
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 February 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00238
The small RNA ErsA of Pseudomonas aeruginosa was previously suggested to be involved in biofilm formation via negative post-transcriptional regulation of the algC gene that encodes the virulence-associated enzyme AlgC, which provides sugar precursors for the synthesis of several polysaccharides. In this study, we show that a knock-out ersA mutant strain forms a flat and uniform biofilm, not characterized by mushroom-multicellular structures typical of a mature biofilm. Conversely, the knock-out mutant strain showed enhanced swarming and twitching motilities. To assess the influence of ErsA on the P. aeruginosa transcriptome, we performed RNA-seq experiments comparing the knock-out mutant with the wild-type. More than 160 genes were found differentially expressed in the knock-out mutant. Parts of these genes, important for biofilm formation and motility regulation, are known to belong also to the AmrZ transcriptional regulator regulon. Here, we show that ErsA binds in vitro and positively regulates amrZ mRNA at post-transcriptional level in vivo suggesting an interesting contribution of the ErsA-amrZ mRNA interaction in biofilm development at several regulatory levels.
Biofilm formation is considered to be an adaptive strategy of the human pathogen Pseudomonas aeruginosa, and the switch from the motile to a sessile mode of growth represents an important step in the virulence of this pathogen (Costerton et al., 1999).
Biofilms are microbial communities assembled in a self-produced matrix of exopolysaccharides, proteins and DNA (Ma et al., 2006), generating conditions that confer resistance and protection against antimicrobial agents and the immune system. The biofilm lifestyle cycle of P. aeruginosa PAO1 develops through coordinated stages. Adhesion to a surface is the first step in the colonization of P. aeruginosa and is followed by cell-to-cell aggregation. Attachment is an irreversible condition characterized by formation of microcolonies that develop in structured and three-dimensional clusters. During these two stages, the bacterial cells display three types of motility: swimming movement in liquid or low-viscosity conditions, swarming on semisolid surface and twitching on a solid surface. Swarming motility is based on flagella and type IV pili as well as on biosurfactants, swimming is flagella-dependent, and twitching relies on extension and retraction of type IV pili (O’Toole and Kolter, 1998; Kohler et al., 2000; Wang et al., 2014). The final stage of biofilm development is bacterial dispersion, in which the bacteria re-enter the planktonic state, spreading and colonizing other surfaces (Flemming et al., 2007; Wang et al., 2014).
As summarized in Figure 1, intertwined regulatory pathways and numerous regulators control transcriptionally and post-transcriptionally biofilm development. Most of these regulators are coordinated by the alternative sigma factor AlgT/U (σ22) (Potvin et al., 2008), a mediator of stress response and a functional homolog of Escherichia coli σE (Yu et al., 1995). AlgU regulates alginate production driving the expression of algD operon, and activating two transcriptional regulators, AlgR and AmrZ, both required for alginate production in multiple mucoid strains (Mohr et al., 1991, 1992; Yu et al., 1995). AmrZ, besides the interaction with algD, also directly affects the P. aeruginosa exopolysaccharides profile. In fact, as shown in Figure 1, AmrZ triggers the expression of the exopolysaccharide Pel interacting with a member of the pel operon (pelB) and represses the expression of the exopolysaccharide Psl binding to the pslA promoter. In addition, AmrZ affects the intracellular levels of the signaling molecule bis (3′–5′)-cyclic diguanylic monophosphate (c-di-GMP) (Jones et al., 2014; Petrova et al., 2014; Xu et al., 2016). Pel and Psl exopolysaccharides are the major contributors to P. aeruginosa biofilm structure and development. Psl supports the cell-to-cell interactions during the initial attachment and adhesion phase, forming a fiber web to constitute a scaffold for the biofilm shaping, and Pel provides structural stability to the global configuration (Ma et al., 2006, 2007; Yang et al., 2011; Jennings et al., 2015).
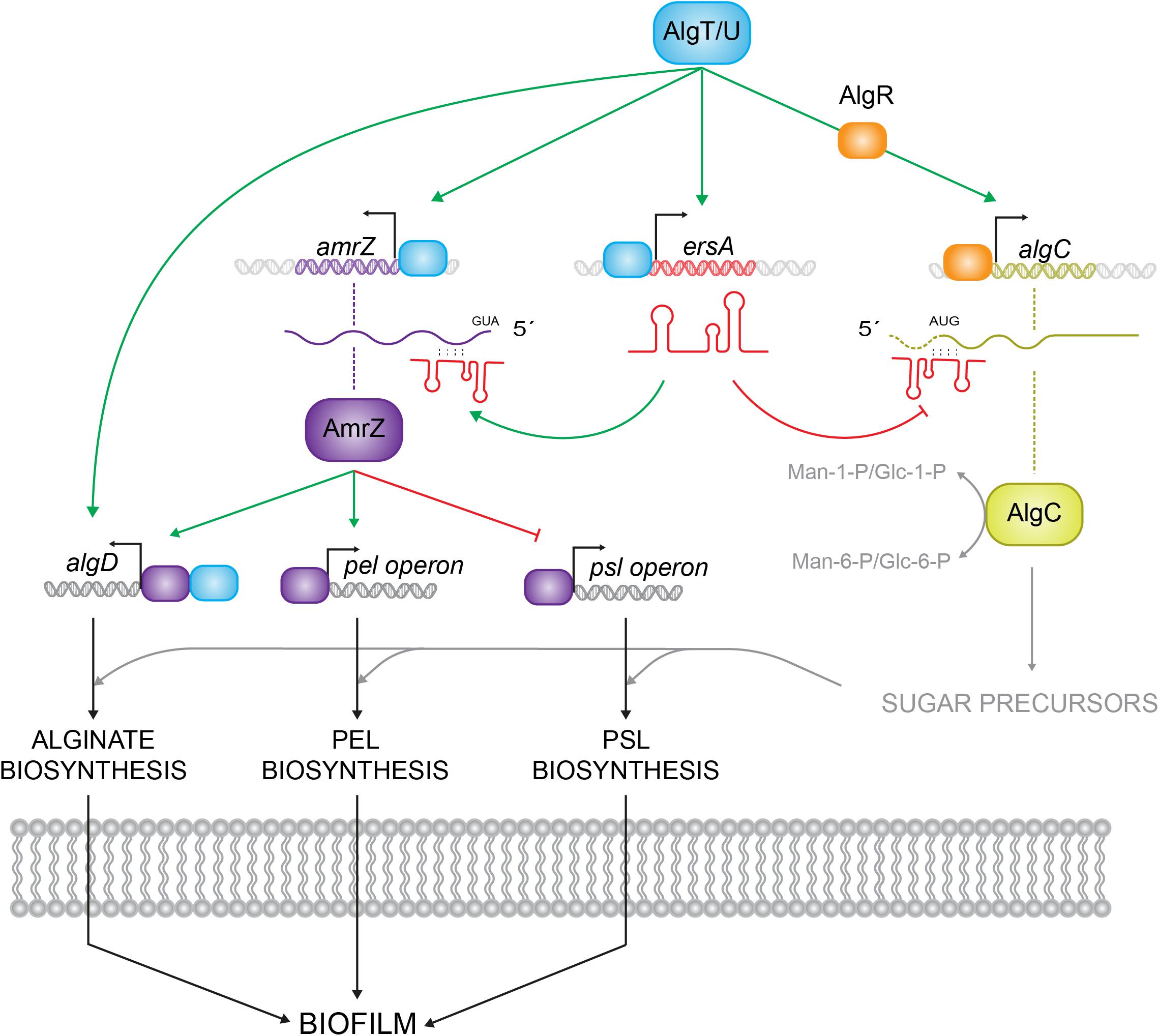
FIGURE 1. Schematic representation of different levels of AlgU-dependent regulatory routes in Pseudomonas aeruginosa. AlgU drives the expression of the alginate biosynthetic operon by activating the expression of algD promoter and it modulates exopolysaccharides (Pel and Psl) production by inducing the expression of transcriptional regulators, as AmrZ and AlgR, and the small RNA ErsA, which regulates algC at the post-transcriptional level. Green arrows represent positive regulation, red arrows negative regulation.
Biosynthesis of Pel, Psl, and LPS uses common sugar precursors supplied by the AlgU-induced AlgC enzyme, which coordinates the levels of exopolysaccharides in the cell, catalyzing the conversion of Man-6-P and glucose-6-P (Glc-6-P) to Man-1-P and Glc-1-P, respectively (Coyne et al., 1994; Ma et al., 2012). AlgC is positively regulated by AlgR at the transcriptional level, and negatively regulated by the small RNA (sRNA) ErsA at the post-transcriptional level (Zielinski et al., 1991; Ferrara et al., 2015). ErsA is a novel sRNA recently characterized in P. aeruginosa whose expression responds to several infection cues such as limited iron availability, temperature shifts from environmental to body temperature and reduced oxygen conditions. The incoherent feed-forward loop settled by ErsA and AlgU to fine-regulate AlgC was supposed to be an additional regulatory route in the complex process of biofilm shaping, in particular balancing the sugar precursors production in the exopolysaccharides biosynthesis (Ferrara et al., 2015).
In a recent study (Zhang et al., 2017), ErsA has been described to bind and regulate at the post-transcriptional level oprD mRNA, coding for a porin which highly contributes to carbapenems sensitivity. The overexpression of ErsA negatively affects translation of oprD mRNA and consequently the OprD protein level, reducing susceptibility to meropenem treatment. These findings contribute to enforce the role of ErsA in P. aeruginosa pathogenesis by regulating different virulence traits.
sRNAs can regulate multiple targets, allowing the cells to have a fast response to stress conditions and adapt in a short time frame to environmental changes (Beisel and Storz, 2010).
ErsA provides a relevant regulatory contribution balancing metabolism and virulence routes by regulating the checkpoint enzyme AlgC and it was conceivable to hypothesize novel ErsA targets in the large landscape of regulatory routes connected to exopolysaccharides production and biofilm formation.
In this study, we scrutinized for the first time the regulatory pattern of ErsA in P. aeruginosa biofilm formation revealing a positive contribution of the sRNA to biofilm maturation and shaping. An RNA-seq approach allowed us to identify several genes involved in this process, whose expression was deregulated in an ErsA deletion mutant. Most of these genes belong to AmrZ regulon, which was shown to be a novel direct target for ErsA (Figure 1).
Bacteria and plasmids used in this study are listed in Supplementary Table S1. E. coli strains were grown at 37°C in Lysogeny Broth (LB). P. aeruginosa strains were grown at 37°C in LB or in Brain Heart Infusion Broth (BHI) or Artificial Sputum Medium (ASM) in flasks at 200 r.p.m.. When required, for E. coli strains the media were supplemented with 10 μg/ml gentamycin, 100 μg/ml ampicillin, 25 μg/ml kanamycin, and for P. aeruginosa strains with 50 μg/ml gentamycin and 300 μg/ml carbenicillin. For monitoring biofilm development in flow-chambers conditions, PAO1 wild-type and PAO1 ΔersA (Ferrara et al., 2015) were chromosomally tagged with green fluorescent protein (GFP) and grown in modified FAB medium (Heydorn et al., 2000) supplemented with 0.3 mM glucose.
ErsA overexpression was obtained from pGM-ersA plasmid (Ferrara et al., 2015) using arabinose 0.2% when required.
Oligonucleotides used in this study are listed in Supplementary Table S2. Translational fusions pBBR1 amrZ::sfGFP, amrZCIS1::sfGFP, amrZΔIS2::sfGFP and amrZCIS1ΔIS2::sfGFP under the PLtetO-1 constitutive promoter were generated as follows. A DNA fragment of 161 bp including 56 nt of UTR-region and 35 codons of the open reading frame (ORF) of amrZ was amplified by PCR with oligos 1/2 (Supplementary Table S2), digested with NsiI-NheI and cloned into the sfGFP reporter vectors pXG10-SF resulting in the plasmid pXG10-amrZ::sfGFP. Likewise for amrZ::sfGFP, 161 bp including 56 nt of UTR-region and 35 codons of the ORF of amrZ were amplified by PCR with oligos 1/2 (Supplementary Table S2) from pUCIDT amrZCIS plasmid, carrying the synthetic and modified sequence of amrZ, digested with NsiI-NheI and cloned into pXG10-SF to generate the translational fusion pXG10-amrZCIS1::sfGFP. The translational fusion pXG10-amrZΔIS2::sfGFP was generated amplifying a fragment of 119 bp including 56 nt of UTR-region and 21 codons of the amrZ ORF with oligos 1/3 (Supplementary Table S2) digested with NsiI-NheI and cloned into pXG10-SF. AmrZCIS1ΔIS2::sfGFP was constructed amplifying a fragment of 119 bp including 56 nt of UTR-region and 21 codons of the amrZ ORF with oligos 1/3 (Supplementary Table S2) from pUCIDT amrZCIS plasmid. All the fragments from the PLtetO-1 promoter to the end of the GFP reporter gene, including the different versions of amrZ, were amplified from pXG10-amrZ::sfGF, amrZCIS1::sfGFP, amrZΔIS2::sfGFP and amrZCIS1ΔIS2::sfGFP, using oligos 9/10, digested with ClaI-XbaI and cloned into the low-copy number shuttle vector pBBR1-MCS5 generating the pBBR1-amrZ::sfGFP, amrZCIS1::sfGFP, amrZΔIS2::sfGFP and amrZCIS1ΔIS2::sfGFP, respectively. All the plasmids were then transformed into P. aeruginosa strains as reported previously (Ferrara et al., 2015).
A PrrB1-gfp-a transposon cassette was inserted into the chromosome of PAO1 wild-type and ΔersA by conjugation using pBK-miniTn7-ΩGm as a delivery plasmid carrying the cassette inserted into NotI site as reported previously (Lambertsen et al., 2004).
A quantity of 200 μl of overnight bacterial cultures grown in BHI or ASM and diluted to OD600 = 0.01, with the addition of carbenicillin 300 μg/ml and arabinose 0.2% when required, was aliquoted into 96-well peg-lid microtiter plates (Nunclon Delta Surface Cat. No.167008, Nunc TSP Cat. No.445497, Thermo Scientific) as reported previously (Harrison et al., 2010). The plates were incubated at 37°C in aerobic conditions with 100 r.p.m. stirring. After 20 h of incubation, growth was monitored by measuring the OD600, and the ability of the P. aeruginosa strains to adhere to the polystyrene peg-lid was tested by crystal violet staining. Briefly, the peg-lid was washed twice with saline solution and then stained with 0.1% crystal violet for 20 min (O’Toole, 2011). Excess of stain was rinsed off by placing the peg-lid in saline solution before to solubilize the dye in absolute ethanol. The optical density of each well was measured at 590 nm. Biofilm formation was expressed in adhesion units as the result of the OD590/OD600 ratio and statistical analysis were performed using T-Test.
Biofilms were grown at 30°C in flow chambers composed of three individual channels as described previously (Møller et al., 1998). PAO1 wild-type and ΔersA overnight cultures diluted to OD600 = 0.01 were inoculated into each flow channel with a small syringe. After 1 h without flow, each channel was supplied with a flow of 3 ml/h of FAB medium with glucose 0.3 mM, using a Watson Marlow 205S peristaltic pump. The mean flow velocity in the flow cells was 0.2 mm/s.
The microscopic analyses were performed using a Zeiss LSM510 confocal laser scanning microscope (CLSM; Carl Zeiss, Jena, Germany) equipped with an Ar/Kr laser and filter sets for GFP detection (excitation, 488 nm; emission, 517 nm). Images were obtained using a 40×/1.3 Plan-Neofluar oil objective.
Simulated shadow projection images and cross sections were generated using the IMARIS software package (Bitplane AG, Zürich, Switzerland).
The experiment was performed in triplicate for each strain acquiring seven random images for each channel every day for 3 days. Thus, 21 images for each time point were employed for the statistical analyses using COMSTAT 2.1 software1 (Heydorn et al., 2000; Vorregaard, 2008).
Swarming assays were performed using Nutrient Broth (Nutrient Broth n°2 Oxoid) medium plates supplemented with 0.5% glucose and 0.5% Bacto-agar (Difco). Overnight cultures normalized at the same OD600 of PAO1 wild-type and ΔersA were spotted on the same plate suitably spaced each other and placed at both 28°C and 37°C for 24 h.
Twitching was performed on LB plates supplemented with 1% Bacto-agar (Difco). The inoculation was performed with a sterile toothpick dipped in the overnight cultures and followed at 37°C for 24 h. Statistical analysis was performed on three independent replicates with GraphPad Prism software.
For RNA-Seq, cultures of wild-type PAO1 and ΔersA strains were grown to early stationary phase (OD600 = 2.7) in BHI medium. For each strain, total RNA was extracted from at least two independent biological replicates using Trizol reagent (Thermo Fisher Scientific Inc.) followed by RNA clean and concentrator kit (Zymo Research, Irvin, CA, United States) accordingly to vendors’ protocols. RNA quality was checked using RNA Nano kit on an Agilent Bioanalyzer 2100 machine. Samples with an RNA integrity number (RIN) greater than 9 were used in downstream analysis. Strand-specific sequencing libraries were prepared using 50 ng of mRNA-enriched samples as input for TruSeq stranded mRNA library preparation kit (Illumina) following vendor’s recommendations. Sequencing was performed on an Illumina NextSeq 500 to a depth of 15–20 million reads per sample. After quality filtering, raw reads were aligned using BWA aligner against P. aeruginosa PAO1 genome (NC_002516.2). Read count for gene relative abundance was obtained using HTSeq-count tool from HTSeq package (Anders et al., 2015), while differential expression analysis and statistical analysis were performed as previously described (Peano et al., 2014). RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI2 under accession number E-MTAB-6247.
Total RNA was extracted as reported previously (Ferrara et al., 2012). RNA for RNA/RNA interaction assays was prepared by T7 RNA polymerase transcription of gel-purified DNA fragments. DNA fragments for ErsA RNA and amrZ mRNAs (amrZ, amrZCIS1, amrZΔIS2, amrZCIS1ΔIS2) preparations were amplified from P.aeruginosa PAO1 genomic DNA with oligo pairs 4/5 or 4/6 and 7/8, respectively. The transcription reactions were performed using the Riboprobe® System-T7 (Promega) with 300 ng of DNA template. DNA probe was 5′-end–labeled with (γ-32P) ATP and T4 polynucleotide kinase (Promega) according to manufacturer’s instruction. Synthesized RNA was precipitated and resuspended in diethylpyrocarbonate-treated water. Purified RNA was checked by denaturing polyacrylamide gel electrophoresis and quantified using a Qubit Fluorometer.
To assess the ErsA/amrZ mRNA interactions in vitro, the binding reactions were set up as described previously (Ferrara et al., 2015). After the electrophoresis, the membrane was UV-crosslinked and hybridized with a [32P]-labeled oligo and the radioactive bands were acquired using a TyphoonTM 8600 variable mode imager scanner (GE Healthcare BioSciences) and visualized with ImageQuant software (Molecular Dynamics).
Non-radioactive EMSA were performed using Mini-Protean® Electrophoresis System (Bio-Rad) at 4°C and 150 V for 45 min. The gel was stained in SYBRTM Gold Nucleic Acid Gel Stain diluted in 0.5 × TBE. Images were acquired by Gel DocTM XR+ (Bio-Rad) imaging system.
Fluorescence measurements of P. aeruginosa strains carrying the reporters pBBR1-amrZ::gfp were carried out as previously reported (Ferrara et al., 2015). Abs595 and fluorescence polarization FP485/535 were measured in a Tecan Infinity PRO 200 reader, using Magellan as data analysis software (Tecan). GFP activities were expressed in Arbitrary Units (AU) as ratio FP485/535/Abs595. Statistical analysis performed on three individual clones per strain using T-test.
We investigated the effects of deleting the ersA gene on biofilm formation using a semi-quantitative microtiter “peg-lid” assay in Brain Heart Infusion medium (BHI). As shown in Figure 2A, the ErsA deletion resulted in decreased biofilm formation in BHI compared to PAO1 wild-type strain, and the complemented strain carrying the plasmid pGM-ersA produces more biofilm than the ersA deletion mutant strain carrying the pGM931 empty vector (Figure 2B).
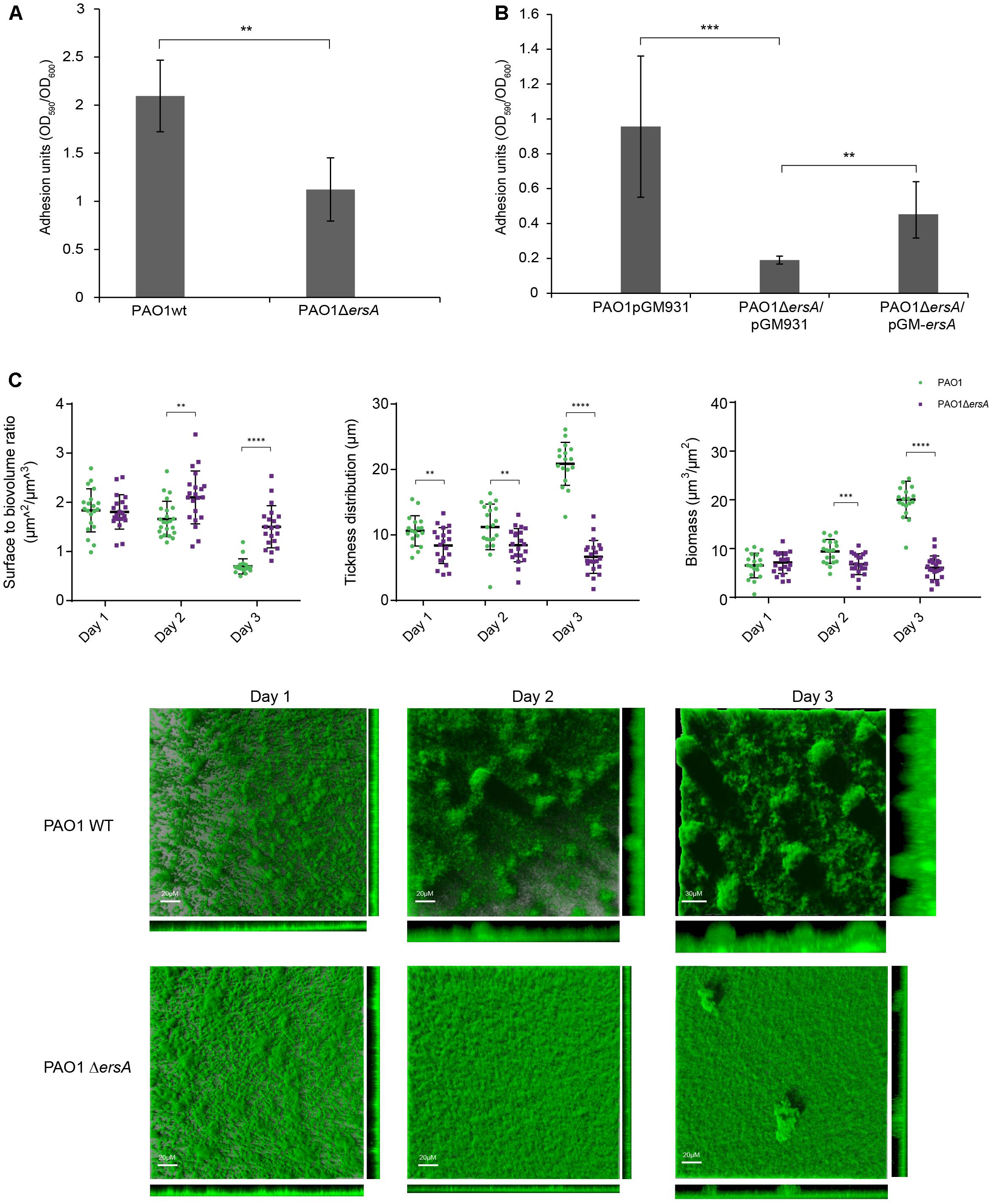
FIGURE 2. Biofilm formation of PAO1 wild-type, ΔersA, wild-type/pGM931, ΔersA/pGM931 and ΔersA/pGM-ersA strains. (A) PAO1 ersA mutant strain produces less biofilm in BHI medium when compared to the wild-type strain. (B) The phenotype is rescued when the ersA mutation is complemented by the pGM-ersA plasmid (four replicates for each strain, 24 h at 37°C. Adhesion units are expressed as the ratio of biofilm formation optical density OD590 normalized for the bacterial growth OD600). T-Test, ∗∗∗p-value < 0.001, ∗∗p < 0.01, ∗p < 0.1. (C) Spatial distribution of 3 days-old flow-chamber-grown biofilms of PAO1 wild-type and ΔersA GFP-tagged strains. The larger central plots are simulated fluorescence projections, in which long shadows indicate large, high micro-colonies. The scale bars shown are also valid for the right and lower frames. Surface to volume ratio, thickness distribution and biomass of PAO1 wild-type and ΔersA values are means of data from 21 image stacks (seven image stacks from three channels). The statistical analysis was performed using GraphPad Prism software (∗∗p-value < 0.01, ∗∗∗∗p < 0.001).
To examine the role of ErsA in P. aeruginosa biofilm architecture development, we cultivated the PAO1 wild-type and the ΔersA GFP-tagged strain, in flow-chambers continuously supplied with modified FAB medium supplemented with glucose. Biofilm development stages were followed and visualized daily for 3 days by Confocal Laser Microscopy (CLSM). In agreement with biofilm formation in the microtiter “peg-lid” assays in BHI medium, the PAO1 ΔersA strain developed less biofilm biomass than the wild-type, which showed the mushroom-like structures typical of 3-days old P. aeruginosa biofilms in flow-cells system (Figure 2C). The statistically significant differences in biomass and spatial structure between PAO1 wild-type and ΔersA biofilms were determined by the COMSTAT 2.1 software (Heydorn et al., 2000; Vorregaard, 2008) as represented in Figure 2C. We further noticed the positive influence of ErsA on adhesion and biofilm formation when overexpressed in PAO1 wild-type and ΔersA strains, grown in ASM (Supplementary Figure S1), which is defined to reflect the chemical environment of CF lungs (Sriramulu et al., 2005; Haley et al., 2012).
Motility is crucial in cell-to-cell adherence and attachment in early biofilm stages and it has been suggested an inverse regulation of motility and biofilm during biofilm development (Caiazza et al., 2007; Wang et al., 2014). Several transcriptional and post-transcriptional regulators are involved in these pathways and some of them coordinate both sessile and motile lifestyles (O’Toole and Kolter, 1998; Ramsey and Whiteley, 2004; Shrout et al., 2006; Gloag et al., 2013). To further investigate the involvement of ErsA on these biofilm-related phenotypes, we performed co-swarming, swimming and co-twitching experiments comparing PAO1 wild-type with ΔersA strain. Our results reveal a negative influence of ErsA on both swarming and twitching motility (Figure 3) and the temperature conditions do not affect ErsA regulation on swarming motility (Figure 3A). No differences between PAO1 wild-type and ΔersA mutant strain were observed for swimming motility (Supplementary Figure S2).
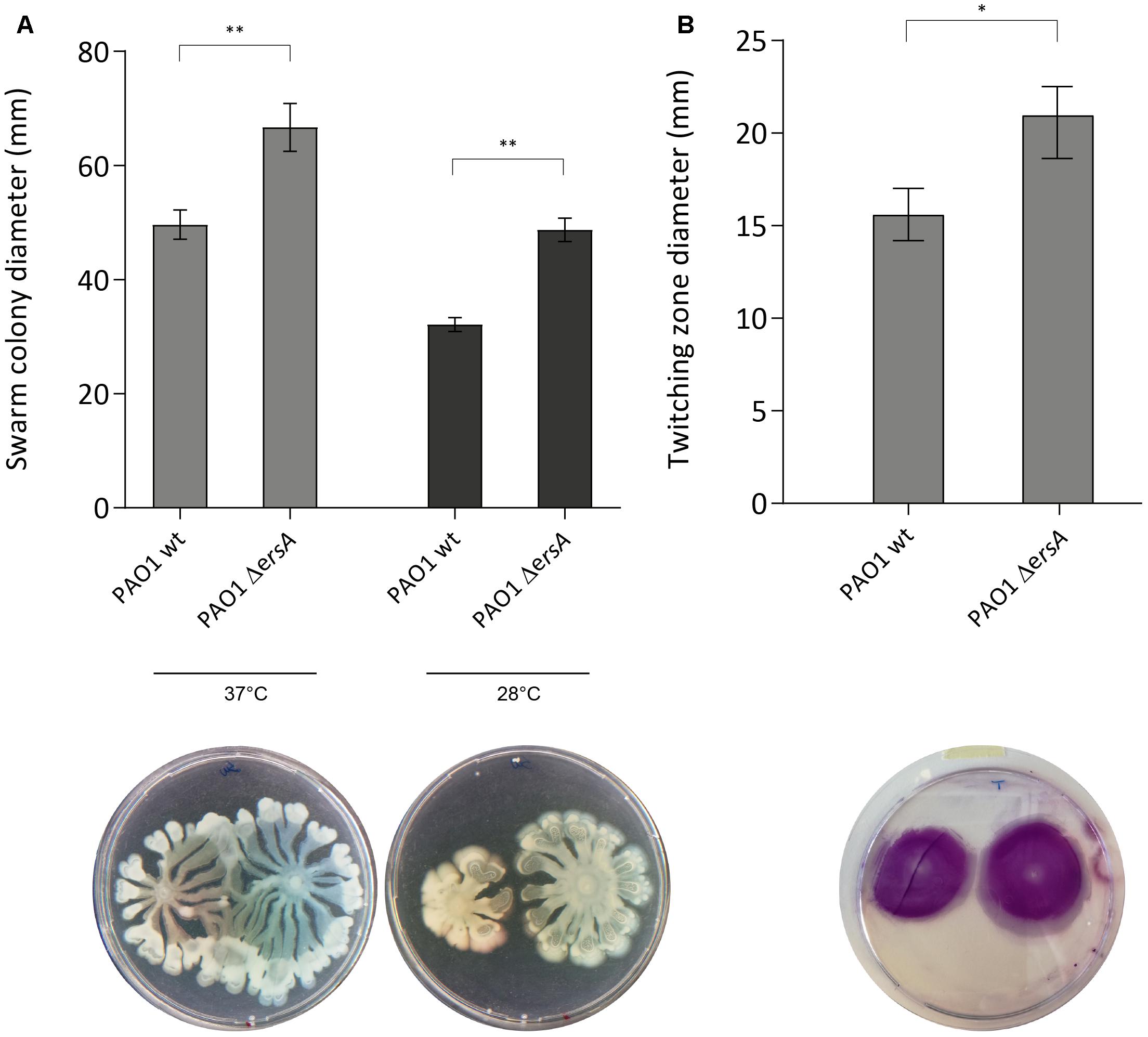
FIGURE 3. PAO1 and PAO1 ΔersA motility. ErsA deletion results in more swarming motility compared to the PAO1 wild-type on 0.5% Nutrient Broth agar plates supplemented with 0.5% glucose at 37°C and 28°C (A), and twitching motility at the plastic-1.0% LB agar interface stained with 0.1% crystal violet (B). Statistical analysis was performed on three independent replicates with GraphPad Prism software (∗p-value < 0.05, ∗∗p < 0.01). The best representative pictures are displayed.
Small RNAs are usually involved in post-transcriptional regulation, and the role of ErsA in biofilm development and motility shown in this study, suggested interference with the translation of transcriptional regulators as AmrZ. Thus, to expand the panel of ErsA targets in P. aeruginosa PAO1, and to have a better view of the effect of ErsA activity on the genome-wide gene expression, we performed an RNA-seq experiment comparing PAO1 wild-type to ErsA deletion mutant strains, grown to late exponential phase (OD600 of 2.7) in BHI medium. We observed 168 genes (Supplementary Table S3 and the most representative genes listed in Table 1) differentially expressed in the ersA deletion mutant when compared to the wild-type strain. Among the 29 genes upregulated in the ersA deletion mutant we identified genes involved in denitrification and nitrate metabolism (narI, narJ, nirN) as well as type VI and III secretion systems effectors (tssA1, tsi4, tse6).
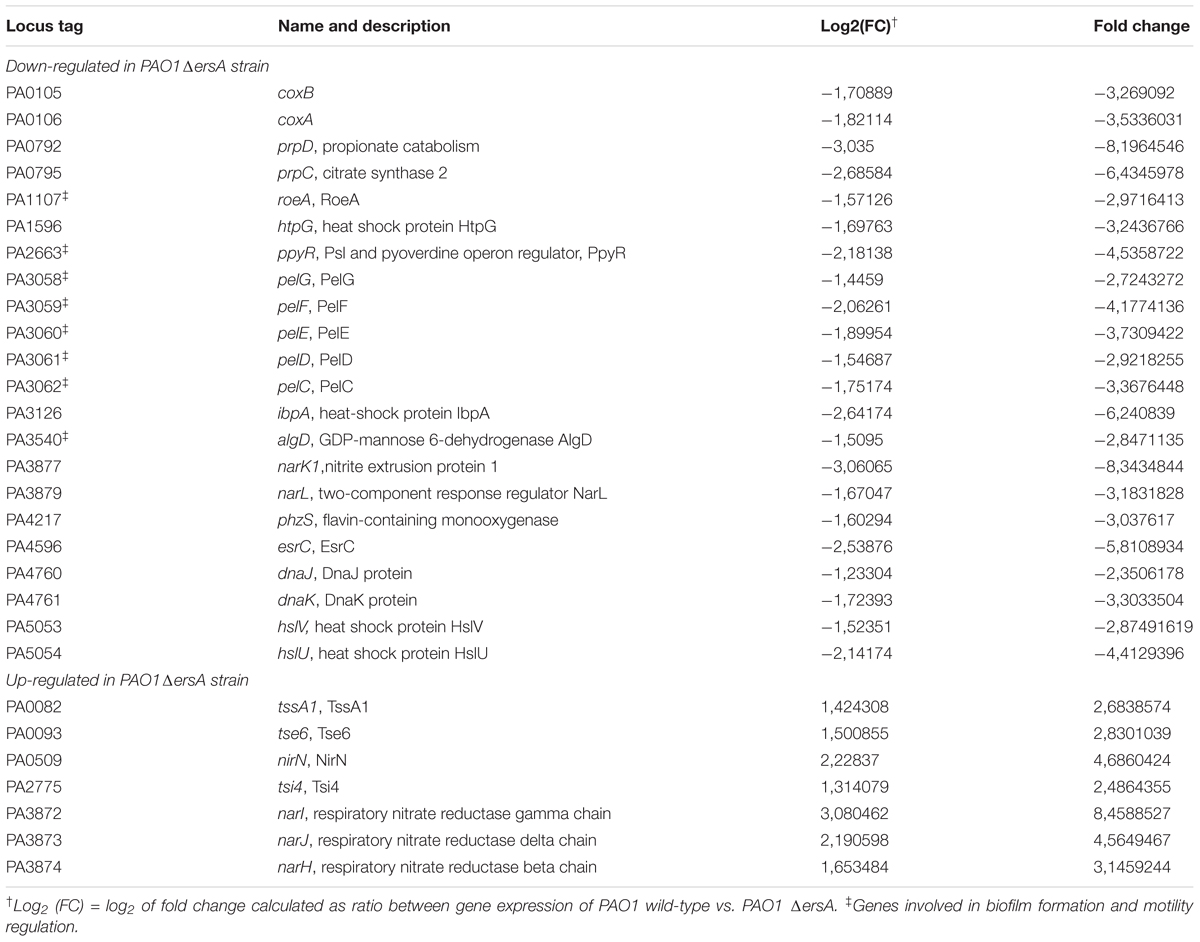
TABLE 1. Selection of the most representative genes differentially expressed in PAO1 ErsA deletion mutant with Log2 (FC) ≤-1 or Log2 (FC) ≥ 1.
The majority of genes were downregulated in absence of ErsA (139 genes); the strongest negative effect was observed for narK1 involved in nitrate transport. The other hits with a change of Log2(FC) ≤-1.5, comprise well described genes involved in biofilm formation and motility (algD, esrC, ppyR, pelCDEFG, roeA), energy and carbon metabolism (prpD, prpC, coxA, coxB), heat-shock proteins (htpG, hslU, hslV, ibpA, dnaK, dnaJ) and phzS involved in pyocyanin production.
In order to investigate the possibility that ErsA regulates biofilm modulating the expression of AmrZ at the post-transcriptional level through direct binding to the amrZ mRNA, we used a plasmid based GFP-reporter system and an electromobility-shift assay for the in vivo and in vitro validation, respectively. Before this, however, we used the full-length ErsA RNA sequence and the amrZ mRNA (including the 5′ untranslated region, 5′-UTR), as inputs in the web tool IntaRNA (Wright et al., 2014) to predict ErsA-amrZ mRNA interactions. The tool identified two putative interaction sites for ErsA on the amrZ mRNA. The interaction site 1 (IS1) involves part of the ErsA U-rich unstructured region, from nt 41 to 52 and is predicted to bind to amrZ mRNA in the region spanning +5 to +14 from the translational starting site AUG (Figure 4A). The ErsA interaction site 2 (IS2) on amrZ mRNA is predicted at positions +65 to +89 and covers a longer region on ErsA unstructured structure, from 26 to 56 nt (Figure 4A).
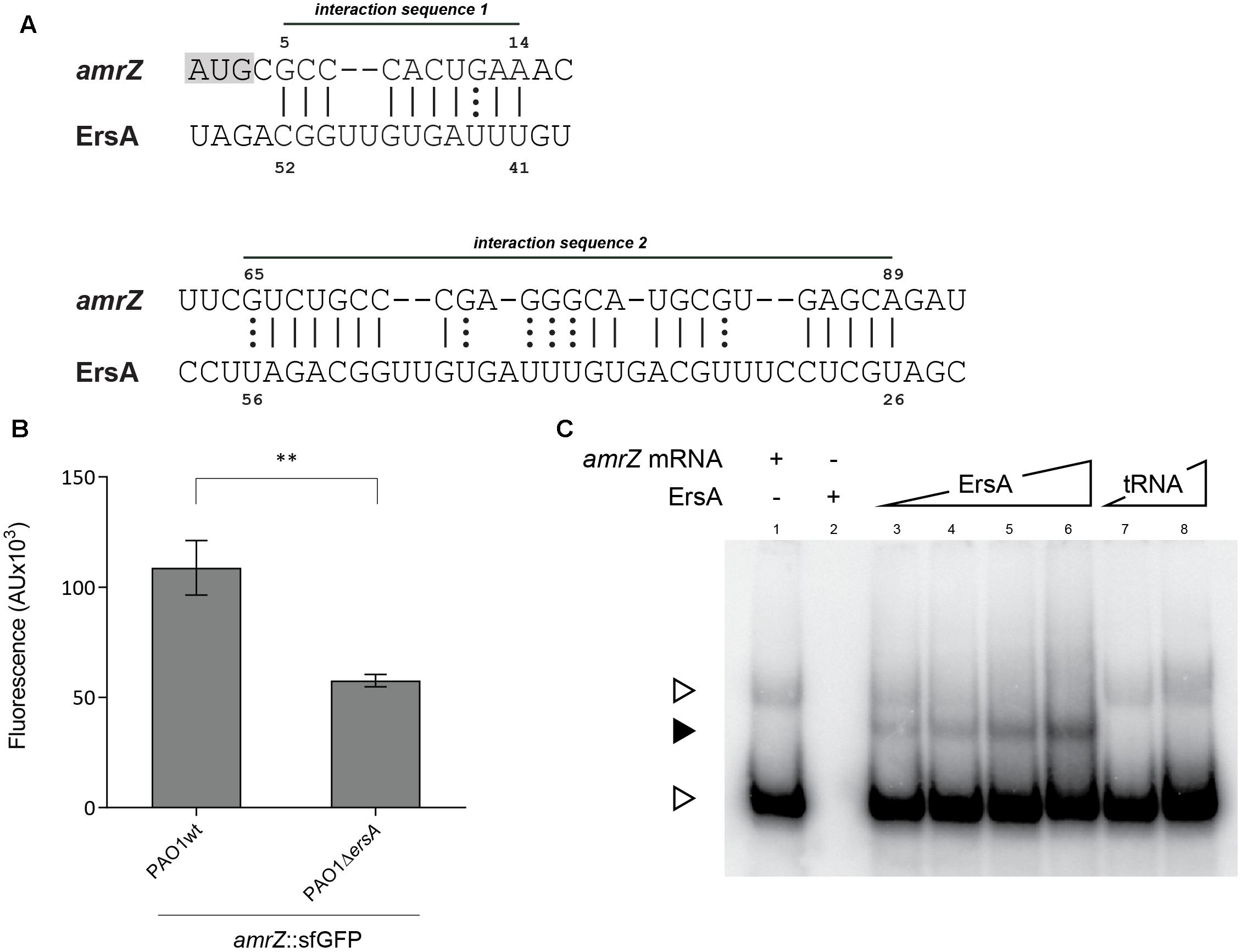
FIGURE 4. Interaction of ErsA with amrZ mRNA. (A) Prediction by IntaRNA software of the two base-pairing interactions between ErsA and amrZ mRNA. ErsA is predicted to bind to amrZ mRNA at two different sites in the ORF; the interaction sequence 1 is close to the ATG (highlighted in gray). (B) Comparison of the fluorescence polarization expressed in arbitrary units (AU) resulting from the translational fusion amrZ::gfp in PAO1 and PAO1 ΔersA. The absence of ErsA results in a reduction of the reporter activity compared to the reference strain (statistical analysis performed on three individual clones per strain using T-test, ∗∗p-value < 0.01). (C) In vitro interaction between ErsA RNA and amrZ mRNA by electrophoretic mobility shift assay. Increasing amounts of ErsA RNA (0.15, 0.3, 0.6, and 1.2 pmol; lanes 3–6) or, as a negative control, yeast tRNA (0.89 and 8.9 pmol; lanes 7 and 8) were incubated with 0.3 pmol of amrZ mRNA at 37°C for 20 min and loaded onto a native 6% polyacrylamide gel. Nucleic acids were transferred onto Hybond N+ nylon membranes. After blots, the ErsA-mRNA interactions were tested using oligonucleotide probes for the mRNA target. Free target mRNA is indicated with open arrowheads, sRNA/mRNA complex with filled arrowheads.
To test the ErsA post-transcriptional regulation on amrZ mRNA, we generated a translational fusion between the 5′-UTR along with the first 35 codons of amrZ mRNA and the superfolder variant gene of the green fluorescent protein (sfGFP) under the control of the heterologous constitutive promoter PLtetO-1. This GFP reporter fusion was transformed into P. aeruginosa PAO1 wild-type and ΔersA strains, respectively. As shown in Figure 4B, the ErsA deletion caused a reduction in GFP activity of the amrZ::sfGFP translational fusion compared to the wild-type and it was possible to increase the amrZ::sfGFP translational levels in ΔersA mutant strain by inducing with arabinose the expression of ersA from the pGM-ersA plasmid (Supplementary Figure S3), suggesting a direct effect of ErsA on amrZ translation efficiency. The lack of a full genetic complementation could be explained by the fact that we observed by Northern blot that the ErsA levels expressed by pGM-ersA in a ΔersA strain are lower than those expressed by the chromosomal copy of ersA gene (data not shown). This scenario is different from the one observed for the expression of ErsA from pGM-ersA in a wild-type background where the ErsA levels resulted to be five–sixfold higher than those expressed by the chromosomal copy of ersA gene (Ferrara et al., 2015). This would suggest a higher ErsA degradation in a ΔersA background.
Interactions of ErsA with the GFP ORF were previously controlled using a plasmid carrying exclusively the gfp gene (Ferrara et al., 2015). These results strongly suggested a positive regulation by ErsA on translation of the amrZ gene. This regulation does not depend on Hfq (data not shown). Furthermore, to document the predicted ErsA–amrZ mRNA interaction also in vitro, ErsA RNA and amrZ mRNA were synthesized, mixed and analyzed by electrophoresis on native polyacrylamide gels. As shown in Figure 4C, ErsA specifically formed a complex with the amrZ mRNA.
To further document the specific ErsA-amrZ mRNA interactions, we generated three amrZ mRNA fragments, (i) amrZCIS1, in which the interaction site 1 has been substituted with its complementary sequence, (ii) amrZΔIS2 characterized by the deletion of the interaction site 2 and (iii) amrZ CIS1ΔIS2 containing both the modifications present in amrZCIS1 and in amrZΔIS2. The in vitro analysis showed that ErsA forms a complex with both amrZCIS1 and amrZΔIS2 (Figures 5A,B), and it does not bind to amrZ CIS1ΔIS2 mRNA (Figure 5C). This suggested that both interaction sequences are involved in ErsA-amrZ binding (Supplementary Figure S4). In vitro results were corroborated by in vivo experiments, measuring the translational levels of amrZCIS1::sfGFP, amrZΔIS2::sfGFP and amrZCIS1ΔIS2::sfGFP in PAO1 wild-type and ΔersA strains. The absence of the interaction sites for ErsA causes a reduction of translational fusions activity in both genetic backgrounds (Figure 5D), associated also to a transcriptional instability (data not shown).
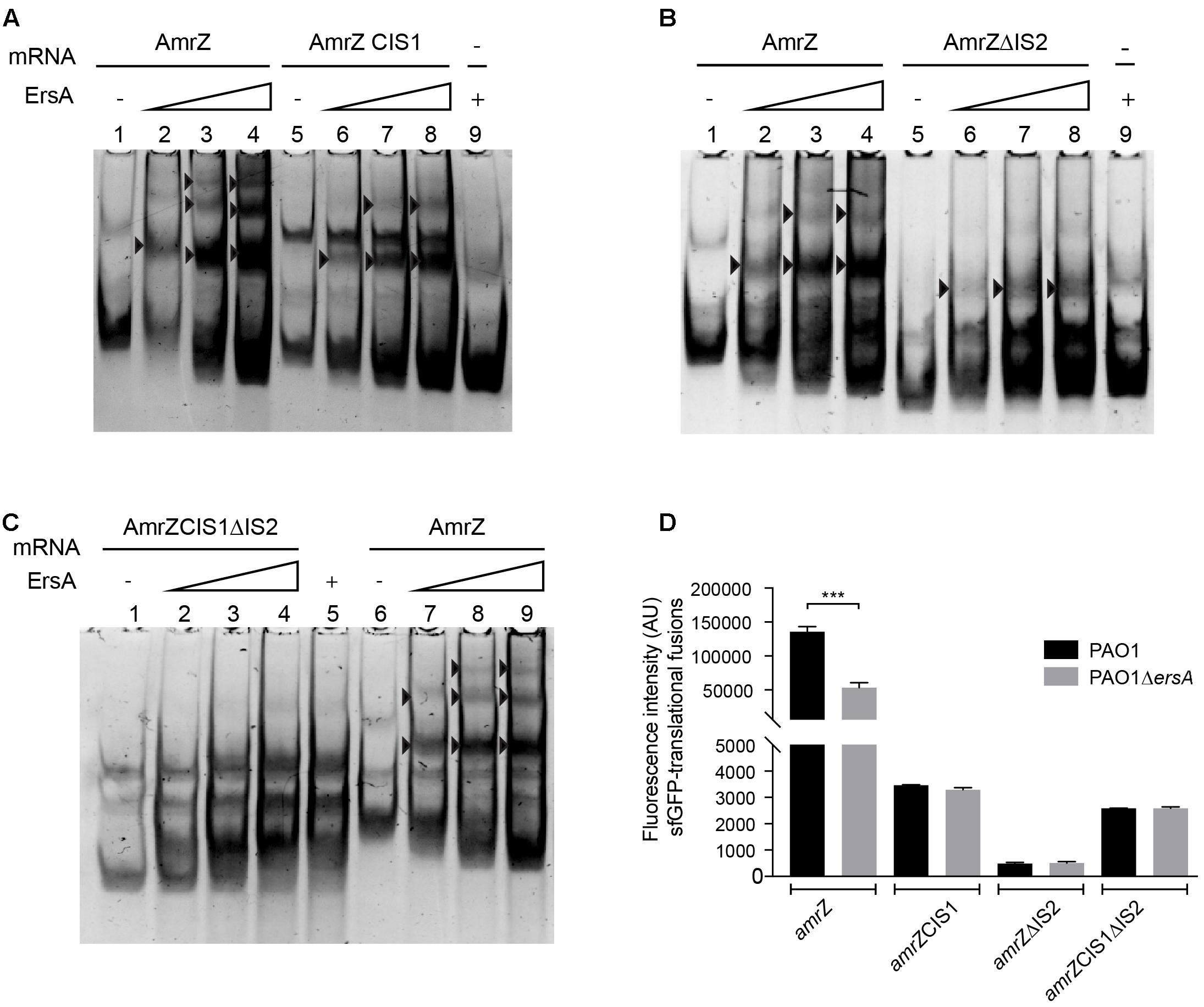
FIGURE 5. In vitro (non-radioactive EMSA) and in vivo analysis of ErsA interactions with amrZ and amrZ modified transcripts. (A) Interactions between ErsA-amrZ mRNA and ErsA- amrZCIS1 mRNA generated by substitution of IS1 with its complement sequence. (B) Interactions between ErsA-amrZ mRNA and ErsA- amrZΔIS2 mRNA carrying the deletion of IS2. (C) Interactions between ErsA-amrZ mRNA and ErsA- amrZCIS1ΔIS2 mRNA characterized by both the modifications present in amrZCIS1 and amrZΔIS2. ErsA specifically binds amrZ, amrZCIS1, and amrZΔIS2 mRNAs (black arrows) but no complex is formed when combined to amrZCIS1ΔIS2 mRNA. Binding reactions were performed mixing the amrZ mRNAs (5 pmol) with increasing amount of ErsA RNA (ratio 1:0.5, 1:1, 1:2). ErsA RNA free form 10 pmol (A lane 9, B lane 9, C lane 5), amrZ mRNA free form 5 pmol (A lane 1, B lane 1, C lane 6). (D) Comparison of the fluorescence intensity expressed in arbitrary units (AU) deriving from amrZ::sfGFP, amrZCIS1::sfGFP, amrZΔIS2::sfGFP and amrZCIS1ΔIS2::sfGFP in PAO1 wild-type and ΔersA strains. Modification in ErsA interaction site IS1 and/or IS2 causes a reduction in the translational levels of amrZ mRNA in PAO1 wild-type, comparable to those measured in ersA deleted strain. T-test ∗∗∗p-value < 0.001.
ErsA is a 132 nt long sRNA expressed in P. aeruginosa in concert with other stress-induced genes. We have previously reported that ErsA regulates exopolysaccharide production, negatively affecting at the post-transcriptional level algC mRNA translation in an incoherent feed-forward loop driven by the alternative sigma factor σ22 (Ferrara et al., 2015). Several sRNAs can regulate a broad spectrum of mRNA targets, usually governing similar or correlated cellular processes (Storz et al., 2011). In this work, we expanded the target spectrum of ErsA, validating its direct interactions with the transcriptional regulator AmrZ, which is involved in biofilm and motility, in particular by promoting multicellular colony formation and repressing swarming and twitching motility.
Pseudomonas aeruginosa strains exhibiting increased swarming phenotype generally develop flat and uniform biofilm in flow cell experiments (Shrout et al., 2006). Likewise, twitching motility is suggested to be required for monolayer creation during the initial stages of biofilm development (Shrout et al., 2006; Guttenplan and Kearns, 2013). In addition, in Gram-negative bacteria, biofilm formation and cellular motility are inversely regulated (O’Toole and Kolter, 1998; Wang et al., 2014). According to these observations, inactivation of ersA gene results in increased twitching and swarming motility leading to a less structured biofilm matrix resulting in development of homogeneous monolayers with high surface to volume ratios compared to the wild-type strain.
These phenotypes were supported by genome-wide expression analysis, showing that inactivation of ErsA affects expression of several genes involved in biofilm development and motility regulation, such as pelCDEFG, algD, ppyR, and roeA. All these genes are known to be directly or indirectly regulated by the transcriptional regulator AmrZ (Jones et al., 2014; Xu et al., 2016).
Small RNAs can positively or negatively affect translation of transcriptional regulators. For example, three sRNAs, DsrA, MicF, and GcvB, inhibit translation of the lrp gene, coding for a transcriptional regulator involved in amino acid transport and utilization (Ottesman et al., 1998; Majdalani et al., 2002; Massé et al., 2005; Prévost et al., 2007). The results of this work strongly suggest that ErsA positively affects amrZ translation through direct binding to amrZ mRNA at two different segments located on the mRNA, IS1 and IS2, with the former positioned close to the translational starting site. ErsA binds to these two regions with the same segment as involved in the algC interaction (Ferrara et al., 2015). Likewise ErsA, other sRNAs are known to regulate target expression via multiple interactions. SgrS, a regulator of the manXYZ operon binds two different sites, both involved in RNaseE-dependent degradation of the mRNA (Rice et al., 2012); the aforementioned GcvB sRNA, interacts with two independent regions on the lrp mRNA (Lee and Gottesman, 2016); and RyhB is suggested to repress expression of msrB, a methionine oxidase gene, interacting with two sites on the same mRNA (Bos et al., 2013).
It is possible that concomitant binding of two ErsA RNAs to the amrZ mRNA, is required to remodel amrZ mRNA secondary structure in order to release the AUG from the interaction with the anti-AUG sequence present in amrZ mRNA in its unbound form (Supplementary Figure S4). These interactions would expose the translational starting site and improve the efficiency of translation of amrZ transcript, thus explaining the positive contribution of ErsA at the post-transcriptional level.
Even though we identified biofilm genes being part of the AmrZ regulon and therefore differentially expressed in the absence of ErsA, the transcriptomics data does not reflect in all cases the known regulation exerted by AmrZ. For example, the roeA and ppyR genes, suggested to be positively regulated by ErsA, are known to be repressed by AmrZ (Sternberg et al., 2008; Merritt et al., 2010; Jones et al., 2014). We cannot exclude that ErsA may also stabilize directly these transcripts, for instance protecting them from degradation, or that these effects depend on the activity of other regulators affecting roeA and ppyR expression. Therefore, ErsA seems to overlap with the AmrZ regulon in guiding the switch from a motile life-style into the biofilm mode, extending our previous findings of its involvement in extracellular matrix production (Ferrara et al., 2015). ErsA, thus stimulates indirectly exopolysaccharide production through its control of AmrZ translation; acting on AlgC, it may redirect the sugar precursor fluxes providing more building blocks for extracellular polysaccharides biosynthesis (Figure 1). ErsA, in this sense, may be part of a mixed-regulatory circuit, like that involved in high osmolarity response in E. coli (Guillier et al., 2006).
This mixed-regulatory circuit could be used to take advantage of ErsA in order to have a more rapid and enhanced response compared to transcriptional regulators, in particular in stress conditions (Shimoni et al., 2007) or for niche-competition in case of mixed-species biofilms. Indeed, ErsA has recently been described to be overexpressed in P. aeruginosa biofilm grown with Staphylococcus aureus. However, the role of ErsA in neutralizing S. aureus agents has to be investigated (Miller et al., 2017).
Thus, ErsA may be employed as a “fast switcher” in the regulation of biofilm development at multiple stages and regulatory levels, fine-tuning the main routes controlled by the alternative sigma factor σ22 in the transition between acute and chronic infection of P. aeruginosa.
GB, SF, and SM conceived and designed the study. MF, SF, GB, and SM conceived the experiments. MF, SF, and ER designed and performed the experiments. MF, GB, SF, SM, ER, and HJ analyzed the data. GB, SM, and HJ contributed reagents, materials and analysis tools. MF, GB, and SM wrote the paper.
This work has been supported by the European Commission (NABATIVI-223670 and EU-FP7-HEALTH-2007-B) and Italian Cystic Fibrosis Research Foundation (FFC#13/2015) with the contribution of Gruppo di Sostegno FFC di Sassari Castelsardo and Delegazione FFC di Boschi Sant’Anna Minerbe and (FFC#14/2016) with the contribution of Delegazione FFC di Reggio Calabria and Gruppo di Sostegno FFC di Vigevano.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CN and handling Editor declared their shared affiliation.
The authors are grateful to Sara Carloni and Janus Anders Juul Haagensen for the helpful contribution in phenotypic experiments design.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00238/full#supplementary-material
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Beisel, C. L., and Storz, G. (2010). Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 34, 866–882. doi: 10.1111/j.1574-6976.2010.00241.x
Bos, J., Duverger, Y., Thouvenot, B., Chiaruttini, C., Branlant, C., Springer, M., et al. (2013). The sRNA RyhB regulates the synthesis of the Escherichia coli methionine sulfoxide reductase MsrB but not MsrA. PLOS ONE 8:e63647. doi: 10.1371/journal.pone.0063647
Caiazza, N. C., Merritt, J. H., Brothers, K. M., and O’Toole, G. A. (2007). Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 3603–3612. doi: 10.1128/JB.01685-06
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Coyne, M. J., Russell, K. S., Coyle, C. L., and Goldberg, J. B. (1994). The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176, 3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994
Ferrara, S., Brugnoli, M., De Bonis, A., Righetti, F., Delvillani, F., Dehò, G., et al. (2012). Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs. PLOS ONE 7:e36553. doi: 10.1371/journal.pone.0036553
Ferrara, S., Carloni, S., Fulco, R., Falcone, M., Macchi, R., and Bertoni, G. (2015). Post-transcriptional regulation of the virulence-associated enzyme AlgC by the σ22-dependent small RNA ErsA of Pseudomonas aeruginosa. Environ. Microbiol. 17, 199–214. doi: 10.1111/1462-2920.12590
Flemming, H. C., Neu, T. R., and Wozniak, D. J. (2007). The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 189, 7945–7947. doi: 10.1128/JB.00858-07
Gloag, E. S., Turnbull, L., Huang, A., Vallotton, P., Wang, H., Nolan, L. M., et al. (2013). Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. U.S.A. 110, 11541–11546. doi: 10.1073/pnas.1218898110
Guillier, M., Gottesman, S., and Storz, G. (2006). Modulating the outer membrane with small RNAs. Genes Dev. 20, 2338–2348. doi: 10.1101/gad.1457506
Guttenplan, S. B., and Kearns, D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37, 849–871. doi: 10.1111/1574-6976.12018
Haley, C. L., Colmer-Hamood, J. A., and Hamood, A. N. (2012). Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol. 12:181. doi: 10.1186/1471-2180-12-181
Harrison, J. J., Stremick, C. A., Turner, R. J., Allan, N. D., Olson, M. E., and Ceri, H. (2010). Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat. Protoc. 5, 1236–1254. doi: 10.1038/nprot.2010.71
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. doi: 10.1099/00221287-146-10-2395
Jennings, L. K., Storek, K. M., Ledvina, H. E., Coulon, C., Marmont, L. S., Sadovskaya, I., et al. (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 112, 11353–11358. doi: 10.1073/pnas.1503058112
Jones, C. J., Newsom, D., Kelly, B., Irie, Y., Jennings, L. K., Xu, B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLOS Pathog. 10:e1003984. doi: 10.1371/journal.ppat.1003984
Kohler, T., Curty, L. K., Barja, F., Van Delden, C., and Pechere, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182, 5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000
Lambertsen, L., Sternberg, C., and Molin, S. (2004). Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726–732. doi: 10.1111/j.1462-2920.2004.00605.x
Lee, H. J., and Gottesman, S. (2016). SRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 44, 6907–6923. doi: 10.1093/nar/gkw358
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/JB.01202-06
Ma, L., Lu, H., Sprinkle, A., Parsek, M. R., and Wozniak, D. J. (2007). Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 189, 8353–8356. doi: 10.1128/JB.00620-07
Ma, L., Wang, J., Wang, S., Anderson, E. M., Lam, J. S., Parsek, M. R., et al. (2012). Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14, 1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x
Majdalani, N., Hernandez, D., and Gottesman, S. (2002). Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46, 813–826. doi: 10.1046/j.1365-2958.2002.03203.x
Massé, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Merritt, J. H., Ha, D. G., Cowles, K. N., Lu, W., Morales, D. K., Rabinowitz, J., et al. (2010). Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10
Miller, C. L., Van Laar, T. A., Chen, T., Karna, S. L. R., Chen, P., You, T., et al. (2017). Global transcriptome responses including small RNAs during mixed-species interactions with methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. Microbiologyopen 6:e00427. doi: 10.1002/mbo3.427
Mohr, C. D., Hibler, N. S., and Deretic, V. (1991). AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol. 173, 5136–5143. doi: 10.1128/JB.173.16.5136-5143.1991
Mohr, C. D., Leveau, J. H., Krieg, D. P., Hibler, N. S., and Deretic, V. (1992). AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 174, 6624–6633. doi: 10.1128/JB.174.20.6624-6633.1992
Møller, S., Sternberg, C., Andersen, J. B., Christensen, B. B., Ramos, J. L., Givskov, M., et al. (1998). In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64, 721–732.
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. doi: 10.3791/2437
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Ottesman, S. U. G., Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. (1998). DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 95, 12462–12467. doi: 10.1073/pnas.95.21.12462
Peano, C., Chiaramonte, F., Motta, S., Pietrelli, A., Jaillon, S., Rossi, E., et al. (2014). Gene and protein expression in response to different growth temperatures and oxygen availability in Burkholderia thailandensis. PLOS ONE 9:e93009. doi: 10.1371/journal.pone.0093009
Petrova, O. E., Cherny, K. E., and Sauer, K. (2014). The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 196, 2827–2841. doi: 10.1128/JB.01628-14
Potvin, E., Sanschagrin, F., and Levesque, R. C. (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55. doi: 10.1111/j.1574-6976.2007.00092.x
Prévost, K., Salvail, H., Desnoyers, G., Jacques, J.-F., Phaneuf,É., and Massé, E. (2007). The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 64, 1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x
Ramsey, M. M., and Whiteley, M. (2004). Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 53, 1075–1087. doi: 10.1111/j.1365-2958.2004.04181.x
Rice, J. B., Balasubramanian, D., and Vanderpool, C. K. (2012). Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc. Natl. Acad. Sci. U.S.A. 109, E2691–E2698. doi: 10.1073/pnas.1207927109
Shimoni, Y., Friedlander, G., Hetzroni, G., Niv, G., Altuvia, S., Biham, O., et al. (2007). Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 3:138. doi: 10.1038/msb4100181
Shrout, J. D., Chopp, D. L., Just, C. L., Hentzer, M., Givskov, M., and Parsek, M. R. (2006). The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62, 1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x
Sriramulu, D. D., Lünsdorf, H., Lam, J. S., and Römling, U. (2005). Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676. doi: 10.1099/jmm.0.45969-0
Sternberg, C., Attila, C., Ueda, A., and Wood, T. K. (2008). PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl. Microbiol. Biotechnol. 78, 1–32. doi: 10.1007/s00253-007-1308-y
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Vorregaard, M. (2008). Comstat2 - A Modern 3D Image Analysis Environment for Biofilms. Ph.D. thesis, Kongens Lyngby, Technical University of Denmark.
Wang, S., Yu, S., Zhang, Z., Wei, Q., Yan, L., Ai, G., et al. (2014). Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 80, 6724–6732. doi: 10.1128/AEM.01237-14
Wright, P. R., Georg, J., Mann, M., Sorescu, D. A., Richter, A. S., Lott, S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42, W119–W123. doi: 10.1093/nar/gku359
Xu, B., Soukup, R. J., Jones, C. J., Fishel, R., and Wozniak, D. J. (2016). Pseudomonas aeruginosa AmrZ binds to four sites in the algD promoter, inducing DNA-AmrZ complex formation and transcriptional activation. J. Bacteriol. 198, 2673–2681. doi: 10.1128/JB.00259-16
Yang, L., Hu, Y., Liu, Y., Zhang, J., Ulstrup, J., and Molin, S. (2011). Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13, 1705–1717. doi: 10.1111/j.1462-2920.2011.02503.x
Yu, H., Schurr, M. J., and Deretic, V. (1995). Functional equivalence of Escherichia coli σE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177, 3259–3268.
Zhang, Y.-F., Han, K., Chandler, C. E., Tjaden, B., Ernst, R. K., and Lory, S. (2017). Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 6, 919–937. doi: 10.1111/mmi.13857
Keywords: Pseudomonas aeruginosa, small regulatory RNA, post-transcriptional regulation, biofilm, virulence
Citation: Falcone M, Ferrara S, Rossi E, Johansen HK, Molin S and Bertoni G (2018) The Small RNA ErsA of Pseudomonas aeruginosa Contributes to Biofilm Development and Motility through Post-transcriptional Modulation of AmrZ. Front. Microbiol. 9:238. doi: 10.3389/fmicb.2018.00238
Received: 21 November 2017; Accepted: 31 January 2018;
Published: 15 February 2018.
Edited by:
Christian Sohlenkamp, Universidad Nacional Autónoma de México, MexicoReviewed by:
David Salvador Zamorano Sanchez, University of California, Santa Cruz, United StatesCopyright © 2018 Falcone, Ferrara, Rossi, Johansen, Molin and Bertoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Bertoni, Z2lvdmFubmkuYmVydG9uaUB1bmltaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.