- 1Department of Plant Physiology, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich-Schiller-University Jena, Jena, Germany
- 2Systems Biology and Bioinformatics Group, Leibniz Institute for Natural Product Research and Infection Biology—Hans-Knöll-Institute, Jena, Germany
- 3Department of Biochemistry, Max-Planck Institute for Chemical Ecology, Jena, Germany
Verticillium dahliae is a soil-borne vascular pathogen that causes severe wilt symptoms in a wide range of plants. Co-culture of the fungus with Arabidopsis roots for 24 h induces many changes in the gene expression profiles of both partners, even before defense-related phytohormone levels are induced in the plant. Both partners reprogram sugar and amino acid metabolism, activate genes for signal perception and transduction, and induce defense- and stress-responsive genes. Furthermore, analysis of Arabidopsis expression profiles suggests a redirection from growth to defense. After 3 weeks, severe disease symptoms can be detected for wild-type plants while mutants impaired in jasmonate synthesis and perception perform much better. Thus, plant jasmonates have an important influence on the interaction, which is already visible at the mRNA level before hormone changes occur. The plant and fungal genes that rapidly respond to the presence of the partner might be crucial for early recognition steps and the future development of the interaction. Thus they are potential targets for the control of V. dahliae-induced wilt diseases.
Introduction
Vascular wilts caused by members of the genus Verticillium are among the most devastating fungal diseases worldwide, and these soil-borne ascomycete fungi attack a large variety of plant hosts in many parts of the world (Decetelaere et al., 2017) which leads to massive yield losses (Pegg and Brady, 2002). Among the 10 species within the Verticillium genus, Verticillium dahliae has the broadest host range with the ability to infect >200 plant species worldwide (Agrios, 1997; Inderbitzin et al., 2011; Inderbitzin and Subbarao, 2014). Verticillium species produce microsclerotia, which can survive in soil or dead plant material for more than 10 years, but they also form resting mycelia which survive in dead plant material. Newly growing hyphae rapidly penetrate the roots of their hosts, reach the vascular tissue and ultimately propagate in the xylem (Puhalla and Bell, 1981; Schnathorst, 1981). Initially, these hemibiotrophic fungi show biotrophic behavior that does not lead to severe reductions in plant performance. However, at later stages, they shift to a necrotrophic interaction characterized by the reprogramming of phytohormone metabolism (Veronese et al., 2003; Thaler et al., 2004; Tjamos et al., 2005), synthesis of hydrogen peroxide (H2O2) and nitric oxide (NO, Yao et al., 2011, 2012), and defense gene activation which ultimately induces host cell death (Reusche et al., 2012; Zhang et al., 2016). A typical symptom of these pathogens is wilting, which occurs as a consequence of impaired vascular transportation (Reusche et al., 2012).
Arabidopsis is an ideal model plant to study the Verticilllium infection process at the molecular level. Sun et al. (2014) showed that jasmonate phytohormones are highly induced upon V. dahliae infection, including the jasmonic acid (JA) pre-cursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA) and the jasmonic acid-isoleucine conjugate (JA-Ile). JA-Ile, synthesized by the enzyme JASMONATE RESISTANT 1 (JAR1), is the active signaling compound that binds to the JA receptor CORONATINE INSENSITIVE 1 (COI1) to initiate the downstream signaling cascade (Xie et al., 1998; Staswick and Tiryaki, 2004; Fonseca et al., 2009). Hydroxylation of JA-Ile by a P450 enzyme leads to the deactivation of the molecule (Koo et al., 2011). Previous observations on JA-deficient def1 tomato plants showed decreased fitness after V. dahliae treatment (Thaler et al., 2004), while external application of methyl JA (MeJA) to cotton and tomato plants partially blocked disease development due to the reduced growth of the fungus (Li et al., 1996). Recent RNA-seq analysis in cotton showed that in addition to the biosynthesis of JA, also other components of the JA signaling cascade are targets of Verticillium. For instance, the gene of a repressor protein of COI1, GhJAZ10, was significantly upregulated in a V. dahliae-resistant cotton cultivar (Chini et al., 2007; Thines et al., 2007; Zhang W. W. et al., 2017). On the other hand, Verticillium itself needs an activated COI1 signaling pathway in the plant to cross the root-shoot barrier and to induce disease symptoms in the leaf. Consequently, V. dahliae-infected coi1 mutant plants showed less severe wilt symptoms (Ralhan et al., 2012).
Besides jasmonates, Verticillium-infested plants also accumulate other phytohormones like salicylic acid (SA) and ethylene (ET), (Fradin and Thomma, 2006; Ratzinger et al., 2009; Sun et al., 2014). SA plays an important role in plant defense against pathogens by activation of systemic acquired resistance (SAR) (Metraux, 2001). In Brassica napus the concentration of SA increased after infection with Verticillium longisporum in the xylem sap of the plants (Ratzinger et al., 2009). Also in Arabidopsis, a significant increase of SA could be detected in V. dahliae-infested plants (Sun et al., 2014). Exogenous application of SA protected cotton callus cells against a V. dahliae (VD)-toxin preparation by increase of chitinase and β-1,3-glucanase activities (Li et al., 2003; Zhen and Li, 2004). Interestingly, several Arabidopsis genotypes affected in different steps of SA signaling did not show altered resistance against V. dahliae infection (Veronese et al., 2003).
The role of ET in pathogen responses is still controversial although it was shown that ET can increase resistance and control symptom expression in some hosts. In soybean, tobacco and Aarabidopsis, ET is involved in host resistance against particular classes of pathogens (Knoester et al., 1998; Hoffman et al., 1999; Thomma et al., 1999), while a tomato mutant impaired in ET perception exhibited a significant reduction in disease symptoms after inoculations with different bacterial and fungal pathogens (Lund et al., 1998). Similarly, a tomato mutant with decreased ET biosynthesis showed significantly reduced symptom development (Robinson et al., 2001). The involvement of ET in response to Verticillium infection and VD-toxins has been shown repeatedly (Pegg and Cronshaw, 1976; Mansoori and Smith, 2005; Sun et al., 2014). Also the ET-resistant Arabidopsis mutant etr1-1 was found to display less chlorosis upon Verticillium inoculation (Veronese et al., 2003; Tjamos et al., 2005; Pantelides et al., 2010).
A growing number of other chemical compounds of both plant and fungal origin have been identified that participate in disease development of infected host plants. For Verticillium, a range of peptides, hormones like cytokinins, and other metabolites were shown to confer partial resistance to infection (Reusche et al., 2013; Bu et al., 2014; Gaspar et al., 2014; Roos et al., 2014). In addition, the Ve gene provides resistance against isolates of V. dahliae in tomato and Arabidopsis (Kawchuk et al., 2001; Fradin et al., 2009, 2011) and the VET1 gene was able to convey increased tolerance with milder chlorosis symptoms (Veronese et al., 2003).
Due to the vascular location of the growing fungus, Verticillium wilt of agricultural plants is difficult to control by chemicals or molecular tools. Even after removal of infected plants from agricultural fields, the resting structures (microsclerotia) remain in soil making them a hazard for future plantings (Fradin and Thomma, 2006). Biocontrol studies with bacterial and fungal isolates like Pseudomonas putida B E2, Pseudomonas chlororaphis K15, Serratia plymuthica R12 or Paenibacillus alvei K165 on Solanaceae, Malvaceae, and Brassicaceae have been shown to be an alternative strategy to restrict Verticillium-induced wilts (Berg et al., 2001; Tjamos et al., 2005; Li et al., 2012). Sun et al. (2014) showed that Piriformospora indica, a beneficial endophytic fungus of Sebacinales which colonizes the roots of many plant species, is an efficient biocontrol agent that restricts V. dahliae growth in the model plant Arabidopsis thaliana.
V. dahliae rapidly and efficiently colonizes Arabidopsis roots within 24 h post germination. In order to identify plant and fungal genes involved in these early recognition processes we performed an RNA-seq analysis under standardized co-cultivation conditions. We found a massive reprogramming of both the plant and fungal expression profiles that occurred before pathogen induced, defense-related phytohormone levels changed in the host. This alteration in expression profiles clearly indicates that both plant and pathogen respond very rapidly to the presence of the other.
Materials and Methods
Growth Conditions of Seedlings
A. thaliana wild-type (ecotype Columbia-0) seeds, and seeds of the jar1, coi1-16, and cyp94B3 mutants (kindly provided by Axel Mithöfer, MPI-CE, Jena) were surface-sterilized and placed on Petri dishes with MS media supplemented with 0.3% gelrite (Murashige and Skoog, 1962). After cold treatment at 4°C for 48 h, plates were incubated vertically for 9 or 14 days at 22°C under long day conditions (16 h light/8 h dark; 80 μmol m−2 s−1, for experimental setup, cf. Figure S1).
Growth Conditions of Fungi and Preparation of Spore Solutions
V. dahliae wild-type (FSU-343, Jena Microbial Resource Center, Germany) and a GFP-labeled strain (IPP0860, kindly provided by Prof. Tiedemann, University of Göttingen) were grown for 1–2 weeks on Potato-Dextrose-Agar (PDA) medium (Bains and Tewari, 1987) at 23°C in the dark. To obtain high spore production, the still white mycelia (with low amount of microsclerotia) were transferred to liquid KM medium (Hill and Kaefer, 2001) and incubated for 4–5 days at room temperature (RT) in the dark and 110 rpm. The cultures were filtered through two layers of a nylon membrane (75 μm pore size), pelleted and washed with water. The spore concentration was determined with a hemocytometer and adjusted to 5 × 106 per ml.
Co-cultivation Assays
For short time co-cultivation assays, 9 day-old A. thaliana seedlings of equal sizes were used. Co-cultivation of A. thaliana and the fungus was performed under in vitro culture conditions on a nylon membrane on PNM medium (Johnson et al., 2011). Two days prior to use, 100 μl of the generated spore solutions (in water) or an equal amount of water were plated on six sterile membrane stripes (4 × 1 cm) placed on PDA plates and incubated at room temperature (Figure S1). For the co-cultivation the membrane stripes with fungus or water were placed on the PNM plates together with two plants per membrane. The plants were placed in the way that the roots were in contact with the fungus while the leaves were not (Figure S1). Plates were sealed with 3M™ Micropore tape and incubated for 24 h at 80 μmol m−2 s−1 with light from one side (leaves directed to light). Roots and leaves were harvested separately for further analysis. All experiments were performed three times independently (≈120 seedlings per replicate).
For long term co-cultivation [10 and 20 days post infection (dpi)], 14 day-old A. thaliana seedlings of equal size were used. Plants were grown and infected as described above and transferred to Magenta boxes (Sigma-Aldrich, Germany) after 24 h. One plant was added per box which contained 30 g sterile vermiculite mixed with 100 ml liquid PNM medium. The boxes were incubated at 23°C under short day conditions (9 h light/15 h dark; 80 μmol m−2 s−1). After 10 or 20 dpi, plants were visually examined and photographed. The performance of the plants [n = 6 (control) and 9 (VD-infected)] was quantified based on the efficiency of the photosynthetic electron transfer measured with the Fluorcam as described before (Matsuo et al., 2015).
RNA Isolation and RNA-Seq
RNA of both plant and fungus was isolated from 100 mg root material (≈ 120 seedlings per replicate, n = 3). The frozen roots were finely ground with mortar and pistil and weighed. The RNA from the samples was extracted with peqGOLD TriFast™ FL (VWR, Darmstadt, Germany) according to the manufacturer's protocol. The RNA was further processed by use of the PureLink™ RNA Mini Kit (Thermo Fisher Scientific, Dreieich, Germany) with on-column DNAse treatment. RNA was dissolved in water and checked for quality. The isolated RNA was shipped to GeneCore (Heidelberg, Germany) where library construction was performed using the mRNA sequencing Sample Preparation Guide (Illumina, Cat#RS-930-1001), followed by a validation of the library and RNA sequencing (Hi Seq 2000, single-end 75 bp, ~50 Mill. reads/sample). The removal of low quality reads and Illumina adapters was performed using Trimmomatic (Bolger et al., 2014). The remaining reads were then aligned to the A. thaliana (TAIR10.33) and V. dahliae (ASM15067v2.37) reference genomes using the RNA-seq aligner STAR (Dobin et al., 2013). Differential gene expression analysis was performed using DESeq2 (Love et al., 2014) with the raw counts obtained from FeatureCounts (Liao et al., 2014). Differentially Expressed Genes (DEGs) of both interaction partners were analyzed [FoldChange ≥ 2 and p-Value (FDR) ≤ 0.05]. The function of the DEGs was analyzed with the TAIR (www.arabidopsis.org) and the Fungi Ensembl (http://fungi.ensembl.org/Verticillium_dahliae) databases. Pathway analysis for the plant and the fungus was executed with the KEGG (Kyoto Encyclopedia of Genes and Genomes) Mapper tool (http://www.genome.jp/kegg/tool/map_pathway2.html; RRID:SCR_012773).
Analysis of Gene Expression and Fungal Colonization
The RNA (n = 3) was isolated as described above. One μg of RNA was transcribed to cDNA using the Omniscript RT Kit (Qiagen, Hilden, Germany). Fifty nanograms of synthesized cDNA was used as template for RT-qPCR in a CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Munich, Germany) with the Brilliant II SYBR® Mastermix (Agilent, Böblingen, Germany). The mRNA levels for each cDNA probe were normalized with respect to the RPS18B (plant) or VD_Actin2 (fungus, e.g., Klimes and Dobinson, 2006; Yang et al., 2013) mRNA levels. The normalized fold expression of GOIs was calculated according to ΔΔCP (Pfaffl, 2001). The primer pairs are given in Table S5. To analyze fungal colonization of different mutant plants, normalized fungal housekeeping gene expression was compared to that in WT plants.
Confocal Microscopy
Samples for confocal laser scanning microscopy were prepared according to the method for the short time co-cultivation described above. The GFP-labeled V. dahliae strain on Arabidopsis seedlings was imaged using a LSM 880 (Zeiss Microscopy GmbH, Jena, Germany) with the 488 nm laser line of an argon multiline laser (11.5 mW). Images were taken with a 40x objective (Plan-Apochromat 40x/0.8). Lambda stacks were created using the 32 channel GaAsP detector followed by Linear Unmixing with ZEN software (Zeiss, Jena, Germany). Z-stacks were taken from specific areas of the sample and Maximum Intensity Projections were produced with ZEN software. Cross-sections of the roots with a width of 14 μm were done with a Microm HM560 Cryostar (Southeast Pathology Instrument Service, Charleston, USA).
Quantification of Phytohormones
Phytohormones were extracted from the green parts of co-cultivated seedlings (1 sample = seedlings from 1 plate, ≈ 50–100 mg, n = 10). Frozen samples were homogenized for 30 s at 1000 rpm in a 2010 Geno/Grinder® (Spex SamplePrep, Stanmore, UK) and mixed with 1 ml methanol containing 40 ng/ml of D6-JA, D6-ABA, D4-SA, and 8 ng/ml of D6-JA-Ile (Scholz et al., 2017). All samples were shaken for 30 min at 4°C and centrifuged at 13,000 rpm for 20 min at 4°C. The supernatants were collected and the sample re-extracted with 500 μl methanol. The combined supernatants were evaporated to dryness at 30°C using a vacuum concentrator. Residues were re-suspended in 200 μl methanol and centrifuged at 13,000 rpm for 10 min. The supernatants were collected and measured with the API 5000 LC-MS/MS system (Applied Biosystems, Framingham, USA) as previously described (Vadassery et al., 2012). Since it was observed that both the D6-labeled JA and JA-Ile contained 40% of the corresponding D5-labeled compounds, both peaks were combined for analysis.
Statistics
Experiments were repeated three times to ensure reproducibility and 120–150 seedlings were used in each treatment for each mutant. Data of all independent experiments were pooled and analyzed. For comparison of two groups, the Mann Whitney U-test was applied. For statistical analyses of multiple groups, one-way analysis of variance (one-way ANOVA) was used as indicated in the figure legends. Different letters indicate significant differences between treatments. SigmaPlot13 and Origin Pro were used for data analysis and graph composition.
Results
Co-cultivation of Arabidopsis Seedlings with V. dahliae for 24 H
Verticillium species are considered as hemibiotrophs, where a biotrophic phase—within the root xylem without a visible disease phenotype—is followed by a necrotrophic phase in the aerial parts of the plant (Reusche et al., 2012). We focused on the very early phase of infection, the pre-vascular growth phase, and analyzed the plant and fungal expression profiles during the first 24 h of fungal root colonization in Arabidopsis. By developing a stable co-cultivation method (scheme in Figure S1), a reproducible colonization of the seedlings by the fungus was achieved. Confocal microscopic pictures taken 24 h after co-culture demonstrate that Arabidopsis roots are already heavily colonized by the GFP-labeled V. dahliae (Figure 1). The hyphae of the fungus form a net over the root surface and root tip while the first spores can be observed at the site of lateral root formation (Figures 1G–I). In cross-sections of the colonized root (Figures 1J–O) it can be observed that the fungus penetrates the root surface, but did not yet invade the vascular tissue after 24 h of co-culture.
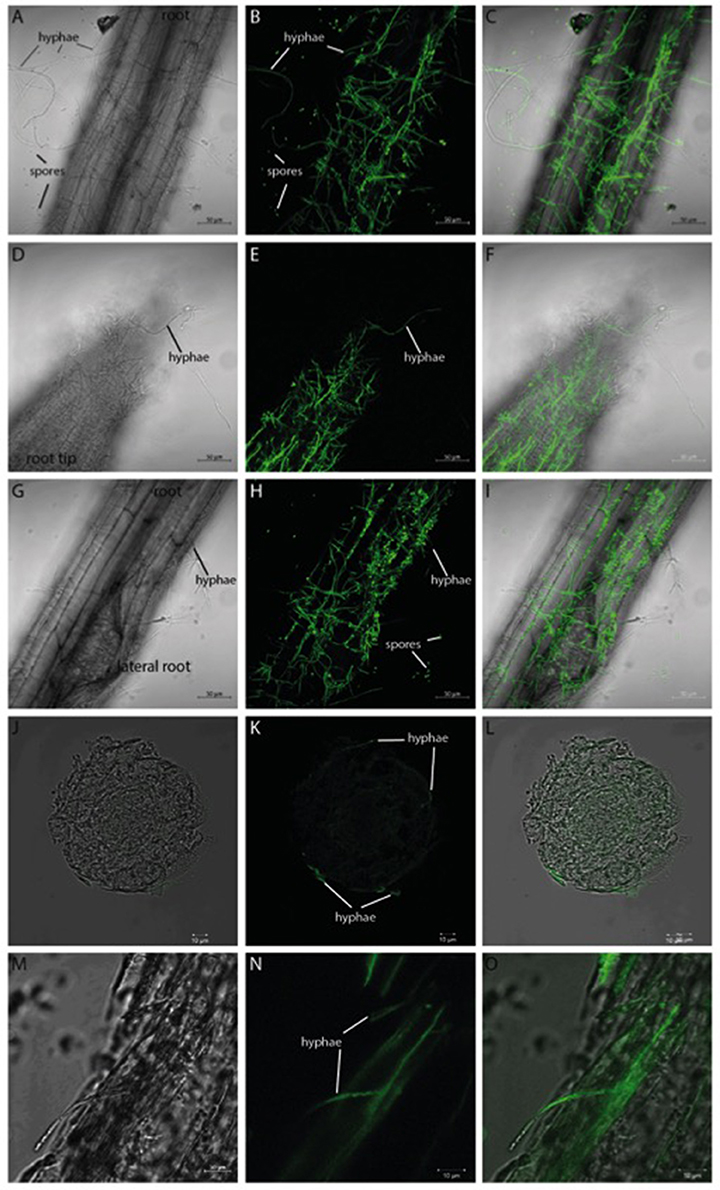
Figure 1. GFP-labeled Verticillium dahliae colonizing roots of 10 day-old seedlings of Arabidopsis thaliana after 24 h co-culture. Shown are pictures (obtained from confocal microscopy) of the colonized Arabidopsis root surface (A–C), root tip (D–F), area around lateral roots (G–I) as well as a cross-section (J–L) and longitudinal section (M–O). A bright field image (left), the GFP image (middle), and the overlay of both (right) are shown for each analyzed area. Both the hyphae and the spores of V. dahliae show a GFP fluorescence.
Dual-RNA-Seq of Arabidopsis-Verticillium Co-culture Reveals High Number of Differentially-Expressed Genes (DEGs)
Recent studies of plant-Verticillium co-cultures focus on analysis of the plant transcriptome by RNA-seq and indicate major changes in nearly 19% of pathways in early phases of infection (1 or 4 dpi, e.g., Faino et al., 2012; Zhang W. W. et al., 2017). The RNA-seq data generated in this study 24 h after co-cultivation was analyzed for changes in both the plant and fungal transcriptomes and revealed a total number of 4432 DEGs for both organisms (Table 1). Compared to Arabidopsis seedlings grown alone, co-cultivation with Verticillium results in 1143 DEGs, 903 of them are significantly up-regulated and 240 down-regulated (Tables S1, S2). For the fungus, 3289 DEGs were detected with 1695 significantly up-regulated (Table S3) and 1594 significantly down-regulated genes (Table S4). This suggests that the fungal expression profile responds more strongly to the presence of a host than the plant genome to a pathogen. The results for selected genes were verified by RT-qPCR (Figures 7, 8, WT).

Table 1. Overview of differentially regulated genes in A. thaliana (ATH) and V. dahliae (VDA) co-culture compared to organisms grown separately.
Multiple Arabidopsis Pathways Are Affected by V. dahliae Infection
Previous studies during longer co-cultivation times have shown that infection of Arabidopsis plants with Verticillium species results in disruption of water transport and massive accumulation of phytohormones like jasmonates (JAs), ethylene (ET), salicylic acid (SA), or abscisic acid (ABA), as well as stomata closure (Pegg and Brady, 2002; Fradin and Thomma, 2006; Klosterman et al., 2009; Ralhan et al., 2012; Sun et al., 2014). To identify early targets of the fungus in Arabidopsis and to analyze and classify the plant DEGs, we chose the 24 h co-cultivation time point and mapped the identified plant DEGs to distinct pathways in the KEGG database (Kanehisa, 1997). In total, 78 pathways were affected by the fungus (Table 2 and Figure 2A). Metabolic pathways involved in carbon and amino acid metabolism seemed to be the major targets, such as SWEET genes and sugar efflux transporters, which often respond to pathogens and symbionts for nutritional gain (Chen et al., 2010). In our study, SWEET11, −3, and −12 were down-regulated. Genes involved in plant development were also affected. So was ROOT CAP POLYGALACTURONASE28, a crucial player in root tip growth, which was also down-regulated at the mRNA level (Kamiya et al., 2016). Defense-related genes, genes involved in the synthesis and propagation of defense signals like JA, SA, NO, and reactive oxygen species (ROS), membrane-associated receptor kinases are up-regulated while pathways involved in or related to photosynthesis, carotenoid and flavonoid biosynthesis are down-regulated. We observed the activation of genes for various defense-related WRKY transcription factors (WRKY18, −28, −30, −33, −41, −45, −53, −55, −71). It has been shown that WRKY71 promotes shoot branching, acceleration of flowering and cell death (cf. references in TAIR, www.Arabidopsis.org); WRKY33 is a target of Botrytis to repress defense in Arabidopsis (Liu et al., 2017), WRKY53 is involved in disease resistance against Verticillium longisporium (Reusche et al., 2013), and WRKY30 confers general abiotic stress response to the plant (Scarpeci et al., 2013). Upregulation of these genes suggest that the plant reprograms its metabolism for defense. Also candidate genes for the perception of pathogen-associated molecular patterns, and enzymes as well as signaling components which might lead to the activation of defense compounds in the host were upregulated in the presence of the fungus. This includes six genes for receptor-like protein kinases (RLP7, −15, −19, −20, −35, −38) and seven genes for proteins with TIR-NBS-LRR domains involved in disease resistance responses (At1g57630; At1g66090; At5g45240; At5g41750; At1g63750; At1g72900, At1g72920). For the RLP genes, no clear function has been described yet. The mRNAs for the Toll and Interleukin-1 Receptor (TIR)-Nucleotide-Binding Site (NBS)-Leucine-Rich Repeat (LRR) proteins (TIR-NBS-LRR), At1g66090 and At1g72920, have been shown to travel to distant tissue upon stress and thus might be involved in systemic signal propagation (Thieme et al., 2015). Furthermore, numerous genes for cytochrome P450 enzymes responded to the fungus. Of these, 17 are up-regulated and 11 have a described role in plant defense (cf. TAIR). Two of the three down-regulated genes (CYP87A2 and CYP705A12) are involved in cytokinin signaling (cf. TAIR). Many of the cyp mRNAs are also known to be mobile within the plant. Finally, defense gene activation is often mediated via Ca2+ signaling and Ca2+-binding proteins. The majority of genes that code for proteins with Ca2+-related functions are involved in signaling, ion uptake and distribution. Many of these are also up-regulated in Arabidopsis upon V. dahliae infection. At an early phase of interaction with the pathogen, the plant appears to shift its resources from primary metabolism to defense processes (cf. Discussion).
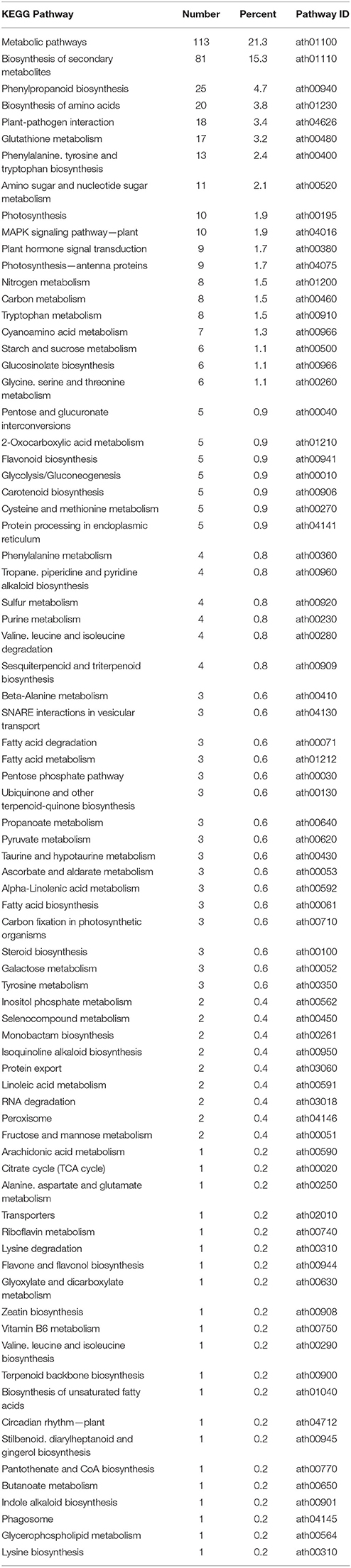
Table 2. KEGG pathway classification of genes differentially expressed in Arabidopsis alone vs. co-culture with V. dahliae.
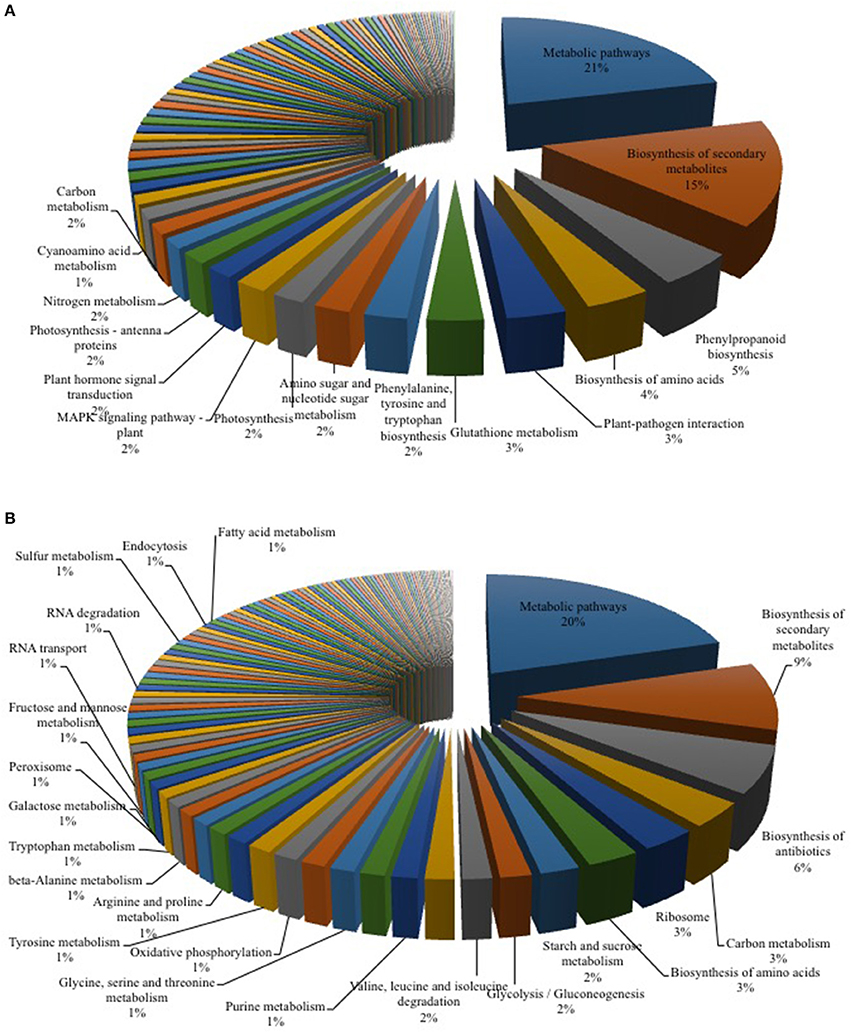
Figure 2. KEGG Mapper analysis of RNA-seq data for Arabidopsis-Verticillium co-culture. Shown are the mapped pathways (n = 3) for differentially expressed genes in Arabidopsis alone vs. co-culture (A) and Verticillium alone vs. co-culture (B). A full list of the other regulated pathways is shown in Tables 2, 3.
Interestingly, two genes for SNARE proteins (soluble N-ethylmaleimide-sensitive-factor attachment receptor, At5g39630, and At1g16225) also responded to the fungus. SNARE proteins are involved in vesicle trafficking and membrane fusion and deliver defense products to infection sites during exocytosis-associated immune responses (Wang et al., 2017, and references cited therein). Stimulation of EXO70H4 and EXO70A3 for EXOCYST subunits is consistent with the stimulation of exocytosis-mediated callose deposition and cell wall maturation by the fungus (Li et al., 2010; Kulich et al., 2015).
V. dahliae Pathways Respond to the Interaction with Arabidopsis
Many genes annotated in the 117 fungal pathways that were affected by co-cultivation with Arabidopsis (Table 3 and Figure 2B), are still uncharacterized or with unknown function, which limits our analysis on the fungal side. However, like on the plant side, the majority of the affected pathways involved in primary metabolism, e.g., genes for the amino acid metabolism, were highly down-regulated, while those for sugar metabolism and sugar transport processes were up-regulated. This suggests that the fungus starts quite early to reprogram its primary metabolism and adapts amino acid and sugar metabolism to being inside a host. mRNA for several enzymes employed in degradation of the plant cell wall, such as the cell wall glycosyl-hydrolase YteR (Moore et al., 2016), were up-regulated as shown previously. Genes involved in processes such as oxidative phosphorylation, ribosome formation, RNA transport and degradation or proteasome functions were preferentially down-regulated in the co-cultivated fungus. Closer inspection of the data supports the idea that the fungus down-regulates genes for essential processes (primary sucrose, N and P metabolism, ion homeostasis, redox processes, defense, and secondary metabolites) perhaps because of its increasing reliance on the plant. Reduced defense gene activation may indicate that the fungus prevents the synthesis of compounds that restrict its growth and propagation in the host.
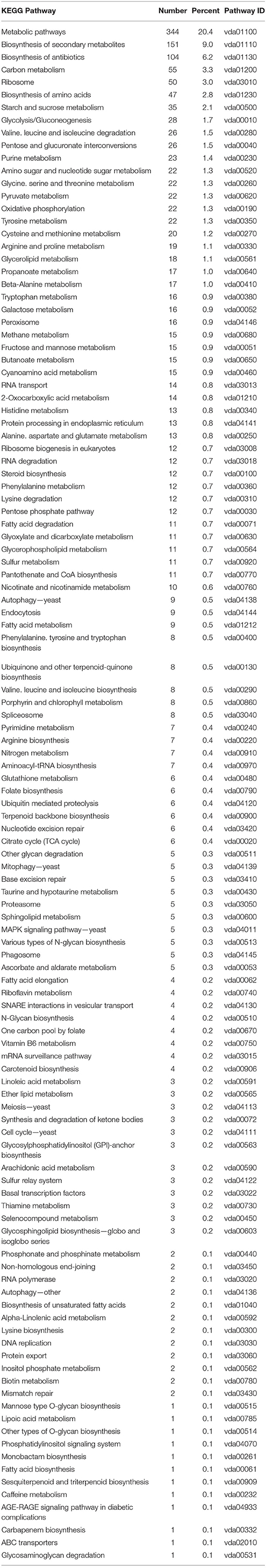
Table 3. KEGG pathway classification of genes differentially expressed in Verticillium alone vs. co-culture.
V. dahliae Growth Is Reduced in Arabidopsis JA Mutants
Former studies indicate that the accumulation and perception of jasmonates are key events for both plant and fungal responses to the interaction. The plant defense machinery is activated by an elevation of jasmonates and activation of the receptor COI1, while the fungus activates the plant COI1-dependent JA pathway to induce disease symptom development in the host (Feys et al., 1994; Xie et al., 1998; Ralhan et al., 2012). Upregulation of genes involved in JA biosynthesis and responses within the first 24 h of co-cultivation demonstrates that the course is already set even before significant changes in the plant hormone levels can be detected (cf. below). We observed several DEGs involved in the α-linolenic acid pathway as well as in plant hormone signal transduction pathways (Table 2 and Figure 2A). While the expression of growth-associated genes like the auxin-responsive genes ARF5 (Krogan et al., 2016) and several members of the GH3-family (e.g., GH3.17, GH3.4, DFL2, WES1, Staswick et al., 2005) was decreased, an up-regulation of the JA biosynthetic genes LIPOXYGENASE 3 and 4 (LOX3 and 4, Acosta and Farmer, 2010; Umate, 2011) and OXOPHYTODIENOATE-REDUCTASE 3 (OPR3, Müssig et al., 2000) as well as of an ET response factor (ERF1, Fujimoto et al., 2000) was observed.
To further investigate the role of JA in the Arabidopsis-V. dahliae interaction, we analyzed the phenotype and vitality of Verticillium-infected JA mutants jar1 (Staswick et al., 1992; Staswick and Tiryaki, 2004), coi1-16 (Ellis and Turner, 2002), and cyp94B3 (Koo et al., 2011). Although there was no visual difference between the mutants and the WT plants 10 dpi, the WT plants were dead after 20 dpi, while the mutant plants showed disease symptoms in leaves but were still vital and alive (Figure 3). This result was confirmed by analysis of the efficiency of the electron transfer during photosynthesis using chlorophyll fluorescence measurements, a sensitive marker for plant vitality (Figure 4). At 10 dpi, there were only small differences between the mutants and the WT, and chlorophyll fluorescence values around 0.81 indicate that the plants were capable of photosynthesis. At 20 dpi, the JA mutants still possessed a fluorescence value between 0.81 and 0.83, while WT plants were at 0.24 corresponding to very low levels of electron transport and photosynthetic efficiency. Thus, these non-invasive measurements provide an efficient tool to determine and quantify disease development in Verticillium-infected hosts. To obtain further insight into the colonization efficiency of V. dahliae, the content of fungal RNA—in the plants previously analyzed for chlorophyll fluorescence—was determined in roots and shoots separately, and the levels were compared to those in WT plants (Figure S2). The colonization pattern at the two tested time points was very different. While coi1-16 and cyp94B3 roots showed a significantly lower colonization at 10 dpi compared to WT (38 and 30%, respectively), jar1 roots showed an intermediate level with 83% (Figure S2A). The respective shoots of the plants show the same trend (Figure S2B). At 20 dpi, no difference in colonization of the mutant shoots was detectable; all of them showed significantly lower fungal RNA levels compared to the WT (Figure S2D). The levels of fungal RNA in the roots of coi1-16 plants was significantly lower compared to those of all other genotypes.
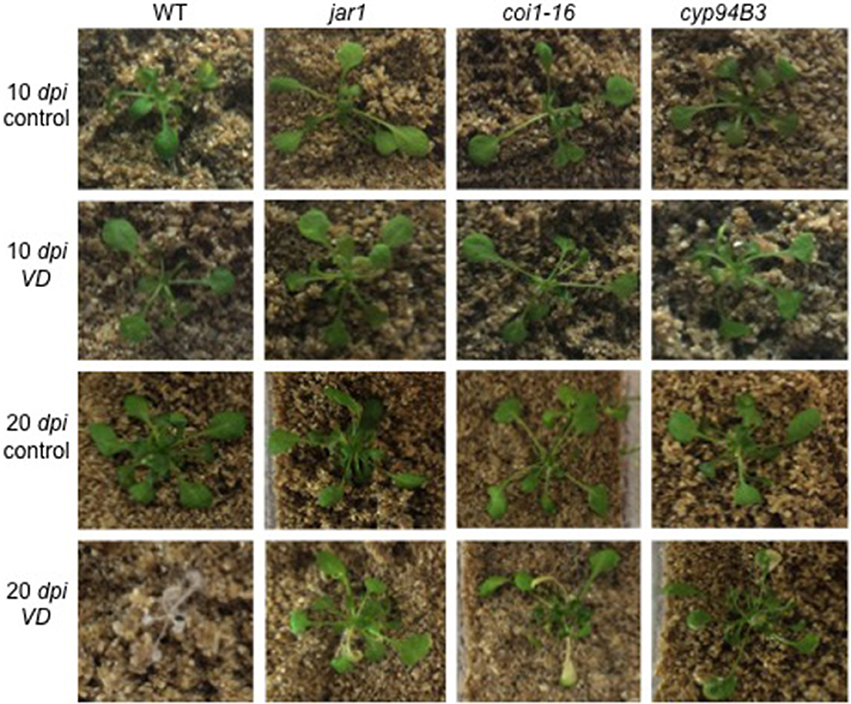
Figure 3. Phenotype of V. dahliae (VD)-infected Arabidopsis WT and JA mutant plants. Shown are representative phenotypes of WT, jar1, coi1-16, and cyp94B3 plants 10 (upper two rows) and 20 dpi (lower two rows) grown in Magenta boxes. 14 day-old plants were treated with water (control, n = 6) or VD (n = 9) for 24 h and then transferred to the boxes.
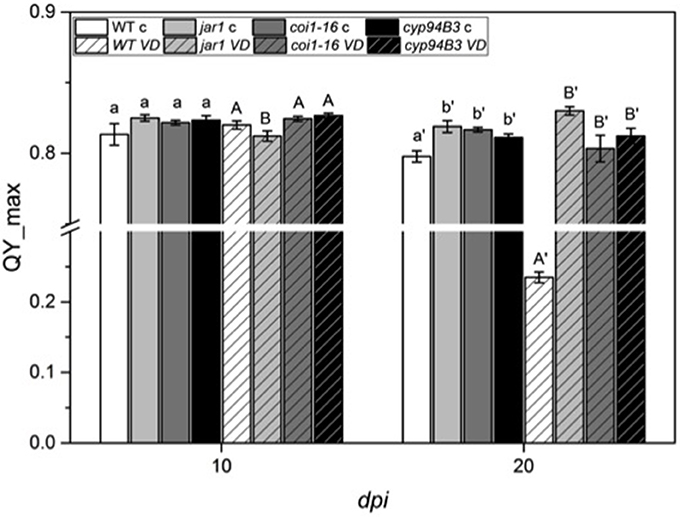
Figure 4. Chlorophyll fluorescence (QY_max) analysis of Arabidopsis WT and JA mutant plants after 24 h of V. dahliae infection. Shown are the QY_max values [± SE, n = 6 (control) and 9 (VD-infected)] in control (plain color) and VD-infected (stripes) plants 10 and 20 dpi. QY_max was analyzed in WT (white), jar1 (light gray), coi1-16 (dark gray) and cyp94B3 (black) plants. Statistically significant differences between controls and between infected plants were analyzed by one-way ANOVA separately, p < 0.05 (Sidak). Different letters indicate a statistically significant difference.
Root Colonization and Plant Hormone Levels Are Not Altered after the First 24h of Co-culture with V. dahliae
To compare the colonization of the mutant plants in the very early phase, the content of fungal RNA was also analyzed 24 h after co-cultivation (Figure 5). All plants showed a similar fungal colonization with no statistically significant differences (Figure 5A). Compared to the WT colonization level (set as 100%), jar1 plants showed a colonization of 90.0%, coi1-16, and cyp94B3 of 121.4 and 129.8%, respectively (Figure 5B). These results suggest that there are also no differences in plant JA levels during the first 24 h of co-culture.
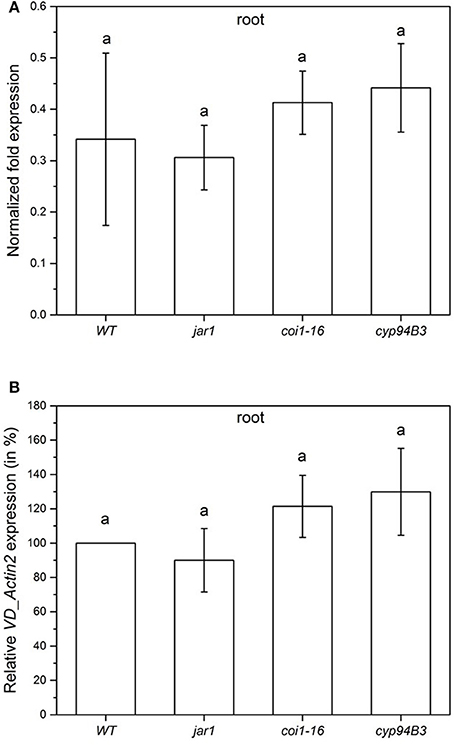
Figure 5. Colonization of Arabidopsis WT and JA mutant plants by V. dahliae after 24 h. Shown is the normalized fold expression (± SE, n = 3) of VD_Actin2 in VD-infected WT, jar1, coi1-16 and cyp94B3 plants (A) and the relative expression in % (B). The expression level of VD_Actin2 in VD-infected WT plants was used as control and set to 1.0. The mRNA levels for each cDNA probe were normalized with respect to the RPS18B mRNA level. Statistically significant differences between the mutants were analyzed by one-way ANOVA, p < 0.05 (Sidak). Different letters indicate a statistically significant difference.
In a previous study we demonstrated that V. dahliae infection leads to an elevation of JA, JA-Ile, and SA 21 dpi (Sun et al., 2014). To analyze whether the accumulation of these phytohormones was already induced in the first 24 h, leaf tissue of WT and the mutants was analyzed, but the content of SA (Figure 6A) and JA-Ile (Figure 6C) were found not to be significantly induced by the fungus. There was also no difference in the JA content in WT, coi1-16 and cyp94B3, while a slight difference was observed in jar1 plants. Accumulation of JA in this mutant is caused by the inactivation of the Ile-conjugating enzyme. These data confirm that the changes in the gene expression profiles described above occur before the fungus induces changes in the plant phytohormone levels. Thus, the identified genes respond most likely to fungal signals and not to fungus-induced phytohormone changes in the plant. JA signaling in the host is necessary for disease development only during later phases of the interaction, which can be strongly repressed or retarded when JA function is impaired.
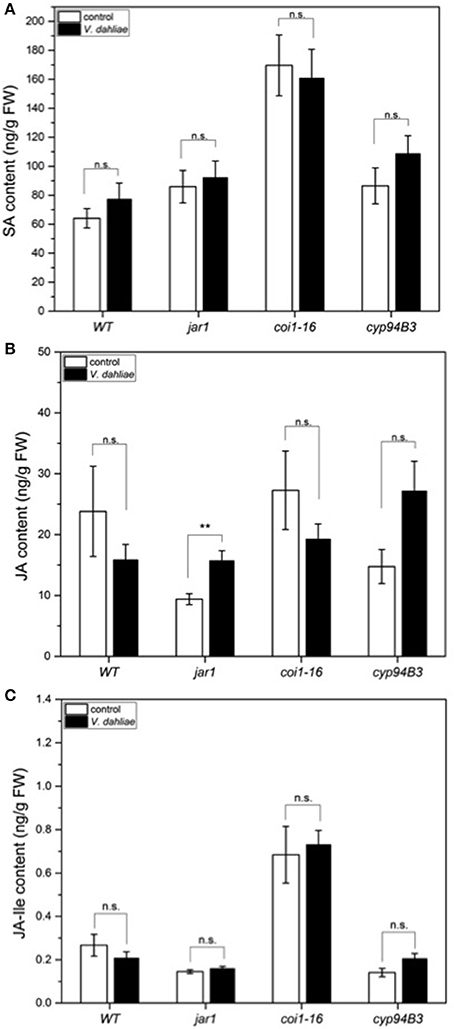
Figure 6. Phytohormone levels in Arabidopsis WT and JA mutant plants after 24 h of V. dahliae infection. Shown is the content (± SE, n = 10) of SA (A), JA (B), and JA-Ile (C) in control (white) and VD-infected (black) WT, jar1, coi1-16, and cyp94B3 plants. Statistically significant differences were analyzed by Mann Whitney U-test, p < 0.05; n.s. not significant, **p < 0.01.
Gene Expression in JA Mutants Is Different to That in the WT in the Early Phase of Colonization
To validate the RNA-seq results obtained, the expression of selected regulated genes in the data set was analyzed by RT-qPCR for co-cultivated WT plants (Figure 7) and for the fungus (Figure 8). Highly regulated members of the SWEET family (Chen et al., 2010) and a Wall-Associated Kinase-like gene (WAKL10, Meier et al., 2010) were chosen for the plant and VDAG_02979 (a glucose transporter) and VDAG_06565 (poly-A-ribonuclease, PARN) for the fungus. In the RNA-seq analysis, the expression of WAKL10 was 56-fold induced in V. dahliae-infested plants compared to plants grown alone. A 10-fold induction was observed in the RT analysis (Figure 7A). SWEET11 and -3 were 20- and 10-fold downregulated in the RNA-seq data set, respectively, while a 9.7- and 6.8-fold decrease was detected by RT-qPCR (Figures 7B,C). Interestingly, the expression of these genes was differently regulated in the JA mutants. The expression of WAKL10 was induced ~100-fold in jar1, i.e., significantly higher than in WT seedlings, while SWEET3, which is down-regulated in the WT, was induced 17-fold in the mutant (Figure 7). Additionally, SWEET11 was less repressed in the mutants: for jar1, the repression was 50% lower compared to WT plants. This clearly indicates that the expression of interaction-specific plant genes is influenced by JA content.
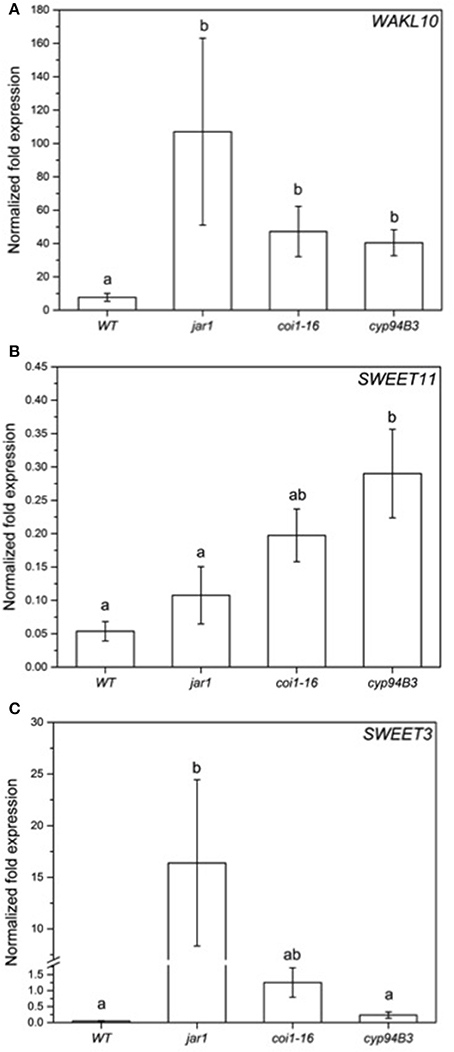
Figure 7. Expression of selected genes in Arabidopsis WT and JA mutant plants after 24 h of V. dahliae infection. Shown is the normalized fold expression (± SE, n = 3) of WAKL10 (A), SWEET11 (B), and SWEET3 (C) in VD-infected WT, jar1, coi1-16, and cyp94B3 plants. The expression level of genes of interest (GOIs) in water-treated plants was used as control and set to 1.0. The mRNA levels for each cDNA probe were normalized with respect to the RPS18B mRNA level. Statistically significant differences between the mutants were analyzed by one-way ANOVA, p < 0.05 (Sidak). Different letters indicate a statistically significant difference.
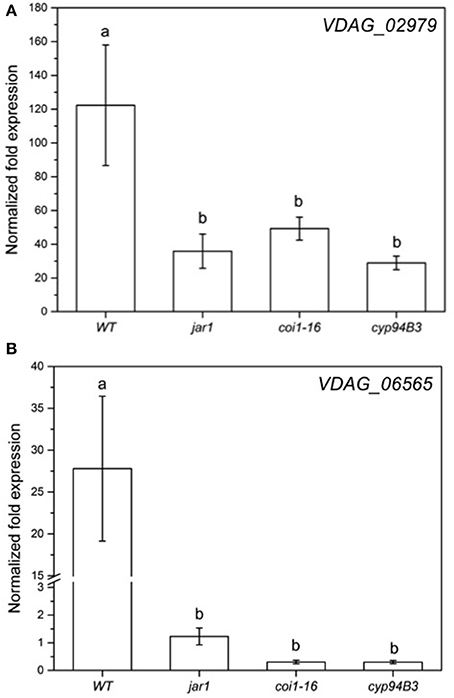
Figure 8. Expression of selected genes in V. dahliae after colonizing Arabidopsis WT and JA mutant plants for 24 h. Shown is the normalized fold expression (± SE, n = 3) of VDAG_02979 (A) and VDAG_06565 (PARN) (B) in VD-colonized WT, jar1, coi1-16, and cyp94B3 plants. The expression level of GOIs in VD alone was used as control and set to 1.0. The mRNA levels for each cDNA probe were normalized with respect to the VD_Actin2 mRNA level. Statistically significant differences between the mutants were analyzed by one-way ANOVA, p < 0.05 (Sidak). Different letters indicate a statistically significant difference.
The RNA-seq results for the fungal genes were also validated by RT-qPCR analysis (Figure 8). The mRNA levels of VDAG_02979 (RNA-seq: + 200-fold; RT-qPCR: + 120-fold) and VDAG_06565 (RNA-seq: + 3-fold; RT-qPCR: + 27-fold) in response to the fungus showed the same trend. Again, when co-cultivation was performed with the JA mutants, the expression of both genes was less induced compared to WT plants (Figure 8). Thus, both partners require plant JA for the typical regulation of genes that respond to the symbiotic interaction.
Discussion
V. dahliae Is in the Pre-vascular Growth Phase 24h after Co-cultivation with Arabidopsis
The propagation of Verticillium species within their hosts is characterized by a biphasic interaction: initially a biotrophic phase allows rapid growth of the microbe in the xylem without major effects on plant performance followed by a necrotrophic phase in which the host is ultimately killed (Reusche et al., 2012). Analysis of the initial contact and early phase of the interaction between both partners could lead to the identification of target genes that will help understand further phases of the interaction and provide useful targets for pest control. Confocal analysis 24 h after co-cultivation demonstrated that the pathogen colonizes the root efficiently (Figure 1) by developing a dense hyphal network at the root surface and root tip. In areas of lateral root formation, an accumulation of conidia spores was detectable (Figures 1G–I). This matches previous observations that showed the lateral root to be a primary area of Verticillium infection in diverse host plants (Zhou et al., 2006; Vallad and Subbarao, 2008; Zhao et al., 2014). Further analyses of root cross-sections demonstrated that the fungus invades the plant tissue but did not reach the vascular system within the first 24 h of co-culture (Figures 1J–O). We conclude that the fungus was still growing in the pre-vascular phase during our experiments, optimal for the identification of genes regulated during the very early interaction phase. During this period we also did not observe significant differences in phytohormone levels, which is consistent with the microscopic information.
Initial Contact of Arabidopsis and V. dahliae Leads to Reprogramming of Primary Metabolism in Both Organisms
Plant-pathogen interactions involve an adaptation of both partners. While the plant typically recognizes the pathogen and induces appropriate defense responses, the fungus manipulates the biology of its host to gain sufficient nutrients for growth and reproduction (Boyd et al., 2013). To identify early targets in the Arabidopsis—V. dahliae interaction, an RNA-seq analysis after 24 h of co-cultivation was performed for both partners. This time point was chosen because no structural changes were detected in plant and fungal tissues under the microscope, and because the phytohormone levels in the plant were not yet altered, thereby avoiding secondary effects due to the response of the plant genome to an altered phytohormone environment. We detected 4432 DEGs for both organisms (Table 1, Tables S1–S4), 1143 DEGs in Arabidopsis belong mainly to primary metabolic pathways (21%, Figure 2A). This result is comparable to a recent study in cotton where 26% of the DEG reads could be assigned to metabolic pathways (Zhang W. W. et al., 2017). Comparably, in both studies, pathways associated with plant-pathogen interaction (3%) and plant hormone signal transduction (2%) were also regulated upon Verticillium stress.
Several members of the plant sucrose efflux transporters of the SWEET family were highly regulated in the analyzed interaction. SWEET11 as well as –3, −12, −15, and –8 were downregulated in decreasing order (Table S2). SWEET11 and −12 are involved in phloem loading with sugars (Chen et al., 2010, 2012; Boyd et al., 2013). By silencing of OsSWEET11 in rice it could be demonstrated that the growth of Xanthomonas oryzae pv. oryzae (Xoo) is decreased which resulted in more resistant plants (Yang et al., 2006). Also SWEET3 was reported to be involved in defense and was downregulated after Geminivirus infection (Ascencio-Ibá-ez et al., 2008). These observations suggest that the plant is trying to prevent export of its sugars to restrict fungal growth.
Besides the defense genes discussed above, members of the WAK family are also highly represented in our data sets. WAK-L10 and WAK3, –5, −1, and −4 responded to Verticillium treatment (in declining order, Table S1). WAK1 is induced upon infection with Ps. maculicola ES4326 and the protein stimulated PR1 expression (He et al., 1998). Additionally, WAK1, −2, −3, and −5 are inducible by SA which accumulated during pathogen infection (He et al., 1999). Likewise, the WAK-like gene WAKL10 is SA-induced and involved in defense against bacteria and fungi (Meier et al., 2010). Among the genes involved in controlling Ca2+ homeostasis, genes for in- and efflux transporters of the cyclic nucleotide-gated channel (CNGC), autoinhibited Ca2+-ATPase (ACA), and glutamate-like receptor (GLR) protein families responded to the fungus, indicating that many signaling events induced during the early phase of the interaction are Ca2+-dependent. Consistent with this idea, genes for Ca2+-binding proteins involved in signal perception and propagation showed increased expression, especially members of the calmodulin-like (CML) protein family. However, closer inspection of the genes did not allow any meaningful conclusion about which pathways are major targets of the fungus. Besides regulation of defense responses, many genes for Ca2+-binding proteins are involved in ion homeostasis, enzyme activity control, and biotrophic plant/microbe interactions (for details compare genes in Table S1 with the TAIR database). An induction of defense genes is always accompanied by a lower investment in plant growth (Huot et al., 2014). This is reflected by the down-regulation of ROOT CAP POLYGALACTURONASE28 and enzymes involved in cytokinin signaling. The phase of reprogramming of plant primary metabolism lasts for several days, as in Verticillium-infected tomato plants where it was shown that both gene expression and protein synthesis of metabolic pathways proteins are still down-regulated at 7 dpi (van Esse et al., 2009; Witzel et al., 2017). The global transcriptome analysis performed in this study provides significant insights into components involved in early phases of the Arabidopsis—V. dahliae interaction and suggests a number of genes and pathways that could be employed as markers in breeding for wilt tolerance.
In contrast to previous studies focusing on analysis of the plant's reaction to the fungus (van Esse et al., 2009; Faino et al., 2012; Witzel et al., 2017; Zhang W. W. et al., 2017), we present also DEGs from the fungus caused by response to the plant. The majority of the 3289 DEGs code for proteins with unknown functions (Tables S3, S4). The up-regulation of VDAG_02979, which encodes for a glucose transporter, suggests that it might play an important role in the early interaction. Besides this initial observation, a huge number of identified genes belong to families which might become important targets after elucidation of their function in V. dahliae. Therefore, this list might provide useful information for genes with interaction-specific functions, in particular since the knowledge about the function of V. dahliae genes is strongly increasing. For example, very recent studies revealed that homeodomain and bZIP transcription factors (Fang et al., 2017; Sarmiento-Villamil et al., 2017), two less characterized transcription factors (Sarmiento-Villamil et al., 2010; Zhang W. Q. et al., 2017), polyketide synthases (Zhang T. et al., 2017), endochitinases (Cheng et al., 2017), a novel V. dahliae protein that targets the plant nucleus (Zhang L. et al., 2017), LysM effectors (Kombrink et al., 2017), a isochorismatase hydrolase (Zhu et al., 2017), a factor involved in the fungal secretory pathway (Xie et al., 2017), a RACK1-like protein involved in root entry (Yuan et al., 2017), pathogenesis-related exudated proteins (Chen et al., 2016), and the mitogen-activated protein kinase 2 (Tian et al., 2016) are important components in controlling V. dahliae-induced disease development in various plant species. Members of all these protein families can be found in the list of V. dahliae genes up-regulated after infection of Arabidopsis.
JA Mutants Are Less Susceptible to Verticillium Infection
Various studies have shown the involvement of several classes of plant hormones in the control of Verticillium growth and propagation in Arabidopsis. While ET perception mutants are more susceptible to Verticillium infection, an elevation of cytokinins enhances plant resistance (Pantelides et al., 2010; Reusche et al., 2013; Sun et al., 2014). Downregulation of plant genes involved in cytokinin signaling might therefore be induced by the fungus. Interestingly, jasmonates are not only accumulated by the plant to induce defense, but the fungus also requires a JA-independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots (Ralhan et al., 2012). To further analyze the role of JA levels in the interaction of Arabidopsis and V. dahliae, JA biosynthesis (jar1), perception (coi1-16) and degradation (cyp94B3) mutants were studied. All of these mutants performed better, showed less severe disease symptom development in the leaves at 20 dpi compared to the WT control, which were already dead at this time point (Figure 3), and had a higher photosynthetic potential, as demonstrated by their QY_max value above 0.80 (Fv/Fm, Figure 4) (e.g., Kim et al., 2009; Sztatelman et al., 2015). The observation, that jar1 plants performed better than WT plants, contradicts earlier findings where jar1 plants were as susceptible as WT plants (Fradin et al., 2011). To analyze this contradiction, the colonization of the mutants used was compared to WT plants (Figure S2). While there was no difference to the WT colonization level after 10 dpi in the roots and the shoots, there was a significant difference at 20 dpi. The colonization level of the root was similar to WT while the colonization in the shoots was significantly lower in jar1 (Figure S2D). This observation could explain the better performance of the aerial parts of the jar1 plants. Taken together, the altered expression of the interaction-specific genes of plant and fungal origin in the JA mutants confirms the important role of this phytohormone also during early phases. Apparently, the altered expression profile occurs before a significant change in phytohormone levels become detectable.
To gain insight into the growth behavior of V. dahliae on JA mutants during the first 24 h of co-cultivation, the colonization of the roots was analyzed. The detected differences in the colonization (Figure 5) were not significant at this time point, but may have a greater impact in later phases where a clear difference is obvious. After 24 h of co-cultivation, there was also no detectable difference in the levels of the phytohormones SA, JA and JA-Ile (Figure 6). Since changes in phytohormone levels upon pathogen attack are normally very rapid in plants, Arabidopsis might have not yet recognized the microbe as friend or foe, in spite of the already initiated reprogramming of its gene expression pattern. It is also conceivable that the penetration rate is still too low to induce the accumulation of JA and JA-Ile, since this is often associated with wounding or pathogen-induced cell disruption, which was not visible in our microscopic studies (e.g., Suza and Staswick, 2008; Koo et al., 2009). The biphasic interaction with an initial biotrophic period followed by a necrotrophic period may also leave the plant undecided whether it responds to the pathogen with SA- or JA-dependent defense strategies. Furthermore, both phytohormones cross-talk (Thaler et al., 2012; Proietti et al., 2013).
The low content of active JA-Ile in jar1 plants and the reduced perception in the receptor mutant coi1-16 lead to a decreased activation of the downstream signaling pathway by the receptor complex SCFCOI1 (Thines et al., 2007). Since Verticillium propagation from the roots to the leaves depends on an activated COI pathway (Ralhan et al., 2012), this could be a reason why Verticillium causes a reduced leaf growth rate of the JA mutant plants (Figure 3). The reduced spread in the green parts of the JA mutants is also reflected in the gene expression analysis of chosen genes for both the plant and the fungus (Figures 7, 8). In the mutant plants, the SA-induced defense gene WAKL10 is more highly expressed than in WT plants and the downregulation of SWEET11 is less pronounced (Figure 7). In the fungus, both target genes VDAG_02979 and VDAG_06565 are more weakly expressed in the mutants than in the WT plants (Figure 8). VDAG_02979 codes for a glucose transporter (http://fungi.ensembl.org/Verticillium_dahliae), which contributes to the nutrient supply of the fungus. The lower growth rate in the mutants may be a consequence of this transporter being less expressed.
In conclusion, biotrophic plant-microbe interactions are characterized by the stimulation of SA, but not JA levels, while the opposite hormone regulation occurs during nectrophophic interactions (reviewed in Chanclud and Morel, 2016). Within the first 24 h of interaction studied here, none of these phytohormone levels increase significantly, while hormone-synthesis related genes, as well as defense-related genes responding to both hormone types are already up-regulated in the host. This suggests that no clear decision has been taken yet about which strategy will follow initial contact. The numerous genes identified during early reprogramming of the fungal and plant development might be crucial for the initiation and propagation of the pest, and thus may be helpful for developing strategies which potentially restrict fungal development after infection. Considering our results, identification of crucial players which control the interaction at early stage is apparently difficult, because many metabolomic pathways are already re-adjusted within the first 24 h of the contact of the two partners.
The strong retardation of disease symptom development in host plants impaired in jasmonate mutants has been attributed to the fact that Verticillium stimulates the host JA functions in order to promote host cell death during the later necrotrophic phase. There is a substantial crosstalk between JA and SA signaling in which each hormone inhibits the accumulation and/or function of the other. Complete or strong inhibition of JA functions in the mutants may favor SA accumulation and/or SA signaling function which—in turn—may prolong the biotrophic phase and thus retard necrosis and disease development.
Author Contributions
SS: designed and performed the experiments; WS-H: analyzed the RNA-seq data and DEGs; AF: prepared, analyzed, and evaluated samples for confocal microscopy; MR: analyzed the phytohormones; RG, JG, and RO: supervised the projects; RO: coordinated the project; SS, JG, and RO: wrote the manuscript; All authors read and approved the final manuscript.
Data Access
Raw and calculated RNA-seq data was submitted to NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE104590.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We like to thank Anna-Sophie Enke for technical assistance and Julia Starke for pre-experiments and set-up of the cultivation method. Special thanks go to Dr. Vladimir Benes and co-workers at GeneCore (http://genecore3.genecore.embl.de/genecore3/index.cfm) for RNA sequencing. This work was supported by the DFG (CRC1127, projects A03 and INF).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00217/full#supplementary-material
References
Acosta, I. F., and Farmer, E. E. (2010). Jasmonates. Arabidopsis Book 8:e0129. doi: 10.1199/tab.0129
Ascencio-Ibá-ez, J. T., Sozzani, R., Lee, T. J., Chu, T. M., Wolfinger, R. D., Cella, R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454, doi: 10.1104/pp.108.121038
Bains, P. S., and Tewari, J. P. (1987). Purification, chemical characterization and host-specificity of the toxin produced by Alternaria brassicae. Physiol. Mol. Plant Pathol. 30, 259–271. doi: 10.1016/0885-5765(87)90039-7
Berg, G., Fritze, A., Roskot, N., and Smalla, K. (2001). Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J. Appl. Microbiol. 91, 963–971. doi: 10.1046/j.1365-2672.2001.01462.x
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyd, L. A., Ridout, C., O'Sullivan, D. M., Leach, J. E., and Leung, H. (2013). Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29, 233–240 doi: 10.1016/j.tig.2012.10.011
Bu, B., Qiu, D., Zeng, H., Guo, L., Yuan, J., and Yang, X. (2014). A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 33, 461–470. doi: 10.1007/s00299-013-1546-7
Chanclud, E., and Morel, J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. doi: 10.1111/mpp.12393
Chen, J. Y., Xiao, H. L., Gui, Y. J., Zhang, D. D., Li, L., Bao, Y. M., et al. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front Microbiol. 7:1709. doi: 10.3389/fmicb.2016.01709
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Cheng, X. X., Zhao, L. H., Klosterman, S. J., Feng, H. J., Feng, Z. L., Wie, F., et al. (2017). The endochitinase VDECH from Verticillium dahliae inhibits spore germination and activates plant defense responses. Plant Sci. 259, 12–23. doi: 10.1016/j.plantsci.2017.03.002
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in JA signalling. Nature 448, 666–674. doi: 10.1038/nature06006
Decetelaere, S., Tyvaert, L., França, S. C., and Höfte, M. (2017). Desirable traits of a good biocontrol agent against Verticillium wilt. Front. Microbiol. 8:1186. doi: 10.3389/fmicb.2017.01186
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Ellis, C., and Turner, J. G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. doi: 10.1007/s00425-002-0787-4
Faino, L., de Jonge, R., and Thomma, B. P. H. J. (2012). The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signal. Behav. 7, 1065–1069. doi: 10.4161/psb.21014
Fang, Y., Xiong, D., Tian, L., Tang, C., Wang, Y., and Tian, C. (2017). Functional characterization of two bZIP transcription factors in Verticillium dahliae. Gene 626, 386–394. doi: 10.1016/j.gene.2017.05.061
Feys, B., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to Methyl JA, and resistant to a bacterial pathogen. Plant Cell. 6, 751–759. doi: 10.1105/tpc.6.5.751
Fonseca, S., Chico, J. M., and Solano, R. (2009). The JA pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. doi: 10.1016/j.pbi.2009.07.013
Fradin, E. F., Abd-El-Haliem, A., Masini, L., van den Berg, G. C., and Joosten, M. H. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Fradin, E. F., Zhang, Z., Ayala, J. C., Castroverde, C. D., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 12, 393–404. doi: 10.1105/tpc.12.3.393
Gaspar, Y. M., McKenna, J. A., McGinness, B. S., Hinch, J., Poon, S., Connelly, A. A., et al. (2014). Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 65, 1541–1550. doi: 10.1093/jxb/eru021
He, Z. H., Cheeseman, I., He, D., and Kohorn, B. D. (1999). A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 39, 1189–1196. doi: 10.1023/A:1006197318246
He, Z.-H., He, D., and Kohorn, B. D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–64. doi: 10.1046/j.1365-313X.1998.00092.x
Hill, T. W., and Kaefer, E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48, 20–21. doi: 10.4148/1941-4765.1173
Hoffman, T., Schmidt, J. S., Zheng, X., and Bent, A. F. (1999). Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 119, 935–950.
Huot, B., Yao, J., Montgomery, B. L., and He, S. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant. 7, 1267–1287. doi: 10.1093/mp/ssu049
Inderbitzin, P., Bostock, R. M., Davis, R. M., Usami, T., Platt, H. W., and Subbarao, K. V. (2011). Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PLoS ONE 6:e28341. doi: 10.1371/journal.pone.0028341
Inderbitzin, P., and Subbarao, K. V. (2014). Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 104, 564–574. doi: 10.1094/PHYTO-11-13-0315-IA
Johnson, J. M., Sherameti, I., Ludwig, A., Nongbri, P. L., Sun, C., Lou, B., et al. (2011). Protocols for Arabidopsis thaliana and Piriformospora indica co-cultivation - a model system to study plant beneficial traits. Endocyt. Cell Res. 21, 101–113.
Kamiya, M., Higashio, S. Y., Isomoto, A., Kim, J. M., Seki, M., Miyashima, S., et al. (2016). Control of root cap maturation and cell detachment by BEARSKIN transcription factors in Arabidopsis. Development 143, 4063–4072. doi: 10.1242/dev.142331
Kanehisa, M. (1997). A database for post-genome analysis. Trends Genet. 9, 375–376. doi: 10.1016/S0168-9525(97)01223-7
Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 6511–6515. doi: 10.1073/pnas.091114198
Kim, E.-H., Li, X.-P., Razeghifard, R., Anderson, J. M., Niyogi, K. K., Pogson, B. J., et al. (2009). The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: a study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 1787, 973–984. doi: 10.1016/j.bbabio.2009.04.009
Klimes, A., and Dobinson, K. F. (2006). A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 43, 283–294. doi: 10.1016/j.fgb.2005.12.006
Klosterman, S. J., Atallah, Z. K., Vallad, G. E., and Subbarao, K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Ann. Rev. Phytopathol. 47, 39–62. doi: 10.1146/annurev-phyto-080508-081748
Knoester, M., Van Loon, L. C., Van Den Heuvel, J., Hennig, J., Bol, J. F., and Linthorst, H. J. M. (1998). Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc. Nat. Acad. Sci. U.S.A. 95, 1933–1937.
Kombrink, A., Rovenich, H., Shi-Kunne, X., Rojas-Padilla, E., van den Berg, G. C., Domazakis, E., et al. (2017). Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol. Plant Pathol. 18, 596–608. doi: 10.1111/mpp.12520
Koo, A. J. K., Cooke, T. F., and Howe, G. A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303. doi: 10.1073/pnas.1103542108
Koo, A. J., Gao, X., Jones, A. D., and Howe, G. A. (2009). A rapid wound signal activates the systemic synthesis of bioactive JAs in Arabidopsis. Plant J. 59, 974–986. doi: 10.1111/j.1365-313X.2009.03924.x
Krogan, N. T., Marcos, D., Weiner, A. I., and Berleth, T. (2016). The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol. 212, 42–50. doi: 10.1111/nph.14107
Kulich, I., Vojtíková, Z., Glanc, M., Ortmannová, J., Rasmann, S., and Žárský, V. (2015). Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 168, 120–131. doi: 10.1104/pp.15.00112
Li, C. H., Shi, L., Han, Q., Hu, H. L., Zhao, M. W., Tang, C. M., et al. (2012). Biocontrol of Verticillium wilt and colonization of cotton plants by an endophytic bacterial isolate. J. Appl. Microbiol. 113, 641–651. doi: 10.1111/j.1365-2672.2012.05371.x
Li, J., Zingen-Sell, I., and Buchenauer, H. (1996). Induction of resistance of cotton plants to Verticillium wilt and of tomato to Fusarium wilt by 3-aminobutyric acid and methyl JA. J. Plant Dis. Plant Prot. 103, 288–299.
Li, S., van Os, G. M., Ren, S., Yu, D., Ketelaar, T., Emons, A. M., et al. (2010). Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type-specific exocytosis. Plant Physiol. 154, 1819–1830. doi: 10.1104/pp.110.164178
Li, Y., Zheng, X., Tang, H., Zhu, J., and Yang, J. (2003). Increase of β-1,3-glucanase and chitinase activities in cotton callus cells treated by salicylic acid and toxin of Verticillium dahliae. Kleb. Acta Bot Sin. 45, 802–808. Available online at: http://www.jipb.net/Abstract_old.aspx?id=2235
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general pur-pose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, S., Ziegler, J., Zeier, J., Birkenbihl, R. P., and Somssich, I. E. (2017). Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ. 40, 2189–2206. doi: 10.1111/pce.13022
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lund, S. T., Stall, R. E., and Klee, H. J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10, 371–382. doi: 10.1105/tpc.10.3.371
Mansoori, B., and Smith, C. J. (2005). Elicitation of ethylene by Verticillium albo-atrum phytotoxins in potato. J. Phytopathol. 153, 143–149. doi: 10.1111/j.1439-0434.2005.00943.x
Matsuo, M., Johnson, J. M., Hieno, A., Tokizawa, M., Nomoto, M., Tada, Y., et al. (2015). High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol. Plant. 8, 1253–1273. doi: 10.1016/j.molp.2015.03.011
Meier, S., Ruzvidzo, O., Morse, M., Donaldson, L., Kwezi, L., and Gehring, C. (2010). The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE. doi: 10.1371/journal.pone.0008904
Metraux, J. P. (2001). Systemic acquired resistance and salicylic acid: current state of knowledge. Eur. J. Plant Pathol. 107, 13–18. doi: 10.1023/A:1008763817367
Moore, G. G., Mack, B. M., Beltz, S. B., and Gilbert, M. K. (2016). Draft genome sequence of an aflatoxigenic aspergillus species, A. bombycis. Genome Biol. Evol. 8, 3297–3300. doi: 10.1093/gbe/evw238
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Müssig, C., Biesgen, C., Lisso, J., Uwer, U., Weiler, E. W., and Altmann, T. (2000). A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between Brassinosteroid-action and Jasmonic-acid synthesis. J. Plant Physiol. 157, 143–152. doi: 10.1016/S0176-1617(00)80184-4
Pantelides, I. S., Tjamos, S. E., and Paplomatas, E. J. (2010). Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol. Plant Pathol.. 11, 191–202. doi: 10.1111/j.1364-3703.2009.00592.x
Pegg, G. F., and Cronshaw, D. K. (1976). Ethylene production in tomato plants infected with Verticillium albo-atrum. Physiol. Plant Pathol. 8, 279–295. doi: 10.1016/0048-4059(76)90022-9
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Proietti, S., Bertini, L., Timperio, A. M., Zolla, L., Caporale, C., and Caruso, C. (2013). Crosstalk between salicylic acid and JA in Arabidopsis investigated by an integrated proteomic and transcriptomic approach. Mol. Biosyst. 9, 1169–1187. doi: 10.1039/c3mb25569g
Puhalla, J. E., and Bell, A. A. (1981). “Genetics and biochemistry of Wilt of pathogens,” in Fungal Wilt Diseases of Plants, eds M. E. Mace, A. A. Bell, and C. H. Beckman (New York, NY: Academic Press Inc.), 146–192.
Ralhan, A., Schöttle, S., Thurow, C., Iven, T., Feussner, I., Polle, A., et al. (2012). The vascular pathogen Verticillium longisporum requires a jasmonic acid-independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots. Plant Physiol. 159, 1192–1203. doi: 10.1104/pp.112.198598
Ratzinger, A., Riediger, N., von Tiedemann, A., and Karlovsky, P. (2009). Salicylic acid and salicylic acid glucoside in xylem sap of Brassica napus infected with Verticillium longisporum. J. Plant Res. 122, 571–579. doi: 10.1007/s10265-009-0237-5
Reusche, M., Klásková, J., Thole, K., Truskina, J., Novák, O., Janz, D., et al. (2013). Stabilization of cytokinin levels enhances Arabidopsis resistance against Verticillium longisporum. Mol. Plant Microbe Interact. 26, 850–860. doi: 10.1094/MPMI-12-12-0287-R
Reusche, M., Thole, K., Janz, D., Truskina, J., Rindfleisch, S., Drübert, C., et al. (2012). Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 24, 3823–3837. doi: 10.1105/tpc.112.103374
Robinson, M. M., Shah, S., Tamot, B., Pauls, K. P., Moffatt, B. A., and Glick, B. R. (2001). Reduced symptoms of Verticillium wilt in transgenic tomato expressing a bacterial ACC deaminase. Mol. Plant Pathol. 2, 135–145. doi: 10.1046/j.1364-3703.2001.00060.x
Roos, J., Bejai, S., Oide, S., and Dixelius, C. (2014). RabGAP22 is required for defense to the vascular pathogen Verticillium longisporum and contributes to stomata immunity. PLoS ONE 9:e88187. doi: 10.1371/journal.pone.0088187
Sarmiento-Villamil, J. L., García-Pedrajas, N. E., Baeza-Monta-ez, L., and García-Pedrajas, M. D. (2010). The APSES transcription factor Vst1 is a key regulator of development in microsclerotium- and resting mycelium-producing Verticillium species. Mol Plant Pathol. 19, 59–76. doi: 10.1111/mpp.12496
Sarmiento-Villamil, J. L., Prieto, P., Klosterman, S. J., and García-Pedrajas, M. D. (2017). Characterization of two homeodomain transcription factors with critical but distinct roles in virulence in the vascular pathogen Verticillium dahliae. Mol Plant Pathol. doi: 10.1111/mpp.12584. [Epub ahead of print].
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., and Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Schnathorst, W. C. (1981). “Life cycle and epidemiology of Verticillium,” in Fungal Wilt Diseases of Plants, eds M. E. Mace, A. A. Bell, and C. H. Beckman (New York, NY: Academic Press Inc.), 81–111.
Scholz, S. S., Malabarba, J., Reichelt, M., Heyer, M., Ludewig, F., and Mithöfer, A. (2017). Evidence for GABA-induced systemic GABA accumulation in arabidopsis upon wounding. Front. Plant Sci. 8:388. doi: 10.3389/fpls.2017.00388
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 16, 2117–2127. doi: 10.1105/tpc.104.023549
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 17, 616–627. doi: 10.1105/tpc.104.026690
Staswick, P. E., Su, W., and Howell, S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl Acad. Sci. U.S.A. 89, 6837–6840. doi: 10.1073/pnas.89.15.6837
Sun, C., Shao, Y., Vahabi, K., Lu, J., Bhattacharya, S., Dong, S., et al. (2014). The beneficial fungus Piriformospora indica protects Arabidopsis from Verticillium dahliae infection by downregulation plant defense responses. BMC Plant Biol. 14, 268–283. doi: 10.1186/s12870-014-0268-5
Suza, W. P., and Staswick, P. E. (2008). The role of JAR1 in Jasmonoyl-L-isoleucine production during Arabidopsis wound response. Planta 227, 1221–1232. doi: 10.1007/s00425-008-0694-4
Sztatelman, O., Grzyb, J., Gabryś, H., and Banaś, A. K. (2015). The effect of UV-B on Arabidopsis leaves depends on light conditions after treatment. BMC Plant Biol. 15, 281. doi: 10.1186/s12870-015-0667-2
Thaler, J. S., Humphrey, P. T., and Whiteman, N. K. (2012). Evolution of JA and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Thaler, J. S., Owen, B., and Higgins, V. J. (2004). The role of the JA response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. doi: 10.1104/pp.104.041566
Thieme, C. J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., et al. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1:15025. doi: 10.1038/nplants.2015.25
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during JA signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Thomma, B. P. H. J., Eggermont, K., Tierens, K. F. M.-J., and Broekaert, W. F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1101.
Tian, L., Wang, Y., Yu, J., Xiong, D., Zhao, H., and Tian, C. (2016). The mitogen-activated protein kinase kinase VdPbs2 of Verticillium dahliae regulates microsclerotia formation, stress response, and plant infection. Front. Microbiol. 7:1532. doi: 10.3389/fmicb.2016.01532
Tjamos, S. E., Flemetakis, E., Paplomatas, E. J., and Katinakis, P. (2005). Induction of resistance to Vericillium dahliae in Arabidopsis thaliana by biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol. Plant Microbe. Interact. 18, 555–561. doi: 10.1094/MPMI-18-0555
Umate, P. (2011). Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal Behav. 6, 335–338. doi: 10.4161/psb.6.3.13546
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithöfer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Vallad, G. E., and Subbarao, K. V. (2008). Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology. 98, 871–885. doi: 10.1094/PHYTO-98-8-0871
van Esse, H. P., Fradin, E. F., de Groot, P. J., de Wit, P. J., and Thomma, B. P. (2009). Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol. Plant Microbe Interact. 22, 245–258. doi: 10.1094/MPMI-22-3-0245
Veronese, P., Narasimhan, M. L., Stevenson, R. A., Zhu, J. K., and Weller, S. C. (2003). Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 35, 574–587. doi: 10.1046/j.1365-313X.2003.01830.x
Wang, P., Zhang, X., Ma, X., Sun, Y., Liu, N., Li, F., et al. (2017). Identification of CkSNAP33, a gene encoding synaptosomal-associated protein from Cynanchum komarovii that enhances Arabidopsis resistance to Verticillium dahliae. PLoS ONE 12:e0178101. doi: 10.1371/journal.pone.0178101
Witzel, K., Buhtz, A., and Grosch, R. (2017). Temporal impact of the vascular wilt pathogen Verticillium dahliae on tomato root proteome. J. Proteomics 169, 215–224. doi: 10.1016/j.jprot.2017.04.008
Xie, C., Li, Q., and Yang, X. (2017). Characterization of VdASP F2 secretory factor from Verticillium dahliae by a fast and easy gene knockout system. Mol. Plant Microbe Interact. 30, 444–454. doi: 10.1094/MPMI-01-17-0007-R
Xie, D.-X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an arabidopsis gene required for JA-regulated defense and fertility. Science 280, 1091–1094.
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yang, X., Ben, S., Sun, Y., Fan, X., Tian, C., and Wang, Y. (2013). Genome-wide identification, phylogeny and expression profile of vesicle fusion components in Verticillium dahliae. PLoS ONE 8:e68681. doi: 10.1371/journal.pone.0068681
Yao, L. L., Pei, B. L., Zhou, Q., and Li, Y. Z. (2012). NO serves as a signaling intermediate downstream of H2O2 to modulate dynamic microtubule cytoskeleton during responses to VD-toxins in Arabidopsis. Plant Signal. Behav. 7, 174–177. doi: 10.4161/psb.18768
Yao, L. L., Zhou, Q., Pei, B. L., and Li, Y. Z. (2011). Hydrogen peroxide modulates the dynamic microtubule cytoskeleton during the defence responses to Verticillium dahliae toxins in Arabidopsis. Plant Cell Environ. 34, 1586–1598. doi: 10.1111/j.1365-3040.2011.02356.x
Yuan, L., Su, Y., Zhou, S., Feng, Y., Guo, W., and Wang, X. (2017). A RACK1-like protein regulates hyphal morphogenesis, root entry and in vivo virulence in Verticillium dahliae. Fungal Genet. Biol. 99, 52–61. doi: 10.1016/j.fgb.2017.01.003
Zhang, H. C., Zhang, W. W., Jian, G. L., Qi, F. J., and Si, N. (2016). The genes involved in the protective effects of phytohormones in response to Verticillium dahliae infection in Gossypium hirsutum. J. Plant Biol. 59, 194–202. doi: 10.1007/s12374-016-0568-4
Zhang, L., Ni, H., Du, X., Wang, S., Ma, X. W., Nürnberger, T., et al. (2017). The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 215, 368–381. doi: 10.1111/nph.14537
Zhang, T., Zhang, B., Hua, C., Meng, P., Wang, S., Chen, Z., et al. (2017). VdPKS1 is required for melanin formation and virulence in a cotton wilt pathogen Verticillium dahliae. Sci. China Life Sci. 60, 868–879. doi: 10.1007/s11427-017-9075-3
Zhang, W. Q., Gui, Y. J., Short, D. P. G., Li, T. G., Zhang, D. D., Zhou, L., et al. (2017). Verticillium dahliae transcription factor VdFTF1 regulates the expression of multiple secreted virulence factors and is required for full virulence in cotton. Mol. Plant Pathol. doi: 10.1111/mpp.12569. [Epub ahead of print].
Zhang, W. W., Zhang, H. C., Liu, K., Jian, G. L., Qi, F. J., and Si, N. (2017). Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis. PLoS ONE 12:e0181609. doi: 10.1371/journal.pone.0181609
Zhao, P., Zhao, Y.-L., Jin, Y., Zhang, T., and Guo, H.-S. (2014). Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell 5, 94–98. doi: 10.1007/s13238-013-0009-9
Zhen, X., and Li, Y. (2004). Ultrastructural changes and location of b-1,3-glucanase in resistant and susceptible cotton callus cells in response to treatment with toxin of Verticillium dahliae and salicylic acid. J. Plant Physiol. 161, 1367–1377. doi: 10.1016/j.jplph.2004.04.007
Zhou, L., Hu, Q., Johansson, A., and Dixelius, C. (2006). Verticillium longisporum and V. dahliae: infection and disease in Brassica napus. Plant Pathol. 55, 137–144. doi: 10.1111/j.1365-3059.2005.01311.x
Keywords: Arabidopsis, calcium, JA, defense, Verticillium dahliae
Citation: Scholz SS, Schmidt-Heck W, Guthke R, Furch ACU, Reichelt M, Gershenzon J and Oelmüller R (2018) Verticillium dahliae-Arabidopsis Interaction Causes Changes in Gene Expression Profiles and Jasmonate Levels on Different Time Scales. Front. Microbiol. 9:217. doi: 10.3389/fmicb.2018.00217
Received: 30 September 2017; Accepted: 30 January 2018;
Published: 13 February 2018.
Edited by:
Katarzyna Turnau, Jagiellonian University, PolandReviewed by:
Sotiris Tjamos, Agricultural University of Athens, GreeceSergio Casas-Flores, Institute for Scientific and Technological Research, Mexico
Copyright © 2018 Scholz, Schmidt-Heck, Guthke, Furch, Reichelt, Gershenzon and Oelmüller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Oelmüller, cmFsZi5vZWxtdWVsbGVyQHVuaS1qZW5hLmRl
 Sandra S. Scholz
Sandra S. Scholz Wolfgang Schmidt-Heck
Wolfgang Schmidt-Heck Reinhard Guthke
Reinhard Guthke Alexandra C. U. Furch
Alexandra C. U. Furch Michael Reichelt
Michael Reichelt Jonathan Gershenzon
Jonathan Gershenzon Ralf Oelmüller1*
Ralf Oelmüller1*