
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 February 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00334
This article is part of the Research TopicMALDI-TOF MS Application for Susceptibility Testing of Microorganisms View all 12 articles
 Annie W. T. Lee1
Annie W. T. Lee1 Johnson K. S. Lam2
Johnson K. S. Lam2 Ricky K. W. Lam2
Ricky K. W. Lam2 Wan H. Ng2
Wan H. Ng2 Ella N. L. Lee1
Ella N. L. Lee1 Vicky T. Y. Lee3
Vicky T. Y. Lee3 Po P. Sze1
Po P. Sze1 Rahim Rajwani1
Rahim Rajwani1 Kitty S. C. Fung4
Kitty S. C. Fung4 Wing K. To5
Wing K. To5 Rodney A. Lee3
Rodney A. Lee3 Dominic N. C. Tsang2
Dominic N. C. Tsang2 Gilman K. H. Siu1*
Gilman K. H. Siu1*Objective: This study evaluated the capability of a MALDI Biotyper system equipped with the newly introduced MBT STAR-BL module to simultaneously perform species identification and β-lactamase-mediated resistance detection in bacteremia -causing bacteria isolated from cultured isolates and patient-derived blood cultures (BCs).
Methods: Two hundred retrospective cultured isolates and 153 prospective BCs containing Gram-negative rods (GNR) were collected and subjected to direct bacterial identification, followed by the measurement of β-lactamase activities against ampicillin, piperacillin, cefotaxime, ceftazidime, and meropenem using the MBT STAR-BL module. The results and turnaround times were compared with those of routine microbiological processing. All strains were also characterized by beta-lactamase PCR and sequencing.
Results: Using the saponin-based extraction method, MALDI-TOF MS correctly identified bacteria in 116/134 (86.6%) monomicrobial BCs. The detection sensitivities for β-lactamase activities against ampicillin, piperacillin, third-generation cephalosporin and meropenem were 91.3, 100, 97.9, and 100% for cultured isolates, and 80.4, 100, 68.8, and 40% for monomicrobial BCs (n = 134) respectively. The overall specificities ranged from 91.5 to 100%. Furthermore, the MBT STAR-BL and conventional drug susceptibility test results were concordant in 14/19 (73.7%) polymicrobial cultures. Reducing the logRQ cut-off value from 0.4 to 0.2 increased the direct detection sensitivities for β-lactamase activities against ampicillin, cefotaxime and meropenem in BCs to 85.7, 87.5, and 100% respectively. The MBT STAR-BL test enabled the reporting of β-lactamase-producing GNR at 14.16 and 47.64 h before the interim and final reports of routine BCs processing, respectively, were available.
Conclusion: The MALDI Biotyper system equipped with the MBT STAR-BL module enables the simultaneous rapid identification of bacterial species and β-lactamase-mediated resistance from BCs and cultured isolates. Adjustment of the logRQ cut-off value to 0.2 significantly increased the detection sensitivities for clinically important drug-resistant pathogens.
Sepsis is a major cause of infectious disease-associated morbidity and mortality (Fleischmann et al., 2016). Proper initial antibiotic therapy is a crucial parameter for improvement of patient outcomes (Kumar et al., 2006; Dellinger et al., 2008). Empirical treatment must be administrated at the time of sepsis diagnosis, and the regimen should be adjusted if necessary when bacterial identification and drug susceptibility results are available (Kang et al., 2005; Dellinger et al., 2008; Kumar, 2011). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) is particularly useful for direct identification of causative agents of bacteremia, which can be inducers of sepsis, within the same day of a positive blood culture (BC) broth result (Schubert et al., 2011; Wuppenhorst et al., 2012; Chen et al., 2013; Clerc et al., 2013). However, predictions of drug susceptibility patterns based on bacterial identification alone have become inaccurate, given the increasing incidence of multidrug-resistance among Gram-negative bacteria (Paterson, 2006; Livermore, 2012).
Recent studies demonstrated the feasibility of using MALDI-TOF MS to predict β-lactam resistance through detection of hydrolytic β-lactam substrates produced by bacterial β-lactamases (Burckhardt and Zimmermann, 2011; Hrabak et al., 2011; Sparbier et al., 2012; Ghebremedhin et al., 2016). Unfortunately, in the absence of automated analysis software, previous studies have used either manual calculations (Sparbier et al., 2012; Ghebremedhin et al., 2016) or self-developed algorithms with ambiguous cut-off values (Jung et al., 2014) to analyze the peak patterns. These techniques introduce intra- and inter-observer variability to the assay and are difficult to implement in routine diagnostic workflows. Recently, Bruker Daltonik launched a software module, the MALDI Biotyper™ Selective Testing for Beta-Lactamase Activity (MBT STAR-BL), for the automatic analysis of drug hydrolysis mass spectra. This module facilitates the simultaneous bacterial identification and detection of β-lactamase-mediated resistance toward ampicillin (AMP), piperacillin (PIP), cefotaxime (CTX), ceftazidime (CAZ), meropenem (MEM), and ertapenem (ETP).
The present study aimed to evaluate the ability of the MALDI Biotyper system equipped with the MBT STAR-BL module to identify bacteremia -causing bacteria and predict β-lactam resistance from plated isolates, as well as BC broths. The time-to-results determined using MBT STAR-BL were also compared with those obtained using a conventional culture-based method.
In the first stage, 200 archived Gram-negative isolates of different species and various drug susceptibility patterns were collected and used to evaluate the ability of the MBT STAR-BL to detect β-lactamase-mediated resistance to all claimed antibiotics except ertapenem, which was not available locally. All strains were isolated from BCs previously collected at four different public hospitals throughout the territory. Escherichia coli strain DH5a was used as a β-lactamase-negative control strain, whereas ATCC E. coli strain BAA-2452, a NDM-1 carbapenemase producer, was used as a positive control in all drug hydrolysis tests.
In the second stage, 153 positive BC broths derived from patients with Gram-negative bacterial bloodstream infections were collected prospectively from January to December 2016. BACTEC™ FX (Becton Dickson, US) and BacT/Alert FA (bioMerieux, France) blood culture system are housed in hospitals. All positive BCs were subjected to direct Gram staining. If Gram-negative rods were found, a 5-mL aliquot of culture broth was transported to our laboratory for direct bacterial identification, followed by the detection of β-lactamase-mediated resistance using MBT STAR-BL within the same day.
On the following day, MALDI-TOF MS analyses were repeated using isolated colonies grown on subculture plates.
For direct bacterial identification from BC broths, bacterial proteins were extracted using saponin-based protocol (Chen et al., 2013). The target plate was then analyzed using the Bruker Microflex LT system and MALDI Biotyper Compass software with the V5.0.0.0 spectra library (5989 spectra).
Solutions of AMP (3 mg/mL), PIP (0.5 mg/mL), CTX (0.5 mg/mL), CAZ (0.25 mg/mL) and MEM (1 mg/mL) were prepared in incubation buffer (10 mM ammonium bicarbonate, 10 μg/mL zinc chloride, pH 8–9).
The antibiotics used for MBT STAR-BL measurement were selected according to the bacterial identification given by MALDI-TOF MS. If Enterobacteriaceae was identified, β-lactamase activity against AMP, CTX/CAZ and MEM were studied. For non-fermentative Gram-negative rods (NFGRs), including Acinetobacter spp. and Pseudomonas spp., β-lactamase activities against PIP, CAZ and MEM were investigated. β-lactamase activities against to all five β-lactam drugs was determined if no reliable ID was obtained.
For plated isolates, sample preparation was done according to Hrabak et al. (2012). For BC broths, a bacterial pellet was isolated using saponin-based extraction method. Briefly, 1 mL of BC broth was treated with 5% saponin and subsequently washed twice with distilled water. After centrifugation, the bacterial pellet was resuspended in 50 μL of the appropriate antibiotic solution and incubated at 37°C under agitation (900 rpm) for 2 h, followed by centrifugation at 2,000 rpm for 2 min to collect the supernatant.
One microliter of supernatant was applied to the MSP96 target plate (reactions were performed in quadruplicate). The dried spots were overlaid with 1 μL of MBT STAR-BL Matrix. Automated mass spectrometric measurements were performed using the STAR-BL module (RUO version) embedded in the MALDI Biotyper Compass software. For instrument calibration, an antibiotic calibration standard (ACS; Bruker Daltonik) was measured in parallel with the samples in each run.
The STAR-BL module automatically calculated the normalized logRQ values for each sample. A value ≤0.2 indicated negative drug hydrolysis (i.e., reported as susceptible), whereas a value ≥0.4 indicated positive β-lactamase activity (i.e., reported as resistant). Normalized logQR values between 0.2 and 0.4 indicated an indeterminate hydrolysis measurement requiring retesting. If the repeated test yielded the same logQR value, the results were reported as indeterminate.
Routine processing of BCs included the subcultivation of positive broths on Columbia Blood agar. The final identification involved a MALDI-TOF MS analysis of single isolated colonies grown on subculture plates.
Interim drug susceptibility patterns were determined by disk diffusion test directly using positive BC broths, whereas final drug susceptibilities relied on the disk diffusion testing patterns obtained from subcultured isolates (Clinical and Laboratory Standards Institute, 2013). Furthermore, the extended-spectrum β-lactamase (ESBL) and carbapenemase phenotypes were confirmed using a combined disk method (Clinical and Laboratory Standards Institute, 2013; Pournaras et al., 2013).
The presence of plasmid-mediated β-lactamases in all identified β-lactam-resistant Enterobacteriaceae was confirmed using multiplex PCR assays, followed by amplicon sequencing-based genotyping (Dallenne et al., 2010; Doyle et al., 2012). PCR sequencing was used to detect the mutations in the ampC promoter/attenuator region as well as in ampD and ampR which are associated with hyperproduction of AmpC in Enterobacteriaceae (Kaneko et al., 2005; Schmidtke and Hanson, 2006; Peter-Getzlaff et al., 2011). For NFGRs, carbapenemases were characterized using a dual-tube multiplex PCR (Kazi et al., 2015), whereas some rare β-lactamases, such as blaPom and blaL1, were detected as previously described (Thaller et al., 2011; Yang et al., 2014). For cases involving discrepancy between the phenotypic DST and MBT STAR-BL assays, the minimal inhibitory concentration (MIC) was determined using an E-test (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's guidelines.
The times-to-results were compared based on a subset of 153 monomicrobial BCs (n = 88) collected from a hospital adjacent to our laboratory that houses the MALDI Biotyper system with STAR-BL module. BCs that were identified as positive between 7:00 a.m. and 4:00 p.m. were collected at the hospital. MBT STAR-BL measurements were performed immediately upon sample arrival in our laboratory. Time-zero was defined as the time at which the primary Gram staining result was reported. For MALDI-TOF MS analysis, the time elapsed between time-zero and the MBT STAR-BL analysis completion time was considered the “Time-to-MBT STAR-BL.” For routine microbiological processing, the total time required to obtain bacterial identification and the results of the direct (interim) disk diffusion test was defined as the “Time-to-interim report,” whereas the total time required to obtain the final drug susceptibility result from subcultured isolates was defined as the “Time-to-final report” (Figure 1).
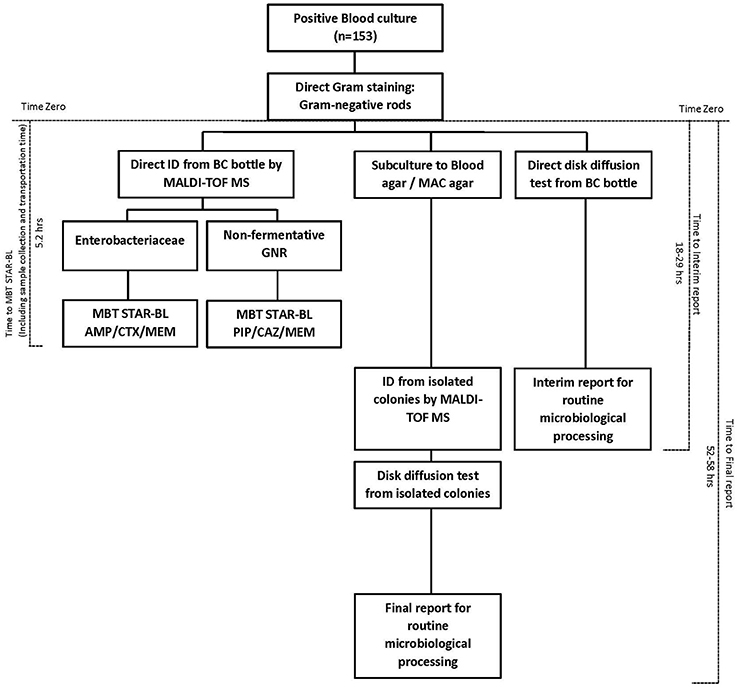
Figure 1. Diagram of the workflow and turnaround time of bacterial identification and β-lactam resistance detection from positive blood cultures, using MBT STAR-BL and routine microbiological processing.
Among 139 Enterobacteriaceae collected in this study, 103 were AMP-resistant, as determined by phenotypic DST, and 94 (91.3%) exhibited positive hydrolysis in the MBT STAR-BL module. Notably, 63/66 (95.5%) AMP-resistant E. coli isolates were correctly identified by MBT STAR-BL with 100% specificity (Table 1).
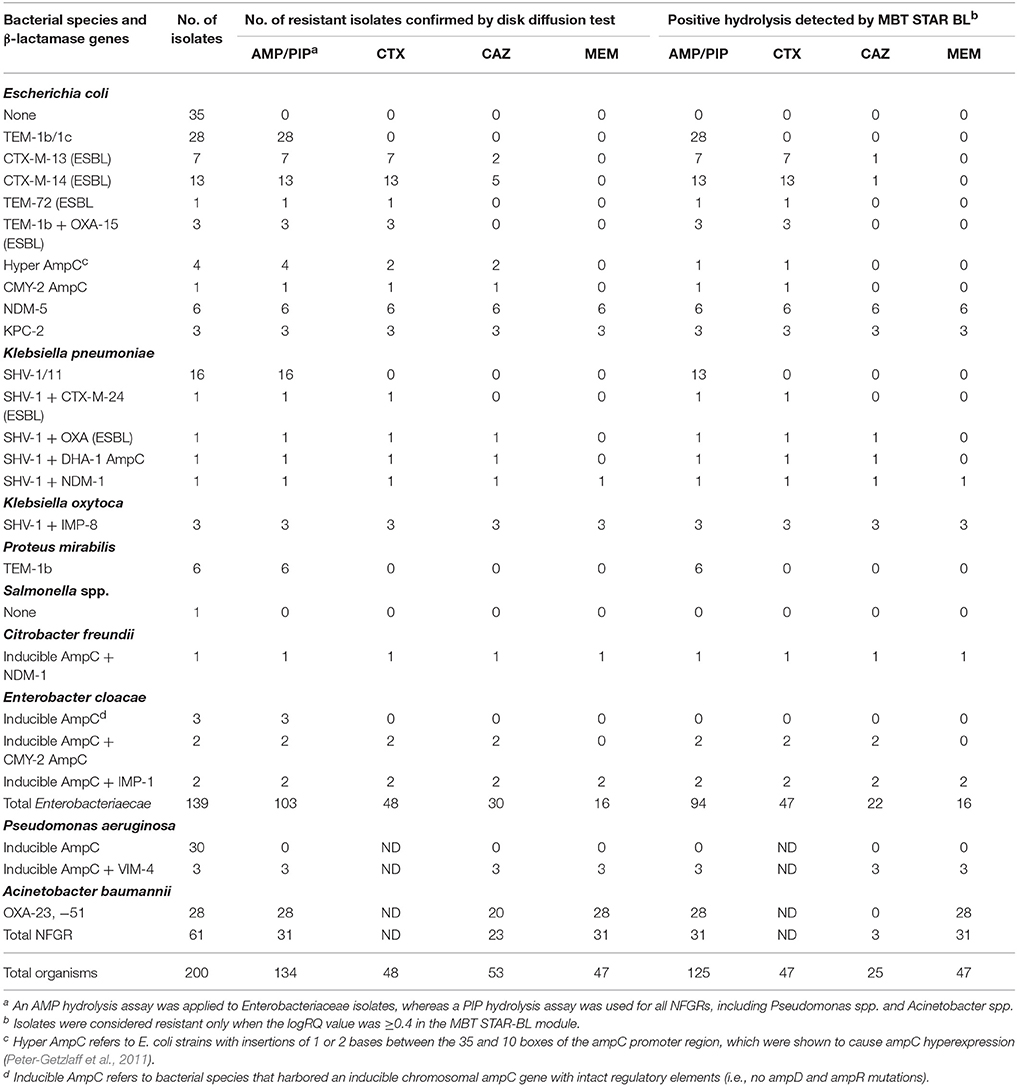
Table 1. Performance of the MBT STAR-BL module for the detection of β-lactamase-mediated resistance in cultured isolates.
MBT STAR-BL successfully identified 47/48 (97.9%) CTX-resistant and 22/30 (73.3%) CAZ-resistant Enterobacteriaceae isolates with no false positivity (Table 1). Interestingly, all 26 ESBL-producing Enterobacteriaceae demonstrated positive CTX hydrolysis, whereas only 3 (11.5%) could hydrolyze CAZ.
Remarkably, all carbapenemase-producing Enterobacteriaceae (n = 16) isolates were correctly identified by MBT STAR-BL with 100% specificity (Table 1).
For NFGRs, MBT STAR-BL successfully detected PIP and MEM hydrolysis in all A. baumannii isolates harboring blaOXA−23 and blaOXA−51 (n = 28) and all P. aeruginosa isolates harboring blaVIM (n = 3) (Table 1). However, all A. baumannii isolates harboring blaOXA−23 and blaOXA−51 failed to exhibit CAZ hydrolysis in MBT-STAR-BL.
The characteristics of the bacterial cultures with discrepant results are shown in Table 4.
A total of 153 positive BC bottles, including monomicrobial (n = 134) and polymicrobial cultures (n = 19), were prospectively collected for this study.
Direct MALDI-TOF MS correctly identified bacteria in 116/134 (86.6%) monomicrobial BCs, and achieved species-level identification in 87/134 (64.9%) (Table S1).
Among 112 monomicrobial BCs harboring AMP-resistant Enterobacteriaceae, 90 (80.4%) exhibited AMP hydrolysis in MBT STAR-BL (Table 2). False negative results were mainly obtained from species that harbored inducible chromosomal ampC (Table 4). For E. coli, 58/63 (92.1%) AMP-resistant cultures were correctly identified by MBT STAR-BL, with the specificity of 92.9% (Table 5).
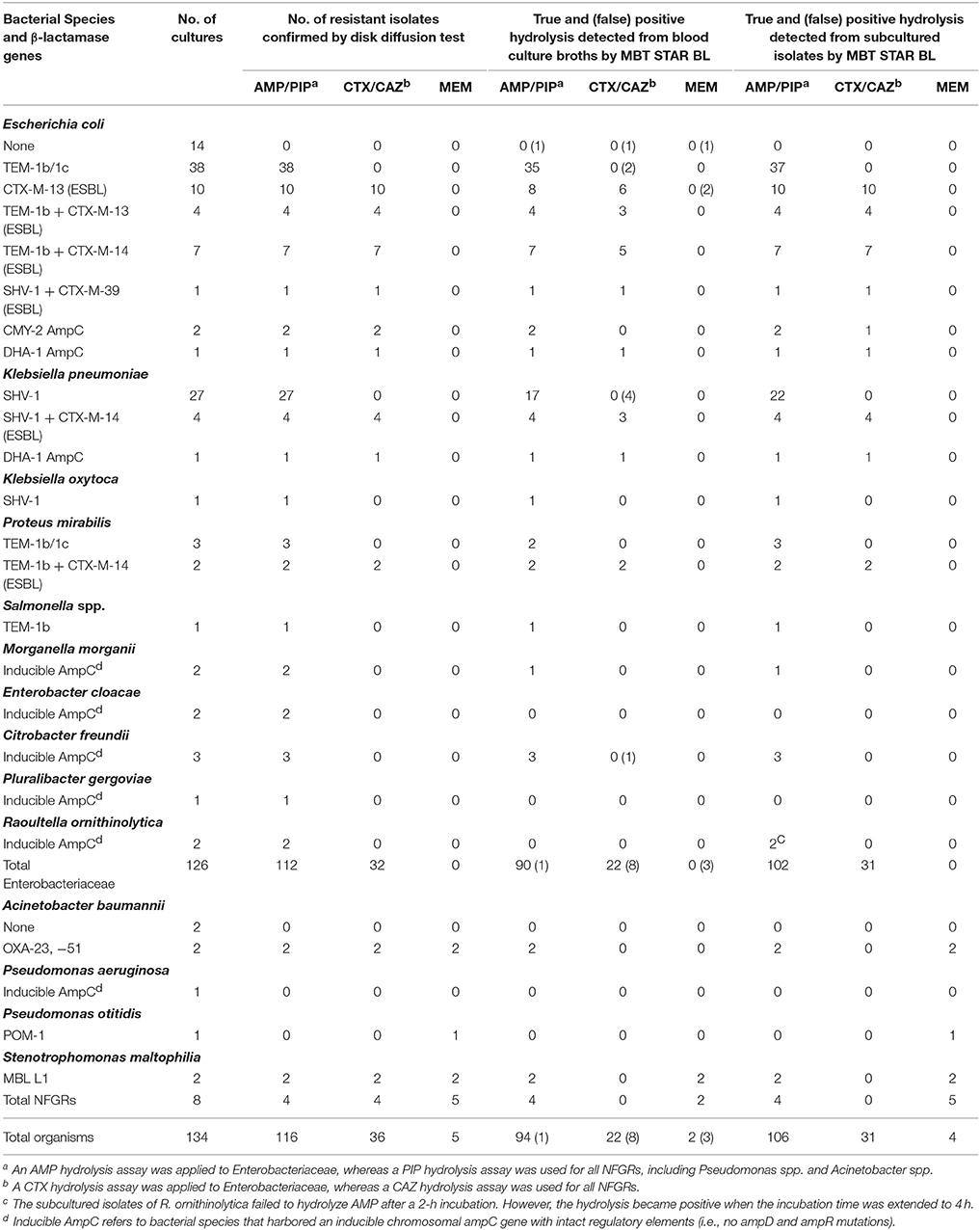
Table 2. Performance of the MBT STAR-BL module in the detection of β-lactamase-mediated resistance in prospectively collected monomicrobial blood cultures.
In addition, MBT STAR-BL successfully detected CTX resistant Enterobacteriaceae in 22/32 (68.8%) BCs (Table 2). Notably, of 28 ESBL-producing Enterobacteriaceae, 20 (71.4%) demonstrated positive CTX hydrolysis (Table 2). The false negative MBT STAR-BL results did not correlate with CTX-M genotypes or MIC levels (Table 4). Moreover, eight BCs containing CTX-susceptible Enterobacteriaceae yielded false positive hydrolysis results, resulting in a specificity of 91.5% (Table 5).
Unfortunately, no carbapenem-resistant Enterobacteriaceae were collected from BCs in this study.
For NFGRs, four BCs were resistant to PIP and CAZ as determined by phenotypic DST. MBT STAR-BL correctly predicted PIP resistance for all of them, but none of the BCs showed detectable CAZ hydrolysis in MBT STAR-BL. In addition, among 5 NFGRs that haboured carbapenemase genes and exhibited MEM resistance in phenotypic DST, MBT STAR-BL only detected MEM hydrolysis in 2 (40%) of the BCs (Table 2).
The characteristics of the BCs with discrepant results are shown in Table 4.
MALDI-TOF MS analyses were repeated for each sample using plated isolates subcultivated from BCs on the following day. The diagnostic sensitivities and specificities of MBT STAR-BL for the subcultured plates resembled those obtained from the archived cultured isolates (Tables 2, 5).
Direct MALDI-TOF MS correctly identified at least 1 bacterial species in 16/19 (84.2%) polymicrobial BCs (Table 3).
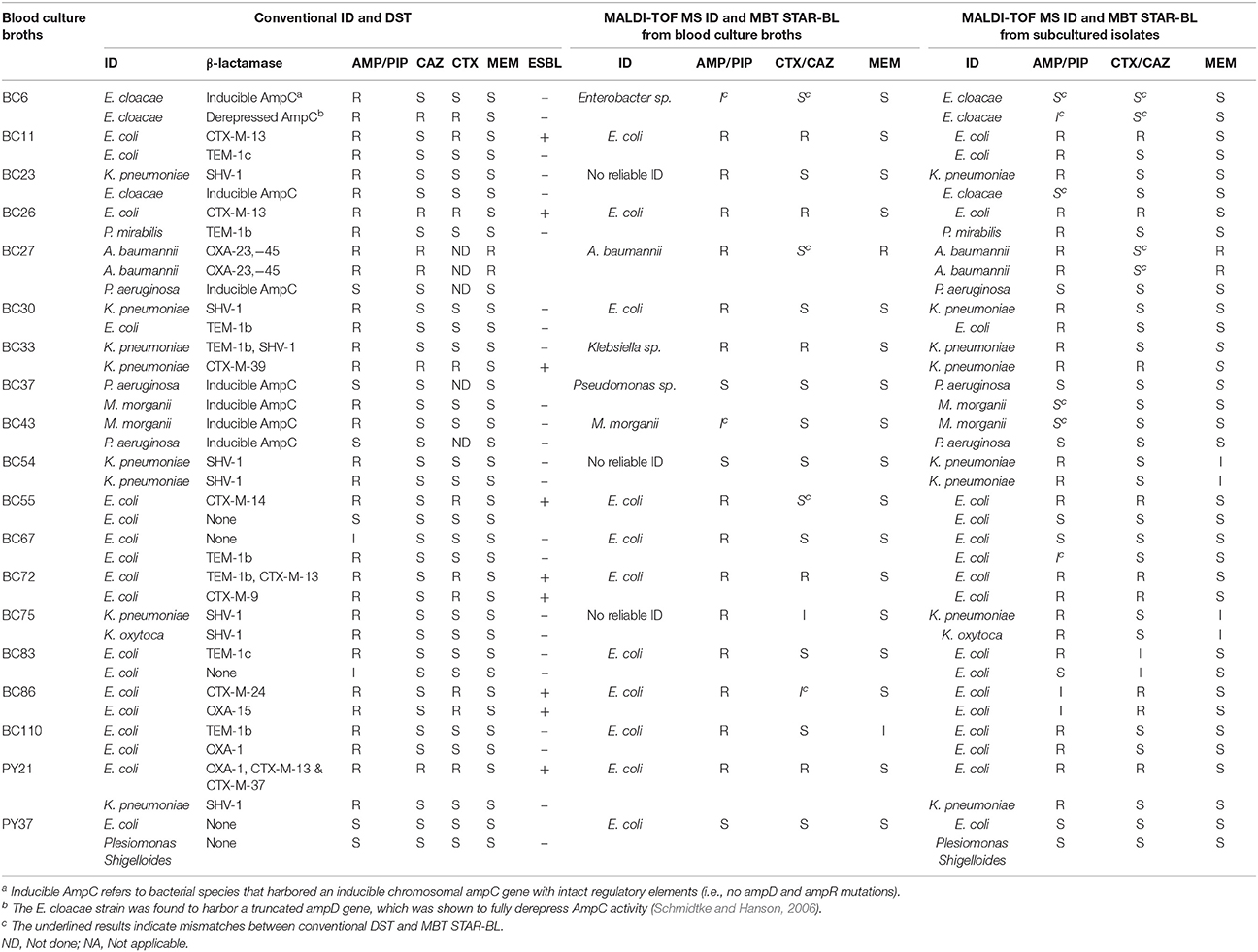
Table 3. Performance of the MALDI-TOF MS workflow for bacterial identification and β-lactamase-mediated resistance detection in polymicrobial blood cultures and respective subcultured isolates.
Additionally, MBT STAR-BL successfully detected AMP hydrolysis in all 10 BCs containing AMP-resistant E. coli strains. Among the 7 polymicrobial BCs containing ESBL-producing Enterobacteriaceae, 5 (71.4%) exhibited CTX hydrolysis in an MBT STAR-BL test (Table 3).
Full concordance was yielded between the MBT STAR-BL and phenotypic DST in prediction of PIP resistance in polymicrobial BCs containing NFGRs (Table 3). For the BCs containing two A. baumannii strains harboring blaOXA−23&51, MBT STAR-BL successfully identified MEM hydrolysis in both the BC broths and subcultured isolates (Table 3). Overall, the β-lactam resistance patterns predicted by MBT STAR-BL were concordant with the phenotypic DST in 14/19 (73.7%) polymicrobial BCs. Details of the mismatches are shown in Table 4.
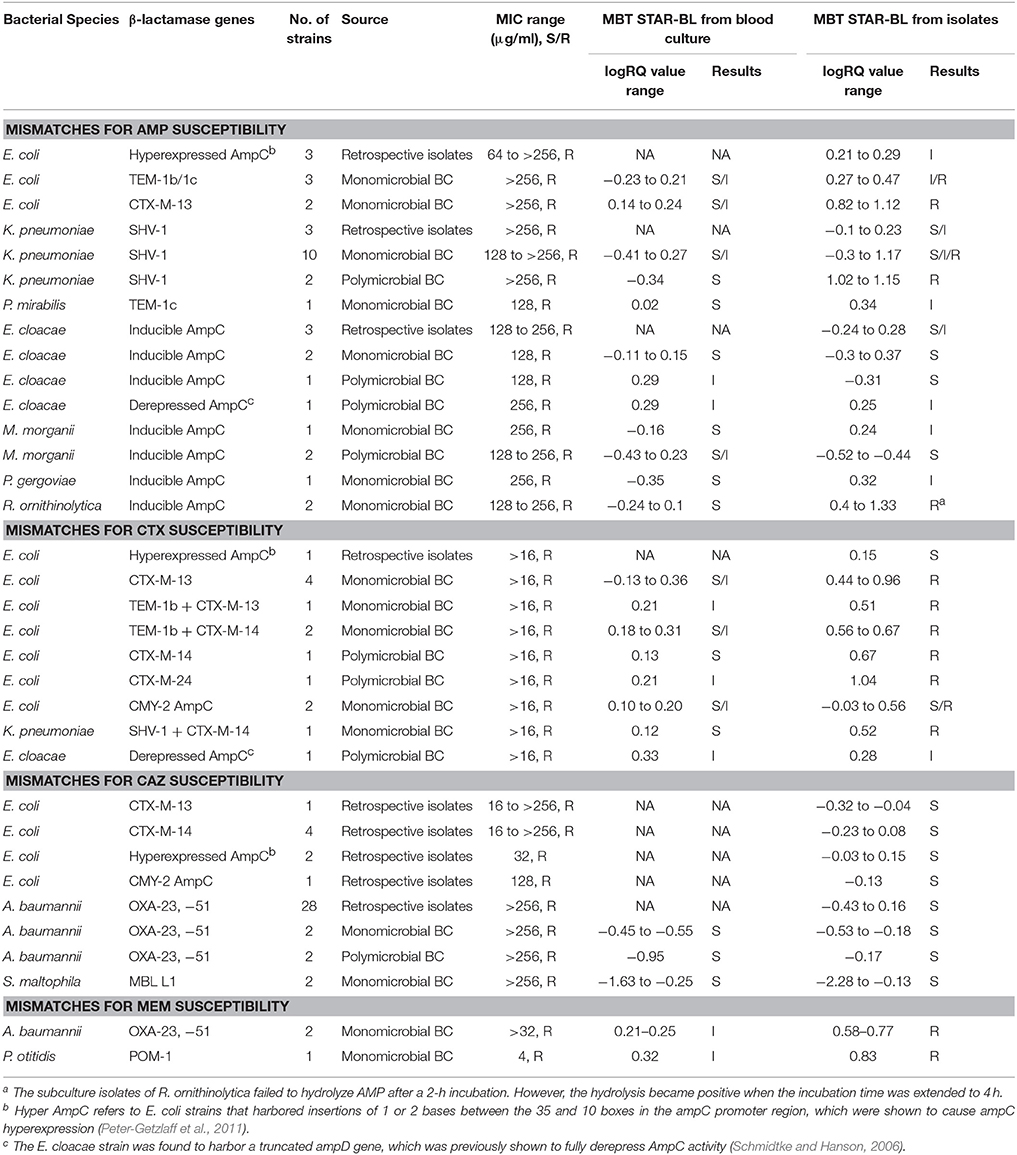
Table 4. The characteristics of bacterial strains with discrepant results from the phenotypic drug susceptibility test and MBT STAR-BL analysis.
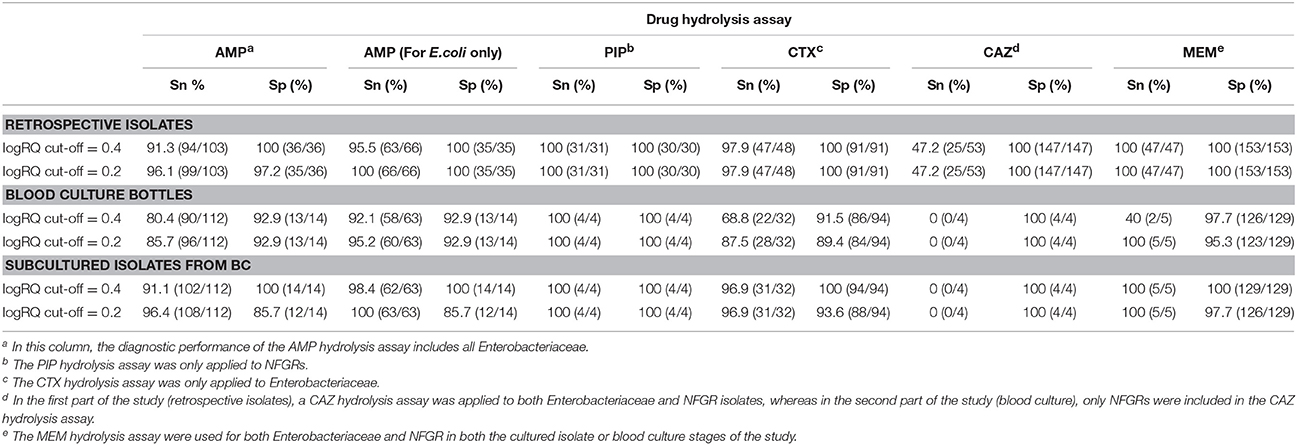
Table 5. The overall diagnostic performances of drug hydrolysis assays for cultured isolates and blood cultures at logRQ cut-off values of 0.4 and 0.2.
The times-to-results based on MBT STAR-BL and routine microbiological processing were investigated for 88 monomicrobial BCs (Table 6). Among the BCs, the average time to MBT STAR-BL was 5.2 h. For Enterobacteriaceae, the MALDI-TOF MS-based workflow allowed the laboratory to identify bacterial species and β-lactamase-mediated resistance to β-lactams at averages of 13.5 and 47.5 h before the interim and final reporting of the routine processing, respectively. For NFGRs, the MBT STAR-BL assay enabled the reporting of bacterial identification and MEM hydrolysis at 23.0 and 49.9 h before the interim and final routine processing, respectively.
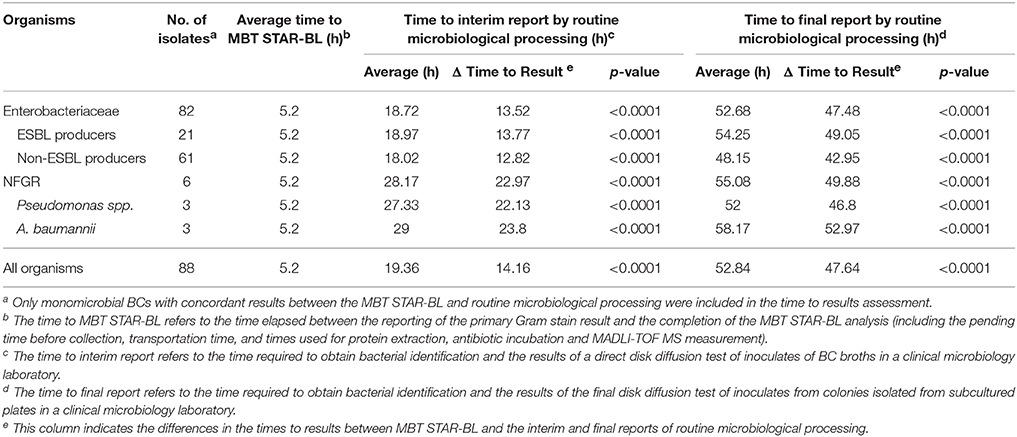
Table 6. Assessment of the times to results, based on MBT STAR-BL and routine microbiological processing.
In this study, we evaluated the practicality of MALDI-TOF MS using the 5989 spectra library, followed by MBT STAR-BL as a diagnostic workflow, for the identification of bacterial species and β-lactamase-mediated resistance in cultured isolates and patient-derived BCs. In contrast to previous studies that mainly investigated origin of carbapenem resistance in Enterobacteriaceae (Hrabak et al., 2011, 2012; Papagiannitsis et al., 2015; Ghebremedhin et al., 2016; Monteferrante et al., 2016; Oviano et al., 2017), this study identified the susceptibilities of a wide spectrum of bacterial species harboring different enzymes. To the best of our knowledge, this is the most comprehensive evaluation of the MBT STAR-BL module since its official launch in 2016.
AMP is used as a surrogate marker of resistance to aminopenicillins in E. coli and other Enterobacteriaceae, such as Proteus mirabilis, Salmonella spp. and Shigella spp. In this study, AMP resistance was detected in E.coli from cultured isolates and BCs at detection sensitivities of 95.5 and 92.1%, respectively (Table 5), consistent with Jung et al. (2014). Although we did not observe a correlation between the logRQ values and MIC levels, we noted that all the strains with high MIC level but low logRQ value are those harboring mutations associated with hyperproduction of AmpC (Table 4). An analysis of the respective mass spectra revealed that the peaks corresponding to hydrolyzed AMP were weak and coexistent with the molecular peaks of intact antibiotics, indicating incomplete drug hydrolysis (Figure 2). We repeated this assay for all chromosomal ampC carriers after extending the incubation time to 4 h. Unfortunately, only two R. ornithinolytica isolates yielded logRQ values ≥ 0.4 whereas most others had values of 0.2–0.4.
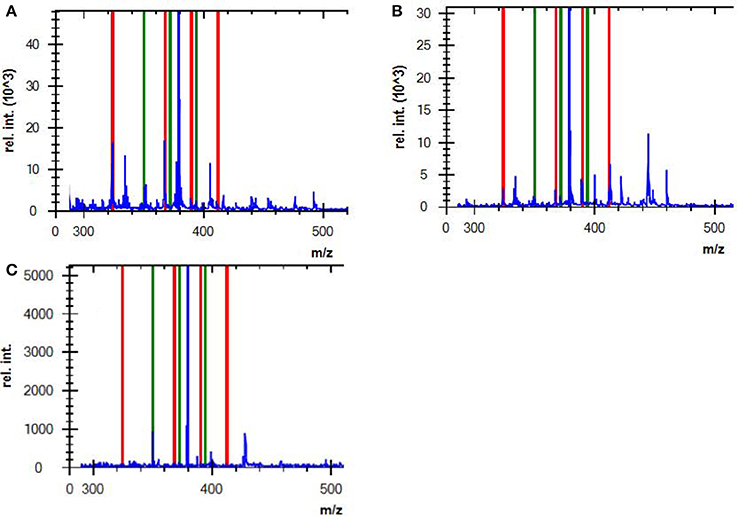
Figure 2. Mass spectra of (A) a complete AMP hydrolysis obtained from a positive control strain (ATCC BAA-2452, NDM-1-producing E. coli) incubated with AMP for 2 h, (B) an incomplete AMP hydrolysis obtained from an E. coli strain that hyperexpressed ampC, and (C) non-hydrolyzed AMP from a negative control strain (DH5α). Red lines indicate the hydrolyzed antibiotic peaks, and green lines indicate intact antibiotic peaks (non-hydrolyzed).
Both CTX and CAZ could be used as indicator drugs for the MBT STAR-BL testing of resistance to third-generation cephalosporin. Consistent with a previous study (Mackenzie et al., 2002), the CAZ hydrolysis assay in our study yielded a high false negative rate in both cultured isolates and BCs, indicating that it might not be appropriate for predicting phenotypic drug resistance. Conversely, the CTX hydrolysis assay successfully identified 100% of ESBL- and 83.3% of AmpC-producing bacterial isolates, suggesting that CTX is a better surrogate marker for the detection of ESBL- and plasmid-mediated AmpC β-lactamase activity. However, the direct detection of ESBL from BCs yielded a CTX hydrolysis sensitivity of only 71.4%, lower than that reported by Jung et al. (2014). We attribute this disagreement to the different logRQ cut-off values used in the two studies (≥0 in Jung et al. vs. ≥0.4 in this study). In fact, 5/7 ESBL producers with false negative results in this study had logRQ values of 0.2–0.4. We note that antibiotic therapy prior to BC collection might also contribute to the occurrence of false negativity in a drug hydrolysis assay. At least two patients from whom the ESBL producers were not detected from BCs by MBT STAR-BL were receiving carbapenem treatment, which might have reduced the viable number of bacterial cells in the BC broths and led to suboptimal CTX and AMP hydrolysis.
Many previous studies involving the mass spectrometric detection of β-lactamase excluded NFGRs, as permeability- and efflux-based resistance mechanisms play major roles in drug resistance in these species (Kumar and Schweizer, 2005). However, NFGRs are important pathogens, and data regarding the ability of MBT STAR-BL to detect drug-resistant NFGR would be useful for routine diagnostic purposes. In our study, the PIP hydrolysis assay correlated perfectly with the phenotypic resistance test for all cultured isolates and BCs. Regarding the MEM hydrolysis assay, MBT STAR-BL correctly identified all carbapenem-resistant NFGRs in cultured isolates. However, in direct BCs, the results for two A. baumannii strains harboring blaOXA−23, −51 and one P. otitidis strain harboring blaPOM−1 were interpreted as indeterminate (logRQ: 0.2–0.4). We attributed the low logRQ values to the low recovery of bacterial cells after extraction and the reported low cell permeability of NFGR (Van Looveren et al., 2004). A previous report suggested that the inclusion of 0.005% SDS in the incubation buffer might have enhanced drug hydrolysis by perforating the membranes of NFGRs (Oviano et al., 2017).
This study also features the unique inclusion of polymicrobial BCs in a full clinical evaluation. The MBT STAR-BL module correctly predicted β-lactam resistance in 14/19 BCs, including those containing ESBL-producing Enterobacteriaceae and carbapenemase-producing NFGRs. These results indicate that the module could help to select the most appropriate antibiotic therapy for patients with polymicrobial bacteremia.
Using the current MBT STAR-BL setting, logRQ values of 0.2–0.4 indicate ambiguous drug hydrolysis and should be reported as indeterminate. However, our findings demonstrate that three AmpC-hyperproducing E. coli isolates, six Enterobacteriaceae harboring blaCTX−M and three carbapenemase-producing NFGRs in BCs had logRQ values within this range. A reduction of the cut-off value to 0.2 could increase the detection sensitivities and identify these clinically important organisms, although the specificities would be slightly compromised. This cut-off value reduction led to increases in the sensitivities of the AMP (E. coli only), CTX and MEM hydrolysis assays from 92.1, 68.8, and 40% to 95.2, 87.5, and 100%, respectively, for BCs (Table 5). Regarding patient safety, the test aims to rapidly provide information that will lead to effective antibiotic therapy. Accordingly, the sensitivity of the test is more important than the specificity. We therefore recommended eliminating the indeterminate range and setting the logRQ cut-off value at 0.2.
We additionally modified the manufacturer's protocol by using 5% saponin instead of the MALDI Sepsityper kit for BC extraction in MBT STAR-BL test. This is the first study to apply this protocol to the direct detection of β-lactamase-mediated resistance in BCs. Our method yielded similar sensitivities and specificities for AMP, CTX and MEM hydrolysis assay as those reported in previous studies using the Sepsityper kit, if the same logRQ cut-off value was applied (Jung et al., 2014; Oviano et al., 2017). Additionally, the use of 5% saponin (<US$1) is a less expensive option for extraction, compared with the Sepsityper kit (US$17).
In this study, the MBT STAR-BL module was not installed in a clinical laboratory, but rather in an adjacent research laboratory. The time required for sample collection and transportation led to longer turnaround times for MBT STAR-BL measurements in this study (5.2 h) than the value claimed by the manufacturer (2.5 h). Nevertheless, the assay still greatly reduced the turnaround times required to identify β-lactamase-producing organisms in patient-derived BCs. Particularly, drug hydrolysis assays could confirm the presence of ESBL-producing Enterobacteriaceae and carbapenemase-producing NFGRs in BCs at an average of 14 and 48 h before the interim and final reports of routine microbiological processing, respectively, were made available. It should be noted that the time saved by MBT STAR-BL might not be as pronounced as we described for laboratories where the final antibiogram is determined by automated microbroth system or disk diffusion test prepared after short-term incubation (e.g., 6 h) on solid media.
Although MBT STAR-BL cannot provide a full antibiogram, elevated logRQ values suggest the likelihood of ESBL and carbapenemase production. On the other hand, owing to high accuracy in prediction of AMP resistance in E. coli, de-escalation of antibiotic therapy might be considered when AMP hydrolysis is negative as determined by MBT STAR-BL. The short assay turnaround time allows the fine-tuning of antibiotic therapy on the same day that a positive blood culture is identified.
There were two weaknesses of this study. First, we could not determine the performance of MBT STAR-BL for the direct detection of carbapenemase-producing Enterobacteriaceae (CPE) in patient-derived BCs, as no CPE were isolated from positive BCs during the study period. Considering that all the archived KPC-, NDM-, and IMP-producing Enterobacteriaceae isolates were correctly detected by MBT STAR-BL in the first part of our study, the analysis module should be capable of identifying CPE regardless of the enzyme type. Second, as it is the first evaluation study of MBT STAR-BL in our locality, the test results were not used to modify the treatment regimen in our hospitals. Therefore we could not determine the impact of the MBT STAR-BL on the patient outcome, such as change in hospital stay, sepsis related mortality, and the cost of care. A large-scale randomized controlled trial study is recommended to further investigate how the implementation of MBT STAR-BL into routine workflow can benefit the patient management.
In conclusion, the MALDI Biotyper system, when equipped with the MBT STAR-BL module, enables the rapid and simultaneous identification of bacterial species and β-lactamase-mediated resistance from BCs and cultured isolates. A reduction of the logRQ cut-off value to 0.2 significantly increased the detection sensitivities for clinically important pathogens. Finally, the low reagent costs and short turnaround time suggest that this test could be used as a tool for early therapeutic guidance in patients with infection.
The biological safety approval was obtained from Health, Safety and Environment Office of The Hong Kong Polytechnic University (Ref. number: RSA15096).
AL, JL, and GS conceived and designed the experiments, performed the experiments, analyzed the data and wrote the paper. RiL, WN, EL, VL, PS, and RR performed the experiments and analyzed the data. KF, WT, RoL, and DT read and approved the final version of the manuscript.
This work was supported by The Hong Kong Polytechnic University Large Equipment Fund (Ref# HTI-4) and HTI Seed Fund (Ref#99TX), which funded the Microflex LT biotyper instrument and reagents respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to express our gratitude to the Bruker Daltonik specialists, Mr. Lee Chi Chung and Dr. Yeung Hoi Sze Betty, for their technical support and advice throughout the project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00334/full#supplementary-material
Burckhardt, I., and Zimmermann, S. (2011). Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49, 3321–3324. doi: 10.1128/JCM.00287-11
Chen, J. H., Ho, P. L., Kwan, G. S., She, K. K., Siu, G. K., Cheng, V. C., et al. (2013). Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J. Clin. Microbiol. 51, 1733–1739. doi: 10.1128/JCM.03259-12
Clerc, O., Prod'hom, G., Vogne, C., Bizzini, A., Calandra, T., and Greub, G. (2013). Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with Gram-negative bacteremia: a prospective observational study. Clin. Infect. Dis. 56, 1101–1107. doi: 10.1093/cid/cis1204
Clinical and Laboratory Standards Institute (2013). “Performance standards for antimicrobial susceptibility testing,” in 23rd Informational Supplemental, CLSI M100-S23 (Wayne, PA: Clinical and Laboratory Standards Institute).
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dellinger, R. P., Levy, M. M., Carlet, J. M., Bion, J., Parker, M. M., Jaeschke, R., et al. (2008). Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit. Care Med. 36, 296–327. doi: 10.1097/01.CCM.0000298158.12101.41
Doyle, D., Peirano, G., Lascols, C., Lloyd, T., Church, D. L., and Pitout, J. D. (2012). Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50, 3877–3880. doi: 10.1128/JCM.02117-12
Fleischmann, C., Scherag, A., Adhikari, N. K., Hartog, C. S., Tsaganos, T., Schlattmann, P., et al. (2016). Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am. J. Respir. Crit. Care Med. 193, 259–272. doi: 10.1164/rccm.201504-0781OC
Ghebremedhin, B., Halstenbach, A., Smiljanic, M., Kaase, M., and Ahmad-Nejad, P. (2016). MALDI-TOF MS based carbapenemase detection from culture isolates and from positive blood culture vials. Ann. Clin. Microbiol. Antimicrob. 15:5. doi: 10.1186/s12941-016-0120-x
Hrabak, J., Studentova, V., Walkova, R., Zemlickova, H., Jakubu, V., Chudackova, E., et al. (2012). Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50, 2441–2443. doi: 10.1128/JCM.01002-12
Hrabak, J., Walkova, R., Studentova, V., Chudackova, E., and Bergerova, T. (2011). Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49, 3222–3227. doi: 10.1128/JCM.00984-11
Jung, J. S., Popp, C., Sparbier, K., Lange, C., Kostrzewa, M., and Schubert, S. (2014). Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid detection of beta-lactam resistance in Enterobacteriaceae derived from blood cultures. J. Clin. Microbiol. 52, 924–930. doi: 10.1128/JCM.02691-13
Kaneko, K., Okamoto, R., Nakano, R., Kawakami, S., and Inoue, M. (2005). Gene mutations responsible for overexpression of AmpC beta-lactamase in some clinical isolates of Enterobacter cloacae. J. Clin. Microbiol. 43, 2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005
Kang, C. I., Kim, S. H., Park, W. B., Lee, K. D., Kim, H. B., Kim, E. C., et al. (2005). Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 49, 760–766. doi: 10.1128/AAC.49.2.760-766.2005
Kazi, M., Shetty, A., and Rodrigues, C. (2015). The carbapenemase menace: do dual mechanisms code for more resistance? Infect. Control Hosp. Epidemiol. 36, 116–117. doi: 10.1017/ice.2014.33
Kumar, A. (2011). Optimizing antimicrobial therapy in sepsis and septic shock. Crit. Care Nurs. Clin. North Am. 23, 79–97. doi: 10.1016/j.ccell.2010.12.005
Kumar, A., Roberts, D., Wood, K. E., Light, B., Parrillo, J. E., Sharma, S., et al. (2006). Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34, 1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9
Kumar, A., and Schweizer, H. P. (2005). Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Deliv. Rev. 57, 1486–1513. doi: 10.1016/j.addr.2005.04.004
Livermore, D. M. (2012). Current epidemiology and growing resistance of gram-negative pathogens. Korean J. Intern. Med. 27, 128–142. doi: 10.3904/kjim.2012.27.2.128
Mackenzie, F. M., Miller, C. A., and Gould, I. M. (2002). Comparison of screening methods for TEM- and SHV-derived extended-spectrum beta-lactamase detection. Clin. Microbiol. Infect. 8, 715–724. doi: 10.1046/j.1469-0691.2002.00473.x
Monteferrante, C. G., Sultan, S., Ten Kate, M. T., Dekker, L. J., Sparbier, K., Peer, M., et al. (2016). Evaluation of different pretreatment protocols to detect accurately clinical carbapenemase-producing Enterobacteriaceae by MALDI-TOF. J. Antimicrob. Chemother. 71, 2856–2867. doi: 10.1093/jac/dkw208
Oviano, M., Ramirez, C. L., Barbeyto, L. P., and Bou, G. (2017). Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J. Antimicrob. Chemother. 72, 1350–1354. doi: 10.1093/jac/dkw579
Papagiannitsis, C. C., Studentova, V., Izdebski, R., Oikonomou, O., Pfeifer, Y., Petinaki, E., et al. (2015). Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J. Clin. Microbiol. 53, 1731–1735. doi: 10.1128/JCM.03094-14
Paterson, D. L. (2006). Resistance in gram-negative bacteria: enterobacteriaceae. Am. J. Infect. Control 34, S20–S28. discussion: S62–S70. doi: 10.1016/j.ajic.2006.05.238
Peter-Getzlaff, S., Polsfuss, S., Poledica, M., Hombach, M., Giger, J., Bottger, E. C., et al. (2011). Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J. Clin. Microbiol. 49, 2924–2932. doi: 10.1128/JCM.00091-11
Pournaras, S., Zarkotou, O., Poulou, A., Kristo, I., Vrioni, G., Themeli-Digalaki, K., et al. (2013). A combined disk test for direct differentiation of carbapenemase-producing enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 51, 2986–2990. doi: 10.1128/JCM.00901-13
Schmidtke, A. J., and Hanson, N. D. (2006). Model system to evaluate the effect of ampD mutations on AmpC-mediated beta-lactam resistance. Antimicrob. Agents Chemother. 50, 2030–2037. doi: 10.1128/AAC.01458-05
Schubert, S., Weinert, K., Wagner, C., Gunzl, B., Wieser, A., Maier, T., et al. (2011). Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagn. 13, 701–706. doi: 10.1016/j.jmoldx.2011.07.004
Sparbier, K., Schubert, S., Weller, U., Boogen, C., and Kostrzewa, M. (2012). Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 50, 927–937. doi: 10.1128/JCM.05737-11
Thaller, M. C., Borgianni, L., Di Lallo, G., Chong, Y., Lee, K., Dajcs, J., et al. (2011). Metallo-beta-lactamase production by Pseudomonas otitidis: a species-related trait. Antimicrob. Agents Chemother. 55, 118–123. doi: 10.1128/AAC.01062-10
Van Looveren, M., Goossens, H., and Group, A. S. (2004). Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10, 684–704. doi: 10.1111/j.1469-0691.2004.00942.x
Wuppenhorst, N., Consoir, C., Lorch, D., and Schneider, C. (2012). Direct identification of bacteria from charcoal-containing blood culture bottles using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2843–2850. doi: 10.1007/s10096-012-1638-2
Keywords: MBT STAR-BL, MALDI-TOF MS, drug resistance, bacterial, drug hydrolysis test, beta-lactamases, blood culture
Citation: Lee AWT, Lam JKS, Lam RKW, Ng WH, Lee ENL, Lee VTY, Sze PP, Rajwani R, Fung KSC, To WK, Lee RA, Tsang DNC and Siu GKH (2018) Comprehensive Evaluation of the MBT STAR-BL Module for Simultaneous Bacterial Identification and β-Lactamase-Mediated Resistance Detection in Gram-Negative Rods from Cultured Isolates and Positive Blood Cultures. Front. Microbiol. 9:334. doi: 10.3389/fmicb.2018.00334
Received: 31 August 2017; Accepted: 12 February 2018;
Published: 23 February 2018.
Edited by:
Karsten Becker, Universität Münster, GermanyReviewed by:
Jaroslav Hrabak, Charles University, CzechiaCopyright © 2018 Lee, Lam, Lam, Ng, Lee, Lee, Sze, Rajwani, Fung, To, Lee, Tsang and Siu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilman K. H. Siu, Z2lsbWFuLnNpdUBwb2x5dS5lZHUuaGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.