, 3955–3964. doi: 10.1128/JB.182.14.3955-3964.2000
Gross, M., Cramton, S. E., Götz, F., and Peschel, A. (2001). Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001
Gupta, R. K., Luong, T. T., and Lee, C. Y. (2015). RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc. Natl. Acad. Sci. U.S.A. 112, 14036–14041. doi: 10.1073/pnas.1509251112
Haag, A. F., and Bagnoli, F. (2016). “The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation,” in Current Topics in Microbiology and Immunology, eds F. Bagnoli, R. Rappuoli, and G. Grandi (Berlin: Springer), 1–54. doi: 10.1007/82_2015_5019
Hogan, S., Stevens, N. T., Humphreys, H., O’Gara, J. P., and O’Neill, E. (2015). Current and future approaches to the prevention and treatment of staphylococcal medical device-related infections. Curr. Pharm. Des. 21, 100–113.
Ingavale, S. S., van Wamel, W., and Cheung, A. L. (2003). Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48, 1451–1466. doi: 10.1046/j.1365-2958.2003.03503.x
Kluytmans, J., van Belkum, A., and Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520.
Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240.
Lasa, I., Toledo-Arana, A., Dobin, A., Villanueva, M., de los Mozos, I. R., Vergara-Irigaray, M., et al. (2011). Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U.S.A 108, 20172–20177. doi: 10.1073/pnas.1113521108
Lasaro, M., Liu, Z., Bishar, R., Kelly, K., Chattopadhyay, S., Paul, S., et al. (2014). Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J. Bacteriol. 196, 1723–1732. doi: 10.1128/JB.01296-13
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Luong, T. T., Newell, S. W., and Lee, C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185, 3703–3710.
Maira-Litrán, T., Kropec, A., Abeygunawardana, C., Joyce, J., Mark, G., Goldmann, D. A., et al. (2002). Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70, 4433–4440.
Mashruwala, A. A., Gries, C. M., Scherr, T. D., Kielian, T., and Boyd, J. M. (2017). SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect. Immun. 85:e00157-17. doi: 10.1128/IAI.00157-17
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. doi: 10.1128/JB.01222-08
Novick, R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x
O’Gara, J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. doi: 10.1111/j.1574-6968.2007.00688.x
O’Neill, E., Pozzi, C., Houston, P., Humphreys, H., Robinson, D. A., Loughman, A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. doi: 10.1128/JB.00167-08
Otto, M. (2012). Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 34, 201–214. doi: 10.1007/s00281-011-0296-2
Pavithra, D., and Doble, M. (2008). Biofilm formation, bacterial adhesion and host response on polymeric implants—issues and prevention. Biomed. Mater. 3, 34003–34014. doi: 10.1088/1748-6041/3/3/034003
Reboul, A., Lemaître, N., Titecat, M., Merchez, M., Deloison, G., Ricard, I., et al. (2014). Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J. Infect. Dis. 210, 1367–1375. doi: 10.1093/infdis/jiu274
Rice, K. C., Mann, E. E., Endres, J. L., Weiss, E. C., Cassat, J. E., Smeltzer, M. S., et al. (2007). The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8113–8118. doi: 10.1073/pnas.0610226104
Ruiz de Los Mozos, I., Vergara-Irigaray, M., Segura, V., Villanueva, M., Bitarte, N., Saramago, M., et al. (2013). Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 9:e1004001. doi: 10.1371/journal.pgen.1004001
Schenk, S., and Laddaga, R. A. (1992). Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73, 133–138.
Schwartz, K., Sekedat, M. D., Syed, A. K., O’Hara, B., Payne, D. E., Lamb, A., et al. (2014). The AgrD N-Terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. Infect. Immun. 82, 3837–3844. doi: 10.1128/IAI.02111-14
Schwartz, K., Syed, A. K., Stephenson, R. E., Rickard, A. H., and Boles, B. R. (2012). Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 8:e1002744. doi: 10.1371/journal.ppat.1002744
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Taglialegna, A., Navarro, S., Ventura, S., Garnett, J. A., Matthews, S., Penadés, J. R., et al. (2016). Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 12:e1005711. doi: 10.1371/journal.ppat.1005711
Thomassin, J.-L., Leclerc, J.-M., Giannakopoulou, N., Zhu, L., Salmon, K., Portt, A., et al. (2017). Systematic analysis of two-component systems in Citrobacter rodentium reveals positive and negative roles in virulence. Infect. Immun. 85:e00654-16. doi: 10.1128/IAI.00654-16
Toledo-Arana, A., Merino, N., Vergara-Irigaray, M., Débarbouillé, M., Penadés, J. R., and Lasa, I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187, 5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005
Trotonda, M. P., Tamber, S., Memmi, G., and Cheung, A. L. (2008). MgrA represses biofilm formation in Staphylococcus aureus. Infect. Immun. 76, 5645–5654. doi: 10.1128/IAI.00735-08
Ulrich, M., Bastian, M., Cramton, S. E., Ziegler, K., Pragman, A. A., Bragonzi, A., et al. (2007). The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65, 1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J.-M., Amorena, B., Penadés, J. R., et al. (2003). SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087.
Vergara-Irigaray, M., Maira-Litran, T., Pier, G. B., Penadés, J. R., and Lasa, I. (2008). Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154, 865–877. doi: 10.1099/mic.0.2007/013292-0
Vergara-Irigaray, M., Valle, J., Merino, N., Latasa, C., García, B., Ruiz de Los Mozos, I., et al. (2009). Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77, 3978–3991. doi: 10.1128/IAI.00616-09
Villanueva, M., García, B., Valle, J., de los Mozos, I. R., Solano, C., Rapun, B., et al. (2018). Sensory deprivation in Staphylococcus aureus. Nat. Commun. 9:523. doi: 10.1038/s41467-018-02949-y
Walker, J. N., Crosby, H. A., Spaulding, A. R., Salgado-Pabón, W., Malone, C. L., Rosenthal, C. B., et al. (2013). The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis. PLoS Pathog. 9:e1003819. doi: 10.1371/journal.ppat.1003819
Wu, Y., Liu, J., Jiang, J., Hu, J., Xu, T., Wang, J., et al. (2014). Role of the two-component regulatory system arlRS in ica operon and aap positive but non-biofilm-forming Staphylococcus epidermidis isolates from hospitalized patients. Microb. Pathog. 76, 89–98. doi: 10.1016/j.micpath.2014.09.013
Wu, Y., Wang, J., Xu, T., Liu, J., Yu, W., Lou, Q., et al. (2012). The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One 7:e40041. doi: 10.1371/journal.pone.0040041
Keywords: two-component systems, Staphylococcus aureus, implants, biofilm, PNAG, arlRS
Citation: Burgui S, Gil C, Solano C, Lasa I and Valle J (2018) A Systematic Evaluation of the Two-Component Systems Network Reveals That ArlRS Is a Key Regulator of Catheter Colonization by Staphylococcus aureus. Front. Microbiol. 9:342. doi: 10.3389/fmicb.2018.00342
Received: 26 November 2017; Accepted: 12 February 2018;
Published: 07 March 2018.
Edited by:
Andrea Guido Oreste Manetti, GlaxoSmithKline, United KingdomReviewed by:
Dipankar Ghosh, Jawaharlal Nehru University, IndiaCopyright © 2018 Burgui, Gil, Solano, Lasa and Valle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaione Valle, amFpb25lLnZhbGxlQHVuYXZhcnJhLmVz Iñigo Lasa, aWxhc2FAdW5hdmFycmEuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.