
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 March 2018
Sec. Virology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00303
This article is part of the Research Topic Genome Invading RNA Networks View all 17 articles
Avian-like H5N1 canine influenza virus (CIV) causes severe respiratory infections in dogs. However, the mechanism underlying H5N1 CIV infection in dogs is unknown. The present study aimed to identify differentially expressed miRNAs and mRNAs in the lungs and trachea in H5N1 CIV-infected dogs through a next-generation sequencing-based method. Eighteen 40-day-old beagles were inoculated intranasally with CIV, A/canine/01/Guangdong/2013 (H5N1) at a tissue culture infectious dose 50 (TCID50) of 106, and lung and tracheal tissues were harvested at 3 and 7 d post-inoculation. The tissues were processed for miRNA and mRNA analysis. By means of miRNA-gene expression integrative negative analysis, we found miRNA–mRNA pairs. Lung and trachea tissues showed 138 and 135 negative miRNA–mRNA pairs, respectively. One hundred and twenty negative miRNA–mRNA pairs were found between the different tissues. In particular, pathways including the influenza A pathway, chemokine signaling pathways, and the PI3K-Akt signaling pathway were significantly enriched in all groups in responses to virus infection. Furthermore, dysregulation of miRNA and mRNA expression was observed in the respiratory tract of H5N1 CIV-infected dogs and notably, TLR4 (miR-146), NF-κB (miR-34c) and CCL5 (miR-335), CCL10 (miR-8908-5p), and GNGT2 (miR-122) were found to play important roles in regulating pathways that resist virus infection. To our knowledge, the present study is the first to analyze miRNA and mRNA expression in H5N1 CIV-infected dogs; furthermore, the present findings provide insights into the molecular mechanisms underlying influenza virus infection.
Although the natural host of influenza A virus is wild aquatic birds, the host barrier is not unbreakable, and the virus can be transmitted to other species, including dogs (Klenk, 2014). Canine influenza virus (CIV) had not been reported until 2004, and it was first discovered in racing Greyhounds in the United States. The first identified CIV was confirmed as subtype H3N8 by sequencing (Crawford et al., 2005). In addition, another subtype of CIV (H3N2) was isolated from sick farmed and pet dogs in South Korea (SK) in 2007 (Lee et al., 2009; Li et al., 2010). Moreover, multiple subtypes of influenza A viruses are reported to infect dogs, including the deadly H5N1 avian influenza virus (Songserm et al., 2006b), influenza A (H1N1) pdm09 virus (Damiani et al., 2012; Su et al., 2014a,b; Yin et al., 2014), H10N8 avian influenza virus (Su et al., 2014b), avian-like H9N2 influenza A virus (Songserm et al., 2006a; Sun et al., 2013), avian-like CIV (Su S. et al., 2013), and H3N2 influenza virus with the PA gene of H9N2 avian influenza virus (Lee et al., 2016). Studies have reported successful experimental infection of CIV in guinea pigs and mice (Tu et al., 2009; Castleman et al., 2010). These findings indicate that dogs may represent a new bridging species for avian and human influenza viruses for interspecies transmission.
In October 2004, highly pathogenic avian influenza (HPAI) H5N1 virus was reported for the first time in Thailand in a dog with severe lung congestion and edema and bloody nasal discharge (Songserm et al., 2006b). Furthermore, previous studies have reported that H5N1 influenza virus infections in dogs have resulted in anorexia, fever, conjunctivitis, labored breathing, and coughing (Songserm et al., 2006b; Maas et al., 2007; Chen et al., 2010; Ashour et al., 2012). Being considered “man’s best friend,” dogs drastically influence human lives. However, influenza viruses are susceptible to mutation; hence, it is important to understand the pathogenesis of H5N1 influenza virus infection among dogs.
MiRNAs are small non-coding RNAs. MiRNA binds on target mRNA to negatively regulate biological processes such as differentiation (Shivdasani, 2006), proliferation (Song et al., 2010), growth (Suh et al., 2015), metabolism (Gomez et al., 2014; Meydan et al., 2016), and apoptosis (Cheng, 2005). Increasing evidence has shown that deregulation of miRNA contributes to antiviral activity during an influenza virus infection. MiR-342-5p participates in macrophage interferon (IFN) antiviral responses against multiple viruses including influenza A (H1N1) (Ploegh et al., 2016). MiR-146a up-regulation significantly decreases H1N1 and H3N2 viral propagation (Terrier et al., 2013). MiR-144 was reported to post-transcriptionally decrease TRAF6 levels to attenuate the antiviral response (Lopez et al., 2017). MiR-485 binds PB1 transcripts in H5N1 to inhibit viral replication (Ingle et al., 2015). These findings suggest that certain miRNAs may play an important role in regulating influenza virus infections in dogs.
Strikingly, recent studies have assumed that miRNAs not only develop functions but also transmit from one species to another, thereby promoting crosstalk (Zhang et al., 2016) and interfering in signal transmission and communication (Hansen et al., 2010). Numerous studies have reported that viruses adapt their own miRNAs based on host miRNA (Pfeffer et al., 2004; Cai et al., 2005; Pfeffer et al., 2005) and establish an environment conducive to viral replication (Grey et al., 2007; Samols et al., 2007; Stern-Ginossar et al., 2007; Choy et al., 2008; Murphy et al., 2008; Nachmani et al., 2009). Hence, understanding the mechanism underlying viral miRNA-mediated adaptations can further our knowledge of the cross-species communication.
When infected with a virus, animals try to defend themselves with transcriptional reprograming of the affected cells. In this process, several genes are key elements and these genes are regulated by miRNAs (Peng et al., 2011; de Cubas et al., 2013). Recently, next-generation sequencing (NGS) technology has been used to obtain comprehensive sequencing data, which was used to detect and study the miRNA and protein expression levels in dogs with influenza virus infection (Zhao et al., 2014; Su et al., 2015). However, no detailed analysis of miRNA and the mRNA transcriptome is available. In this study, we used miRNA and mRNA profiles to perform a deep analysis of critical genes, miRNAs, and pathways related to virus infections.
Canine influenza virus, A/canine/01/Guangdong/2013 (H5N1), was isolated in 2013 from a dog with severe respiratory symptoms. Eighteen 40-day-old beagles were assigned to experimental and control groups. Dogs were housed in the Laboratory Animal Center of South China Agricultural University with number SYXK (YUE) 2014-0136. All study protocols were approved by the ABSL-3 Committee of South China Agricultural University in this study. A hemagglutination inhibition (HI) assay revealed that these dogs were sero-negative for avian-origin CIV (H3N2 and H5N1) and for H1N1, H3N1, and influenza B viruses of seasonal influenza viruses. Nine beagles were randomly divided into each group, i.e., experimentally infected (I) and non-infected (NI). After beagles were anesthetized with tiletamine–zolazepam (Virbac, 10–15 mg/kg), they were inoculated intranasally with H5N1 CIV at a tissue culture infectious dose 50 (TCID50) 106 and the control group similarly received 1.0 ml of phosphate-buffered saline. Nasal samples were collected from all beagles before infection and continuously for 14 d after infection. At 3 and 7 d post-infection (dpi), three beagles from each group were euthanized through a pentobarbital overdose. Lesions in the lungs and trachea were collected and frozen in liquid nitrogen. One section was immediately used for RNA isolation, and the others were stored at -80°C for further use. An RNA library was generated for each group from the total RNAs collected from three dogs. Total RNAs were isolated using TRIZOL (Takara, Otsu, Japan) in accordance with the manufacturer’s protocol. The concentration and purity of RNA were determined by measuring absorbance at 260 nm and the A260/A280 ratio using a microspectrophotometer (Nanophometer, Germany). RNA samples were stored at -80°C until further use.
All procedures in the animal experiments were approved by the South China Agricultural University Experimental Animal Welfare Ethics Committee with a reference number of 2016-07.
Eight total RNA samples were obtained to generate RNA libraries for each sample. After the samples were qualified, using mRNA Capture Beads Enriched eukaryote mRNA, mRNA was fragmented by heating. These short mRNA and random hexamers were used to generate the first cDNA and then the second cDNA was synthesized. The second cDNA was purified using VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and then ligated to sequencing adapters. The fragments were amplified by using polymerase chain reaction (PCR) and purified using VAHTSTM DNA Clean Beads and then sequenced using an Illumina HiSeq (Vazyme, Nanjing, China). Raw sequence data were assessed, and sequences containing adaptor tags and those of low quality were excluded. Filtered reads were used for subsequent analysis, and the unique reads were used to identify differentially expressed genes (DEGs) with |log2Ratio|≥ 1 and q-value |FDR|≤ 0.05.
Eight RNA libraries were generated with total RNA from samples. Total RNA was extracted and different fragments of RNAs by polyacrylamide gel electrophoresis (PAGE) were separated. Polyacrylamide electrophoresis gels were used to purify fragments that were 18–30 nt in length and 5′- and 3′-ends adaptors were ligated. The PCR products were generated after reverse-transcription (RT)-PCR and isolated using PAGE. Then, Illumina HiSeq 2000 (Vazyme, Nanjing, China) was used to sequence the purified cDNAs. After excluding reads with 3′- and 5′-primer contaminants or a poly(A) tails shorter than 18 nt, those with low-quality or those without the insert tag, were compared to clean reads in databases to annotate all known small RNA sequences. The unannotated sequences were searched against known miRNA precursors and mature miRNAs identified as known miRNAs. Differentially expressed (DE) miRNAs between the different samples were measured by |log2Ratio|≥ 1 and q-value |FDR|≤ 0.05.
To elucidate the interaction network of miRNA–mRNA with positive and negative correlations, we constructed an miRNA–mRNA regulatory network. The DE mRNAs and miRNAs were collected from the mRNA list and miRNA list. Then, we used miRanda1 and TargetScan2 to confirm the relationship between miRNA and mRNA. Finally, the relationship between miRNA and mRNA was verified.
Depending on the miRNA–mRNA integrative genomic analysis, we used Gene Ontology (GO) to identify biological themes for each negatively correlated miRNA–mRNA pair. There are three ontologies in GO: molecular function, cellular component, and biological process. Negative miRNA–mRNA correlations were imported into the KEGG database3 for pathway analysis.
cDNA was synthesized with oligo (dt) primer for mRNA, using PrimeScriptTM RT Master Mix (Takara, Otsu, Japan). qPCR was performed using the SYBR Premix Ex TaqTM (Tli RNaseH Plus) (Takara, Otsu, Japan) on the LC480 Real-Time PCR System (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions (the primers are listed in Supplementary Tables 1, 2). cDNA was synthesized with A tail for miRNA, miRcute Plus miRNA Firsr-Strand cDNA Synthesis Kit (Tiangen, Beijing, China), and qPCR was performed using miRcute Plus miRNA qPCR Detection Kit (Tiangen, Beijing, China) on the LC480 Real-Time PCR System (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions. GAPDH was used as an endogenous control gene for mRNA and U6 was used for miRNA. We used the 2-ΔΔCT method to analyze these data. qPCR in each reaction was performed in triplicate, and the data were expressed as the mean ± standard error (n = 3).
Following infection with H5N1 influenza virus, clinical symptoms were observed, including cough, running nose, and an increased temperature. After infection with the virus, nasal swabs were collected from 1 to 14 dpi, and the peak temperature was 40.6°C at the second day after infection. The highest virus titer of a nasal swab was 104.56. Virus replication in lung and trachea tissues was detected at 3 and 7 dpi, and the mean viral titers of 3 and 7 dpi were 105.6 and 102.16 TCID50/ml in lung tissues, respectively, whereas it was 101.3 and 100.5 TCID50/ml, respectively, in the trachea.
Eight gene libraries as shown in Supplementary Data Sheet 1 were collected to identify mRNA differentiation of beagles when infected with H5N1 influenza virus. The beagles in the control group (KL3, KT3, KL7, and KT7) and experiment group (LUN3, TRA3, LUN7, and TRA7) were represented in the eight libraries. The original results of sequencing data and assembling results are shown in Supplementary Tables 3, 4. Among all of these reads, >90% of the clean reads were mapped to the canine reference genome.
Total RNA from the lung and trachea were used to build small RNA libraries to assess the characteristics of miRNAs in the lung and trachea after infection. After removing the contaminant and adaptor sequences and filtering low-quality tags, 10.5–12.4 million clean reads were collected in eight samples (as shown in Supplementary Table 5). Clean reads included rRNAs, tRNAs, snRNAs, and snoRNAs. After other RNAs were removed, 291, 290, 290, 283, 280, 287, 282, and 288 mature miRNAs were found in LUN3, TR3, LUN7, TR7, KL3, KT3, KL7, and KT7, respectively.
A total of 455 miRNAs were identified. The DE miRNAs were identified through |log2Ratio|≥ 1 and q-value |FDR|≤ 0.05 (as shown in Figure 1A). Compared with NI tissues, H5N1-infected tissues displayed differential expression of 19 mature miRNAs (as shown in Figure 2). Only miR-126 was DE on day 3 in lung tissue, miR-126 has been reported to restrict the replication of H5N1 in endothelial cells to ameliorate disease condition (Sant et al., 2017), probably implying that when infected with the virus, miR-126 may play an important role in restricting viral replication. MiR-34c, miR-146b, and miR-34b were DE on day 7 in lung tissue, playing anti-inflammatory (miR-146b) (Comer et al., 2014) and resistant apoptosis (miR-34) (Catuogno et al., 2012) roles at this stage. On the seventh day, the mi-34 family is predicted to protect the lung from undergoing apoptosis to restore lung function to normal. Fifteen DE miRNAs were identified on the seventh day in the tracheal tissue (as shown in Supplementary Table 6). At this stage, miRNAs may restore the state of the organism to normal; these included miR-379, which inhibited cell proliferation invasion (Li et al., 2017), anti-inflammatory-related miR-127 (Zhang et al., 2017), and apoptosis-related miR-410 (Palumbo et al., 2016; Deng H. et al., 2017).
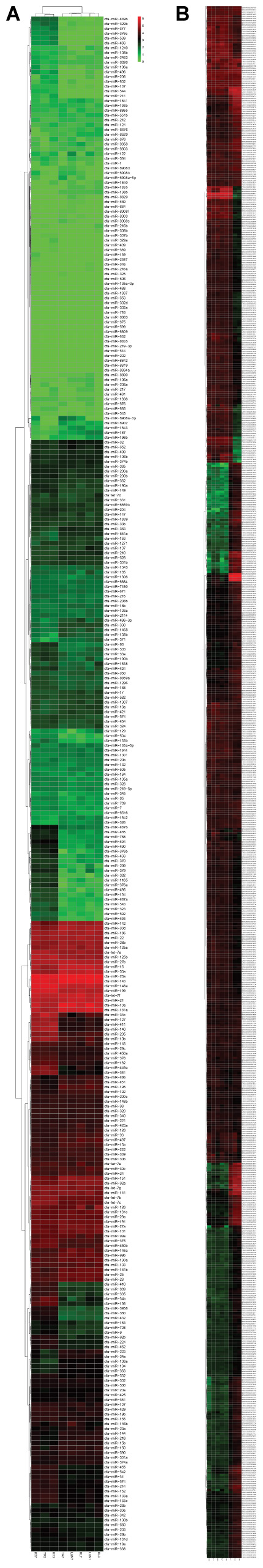
FIGURE 1. (A,B) Hierarchical clustering of DE miRNAs and DE mRNAs among eight RNA libraries. Heatmap of the number of DE miRNAs and DE mRNAs libraries for the DEGs between the infected groups and non-infected groups. MiRNA and mRNA heatmaps of all DE miRNAs and mRNAs are included.
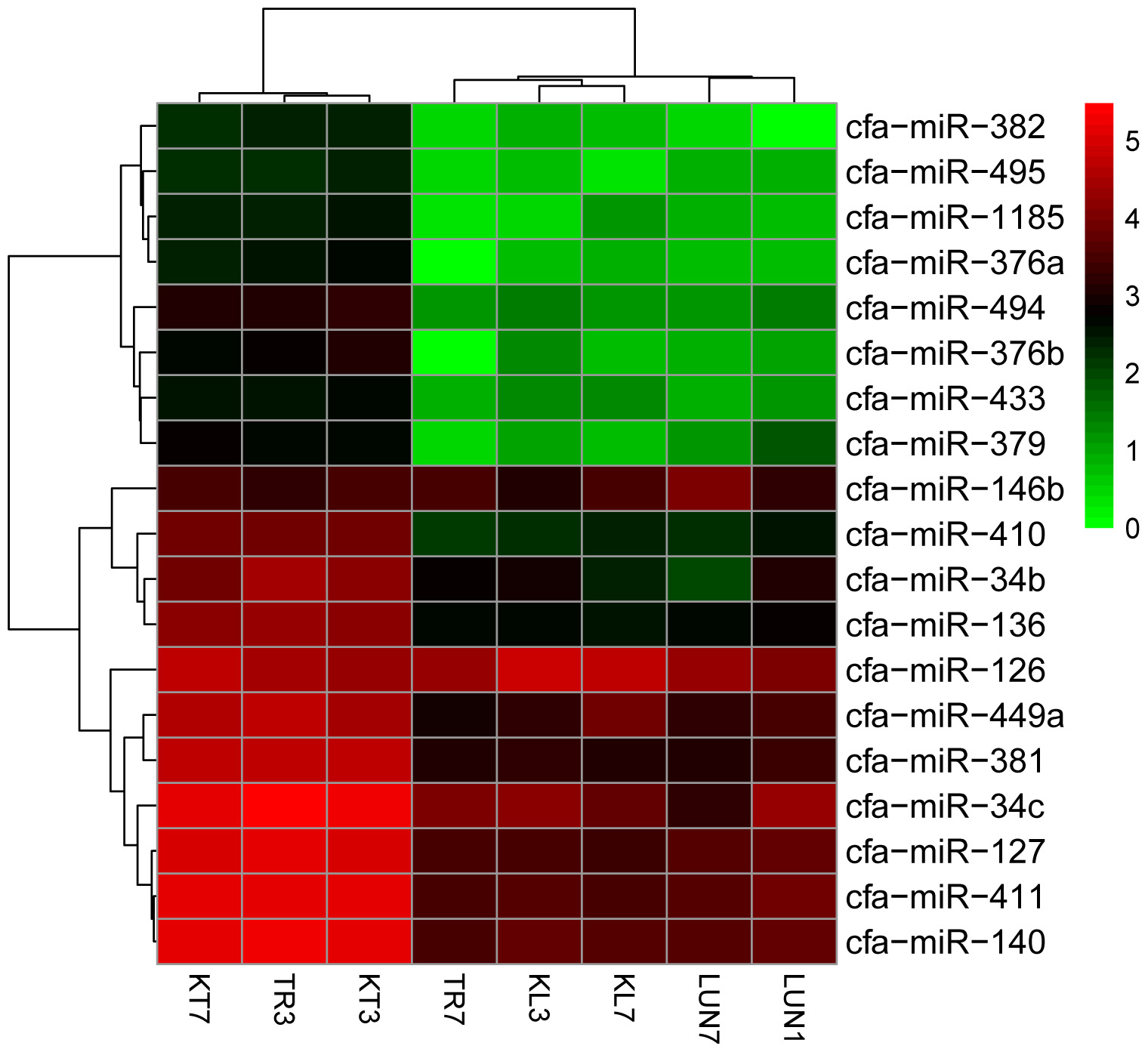
FIGURE 2. Clustering of the 19 up-regulated and down-regulated DE miRNAs for assigning dogs to infected groups and non-infected groups. The colors in the heat map represent the normalized expression values, with lower expression values indicated in shades of green and higher expression values in shades of red.
To determine whether differential miRNA expression was related to differences in post-infection stages, four comparisons were made. Within the tissues, 24 and 132 different miRNAs were identified from lung and tracheal tissues at different times, respectively. Furthermore, 140 and 12 miRNAs were DE at 3 and 7 dpi, respectively (Supplementary Table 7). There were more up-regulated miRNAs (15 of 24 in the lung and 90 of 132 in the trachea) than down-regulated miRNAs in the lung and trachea (Figure 3). Furthermore, three miRNAs, miR-379, miR-34c, and miR-34b, were DE at different times (Figure 4). These results suggest that when infected with H5N1 influenza virus, miR-379 and miR-34 probably play significant roles in resisting viral invasion.
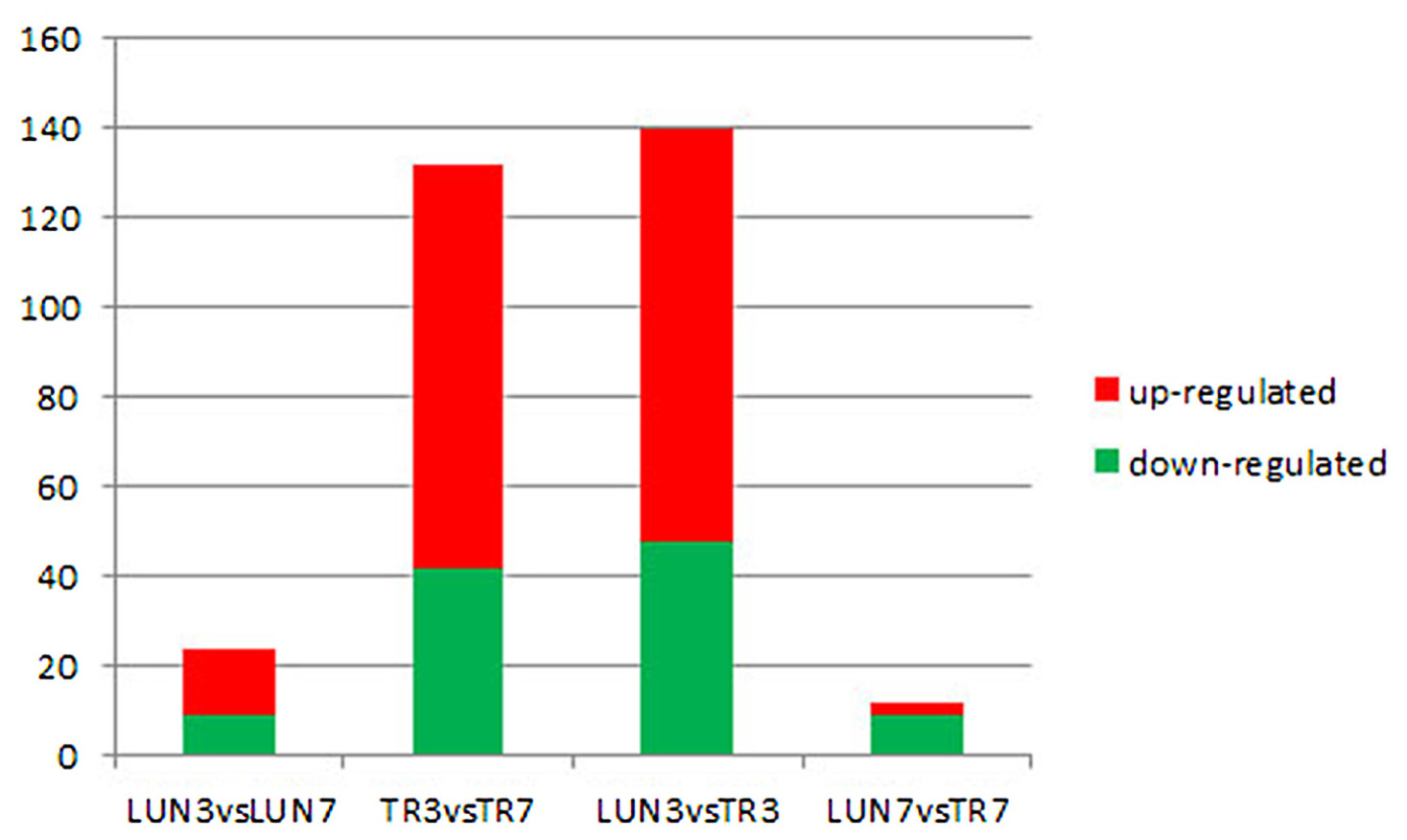
FIGURE 3. Four comparisons of different post-infection stages, miRNAs showing a differential expression pattern, with down-regulation indicated in green and up-regulation indicated in red.
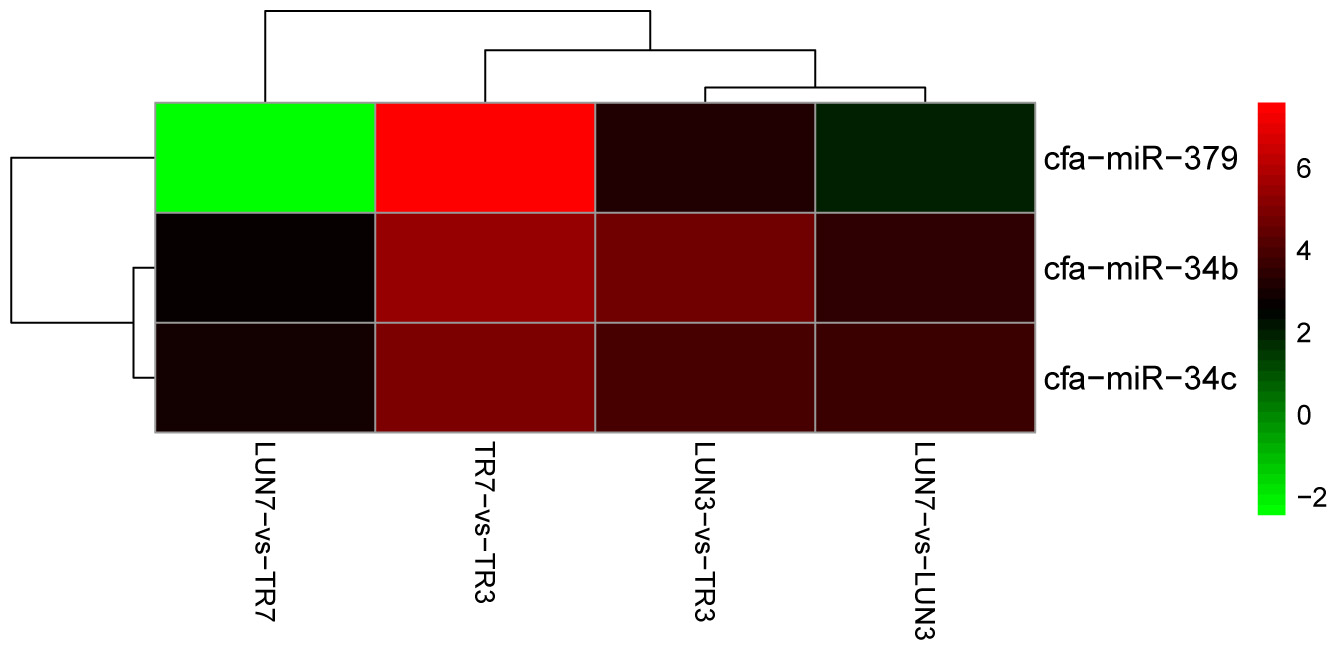
FIGURE 4. MiR-379, miR-34c, and miR-34b were DE at all different post-infection stages in four comparisons (LUN7 vs. TR7, TR7 vs. TR3, LUN3 vs. TR3, and LUN7 vs. LUN3).
Compared with the control group, 1236 significant DEGs (Figure 1B) were identified. There were more down-regulated genes than up-regulated genes (Figures 5A,B). Among the down-regulated genes, 7 were found in the lung at different times, only 1 was found in the trachea at a different time, and 6 and 39 were found at 3 and 7 dpi, respectively; these included immune-related genes IL2R2 (Schliemann et al., 2008), CLEC4D (Steichen et al., 2013), IGFBP2 (Ambrosini-Spaltro et al., 2011), and GPNMB (Schwarzbich et al., 2011) and antiviral genes FOSB (Baumann et al., 2003) and TNIP3 (Zhao et al., 2017).
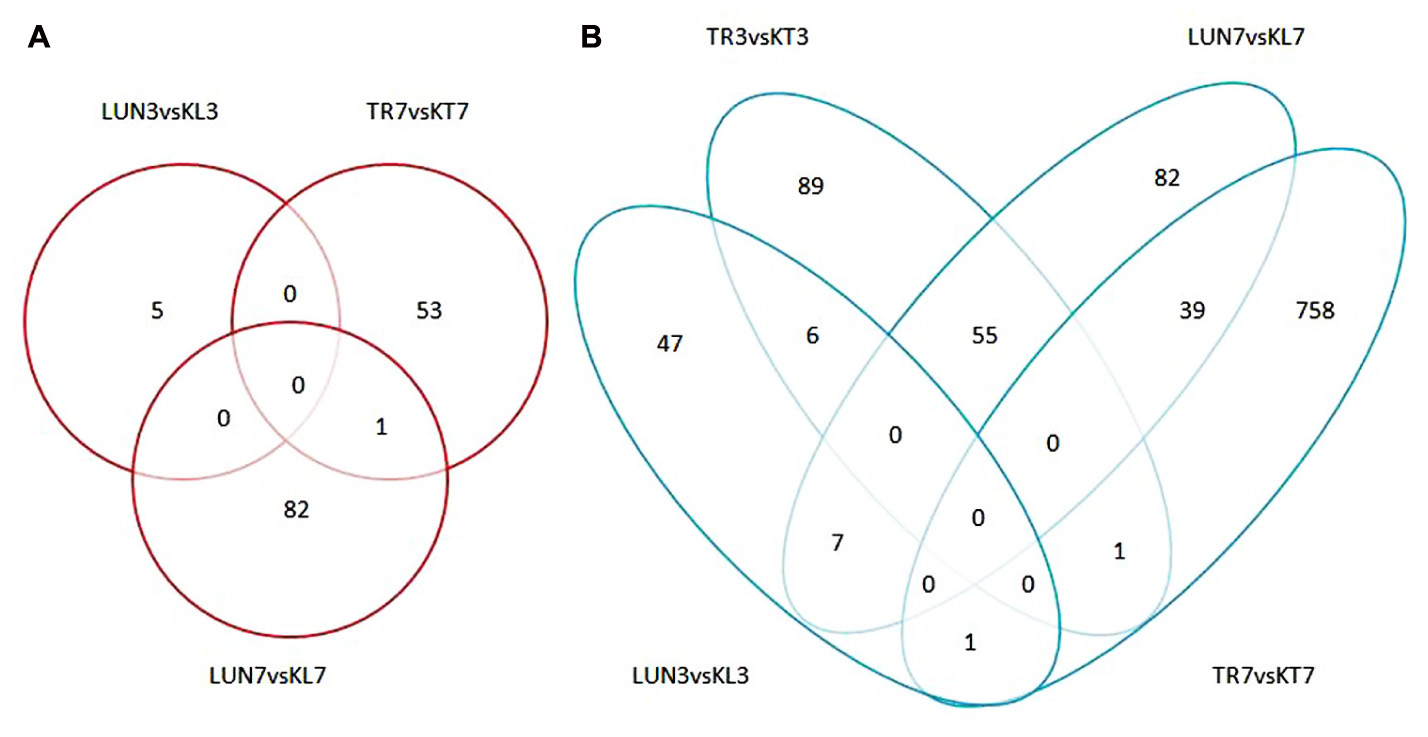
FIGURE 5. (A) Numbers of up-regulated DEGs in infected groups and non-infected groups. (B) Numbers of down-regulated DEGs in infected groups and non-infected groups. An FDR of 0.05 was used to classify genes as DE. The lung and tracheal tissues from beagles in the control group, named KL3 and KT3 on the third day and KL7 and KT7 on the seventh day, and the experimental group named LUN3 and TRA3 on the third day and LUN7 and TRA7 on the seventh day.
To explore the mechanisms underlying resistance to influenza virus infection in dogs, three groups were formed. We identified 919 and 2314 significant DEGs from lung and tracheal tissues, respectively; furthermore, DE genes were also identified under the infected state in the lung and tracheal tissues; 2411 DEGs were collected under the infected state in different tissues. These data were imported into KEGG databases. Significant (p-value < 0.05) pathways were identified (Figure 6 X2, X4, and X6). The signaling pathway and immune related were the two most represented subunits among these groups. The chemokine signaling pathway and hematopoietic cell lineage belonged to the immune system, and all participated in the regulation of these groups. All were involved in the modulation of infected CIV signal pathways including cytokine–cytokine receptor interactions, the PI3K-Akt signaling pathway, cell adhesion molecules (CAMs), and ECM–receptor interactions. Apart from these, infectious diseases were another subclass pathway. Furthermore, influenza A was in the enriched pathway in all of the groups. These results suggested that genes in these pathways may play an important role in response to influenza virus.
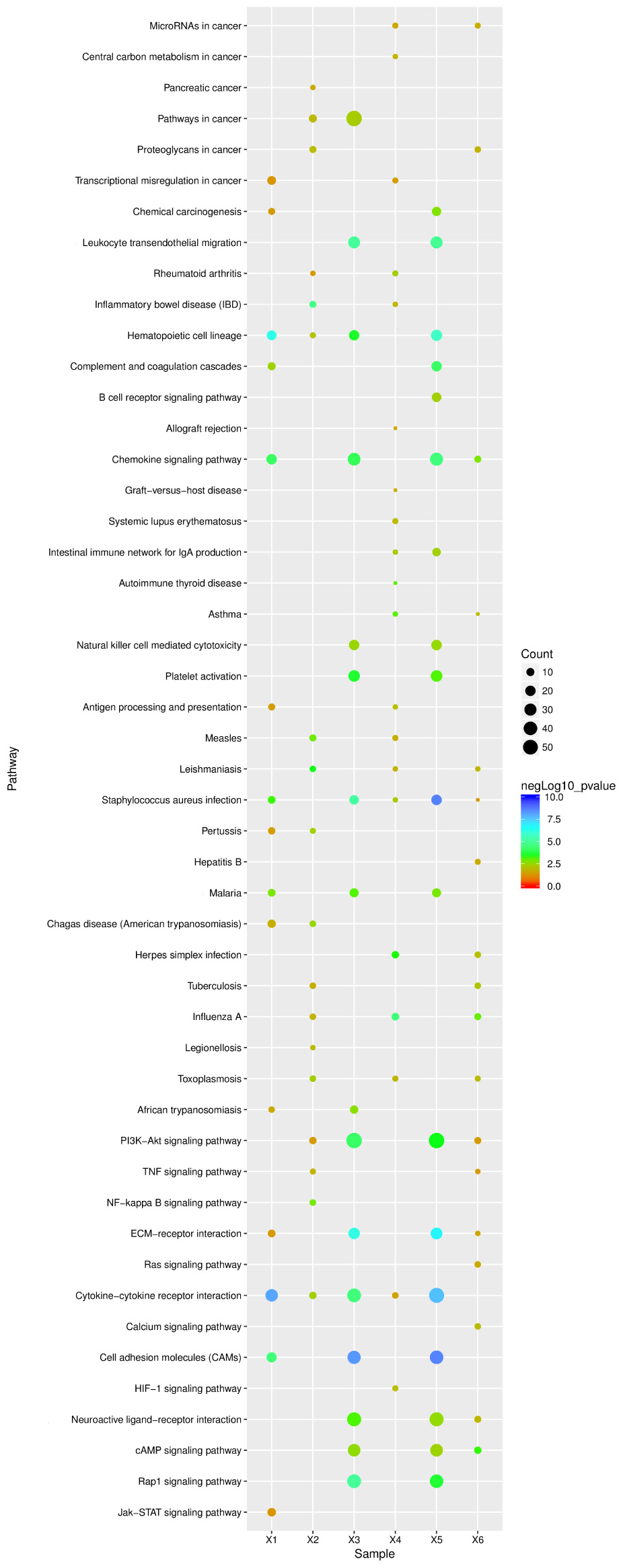
FIGURE 6. KEGG pathway enrichment analysis of DE unigenes of negative miRNA–mRNA and DE unigenes of RNA-seq. X1, 138 negative miRNA–mRNA interactions with 24 mature miRNAs and 201 validated mRNAs DE in the lung (15 pathways); X2, 916 DE unigenes of RNA-seq in lung tissue (18 pathways); X3, 134 negative miRNA–mRNA interactions with 97 mature miRNAs and 135 mRNAs were found in the trachea (16 pathways); X4, 2314 DE unigenes of RNA-seq in trachea tissue (20 pathways); X5, 65 mature miRNAs and 78 mRNA with 120 negative miRNA–mRNA interactions were identified under the infected state with different tissues (18 pathways); X6, 2411 DE unigenes of RNA-seq were identified under the infected state with different tissues (pathways). The ordinate represents the pathway name, the abscissa represents the sample name, the size of the dots indicates the number of DEGs in this pathway, and the point colors correspond to different negLog10 p-value ranges.
To further investigate miRNA–mRNA regulatory information in dogs, transcriptome analyses were performed to identify genes that were co-expressed with miRNAs and miRNA targets were also predicted to identify genes that were bound to miRNA. A total of 268 and 251 miRNA–mRNA pairs with both positive and negative correlations, respectively, were identified in the lung and tracheal tissues, respectively; moreover, under the infected state, miRNA–mRNA pairs were also different between lung and tracheal tissues, and 255 miRNA–mRNA pairs were identified under the infected state in different tissues. However, miRNA generally negatively targeted genes. Therefore, when miRNAs are induced by the influenza virus, their target mRNAs are down-regulated and vice versa. There were 138 negative miRNA–mRNA interactions with 24 mature miRNAs and 201 validated mRNAs were DE in the lung. A total of 134 negative miRNA–mRNA interacted with 97 mature miRNAs and 135 mRNAs were found in the trachea. Moreover, 65 mature miRNAs and 78 mRNAs with 120 negative miRNA–mRNA interactions were identified under the infected state in different tissues. All target genes of the miRNAs were predicted using miRanda and TargetScan.
To identify enriched functional terms of these predicted target genes and to further explore the significant negatively correlated miRNA–mRNA pairs, a GO analysis was carried out on these 105 mRNAs in the lung, 84 mRNAs in the trachea, and 78 mRNAs in different infected tissues, which were up-regulated and down-regulated. GO enrichment analysis was involved in biological processes, cellular components, and molecular functions.
For the 109 mRNAs in the lung, 15 immune-related GO terms in biological process were significantly enriched (p < 0.05) (Figure 7A), and immune response was the highest fold enrichment (eightfold). Inflammatory responses, transcription, and positive regulation of fever generation were also identified. Furthermore, 17 immune-related GO terms in biological processes were significantly enriched (p < 0.05) in the trachea of 84 mRNAs (Figure 7B). Inflammatory responses and innate immune responses had the highest fold enrichment (ninefold). In addition, an infected state with different tissues was associated with 10 immune-related GO terms in biological processes (Figure 7C) including positive regulation of cytosolic calcium ion concentrations, positive regulation of I-kappaB kinase/NF-κB signaling, and positive regulation of the Toll-like receptor (TLR) 2 signaling pathway (p < 0.05). In vivo, different genes coordinate each other to perform biological functions. Pathway analysis helps to gain a better understanding of the biological function of genes.
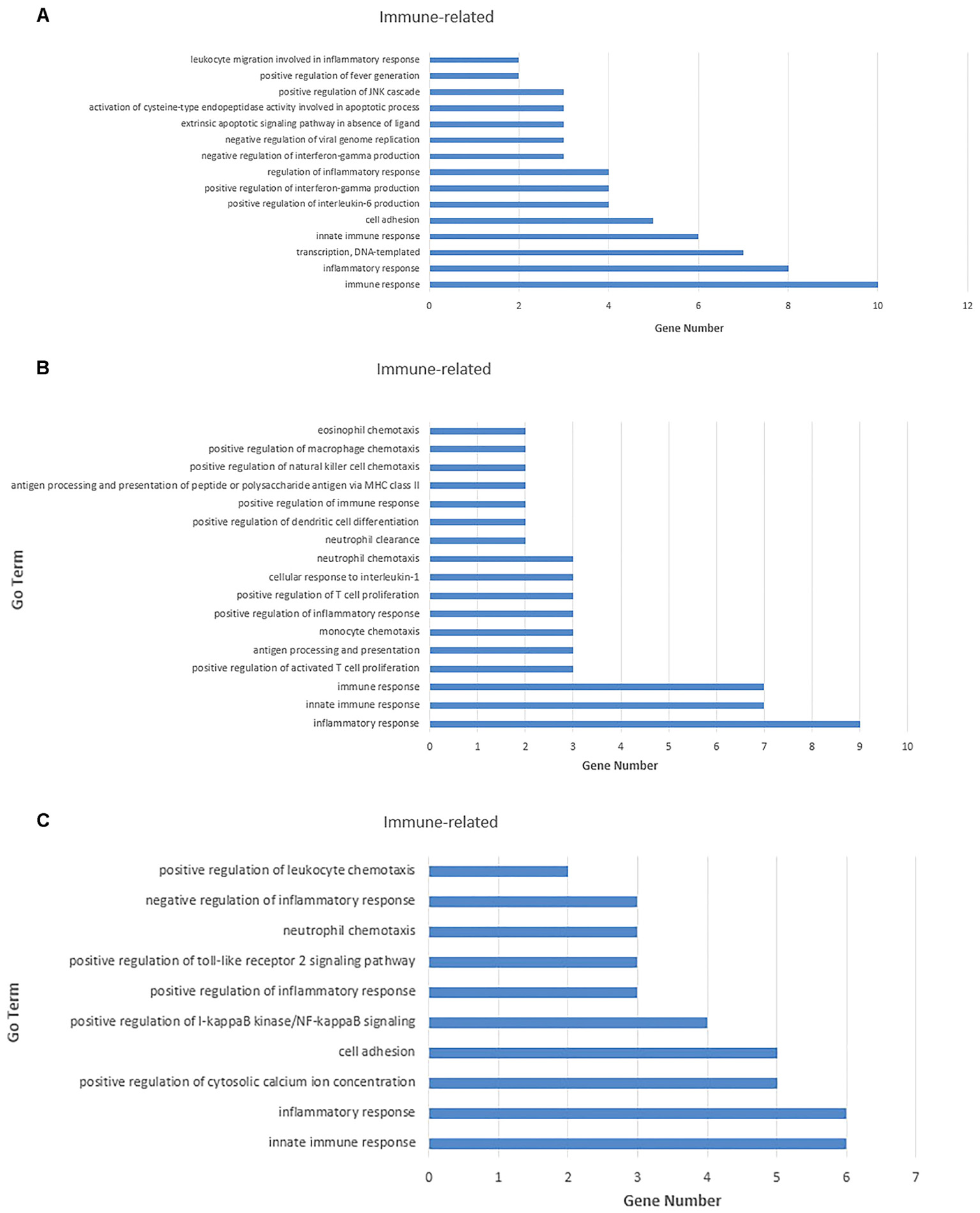
FIGURE 7. Gene ontology (GO) enrichment analysis for negatively correlated miRNA–mRNA. (A) Enriched immune-related GO terms of 109 target genes of repressed DE miRNAs in the lung tissue. (B) Enriched immune-related GO terms of 135 target genes of repressed DE miRNAs in the trachea tissue. (C) Enriched immune-related GO terms of 141 target genes of repressed DE miRNAs under the infected state with different tissues. The abscissa represents the sample name, and the numbers indicate related genes.
Pathway enrichment analysis for 109 mRNAs of 138 negative miRNA–mRNA pairs in the lung, 84 mRNAs of 134 negative pairs in the trachea, and 78 mRNAs of 120 negative miRNA–mRNA pairs in different infected tissues. Further analysis found that among these significant (p < 0.05) pathways, signal- and immune-related pathways were also the most in all of these groups. In addition, infectious diseases were also another enrichment class of the pathways, and the influenza A pathway was also enriched in all groups (Figure 6 X1, X3, and X5).
We also found several genes played roles in multiple pathways. For example, TLR4 (miR-122) and nuclear factor kappa B subunit 1 (NFKB1) (miR-34c) were all involved in the influenza A pathway, as well as the TLR signaling pathway. While tumor necrosis factor (TNF) (miR-331) and nitric oxide synthase 3 (NOS3) (miR-335) participate in the PI3K-Akt signaling pathway and TNF signaling pathway (Figure 8).
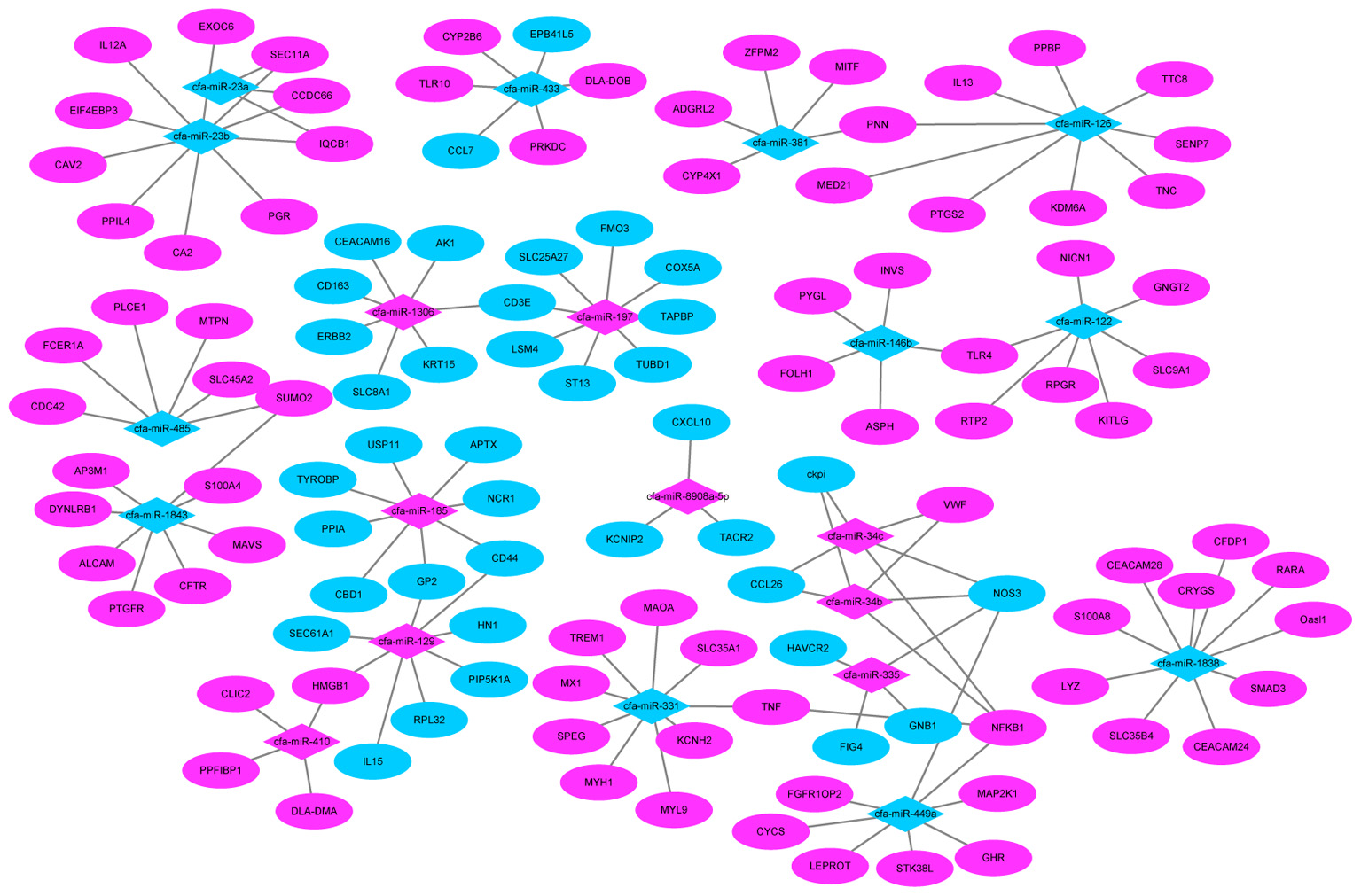
FIGURE 8. MiRNA–mRNA negative correlation network. Red indicates up-regulation and green indicates down-regulation.
Seventeen DE mature miRNAs (cfa-miR-122, cfa-miR-129, cfa-miR-1838, cfa-miR-185, cfa-miR-23a, cfa-miR-331, cfa-miR-34c, cfa-miR-410, cfa-miR-433, cfa-miR-449a, cfa-miR-1843, cfa-miR-8908a-5p, cfa-miR-335, cfa-miR-34b, cfa-miR-485, cfa-miR-126, and cfa-miR-370) and 19 DEGs among the negative miRNA–mRNA interaction network were validated by qRT-PCR. The validation with qPCR was consistent with the sequencing results (as shown in Tables 1, 2).
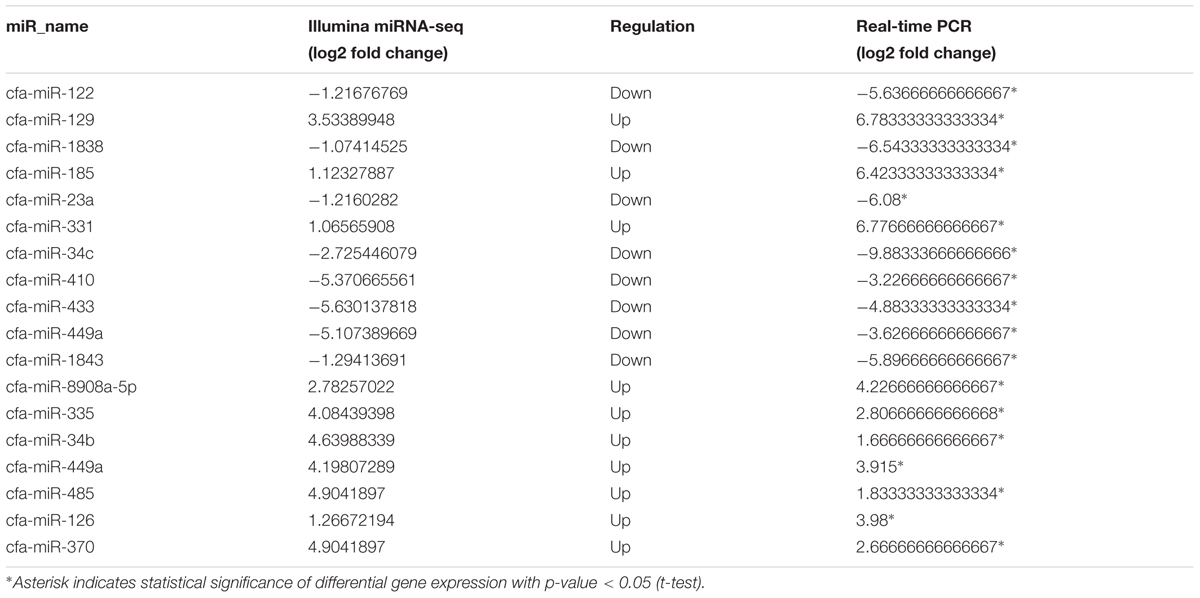
TABLE 1. Relative miRNA expression of selected differentially expressed genes (DEGs) as revealed through miRNA-Seq and quantitative real-time PCR analyses.
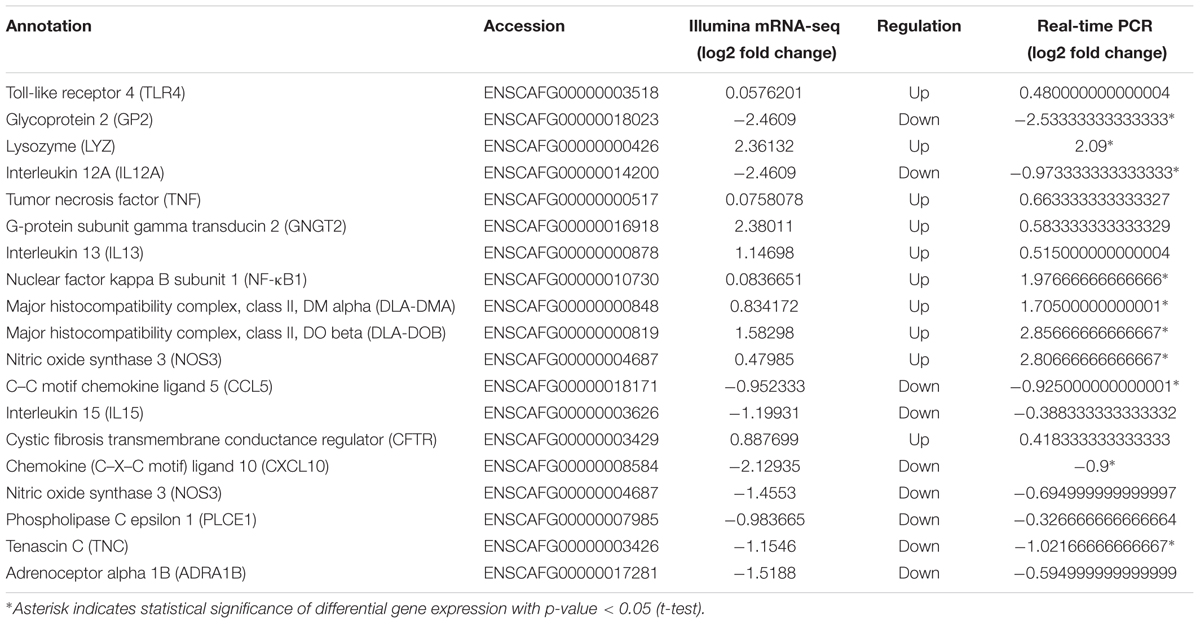
TABLE 2. Relative mRNA expression of 10 selected DEGs as revealed through mRNA-Seq and Quantitative real-time PCR.
H5N1 infections are characterized with a high-fatality rate; hence, in this study, we choose beagles as the animal model of H5N1 infection. We investigated the effect of H5N1 virus infection on miRNA and mRNA, and analyzed the interaction between miRNA and mRNA. Dysregulation of 24 miRNAs and 204 mRNAs with 138 negative miRNA–mRNA pairs was observed in the lung tissue, and dysregulation of 97 miRNAs and 135 mRNAs with 134 negative miRNA–mRNA pairs was observed in the tracheal tissue; moreover, under the infected state, between lung and trachea, miRNAs and mRNAs were also different expressed: 97 miRNAs and 141 mRNAs with 120 negative miRNA–mRNA pairs were found between different tissues. Target mRNA gene functions were analyzed with GO and the KEGG database. Several pathways were activated by relevant target genes of DE miRNAs with a negative miRNA–mRNA correlation.
Infection with H5N1 influenza virus caused differential expression of mRNA in lung and trachea tissues. Comparisons among these infected groups revealed 65 similar up-regulated genes and 7 similar down-regulated genes. These up-regulated genes included antiviral genes, such as cell surface receptor (CD59), calcium ion binding gene (EFHC1 and CAPS2), and protein coding genes (RIBC2, CFAP157, DRC7, CFAP45, CFAP65, and CCDC33). CD59 (Zhang et al., 2010) can inhibit local inflammatory reactions caused by the influenza virus. EFHC1 affects cellular apoptosis (Wang et al., 2002; Katano et al., 2012). CFAP157 (Benos et al., 2012), DRC7 (Yang et al., 2011), and CFAP45 (Li et al., 1999) are involved in the regulation of flagella and cilia motility for viral invasion. Of the down-regulated genes, CD5L combined with any modified low-density lipoprotein (LDL) or other polyanionic ligand and delivered the ligand into the cell via receptor-mediated endocytosis to regulate innate immunity (Peiser et al., 2002). SLC11A1 is a natural resistance-associated macrophage protein (NRAMP1), which is expressed only in immune-related cells (Govoni and Gros, 1998). As a divalent transition metal transporter, it was also involved in iron metabolism and assisted with host resistance to certain pathogens. In conclusion, when infected by H5N1 CIV, immune-related genes and antiviral genes are activated to resist viral infection.
Differential miRNA expression in the lung and trachea revealed that the regulatory mechanism of miRNA on host responses when infected with CIV and that between lung and trachea is different. At different post-infection stages, there were more miRNAs expressed at 3 than 7 dpi. Immunogenicity was enhanced by miR-155 (Izzard et al., 2017), miR-375 (Lin et al., 2017), miR-155, miR-146a (Zhou et al., 2017), and miR-136 (Zhao et al., 2015). Furthermore, miR-146a (Terrier et al., 2013) plays an antiviral role. Moreover, miR-29 activates cyclooxygenase and lambda-1 IFN to resist viral infection (Fang et al., 2011). As a cytolytic virus, influenza A virus induces apoptosis, which results in organ and cellular dysfunction. MiR-15b and miR-451 regulate a series of pro-inflammatory cytokine responses (Chan et al., 2011; Rosenberger et al., 2012). Moreover, miR-29c inhibits BCL2L2 expression to regulate apoptosis induced by influenza A virus (Guan et al., 2012). These data reveal that viral infections typically induce miRNAs that regulate cytokine production and the anti-viral immune response.
The present study reveals novel findings regarding H5N1 influenza virus-infected dogs and lung and tracheal tissue profiling, since these have not been previously performed. MiRNAs and their target genes have multiple relationships (Perez et al., 2009; Fan and Wang, 2016; Nakamura et al., 2016). Dysregulated miRNAs may serve as a diagnostic and prognostic biomarker (Okkenhaug and Vanhaesebroeck, 2003; Hale et al., 2010; Haneklaus et al., 2013; Ingle et al., 2015).
Influenza, caused by influenza virus, is an acute febrile contagious respiratory infectious disease. The innate immune system recognizes invading viruses (Hale et al., 2010; Iwasaki and Pillai, 2014) through multiple mechanisms. The non-structural NS1 inhibits type I IFN production (Haye et al., 2009; Pekosz et al., 2012) by inactivating transcription factors (MacEwan, 2002; Wang et al., 2017) such as IRF3 (Wang et al., 2017), AP1, and NF-κB (miR-34) (Taganov et al., 2006). Pattern recognition receptors (PRRs) recognize viral RNA (Thompson et al., 2011; Okamoto et al., 2017) to activate a specialized immune response at mucosal surfaces to combat viral invasion. TLR4 (miR-146) has been reported to attenuate influenza virus infection with TLR4 antagonists, which may be a novel therapeutic approach to combat infection (Shirey et al., 2013). MiR-146a regulates TRAF6 when infected with H3N2 virus (Deng Y. et al., 2017). Moreover, TRAF6 and MEKK1 are critical genes that IPS-1 activates NF-κB and induces IFNs (Yoshida et al., 2008). In addition, miR-144 targets TRAF4-IRF7 through NF-κB to attenuate the host response to influenza virus (Lopez et al., 2017). Furthermore, miR-34 targets pro-apoptosis Bax through downregulation (Fan and Wang, 2016) during an influenza virus infection. Despite the paucity in the number of studies on dogs, it is important to refer to previous studies. Briefly, NF-κB and TLR4 with their target miR-34c and miR-146 through IPS-1 and TRAF family may have important roles in regulating the innate immune system during an H5N1 CIV infection.
Toll-like receptors activate IFNs and lead to antiviral responses through cytokine–cytokine receptor interactions. As chemokines (Zlotnik and Yoshie, 2000), ccl5 (miR-335) and cxcl10 (miR-8908a-5p) activate their receptors once activated, different downstream pathways are activated and may produce a series of immune responses. In addition, P13K-Akt signaling pathway regulates transcription, growth, proliferation, translation, and survival of fundamental cellular functions (Koyasu, 2003; Okkenhaug and Vanhaesebroeck, 2003; Richardson et al., 2004; Duronio, 2008; Hers et al., 2011). When cells are infected with the influenza A virus, viral NS1 binds to and activates P13K, inducing the beta-IFN and the apoptotic responses (Ehrhardt et al., 2007; Ehrhardt and Ludwig, 2009). In our findings, up-regulated GNGT2 and down-regulated miR-122 activate the P13K-Akt pathway during an influenza virus infection. Despite the paucity in the number of studies on canine microRNA and mRNA, it is important to refer to previous studies to explain the function of microRNAs in dogs and is of great significance in studies on dogs.
Although the expression levels and functions of miRNAs in the lung and trachea have been studied, the effects of beagles infected with influenza virus on mRNA and miRNA expression and influenza virus-related pathways have not been investigated. To our knowledge, this is the first study to assess the effect of beagles on miRNA–mRNA expression when infected with H5N1 influenza virus. Despite the low chance of H5N1 infection in domestic animals, direct contact of H5N1-infected dogs with humans may occur. Our results provide information for understanding the mechanisms of viral pathogenesis in dogs.
CF contributed to the data analysis and the writing of the manuscript. JL contributed to the drafting of the manuscript. SY and ZY contributed to the data collection and the laboratory work. CF and JL contributed to the animal experiment. SL contributed to the conception of the idea and design. All authors read and approved the manuscript.
This project was supported in part by The National Natural Science Foundation of China (31672563), The National Key Research and Development Program of China (2016YFD0501004), and The Guangdong Provincial Key Laboratory of Prevention and Control for Severe Clinical Animal Diseases (2017B030314142).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00303/full#supplementary-material
Ambrosini-Spaltro, A., Farnedi, A., Montironi, R., and Foschini, M. P. (2011). IGFBP2 as an immunohistochemical marker for prostatic adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 19, 318–328. doi: 10.1097/PAI.0b013e3128052936
Ashour, M. M., Khatab, A. M., El-Folly, R. F., and Amer, W. A. (2012). Clinical features of avian influenza in Egyptian patients. J. Egypt Soc. Parasitol. 42, 385–396. doi: 10.12816/0006325
Baumann, S., Hess, J., Eichhorst, S. T., Krueger, A., Angel, P., Krammer, P. H., et al. (2003). An unexpected role for FosB in activation-induced cell death of T cells. Oncogene 22, 1333–1339. doi: 10.1038/sj.onc.1206126
Benos, P. V., Hoh, R. A., Stowe, T. R., Turk, E., and Stearns, T. (2012). Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One 7:e52166. doi: 10.1371/journal.pone.0052166
Cai, X., Lu, S., Zhang, Z., Gonzalez, C. M., Damania, B., and Cullen, B. R. (2005). Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U.S.A. 102, 5570–5575. doi: 10.1073/pnas.0408192102
Castleman, W. L., Powe, J. R., Crawford, P. C., Gibbs, E. P. J., Dubovi, E. J., Donis, R. O., et al. (2010). Canine H3N8 influenza virus infection in dogs and mice. Vet. Pathol. 47, 507–517. doi: 10.1177/0300985810363718
Catuogno, S., Cerchia, L., Romano, G., Pognonec, P., Condorelli, G., and de Franciscis, V. (2012). miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene 32, 341–351. doi: 10.1038/onc.2012.51
Chan, L. Y., Kwok, H. H., Chan, R. W. Y., Peiris, M. J. S., Mak, N. K., Wong, R. N. S., et al. (2011). Dual functions of ginsenosides in protecting human endothelial cells against influenza H9N2-induced inflammation and apoptosis. J. Ethnopharmacol. 137, 1542–1546. doi: 10.1016/j.jep.2011.08.022
Chen, Y., Zhong, G., Wang, G., Deng, G., Li, Y., Shi, J., et al. (2010). Dogs are highly susceptible to H5N1 avian influenza virus. Virology 405, 15–19. doi: 10.1016/j.virol.2010.05.024
Cheng, A. M. (2005). Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33, 1290–1297. doi: 10.1093/nar/gki200
Choy, E. Y.-W., Siu, K.-L., Kok, K.-H., Lung, R. W.-M., Tsang, C. M., To, K.-F., et al. (2008). An Epstein-Barr virus–encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205, 2551–2560. doi: 10.1084/jem.20072581
Comer, B. S., Camoretti-Mercado, B., Kogut, P. C., Halayko, A. J., Solway, J., and Gerthoffer, W. T. (2014). MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L727–L734. doi: 10.1152/ajplung.00174.2014
Crawford, P. C., Dubovi, E. J., Castleman, W. L., Stephenson, I., Gibbs, E. P., Chen, L., et al. (2005). Transmission of Equine to dog. Science 310, 482–485. doi: 10.1126/science.1117950
Damiani, A. M., Kalthoff, D., Beer, M., Müller, E., and Osterrieder, N. (2012). Serological survey in dogs and cats for influenza A(H1N1)pdm09 in Germany. Zoonoses Public Health 59, 549–552. doi: 10.1111/j.1863-2378.2012.01541.x
de Cubas, A. A., Leandro-Garcia, L. J., Schiavi, F., Mancikova, V., Comino-Mendez, I., Inglada-Perez, L., et al. (2013). Integrative analysis of miRNA and mRNA expression profiles in pheochromocytoma and paraganglioma identifies genotype-specific markers and potentially regulated pathways. Endocr. Relat. Cancer 20, 477–493. doi: 10.1530/erc-12-0183
Deng, H., Chu, X., Song, Z., Deng, X., Xu, H., Ye, Y., et al. (2017). MicroRNA-1185 induces endothelial cell apoptosis by targeting UVRAG and KRIT1. Cell Physiol. Biochem. 41, 2171–2182. doi: 10.1159/000475571
Deng, Y., Yan, Y., Tan, K. S., Liu, J., Chow, V. T., Tao, Z.-Z., et al. (2017). MicroRNA-146a induction during influenza H3N2 virus infection targets and regulates TRAF6 levels in human nasal epithelial cells (hNECs). Exp. Cell Res. 352, 184–192. doi: 10.1016/j.yexcr.2017.01.011
Duronio, V. (2008). The life of a cell apoptosis regulation by the PI3/PKB pathway. Biochem. J. 415, 333–344. doi: 10.1042/BJ20081056
Ehrhardt, C., and Ludwig, S. (2009). A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell. Microbiol. 11, 863–871. doi: 10.1111/j.1462-5822.2009.01309.x
Ehrhardt, C., Wolff, T., Pleschka, S., Planz, O., Beermann, W., Bode, J. G., et al. (2007). Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81, 3058–3067. doi: 10.1128/jvi.02082-06
Fan, N., and Wang, J. (2016). MicroRNA 34a contributes to virus-mediated apoptosis through binding to its target gene Bax in influenza A virus infection. Biomed. Pharmacother. 83, 1464–1470. doi: 10.1016/j.biopha.2016.08.049
Fang, J., Hao, Q., Liu, L., Li, Y., Wu, J., Huo, X., et al. (2011). Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J. Virol. 86, 1010–1020. doi: 10.1128/jvi.06169-11
Gomez, I. G., MacKenna, D. A., Johnson, B. G., Kaimal, V., Roach, A. M., Ren, S., et al. (2014). Anti–microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Invest. 125, 141–156. doi: 10.1172/jci75852
Govoni, G., and Gros, P. (1998). Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47, 277–284. doi: 10.1007/s000110050330
Grey, F., Meyers, H., White, E. A., Spector, D. H., and Nelson, J. (2007). A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3:e163. doi: 10.1371/journal.ppat.0030163
Guan, Z., Shi, N., Song, Y., Zhang, X., Zhang, M., and Duan, M. (2012). Induction of the cellular microRNA-29c by influenza virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors BCL2L2. Biochem. Biophys. Res. Commun. 425, 662–667. doi: 10.1016/j.bbrc.2012.07.114
Hale, B. G., Albrecht, R. A., and García-Sastre, A. (2010). Innate immune evasion strategies of influenza virus. Future Microbiol. 5, 23–41. doi: 10.2217/FMB.09.108
Haneklaus, M., Gerlic, M., O’Neill, L. A. J., and Masters, S. L. (2013). miR-223: infection, inflammation and cancer. J. Intern. Med. 274, 215–226. doi: 10.1111/joim.12099
Hansen, A., Henderson, S., Lagos, D., Nikitenko, L., Coulter, E., Roberts, S., et al. (2010). KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24, 195–205. doi: 10.1101/gad.553410
Haye, K., Burmakina, S., Moran, T., Garcia-Sastre, A., and Fernandez-Sesma, A. (2009). The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J. Virol. 83, 6849–6862. doi: 10.1128/jvi.02323-08
Hers, I., Vincent, E. E., and Tavaré, J. M. (2011). Akt signalling in health and disease. Cell. Signal. 23, 1515–1527. doi: 10.1016/j.cellsig.2011.05.004
Ingle, H., Kumar, S., Raut, A. A., Mishra, A., Kulkarni, D. D., Kameyama, T., et al. (2015). The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 8:ra126. doi: 10.1126/scisignal.aab3183
Iwasaki, A., and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328. doi: 10.1038/nri3665
Izzard, L., Dlugolenski, D., Xia, Y., McMahon, M., Middleton, D., Tripp, R. A., et al. (2017). Enhanced immunogenicity following miR-155 incorporation into the influenza A virus genome. Virus Res. 235, 115–120. doi: 10.1016/j.virusres.2017.04.002
Katano, M., Numata, T., Aguan, K., Hara, Y., Kiyonaka, S., Yamamoto, S., et al. (2012). The juvenile myoclonic epilepsy-related protein EFHC1 interacts with the redox-sensitive TRPM2 channel linked to cell death. Cell Calcium 51, 179–185. doi: 10.1016/j.ceca.2011.12.011
Klenk, H. D. (2014). Influenza viruses en route from birds to man. Cell Host Microbe 15, 653–654. doi: 10.1016/j.chom.2014.05.019
Lee, C., Song, D., Kang, B., Kang, D., Yoo, J., Jung, K., et al. (2009). A serological survey of avian origin canine H3N2 influenza virus in dogs in Korea. Vet. Microbiol. 137, 359–362. doi: 10.1016/j.vetmic.2009.01.019
Lee, I. H., Le, T. B., Kim, H. S., and Seo, S. H. (2016). Isolation of a novel H3N2 influenza virus containing a gene of H9N2 avian influenza in a dog in South Korea in 2015. Virus Genes 52, 142–145. doi: 10.1007/s11262-015-1272-z
Li, K., Wang, Y., Zhang, A., Liu, B., and Jia, L. (2017). miR-379 inhibits cell proliferation, invasion, and migration of vascular smooth muscle cells by targeting insulin-like factor-1. Yonsei Med. J. 58, 234–240. doi: 10.3349/ymj.2017.58.1.234
Li, S., Shi, Z., Jiao, P., Zhang, G., Zhong, Z., Tian, W., et al. (2010). Avian-origin H3N2 canine influenza A viruses in Southern China. Infect. Genet. Evol. 10, 1286–1288. doi: 10.1016/j.meegid.2010.08.010
Li, Z., Yao, K., and Cao, Y. (1999). Molecular cloning of a novel tissue-specific gene from human nasopharyngeal epithelium. Gene 237, 235–240. doi: 10.1016/S0378-1119(99)00234-6
Lin, J., Xia, J., Tu, C. Z., Zhang, K. Y., Zeng, Y., and Yang, Q. (2017). H9N2 avian influenza virus protein PB1 enhances the immune responses of bone marrow-derived dendritic cells by down-regulating miR375. Front. Microbiol. 8:287. doi: 10.3389/fmicb.2017.00287
Lopez, C. B., Rosenberger, C. M., Podyminogin, R. L., Diercks, A. H., Treuting, P. M., Peschon, J. J., et al. (2017). miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis. PLoS Pathog. 13:e1006305. doi: 10.1371/journal.ppat.1006305
Maas, R., Tacken, M., Ruuls, L., Koch, G., van Rooij, E., and Stockhofe-Zurwieden, N. (2007). Avian influenza (H5N1) susceptibility and receptors in dogs. Emerg. Infect. Dis. 13, 1219–1221. doi: 10.3201/eid1308.070393
MacEwan, D. J. (2002). TNF receptor subtype signalling: differences and cellular consequences. Cell. Signal. 14, 477–492. doi: 10.1016/S0898-6568(01)00262-5
Meydan, C., Shenhar-Tsarfaty, S., and Soreq, H. (2016). MicroRNA regulators of anxiety and metabolic disorders. Trends Mol. Med. 22, 798–812. doi: 10.1016/j.molmed.2016.07.001
Murphy, E., Vanicek, J., Robins, H., Shenk, T., and Levine, A. J. (2008). Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U.S.A. 105, 5453–5458. doi: 10.1073/pnas.0711910105
Nachmani, D., Stern-Ginossar, N., Sarid, R., and Mandelboim, O. (2009). Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5, 376–385. doi: 10.1016/j.chom.2009.03.003
Nakamura, S., Horie, M., Daidoji, T., Honda, T., Yasugi, M., Kuno, A., et al. (2016). Influenza A virus-induced expression of a GalNAc transferase, GALNT3, via microRNAs is required for enhanced viral replication. J. Virol. 90, 1788–1801. doi: 10.1128/jvi.02246-15
Okamoto, M., Tsukamoto, H., Kouwaki, T., Seya, T., and Oshiumi, H. (2017). Recognition of viral RNA by pattern recognition receptors in the induction of innate immunity and excessive inflammation during respiratory viral infections. Viral Immunol. 30, 408–420. doi: 10.1089/vim.2016.0178
Okkenhaug, K., and Vanhaesebroeck, B. (2003). PI3K in lymphocyte development, differentiation and activation. Nat. Rev. Immunol. 3, 317–330. doi: 10.1038/nri1056
Palumbo, T., Poultsides, G. A., Kouraklis, G., Liakakos, T., Drakaki, A., Peros, G., et al. (2016). A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer 16:353. doi: 10.1186/s12885-016-2384-0
Peiser, L., Mukhopadhyay, S., and Gordon, S. (2002). Scavenger receptors in innate immunity. Curr. Opin. Immunol 14, 123–128. doi: 10.1016/S0952-7915(01)00307-7
Pekosz, A., Rajsbaum, R., Albrecht, R. A., Wang, M. K., Maharaj, N. P., Versteeg, G. A., et al. (2012). Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 8:e1003059. doi: 10.1371/journal.ppat.1003059
Peng, X., Gralinski, L., Ferris, M. T., Frieman, M. B., Thomas, M. J., Proll, S., et al. (2011). Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. mBio 2:e198-11. doi: 10.1128/mBio.00198-11
Perez, J. T., Pham, A. M., Lorini, M. H., Chua, M. A., Steel, J., and tenOever, B. R. (2009). MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 27, 572–576. doi: 10.1038/nbt.1542
Pfeffer, S., Sewer, A., Lagos-Quintana, M., Sheridan, R., Sander, C., Grässer, F. A., et al. (2005). Identification of microRNAs of the herpesvirus family. Nat. Methods 2, 269–276. doi: 10.1038/nmeth746
Pfeffer, S., Zavolan, M., Grässer, F. A., Chien, M., Russo, J. J., Ju, J., et al. (2004). Identification of virus-encoded microRNAs. Science 304, 734–736. doi: 10.1126/science.1096781
Ploegh, H. L., Robertson, K. A., Hsieh, W. Y., Forster, T., Blanc, M., Lu, H., et al. (2016). An interferon regulated microRNA provides broad cell-intrinsic antiviral immunity through multihit host-directed targeting of the sterol pathway. PLoS Biol. 14:e1002364. doi: 10.1371/journal.pbio.1002364
Richardson, C. J., Schalm, S. S., and Blenis, J. (2004). PI3-kinase and TOR: PIKTORing cell growth. Semin. Cell Dev. Biol. 15, 147–159. doi: 10.1016/j.semcdb.2003.12.023
Rosenberger, C. M., Podyminogin, R. L., Navarro, G., Zhao, G. W., Askovich, P. S., Weiss, M. J., et al. (2012). miR-451 regulates dendritic cell cytokine responses to influenza infection. J. Immunol. 189, 5965–5975. doi: 10.4049/jimmunol.1201437
Samols, M. A., Skalsky, R. L., Maldonado, A. M., Riva, A., Lopez, M. C., Baker, H. V., et al. (2007). Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. doi: 10.1371/journal.ppat.0030065
Sant, A. J., Tundup, S., Kandasamy, M., Perez, J. T., Mena, N., Steel, J., et al. (2017). Endothelial cell tropism is a determinant of H5N1 pathogenesis in mammalian species. PLoS Pathog. 13:e1006270. doi: 10.1371/journal.ppat.1006270
Schliemann, C., Palumbo, A., Zuberbuhler, K., Villa, A., Kaspar, M., Trachsel, E., et al. (2008). Complete eradication of human B-cell lymphoma xenografts using rituximab in combination with the immunocytokine L19-IL2. Blood 113, 2275–2283. doi: 10.1182/blood-2008-05-160747
Schwarzbich, M.-A., Gutknecht, M., Salih, J., Salih, H. R., Brossart, P., Rittig, S. M., et al. (2011). The immune inhibitory receptor osteoactivin is upregulated in monocyte-derived dendritic cells by BCR–ABL tyrosine kinase inhibitors. Cancer Immunol. Immunother. 61, 193–202. doi: 10.1007/s00262-011-1096-1
Shirey, K. A., Lai, W., Scott, A. J., Lipsky, M., Mistry, P., Pletneva, L. M., et al. (2013). The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature 497, 498–502. doi: 10.1038/nature12118
Shivdasani, R. A. (2006). MicroRNAs regulators of gene expression and cell differentiatio. Blood 108, 3646–3653. doi: 10.1182/blood-200601-030015
Song, G., Sharma, A. D., Roll, G. R., Ng, R., Lee, A. Y., Blelloch, R. H., et al. (2010). MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51, 1735–1743. doi: 10.1002/hep.23547
Songserm, T., Amonsin, A., Jam-on, R., Sae-Heng, N., Pariyothorn, N., Payungporn, S., et al. (2006a). Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect. Genet. Evol. 12, 1744–1747. doi: 10.3201/eid1211.060542
Songserm, T., Amonsin, A., Jam-on, R., Sae-Heng, N., Pariyothorn, N., Payungporn, S., et al. (2006b). Fatal avian influenza A H5N1 IN a dog. Emerg. Infect. Dis. 12, 1744–1747. doi: 10.3201/eid1211.060542
Steichen, A. L., Binstock, B. J., Mishra, B. B., and Sharma, J. (2013). C-type lectin receptor Clec4d plays a protective role in resolution of Gram-negative pneumonia. J. Leukoc. Biol. 94, 393–398. doi: 10.1189/jlb.1212622
Stern-Ginossar, N., Elefant, N., Zimmermann, A., Wolf, D. G., Saleh, N., Biton, M., et al. (2007). Host immune system gene targeting by a viral miRNA. Science 317, 376–381. doi: 10.1126/science.1140956
Su, S., Chen, J., Jia, K., Khan, S. U., He, S., Fu, X., et al. (2014a). Evidence for subclinical influenza A(H1N1)pdm09 virus infection among dogs in Guangdong Province, China. J. Clin. Microbiol. 52, 1762–1765. doi: 10.1128/jcm.03522-13
Su, S., Chen, Y., Zhao, F.-R., Chen, J.-D., Xie, J.-X., Chen, Z.-M., et al. (2013). Avian-origin H3N2 canine influenza virus circulating in farmed dogs in Guangdong, China. Infect. Genet. Evol. 19, 251–256. doi: 10.1016/j.meegid.2013.05.022
Su, S., Qi, W., Zhou, P., Xiao, C., Yan, Z., Cui, J., et al. (2014b). First evidence of H10N8 avian influenza virus infections among feral dogs in live poultry markets in Guangdong province, China. Clin. Infect. Dis. 59, 748–750. doi: 10.1093/cid/ciu345
Su, S., Tian, J., Hong, M., Zhou, P., Lu, G., Zhu, H., et al. (2015). Global and quantitative proteomic analysis of dogs infected by avian-like H3N2 canine influenza virus. Front. Microbiol. 6:228. doi: 10.3389/fmicb.2015.00228
Suh, Y. S., Bhat, S., Hong, S.-H., Shin, M., Bahk, S., Cho, K. S., et al. (2015). Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat. Commun. 6:7693. doi: 10.1038/ncomms8693
Sun, X., Xu, X., Liu, Q., Liang, D., Li, C., He, Q., et al. (2013). Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect. Genet. Evol. 20, 471–475. doi: 10.1016/j.meegid.2013.10.012
Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D. (2006). NF- B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486. doi: 10.1073/pnas.0605298103
Terrier, O., Textoris, J., Carron, C., Marcel, V., Bourdon, J. C., and Rosa-Calatrava, M. (2013). Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J. Gen. Virol. 94(Pt 5), 985–995. doi: 10.1099/vir.0.049528-0
Thompson, M. R., Kaminski, J. J., Kurt-Jones, E. A., and Fitzgerald, K. A. (2011). Pattern recognition receptors and the innate immune response to viral infection. Viruses 3, 920–940. doi: 10.3390/v3060920
Tu, J., Zhou, H., Jiang, T., Li, C., Zhang, A., Guo, X., et al. (2009). Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch. Virol. 154, 887–890. doi: 10.1007/s00705-009-0381-1
Wang, L., Fu, X., Zheng, Y., Zhou, P., Fang, B., Huang, S., et al. (2017). The NS1 protein of H5N6 feline influenza virus inhibits feline beta interferon response by preventing NF-κB and IRF3 activation. Dev. Comp. Immunol. 74, 60–68. doi: 10.1016/j.dci.2017.04.003
Wang, S., Chen, J.-Z., Zhang, Z., Huang, Q., Gu, S., Ying, K., et al. (2002). Cloning, characterization, and expression of calcyphosine 2, a novel human gene encoding an EF-Hand Ca2+-binding protein. Biochem. Biophys. Res. Commun. 291, 414–420. doi: 10.1006/bbrc.2002.6461
Yang, Y., Cochran, D. A., Gargano, M. D., King, I., Samhat, N. K., Burger, B. P., et al. (2011). Regulation of flagellar motility by the conserved flagellar protein CG34110/Ccdc135/FAP50. Mol. Biol. Cell 22, 976–987. doi: 10.1091/mbc.E10-04-0331
Yin, X., Zhao, F.-R., Zhou, D.-H., Wei, P., and Chang, H.-Y. (2014). Serological report of pandemic and seasonal human influenza virus infection in dogs in southern China. Arch. Virol. 159, 2877–2882. doi: 10.1007/s00705-014-2119-y
Yoshida, R., Takaesu, G., Yoshida, H., Okamoto, F., Yoshioka, T., Choi, Y., et al. (2008). TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J. Biol. Chem. 283, 36211–36220. doi: 10.1074/jbc.M806576200
Zhang, C., Xu, Y., Jia, L., Yang, Y., Wang, Y., Sun, Y., et al. (2010). A new therapeutic strategy for lung tissue injury induced by influenza with CR2 targeting complement inhibitor. Virol. J. 7:30. doi: 10.1186/1743-422x-7-30
Zhang, H., Li, Y., Liu, Y., Liu, H., Wang, H., Jin, W., et al. (2016). Role of plant MicroRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 10:60. doi: 10.1186/s12918-016-0292-1
Zhang, Z., Wan, F., Zhuang, Q., Zhang, Y., and Xu, Z. (2017). Suppression of miR-127 protects PC-12 cells from LPS-induced inflammatory injury by downregulation of PDCD4. Biomed. Pharmacother. 96, 1154–1162. doi: 10.1016/j.biopha.2017.11.107
Zhao, F.-R., Su, S., Zhou, D.-H., Zhou, P., Xu, T.-C., Zhang, L.-Q., et al. (2014). Comparative analysis of microRNAs from the lungs and trachea of dogs (Canis familiaris) infected with canine influenza virus. Infect. Genet. Evol. 21, 367–374. doi: 10.1016/j.meegid.2013.11.019
Zhao, H., Wang, L., Luo, H., Li, Q.-Z., and Zuo, X. (2017). TNFAIP3 downregulation mediated by histone modification contributes to T-cell dysfunction in systemic lupus erythematosus. Rheumatology 56, 835–843. doi: 10.1093/rheumatology/kew508
Zhao, L., Zhu, J., Zhou, H., Zhao, Z., Zou, Z., Liu, X., et al. (2015). Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci. Rep. 5:14991. doi: 10.1038/srep14991
Zhou, D., Bitar, A., De, R., Melgar, S., Aung, K. M., Rahman, A., et al. (2017). Induction of immunomodulatory miR-146a and miR-155 in small intestinal epithelium of Vibrio cholerae infected patients at acute stage of cholera. PLoS One 12:e0173817. doi: 10.1371/journal.pone.0173817
Keywords: canine influenza virus, H5N1, mRNA–miRNA integrate analysis, negative, KEGG
Citation: Fu C, Luo J, Ye S, Yuan Z and Li S (2018) Integrated Lung and Tracheal mRNA-Seq and miRNA-Seq Analysis of Dogs with an Avian-Like H5N1 Canine Influenza Virus Infection. Front. Microbiol. 9:303. doi: 10.3389/fmicb.2018.00303
Received: 13 October 2017; Accepted: 09 February 2018;
Published: 05 March 2018.
Edited by:
Akio Adachi, Tokushima University, JapanReviewed by:
Luis Villarreal, University of Califiornia, Irvine, United StatesCopyright © 2018 Fu, Luo, Ye, Yuan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoujun Li, c2hvdWp1bmxpQHNjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.